Abstract
Background and Aims
There is increasing interest in machine learning-based prediction models in inflammatory bowel diseases [IBD]. We synthesised and critically appraised studies comparing machine learning vs traditional statistical models, using routinely available clinical data for risk prediction in IBD.
Methods
Through a systematic review till January 1, 2021, we identified cohort studies that derived and/or validated machine learning models, based on routinely collected clinical data in patients with IBD, to predict the risk of harbouring or developing adverse clinical outcomes, and reported its predictive performance against a traditional statistical model for the same outcome. We appraised the risk of bias in these studies using the Prediction model Risk of Bias ASsessment [PROBAST] tool.
Results
We included 13 studies on machine learning-based prediction models in IBD, encompassing themes of predicting treatment response to biologics and thiopurines and predicting longitudinal disease activity and complications and outcomes in patients with acute severe ulcerative colitis. The most common machine learning models used were tree-based algorithms, which are classification approaches achieved through supervised learning. Machine learning models outperformed traditional statistical models in risk prediction. However, most models were at high risk of bias, and only one was externally validated.
Conclusions
Machine learning-based prediction models based on routinely collected data generally perform better than traditional statistical models in risk prediction in IBD, though frequently have high risk of bias. Future studies examining these approaches are warranted, with special focus on external validation and clinical applicability.
Keywords: Machine learning, prediction, big data, Crohn’s disease, ulcerative colitis
1. Introduction
Inflammatory bowel diseases [IBD] are chronic, debilitating, autoimmune diseases with rising global incidence and prevalence, and with a lifelong unpredictable relapsing-remitting course, leading to substantial morbidity, diminished quality of life, and disability. IBD is one of the top five most expensive gastrointestinal conditions, with annual costs exceeding $25 billion.1,2 Over the past two decades, with improved understanding of disease pathophysiology, diagnosis, new therapies, and evolving management strategies, we have made substantial advancements in management of IBD which have led to higher rates of remission and decrease in the risk of surgery.3 Personalized, treatment-management strategies are critically needed to improve clinical care.
Conventional approaches to risk stratification have relied on traditional statistical methods. However over the past decade, advanced computational methods like machine learning, which are able to use the wealth of unrelated data from diverse sources, have been used for risk prediction and prognostication in multiple conditions. Artificial intelligence approaches for image analysis, such as for endoscopy, radiology, or histology, have received most attention in the management of IBD, and it remains unclear whether machine learning-based prediction models using routinely available clinical data offer any advantages over conventional risk prediction approaches.4–10
Hence, we sought to systematically synthesise the performance of machine learning-based prediction models based on typically available clinical data vs conventional risk prediction models, for diagnosis and prognosis in patients with IBD. With rapid expansion in the number of publications applying machine learning techniques for clinical research, we also critically appraised risk of bias in these studies, using a standardised approach.
2. Methods
Our systematic review was conducted and reported based on the Preferred Reporting Items for Systematic Reviews and Meta-analysis [PRSMA] statement, and the process followed an a priori established protocol.
2.1. Selection criteria
We included cohort studies that derived or validated machine learning [ML] models based on routinely collected clinical data in patients with IBD to predict the risk of harbouring or developing adverse clinical outcomes, and reported predictive performance against a traditional statistical model for the same outcome.
We excluded studies that reported the derivation and/or validation of ML models based on variables not routinely collected or reported in usual practice, such as -omics predictors, as well as studies based on use of artificial intelligence for identifying endoscopic, imaging, and/or histological disease activity. Studies using the same cohort could be included in our systematic review if they developed and/or validated different models to prevent overlap.
2.2. Search strategy
Our search strategy was designed and conducted by an experienced medical librarian with input from the study’s investigators [SS and PSD], using a controlled vocabulary supplement with keywords, expanded terminology, and different algorithms for studies on the derivation and/or validation of ML models for clinical outcomes in patients with chronic conditions [see online Supplement, available as Supplementary data at ECCO-JCC online]. Our initial search was broad, evaluating these ML models in diverse chronic diseases including cancer, cardiovascular disease cirrhosis, diabetes, hypertension, arthritis, inflammatory bowel diseases, etc. The databases included Ovid MEDLINE[R] and Epub Ahead of Print, In-Process & Other Non-Indexed Citations, and Daily, Ovid EMBASE, Ovid Cochrane Central Register of Controlled Trials, Ovid Cochrane Database of Systematic Reviews, Scopus, and Web of Science, and dates of search ranged from January 1, 2000 to \august 27, 2018 [see online Supplement]. Subsequently, a focused literature search was updated in PubMed on January 1, 2021. We did not include conference proceedings due to limited information on details of the ML models.
2.3. Data abstraction and model assessment
We collected data on the following study-, patient-, and model-related characteristics, using a standardised case report form: 1] study characteristics including primary author[s], time period of study or year of publication, location of population studied, study design, number of centres, and source of primary data; 2] patient characteristics including IBD subtype, age, proportion of male patients, proportion of smokers, proportion with Crohn’s disease, location and behaviour of IBD, medications at time of enrolment [e.g,. steroids, immunomodulators, biologics]; and 3] model-related characteristics including primary and secondary outcomes, type of ML model [e.g., classification, regression], diagnostic or prognostic model, derivation or validation model, type of machine learning models, model performance [e.g., metrics such as area under the curve receiver operating characteristic [AUC ROC], sensitivity, specificity, and others]. Comparative accuracy of different models [machine learning vs conventional regression-based statistical model] was determined based on ROC curves.
2.4. Critical appraisal of individual studies
To assess the quality of included studies, we performed an in-depth assessment for the risk of bias [ROB] using Prediction model Risk of Bias ASsessment [PROBAST] tool.11,12 PROBAST was developed based on a four-stage approach for developing health research reporting guidelines and was designed to evaluate studies of clinical prediction model development and model validation. This consists of four domains [participants, predictors, outcome, and analysis] containing 20 signalling questions for ROB assessment and evaluation for concerns regarding applicability [Supplementary Table, available as Supplementary data at ECCO-JCC online]. Each domain focuses on an important aspect of model development and validation including: 1] participants: appropriate recruitment of patients and use of data sources; 2] predictors: appropriate definition and assessment of predictors for model development/validation; 3] outcome: appropriate definition and assessment, in relation to the predictors, to ensure standard definitions and appropriate timing between predictors and outcome variable[s]; and 4] analysis: analytical approach that appropriately includes a reasonable number of participants with the outcome, evaluation and assessment of categorial and continuous variables, evaluation of missing data, adjustment for complexities in the data, and accounting for model performances by evaluation and assessment for model overfitting, underfitting, and optimism. Signalling questions are answered as ‘yes’, ‘probably yes’, ‘probably no’, ‘no’, or ‘no information’. Risk of bias is evaluated based on a scale of low, high, or unclear, with all signalling questions inclusive of ‘yes’ indicating absence of bias, and any signalling questioned answered as ‘no’ or ‘probably no’ indicating potential bias. Concerns regarding applicability questions were developed to assess whether the population, predictors, or outcomes of a primary study differ from those detailed in the review question. Concerns regarding applicability are rated on a similar scale to ROB [low, high, or unclear] but without signalling questions.11,12 Whereas studies could be judged to be low risk of bias or low concern regarding applicability from signalling questions, the ultimate judgment was determined by the evaluators after evaluating the totality of information and the context of the review question.
Each study was evaluated for ROB and concerns regarding applicability using a word template developed for PROBAST. The Supplementary files, available as Supplementary data at ECCO-JCC online, contain the detailed ROB assessment and concerns regarding applicability for each study.
