Abstract
An updated review of emerging plant proteases with potential biotechnological application is presented. Plant proteases show comparable or even greater performance than animal or microbial proteases for by‐product valorization through hydrolysis for, for example, cheese whey, bird feathers, collagen, keratinous materials, gelatin, fish protein, and soy protein. Active biopeptides can be obtained as high added value products, which have shown numerous beneficial effects on human health. Plant proteases can also be used for wastewater treatment. The production of new plant proteases is encouraged for the following advantages: low cost of isolation using simple procedures, remarkable stability over a wide range of operating conditions (temperature, pH, salinity, and organic solvents), substantial affinity to a broad variety of substrates, and possibility of immobilization. Vegetable proteases have enormous application potential for the valorization of industrial waste and its conversion into products with high added value through low‐cost processes.
Keywords: application of plant proteases, by-product valorization, hydrolysis, plant proteases production, sustainability
Plant proteases are increasingly used in industrial applications. Potential application fields are continuously broadening, ranging from the production of bioactive peptides to the treatment of liquid effluents, including the recovery of by‐products with high protein content and many other interesting applications which are reviewed.
1. Introduction
Proteases (also called proteinases, or proteolytic enzymes) are enzymes able to hydrolyze the peptide linkages in proteins. In biological environments, proteases are essential for the normal cellular metabolism, including mitochondrial process. [1] Proteases also play a vital role during various biochemical processes, controlling the size, structure, and composition of key proteins. [2]
From an economic perspective, the research of new proteases is in continuous growth because they represent 60 % of all commercialized enzymes in the world. The overall protease enzyme's market has been estimated above 3 billion US$, showing a compound annual growth rate (CAGR) of 6.1 % by 2024. [3] Nowadays, the industrial applications of proteases cover a wide range of industries, such as leather and detergent industries, food technology and pharmaceutical manufacturing. [4]
The proteases can be classified attending the following criteria:
The catalyzed reaction; according to this category the proteases are included in hydrolases (group 3), and subgroup 4 (hydrolases of peptide bonds). Subclass 3.4 can be also subdivided into exo‐ or endopeptidases. The first ones hydrolyze external peptidic linkages (amino‐terminal or carboxyl‐terminal), meanwhile, the endopeptidases hydrolyze the internal peptide bonds in the protein. [5]
Nature of the active site; following this criterion the proteases are classified within seven categories: cysteine endopeptidases (also known as thiol proteases), serine endopeptidases, aspartic endopeptidases (first called acid proteases), glutamic endopeptidases, metalloendopeptidases, threonine endopeptidases, and peptidases with unknown action mechanism. They are grouped if a covalent complex between the enzyme and the substrate is generated during the enzymatic mechanism (cysteine, threonine, and serine endopeptidases) or not (aspartic, glutamic, and metalloendopeptidases). The formation of the covalent enzyme‐substrate complex can be understood considering if the nucleophile is an amino acid in serine, threonine, and cysteine proteases, meanwhile, in other peptidases, the nucleophile is activated by water molecules. [6]
Structure‐based evolutive relationships; where the proteins that share sufficient sequence homology (amino acidic sequence) are included within the same family, while a clan is integrated by families with a common ancestor protein. The MEROPS database (http://merops.sanger.ac.uk/index.htm ) contains 244 families and 55 clans. The names of clans and families are designed with basis on the letters S, C, T, A, G, M, and U, in concordance with the specific catalyst mechanism. Some clans contain families with different mechanisms, which are designed with the letter “P”. [7]
Proteases are found from prokaryotes to complex organisms (plants and animals). The type of protease, as well its functional properties, directly depend on its particular function and the organism conditions of the host. [6] Proteases are involved in the physiology of the plants during their entire cycle life (chloroplast photoinhibition, defense mechanisms, photomorphogenesis, and seed germination, among others) The most abundant type of protease in plants is cysteine (CPs), followed by serine proteases (SPs) and aspartic proteases (APs).
In the cysteine proteases, the nucleophile is the sulfhydryl group of a cysteine residue and the proton donor is a histidine residue (a feature shared with serine proteases). They are active over a wide range of temperatures and pHs. The thiol group of the enzyme has to be in the reduced form for catalytic activity. Thus, the cysteine proteases require a rather reducing environment to be active. Most cysteine proteases have molecular weights between 25 and 30 kDa, and show an optimum pH of 5 to 8. According to the MEROPS database, CPs are divided into ten clans: CA, CD, CE, CF, CH, CL, CM, CN, CO, and C−, and to date, plant CPs have been described as belonging to five of these clans (CA, CD, CF, CO, and CE). The structure of these proteases reveals an α‐helix and a β‐barrel‐like separated from the active site (Cys‐25 and His‐159), which are located at each side of a groove and are well conserved in all members of the family.
Plant CPs play an important role in protein mobilization and defense against biotic and abiotic agents. Plants CPs are inhibited by Iodoacetamide (IAA) and p‐chloromercury benzoate (PCMB). [8]
Plant SPs have a Ser residue as the nucleophile. The catalytic mechanism involves a proton donor in addition to the serine that carries the nucleophile. In 4 clans, the proton donor is a histidine residue; with a third residue present: aspartate in 3 clans and another histidine in the fourth one. In the other 2 clans, the proton donor is a lysine residue and a third catalytic residue is not required. In one of these, there is a Ser/His catalytic dyad. Three clans, (SA, SB, and SC) share a catalytic triad of serine (S) aspartate (D), and histidine (H) in different orders (e. g., HDS (SA), DHS (SB), and SDH (SC)).
Plant SPs are involved in numerous physiological processes, such as microsporogenesis, pathogen attack, and specialization of tissues. They have a molecular weight between 20 and 120 kDa (most of them lies within the 60–80 kDa range). The optimum conditions for their highest activity are alkaline medium (pH: 7–11), and temperature up to 50 °C. Some exceptions are Ara12 from A. thaliana, RSIP from maize, and C1 from soybean, showing an optimum pH between 3.5 and 6.5. [9] Furthermore, diisopropyl‐fluorophosphate and phenylmethylsulfonyl fluoride are serine peptidase inhibitors.
Plant aspartic proteases are found in seeds, flowers, and leaves, differing from cysteine and serine proteases the nucleophile is an activated water molecule rather than the nucleophilic side chain of an amino acid. According to the MEROPS database, APs are grouped in AA (A1, A3, A11, and A12 families) and AD clan (A22 family), being most of them belonging to the A1 family. The primary structure of APs contains a signal sequence at the amino terminus for translocation, a prosegment responsible for correct folding and stability, and a plant‐specific insert (PSI, also known as swaposin domain). PSI contain about 100 amino acids, being responsible for the vacuolar transport of the enzyme. Regarding the second structure, APs mainly contain β‐sheets with a biloval conformation. [10]
Most of APs have two aspartic residues, which are responsible for the activity of the enzyme, being inhibited by pepstatin A. The active site is formed by two Asp residues responsible for water activation. APs exhibit high specificity towards the cleavage at peptide bonds between hydrophobic amino acid residues. The majority of APs are active in acidic conditions, showing an optimum pH of 4–6.5 and a temperature up to 55 °C. APs show a molecular weight range of 35–65 kDa. The enzyme mass depends on whether the PSI fragment was removed by proteolysis previous to activation (heterodimeric) or not (monomeric). The majority of plant APs are heterodimeric with a mass from 35 to 51 kDa.
Plant proteases have been traditionally used for decades; highlighting papain, bromelain, ficin, and actinidin (see Table 1). Over recent decades, new plants have emerged as a valuable raw source of proteases with competitive commercial potential. Table 2 shows emerging serine and aspartic endopeptidases isolated from plants with industrial applications reported in recent years. Proteases derived from plants are gaining attention due to their high proteolytic activity and affinity for specific substrates, remarkable stability over a wide range of operating conditions (pH: 4–10 and temperature up to 60 °C), and low cost associated with the raw source.
Table 1.
Traditional plant proteases with industrial applications.
|
Protein |
Source |
pH[a] |
Temp[a] [°C] |
Molecular weight [kDa] |
Uses |
Ref. |
|---|---|---|---|---|---|---|
|
Papain |
Carica papaya (fruit, root and leaves) |
5–9 |
65 |
23.4 |
• Meat tenderization: connective tissue and myofibrillar proteins hydrolysis. • Dairy industry: production of semisoft cheese or cream cheese), protein hydrolysates production. • Baking industry: reduction of the allergenic protein content of cereals. • Animal feed: bioactive peptides production. • Brewing and wine industry: protein aggregates solubilization. • Bioethanol production: deflocculating agent. • Tooth whitening and biomedicine: tissue repairing of venous ulcers and antibacterial activity. |
[14] |
|
Bromelain |
Ananas comosus (stem and juices) |
5–10 |
70 |
28–32.5 |
• Meat tenderization: myofibrillar proteins hydrolysis. • Fish industry: biopeptides hydrolysates production. • Alcohol production: protein aggregates solubilization. • Animal feed: protein degradation in ruminant feed degradation. • Textile industry: protein solubilization. |
[15] |
|
|
|
|
|
• Biomedicine: blood coagulation, fibrinolysis, antibacterial activity, tissue recovering, anti‐inflammatory agent. |
|
|
|
Ficin |
Ficus carica |
8 |
60 |
23.8 |
• Meat tenderization: myofibrillar proteins hydrolysis. • Milk clotting. • Bioactive peptides production. • Synthetic fibers hydrolysis. • Biomedicine: Production of active antibodies fragments by proteolysis, hemostatic. • Brew industry: protein extraction from barley and malt. • Meat tenderization: myofibrillar proteins hydrolysis. |
[16] |
|
Actinidin |
Actinidia deliciosa (Kiwi fruit) |
6 |
40 |
24.5 |
• Chicken and fish protein hydrolysis. • Alcohol production: protein aggregates solubilization. |
[17] |
[a] Optimal pH or temperature.
Table 2.
Emerging serine and aspartic plant endopeptidases with industrial applications reported in recent years.
|
Protease type |
Name |
Source |
pH[a] |
Temp[a] [°C] |
Molecular weight [kDa] |
Uses or activity |
Ref. |
|---|---|---|---|---|---|---|---|
|
Serine |
Milii |
Latex of Euphorbia milii |
9.0 |
35 |
80 |
Molluscicidal agent and antithrombic drugs |
[18] |
|
Wrightin |
Wrightia tinctoria latex |
7.5–10 |
70 |
58 |
Food industry |
[19] |
|
|
Carnein |
Latex of Ipomea carnea |
6.5 |
65 |
80 |
Brewing wine |
[20] |
|
|
Neriifolin |
Euphorbia neriifolia latex |
9.5 |
50 |
94 |
Dairy industry |
[21] |
|
|
Dubiumin |
Solanum dubium Fresen |
11.0 |
70 |
66 |
Milk‐clotting |
[22] |
|
|
Milin |
Euphorbia mill |
8.0 |
60 |
51 |
Milk‐clotting |
[23] |
|
|
Religiosin |
Ficus religiosa |
8.0–8.5 |
50 |
43 |
Milk‐clotting |
[24] |
|
|
LGP (Latex glycoprotein) |
Synadenium grantii |
7–8 |
37 |
34 |
Casein hydrolysis |
[25] |
|
|
Aspartic |
Asteraceae |
Silybum marianum flowers |
4.5–6.8 |
37 |
27 |
Casein hydrolysis |
[26] |
|
Purified extract (APs) |
Centaurea calcitrapa |
5.1 |
52 |
45–67 |
Milk‐clotting |
[27] |
|
|
Protein extract (APs) |
Ficus racemosa latex |
4.5–6.5 |
60 |
45–70 |
Dairy industry |
[28] |
|
|
Arctiumisin |
Arctium minus (Hill) Bernh |
7.0–8.2 |
50 |
30 |
Bioactive peptides production |
[29] |
|
|
Cardosin |
Cynara cardunculus latex |
5.5 |
– |
55 |
Milk‐clotting |
[30] |
|
|
Onopordosin |
Onopordum acanthium |
2.5 |
30 |
47 |
Bovine milk‐clotting |
[31] |
[a] Optimal pH or temperature.
This work provides an update and critical review of the production techniques, isolation procedures, and immobilization of plant proteases. Emerging applications of plant proteases are discussed in detail, including valorization of products and waste from the food industry (e. g., bird feathers, fish, dairy, gluten, soy), hydrolysis of keratinous materials, organic chemistry, water remediation, biopeptide production, and therapeutic uses. These applications could provide new alternatives for the use of plant proteases to enhance the feasibility of the food and biomedical industries.
1.1. Catalytic Mechanism
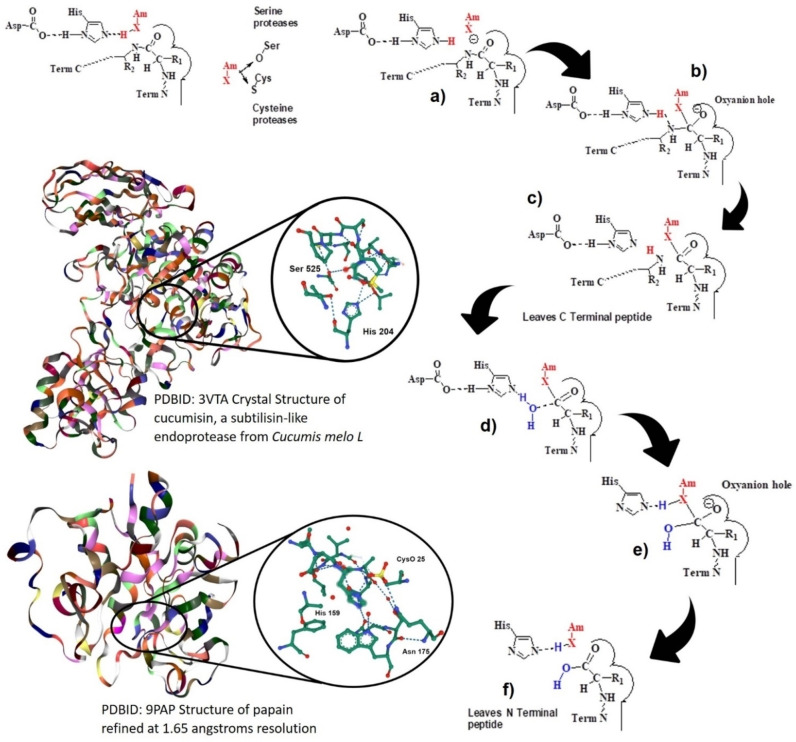
As previously mentioned, the most abundant plant proteases are cysteine proteases, followed by serine proteases and then aspartic proteases. The catalytic mechanism for these major plant proteases is described below. Figure 1 shows the mechanism for the serine/cysteine protease catalyzed protein hydrolysis reaction. The mechanism of action of serine proteases is related to a catalytic triad where the acidic Asp is required to stabilize the positively charged His, which enhances the nucleophilicity of the serine oxygen. In the case of cysteine proteases, the Asp is not needed and the catalytic site is a dyad Cys‐His. [11] The nucleophilic attack occurs by the negatively polarized X (O or S) (Figure 1a), resulting in a tetrahedral intermediary where the negatively charged X is bound to the oxyanion hole (Figure 1 b). Protonation of the NH‐ breaks the bond with the release of the product 1 (C‐terminal peptide (Figure 1c)) and formation of the acyl intermediate, which is attacked by catalytic water (Figure 1d). A second intermediate is formed (Figure 1e). Finally, product 2 is released (N‐terminal peptide) and the active site is regenerated (Figure 1f).
Figure 1.
Mechanism of protein hydrolysis catalyzed by plant serine/cysteine proteases.
Aspartate proteases are different: the catalytic site is a dyad with two Asp. With water present, the reaction involves three main steps: 1. Nucleophilic attack by an activated water molecule with a first transition state and formation of a tetrahedral intermediate. 2. Nitrogen protonation. 3. Fission of the scissile bond and release of the products with new C‐ and N‐terminal peptides. [11] Figure 1 includes the PDB figure for papain (https://www.rcsb.org/structure/9pap) and cucumisin (https://www.rcsb.org/3d‐view/ngl/3vta).[ 12 , 13 ]
2. Production of Plant Proteases
During the two last decades, new strategies have been developed to decrease the cost and the growth time of plants. The plant proteases can be obtained in vivo (using a biological vector to express a specific vegetable protein), through in vitro cultures (employing cell, tissues, and organs), or directly from vegetable biomass (latex, flowers, fruits, roots) using traditional cultivation. From an overview, in vivo and in vitro strategies involve higher costs compared to protein production from crops protein. However, they ensure continuous production of protease with standard parameters. On the other side, proteases derived from traditional crops are more affordable from an economic perspective. However, the homogeneity of target protease depends on the plant response to changes in climatic conditions, modifications in mineral concentrations in the soil, or the presence of other plants. Besides, other factors need to be considered to analyze the feasibility of large‐scale plant protease production, for example, availability of agricultural land for cultivation, labor costs, and plant growth time. [4] Figure 2 summarizes the main advantages and disadvantages of plant protease production using in vivo and in vitro methodology, and traditional cultivation.
Figure 2.
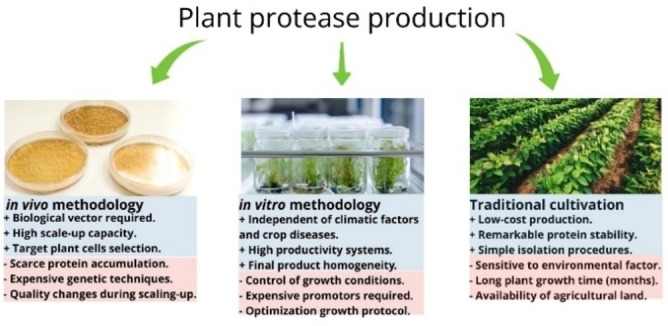
Methodologies for plant protease production.+Advantages, ‐Disadvantages.
2.1. In vivo Protease Production
Protease production via in vivo methodology is a complex process, which can also involve post‐translational modifications (proper folding, glycosylation, phosphorylation) of the proteins to enhance their proteolytic activity and stability. The proteins produced through in vivo strategy (also known as heterologous proteins) are grouped within three categories according to their application: therapeutic proteins, reagents proteins for research and proteins with various industrial uses. A gene or cDNA encoding desired protein and an efficient biological vector with the capacity to transcribe the transgene into the desired protein are required by in vivo methodology. [32] Thus, the target protease can be expressed and isolated.
The requirements for the application of a viable expression system on a large scale are high productivity, easy handling and maintenance, and inexpensive and simple post‐processing. [46] The use of plants as expression systems has exhibited a growing interest in recent years considering its following advantages: low‐cost production compared to mammalian cell culture and microbial fermentation, targeted cells selection (seed, tubers) to express the desired protein, and affordable scale‐up capacity (cultivation of more plants). [33] Furthermore, plants as expression systems show a low risk of contamination with human pathogenic microorganisms.
In contrast, the main limitations of plants as expression systems are low level of protein expression/biomass production ratio, low level of accumulation of expressed protein, changes in protein quality during scale transition from laboratory to agricultural scale, and long production periods (months or even years). [34] The protein accumulation can be increased using promoters to transcribe sequences encoding recombinant proteins, protein fusion to increase product stability, or targeting specific subcellular sites and tissues.[ 33 , 34 ]
The in vivo protein production demands in advance extensive research at laboratory scale to determine the optimum vector and procedures to achieve a feasible protein accumulation yield in the plant, including the use of genetic engineering techniques, thus increasing the protein production cost.[ 35 , 36 ] In consequence, the application of in vivo methodologies in plants is frequently reserved for the production of therapeutic or reagent proteins with high added‐value, whose commercial prices permit the use of complex and expensive genetic techniques.[ 34 , 37 ] An extensive recompilation of genetic technique used for in vivo methodology is presented by Desai et al. [34]
Aspartic, cysteine, and serine proteases have been successfully produced using in vivo methodology from a broad variety of tissues, such as seed (Arabidopsis thaliana, rice, and barley), leaves (tomato and potato), and flowers (cardoon). Cardosin A (Cynara cardunculus) is an aspartic protease whose accumulation in pistils tissue has been increased through in vivo production. In this way, the proteases show higher milk‐clotting activity and their specificity could be also enhanced to the substrate target (k‐casein). [38] Regarding cysteine proteases, papain, bromelain and ficin have been produced through in vivo techniques, showing an optimum activity at 65 °C and a slight enhance in their thermal stability compared with the isolated from crops. In the case of serine proteases, taraxilisin (M. pomifera) and cucumisin (Cucumis melo) increased their specificity (myofibrillar proteins) through in vivo methodologies.[ 39 , 40 ]
In summary, the use of in vivo techniques for plant protease production is applied to improve their specificity, thermal stability and activity. However, the methodologies are reserved for proteases with a wide reported characterization, cases in which the use of complex genetic procedures is feasible.
2.2. In vitro Protease Production
In vitro cell culture methodology has emerged as an attractive alternative, allowing the relative homogenization of large batch of proteases with competitive costs for the production of traditional plant proteases, for instance; papain, bromelain, ficin, and cardosin. The grown of cells and tissues in vitro is independent of climatic factors, crop diseases, heterogeneity in source material, pesticides, remote geographical location and economic problems associated with unstable cultivation policies. [41]
The application of bioreactors for tissue and cell suspension culture grown exhibits major advantages, such as high ratio extract protein volume/amount of biomass in the medium, and rigorous control of environmental conditions. Controlling these variables contributes to obtaining homogeneous compounds and reduces erratic variations due to climatic and seasonal factors. These advantages contribute to achieving a feasible yield in the elaboration of plant proteases whose production by traditional plant agriculture is not economically viable.
The in vitro tissue culture can be carried out using genetically modified plants to maximize the protease volumetric productivity. Relevant grown culture parameters are the selection of transgenic cell lines, temperature and pH, oxygen demand, and bioreactor type (stirred‐tank, pneumatic, wave, membrane, and scale‐down). [42] In recent years, there has been a growing interest by roots culture (also known as “hairy roots”), which offer in contrast to normal roots, high genetic stability, and fast growth rates. These benefits are especially important for recombinant protein production, phytoremediation, enhanced secondary metabolite production, and plant breeding. [43] The in vitro methodologies can be distinguished into two categories: (i) micropropagation and somatic embryogenesis and ii) callus and cell suspension cultures.
2.2.1. Micropropagation and Somatic Embryogenesis
The micropropagation techniques make it possible to reduce the growth periods of large volumes of clonal plants from years to months, producing pathogen free specimens. Micropropagation methodology has been successfully applied to Ficus carica, Carica Papaya, Taxus canadensis, Coleus forskohlii, Hypericum perforatum, and Catharanthus roseus. Micropropagation can be carried out using nodal explants, shoot tips, or leaf segments, which are immersed within a specific growth media. BAP (6‐benzylaminopurine) and NAA (naphthaleneacetic acid) are commonly used as regulators to promote shoot proliferation, thus increasing the enzyme production rate. [44]
The costs associated to regulators compounds are crucial to scale up of micropropagation techniques to produce high‐quality roots. An optimized growth medium is required for each particular enzyme according to the explant types in order to achieve a feasible yield. [45] Specific protocols have been developed for the production of commercial proteases on a large scale. Regarding papaya, the micropropagation is performed using shoots immersed in agar medium with low concentration of IBA (indole‐3‐butyric acid). Once the induction stage was accomplished, the root development is carried out using vermiculite medium with MS (Murashige and Skoog) under aerated conditions. In the case of bromelain, its proteolytic activity could be increased when the medium consists of ANA (α‐naphthaleneacetic acid) and 0.5 ppm of BA (benzyladenine) at 24 °C in 16 h light/8 h dark photoperiod using nodus callus. [41]
On the other hand, somatic embryogenesis process makes it possible to achieve massive propagation using ovular tissue, immature zygotic embryos, mature zygotic embryos, anthers, apical meristems or stems. 2,4‐Dichlorophenoxyacetic acid and adenine hemisulphate are growth regulators commonly used to promote the multiplication of healthy plants, which are after transplanted to the field. [46] The Morishige and Skoog medium (inclusive in combination with kinetin and indolebutyric acid) is broadly used for the cultivation of plant cell culture.[ 45 , 46 ] As mentioned for micropropagation techniques, a specific protocol for plant production using somatic embryogenesis is also required to achieve an efficient large‐scale performance. The application of solvents and regulators with low ecotoxicity and competitive purchase cost is a continuous challenge to achieve sustainable plant production through micropropagation and somatic embryogenesis.
2.2.2. Callus and Cell Suspension Cultures
Callus and cell suspension favor the synthesis of a specific metabolite through the control of growth conditions (temperature, pH, and regulators concentration). Only the specimens with major induction capacity are selected to be transferred into the growth medium, where the callus formation begins. This represents a significant advantage in terms of culture periods compared to micropropagation, ensuring the continuous availability of enzymes with high homogeneity levels.
Callus and cell suspension cultures methodologies have been successfully applied for the production of cardosin (Cynara cardunculus) and silymarin (Silybum marianum L.). [47] The protocol used during growth periods has a predominant effect on proteolytic activity.[ 48 , 49 ] Fernandez and Pomilio optimized the growth protocol for bromelain production from Ananas comosus, observing that protease obtained through callus culture exhibited higher specific activity compared to those obtained from traditional cultivation. [41] In the case of Ficus carica, Dini et al. documented the production of a protein extract using cell cultures with dermatological applications. [50] The authors observed that the specific activity of the extract is sensitive to the concentration of regulators in the medium. The correct selection of regulators and growing conditions is crucial to achieve a high yield of the target enzyme or protein extract. Protein‐free media can be also used for metabolites production, containing only salts, sugars, and specific growth‐regulating hormones. This strategy involves a low cost of production and contamination risk. [51]
The cell suspension cultures using large‐scale bioreactors commonly show a high protein accumulation/amount of biomass ratio. The use of bioreactors for plant cells cultivation overcomes the problems associated with climatic and seasonal factors, showing a simplified purification process when the product is secreted into the extracellular medium. On the other hand, cell suspension cultures require the application of promotors systems (similar to those used in micropropagation and somatic embryogenesis), and rigorous control of the healthy specimens and growth conditions. [52]
2.3. Protein Obtained Directly from Plants
The main benefit of traditional cultivation is its low‐cost production, without the need to employ complex genetic engineering techniques. Similar to in vivo methodology, the production of proteases from crops is sensitive to environmental conditions changes (floods or droughts), fertilizers, and land‐use patterns (crops rotation), which can affect the homogeneity of the product. Furthermore, the long growing periods between planting and harvesting must be taken into account for the techno‐economic evaluation.
Protein extracts can be obtained from different plant organs (roots, fruits, flowers, peels, or latex) as is indicated in the work of Sun et al. The authors carried out extensive comparative analysis on proteolytic activity among 90 plant resources, using casein as substrate at pH from 3.0 to 10.5. [53] Remarkable proteolytic activity (1 % casein substrate) was observed in that extracts derived from kiwifruit (28.8 U g−1), broccoli (16.9 U g−1), ginger (16.6 U g−1), leek (32.7 U g−1), and red pepper (15.8 U g−1). The authors noted that there are promising plants with potential for the production of proteases.
Numerous proteases present in the latex of plants belonging to families such as Caricaceae (mexicain, papain, clycylendopeptidase), Moraceae (ficain, ficain P I, macluralisin), Asclepiadaceae (funastrains CII and CI, morrenains BI and BII, Morrenain O II, asclepain CI), Asteraceae (taraxalisin), Convolvulaceae (carnein), Apocynaceae (cryptolepain, ervatamins A, B, C, heynein, philibertain G I), and Euphorbiaceae (euphorbains Y‐1, Y‐2, Y‐3, P, La1, La2, La3 Lc, T1, T2, T3, T4, milin) have been identified. [54] Serine and cysteine proteases obtained from latex are stable in a broader range of pH and temperature compared with aspartic proteases. However, APs have fewer inhibitors. Table 3 shows the common properties of PPs isolated from laticifers plants.
Table 3.
|
Protease |
Molecular weights [kDa] |
Stability range |
Optimal conditions |
Common inhibitors |
||
|---|---|---|---|---|---|---|
|
pH |
Temp. [°C] |
pH |
Temp. [°C] |
|||
|
Serine |
33–117 (majority 60–80) |
2.5–11 |
<80 |
5.2–10 |
40–75 (most of them 60–70) |
Diisopropyl fluorophosphate (DFP), Phenylmethanesulfonyl fluoride (PMSF), Pamidinomethanesulfonyl fluoride (APMSF), Chymostatin and Ciethyl pyrocarbonate (DEPC). |
|
Aspartic |
45–60 |
4.0–7.5 |
<70 |
4.5–6.5 |
60 |
Pepstatin A |
|
Cysteine |
21–29 |
3–12 |
<80 |
7–9 |
60 |
Iodoacetamide (IAA), p‐chloromercury benzoate (PCMB), sodium tetrathionate, mercuric chloride, transepoxysuccinyl‐ l‐leucylamido‐(4‐guanidino)butane (E‐64). |
Flowers are also a suitable source of PPs.[ 55 , 56 , 57 ] For example, Cardosin and cyprosins proteases obtained from fresh flowers (Cynara cardunculus) have been traditionally used on cheese elaboration (optimum temperature 65 °C and pH 5–7). In recent years, peel and kernel (food processing wastes) have been considered valuable vegetable protease sources. Within this context, Mehrnoush et al. optimized the extraction and purification of serine protease from mango peel using an aqueous two‐phase polyethylene glycol/dextran system, reaching 97.3 % yield under optimal conditions (8000 g mol−1 of polyethylene glycol (PEG) and 4.5 % of NaCl at pH 7.5). [58] The proteases showed to be active for azocasein hydrolysis, indicating that mango peel could be an effective source of natural enzymes considering the large amounts of peel and kernel generated during mango processing.
