SUMMARY
Lysosomes promote cellular homeostasis through macromolecular hydrolysis within their lumen and metabolic signaling by the mTORC1 kinase on their limiting membranes. Both hydrolytic and signaling functions require precise regulation of lysosomal cholesterol content. In Niemann-Pick type C (NPC), loss of the cholesterol exporter, NPC1, causes cholesterol accumulation within lysosomes, leading to mTORC1 hyperactivation, disrupted mitochondrial function and neurodegeneration. The compositional and functional alterations in NPC lysosomes, and how aberrant cholesterol-mTORC1 signaling contributes to organelle pathogenesis are not understood. Through proteomic profiling of NPC lysosomes, we find pronounced proteolytic impairment compounded with hydrolase depletion, enhanced membrane damage and defective mitophagy. Genetic and pharmacologic mTORC1 inhibition restores lysosomal proteolysis without correcting cholesterol storage, implicating aberrant mTORC1 as a pathogenic driver downstream of cholesterol accumulation. Consistently, mTORC1 inhibition ameliorates mitochondrial dysfunction in a neuronal model of NPC. Thus, cholesterol-mTORC1 signaling controls organelle homeostasis and is a targetable pathway in NPC.
eTOC Blurb:
Niemann-Pick type C is a devastating neurodegenerative disease caused by cholesterol buildup in lysosomes. Through organelle proteomics, Davis et al identify degradative and structural defects of NPC lysosomes. Aberrant mTORC1 signaling drives lysosomal dysfunction downstream of cholesterol accumulation, and chemical and genetic mTORC1 inhibition restores organelle homeostasis in NPC cells.
Graphical Abstract

INTRODUCTION
Lysosomes are key degradative organelles for macromolecular turnover and recycling of cellular building blocks, including lipids. Through autophagy, lysosomes also help detoxify potentially harmful cellular components, such as damaged mitochondria. Along with these catabolic roles, lysosomes have been recently recognized as signaling compartments that support the activity of the master growth regulator, mechanistic Target of Rapamycin Complex 1 (mTORC1) (Liu and Sabatini, 2020; Perera and Zoncu, 2016).
The degradative and signaling roles of lysosomes are highly integrated. Intracellular nutrients drive mTORC1 translocation from the cytosol to the lysosomal limiting membrane, where growth factor signals relayed by the phosphatidylinositol 3-kinase (PI3K)-AKT pathway trigger mTORC1 kinase function (Castellano et al., 2017; Menon et al., 2014; Sancak et al., 2010; Wyant et al., 2017; Zoncu et al., 2011). In turn, active mTORC1 suppresses initiation of autophagy, steering the cell toward net mass accumulation (Düvel et al., 2010; Kim et al., 2011; Puente et al., 2016; Settembre et al., 2012). The tight integration of lysosomal catabolism and signaling suggests that, in diseases driven by lysosomal dysfunction, their aberrant cross-talk may synergize to disrupt cellular homeostasis (Perera and Zoncu, 2016).
Niemann-Pick type C (NPC) is one of a family of approximately 60 diseases known as lysosomal storage disorders (LSDs), in which genetic inactivation of lysosomal hydrolases or transporters triggers massive and pathogenic accumulation of their respective substrates within the lysosome (Ballabio and Gieselmann, 2009; Platt et al., 2018). NPC is triggered by inactivating mutations in NPC1, a polytopic transmembrane cholesterol transporter located on the lysosomal limiting membrane (Gong et al., 2016; Kwon et al., 2009; Li et al., 2016; Winkler et al., 2019). In conjunction with NPC2, a cholesterol-binding protein of the lysosomal lumen, NPC1 exports cholesterol released from Low-Density Lipoprotein (LDL) to acceptor compartments such as the endoplasmic reticulum (ER), the Golgi and the plasma membrane (Feltes et al., 2020; Infante and Radhakrishnan, 2017; Infante et al., 2008; Pfeffer, 2019).
Heritable mutations in the NPC1 gene lead to an unstable NPC1 protein, which is degraded in the ER (Schultz et al., 2018). In cells lacking NPC1, cholesterol accumulates massively within the lysosomal lumen, as well as on its limiting membrane. Cholesterol storage results in enlarged lysosomes that exhibit morphological, trafficking and functional defects. Moreover, the primary lysosomal phenotype is accompanied by dysfunction in other cellular compartments, including autophagosomes (Elrick et al., 2012; Ordonez et al., 2012; Sarkar et al., 2013), mitochondria (Kennedy et al., 2014; Ordonez, 2012; Yambire et al., 2019a; Yu et al., 2005) and peroxisomes (Schedin et al., 1997).
Mechanistically, how cholesterol accumulation caused by loss of NPC1 leads to lysosomal dysfunction remains poorly understood. There is evidence of defective degradation of autophagosomal cargo (Elrick et al., 2012; Ordonez et al., 2012), as well as defective trafficking of autophagic vesicles to lysosomes (Sarkar et al., 2013). However, the mechanistic basis for impaired lysosomal catabolism remains to be fully elucidated. Moreover, the molecular processes that connect the primary lysosomal dysfunction to the impairment of other organelle populations remain largely mysterious.
Limiting our understanding of the pathogenic processes that drive NPC is the lack of a comprehensive view of the structural and functional alterations that occur in NPC-defective lysosomes. The recent development of techniques for rapid immunoisolation and mass spectrometry-based profiling of intact organelles presents with an opportunity to address this critical point (Abu-Remaileh et al., 2017; Castellano et al., 2017; Sleat et al., 2013; Wyant et al., 2017; Zoncu et al., 2011).
An especially important question is whether disruption of lysosomal signaling pathways may contribute to the multi-organellar defects of NPC cells. Cholesterol was recently identified as a nutrient input that promotes mTORC1 activation at the lysosome. Cholesterol stimulates mTORC1 activity via a protein complex also involved in amino acid-dependent mTORC1 activation: the heterodimeric Rag guanosine triphosphatases (GTPases), their membrane anchor Ragulator, and the lysosomal amino acid permease SLC38A9 (Castellano et al., 2017; Sancak et al., 2010; Wang et al., 2015; Wyant et al., 2017). This complex responds to both extracellular cholesterol, carried by low-density lipoprotein (LDL) and to intracellular cholesterol transferred across ER-lysosome contacts by oxysterol binding protein (OSBP) and its ER anchors, VAPA and B (Castellano et al., 2017; Dong et al., 2016; Lim et al., 2019; Mesmin et al., 2013).
In contrast to these positive activators, NPC1 appears to antagonize cholesterol-dependent mTORC1 signaling. In NPC1-defective cells, mTORC1 signaling is elevated compared to wild-type cells and is immune to inhibition by cholesterol-depleting agents. mTORC1 dysregulation may result from massive accumulation of lysosomal cholesterol resulting from loss of NPC1-dependent export. NPC1 may also function as a sensor, regulating the mTORC1-scaffolding complex via cholesterol-dependent physical interaction (Castellano et al., 2017).
mTORC1 signaling has profound effects on organelle composition and function. mTORC1 inhibits the master regulators of lysosomal biogenesis and autophagy, MiT-TFE transcription factors (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Sardiello et al., 2009; Settembre et al., 2012). mTORC1 drives mitochondrial biogenesis and function at the transcriptional and translational level (Bentzinger et al., 2008; Cunningham et al., 2007; Morita et al., 2013), and stimulates mitochondrial programs that support cell growth and proliferation (Ben-Sahra et al., 2016; Csibi et al., 2013). Thus, dysregulation of mTORC1 signaling due to loss of NPC1 could affect organelle homeostasis via multiple mechanisms.
To shed light into the relationship between dysregulated mTORC1 signaling and organelle homeostasis in NPC, we carried out mass spectrometry-based profiling of immunopurified lysosomes combined with functional assays for mTORC1 signaling and lysosome function in both engineered cell lines and in iPSC-derived neuronal cultures lacking NPC1. We uncover a more profound disruption of lysosomal composition and function, mitochondrial homeostasis and cholesterol-mTORC1 signaling associated with NPC than previously anticipated. Importantly, pharmacological suppression of mTORC1 signaling was able to correct several aspects of organelle function independent of the primary cholesterol storage defect, thus implicating dysregulated mTORC1 signaling as an important pathogenic driver in Niemann-Pick type C.
RESULTS
NPC1-null lysosomes display extensive proteolytic defects
To investigate the molecular basis for lysosomal dysfunction in NPC, we conducted lysosome immunoprecipitation (lyso-IP) followed by label-free proteomics-based profiling from HEK-293T cells that either have intact NPC1 function or in which the NPC1 gene was targeted using CRISPR/Cas9, resulting in near-complete (~93%) loss of NPC1 protein (Table S1) and establishment of an NPC-like phenotype characterized by massive cholesterol accumulation in lysosomes (Figure S1A) (Abu-Remaileh et al., 2017; Castellano et al., 2017; Lim et al., 2019; Wyant et al., 2017; Zoncu et al., 2011). To identify cargo proteins being actively degraded, additional samples were treated with the broad-spectrum hydrolase inhibitors, leupeptin and pepstatin (Figure S1B and S1C). In lysosomes purified from control (‘sgNT’) cells, the abundance of resident lysosomal proteins (hydrolases, permeases, signaling components) remained unchanged. In contrast, lysosomal substrates were enriched 2-fold or more in the leupeptin:pepstatin-treated samples (Figure 1A). These included autophagic adaptors, delivered to the lysosome via fusion with autophagosomes (Khaminets et al., 2016). Unlike lysosomes from control cells, NPC1-defective lysosomes (‘sgNPC1’) showed marked accumulation of numerous bona fide substrates in the non-leupeptin:pepstatin condition, suggesting defective proteolysis in NPC lysosomes (Figure 1B and 1C).