3. Results
No new data were generated or analysed in support of this research.
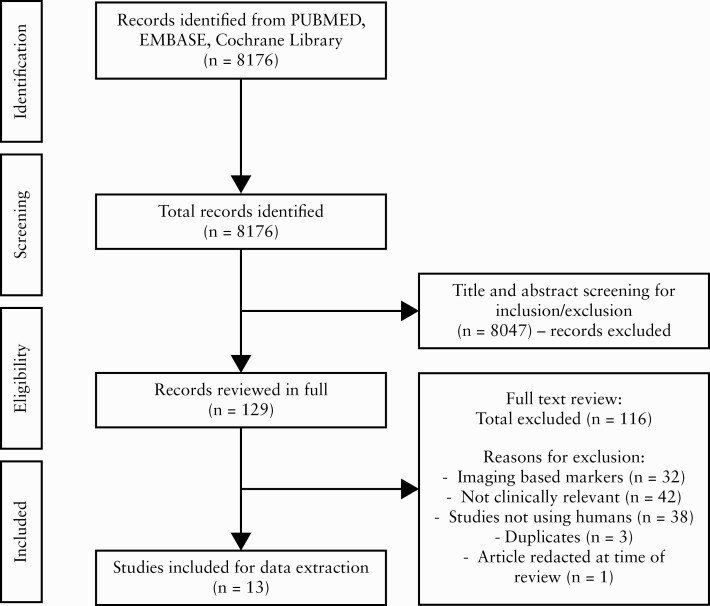
Our systematic search identified 8176 articles, of which 8047 were excluded based on title or abstract. One article was excluded as it was redacted at the time of full-text screening. Three duplicate studies were identified and removed. A total of 116 articles were excluded during full-text screening. A total of 13 full-length articles were included into the final analysis [Figure 1].7,13–26
Figure 1.
Study selection flowsheet.
Table 1 summarises key findings on model-related characteristics, outcomes, and study findings. Table 2 reports detailed study-, patient-, and treatment-related characteristics in the included studies. We evaluated the studies based on themes: 1] predicting treatment response to biologics; 2] predicting response to thiopurines; 3] longitudinal disease activity and complications (e.g., risk of surgery, changes in C-reactive protein [CRP] as a biochemical endpoint, presence of extra-intestinal manifestations [EIMs], and seasonal association with relapses); and 4] outcomes in patients with acute severe ulcerative colitis [ASUC].7,13–26 All studies except one19 validated their ML prediction model, though only one study performed external validation.14 Three studies included data from international, multicentre, randomised controlled trials and developed prognostic models to predict treatment outcomes in patients who received biologic therapies.23,24,26
Table 1.
Summary of findings from included studies.
| Study, year of publication | Aim, outcome, type of machine learning model, derivation/validation study, type of validation | Machine learning model[s] used; comparator | Performance [AUC, 95% CI] | Observations |
|---|---|---|---|---|
| Bottigliengo, 2019 | Aim: role of genetic factors in identifying extra-intestinal manifestations in CD patients Outcome: patients were classified according to the presence or absence of EIMs Type of ML model: classification Derivation or validation: both Validation: internal [bootstrap] |
Three models from the Bayesian machine learning family: Naïve Bayes [NB], Bayesian Network [BN] and Bayesian Additive Regression Trees [BART] Comparators: models used in the study by Giachino 2007 |
Model without genetic variables • LR: 0.72 • GAM: 0.72 • PPR: 0.82 • LDA: 0.70 • QDA: 0.67 • ANN: 0.79 • NB: 0.71 • BN: 0.50 • BART: 0.76 Model with genetic variables • LR: 0.77 • GAM: 0.77 • PPR: 0.94 • LDA: 0.77 • QDA: 0.88 • ANN: 0.87 • NB: 0.75 • BN: 0.67 • BART: 0.78 |
1. Bayesian machine learning family [NB, BN, and BART] were not better than methods used in Giachino 2007 in classification accuracy 2. Bayesian machine learning family did worst in classification when genetic information was added 3. Models with genetic variables did better than models without genetic variables |
| Dias, 2017 | Aim: predict disabling disease and/or re-operation using tree augmented Naïve Bayes Classifier [TAN] to build risk matrices Outcome: primary was disabling disease, while secondary re-operation Disabling disease was defined as one or more surgeries in the first 5 years after diagnosis, more than one surgery during follow-up, more than two hospitalisations, at least two courses of corticosteroids, need to switch immunosuppression and/or anti-TNF drugs, stenosis, penetrating disease, or anal disease Type of ML model: classification Derivation or validation: both Validation: external |
Tree Augmented Naïve Bayes [TAN] classifier; none | For disabling disease: • Derivation: 0.79 [0.75–0.83] • Validation: 0.78 [0.69–0.86] For re-operation: • Derivation: 0.86 [0.82–0.89] • Validation: 0.86 [0.80–0.93] |
1. The authors utilized TAN to identify key variables to include into risk matrices. Variables identified using TAN were then entered into logistic regression models to build risk matrices 2. For disabling disease, an association between perianal disease and gender was observed 3. For re-operation, an association between gender and upper tract was observed |
| Giachino, 2007 | Aim: role of genetic factors on identifying extra-intestinal manifestations in CD patients | Artificial Neural Network [ANN]: 1. Generalised additive model [GAM]; 2. projection pursuit regression [PPR]; 3. linear discriminant | Model without genetic variables: • LR: 0.72 • GAM: 0.72 • PPR: 0.82 • LDA: 0.70 • QDA: 0.67 • ANN: 0.79 |
The best fitting model was PPR. Model without genetic factors had AUC of 0.82 and increased to 0.94 by including genetic variables |
|
Outcome: patients were classified according to the presence or absence of EIMs Type of ML model: classification Derivation or validation: both Validation: internal [bootstrap] |
analysis [LDA]; 4. quadratic discriminant analysis [QDA]; 5. logistic regression [LR] |
Model with genetic variables: • LR: 0.727 • GAM: 0.77 • PPR: 0.94 • LDA: 0.77 • QDA: 0.88 • ANN: 0.87 |
||
| Peng, 2015 | Aim: assess for a seasonal pattern for the frequency of onset and relapse of IBD Outcome: clinical relapse Type of ML model: regression Derivation or validation: both Validation: internal |
Artificial Neural Network [ANN]; none | Did not use AUC, used Mean Square Error [MSE] and Mean Absolute Percentage Error [MAPE] Training: • Onset fq: MSE 1.14 × 10−3; MAPE 0.375 • Fq of relapse: MSE 3.82 × 10−3; MAPE 0.0487 Validation: • Onset fq: MSE 0.076; MAPE 0.06 • Fq of relapse: MSE 0.009; MAPE 0.171 |
The ANN model predicted a seasonal association in the frequency of onset and relapse in patients with CD but not in patients with UC |
| Reddy, 2018 | Aim: predict inflammation severity among patients with CD Outcome: inflammation severity [defined as 100% change in CRP value from baseline] Type of model: classification Derivation or validation: both Validation: internal |
Gradient Boosting Machine [GBM]; LR and Regularised Regression | Internal validation [mean AUC values across 10 times 10-fold]: • LR: 0.813 • Regularised Regression: 0.827 • GBM: 0.928 |
The authors compared three different models to predict CRP level/severity: GBM, LR, and regularised regression. The GBM model performed the best with a median AUC of 93.43 followed by regularised regression and logistic regression |
| Saito, 2012 | Aim: response to intravenous ciclosporin for severe UC. Outcome: 3-month colectomy Type of model: classification Derivation or validation: derivation Validation: none |
Decision Tree Analysis [chi square Automatic Interaction Detection, CHAID model]; logistics regression [LR] | Did not use AUC. Reported positive predictive value [PPV], negative predictive value [NPV] and accuracy CHAID • PPV: 0.72 • NPV: 1.0 • Accuracy: 0.90 LR • PPV: 0.83 • NPV: 0.91 • Accuracy: 0.88 |