Seeds and rhizomes of plants are important cysteine storage and play an important role during germination. [62] Research on the extraction of proteases from seeds and rhizomes focused on the obtention of new enzymes for milk‐clotting. Cysteine proteases obtained from traditional cultivation show particular specificity towards casein substrate and they commonly present stability higher than 75–80 % over a wide range of pH (4–10) and temperature (60–70 °C).[ 63 , 64 , 65 ]
As mentioned previously, plant latices are an extremely interesting source of proteases due to their easy extraction, high enzymatic activities, and relatively simple purification processes. These proteases protect ripening fruits against plant pathogens such as insects and fungi and most belong to the cysteine and serine proteases family, only one is a member of an aspartic proteases and none is a metalloprotease (at least for now). Table 4 shows the main structural facts of the most important Serine proteases found in plant latices.
Table 4.
Some serine proteases in plant latices.
|
Protease |
Plant |
MW [kDa] |
Isoelectric point |
pH; T [°C] opt. |
|---|---|---|---|---|
|
Macluralisin |
Maclura pomifera |
65 |
– |
8.5; 58 |
|
Taraxalisin |
Taraxacum officinale |
65 |
4.5 |
8; 40 |
|
Euphorbain L, Y1, Y2, Y3, P La1, La2, L3, Lc, T1, T2, T3, T4 |
Euphorbia, different |
33–74 |
4.5–7 |
5.5–8.3 |
|
Hevains A, B, L |
Hevea brasilensis |
58–80 |
4.3–6.9 |
6–7 |
|
Parthenain |
Parthenium argentatum |
63 |
6.3 |
7–8 |
|
Ficin E Other A, B and C |
Ficus elastica |
50 |
3.7 |
6 |
|
Carnein |
Ipomoea carnea ssp fistulosa |
80.2 |
6.5 |
60 |
|
Artocarpin |
Artocarpus heterophyllus |
79.5 |
6.3 |
8; 60 |
|
Milin |
Euphoris milii |
51.4 |
7.2 |
8; 60 |
Some of the serine proteases have homology with subtilisin‐like proteases. Subtilisin‐like proteases (or subtilases) are serine peptidases present mostly in plants. Plant subtilisins (plant SBTs) are homologs of the bacterial subtilisins that were first identified in the Carlsberg and BPN strains of Bacillus subtilis. [66] Macluralisin's N‐terminal sequence shares 30 % homology with the sequence of subtilisin‐like proteinase K from Tritirachium album whereas Taraxalisin's has 40 % of its residues identical to those of Subtilisin Carlsberg. Subtilisin Carlsberg from B. licheniformis (also known as subtilisin A, subtilopeptidase A, alcalase Novo), was discovered by Linderstrøm‐Lang and Ottensen while studying the conversion of ovalbumin to plakalbumin. The similarity of catalytic and binding site geometries for subtilisin and chymotrypsin, despite their different tertiary structures, gave rise to the notion of convergent molecular evolution. Studies about these enzymes have been crucial in understanding the mechanism of serine protease activity. [67]
Protease B from the latex of E. supina Rafin is a Cucumisin‐like serine protease, whereas Carnein's N‐terminal sequence showed a high degree of identity with that of Subtilisin‐like serine protease. Cryptlepain and Milin sequences do not match with any sequence of known plant serine proteases.
Latex cysteine proteases are in the range from 21 to 29 kDa in molecular weight, are stable in the range 3–12 of pH and up to 80 °C. The manuscript from Domsalla and Melzig presents a complete description of properties of several latex proteases. [54] Here some of them are presented in Table 5.
Table 5.
Cysteine proteases from plant latices.
|
Protease |
Plant |
MW [kDa] |
Isoelectric point |
pH; T [°C] opt. |
|---|---|---|---|---|
|
Ervatamin A |
Ervatamia coronaria |
27.6 |
8.37 |
8–8.5; 50–55 |
|
Ervatamin B |
Ervatamia coronaria |
26 |
9.35 |
6–6.5; 50–55 |
|
Ervatamin C |
Ervatamia coronaria |
23 |
9.54 |
7.5–8; 50 |
|
Funastrain CII |
Funastrum clausum |
23.6 |
>9.3 |
9–10; up to 70 |
|
Asclepain S |
Asclepias speciosa Torr. |
– |
– |
7–8; 65–75 |
|
Calotropins DI DII |
Calotropis gigantea |
23.8‐24.2 |
9.60 |
|
|
Araujiain HI, HII and HIII |
Araujia hortorum |
24.03 |
>9.3 |
8–9.5; 60 |
|
Papain |
Carica papaya |
23.43 |
8.75 |
5.5–7 |
|
Caricain |
Carica papaya |
23.28 |
11.7 |
7 |
Ervatamin C has a similarity of 66 % to Ervatamin B and 50 % to Papain. Funastrain CII shows remarkable stability of its caseinolytic activity. The N‐terminal sequence of it shows a high degree of homology with Asclepain F. Another group is the Morrenains BI, BII, and OII. Asclepain CI is the major purified protease from latex of stems of Scarlet milkweed Asclepias crussasiva L. and shows a high identity with Funastrain CII (87 %) and Asclepain F (86 %). Araujiain HI, HII, and HIII show partial homology (36–48 %) with other plant cysteine proteinases.
There is an important group of proteases: papain‐like cysteine proteases (PLCP). Papain‐like cysteine proteases genes belong to a large multigenic family with 31, 43, 40, 26, 40, and 24 PLCP family members were identified in Arabidopsis, rubber, cassava, castor, poplar, and grapevine, respectively, divided into 9 subfamilies based on structural characteristics. [68]
Homology in plant proteases is very heterogeneous: from no homology at all to up to 90 % homology. Within this context, besides the examples cited above, ficins are glycoproteins and have high homology with bromelain. [16]
Table 6 presents a comparison of the activity of different latex proteases with some of the more recognized plant proteases. In general, the characterization is performed using a model reaction with a model substrate (azo) casein, (azo) collagen, or (azo) albumin. A typical reaction with casein involves 100 μL of enzyme solution added to 900 μL of substrate solution (2 % casein in 10 mm. Tris‐Cl buffer pH 8.0) incubated at 50 °C for 20 min. The reaction is terminated by the addition of an equal volume of 10 % chilled trichloroacetic acid (TCA). One protease unit is defined as the amount of enzymes that release 1 μmol of tyrosine per mL per minute under the above assay condition. The specific activity is expressed in unit of enzymes activity per milligram of protein. Other ways of determining proteolytic activity have been used. For example, to 10 mL solution containing 1 % (w/v) casein, 0.05 m acetate, 0.05 m phosphate, and 5.0×10 m EDTA 0,05 mL of enzyme were added. Two‐milliliter aliquots were removed at 0, 10, 20, and 30 minutes and added to 3.0 mL of 5 % trichloroacetic acid solution. One unit of activity is defined as a change of 0.001 in absorbance at 280 nm in one minute.
Table 6.
Examples of degree of hydrolysis (%) or Units mg−1 protein for different proteases with casein, whey concentrate, or milk as substrates.
|
Protease |
Protein |
pH |
Temperature [°C] |
Reaction time |
Degree of hydrolysis |
|
|---|---|---|---|---|---|---|
|
Reference |
Enzyme |
|
|
|
|
|
|
[71] |
Papain |
Whey concentrate |
7 |
37 |
0.2–24 h |
13.3–22 % |
|
|
Commercial Papain |
|
|
|
|
10 U mg−1 |
|
[72] |
Actinidin |
Whey concentrate |
– |
15–70 |
5 h |
3.7–15 % |
|
[73] |
Bromelain |
Casein 1 % |
5.1 |
35 |
30 min |
41.7 U mg−1 |
|
|
Commercial bromelain |
|
|
|
|
3–15 U mg−1 |
|
[69] |
From latex |
|
|
|
|
|
|
|
Euphorbia synudenium |
Casein 2 % |
8 |
50 |
20 min |
9.44 U mg−1 |
|
|
Carica papaya |
Casein 2 % |
8 |
50 |
20 min |
0.935 U mg−1 |
|
|
Calotropis gigantea |
Casein 2 % |
8 |
50 |
20 min |
0.618 U mg−1 |
|
|
Calotropis procera |
Casein 2 % |
8 |
50 |
20 min |
0.82 U mg−1 |
|
|
Plumberia Rubera |
Casein 2 % |
8 |
50 |
20 min |
1.03 U mg−1 |
|
|
Ficus religiosa |
Casein 2 % |
8 |
50 |
20 min |
2.17 U mg−1 |
|
[74] |
Bromelain 5 % |
Cow milk |
6 |
45 |
340 min |
22.6 % |
|
[74] |
Papain 5 % |
Cow milk |
6 |
45 |
340 min |
17 % |
|
[70] |
Araujia hortorum latex |
Casein 1 % |
8 |
45 |
2 min |
12.8 U mg−1 |
|
|
Araujiain |
Casein 1 % |
8 |
45 |
2 min |
77.9 U mg−1 |
From the reference of Borde et al. it is clear that the specific activity for different latex proteases is heterogeneous in casein hydrolysis and from 0.61 to 9.44 U mg−1 latex. [69] However, the purified enzymes have very high activity, reaching 77.9 U mg−1 for araujiain. [70]
Depending on the source of the protease and its degree of purity, the enzymatic activity can range from less than 1 U mg−1 to about 80 U mg−1. However, the values depend on the experimental conditions and the additives (cysteine, group 2 ions, metal ions, other).
3. Isolation and Purification of Plant Proteases
Isolation and purification of proteases involve consecutive steps that could lead to loss of product yield. Nowadays, the industry demands efficient downstream process for enzymes purification, highlighting the following requirements: application of mild conditions, easy scale‐up, low material costs and minimization of protein denaturation.
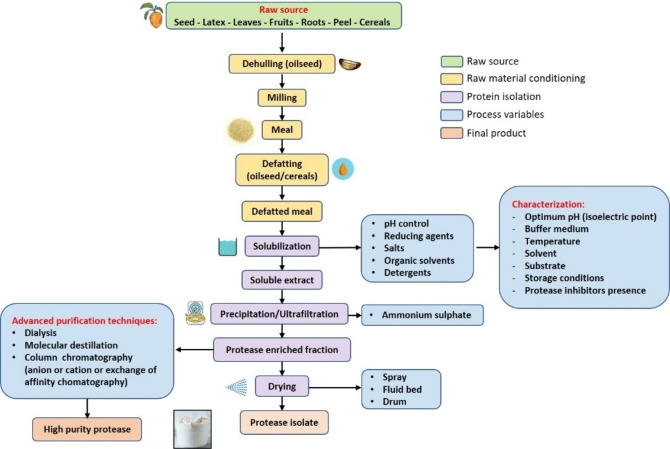
From an economic as well as an environmental perspective, an effective extraction method contributes to favor the commercial possibilities of emerging proteases. There is a continuous development of isolation and purification techniques to achieve sustainable plant proteases production. The amount of protein required, the degree of final purity of the product, whether loss of activity is acceptable, time required for isolation and purification costs should be considered to design a feasible isolation process before scaling up. When the protease extract is obtained from crops, additional considerations should be taken into account, such as plant species, seasonal and environmental variations, application of fertilizer during growth stage, structure and properties of raw material (leaves, seed, roots, latex, and fruits, among others), and previous processing. [75] Figure 3 shows the general procedure for isolation and purification of plant proteases.
Figure 3.
Plant protease purification methodologies.
The first stage of protease isolation process involves the disruption of the tissues where the proteases are accumulated during milling procedure. In the case of seeds, the previous dehulling is required to achieve a feasible homogenization of raw material powder. [76] An extra pre‐treatment of raw material is the defatting of the flours. This stage is only performed for oilseed and cereals, which have high level of fat/oil. [40]
Once the material was conditioned, the solubilization process is performed, obtaining an extract with the target protease in a soluble form. The efficiency of the solubilization process and the distribution coefficient predominantly depend on material composition (protein, lipids, carbohydrates, pigments, fibers, polyphenols, gums, polysaccharides among other compounds), solvent buffer, extraction time, pH, ionic strength, reducing agents, and temperature. [77] During the solubilization process, rigorous control of operating conditions is required to prevent protein denaturation and functional adverse reactions. For most plant proteases, the extraction is carried out at pH between 4 and 9 because solubilization at extreme basic pH could produce the racemization of amino acids.
The protein solubility on the selected solvent is a key parameter. Commonly, previous studies at laboratory scale are developed before scaling‐up, considering the large volume of solvents required and their large‐scale handling. [16] The solvents or buffers used in proteases extraction generally are high purity quality with remarkable costs associated. Consequently, a preliminary techno‐economic assessment of solvent recovery design should be carried out. The solvent selection also depends on raw material sources. Proteases obtained from oilseeds and legumes generally present a considerable solubility in aqueous media, whereas cereal proteases exhibit the opposite behavior. During the extraction step, surfactants, salts, or organic solvents (ethanol) are usually used to increase the solubility of cereal proteases. [75]
Acetate or citrate buffers are commonly used during aspartic proteases extraction to increase their activity and stability. [78] On the other hand, tris(hydroxymethyl)aminomethane buffer is broadly used on serine and cysteine protease extraction. Also reducing agents could be used to improve the cysteine proteolytic activity, for instance, dithiothreitol (DTT). Ethylendiaminetetraacetic acid (EDTA) and polyvinylpyrrolidone (PVP) are commonly used as chelating agents, preventing protein oxidation and tannin‐protein aggregation during isolation processing. [55] In recent years, there is a growing interest in protease extraction using neutral polymers, such as dextrose and PEG. The application of neutral polymers is efficient when flowers and leaves are used as raw materials. The process is commonly carried out at low temperature (4 °C) using buffer (Tris‐HCl) at pH from 6 to 8, with short contact time (30–60 s). After blending, filtration and subsequent centrifugation stages are performed. [79]
Once the soluble extract is obtained, the clarification step is performed to separate the protease from unwanted material using precipitation, ultracentrifugation, or ultrafiltration techniques. Ultrafiltration shows higher performance than precipitation, obtaining a final product with better functional quality and higher proteolytic activity. On the other hand, the application of ultrafiltration methodology involves higher cost and membrane maintenance compared to other technologies. [80] Precipitation is performed through several methods, such as isoelectric precipitation, organic solvents applications, addition of salts (reducing ionic strength), or use of exclusion polymers.
Isoelectric precipitation is the predilect technique due to its low operating cost. In contrast, this methodology could modify the protease solubility when concentrated acids or alkalis are used, generating the partial protease denaturation by local extreme pH values. [81] In the case of vegetable protease, pigments, carbohydrates, fibers, and lipids are removed from protein extract using a mixture of water/alcohol. The application of organic solvents should be restricted to prevent protein denaturation and the modification of the active site. Another concentration methodology is heat coagulation, which is scarcely used in view of the high level of denaturation achieved during the processing. The use of heat coagulation is reserved for protein extracts with nutritional applications or to increase the selective denaturalization of specific proteins.
Isolation methods by adding salts are scarcely used because of the high cost associated with salt recovery. These methods show comparable performance to isoelectric precipitation, with low denaturation rate. [82] The use of specific salts introduces modifications of the ionic strength of the medium, resulting in alterations in protein solubility without heating, thus conserving the properties of the final product.
Three‐phase partitioning system (TPP) provides a simple and effective protein purification. This technique also can be applied to complex mixtures, involving the blending of crude plant extract with ammonium sulfate ((NH4)2SO4), in combination with organic solvent, commonly tert‐butanol (t‐BuOH). During the procedure, three phases are formed: aqueous phase (rich in carbohydrates and polar compounds) and solvent layer (rich in pigments, lipids, fibers, and nonpolar compounds), while the proteins are selectively partitioned as a precipitate. [83]
After the protease‐enriched fraction was obtained, the drying is carried out. Rigorous temperature control is carried out at this stage to avoid denaturation of the protein. Most vegetable proteases are stable at temperatures up to 70 °C. Drying methods used on an industrial scale are drum drying, spray drying and fluidized bed drying. Spray drying is broadly used owing to its high efficiency to produce protease preparations without denaturalization. However, irreversible aggregation could occur during the process, in which the protein‐enriched solution is atomized and dried with flow hot air. In contrast, during drum drying, the protein extract is applied as a thin layer on a hot surface. This methodology produces the evaporation of large amount of water using short contact periods. [84]
Pre‐concentrated enzymes can be further subjected to affinity chromatographic, size‐exclusion, and ion‐exchange (anionic or cationic) procedures. These techniques are used when a high purity protein preparation is required, for example for biopharmaceutical industry or small quantities production.
4. Plant Protease Applications
4.1. Traditional Applications of Proteases (Cheese Making, Meat Tenderizing, Detergents, Food and Beverages)
Vegetable proteases have been used in manufacturing processes for decades, such as cheese‐making, dairy industry, meat tenderization, brew industry, tanning of leather, and peptide production. An enormous application field of plant proteases is based on hydrolysis of protein‐based substrate from vegetal and animal origin.
4.1.1. Leather Industry: Tanning
Conventional leather processing involves consecutive unit operations (soaking, liming, hair removal, deliming, bating, degreasing, and pickling) where harmful chemicals are used (lime, solvents, sodium sulfide, and ammonium salts). The effluents from leather factories generate serious pollution problems when they are discharged without the correct treatment.
Within this context, plant proteases are an eco‐friendly alternative to replace the use of sodium sulfide for dehairing and tanning leather. [85] The mechanism of hair removal using enzymes is a complex process and currently is not fully understood. Sodium sulfide acts over the hair shaft outside the skin, reducing the S−S bond and increasing protein solubilization. Proteases dehairing act over the active sites of the basement membrane and the specific cells of the outer root sheath and follicle bulb (see Figure 4). Collagen is the most abundant protein found in skin and hides, followed by globular proteins and other fibrous proteins. [86] The glycoconjugates proteins, including glycoprotein (GPs), proteoglycan (PGs), and glycosaminoglycan (GAGs) play an important role in forming collagen fibrils and fixation of hairs in hair follicles. Enzymes attack the glycosidic bonds of GAGs chains and the link between the core protein and GAG. [87] Recently was proposed that enzymes attack β‐1,4‐bonds between N‐acetylglucosamine moieties in the glycosylation of serine and threonine (non‐coiled head structure of the keratin filaments), causing the de‐assembling of the root. [88]
Figure 4.
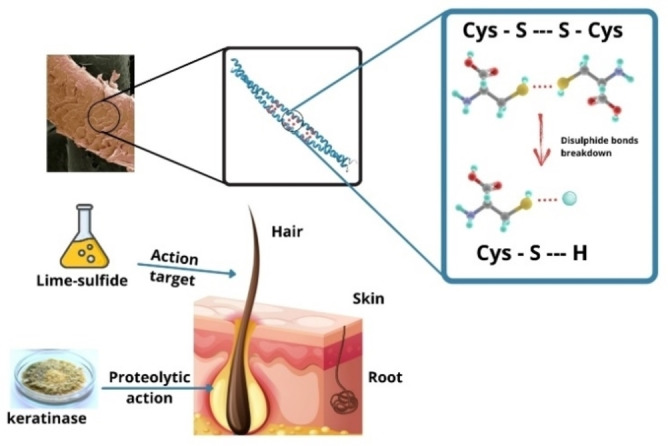
Lime‐sulfide and keratinase action during leather dehairing stage.
High collagenase activity is undesirable in order to obtain leathers with substantial‐quality. [89] A keratinolytic/caseinolytic activity (K/C) ratio lower than 0.5 is required to prevent leather degradation, this requirement is satisfied by plant proteases. In addition, plant proteases show remarkable stability under the operative conditions of leather processing (temperature: 30–50 °C, pH: 4–8, and high salt concentration). [90]
Few reports of vegetable proteases in leather tanning are found in the literature. However, proteases extracted isolated from different plants (Apocynaceae, Bromiliaceae, Anana comosus and Euphorbia nivulia) have shown potential to partially substitute the environmentally toxic lime‐sulfide treatment.
Lopéz et al. investigated the application of peptidases from Calotropis procera latex (CpLP) and Cryptostegia grandiflora latex (CgPL), plants belonging to the Apocynaceae family, for dehairing process. [91]
The latex was submitted to centrifugation and dialysis process to obtain protein enriched‐fraction. The dehairing tests were performed at 25 °C for 24 h at a pilot scale with 0.05 % w/v enzyme. Keratin azure and epidermis substrates were used (hide powder azure (HPA) and azocollagen as keratin, and Elastin‐Congo Red as elastin substrates). CpLP showed twice the proteolytic activity on all substrates compared to CgPL, except Elastin‐Congo Red, which the activity was not detected. In contrast, CgPL showed an elastin activity of 0.12 U mg−1. CpLP exhibited the following activities: 13.4 U mg−1 keratin azure, 36.3 U mg−1 epidermis, 93.0 U mg−1 HPA, and 403.3 U mg−1 azocollagen. From these results, it can be inferred that CpLP presented the highest ratio of keratinolytic/collagenolytic activities.
The activity of CpLP was enhanced by adding sodium sulfite (optimum conditions: 0.05 % w/v enzyme, 0.6 % w/v sodium sulfite at 30 °C for 24 h) without damaging the collagen layer of skin (incomplete depilation was reached using only CpLP). This synergistic effect was based on the activation of cysteine peptidases and the cleavage of keratin disulfide bonds promoted by sodium sulfite.
Proteolytic extract derived from Agave americana serine protease has been isolated and tested on leather liming. Bouhlel et al. optimized the extraction and purification of protease of Agave americana (PPA). [92] The protein‐enriched fraction was initially obtained from leaves using Tris‐HCl buffer (pH 7.8), incubated at 4 °C overnight using CaCl2 as activity stabilizer agent, and subsequent centrifugation. The purification of PAA was carried out using ammonium sulfate precipitation at the level of 80 %, followed by ethanol fractionation and finally, gel filtration coupled to HPLC system. PPA (with a molecular weight of 35 kDa) showed to be stable up to 70 °C and pH from 6.5 to 8.5, exhibiting the highest activity at 60 °C. Wool remotion from sheepskin essays using PPA at 40 °C for 24 h were performed. The authors observed that PPA efficiently unhaired the sheepskin (no major data was reported).
The use of crude bromelain derived from Anana comosus for cowhide pieces dehairing was also studied. The crude extract showed a casein activity of 4.71 U mL−1 casein at pH 4.5. [93] The extract was immersed in sodium acetate buffer solution (pH 4.5) with cowhide using an enzyme concentration of 100 % w/v of skin distilled water. After 24 h, around 50 % hair was removed, probably due to the reaction process was limited by the enzyme diffusion through the skin matrix.
In recent work, the potential application of Carica papaya (Cp) and Vasconcellea quercifolia (Vq) obtained, using simple procedures, from latex on the dehairing process was documented. [94] The activity of both protease extracts was measured using HPA, keratin azure, and elastin‐congo red epidermis substrate under eco‐friendly conditions (35 °C and pH 8), and compared to commercial dehairing enzyme. Cp showed the double of keratinolytic activity (10 keratin/casein units) compared to Vq and the commercial dehairing enzyme (5 keratin/casein units). However, an excessive keratinolytic activity could contribute to skin damage. Besides, three enzymes had a similar collagen activity (around 2300 HPA/casein unit). The result most notable was observed in elastinolytic activity, where Cp and Vq exhibited lower values (20 and 10 elastin/casein activity, respectively) than those the commercial dehairing enzyme (98 elastin/casein activity). In this way, the hide treated with Cp and Vq shows higher softness and flexibility. Regarding epidermis activity, Cp and Vq showed almost 70 % of that of the commercial enzyme activity, indicating that the dehairing process was less efficient. Comparing both plant proteases, Cp was more efficient than the Vp preparation, producing hair‐free hides with clean pores and without significant damage on the grain surface. Moreover, cowhide treated with Vq was rougher than that treated with Cp. Further studies are required to understand the relationship between the different substrate activities (all increased with temperature) to improve the efficiency of the dehairing process using Cp and Vq.
Another protein extract with promising results is those derived from Euphorbia nivulia. [95] This protease preparation is stable between pH 5 to 8 and up to 60 °C (75 % residual activity), showing high compatibility with metal ions, detergents, oxidizing agents, surfactants, and organic solvents. A preliminary dehairing study was performed at 30 °C and pH 7 for 18 h without sodium sulfate. The results indicated that the protease extract completely removed of fine hairs of goat hide (no major data was reported). Only a few reports with quantitative data of plant proteases on the unhairing process are found in literature CpLP and Cp are the most promising plant proteases. However, supplementary researches are needed to understand the crucial relationship between the selectively towards keratin, collagen, and elastin substrates in the skin to develop a potential enzyme procedure substitute of lime‐sulfide treatment.
4.1.2. Hydrolysis of Protein‐Based Substrates
A huge amount of waste rich in protein substrates is wasted in the fishing, dairy, food, and bakery industries. Plant proteases have emerged as an affordable technology to valorize these wastes via hydrolysis in order to obtain high value‐added products such as bioactive peptides. An updated review of the hydrolysis of cheese whey, keratinous materials from poultry feathers (keratin, collagen, and gelatin), gluten and soy and legume proteins, and fish is provided in the next section.
4.1.2.1. Dairy Industry: Valorization of Cheese Whey, Milk Hydrolysates, and Milk‐Clotting
Cheese production has grown at a rate of 3 % per year in the last 15 years, reaching a production volume around of 180–190 million tons/year. [96] Cheese production involves the generation of several byproducts, one of the most abundant is cheese whey (CW). An average of 90 % of the mass of milk used in cheese making is extracted in form of the whey and approximately half of total volume is discarded without the treatment required, thus generating serious pollution problems and environmental concerns. The discharge of these effluents into bodies of water without the necessary treatment has a negative impact on aquatic life due to dissolved oxygen depletion. [97]
The wastewater whey volume also depends on the animal milk used, with cow‘s milk showing the highest effluent compared to sheep and goat milk (Table 7). Milk coagulation can produce two types of whey. Acid whey (pH <5) is obtained from fermentation or addition of organic or mineral acids, and sweet whey (pH 6–7), which is produced during the addition of proteolytic enzymes like chymosin. [98] Table 8 shows the protein composition of acid and sweet whey, respectively.
Table 7.
Volume of whey generated and yield to cheese for milks from cow, goat, sheep and camel. [100]
|
Parameter |
Milk |
|||
|---|---|---|---|---|
|
Cow |
Goat |
Sheep |
Camel |
|
|
Cheese yield (kg/100 kg milk) |
9.86 |
14.78 |
9.84 |
10.12 |
|
Whey volume (L L−1) |
0.873 |
0.822 |
0.873 |
0.834 |
Table 8.
Protein composition of acid and sweet whey. [96]
|
Protein |
Sweet whey[a] |
Acid whey[b] |
|---|---|---|
|
β‐Lactoglobulin (β‐Lg) |
46 % |
44 % |
|
Peptide fraction |
30 % |
40 % |
|
α‐Lactalbumin (α‐La) |
16 % |
11 % |
|
Immunoglobulin (IgG) |
5 % |
3 % |
|
Others |
4 % |
3 % |
[a] pH 6.5, [b] pH 3.5.
Cheese whey is a heterogeneous mixture of different proteases: β‐lactoglobulin (β‐LG), α‐lactalbumin (α‐LA), immunoglobulins (Igs), bovine serum albumin (BSA), lactoferrin (Lf), and peptide‐fraction. Cheese whey also can contain lactoperoxidase (LP), proteose‐peptone, and glycomacropeptide (GMP). The composition of whey depends on procedures used for casein removal from milk. The studies related to the kinetics of hydrolysis have been focus on the major substrates (β‐LG and α‐LA). A simplified mechanism has been proposed with satisfactory reproducibility of experimental data adopting the Michaelis–Menten model. First, the plant protease bond to the substrate forming an intermediate in equilibrium with milk protein and the enzyme. Then, the enzyme cleaves the protein chain according to the nature of its active site. The hydrolysis β‐LG and α‐LA occur simultaneously. [99] It was also found that the kinetic of whey protein is sensitive to enzyme concentration, the composition of whey, pH, and temperature.
Approximately 55 % of the total organic and inorganic compounds in milk are retained in cheese whey, which make it an attractive raw material for the production of economically valuable products.