Figure 1. NPC1-deficient lysosomes have reduced degradative capacity relative to sgNT lysosomes.
(A-C) Proteomic analysis of Lyso-IP samples from sgNT and sgNPC1 293Ts. Volcano plots of the ratio of leupeptin+pepstatin(“L+P”) to untreated (DMSO) for sgNT (A) and sgNPC1 (B) 293Ts. Proteins with statistically significant (p-value≥0.5, two-tailed unpaired t-test) fold change L+P/DMSO>2 (“sgNT cargo”) in (A) are displayed as red circles. The sgNT cargo proteins identified in (A) are also displayed as red circles in (B). (C) Average peptide counts (raw) for selected autophagy-related proteins. Data are averaged from three independent biological replicates for all conditions except DMSO-treated sgNPC1 samples which are from two biological replicates.
(D) Immunoblots of Lyso-IP samples and corresponding post-nuclear supernatant (PNS) from sgNT or sgNPC1 293Ts treated with leupeptin and pepstatin for 24h.
(E and F) sgNT and sgNPC1 293Ts were treated with bafilomycin A1 (BafA1) for 4h before immunostaining for LC3B and LAMP2. (E) Representative confocal micrographs. (F) Quantification of LC3B and LAMP2 co-localization from 10 non-overlapping fields containing at least 3 cells per field; ****P<0.0001, ns=not significant, ANOVA with Tukey’s multiple comparisons test.
(G and H) sgNT and sgNPC1 293Ts expressing the indicated NPC1-FLAG cDNA were immunostained for LC3B and LAMP2. (I) Representative confocal micrographs. (J) Quantification of LC3B and LAMP2 co-localization from 10 non-overlapping fields containing at least 3 cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
(I) Volcano plot of Lyso-IP proteomic data (from A-C) for the ratio of untreated sgNT/sgNPC1 LAMP1-normalized peptide counts. Proteins that are classified by GO:0005775 “vacuolar lumen” are depicted as orange circles. Statistical analysis was performed using two-tailed unpaired t-test.
(J) Relative galactosylceramidase (GALC) activity in sgNT or sgNPC1 293Ts from cells labeled with a GALC activity probe (GalGreen). GalGreen fluorescence is normalized by total lysosomal content, as determined by LysoTracker Red fluorescence; nine independent replicate measurements were performed per condition; ***P=0.0004, two-tailed unpaired t-test.
All bar graphs depict mean ± SD. Scale bars are 10μm. See also Figure S1–S3 and Supplemental Table 1.
Direct immunoblotting of lysosomal immunoprecipitates from both control and sgNPC1 HEK293T cells confirmed that the steady-state levels of several autophagic adaptor proteins, including LC3, p62/SQSTM1, TAX1BP1 and NCOA4 were significantly elevated in NPC1-null lysosomes compared to control lysosomes (Figure 1D, S1D). Blocking lysosomal proteolysis with leupeptin:pepstatin or with the vacuolar H+ATPase (v-ATPase) inhibitor, bafilomycin A1 (BafA1) caused further buildup of autophagic substrates in both genetic backgrounds, however this buildup was significantly reduced in NPC1-null lysosomes (Figure 1D, S1D). Immunoblotting of lysosomal immunoprecipitates from NPC1-knock out mouse embryonic fibroblasts (MEFs) also confirmed increased amounts of undigested autophagic substrates (Figure S1E).
We further confirmed accumulation of undigested autophagic material in NPC1-defective lysosomes via immunofluorescence staining for endogenous LC3B and LAMP2, which showed strong LC3B signal within the lumen of LAMP2-positive vesicles in sgNPC1 but not sgNT cells (Figure 1E–1F, and S1F–S1G). Consistent with defective proteolysis, BafA1 treatment caused a 5-fold increase of LC3B signal in control lysosomes, but no significant change in NPC1-defective lysosomes (Figure 1E–1F).
Accumulation of undigested cargo in NPC lysosomes was dependent on autophagic flux. In cells deleted for the essential autophagic regulator, ATG7, blocking NPC1 function via the specific NPC1 inhibitor, U-18666A (Lu et al., 2015) did not result in accumulation of LC3 and TAX1BP1 within LAMP2-positive lysosomes, whereas U-18666A led to lysosomal buildup of these proteins in cells with intact autophagic function (Figure S2A–S2D).
To causally link the cholesterol-exporting activity of NPC1 to lysosomal proteolysis, we carried out reconstitution experiments in NPC1-defective cells. Either wild-type NPC1 or with a mutant (D786N) predicted to have enhanced transport activity based on homology to the cholesterol sensor SCAP (Gao et al., 2017; Yabe et al., 2002) reversed lysosomal LC3B accumulation (Figure 1G and 1H). In contrast expression of several NPC1 mutant variants that fail to export cholesterol due to either impaired binding to NPC2 (F503/504A) or impaired cholesterol binding to the sterol-sensing domain (SSD: P691S) (Gong et al., 2016; Li et al., 2016; Millard et al., 2005) failed to clear LC3B-positive material from the lysosomal lumen (Figure 1G and 1H). F503/504A and SSD mutants were correctly targeted to the lysosome membrane (Figure S3A) but failed to correct lysosomal cholesterol buildup, as shown by unchanged staining with filipin (Figure S3B) and with the recombinant sterol probe, mCherry-D4H*, which binds to cholesterol on the limiting membrane of the lysosome (Lim et al., 2019; Maekawa and Fairn, 2015) (Figure S3C and S3D).
Consistent with the pronounced proteolysis defect of NPC1-defective lysosomes, several lumenal hydrolases were decreased or undetectable in the NPC-null compared to control lysosomal samples. These included the Cathepsin Z protease (CTSZ), the acid lipase (LIPA), which de-esterifies LDL-derived cholesterol, and several enzymes involved in degradation of glycans and glycosphingolipids such as galactosylceramidase (GALC) and N-acetyl-alpha-glucosaminidase (NAGLU) (Figure 1I and S3E). Accordingly, a fluorescence-based reporter assay for the activity of GALC showed a marked reduction in NPC1-depleted cells compared to control cells (Figure 1J). Thus, reduced abundance of resident hydrolases in NPC lysosomes may decrease their degradative capacity and cause autophagic cargo buildup.
NPC1-null lysosomes display increased susceptibility to membrane damage.
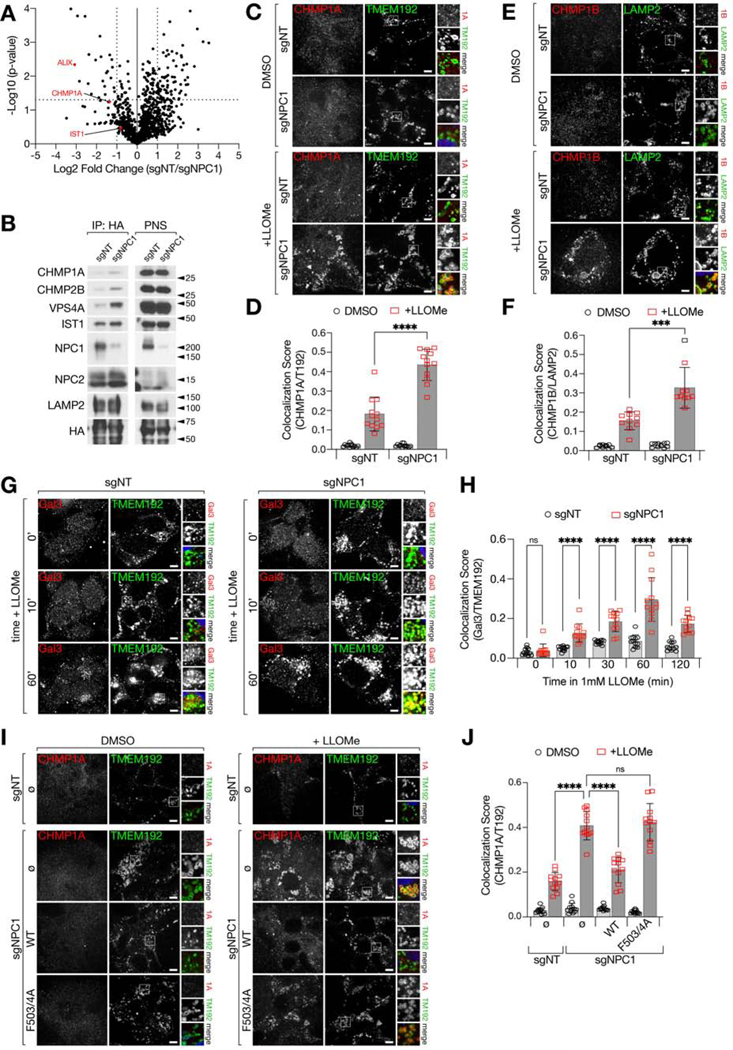
Recently, it has emerged that the lysosomal limiting membrane is susceptible to damage resulting from undigested/undigestible substrates that accumulate within the lumen (Jia et al., 2020; Maejima et al., 2013; Radulovic et al., 2018; Skowyra et al., 2018). An early detection system provided by the Endosomal Sorting Complex Required for Transport (ESCRT) III proteins detects ‘microtears’ in the lysosomal membrane and repairs them via a scission process thought to topologically resemble intraluminal vesicle budding during endosomal maturation (Nguyen et al., 2020; Radulovic et al., 2018; Schöneberg et al., 2017; Skowyra et al., 2018). Suggestive of ongoing damage to NPC lysosome membranes, our proteomics analysis of NPC lysosomes found a significant enrichment of the ESCRT III components CHMP1A and IST1, along with ALIX/ PDCD6IP, which provides a recruiting platform for the assembly of coiled ESCRT III filaments (Radulovic et al., 2018; Skowyra et al., 2018) (Figure 2A and Table S1).