The authors used the LR model to identify four predictors associated with response to intravenous ciclosporin. They then included these four predictors into the CHAID model to build a multivariable decision tree model to predict 3-month colectomy after receiving intravenous ciclosporin. This model had an accuracy of 90.4%, PPV of 72.2%, and NPV of 100.0% |
| Takayama, 2015 | Aim: need for operation after cytoapheresis [CAP] therapy Outcomes: need of operation after CAP therapy Type of ML model: classification |
Artificial Neural Network [ANN]; none | Did not use AUC. Authors reported sensitivity and specificity: Using all 13 variables: • Sens: 0.96 • Spec: 0.97 |
The authors observed that using ANN, they could predict the requirement of operation after CAP therapy. Sensitivity and specificity were 0.96 and 0.97. The authors did not report sensitivity and specificity in validation cohort. |
|
Derivation or validation: both Validation: internal |
With 2 factors removed [events of operation & history of admission]: • Sens: 0.87 • Spec: 0.75 With 4 factors removed [events of operation, history of admission, combination use of immunomodulators, and effect of CAP to the requirement of operations after CAP therapy]: • Sens: 0.60 • Spec: 0.71 |
Significant bias due to unclear definition/assessment/timing of predictors and outcome and also a mix of patients from outpatient and inpatient settings | ||
| Waljee, 2010 | Aim: predict outcomes in IBD patients on thiopurine therapy based on a machine learning approach Three outcome variables: 1] clinical response to thiopurines; 2] thiopurine non-response; 3] patients who receive thiopurines and shunt from 6-thioguanine nucleotide [6-TGN] to 6-methylmercaptopurine [6-MMP] metabolites Type of ML model: classification Derivation or validation: both Validation: internal. Calibration of the models was assessed with Hosmer‐Lemeshow tables |
Random Forest [RF] algorithms; Logistic Regression [LR] | Area under the receiver operating characteristic curve [AUROC] and 95% confidence intervals [CI] RF: • Clinical response: 0.856, 95% CI: 0.793–0.919 • Excluding patients on steroids: 0.807, 95% CI: 0.721–0.893 • Adding 6-TGN as independent variable to RF: 0.862, 95% CI: 0.800–0.924 • Non-adherence with RF: 0.813, 95% CI: 0.763–0.863 • Shunting with RF: 0.797, 95% CI: 0.743–0.850 LR: • Clinical response: 0.715, 95% CI: 0.597–0.833 • Non-adherence: 0.704, 95% CI: 0.637–0.771 • Shunting with LR: 0.613, 95% CI: 0.561–0.665 |
The authors demonstrated that RF models were superior to LR models when evaluating three outcome variables: clinical response, non-adherence, and preferential shunting to 6-MMP rather than metabolism to 6-TGN. The authors performed multiple sensitivity analyses with their ML models by adjusting certain features in their models: 1] adding 6-TGN metabolite value; and 2] excluding patients who are on steroids, since steroids can affect lab values [such as glucose and eosinophils] |
| Waljee, Sauder, et al., 2017 | Aim:predict objective evidence of remission in patients with IBD Three outcomes: objective evidence of remission [defined as absence of intestinal inflammation], non-adherence to thiopurines, and shunting to 6-metyhlmercaptopurine [6-MMP] |
Random Forest [RF] algorithms; none | The authors used out-of-bag [OOB] predictions on all observations and reported area under the receiver operating characteristic curve [AUROC]. Confusion matrix and Brier scores were also calculated. They also performed sensitivity analyses. | The authors built on their original cohort reported in Waljee 2010. They used objective remission instead of clinical response as their main outcome measurement. They also performed OOB evaluation to account for overfitting and performed sensitivity analysis based on the visit time |
|
Objective remission defined as: absence of C-reactive protein [CRP], erythrocyte sedimentation rate [ESR] or faecal calprotectin Type of ML model: classification Derivation or validation: both Validation: internal |
RF: • Objective remission: 0.79, 95% CI: 0.78–0.81 • Non-adherence with RF: 0.84, 95% CI: 0.79–0.89 • Shunting with RF: 0.78, 95% CI: 0.74–0.82 |
The authors excluded patients on biologics, anti-TNF therapy The authors demonstrated that an RF had good performance for all three outcomes based on Brier scores and AuROC The authors performed multiple sensitivity analyses with their ML models by adjusting certain features in their models: 1] adding 6-TGN metabolite value; and 2] excluding patients who are on steroids, since steroids can affect lab values [such as glucose and eosinophils]. They performed sensitivity analyses examining the distribution of the criteria that defined their objective remission outcome to assess the performance of their model The authors also conducted a time-based analysis to examine how well their model did over time in predicting steroid prescriptions, hospitalisations, and abdominal surgeries |
||
| Waljee, Liu et al., 2018 [CD] | Aim: predict corticosteroid-free biologic remission at Week 52 using baseline or Week 6 laboratory data in patients with CD who received vedolizumab [VDZ] Outcome: composite of no use of corticosteroid medications at Week 52 and reduction of C-reactive protein from >5 mg/L at baseline to ≤5 mg /L at Week 52 Type of ML model: classification Derivation or validation: both Validation: internal |
Random Forest [RF] models; none | receiver operating characteristic [AUROC]. The authors also included accuracy, sensitivity, specificity, and misclassification tables RF model with baseline data: • AuROC 0.65, 95% CI: 0.53–0.77 • Accuracy: 0.58 • Sensitivity: 0.64 • Specificity: 0.56 Simplified model using important variables identified from RF model through Week 6: Area under the curve • AuROC 0.75, 95% CI: 0. 64–0.86 • Accuracy: 0.72 • Sensitivity: 0.76 • Specificity: 0.71 Simplified Week 6 model using RF: |
The authors included all patients: those on every 4-week and every 8-week schedule [those on every 4-week most likely had more severe disease]. Potential selection bias in that ~40% of the cohort was not used for building the prediction models and 228 patients of 472 patients were already corticosteroid-free at baseline The authors demonstrated better performance with RF model that included longitudinal data [data through Week 6] to predict their composite outcome compared with a model that only included baseline variables |
|
• AuROC 0.75, 95% CI: 0.70–0.81 • Accuracy: 0.69 • Sensitivity: 0.73 • Specificity: 0.69 |
The authors also created a simplified model using predictors that were identified by a variable importance plot at Week 6 and identified that a single quotient [haemoglobin * albumin * vedolizumab level] / [C-reactive protein * weight in kg] value allowed for accurate prediction of their composite outcome |