Based on the absence of any toxic agent, the following approaches have been used for cheese whey valorization: prebiotic galacto‐oligosaccharides from lactose, generation of bioethanol, production of high‐grade lactose for pharmaceutical purposes and hydrolysates production, protein concentrates, lactose and organic acids, among others. Cheese whey is also used in sports drinks, nutritional products, and high protein preparation. [101]
Cheese whey valorization appears as a huge field of application for plant proteases. Plant proteases, especially cysteine, are selective to casein substrates and many of them have been traditionally used for milk‐clotting. The use of plant proteases for cheese whey hydrolysis permit to obtain hydrolysates with functional properties (emulsifiers and foaming agents), nutritional foods and bioactive peptides with beneficial properties. [40] Madureira et al. reviewed the main biological properties of whey proteins and peptides, including benefit on the immune system (e. g., antimicrobial, immunomodulation, and cytomodulation activities), on the cardiovascular system (e. g., antihypertensive, antioxidant, and antithrombotic activities), on the nervous system (e. g., opioid agonists and antagonists) and on the gastrointestinal system, (e. g., anti‐appetitive and mineral‐binding vectors).[ 102 , 103 ]
The obtention of biopeptides for health care through whey protein hydrolysis using is gaining interest. Especially plant proteases isolated through simple procedures represent an affordable technology to obtain these biopeptides. Protein extracts from melon fruit (Cucumis melo), trompillo berries (Solanum elaeagnifolium), and citrus flowers (Citrus aurantium) have been tested for the production of bioactive peptides with the capacity to inhibit angiotensin‐converting enzyme (ACE) for hypertension treatment. [104] It has been observed that enzymatic crude extract from trompillo and melon fruit are selective towards β‐Lg and α‐La hydrolysis in sweet whey (pH 6.5, 60 °C, 5 : 95 (v/v) enzyme/substrate ratio). Trompillo almost completely degraded β‐Lg and α‐La after 24 h, whereas, only 30 % β‐Lg was hydrolyzed by melon proteases at the same time.
Lactoferrin (Lf), bovine serum albumin (BSA), and immunoglobulin (IgG) are minimally hydrolyzed for both enzymes. The similar enzymatic behavior exhibited by both plant protease extracts (trompillo and melon) may be attributed to the high concentration of serine proteases in each extract. On the other hand, citrus proteases showed minimum hydrolysis of sweet whey, while they exhibited higher bioactivity for acid whey, favoring the degradation of Lf, BSA, α‐La, and IgG under the same test conditions (no quantitative hydrolysis data was reported). This enzymatic behavior can be understood in view of the fact that citrus proteases have optimal proteolytic activity under acidic conditions (3.0–4.5). The ACE ‐ inhibitory activity of peptide sequences derived from CW hydrolysis using melon, trompillo, and citrus flower proteases was in the range of 85–90 %, 75–80 %, and 32–43 %, respectively. The results indicated that these proteases show potential for the production of hydrolysates for hypertension treatment.
Promising results were also documented using Maclura pomifera proteases latex. [105] The enzymatic preparation (also called pomiferin) showed a caseinolytic activity of 14.1 U mL−1, with a protein concentration of 1.5 mg mL−1. Ucas is an enzyme unit (caseinolytic unit) defined as the amount of protease producing an increment of one absorbance unit per min. The extract exhibited the highest proteolytic activity towards whey hydrolysis at 45 °C and pH 6. A linear correlation was observed between the hydrolysis time and the enzyme dissolution from 30 to 45 °C. When no dissolution was performed (10 (v/v) enzyme/substrate) the reaction took place almost instantly. Meanwhile, when the factor dissolution was 20 the reaction time was between 40 (45 °C) and 190 s (30 °C). Pomiferin hydrolyzed milk protein into small peptides (19–25 kDa), which exhibited antioxidant activity and ACE‐inhibitory activity of 57 % and 11 %, respectively. Thus, milk hydrolysates could be used for therapeutic applications.
In literature, a few reports of milk hydrolysates production using vegetable enzymes at pilot scale were found. Tavares et al. investigated the valorization of cow whey using selective filtration techniques (from ultrafiltration to nanofiltration), in combination with hydrolysis using proteolytic enzymes from Cynara cardunculus aqueous extracts. [106] The optimal hydrolysis conditions were: enzyme/substrate ratio of 1.6 % v/v, pH 5.2, 55 °C, and incubation time of 7 h. The results indicated that 87 % of α‐La was hydrolyzed, while no degradation of β‐Lg was registered. The final products of the process were a bioactive peptide concentrate, an hydrolysates fraction with molecular weight lower than 3 kDa and β‐Lg rich fraction, containing 73, 43, and 91 % w/w protein, respectively. All these fractions were low in lactose and salt. Furthermore, the small peptide fraction showed potential antihypertensive activity (ACE‐inhibitory activity of 710 μg mL−1 according to the IC50 method).
Highly hydrolyzed milk protein also could be used ingredients for infant preparations and therapeutic baby foods. From a nutritional perspective, the absorption of milk proteins hydrolysates and short‐chain peptides in the gastrointestinal tract is better compared to the absorption of high‐molecular‐weight proteins contained in milk. Thus, milk hydrolysates contribute to reducing the residual antigenicity (amount of undigested protein with the capacity to interact with antibodies) in foods. [107] In general, hydrolysates products are between 103 to 106 times less antigenic than the native proteins. [108]
Antimicrobial peptides are another valuable product derived from milk hydrolysis. These peptides commonly contain from 12 to 50 residues, including at least two positively charged residues and a remarkable proportion of hydrophobic residues (generally >50 %). The cationic side chains of basic residues (e. g., arginine, lysine, and histidine) interact with the negatively charged membranes cell of bacteria, including lipopolysaccharide. In comparison to traditional antibiotics to treat microbial infections, antimicrobial peptides derived from casein and milk hydrolysis show the capacity to attack target cells rapidly over different types of bacteria. [109] The peptides derived from bovine milk hydrolysis employing root latex proteases of Jacaratia corumbensis (Caricaceae family) have been shown potential antimicrobial. [110] The proteases exhibited high proteolytic activity (2.5 ⋅ 10−4 U mg−1) at room temperature and pH 7.6, leading to the formation of hydrolysates peptides, which were consecutively purified. The hydrolysates obtained after 2 h hydrolysis showed efficient antimicrobial activity against Enterococcus faecalis, Bacillus subtilis, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae and Staphylococcus aureus. The antimicrobial test was performed following the Minimum Inhibitory Concentration method (MIC). From the study was obtained a MIC of 50 mg mL−1 for all bacteria previously cited, except for Staphylococcus aureus, with a value of 40 mg mL−1 (107 Colony Forming Units/mL).
Similar therapeutic uses could be reached from hydrolysates obtained using proteases from Araujia hortorum (Asclepiadaceae). These proteases have simple and economic procedure extraction and high caseinolytic activity.[ 111 , 112 ] Quiroga et al. studied the synthesis of peptides in aqueous‐organic media catalyzed by proteases from Araujia hortorum latex at pH 9. [113] The activity of enzymatic preparation in 50 % (v/v) ethyl acetate, 50 % (v/v) hexane, 50 % (v/v) propanone and N,N‐dimethylformamide was 1.96, 7.84, 9.52, and 40.9 Ucas mg−1 of protein, respectively. The protease extracts were selective for generating short peptides, especially Z−Ala−Phe−OMe, which could be used in therapeutic applications.
The composition of milk hydrolysates peptides, and consequently their potential uses, depends on protease nature and reaction conditions, as well the composition of raw material. Table 9 shows the composition of cow, goat, sheep, and camel milk. Cows are the main source of milk worldwide, meanwhile, small ruminants (sheep and goat) produce around 3.5 % of the world's milk. In some cases, sheep and goat milk are suitable alternatives for persons with cow milk allergies. Furthermore, goat milk contains larger proteins, smaller fat globules, and shorter fatty acids than cow milk. On the other hand, sheep milk is rich in proteins and minerals (e. g., calcium, phosphate, and magnesium), showing higher amount of polyunsaturated fatty acids (PUFA) and conjugated linoleic acids (CLAs) compared to goat milk. [114] Camel milk exhibits less amount of lactose and higher level of β‐CN, α‐La, and serum albumin compared to cow milk. Besides, camel milk is rich in iron, magnesium, riboflavin (vitamin B2), folic acid (vitamin B9), vitamin B11, vitamins C and D, and fatty acids. Furthermore, caseins represent between 75–80 % of the total protein of camel milk.[ 114 , 115 ]
Table 9.
|
Compound [g/100 g] |
Milk |
|||
|---|---|---|---|---|
|
Cow |
Goat |
Sheep |
Camel |
|
|
Moisture |
87.9 |
87.6 |
82.9 |
86.9 |
|
Fat |
3.7 |
4.3 |
6.1 |
4.1 |
|
Lactose |
4.7 |
4.1 |
4.8 |
4.1 |
|
Protein |
3.4 |
3.7 |
5.5 |
3.4 |
|
Casein |
3.0 |
2.4 |
4.7 |
2.1 |
|
Ash |
0.7 |
0.8 |
0.9 |
0.7 |
|
Minerals [mg/100 g] |
|
|||
|
Potassium |
145.0 |
185.0 |
138.0 |
60 |
|
Calcium |
112.0 |
130.0 |
197.5 |
117 |
|
Phosphorous |
91.0 |
109.0 |
141.0 |
51.5 |
|
Sodium |
42.0 |
39.5 |
39.0 |
69 |
|
Magnesium |
11.0 |
14.5 |
19.5 |
14.2 |
|
Zinc |
0.40 |
0.43 |
0.60 |
Trace |
|
Iron |
0.10 |
0.06 |
0.10 |
0.39 |
|
Copper |
Trace |
0.04 |
0.10 |
Trace |
|
Vitamins [mg/100 g] |
|
|||
|
Vitamin A |
37.0 |
54.3 |
64.0 |
35.4 |
|
Riboflavin |
0.20 |
0.17 |
0.30 |
0.57 |
|
Vitamin E |
0.08 |
0.04 |
0.11 |
0.07 |
|
Thiamin |
0.04 |
0.06 |
0.07 |
0.04 |
|
Retinol |
35.0 |
0.04 |
64.0 |
27.0 |
|
Carotenoids |
16.0 |
Trace |
Trace |
Trace |
|
Folic acid |
8.5 |
1.0 |
6.0 |
8.7 |
|
Vitamin C |
1.0 |
1.3 |
4.6 |
4.5 |
|
Vitamin D |
0.2 |
0.15 |
0.2 |
0.5 |
|
Vitamin B12 |
0.50 |
0.06 |
0.66 |
0.85 |
|
Fatty acids [g/100 g] |
|
|||
|
C16 : 0 |
27.9 |
28.2 |
25.9 |
29.8 |
|
C18 : 0 |
12.2 |
8.8 |
9.6 |
12.9 |
|
C18 : 2 (n‐6) |
1.4 |
3.2 |
3.2 |
3.1 |
|
C18 : 3 (n‐3) |
1.0 |
0.42 |
0.80 |
1.4 |
|
Total SFA[a] |
68.7 |
68.7 |
64.2 |
58.9 |
|
Total PUFA[b] |
4.0 |
3.7 |
4.8 |
4.5 |
|
Total CLAs[c] |
1.1 |
0.70 |
1.6 |
3.1 |
[a] SFA=saturated fatty acids, [b] PUFA=polyunsaturated fatty acids, [c] CLAs=conjugated linoleic acids.
The protein content and vitamin composition of camel milk have encouraged the production of small bioactive peptides with, antioxidant and antimicrobial properties. Al‐Shamsi et al. explore the enzymatic hydrolysis of camel milk hydrolysates using bromelain and papain. [116] The protein hydrolysis was performed at 50 °C under constant stirring in a water bath using an enzyme/substrate ratio of 1 : 100 (wt/wt) at neutral pH. After 6 h of reaction, a hydrolysis degree of 24 % and 40 % was reached using bromelain and papain, respectively. The biopeptides (<14 kDa) showed notable antioxidant improvement compare to the camel milk source. DPPH (2,2‐diphenyl‐1‐picrylhydrazyl), ABTS (2,2‐azinobis 3‐ethylbenzthiazoline‐6‐sulfonic acid) radical scavenging, and ferrous Iron‐Chelating activity activities were increased 50 % and 33 %, 4.75 and 11.75 times, and 21 % and 2 %, using bromelain and papain, respectively. Similar observations were reported by Wali et al., who isolated specific antioxidant peptides produced during camel milk hydrolysis using papain. [117] The optimal conditions reported were: pH 5.8, 50 °C and an enzyme concentration of 5 kU g−1 protease. In addition, the isolated peptides showed comparable ABTS radical scavenging activity to the results reported by Al‐Shamsi et al. [116]
Cheese production is considered one of the most important activities in the dairy industry. About 80 % of world cheese production (20 million tons) is derived from cow‘s milk. [121] Cheese manufacturing from milk is a complex process, being enzymatic coagulation, the first step. [122] During milk clotting, caseins are separated as a coagulum and the whey is released (fat trapped in the gel network). Plant proteases have been traditionally used for milk clotting in cheesemaking manufacturing. [122] The presence of enzymes in coagulants preparation determines the final gel firmness, curd draining properties, moisture content, texture, and flavor of the final product. The catalytic mechanism of most plants extracts with milk‐clotting activity initiate with the hydrolysis of the casein micelle‐protective protein (k‐CN), in particular with the breakdown of the Phe105‐Met106 peptide bond. The glycomacropeptide (f106‐169, hydrophilic portion) is then released from the casein micelle surface, decreasing the electrostatic and steric repulsion between the micelles, thus causing casein aggregation and clot formation. The clot formation is commonly observed when 70–80 % of κ‐CN is hydrolyzed. The milk‐clotting activity depends on protease type or extract composition, specificity, and optimum conditions (enzyme concentration, pH, temperature, and calcium ions). Commonly, calcium ions are added to the milk in the form of CaCl2 (10–40 g CaCl2 100 kg−1 milk) to favor the formation of the gel matrix. [122]
A paramount parameter of enzymatic coagulation is the milk‐clotting/proteolytic activity ratio (MCA/PA). Proteases with high values of MCA/PA ratio favor the production of cheese with suitable textural and flavor features. In contrast, proteases with a low MCA/PA ratio are associated with weak curds, higher protein losses in the whey, soft texture, and bitter cheeses.[ 10 , 122 ] Industrial milk coagulation traditionally is carried out at 32–37 °C and pH 6.3‐6.8 using the chymosin enzyme. [10] Crude extracts from kiwifruit, melon mesocarp and ginger rhizome exhibited a higher MCA/PA ratio than chymosin, favoring the rheological properties of the final product. [104] Table 10 shows the milk‐clotting activity and the MCA/PA ratio of crude extracts and plant purified enzymes in comparison with the chymosin enzyme. The higher MCA/PA ratio shown by different plant proteases compared to chymosin represent their potential to be applied for milk coagulation at an industrial scale. In the case of Bromelia penguin, their extracts are able to hydrolyze selectively the κ‐CN in the Phe105‐Met106 peptide, leading to the formation of amino para‐kappacasein fragment (F1‐105 κ‐CN) and glycomacropeptide (F106‐169 κ‐CN). In contrast, cardosin A and B are selective towards the hydrophilic sections αs1‐CN (Phe23‐Phe24, Phe153‐Tyr154, and Typ164‐Tyr165) and β‐CN (Leu127‐Thr128, Leu165‐Ser166, and Leu192‐Tyr193).
Table 10.
Milk‐clotting activity and the MCA/PA ratio of plant preparations in comparison with chymosin enzyme.[ 122 , 124 , 125 ]
|
Enzyme or extract |
Milk‐clotting activity (MCA) |
MCA/PA[a] |
Optimum conditions |
|
|---|---|---|---|---|
|
T [°C] |
pH |
|||
|
Crude extracts | ||||
|
Kiwifruit mesocarp (A. deliciosa) |
2.7 U mg−1 |
5 (104) |
40 |
7 |
|
Rhizome (Z. officinale) |
2.3 U mg−1 |
3.2 (162) |
60 |
7 |
|
Melon mesocarp (C. melo) |
1.5 U mg−1 |
2.5 (208) |
40 |
7 |
|
Cardoon flowers (C. cardunculus) |
61 IMCU mL−1 |
NA |
50 |
6.5 |
|
Fruit mesocarp (B. pinguin) |
2.59 U mg−1 |
1.3 (162) |
50 |
7–9 |
|
Purified enzymes | ||||
|
Cardosin A (C. cardunculus) |
1160 IMCU g−1 |
NA |
40 |
6.6 |
|
Cardosin B (C. cardunculus) |
7556 IMCU g−1 |
NA |
40 |
6.6 |
|
Actinidin (A. deliciosa) |
1 RU mg−1 |
0.46 (10.2) |
37 |
6.5 |
|
Cynarase C (C. scolymus) |
43,000 IMCU g−1 |
34.87 (2.7) |
60 |
5.5 |
|
Dubiumin (S. dubium) |
3520 U mg−1 |
2490 (2) |
70 |
11 |
[a] Values in parenthesis in MCA/PA represent the relative ratio of MCA/PA between the plant enzyme and chymosin. NA: not available.
Plant extracts can exhibit high proteolytic activity. From an overview, a high rate of κ‐CN involves a lower clotting time; however, an excessive activity could generate the degradation of the casein network, thus decreasing the curd yield. Similar observations have been made when milk clotting temperature is near 70 °C, where the over‐proteolysis negatively affects the fat transfer from milk to casein curd. Enzyme concentration, temperature, and coagulation time are relevant parameters to be controlled to guarantee the quality during cheesemaking. [123]
Plant proteases for milk clotting can be obtained directly from organs of the plant as is the case of C. scolymus (stigmas), Moringa oleifera (dried flowers), Silybum marianum (fresh flowers), Solanum dubium (seeds), and Solanum elaeagnifolium (berries). [122] Proteases for milk‐clotting produced via in vitro techniques are an alternative to obtention of proteases from organs. As well callus suspension (Cynara cardunculus and Centaurea calcitrapa), as callus (Mirabilis jalapa, Silybum marianum, Cynara cardunculus, and Mirabilis jalapa) can be used. [10]
Promising results have been obtained using new plant proteases. Proteases derived from Solanum elaeagnifolium and Solanum dubium have been demonstrated high milk‐clotting activity with 39.4 and 880 milk‐clotting units (MCU) at 32 °C and 37 °C, respectively. [126] Dubiumin protease showed high resistance to different salt concentrations, retaining its activity at 60 °C after 1 h. Hieronymain (Bromelia hieronymi) efficiently degraded κ‐casein, α‐lactalbumin and β‐lactoglobulin at 60 °C after 10, 30 and 60 min, respectively. [22] Onopordosin (Onopordum acanthium) exhibited a similar degradation behavior to chymosin over αs1‐ casein and β‐casein (pH 2.5). [31]
In summary, new plant proteases show comparable or even superior milk‐clotting activity and MCA/PA ratio compared to chymosin. Further studies at larger scales are required to analyze the feasibility of their use in cheese making.
4.1.2.2. Hydrolysis of Gluten and Obtention of Bioactive Peptides
Over the last decades, there has been a growing interest in the obtention of antioxidant compounds from vegetable sources using efficient and low‐cost procedures. Corn and wheat gluten hydrolysis have emerged as promising routes for obtaining antioxidants. The bakery industry demands around 60 % gluten worldwide market; besides, gluten is also used for meat processing, dry breakfast, dairy products, and seafood preparation. The large volumes of gluten wasted in the bakery industry have encouraged the research for new applications. [127]
Gluten is generally isolated during starch manufacture. After drying, gluten is a powder with a neutral taste, containing around 75–80 % protein, 5–10 % moisture, and minorities compounds as starch, fats, and non‐starch polysaccharides.
Wheat gluten contains proteins that form either monomers or oligo‐ and polymers linked by disulfide and non‐covalent bonds. The proteins are categorized according to their solubility in water‐alcohol solution into gliadins (50‐60 %) and glutenins (40‐50 %). Gliadins are single‐chain proteins (or monomeric), which are grouped into α‐, β‐, γ‐, and ω‐fraction on the basis of N‐terminal amino acid sequence. Gliadin fractions, with exception of ω‐class, contain cysteine proteins, which forms disulfide bonds with other peptide chains. Regarding molecular weight, ω‐gliadin contains proteins between 46 and 75 kDa, whereas α‐, β‐, γ‐fraction exhibits from 30 to 45 kDa. On the other hand, glutenins are made up of protein chains linked by disulfide bonds forming polymers. Three groups of glutenin are distinguished: high molecular weight group (HMW, 65–90 kDa), medium molecular weight group (MMW, 50–65 kDa), and low molecular weight subunit (LMS, 30–60 kDa). The gluten commonly exhibits a high and low molecular weight subunits (HMS and LMS) with an HMS/LMS ratio of 0.25. Gluten contains a complex mixture of HMS and globular (LMS and monomeric gliadins) proteins. [127]
The average amino acid composition of commercial gluten is given in Table 11. Gluten is rich in glutamine, which favor the formation of hydrogen bonds between the protein chains, thus reducing the water solubility because of the presence of hydrophobic amino acids on the proteins surface, especially proline. The low solubility of gluten protein in water can be increased using urea, lactic acid, acetic acid, sodium hydroxide or ethanol (70 %). When gluten comes into contact with an aqueous medium, the interaction between glutenins and gliadins leads to the formation of a similar sponge‐net structure, with particular functional properties (viscoelasticity, water‐holding, gelling, foaming, and fat emulsifying). [128] Furthermore, in aqueous medium when gluten proteins are heated, cysteine residues are generated due to protein denaturation. Even glutenins and gliadins can form cross‐links at temperatures above 70 and 90 °C. The gluten hydrolysis leads to the formation of peptides with substantial capacity of foaming, emulsification, and rheological properties modifications of the dough. With this purpose, the hydrolysis of gluten needs to be carried out under strictly defined conditions to reach the desired organoleptic and functional properties (pH 6.5 and 0.2 % wt. NaCl). [129]
Table 11.
Amino acid composition of commercial gluten (adapted from Ref. [130]).
|
Amino acid |
wt.% |
Amino acid |
wt.% |
|---|---|---|---|
|
Glutamine/glutamic acid |
37.5 |
Arginine |
3.6 |
|
Proline |
12.0 |
Threonine |
2.5 |
|
Leucine |
6.7 |
Cysteine |
2.5 |
|
Phenylalanine+tyrosine |
8.3 |
Alanine |
2.3 |
|
Serine |
4.7 |
Methionine |
2.4 |
|
Valine |
3.9 |
Histidine |
2.1 |
|
Isoleucine |
3.8 |
Lysine |
1.6 |
|
Asparagine/aspartic acid |
3.1 |
Tryptophan |
1.1 |
|
Glycine |
2.8 |
Plant proteases exhibit a high affinity towards gluten protein, considering that, during germination, gluten proteins are degraded to supply the developing embryo with amino acids and nitrogen. Plant protease shows the following upsides for gluten hydrolysis: (i) high stability (pH: 4–9 and T: 30–60 °C) and bioactivity, (ii) cleavage specificity naturally optimized, (iii) they are food‐grade, (iv) isolation using simple procedures (e. g., ammonium sulfate precipitation), (v) they can be integrated into the production process in a relatively simple manner (by adding to the gluten manufacturing), (vi) and low production cost. [131] The hydrolysis mechanism of gliadins and glutenin proteins remains unclear. It has been postulated that, in the first stage, the depolymerization of glutenin macropolymers occurs through the breaking of disulfide bonds, which increases the solubility of the substrate. In secondary proteolysis, the exposed gliadin and glutenin protein are attacked, forming larger protein chains. Finally, the chains are converted into small peptides and amino acids. The cleavage amino acid depends on enzyme specificity.[ 128 , 129 ]
Health concerns have also encouraged the development of new alternatives for gluten hydrolysis. Celiac disease (CD) is an inflammatory disorder of the digestive system caused by the intolerance of the immune system against gluten and gliadin proteins (glutamine and proline) in products (wheat, barley, and rye).[ 132 , 133 ] It is estimated that 1.4 % of the world population is affected by CD, which is characterized by severe immune damage to the intestinal mucosa with gluten consumption. [134]
Promising results to reduce the gluten allergenicity through enzymatic hydrolysis using Nigella sativa proteases have been documented. Gabr tested the activity of protease extract purified from Nigella Sativa in gluten and gliadin hydrolysis. [135] The enzymatic extraction was carried out using ammonium sulfate, acetone and trichloroacetic acid as solvents. According to the results obtained, the highest protein purity was reached using acetone 80 % and 0.2 m trichloroacetic acid. The enzymatic tests were performed at pH 6.9 (phosphate‐citrate buffer), 31 °C, and 2 % by weight of gluten or gliadin, with a reaction time of 2 h. The protease preparation showed to be active for gluten and gliadin hydrolysis, reaching a conversion of 28.5 % and 32.0 % for gluten, and 22.7 % and 16.8 % for gliadin, using an enzymatic concentration of 1 and 2 mg mL−1, respectively. Bellir et al. studied the performance of Nigella sativa seed crude extract on Triticum aestivum and Triticum durum gliadin hydrolysis. [136] The extract exhibited a maximum casein activity of 84.26 U g−1 (50 °C and pH 1.5). Meanwhile, the enzymatic hydrolysis was carried out at pH 7.5 and 37 °C with 2 h incubation. Under these conditions, the activity reported for Triticum durum were 19.7 and 73.1 U g−1 and for Triticum aestivum 69.3 and 101.8 U g−1, using crude enzyme extract and partially purified enzyme, respectively. The results indicated that Nigella sativa could be used to detoxify gliadin protein in gluten.
Another protease with remarkable selectivity towards gliadin hydrolysis is Neprosin, which is obtained from Nepenthes pitcher. Only preliminary studies have been performed, reaching gliadin hydrolysis around 58 % after 80 min (37 °C, pH 7.5 and enzyme concentration 10 mg mL−1) showing the capacity to detoxify gluten under mild conditions. [137] It was also reported that cumin (Cuminum cyminum latex) could increase the proteolytic activity of pepsin (up to 400 %) in hydrolysis reaction, however, no experiments with gluten protein have been reported.
Caricain (Carica Papaya latex) is a potential alternative protease for gluten degradation. Buddrick et al. studied the ability of caricain preparation (crude and enriched through ion‐exchange chromatography) to detoxify gliadin in whole wheat flour. [138] The enzymatic formulations were incorporated during bread dough preparation. The reduction of gliadin in bread samples treated with crude enzyme was 65.7 %, using an enzymatic concentration of 0.03 % wt. at 37 °C after 5 h. Meanwhile, a gliadin reduction of 97.8 % was reached using purified caricain under the same operating conditions. Thus, caricain protease extracts could be directly incorporated during bread dough preparation to develop products suitable for celiac and gluten intolerant individuals.
The combination of heat treatment with enzymatic hydrolysis could have a synergic effect to reach a higher gluten hydrolysis degree. The gluten hydrolysis using papain with heat treatment was investigated by Wang et al. [139] First, wheat gluten was incubated in a water bath with a temperature between 50 and 90 °C for 10 to 60 min. The heat treatment produced the denaturation of the gluten protein due to the breaking of the sulfide bonds, reducing the free SH‐contents up to 60 % (90 °C for 60 min). In a second step, hydrolysis was carried out using papain for 6 h at 45 °C and pH 6.5 with a ratio of enzyme to gluten proteins of 1500 U g−1 (optimal conditions). The heat treatment at 70 °C for 30 min in combination with the hydrolysis under optimal conditions produced peptides with different molecular weights. Peptides with >1 kDa (68 %), between 5 and 10 kDa (27 %) and the rest <5 kDa. The approach of employing plant proteases in combination with heat treatment is a valuable alternative to produce protein hydrolysates for nutritional supplements, functional ingredients, flavor enhancers, coffee whiteners, cosmetics, personal care products, fortification of soft drinks and juices and improving the functionalities of food proteins.
In addition to caricain, papain (also derived from Carica Papaya) has been demonstrated potential on gluten hydrolysis. Xue et al. reported that gluten hydrolysis using papain leads to the modification of the conformational structures of gliadins, especially the secondary structure. [140] The enzymatic tests were carried out at 60 °C, pH 4.6, substrate concentration of 80 g gluten L−1, and a papain/gluten ratio of 200 U g−1. The authors observed a reduction of β‐sheet (22 %), a slight increase in β‐turn (3 %) α‐helix (2) for a hydrolysis degree of 20 %. The gluten hydrolysates showed a significant allergenicity reduction in different tests (Index of Spleen: 6 mg g−1, concentration of serum histamine: 15 ng mL−1, and serum gliadin‐specific IgE level: 0.9). On the other hand, Li et al. carried out the optimization of wheat gliadin hydrolysis using papain, alcalase (B. licheniformis), flavourzyme (Aspergillus oryzae), α‐chymotrypsin (bovine pancreas), trypsin (bovine pancreas), and pepsin (porcine gastric mucosa). [141] The optimal conditions for papain were: enzyme/substrate ratio of 0.26 %, hydrolysis time of 32 min, pH 7.0 and 48 °C, reaching almost 100 % reduction of gliadin. Papain exhibited higher activity compared to the other enzyme. The combination of alcalase‐papain treatment was the most effective procedure to reduce allergen of wheat flour (IgE‐Binding Inhibition: 80 %. Alcalase and papain have different preferred cleaving sites, producing more peptide bonds than a single enzyme.
