ABSTRACT
Decades of research, much of it in Escherichia coli, have yielded a wealth of insight into bacterial cell division. Here, we provide an overview of the E. coli division machinery with an emphasis on recent findings. We begin with a short historical perspective into the discovery of FtsZ, the tubulin homolog that is essential for division in bacteria and archaea. We then discuss assembly of the divisome, an FtsZ-dependent multiprotein platform, at the midcell septal site. Not simply a scaffold, the dynamic properties of polymeric FtsZ ensure the efficient and uniform synthesis of septal peptidoglycan. Next, we describe the remodeling of the cell wall, invagination of the cell envelope, and disassembly of the division apparatus culminating in scission of the mother cell into two daughter cells. We conclude this review by highlighting some of the open questions in the cell division field, emphasizing that much remains to be discovered, even in an organism as extensively studied as E. coli.
KEYWORDS: Escherichia coli, FtsZ, cell division, divisome, peptidoglycan
OVERVIEW OF E. COLI DIVISION
In Escherichia coli and other bacteria, cell division is executed by a multiprotein transenvelope complex called the “divisome” (1–5). The first protein to localize to the midcell division site—the cytoplasmic tubulin-like GTP hydrolase FtsZ—plays a foundational role in divisome organization, and dynamic treadmilling behavior of FtsZ polymers ensures uniform assembly of the peptidoglycan (PG) cross wall (6–9). Conserved in bacteria, archaea, and chloroplasts, FtsZ is also found in mitochondria from diverse single-celled eukaryotes but not in animal or plant nuclear genomes (10). FtsZ associates with the cytoplasmic membrane in a ring-like arrangement, the eponymous “Z ring.” Membrane association is mediated by two essential proteins, FtsA and ZipA (4). The intrinsic properties of FtsZ together with the concerted actions of several FtsZ modulatory proteins help corral diffuse FtsZ polymers to the nascent septal site. Additional essential proteins that traverse the cytoplasmic membrane are recruited in a genetically defined sequence: first FtsK, then FtsQ, FtsL, and FtsB, followed by FtsW with FtsI, and finally, FtsN (11, 12). These factors are involved in structural, modulatory, and PG-remodeling functions. The assembly of the divisome at the nascent division site is coordinated with growth and cell cycle progression. Once core proteins reach optimal concentrations within the divisome, an apparent conformational change stimulates activation of cell wall synthesis via specialized septal PG synthases. Treadmilling behavior within FtsZ polymers guides the distribution of the cell wall synthesis enzymes uniformly around the septum (8). Cross wall synthesis is a major driver of constriction, and other transenvelope divisome components ensure the coordinated invagination of the outer membrane (OM). Division culminates with divisome disassembly and daughter cell separation (13). Focusing on the molecular details and recent literature, below, we review the mechanisms ensuring temporal and spatial fidelity during the course of E. coli division.
BRIEF HISTORY OF THE DISCOVERY OF FTSZ
The earliest mention of nondividing E. coli cell “filaments” or “snakes” in response to low doses of UV radiation can be traced back to reports in the 1930s and early 40s (14). In the 1960s, phenotypic analysis of libraries of heat-sensitive mutants identified a subset that formed long, filamentous cells at elevated growth temperatures of 42°C. Termed “filamentation temperature sensitive (fts)” genes based on this phenotype, this nomenclature persists to the present day as a descriptor for essential divisome components (15–17) (Fig. 1A). Pioneering studies by Lutkenhaus and Donachie led to the cloning of ftsA and ftsZ (18–20).
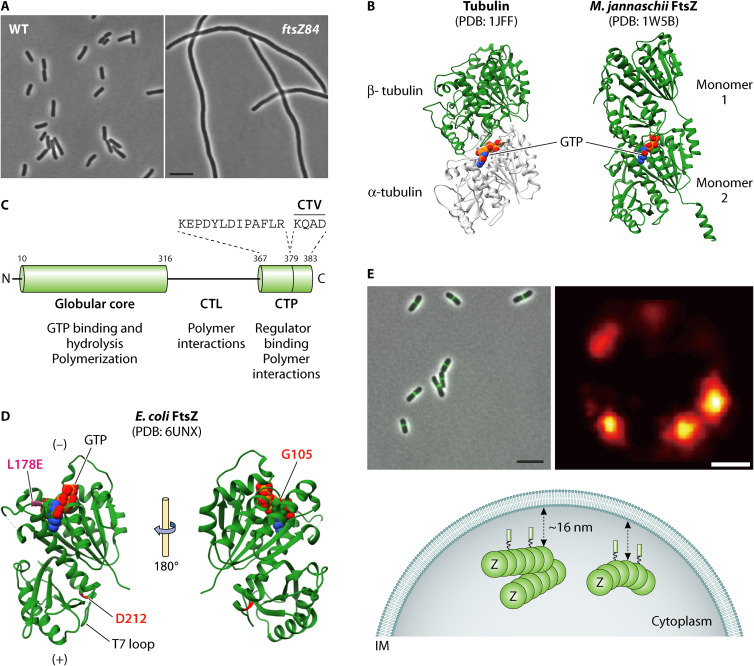
FIG 1.
The tubulin-like FtsZ plays a foundational role in E. coli division. (A) Phase images of wild-type E. coli under permissive (30°C) and ftsZ84 (Ts) mutant cells under restrictive (42°C) growth temperatures (M. Buczek, unpublished data). FtsZ84 fails to localize to the nascent division site under nonpermissive growth conditions. Bar, 5 μm. (B) Three-dimensional structures of Tubulin and FtsZ: an α/β heterodimer of tubulin (PDB 1JFF) (left); Methanococcus jannaschii FtsZ (PDB 1W5B) dimer made up of two identical monomers (right). The C-terminal linker (CTL) and C-terminal peptide (CTP) domains are not present in the structure. Sandwiched in the dimeric units of each protein is a space-filled model of GTP. The two structures were modeled using UCSF Chimera (https://www.cgl.ucsf.edu/chimera/). (C) The domain architecture of FtsZ consists of a short disordered N-terminal end followed by the globular core, which contains the tubulin-signature motif responsible for binding GTP and GTP hydrolysis residues. The core is linked to the C-terminal peptide (CTP) by an intrinsically disordered linker region (CTL). The 14 aa CTP serves as a common binding site for various FtsZ binding proteins and can be further split into conserved and variable (CTV) regions. (D) The structure of E. coli FtsZ (ftsZL178E; residue L178 is shown in pink; PDB 6UNX) monomer bound to GTP is shown and was generated using UCSF Chimera. FtsZ L178E is incompetent for assembly in the presence of GTP (32). Other critical residues D212 and G105 are labeled and are mutated in strains ftsZ2 (D212G) and ftsZ84 (G105S) alleles referred to in the main text. In the polymer, the T7 synergy loop at the bottom surface is inserted into the nucleotide binding pocket of the second subunit resulting in GTP hydrolysis. Monomers are added toward the T7 loop-end of the growing polymer referred to as the “+” end. Monomers disperse from the end to which GTP is bound, which is referred to as the “−” end of a treadmilling FtsZ polymer. (E) Conventional fluorescence images of FtsZ polymers at midcell in wild-type E. coli cells. ZapA-GFP is used as a proxy for Z ring localization in these cells (A. Cardenas Arevalo, unpublished data). Bar, 5 μm; super resolution PALM microscopy image of FtsZ polymer assemblies in a cross section of the “ring” in a wild-type cell reveals a heterogeneous wreath-like arrangement (image courtesy of Z. Jason Lyu and Jie Xiao). Bar, 200 nm. A diagrammatic interpretation of cytoplasmic FtsZ polymers arranged in a random, heterogenous structure 16 nm away from the inner membrane (IM). The FtsZ polymers are tethered to the IM by FtsA or ZipA (not shown).
The 1990s saw an explosion of data establishing FtsZ as a bacterial cytoskeletal protein. These efforts were led by the groundbreaking 1991 study of Bi and Lutkenhaus which combined electron microscopy and immunogold labeling to reveal FtsZ localization at the leading edge of the invaginating cytoplasmic membrane (6). Fluorescence microscopy, known for its higher sensitivity but lower resolution compared to immunogold electron microscopy, displayed FtsZ in a continuous ring-like configuration at midcell, the Z ring (21, 22). This advance, in turn, permitted experiments illuminating the hierarchy of divisome assembly taking advantage of conditional mutations in essential cell division proteins (12). Together with the high degree of conservation of FtsZ and data indicating that it was the target of both the SOS (DNA damage response system) and the positional regulator of cell division, Min, these findings firmly established FtsZ as a foundational component of the bacterial division machinery (6, 23–26). Biochemical analysis from the de Boer, RayChaudhuri, and Lutkenhaus groups confirmed that FtsZ functioned as a GTPase (27–29). Like the eukaryotic cytoskeletal protein tubulin, FtsZ polymerization is dependent on GTP and can form single-stranded polymers/filaments, sheets, bundles, and rings, depending on buffer conditions. FtsZ does not, however, form the 13-stranded microtubule structure typical of α/β tubulin dimers (29–31). Structural data indicating that FtsZ and tubulin share common core domains despite limited primary sequence similarity unequivocally established FtsZ as an ancestral homolog of tubulin (32, 33) (Fig. 1B).
Collectively, these early studies contributed to the paradigmatic shift in our understanding of subcellular organization in bacteria and contributed to the emergence of an entirely new field, bacterial cell biology. Enabled by ingenious genetic approaches, cell-free reconstitution studies, superresolution microscopy, and high-resolution structural analyses, the last 20 years have revealed basic principles underlying assembly and activation of the division machinery. We summarize key findings in these areas focusing on E. coli and include data from other bacterial species where appropriate.
THE PROPERTIES OF FTSZ
FtsZ, the protein.
The FtsZ polypeptide can be subdivided into five domains (Fig. 1C). First is an unstructured N terminus of variable length ranging from 10 amino acid (aa) residues in E. coli to 68 aa in cyanobacteria (34, 35). The role of the N terminus is unknown, although in cyanobacteria, it is considered to influence FtsZ polymerization (34). Following the N terminus is a large tubulin-like core domain that contains GTP-binding residues and is sufficient for polymerization in vitro but cannot support division in vivo (32, 35). Next is the C-terminal linker (CTL), an intrinsically disordered region ranging between ∼50 aa in the Gammaproteobacteria and firmicutes to over 250 aa in some Alphaproteobacteria (36). Poorly conserved at the primary sequence level, differences in CTL composition affect FtsZ polymer morphologies and function in vivo (36–38). The CTL is followed by a highly conserved patch of 14 aa (the C-terminal peptide [CTP]) that is involved in mediating FtsZ interactions with several essential and accessory proteins in division (39, 40). The CTL provides the CTP the conformational freedom to interact with a variety of protein partners (41). Recent data suggest that the CTL and CTP can also influence FtsZ-FtsZ interactions within polymers and polymer bundles, although this has yet to be thoroughly explored (42). Finally, the extreme C terminus of FtsZ, the C-terminal variable (CTV) region, differs widely in length and composition between species, the latter dictating its influence on lateral interactions between FtsZ polymers (43).
FtsZ, the dynamic polymer.
FtsZ assembles into single-stranded homopolymers by a tail-to-head association of monomers. Although monomers are capable of binding GTP on their own, the catalytic site for GTP hydrolysis forms exclusively at the intermonomer interface (27–29, 32, 44) (Fig. 1D). FtsZ polymerization is thus dependent on GTP binding but not hydrolysis. GTP hydrolysis destabilizes the FtsZ polymer leading to disassembly. The available intracellular concentration of FtsZ (∼4 to 6 μM) far exceeds that of the critical concentration of assembly in vitro (7, 45). Curiously, formation of even single-stranded FtsZ polymers exhibits cooperativity with a critical concentration of 1 μM (46). A plausible mechanism to reconcile this apparent contradiction resides in the ability of FtsZ to adopt two conformations: open, residing primarily in the polymer, and closed, present primarily in the monomer pool. A switch between the closed and open conformations upon polymerization exposes a different interface, which in turn promotes additional polymerization (47, 48). Mathematical models predict that such polymerization-associated conformational change would allow for higher-affinity associations among the subunits of the polymer, explaining why assembly is cooperative (49, 50).