Figure 2. NPC1-deficient lysosomes display an increased propensity for membrane damage.
(A) Volcano plot of Lyso-IP proteomic data (from Fig. 1A–1C) for the ratio of untreated sgNT/sgNPC1 LAMP1-normalized peptide counts. Selected ESCRT proteins are depicted as red circles. Statistical analysis was performed using two-tailed unpaired t-test.
(B) Immunoblots of Lyso-IP samples from sgNT or sgNPC1 293Ts.
(C-F) sgNT or sgNPC1 293Ts were treated with LLOMe or vehicle for 10m before immunostaining with the indicated antibodies. Representative confocal micrographs for cells stained with CHMP1A and TMEM192 (C) or CHMP1B and LAMP2 (E). Quantification of CHMP1A and TMEM192 co-localization (D) or CHMP1B and LAMP2 (F) from 12 (F) or 10 (D) non-overlapping fields containing at least 3 cells per field; ****P<0.0001, ***P=0.0005, two-tailed unpaired t-test with Welch’s correction.
(G and H) sgNT or sgNPC1 293Ts were treated with LLOMe for the indicated amounts of time before immunostaining for Galectin-3 (Gal3) and TMEM192. (G) Representative confocal micrographs for selected time points. (H) Quantification of Gal3 and TMEM192 co-localization from 12 non-overlapping fields containing at least 3 cells per field; ****P<0.0001, ns=not significant, two-tailed unpaired t-test with Welch’s correction.
(I and J) sgNT and sgNPC1 293Ts expressing the indicated NPC1-FLAG cDNA were immunostained for CHMP1A and TMEM192. (I) Representative confocal micrographs. (J) Quantification of CHMP1A and TMEM192 co-localization from 12 non-overlapping fields containing at least 3 cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
All bar graphs depict mean ± SD. Scale bars are 10μm. See also Figure S3–S4, and Supplemental Table 1.
Consistent with the proteomic data, immunoblotting of lysosomal immunoprecipitates from NPC1-defective HEK-293Ts confirmed increased lysosomal accumulation of several ESCRT III components, including CHMP1A, CHMP2B and IST1, as well as the AAA+ ATPase, VPS4A, which mediates ESCRT III polymer remodeling and disassembly (Chiaruttini et al., 2015; Schöneberg et al., 2017) (Figure 2B). In contrast to the clear proteomics and immunoblotting results, ESCRT III accumulation in unperturbed NPC cells was not readily visible by double immunofluorescence staining for CHMP1A or CHMP1B and LAMP2, possibly due to limitations of the available antibodies. However, enhanced ESCRT III accumulation became evident upon induction of lysosomal damage using the membrane destabilizing agent L-leucyl-L-leucine methyl ester (LLOMe), both in NPC1-defective HEK293Ts and in NPC1-null MEFs (Figure 2C–2F and Figure S4A–S4D).
To confirm that accumulation of ESCRT III subunits signals ongoing membrane damage, we subjected both sgNT and sgNPC1 HEK-293T cells to a time-course of LLOMe treatment and stained cells for Galectin-3 (Gal3), a cytosolic lectin that recognizes and binds to glycan chains on the luminal side of endolysosomal membrane proteins, when these become exposed upon membrane permeabilization (Jia et al., 2020; Maejima et al., 2013). Prolonged LLOMe treatment led to stronger Gal-3 accumulation in NPC1-deleted over control lysosomes at all time points, indicating a higher propensity for lysosomal membrane rupture in the absence of NPC1 function (Figure 2G and 2H). Moreover, LLOMe treatment enhanced leakage of the lysosomotropic dye, LysoTracker red, from NPC1-defective compared to control lysosomes (Figure S4E and S4F).
Given that NPC lysosomes maintain their luminal acidity (Elrick et al., 2012), the enrichment of ESCRT III components likely reflects not unrecoverable membrane permeabilization, but higher propensity to damage compensated, at least in part, by local ESCRT III polymerization (Radulovic et al., 2018; Skowyra et al., 2018).
Reconstituting NPC1-defective HEK293T cells with wild-type NPC1 reduced LLOMe-induced lysosomal CHMP1A and CHMP1B accumulation to control levels, whereas the transport-defective F503/4A mutant failed to do so (Figure 2I–2J and Figure S4G–S4H). Thus, the higher damage propensity of NPC lysosomes directly results from loss of NPC1-dependent cholesterol export.
NPC1 regulates mTORC1 via its cholesterol-exporting function
To begin to establish a mechanistic link between the lysosomal defects described above and faulty mTORC1 regulation, we tested the ability of several sterol transport-defective NPC1 mutants to restore mTORC1 regulation by cholesterol. Unlike wild-type NPC1, NPC2-binding defective NPC1F503/504A and cholesterol-binding defective NPC1P202/203A and NPC1P691S failed to restore mTORC1 regulation by cholesterol depletion-refeed (Figure 3A, S3A–S3B and S5A–S5B). In agreement with the signaling results, the transport defective NPC1 isoforms failed to re-establish regulation of mTORC1 localization to LAMP2-positive lysosomes in response to changes in cellular cholesterol levels (Figure 3B and 3C).
Figure 3. Cholesterol transport by NPC1 controls mTORC1 activity in response to lysosomal cholesterol.
(A) Immunoblots from sgNT and sgNPC1 293Ts expressing the indicated NPC1-FLAG cDNA. Cells were depleted of sterols using methyl-β-cyclodextrin (MCD), followed by re-feeding with cholesterol in complex with MCD, as indicated.
(B and C) Cells were starved for and re-fed with cholesterol as in (A) before immunostaining for mTOR and LAMP2. (B) Representative confocal micrographs. (C) Quantification of mTOR and LAMP2 co-localization from 12 non-overlapping fields containing at least 5 cells per field; ****P<0.0001, *P=0.0121, ns=not significant, two-tailed unpaired t-test with Welch’s correction.
(D) Immunoblots from sgNT and sgNPC1 293Ts expressing the indicated NPC1-FLAG cDNA. Cells were pre-treated with OSW-1, followed by sterol depletion and re-feeding as in (A) in the presence of OSW-1, as indicated.
All bar graphs depict mean ± SD. Scale bars are 20μm. See also Figure S3.
In cells expressing the transport-defective NPC1 mutants, constitutive mTORC1 signaling still required cholesterol, as it was abolished by treatment with OSW-1, a natural product inhibitor of OSBP that causes cholesterol sequestration in the ER and its depletion from the lysosomal limiting membrane (Burgett et al., 2011; Lim et al., 2019) (Figure 3D).
In summary, these results show that failure of NPC1 to transport cholesterol underlies both dysregulated mTORC1 signaling and the defects in lysosomal proteolysis and membrane integrity described above.
mTORC1 inhibition restores integrity and proteolytic function of NPC lysosomes downstream of cholesterol storage.
We next investigated the cellular effects of aberrant mTORC1 regulation in NPC. Due to its ability to stimulate lipogenic programs (Düvel et al., 2010; Li et al., 2010; Peterson et al., 2011), hyperactive mTORC1 could contribute to lysosomal cholesterol buildup in NPC1-null cells. However, treating NPC cells with the ATP-competitive mTOR inhibitor, Torin1 (Thoreen et al., 2009), did not significantly reduce cholesterol accumulation in the lysosomal lumen or limiting membrane, as indicated by unchanged staining with filipin and mCherry-D4H*, respectively (Figure 4A and 4B). Moreover, lipidomic profiling of lysosomes immunopurified from sgNT and sgNPC1 HEK-293T cells revealed that, while NPC lysosomes had the expected increased content of cholesterol and sphingolipids, the levels of these lipids were not significantly changed by mTORC1 inhibition (Figure 4C, Figure S6A–S6B and Table S2). Thus, mTORC1 dysregulation occurs downstream of cholesterol storage and does not appear to contribute significantly to its establishment.
Figure 4. Inhibition of mTORC1 activity alleviates lysosomal pathologies associated with loss of NPC1.
(A and B) sgNPC1 293Ts were treated with Torin1, OSW-1, or vehicle before being fixed and semi-permeabilized with a liquid N2 pulse, followed by cholesterol labeling with D4H*-mCherry and filipin, and immunostaining for LAMP2. (A) Representative confocal micrographs. Scale bars are 20μm. (B) Quantification of D4H*-mCherry and filipin co-localization from 10 non-overlapping fields containing at least 3 cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
(C) Lysosomes from sgNT and sgNPC1 cells, treated with Torin1 or vehicle, were isolated by Lyso-IP and free cholesterol content was measured by LC-MS/MS. Measurements of cholesterol peak intensity from three independent biological replicates are shown. ***P(adj.)=0.0002, ns=not significant, ANOVA with Dunnett’s multiple comparisons test.
(D and E) sgNT or sgNPC1 293Ts were treated with Torin1, rapamycin, or vehicle as indicated before being immunostaining for LC3B and LAMP2. (D) Representative confocal micrographs. Scale bars are 10μm. (E) Quantification of LC3B and LAMP2 co-localization from 10 non-overlapping fields containing at least 3 cells per field; ****P(adj.)<0.0001, **P(adj.)=0.0014, ANOVA with Dunnett’s multiple comparisons test.
(F) Immunoblots of Lyso-IP samples from sgNT or sgNPC1 293Ts treated as in (D).
(G and H) sgNT or sgNPC1 293Ts expressing control (shLuciferase) or Ragulator-specific (shLAMTOR5) shRNAs were immunostained for LC3B and LAMP2. (G) Representative confocal micrographs. Scale bars are 10μm. (H) Quantification of LC3B and LAMP2 co-localization from 12 non-overlapping fields containing at least 3 cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
(I) Immunoblots of Lyso-IP samples from sgNT or sgNPC1 293Ts treated as in (D).
