Aminoglycosides are highly potent, broad-spectrum antibiotics with many desirable properties for the treatment of life-threatening infections (28). Their history begins in 1944 with streptomycin and was thereafter marked by the successive introduction of a series of milestone compounds (kanamycin, gentamicin, and tobramycin) which definitively established the usefulness of this class of antibiotics for the treatement of gram-negative bacillary infections. In the 1970s, the semisynthetic aminoglycosides dibekacin, amikacin, and netilmicin demonstrated the possibility of obtaining compounds which were active against strains that had developed resistance mechanisms towards earlier aminoglycosides as well as displaying distinct toxicological profiles (65). Since then, however, the pace of development of new aminoglycosides has markedly slowed down. Conversely, we have witnessed a period of extensive basic and clinical research which has made us view these drugs very differently from what was commonly accepted when they were first introduced in the clinic. We attempt to present and discuss these developments, not to ascertain whether there is a likelihood that new molecules or effective means to avoid bacterial resistance and drug-induced toxicity will eventually reach the clinical arena, but to foster continuing research on aminoglycosides and to make the clinician aware of the pertinent progress made in this area. The present paper is focused on activity and resistance, whereas the companion review (65) deals with nephrotoxicity (ototoxicity has been reviewed earlier in this journal [4]). In both reviews, we did not attempt to be exhaustive in any of these domains, and the material presented has been selected on the basis of its interest in terms of new concepts or because it deals directly with the design of new aminoglycosides or an improved use of the available agents.
BASIS OF ANTIMICROBIAL ACTION
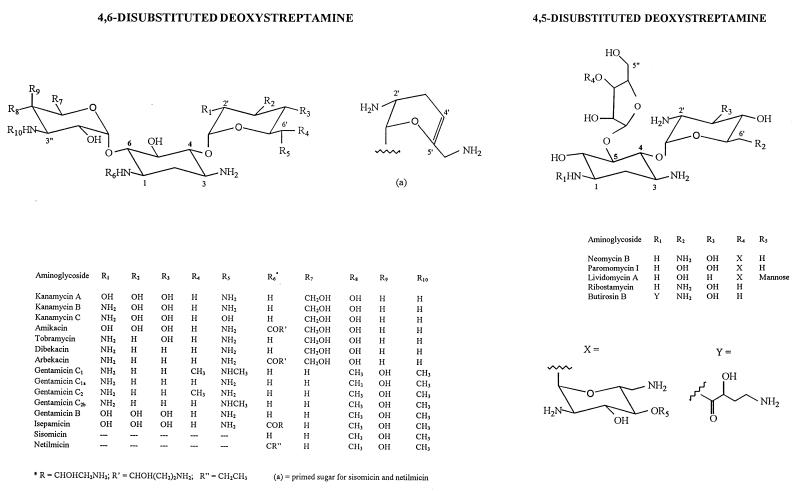
The basic chemical structure required for both potency and the spectrum of antimicrobial activity of aminoglycosides is that of one or several aminated sugars joined in glycosidic linkages to a dibasic cyclitol. In most clinically used aminoglycosides the latter is 2-deoxystreptamine, and it is streptidine in streptomycin and derivatives and fortamine in the fortimicin series (Fig. 1 and 2). Aminoglycosides act primarily by impairing bacterial protein synthesis through binding to prokaryotic ribosomes. Passage of these highly polar molecules across the outer membrane of gram-negative bacteria (31, 32) is a self-promoted uptake process involving the drug-induced disruption of Mg2+ bridges between adjacent lipopolysaccharide molecules (108). Penetration through porin channels is unlikely because of the large size of aminoglycosides (approximately 1.8 by 1.0 by 1.0 nm [11]). Subsequent transport of aminoglycosides across the cytoplasmic (inner) membrane is dependent upon electron transport and is termed energy-dependent phase I (EDP-I [5]). It is rate limiting and is blocked or inhibited by divalent cations, hyperosmolarity, low pH (114), and anaerobiosis. In the cytosol, aminoglycosides bind to the 30S subunit of ribosomes, again through an energy-dependent process (energy-dependent phase II [EDP-II] [5]). While this binding does not prevent formation of the initiation complex of peptide synthesis (binding of mRNA, fMetRNA, and association of the 50S subunit), it perturbs the elongation of the nascent chain by impairing the proofreading process controlling translational accuracy (misreading and/or premature termination [60]). The aberrant proteins may be inserted into the cell membrane, leading to altered permeability and further stimulation of aminoglycoside transport (7). It is the 2-deoxystreptamine and the primed amino sugar (on the right in the structures shown in Fig. 1) which are essential for causing the lack of fidelity in the translation process (23). The nucleotides responsible for aminoglycoside binding form an asymmetrical internal loop caused by noncanonical base pairs (23, 67). These key structural features are also found in the rev-binding site of human immunodeficiency virus type 1 (HIV-1) (10, 110), but aminoglycosides are unlikely to become anti-HIV drugs, as was originally hoped (116), without thorough chemical optimization and/or screening (51, 78) because of their lack of specificity.
FIG. 1.
Structural formulae of the 2-deoxystreptamine-containing aminoglycosides cited in the text. The numbering of the atoms shown in the figure follows the recommendations in reference 73 with the primed numbers (′) being ascribed to the sugar attached to the C-4 of the 2-deoxystreptamine (since this C is of the R configuration) and the doubly primed numbers (") being ascribed to the sugar attached on either the C-6 (S configuration) for the 4,6-disubstituted deoxystreptamine or the C-5 (R configuration) for the 4,5-disubstituted-2-deoxystreptamine. Note that sisomicin and netilmicin have a particular primed sugar with a double bond between C-4′ and C-5′.
FIG. 2.
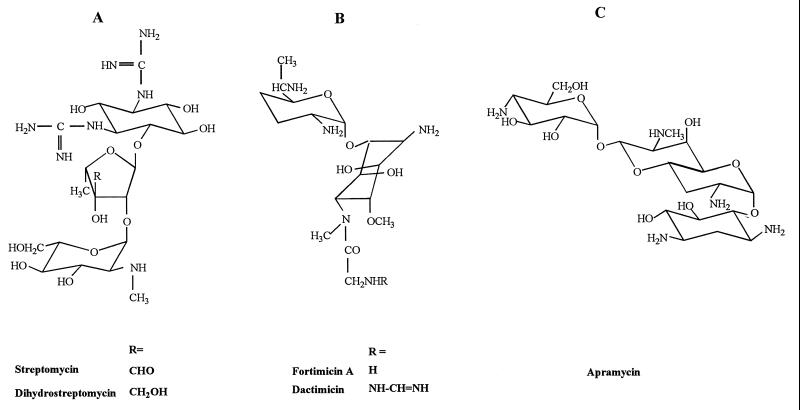
Structural formulae of the main aminoglycosides containing an aminocyclitol other than 2-deoxystreptamine. (A) streptomycin and derivatives; (B) fortimicin and derivatives; (C) apramycin.
Whereas all clinically used aminoglycosides are inhibitors of prokaryotic protein synthesis at commonly accepted therapeutic concentrations (≤25 μg/ml), they may also affect the protein synthesis of mammalian cells at larger concentrations (96), probably through nonspecific binding to eukaryotic ribosomes and/or nucleic acids. Molecules with a hydroxyl function at C-6′ in place of an amino function, such as in kanamycin C, gentamicin A factors, and antibiotic G418 (3′,4′-dihydroxy-6′-hydroxy-6′-deamino-gentamicin C1, also called geneticin [30]), are more effective inhibitors of eukaryotic protein synthesis (22, 52). Geneticin and hygromycin B (an atypical aminoglycoside with properties similar to G418) are now commonly used in molecular biology to select eukaryotic cells transfected with a gene coding for an appropriate aminoglycoside-modifying enzyme (17, 88). Antibiotic G418 was actually shown to bind directly to the eukaryotic 80S ribosomal complex (2), and its mode of action, and probably that of the other 6′-OH aminoglycosides, is therefore expected to be quite different from that of conventional aminoglycosides. It is not clear, however, how antibiotic G418 and hygromycin B reach their target in the cytosol in sufficient amounts (6), since aminoglycosides in general are too polar to cross mammalian cell membranes by diffusion and are mainly stored in lysosomes of mammalian cells grown in their presence (103).
RESISTANCE MECHANISMS
The emergence of resistant strains has somewhat reduced the potential of aminoglycosides in empiric therapies (71). Alteration of the ribosomal binding sites will not be reviewed here since it causes significant resistance to streptomycin only. The other main mechanisms which may affect all aminoglycosides are (i) a decreased uptake and/or accumulation of the drug in bacteria and (ii) the bacterial expression of enzymes which modify the antibiotic and thereby inactivate it (18, 89).