In vitro, FtsZ polymers exhibit continuous chiral treadmilling, a motion defined by elongation of a polymer at one end and shortening of the same polymer at the opposite end. Subunits are added to the “plus” end and fall off the polymer at the “minus” end, with individual subunits within the polymer remaining stationary (51). In vivo, only ∼30% of cellular FtsZ is present in the Z ring at a given time and FtsZ molecules from cytoplasmic and membrane-bound pools are in rapid dynamic exchange with a lifetime of ∼8 s (52, 53). Consistent with treadmilling, single-molecule tracking analysis indicates that individual FtsZ molecules within the cytokinetic ring are largely immobile (54). Seminal superresolution imaging studies in E. coli and Bacillus subtilis concurrently revealed that FtsZ treadmilling is powered by GTP hydrolysis; mutants that perturb nucleotide hydrolysis alter the rate of treadmilling in a proportional fashion (55, 56). Further, In B. subtilis, FtsZ treadmilling coalesces FtsZ polymers into a dense, tight-pitched ring at the nascent septal site (57). Treadmilling FtsZ polymers are oriented in both directions around the division plane and contribute to the uniform distribution of the PG synthesis machinery at the midcell septal site in E. coli and B. subtilis (55, 56) (discussed further in “Coupling of septal PG synthesis to FtsZ treadmilling”).
The treadmilling behavior of FtsZ implies a defined polarity for FtsZ filaments with regard to subunit-subunit interactions, raising the question of the identity of the plus end (58). Elegant genetic work from the Lutkenhaus group suggests that the plus end resides proximal to the T7 synergy loop involved in GTP hydrolysis at the base of the subunit rather than closer to the GTP-binding site on top of the subunit (59) (Fig. 1D). Mutations proximal to the T7 synergy loop are not tolerated in vivo, suggesting that new polymers are added to the FtsZ filament at this interface (59). Notably, the polarity exhibited by FtsZ is the reverse of that of microtubules (59).
Although recent work suggests that FtsZ primarily serves as a guide for septal cross wall synthesis, a longstanding controversy in the field centers on the potential for FtsZ to generate the force necessary for constriction. This model, which implicates GTP hydrolysis as a driver of constriction via a change in the morphology of FtsZ polymers, is supported by the ability of reconstituted membrane-anchored FtsZ to deform tubular liposomes (60–62). Substantial data, however, argue against FtsZ as a major force-generating mechanism after constriction initiation. A mutation in ftsZ, ftsZ26, that shifts the geometry of the divisome from a ring to an extended spiral results in the formation of skewed cross walls that contract in a twisted formation, indicating that constriction of an annular cytokinetic ring is dispensable for division (63, 64). Likewise, FtsZ mutants defective in GTP binding and/or hydrolysis in vitro (FtsZ84 [G105S] and FtsZ2 [D212G]) retain the ability to support septation in vivo, suggesting that nearly all of GTPase activity of FtsZ is dispensable for division (27, 28, 55, 63, 65–68). Instead, nucleotide hydrolysis appears to be required primarily for controlling FtsZ-treadmilling dynamics and the uniform circumferential distribution of septal cell wall synthases (55, 56). This conclusion is underscored by recent findings in B. subtilis: FtsZ treadmilling condenses FtsZ polymers into a dense midcell ring that recruits cell wall synthases more efficiently, concentrating their enzymatic activity into a tight-pitched midcell zone (57, 69).
FtsZ, as part of the cytokinetic ring.
While the precise organization of FtsZ filaments within the cytokinetic ring is still unsettled, recent advances in high-resolution imaging and single-molecule analyses have provided important insights into the in vivo architecture of the Z ring. In cells at the beginning of visible constriction, the ring is estimated to have a cross-sectional diameter of ∼550 to 600 nm with a width (along the cell length axis) and radial thickness of ∼100 and ∼60 nm, respectively (68). Increasing intracellular FtsZ concentration has little, if any, impact on the width of the Z ring, suggesting that it is a defined parameter (70). Conventional fluorescence microscopy suggests that the Z ring is continuous, while superresolution imaging supports a discontinuous, heterogeneous structure consisting of short, randomly overlapping filaments arranged into a wreath-like array with nodes of higher density scattered throughout (21, 70, 71) (Fig. 1E). The peak intensity nodes likely correspond to clusters of FtsZ polymers at a spatial resolution limit of ∼35 nm (8, 70). Cryoelectron tomograms in E. coli and other species indicate the presence of long overlapping polymers of FtsZ positioned a short distance (∼16 nm) away from the cytoplasmic membrane resembling a more continuous structure (72) (Fig. 1E).
CORRALLING FTSZ POLYMERS TO MIDCELL
The topological factors.
The prevailing view is that the timing and position of FtsZ assembly are governed primarily by modulatory factors (73–75). Nearly 94% of cells under nutrient-rich conditions supporting fast growth and ∼60% of cells under nutrient-poor conditions supporting slow growth contain Z rings (73, 76). FtsZ concentration remains largely unchanged over the course of the cell cycle in rich medium. Although FtsZ concentration increases by ∼20% during the lifetime of a cell cultured in nutrient-poor medium, the correlation between Z ring assembly and intracellular concentration remains weak under these conditions (73, 76).
Dispensable for growth individually and largely in combination, a few proteins control the location of FtsZ assembly in E. coli (Fig. 2A). The predominant function of these proteins appears to be inhibitory, effectively confining FtsZ polymers to midcell to ensure high local concentration at the nascent division site. Best known is the Min system, which inhibits FtsZ assembly specifically at cell poles (77–79). Additional factors include those that help prevent FtsZ assemblies over unsegregated chromosomal material (e.g., SlmA) and those that coordinate FtsZ assembly with the termination of DNA replication (e.g., Ter-MatP linkage) (80, 81). The activity of these dedicated topological systems is facilitated by general-acting factors, including the Zaps (FtsZ-associated proteins) that promote FtsZ assembly within the context of the ring (39). It is unclear whether the nonessential nature of these systems foretells the existence of an as-yet-unidentified positioning factor(s) or if FtsZ assembly at the nascent division site is promoted when the local concentration of the protein reaches high enough levels by the collective action of these factors that the system becomes self-reinforcing.
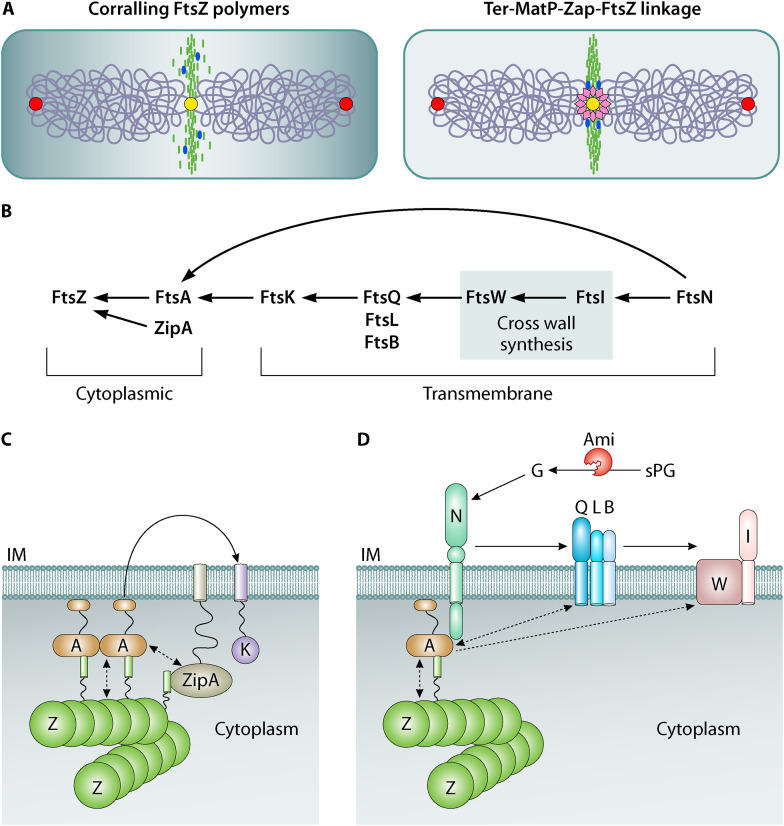
FIG 2.
Assembly and activation of the divisome. (A) Left, FtsZ polymers (green) are corralled at midcell by a number of factors. Negative positional factors Min and nucleoid occlusion (NO), which together generate a high to low gradient of inhibitor concentration from poles to the cell center (gray), prevent FtsZ assembly elsewhere in the cell. The replicating ori regions (red ovals) and the ter region (yellow) on a duplicating chromosome are marked. FtsZ cross-linking proteins, including ZapA, ZapC, and ZapD (blue), are postulated to condense FtsZ polymers through lateral interactions to ensure efficient recruitment of cell wall synthesis enzymes to midcell. The FtsZ CTL domain is also considered to contribute to FtsZ lateral interactions. Right, the Ter-MatP linkage provides a positive positional signal that possibly links chromosome replication and segregation with FtsZ placement. The DNA-binding protein MatP (pink diamonds) binds both the terminus (ter) region on the chromosome and the ZapB-ZapA complex. (B) The genetically defined assembly of the essential E. coli divisome proteins is shown. The direction of the arrows represents the presence of a protein that helps in the recruitment of the next protein in the sequence. In this scheme, the proteins that constitute the cytokinetic ring at the cytoplasmic face of the IM can be visualized initially followed by the integral membrane proteins which predominantly play structural, regulatory, and synthesis roles in cell wall assembly. FtsN, the last protein to be recruited, can in certain genetically defined backgrounds arrive early to the divisome and through its interactions with FtsA (arrow) back recruit the other proteins. (C) The cytoplasmic core of the cytokinetic ring is composed of three proteins: FtsZ, FtsA, and ZipA. Together, FtsA and ZipA anchor FtsZ filaments to the membrane. Of the two, FtsA plays the primary role in division progress. ZipA influences FtsA polymerization (dotted double arrow) and enhances the recruitment of other division proteins to the midcell site (curved arrow; only FtsK is included in this figure for clarity). FtsZ and FtsA polymer architecture are considered to be influenced by each other (dotted double arrow). (D) In dividing cells, high levels of FtsN accumulate at the septum where it interacts with FtsA on the cytoplasmic side. The periplasmic essential (E) subdomain of FtsN is involved in activating (arrow) the FtsQLB complex. The resulting conformational changes in FtsQLB allow it to transition from a recruitment state to an activated form. At this point, FtsL interacts with FtsI to stimulate the transglycosylase and transpeptidase activities of FtsW and FtsI, respectively, leading to septal PG (sPG) synthesis. The periplasmic SPOR domain of FtsN also interacts with denuded glycan strands (G) at the septum, formed due to the action of amidases (Ami) cleaving the sPG. This invokes a septal PG feedback loop and the recruitment of additional FtsN molecules to midcell. Signals from both cytoplasmic (FtsA-FtsN) and periplasmic (FtsN-PG and FtsN-FtsQLB) compartments lead to the activation of cross wall synthesis. FtsA and FtsQ interactions may help stabilize FtsQLB directly or indirectly (dotted double arrow). FtsA oligomerization state may also influence FtsWI activity (dotted arrow). IM, inner membrane; PG, peptidoglycan; OM, outer membrane.
Min, a negative positional regulator.
The classic min mutant phenotype of heterogeneously sized and anucleate “mini-cells” resulting from midcell and polar divisions was first described by Adler in 1967 (82). Cloned by de Boer and colleagues in the late 1980s, the E. coli min locus encodes three proteins: MinC, the membrane-associated ATPase MinD, and MinE. MinD and MinE provide topological specificity to MinC, which interacts directly with FtsZ to inhibit assembly (24, 77). Data suggest that MinC binds to the FtsZ CTP, placing its inhibitory domain at the intermonomer interface of an FtsZ subunit occluding polymer assembly (83–85). MinC is activated by MinD-mediated recruitment to the membrane and, potentially, additional factors (86–88). Fluorescence microscopy, genetics, and biochemistry indicate that Min-mediated inhibition of FtsZ is a dynamic process where MinC oscillates on and off the cell membrane driven by repeating cycles of ATP hydrolysis of the membrane-associated MinD and the activity of MinE (41, 78, 89–92). MinE stimulates the ATPase activity of MinD at cell poles, thereby promoting disassociation of the MinCD complex (93). Subsequently, MinD reassembles on the membrane at the other half of the cell and recruits MinC afresh from the cytoplasmic pool. Data indicating that MinE interferes with the MinC-MinD interaction on the membrane even when ATP hydrolysis by MinD is blocked and MinD remains associated with membrane provide further support for this model (92). Together, oscillations of MinC and MinD on and off membrane at alternate poles establish a time-averaged gradient of FtsZ inhibition at the cytoplasmic membrane with a minimum at midcell, favoring FtsZ assembly at this position (78, 90) (Fig. 2A).
The chromosome as a positional regulator.