(J and K) sgNT and sgNPC1 293Ts were pre-treated with Torin1 or vehicle for 24h before being treated with LLOMe, as indicated, followed by immunostaining for CHMP1A and TMEM192. (J) Representative confocal micrographs. Scale bars are 10μm. (K) Quantification of CHMP1A and TMEM192 co-localization from 12 non-overlapping fields containing at least 3 cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
All bar graphs depict mean ± SD. See also Figure S5 and Supplemental Table 2.
Due to the ability of mTORC1 to control lysosomal biogenesis and catabolism (Kim et al., 2011; Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012), we next tested whether mTORC1 inhibition could alleviate at least some aspects of lysosomal dysfunction identified by our lysosomal proteomic analysis. This was clearly the case. Treating NPC1-null cells with Torin1 promoted clearance of autophagic material from the lysosomal lumen, as judged by both LC3B-LAMP2 double immunofluorescence (Figure 4D–4E and S7A–S7B) and by direct immunoblotting of immunopurified lysosomal samples from NPC1-deleted HEK-293T and MEFs (Figure 4F and S7C). Also, inhibiting mTORC1 signaling via shRNA-mediated knock down of LAMTOR5, which is essential for mTORC1 recruitment to and activation at the lysosome (but is not required for mTORC2-dependent signaling) (Anandapadamanaban et al., 2019; de Araujo et al., 2017; Bar-Peled et al., 2012; Rogala et al., 2019; Su et al., 2017), led to pronounced clearance of accumulated LC3 from the lumen of LAMP2-positive NPC lysosomes (Figure 4G–4H and S7D).
In contrast to mTORC1 inhibition via Torin1 or Lamtor5 knock down, little effect on lysosomal clearance was observed when cells were treated with a less complete mTORC1 inhibitor, Rapamycin (Figure 4D–4F and S7A–S7C), suggesting that inhibition of protein synthesis and activation of the autophagy-lysosome system are both key for restoration of lysosomal proteolysis downstream of mTORC1 inhibition (Lawrence and Zoncu, 2019; Liu and Sabatini, 2020).
mTORC1 inhibition via Torin1 also corrected the higher propensity for damage of NPC1 lysosomes, as shown by decreased recruitment of ESCRT III subunits both by immunoblotting of purified lysosomal samples and by double immunofluorescence for CHMP1A or CHMP1B and lysosomal markers (Figure 4I–4K and S7E–S7G). Given that mTORC1 inhibition reversed proteolytic failure but not cholesterol accumulation within NPC lysosomes, we conclude that the higher propensity of NPC lysosomes to undergo membrane damage primarily results from accumulation of undigested substrates within the lumen and not from altered fluidity of the membrane that may result from increased cholesterol content.
mTORC1 inhibition restores lysosomal function in iPSC-derived NPC neuronal cultures
The cell type most compromised by loss of NPC1 are postmitotic neurons in the cerebellum and cerebral cortex (Walkley and Suzuki, 2004). To characterize the role of aberrant mTORC1 signaling in a neuronal model of NPC, we deleted the NPC1 gene in induced pluripotent stem cells (iPSCs) using CRISPR/Cas9, and differentiated this population into neurons as previously described (Ordonez et al., 2012) (Figure S8A-S8B). Neural stem cells and neurons thus derived express neuronal lineage markers (nestin, MAP2, ßIII tubulin) (Figure S8B-S8C) and have been shown to be electrophysiologically active (Israel et al., 2012).
Consistent with the results in HEK-293T and MEFs and with previous reports (Elrick et al., 2012), iPSC-derived NPC neurons showed accumulation of undigested autophagic adaptors TAX1BP1 and GABARAP both within and outside LAMP2-positive lysosomes (Figure 5A–5D). Overnight treatment of NPC neurons with Torin1 largely cleared intracellular TAX1BP1 and GABARAP aggregates, suggesting that mTORC1 inhibition is sufficient to restore the function of the autolysosomal system of neuronal cells (Figure 5A–5D).
Figure 5. Inhibition of mTORC1 corrects lysosomal defects associated with loss of NPC1 in and iPSC-derived neuronal cell model.

(A and B) Control or NPC1−/− iPSC-derived neural lineage cells were treated with Torin1, BafA1, or vehicle before immunostaining for TAX1BP1, LAMP2, and MAP2 (not shown). (A) Representative confocal micrographs. (B) Quantification of the number of TAX1BP1 spots per cell area (defined by MAP2, see Figure S6C) from 10 non-overlapping fields containing at least 3 MAP2-positive cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
(C and D) Control or NPC1−/− iPSC-derived neuronal lineage cells were treated as in (A) before immunostaining for GABARAP, LAMP2, and MAP2 (not shown). (C) Representative confocal micrographs. (D) Quantification of GABARAP and LAMP2 co-localization from 10 non-overlapping fields containing at least 3 MAP2-positive cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
(E and F) Control or NPC1−/− iPSC-derived neuronal lineage cells were pre-treated with Torin1 or vehicle for 24h before being treated with LLOMe, as indicated, followed by immunostaining for CHMP1A, TMEM192, and MAP2 (not shown). (E) Representative confocal micrographs. (F) Quantification of CHMP1A and TMEM192 co-localization from 12 non-overlapping fields containing at least 3 MAP2-positive cells per field; ****P(adj.)<0.0001, ANOVA with Dunnett’s multiple comparisons test.
All bar graphs depict mean ± SD. Scale bars are 20μm. See also Figure S6.
Similar to NPC1-depleted HEK-293T cells, iPSC-derived NPC neurons also had increased sensitivity to membrane damage, as shown by enhanced recruitment of ESCRT III upon treatment with LLOMe (Figure 5E5F), and this prenotype was reversed by Torin1 treatment (Figure 5E–5F).
Thus, the compositional and functional defects revealed by our lysosomal proteomics in HEK-293T cells extend to NPC1-null neurons and are similarly corrected by mTORC1 inhibition.
mTORC1 inhibition restores defective mitochondrial function in NPC1-null cells
Mitochondria are especially dependent on efficient lysosome-autophagy function for their morphological and functional homeostasis; accordingly, severe mitochondria defects have been reported in NPC1-null cell lines and in neuronal cultures derived from human embryonic stem cells (hESCs) (Ordonez et al., 2012; Yambire et al., 2019a). Dysregulated mTORC1 activity could contribute to mitochondrial dysfunction via increased translational burden, production of reactive intermediates and morphological alterations (Ebrahimi-Fakhari et al., 2016; Morita et al., 2013, 2017).
We analyzed our lysosomal proteomic datasets for the relative representation of mitochondrial proteins, defined by MitoCarta (Calvo et al., 2016) or Gene Ontology classification. Surprisingly, we discovered that mitochondrial proteins were significantly under-represented in NPC1-defective compared to control lysosomes (Figure 6A). When proteins were ranked based on their preferential enrichment in wild-type over NPC1 lysosomes, approximately 50% of the proteins in the top quartile were classified as mitochondrial, whereas only 23.6% of proteins in the other three quartiles were mitochondrial (Figure 6B). About half of the proteins classified as mitochondrial displayed the expected behavior of substrates, where their peptide count was higher in leupeptin:pepstatin than in DMSO (“mito substrates”, Figure 6A–6B). Thus, unlike other substrates that reach the lumen of NPC lysosomes but fail to be degraded, lysosomal delivery of mitochondria appears hampered in NPC, possibly reflecting disruption of the process of mitophagy (Ebrahimi-Fakhari et al., 2016; Khaminets et al., 2016; Ordonez, 2012).
Figure 6. Mitochondrial morphology and function are disrupted by loss of NPC1 and is restored by inhibition of mTORC1.
(A) Volcano plot of Lyso-IP proteomic data (as in Figure 2A). Proteins identified as mitochondrial in the human MitoCarta 2.0 database (Calvo et al., 2016) are shown as red circles. Mitochondrial proteins that behave as substrates for lysosomal proteolysis are highlighted with a green outline.
(B) Percentages of MitoCarta or “mito substrates” proteins that are in the top quartile (>75% enrichment) or remaining three quartiles (<75% enrichment) of proteins enriched in sgNT over sgNPC1 lysosomes. Total number of proteins is 1254, total number in top quartile is 62.
(C) Immunoblots of Lyso-IP samples and corresponding PNS from sgNT or sgNPC1 293Ts treated with vehicle or leupeptin and pepstatin, and/or treated with 10μM CCCP for 5h.
(D and E) Control or NPC1−/− iPSC-derived neuronal lineage cells were treated with Torin1, BafA1, or vehicle before immunostaining for TOM20. (D) Representative confocal micrographs. (E) Lengths of individual mitochondria were measured and quantified.
(F and G) Control or NPC1−/− iPSC-derived neuronal lineage cells were treated with Torin1, BafA1, or vehicle before being stained with the ratiometric MMP dye JC-10. (F) Dot plots showing MMP-independent (“green”) and MMP-dependent (“red”) fluorescence distribution of individual cells from one representative experiment. (G) Percentages of cells classified as depolarized (Q3: greenhigh, redlow) or polarized (Q3: greenhigh, redhigh). Values from six individual replicates are shown as points, bars represent average values across all replicates.
All bar graphs depict mean ± SD. Scale bars are 20μm. See also Figure S6–S7.
To gain insight into a possible mitophagy defect downstream of NPC1 loss, we treated sgNT and sgNPC1 cells with the mitochondrial depolarizing agent carbonyl cyanide m-chlorophenyl hydrazone (CCCP), which triggers stabilization of PTEN-induced kinase 1 (PINK1) at the mitochondrial outer membrane (Jin et al., 2010; Lazarou et al., 2015). Stabilized PINK1 phosphorylates ubiquitin at Ser65, which in turn allosterically activates the E3 ligase Parkin, leading to ubiquitylation of mitochondrial outer membrane proteins and capture of ubiquitylated mitochondria into nascent autophagosomes (Kane et al., 2014; Kazlauskaite et al., 2014; Wauer et al., 2015).