Decrease in drug uptake and accumulation.
Reduced drug uptake, mostly seen in Pseudomonas spp. and other non-fermenting gram-negative bacilli, is likely to be due to membrane impermeabilization, but the underlying molecular mechanisms are largely unknown (8). It is highly significant in the clinic since it affects all aminoglycosides, is a stable characteristic, and results in a moderate level of resistance (intermediate susceptibility). Aerobic gram-negative bacilli in general also show a phenomenon of adaptive resistance (transiently reduced antimicrobial killing in originally susceptible bacteria [42, 115]). Membrane protein changes and alteration in the regulation of genes of the anaerobic respiratory pathway in bacteria exposed to aminoglycosides (42) are probably responsible for this phenomenon (55), which gives a pharmacodynamic rationale for high dosages associated with long intervals between successive administrations (16). Active efflux has been evidenced for neomycin, kanamycin, and hygromycin A in Escherichia coli (21, 33) (protein Mdfa, a member of the family of multidrug resistance proteins), but its clinical significance is still uncertain compared to that of other antibiotics (54, 75).
Aminoglycoside-modifying enzymes.
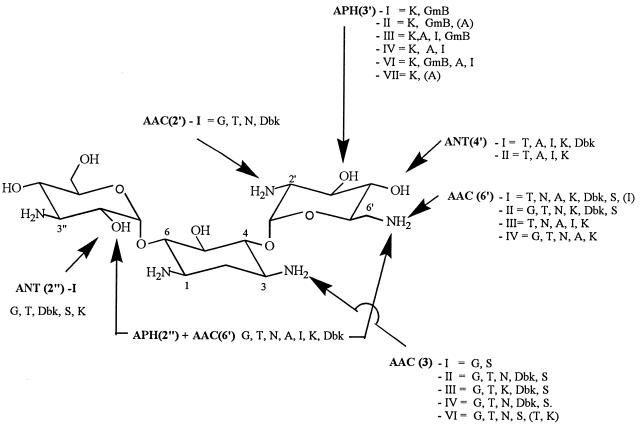
Aminoglycoside-modifying enzymes catalyze the covalent modification of specific amino or hydroxyl functions, leading to a chemically modified drug which binds poorly to ribosomes and for which the EDP-II of accelerated drug uptake also fails to occur, thereby resulting most often in high-level resistance. The enzymes modifying aminoglycosides are N-acetyltransferases (AAC), which use acetyl-coenzyme A as donor and affect amino functions, and O-nucleotidyltransferases (ANT) and O-phosphotransferases (APH), which both use ATP as donor and affect hydroxyl functions. The functions affected in typical aminoglycosides (kanamycin and gentamicin derivatives) are on positions 3, 2′, and 6′ for AAC, positions 4′ and 2" for ANT; and positions 3′ and 2" for APH (Fig. 3). Aminoglycoside-modifying enzymes are often plasmid encoded but are also associated with transposable elements. Plasmid exchange and dissemination of transposons facilitate the rapid acquisition of a drug-resistance phenotype not only within a given species but among a large variety of bacterial species. In epidemiological surveys, aminoglycoside resistance mechanisms have first been ascertained by examining the susceptibility of the isolates to a panel of clinically used and experimental aminoglycosides with specific susceptibilities to these enzymes (phenotypic characterization [89]). Such studies quickly led to the recognition of a large diversity of phenotypes with almost every susceptible position in each drug being modified by several distinct enzymes. With the development of molecular biology techniques, a considerably larger number of genes have been characterized so that each phenotype has now been associated with the expression of several distinct proteins with the same aminoglycoside-modifying activity (64a, 89). Large variations in substrate specificity may develop from a few and sometimes a single amino acid change in the protein (83, 113). Moreover, several genes could derive from one or a few single common ancestors, suggesting a large plasticity in the type of activities a bacterium may express. It is therefore anticipated that bacteria will quickly catch up to or defeat our efforts at making a given aminoglycoside resistant to inactivation by a specific enzyme. This insight also places the existence of aminoglycoside-modifying enzymes in a new perspective. It is indeed likely that these enzymes have physiological functions in bacteria (acetylation, nucleotidylation, or phosphorylation of natural substrates) and that their activities against aminoglycosides in the wild-type strains are simply too weak to confer a phenotype of resistance. Yet, these proteins may be overexpressed under the pressure of aminoglycosides (84, 112). In some cases, aminoglycosides may still remain poor substrates compared to the natural ones, but a moderate overexpression will nevertheless be enough to cause low-level resistance and further selection of the so-called intermediate susceptibility strains. These strains often fail to be clearly recognized in routine microbiological testing unless their MICs are determined, but their presence may explain clinical failures seen with patients with low peak levels (70) of aminoglycosides in serum.
FIG. 3.
Major aminoglycoside-modifying enzymes acting on kanamycin B (this aminoglycoside is susceptible to the largest number of enzymes). Each group of enzymes inactivates specific sites, but each of these sites can be acted upon by distinct isoenzymes (roman numerals) with different substrate specificities (phenotypic classification; each phenotype comprises several distinct gene products [denoted by lowercase letters after the roman numeral in the text]); at least one enzyme is bifunctional and affects both positions 2" (O-phosphorylation) and 6′ (N-acetylation)). The main clinically used aminoglycosides on which these enzymes act are as follows: amikacin (A), dibekacin (Dbk), commercial gentamicin (G) (see text), gentamicin B (GmB), kanamycin A (K), isepamicin (I), netilmicin (N), sisomicin (S), and tobramycin (T) (see text for discussion of arbekacin, sagamicin, and dactimicin). The drug abbreviations which appear in parentheses are those for which resistance was detectable in vitro even though clinical resistance was not conferred. Based on the data of Shaw et al. (89).
All these observations most probably account for the fact that while early aminoglycoside resistance surveys (19, 82, 90) showed resistant strains to be characterized by a single modifying enzyme, subsequent surveys in the same regions began to show the emergence of complex phenotypes and a multiplicity of genotypes (20, 62). Most interestingly, some changes in the pattern of mechanisms in certain species of Enterobacteriaceae were found to correlate with modifications in aminoglycoside usage at the level of either geographic region or individual hospital (62–64) (for a web site with regular updates on aminoglycoside-inactivating enzymes and on methods for their identification see reference 88a). Such phenomena explain why the choice of an aminoglycoside has become so much more complex today and must be largely individualized on the basis of the local epidemiology and appropriate determination of effective susceptibility. An additional source of complexity in human therapy is also the spreading of resistance genes from bacteria exposed to aminoglycosides used more specifically in animals, such as apramycin (9, 41), causing selection pressure even in the absence of drug usage in the hospital setup. Pseudomonas is perhaps an exception to this evolution towards more complexity since the main change observed from the mid-1970s to now seems to be an increase in the frequency of the so-called decreased permeability type of resistance. Yet, because this mechanism is often not characterized in epidemiological surveys, we cannot exclude the undetected emergence of enzymatically mediated low-level resistance mechanisms.
DESIGN AND CHARACTERIZATION OF COMPOUNDS ACTIVE AGAINST ORGANISMS PRODUCING AMINOGLYCOSIDE-MODIFYING ENZYMES
So far, the most successful approaches to obtaining derivatives with activity against strains producing aminoglycoside-modifying enzymes have used the conventional pharmacochemical methodology of structure-activity relationship studies and medicinal chemistry, starting from existing compounds and modifying them to decrease their recognition by one or several enzymes while retaining most of their intrinsic activity. Approaches by using the knowledge of the aminoglycoside-modifying enzymes to produce totally new compounds are recent and still largely unproven.
PHARMACOCHEMICAL METHODOLOGY APPROACHES
Most of these approaches have been applied to the 2-deoxystreptamine derivatives, and these will be therefore discussed in detail (Fig. 4). Indeed, among the non-2-deoxystreptamine aminoglycosides only fortimicins, made of a diaminocyclitol (fortamine) and only one aminated sugar (see Fig. 2), have been studied [fortimicins lack the functions modified by APH(3′), ANT(2"), and ANT(4′) and are therefore sensitive only to AAC(6′) and AAC(3); dactimicin, a natural derivative of fortimicin A carrying a formimidoyl substitution on the amino group of the glycyl residue (76) (Fig. 2) is protected against AAC(6′)-I and -II but not against the bifunctional enzyme AAC(6′)-APH(2") (29); it remains sensitive to AAC(3)].
FIG. 4.