In the late 1980s, Conrad Woldringh observed that the site of cell division rarely overlaps with the chromosome (nucleoid), even when DNA replication is perturbed, a phenomenon later termed “nucleoid occlusion” (NO) (94, 95). The chromosome occupies a majority of the cell; thus, together with the Min system, NO may be sufficient to prevent aberrant septation (96). In E. coli, genetic data suggest that the FtsZ inhibitor and DNA-binding protein SlmA contributes to NO by binding to DNA throughout the cell (97, 98). SlmA interacts more or less equally with all regions of the chromosome but the terminus, which is positioned at the cell center prior to division, leaving a SlmA-free zone for FtsZ assembly (98) (Fig. 2A).
At the same time, defects in slmA have no detectable impact on the timing or position of Z ring formation, suggesting either that the Min system alone is sufficient to regulate FtsZ assembly at midcell during standard growth and SlmA is needed when DNA integrity is compromised or that additional NO factors exist (99). While loss of slmA is synthetically lethal with defects in min, the viability of min slmA double mutants is restored by increasing FtsZ levels 2- to 3-fold or by growth in minimal medium when the cell cycle is slowed down (97). These data suggest that SlmA together with Min helps increase the local concentration of FtsZ, facilitating assembly at midcell. If correct, this function would be shared with the unrelated B. subtilis NO factor, Noc, which condenses FtsZ polymers into a tight-pitched ring at the nascent septum in conjunction with its native Min system (100).
Ter-MatP linkage, a positive positional regulator.
The Ter-MatP linkage system promotes FtsZ assemblies at midcell (80), making it the only positive topological regulator of FtsZ assembly identified to date in E. coli. The E. coli chromosome is spatially organized into four macrodomains, large stretches of DNA that occupy specific regions of the cell during DNA replication. These domains include Ori, which encodes the origin of DNA replication, the right and left lateral domains, and the Ter macrodomain, which encodes the terminus of replication (101). The Ter region is organized into a macrodomain by MatP, a ter-binding protein (101). DNA-bound MatP recruits ZapB and, indirectly, its binding partner, ZapA (102). In vitro, ZapA interacts directly with FtsZ polymers promoting lateral interactions, and genetic and cytological data support a similar function in vivo (81, 103–105). The MatP-ZapB-ZapA protein complex aids FtsZ assembly at midcell by anchoring the terminus of the chromosome to the division site, providing a possible link in coordinating division with DNA replication (102) (Fig. 2A).
ASSEMBLY OF THE CYTOKINETIC RING
Association of FtsZ with the membrane.
In E. coli, FtsZ associates with the cytoplasmic membrane by the aid of two proteins, FtsA and ZipA, and together they form the essential core of the cytokinetic ring at midcell (106, 107). FtsA and ZipA both interact with the FtsZ CTP, and while either can anchor FtsZ polymers to the membrane and support FtsZ assembly in vivo, both are essential for division to proceed (107–112). A critical ratio of FtsA:FtsZ and ZipA:FtsZ levels is required for division; excess FtsA/ZipA inhibits division, but a compensatory increase in FtsZ levels overcomes this inhibition (106, 113).
A peripheral membrane protein, FtsA, is widely conserved, and accumulating data suggest that in addition to mediating interaction of FtsZ polymers to the cytoplasmic membrane, it also coordinates recruitment of downstream divisome components. FtsA belongs to the actin superfamily of ATPases but possesses a different domain architecture (114, 115). Electron tomography of E. coli cells overexpressing ftsA and ftsZ reveals FtsA-FtsZ polymer complexes with FtsZ polymers situated ∼16 nm away from the cytoplasmic membrane and partial FtsA polymers present nearly halfway between the two (72). FtsZ polymer dynamics on a lipid bilayer containing FtsA is driven by GTP-dependent treadmilling of FtsZ (51). A role for the ATP hydrolytic function of FtsA in division is not clear. Mutations located near the ATP-binding pocket of FtsA fail to support cell division at higher temperatures (116). It is plausible that ATP hydrolysis is involved in FtsA oligomer disassembly or turnover, as mutants that reduce FtsA oligomerization suppress defects in ATP binding/hydrolysis and ATP hydrolysis rates are reduced in oligomerization-defective FtsA (116, 117).
Restricted to the Gammaproteobacteria, the integral membrane protein ZipA anchors FtsZ polymers to the cytoplasmic membrane (106). ZipA promotes the stability of the FtsZ cytokinetic ring in vivo, likely by corralling FtsZ polymers at midcell, a notion bolstered by the ability of ZipA to cross-link FtsZ polymers in vitro (112, 118). The requirement for ZipA can be bypassed by gain-of-function mutations in ftsA (e.g., ftsA* [R286W]), suggesting that the primary function of ZipA is to serve as a supplementary membrane anchor of FtsZ (119, 120) (Table 1) (discussed in “Role of FtsA in recruitment of late division proteins”).
TABLE 1.
The essential divisome of E. coli under standard laboratory growth conditions
| Protein (gene) | Function | Location (size in kDa)a | Coreb | Protein-protein interactionsc | Bypass mutations or multicopy suppressorsd | Source |
|---|---|---|---|---|---|---|
| FtsZ (ftsZ) | Tubulin-like principal regulator of septal PG synthesis | C (40.3) | + | FtsZ, FtsA, and ZipA | ||
| FtsA (ftsA) | Primary membrane tether of FtsZ; recruitment of other division proteins | C and IM (45.3) | + | FtsZ, FtsA, FtsQ, FtsI, and FtsN | ftsL* (E88K) partial bypass; multicopy ftsN [ftsA12 (Ts)] | 152, 177 |
| ZipA (zipA) | Alternate membrane tether of FtsZ; enhances recruitment of other division proteins; preseptal PG synthesis | C and IM (36.5) | FtsZ and FtsA | ftsA* (R286W); ftsA*-like alleles; ftsA (I143L); multicopy ftsN; ftsB* (E56A/K); ftsL*; ftsZ* (L169R); ftsW (M269I) | 119, 120, 136, 152, 178, 223, 224 | |
| FtsK (ftsK) | Recruitment of other division proteins; chromosome segregation; PG synthesis | IM (146.7) | FtsZ, FtsK, FtsQ, FtsL, and FtsI | ftsA*; ftsA (I143L); multicopy ftsB; ftsL*; multicopy ftsN or ftsZ* [ftsK44(Ts)] | 119, 136, 152, 171, 178, 181, 223, 225 | |
| FtsQ (ftsQ) | Recruitment of other division proteins; structural and regulatory role in PG synthase activity | IM (31.4) | + | FtsA, FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI, and FtsN | Multicopy ftsN or ftsZ* [ftsQ1 (Ts)]; ftsA (I143L) [localization of ftsQ (V92D)] | 136, 177, 223 |
| FtsL (ftsL) | Recruitment of other division proteins; structural and regulatory role in PG synthase activity | IM (13.6) | + | FtsK, FtsQ, FtsL, FtsB, FtsW, and FtsI | ||
| FtsB (ftsB) | Recruitment of other division proteins; structural and regulatory role in PG synthase activity | IM (11.6) | + | FtsQ, FtsL, FtsB, and FtsI | ||
| FtsW (ftsW) | Septal transglycosylase | IM (46) | + | FtsQ, FtsL, and FtsI | ||
| FtsI (ftsI; pbp3) | Septal transpeptidase | IM (63.9) | + | FtsA, FtsK, FtsQ, FtsL, FtsB, FtsW, and FtsI | Multicopy ftsN [ftsI23(Ts)] | 177 |
| FtsN (ftsN) | Regulation of septal PG synthesis activity in response to cytoplasmic and periplasmic signals | IM (35.8) | FtsA, FtsQ, FtsI, and FtsN | ftsA*; multicopy ftsA (E124A or I143L); ftsL*; ftsB*; multicopy ftsN (E) peptide; multicopy ftsW (M269I) partial rescue of FtsN function; ftsW (E289G) | 151, 152, 167, 178–180 |
C, cytoplasmic; IM, inner membrane; predicted sizes as reported on Uniprot (https://www.uniprot.org).
The essential functions of these genes cannot be completely bypassed.
Based on bacterial two-hybrid and coimmunoprecipitation assays as described in references 226 and 227.
Mutant or multicopy suppression is defined by the ability to bypass lethal defects in essential components of the divisome, partially or completely restoring growth and viability.
Lateral interactions of FtsZ polymers in the cytokinetic ring.
Note, for the purpose of this review, that we refer to FtsZ assemblies with lateral contacts directly between polymers as “bundles” and close polymer associations facilitated by protein partners as “cross-links.”
The relative ability of purified FtsZ to bundle in vitro differs substantially between bacterial species (43). E. coli FtsZ exists primarily as single-stranded polymers under standard assembly conditions (50 mM morpholineethanesulfonic acid [MES; pH 6.5], 50 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, and 1 mM GTP), while B. subtilis FtsZ forms large sheets and bundles in the same buffer conditions (43). Differences in intrinsic lateral interactions between the two species of FtsZ appear to be mediated through the core and CTV domains, potentially influenced by the CTL (36, 43). In vitro, bundling is enhanced in FtsZ proteins derived from various species when FtsZ concentration is increased or in the presence of crowding agents, such as DEAE-dextran, and divalent cations (31, 121–123).
Outside intrinsically driven lateral interactions, at least three factors, the ZapAB complex, ZapC, and ZapD, bind FtsZ directly and promote FtsZ polymer associations within the cytokinetic ring (39) (Fig. 2A). Although individually dispensable for division, cells lacking zapA, zapB, or zapC exhibit diffuse Z rings and those deleted of two or more Zaps show moderate filamentation, suggesting overlapping roles in FtsZ assembly (124–127). This redundancy probably lies in corralling loosely assembled clusters of FtsZ polymers, increasing local concentration of the protein at the division site. Only ZapA is widely conserved, while ZapC and ZapD are restricted to the gammaproteobacterial class (103, 125–127). Biochemistry indicates that all three Zaps cross-link FtsZ polymers by distinct mechanisms. Tetramers of ZapA cooperatively link FtsZ filaments through interactions with the FtsZ core domain while dimers of ZapD cross-link via interactions with the CTP (41, 128, 129). At least two FtsZ binding sites have been identified in ZapC, supporting a model where ZapC monomers cross-link adjacent FtsZ polymers through interactions with the core domain (130, 131). Of note, ZipA also cross-links FtsZ polymers and in this aspect appears to be functionally redundant to the Zap proteins (118).
Lateral interactions and polymer treadmilling, a conundrum.
It is not trivial to reconcile increases in lateral interactions between FtsZ filaments with the treadmilling behavior of FtsZ in the cell. In the simplest sense, increasing contacts between adjacent polymers should stabilize subunits within both polymers, inhibiting turnover. Consistent with this prediction, GTP hydrolysis rates are reduced in laterally associated FtsZ polymers in vitro (7, 122). Treadmilling dynamics, however, appear to differ between bundled and cross-linked FtsZ polymers. While FtsZ CTL domain mutations that enhance bundling in vitro reduce treadmilling velocity in a reconstitution assay, the dynamic properties of ZapA-cross-linked FtsZ polymers are essentially identical to “naked” wild-type FtsZ polymers (55, 132, 133). This suggests that the CTL and ZapA control the distance between FtsZ polymer lateral associations without strongly affecting the dynamics of the single-stranded polymer. The weak affinity (micromolar range) of ZapA for FtsZ and the competition for binding with other FtsZ modulatory proteins in the context of the divisome likely render the ZapA-FtsZ interaction transient (133). Individual polymer dynamics may thus be less affected by cross-links than first principles would suggest. The physiological relevance of FtsZ lateral associations in Z ring assembly therefore appears to lie in funneling FtsZ polymers to the nascent septal site and propagating more treadmilling filaments, the latter potentially a consequence of increased polymer collisions at high local concentrations of the protein (57, 134).
A self-reinforcing corralling model for midcell FtsZ assembly.
Accumulating data support a model in which the topological factors together with the Zaps drive assembly of the septal cytokinetic ring by corralling FtsZ polymers at midcell (69, 100, 132). In agreement with this “self-reinforcing corral” model, accrual of FtsZ to threshold numbers at the nascent septal site appears to be critical for coordinating division with cell size (135). Genetic data are also consistent with this view. Increasing concentration of wild-type FtsZ or expressing a hypermorphic variant, ftsZ*, that exhibits increased lateral interactions in vitro suppresses the filamentation associated with double deletions in min and slmA or zapA and zapC (97, 136, 137). Finally, advanced microscopy in B. subtilis cells supports the idea that FtsZ treadmilling-dependent lateral associations corral FtsZ polymers at the nascent septal site and enhance the recruitment of cell wall synthesis enzymes (69).