CCCP-induced stabilization of PINK1 appeared nearly identical between sgNT and sgNPC1 HEK-293T cells, as were cellular levels of Parkin (Figure S9A). Moreover, total cellular levels of Ser65-phosphorylated ubiquitin, which decorated numerous proteins over a wide range of molecular weights, were very similar between CCCP-treated control and NPC1-defective cells (‘PNS’ in Figure 6C). Thus, initiation of PINK1- and Parkin-dependent mitophagy appears to be independent of NPC1 status.
In contrast, immunoblotting of lysosomal immunoprecipitates showed a striking reduction of pSer65-ubiquitylated proteins in NPC1-defective versus control lysosomes (Figure 6C). This reduction paralleled that of other mitochondrial proteins such as TOM20, TOM40, VDAC and PINK1 itself, in line with the proteomic results of Figure 6A, and was in contrast to the accumulation of canonical autophagic substrates SQSTM1 and TAX1BP1.
These results support a model in which lack of NPC1 does not impact the initial response to mitochondrial stress, including PINK1-dependent phosphorylation of ubiquitin at Ser65 and the attachment of pSer-ubiquitin to mitochondrial proteins, whereas it appears to severely impair subsequent steps in mitophagy, such as recognition of Ser65-Ub-modified mitochondrial proteins by autophagic adaptors, or the fusion of mitochondria-loaded autophagic vesicles with NPC lysosomes.
Consistent with defective mitophagy, the cytoplasm of iPSC-derived NPC neurons was disseminated with fragmented mitochondria, as shown by Tom20 immunostaining (Figure 6D–6E). Treatment with the v-ATPase inhibitor, BafA1, only modestly increased the already high degree of mitochondrial fragmentation observed in NPC1-null cells, implicating lysosomal dysfunction as the main driver of this process (Ordonez et al., 2012; Weber et al., 2020; Yambire et al., 2019b) (Figure 6D–6E). Further supporting this idea, overnight treatment with Torin1 restored mean mitochondrial length of NPC1-null neurons to wild-type levels, whereas it caused no change to mitochondria of their wild-type counterparts (Figure 6D–6E). Similar to NPC neurons, mitochondria of NPC1-defective HEK293T cells were highly fragmented, and their morphology was corrected by Torin1 but not rapamycin (Figure S9B-S9E).
Loss of mitochondrial integrity is often indicative of decreased mitochondrial membrane potential (MMP). Indeed, mitochondria from NPC1-null iPSC-derived neurons showed significantly reduced MMP compared to WT cells, as determined by flow cytometry analysis of the ratiometric MMP indicator, JC-10 (Figure 6F–6G). BafA1 treatment decreased the MMP of wild-type neurons, whereas it had a smaller effect on the MMP of NPC cells. Similar to mitochondrial fragmentation, Torin1 significantly rescued the MMP of NPC neurons, whereas it had a negligible effect on wild-type cultures.
To gain mechanistic insight into how mTORC1 inhibition improves the mitochondrial morphology and function of NPC cells, we measured CCCP-dependent mitophagy induction in DMSO- and Torin1-treated control and NPC1-defective cells. Interestingly, Torin1 treatment inhibited CCCP-dependent PINK1 stabilization and reduced the amount of CCCP-induced, pS65-ubiquitin-decorated proteins both in whole lysates and in immunoprecipitated lysosomes from sgNT and sgNPC1 HEK293T cells (Figure S10), possibly reflecting increased ability of Torin1-treated mitochondria to maintain their MMP upon CCCP challenge
Finally, in the lyso-IP samples the amount of pSer65-ubiquitin-modified proteins was lower overall in the Torin1-treated condition, but more comparable between sgNT and sgNPC1 than in vehicle-treated samples (Figure S10). Thus, mTORC1 inhibition appears to both improve mitochondrial health and stress resilience, and possibly correct the defective autophagic capture and lysosomal delivery of ubiquitylated mitochondrial material.
DISCUSSION
Taken together, our results support a central role for the NPC1-cholesterol-mTORC1 signaling axis in the maintenance of organelle function and cellular homeostasis (Figure 7). Export of lysosomal cholesterol by NPC1 is essential for optimal regulation of mTORC1 signaling outputs. In turn, mTORC1 signaling plays a critical role in sustaining the hydrolytic activities of the lysosome, the integrity of its limiting membranes, and the morphology and polarization of mitochondrial.
Figure 7.
Model illustrating the relationship between NPC1, lysosomal cholesterol, mTORC1 signaling and organelle homeostasis in both normal and NPC cells.
A key point that we address concerns the mechanisms via which NPC1 regulates mTORC1. Hyperactive mTORC1 signaling in NPC cells is consistent with both a cholesterol-transporting and a cholesterol-sensing role for NPC1 (Castellano et al., 2017; Lim et al., 2019). To distinguish between these models, we systematically rescued NPC1-defective cells with NPC1 mutants that lack cholesterol-exporting activities. The tight correlation between the cholesterol transporting function of NPC1 and its ability to regulate mTORC1 signaling support a model in which NPC1 functions upstream of cholesterol, not downstream of it.
In turn, dysregulated mTORC1 signaling emerges as a key driver of organelle dysfunction downstream of NPC1 loss and the resulting lysosomal cholesterol buildup. This is shown by genetic and pharmacologic inhibition of mTORC1, which failed to correct cholesterol storage in the lysosomal lumen or on its limiting membrane but effectively corrected compositional and functional defects of the lysosome. How the lysosomal defects that we uncovered relate to each other will require further investigation. The depletion of several hydrolases (including proteases such as Cathepsin Z) provides a likely explanation for the profound proteolytic impairment of NPC lysosomes, and could stem from transcriptional inhibition or from their defective trafficking (Kobayashi et al., 1999; Saftig and Klumperman, 2009; Sleat et al., 2013). Alternatively, the increased propensity of NPC lysosomes to undergo membrane damage, as shown by our data and by a recent report (Liu et al., 2020), could trigger leakage of resident hydrolases, a proposed phenomenon in NPC and other pathophysiological contexts (Chung et al., 2016; Hämälistö et al., 2020; Maejima et al., 2013; Sakamachi et al., 2017).
The factors driving membrane damage in NPC remain to be determined. Alterations in limiting membrane fluidity caused by massive cholesterol accumulation could increase the propensity for membrane rupture through discontinuities between cholesterol-rich, crystalline-like microdomains and the surrounding membrane (Toulmay and Prinz, 2013; Tsuji et al., 2017). Damage could also result from undigested cargo accumulating within the lumen, a mechanism similar to that induced by LLOMe, which assembles into membrane-piercing polymers within the acidic lysosomal lumen (Maejima et al., 2013; Radulovic et al., 2018; Skowyra et al., 2018). The observation that mTORC1 inhibition corrects membrane damage and defective proteolysis without altering the cholesterol content of NPC lysosomes favors the latter possibility.
The observation that hydrolases mutated in other lysosomal storage disorders, such as acid lipase, beta-glucosidase and galactosidase are depleted from NPC lysosomes suggests that NPC may share pathogenic traits with diseases such as Wolman, Krabbe and Mucopolysaccharidosis III (Ballabio and Gieselmann, 2009), and could provide a mechanistic basis for the accumulation of neurofibrillary tangles, a hallmark of Alzheimer Disease, in NPC brains (Walkley and Suzuki, 2004). More generally, hydrolase loss may be a common trait of several LSDs (Danyukova et al., 2018; Platt et al., 2018), a possibility that can be tested through lysosomal profiling in disease-specific models.
Given that mTORC1 inhibition decouples cholesterol storage from proteolytic failure and lysosomal membrane damage, an outstanding question is how NPC1-mTORC1 signaling controls these important lysosomal properties. mTORC1 drives anabolic programs that could increase the proteolytic load of the lysosome, such as protein synthesis and ribosome biogenesis, while actively suppressing catabolic programs that could help restore lysosomal function (Düvel et al., 2010; Perera and Zoncu, 2016; Settembre et al., 2012). It is likely that simultaneous inhibition of biosynthetic programs and activation of catabolic ones by mTORC1 inhibitors may synergize in restoring autophagic function in NPC.
Similarly, the ability of catalytic mTORC1 inhibitors to restore mitochondrial morphology and membrane potential likely stems from a combination of decreased translational burden and increased repair due to restoration of lysosomal proteolysis. mTORC1 stimulates translation of multiple nuclear-encoded mitochondrial transcripts (Morita et al., 2013), stimulates energy production as well as generation of anabolic intermediates in mitochondria (Ben-Sahra et al., 2016; Cunningham et al., 2007; Morita et al., 2013) and can indirectly promote mitochondrial fission (Morita et al., 2017). In turn, mTORC1-driven remodeling of mitochondria may render them more susceptible to stress, consistent with our observation of reduced CCCP-dependent mitophagy initiation in Torin1-treated cells. Moreover, by inhibiting autophagic initiation, elevated mTORC1 signaling can suppress the capture and clearance of damaged mitochondria, in line with our lysosomal proteomic data (Ebrahimi-Fakhari et al., 2016; Perera and Zoncu, 2016).