|||
| Waljee, Liu et al., 2018 [UC] | Aim: predict corticosteroid-free endoscopic remission at Week 52 using baseline or Week 6 laboratory data in patients with UC who received vedolizumab [VDZ] Outcome: composite outcome of no use of corticosteroid medications at Week 52 and a Mayo Sigmoidoscopy Score of 0 or 1 at Week 52 Type of ML model: classification Derivation or validation: both Validation: internal |
Random Forest [RF] models; none | Area under the curve receiver operating characteristic [AUROC], sensitivity, and specificity Baseline RF model: • 0.62 [95% CI: 0.53–0.72] • Sensitivity: 0.63 • Specificity: 0.62 Using faecal calprotectin [FCP] before first dose of VDZ • 0.58 [95% CI: 0.52–0.63] • Sensitivity: 0.57 • Specificity: 0.57 Week 6 model using RF • 0.73 [95% CI: 0.65–0.82] • Sensitivity: 0.72 • Specificity 0.68: Simplified model using Week 6 FCP/VDZ level ratio [best cut-off 12.35]: • 0.71 [95% CI: 0.67–0.76] • Sensitivity: 0.68 • Specificity: 0.66 Simplified with Week 6 FCP value alone [best cut-off 233.67]: • 0.71 [95% CI: 0.66–0.76] • Sensitivity: 0.58 • Specificity: 0.73 |
Potential selection bias in that ~21% of the cohort was not used for building the prediction models and 224 patients of 491 patients were already corticosteroid-free at baseline The best model was a Week 6 model with AUROC of 0.73 [95% CI: 0.65–0.82]; this model incorporated the change in faecal calprotectin over time, VDZ levels, and the slope of the VDZ concentration and the lab results at Week 6 |
| Waljee, Lipson et al., 2017 | Aim: predict composite outcome of hospitalisation and/or corticosteroid use in patients with IBD within 6 months Outcome: composite of hospitalisation and/or corticosteroid use within 6 months |
Random Forest [RF]; Logistic Regression using baseline data [LR] | Area under the curve receiver operating characteristic [AUROC] RF without previous events [defined as prior hospitalisation and/or corticosteroid use]: • Overall: 0.85 [95% CI, 0.84–0.85] • CD: 0.84 [95% CI,0.83–0.85] • UC: 0.85 [95% CI, 0.84–0.86] |
The authors used two RF models with different data and demonstrated better performance with both algorithms compared with a logistic regression using only baseline data |
|
Type of ML model: classification Derivation or validation: both Validation: internal |
• IC: 0.82 [95% CI, 0.81–0.83] RF including previous events: • Overall: 0.87 [95% CI, 0.87- 0.88] • CD: 0.87 [95% CI, 0.87–0.88]: • UC: 0.88 [0.87–0.88] IC: 0.85 [0.84–0.86] LR [with baseline variables]: • Overall: 0.68 [95% CI, 0.67–0.68] LR [with longitudinal data]: • Overall: 0.79 [95% CI, 0.79- 0.80] |
The authors performed many sensitivity analyses and demonstrated excellent performance of their ML algorithms: predicting outpatient corticosteroid use only, predicting 12-month outcomes, UC patients only, CD patients only, and indeterminate colitis patients only When the authors used a longitudinal LR, this model performed better than the baseline LR model: 1] with immunosuppressive medication predictor, AUROC 0.79 [95% 0.79–0.80]; 2] model without immunosuppressive medication, AuROC 0.79 [95% CI 0.79–0.80] |
||
| Waljee, 2019 | Aim: to predict biological remission beyond Week 42 of ustekinumab [UST] treatment in patients with CD Outcome: biological remission [defined as a C-reactive protein level <5 mg/dL] Type of ML model: classification Derivation or validation: both Validation: internal |
Random Forest [RF] models]; none | Area under the curve receiver operating characteristic [AUROC] and confusion matrices. Brier scores reported for baseline and Week 8 models RF baseline model: • 0.59, [95% CI, 0.46–0.72] • Sensitivity: 0.63 • Specificity: 0.64 RF Week 8 model: • 0.78, [95% CI, 0.69–0.87] • Sensitivity: 0.79 • Specificity: 0.67 Simplified model with Week 6 albumin/CRP ratio: • 0.76 [95% CI, 0.71–0.82] • Sensitivity: 0.77 • Specificity: 0.68 |
The authors performed two RF models, one at baseline and one at Week 8, to predict biological remission using CRP as a surrogate marker. Their Week 8 model performed better than their baseline model. Using a simplified Week 6 albumin-to-CRP ratio, their model had an AUROC of 0.77 [95% CI, 0.71–0.82] |
CD, Crohn’s disease; UC, ulcerative colitis; EIM, extra-intestinal manifestations; ML, machine learning; TNF, tumour necrosis factor.
Table 2.
Characteristics of patients in included studies.
| Author, year | Study, location, patient population, centres, source | Groups, study participants | Age, % males, % smokers, % with CD | Disease type | Medications at time of index admission | |||
|---|---|---|---|---|---|---|---|---|
| Crohn’s disease: Location [%] Ileal, colonic, ileocolonic, upper GI tract Disease phenotype [%] inflammatory, obstructive, penetrating, perianal disease |
Ulcerative colitis Location [%] Proctitis, left-sided, extensive |
Steroids | Immuno-modulators [%] | Biologics [%] | ||||
| Bottigliengo, 2019 | Retrospective cohort, Italy, adults, multicentre *Same cohort as Giachino, 2007 |
152 patients with CD and extra-intestinal manifestations [EIMs] | Overall: <40 [68%], 45, 39%, 100% | Overall Location 18%, 15%, 32%, 5% Behaviour 33%, 23%, 15% |
NA | NA | NA | NA |
| Dias, 2017 | Retrospective cohort, Portugal, adults, multicentre | Patients with CD who underwent medical therapy or surgeries Derivation: 489 [64% with disabling disease and 18% with re-operation]; validation: 129 [similar proportion as derivation cohort] |
Overall: <40 [79%], 46%, NA, 100% | Overall Location 47%, 10%, 43%, 12% Behaviour 32%, 36%, 32%, 26% |
NA | 67% | NA | 37% |
| Giachino, 2007 | Retrospective cohort, Italy, adults, multicentre | Patients with CD and extra-intestinal manifestations [EIMs]; 152 patients | Overall: <40 [68%], 45%, 39%, 100% | Overall Location 18%, 15%, 32%, 5% Behaviour 33%, 23%, 15% |
NA | NA | NA | NA |
| Peng, 2015 | Retrospective cohort, China, single centre; electronic health records | 901 patients were diagnosed as UC or CD at a single centre | Overall: Mean age 39.8 [IQR 26–51], 49%, NA, NA | Overall Location NA, NA, NA, NA Behaviour NA, NA, NA, NA |
NA | NA | NA | NA |
| Reddy, 2018 | Retrospective, USA, multicentre; electronic health records [Cerner] | 82 CD patients based on administrative codes using Cerner | Overall: NA, NA, NA, 100% | Overall Location NA, NA, NA, NA Behaviour NA, NA, NA, NA |
NA | NA | NA | NA |
| Saito, 2012 | Retrospective, Japan, single-centre; electronic health records | 52 patients with severe ulcerative colitis flare refractory to corticosteroids at a single centre | Overall: 40.15 ± 15.6, 37%, NA, 0% | Overall Location NA, NA, NA, NA Behaviour NA, NA, NA, NA |
Overall Location 0%, 31%, 67%, [1 patient had right-sided inflammation] |
100% | NA | NA |