THE BUSINESS END OF THE DIVISOME
The transenvelope divisome, a cross wall synthesis machine.
Composed of scaffolding proteins, regulatory factors, and PG synthesis enzymes, transenvelope components of the divisome play an essential and direct role in cross wall synthesis. Despite being commonly referred to as “late” proteins, transenvelope divisome components assemble in concert with cytoplasmic components, guided in part via interactions with FtsA (138–140). The late protein moniker is historical, stemming from genetic-hierarchy findings (12). Like those of FtsZ, the levels of transenvelope proteins are largely unchanged throughout the cell cycle, their absolute numbers increasing but concentration remaining unaffected as the cell grows (73, 75). Analysis of a fluorescent FtsZ fusion protein indicates that division initiates when FtsZ, and likely other cell division proteins, reaches threshold numbers at the nascent septal site, prompting a conformational change that stimulates PG synthesis at midcell (141). Below, we describe the function of individual essential components of the septal ring, for simplicity tackling them according to their hierarchy of assembly as established by genetic analysis (FtsK < FtsQ, FtsL, FtsB < FtsW, < FtsI, < and FtsN) (Fig. 2B).
Cellular roles of transenvelope divisome components. (i) FtsK.
A four-pass transmembrane protein whose C-terminal domain shares homology with proteins involved in chromosome partitioning, FtsK, a DNA translocase, corrects aberrant division across unsegregated chromosomal material by “pumping” DNA away from the invaginating septum (142–144). The chromosome partitioning domain is dispensable for E. coli growth (145, 146), arguing against a major role for FtsK as a checkpoint linking division and chromosome segregation. Instead, the essentiality of FtsK appears to lie in its ability to recruit other transenvelope proteins, particularly the FtsQLB complex.
(ii) FtsQLB.
The widely conserved bitopic membrane proteins, FtsQ, FtsL, and FtsB, play an important structural role in transenvelope assembly of the divisome (147–153). FtsQ contains a large periplasmic domain required for recruitment of FtsL and FtsB, and the three proteins localize as a FtsQLB complex to midcell (154, 155). FtsQ is considered to interact with multiple division proteins, including FtsN, the last protein in the genetically defined pathway (149, 156). FtsL and FtsB interact with each other through periplasmic leucine-zipper motifs and are critical for recruitment of PG synthesis enzymes FtsW and FtsI (PBP3), in addition to FtsN (157). Genetic data suggest that beyond its role as a scaffold for transenvelope protein assembly, FtsQLB promotes septal PG synthesis following recruitment of threshold levels of FtsN (151–153).
(iii) FtsW and FtsI.
Together, FtsW and FtsI mediate synthesis of the PG cross wall. A 10-pass transmembrane protein, FtsW is a monofunctional transglycosylase responsible for covalently linking N-acetylglucosamine and N-acetylmuramic acid molecules that make up the backbone of PG (158–160). Encoded in the same “2-min” division and cell wall (dcw) region of the chromosome as ftsW, ftsI encodes the class B penicillin-binding protein, PBP3, a monofunctional transpeptidase that catalyzes linkages between the peptide stems of PG subunits (161). Its name is derived from its nature as a target of penicillin and other beta-lactam antibiotics; treatment of E. coli with the PBP3-specific beta-lactam, cephalexin, inhibits division entirely (162–164).
(iv) FtsN.
Last in the genetically derived divisome assembly hierarchy, FtsN consists of a short, N-terminal cytoplasmic domain (165, 166), a 19-residue periplasmic subdomain (E) essential for activation of cross wall synthesis (167), and a C-terminal SPOR (sporulation related repeat) domain (168). The latter interacts with so-called “denuded” glycans (G), the product of cell wall hydrolases that remove the peptide stem from the glycan backbone as part of PG remodeling during growth and cell separation (169, 170). Within the divisome, FtsN interacts not only with its immediately upstream assembly partner, FtsI, but also with FtsQ and FtsA, providing a connection between periplasmic, membrane, and cytoplasmic components of the divisome (171–175). Interaction between FtsA and FtsN is considered to propagate the signal that activates division once assembly of the cytokinetic ring is complete (151, 176). Consistent with a role as a mediator of divisome integrity and function, ftsN was first identified as a multicopy suppressor of an ftsA (Ts) mutant (177). Overexpression of FtsN also suppresses conditional alleles of ftsK, ftsQ, and ftsI and can bypass the requirement for the conditionally essential division regulator, FtsEX (139, 176) (Table 1). Although frequently referred to as the “trigger” for division, suggesting a transient but essential role, FtsN activity is bypassed by gain-of-function mutations in ftsA, ftsL, ftsB, ftsW, and ftsI, suggesting a more nuanced role (151–153, 178–180) (Table 1) (discussed in “Activation of cross wall synthesis and cell separation”).
Role of FtsA in recruitment of late division proteins.
The last decade of work points to a critical role for FtsA in bridging the transition between cytoplasmic and periplasmic components of the divisome. Several hypermorphic ftsA alleles bypass the requirement for ZipA, the best characterized of which is ftsA* (R286W), suggesting that they mimic the physiological role of ZipA (119, 120). Notably, FtsA* and FtsA*-like mutants are also able to suppress the requirement for ftsN, ftsK, and ZapC overexpression-related FtsZ assembly defects (178, 181, 182). Consistent with accelerated divisome assembly and activation in ftsA*-bearing cells, they are ∼20% shorter than wild-type cells (183). Cytological and biochemical data suggest that FtsA* and FtsA*-like proteins form shorter oligomers, which may explain the basis of the gain of function (115, 120, 184, 185). Shorter FtsA oligomers expose more sites of interactions with downstream proteins that would normally be occluded in the longer FtsA polymer. Together, these data support a model in which ZipA enhances interactions between FtsA and downstream division proteins by competing for the common FtsA/ZipA-binding site on the FtsZ CTP and disrupting FtsA polymers into shorter fragments (111, 120) (Fig. 2C). FtsA and ZipA interact in an in vivo cross-linking assay, suggesting that ZipA may also function as a direct modulator of FtsA activity (186).
ACTIVATION OF THE DIVISOME
Activation of cross wall synthesis and cell separation.
To ensure the production of viable daughter cells, division must be coordinated with both DNA replication and cell growth. As discussed above, part of this coordination is achieved through the Min system and NO (see “Min, a negative positional regulator” and “The chromosome as a positional regulator”), through interactions between FtsZ and ter-binding protein MatP (see “Ter-MatP linkage, a positive positional regulator”), and by the chromosome pumping activity of FtsK (see “Cellular roles of transenvelope divisome components”). The mechanisms coordinating division with cell growth, however, are only now beginning to emerge. Recent work suggests that two factors—accumulation of division proteins to threshold numbers within the divisome and related changes in the nature of interactions between divisome components—underlie activation of cross wall synthesis by FtsW and FtsI (141, 151–153). Activation of FtsW and FtsI, in turn, shifts PG synthesis from elongation of the cylindrical wall to cross wall formation by an as-yet-unknown mechanism (161).
Interactions between FtsN and FtsQLB are responsible for the initial activation of FtsW and FtsI. Early in the cell cycle, the concentration of FtsN, FtsQLB, and other transenvelope components at the nascent division site is low, a consequence of reduced numbers of FtsZ polymers at this position (141), their own growth-dependent accumulation, and limited availability of the FtsN binding site of FtsA (187). In this state, FtsQLB is structurally “off,” unable to activate FtsWI-mediated septal PG synthesis. As the cell grows, numbers of FtsQLB, FtsWI, and FtsN molecules increase within the divisome as their recruitment is enhanced by ZipA- and FtsEX-mediated exposure of the FtsN binding site of FtsA (see discussion of FtsEX in the next section). Increases in their local concentration facilitate interaction between the periplasmic essential subdomain (E) of FtsN and FtsQLB, shifting the latter to an “on” state (167). Interaction of the cytoplasmic N-terminal domain of FtsN with FtsA is thought to reinforce the FtsQLB “on” state (188). Once on, FtsL stimulates FtsW and FtsI activity, initiating cross wall synthesis (151–153) (Fig. 2D). This “threshold activation” model is supported by the reduced cell lengths of ftsA, ftsL, ftsB, ftsW, and ftsI hypermorphs and cells overexpressing ftsN (151, 152, 179, 180, 183, 189). Overproduction of the region containing the FtsN (E) subdomain alone bypasses the requirement for full-length FtsN for division, further highlighting the critical nature of this region to cross wall synthesis (167). Purified Pseudomonas aeruginosa FtsQLB activates FtsWI-mediated PG synthesis in the presence of PG precursors, providing additional support for this model (190). Notably, in this scheme, one can envision the role of FtsN somewhat akin to a “clamp,” holding the divisome in the appropriate conformation and thus promoting continued cell wall synthesis via FtsWI.
Cross wall synthesis is followed by cross wall splitting, which in turn reinforces FtsN recruitment and activation of FtsWI (151). Synthesis of new PG at the leading edge of the septum stimulates splitting of the PG macromolecule, a consequence of cell wall hydrolase (amidase) activity proximal to the OM. As mentioned, amidases clip peptide stems from the glycan backbone, facilitating cell separation (161). The strong affinity of FtsN for the products of amidase activity (denuded glycans [G]) enhances its recruitment to the septum, further potentiating the FtsQLB-mediated activation of FtsWI (Fig. 2D). Accessory SPOR domain-containing proteins also localize to the midcell (167, 188, 191). Of these, DedD interacts with the FtsQLB complex and stimulates FtsWI activity in wild-type cells in parallel with FtsN (188). DedD becomes essential when FtsN activity is reduced, suggesting that other SPOR proteins may play a critical septal PG synthesis role under specific genetic conditions (188).
FtsEX, the conditionally essential division regulators.
An important modulator of divisome activity, FtsEX localizes to midcell dependent on the interaction of FtsE with FtsZ (192–195). FtsEX contributes to the recruitment of FtsK and the nonessential amidase activator, EnvC (196). During division, the role of FtsEX is twofold. FtsX interacts with FtsA, helping expose the FtsA interface, facilitating assembly of additional division proteins, a function analogous to that proposed for ZipA (179) (discussed in “Role of FtsA in recruitment of late division proteins”). FtsEX also enhances the septal PG feedback cycle indirectly by promoting amidase activity via interactions with activator EnvC (188, 196). Despite its contribution to important aspects of division, increases in the osmotic strength of the medium, the presence of hyperactive ftsA*, ftsB*, ftsL*, or ftsW* alleles, or the overexpression of FtsN can bypass the essentiality of FtsEX (179).
CONSTRICTION OF THE DIVISION SEPTUM AND CELL SEPARATION
FtsZ, a guiding force.
Constriction of the multilayered Gram-negative septum requires coordinated synthesis of new cytoplasmic membrane, PG, and OM. As touched on earlier, cytoplasmic membrane invagination is largely a consequence of PG material “pushing” it toward the center like the closing of an annular ring rather than constriction of the Z ring itself (Fig. 3A). Consistent with this idea, nearly 50 years ago, experiments targeting FtsI (PBP3) demonstrated that PG synthesis is rate limiting for division (164). This finding was refined by the observation of twisted septa in an ftsZ GTPase mutant (ftsZ26) with otherwise no changes to the timing of division or cell growth, pointing to a role for FtsZ GTP hydrolysis activity in largely guiding the formation of a symmetrical septum (63) (Fig. 3B). More recently, quantitative superresolution imaging in various species buttresses the conclusion that constriction is limited by the rate of septal PG synthesis and is independent of FtsZ-treadmilling dynamics (57, 68, 197). Most important to the argument in favor of cell wall synthesis as the driver of septation is the observation that FtsZ disassembles from the divisome prior to the closure of the cytoplasm. This finding suggests that FtsZ is dispensable, at least in the final stages of division (198, 199). Notwithstanding, the ability of FtsZ GTP hydrolysis-driven polymer dynamics to deform liposomes and membranes may support a role for “pulling” forces on the membrane as a signal to initiate early stages of constriction (133, 200–202).