The restorative effects of mTORC1 inhibition on organelle functions suggests the attractive possibility that inhibiting this pathway could have therapeutic value in NPC and other lysosomal disorders characterized by dysregulated mTORC1 signaling (Andrzejewska et al., 2016; Bartolomeo et al., 2017; Brown et al., 2019; Ivanova et al., 2016). Our data indicate that clinical derivatives of rapamycin (rapalogues) are unlikely to be beneficial, whereas the more potent and complete ATP-competitive inhibitors may be effective. However, the applicability of these inhibitors is limited by poor bioavailability and toxic off-target inhibition of mTORC2 blockade (Lamming et al., 2012; Liu and Sabatini, 2020). The development of new-generation, mTORC1-specific inhibitors with more complete inhibitory profiles than rapamycin may provide an avenue for safe and effective mTORC1 modulation in NPC and other lysosomal disorders (Chung et al., 2019; Mahoney et al., 2018; Rodrik-Outmezguine et al., 2016; Schreiber et al., 2019)
RESOURCE AVAILABILITY
Lead Contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact, Roberto Zoncu (rzoncu@berkeley.edu)
Materials Availability
Plasmids and cell lines generated in this study are available upon request from the lead contact.
Data and Code Availability
The supplemental tables accompanying the published article includes all datasets generated in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HEK293T sgNT and sgNPC1 cells were generated using Cas9/sgRNA RNP transfection as previously described (Castellano, et al. 2017). Briefly, cells were transfected by electroporation with recombinant Cas9 protein complexed with sgRNA targeting the first exon of the NPC1 gene, individual cells were seeded into wells of a 96-well plate, and clonal cell populations were screened for cholesterol accumulation by filipin staining. Clones showing endosomal filipin staining were selected for further validation of NPC1 expression by immunoblotting. NPC+/+ (WT) and NPC1−/− MEFs were a gift from Dr. Sovan Sarkar (University of Birmingham). HEK293T and MEF cells were maintained in DMEM (Gibco, 11995) supplemented with 10% (v/v) fetal bovine serum (Sigma, F0926) and 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco, 15140–122). All cells were cultured at 37°C in 5% CO2. Cells were free of mycoplasma and routinely tested using MycoAlert Mycoplasma Detection Kit (Lonza, LT07-318).
hIPSC generation and neuronal differentiation
Control hIPSC lines were derived from fibroblasts obtained from one healthy adult (J. Craig Venter) whose genome is fully sequenced and published. hIPSC lines are generated by four-factor reprogramming (Israel et al., 2012), and were examined for pluripotency by labeling with lineage specific markers Tra 1–81 (BD Bioscience), Oct-4 and Nanog (Santa Cruz). Pluripotency is assessed by embryoid body formation and staining for the 3 germ layers, endoderm (Alpha-fetoprotein, DAKO), mesoderm (smooth muscle actin, Millipore) and ectoderm (Nestin, Millipore). NSC and neurons were generated using previously described protocols and purified by FACS NSCs and neurons were purified by FACS, where NSCs were stained for Nestin (Millipore) Sox1, Sox2 and Pax 6, and neurons were stained for MAP2 and TUJ (Yuan et al., 2011). For each experiment involving hIPSC derived neurons, neurons from at least two independent differentiations were examined in duplicate or triplicate format.
Generation of NPC1 knock-out hIPSCs
CRISPR/Cas9 gene editing was used to generate an NPC1 knock-out (KO) in the Craig Venter control hIPSC line. We inserted a frame-shift mutation in Exon 4 that engineered a premature stop codon leading to complete ablation of NPC1. Transfected hIPSCs were sorted based on GFP and Tra181 expression and sparsely plated onto 10cm MEF plates. Individual colonies were picked manually and transferred to 96-well plates. Candidates were screened by PCR and TOPO cloning, and positive hits were karyotyped to ensure genetic stability. Digital karyotype was normal. A microamplification on chromosome 1p was visually observed in parental CV line and NPC1 KO line but this was below threshold and considered to be an artifact. Ablation of NPC1 was confirmed by RTqPCR and Western Blot.
METHOD DETAILS
Cell Line Generation
HEK293T and MEF cells stably expressing recombinantly tagged cDNAs or shRNAs were generated as follows. A synthetic cDNA encoding TMEM192-RFP-3xHA was cloned into the pLJM1 lentiviral vector (Lim et al., 2019). TMEM192-FLAG was cloned also cloned in the pLJM1 vector by amplifying the TMEM192 cDNA using primers to append a DYKDDDK (“FLAG”) peptide to the c-terminus of the resulting protein. Codon-optimized NPC1 cDNA containing a FLAG tag (Castellano et al., 2017) was subcloned into the pLVX lentiviral vector (Clontech). All NPC1 mutants were generated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent, 200524).
Short-hairpin oligonucleotides (shRNAs) directed against LAMTOR5 (TRCN0000153443) or Luciferase (TRCN0000072243, used as a non-targeting control) were cloned into the pLKO.1 lentiviral vector (The RNAi Consortium, Broad Institute) according to the manufacturer’s instructions.
Expression of protein-encoding cDNAs or shRNA constructs was performed by stable lentiviral transduction. Lentivirus was generated by co-transfection of lentiviral vector carrying the construct of interest with lentiviral packaging plasmids (pMD2.G, Addgene 12259; and psPAX2, Addgene 12260) in a 5:3.75:1.25 ratio, respectively, using polyethylenimine (PEI). Viral supernatant was harvested 48 h after transfection, cleared by centrifugation, and concentrated using Lenti-X Concentrator (Clontech, 631231) according to the manufacturer’s instructions. Target cells were plated in 6-well plates in media supplemented with 8 μg/ml polybrene (Millipore, TR-1003-G) and concentrated virus. Virus-containing media was removed after 24 h and replaced with media containing 1.5 μg/ml puromycin. Protein expression or knockdown was confirmed by immunoblotting. For shRNA knockdown, cells were maintained in selective media for 3 days before use in assays in order to ensure complete knockdown.
Drug Treatments
Drug treatments were performed as follows unless otherwise specified. Leupeptin (Alfa Aesar, J61188) and pepstatin A (MP Biologicals, 195368) were used at 20 μM each for 24 h. Bafilomycin A1 (Alfa Aesar, J61835) was used at 500 nM for 4 h. LLOMe (Sigma, L7393) was used at 1 mM for 10 m unless otherwise indicated in the figure legend. Torin1 (Tocris, 4247) was used at 250 nM for 24 h, and rapamycin (Calbiochem, 553210) was used at 100 nM for 24 h. OSW-1 (a generous gift from Matthew Shair, Harvard University) was used at 10 nM for 8 h. CCCP (Sigma, C2759) was used at 10μM for 5 h. U18666A (Tocris, 1638) was used at 5 μg/ml for 24 h.
Lysosome Immunoprecipitation (Lyso-IP)
Lysosomes from cells expressing TMEM192-RFP-3xHA were purified as previously described (Lim et al., 2019). Briefly, cells were seeded in a 15cm at a density appropriate for them to reach confluency after 24h. All subsequent steps were performed on ice or at 4°C unless otherwise noted. Media was removed, cell monolayers were rinsed with ice-cold KPBS buffer (136 mM KCl, 10m M KH2PO4, pH 7.25, supplemented with Pierce Protease Inhibitor Tablets (Thermo, A32965)), scraped into 10 ml of KPBS and collected by centrifugation at 1500 rpm for 5 min. Pelleted cells were resuspended in a total volume of 1ml KPBS (supplemented with 3.6% (w/v) OptiPrep (Sigma, D1556)) and fractionated by passing through a 23G syringe 5 times followed by centrifugation at 2700 rpm for 10 min. Post-nuclear supernatant was harvested and incubated with 40 μl of anti-HA magnetic beads (Thermo, 88836, prewashed with KPBS buffer) with end-over-end rotation for 10 min. Lysosome-bound beads were washed two times with KPBS(+ OptiPrep) and two times with KPBS. For immunoblotting, samples were incubated with a 1:1 mixture of KPBS and 2x urea sample buffer (150 mM Tris, pH 6.5, 6 M urea, 6% SDS, 25% glycerol, 5% β-mercaptoethanol, 0.02% bromophenol blue) for 30 min at 37°C. For proteomics experiments, lysosomal immunoprecipitates were eluted from beads using 0.1% NP-40 in PBS for 30 min at 37°C, beads were removed and the resulting eluate was snap-frozen with LN2.
Proteomics Analysis
For comparative analysis between treatment conditions and genotypes, minimum peptide abundance was set to 1 for all replicates. Experimental datasets were first compared to the proteomic dataset generated from anti-HA Lyso-IP performed on cells expressing TMEM192-FLAG (“blank” samples). Only proteins present with a combined average peptide abundance across both experimental samples >1.5-fold enrichment over blank samples were included in further analysis. Fold changes between experimental samples were then calculated, and the significance of these fold changes were calculated using a two-tailed unpaired t-test. For comparative analysis between genotypes, peptide counts from each replicate were additionally normalized to the peptide abundance of LAMP1 within each replicate, before calculation of fold changes and significance values. Data in all volcano plots are displayed as the log2 of the fold change, and the -log10 of the p-value.
The list of “cargo” proteins was generated by identifying all proteins whose abundance increased ≥2-fold upon inhibition of lysosomal proteolysis (leupeptin/pepstatin treatment). This list was cross-referenced to the dataset comparing leupeptin/pepstatin-treated to vehicle-treated sgNPC1 cells.
To analyze mitochondrial proteins present in Lyso-IP samples, the filtered datasets were cross-referenced with the Human MitoCarta 2.0 database (Calvo et al., 2016; Pagliarini et al., 2008). This list was further refined by eliminating proteins that did not obey expected behavior upon lysosomal proteolysis inhibition (i.e. any protein whose abundance decreased upon leupeptin/pepstatin treatment in either genotype was excluded) to generate the “mito substrates” subset. Quartile analysis is based on the enrichment of proteins in sgNT samples over sgNPC1 samples, where “top quartile” are proteins with an enrichment of >75% in sgNT samples, and “bottom three quartiles” are all other proteins. Percentages shown in pie charts represent the fraction of proteins identified in MitoCarta 2.0 database, or as “mito substrates”.