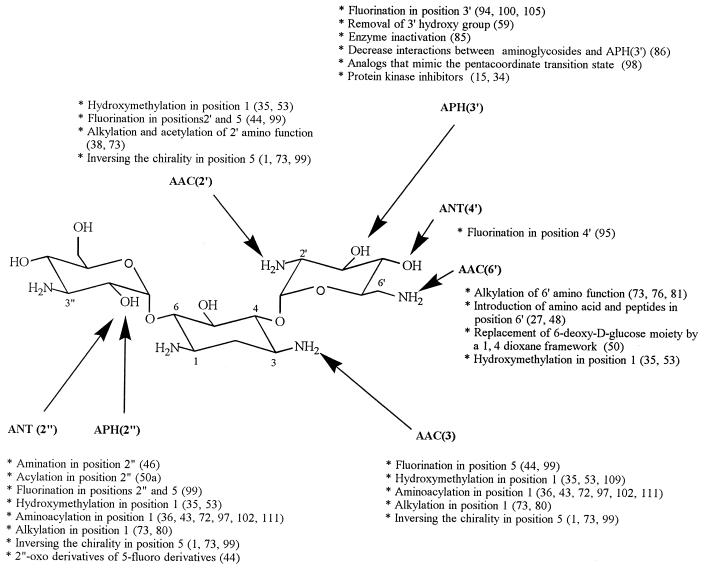
Chemical modifications performed on kanamycin B to obtain compounds capable of resisting inactivation by specific aminoglycoside-modifying enzymes (see Fig. 3 for the types of enzymes acting on each of these positions).
Generally speaking, two types of leads have been followed. In the first one, resistance to specific enzymes has been sought for by introducing minor but often very efficient modifications of one given function. In the second one, more extensive changes have been introduced which confer resistance to several aminoglycoside-modifying enzymes acting on distinct functions in the parent molecule because its overall properties are significantly modified.
Protection against specific aminoglycoside-modifying enzymes. (i) Phosphotransferases.
This line of research was pioneered in the late 1960s-early 1970s by H. Umezawa and colleagues, who aimed at protecting kanamycins against APH enzymes and especially APH(3′), which was most prevalent at that time (68). This led to the synthesis of 3′-deoxykanamycin A (107), with properties similar to those of tobramycin in this context (66), and to that of 3′,4′-dideoxykanamycin B (dibekacin) (106), which resists both APH(3′) and ANT(4′) enzymes (note that a similar protection is observed in the naturally occurring gentamicins C and sisomicin (and its derivative netilmicin). Insensitivity of the 3′-deoxyaminoglycosides to phosphorylation is, however, only partial because one of the APH(3) enzymes [APH(3′)-I] tightly binds to these molecules, even though they are not substrates, resulting in a phenotype of resistance (61).
(ii) Acetyltransferases.
Among the acetyltransferases the 6′-N-acetyltransferases, and particularly AAC(6′)-I, have attracted the greatest attention because these enzymes confer resistance not only to the naturally occurring kanamycins and tobramycin but also to the 1-N-substituted derivatives discussed below (amikacin and netilmicin). Commercial, clinically used gentamicin appears to be resistant to AAC(6′)-I enzymes because of its content of gentamicin C1 (approximately one-third). Indeed, this component of the gentamicin complex carries a methyl group on N-6′ (as well as on C-6′) giving rise to a secondary amine which is far less susceptible to AAC(6′) enzymes. Sagamicin or gentamicin C2b, a minor component of the gentamicin complex which is methylated in N-6′ only (77), is also resistant to AAC(6′) enzymes. Reduction of the sensitivity of the other aminoglycosides towards AAC(6′) enzymes was therefore sought by a similar substitution of the N-6′ either as such (e.g., 6′-N-methyl-sisomicin [G52]) or in combination with the 1-N-substitutions discussed below (6′-N-ethyl-netilmicin, 6′-N-methyl-amikacin [BB-K 28], and with the deoxygenation of position 4′ already discussed, 4′-deoxy-6′-N-methyl-amikacin [BB-K 311] [73, 81]). All these semisynthetic compounds, however, have lower intrinsic activities against sensitive isolates with no or little commensurate decrease in toxicity, so that none of them has been developed so far for clinical use. Following the observation that 6′-N-amino acid and peptide derivatives of neamine retain the weak but nevertheless measurable microbiological activity of the parent compound (27), 6′-N-amino acid- and peptides derivatives of kanamycin A and netilmicin have been synthesized (48). Unfortunately, these compounds lacked activity against both susceptible and resistant strains, demonstrating that the 6′-N atom must remain ionizable, as was already deduced from the lack of significant activity of 6′-N-hydroxyaminobutyryl-kanamycin A, a positional isomer of amikacin (74).
Synthesis of compounds designed for specific resistance to other acetyltransferases [AAC(2′) and AAC(3)] has been much more limited, with the exception of 2′-N-ethyl-netilmicin (73), which has not been developed for clinical use.
Protection against several aminoglycoside-modifying enzymes. (i) 1-N-substituted derivatives.
The strategy of developing compounds by substituting a short amino acyl or alkyl chain for the N-1 amino function has been highly successful. The basis of this strategy was the recognition in the late 1960s that the naturally occurring butirosins possessing an amino acyl chain on N-1 are resistant to many inactivating enzymes while retaining most of the intrinsic activity of the unsubstituted molecules against susceptible strains (102, 111). While butirosins themselves are of little interest, this observation led to the semisynthesis of analogous 1-N-acyl derivatives of the kanamycins and gentamicins yielding, successfully, amikacin {1-N-[(S)-4-amino-2-hydroxybutyryl]-kanamycin A (36, 43)}, isepamicin {1-N-[(S)-4-amino-2-hydroxypropionyl]-gentamicin B (72)}, and arbekacin {1-N-[(S)-4-amino-2-hydroxybutyryl-3′,4′-dideoxykanamycin B, formerly called habekacin] (39, 45, 97)}, all three of which have been clinically developed (similar compounds were made with other naturally occurring aminoglycosides such as sisomicin [73] but were not brought to the clinics). In these derivatives, the N-1 atom is no longer basic or ionizable, and the basic amino function is actually displaced to the end of the side chain. These compounds are not only protected against enzymes acting on position 2" [ANT(2")-I enzymes], which most likely results from steric hindrance, but also on position 3 [AAC(3) enzymes], and for amikacin and isepamicin, to some of the enzymes acting on position 3′. The explanation for this wider-than-expected protection as well as for the maintenance of activity towards susceptible strains is probably that the side chain folds in such a way as to present its terminal amino function at a position which, compared to that of the N-1 function in the parent compound, is sufficiently close for efficient binding to the ribosomes. Yet, it also remains sufficiently remote to circumvent the AAC(3) and APH(3′) enzymes by slightly misaligning them on the drug molecule (73). This useful feature explains the wide success of the 1-N-substituted derivatives in situations of resistance to kanamycin, gentamicin, or tobramycin, with amikacin being the lead compound nowadays in North America and Europe (isepamicin, largely used in Japan, is now also available in Europe).
1-N-aminoacyl derivatives, however, remain largely susceptible to enzymes acting on position 4′ unless that position is made unreactive by deoxygenation such as in BB K-311 (discussed above) or arbekacin. Arbekacin, having no hydroxyl function at C-3′ and C-4′, is not modified by APH(3′) and ANT(4′) enzymes. It is also not inactivated by the bifunctional enzymes APH(2")-AAC (6′) and ANT(4′)-ANT(4"), produced by Staphylococcus aureus and Staphylococcus epidermidis (104). Since methicillin-resistant Staphylococcus aureus often harbors these bifunctional enzymes (46, 47), arbekacin has found an interesting application in the treatment of infections caused by these organisms (39, 40). Arbekacin carries a 2′-amino function and is therefore slowly modified by AAC(2′) enzymes (49). Yet, the resulting 2′-N-acetyl derivative retains substantial antibiotic activity against a variety of bacterial species (38).
The 1-N substitution in amikacin and arbekacin does not confer protection against the enzymes causing the AAC(6′)-I phenotype. Yet, surprisingly, isepamicin is resistant to many of them, perhaps due to the fact that it contains a secondary, methylated amine at position 3", an important determinant for binding and correct positioning of the AAC(6′) enzymes (63). This explanation, however, does not account for why netilmicin remains fully susceptible to the same enzymes since it shares the same doubly primed sugar as isepamicin.
A parallel direction of research has been the substitution of the N-1 position of aminoglycosides by an alkyl group, a strategy that results in the introduction of substituents which also protect the aminoglycoside from the ANT(2") and AAC(3) enzymes but which leaves the N-1 group as a secondary ionizable amine. These compounds more fully maintain their intrinsic antibacterial activity than the 1-N-amino acyl derivatives. Yet, for the same reason, they are not protected against enzymes acting further away from positions 2" and 3. This approach yielded a large number of drug candidates such as 1-N-butyl-kanamycin A, 1-N-butyl-kanamycin B, and 1-N-ethyl-sisomicin (80). Only the latter, under the name of netilmicin, has been clinically developed, apparently mainly on grounds of toxicological considerations (73). Since netilmicin is built from sisomicin, which has no hydroxyl function in C-3′ and C-4′, it is not modified by APH(3′) and ANT(4′) enzymes. Conversely, it remains sensitive to AAC(2′) and to all AAC(6′) enzymes. Amino alkyl rather than amino acyl or alkyl groups have also been used to substitute the N-1 position of aminoglycosides [e.g., 1-N-(4-aminobutyl) sisomicin (73)], an approach that would not only leave this N-1 ionizable (as in netilmicin) but would also place a terminal ionizable function on the side chain (as in amikacin, isepamicin, and arbekacin). Apparently, this lead was not followed.