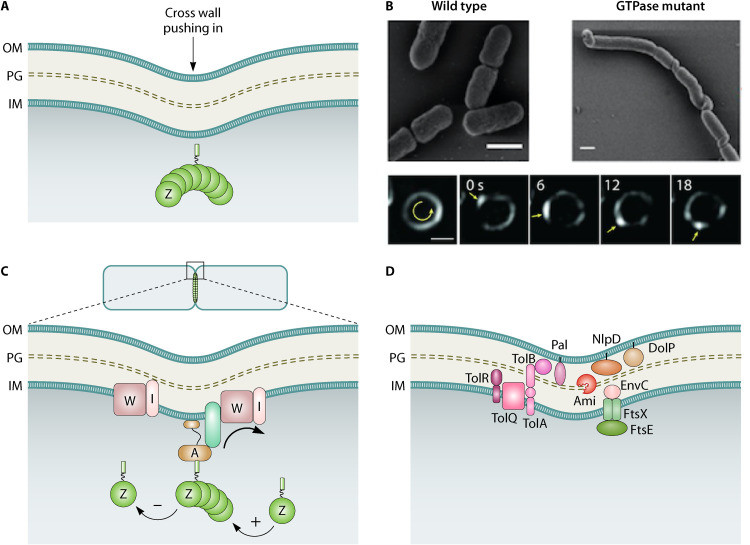
FIG 3.
FtsZ treadmilling drives uniform distribution of cross wall synthases leading to septal PG (sPG) synthesis, constriction, and scission. (A) Accumulating evidence shows that the “pushing” force of sPG synthesis is the major driver of constriction and has long been known to be the rate-limiting step. FtsZ membrane anchors FtsA and ZipA are not shown. (B) Super resolution microscopy images of E. coli FtsZ treadmilling across the septum at different time points is shown (reprinted from reference 55 with permission of the publisher). Treadmilling is GTP dependent and influences the distribution of septal cross wall synthesis enzymes (reprinted from reference 55 with permission of the publisher). (C) Cartoon of treadmilling FtsZ polymers guiding sPG synthesis. The polarity of the FtsZ polymer is shown with growing (+) and shrinking (−) ends. The treadmilling FtsZ polymer is linked (via essential divisomal factors represented as a green rod) to sPG synthases and can move bidirectionally around the septal plane. The essential septal transglycosylase FtsW and transpeptidase FtsI are shown in two tracks. The FtsZ track (shown associated via other divisiomal proteins in green) promotes uniform distribution of the two synthases around the septum (arrow). Independent of FtsZ (shown alone), the enzymatically active synthases generate new cross wall exhibiting slower directional movements. (D) PG and OM remodeling is coordinated during division. The activity of the periplasmic amidases (Ami) involved in PG hydrolysis is controlled from the IM by FtsEX-linked activation of the periplasmic EnvC and from the OM by lipoproteins NlpD-DolP. The Tol-Pal complex spanning the cell envelope layers is energized by the proton motive force across the IM and plays a major role in constriction. This protein complex coordinates the constriction of the OM with the restructuring of the PG as well. IM, inner membrane; PG, peptidoglycan; OM, outer membrane.
Coupling of septal PG synthesis to FtsZ treadmilling.
While it is established that FtsZ-treadmilling velocity is dependent on GTP hydrolysis and directs PG synthesis enzymes circumferentially across the septum, precisely how FtsZ treadmilling at the cytoplasmic face of the membrane controls septal PG synthesis at the periplasmic face of the membrane is just beginning to be elucidated (55) (Fig. 3C). Truncated FtsQ and FtsN constructs bound to supported lipid bilayers exhibit dynamics similar to those of FtsZ in the presence of both FtsZ and FtsA (175). Notably, these motions are observed only in association with FtsZ (175). On their own, the peptides displayed random stop and go motions, suggesting that FtsQ, FtsN, and associated proteins form transient associations with treadmilling FtsZ polymers via a “diffusion and capture” mechanism. While single-molecule analysis supports such dynamics for FtsN and FtsWI (180), fluorescence recovery after photobleaching (FRAP) and superresolution imaging reveal a spatial separation between FtsZ and FtsN in cells (203). Such separation makes it unlikely that a majority of FtsN movement in the cells is directly affected by FtsZ dynamics.
FtsW and FtsI exist in two states: a population of “fast-track” molecules coupled to FtsZ-treadmilling dynamics and a “slow-track” population that exhibits directional motion independent of FtsZ (180). Strikingly, only the slow-moving population is sensitive to inhibition of septal transglycosylase or transpeptidase activities, suggesting that these are active in septal PG synthesis (180). The model emerging from these experiments is that inactive FtsWI molecules traverse the septum directionally by end-tracking with treadmilling FtsZ polymers. While FtsZ subunits within the polymer remain stationary, the plus and minus ends grow and shrink, respectively. End-tracking thus allows for even distribution of FtsWI synthases at the division plane. Accordingly, an E. coli ftsZ GTPase mutant is affected in its ability to evenly distribute septal cell wall synthases but not in overall PG synthesis activity (55). Conformational changes in divisome regulators such as FtsN and FtsQLB lead to a switch in FtsW and FtsI from an enzymatically inactive state to an active one. This switch coincides with dissociation of the synthases from a treadmilling FtsZ polymer to slower directional motions where the enzymes are actively involved in cross wall assembly and setting the rate of constriction.
PG synthesis and cleavage.
Synthesis and cleavage of septal PG have to be carefully regulated to ensure the fidelity of the cell envelope during division. Among the key enzymatic activities responsible for PG synthesis and breakdown are transglycosylation, transpeptidation, and the action of hydrolases and glycosylases, involved in PG cleavage (161). In E. coli, cell wall synthesis is largely segregated into two machines: the divisome and the elongasome. Responsible for lateral cell wall synthesis, the elongasome is scaffolded by the actin-like protein MreB and encodes its own primary transglycosylase, RodA, and transpeptidase, PBP2 (161). Both the divisome and elongasome rely on an overlapping set of proteins to modify and remodel the cell wall, allowing for insertion of new material and repair of damaged sections (161). Notably, accumulating data suggest that the activities of many of these enzymes are modulated by the external environment to which they are constantly exposed (204). Below, we review the major PG synthesis and modification proteins involved in cell division.
(i) Additional septal PG synthesis by PBP1b.
While FtsW and FtsI are essential for septal PG synthesis, the bifunctional class A penicillin-binding protein, PBP1b, is thought to fortify their activity (205). Regulated from the cytoplasmic membrane by FtsW and FtsI and from the OM by a lipoprotein-periplasmic linkage LpoB-CpoB, PBP1b carries both transpeptidase and transglycosylase activities (206). While PBP1b localizes throughout the cell envelope, it is enriched at the division site (205, 207). Recent evidence suggests that nonessential SPOR domain-containing proteins, including DedD, stimulate the functionality of PBP1b (208). Although dispensable under normal laboratory growth conditions, defects in PBP1b increase sensitivity to beta-lactam antibiotics in general (209).
(ii) Septal PG hydrolysis by amidases.
As touched on above (“Activation of cross wall synthesis and cell separation”), the outer layer of the septal PG is cleaved by a set of periplasmic amidases, AmiA, AmiB, and AmiC, in parallel with PG synthesis. The amidases AmiB and AmiC are recruited to the divisome dependent on the prior localization of FtsN (210, 211). AmiA is localized throughout the periplasm during all stages of the cell cycle (210). Deletion of all three amidases results in chains of cells attached to one another by septal PG layers that fail to split and interfere with OM invagination (212, 213). To prevent discordant PG hydrolytic activity, the amidases are tightly regulated. AmiA and AmiB activity is mediated by EnvC on the cytoplasmic membrane (196, 213, 214) (Fig. 3D). EnvC itself is allosterically controlled by ATP-dependent changes in FtsEX conformation (discussed in “FtsEX, the conditionally essential division regulators”). AmiC, on the other hand, is activated by the OM proteins NlpD-YraP/DolP coincident with constriction via the Tol-Pal system (211) (Fig. 3D) (discussed below).
Invagination of the OM.
Spanning the cell envelope, the Tol-Pal complex coordinates OM invagination with PG synthesis. Five proteins, TolQ, TolA, and TolR at the cytoplasmic membrane, TolB in the periplasm, and the lipoprotein Pal in the OM, link the different layers of the cell envelope through protein-protein and protein-PG interactions (215). Mutants defective in tol-pal display multiple phenotypes, including chaining, OM vesiculation, defects in membrane integrity, and a delay in OM invagination (216). Interactions between Pal and TolB or Pal and the PG are critical for efficient OM invagination (217, 218). Prior to division, TolB-Pal complexes diffuse through the OM, while TolQR and TolA are associated with the cytoplasmic membrane. In dividing cells, TolQAR accumulates at the septum and proton motive force-driven conformational changes in TolR and TolA promote the association of TolA with TolB at the OM. Consequently, the OM lipoprotein Pal is liberated from TolB and interacts with the septal PG (216, 218). Tol-Pal also functions in activating PG-remodeling enzymes to complete the processing of remaining denuded glycan strands at the septum. Together, Tol-Pal activities help coordinate completion of septal PG remodeling with the inward folding of the OM (219) (Fig. 3D).
Disassembly and closing the pore.
Once septal PG synthesis is initiated, constriction proceeds at a relatively steady pace, taking ∼12 min in nutrient-rich medium at 37°C (68). As mentioned earlier, the rate of septum closure is driven primarily by cell wall synthesis but is partially influenced by chromosome segregation; for instance, matP mutant cells display higher rates of pore closure (68). The cytoplasm of the two daughter cells is separated first, potentially by membrane fusion, inward growth of PG, or a combination of both (198). The OM is sealed in the final step subsequent to the amidase-mediated separation of septal PG. The mechanisms governing these two membrane fusion events are unclear.
The few details we do know about the final stage of division come from the work conducted by Daley, Söderström, and colleagues. Combining superresolution microscopy with FRAP analyses they determined that division proteins disassemble in essentially the reverse order of the genetically determined assembly hierarchy (13). As discussed earlier (see “FtsZ, a guiding force”), FtsZ is not required for completion of constriction as it exits the septum prior to the sealing of the cytoplasm at a pore diameter of ∼250 nm (199). Other division proteins follow with FtsA/ZipA dispersing first, followed by FtsQL, FtsI, and finally FtsN. Strikingly, some FtsN molecules persist at the closed pore, dependent on the FtsN SPOR domain, suggesting that dispersal of all FtsN molecules from the closing septum takes place only when the denuded glycan (G) strands are completely processed (203). The signal that triggers divisome disassembly is unknown. Biophysical (e.g., membrane curvature) and biochemical properties of the closing septal pore and/or transient interactions with as-yet-unidentified protein regulators may affect this process (199). In support of the latter, ZapB and ZapE are considered to contribute to disassembly (174, 220).
CONCLUDING REMARKS
The divisome conjures up an image of a single macromolecular complex that enables a bacterial cell to build a midcell septum and divide into two new daughter cells. However, it is clear that the bacterial divisome is not a monolith but instead is a multipartite machine distributed across cellular compartments and membrane layers finely tuned to respond to cytoplasmic and extracellular stimuli that ensure the spatiotemporal precision of division.
Since FtsZ was first visualized at the septum in 1991, we have made tremendous strides toward elucidating the mechanistic details underlying cell division in E. coli. Even so, questions remain. These include, but are not limited to, the spatial architecture of the divisome through the different stages of division, the contribution of septal cell wall precursor availability to the timing and/or rate of division, and the nature of the signals stimulating cell wall constriction and disassembly of the division machinery. Further, precise molecular details of how division is coordinated with the cell cycle and how different environmental conditions shape these processes are difficult problems that are currently being investigated.
Finally, while this chapter has focused on the molecular mechanisms underlying FtsZ-based division, it would be amiss on our part not to mention atypical modes of growth and proliferation exhibited by E. coli variants such as the “L-forms” that lack PG. The existence of L-form variants has been known for nearly a century, but details of how they divide were sparse (221). Pioneering work from the Errington group have provided some insights into division mechanisms in these variants, even in the absence of FtsZ (222). These findings illustrate the awe-inspiring evolutionary plasticity present in bacterial division and emphasize that there is plenty left to discover, even in a model organism as richly investigated and understood as E. coli. With the advances in several techniques, including live-cell single-molecule imaging, in situ cryotomography, microfluidics, and whole-genome sequencing, to name a few, it is perhaps not an exaggeration to state that the versatile toolkit that is available to understand the mechanistic strategies of bacterial division is greater than it has ever been before. The lessons we have learned, and continue to learn, from E. coli division will therefore remain an invaluable reference in understanding division in other species, including in traditionally understudied ones.
ACKNOWLEDGMENTS
It is difficult to do justice to the extensive body of work in the area of E. coli cell division. We apologize to the many authors who have made significant contributions to our knowledge in this area and whose publications we have failed to include here. We thank the members of our labs for lively and helpful conversations over the course of writing this article, the reviewers whose comments and suggestions helped improve the final manuscript, and Patrick Lane for help with the figures. Research in the Levin lab is supported by NIH GM127331 and Janakiraman lab by NSF MCB 1615858.
Contributor Information
Petra Anne Levin, Email: plevin@wustl.edu.