| Takayama, 2015 | Retrospective, Japan, single-centre; electronic health records | 90 patients with UC who underwent treatment with cytoapheresis. Training: 54; testing 36 |
Overall: 38.5 ± 15.8. 58%, NA, 0% |
Overall Location NA, NA, NA, NA Behaviour NA, NA, NA, NA |
Overall: Location 6%, 29%, 61%, [4 patients described as other] |
NA | 16 | NA |
| Waljee, 2010 | Retrospective, USA, single-centre; electronic medical records | 395 patients, 774 cases. Mix of UC and CD patients who all received thiopurine. Patients on biologic therapy were excluded Clinical response sample: 395 cases Adherence sample: 774 cases Shunting sample: 716 cases |
Overall: Median 23 [16–40], 51%, NA, 63% |
Overall Location Could not calculate due to presentation of data [26 had small bowel CD, 24 had large bowel CD, and 168 had both small and large bowel CD] Behaviour NA |
Overall: Location Only 93 patients had data reported [2 had proctitis, 15 had left-sided, and 76 had pancolitis] |
29% | 100% | 0% |
| Waljee, Sauder, et al., 2017 | Retrospective, USA, single-centre; electronic medical records *Built on the initial cohort reported in Waljee 2010 |
1080 patients, 3263 cases. Mix of UC and CD patients who all received thiopurine. Patients on biologic therapy were excluded Objective remission: 3263 cases Adherence/shunting samples: 509 cases for both samples *The authors supplemented their non-adherence study sample with 608 IBD patients with measurements of thiopurine metabolites from a previous ThioMon dataset [ThioMon v1] for a total of 1117 cases and 603 patients |
Overall: Median 29.3 [17.3–45.8], 52%, NA, 57% |
Overall Location Could not calculate due to presentation of data [155 had small bowel CD, 100 had large bowel CD, 352 had both small and large bowel CD, 9 had unknown location] |
Overall: Location 3%, 32%, 61%, [4% had unknown location] |
35% | 100% [82% azathioprine; 18% 6-mercaptopurine] | 0% |
| Waljee, Liu, et al., 2018 [CD] | Retrospective, international, multicentre, used data from a randomised controlled trial [GEMINI II] | Original cohort included 813 patients. A total of 472 were included after applying inclusion/exclusion criteria | Overall [final model cohort]: 34.2 ± 11.4, 49%, NA, 100% |
Overall Location 17%, 28%, 54%, NA Behaviour NA, NA, NA, NA |
Overall: Location NA, NA, NA |
51% | 22% | 100% |
| Waljee, Liu, et al., 2018 [UC] | Retrospective, international, multicentre, used data from a randomised controlled trial [GEMINI I] | Original cohort included 895 patients. A total of 491 were included in the final analysis | Overall [final model cohort]: 40.2 ± 13.4, 57%, NA, NA |
Overall Location NA, NA, NA, NA Behaviour NA, NA, NA, NA |
Overall: Location 14%, 38%, 48% |
55% | 34% | 100% |
| Waljee, Lipson, et al., 2017 | Retrospective, USA, multicentre, claims-based using national data from Veterans Health Administration database | 20 369 patients from the Veterans Health Administration database | Overall: 59.1 ± 14.9, 93%, NA, 35% |
Overall Location NA, NA, NA, NA Behaviour NA, NA, NA, NA |
Overall: Location NA, NA, NA |
NA | 18% *The authors defined immunosuppressive medications as thiopurine, methotrexate, anti-TNF, or combination therapy |
18% *The authors defined immunosuppressive medications as thiopurine, methotrexate, anti-TNF, or combination therapy |
| Waljee, 2019 | Retrospective, international, multicentre, used data from three randomised controlled trials [UNITI-1, UNITI-2, and IM-UNITI] | Original cohort included 1409 patients, 668 were either randomised to placebo or lost to follow-up by Week 8 so were excluded. 401 included in the Week 8 model and 371 were included in baseline model | Overall: 37.7 ± 12.8, 42%, NA, 100% |
Overall Location 20%, 17%, 62%, 18% Behaviour NA, NA, NA, NA |
Overall: Location NA, NA, NA |
56% | 42% | 100% |
CD, Crohn’s patients; UC, ulcerative colitis patients; IBD, inflammatory bowel disease; GI, gastro-intestinal; NA, not available; IQR, interquartile range.
*Waljee and colleagues published two studies in 2017/2018 using the same dataset but different cohort.
3.1. Predicting treatment response to biologics
Three studies developed prediction models using data collected from international, multicentre, randomised controlled trials in patients who received biologic therapy with either vedolizumab or ustekinumab.23,24,26 In one study, Waljee and colleagues developed random forest [RF] models to predict corticosteroid-free biological remission [composite of no corticosteroid use and CRP reduction from 5 mg/L at baseline to ≤5 mg/L] at Week 52, using baseline or Week 6 laboratory data in CD patients who were treated with vedolizumab [GEMINI II].23 The authors also attempted to create a simplified model [haemoglobin * albumin * vedolizumab level] / [C-reactive protein * weight in kg] using predictors that were identified by a variable importance plot from the ML algorithm to develop an equation to accurately predict their composite outcome. Of the three models [baseline model without Week 6 data, Week 6 model, and simplified model using Week 6 data], the authors demonstrated that the Week 6 model (AUC 0.75; 95% confidence interval [CI] 0.64–0.86) and the simplified Week 6 model [AUC 0.75; 95% CI 0.70–0.81] had the best performance in predicting the combined outcome. Overall, the study was judged to be low ROB and low concern for applicability. In a similar study, Waljee and colleagues developed RF models to predict corticosteroid-free endoscopic remission [composite of no corticosteroid use and a Mayo sigmoidoscopy score of 0 or 1] at Week 52, using baseline or Week 6 laboratory data in UC patients who were treated with vedolizumab [GEMINI I].24 The authors also developed a simplified Week 6 model with a faecal calprotectin cut-off of <234 µg/g to predict the composite outcome. The authors demonstrated that the best model was the Week 6 model, which was numerically more accurate and with an AUC of 0.73 [95% CI: 0.65–0.82] compared with the baseline model and simplified Week 6 model with faecal calprotectin cut-off. The study was judged to be low ROB and low concern for applicability. In a recent study using clinical trial data of patients with CD treated with ustekinumab [UNITI-1, UNITI-2, and IM-UNITI], Waljee and colleagues developed two RF models [one at baseline and one at Week 8] to predict biological remission [defined as a C-reactive protein level <5 mg/dL] in CD patients who were treated with ustekinumab. The authors also developed a simplified Week 6 albumin-to-CRP ratio to predict their primary outcome. The Week 8 model had an AUC of 0.78 [95% CI 0.69–0.87] and the simplified Week 6 model had an AUC of 0.76 [95% CI 0.71–0.82] in predicting biological remission. Overall, the study was judged to be at low ROB and low concern for applicability.