(ii) Derivatives with inverted chirality.
Inverting the stereochemistry of the substituents of a carbon atom carrying a functional group which is a target for a modifying enzyme was thought to be sufficient to make this function insensitive to the corresponding enzyme. An inversion of stereochemistry may have broader effects. Indeed, as demonstrated by the studies made in the sisomicin series, changing the orientation of the 5-OH function from equatorial to axial (yielding 5-episisomicin) makes the molecule resistant to ANT(2"), AAC(2′), and most AAC(3) enzymes. It is thought that the change of orientation of the hydroxyl group allows for more rotational freedom of the two sugars around the corresponding carbon atoms of the aglycone (C-4 and C-6) which are flanking the C-5 (73). This interesting lead was also followed for amikacin or arbekacin (1, 99) but has so far not yielded clinically usable compounds.
(iii) 1-C-substituted derivatives.
In a search for alternatives to the N-1 substitutions, a direct linking of various side chains to the C-1 of the 2-deoxystreptamine has been attempted. Introduction of a hydroxymethyl substituent at that position in gentamicin C2 yielded a compound (S87351) that overcomes all clinically relevant types of enzymatically-mediated resistance of importance for gentamicin (35, 53) and specifically resists the enzymes acting at positions 2", 3, 2′, and 6′. The same substitution, however, was ineffective in the kanamycin series (93a, 109) with no factual explanation given so far for this unsuspected but most frustrating limitation beyond considerations on the comparative structures of gentamicins versus kanamycins.
(iv) Halogenated derivatives.
More recently, halogenation of specific positions was attempted not only to render the function no longer susceptible to specific enzymes but also to confer protection of the neighboring functions thanks to the electron-withdrawing properties of the halogen. This was indeed observed for kanamycins carrying one or two fluorine atoms in position 5 of the 2-deoxystreptamine [protection against ANT(2"), APH(3′), AAC(3), and partly against AAC(2′) [(91)]. Yet, this approach has clear limitations since the introduction of a fluorine in positions 2′, 3′, and 4′ (95, 100, 101, 105) or in positions 2" and 6" (44) protects only against the enzymes acting on the modified positions. Chlorine has also been introduced into positions 3′ and 6" of kanamycin A (105) as well as in position 6" of amikacin. Although 3′-deoxy-3′-chlorokanamycin was active against resistant bacteria, the activity of this compound was approximately one sixth of that of 3′-deoxy-3′-fluorokanamycin A against sensitive strains, discouraging further developments in this direction.
(v) Other modifications.
Another original modification of the primed sugar of kanamycin to reduce its susceptibility to AAC(6′) enzymes has been the replacement of the endocyclic C-3′ by an oxygen atom (yielding a dioxane), causing a major change of conformation and a marked decrease of the polarity of this moiety by the removal of the 3′-OH group. The corresponding derivative compounds, unfortunately, are substantially less active than kanamycins (50).
ENZYME KNOWLEDGE-BASED APPROACHES
Phosphotransferases.
Most efforts have been directed towards the APH(3′) enzymes, which inactivate kanamycin and other aminoglycosides carrying a 3′-OH function such as amikacin and isepamicin. They make up to 2% of the total bacterial protein (92, 93) and exist in several subgroups. Purification procedures and kinetic mechanisms have been worked out in detail for three of these subgroups [APH(3′)-Ia (93); APH(3′)-IIa (92), and APH(3′)-IIIa (12, 13, 56–59, 98)]. Four strategies, illustrated in Fig. 5, have been suggested to protect against APH(3′) enzyme. The first strategy (Fig. 5A) (86) consists in the deamination of position 1, 6′, or 3" of neamine or of kanamycin A, leading to a 104 to 106 decrease of the catalytic constants of APH(3′)-IIa towards 1-deaminokanamycin A and of APH(3′)-Ia towards 3"-deaminokanamycin A, respectively. Unfortunately, the intrinsic activity of these deaminated derivatives was too low to warrant clinical applications. A second and novel strategy in the aminoglycoside resistance field has been the development of suicide substrates for the APH(3′) enzymes. This has been attempted with 2′-nitroderivatives of neamine and kanamycin B (Fig. 5B) (85). These compounds are excellent substrates of APH(3′) but are poor antibiotics. After the compounds have been modified by APH(3′), the 2′-nitrofunction becomes vicinal to the phosphogroup. The latter being an excellent leaving group, it will give rise, after facile and spontaneous elimination, to a nitroalkene function. The electrophilic intermediate which is thereby generated right in the active site of the enzyme will react with the nucleophilic side chain(s) of the neighboring aminoacids, resulting in an irreversible inactivation of the enzyme. The third strategy (Fig. 5C) is based on the mechanism of phosphate transfer (98) which involves the participation of a pentacoordinate transition state intermediate. Analogues of this compound unable to yield a final product should block the enzyme and, therefore, be good inhibitors. The last strategy (Fig. 5D) uses the knowledge of the enzymes tridimensional structure based on X-ray crystallographic data (37). Because there are many similarities between APH(3′)-IIIa and eukaryotic protein kinases, a large series of inhibitors of the latter enzymes have been tested (15), among which the isoquinoline sulfonamides, which act through competition with ATP (34), have proved effective against not only APH(3′)-IIIa but also against the bifunctional APH(2′)-AAC(6′) enzyme. This observation is of major clinical interest since, as mentioned above, methicillin-resistant Staphylococcus aureus often harbor this enzyme making it resistant to most available aminoglycosides except arbekacin.
FIG. 5.
Potential strategies to overcome or avoid inactivation of aminoglycosides by APH(3′) enzymes (see Fig. 3 for the site of action these enzymes). 5A, reduction of the electrostatic binding of the drug to the enzymes by deamination of the 6′ position and/or the 1 and 3" positions; 5B, design of a suicide substrate (2′-nitro derivative allowing for the formation of a 2′-nitro, 3′-phospho derivative which then gives rise to a highly reactive 2′-nitroalkene capable of inactivating the enzyme by chemical binding to amino acids close to the active site); 5C, design of analogues of the pentacoordinate transition state compound involved in the enzymatic phosphorylation of the drug; 5D, use of inhibitors of protein kinases based on the similarities between these enzymes and APH(3′)-IIIa.
Nucleotidyltransferases.
Two enzymes in this group, ANT(2")-I and ANT(4′)-I, have been studied in great detail. Both are of clinical interest since the first inactivates gentamicin and tobramycin while the latter inactivates tobramycin, amikacin, isepamicin, and all other 4′-OH aminoglycosides. The molecular mechanisms involved in ANT(2")-I action appear to be very complex, however, with at least eight steps in the reaction itself plus additional critical steps for drug recognition (24, 26). It is therefore unlikely that effective inhibitors can be easily designed on the basis of these complex mechanisms. It must also be remembered that the 1-N-alkyl or 1-N-acyl derivatives discussed above are effectively protected against this enzyme. Moreover, there is a rather striking parallelism for a large series of aminoglycosides between their antibacterial activity and their susceptibility to nucleotidylation by ANT(2")-I enzymes (25). Both properties indeed show a marked dependence on the number and position of the amino groups in the primed sugar (2′,6′-diamino- > 6′-amino- > 2′-amino- > deamino sugar [3, 25]) and are enhanced if the same primed sugar is not hydroxylated (thus gentamicins C, which have no hydroxyl group on their primed sugar, are generally better antibiotics than kanamycins [69] and at the same time are better substrates for ANT(2")-I [23]). ANT(4′)-I has also been extensively characterized down to the determination of the amino acids involved in its antibiotic binding domain (79, 87). This showed that the 4′-hydroxyl group of the antibiotic lies at a distance of only approximately 5 Å from the a-phosphorus of the nucleotide and in the proper orientation for a single in-line displacement attack at the phosphorus. Glu 145, which lies within hydrogen-bonding distance of the 4′-hydroxyl group, is proposed to be the active-site base, giving clear suggestion for the design of specific inhibitors targeted towards this amino acid.