The consumption of kiwifruit could contribute to reducing gluten allergenicity. Kaur et al. reported in an in vitro study that actinidin protease from green kiwifruit enhances the gluten protein degradation in the small intestine. [142] In subsequent work, Jayawardana et al. investigated the effect of actinidin on the hydrolysis of gluten proteins and digestion‐resistant gluten peptides (synthetic 33‐mer peptide and pentapeptide epitopes) under static simulated gastrointestinal tract conditions (37 °C, pH 4). [143] The results indicated that actinidin efficiently hydrolyzed gluten protein and resistant peptide bonds adjacent to proline residues in the 33‐mer peptide. The hydrolysis degree was 25 % for gluten and 21 % for gliadin under optimal conditions (pH 6 and actinidin concentration of 4.4 U mL−1 for 60 min). The gastric degree rate of gliadin hydrolysis by actinidin (0.8 %/min) was higher compared to papain or bromelain (on average 0.4 % min−1). These results are in concordance with the observations reported by Jayawardana et al. in a posterior work, where it was stated that gluten intolerance could be minimized through co‐consumption of papaya, pineapple, and green kiwifruit. [144]
Previous studies have reported that oral intake of gluten hydrolysates has beneficial effects on therapeutic treatments, for instance, muscular injuries. In this way, Taga et al. explored the production of a novel type of wheat gluten hydrolysate using ginger protease. [145] The hydrolysis test was carried out at 50 °C, pH 5.2, and with an enzyme concentration of 30 mg mL−1 (100 mm sodium acetate buffer) for 16 h. Under these conditions, ginger protease efficiently hydrolyzed gluten, producing peptides with an average molecular weight lower than 600 Da. The reaction pH had a negative effect on hydrolysates solubility (54.1 % at pH 5.6, 34.4 % at pH 6.0, and 25.7 % at pH 6.4) compared with that at acidic pH (65.2–67.7 % at pH 3.6–5.2). Besides, gluten hydrolysate contained considerable amounts of tripeptides, including Gln‐Pro‐Gln, Gln‐Pro‐Gly, Gln‐Pro‐Phe, Leu‐Pro‐Gln, and Ser‐ Pro‐Gln (40.7 mg g−1 at pH 5.2), which showed remarkable inhibitory activity on dipeptidyl peptidase‐IV with IC50 values of 79.8, 70.9, 71.7, 56.7, and 78.9 μm, respectively. Thus, the hydrolysates produced using ginger protease could be applied in type II diabetes treatment.
Another important source of gluten is corn, which is one of the most important crops in the world with a protein content of around 10–15 %. The corn seeds are rich in zein (68 %) and glutelin (28 %), meanwhile, Lys and Trp amino acids are presented in low proportion. Corn gluten meal (CGM) is a high protein co‐product (60‐70 %) generated during the wet‐milling of corn. Corn gluten meal is a suitable source of antioxidative hydrolysates taking into account its high protein content. Hu et al. investigated the hydrolysis of this substrate employing papain, ficin, and bromelain. [146] The hydrolysis assays were performed at 50 °C, using a pH between 5 and 6.5, with an enzyme amount/corn protein ratio of 40, 225.5, and 150 mg g−1, for papain, ficin, and bromelain, respectively. The highest hydrolysis degree was 16 % for papain (4 h) and 12 % (5 h) for ficin and bromelain. The hydrolysates fractions presented a molecular weight from 1 to 10 kDa, showing high antioxidant activity. The antioxidant yield reached after 5 h was 80 %, 73 %, and 31 % for ficin, bromelain and papain, respectively. Besides, DPPH scavenging rate of the hydrolysates obtained using ficin, bromelain and papain (reaction time 5 h) was 68 %, 78 %, and 74 %, respectively. According to the results, CGM hydrolysates could be applied as antioxidants in food preparations to prevent lipid oxidation and to enhance the storage stability of the product.
Plant proteases could also be used to hydrolyze rice bran, which contains around 12–20 % high‐quality protein, showing hypoallergenic properties. Enzymatic hydrolysis of rice bran protein is an alternative to enhance the functional properties of rice bran, increasing the protein solubility in water medium, and consequently, their extractability. Apinunjarupong et al. investigated the rice bran hydrolysis using bromelain enzyme. [147] The optimal conditions were: 6 % bromelain concentration, pH 9 and 50 °C, achieving a hydrolysis degree of 36.5 % after 30 min. Thus, bromelain could be used under alkaline conditions for the production of biopeptides with hypoallergenic functions for food preparations.
Plant proteases have high proteolytic activity towards gluten protein hydrolysis (wheat, corn, and rice) under mild conditions, being possible to obtain peptides with great antioxidant capacity and nutritional value.
4.1.2.3. Bird Feathers Valorization
The chicken meat processing industry shows a rapid growth rate around the world. This expansion is due to the fast growth rate of chickens, nutritional supply for human consumption, short production time and lower price than beef. [148] Feathers are a major component of the inedible by‐products obtained during chicken processing are considered environmentally polluting waste. The disposal techniques commonly used are incineration or burial and controlled landfilling.
Chicken feathers are an important protein resource (91 % keratin) in combination com other compounds as water (8 %) and lipids (1 %). The fibrous structure of keratin protein forms intermediate filament and filamentous polymers conforming long fibers. For this reason, keratin fiber extraction could be spun into: filamentous regenerated fibers using electro‐spinning techniques (manufacturing plastics), technical materials, textile fibers, biodegradable plastics, packaging materials, filtration and paper applications, and biofertilizers, among others. [149] The application of chicken feathers on biofertilizers composite is gaining importance considering the significant benefits in promoting plant growth promoting. [150]
The protein chains in chicken feathers are mainly composed of cysteine, glutamine, proline, leucine, and serine. [151] Table 12 shows the relative amino acid composition of keratin fibers in chicken feathers. Keratin protein exhibits low solubility and remarkable resistance to proteolytic hydrolysis because of the presence of disulfide bonds, hydrogen bonds, salt linkage and cross‐linkages. The secondary structure of keratin protein in chicken feathers contains α‐helical and β‐sheet conformations, showing higher cysteine content than α‐helix. [152] Hydrogen bonds and hydrophobic interactions generate a three‐dimensional protein structure with great stability and difficult to hydrolyze. Furthermore, post‐translational modifications, such as phosphorylation and glycosylation, favor the formation of keratin filaments, thus modifying the accessibility of the substrate to the active site during the hydrolysis process. [88]
Table 12.
Relative content of amino acids in chicken feathers (adapted from [150]).
|
Functional group |
Amino acid |
wt.% |
||
|---|---|---|---|---|
|
Positively charged |
Arginine |
4.3 |
6.57 |
6.16 |
|
Negatively charged |
Aspartic acid |
6 |
4.76 |
– |
|
Glutamic acid |
– |
9.18 |
8.76 |
|
|
Glutamine |
7.6 |
– |
– |
|
|
Hydrophobic |
Tyrosine |
– |
– |
2.43 |
|
Leucine |
2.62 |
7.48 |
7.38 |
|
|
Isoleucine |
3.32 |
4.93 |
4.28 |
|
|
Valine |
1.6 |
7.2 |
6.1 |
|
|
Glycine |
– |
7.57 |
6.31 |
|
|
Cysteine |
– |
– |
7.16 |
|
|
Alanine |
3.4 |
3.6 |
4 |
|
|
Phenylalanine |
0.86 |
4.11 |
4.4 |
|
|
Methionine |
1.02 |
0.03 |
0.025 |
|
|
Lysine |
– |
0.57 |
1.1 |
|
|
Histidine |
– |
0.02 |
0.4 |
|
|
Hygroscopic |
Threonine |
4 |
4.11 |
3.76 |
|
Serine |
16 |
13.57 |
8.92 |
|
|
Special |
Proline |
12 |
1.01 |
8.84 |
|
Asparagine |
4 |
– |
5.23 |
|
|
Tryptophan |
1 |
1.85 |
0.97 |
|
|
Reference |
|
[151] |
[154] |
[155] |
Considering the complex structure of keratins, there is increasing evidence that a mixture of enzymes is required to achieve a feasible yield and selectivity on feather degradation. The mechanism of feather hydrolysis involves the disulfide bond reduction (sulfitolysis) by keratinase in a first stage. It has been reported that the addition of 80 mm Na2SO3 addition enhances between 11 and 15 % the efficiency of the sulfitolysis process. Also, cysteinyl groups could be used as reducing agents. The breaking down of disulfide bonds causes instability in the three‐dimensional protein structure. In a second step, the proteases act over the hydrogen bonds and cleave the protein chain, producing peptides and free amino acids. The specific mechanism depends strictly on the active site of enzymes. [153] Plant proteases could be a valuable complement on enzyme preparations, mainly acting during the second step of the feather degradation mechanism. There are few works on this topic in the literature.
Mahajan et al. investigated the chicken feather degradation using latex from medicinal plants of Euphorbiaceae family, mainly Euphorbia tirucalli, Euphorbia nivulia, Euphorbia nerifolia, and Pedilanthus tithymaloides. [156] The proteolytic essays were carried out at 50 °C using phosphate buffer (0.1 m with pH 7.4). A feather degradation activity of 15 % and 25 % after 12 h was reached using crude enzyme of E. nerifolia and papain, respectively. These yields are comparable with those obtained using bacteria or fungi. [157] Similar results were reported by Jin et al., who compared the proteolytic activity of crude extracts from Fervidobacterium islandicum AW‐1, proteinase K, trypsin, and papain. [158] The test using papain was performed at pH 8 (Tris‐HCl buffer), 70 °C, with an enzyme concentration from 0.001 to 0.02 mg mL−1. Papain exhibited remarkable affinity towards casein, with a free amino acid generation proportional to protein concentration. However, the activity towards soluble keratins was negligible, showing a keratinolytic/caseinolytic activity ratio of papain was 1/10 (both Chr2_FK4 and Chr27_FK12 β keratins). The preliminary data reported indicated that plant proteases are suitable to be applied in combination with other enzymes (trypsin and proteinase K) to achieve feasible yields in keratin degradation has been reported. [159]
4.1.2.4. Hydrolysis of Collagen, Keratin, and Gelatin
Keratin is an important structural protein produced by mammals, birds, amphibians and reptiles, acting as protective barrier or mechanical support. Keratin is found in feathers, hair, nails, horns, hooves, bones, furs, claws, hides, bird beaks, skin, wool, scales, bristles, and epithelial cells of digestive organs (liver, pancreas, intestine, and gallbladder). [160] The amount of keratin depends on the source of raw material. For instance, wool contains up to 95 % of keratin, while hair and nail contain about 80 % keratin. On the other hand, bird feathers commonly contain 90 % keratin, chicken feathers stand out due to the large volume generated each year (see 4.1.2.3 section). Hooves and horns are composed of keratinous material arranged in tubular form, which is surrounded by a keratin cortex with a high content of cysteine amino acids (22 %). [161]
Keratin structure is stabilized through crosslinking of disulfide bonds, hydrogen bonding, or hydrophobic interactions. Three types of keratin proteins can be distinguished: α‐keratins (alpha helix), β‐keratin (beta sheets), and γ‐keratin. α‐Keratins are commonly found in hair, wool, horns, nails, bristles, claws, and hooves of mammals, whereas β‐keratins are found in bird feathers, beaks, and claws. [162] α‐Keratins have low sulfur content and they can be grouped into two subfamilies, type I (acidic keratins, 40–50 kDa) and type II (neutral or basic keratins 55–65 kDa). β‐keratins have a molecular weight of 10–22 kDa and higher sulfur content than α‐keratins. γ‐keratins present a globular structure with high sulfur content and a molecular weight close to 15 kDa. The aforementioned types of keratin proteins interact with each other to form tetramers, which in turn form intermediate filaments (75–90 Å). Eight tetramers form micro‐fibril, which could conform macro‐fibril (0.1–0.4 mm). [163]
The large volumes of keratinous wastes generated throughout the world each year during food manufacturing have promoted the valorization of these wastes through eco‐friendly alternatives. Keratin wastes are commonly incinerated or disposed into landfills or rendering. However, recent studies have demonstrated that they are valuable sources of protein for animal nutrition, fertilizers, glues, biodegradable films, and foaming agents for fire extinguishers. Hydrothermal treatments or enzymatic hydrolysis can be used to perform appropriate hydrolysis of keratinous material. Hydrothermal treatments involve the use of acids or bases (commonly HCl or NaOH) as hydrolytic agents at high temperatures (100–150 °C) and pressure (1.5 atm). Acidic hydrolysis generally shows substantial efficiency; however, additional neutralization is required, the amount of ash in the final product is high and extreme acidic conditions could lead to amino acids denaturation (e. g., tryptophan). During alkaline hydrolysis, the loss of amino acids is avoided, however, this process is slower than acidic hydrolysis.[ 164 , 165 ]
Another alternative is enzymatic hydrolysis, which is performed under mild conditions, reaching yields as high as 90 %. Enzymatic hydrolysis is performed using keratinases produced extracellularly by bacteria (Bacillus and Vibrio) or fungi (Paecilomyces marquandii). This procedure consists of two stages. During the first step, keratinases are produced from the action of microbial or fungi on keratinous waste. Meanwhile, in the second step, keratinases hydrolyze the cross‐links, generating keratin hydrolysates. Keratinases expressed by organisms commonly are serine or organo‐metallic proteases, which act over a wide pH (7.0–9.5) and temperature range (40‐50 °C). [162]
As it was previously stated for feather degradation, the hydrolysis of keratinous materials first involves the breaking down of disulfide bonds, and then the hydrolysis of the protein chain. The use of plant proteases on active enzymatic preparation is an attractive alternative to reach feasible keratin degradation yield. The reports of vegetable proteases on keratin degradation are scarce. [166] A potential application is based on wool hydrolysis. Wang et al. carried out a comparative study of cutinase‐pretreated wool hydrolysis using savinase (serine proteases) and papain. [167] Both enzymes showed to be active at 55 °C, and pH 7.0 for savinase, and pH 8.5 for papain, resulting in a weight loss of 2 % (200 U g−1), and preventing undesirable fiber damages during the hydrolysis. Furthermore, the proteolytic activity was increased when the enzymes were combined (3.4 % weight loss). This can be understood considering the hydrolysis mechanisms of each enzyme. Papain was selective towards peptide bonds acting on hydrophobic residues in particular positions from the underlying protein structure (especially Cys, His, and Asn amino acids), while savinase acted on the filaments of keratin.
In recent work, Yoshida‐Yamamoto et al. studied the keratinolytic activity of proteases derived from Cucumis melo, Pyrus communis, Actinidia chinensis, Ficus carica, Ananas Comosus, and Carica Papaya, on nail clippings valorization. [168] The enzymatic tests were performed at temperature from 37 °C (papain) to 80 °C (ficin) and pH between 6 and 10. Papain and Cucumis melo protease showed higher keratinolytic activity than proteinase K, which is commonly used for keratin digestion. Cucumis melo protease exhibited the highest activity during keratin hydrolysis, approximately 1.78 times higher compared to proteinase K. On the other hand, the keratinolytic activity observed for the other proteases relative to the proteinase K activity were: papain (1.2 times), bromelain (0.75 times), Pyrus communis (0.4 times), Actinidia chinensis (0.65 times) and Ficus carica (0.62 times). Based on the results, proteases derived from Cucumis melo could be promising enzymes for keratin degradation applications.
The keratinolytic activity of papain could be enhanced using green reagents. Shi et al. studied the synergistic effect of H2O2 and papain (100,000 U g−1) during keratinous protein fiber hydrolysis (wool fiber). [169] First, the wool fabric (2 g) was incubated in a solution with H2O2 (50 mL L−1) for 30 min at 50 °C. Then, the hydrolysis was carried out at 50 °C for 60 min in a 60 mL solution of papain (0.12 g). Hydrogen peroxide breaks the S−S bonds of the keratin and converts them into S−O bonds, thus improving the capacity of papain to act on amide bonds (no hydrolysis degree was reported). Further studies on keratin degradation using plant proteases are required, focusing on serine plant proteases, which commonly show higher keratinolytic activity than cysteine proteases. Furthermore, green reagents could be applied in combination with serine plant proteases to provide the disulfide bond hydrolyzing function of keratinases.
Collagen is a fibrous structural protein found in the extracellular matrix and fibrous tissues, such as skin, ligament, tendon (elongated fibrils), cornea, blood vessels, bone, cartilage, and even in the gut. Collagen proteins are abundant in mammals, makeup about 30 % of the total proteins of the body.
Collagen presents a complex structure, showing a unique tertiary structure, triple helix. The structure contains three polypeptide chains (identical or non), where each chain is composed of about 1000 amino acids. The interaction of these three protein chains leads to the formation of supercoiling in a left‐handed manner around a common axis with a triple‐helical conformation. The close packing of the three chains is permitted because glycine is repeated at every third location. Glycine is the only amino acid that can be accommodated within the helix without generating chain distortions or major positional modifications. Besides, approximately 35 % of non‐glycine positions are occupied by proline. [170] Collagen also contains around 10 % of hydroxyproline (generated by post‐translational hydroxylation of proline), based on amino acid composition. Approximately 28 collagen types have been identified in vertebrates, which are composed of 46 distinct polypeptide chains. All collagen types present the characteristic triple helicoidal helix, but they differ on their length, the most common being the following: [171]
Collagen I: main constituent of the organic part of bones. Also, it is found in skin, bone, teeth, tendon, ligament, and vascular ligature.
Collagen II: main component of cartilage (eyes and cartilage).
Collagen III: main constituent of reticular fibers (skin, muscle, blood vessels).
Collagen IV: main compound of the epithelium‐secreted layer of the basement membrane and the basal lamina.
Collagen V: it is commonly found in hair, cell surfaces, and placenta.
A significant amount of collagen waste is produced during food manufacturing around the world each year. These wastes commonly contain body parts of animals, such as: head, skin, bones, cartilage, teeth, tendon, ligament, and scales, among others.
The collagen used in industrial products is generally obtained from mammalian sources (bovine and porcine). Collagen is used as biomaterial in a wide spectrum of applications, mainly in pharmaceutical (mini pellets, tissue engineering, wound dressing, sponges, films, membranes, and composites) and food industry (dietary supplements, foaming agents, emulsifiers, stabilizers, microencapsulation, and biodegradable films). Collagen is broadly used as enhancer in supplementation preparations, helping to meet the body‘s needs for collagen, resulting in significant health benefits. [172] It has been also reported that collagen could be used in drug delivery, tissue recovery, cosmetic ingredient, disease treatment, and skin restoration (hydrolyzed collagen is incorporated into the formulation to counteract skin dryness as well as prevent skin damage).[ 173 , 174 ]
Collagenases are proteases with the capacity to hydrolyze various types of collagens, which are resistant to degradation due to the rigid triple helical structure. These proteases are categorized into two main groups: metallo‐collagenases and serine‐collagenases. Metallo‐collagenases contain zinc and commonly require calcium for optimal activity and stability. Meanwhile, serine‐collagenases generally do not require cofactors.
In view of the potential uses of collagenases and their increasing demand, there is a growing interest in finding new sources for these proteases. After an extensive review of the literature, few articles reporting the hydrolysis of collagen using plant proteases were found. Most of the research has focused on hydrolysis using zingibain, bromelain, papain, actinidin, and ficin, over different substrates. Kim et al. investigated the application of cysteine protease obtained from ginger (Zingiber officinale) and papain on collagen type I hydrolysis. [175] The enzymatic tests were performed at 20 °C, pH 5.5 (0.1 m acetate buffer), and an enzyme concentration of 0.5 mg mL−1 for 64 h. Ginger protease hydrolyzed native type I collagen with at least 10‐fold higher efficiency of hydrolysis than papain, showing preference for Pro in the P2 position. The products were collagen hydrolysates with a molecular weight between 36 and 210 kDa. The β conformations of collagen were completely degraded due to the hydrolysis in the non‐helical, cross‐linked ends of the protein chains. The authors attributed the remarkable collagenolytic activity of ginger protease to their biological function in the rhizome to defend the plant against nematodes. In a posterior work, Ketnawa et al. studied the collagen hydrolysis using bromelain extract obtained from pineapple peel (Nang Lae cultivar). [176] Collagen from bovine Achilles’ tendon and farmed giant catfish skins were used as substrates. The optimal enzymatic conditions were 55 °C and pH 7.0, generating hydrolysates from 39 to 120 kDa. From an overview, higher activity was observed by increasing the enzyme concentration. A similar hydrolyzed pattern between bovine and giant catfish skin was found for a specific activity of 0.18 U mg−1. However, bovine collagen was more resistant to bromelain hydrolysis. All collagen compounds were hydrolyzed when the specific activity of bromelain was 0.3 and 0.12 U mg−1 for bovine and giant catfish skin collagens, respectively. Furthermore, the β, α1, and α2 collagen of giant catfish skin were hydrolyzed using and specific activity of 0.02 U mg−1.
Commercial plant proteases can be combined to achieve higher yields, as proposed Ha et al. [177] The authors evaluated the activity of commercial plant protease preparations (papain, bromelain, actinidin, and zingibain) on the hydrolysis proteins of beef connective tissue and topside myofibril extracts. The hydrolysis conditions were: 55 °C, pH 6.0, reaction volume of 200 μL, and an enzyme amount of 0.07, 0.5, 2.0, and 17.9 μg for papain, bromelain, actinidin and zingibain, respectively. In the case of zingibain, most collagen hydrolysis occurred during the first hour with a gradual increase over the first three hours. The specific activity of the four protease preparations tested was 0.02, 0.7, 0.05, and 0.03 A520 min−1 mg−1 Azocoll for papain, bromelain, actinidin, and zingibain, respectively. Azocoll is an insoluble collagen to which a bright‐red dye is attached, 1 mg mL−1 of azocoll yielded an Absorbance of 0.593 if totally digested. The four enzymes preparation also were tested for connective tissue and meat myofibril extracts hydrolysis. The actinidin protease was the most effective at hydrolyzing beef myofibril proteins, while zingibain showed the highest efficiency at hydrolyzing connective tissue proteins. The hydrolysates produced after 24 h incubation presented a molecular weight range between 18 kDa (Troponin C) and 138.8 kDa (Collagen type I α‐1) The authors indicated that the bioactivity of plant proteases depends on the natural collagen source. The actinidin activity (obtained from kiwifruit by salt precipitation and purified through ion‐exchange chromatography) on collagen hydrolysis was also reported by Mostafaie et al. [178] The collagen test was carried out at 37 °C with an actinidin concentration of 1 mg mL−1, incubation time of 1 h or 2 h, and different pH. Actinidin hydrolyzed collagen type I and II using neutral (20 mm phosphate (pH 7)) and alkaline (20 mm Tris‐HCl (pH 8.5) buffers) buffers, while in acidic conditions (20 mm acetate (pH 4) and 20 mm citrate (pH 5.5)) the enzyme did not hydrolyze the substrate. The highest bioactivity was observed using Tris‐HCl (pH 8.5) buffer, hydrolyzing almost completely type I collagen after 1 h. Meanwhile, using phosphate (pH 7) buffer, only around 50 % type I collagen was hydrolyzed. On the other hand, most of the type II collagen was hydrolyzed using Tris‐HCl and phosphate buffers after 2 h (no hydrolysis degree was reported). At pH 8.5, the hydrolysis products presented molecular weights 116 and 230 kDa. The results addressed that actinidin could be efficiently applied for hydrolysis of collagen type I and II.
Recently, the purification and characterization of a novel collagenolytic serine protease derived from Ficus carica (var. Brown Turkey) latex was reported. [179] The enzyme is a monomeric protein with a molecular weight of 41 kDa, with optimal pH of 8.0–8.5 and temperature of 60 °C. The serine protease was stable under a wide range of pH (5–9) and temperature (up to 80 °C) without loss of activity. The purified enzyme showed a proteolytic activity of 28.4 CDU mg−1, reaching 94 % yield and generating hydrolysates with a mass from 14.4 and 116 kDa. The high proteolytic activity shown by the new serine protease from Ficus carica is in good agreement with the activity of other serine proteases documented by Gomes et al., highlighting cryptolepain (Cryptolepis buchanani) and pedilanthin (Vedilanthus tithymaloids). [180] Serine protease from Wrightia tinctoria was recently isolated and characterized. [181] The protease exhibits high stability up to 70 °C, and pH from 5 to 10. However, the activity of the enzyme was inhibited by PMSF (a dimethylamino benzaldehyde). The enzyme presents caseinolytic, gelatinolytic, and collagenolytic activity at 37 °C and pH 8.5 (no hydrolysis degree of gelatin and collagen was reported). The caseinolytic activity of the protease was 22 U h−1 (10 μg). The enzyme hydrolyzes type I collagen to a high degree, generating small molecular weight protein and peptide fragments (50–80 kDa). On the other hand, cardosin A obtained from Cynara cardunculus latex is a promising enzyme for collagen hydrolysis according to Duarte et al. [182] Cardosin A was able to hydrolyze the fibrillar type I collagen, showing high selectivity towards α‐chains. After 24 h incubation at 37 °C (enzyme/substrate mass ratio of 1/50), collagen was efficiently hydrolyzed forming small peptides with a molecular weight from 20 to 50 kDa. Under these conditions, a hydrolysis degree of 30 % and 55 % was reached for α1 and α2 chains of collagen type I, respectively. The cleavage site was Phe464‐Gln465 in the α2 chains of collagen I. The high specificity of cardosin A towards collagen indicated a potential use for extracellular matrix remodeling (EMC). Another important protein for bioactive peptide production is gelatin, which is a heterogeneous mixture of peptides obtained from the breakdown of cross‐linking between the protein chains and some breakage of polypeptide bonds during collagen hydrolysis. The primary structure of gelatin is similar to collagen, while the secondary structure is made up of polypeptide chains: α‐chains, β (dimers of α‐chain) and γ (trimers of α‐chain).
Common sources of gelatin are pig skin (46 %), bovine hide (30 %), and pork and cattle bones (24 %). Meanwhile, the industrial production of gelatin derived from nonmammalian species is gaining attention. [183] Two types of gelatins can be obtained from collagen hydrolysis depending on the pre‐treatment procedure: type A and type B (both types are soluble in water). Type A gelatin has an isoelectric point at pH ≈8‐9 and it is obtained using an acidic pretreatment, whereas, type B presents an isoelectric point at pH ≈4‐5 employing alkaline conditions in pretreatment.
Commonly, gelatin hydrolysates have a molecular weight between 1 and 24 kDa, depending on gelatin source, enzyme used (alcalase, pepsin, trypsin, α‐chymotrypsin, neutrase, papain, properase E, protamex, savinase, and NS37005), and assay conditions (pH, temperature, and extraction time). [184] The structure of gelatin‐derived peptides depends on the hydrolysis degree, however, Gly, Pro, and Hyp together represent more than half of residues, where Gly‐Pro‐Hyp sequence conforms about 10 % of the chain. A complete review of amino acid sequence on gelatin‐derived peptides was documented by Liu et al. [185]
Gelatin hydrolysis is growing in importance taking into account the functional properties of its biopeptides. Gelatin‐derived peptides show potential biological benefits, such as antioxidant, antihypertensive, anticancer, antiphotoaging, and cholesterol‐lowering effects. [186] Gelatin hydrolysates can be used as enhancers of antioxidative potential and cryoprotective properties on functional foods. On the other hand, gelatin‐derived peptides have been shown to be useful as anti‐aging agents.[ 187 , 188 ]
Traditionally, gelatin hydrolysis has been studied using microbial gelatinase, involving the use of complex recombinant techniques. An extensive review of microbial collagenases was reported by Pal and PV. [189] The mechanism of collagenase for the degradation of collagen is still unclear. In recent years, preliminary studies for gelatin hydrolysis using plant proteases with satisfactory productivity have been documented. Gautam et al. carried out a comparative study for gelatin hydrolysis at 45 °C and pH 6.0 using crude and purified bromelain extract (Ananas comosus). [190] Stem bromelain showed higher bioactivity (16 U mg−1) compared to bromelain from fruit (2 U mg−1). It was also observed that stability and activity of purified bromelain extract is important to gelatin hydrolysis. Comparable results for gelatin hydrolysis using bromelain extract were documented by Kaur et al., indicating that the activity of bromelains over gelatin substrate is sensitive to the organ source of the plant. [191]
The gelatinolytic activity of ficin enzyme was reported by Raskovic et al. [179] The authors tested the plant protease using an enzyme enzymatic concentration of 20 mg mL−1 at 37 °C and pH 8.1 (50 mm Tris‐HCl buffer). Ficin showed a proteolytic activity of 24.8 U mg−1, producing biopeptides with a molecular weight between 35 and 45 kDa after 15 min. The protease maintained 80 % of its activity when it was incubated at the range of pH 4‐9 and temperature 20–80 °C. The high stability of enzyme address further works to optimize the gelatin hydrolysis.