Anuradha Janakiraman, Email: anuj@ccny.cuny.edu.
James M. Slauch, University of Illinois at Urbana Champaign
REFERENCES
- 1.Nanninga N. 1991. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol 5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 2.de Boer PAJ. 2010. Advances in understanding E. coli cell fission. Curr Opin Microbiol 13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haeusser DP, Margolin W. 2016. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14:305–319. doi: 10.1038/nrmicro.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du S, Lutkenhaus J. 2019. At the heart of bacterial cytokinesis: the Z ring. Trends Microbiol 27:781–791. doi: 10.1016/j.tim.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Söderström B, Daley DO. 2017. The bacterial divisome: more than a ring? Curr Genet 63:161–164. doi: 10.1007/s00294-016-0630-2. [DOI] [PubMed] [Google Scholar]
- 6.Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 7.Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McQuillen R, Xiao J. 2020. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu Rev Biophys 49:309–341. doi: 10.1146/annurev-biophys-121219-081703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrows JM, Goley ED. 2021. FtsZ dynamics in bacterial division: what, how, and why? Curr Opin Cell Biol 68:163–172. doi: 10.1016/j.ceb.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leger MM, Petrů M, Žárský V, Eme L, Vlček Č, Harding T, Lang BF, Eliáš M, Doležal P, Roger AJ. 2015. An ancestral bacterial division system is widespread in eukaryotic mitochondria. Proc Natl Acad Sci U S A 112:10239–10246. doi: 10.1073/pnas.1421392112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 12.Goehring NW, Beckwith J. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol 15:R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 13.Söderström B, Mirzadeh K, Toddo S, von Heijne G, Skoglund U, Daley DO. 2016. Coordinated disassembly of the divisome complex in Escherichia coli. Mol Microbiol 101:425–438. doi: 10.1111/mmi.13400. [DOI] [PubMed] [Google Scholar]
- 14.Witkin EM. 1947. Genetics of resistance to radiation in Escherichia coli. Genetics 32:221–248. doi: 10.1093/genetics/32.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van De Putte P, Van D, Roersch A. 1964. The selection of mutants of Escherichia Coli with impaired cell division at elevated temperature. Mutat Res 106:121–128. doi: 10.1016/0027-5107(64)90014-4. [DOI] [PubMed] [Google Scholar]
- 16.Hirota Y, Ryter A, Jacob F. 1968. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harbor Symp Quant Biol 33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- 17.Wijsman HJ. 1972. A genetic map of several mutations affecting the mucopeptide layer of Escherichia coli. Genet Res 20:65–74. doi: 10.1017/s0016672300013598. [DOI] [PubMed] [Google Scholar]
- 18.Donachie WD, Begg KJ, Lutkenhaus JF, Salmond GP, Martinez-Salas E, Vincente M. 1979. Role of the ftsA gene product in control of Escherichia coli cell division. J Bacteriol 140:388–394. doi: 10.1128/jb.140.2.388-394.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutkenhaus JF, Donachie WD. 1979. Identification of the ftsA gene product. J Bacteriol 137:1088–1094. doi: 10.1128/jb.137.3.1088-1094.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutkenhaus JF, Wolf-Watz H, Donachie WD. 1980. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol 142:615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X, Ehrhardt DW, Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci U S A 93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin PA, Losick R. 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev 10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 23.Huisman O, D'Ari R, Gottesman S. 1984. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci U S A 81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Boer PA, Crossley RE, Rothfield LI. 1988. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol 170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Boer PA, Crossley RE, Rothfield LI. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol 174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi E, Lutkenhaus J. 1993. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol 175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Boer PAJ, Crossley RE, Rothfield LI. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 28.RayChaudhuri D, Park JT. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee A, Lutkenhaus J. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol 176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bramhill D, Thompson CM. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc Natl Acad Sci U S A 91:5813–5817. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popp D, Iwasa M, Narita A, Erickson HP, Maéda Y. 2009. FtsZ condensates: an in vitro electron microscopy study. Biopolymers 91:340–350. doi: 10.1002/bip.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Löwe J, Amos LA. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 33.Nogales E, Downing KH, Amos LA, Löwe J. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 34.Corrales-Guerrero L, Camargo S, Valladares A, Picossi S, Luque I, Ochoa de Alda JAG, Herrero A. 2018. FtsZ of filamentous, heterocyst-forming cyanobacteria has a conserved N-terminal peptide required for normal FtsZ polymerization and cell division. Front Microbiol 9:2260. doi: 10.3389/fmicb.2018.02260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher MA, Ohashi T, Corbin L, Erickson HP. 2020. High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP. Acta Crystallogr F Struct Biol Commun 76:94–102. doi: 10.1107/S2053230X20001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardner KAJA, Moore DA, Erickson HP. 2013. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol Microbiol 89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buske PJ, Levin PA. 2013. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol 89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundararajan K, Miguel A, Desmarais SM, Meier EL, Casey Huang K, Goley ED. 2015. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat Commun 6:7281. doi: 10.1038/ncomms8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang K-H, Durand-Heredia JM, Janakiraman A. 2013. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol 195:1859–1868. doi: 10.1128/JB.02157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortiz C, Natale P, Cueto L, Vicente M. 2016. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol Rev 40:57–67. doi: 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 41.Schumacher MA, Huang KH, Zeng W, Janakiraman A. 2017. Structure of the Z ring-associated protein, ZapD, bound to the C-terminal domain of the tubulin-like protein, FtsZ, suggests mechanism of Z ring stabilization through FtsZ cross-linking. J Biol Chem 292:3740–3750. doi: 10.1074/jbc.M116.773192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohan MC, Eddelbuettel AMP, Levin PA, Pappu RV. 2020. Dissecting the functional contributions of the intrinsically disordered C-terminal tail of Bacillus subtilis FtsZ. J Mol Biol 432:3205–3221. doi: 10.1016/j.jmb.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buske PJ, Levin PA. 2012. Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J Biol Chem 287:10945–10957. doi: 10.1074/jbc.M111.330324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheffers DJ, de Wit JG, den Blaauwen T, Driessen AJ. 2002. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41:521–529. doi: 10.1021/bi011370i. [DOI] [PubMed] [Google Scholar]
- 45.Romberg L, Simon M, Erickson HP. 2001. Polymerization of FtsZ, a bacterial homolog of tubulin. J Biol Chem 276:11743–11753. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- 46.Caplan MR, Erickson HP. 2003. Apparent cooperative assembly of the bacterial cell division protein FtsZ demonstrated by isothermal titration calorimetry. J Biol Chem 278:13784–13788. doi: 10.1074/jbc.M300860200. [DOI] [PubMed] [Google Scholar]
- 47.Matsui T, Han X, Yu J, Yao M, Tanaka I. 2014. Structural change in FtsZ induced by intermolecular interactions between bound GTP and the T7 loop. J Biol Chem 289:3501–3509. doi: 10.1074/jbc.M113.514901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagstaff JM, Tsim M, Oliva MA, García-Sanchez A, Kureisaite-Ciziene D, Andreu JM, Löwe J. 2017. A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. mBio 8. doi: 10.1128/mBio.00254-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miraldi ER, Thomas PJ, Romberg L. 2008. Allosteric models for cooperative polymerization of linear polymers. Biophys J 95:2470–2486. doi: 10.1529/biophysj.107.126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbin LC, Erickson HP. 2020. A unified model for treadmilling and nucleation of single-stranded FtsZ protofilaments. Biophys J 119:792–805. doi: 10.1016/j.bpj.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loose M, Mitchison TJ. 2014. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol 16:38–46. doi: 10.1038/ncb2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson DE, Gueiros-Filho FJ, Erickson HP. 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol 186:5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y, Erickson HP. 2005. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J Biol Chem 280:22549–22554. doi: 10.1074/jbc.M500895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niu L, Yu J. 2008. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J 95:2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. 2017. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355:744–747. doi: 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bisson-Filho AW, Hsu Y-P, Squyres GR, Kuru E, Wu F, Jukes C, Sun Y, Dekker C, Holden S, VanNieuwenhze MS, Brun YV, Garner EC. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355:739–743. doi: 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitley KD, Jukes C, Tregidgo N, Karinou E, Almada P, Cesbron Y, Henriques R, Dekker C, Holden S. 2021. FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nat Commun 12:2448. doi: 10.1038/s41467-021-22526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Redick SD, Stricker J, Briscoe G, Erickson HP. 2005. Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J Bacteriol 187:2727–2736. doi: 10.1128/JB.187.8.2727-2736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du S, Pichoff S, Kruse K, Lutkenhaus J. 2018. FtsZ filaments have the opposite kinetic polarity of microtubules. Proc Natl Acad Sci U S A 115:10768–10773. doi: 10.1073/pnas.1811919115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu C, Erickson HP. 1999. The straight and curved conformation of FtsZ protofilaments-evidence for rapid exchange of GTP into the curved protofilament. Cell Struct Funct 24:285–290. doi: 10.1247/csf.24.285. [DOI] [PubMed] [Google Scholar]
- 61.Osawa M, Anderson DE, Erickson HP. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osawa M, Erickson HP. 2013. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci U S A 110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bi E, Lutkenhaus J. 1992. Isolation and characterization of ftsZ alleles that affect septal morphology. J Bacteriol 174:5414–5423. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Addinall SG, Lutkenhaus J. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol 22:231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 65.RayChaudhuri D, Park JT. 1994. A point mutation converts Escherichia coli FtsZ septation GTPase to an ATPase. J Biol Chem 269:22941–22944. doi: 10.1016/S0021-9258(17)31600-9. [DOI] [PubMed] [Google Scholar]
- 66.Mukherjee A, Saez C, Lutkenhaus J. 2001. Assembly of an FtsZ mutant deficient in GTPase activity has implications for FtsZ assembly and the role of the Z ring in cell division. J Bacteriol 183:7190–7197. doi: 10.1128/JB.183.24.7190-7197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arjes HA, Lai B, Emelue E, Steinbach A, Levin PA. 2015. Mutations in the bacterial cell division protein FtsZ highlight the role of GTP binding and longitudinal subunit interactions in assembly and function. BMC Microbiol 15:209. doi: 10.1186/s12866-015-0544-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coltharp C, Buss J, Plumer TM, Xiao J. 2016. Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci U S A 113:E1044–1053. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Squyres GR, Holmes MJ, Barger SR, Pennycook BR, Ryan J, Yan VT, Garner EC. 2021. Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat Microbiol 6:553–562. doi: 10.1038/s41564-021-00878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu G, Huang T, Buss JA, Coltharp C, Hensel Z, Xiao J. 2010. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS One 5:e12680. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowlett VW, Margolin W. 2014. 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J 107:L17–L20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szwedziak P, Wang Q, Bharat TAM, Tsim M, Löwe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife 3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weart RB, Levin PA. 2003. Growth rate-dependent regulation of medial FtsZ ring formation. J Bacteriol 185:2826–2834. doi: 10.1128/JB.185.9.2826-2834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rueda S, Vicente M, Mingorance J. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J Bacteriol 185:3344–3351. doi: 10.1128/JB.185.11.3344-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vischer NO, Verheul J, Postma M, van den Berg van Saparoea B, Galli E, Natale P, Gerdes K, Luirink J, Vollmer W, Vicente M, den Blaauwen T. 2015. Cell age dependent concentration of Escherichia coli divisome proteins analyzed with ImageJ and ObjectJ. Front Microbiol 6:586. doi: 10.3389/fmicb.2015.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Männik J, Walker BE, Männik J. 2018. Cell cycle-dependent regulation of FtsZ in Escherichia coli in slow growth conditions. Mol Microbiol 110:1030–1044. doi: 10.1111/mmi.14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Boer PA, Crossley RE, Rothfield LI. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 78.Lutkenhaus J. 2008. Min oscillation in bacteria. Adv Exp Med Biol 641:49–61. doi: 10.1007/978-0-387-09794-7_4. [DOI] [PubMed] [Google Scholar]