3.2. Predicting treatment response to thiopurines
In three different studies, Waljee and colleagues developed multiple random forest [RF] models to predict treatment-related outcomes in patients with IBD, using real-world cohorts from a single-centre electronic health record or the national Veterans Health Administration database warehouse.21,22,25 In the first study, Waljee and colleagues developed and compared three RF models with a traditional logistic regression [LR] model in identifying three different outcomes in patients with IBD treated with thiopurines: 1] clinical response to thiopurines; 2] thiopurine non-adherence; and 3] patients who were more likely to shunt from 6-thioguanine nucleotide [6-TGN] to 6-methylmercaptopurine [6-MMP] metabolites.21 The authors demonstrated that the RF models had excellent performance in predicting clinical response [AUC 0.86; 95% CI 0.79–0.92], non-adherence [AUC 0.81; 95% CI 0.76–0.86], and shunting [AUC 0.80; 95% CI 0.74–0.85] and performed better than LR models for each outcome. The study was judged to have low concern for applicability but high ROB, since the authors performed a random-split of their data for validation instead of relying on other techniques [e.g., k-fold cross-validation, internal bootstrapping, out-of-bag, etc.] to account for overfitting. In an updated study using the same cohort, Waljee and colleagues evaluated RF models in predicting three similar outcomes related to thiopurine therapy but focused on objective remission [defined as absence of intestinal inflammation], which has been shown to be superior to clinical activity indices in predicting treatment-related outcomes, instead of clinical response.25 The authors demonstrated excellent performance of their ML models in predicting objective remission [AUC 0.79; 95% CI 0.78–0.81], non-adherence to thiopurine [AUC 0.84; 95% CI 0.79–0.89], and shunting of thiopurine metabolites [AUC 0.78; 95% CI 0.74–0.82]. The study was rated to have low ROB and low concern for applicability, since the authors relied on out-of-bag predictions to account for overfitting and objective assessment of disease activity. In a recent study evaluating the effect of treatment (immunomodulators and/or anti-tumour necrosis factor [TNF] therapy) in patients with IBD from the Veterans Health Administration database, Waljee and colleagues developed two RF models [one without longitudinal data and one with longitudinal data] to predict a composite outcome of IBD-related hospitalisation and outpatient corticosteroid use. The authors demonstrated that incorporating longitudinal data with previous IBD-related flares improved the performance of their RF model [AUC 0.87; 95% CI 0.87–0.88] compared with relying on longitudinal data alone [AUC 0.85; 95% CI 0.84–0.85] in predicting the composite outcome. Overall, the study was judged to be low ROB and low concern for applicability.
3.3. Longitudinal disease activity and complications
Four studies developed diagnostic prediction models to identify complications of IBD or severity of inflammation.7,13,15,18 Three studies developed prediction models in patients with CD only.13,15,18 None of the studies sought to detect the presence of IBD or differentiate UC from CD, in patients with suggestive symptomatology and biochemical abnormalities.
3.3.1. Predicting clinical phenotypes
Giachino and colleagues compared six different ML and regression models to identify patients with CD with extra-intestinal manifestation. They used an artificial neural network, generalised additive model, projection pursuit regression, linear discriminant analysis, quadratic discriminant analysis, and traditional logistic regression to identify the best model. Projection pursuit regression model had the best discriminatory performance, with an AUC of 0.82 without genetic factors and AUC of 0.94 with genetic factors [Table 1].15 Risk of bias and concern for applicability were determined to be high, since EIMs used non-standardised definitions and dates of enrolment of participants were not clearly stated [Table 3]. In an updated analysis of the same cohort, Bottigliengo and colleagues build newer models from the Bayesian machine learning family [Naïve Bayes, Bayesian Network, and Bayesian Additive Regression Trees] to evaluate whether these models would perform better than the original models.13 Newer Bayesian models performed worse than previous models in classifying the presence of EIMs [see Table 1].
Table 3.
Risk of bias in included studies based on PROBAST tool
| Study | Risk of bias | Applicability | Overall | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Participants | Predictors | Outcome | Analysis | Participants | Predictors | Outcome | Risk of bias | Applicability | |
| Bottigliengo, 2019 | - | + | - | + | - | + | - | - | - |
| Dias, 2017 | + | + | + | + | + | + | + | + | + |
| Giachino, 2007 | - | + | - | + | - | + | - | - | - |
| Peng, 2015 | ? | - | - | + | - | - | - | - | |
| Reddy, 2018 | - | - | - | - | - | - | - | - | - |
| Saito, 2012 | - | + | + | - | + | + | + | - | + |
| Takayama, 2015 | - | - | ? | - | - | - | ? | - | - |
| Waljee, 2010 | ? | + | - | - | ? | + | - | - | - |
| Waljee, Sauder, et al., 2017 | ? | + | + | + | ? | + | + | + | + |
| Waljee, Liu et al. 2018 [CD] | ? | + | + | + | ? | + | + | + | + |
| Waljee, Liu et al. 2018 [UC] | ? | + | + | + | ? | + | + | + | + |
| Waljee, Lipson, et al., 2017 | + | + | + | + | + | + | + | + | + |
| Waljee, 2019 | + | + | + | + | + | + | + | + | + |
Waljee and colleagues published two studies in 2017/2018 using the same dataset but different cohort.
PROBAST, Prediction model of Risk Of Bias ASsessment Tool; CD, Crohn’s patient; UC, ulcerative colitis patient.
* + indicates low risk of bias or low concern regarding applicability; - indicates high risk of bias or high concern regarding applicability; and? indicates unclear risk of bias or risk of concern regarding applicability.
Dias and colleagues used a tree-augmented Naïve Bayes classifier to identify predictive factors of disabling disease [defined as one or more surgeries in the 5 years after diagnosis, more than one surgery during follow-up, more than two hospitalisations, at least two courses of corticosteroids, need to switch immunosuppression and/or anti-TNF drugs, stenosis, penetrating disease, or anal disease] to include into treatment-stratified risk matrices to provide differential probabilities of patients with CD for developing disabling disease.14 A secondary outcome was to identify patients at risk for re-operation. The models had excellent AUC in the external validation cohort for risk of disabling disease [AUC 79%; 95% CI 75–83%] and re-operation [AUC 86%; 95% CI 90–93%]. Overall, the study was rated as low ROB and low concern for applicability.
3.3.2. Identify disease activity
Reddy and colleagues compared traditional regression models [logistic regression and regularised regression] to a gradient boosting machine [GBM] learning model, which is an ensemble approach to making predictions, to identify CD patients with 100% or more increase in the level of C-reactive protein [CRP] from baseline to a subsequent visit.18 They leverage a large electronic medical record [EMR], Cerner EMR, to build a decision support tool using ML approaches to allow for real-time diagnosis of patients with inflammation. The GBM model was the best performing model, with a median AUC of 0.93 across the 10 folds, at identifying patients with at least a doubling of their CRP value from baseline to a subsequent visit. This study was also deemed to have high risk of bias and concern for applicability due to missing data—there were initially 3335 unique patients with CD but, after preprocessing laboratory, encounter, procedure, medication, and clinical data, only 82 patients were available for analysis and model development.
In a novel approach to identify potential environmental factors in predicting clinical relapse in patients with IBD, Peng and colleagues developed an artificial neural network [ANN] to predict a seasonal association in the frequency of onset and relapse in patients with IBD, by collecting monthly temperature, air pressure, and humidity data from 2003 to 2011.17 Using an ANN model, the authors found a significant seasonal variation in the onset and relapse of CD, with a peak in July and August, but no significant seasonal variation for patients with UC. The study was rated as high ROB and high concern for applicability from: 1] unclear number of predictors; 2] unclear definition of clinical relapse; and 3] not all temperature, air pressure, and humidity values reported for each month in their study period.
3.4. Acute severe ulcerative colitis
In a study that evaluated hospitalised patients with IBD, Saito and colleagues attempted to use a combination of logistic regression and decision tree analysis [chi square automatic interaction detection, CHAID] to identify predictors associated with response [defined as being colectomy-free] in patients with acute severe UC who were steroid-refractory and received intravenous ciclosporin as second-line therapy.19 The authors found that a decision tree using a combination of age at hospitalisation, platelet count on the first day of admission, Lichtiger score on the third day, and total protein on the third day minus total protein on the first day, predicted colectomy with an accuracy of 90.4%, positive predictive value of 72.2% and negative predictive value of 100.0%. Overall, the study was rated at high ROB [due to small sample size and lack of model validation] and low concern for applicability. In a similar study evaluating the outcome of colectomy in patients with moderate-severe UC, Takayama and colleagues evaluated the performance of ANN in predicting patients with UC who would respond to cytoapheresis, a standard of care treatment in Japan.20 The authors reported high sensitivity and specificity [96% and 0.97, respectively] in predicting response to cytoapheresis. Overall, the study was rated at high ROB and high concern for applicability due to the: small sample size; unclear definition, assessment, and timing of the predictors and outcome; and mix of patients from inpatient and outpatient settings.