Recently, the possibility of hydrolysis of fish gelatin to obtain antioxidant peptides using papain was documented. Kittiphattanabawon et al. investigated the hydrolysis of gelatin from blacktip shark skin using papaya latex enzyme. [192] The hydrolysis assays were performed at pH 7.5 and 40 °C with an enzyme concentration of 3 % (w/v). The antioxidant activity of gelatin hydrolysates increased with hydrolysis degree. When the hydrolysis degree was 40 %, the oxygen radical absorbance capacity (ORAC) and chelating activity remained constant or slightly increased from pH 1 to 9 and during heating (100 °C) for 240 min. The peptides in low concentration (500‐1000 ppm) could be applied as natural antioxidants to prevent the oxidation of β‐carotene linoleate (chelating activity of Fe+3: 3.2 μmol EE g−1 solid, and HOCl scavenging capacity: 170 μmol TE g−1 solid). On the other hand, You et al. carried out the optimization of fish gelatin hydrolysis employing papain for the production of antioxidant peptides using response surface methodology. [184] The optimal conditions were: enzyme to substrate ratio of 2 %, 56.8 °C, 2.11 h reaction time, and pH 7.4, reaching a hydrolysis degree around 50 %. Thus, papain could be used for the production of fish gelatin‐derived peptides with antioxidant properties.
4.1.2.5. Fish Protein Hydrolysis
Fish processing plants generate large amounts of by‐products that are discarded in most cases. About 60 % of fish raw material is discarded, including head, bone, skin, fin, meat, scales, trimmings, and roe, among others. In the case of shrimp production, 3.4 million tons per year worldwide are processed, which 48–56 % results in other products.[ 193 , 194 ] It is estimated that the annual production of fish will achieve around 201 million tons in 2030.
Fish wastes contain protein‐rich fractions, essential omega‐3 fatty acids, macronutrients (phosphorus and calcium), micronutrients (zinc, iron, and selenium) and important bioactive molecules, which have potential applications for human health.
Fish by‐products hydrolysis can be carried out employing chemical or enzymatic methodologies. Chemical strategies are classified as acidic or alkaline hydrolysis based on the pH of the medium. A high hydrolysis degree is achieved during chemical hydrolysis, however, these methods show low selectivity towards a specific substrate, producing the destruction of some individual amino acids and generating toxic substances. On the other hand, enzymatic hydrolysis is performed under mild reaction conditions (pH and temperature), showing remarkable substrate affinity with precise control of hydrolysis degree, thus conserving the nutritional value of the source protein. Commercial proteolytic enzymes are used for fish hydrolysates production, including alcalase, papain, pepsin, trypsin, alpha‐chymotrypsin, pancreatin, flavourzyme, pronase, neutrase, protamex, bromelain, cryotin F, protease N, protease A, orientase, thermolysin, and validase. [195]
Fish protein hydrolysates are one of the best protein sources due to their nutritional value, amino acid balance, and high digestibility. The bioactive peptides found in fish hydrolysates can act as anticoagulants, antioxidants, high blood pressure controllers (reducing the risk of cardiovascular disease) and used as anti‐cancer and anti‐bacterial compounds. [196]
Fish protein hydrolysates commonly contain between 60 % and 90 % of proteins. The moisture content in fish hydrolysates commonly is less than 10 %, which is caused by the high temperature used during the evaporation process and spray drying. [197] The ash content of fish hydrolysates ranges between 0.45 % and 27 % of the total composition. The wide range of ash content is consequence of the use of acids or bases to adjust the pH medium.[ 198 , 199 ] Protein hydrolysates contain free amino acids and short peptides chains. Besides, aspartic acid and glutamic acid are found in high proportion in fish hydrolysates. [200] All essential and non‐essential amino acids for human health are found in fish muscle, head, skin, and visceral hydrolysates, whereas aromatic amino acids are in low proportion. Chalamaiah et al. documented an extensive recompilation of amino acid composition of protein hydrolysates prepared from various fish processing wastes. [194]
The mechanism of fish protein hydrolysis is not fully understood due to the substrate diversity, multiple reactions, and medium conditions. However, it has been postulated that the hydrolysis of fish protein occurs in three consecutive reactions: (i) formation of the Michaelis complex between the substrate and the enzyme, (ii) cleavage of the peptide bond releasing one of the two peptides, and (iii) a nucleophilic attack on the remains peptide bonds of the complex to split off the other peptide, thus regenerating the protease. The hydrolysis of peptide bonds generates ionizable groups (NH3+ and COO−), while simultaneously the size of the polypeptide chain decrease. These factors contribute to exposure to the hydrophobic interior of the original protein, favoring the production of small peptides. [201]
In this context, plant proteases emerge as an affordable enzyme source on fish hydrolysis to obtain add‐value products, such as beneficial fish hydrolysates or nutritional preparations for plants. A case study of the last one was reported by Ranasinghe et al., who investigated the hydrolysis of tuna fish waste using fruit wastes (Ananas comosus and Carica papaya) for the production of liquid fertilizers. [202] The proteases were extracted from leaves, ripe fruit peels, and pulp. The optimal conditions of the enzymatic tests were 37 °C and pH 7.5, with bromelain obtained from ripe fruit peels showing greater activity (0.34 U mL−1) than papain (0.3 U mL−1) after 24 h. The protease fraction extracted from Ananas comosus leaves generated the highest hydrolyzed protein amount (0.33 g g−1 enzyme) after 5 h. Meanwhile, the major content of fish protein hydrolyzed after 24 h was obtained using protease fraction derived from ananas comosus crown. The liquid fertilizers with fish protein hydrolysates were tested on the growth of Basella alba. The fertilizer derived from the hydrolysates obtained using bromelain or papain exhibited the same performance that a fertilizer liquid. Thus, the high nitrogen content on fish protein could be converted into natural fertilizer.
Seafish are also a valuable source for biopeptide production with antimicrobial properties. Liu et al. investigated the production of cysteine‐rich antimicrobial peptides from hydrolysis of oysters (Crassostrea gigas). [203] The raw material was first hydrolyzed using alcalase (3 wt.% enzyme, pH 8.5, 50 °C and 3 h), and then using bromelain (under the same operating conditions but at pH 5.5). The combination of both enzymes produced hydrolysates from 5 to 10 kDa (no hydrolysis degree was reported). The peptide CgPep33 (rich in cysteine) inhibited the growth of the studied bacteria (Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis, and Staphylococcus aureus) and fungi (Botrytis cinerea, and Penicillium expansum). The IC50 (effective concentration for 50 % growth inhibition) values of CgPep33 against all tested bacteria and fungi were from 18.6 to 48.2 μg mL−1. Meanwhile, Gram‐positive bacteria were the most sensitive, showing MIC (minimal inhibitory concentrations) values ranging between 40 and 60 μg mL−1. The authors remarked that safe antimicrobial peptides from marine bivalved mollusks can be obtained.
The production of peptides from salmon skin has been also documented. Ahn et al. studied the production of Angiotensin I peptide from the collagen extracted from Atlantic salmon (Salmo salar) skin. [204] The collagen was hydrolyzed with alcalase and papain (a similar procedure was described by Liu et al. [203] ). The hydrolysates were then isolated by multistage separation. The collagen hydrolysates from salmon skin had high protein content (91.2 %), with low molecular weight (91 % of peptides had less than 1 kDa). Eleven peptide fractions were identified and tested for Angiotensin I‐converting enzyme (ACE) inhibition. The highest IC50 values against ACE activity found were 9.10 μm, 10.77 μm, and 7.72 μm, for Val‐Trp‐Asp‐Pro‐Pro‐Lys‐Phe‐Asp, Phe‐Glu‐Asp‐Tyr‐Val‐Pro‐Leu‐Ser‐Cys‐Phe, and Phe‐Asn‐Val‐Pro‐Leu‐Tyr‐Glu peptide, respectively. These peptides could be applied for hypertension treatment.
Threadfin breams are a common waste on fish processing, being a potential peptide source. Gajanan et al. studied the peptide production from the waste of threadfin breams (Nemipterus japonicus) using papain and bromelain. [205] The optimal conditions for both enzymes were 50 °C and pH 6.8, reaching a hydrolysis degree of 15 % after 60 min. However, an enzyme to substrate ratio of 0.70 and 3.79 was required for papain and bromelain, respectively. Three peptide fractions with a molecular weight ranging between 812 and 7562 Da were obtained. According to the results, papain exhibited higher activity for hydrolysis of threadfin breams, however, the peptides generated using bromelain had higher antioxidant potential and ACE inhibitory activity. For a hydrolysis degree of 15 %, the purified peptide fractions obtained using bromelain exhibited the following values on antioxidant tests: 75 % DPPH free radical scavenging activity and 65 % linoleic acid peroxidation inhibition activity. Liu et al. also explored the production of peptides with ACE inhibitory activity from jellyfish (Rhopilema esculentum). [206] The optimal conditions using papain were: 37 °C and pH 6, with an enzyme/substrate ratio of 2.8 %, reaching a hydrolysis degree of 20.6 % (0.3 g substrate mL−1). The proteolytic activity was increased when papain was used in combination with other proteases (pepsin, protamex, and alcalase), producing four novel ACE inhibitory peptides, which were further purified. The IC50 values for Val‐Gly‐Pro‐Tyr, Phe‐Thr‐Tyr‐Val‐Pro‐Gly, Phe‐Thr‐Tyr‐Val‐Pro‐Gly‐Ala, and Phe‐Gln‐Ala‐Val‐Trp‐Ala‐Gl were 8.40, 23.42, 21.15, and 19.11 μm. In concordance with these, the synergic effect between plant proteases with animal enzymes for fish waste hydrolysis is reported in the literature. [204]
Promising results have been published using emerging plant proteases as reported by Romero‐Garay et al. [207] The authors evaluated the application of plant proteases derived from Bromelia karatas (BK) and Bromelia pinguin (BP) for antioxidant hydrolysates/bioactive peptides production through fish waste hydrolysis. The plant proteases were extracted and semi‐purified using simple procedures. The highest yield (54.8 %) was reached using bromelain (37 °C, pH 7.0 and 4 h), followed by BP fraction (54.3 %; pH 6.5, 40 °C and 30 min), and by BK extract (49.7 %; pH 6.5, 40 °C and 4 h). BP protease exhibited higher proteolytic activity compared to bromelain. Meanwhile, the hydrolysates generated using BK proteases showed hydrophilic character with more than 50 % of peptides with less than 17.5 kDa, exhibiting great antioxidant capacity (DPPH activity: 380 μm ET mL−1, ABTS: 37 μm ET mL−1, and FRAP: 600 μm ET mL−1). It was also observed that the biopeptide production does not necessarily increase with hydrolysis degree.
The use of ficin on fish by‐products hydrolysis has been scarcely documented. Tan et al. carried out a kinetic study of the hydrolysis of channel catfish (Ictalurus punctatus) by‐products (heads and frames) using papain, ficin, bromelain, neutrase, alcalase, protamex, novo‐proD, and thermolysin at pH 7.2 varying the concentration of enzyme from 5 to 80 AzU g−1 of protein in the substrate and the temperature between 30 and 70 °C. [208] The highest hydrolysis degree (71 %) was obtained employing ficin at 30 °C and after 120 min. The authors also observed that the functional properties of hydrolysates were affected by the pH medium.
Plant proteases have shown promising results in combination with several bacteria or fungi proteases, producing hydrolysates with active biological properties from a wide spectrum of fish by‐products.[ 209 , 210 , 211 , 212 ] Further studies should focus on optimizing hydrolysis and kinetic studies are required to exploit the potential of plant proteases in this field.
4.1.2.6. Valorization of Soy Proteins and Other Legumes
Functional food preparations are an important sector of the food industry. New food formulations are continually being developed to improve the functional properties of the products and their nutritional value. Vegetable proteins are major compounds commonly used in the formulation of functional foods, acting as amino acids resource to regulate the physicochemical properties of the final product.
The application of hydrolyzed vegetable proteins improves the physiological and functional properties of food formulations compared to crude protein due to the release of biological peptides during enzymatic hydrolysis. These biopeptides favor the regulation of chronic diseases, for instance, oxidative stress, diabetes, and hypertension. [213] Nevertheless, the peptides derived from soybean, rice, canola, peas, wheat, and walnut proteins present remarkable antioxidant capacity. Peptides obtained from chickpea and pea protein hydrolysates showed an inhibitory effect on angiotensin I‐converting enzyme by gastrointestinal simulation. [214] In contrast, potato protein hydrolysates had the capacity to diminish the oxidation of myofibril proteins, indicating that they could be used as natural antioxidant additive for muscle foods preparation. [215] Furthermore, it was reported that compounds produced from peanut protein hydrolysis have proven antioxidant and functional properties, and they could be incorporated into food supplement. [216] On the other hand, walnut (Juglans regia L.) protein hydrolysates have anti‐atherogenic, anti‐mutagenic, anti‐inflammatory, and antioxidant properties. [217]
In recent years, the application of biopeptides derived from vegetable protein hydrolysates for anti‐cancer treatment has been documented. Fenugreek (Trigonella foenum graecum) protein hydrolysates can be applied as nutraceuticals for treating colon cancer, exhibiting a similar mechanism to antimicrobial peptides. [218] Peptides derived from beans (Vicia faba) hydrolysis were suitable for cancer treatment. They can be used directly, added as ingredients into functional foods, or incorporated into pharmaceuticals.[ 219 , 220 ]
Soybean is one of the most important oilseeds in the world. Soybean manufacture produces soy defatted flour, which is rich in proteins with higher nutritional value compared to other vegetable proteins. [221] The use of peptides derived from soy protein is gaining attention due to their substantial level of glycine (4.2 % wt.). The oral administration of biological peptides obtained from soy protein hydrolysis has antioxidant effects, antimicrobial, antihypertensive, reducing cholesterol, anti‐osteoporosis, anti‐cancer, and immunization properties. [222] Long‐term consumption of soy protein hydrolysates could delay the increase of the blood pressure due to the inhibitory effect of angiotensin converting enzyme. It was also reported that soy proteins could prevent renal insufficiency, decreasing kidney TNF‐a levels. [223]
The hydrolysis of soybean protein involves the handling of three challenges. [224] First, the extension of the hydrolysis degree requires to be carefully controlled because soybean develops bitter flavor when is hydrolyzed. The flavor of soybean hydrolysates depends on free amino acids, smaller peptides, and volatile compounds composition. Within this context, the enzymatic hydrolysis under mild conditions provides precise control on hydrolysis degree. Second, the costs of enzymes commonly used for soybean hydrolysis (trypsin and alcalase) are high. Regarding this point, plant proteases offer a significant advantage, involving lower production costs. Third, soy proteins could coagulate during enzymatic hydrolysis due to the interaction between the protein fragments in aqueous medium. The coagulum is insoluble and decreases the yield of the soluble hydrolysates. The determination of optimal conditions to prevent the coagulation process is required for each particular enzyme.
Two methods can be applied to hydrolyze soy proteins: acid hydrolysis and enzymatic hydrolysis. Acid hydrolysis involves the use of concentrated hydrochloric acid, leading to the formation of chlorohydrins, like 3‐chloro‐1, 2‐propanediol (MCPD) and 1,3‐dichloro‐2‐propanol (DCP) during the process, which generates environmental concerns. Furthermore, acid hydrolysis shows low specificity towards protein substrate. [225] On other hand, enzymatic hydrolysis is performed under mild reaction conditions and the undesired reactions are suppressed. Trypsin, alcalase, pepsin and chymotrypsin enzyme are commonly used due to their high bioactivity, however, these enzymes offer low affinity towards soy protein substrates. [224] In contrast, soy proteins have high biocompatibility with plant proteases, being this, their main advantage compared to bacteria or fungi enzymes. Soybean flour contains a wide spectrum of proteins, and the enzymatic hydrolysis mechanism is still unclear. The Linderstrom–Lang model has been proposed to explain the product distribution of soybean protein hydrolysis using vegetable proteases. The model describes two pure mechanisms: zipper and one‐by‐one. The zipper mechanism occurs through the protein structure destabilization, exposing peptide bonds, which result in various intermediate products. The one‐by‐one mechanism proposes a first slow step in which soybean proteins are hydrolyzed one by one to the final products without the detection of intermediate products. In most cases, hydrolysis using plant proteases occurs through a combination of both mechanisms. For instance, hydrolysis using pomiferin or bromelains has the prevalence of zipper mechanism, while employing papain or hieronymin, one‐by‐one mechanism occurs in higher extension. The protein solubility, pH, substrate composition, and temperature affect the kinetics of the mechanism. [226]
New plant proteases with remarkable specificity towards soy protein have been recently documented. Mesa‐Espinoza et al. tested the antioxidant capacity of hydrolysate fractions derived from soy protein using enzymes extracted from Bromelia pinguin and Bromelia karatas fruits. [227] The optimal hydrolysis conditions were pH 7, 60 °C, incubation time of 35 min, and enzyme concentration of 0.46 g mL−1. The results indicated that both enzymes were active, showing B. pinguin and B. karatas proteases a specific activity of 7.91 and 3.19 U mg−1, respectively. B. karatas proteases retained 68–95 % of their activity after incubation at 37–60 °C for 60 min. Meanwhile, B. pinguin exhibited a residual activity between 70 and 93 % under the same conditions. The hydrolysis products were biopeptides fractions with molecular weight up to 30 kDa.
Promising results were found employing peptidases from the latex of Maclura pomifera fruits. [228] The enzymatic essays were performed at 45 °C and pH 8.0 using soybean flour as substrate (4.2 mg mL−1), reaching a hydrolysis degree of 36.2 % after 180 min of reaction. The soy hydrolysates obtained at 90 min (28.5 % hydrolysis degree) had great antioxidant capacity, presenting an IC50 of 31.6 μg mL−1, and a Trolox equivalent antioxidant capacity of 157.6 and 176.9 μmol TE g−1 peptide. The authors remarked that the 90‐min peptides could be applied for designing functional foods.
Lattices proteases are recognized for their high bioactivity towards vegetable substrates. In this way, Torres et al. tested the proteolytic activity of various plant proteases, including papain, bromelain, ficin, and a new peptidase derived from Asclepias fruticosa latex (asclepain f). [229] The Michaelis constant (Km) value for asclepain f was 6 to 8 times higher than those reached for papain, bromelain, and ficin. The optimal enzymatic conditions using asclepain f were: 45 °C, pH 10, and an enzyme/substrate ratio of 0.2 % (w/w), reaching a hydrolysis degree around 7 %. The hydrolyzed soy protein showed an increment of 60 % in water solubility compared to the original proteins. Meanwhile, the water‐holding capacity (WHC) was increased by 71 % at 20 °C and by 134 % at 45 °C after hydrolysis. Asclepain f could be applied to increase the solubilization of soy protein at a low hydrolysis degree.
The combination of heat treatment followed by enzymatic hydrolysis is a new approach. Zhang et al. evaluated the combination of both procedures for the production of black bean hydrolysates. [230] First, the heat treatment of black beans was carried out at 90 °C for 15 min, reaching a hydrolysis degree of 26.6 %. The heat treatment could break the disulfide bonds, however, if the temperature is too high, the black bean protein chains could form network polymers, thus reducing the enzymatic activity in the next step. Then, the enzymatic hydrolysis was performed at 50 °C, pH 8.0, and enzyme/substrate ratio of 0.05 (w/w), achieving a hydrolysis degree of 22 % in this stage. Pretreatment of black bean substrate with thermal methods before enzymatic essay generated a higher hydrolysis degree. However, heat treatment exhibits low substrate specificity.
Soy protein with antioxidant properties can be obtained using plant proteases, being possible to increase their activity in combination with animal proteases or heat treatments. The mild conditions and short incubation time are significant advantages of plant proteases. Further studies focusing on kinetics and optimization assays are required to provide a deeper understanding of the reaction mechanism.
4.2. Non‐Traditional Applications
The research of plant proteases with biotechnical applications has been encouraged in the last decades, considering their bioactivity and stability on a wide range of pH and temperature. The number of publications referred to extraction and purification of plant protease is in continuous growth, which displays a wide spectrum of opportunities focused on the by‐product valorization and chemical compounds substitution in traditional industries.
In the present review, traditional and non‐traditional applications of plant proteases are described. Traditional applications are those in which the action of plant proteases has been widely explored. The traditional uses covered in this work (see section 4.1) include the valorization of whey, milk hydrolysates and cheese making, gluten hydrolysis and obtaining bioactive peptides, poultry feather valorization, and collagen, keratin and gelatin hydrolysis. In addition, the application of plant proteases in the leather industry as an alternative to the conventional process, hydrolysis of fish proteins, recovery of soy proteins and other legumes were considered.
In this section those applications in which the use of plant proteases has been least explored will be addressed; these applications can be considered as non‐traditional. Effluent treatment and waste management, therapeutic applications, cosmetic medicine and beauty products, and pharmaceutical and medical uses are included.
Figure 5 summarizes the main applications for proteases of plant origin.
Figure 5.
Main applications of plant proteases.
4.2.1. Effluent Treatment and Waste Management
The impact of industrial activity on the environment generates serious concerns around the world. Enormous volumes of wastewater are generated for industries each year, which required adequate treatment before being discharged to minimize their environmental impact. A wide spectrum of methods can be used for water management, such as filtration, adsorption, coagulation and precipitation, membrane separation, advanced oxidation process (AOPs), and bioremediation. Commonly, a combination of the above methods is used to achieve the desired water remediation, for example, precipitation‐AOP, coagulation‐precipitation‐adsorption, and adsorption‐bioremediation. The sequence of treatment methodologies and the operating conditions on each step depend on concentration and type of harmful compounds, chemical and biological specification determined by environmental legislation, costs, maintenance labor, technological availability, and selectivity of the method, among others. [231]
AOPs and bioremediation involve the degradation of harmful compounds into less toxic substances, meanwhile, the other methods involve the generation of a by‐product rich in contaminants, which requires further treatment. In the bioremediation process, biological organisms are used to degrade organic pollutants. However, rigorous control of operating conditions (pH, temperature, alkalinity, and oxygen concentration) is necessary to prevent the deterioration of the organism colony. [232] Industrial wastewater commonly shows considerable physicochemical variations, which difficult the implementation of efficient monitoring during the bioremediation process.
In recent years, the application of enzymatic process for wastewater remediation has been gaining attention, especially plant proteases in view of their low cost and stability over a wide range of pH. The use of enzymes for wastewater treatment has been focused on effluents rich in polyphenols, oils and greases, sugar, and whey. [233] Few reports associated with the use of plant proteases on effluent treatment are found in the literature. In most of cases, plant proteases are unable to hydrolyze the harmful compounds, and they are applied as flocculant and chelating agents.
Lea studied the application of seed protein extract from Moringa oleifera tree as a low‐cost clarification agent for highly turbid and untreated pathogenic domiciliary wastewater. [84] The seed extract was obtained using simple procedures, reaching a content protein of 34 % with a molecular weight between 6 and 16 kDa (isoelectric pH of 10 to 11). The protein extract acted as flocculant, binding suspended particles in the colloidal suspension, producing a turbidity reduction between 80.0 % and 99.5 % at natural pH, accompanied by 90.00 % to 99.99 % bacterial reduction. This can be understood considering that bacteria are commonly attached to solid particles. Thus, Moringa oleifera seed extract is an affordable method for wastewater treatment with high organic matter content, as domiciliary effluent. This observation is in agreement with the findings documented by Dzuvor et al. [234] Seed extracts of Moringa oleifera could be also applied to remove microorganisms and metals (4‐6 times higher efficiency compared with alum) from wastewater owing to their coagulation properties.
Heavy metals are considered one of the most toxic compounds. Several industries have restricted the use of chemical reagents with mercury, chrome, arsenic and lead, however, large volumes of effluents with these toxic compounds are discharged into bodies of water. Efficient heavy metals removal using plant proteases has been reported. Plant proteases act as chelating agents during heavy metal removal, forming a complex with the metal, however, this commonly leads to enzyme inhibition.
Dutta et al. investigated the use of papain immobilized on charcoal by a physical adsorption method for mercury remotion from aqueous systems. [235] The optimal conditions for papain immobilization were: initial papain concentration of 40 g L−1, activated charcoal amount of 0.5 g, and pH 7. The adsorption equilibrium data was successfully described adopting the Langmuir isotherm model. The immobilized papain sample showed maximum mercury (HgCl2) removal of 99.4 % (20 mg L−1 metal, pH 7, 0.03 g immobilized papain). Immobilized papain exhibited a high affinity for mercury removal; thus, it could be used for rigorous separation at low concentrations of mercury.
Chatterjee et al. studied the removal of lead(II) and chromium(VI) by adsorption using bromelain immobilized on activated charcoal. [236] The optimal conditions for immobilization were: 20 g L−1 initial concentration of bromelain, 0.3 g of charcoal, pH 7, and 35 °C. The enzymatic tests were carried out at 35 °C and pH 7 to make the process more affordable. The removal of lead(II) and chromium(VI) was 99.9 % and 96.8 %, respectively, when the initial metal concentration was 10 mg L−1 and 0.3 g enzyme immobilized were used. According to the kinetic study, major removal took place in the first 5 min, reaching the equilibrium after 10 min. The authors also observed that the metal was linked to the sulfhydryl group of bromelains during the adsorption process.
Polyphenol effluents are commonly produced in pharmaceutical manufacturing. Polyphenols are toxic compounds resistant to degradation, which generate serious concerns. In this context, plant proteases could be used in combination with other proteases to provide an active enzymatic extract capable to degrade phenols substances. Cheriyan and Abraham investigated the enzymatic treatment at neutral pH of cashew nut shell liquid (CNSL) obtained as by‐product of cashew kernel manufacture. [237] The effluent had caustic properties and contained phenolic compounds, mainly cardanol (60‐65 %). The enzymatic treatment was carried out in two steps. First, crude protein fraction derived from Eupatorium odoratum leaves was used for decolorization. In the second step, laccase and papain immobilized on starch‐alginate beads were used in equal proportion to degrade cardanol. Laccase showed higher degradation ratio (28.6 % in 2 h) compared to papain (43.28 % in 73 h). The combination of laccase and papain immobilized resulted in the efficient degradation of cardanol.
Textile manufacture is considered one of the most polluting industries based on the large volume of colored liquid effluents generated. Dyestuffs require powerful oxidant conditions for their degradation, commonly peroxide radicals are generated by adding H2O2 and chemical agents (Fe+2/Fe+3 in Fenton process). These radicals form peroxide complexes with the aromatic rings of the dyestuff molecule, inducing its instability and breaking down the C−C bonds. Regarding textile effluent, plant proteases could provide an affordable and sustainable technology treatment. It was found that plants extract from Blumea malcolmii have the capacity to decolorize colored effluents. [238] The root extract contained a significant amount of lignin peroxidase, tyrosinase, DCIP (2,6‐dichlorophenol‐indophenol) reductase, azoreductase and riboflavin reductase, which degraded the aforementioned dye compounds. The enzymatic tests were carried out at 30 °C and pH 6.8, with a dye concentration of 20 mg L−1. After three days the dyes were efficiently degraded: 96.7 % malachite green, 87.9 % red HE8B, 80.2 % methyl orange, 42 % reactive red 2, and 76.5 % direct Red 5B after three days. Plant extract could be applied for water remediation, offering a sustainable alternative to chemical methods.
4.2.2. Cosmetic Medicine and Beauty Products
The cosmetic market is in continuous growth worldwide. Cosmetics are articled to promote cleansing, beautifying, and attractiveness without introducing modifications to body structure. Cosmetic preparations also are used to prevent diseases or protect the body, such as sunscreen or antidandruff shampoos. Cosmetics could contain a broad diversity of compounds according to their purpose. Creams, lotions, and ointments are common ingredients in skincare products. On the other hand, ingestible beauty products or nutricosmetics are commercialized as pills, liquids, or functional foods. Nutricosmetics contain vitamins, minerals, botanical extracts, and antioxidants, promoting and favoring the health of skin, hair, lips, and nails at cellular levels. [239] The content of enzymes in cosmetic formulations is rigorously monitored during process manufacturing to reduce the risk of allergy of most individuals. The use of plant proteases highlights by their high proteolytic activity at pH of cosmetic products (6.8‐7.8).
The application of plant proteases for cosmetic medicine and beauty products preparation is based on two approaches: (i) obtention of bioactive compounds with functional properties (e. g., biopeptides, antioxidant hydrolysates) through protein‐based substrates (see section 4.1.2.), and (ii) as an active ingredient in cosmetic formulation. Regarding the first approach, hydrolyzed collagen is incorporated in skin regenerating products to prevent skin dryness and damage. On the other hand, plant proteases are primarily used to promote skin exfoliation (scaling of the keratinized superficial corneal layer), increasing the absorption of water and other cosmetic ingredients, treating skin ulcers, and healing burn wounds.