CONCLUSIONS
Extensive structural information has now been assembled over the last 20 years not only on aminoglycoside-modifying enzymes but also on the fine mechanisms governing the activity of these drugs at the molecular level. In parallel, the various steps in drug uptake and disposition by bacteria have now been almost entirely uncovered. Together with the extensive progress made in the characterization of the key pharmacodynamic parameters of governing aminoglycoside activity (dose dependency, first-exposure and post-antibiotic effects) and toxicity (saturation of uptake by kidney and cochlear cells) (see references 14, 65, and 117 for reviews), this large body of knowledge could now be put into practice for the effective design and evaluation of new aminoglycosides or for the design of inhibitors of aminoglycoside-modifying enzymes. Both approaches appear feasible and could lead to substantial improvements in aminoglycoside-based chemotherapy.
ACKNOWLEDGMENTS
We thank G. H. Miller from the Schering-Plough Corp. (Kenilworth, N.J.) for helpful discussions and suggestions during the preparation of this review.
M.-P. M. is Chercheur Qualifié of the Belgian Fonds National de la Recherche Scientifique. Support was received from the Belgian Fonds de la Recherche Scientifique Médicale (grant no. 3.4589.96, 3.4516.94, 9.4541.95.F, and 9.4514.92), the Fonds National de la Recherche Scientifique (grant no. 9.4546.94), the Actions de Recherches Concertées 94/99-172 of the Direction Générale de la Recherche Scientifique—Communauté Française de Belgique, Belgium, and the French nonprofit organization (Association-loi 1901) Vaincre les Maladies Lysosomales.
REFERENCES
- 1.Albert R, Dax K, Stũtz A E, Hildebrandt J. Chemical modification of amikacin at C-4" with inversion of configuration. J Antibiot (Tokyo) 1985;38:275–278. doi: 10.7164/antibiotics.38.275. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Nun S, Shneyour Y, Beckman J S. G-418, an elongation inhibitor of 80 S ribosomes. Biochim Biophys Acta. 1983;741:123–127. doi: 10.1016/0167-4781(83)90018-0. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste R, Davies J. Structure-activity relationships among the aminoglycoside antibiotics: role of hydroxyl and amino groups. Antimicrob Agents Chemother. 1973;4:402–409. doi: 10.1128/aac.4.4.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brummett R, Fox K. Aminoglycoside-induced hearing loss in humans. Antimicrob Agents Chemother. 1989;33:797–800. doi: 10.1128/aac.33.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan L E, Kwan S. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob Agents Chemother. 1983;23:835–845. doi: 10.1128/aac.23.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan J H, Stevens A, Sidhu J. Aminoglycoside antibiotic treatment of human fibroblasts: intracellular accumulation, molecular changes and the loss of ribosomal accuracy. Eur J Cell Biol. 1987;43:141–147. [PubMed] [Google Scholar]
- 7.Busse H J, Wostmann C, Bakker E P. The bactericidal action of streptomycin: membrane permeabilization caused by the insertion of mistranslated proteins into the cytoplasmic membrane of Escherichia coli and subsequent caging of the antibiotic inside the cells: degradation of these proteins. J Gen Microbiol. 1992;138:551–561. doi: 10.1099/00221287-138-3-551. [DOI] [PubMed] [Google Scholar]
- 8.Chambers H F, Sande M A. Antimicrobial agents: the aminoglycosides. In: Hardman J G, Limbird L E, Molinoff P B, Ruddon R W, Goodman Gilman A, editors. The pharmacological basis of therapeutics. New York, N.Y: McGraw-Hill; 1995. pp. 1103–1121. [Google Scholar]
- 9.Chaslus-Dancla E, Glupczynski Y, Gerbaud G, Lagorce M, Lafont J P, Courvalin P. Detection of apramycin resistant Enterobacteriaceae in hospital isolates. FEMS Microbiol Lett. 1989;61:261–266. doi: 10.1016/0378-1097(89)90208-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen Q, Shafer R H, Kuntz I D. Structure-based discovery of ligands targeted to the RNA double helix. Biochemistry. 1997;36:11402–11407. doi: 10.1021/bi970756j. [DOI] [PubMed] [Google Scholar]
- 11.Chung L, Kaloyanides G, McDaniel R, McLaughlin A, McLaughin S. Interaction of gentamicin and spermine with bilayer membranes containing negatively-charged phospholipids. Biochemistry. 1985;24:442–452. doi: 10.1021/bi00323a030. [DOI] [PubMed] [Google Scholar]
- 12.Cox J R, Serpersu E H. Biologically important conformations of aminoglycoside antibiotics bound to an aminoglycoside 3′-phosphotransferase as determined by transferred nuclear Overhauser effect spectroscopy. Biochemistry. 1997;36:2353–2359. doi: 10.1021/bi9626822. [DOI] [PubMed] [Google Scholar]
- 13.Cox J R, McKay G A, Wright G D, Serpersu E H. Arrangement of substrate at the active site of an aminoglycoside antibiotic 3′-phosphotransferase as determined by NMR. J Am Chem Soc. 1996;18:1295–1301. [Google Scholar]
- 14.Craig W A. Post-antibiotic effects in experimental infection models: relationship to in-vitro phenomena and to treatment of infections in man. J Antimicrob Chemother. 1993;31(Suppl. D):149–158. doi: 10.1093/jac/31.suppl_d.149. [DOI] [PubMed] [Google Scholar]
- 15.Daigle D M, McKay G A, Wright G D. Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors. J Biol Chem. 1997;272:24755–24758. doi: 10.1074/jbc.272.40.24755. [DOI] [PubMed] [Google Scholar]
- 16.Daikos G L, Lolans V T, Jackson G G. First-exposure adaptive resistance to aminoglycoside antibiotics in vivo with meaning for optimal clinical use. Antimicrob Agents Chemother. 1991;35:117–123. doi: 10.1128/aac.35.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies J, Jimenez A. A new selective agent for eukaryotic cloning vectors. Am J Trop Med Hyg. 1980;29:1089–1092. doi: 10.4269/ajtmh.1980.29.1089. [DOI] [PubMed] [Google Scholar]
- 18.Davies J, Wright G D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 19.Dornbusch K. European study group on antibiotic resistance. In vitro susceptibility to aminoglycoside antibiotics in blood and urine isolates consecutively collected in twenty-nine European laboratories. Eur J Clin Microbiol. 1987;6:378–385. doi: 10.1007/BF02013090. [DOI] [PubMed] [Google Scholar]
- 20.Dornbusch K, Miller G H, Hare R S, Shaw K J. Resistance to aminoglycoside antibiotics in gram-negative bacilli and staphylococci isolated from blood. Report from a European collaborative study. J Antimicrob Chemother. 1990;26:131–144. doi: 10.1093/jac/26.1.131. [DOI] [PubMed] [Google Scholar]
- 21.Edgar R, Bibi E. Mdfa, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eustice D C, Wilhelm J M. Mechanisms of action of aminoglycoside antibiotics in eukaryotic protein synthesis. Antimicrob Agents Chemother. 1984;26:53–60. doi: 10.1128/aac.26.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fourmy D, Recht M I, Blanchard S C, Puglisi J D. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 24.Gates C A, Northrop B. Alternative substrate and inhibition kinetics of aminoglycoside nucleotidyltransferase 2"-I in support of a Theorell-Chance kinetic mechanism. Biochemistry. 1988;27:3826–3833. doi: 10.1021/bi00410a046. [DOI] [PubMed] [Google Scholar]
- 25.Gates C A, Northrop D B. Substrate specificities and structure-activity relationships for the nucleotidylation of antibiotics catalyzed by aminoglycoside nucleotidyltransferase 2"-I. Biochemistry. 1988;27:3820–3825. doi: 10.1021/bi00410a045. [DOI] [PubMed] [Google Scholar]