4. Discussion
In this systematic review of 13 studies of ML learning models in patients with IBD, we noted the broad application of different ML algorithms across multiple health care settings and from various data sources to accurately predict disease- and treatment-related outcomes. With the rapid development of large volumes of data from variable sources, computational-based approaches such as artificial intelligence and ML learning allow clinicians and researchers a unique opportunity to achieve the goal of personalised medicine, compared with traditional data mining and statistical approaches.4,6 In studies where ML models were compared with traditional statistical approaches, ML models were shown to be more effective at disease monitoring and predicting disease complications, treatment response to immunosuppression and biologics, corticosteroid use, and hospitalisation.15,16,18,21,22 ML models are attractive in the field of IBD, given the exponential growth of big data in genomics, transcriptomics, proteomics, imaging, therapeutics, and electronic health information, where technological advances are needed to analyse and interpret large amounts of complex and inter-related data. The most common ML models used in this systematic review were tree-based ML algorithms, which are classification approaches achieved through supervised learning and are easy to interpret compared with other ML techniques.5,6 Decision trees, compared with other classifiers such as neural networks and support vector machines, are frequently used because they can incorporate large amounts of complex data and condense the findings into easily interpretable results, by combining simple questions about the data in an understandable and intuitive approach that mimics human decision making.27
Despite the rapid development and introduction of new therapeutics to the market, the optimal positioning of therapies is elusive, as current prediction models have modest performance in accurately risk-stratifying patients who may benefit from treatment.5,6,28 Studies by Waljee and colleagues have demonstrated that ML models can be helpful in monitoring response to thiopurine therapy based on a combination of clinical and laboratory data, which is a more cost-effective and less labour-intensive approach compared with traditional laboratory monitoring.21,25 The authors have also demonstrated the effectiveness of ML algorithms in predicting treatment response in patients who receive newer and more expensive agents, such as vedolizumab and ustekinumab.23,24,26
Although a major strength of ML models is their ability to identify non-linear relationships in large and complex datasets, there has been limited adoption of these models in clinical practice due to the concern of lack of interpretability of ML models. Waljee and colleagues have attempted to address this concern by developing simplified models of their ML algorithms and demonstrated comparable performances between the simplified and full ML models. The authors developed a Week 6 quotient [haemoglobin * albumin * vedolizumab level] / [C-reactive protein * weight in kg] to predict corticosteroid-free biological remission at Week 52 in patients with CD treated with vedolizumab [AUC 0.75; 95% CI 0.70–0.81] and a Week 6 albumin-to-CRP ratio in patients with CD treated with ustekinumab [AUC 0.76; 95% CI 0.71–0.82] all with excellent AUCs.23,26
Whereas this systematic review highlights the potential benefits of applying ML techniques in building prediction models for clinical outcomes in patients with IBD, there are important limitations that need to be highlighted. All studies attempted to account for potential model overfitting by performing internal validation of their models, but only one study attempted to externally validate their models by relying on an external cohort of patients. Second, ML models are only as good as their input and outcome variables, and there are potential biases when the input and outcome variables are not clearly measured and validated. In some of the studies, outcome measurements could have introduced significant bias in model performance, especially when the primary outcomes were endoscopic measures and/or biochemical measures. Not all studies that evaluated endoscopic outcomes relied on blinded centralised endoscopy reading, which has been shown to reduce site-related scoring biases.29,30 Additionally, there is potential for bias in studies that relied on CRP values to differentiate remission from active disease, since previous studies have noted that CRP can be falsely low despite evidence of active mucosal inflammation.31 Although PROBAST is an excellent method for uniformly addressing potential bias, the tool is not without limitation in terms of bias assessment and concern regarding applicability. There are signalling questions available for bias assessment, but the tool allows for evaluators to make the final and subjective decision on risk of bias assessment outside the signalling questions. Furthermore, concern regarding applicability does not have signalling questions, which also introduces subjective assessments by the evaluators. Future systematic reviews of prediction models using the PROBAST tool should provide direct reviewer commentary to expand on the reasons for bias judgement, and a summary statement regarding concern for applicability, to provide detailed explanations of the reviewer’s assessments to the reader.
Big data and artificial intelligence offer a unique opportunity to enable integration of big data and discovery of novel clinical knowledge to guide both IBD research and clinical practice. With the continued growth of big data and need for computational methods to handle large and complex datasets, artificial intelligence and ML will ultimately guide the field of IBD closer to the goal of personalised medicine.
Supplementary Material
Contributor Information
Nghia H Nguyen, Division of Gastroenterology, Department of Medicine, University of California San Diego, La Jolla, CA, USA.
Dominic Picetti, Division of Gastroenterology, Department of Medicine, University of California San Diego, La Jolla, CA, USA.
Parambir S Dulai, Division of Gastroenterology, Department of Medicine, University of California San Diego, La Jolla, CA, USA.
Vipul Jairath, Department of Epidemiology and Biostatistics, Western University, London, ON, Canada; Division of Gastroenterology, Western University, London, ON, Canada.
William J Sandborn, Division of Gastroenterology, Department of Medicine, University of California San Diego, La Jolla, CA, USA.
Lucila Ohno-Machado, Division of Biomedical Informatics, University of California San Diego, La Jolla, CA, USA.
Peter L Chen, HyperPlanar, San Diego, CA, USA.
Siddharth Singh, Division of Gastroenterology, Department of Medicine, University of California San Diego, La Jolla, CA, USA; Division of Biomedical Informatics, University of California San Diego, La Jolla, CA, USA.
Funding
No direct funding was used for this project. NHN is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award number T32DK007202 and T15LM01127WJS is supported in part by NIDDK-funded San Diego Digestive Diseases Research Centre [P30 DK120515]. SS is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award number K23DK117058.
Conflict of Interest
PSD: research support and consulting for Takeda, Abbvie, Janssen, Pfizer. WJS: research grants from Atlantic Healthcare, Amgen, Genentech, Gilead Sciences, Abbvie, Janssen, Takeda, Lilly, Celgene/Receptos,Pfizer, Prometheus Laboratories; consulting fees from Abbvie, Allergan, Amgen, Arena Pharmaceuticals, Avexegen Therapeutics, BeiGene, Boehringer Ingelheim, Celgene, Celltrion, Conatus, Cosmo, Escalier Biosciences, Ferring, Forbion, Genentech, Gilead Sciences, Gossamer Bio, Incyte, Janssen, Kyowa Kirin Pharmaceutical Research, Landos Biopharma, Lilly, Oppilan Pharma, Otsuka, Pfizer, Precision IBD, Progenity, Prometheus Laboratories, Reistone, Ritter Pharmaceuticals, Robarts Clinical Trials [owned by Health Academic Research Trust, HART], Series Therapeutics, Shire, Sienna Biopharmaceuticals, Sigmoid Biotechnologies, Sterna Biologicals, Sublimity Therapeutics, Takeda, Theravance Biopharma, Tigenix, Tillotts Pharma, UCB Pharma, Ventyx Biosciences, Vimalan Biosciences, Vivelix Pharmaceuticals; and stock or stock options from BeiGene, Escalier Biosciences, Gossamer Bio, Oppilan Pharma, Precision IBD, Progenity, Ritter Pharmaceuticals, Ventyx Biosciences, Vimalan Biosciences. Spouse: Opthotech, consultant; stock options: Progenity—consultant, stock; Oppilan Pharma—employee, stock options; Escalier Biosciences—employee, stock options; Precision IBD—employee, stock options; Ventyx Biosciences—employee, stock options; Vimalan Biosciences—employee, stock options. PLC: founder, HyperPlanar. SS: research grants from AbbVie and Janssen, personal fees from Pfizer for ad-hoc grant review.