- 79.Rowlett VW, Margolin W. 2013. The bacterial Min system. Curr Biol 23:R553–556. doi: 10.1016/j.cub.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 80.Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Männik J. 2014. Evidence for divisome localization mechanisms independent of the min system and SlmA in Escherichia coli. PLoS Genet 10:e1004504. doi: 10.1371/journal.pgen.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Buss JA, Peters NT, Xiao J, Bernhardt TG. 2017. ZapA and ZapB form an FtsZ-independent structure at midcell. Mol Microbiol 104:652–663. doi: 10.1111/mmi.13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adler HI, Fisher WD, Cohen A, Hardigree AA. 1967. Miniature Escherichia coli cells deficient in DNA. Proc Natl Acad Sci U S A 57:321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci U S A 96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen B, Lutkenhaus J. 2009. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD. Mol Microbiol 72:410–424. doi: 10.1111/j.1365-2958.2009.06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. 2008. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol 18:235–244. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 86.Hu Z, Saez C, Lutkenhaus J. 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J Bacteriol 185:196–203. doi: 10.1128/JB.185.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou H, Lutkenhaus J. 2004. The switch I and II regions of MinD are required for binding and activating MinC. J Bacteriol 186:1546–1555. doi: 10.1128/JB.186.5.1546-1555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Szwedziak P, Ghosal D. 2017. FtsZ-ring architecture and its control by MinCD, p 213–244. In Löwe J, Amos LA (ed), Prokaryotic cytoskeletons: filamentous protein polymers active in the cytoplasm of bacterial and archaeal cells. Springer International Publishing, Cham, Switzerland. [DOI] [PubMed] [Google Scholar]
- 89.de Boer PA, Crossley RE, Hand AR, Rothfield LI. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J 10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raskin DM, de Boer PA. 1999. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181:6419–6424. doi: 10.1128/JB.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hale CA, Meinhardt H, de Boer PA. 2001. Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J 20:1563–1572. doi: 10.1093/emboj/20.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lackner LL, Raskin DM, de Boer PAJ. 2003. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol 185:735–749. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park KT, Villar MT, Artigues A, Lutkenhaus J. 2017. MinE conformational dynamics regulate membrane binding, MinD interaction, and Min oscillation. Proc Natl Acad Sci U S A 114:7497–7504. doi: 10.1073/pnas.1707385114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cook WR, de Boer PA, Rothfield LI. 1989. Differentiation of the bacterial cell division site. Int Rev Cytol 118:1–31. doi: 10.1016/s0074-7696(08)60871-2. [DOI] [PubMed] [Google Scholar]
- 95.Woldringh CL, Mulder E, Huls PG, Vischer N. 1991. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol 142:309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- 96.Yu XC, Margolin W. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 32:315–326. doi: 10.1046/j.1365-2958.1999.01351.x. [DOI] [PubMed] [Google Scholar]
- 97.Bernhardt TG, de Boer PA. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cho H, Bernhardt TG. 2013. Identification of the SlmA active site responsible for blocking bacterial cytokinetic ring assembly over the chromosome. PLoS Genet 9:e1003304. doi: 10.1371/journal.pgen.1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mannik J, Wu F, Hol FJ, Bisicchia P, Sherratt DJ, Keymer JE, Dekker C. 2012. Robustness and accuracy of cell division in Escherichia coli in diverse cell shapes. Proc Natl Acad Sci U S A 109:6957–6962. doi: 10.1073/pnas.1120854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu Y, Zhou J, Gueiros-Filho FJ, Kearns DB, Jacobson SC. 2021. Noc corrals migration of FtsZ protofilaments during cytokinesis in Bacillus subtilis. mBio 12:e02964-20. doi: 10.1128/mBio.02964-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O. 2008. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135:475–485. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 102.Espeli O, Borne R, Dupaigne P, Thiel A, Gigant E, Mercier R, Boccard F. 2012. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J 31:3198–3211. doi: 10.1038/emboj.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16:2544–2556. doi: 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mohammadi T, Ploeger GE, Verheul J, Comvalius AD, Martos A, Alfonso C, van Marle J, Rivas G, den Blaauwen T. 2009. The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48:11056–11066. doi: 10.1021/bi901461p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Galli E, Gerdes K. 2012. FtsZ-ZapA-ZapB interactome of Escherichia coli. J Bacteriol 194:292–302. doi: 10.1128/JB.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hale CA, de Boer PA. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175–185. doi: 10.1016/S0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 107.Pichoff S, Lutkenhaus J. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hale CA, de Boer PA. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol 181:167–176. doi: 10.1128/JB.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu Z, Mukherjee A, Lutkenhaus J. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol 31:1853–1861. doi: 10.1046/j.1365-2958.1999.01322.x. [DOI] [PubMed] [Google Scholar]
- 110.Ma X, Margolin W. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol 181:7531–7544. doi: 10.1128/JB.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haney SA, Glasfeld E, Hale CA, Keeney D, He Z, de Boer PAJ. 2001. Genetic analysis of the Escherichia coli FtsZ·ZipA interaction in the yeast two-hybrid system. J Biol Chem 276:11980–11987. doi: 10.1074/jbc.M009810200. [DOI] [PubMed] [Google Scholar]
- 112.RayChaudhuri D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J 18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dai K, Lutkenhaus J. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol 174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.van den Ent F, Lowe J. 2000. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J 19:5300–5307. doi: 10.1093/emboj/19.20.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Szwedziak P, Wang Q, Freund SM, Lowe J. 2012. FtsA forms actin-like protofilaments. EMBO J 31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Herricks JR, Nguyen D, Margolin W. 2014. A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Mol Microbiol 94:713–727. doi: 10.1111/mmi.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Conti J, Viola MG, Camberg JL. 2018. FtsA reshapes membrane architecture and remodels the Z-ring in Escherichia coli. Mol Microbiol 107:558–576. doi: 10.1111/mmi.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hale CA, Rhee AC, de Boer PA. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J Bacteriol 182:5153–5166. doi: 10.1128/JB.182.18.5153-5166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Geissler B, Elraheb D, Margolin W. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci U S A 100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pichoff S, Shen B, Sullivan B, Lutkenhaus J. 2012. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol 83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mukherjee A, Lutkenhaus J. 1998. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J 17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mukherjee A, Lutkenhaus J. 1999. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol 181:823–832. doi: 10.1128/JB.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yu XC, Margolin W. 1997. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J 16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Buss JA, Coltharp C, Huang T, Pohlmeyer C, Wang S-C, Hatem C, Xiao J. 2013. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol 89:1099–1120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Durand-Heredia JM, Yu HH, De Carlo S, Lesser CF, Janakiraman A. 2011. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J Bacteriol 193:1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Durand-Heredia JM, Rivkin E, Fan G, Morales J, Janakiraman A. 2012. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J Bacteriol 194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hale CA, Shiomi D, Liu B, Bernhardt TG, Margolin W, Niki H, de Boer PAJ. 2011. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J Bacteriol 193:1393–1404. doi: 10.1128/JB.01245-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang K-H, Mychack A, Tchorzewski L, Janakiraman A. 2016. Characterization of the FtsZ C-terminal variable (CTV) region in Z-ring assembly and interaction with the Z-ring stabilizer ZapD in E. coli cytokinesis. PLoS One 11:e0153337. doi: 10.1371/journal.pone.0153337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roseboom W, Nazir MG, Meiresonne NY, Mohammadi T, Verheul J, Buncherd H, Bonvin A, de Koning LJ, de Koster CG, de Jong L, den Blaauwen T. 2018. Mapping the contact sites of the Escherichia coli division-initiating proteins FtsZ and ZapA by BAMG cross-linking and site-directed mutagenesis. Int J Mol Sci 19:2928. doi: 10.3390/ijms19102928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Buczek MS, Cardenas Arevalo AL, Janakiraman A. 2016. ClpXP and ClpAP control the Escherichia coli division protein ZapC by proteolysis. Microbiology (Reading) 162:909–920. doi: 10.1099/mic.0.000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schumacher MA, Zeng W, Huang K-H, Tchorzewski L, Janakiraman A. 2016. Structural and functional analyses reveal insights into the molecular properties of the Escherichia coli Z ring stabilizing protein, ZapC. J Biol Chem 291:2485–2498. doi: 10.1074/jbc.M115.697037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Caldas P, López-Pelegrín M, Pearce DJG, Budanur NB, Brugués J, Loose M. 2019. Cooperative ordering of treadmilling filaments in cytoskeletal networks of FtsZ and its crosslinker ZapA. Nat Commun 10:5744. doi: 10.1038/s41467-019-13702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Garcia-Soriano DA, Heermann T, Raso A, Rivas G, Schwille P. 2020. The speed of FtsZ treadmilling is tightly regulated by membrane binding. Sci Rep 10:10447. doi: 10.1038/s41598-020-67224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Walker BE, Männik J, Männik J. 2020. Transient membrane-linked FtsZ assemblies precede Z-ring formation in Escherichia coli. Curr Biol 30:499–508. doi: 10.1016/j.cub.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S. 2015. Cell-size control and homeostasis in bacteria. Curr Biol 25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Haeusser DP, Rowlett VW, Margolin W. 2015. A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol Microbiol 97:988–1005. doi: 10.1111/mmi.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Krupka M, Margolin W. 2018. Unite to divide: oligomerization of tubulin and actin homologs regulates initiation of bacterial cell division. F1000Res 7:235. doi: 10.12688/f1000research.13504.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Du S, Lutkenhaus J. 2017. Assembly and activation of the Escherichia coli divisome. Mol Microbiol 105:177–187. doi: 10.1111/mmi.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Goehring N, Gonzalez M, Beckwith J. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol Microbiol 61:33–45. doi: 10.1111/j.1365-2958.2006.05206.x. [DOI] [PubMed] [Google Scholar]
- 140.Alexeeva S, Gadella TW, Jr, Verheul J, Verhoeven GS, den Blaauwen T. 2010. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol 77:384–398. doi: 10.1111/j.1365-2958.2010.07211.x. [DOI] [PubMed] [Google Scholar]
- 141.Si F, Le Treut G, Sauls JT, Vadia S, Levin PA, Jun S. 2019. Mechanistic origin of cell-size control and homeostasis in bacteria. Curr Biol 29:1760–1770.e7. doi: 10.1016/j.cub.2019.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Begg KJ, Dewar SJ, Donachie WD. 1995. A new Escherichia coli cell division gene, ftsK. J Bacteriol 177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dubarry N, Possoz C, Barre F-X. 2010. Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol Microbiol 78:1088–1100. doi: 10.1111/j.1365-2958.2010.07412.x. [DOI] [PubMed] [Google Scholar]
- 144.Jean NL, Rutherford TJ, Lowe J. 2020. FtsK in motion reveals its mechanism for double-stranded DNA translocation. Proc Natl Acad Sci U S A 117:14202–14208. doi: 10.1073/pnas.2001324117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu XC, Weihe EK, Margolin W. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J Bacteriol 180:6424–6428. doi: 10.1128/JB.180.23.6424-6428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang L, Lutkenhaus J. 1998. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol 29:731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- 147.Begg KJ, Hatfull GF, Donachie WD. 1980. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell division gene ftsQ. J Bacteriol 144:435–437. doi: 10.1128/jb.144.1.435-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Buddelmeijer N, Beckwith J. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol Microbiol 52:1315–1327. doi: 10.1111/j.1365-2958.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- 149.D'ulisse V, Fagioli M, Ghelardini P, Paolozzi L. 2007. Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology (Reading) 153:124–138. doi: 10.1099/mic.0.2006/000265-0. [DOI] [PubMed] [Google Scholar]
- 150.Scheffers D, Robichon C, Haan GJ, Den Blaauwen T, Koningstein G, van Bloois E, Beckwith J, Luirink J. 2007. Contribution of the FtsQ transmembrane segment to localization to the cell division site. J Bacteriol 189:7273–7280. doi: 10.1128/JB.00723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu B, Persons L, Lee L, de Boer PAJ. 2015. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol Microbiol 95:945–970. doi: 10.1111/mmi.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tsang M-J, Bernhardt TG. 2015. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol Microbiol 95:925–944. doi: 10.1111/mmi.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Park K-T, Du S, Lutkenhaus J. 2020. Essential role for FtsL in activation of septal PG synthesis. mBio 11:e03012-20. doi: 10.1128/mBio.03012-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ghigo JM, Weiss DS, Chen JC, Yarrow JC, Beckwith J. 1999. Localization of FtsL to the Escherichia coli septal ring. Mol Microbiol 31:725–737. doi: 10.1046/j.1365-2958.1999.01213.x. [DOI] [PubMed] [Google Scholar]
- 155.Van Den Ent F, Vinkenvleugel TM, Ind A, West P, Veprintsev D, Nanninga N, Den Blaauwen T, Löwe J. 2008. Structural and mutational analysis of the cell division protein FtsQ. Mol Microbiol 68:110–123. doi: 10.1111/j.1365-2958.2008.06141.x. [DOI] [PubMed] [Google Scholar]