Exfoliants remove the dead cell layer (stratum corneum) of the skin, and they are commonly used in aging skin, photo‐damaged skin, acne, and dry skin treatments. Commercial exfoliants contain alpha hydroxy acids, beta hydroxy acids, and retinoids, which may cause allergic reactions. An alternative to these compounds is papain, which is a non‐irritating exfoliant (specially for individuals with sun‐damaged skin). Papain also contributes to attenuating freckles and brown spots due to exposure to sunlight and smoothing the skin (peeling and soothing). Papain acts over Type‐I collagen, which is the major structural component of the extracellular matrix (ECM). The accumulation of damaged proteins and connective tissue gives an aged appearance to the skin. Papain has the capacity to hydrolyze around 15 % of connective tissue protein, thus enhancing the appearance of the skin. [240] The addition of papain in cosmetic preparation also could improve stratum corneum hydration level and water holding capacity of the skin, which is attributed to pH increase. Linked papain (papain carbomer) is found in numerous cosmetic formulations with an enzyme concentration between 2 and 5 %. The optimal activity of papain is observed at pH 6, however; the protease maintains 75 % of their activity between pH 5 to 7. The capacity of papain to act as exfoliant in the majority of cosmetic products (pH 7) favors the penetration of different medicine agents (e. g., biological additives and moisturizers). Some common herbal extracts used are those from Echinacea angustifolia, Mimosa tenuiflora, Hydrocotyl, Gingko biloba, tea tree oil, Matricaria chamomila, Hypericum perforatum, and Aloe barbadensis and caterndiele. Papain is also used in combination with non‐enzymatic compounds for chemical debridement, removing the devitalized extracellular material of wounds. In chemical debridement agents, the enzyme exfoliates the necrotic tissue, promoting the liquification of slough in acute and chronic lesions. Papain is ingredient of the following enzymatic debriding preparations: Accuzyme, Panafil, Kovia, Ethenzyme, Galadase, and Ziox. [241]
Bromelain also has been used in cosmetic applications, such as tooth whitening, acne, wrinkles, dry skin treatment, and post‐injection bruising and swelling reduction. [242] Bromelain stability in cosmetic preparation is a major challenge. A recent study reported that bromelain protease on anhydrous gel presents physical instability, which is increased with enzyme content (up to 10 % w/w). The instability was attributed to the absence of water in the formulation. In contrast, cream‐gel with 1 % bromelain has no modification during 90 days of testing, showing optimal temperature conservation of 4 °C. [243] Bromelain is also an attractive nutraceutical for cosmetic dental applications, especially for tooth whitening. The formulation of tooth bleaching gels containing bromelain (1 % wt.) shows a potent effect on peroxide‐free tooth whitening gels. The whiteness index alteration was 15 and the color difference standard deviations was 4.3. Similar findings were found for papain and ficin, indicating that these plant proteases have the potential to be used as active ingredients of peroxide‐free whitening products. [244]
Oxidative stress contributes to skin aging. This process is caused by the hydrolysis of melanin synthesis by tyrosinase. In this way, novel protein extracts derived from plants have been reported for tyrosinase inhibition. Wang et al. examined 25 traditional Chinese herbal medicines with potential uses for skin‐whitening. [245] The extracts (100 μg mL−1) from Pharbitis nil, Sophora japonica, Spatholobus suberectus, and Morus alba, exhibited an efficient inhibitory effect on tyrosine, showing an IC50 value of 24.9, 95.6, 83.9, and 78.3 μg mL−1, respectively. Specially Sophora japonica and Spatholobus suberectus showed high activity in human epidermal melanocytes (HEMn) in terms of free radical scavenging effects and high phenolic content. The values reported of DPPH were 1.95 and 4.36 μg mL−1 for Sophora japonica and Spatholobus suberectus, respectively. Both plant extracts were the most promising candidates for cosmetic application, reaching a tyrosinase inhibition of around 55 %.
Haircare is a major field of cosmetics products. Plant proteases show attractive properties for these hair products. Choi et al. investigated the feasibility of using kiwifruit protease for human hair care (optimal conditions: 45–50 °C and pH 7). [246] The hair keratin was partially hydrolyzed, removing the dirty adsorbed on the hair and the samples becoming thinner. The tensile strength of hairs was reduced by less than 15 % after 48 h (0.0820 kgf). These modifications enhance the quality of the hair by refining the hair cuticles, being this the major attribute of kiwifruit protease to be used in shampoo products.
In recent decades, the use of extracts obtained from herbal stem cells (in vitro production) is gaining attention. These extracts present the following benefits for human the body: prolongation of fibroblasts life, increased epidermis flexibility, regulation of cell division, reconstruction of the damaged epidermis, activation of cell DNA repair, and protection against UV radiation. [247] Some examples of plant stem cells are those corresponding to Malus domestica, Saponaria pumila Argania spinosa, Syringa vulgaris (skin anti‐inflammatory and anti‐aging), Lycopersicon esculentun (protecting skin cells from oxidative stress), Coffea bengalensis and Nicotiana Sylvestris (fibroblasts collagen production stimulation), and Zingiber officinale (skin pores reduction). [248]
4.2.3. Pharmaceuticals and Medical Applications
Medical applications using enzymes are in continuous development to treat human diseases or disorders with non‐invasive methods. The type and specificity of the enzyme have a predominant effect on the success rate of the treatment, acting selectively over the target substrate without harming the surrounding cells. Besides, therapeutic enzymes should be active at low concentrations in view of the high purity degree required for therapeutic applications, which commonly involves a remarkable purification cost. In this context, proteases derived from plant resources have emerged as an affordable alternative to microbial proteases due to simple extraction procedures that produce proteases with high selectivity at a lower cost and without pathogenic potential for humans or animals. In general, plant proteases with therapeutic potential are not heterologous proteins, which can be obtained from plant organs (fruit, stem, flower, and leaf) or from latex, without affecting the viability of the plant. An extensive review of therapeutic proteases isolated from plant latex was documented by Urs et al. [249] The data of plant proteases documented with medical applications has been increasing in the last 10 years. [250] Table 13 summarizes plant proteases with proven therapeutic applications.
Table 13.
Therapeutic uses of plant proteases.
|
Protease |
Source |
Therapeutic applications |
Ref. |
|---|---|---|---|
|
ZCPG |
Zinger (Zingiber montanum) |
Powerful anti‐oxidant |
[261] |
|
Zingipain |
Zinger (Zingiber officinale) |
Anti‐proliferative agent for cancer treatment |
[262] |
|
SgCDF |
Sesbania grandiflora |
Hemostatic and fibrinogenolytic activity |
[263] |
|
Actidin |
Kiwifruit (Actinidia deliciosa) |
Healing of neuropathic diabetic foot ulcer, increase protein digestion and ameliorate of constipation. |
[264] |
|
Bromelain |
Pineapple (Ananas comosus) |
Treatment of thrombosis, rheumatoid arthritis, wounds, cancer, asthma, angina, bronchitis, sinusitis, osteoarthritis, surgical traumas, pyelonephritis and inflammatory diseases in general. |
[252] |
|
Cardosin |
Cardoon (Cynara cardunculus) |
Drug delivery |
[265] |
|
Cucumisin |
Melon (Cucumis melo) |
Fibrin clotting lysis for thrombotic disorders treatment. |
[266] |
|
Ficin |
Fig (Ficus genus) |
Tissue recovery, immunoglobulin G cleavage, antimicrobial and digestive disorders treatment |
[267] |
|
Papain |
Papaya (Carica papaya) |
Treatment of edemas, sinusitis, gluten intolerance, hypochlorhydria, digestive disorders, caries removal, healing burn wound and infections. |
[257] |
|
Wrightin |
Wrightia tinctoria |
Antifungal, antioxidant and hepatoprotective, and vasculoprotective agent. |
[268] |
|
Protein extract |
Moringa Oleifera |
Antimicrobial |
[269] |
Bromelain could be also used in combination with other active ingredients (lutein, zeaxanthin, N‐acetylcysteine, vitamin B12, vitamin D3, alpha‐lipoic acid, rutin, vitamin C, zinc oxide, Vaccinium myrtillus 36 % anthocyanosides, and Ganoderma lucidum) for the treatment of patients with intermediate age‐related macular degeneration. The preparation contributes to increase the function of macular pre‐ganglionic elements. [251]
Bromelain has proved benefits for cardiovascular diseases treatment, osteoarthritis treatment, immunogenicity, blood coagulation and fibrinolysis. [252]
Recent studies highlight the anti‐inflammatory properties of bromelain, in spite of the action mechanism is not fully understood. Three different pathways have been established: (i) kallikrein‐kinin, (ii) arachidonic acid, and (iii) cell‐migration immunity. In the kallikrein‐kinin pathway, bromelain regulates the plasma fibrinogen levels and blood levels of bradykinin, thus improving serum fibrinolytic activity by activating factor XI, activating plasma prekallikrein. In the second pathway, bromelain regulates pro‐inflammatory prostaglandins (through the inhibition of prostaglandin E2 and thromboxane A2). This improves the anti‐inflammatory mediators, thus increasing platelet cyclic adenosine monophosphate (cAMP), and consequently, the levels of prostaglandin (PG) I2 and PGE1. Finally, bromelain regulates the cell‐migration immunity, acting on the migration of neutrophils to inflammation sites and removing cell‐surface molecules (e. g., CD128a/CXCR1, CD128b/CXCR2, CD14, CD44, CD16, and CD21). The three pathways confer bromelain a potent effect in disorders associated with inflammation and blood coagulation (160 mg/day oral administration). Regarding cardiovascular diseases, bromelain could reduce the risk of angina and transient ischemic attack. It was also documented that the oral consumption of bromelain significantly reduces the risk of acute thrombophlebitis. [253] An extensive recompilation of bromelain applications (sports injuries, perioperative, and osteoarthritis, among others) was reported by Colleti et al. [254]
Papain is used for the treatment of edemas, gluten intolerance, hypochlorhydria, digestive disorders, and infections. Papain also shows potent activity against the following bacteria: Alicyclo bacillus, Bacillus subtilis, Enterobacter cloacae, Escherichia coli, Listeria monocytogenes, Salmonella typhimurium, Staphylococcus aureus, and Proteus vulgaris. [255] Antihelmintic action of papain was documented against Ascaris suum, Haemonchus contortus, Heligmosomoides polygyrus, Trichuris muris, Protospirura muricola, and Strongyloides venezuelensis. [256] Papain is able to the cleavage protein content of glycoproteins in the membrane cell of fungi, demonstrating antifungal activity (Aspergillus niger, Candida albicans, Mucor spp., and Rhizopus spp). Gels based on papain with antibacterial and anti‐inflammatory could be a non‐invasive procedure for caries removal. Liposome‐containing papain formulations have proven benefits on hypertrophic scar treatment. The action of papain stimulates the production of cytokines that promote cell repair. Although papain is the most studied cysteine protease, the reports about therapeutic applications and toxicological data are scarce. [257]
Ficin is used as hemostatic agent. [16] Regarding other medical applications, no toxicological and clinical trials have been performed in extension. The main know pharmaceutical use of ficin is for intestinal worm treatment. Ficin also exhibits potential anthelmintic activity against Syphacia obvelata, Aspiculuris tetraptera, and Vampirolepis nana. The daily dose reported in tests for the treatment is 1–4 mL kg−1 for three days and repeated three months later. [258]
New plant proteases have been recently documented with potential therapeutic uses as actinidin (kiwifruit) and cucumisin family proteases (Cucumis melo). [84] Promising studies have been indicated that actinidin could be used for diabetes mellitus due to its ability to degrade α‐amylase. [259] Furthermore, the topical administration of kiwifruit for 4 weeks could heal diabetic ulcers in patients. [260] StSBTc‐3 is a cucumisin‐like protease obtained from Solanum tuberosum with the capacity to inhibit platelet aggregation without cytotoxic activity on human erythrocytes agent. Therefore, StSBTc‐3 shows potential to be used in the treatment of thromboembolic disorders such as strokes, pulmonary embolism, and deep vein thrombosis. No clinical or toxicological studies about cucumisin have been reported.
Plant extracts with high content of proteases have been used over time for the treatment of digestive disorders, wound debridement, inflammation, and immune modulation. Protease inhibitors (PIs) formulation is an important field of application for plant proteases. PIs are regulatory proteins that reduce and inhibit the exacerbated activity of the target proteases. PIs are effective tools for the treatment of arthritis, pancreatitis, hepatitis, cancer, AIDS, thrombosis, emphysema, hypertension, and muscular dystrophy, among others. [84] Plant PIs (small peptides rich in cysteine residues and disulfide bonds) show remarkable resistance to heat treatment, shifts in pH, denaturing agents, and ionic strength. Thus, plant PIs could be used as antimicrobial, anticoagulant, or antioxidant agents.
Although plant cysteine proteases are commonly used in the medical industry, their activity in biological environment is affected by oxidation, by the presence of metal ions, or chelating agents. On the other hand, serine proteases obtained from vegetable sources do not require any co‐factors. Other variables that influence on therapeutic efficacy are protein specificity, temperature, and the presence of inhibitors in the medium.
An important challenge for the development of therapeutic proteases is related to their denaturalization by oral administration. The low pH of the stomach could affect the activity of the protease and its selectivity towards the target substrate. Regarding this, cysteine proteases, which are commonly found in plants, exhibit remarkable resistance to high temperatures, extreme pH values, and high salinity. This can be understood considering the stable structure of cysteine proteases linked by numerous disulfide bonds. [256] The substantial resistance to extreme values and abrupt pH shifts exhibited by cysteine plant proteases is a valuable advantage for therapeutic applications compared to microbial proteases, thus favoring the action of the enzyme on the target substrate.
5. Biotechnological Applications of Plant Proteases: Perspectives and Challenges
The application of enzymes on an industrial scale is a major technological challenge that requires previous studies to determine the optimal operating conditions. Reaction time, reuse strategies, enzyme purity, production volume, specificity substrate, and enzymatic test conditions (temperature, pH, and use of additives or co‐factors) are relevant parameters to analyze the feasibility of an enzyme on an industrial scale. The commercial price of the final product could limit the industrial application of an enzyme. In the case of therapeutic enzymes, the remarkable cost associated with their isolation and high degree purification is aligned with the obtention of high‐value products. Meanwhile, reuse strategies for enzymatic wastewater treatment are required to achieve a sustainable application.
The implementation of immobilized enzymes is a valuable strategy to improve the enzymatic performance in the pharmaceutical, chemical, cosmetic and food industries in the long terms. Table 14 shows the advantages and disadvantages of the application of immobilized enzymes in a heterogeneous form. The final decision to use immobilized protease depends on the technical‐economic evaluation, where the costs of immobilization process are compared with its benefits. It is also noteworthy that the application of immobilized enzymes on an industrial scale is increasing. [270]
Table 14.
Advantages and disadvantages of immobilized enzymes application (adapted from Ref. [279]).
|
Advantages |
Disadvantages |
|---|---|
|
Simple biocatalyst recovery. |
Lower proteolytic activity compared to native protein caused by undesired conformation. |
|
Reduced costs of downstream separation. |
Additional costs associated to carries and immobilization procedure. |
|
Reusability of enzyme. |
Biocatalyst fouling. |
|
Improved stability over broad range of operating conditions (organic solvents, pH, salinity and temperature) |
Mass transfer restrictions. |
|
Possibility of use fixed bed or batch reactors without need of membrane to isolate biocatalyst from the reaction medium. |
Disposal of exhausted immobilized enzyme. |
|
Functionality in continuous operations. |
Laborious and time‐consuming for immobilization processes |
Enzyme immobilization arose from the need to recover and reuse the biocatalyst, reducing the costs associated with the use of enzymes and avoiding the presence of the enzyme in the final product.[ 271 , 272 ] In addition, the immobilization of proteases, as with other enzymes, can lead to other benefits. The enzymatic activity can be increased due to conformational changes that occur during the immobilization process; these conformational changes can also generate changes in the selectivity/specificity of the protease. [273] In general, immobilization (depending on the methodology) can confer greater rigidity to the protein structure, thus, the immobilized enzyme shows greater thermoresistance, tolerance to solvents and/or changes in the pH of the medium.[ 272 , 274 , 275 ] Even more, the immobilization of enzymes can be coupled with the purification of the enzyme itself by simple procedures. [275] Additionally, in the case of proteases, immobilization could prevent autolysis. [264] However, due to the size of the protease substrates, immobilization must be properly evaluated. Limitations in substrate diffusion, orientation of the immobilized protease, and even high loading of immobilized protease (in porous materials) can lead to low enzyme activities.[ 276 , 277 , 278 ]
Three immobilization techniques can be used: carrier binding attachment, encapsulation or entrapment, and the formation of crosslinked enzyme aggregates. Carrier binding immobilization is characterized by the attachment of the enzyme onto a prefabricated solid material (ceramic, metal oxides, nanomaterials, polymers, or silica gel) by physisorption or chemisorption binding to reduce the solubility of enzyme in the reaction medium. Carrier binding immobilization involves low cost, low loss of activity and easy preparation.
Enzyme entrapment represents the immobilization of the enzyme into carriers with a degree of porosity commonly between 41 and 59 %, and permeability from 10 nm to 5 μm, or even more, enhancing the protein stability due to the control of its microenvironment. Sol gels, hydrogels, polymers, nanomaterials, and alginate beads are commonly used as carriers. Enzyme entrapment requires expense preparation; however, it offers a high affinity to the substrate.
On the other hand, enzyme immobilization can be carried out through the formation of cross‐linked enzyme aggregates (CLEAs). First, the precipitating stage is performed, where numerous precipitating reagents can be used (e. g., ammonium sulfate, acetone, acetone, ethanol, or tert‐butanol). In a second step, copolymerization of enzyme aggregates with a cross‐linking agent, commonly glutaraldehyde, is performed. CLEAs show high stability under a wide range of operating conditions, reusability, and low leaching in aqueous medium. An optimization process of synthesis conditions is required for each target biocatalyst using immobilization through the cross‐linking aggregate technique.
The immobilization of plant proteases through binding to supports is commonly performed considering the low cost and high stability of the immobilized enzyme. The selection of material support depends on the protease type and application conditions. In recent years, promising immobilization results have been documented using novel supports. Table 15 summarizes support materials commonly used for plant protease immobilization. In general, immobilized enzymes exhibit better adaptability to alkaline conditions, reusability and higher resistance to thermal inactivation and extreme pH values than the soluble ones.[ 4 , 280 ] The selection of optimal support and activation methods depends on affinity to enzyme, biocompatibility, availability and price, presence of reactional functional groups, regeneration and reusability, insolubility under reaction conditions and chemical and thermal stability. The procedures for plant proteases immobilization should provide precise control of enzyme orientation to preserve the functional properties of the active site during the process.
Table 15.
|
Support |
Features |
Examples |
|---|---|---|
|
Agarose beads |
• Commercial availability (well‐defined pore size: 45 to 620 nm), inert support. • Compatible with mechanical stirring. • Transparent support (enzyme fluorescence). • Possibility of rigorous control during immobilization process. |
• Papain [282] • Ficin [283] • Cardosin A [284] • S. origanifolia protease [285] |
|
Cellulose beads |
• Biological polymer (non‐porous nanoparticles or macroporous particles). • Support activation by direct oxidation (sodium periodate) forming di‐aldehyde that reacts with primary amino groups of proteins. • Immobilization process at 45 °C and pH 7. • High proteolytic stability. • Glutaraldehyde or succinic anhydride are used to covalently immobilize the enzyme to the support. |
|
|
Cotton fabric |
• Previous oxidation with sodium periodate is required. • Low loss of enzyme activity. • Low stability through consecutive uses, specially at alkaline conditions or in the presence of detergents. • Optimal pH shifted compared to native enzyme (commonly optimal pH is increased during immobilization). |
• Papain [288] |
|
Chitosan |
• Polysaccharide derived from chitin rich is hydroxyl groups and glucosamine (weak anion exchanger). • Activating agents are required to obtain a covalent enzyme immobilization (e. g., glutaraldehyde, epichlorohydrin, divinylsulfone, genipin). • High enzyme stability. • High microbial resistance. • Chitosan matrix acts as enzyme photoprotector. |
• Mungbean protease [289] • Procerain B [290] • Papain [291] • Bromelain [292] • Ficin [293] |
|
Alginate |
• Small pore size of alginate beads of enzyme immobilization is required (low commercial availability). • Alginate can adsorb metal ions from the reaction medium, protecting the enzyme properties. • Low loss of catalytic activity. |
• Papain [294] • Araujiain [295] • N. tabacum protease [296] • M. oleifera protease [296] • M. koenigii protease [296] • C. sativum protease [296] |
|
Synthetic organic supports |
• The chemical structure of the matrix could be designed (e. g., nylon grafted with polyacrylamide, monofunctional acrylate, polyoxyethylene dimethacrylate, ionic resin exchange, poly‐L‐lactic acid polymeric beads, polyacrylamide) • Activating agents are required (glutaraldehyde, polyethylene glycol, succinic anhydride) • High proteolytic activity. • High thermal stability of immobilized enzyme with remarkable microbial resistance. |
• Papain (p(HEMA‐EGDMA)) [297] • Ficin (PVA) [298] • Ficin (Poly(α‐hydroxyacids)) [299] |
|
Polymeric membranes |
• Commonly used for industrial applications: pharmaceutics (drug preparation), wastewater treatment (toxic compounds adsorption), biorefinery (hydrolysis reaction), biomedicine (drug delivery) and food processing (food preservation). • Great design reactor flexibility. • Materials support: vinyl alcohol/vinyl butyral copolymer (PVAB), hydroxyethyl cellulose coated with polyethersulfone (PES) hollow fibers • Higher specificity towards substrate than free enzyme. • High thermal stability over a wide range of pH. • Remarkable storage properties and stability. • High‐cost immobilization. |
|
|
Smart polymers |
• These materials change the solubility/insolubility status depending on the reaction conditions (pH and temperature). • Easy enzyme recovery and reuse. • Support materials: polymethyl methacrylate/N‐isopropylacrylamide/methacrylic acid, poly(styrene/N‐isopropylacrylamide/methacrylic acid. • High bioactivity and affinity towards substrate. • Remarkable enzyme stability after consecutive uses. |
• Papain (polymethyl metha crylate/N‐isopropylacrylamide/ methacrylic acid) [302] |
|
Mesoporous silicates |
• Well‐defined pore geometry, narrow pore size distribution and large surface area (spherosil, aminoorganosilica activated and porous silica). • High thermal and mechanical stability. • Remarkable dispersion in water and storage stability. • Abundant amount of hydroxyl groups on the surface, thus facilitating the binding of enzymes. • Surface modifying agents are required (e. g., trimethoxy‐derivatives, glutaraldehyde). • Better pH and thermal stability of immobilized protein than the source enzyme. • Low costs associated to materials purchase. |
• S. melongena (ZSM‐5 zeolite) [303] • A. curassavica protease (octyl‐glyoxyl‐silica) [304] • Antiacanthain (glyoxyl‐silica) [305] • Granulosain f (glyoxyl‐silica) [305] • Papain (mesoporous silica) [306] |
|
Inorganic oxide supports |
• Oxide‐based materials: titanium, aluminum, and zirconium oxides are commonly used for enzyme immobilization. • Attractive material properties, such as: high stability, resistant mechanical strength, good adsorption capacity and high hydrophilicity. • Low‐cost processing. |
• Araujiain (TiO2) [307] • C. Linamarase (clay of kaolin) [308] • Papain (Al2O3) [309] |
|
Magnetic particles |
• Easy recovery of the biocatalyst after the enzymatic tests using a magnetic field. • Large particles could lead to considerable diffusional limitations. • The surface of ferrite particles (Fe3O4) is commonly activated using thionylchloride to generate reactive chloride groups that interact with the free amino groups of enzymes to form amide bonds. • The immobilized biocatalysts exhibit higher pH, thermal and storage stabilities as well as environmental adaptability and reusability compared to the free enzyme. • Low immobilization costs. • Slight activity loss of biocatalyst during consecutive uses. • Valuable alternative for substrate hydrolysis in suspension. |
• Bromelain [310] • C. cardunculus extract (CoFe2O4) [311] • Papain (Ag/CuFe2O4) [312] |
From an overview, the implementation of an optimized biocatalytic process on an industrial scale involves the application of the following disciplines:
Process engineering: bioreactor design, optimization of operating conditions (solvent, concentration, temperature, pH, co‐enzymes, among others), and techno‐economic analysis of life cycle assessment.
Enzyme immobilization: stability of activity, reusability, biocatalyst recovery, mass transfer limitations, and molecular simulation for the optimal design.
Protein engineering: solvent‐ and thermostability, isolation and purifications optimization, and enhancement of enzyme‐substrate affinity using co‐factors and surfactants. [271]
A preliminary optimization process should be carried out for each particular plant protease in order to determine the optimal operating conditions of the biocatalyst before scaling up.
6. Conclusions
Plant proteases show enormous potential for different applications, from the most traditional to the least explored. Emerging vegetable proteases with attractive functional properties and remarkable proteolytic activity on a broad spectrum of substrates (e. g., keratin, gelatin, collagen, gliadin, fish protein, whey protein, soy protein, glutenins) are documented every year. The main advantages of plant proteases obtained from crops in comparison to animal and microbial proteases are low productions cost, the use of genetic engineering techniques is not required, abundant protein sources (flowers, latex, node, stem, roots, and fruits), broad substrate specificity, and substantial stability over a wide range of operating conditions (pH, temperature, salinity, and organic solvents). Large‐scale plant proteases production can be significatively increased using in vitro methodologies (micropropagation, somatic embryogenesis, callus, and cell suspension cultures), which are independent of climatic factors, crop diseases, heterogeneity in the source material, and the application of pesticides. However, rigorous control of environmental conditions is required. In contrast, plant proteases derived from crops can be isolated using simple procedures (e. g., solubilization with mild solvents, ammonium precipitation, and centrifugation), followed by final conditioning (spray drying).
In recent years the use of plant proteases has expanded to new potential industrial applications. Promising results for the valorization of cheese whey (rich protein source) through hydrolysis using plant proteases to produce therapeutic biopeptides have been documented, especially using Maclura pomifera, Cynara cardunculus extracts, and ficin. Even proteases obtained from Araujia hortorum latex are an attractive alternative. Plant proteases present a high affinity towards gluten as a substrate, which can be hydrolyzed to obtain bioactive peptides with antioxidant properties. Substantial reduction of gluten protein allergenicity by hydrolysis has been documented in the literature. Besides, plant proteases could be directly added to bakery preparations.
Regarding bird feather valorization, scarce studies have been reported. Vegetable proteases should be used in combination with other proteases (proteinase K, trypsin) to achieve a feasible yield on keratin degradation. Similar observations can be made for the hydrolysis of keratinous material, collagen, and gelatin. Keratinase and collagenase proteins are not abundant in plants. Favorable results for keratin and gelatin hydrolysis under alkaline conditions have been reported using zingibain (Zingiber officinale), Cucumis melo protease, papain, actinidin, cardosin, and ficin.
Another emerging application for plant proteases is in the leather industry. Papain, bromelain, and proteases from Calotropis procera and Agave americana show suitable properties for the dehairing process without damaging the hide, offering an alternative to the use of harmful chemicals. Furthermore, vegetable proteins extracts have demonstrated stability higher than 60 % at operating conditions commonly used in the leather industry (45 °C, pH between 6 and 10).
Fish hydrolysis using commercial plant proteases (papain, ficin, bromelains) has been studied. Vegetable proteases have remarkable performance when are combined with other proteases (neutrase, alcalase, protamex). Plant proteases are selective to produce biopeptides with therapeutic uses such as antioxidant effects, antimicrobial, antihypertensive, reducing cholesterol, anti‐osteoporosis, anti‐cancer, and immunization properties. Soy proteins are another valuable source for obtaining biopeptides. For this raw material, plant proteases exhibit higher affinity compared to animal or microbial proteases. Besides commercial proteases, emerging peptidases derived from plant latex have shown high selective towards biopeptide production, such as Maclura pomifera and Asclepias fruticosa.
There are few reports of water treatment using plant proteases. The studies focus on metal removal employing immobilized plant proteases on charcoal through physical adsorption procedures. Vegetable proteases could be used for mercury, lead, and chromium removal at low concentrations. However, the regeneration of the respective biocatalyst is a major challenge. Further studies are required for the applications of plant peroxidases and proteases on the degradation of organic compounds.
The application of plant proteases at industrial scale is an important technological challenge. Optimization of operating conditions, reuse strategies, and enzyme purity should be studied before scaling up. Protein immobilization by binding to a support is an affordable methodology to increase the stability of plant protease in aqueous medium, permitting an easy recovering of the biocatalyst. This is significant to decrease the processing cost on the valorization of industrial by‐products rich in proteins and amino acids.
Conflict of interest
The authors declare no conflict of interest.
7.
Biographical Information
Franco Troncoso obtained his PhD. in chemical engineering in 2020 from the Universidad Nacional del Sur (Argentina). In 2021 he joined Prof. María Luján Ferreira's group as postdoctoral researcher in PLAPIQUI. His research interests include green chemistry, with special interest in the application of biocatalysts. He has experience in the field of heterogeneous catalysis, with emphasis on structured catalysts, continuous flow reactions, and process simulations. He has also acted in the following topics: biofuel production, process engineering design, and industrial innovation.

Biographical Information
Ph.D. in chemical engineering Daniel Sanchez is an Assistant Scientific Researcher of the Consejo Nacional de Investigaciones Cinetíficas y Técnicas (CONICET, Argentina). He obtained his doctorate degree from the Universidad Nacional del Sur (Argentina) in 2016. His research interests are focused on biocatalysis, ranging from obtaining biocatalysts based on new sources of enzymes to the application of biocatalysts in the synthesis of products with high added value.