Immunofluorescence
Cells were seeded on fibronectin-coated glass coverslips in 12-well plates at 150,000–300,000 cells per well, and allowed to attach overnight. Cells were treated with compounds at the specified concentrations and length of time as indicated before being fixed and stained. For LC3B, GABARAP, and TAX1BP1 staining cells were first fixed and permeabilized with ice-cold 100% methanol for 5 min at −20°C and then rinsed three times with PBS. For Galectin-3 staining, cells were first fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature, rinsed three times with PBS, permeabilized with 0.3% Triton X-100 (v/v) in PBS for 10 min at room temperature, and rinsed three times with PBS. For all other stainings, cells were first fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature, rinsed three times with PBS, permeabilized with 0.1% (w/v) saponin in PBS for 10 min at room temperature, and rinsed three times with PBS. Primary antibodies were diluted into 5% normal donkey serum (Jackson ImmunoResearch, 017-000-121) and coverslips were labeled with this solution overnight at 4°C. Coverslips were rinsed three times with PBS and then labeled with fluorescently-conjugated secondary antibodies (diluted 1:400 in 5% normal donkey serum, PBS) for 45 min at room temperature, protected from light. Coverslips were rinsed with PBS six times (incubating in every other wash for 5 min at room temperature) and then mounted on glass slides using VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratories, H-1200).
Microscopy
All confocal microscopy was performed on a spinning-disk Nikon Ti-E inverted microscope (Nikon Instruments) system using a Plan Apo 60x oil objective. Images of fine cellular detail were acquired with an additional 1.5x magnifier. All images were acquired with an Andor Zyla-4.5 scientific complementary metal-oxide-semiconductor camera (Andor Technology) using iQ3 acquisition software (Andor Technology).
Image analysis
For quantification of co-localization, 10–12 non-overlapping images were acquired from each coverslip. Raw, unprocessed images were imported into FIJI v.2.0.0-rc-69/1.52i and converted to 8-bit images, and images of individual channels were thresholded independently to exclude background and non-specific staining noise and converted to binary masks. Co-localization between lysosomes and the marker of interest was determined using the “AND” function of the image calculator. Data are plotted as the fraction of lysosomes that are positive for the marker of interest (the “Colocalization Score”).
For quantification of TAX1BP1 aggregates in hiPSC-derived neuronal cell cultures, thresholded images of the channel corresponding to MAP2 staining were used to generate a binary mask to define and measure the total area occupied by MAP2+ cells. Masks were then applied to independently thresholded images of channel corresponding to TAX1BP1 staining to exclude signal outside the defined cell area. Individual TAX1BP1 aggregates in the resulting image were counted using the “Analyze Particles” function, and data from individual frames are plotted as the average number of TAX1BP1 spots per MAP2+ cell area.
For mitochondrial perimeter measurements, individual cells were isolated into separate images and blinded before analysis. Each image was individually thresholded and converted to binary masks. The “Analyze Particles” function was used to identify and measure the perimeter of every particle in the resulting mask. Data are plotted as the average mitochondrial perimeter per cell analyzed. For mitochondrial length measurements, images were first blinded and then the length of individual mitochondria was measured manually. Data are plotted as the length of every individual mitochondria in each condition.
Measurement of GALC activity
HEK293T cells were seeded at 10,000 cells per well in fibronectin-coated flat-bottom black 96-well plates with clear bottom (Greiner, 655090) and allowed to adhere overnight. Media was aspirated and then replaced with fresh complete growth media supplemented with 15 μM LysoLive GalGreen fluorogenic substrate (MarkerGene Technologies, M2776) and 50 nM LysoTracker Red DND-99 (Thermo Scientific, L7528) and incubated at 37°C for 2 h. Media was aspirated, wells were rinsed once with warm PBS, and then replaced with Imaging Buffer (136 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.3 mM MgCl2, 10 mM HEPES, pH 7.4). Endpoint fluorescence was measured on a Bio-Tek Synergy HT Multi-Mode Microplate Reader, using 485 nm/20 nm excitation with 528 nm/20 nm emission, and 570 nm/9 nm excitation with 590 nm/9 nm emission, read through the bottom of the plate. GalGreen fluorescence was normalized per well to LysoTracker Red fluorescence, and values from individual wells are plotted as points.
Cholesterol starvation and replenishment
HEK293T cells were seeded in fibronectin coated culture dishes so they would reach 80–90% confluency at the start of the assay. For cholesterol depletion, cells were incubated in DMEM supplemented with 0.75% (w/v) methyl-ß-cyclodextrin (MCD, Sigma C4555) and 0.5% (v/v) lipid-depleted serum (LDS) for 2 h. For cholesterol re-feeding cells were incubated with DMEM supplemented with 0.1% MCD and 0.5% LDS containing either 50 μM cholesterol (Sigma, C3045) or 100 mg/ml human LDL (Alfa Aesar, J65039), as indicated.
Cell lysis and immunoblotting
Cells were incubated in lysis buffer (1% Triton X-100, 10 mM β-glycerol phosphate, 10 mM sodium pyrophosphate, 4 mM EDTA, 40 mM HEPES, pH 7.4, supplemented with Pierce protease inhibitor tablets) for 30 min at 4°C with rocking to ensure complete lysis. Lysates were harvested and cleared by centrifuging at 17,000g for 10 min at 4°C, protein concentration in the supernatant was measured by Bradford assay. Samples of equalized concentration were prepared for SDS-PAGE by addition of 2x Urea samples buffer or 5x sample buffer (235 mM Tris, pH 6.8, 10% SDS, 25% glycerol, 25% β-mercaptoethanol, 0.1% bromophenol blue). 5 μg of total protein from per sample was loaded per lane in a 12% Tris-Glycine gel (Thermo Scientific, XP00122) and resolved by electrophoresis in a Tris-Glycine running buffer (25 mM Tris, 190 mM glycine, 0.1% (w/v) SDS). For Lyso-IP samples 10% of the total immunoprecipitated material was loaded per lane, and 0.5% of total PNS was loaded per lane. Proteins were transferred to a PVDF membrane (Millipore IPVH00010), blocked with 5% non-fat milk in TBS-T, and incubated in primary antibodies (diluted in 5% milk in TBS-T) overnight at 4°C. Membranes were rinsed with TBS-T and incubated with horseradish peroxidase conjugated anti-rabbit or anti-mouse secondary antibodies (diluted in 5% milk in TBS-T) for 1 h at room temperature. Membranes were washed again with TBS-T and incubated with Pierce ECL Western Blotting Substrate (Thermo Scientific, 32109) before being exposed to ProSignal ECL Blotting Film (Genesee Scientific, 30–810L). For phosphorylation site specific antibodies, PBS-T was used in place of TBS-T for all steps, and antibodies were diluted in 5% BSA in PBS-T.
Cholesterol labeling in situ with D4H*-mCherry and filipin
Recombinantly expressed GST-D4H*-mCherry was purified from BL21 E. coli as previously described (Lim et al., 2019). Briefly, bacterial expression was induced with 0.4mM IPTG for 20h at 18°C. Bacteria were lysed by sonication at 4°C and subsequent incubation in 0.5% Triton X-100 at 4°C for 15 min. Cleared lysate was incubated with Glutathione Sepharose 4B beads for 2h, bound protein was sequentially eluted with 25mM L-glutathione, eluted fractions were pooled and filtered sequentially with 30kDa and 3kDa centrifugal filter units. Concentrated protein solution was supplemented with 20% sucrose and snap frozen in liquid nitrogen. Labeling with D4H*-mCherry and filipin was also performed as previously detailed (Lim et al., 2019). Briefly, cells were plated on fibronectin-coated coverslips and treated as indicated before being fixed with 4% PFA in PBS for 15 min at room temperature. Coverslips were rinsed in PBS, and then selectively permeabilized by immersion in LN2 for 25 sec. Coverslips were then blocked with 1% BSA in PBS for 1 h at room temperature, followed by incubation in D4H*-mCherry, diluted 1:50 in 1% BSA/PBS, for 2 h at room temperature, protected from light. Coverslips were rinsed with PBS and then fixed again with 4% PFA/PBS for 10 min at room temperature. Coverslips were rinsed with PBS and filipin labeling was performed simultaneously with immunofluorescent staining of LAMP2. Primary and secondary antibodies were diluted in 1% BSA in PBS supplemented with 0.5 mg/ml filipin (Sigma, F9765) and performed each for 1 h at room temperature, rinsing the coverslips with PBS after each incubation. Coverslips were mounted on glass slides in VECTASHEILD Antifade Mounting Media (Vector Laboratories, H-1000).
Lipidomics
Lysosomes were purified from treated cells by Lyso-IP. After washing, lysosome-bound beads were resuspended in 400 μl of ice-cold Methanol and vortexed vigorously. 4 μl of a 15:0–18:1 phosphocholine (Avanti, 330716) internal standard were then added to each sample and vortexed vigorously. 500 μl of Tert-butyl methyl ether (TBME, Sigma, 306975) was added to each sample, followed by 500 μl of dH2O to induce phase separation. Samples were vortexed vigorously again, and then centrifuged at 2000xg, 4°C, for 5 min to complete phase separation. 400 μl of the upper phase was collected and 75 μl of was transferred into each of two amber glass LCMS vials (Sigma, 29398).