Author Contributions
Study concept and design: NHN, DP, PSD, SS. Acquisition of data: NHN, DP. Analysis and interpretation of data: NHN, DP, SS. Drafting of the manuscript: NHN, DP, SS. Critical revision of the manuscript for important intellectual content: PSD, WJS, LOM, PLC. Approval of the final manuscript: NHN, DP, PSD, WJS, LOM, PLC, SS. Guarantor of article: SS.
References
- 1. Dieleman JL, Squires E, Bui AL, et al. Factors associated with increases in US health care spending, 1996-2013. JAMA 2017;318:1668–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dieleman JL, Cao J, Chapin A, et al. US health care spending by payer and health condition, 1996-2016. JAMA 2020;323:863–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsai L, Ma C, Dulai PS, et al. Contemporary risk of surgery in patients with ulcerative colitis and Crohn’s disease: a meta-analysis of population-based cohorts. Clin Gastroenterol Hepatol 2020:S1542-3565(20)31497-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kohli A, Holzwanger EA, Levy AN. Emerging use of artificial intelligence in inflammatory bowel disease. World J Gastroenterol 2020;26:6923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olivera P, Danese S, Jay N, Natoli G, Peyrin-Biroulet L. Big data in IBD: a look into the future. Nat Rev Gastroenterol Hepatol 2019;16:312–21. [DOI] [PubMed] [Google Scholar]
- 6. Seyed Tabib NS, Madgwick M, Sudhakar P, Verstockt B, Korcsmaros T, Vermeire S. Big data in IBD: big progress for clinical practice. Gut 2020;69:1520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stidham RW, Liu W, Bishu S, et al. Performance of a deep learning model vs human reviewers in grading endoscopic disease severity of patients with ulcerative colitis. JAMA Netw Open 2019;2:e193963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takenaka K, Ohtsuka K, Fujii T, et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology 2020;158:2150–7. [DOI] [PubMed] [Google Scholar]
- 9. Klang E, Barash Y, Margalit RY, et al. Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest Endosc 2020;91:606–13.e2. [DOI] [PubMed] [Google Scholar]
- 10. Ellmann S, Langer V, Britzen-Laurent N, et al. Application of machine learning algorithms for multiparametric MRI-based evaluation of murine colitis. PLoS One 2018;13:e0206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moons KGM, Wolff RF, Riley RD, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med 2019;170:W1–33. [DOI] [PubMed] [Google Scholar]
- 12. Wolff RF, Moons KGM, Riley RD, et al. ; PROBAST Group† . PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med 2019;170:51–8. [DOI] [PubMed] [Google Scholar]
- 13. Bottigliengo D, Berchialla P, Lanera C, et al. The role of genetic factors in characterizing extra-intestinal manifestations in Crohn’s disease patients: are bayesian machine learning methods improving outcome predictions? J Clin Med 2019;8:865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dias CC, Rodrigues PP, Coelho R, et al. ; on behalf of GEDII. Development and validation of risk matrices for Crohn’s disease outcomes in patients who underwent early therapeutic interventions. J Crohns Colitis 2017;11:445–53. [DOI] [PubMed] [Google Scholar]
- 15. Giachino DF, Regazzoni S, Bardessono M, De Marchi M, Gregori D; Piedmont Study Group on the Genetics of IBD . Modeling the role of genetic factors in characterizing extra-intestinal manifestations in Crohn’s disease patients: does this improve outcome predictions? Curr Med Res Opin 2007;23:1657–65. [DOI] [PubMed] [Google Scholar]
- 16. Klein A, Mazor Y, Karban A, Ben-Itzhak O, Chowers Y, Sabo E. Early histological findings may predict the clinical phenotype in Crohn’s colitis. United European Gastroenterol J 2017;5:694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng JC, Ran ZH, Shen J. Seasonal variation in onset and relapse of IBD and a model to predict the frequency of onset, relapse, and severity of IBD based on artificial neural network. Int J Colorectal Dis 2015;30:1267–73. [DOI] [PubMed] [Google Scholar]
- 18. Reddy BK, Delen D, Agrawal RK. Predicting and explaining inflammation in Crohn’s disease patients using predictive analytics methods and electronic medical record data. Health Informatics J 2019;25:1201–18. [DOI] [PubMed] [Google Scholar]
- 19. Saito K, Katsuno T, Nakagawa T, et al. Predictive factors of response to intravenous ciclosporin in severe ulcerative colitis: the development of a novel prediction formula. Aliment Pharmacol Ther 2012;36:744–54. [DOI] [PubMed] [Google Scholar]
- 20. Takayama T, Okamoto S, Hisamatsu T, et al. Computer-aided prediction of long-term prognosis of patients with ulcerative colitis after cytoapheresis therapy. PLoS One 2015;10:e0131197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Waljee AK, Joyce JC, Wang S, et al. Algorithms outperform metabolite tests in predicting response of patients with inflammatory bowel disease to thiopurines. Clin Gastroenterol Hepatol 2010;8:143–50. [DOI] [PubMed] [Google Scholar]
- 22. Waljee AK, Lipson R, Wiitala WL, et al. Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients using machine learning. Inflamm Bowel Dis 2017;24:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Waljee AK, Liu B, Sauder K, et al. Predicting corticosteroid-free biologic remission with vedolizumab in Crohn’s disease. Inflamm Bowel Dis 2018;24:1185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waljee AK, Liu B, Sauder K, et al. Predicting corticosteroid-free endoscopic remission with vedolizumab in ulcerative colitis. Aliment Pharmacol Ther 2018;47:763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waljee AK, Sauder K, Patel A, et al. Machine learning algorithms for objective remission and clinical outcomes with thiopurines. J Crohns Colitis 2017;11:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Waljee AK, Wallace BI, Cohen-Mekelburg S, et al. Development and validation of machine learning models in prediction of remission in patients with moderate to severe Crohn disease. JAMA Netw Open 2019;2:e193721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kingsford C, Salzberg SL. What are decision trees? Nat Biotechnol 2008;26:1011–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noor NM, Verstockt B, Parkes M, Lee JC. Personalised medicine in Crohn’s disease. Lancet Gastroenterol Hepatol 2020;5:80–92. [DOI] [PubMed] [Google Scholar]
- 29. Khanna R, Ma C, Jairath V, et al. Endoscopic assessment of inflammatory bowel disease activity in clinical trials. Clin Gastroenterol Hepatol 2020:S1542-3565(20)31674-8. [DOI] [PubMed] [Google Scholar]
- 30. Gottlieb K, Travis S, Feagan B, Hussain F, Sandborn WJ, Rutgeerts P. Central reading of endoscopy endpoints in inflammatory bowel disease trials. Inflamm Bowel Dis 2015;21:2475–82. [DOI] [PubMed] [Google Scholar]
- 31. Chang S, Malter L, Hudesman D. Disease monitoring in inflammatory bowel disease. World J Gastroenterol 2015;21:11246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.