Biographical Information
Ph.D. in chemistry María Luján Ferreira is a Senior Researcher of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina). She has more than 25 years of experience as a Researcher. Her interests include from molecular modeling to applied research in biotechnology and nanotechnology. Her research has been focused especially on lipases and more recently in looking at vegetable sources of enzymes, with applications in fields of food, cosmetics, medicine, and pharmaceuticals, among others.

Acknowledgements
The authors thank the Agencia Nacional de Promoción Científica y Tecnológica (National Agency of Scientific and Technological Promotion, Argentina) (PICT 2018‐03425) and the Consejo Nacional de Investigaciones Científicas y Técnicas (National Council for Scientific and Technological Research, CONICET) for the financial support
F. David Troncoso, D. Alberto Sánchez, M. Luján Ferreira, ChemistryOpen 2022, 11, e202200017.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Quirós P. M., Langer T., López-Otín C., Nat. Rev. Mol. Cell Biol. 2015, 16, 345–359. [DOI] [PubMed] [Google Scholar]
- 2. De Strooper B., Annaert W., J. Cell Sci. 2000, 113, 1857–1870. [DOI] [PubMed] [Google Scholar]
- 3. Naveed M., Nadeem F., Mehmood T., Bilal M., Anwar Z., Amjad F., Catal. Lett. 2021, 151, 307–323. [Google Scholar]
- 4. Feijoo-Siota L., Villa T. G., Food Bioproc. Technol. 2011, 4, 1066–1088. [Google Scholar]
- 5. Barrett A. J., Methods Enzymol. 1994, 244, 1–15. [DOI] [PubMed] [Google Scholar]
- 6.M. G. Guevara, G. R. Daleo, Biotechnological Applications of Plant Proteolytic Enzymes, Springer International Publishing, 2018.
- 7. Rawlings N. D., Bateman A., Protein Sci. 2021, 30, 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martínez M., Cambra I., González-Melendi P., Santamaría M. E., Díaz I., Physiol. Plant. 2012, 145, 85–94. [DOI] [PubMed] [Google Scholar]
- 9. Van Der Hoorn R. A. L., Klemenčič M., J. Exp. Bot. 2021, 72, 3337–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shah M. A., Mir S. A., Paray M. A., Dairy Sci. Technol. 2014, 94, 5–16. [Google Scholar]
- 11. Elsässer B., Goettig P., Int. J. Mol. Sci. 2021, 22, 3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murayama K., Kato-Murayama M., Hosaka T., Sotokawauchi A., Yokoyama S., Arima K., Shirouzu M., J. Mol. Biol. 2012, 423, 386–396. [DOI] [PubMed] [Google Scholar]
- 13. Kamphuis I. G., Kalk K. H., Swarte M. B. A., Drenth J., J. Mol. Biol. 1984, 179, 233–256. [DOI] [PubMed] [Google Scholar]
- 14. Fernández-Lucas J., Castañeda D., Hormigo D., Trends Food Sci. Technol. 2017, 68, 91–101. [Google Scholar]
- 15. Arshad Z. I. M., Amid A., Yusof F., Jaswir I., Ahmad K., Loke S. P., Appl. Microbiol. Biotechnol. 2014, 98, 7283–7297. [DOI] [PubMed] [Google Scholar]
- 16. Morellon-Sterling R., El-Siar H., Tavano O. L., Berenguer-Murcia Á., Fernández-Lafuente R., Int. J. Biol. Macromol. 2020, 162, 394–404. [DOI] [PubMed] [Google Scholar]
- 17. Homaei A., Etemadipour R., Int. J. Biol. Macromol. 2015, 72, 1176–1181. [DOI] [PubMed] [Google Scholar]
- 18. Moro L. P., Cabral H., Okamoto D. N., Hirata I., Juliano M. A., Juliano L., Bonilla-Rodriguez G. O., Process Biochem. 2013, 48, 633–637. [Google Scholar]
- 19. Tomar R., Kumar R., Jagannadham M. V., J. Agric. Food Chem. 2008, 56, 1479–1487. [DOI] [PubMed] [Google Scholar]
- 20. Patel A. K., Singh V. K., Jagannadham M. V., J. Agric. Food Chem. 2007, 55, 5809–5818. [DOI] [PubMed] [Google Scholar]
- 21. Yadav R. P., Patel A. K., Jagannadham M. V., Food Chem. 2012, 132, 1296–1304. [DOI] [PubMed] [Google Scholar]
- 22. Mohamed Ahmed I. A., Morishima I., Babiker E. E., Mori N., Phytochemistry 2009, 70, 483–491. [DOI] [PubMed] [Google Scholar]
- 23. Yadav S. C., Pande M., Jagannadham M. V., Phytochemistry 2006, 67, 1414–1426. [DOI] [PubMed] [Google Scholar]
- 24. Kumari M., Sharma A., Jagannadham M. V., J. Agric. Food Chem. 2010, 58, 8027–8034. [DOI] [PubMed] [Google Scholar]
- 25. Rajesh R., Nataraju A., Gowda C. D. R., Frey B. M., Frey F. J., Vishwanath B. S., Biochimie 2006, 88, 1313–1322. [DOI] [PubMed] [Google Scholar]
- 26. Vairo Cavalli S., Silva S. V., Cimino C., Malcata F. X., Priolo N., Food Chem. 2008, 106, 997–1003. [Google Scholar]
- 27. Raposo S., Domingos A., Process Biochem. 2008, 43, 139–144. [Google Scholar]
- 28. Devaraj K. B., Gowda L. R., Prakash V., Phytochemistry 2008, 69, 647–655. [DOI] [PubMed] [Google Scholar]
- 29. Cimino C. V., Colombo M. L., Liggieri C., Bruno M., Vairo-Cavalli S., J. Med. Food 2015, 18, 856–864. [DOI] [PubMed] [Google Scholar]
- 30. Oliveira A., Pereira C., da Costa D. S., Teixeira J., Fidalgo F., Pereira S., Pissarra J., Plant Sci. 2010, 178, 140–146. [Google Scholar]
- 31. Brutti C. B., Pardo M. F., Caffini N. O., Natalucci C. L., LWT-Food Sci. Technol. 2012, 45, 172–179. [Google Scholar]
- 32. Streatfield S. J., Plant Biotechnol. J. 2007, 5, 2–15. [DOI] [PubMed] [Google Scholar]
- 33. Khan M. S., Joyia F. A., Mustafa G., Protein Pept. Lett. 2019, 27, 89–104. [DOI] [PubMed] [Google Scholar]
- 34. Desai P. N., Shrivastava N., Padh H., Biotechnol. Adv. 2010, 28, 427–435. [DOI] [PubMed] [Google Scholar]
- 35. Egelkrout E., Rajan V., Howard J. A., Plant Sci. 2012, 184, 83–101. [DOI] [PubMed] [Google Scholar]
- 36. Porto M. S., Pinheiro M. P. N., Batista V. G. L., Dos Santos R. C., De Albuquerque Melo Filho P., De Lima L. M., Mol. Biotechnol. 2014, 56, 38–49. [DOI] [PubMed] [Google Scholar]
- 37. Lico C., Santi L., Twyman R. M., Pezzotti M., Avesani L., Plant Cell Rep. 2012, 31, 439–451. [DOI] [PubMed] [Google Scholar]
- 38. Castanheira P., Samyn B., Sergeant K., Clemente J. C., Dunn B. M., Pires E., Van Beeumen J., Faro C., J. Biol. Chem. 2005, 280, 13047–13054. [DOI] [PubMed] [Google Scholar]
- 39. González-Rábade N., Badillo-Corona J. A., Aranda-Barradas J. S., del C. Oliver-Salvador M., Biotechnol. Adv. 2011, 29, 983–996. [DOI] [PubMed] [Google Scholar]
- 40. Mazorra-Manzano M. A., Ramírez-Suarez J. C., Yada R. Y., Crit. Rev. Food Sci. Nutr. 2018, 58, 2147–2163. [DOI] [PubMed] [Google Scholar]
- 41. Fernández G., Pomilio A. B., Mol. Med. Chem. 2003, 1, 39–49. [Google Scholar]
- 42. Huang H. L., Hsing H. W., Lai T. C., Chen Y. W., Lee T. R., Chan H. T., Lyu P. C., Wu C. L., Lu Y. C., Lin S. T., Lin C. W., Lai C. H., Chang H. T., Chou H. C., Chan H. L., J. Biomed. Sci. 2010, 17, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gutierrez-Valdes N., Häkkinen S. T., Lemasson C., Guillet M., Oksman-Caldentey K. M., Ritala A., Cardon F., Front. Plant Sci. 2020, 11, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gupta N., Jain V., Joseph M. R., Devi S., Asian J. Pharm. Res. Dev. 2020, 8, 86–93. [Google Scholar]
- 45. Phulwaria M., Rai M. K., Harish, Gupta A. K., Ram K., Shekhawat N. S., Acta Physiol. Plant. 2012, 34, 299–305. [Google Scholar]
- 46. Nuño-Ayala A., Rodríguez-Garay B., Gutiérrez-Mora A., Plant Cell Tissue Organ Cult. 2012, 109, 33–39. [Google Scholar]
- 47. Anandan R., Sudhakar D., Balasubramanian P., Gutieórrez-Mora A., Sci. Hortic. 2012, 136, 43–49. [Google Scholar]
- 48. Folgado A., Pires A. S., Figueiredo A. C., Pimentel C., Abranches R., Plant Cell Rep. 2020, 39, 89–100. [DOI] [PubMed] [Google Scholar]
- 49. Elateeq A. A., Sun Y., Nxumalo W., Gabr A. M. M., Biocatal. Agric. Biotechnol. 2020, 29, 101775. [Google Scholar]
- 50. Dini I., Falanga D., Di Lorenzo R., Tito A., Carotenuto G., Zappelli C., Grumetto L., Sacchi A., Laneri S., Apone F., Antioxidants 2021, 10, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hussain M. S., Fareed S., Ansari S., Rahman M. A., Ahmad I. Z., Saeed M., J. Pharm. BioAllied Sci. 2012, 4, 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Corbin J. M., McNulty M. J., Macharoen K., McDonald K. A., Nandi S., Biotechnol. Bioeng. 2020, 117, 3053–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sun Q., Zhang B., Yan Q. J., Jiang Z. Q., Food Chem. 2016, 213, 708–713. [DOI] [PubMed] [Google Scholar]
- 54. Domsalla A., Melzig M. F., Planta Med. 2008, 74, 699–711. [DOI] [PubMed] [Google Scholar]
- 55. Mazorra-Manzano M. A., Moreno-Hernández J. M., Ramírez-Suarez J. C., de J. Torres-Llanez M., González-Córdova A. F., Vallejo-Córdoba B., LWT-Food Sci. Technol. 2013, 54, 325–330. [Google Scholar]
- 56. Shi Y., Wang X., Huang A., Int. J. Biol. Macromol. 2018, 115, 883–890. [DOI] [PubMed] [Google Scholar]
- 57. Esposito M., Di Pierro P., Dejonghe W., Mariniello L., Porta R., Food Chem. 2016, 204, 115–121. [DOI] [PubMed] [Google Scholar]
- 58. Mehrnoush A., Mustafa S., Sarker M. Z. I., Yazid A. M. M., Int. J. Mol. Sci. 2012, 13, 3636–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Badgujar S. B., J. Ethnopharmacol. 2014, 151, 733–739. [DOI] [PubMed] [Google Scholar]
- 60. Fonseca K. C., Morais N. C. G., Queiroz M. R., Silva M. C., Gomes M. S., Costa J. O., Mamede C. C. N., Torres F. S., Penha-Silva N., Beletti M. E., Canabrava H. A. N., Oliveira F., Phytochemistry 2010, 71, 708–715. [DOI] [PubMed] [Google Scholar]
- 61. Schaaf A., Reski R., Decker E. L., Eur. J. Cell Biol. 2004, 83, 145–152. [DOI] [PubMed] [Google Scholar]
- 62. Benchabane M., Schlüter U., Vorster J., Goulet M. C., Michaud D., Biochimie 2010, 92, 1657–1666. [DOI] [PubMed] [Google Scholar]
- 63. Bruno M. A., Lazza C. M., Errasti M. E., López L. M. I., Caffini N. O., Pardo M. F., LWT-Food Sci. Technol. 2010, 43, 695–701. [Google Scholar]
- 64. Ben Amira A., Besbes S., Attia H., Blecker C., Int. J. Food Prop. 2017, 20, 76–93. [Google Scholar]
- 65. Ahmed I. A. M., Morishima I., Babiker E. E., Mori N., Food Chem. 2009, 116, 395–400. [Google Scholar]
- 66. Schaller A., Handb. Proteolytic Enzym. 2013, 3, 3247–3254. [Google Scholar]
- 67. Graycar T. P., Bott R. R., Power S. D., Estell D. A., Handb. Proteolytic Enzym. 2013, 3, 3148–3155. [Google Scholar]
- 68. Liu H., Hu M., Wang Q., Cheng L., Zhang Z., Front. Plant Sci. 2018, 871, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Borde V., Pawar D., Thorat D., Int. J. Curr. Microbiol. Appl. Sci. 2013, 2, 369–375. [Google Scholar]
- 70. Priolo N., del Valle S. M., Arribére M. C., López L., Caffini N., J. Protein Chem. 2000, 19, 39–49. [DOI] [PubMed] [Google Scholar]
- 71. de Castro R. J. S., Bagagli M. P., Sato H. H., Curr. Opin. Food Sci. 2015, 1, 64–69. [Google Scholar]
- 72. Kaur S., Huppertz T., Vasiljevic T., Int. Dairy J. 2021, 118, 105029. [Google Scholar]
- 73. Fadhilah Y., Shoobihah A., Setiasih S., Handayani S., Hudiyono S., AIP Conf. Proc. 2018, 2049, 020029. [Google Scholar]
- 74. Shu G., Huang J., Bao C., Meng J., Chen H., Cao J., Biomol. 2018, 8, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rodrigues I. M., Coelho J. F. J., Carvalho M. G. V. S., J. Food Eng. 2012, 109, 337–346. [Google Scholar]
- 76. Nehete J. Y., Bhambar R. S., Narkhede M. R., Gawali S. R., Pharmacogn. Rev. 2013, 7, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rajagopalan A., Sukumaran B. O., Int. J. Biol. Macromol. 2018, 118, 279–288. [DOI] [PubMed] [Google Scholar]
- 78. Pontual E. V., Carvalho B. E. A., Bezerra R. S., Coelho L. C. B. B., Napoleão T. H., Paiva P. M. G., Food Chem. 2012, 135, 1848–1854. [DOI] [PubMed] [Google Scholar]
- 79. Mehrnoush A., Mustafa S., Sarker M. Z. I., Yazid A. M. M., Molecules 2011, 16, 9245–9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Alfaro-Diaz A., Urías-Silvas J. E., Loarca-Piña G., Gaytan-Martínez M., Prado-Ramirez R., Mojica L., LWT 2021, 136, 110296. [Google Scholar]
- 81. Zhang D. Q., Mu T. H., Sun H. N., Chen J. W., Zhang M., Int. J. Food Prop. 2017, 20, 2113–2127. [Google Scholar]
- 82. Duong-Ly K. C., Gabelli S. B., Methods Enzymol. 2014, 541, 85–94. [DOI] [PubMed] [Google Scholar]
- 83. Gagaoua M., Ziane F., Nait Rabah S., Boucherba N., Ait Kaki El-Hadef El-Okki A., Bouanane-Darenfed A., Hafid K., Int. J. Biol. Macromol. 2017, 102, 515–525. [DOI] [PubMed] [Google Scholar]
- 84. Abdul-Fattah A. M., Kalonia D. S., Pikal M. J., J. Pharm. Sci. 2007, 96, 1886–1916. [DOI] [PubMed] [Google Scholar]
- 85. Saran S., Mahajan R. V., Kaushik R., Isar J., Saxena R. K., J. Cleaner Prod. 2013, 54, 315–322. [Google Scholar]
- 86. Sujitha P., Kavitha S., Shakilanishi S., Babu N. K. C., Shanthi C., Int. J. Biol. Macromol. 2018, 118, 168–179. [DOI] [PubMed] [Google Scholar]
- 87. Luo F., Zhong X., Gao M., Peng B., Long Z., J. Leather Sci. Eng. 2020, 2, 1–16. [Google Scholar]
- 88. Lange L., Huang Y., Busk P. K., Appl. Microbiol. Biotechnol. 2016, 100, 2083–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Senthilvelan T., Kanagaraj J., Mandal A. B., Clean Technol. Environ. Policy 2012, 14, 889–897. [Google Scholar]
- 90. Fang Z., Yong Y. C., Zhang J., Du G., Chen J., Appl. Microbiol. Biotechnol. 2017, 101, 7771–7779. [DOI] [PubMed] [Google Scholar]
- 91. Lopéz L. M. I., Viana C. A., Errasti M. E., Garro M. L., Martegani J. E., Mazzilli G. A., Freitas C. D. T., Araújo Í. M. S., Da Silva R. O., Ramos M. V., Bioprocess Biosyst. Eng. 2017, 40, 1391–1398. [DOI] [PubMed] [Google Scholar]
- 92. Bouhlel M., Brini F., Saibi W., Biochem. Mol. Biol. 2021, 7, 15. [Google Scholar]
- 93. Mohan R., Sivakumar V., Rangasamy T., Muralidharan C., Am. J. Biochem. Biotechnol. 2016, 12, 188–195. [Google Scholar]
- 94. Errasti M. E., Torres M. J., Mercerat J. R., Caffini N. O., López L. M. I., Biocatal. Biotransform. 2020, 38, 357–366. [Google Scholar]
- 95. Badgujar S. B., Mahajan R. T., Sci. World J. 2013, 2013, 716545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mazorra-Manzano M. A., Mora-Cortes W. G., Leandro-Roldan M. M., González-Velázquez D. A., Torres-Llanez M. J., Ramírez-Suarez J. C., González-Córdova A. F., Vallejo-Córdoba B., Biocatal. Agric. Biotechnol. 2020, 28, 101724. [Google Scholar]
- 97. Bacenetti J., Bava L., Schievano A., Zucali M., J. Food Eng. 2018, 224, 139–147. [Google Scholar]
- 98. Ryan M. P., Walsh G., Rev. Environ. Sci. Bio/Technol. 2016, 15, 479–498. [Google Scholar]
- 99. Barros R. M., Malcata F. X., Food Chem. 2004, 88, 351–359. [Google Scholar]
- 100. Carvalho F., Prazeres A. R., Rivas J., Sci. Total Environ. 2013, 445–446, 385–396. [DOI] [PubMed] [Google Scholar]
- 101. Addai F. P., Lin F., Wang T., Kosiba A. A., Sheng P., Yu F., Gu J., Zhou Y., Shi H., Food Funct. 2020, 11, 8407–8423. [DOI] [PubMed] [Google Scholar]
- 102. Madureira A. R., Tavares T., Gomes A. M. P., Pintado M. E., Malcata F. X., J. Dairy Sci. 2010, 93, 437–455. [DOI] [PubMed] [Google Scholar]
- 103. Madureira A. R., Pereira C. I., Gomes A. M. P., Pintado M. E., X–Malcata F., Food Res. Int. 2007, 40, 1197–1211. [Google Scholar]
- 104. Mazorra-Manzano M. A., Perea-Gutiérrez T. C., Lugo-Sánchez M. E., Ramirez-Suarez J. C., Torres-Llanez M. J., González-Córdova A. F., Vallejo-Cordoba B., Food Chem. 2013, 141, 1902–1907. [DOI] [PubMed] [Google Scholar]
- 105. Corrons M. A., Bertucci J. I., Liggieri C. S., López L. M. I., Bruno M. A., LWT-Food Sci. Technol. 2012, 47, 103–109. [Google Scholar]
- 106. Tavares T. G., Amorim M., Gomes D., Pintado M. E., Pereira C. D., Malcata F. X., J. Food Eng. 2012, 110, 547–552. [Google Scholar]
- 107. Iwaniak A., Minkiewicz P., Hrynkiewicz M., Bucholska J., Darewicz M., Polish J. Food Nutr. Sci. 2020, 70, 139–150. [Google Scholar]
- 108. Aider M., JDS Commun. 2021, 5, 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Salami M., Yousefi R., Ehsani M. R., Dalgalarrondo M., Chobert J. M., Haertlé T., Razavi S. H., Saboury A. A., Niasari-Naslaji A., Moosavi-Movahedi A. A., Int. Dairy J. 2008, 18, 1097–1102. [Google Scholar]
- 110. Arruda M. S., Silva F. O., Egito A. S., Silva T. M. S., Lima-Filho J. L., Porto A. L. F., Moreira K. A., LWT 2012, 49, 73–79. [Google Scholar]
- 111. Obregón W. D., Arribére M. C., Morcelle del Valle S., Liggieri C., Caffini N., Priolo N., J. Protein Chem. 2001, 20, 317–325. [DOI] [PubMed] [Google Scholar]
- 112. Obregón W. D., Liggieri C. S., Trejo S. A., Avilés F. X., Vairo-Cavalli S. E., Priolo N. S., Biochimie 2009, 91, 1457–1464. [DOI] [PubMed] [Google Scholar]
- 113. Quiroga E., Priolo N., Obregón D., Marchese J., Barberis S., Biochem. Eng. J. 2008, 39, 115–120. [Google Scholar]
- 114. Zhao D. B., Bai Y. H., Niu Y. W., Small Rumim. Res. 2015, 127, 58–67. [Google Scholar]
- 115. Park Y. W., Juárez M., Ramos M., Haenlein G. F. W., Small Rumin. Res. 2007, 68, 88–113. [Google Scholar]
- 116. Al-Shamsi K. A., Mudgil P., Hassan H. M., Maqsood S., J. Dairy Sci. 2018, 101, 47–60. [DOI] [PubMed] [Google Scholar]
- 117. Wali A., Yanhua G., Ishimov U., Yili A., Aisa H. A., Salikhov S., Int. J. Pept. Res. Ther. 2019, 26, 641–650. [Google Scholar]
- 118. Landi N., Ragucci S., Di Maro A., Food 2021, 10, 2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Navarrete-Rodríguez E. M., Ríos-Villalobos L. A., Alcocer-Arreguín C. R., Del-Rio-Navarro B. E., Del Rio-Chivardi J. M., Saucedo-Ramírez O. J., Sienra-Monge J. J. L., Frias R. V., Allergol. Immunopathol. 2018, 46, 149–154. [DOI] [PubMed] [Google Scholar]
- 120. Faye B., Konuspayeva G., Bengoumi M., J. Camelid Sci. 2019, 12, 17–32. [Google Scholar]
- 121. Pires A. F., Marnotes N. G., Rubio O. D., Garcia A. C., Pereira C. D., Food 2021, 10, 1067. [Google Scholar]
- 122.M. A. Mazorra-Manzano, J. M. Moreno-Hernández, J. C. Ramírez-Suarez, in Biotechnological Applications of Plant Proteolytic Enzymes. Milk-Clotting Plant Proteases for Cheesemaking, Proceedings (Eds.:M. G. Guevara, G. R. Daleo), Springer, Cham, 2018, pp. 21–41.