- 26.Gates C A, Northrop D B. Determination of the rate-limiting segment of aminoglycoside nucleotidyltransferase 2"-I by pH- and viscosity-dependent kinetics. Biochemistry. 1988;27:3834–3842. doi: 10.1021/bi00410a047. [DOI] [PubMed] [Google Scholar]
- 27.Georgiadis M P, Constantinou-Kokotou V. Synthesis of amino acid derivatives of neamine and 2-deoxystreptamine to be used as mutasynthons. J Carbohydr Chem. 1991;10:739–748. [Google Scholar]
- 28.Gilbert D N. Aminoglycosides. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 4th ed. New York, N.Y: Churchill Livingstone; 1995. pp. 279–306. [Google Scholar]
- 29.Gomez-Lus R, Gomez-Lus S, Goni M P, Rivera M J, Martin C, Rubio-Calvo M C. Stability of dactimicin to aminoglycoside-modifying enzymes produced by 341 bacterial clinical isolates. Drugs Exp Clin Res. 1989;15:129–132. [PubMed] [Google Scholar]
- 30.Halaban R, Alfano F D. Selective elimination of fibroblasts from cultures of normal human melanocytes. In Vitro. 1984;20:447–450. doi: 10.1007/BF02619590. [DOI] [PubMed] [Google Scholar]
- 31.Hancock R E W. Aminoglycoside uptake and mode of action—with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J Antimicrob Chemother. 1981;8:249–276. doi: 10.1093/jac/8.4.249. [DOI] [PubMed] [Google Scholar]
- 32.Hancock R E, Farmer S W, Li Z S, Poole K. Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1309–1314. doi: 10.1128/aac.35.7.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi S F, Norcia L J, Seibel S B, Silvia A M. Structure-activity relationships of hygromycin A and its analogs: protein synthesis inhibition activity in a cell free system. J Antibiot (Tokyo) 1997;50:514–521. doi: 10.7164/antibiotics.50.514. [DOI] [PubMed] [Google Scholar]
- 34.Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Isoquinolinesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- 35.Hildebrandt J, Loibner H, Schütze E, Streicher W, Stütz P, Wenzel A. Program and abstracts of the 24th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1984. Synthesis and biological properties of the aminoglycoside 87-351, a new standard in overcoming enzyme-mediated aminoglycoside resistance, abstr. 310. [Google Scholar]
- 36.Holm S E, Hill B, Lowestad A, Maller R, Vikefors A. A prospective, randomized study of amikacin and gentamicin in serious infections with focus on efficacy, toxicity and duration of serum levels above the MIC. J Antimicrob Chemother. 1983;12:393–402. doi: 10.1093/jac/12.4.393. [DOI] [PubMed] [Google Scholar]
- 37.Hon W C, McKay G A, Thompson P R, Sweet R M, Yang D S, Wright G D, Berghuis A M. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell. 1997;89:887–895. doi: 10.1016/s0092-8674(00)80274-3. [DOI] [PubMed] [Google Scholar]
- 38.Hotta K, Zhu C B, Ogata T, Sunada A, Ishikawa J, Mizuno S, Kondo S. Enzymatic 2′-N-acetylation of arbekacin and antibiotic activity of its product. J Antibiot (Tokyo) 1996;49:458–464. doi: 10.7164/antibiotics.49.458. [DOI] [PubMed] [Google Scholar]
- 39.Inoue M, Nonoyama M, Okamoto R, Ida T. Antimicrobial activity of arbekacin, a new aminoglycoside antibiotic, against methicillin-resistant Staphylococcus aureus. Drugs Exp Clin Res. 1994;20:233–239. [PubMed] [Google Scholar]
- 40.Inouye Y, Hashimoto M, Sugiyama M, Takesue Y, Santo T, Yokoyama T. Susceptibility of methicillin-resistant Staphylococcus aureus clinical isolates to various antimicrobial agents. IV. Aminoglycoside-modifying enzyme AAC(6′)-APH(2") is responsible for arbekacin-resistance enhanced by bleomycin. Hiroshima J Med Sci. 1994;43:87–92. [PubMed] [Google Scholar]
- 41.Johnson A P, Malde M, Woodford N, Cunney R J, Smyth E G. Urinary isolates of apramycin-resistant Escherichia coli and Klebsiella pneumoniae from Dublin. Epidemiol Infect. 1995;114:105–112. doi: 10.1017/s0950268800051955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karlowsky J A, Hoban D J, Zelenitsky S A, Zhanel G G. Altered denA and anr gene expression in aminoglycoside adaptive resistance in Pseudomonas aeruginosa. J Antimicrob Chemother. 1997;40:371–376. doi: 10.1093/jac/40.3.371. [DOI] [PubMed] [Google Scholar]
- 43.Kawaguchi H, Naito T, Nakagawa S, Fujisawa K. BB-K 8, a new semisynthetic aminoglycoside. J Antibiot (Tokyo) 1972;25:695–708. doi: 10.7164/antibiotics.25.695. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi Y, Tsuchiya T. Synthesis of 2"-oxidized derivatives of 5-deoxy-5-epi-5-fluoro-dibekacin and -arbekacin, and study on structure-chemical shift relationships of urethane (or amide)-type NH protons in synthetic intermediates. Carbohydr Res. 1997;298:261–277. doi: 10.1016/s0008-6215(96)00318-7. [DOI] [PubMed] [Google Scholar]
- 45.Kondo S, Jinuma K, Yamamoto H, Maeda K, Umezawa H. Syntheses of 1-N-(S)-4-amino-2-hydroxybutyryl-kanamycin B and-3′,4′-dideoxykanamycin B active against kanamycin-resistant bacteria. J Antibiot (Tokyo) 1973;26:412–415. doi: 10.7164/antibiotics.26.412. [DOI] [PubMed] [Google Scholar]
- 46.Kondo S, Shibahara S, Usui T, Kudo T, Tamura A, Gomi S, Takeuchi T. New 2"-amino derivatives of arbekacin, potent aminoglycoside antibiotics against methicillin-resistant Staphylococcus aureus. J Antibiot (Tokyo) 1993;46:531–534. doi: 10.7164/antibiotics.46.531. [DOI] [PubMed] [Google Scholar]
- 47.Kondo S, Tamura A, Gomi S, Ikeda Y, Takeuchi T, Mitsuhashi S. Structures of enzymatically modified products of arbekacin by methicillin-resistant Staphylococcus aureus. J Antibiot (Tokyo) 1993;46:310–315. doi: 10.7164/antibiotics.46.310. [DOI] [PubMed] [Google Scholar]
- 48.Kotretsou S, Mingeot-Leclercq M-P, Constantinou-Kokotou V, Brasseur R, Georgiadis M P, Tulkens P M. Synthesis and antimicrobial and toxicological studies of amino acid and peptide derivatives of kanamycin A and netilmicin. J Med Chem. 1995;38:4710–4719. doi: 10.1021/jm00023a011. [DOI] [PubMed] [Google Scholar]
- 49.Kurebe M, Yokota M, Niizato T, Kai F, Yoshida T, Okamoto R, Mitsuhashi S. Antibacterial activity and ototoxicity in guinea pigs, and nephrotoxicity in rats of arbekacin. Arzneim-Forsch. 1986;36:1511–1517. [PubMed] [Google Scholar]
- 50.Kuwahara R, Tsuchiya T. Synthesis of dibekacin analogs containing 3-oxa- and 3-aza-2,3,4,-trideoxy-d-glycero-hexopyranose. Carbohydr Res. 1996;293:15–20. doi: 10.1016/0008-6215(96)00176-0. [DOI] [PubMed] [Google Scholar]
- 50a.Kuwahara R, Tsuchiya T. Synthesis of 2′-acylamido derivatives of 2′-amino-5,2′-dideoxy-5-epi-5-fluorodibekacin and a study on the structures of 5-fluorinated dibekacin analogs by 13C NMR. Carbohydr Res. 1997;299:271–279. [Google Scholar]
- 51.Li K, Fernandez-Saiz M, Rigl C T, Kumar A, Ragunathan K G, McConnaughie A W, Wilson W D. Design and analysis of molecular motifs for specific recognition of RNA. Bioorg Med Chem. 1997;5:1157–1172. doi: 10.1016/s0968-0896(97)00054-0. [DOI] [PubMed] [Google Scholar]
- 52.Loebenberg D, Counelis M, Waitz J A. Antibiotic G-418, a new Micromonospora-produced aminoglycoside with activity against protozoa and helminths: antiparasitic activity. Antimicrob Agents Chemother. 1975;7:811–815. doi: 10.1128/aac.7.6.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loibner, H., W. Streicher, and P. Stütz. 5 March 1985. European patent application US1982000404817.