- 156.Guzman LM, Weiss DS, Beckwith J. 1997. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol 179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gonzalez MD, Akbay EA, Boyd D, Beckwith J. 2010. Multiple interaction domains in FtsL, a protein component of the widely conserved bacterial FtsLBQ cell division complex. J Bacteriol 192:2757–2768. doi: 10.1128/JB.01609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Khattar MM, Begg KJ, Donachie WD. 1994. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol 176:7140–7147. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boyle DS, Khattar MM, Addinall SG, Lutkenhaus J, Donachie WD. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol 24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 160.Taguchi A, Welsh MA, Marmont LS, Lee W, Sjodt M, Kruse AC, Kahne D, Bernhardt TG, Walker S. 2019. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat Microbiol 4:587–594. doi: 10.1038/s41564-018-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Egan AJF, Errington J, Vollmer W. 2020. Regulation of peptidoglycan synthesis and remodelling. Nat Rev Microbiol 18:446–460. doi: 10.1038/s41579-020-0366-3. [DOI] [PubMed] [Google Scholar]
- 162.Wang L, Khattar MK, Donachie WD, Lutkenhaus J. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J Bacteriol 180:2810–2816. doi: 10.1128/JB.180.11.2810-2816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. doi: 10.1128/JB.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Spratt BG. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A 72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Addinall SG, Cao C, Lutkenhaus J. 1997. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol 25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 166.Weiss DS. 2015. Last but not least: new insights into how FtsN triggers constriction during Escherichia coli cell division. Mol Microbiol 95:903–909. doi: 10.1111/mmi.12925. [DOI] [PubMed] [Google Scholar]
- 167.Gerding MA, Liu B, Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. 2009. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol 191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Duncan TR, Yahashiri A, Arends SJ, Popham DL, Weiss DS. 2013. Identification of SPOR domain amino acids important for septal localization, peptidoglycan binding, and a disulfide bond in the cell division protein FtsN. J Bacteriol 195:5308–5315. doi: 10.1128/JB.00911-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ursinus A, van den Ent F, Brechtel S, de Pedro M, Holtje JV, Lowe J, Vollmer W. 2004. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J Bacteriol 186:6728–6737. doi: 10.1128/JB.186.20.6728-6737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yahashiri A, Jorgenson MA, Weiss DS. 2015. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc Natl Acad Sci U S A 112:11347–11352. doi: 10.1073/pnas.1508536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Goehring N, Robichon C, Beckwith J. 2007. Role for the nonessential N terminus of FtsN in divisome assembly. J Bacteriol 189:646–649. doi: 10.1128/JB.00992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Busiek KK, Eraso JM, Wang Y, Margolin W. 2012. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol 194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rico AI, Garcia-Ovalle M, Mingorance J, Vicente M. 2004. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol Microbiol 53:1359–1371. doi: 10.1111/j.1365-2958.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- 174.Pazos M, Natale P, Margolin W, Vicente M. 2013. Interactions among the early Escherichia coli divisome proteins revealed by bimolecular fluorescence complementation. Environ Microbiol 15:3282–3291. doi: 10.1111/1462-2920.12225. [DOI] [PubMed] [Google Scholar]
- 175.Baranova N, Radler P, Hernández-Rocamora VM, Alfonso C, López-Pelegrín M, Rivas G, Vollmer W, Loose M. 2020. Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins. Nat Microbiol 5:407–411. doi: 10.1038/s41564-019-0657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pichoff S, Du S, Lutkenhaus J. 2018. Disruption of divisome assembly rescued by FtsN-FtsA interaction in Escherichia coli. Proc Natl Acad Sci U S A 115:E6855–E6862. doi: 10.1073/pnas.1806450115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dai K, Xu Y, Lutkenhaus J. 1993. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts). J Bacteriol 175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bernard CS, Sadasivam M, Shiomi D, Margolin W. 2007. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol Microbiol 64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Du S, Pichoff S, Lutkenhaus J. 2016. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc Natl Acad Sci U S A 113:E5052–5061. doi: 10.1073/pnas.1606656113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang X, McQuillen R, Lyu Z, Phillips-Mason P, De La Cruz A, McCausland JW, Liang H, DeMeester KE, Santiago CC, Grimes CL, de Boer P, Xiao J. 2021. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat Microbiol 6:584–593. doi: 10.1038/s41564-020-00853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Geissler B, Margolin W. 2005. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol 58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ortiz C, Casanova M, Palacios P, Vicente M. 2017. The hypermorph FtsA* protein has an in vivo role in relieving the Escherichia coli proto-ring block caused by excess ZapC. PLoS One 12:e0184184. doi: 10.1371/journal.pone.0184184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Geissler B, Shiomi D, Margolin W. 2007. The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology (Reading) 153:814–825. doi: 10.1099/mic.0.2006/001834-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Krupka M, Rowlett VW, Morado D, Vitrac H, Schoenemann K, Liu J, Margolin W. 2017. Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat Commun 8:15957. doi: 10.1038/ncomms15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schoenemann KM, Krupka M, Rowlett VW, Distelhorst SL, Hu B, Margolin W. 2018. Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol Microbiol 109:676–693. doi: 10.1111/mmi.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Vega DE, Margolin W. 2019. Direct interaction between the two Z ring membrane anchors FtsA and ZipA. J Bacteriol 201:305–317. doi: 10.1128/JB.00579-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Corbin BD, Geissler B, Sadasivam M, Margolin W. 2004. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol 186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu B, Hale CA, Persons L, Phillips-Mason PJ, de Boer PAJ. 2019. Roles of the DedD protein in Escherichia coli cell constriction. J Bacteriol 201:e00698-18. doi: 10.1128/JB.00698-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mueller EA, Westfall CS, Levin PA. 2020. pH-dependent activation of cytokinesis modulates Escherichia coli cell size. PLoS Genet 16:e1008685. doi: 10.1371/journal.pgen.1008685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Marmont LS, Bernhardt TG. 2020. A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc Natl Acad Sci U S A 117:23879–23885. doi: 10.1073/pnas.2004598117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Arends SJR, Williams K, Scott RJ, Rolong S, Popham DL, Weiss DS. 2010. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J Bacteriol 192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol 186:785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Corbin BD, Wang Y, Beuria TK, Margolin W. 2007. Interaction between cell division proteins FtsE and FtsZ. J Bacteriol 189:3026–3035. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Reddy M. 2007. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J Bacteriol 189:98–108. doi: 10.1128/JB.01347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Du S, Henke W, Pichoff S, Lutkenhaus J. 2019. How FtsEX localizes to the Z ring and interacts with FtsA to regulate cell division. Mol Microbiol 112:881–895. doi: 10.1111/mmi.14324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. 2011. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci U S A 108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, Tavares AC, Santos M, Ferreira MT, Macario V, VanNieuwenhze MS, Filipe SR, Pinho MG. 2018. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554:528–532. doi: 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Skoog K, Soderstrom B, Widengren J, von Heijne G, Daley DO. 2012. Sequential closure of the cytoplasm and then the periplasm during cell division in Escherichia coli. J Bacteriol 194:584–586. doi: 10.1128/JB.06091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Söderström B, Skoog K, Blom H, Weiss DS, von Heijne G, Daley DO. 2014. Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol Microbiol 92:1–9. doi: 10.1111/mmi.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Erickson HP, Osawa M. 2017. FtsZ constriction force - curved protofilaments bending membranes. Subcell Biochem 84:139–160. doi: 10.1007/978-3-319-53047-5_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ganzinger KA, Merino-Salomon A, Garcia-Soriano DA, Butterfield AN, Litschel T, Siedler F, Schwille P. 2020. FtsZ reorganization facilitates deformation of giant vesicles in microfluidic traps. Angew Chem Int Ed Engl 59:21372–21376. doi: 10.1002/anie.202001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Osawa M, Erickson HP. 2018. Turgor pressure and possible constriction mechanisms in bacterial division. Front Microbiol 9:111. doi: 10.3389/fmicb.2018.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Söderström B, Chan H, Shilling PJ, Skoglund U, Daley DO. 2018. Spatial separation of FtsZ and FtsN during cell division. Mol Microbiol 107:387–401. doi: 10.1111/mmi.13888. [DOI] [PubMed] [Google Scholar]
- 204.Mueller EA, Levin PA. 2020. Bacterial cell wall quality control during environmental stress. mBio 11. doi: 10.1128/mBio.02456-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, von Rechenberg M, Nguyen-Disteche M, den Blaauwen T, Holtje JV, Vollmer W. 2006. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol 61:675–690. doi: 10.1111/j.1365-2958.2006.05280.x. [DOI] [PubMed] [Google Scholar]
- 206.Gray AN, Egan AJ, Van't Veer IL, Verheul J, Colavin A, Koumoutsi A, Biboy J, Altelaar AF, Damen MJ, Huang KC, Simorre JP, Breukink E, den Blaauwen T, Typas A, Gross CA, Vollmer W. 2015. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. Elife 4. doi: 10.7554/eLife.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Egan AJF, Jean NL, Koumoutsi A, Bougault CM, Biboy J, Sassine J, Solovyova AS, Breukink E, Typas A, Vollmer W, Simorre JP. 2014. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc Natl Acad Sci U S A 111:8197–8202. doi: 10.1073/pnas.1400376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pazos M, Peters K, Boes A, Safaei Y, Kenward C, Caveney NA, Laguri C, Breukink E, Strynadka NCJ, Simorre JP, Terrak M, Vollmer W. 2020. SPOR proteins are required for functionality of class A penicillin-binding proteins in Escherichia coli. mBio 11. doi: 10.1128/mBio.02796-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cho H, Uehara T, Bernhardt TG. 2014. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159:1300–1311. doi: 10.1016/j.cell.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bernhardt T, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tsang M-J, Yakhnina AA, Bernhardt TG. 2017. NlpD links cell wall remodeling and outer membrane invagination during cytokinesis in Escherichia coli. PLoS Genet 13:e1006888. doi: 10.1371/journal.pgen.1006888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Höltje JV. 2001. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol 41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 213.Uehara T, Dinh T, Bernhardt TG. 2009. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol 191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bernhardt T, de Boer PA. 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol 52:1255–1269. doi: 10.1111/j.1365-2958.2004.04063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Szczepaniak J, Press C, Kleanthous C. 2020. The multifarious roles of Tol-Pal in Gram-negative bacteria. FEMS Microbiol Rev 44:490–506. doi: 10.1093/femsre/fuaa018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PAJ. 2007. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol 63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Petiti M, Serrano B, Faure L, Lloubes R, Mignot T, Duché D. 2019. Tol energy-driven localization of Pal and anchoring to the peptidoglycan promote outer-membrane constriction. J Mol Biol 431:3275–3288. doi: 10.1016/j.jmb.2019.05.039. [DOI] [PubMed] [Google Scholar]
- 218.Szczepaniak J, Holmes P, Rajasekar K, Kaminska R, Samsudin F, Inns PG, Rassam P, Khalid S, Murray SM, Redfield C, Kleanthous C. 2020. The lipoprotein Pal stabilises the bacterial outer membrane during constriction by a mobilisation-and-capture mechanism. Nat Commun 11:1305. doi: 10.1038/s41467-020-15083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yakhnina AA, Bernhardt TG. 2020. The Tol-Pal system is required for peptidoglycan-cleaving enzymes to complete bacterial cell division. Proc Natl Acad Sci U S A 117:6777–6783. doi: 10.1073/pnas.1919267117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Marteyn BS, Karimova G, Fenton AK, Gazi AD, West N, Touqui L, Prevost MC, Betton JM, Poyraz O, Ladant D, Gerdes K, Sansonetti PJ, Tang CM. 2014. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio 5:e00022-14. doi: 10.1128/mBio.00022-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Errington J. 2017. Cell wall-deficient, L-form bacteria in the 21st century: a personal perspective. Biochem Soc Trans 45:287–295. doi: 10.1042/BST20160435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mercier R, Kawai Y, Errington J. 2014. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. Elife 3. doi: 10.7554/eLife.04629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Goehring NW, Petrovska I, Boyd D, Beckwith J. 2007. Mutants, suppressors, and wrinkled colonies: mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J Bacteriol 189:633–645. doi: 10.1128/JB.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Potluri LP, Kannan S, Young KD. 2012. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol 194:5334–5342. doi: 10.1128/JB.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Draper GC, McLennan N, Begg K, Masters M, Donachie WD. 1998. Only the N-terminal domain of FtsK functions in cell division. J Bacteriol 180:4621–4627. doi: 10.1128/JB.180.17.4621-4627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Maggi S, Massidda O, Luzi G, Fadda D, Paolozzi L, Ghelardini P. 2008. Division protein interaction web: identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology (Reading) 154:3042–3052. doi: 10.1099/mic.0.2008/018697-0. [DOI] [PubMed] [Google Scholar]