- 123. Beigomi M., Mohammadifar M. A., Hashemi M., Rohani M. G., Senthil K., Valizadeh M., Food Sci. Biotechnol. 2014, 23, 1805–1813. [Google Scholar]
- 124. Grozdanovic M. M., Burazer L., Gavrovic-Jankulovic M., Int. Dairy J. 2013, 32, 46–52. [Google Scholar]
- 125. Jacob M., Jaros D., Rohm H., Int. J. Dairy Technol. 2011, 64, 14–33. [Google Scholar]
- 126. Néstor G. M., Dely Rubí C. G., Héctor J. C., J. Food Sci. 2012, 77, C89–C94. [DOI] [PubMed] [Google Scholar]
- 127. Jin J., Ma H., Zhou C., Luo M., Liu W., Qu W., He R., Luo L., Yagoub A. E. G. A., J. Sci. Food Agric. 2015, 95, 2501–2509. [DOI] [PubMed] [Google Scholar]
- 128. Asrarkulova A. S., Bulushova N. V., Appl. Biochem. Microbiol. 2019, 54, 825–833. [Google Scholar]
- 129. Popineau Y., Huchet B., Larré C., Bérot S., J. Cereal Sci. 2002, 35, 327–335. [Google Scholar]
- 130. Rombouts I., Lamberts L., Celus I., Lagrain B., Brijs K., Delcour J. A., J. Chromatogr. A 2009, 1216, 5557–5562. [DOI] [PubMed] [Google Scholar]
- 131. Knorr V., Kerpes R., Wieser H., Zarnkow M., Becker T., Koehler P., Eur. Food Res. Technol. 2016, 242, 585–597. [Google Scholar]
- 132. Gänzle M. G., Loponen J., Gobbetti M., Trends Food Sci. Technol. 2008, 19, 513–521. [Google Scholar]
- 133. Selinheimo E., Autio K., Kruus K., Buchert J., J. Agric. Food Chem. 2007, 55, 6357–6365. [DOI] [PubMed] [Google Scholar]
- 134. Scherf K. A., Curr. Opin. Food Sci. 2019, 25, 35–41. [Google Scholar]
- 135. Gabr G. A., Asian J. Biotechnol. Bioresour. Technol. 2018, 8, 1–9. [Google Scholar]
- 136. Bellir N., Bellir M. N., Rouabah L., World J. Pharm. Sci. 2014, 12, 1555–1571. [Google Scholar]
- 137. Ravee R., Salleh F. I. M., Goh H., PeerJ 2018, 6, e4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Buddrick O., Cornell H. J., Small D. M., Food Chem. 2015, 170, 343–347. [DOI] [PubMed] [Google Scholar]
- 139. Wang J. S., Wei Z. Y., Li L., Bian K., Zhao M. M., J. Cereal Sci. 2009, 50, 205–209. [Google Scholar]
- 140. Xue L., Li Y., Li T., Pan H., Liu J., Fan M., Qian H., Zhang H., Ying H., Wang L., J. Agric. Food Chem. 2019, 67, 6313–6323. [DOI] [PubMed] [Google Scholar]
- 141. Li Y., Yu J., Goktepe I., Ahmedna M., Food Chem. 2016, 196, 1338–1345. [DOI] [PubMed] [Google Scholar]
- 142. Kaur L., Rutherfurd S. M., Moughan P. J., Drummond L., Boland M. J., J. Agric. Food Chem. 2010, 58, 5074–5080. [DOI] [PubMed] [Google Scholar]
- 143. Jayawardana I. A., Boland M. J., Higgs K., Zou M., Loo T., Mcnabb W. C., Montoya C. A., Food Chem. 2021, 341, 128239. [DOI] [PubMed] [Google Scholar]
- 144. Jayawardana I. A., Montoya C. A., McNabb W. C., Boland M. J., Trends Food Sci. Technol. 2019, 94, 91–97. [Google Scholar]
- 145. Taga Y., Hayashida O., Kusubata M., Ogawa-Goto K., Hattori S., Biosci. Biotechnol. Biochem. 2017, 81, 1823–1828. [DOI] [PubMed] [Google Scholar]
- 146. Hu R., Chen G., Li Y., Molecules 2020, 25, 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Apinunjarupong S., Lapnirun S., Theerakulkait C., Prep. Biochem. Biotechnol. 2009, 39, 183–193. [DOI] [PubMed] [Google Scholar]
- 148. Peng Z., Mao X., Zhang J., Du G., Chen J., Microb. Cell Fact. 2019, 18, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Tesfaye T., Sithole B., Ramjugernath D., Clean Technol. Environ. Policy 2017, 19, 2363–2378. [Google Scholar]
- 150. Bhari R., Kaur M., Singh R. S., Curr. Microbiol. 2021, 78, 2212–2230. [DOI] [PubMed] [Google Scholar]
- 151. Saravanan K., Dhurai B., J. Text. Apparel, Technol. Manag. 2012, 7, 1–6. [Google Scholar]
- 152. Korniłłowicz-Kowalska T., Bohacz J., Waste Manage. 2011, 31, 1689–1701. [DOI] [PubMed] [Google Scholar]
- 153. Li Z. W., Liang S., Ke Y., Deng J. J., Zhang M. S., Lu D. L., Li J. Z., Luo X. C., Commun. Biol. 2020, 3, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Gupta A., Kamarudin N. B., Chua G. K., Yunus R. M., Yeo C., Kee G., Bin R., Yunus M., J. Chem. Chem. Eng. 2012, 6, 732–737. [Google Scholar]
- 155. Yusuf I., Garba L., Shehu M. A., Oyiza A. M., Kabir M. R., Haruna M., Int. Microbiol. 2019, 23, 189–200. [DOI] [PubMed] [Google Scholar]
- 156. Mahajan R., Chaudhari G., Chopadaa M., J. Appl. Biotechnol. Rep. 2015, 2, 333–337. [Google Scholar]
- 157. Callegaro K., Brandelli A., Daroit D. J., Waste Manage. 2019, 95, 399–415. [DOI] [PubMed] [Google Scholar]
- 158. Jin H. S., Park S. Y., Kim K., Lee Y. J., Nam G. W., Kang N. J., Lee D. W., PLoS One 2017, 12, e172712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Ren Y., Luo H., Huang H., Hakulinen N., Wang Y., Wang Y., Su X., Bai Y., Zhang J., Yao B., Wang G., Tu T., Int. J. Biol. Macromol. 2020, 154, 1586–1595. [DOI] [PubMed] [Google Scholar]
- 160. Mi X., Chang Y., Xu H., Yang Y., Food Chem. 2019, 300, 125181. [DOI] [PubMed] [Google Scholar]
- 161. Chojnacka K., Górecka H., Michalak I., Górecki H., Waste Biomass Valori. 2011, 2, 317–321. [Google Scholar]
- 162. Holkar C. R., Jain S. S., Jadhav A. J., Pinjari D. V., Process Saf. Environ. Prot. 2018, 115, 85–98. [Google Scholar]
- 163. Martinez J. P. D. O., Cai G., Nachtschatt M., Navone L., Zhang Z., Robins K., Speight R., Catal. 2020, 10, 184. [Google Scholar]
- 164. Sinkiewicz I., Śliwińska A., Staroszczyk H., Kołodziejska I., Waste Biomass Valori. 2016, 8, 1043–1048. [Google Scholar]
- 165. Nurdiawati A., Nakhshiniev B., Gonzales H. B., Yoshikawa K., Appl. Soil Ecol. 2019, 134, 98–104. [Google Scholar]
- 166. Nanda R. F., Bahar R., Syukri D., Thu N. N. A., Kasim A., Andalasian Int. J. Agric. Nat. Sci. 2020, 1, 33–44. [Google Scholar]
- 167. Wang P., Wang Q., Cui L., Fan X., Yuan J., Gao M., Fibers Polym. 2010, 11, 586–592. [Google Scholar]
- 168. Yoshida-Yamamoto S., Nishimura S., Okuno T., Rakuman M., Takii Y., Mol. Biotechnol. 2010, 46, 41–48. [DOI] [PubMed] [Google Scholar]
- 169. Shi F., Liu Q., Zhao H., Fang K., Xie R., Song L., Wang M., Chen W., ACS Sustainable Chem. Eng. 2021, 9, 10361–10369. [Google Scholar]
- 170. Bhagwat P. K., Dandge P. B., Biocatal. Agric. Biotechnol. 2018, 15, 43–55. [Google Scholar]
- 171. Silvipriya K. S., Kumar K. K., Bhat A. R., Dinesh Kumar B., John A., James S., J. Appl. Pharmacol. 2015, 5, 123–127. [Google Scholar]
- 172. Barati M., Jabbari M., Navekar R., Farahmand F., Zeinalian R., Salehi-Sahlabadi A., Abbaszadeh N., Mokari-Yamchi A., Davoodi S. H., J. Cosmet. Dermatol. 2020, 19, 2820–2829. [DOI] [PubMed] [Google Scholar]
- 173. Rodríguez M. I. A., Barroso L. G. R., Sánchez M. L., J. Cosmet. Dermatol. 2018, 17, 20–26. [DOI] [PubMed] [Google Scholar]
- 174. Parenteau-Bareil R., Gauvin R., Berthod F., Mater. 2010, 3, 1863–1887. [Google Scholar]
- 175. Kim M., Hamilton S. E., Guddat L. W., Overall C. M., BBA-Gen. Subjects. 2007, 1770, 1627–1635. [DOI] [PubMed] [Google Scholar]
- 176. Ketnawa S., Rawdkuen S., Chaiwut P., Biochem. Eng. J. 2010, 52, 205–211. [Google Scholar]
- 177. Ha M., Bekhit A. E. D. A., Carne A., Hopkins D. L., Food Chem. 2012, 134, 95–105. [Google Scholar]
- 178. Mostafaie A., Bidmeshkipour A., Shirvani Z., Mansouri K., Chalabi M., Appl. Biochem. Biotechnol. 2008, 144, 123–131. [DOI] [PubMed] [Google Scholar]
- 179. Raskovic B., Bozovic O., Prodanovic R., Niketic V., Polovic N., J. Biosci. Bioeng. 2014, 118, 622–627. [DOI] [PubMed] [Google Scholar]
- 180. Gomes M. T. R., Oliva M. L., Lopes M. T. P., Salas C. E., Curr. Protein Pept. Sci. 2011, 12, 417–436. [DOI] [PubMed] [Google Scholar]
- 181. Yariswamy M., Shivaprasad H. V., Joshi V., Nanjaraj Urs A. N., Nataraju A., Vishwanath B. S., J. Ethnopharmacol. 2013, 149, 377–383. [DOI] [PubMed] [Google Scholar]
- 182. Duarte A., Pereira A., Cabrita A., Moir A., Pires E., Barros M., Curr. Drug Discov. Technol. 2006, 2, 37–44. [DOI] [PubMed] [Google Scholar]
- 183. Gomez-Guillen M. C., Gimenez B., Lopez-Caballero M. E., Montero M. P., Food Hydrocolloids 2011, 25, 1813–1827. [Google Scholar]
- 184. You L., Regenstein J. M., Liu R. H., J. Food Sci. 2010, 75, C582–C587. [DOI] [PubMed] [Google Scholar]
- 185. Liu D., Nikoo M., Boran G., Zhou P., Regenstein J. M., Annu. Rev. Food Sci. Technol. 2015, 6, 527–557. [DOI] [PubMed] [Google Scholar]
- 186. Gao R., Yu Q., Shen Y., Chu Q., Chen G., Fen S., Yang M., Yuan L., McClements D. J., Sun Q., Trends Food Sci. Technol. 2021, 110, 687–699. [Google Scholar]
- 187. Al-Nimry S., Dayah A. A., Hasan I., Daghmash R., Mar. Drugs 2021, 19, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Alfaro A. T., Balbinot E., Weber C. I., Tonial I. B., Machado-Lunkes A., Food Eng. 2014, 7, 33–44. [Google Scholar]
- 189. Pal G. K., RSC Adv. 2016, 6, 33763–33780. [Google Scholar]
- 190. Gautam S. S., Mishra S. K., Dash V., Goyal A. K., Rath G., Thai J. Pharm. Sci. 2010, 34, 67–76. [Google Scholar]
- 191. Kaur T., Kaur A., Grewal R. K., J. Food Sci. Technol. 2014, 52, 5954–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Kittiphattanabawon P., Benjakul S., Visessanguan W., Shahidi F., Food Chem. 2012, 135, 1118–1126. [DOI] [PubMed] [Google Scholar]
- 193. Sachindra N. M., Bhaskar N., Mahendrakar N. S., J. Sci. Food Agric. 2005, 85, 167–172. [Google Scholar]
- 194. Chalamaiah M., Dinesh Kumar B., Hemalatha R., Jyothirmayi T., Food Chem. 2012, 135, 3020–3038. [DOI] [PubMed] [Google Scholar]
- 195. Zamora-Sillero J., Gharsallaoui A., Prentice C., Mar. Biotechnol. 2018, 20, 118–130. [DOI] [PubMed] [Google Scholar]
- 196. Picot L., Ravallec R., Fouchereau-Péron M., Vandanjon L., Jaouen P., Chaplain-Derouiniot M., Guérard F., Chabeaud A., LeGal Y., Alvarez O. M., Bergé J. P., Piot J. M., Batista I., Pires C., Thorkelsson G., Delannoy C., Jakobsen G., Johansson I., Bourseau P., J. Sci. Food Agric. 2010, 90, 1819–1826. [DOI] [PubMed] [Google Scholar]
- 197. Chalamaiah M., Rao G. N., Rao D. G., Jyothirmayi T., Food Chem. 2010, 120, 652–657. [Google Scholar]
- 198. Pacheco-Aguilar R., Mazorra-Manzano M. A., Ramírez-Suárez J. C., Food Chem. 2008, 109, 782–789. [DOI] [PubMed] [Google Scholar]
- 199. Choi Y. J., Hur S., Choi B.-D., Konno K., Park J. W., J. Food Sci. 2009, 74, C17–C24. [DOI] [PubMed] [Google Scholar]
- 200. Ghassem M., Fern S. S., Said M., Ali Z. M., Ibrahim S., Babji A. S., J. Food Sci. Technol. 2011, 51, 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Kristinsson H. G., Rasco B. A., Crit. Rev. Food Sci. 2000, 40, 43–81. [DOI] [PubMed] [Google Scholar]
- 202. Ranasinghe R., Kannagara B., Ratnayake R., Int. J. Recycl. Org. Waste Agric. 2021, 10, 129–143. [Google Scholar]
- 203. Liu Z., Dong S., Xu J., Zeng M., Song H., Zhao Y., Food Control 2008, 19, 231–235. [Google Scholar]
- 204. Ahn C. B., Jeon Y. J., Kim Y. T., Je J. Y., Process Biochem. 2012, 47, 2240–2245. [Google Scholar]
- 205. Gajanan P. G., Elavarasan K., Shamasundar B. A., Environ. Sci. Pollut. Res. Int. 2016, 23, 24901–24911. [DOI] [PubMed] [Google Scholar]
- 206. Liu X., Zhang M., Shi Y., Qiao R., Tang W., Sun Z., J. Sci. Food Agric. 2016, 96, 3240–3248. [DOI] [PubMed] [Google Scholar]
- 207. Romero-Garay M. G., Martínez-Montaño E., Hernández-Mendoza A., Vallejo-Cordoba B., González-Córdova A. F., Montalvo-González E., De L. García-Magaña M., Appl. Biol. Chem. 2020, 63, 1–11. [Google Scholar]
- 208. Tan Y., Chang S. K. C., Meng S., LWT 2019, 111, 809–820. [Google Scholar]
- 209. Chi C. F., Wang B., Wang Y. M., Zhang B., Deng S. G., J. Funct. Foods 2015, 12, 1–10. [Google Scholar]
- 210. Sheriff S. A., Sundaram B., Ramamoorthy B., Ponnusamy P., Saudi J. Biol. Sci. 2014, 21, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Choonpicharn S., Jaturasitha S., Rakariyatham N., Suree N., Niamsup H., J. Food Sci. Technol. 2014, 52, 3134–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Li-Chan E. C. Y., Hunag S. L., Jao C. L., Ho K. P., Hsu K. C., J. Agric. Food Chem. 2012, 60, 973–978. [DOI] [PubMed] [Google Scholar]
- 213. Udenigwe C. C., Aluko R. E., J. Food Sci. 2012, 77, R11–R24. [DOI] [PubMed] [Google Scholar]
- 214. Barbana C., Boye J. I., Food Res. Int. 2010, 43, 1642–1649. [Google Scholar]
- 215. Wang L. L., Xiong Y. L., J. Food Sci. 2008, 73, C482–C487. [DOI] [PubMed] [Google Scholar]
- 216. Tang L., Sun J., Zhang H. C., Zhang C. S., Yu L. N., Bi J., Zhu F., Liu S. F., Yang Q. L., PLoS One 2012, 7, e37863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Jahanbani R., Ghaffari S. M., Salami M., Vahdati K., Sepehri H., Sarvestani N. N., Sheibani N., Moosavi-Movahedi A. A., Plant Foods Hum. Nutr. 2016, 71, 402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Allaoui A., Gascón S., Benomar S., Quero J., Osada J., Nasri M., Rodríguez-Yoldi M. J., Boualga A., Nutr. 2019, 11, 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. González-Montoya M., Cano-Sampedro E., Mora-Escobedo R., Int. J. Cancer Clin. Res. 2017, 4, 81. [Google Scholar]
- 220. León-Espinosa E. B., Sánchez-Chino X., Garduño-Siciliano L., Álvarez-González R. I., Dávila-Ortiz G., Madrigal-Bujaidar E., Téllez-Medina D. I., Jiménez-Martínez C., Nutr. Cancer 2016, 68, 856–864. [DOI] [PubMed] [Google Scholar]
- 221. Xie Y., Liang X., Wei M., Zhao W., He B., Lu Q., Huo Q., Ma C., Int. J. Mol. Sci. 2012, 13, 7483–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Chatterjee C., Gleddie S., Xiao C. W., Nutr. 2018, 10, 1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Yang H. Y., Chen J. R., Chang L. S., Hypertens. Res. 2008, 31, 957–963. [DOI] [PubMed] [Google Scholar]
- 224. Sun X. D., Int. J. Food Sci. Technol. 2011, 46, 2447–2459. [Google Scholar]
- 225. Ashaolu T. J., Int. J. Food Sci. Technol. 2020, 55, 421–428. [Google Scholar]
- 226. Molina Ortiz S. E., Añón M. C., J. Am. Oil Chem. Soc. 2000, 77, 1293–1301. [Google Scholar]
- 227. Meza-Espinoza L., de los Ángeles Vivar-Vera M., de Lourdes García-Magaña M., Sáyago-Ayerdi S. G., Chacón-López A., Becerrea-Verdín E. M., Montalvo-González E., Food Sci. Biotechnol. 2017, 27, 509–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Jara A. M. R., Liggieri C. S., Bruno M. A., Food Chem. 2018, 264, 326–333. [DOI] [PubMed] [Google Scholar]
- 229. Torres M. J., Natalucci C., López L. M. I., Trejo S. A., Biotechnol. Lett. 2019, 41, 1043–1050. [DOI] [PubMed] [Google Scholar]
- 230. Zhang H. Y., Tian T., Chen H., Adv. Mater. Res. 2013, 781–784, 875–879. [Google Scholar]
- 231. Prasse C., Stalter D., Schulte-Oehlmann U., Oehlmann J., Ternes T. A., Water Res. 2015, 87, 237–270. [DOI] [PubMed] [Google Scholar]
- 232. Shah A., Shah M., Groundw. Sustain. Dev. 2020, 11, 100383. [Google Scholar]
- 233. Alshabib M., Onaizi S. A., Sep. Purif. Technol. 2019, 219, 186–207. [Google Scholar]
- 234. Dzuvor C. K. O., Pan S., Amanze C., Amuzu P., Asakiya C., Kubi F., Crit. Rev. Biotechnol. 2021, 1, 23. [DOI] [PubMed] [Google Scholar]
- 235. Dutta S., Bhattacharyya A., De P., Ray P., Basu S., J. Hazard. Mater. 2009, 172, 888–896. [DOI] [PubMed] [Google Scholar]
- 236. Chatterjee S., Basu S., Dutta S., Chattaraj R., Banerjee D., Sinha S., Mater. Today: Proc. 2016, 3, 3258–3268. [Google Scholar]
- 237. Cheriyan S., Abraham E. T., J. Hazard. Mater. 2010, 176, 1097–1100. [DOI] [PubMed] [Google Scholar]
- 238. Kagalkar A. N., Jagtap U. B., Jadhav J. P., Bapat V. A., Govindwar S. P., Bioresour. Technol. 2009, 100, 4104–4110. [DOI] [PubMed] [Google Scholar]
- 239.K. Sunar, U. Kumar, S. K. Deshmukh, in Agro-Industrial Wastes as Feedstock for Enzyme Production. Recent Applications of Enzymes in Personal Care Products, Proceedings (Eds.: G. S. Dhillon, S. Kaur), Academic Press, 2016, pp. 279–298.
- 240. Norlén L., Int. J. Cosmet. Sci. 2006, 28, 397–425. [DOI] [PubMed] [Google Scholar]
- 241. Banchhor M., Saraf S., Pharmacogn. Rev. 2008, 2, 266. [Google Scholar]
- 242. Levy L. L., Emer J. J., J. Cutan. Aesthet. Surg. 2012, 5, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Spir L. G., Ataide J. A., De Lencastre Novaes L. C., Moriel P., Mazzola P. G., De Borba Gurpilhares D., Silveira E., Pessoa A., Tambourgi E. B., Biotechnol. Prog. 2015, 31, 937–945. [DOI] [PubMed] [Google Scholar]
- 244. Ribeiro J. S., da S. Barboza A., Cuevas-Suárez C. E., da Silva A. F., Piva E., Lund R. G., Sci. Reports 2020, 10, 10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Wang K. H., Lin R. D., Hsu F. L., Huang Y. H., Chang H. C., Huang C. Y., Lee M. H., J. Ethnopharmacol. 2006, 106, 353–359. [DOI] [PubMed] [Google Scholar]
- 246. Choi W. J., Kang S. M., Koh J., Fibers Polym. 2012, 13, 1058–1064. [Google Scholar]
- 247. Georgiev V., Slavov A., Vasileva I., Pavlov A., Eng. Life Sci. 2018, 18, 779–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Trehan S., Michniak-Kohn B., Beri K., Future Sci. 2017, 3, FSO226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.A. P. Urs, V. N. Manjuprasanna, G. V. Rudresha, M. Yariswamy, B. S. Vishwanath, In Proteases in Physiology and Pathology. Plant Latex Proteases: Natural Wound Healers, Proceedings (Eds: S. Chakraborti, N. Dhalla), Springer, Singapore, 2017, pp 297–323.
- 250. Gurumallesh P., Alagu K., Ramakrishnan B., Muthusamy S., Int. J. Biol. Macromol. 2019, 128, 254–267. [DOI] [PubMed] [Google Scholar]
- 251. Parravano M., Tedeschi M., Manca D., Costanzo E., Di Renzo A., Giorno P., Barbano L., Ziccardi L., Varano M., Parisi V., Adv. Ther. 2019, 36, 2493–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Pavan R., Jain S., Kumar A., Biotechnol. Res. Int. 2012, 1, 976203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Ley C. M., Tsiami A., Ni Q., Robinson N., J. Chin. Integer. Med. 2011, 9, 702–710. [DOI] [PubMed] [Google Scholar]
- 254. Colletti A., Li S., Marengo M., Adinolfi S., Cravotto G., Appl. Sci. 2021, 11, 8428. [Google Scholar]
- 255. dos Anjos M. M., da Silva A. A., de Pascoli I. C., Mikcha J. M. G., Machinski M., Peralta R. M., de Abreu Filho B. A., Int. J. Food Microbiol. 2016, 216, 121–126. [DOI] [PubMed] [Google Scholar]
- 256. Cotabarren J., Lufrano D., Parisi M. G., Obregón W. D., Plant Sci. 2020, 292, 110398. [DOI] [PubMed] [Google Scholar]
- 257. Silva-López R. E., Gonçalves R. N., J. Appl. Biotechnol. Bioeng. 2019, 6, 101–109. [Google Scholar]
- 258. Wanderley L. F., Soares A. M. D. S., E Silva C. R., de Figueiredo I. M., da S. Ferreira A. T., Perales J., de O. Mota H. R., Oliveira J. T. A., Costa Junior L. M., Rev. Bras. Parasitol. Vet. 2018, 27, 473–480. [DOI] [PubMed] [Google Scholar]
- 259. Dhiman V. K., Chauhan V., Kanwar S. S., Singh D., Pandey H., Bull. Natl. Res. Cent. 2021, 45, 1–9. [Google Scholar]
- 260. Kardoust M., Salehi H., Taghipour Z., Sayadi A., Int. J. Low. Extrem. Wounds 2021, 20, 104–110. [DOI] [PubMed] [Google Scholar]
- 261. Jamir K., Seshagirirao K., Int. J. Biol. Macromol. 2018, 106, 719–729. [DOI] [PubMed] [Google Scholar]
- 262. Shukla Y., Singh M., Food Chem. Toxicol. 2007, 45, 683–690. [DOI] [PubMed] [Google Scholar]
- 263. Shivamadhu M. C., Balaji K. S., Jayarama S., Biocatal. Agric. Biotechnol. 2017, 12, 10–14. [Google Scholar]
- 264. Tavano O. L., Berenguer-Murcia A., Secundo F., Fernandez-Lafuente R., Compr. Rev. Food Sci. Food Saf. 2018, 17, 412–436. [DOI] [PubMed] [Google Scholar]
- 265. Pereira A., Cartucho D., Duarte A., Gil M., Cabrita A., Patricio J., Barros M., Curr. Drug Discovery Technol. 2005, 2, 231–238. [DOI] [PubMed] [Google Scholar]
- 266. Hendryanti D. N., Jeong H., Kim J. Y., Kwon O., J. Funct. Foods 2020, 68, 103925. [Google Scholar]
- 267. Baidamshina D. R., Koroleva V. A., Trizna E. Y., Pankova S. M., Agafonova M. N., Chirkova M. N., Vasileva O. S., Akhmetov N., Shubina V. V., Porfiryev A. G., Semenova E. V., Sachenkov O. A., Bogachev M. I., Artyukhov V. G., Baltina T. V., Holyavka M. G., Kayumov A. R., Int. J. Biol. Macromol. 2020, 164, 4205–4217. [DOI] [PubMed] [Google Scholar]
- 268. Jamshed H., Siddiqi H. S., Gilani A.-H., Arslan J., Qasim M., Gul B., Phyther. Res. 2019, 33, 2310–2318. [DOI] [PubMed] [Google Scholar]
- 269. Abd-ElKhalek A. M., Seoudi D. M., Ibrahim O. A., Abd-Rabou N. S., Abd ElAzeem E. M., J. Appl. Biotechnol. Reports 2020, 7, 243–250. [Google Scholar]
- 270. Basso A., Serban S., J. Mol. Catal. 2019, 479, 110607. [Google Scholar]
- 271. Nguyen H. H., Kim M., Appl. Sci. Converg. Technol. 2017, 26, 157–163. [Google Scholar]
- 272. Guzik U., Hupert-Kocurek K., Wojcieszynska D., Mol. 2014, 19, 8995–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Secundo F., Chem. Soc. Rev. 2013, 42, 6250–6261. [DOI] [PubMed] [Google Scholar]
- 274. Hermanová S., Zarevúcká M., Bouša D., Pumera M., Sofer Z., Nanoscale 2015, 7, 5852–5858. [DOI] [PubMed] [Google Scholar]
- 275. Rodrigues R. C., Berenguer-Murcia Á., Carballares D., Morellon-Sterling R., Fernandez-Lafuente R., Biotechnol. Adv. 2021, 52, 107821. [DOI] [PubMed] [Google Scholar]
- 276. Garcia-Galan C., Berenguer-Murcia Á., Fernandez-Lafuente R., Rodrigues R. C., Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar]
- 277. Rodrigues R. C., Ortiz C., Berenguer-Murcia Á., Torres R., Fernández-Lafuente R., Chem. Soc. Rev. 2013, 42, 6290–6307. [DOI] [PubMed] [Google Scholar]
- 278. Hernandez K., Fernandez-Lafuente R., Enzyme Microb. Technol. 2011, 48, 107–122. [DOI] [PubMed] [Google Scholar]
- 279. Chapman J., Ismail A. E., Dinu C. Z., Catal. 2018, 8, 238. [Google Scholar]
- 280. Tacias-Pascacio V. G., Morellon-Sterling R., Castañeda-Valbuena D., Berenguer-Murcia Á., Kamli M. R., Tavano O., Fernandez-Lafuente R., Int. J. Biol. Macromol. 2021, 188, 94–113. [DOI] [PubMed] [Google Scholar]
- 281. Zdarta J., Meyer A. S., Jesionowski T., Pinelo M., Catal. 2018, 8, 92. [Google Scholar]
- 282. Homaei A., Int. J. Biol. Macromol. 2015, 75, 373–377. [DOI] [PubMed] [Google Scholar]
- 283. Siar E. H., Zaak H., Kornecki J. F., Zidoune M. N., Barbosa O., Fernandez-Lafuente R., Process Biochem. 2017, 58, 98–104. [Google Scholar]
- 284. Barros R. M., Extremina C. I., Gonçalves I. C., Braga B. O., Balcão V. M., Malcata F. X., Enzyme Microb. Technol. 2003, 33, 908–916. [Google Scholar]
- 285. Rocha G. F., Kise F., Rosso A. M., Parisi M. G., Food Chem. 2017, 237, 350–355. [DOI] [PubMed] [Google Scholar]
- 286. Vasconcelos N. F., Cunha A. P., Ricardo N. M. P. S., Freire R. S., de A. P. Vieira L., Brígida A. I. S., de F. Borges M., de F. Rosa M., Vieira R. S., Andrade F. K., Int. J. Biol. Macromol. 2020, 165, 3065–3077. [DOI] [PubMed] [Google Scholar]
- 287. Islam M. R., Kwak J. W., Lee J. S., Hong S. W., Khan M. R. I., Lee Y., Lee Y., Lee S. W., Hwang I., Plant Biotechnol. J. 2019, 17, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Xue Y., Nie H., Zhu L., Li S., Zhang H., Appl. Biochem. Biotechnol. 2009, 160, 109–121. [DOI] [PubMed] [Google Scholar]
- 289. Bhandari S., Gupta V. K., Singh H., Biocatal. Biotransform. 2009, 27, 71–77. [Google Scholar]
- 290. Singh A. N., Singh S., Suthar N., Dubey V. K., J. Agric. Food Chem. 2011, 59, 6256–6262. [DOI] [PubMed] [Google Scholar]
- 291. Baidamshina D. R., Koroleva V. A., Olshannikova S. S., Trizna E. Y., Bogachev M. I., Artyukhov V. G., Holyavka M. G., Kayumov A. R., Mar. Drugs 2021, 19, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Holyavka M., Pankova S., Koroleva V., Vyshkvorkina Y., Lukin A., Kondratyev M., Artyukhov V., J. Photochem. Photobiol. B 2019, 201, 111681. [DOI] [PubMed] [Google Scholar]
- 293. Holyavka M., Faizullin D., Koroleva V., Olshannikova S., Zakhartchenko N., Zuev Y., Kondratyev M., Zakharova E., Artyukhov V., Int. J. Biol. Macromol. 2021, 180, 161–176. [DOI] [PubMed] [Google Scholar]
- 294. Moreira Filho R. N. F., Vasconcelos N. F., Andrade F. K., de F. Rosa M., Vieira R. S., Colloids Surf. B 2020, 194, 111222. [DOI] [PubMed] [Google Scholar]
- 295. Quiroga E., Illanes C. O., Ochoa N. A., Barberis S., Process Biochem. 2011, 46, 1029–1034. [Google Scholar]
- 296. Rebecca J., Sharmila S., Rebecca L. J., Saduzzaman M., J. Chem. Pharm. Res. 2012, 4, 4484–4488. [Google Scholar]
- 297. Bayramoglu G., Filiz Senkal B., Yilmaz M., Yakup Arica M., Bioresour. Technol. 2011, 102, 9833–9837. [DOI] [PubMed] [Google Scholar]
- 298. Rojas-Mercado A. S., Moreno-Cortez I. E., Lucio-Porto R., Pavón L. L., Int. J. Biol. Macromol. 2018, 118, 2287–2295. [DOI] [PubMed] [Google Scholar]
- 299. Lipsa R., Tudorachi N., Vasile C., E-Polymers 2010, 10, 087. [Google Scholar]
- 300. Han J. H., Taylor J. D., Kim D. S., Kim Y. S., Kim Y. T., Cha G. S., Nam H., Sens. Actuators B 2007, 123, 384–390. [Google Scholar]
- 301. Ganapathi S., Butterfield D. A., Bhattacharyya D., J. Chem. Technol. Biotechnol. 1995, 64, 157–164. [Google Scholar]
- 302. Kondo A., Imura K., Nakama K., Higashitani K., J. Ferment. Bioeng. 1994, 78, 241–245. [Google Scholar]
- 303. Kumari A., Kaur B., Srivastava R., Sangwan R. S., Biochem. Biophys. Rep. 2015, 2, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Origone A., Barberis S., Illanes A., Guzmán F., Camí G., Liggieri C., Martínez R., Bernal C., Process Biochem. 2020, 95, 36–46. [Google Scholar]
- 305. Adaro M., Bersi G., Talia J. M., Bernal C., Guzmán F., Vallés D., Barberis S., Front. Nutr. Sci. 2021, 8, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Solís S., Paniagua J., Martínez J. C., Asomoza M., J. Sol-Gel Sci. Technol. 2006, 37, 125–127. [Google Scholar]
- 307. Llerena-Suster C. R. F., Foresti M. L., Briand L. E., Morcelle S. R., Colloids Surf. B 2009, 72, 16–24. [DOI] [PubMed] [Google Scholar]
- 308. Ajayi O. A., Nok J., Adefila S. S., J. Nat. Sci. Res. 2012, 2, 55–62. [Google Scholar]
- 309. Hyndman D., Burrell R., Lever G., Flynn T. G., Biotechnol. Bioeng. 1992, 40, 1328–1336. [DOI] [PubMed] [Google Scholar]
- 310. Song M. M., Nie H. L., Zhou Y. T., Zhu L. M., Bao J. Y., Sep. Sci. Technol. 2011, 46, 473–482. [Google Scholar]
- 311. Liburdi K., Emiliani Spinelli S., Benucci I., Lombardelli C., Esti M., Food Chem. 2018, 239, 157–164. [DOI] [PubMed] [Google Scholar]
- 312. Atacan K., Özacar M., Özacar M., Int. J. Biol. Macromol. 2018, 109, 720–731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.