- 54.Lynch C, Courvalin P, Nikaido H. Active efflux of antimicrobial agents in wild-type strains of Enterococci. Antimicrob Agents Chemother. 1997;41:869–871. doi: 10.1128/aac.41.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacArthur R D, Lolans V T, Zar F A, Jackson G G. Biphasic, concentration-dependent and rate-limited, concentration-independent bacterial killing by an aminoglycoside antibiotic. J Infect Dis. 1984;150:778–779. doi: 10.1093/infdis/150.5.778. [DOI] [PubMed] [Google Scholar]
- 56.McKay G A, Wright G D. Kinetic mechanism of aminoglycoside phosphotransferase type IIIa. J Biol Chem. 1995;270:24686–24692. doi: 10.1074/jbc.270.42.24686. [DOI] [PubMed] [Google Scholar]
- 57.McKay G A, Robinson R A, Lane W S, Wright G D. Active-site labeling of an aminoglycoside antibiotic phosphotransferase APH(3′)-IIIa. Biochemistry. 1994;33:14115–14120. doi: 10.1021/bi00251a021. [DOI] [PubMed] [Google Scholar]
- 58.McKay G A, Roestamadji J, Mobashery S, Wright G D. Recognition of aminoglycoside antibiotics by enterococcal-staphylococcal aminoglycoside 3′-phosphotransferase type IIIa: role of substrate amino groups. Antimicrob Agents Chemother. 1996;40:2648–2650. doi: 10.1128/aac.40.11.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKay G A, Thompson P R, Wright G D. Broad spectrum aminoglycoside phosphotransferase type III from Enterococcus: overexpression, purification and substrate specificity. Biochemistry. 1994;33:6936–6944. doi: 10.1021/bi00188a024. [DOI] [PubMed] [Google Scholar]
- 60.Melancon P, Tapprich W E, Brakier-Gingras L. Single-base mutations at position 2661 of Escherichia coli 23S rRNA increase efficiency of translational proofreading. J Bacteriol. 1992;174:7896–7901. doi: 10.1128/jb.174.24.7896-7901.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menard R, Molinas C, Arthur M, Duval J, Courvalin P, Leclercq R. Overproduction of 3′-aminoglycoside phosphotransferase type I confers resistance to tobramycin in Escherichia coli. Antimicrob Agents Chemother. 1993;37:78–83. doi: 10.1128/aac.37.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miller G H, Sabatelli F J, Hare R S, Glupczynski Y. The most frequent aminoglycoside resistance mechanism—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Clin Infect Dis. 1997;24:S46–S62. doi: 10.1093/clinids/24.supplement_1.s46. [DOI] [PubMed] [Google Scholar]
- 63.Miller G H, Sabatelli F J, Naples L, Hare R S, Shaw K J. The changing nature of aminoglycoside resistance mechanisms and the role of isepamicin—a new broad-spectrum aminoglycoside. J Chemother. 1995;7:31–44. [PubMed] [Google Scholar]
- 64.Miller G H, Sabatelli F J, Naples L, Hare R S, Shaw K J. The most frequently occurring aminoglycoside resistance mechanisms—combined results of surveys in eight regions of the world. J Chemother. 1995;7:17–30. [PubMed] [Google Scholar]
- 64a.Miller, G. H. Personal communication.
- 65.Mingeot-Leclercq M P, Tulkens P M. Aminoglycosides: Nephrotoxicity. 1999. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitsuhashi S, Kawade H. Aminoglycoside antibiotic resistance in bacteria. In: Whelton A, Neu H C, editors. The aminoglycosides. Microbiology, clinical use and toxicity. New York, N.Y: Marcel Dekker, Inc.; 1982. pp. 97–122. [Google Scholar]
- 67.Miyaguchi H, Narita H, Sakamoto K, Yokoyama S. An antibiotic-binding motif of an RNA fragment derived from the A-site-related region of Escherichia coli 16S rRNA. Nucleic Acids Res. 1996;24:3700–3706. doi: 10.1093/nar/24.19.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyake T, Tsuchiya T, Umezawa S, Umezawa H. Synthesis of 3′,4′-dideoxykanamycin B. Carbohydr Res. 1976;49:141–151. doi: 10.1016/s0008-6215(00)83132-8. [DOI] [PubMed] [Google Scholar]
- 69.Moellering R C., Jr . Clinical microbiology and the in vitro activity of aminoglycosides. In: Whelton A, Neu H C, editors. The aminoglycosides. New York, N.Y: Marcel Dekker, Inc.; 1982. pp. 65–95. [Google Scholar]
- 70.Moore R D, Lietman P S, Smith C R. Clinical response to aminoglycoside therapy: importance of the ratio of peak concentration to minimal inhibitory concentration. J Infect Dis. 1987;155:93–99. doi: 10.1093/infdis/155.1.93. [DOI] [PubMed] [Google Scholar]
- 71.Murray B E. New aspects of antimicrobial resistance and the resulting therapeutic dilemmas. J Infect Dis. 1991;163:1185–1194. [PubMed] [Google Scholar]
- 72.Nagabhushan T L, Cooper A B, Tsai H, Daniels P J L, Miller G H. The syntheses and biological properties of 1-N-((S)-4-amino-2-hydroxybutyryl)-gentamicin B and 1-N-((S)-3-amino-2-hydroxypropionyl)-gentamicin B. J Antibiot (Tokyo) 1978;31:681–687. doi: 10.7164/antibiotics.31.681. [DOI] [PubMed] [Google Scholar]
- 73.Nagabhushan T L, Miller G H, Weinstein M J. Structure-activity relationships in aminoglycoside-aminocyclitol antibiotics. In: Whelton A, Neu H C, editors. The aminoglycosides. New York, N.Y: Marcel Dekker, Inc.; 1982. pp. 3–27. [Google Scholar]
- 74.Naito T, Nakagawa S, Abe Y, Toda S, Fujisawa K, Miyaki T, Kawaguchi H. Aminoglycoside antibiotics. II. Configurational and positional isomers of BB-K 8. J Antibiot (Tokyo) 1973;26:297–301. [PubMed] [Google Scholar]
- 75.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohba K, Tsuruoka T, Mizutani K, Kato N, Omoto S, Ezaki N, Watanabe K. Studies on a new aminoglycoside antibiotic, dactimicin. II. Isolation, structure and chemical degradation. J Antibiot (Tokyo) 1981;34:1090–1100. doi: 10.7164/antibiotics.34.1090. [DOI] [PubMed] [Google Scholar]
- 77.Okachi R, Kawamoto I, Takasawa S, Yamamoto M, Sato S, Sato T, Nara T. A new antibiotic XK-62-2 (sagamicin). I. Isolation, physicochemical and antibacterial properties. J Antibiot (Tokyo) 1974;27:793–800. doi: 10.7164/antibiotics.27.793. [DOI] [PubMed] [Google Scholar]
- 78.Pearson N D, Prescott C D. RNA as a drug target. Chem Biol. 1997;4:409–414. doi: 10.1016/s1074-5521(97)90192-7. [DOI] [PubMed] [Google Scholar]
- 79.Pedersen L C, Benning M M, Holden H M. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry. 1995;34:13305–13311. doi: 10.1021/bi00041a005. [DOI] [PubMed] [Google Scholar]
- 80.Price K E. Aminoglycoside research 1975–1985: prospects for development of improved agents. Antimicrob Agents Chemother. 1986;29:543–548. doi: 10.1128/aac.29.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Price K E, Godfrey J C, Kawaguchi H. Effect of structural modifications on the biological properties of aminoglycoside antibiotics containing 2-deoxystreptamine. In: Perlman D, editor. Structure-activity relationships among the semi-synthetic antibiotics. New York, N.Y: Academic Press; 1977. pp. 239–355. [Google Scholar]
- 82.Price K E, Kresel P A, Farchione L A, Siskin S B, Karpow S A. Epidemiological studies of aminoglycoside resistance in the USA. J Antimicrob Chemother. 1981;8(Suppl. A):89–105. doi: 10.1093/jac/8.suppl_a.89. [DOI] [PubMed] [Google Scholar]
- 83.Rather P N, Munayyer H, Mann P A, Hare R S, Miller G H, Shaw K J. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase-Ib and IIa proteins. J Bacteriol. 1992;174:3196–3203. doi: 10.1128/jb.174.10.3196-3203.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rather P N, Parojcic M M, Paradise M R. An extracellular factor regulating expression of the chromosomal aminoglycoside 2′-N-acetyltransferase of Providencia stuartii. Antimicrob Agents Chemother. 1997;41:1749–1754. doi: 10.1128/aac.41.8.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roestamadji J, Grapsas I, Mobashery S. Mechanism-based inactivation of bacterial aminoglycoside 3′-phosphotransferases. J Am Chem Soc. 1995;117:80–84. [Google Scholar]
- 86.Roestamadji J, Grapsas I, Mobashery S. Loss of individual electrostatic interactions between aminoglycoside antibiotics and resistance enzymes as an effective means to overcoming bacterial drug resistance. J Am Chem Soc. 1995;117:11060–11069. [Google Scholar]
- 87.Sakon J, Liao H H, Kanikula A M, Benning M M, Rayment I, Holden H M. Molecular structure of kanamycin nucleotidyltransferase determined to 3.0-Å resolution. Biochemistry. 1993;32:11977–11984. doi: 10.1021/bi00096a006. [DOI] [PubMed] [Google Scholar]
- 88.Santerre R F, Allen N E, Hobbs J N, Rao R N, Schmidt R J. Expression of prokaryotic genes for hygromycin B and G418 resistance as dominant-selection markers in mouse L cells. Gene. 1984;30:147–156. doi: 10.1016/0378-1119(84)90115-x. [DOI] [PubMed] [Google Scholar]
- 88a.Schindler, J. 26 July 1997, posting date. Aminoglycoside-inactivating enzymes. [Online.] http://www.warn.cas.cz. [26 February 1999, last date accessed.]
- 89.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shimizu K, Kumada T, Hare R S, Miller G H, Sabatelli F J, Howard G. Comparison of aminoglycoside resistance patterns in Japan, Formosa, and Korea, Chile and the United States. Antimicrob Agents Chemother. 1985;28:282–288. doi: 10.1128/aac.28.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shitara T, Kobayashi Y, Tsuchiya T, Umezawa S. Synthesis of 5-deoxy-5-fluoro and 5-deoxy-5,5-difluoro derivatives of kanamycin B and its analogs. Study on structure-toxicity relationships. Carbohydr Res. 1992;232:273–290. doi: 10.1016/0008-6215(92)80060-e. [DOI] [PubMed] [Google Scholar]
- 92.Siregar J J, Lerner S A, Mobashery S. Purification and characterization of aminoglycoside 3′-phosphotransferase type IIa and kinetic comparison with a new mutant enzyme. Antimicrob Agents Chemother. 1994;38:641–647. doi: 10.1128/aac.38.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Siregar J J, Miroshnikov K, Mobashery S. Purification, characterization, and investigation of the mechanism of aminoglycoside 3′-phosphotransferase type Ia. Biochemistry. 1995;34:12681–12688. doi: 10.1021/bi00039a026. [DOI] [PubMed] [Google Scholar]
- 93a.Stütz, P. Personal communication.
- 94.Takahashi Y, Tsuchiya T, Umezawa S, Umezawa H. Synthesis of 3′-deoxy-3′-fluorokanamycin A and 3′,4′-dideoxy-3′-fluorokanamycin A. Carbohydr Res. 1991;210:221–232. doi: 10.1016/0008-6215(91)80124-6. [DOI] [PubMed] [Google Scholar]
- 95.Takahashi Y, Tsuneda S, Tsuchiya T, Koyama Y, Umezawa S. Synthesis of 4′-deoxy-4′-fluorokanamycin A and B. Carbohydr Res. 1992;232:89–105. doi: 10.1016/s0008-6215(00)90996-0. [DOI] [PubMed] [Google Scholar]
- 96.Takano M, Okuda M, Yasuhara M, Hori R. Cellular toxicity of aminoglycoside antibiotics in G418-sensitive and -resistant LLC-PK1 cells. Pharm Res. 1994;11:609–615. doi: 10.1023/a:1018999423464. [DOI] [PubMed] [Google Scholar]
- 97.Tanaka N, Matsunaga K, Hirata A, Matsuhisa Y, Nishimura T. Mechanism of action of habekacin, a novel amino acid-containing aminoglycoside antibiotic. Antimicrob Agents Chemother. 1983;24:797–802. doi: 10.1128/aac.24.5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thompson P R, Hughes D W, Wright G D. Mechanism of aminoglycoside 3′-phosphotransferase type IIIa: HIS188 is not a phosphate accepting residue. Chem Biol. 1996;3:747–755. doi: 10.1016/s1074-5521(96)90251-3. [DOI] [PubMed] [Google Scholar]
- 99.Tsuchiya T, Shitara T, Umezawa S, Takeuchi T, Hamada M, Tomono N, Umemura E. Synthesis of low-toxicity, 5-deoxy-5-fluoro and 5-deoxy-5,5-difluoro derivatives of arbekacin and its analogs, and study of structure-toxicity relationships. Carbohydr Res. 1993;240:307–312. doi: 10.1016/0008-6215(93)84194-b. [DOI] [PubMed] [Google Scholar]
- 100.Tsuchiya T, Takahashi Y, Kobayashi Y, Umezawa S, Umezawa H. Synthesis of 3′-deoxy-3′-fluorokanamycins A and B active against resistant bacteria. J Antibiot (Tokyo) 1985;38:1287–1290. doi: 10.7164/antibiotics.38.1287. [DOI] [PubMed] [Google Scholar]
- 101.Tsuchiya T, Takahashi Y, Kobayashi Y, Umezawa S, Umezawa H. Synthesis of 2′,3′-dideoxy-2′-fluorokanamycin A. J Carbohydr Chem. 1985;4:587–611. [Google Scholar]
- 102.Tsukiura H, Fujisawa K, Konishi M, Saito K, Numata K, Ishikawa H, Kawaguchi H. Aminoglycoside antibiotics. III. Bio-active degradation products from butirosins and semi-synthesis of butirosin analogs. J Antibiot (Tokyo) 1973;26:351–357. [Google Scholar]
- 103.Tulkens P, Trouet A. The uptake and intracellular accumulation of aminoglycoside antibiotics in lysosomes of cultured rat fibroblasts. Biochem Pharmacol. 1978;27:415–424. doi: 10.1016/0006-2952(78)90370-2. [DOI] [PubMed] [Google Scholar]
- 104.Ubukata K, Yamashita N, Gotoh A, Konno M. Purification and characterization of aminoglycoside-modifying enzymes from Staphylococcus aureus and staphylococcus epidermidis. Antimicrob Agents Chemother. 1984;25:754–759. doi: 10.1128/aac.25.6.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Umemura E, Tsuchiya T, Kobayashi Y, Tanaka K. A synthetic study of methyl 3-deoxy-3-fluoro-alpha-d-glucopyranosides from methyl 2,3-anhydro-alpha-d-allopyranosides, and synthesis of 3′-deoxy-3′-fluorokanamycin A and 3′-chloro-3′-deoxykanamycin A. Carbohydr Res. 1992;224:141–163. doi: 10.1016/0008-6215(92)84101-w. [DOI] [PubMed] [Google Scholar]
- 106.Umezawa H, Umezawa S, Tsuchiya T, Okazaki Y. 3′,4′-dideoxykanamycin B active against kanamycin resistant Escherichia coli and Pseudomonas aeruginosa. J Antibiot (Tokyo) 1971;24:485–487. doi: 10.7164/antibiotics.24.485. [DOI] [PubMed] [Google Scholar]
- 107.Umezawa S, Tsuchiya T, Muto R, Nishimura Y, Umezawa H. Synthesis of 3′-deoxykanamycin effective against kanamycin resistant Escherichia coli and Pseudomonas aeruginosa. J Antibiot (Tokyo) 1971;24:274–275. doi: 10.7164/antibiotics.24.274. [DOI] [PubMed] [Google Scholar]
- 108.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Schepdael A, Busson R, Vanderhaeghe H J, Verbist L, Claes P J, Mingeot-Leclercq M P, Tulkens P M. Synthesis, antibacterial activity and toxicological evaluation (in vitro and computer-aided) of 1-C-hydroxymethyl kanamycin B and 1-C-hydroxymethyl, 6" chlorokanamycin B. J Med Chem. 1991;34:1483–1492. doi: 10.1021/jm00108a037. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Hamasaki K, Rando R R. Specificity of aminoglycoside binding to RNA constructs derived from the 16S rRNA decoding region and the HIV-RRE activator region. Biochemistry. 1997;36:768–779. doi: 10.1021/bi962095g. [DOI] [PubMed] [Google Scholar]
- 111.Woo P W K, Dion W, Bartz Q R. Tetrahedron Lett. 1971. Butirosins A and B, aminoglycoside antibiotics. III. Structures; pp. 2625–2628. [Google Scholar]
- 112.Wright G D, Ladak P. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob Agents Chemother. 1997;41:956–960. doi: 10.1128/aac.41.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu H Y, Miller G H, Blanco M G, Hare R S, Shaw K J. Cloning and characterization of an aminoglycoside 6′-N-acetyltransferase gene from Citrobacter freundii which confers an altered resistance profile. Antimicrob Agents Chemother. 1997;41:2439–2447. doi: 10.1128/aac.41.11.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiong Y Q, Caillon J, Drugeon H, Potel G, Baron D. Influence of pH on adaptive resistance of Pseudomonas aeruginosa to aminoglycosides and their postantibiotic effects. Antimicrob Agents Chemother. 1996;40:35–39. doi: 10.1128/aac.40.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xiong Y-Q, Caillon J, Kergueris M F, Drugeon H, Baron D, Potel G, Bayer A S. Adaptive resistance of Pseudomonas aeruginosa induced by aminoglycosides and killing kinetics in a rabbit endocarditis model. Antimicrob Agents Chemother. 1997;41:823–826. doi: 10.1128/aac.41.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zapp M L, Stern S, Green M R. Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell. 1993;74:969–978. doi: 10.1016/0092-8674(93)90720-b. [DOI] [PubMed] [Google Scholar]
- 117.Zhanel G G, Craig W A. Pharmacokinetic contributions to postantibiotic effects. Focus on aminoglycosides. Clin Pharmacokinet. 1994;27:377–399. doi: 10.2165/00003088-199427050-00005. [DOI] [PubMed] [Google Scholar]