Lipids were analyzed using a Shimadzu 10A HPLC system and a Shimadzu SIL-20AC HT auto-sampler coupled to a Thermo Scientific TSQ Quantum Ultra triple quadrupole mass spectrometer. Lipids analyzed in positive and negative ionization mode were separated using a Luna® C5 column, 5 μm particle size, 100 Å, 50 × 4.6 mm. For positive mode, mobile phase A was 95:5 water:methanol, 0.1 % formic acid, 5 mM ammonium formate; mobile phase B was 60:35:5 isopropanol:methanol:water, 0.1 % formic acid, 5 mM ammonium formate. For negative mode, mobile phase A was 95:5 water:methanol, 0.1 % ammonium hydroxide, 5 mM ammonium formate; mobile phase B was 60:35:5 isopropanol:methanol:water, 0.1 % ammonium hydroxide, 5 mM ammonium formate. For both positive and negative mode, the gradient was t = 0, 0% B, 100 μL/min; t = 5.00, 0% B, 100 μL/min; t = 5.01, 0% B, 400 μL/min; t = 45.00, 100% B, 400 μL/min; t = 45.01, 100% B, 500 μL/min; t = 55.00, 100% B, 500 μL/min; t = 55.01, 0% B, 400 μL/min. Peak identify and intergration were performed using Aglient MassHunter Software version 8.06.00 (Aglient Technologies, Inc.).
Measurement of lysosomal permeability
LLOMe treatment, LysoTracker Red staining, and flow cytometry were performed according to previously published protocols (Repnik et al., 2016). Briefly, HEK293T cells were harvested and transferred to 5 mL polystyrene tubes and treated with the indicated concentration of LLOMe for 10 minutes at 37°C. Cells were then centrifuged to pellet, resuspended in complete media containing 50 nM LysoTracker Red DND-99, and incubated at 37°C for 15 minutes. Cells were centrifuged again and resuspended in 1X PBS before flow cytometry analysis. Cytometry was performed on an LSR Fortessa instrument (BD Biosciences). Forward and side scatter parameters were collected, and LysoTracker Red was excited using a 561 nm laser line and collected with a 586/15 nm emission filter. Cell aggregates were discriminated by gating FSC area against FSC height, and 100,000 single cell events were collected per condition.
QUANTIFICATION AND STATISTICAL ANALYSIS
All graphs were assembled and statistics were performed using Prism 8 (GraphPad) with the exception of flow cytometry histograms, which were generated using FlowJo version 10 (BD). Error bars on all graphs are shown as the mean ± SD. The details of each statistical test performed are given in the legend accompanying each figure. Unless otherwise indicated, all co-localization analysis was performed on 10 non-overlapping fields that contain a minimum of 3 cells per field. Unless otherwise indicated, all proteomic and lipidomic measurements were performed on three independent biological replicates for each sample.
Supplementary Material
Table S1. List of proteins and spectral abundances identified from Lyso-IP proteomics. Related to Figures 1, 2, and 6.
Table S2. List of LC-MS peak intensities for lipid species extracted from purified lysosomes. Related to Figure 4.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-HA-Tag | Cell Signaling Technology | Cat#3724; RRID:AB_1549585 |
| Rabbit monoclonal anti-LC3B | Cell Signaling Technology | Cat#3868; RRID:AB_2137707 |
| Rabbit monoclonal anti-SQSTM1/p62 | Cell Signaling Technology | Cat#39749; RRID:AB_2799160 |
| Rabbit monoclonal anti-PEX5 | Cell Signaling Technology | Cat#83020; RRID:AB_2800006 |
| Rabbit monoclonal anti-GM130 | Cell Signaling Technology | Cat#12480; RRID:AB_2797933 |
| Rabbit monoclonal anti-PDI | Cell Signaling Technology | Cat#3501; RRID:AB_2156433 |
| Rabbit monoclonal anti-TAX1BP1 | Cell Signaling Technology | Cat#5105; RRID:AB_11178939 |
| Rabbit monoclonal anti-GABARAP | Cell Signaling Technology | Cat#13733; RRID:AB_2798306 |
| Rabbit monoclonal anti-VDAC | Cell Signaling Technology | Cat#4661; RRID:AB_10557420 |
| Rabbit polyclonal anti-LC3B | Cell Signaling Technology | Cat#2775; RRID:AB_915950 |
| Mouse monoclonal anti-LAMP1 | Cell Signaling Technology | Cat#15665; RRID:AB_2798750 |
| Rabbit monoclonal anti-DYKDDDK Tag | Cell Signaling Technology | Cat#2368 |
| Rabbit monoclonal anti-phospho-p70 S6 Kinase (Thr389) | Cell Signaling Technology | Cat#9234; RRID:AB_2269803 |
| Rabbit monoclonal anti-p70 S6 Kinase | Cell Signaling Technology | Cat#2708; RRID:AB_390722 |
| Rabbit polyclonal anti-phospho-4E-BP1 (Ser65) | Cell Signaling Technology | Cat#9451 |
| Rabbit monoclonal anti-4E-BP1 | Cell Signaling Technology | Cat#9644; RRID:AB_2097841 |
| Rabbit monoclonal anti-mTOR | Cell Signaling Technology | Cat#2983; RRID:AB_2105622 |
| Rabbit monoclonal anti-β-Actin | Cell Signaling Technology | Cat#4970 |
| Rabbit monoclonal anti-NBR1 | Cell Signaling Technology | Cat#9891; RRID:AB_10949888 |
| Rabbit monoclonal anti-Raptor | Cell Signaling Technology | Cat#2280; RRID:AB_5612451 |
| Rabbit monoclonal anti-LAMTOR5/HBXIP | Cell Signaling Technology | Cat#14633; RRID:AB_2798547 |
| Rabbit monoclonal anti-phospho-Ubiquitin (Ser65) | Cell Signaling Technology | Cat#62802; RRID:AB_2799632 |
| Rabbit monoclonal anti-PINK1 | Cell Signaling Technology | Cat#6946; RRID:AB_11179069 |
| Rabbit monoclonal anti-Citrate Synthase | Cell Signaling Technology | Cat#14309; RRID:AB_2665545 |
| Rabbit monoclonal anti-Mitofusin-1 | Cell Signaling Technology | Cat#14739; RRID:AB_2744531 |
| Rabbit monoclonal anti-NPC1 | abcam | Cat#ab134113; RRID:AB_2734695 |
| Rabbit polyclonal anti-CHMP2B | abcam | Cat#ab33174; RRID:AB_2079471 |
| Rabbit monoclonal anti-TMEM192 | abcam | Cat#ab185545 |
| Rabbit polyclonal anti-ARA70 (NCOA4) | Bethyl Laboratories | Cat#A302-272A; RRID:AB_1850160 |
| Rabbit polyclonal anti-NPC2 | Proteintech | Cat#19888-1-AP; RRID:AB_10639363 |
| Rabbit polyclonal anti-CHMP1B | Proteintech | Cat#14639-1-AP; RRID:AB_2079353 |
| Rabbit polyclonal anti-hIST1 | Proteintech | Cat#19842-1-AP |
| Rabbit polyclonal anti-Parkin | Proteintech | Cat#14060-1-AP |
| Rabbit polyclonal anti-MTCO2 (COXII) | Proteintech | Cat#55070-1-AP; RRID:AB_10859832 |
| Rabbit polyclonal anti-GALC | Proteintech | Cat#11991-1-AP; RRID:AB_10641987 |
| Mouse monoclonal anti-LAMP-2 | Santa Cruz Biotechnology | Cat#sc-18822; RRID:AB_626858 |
| Mouse monoclonal anti-CHMP1A | Santa Cruz Biotechnology | Cat#sc-271617; RRID:AB_10707967 |
| Mouse monoclonal anti-TSG101 | Santa Cruz Biotechnology | Cat#sc-7964; RRID:AB_671392 |
| Rabbit polyclonal anti-TOM40 | Santa Cruz Biotechnology | Cat#sc-11414; RRID:AB_793274 |
| Mouse monoclonal anti-CLN1 (PPT1) | Santa Cruz Biotechnology | Cat#sc-517323 |
| Mouse monoclonal anti-CLN2 (TPP1) | Santa Cruz Biotechnology | Cat#sc-393961 |
| Rat monoclonal anti-LAMP1 | Development Studies Hybridoma Bank | Cat#1D4B; RRID:AB_2134500 |
| Mouse monoclonal anti-VPS4A | Sigma Aldrich | Cat# SAB4200215; RRID:AB_10669419 |
| Mouse monoclonal anti-Alix | Biolegend | Cat# 634502; RRID:AB_2162471 |
| Chicken polyclonal anti-MAP2 | Encor Biotechnology | Cat#CPCA-MAP2; RRID:AB_2138173 |
| Mouse monoclonal anti-Galectin-3 | BD Biosciences | Cat#556904; RRID:AB_396531 |
| Goat polyclonal anti-Cathepsin Z | R&D Systems | Cat#AF934; RRID:AB_2087676 |
| Mouse monoclonal anti-GBA | R&D Systems | Cat#MAB7410 |
| Goat anti-Rabbit IgG, peroxidase labeled | Vector Laboratories | Cat#PI-1000; RRID:AB_2336198 |
| Goat anti-Mouse IgG, peroxidase labeled | Sigma Aldrich | Cat#A4416; RRID:AB_258167 |
| Goat anti-Chicken IgY, AlexaFluor647 labeled | ThermoFisher | Cat#A21449; RRID:AB_2535866 |
| Goat anti-Mouse IgG, AlexaFluor488 labeled | ThermoFisher | Cat#A11001; RRID:AB_2534069 |
| Goat anti-Mouse IgG, AlexaFluor568 labeled | ThermoFisher | Cat#A11004; RRID:AB_2534072 |
| Goat anti-Rabbit IgG, AlexaFluor488 labeled | ThermoFisher | Cat#A11011; RRID:AB_143157 |
| Goat anti-Rabbit IgG, AlexaFluor568 labeled | ThermoFisher | Cat#A11008; RRID:AB_143165 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Leupeptin hemisulfate | Alfa Aesar | Cat#J61188; CAS: 103476-89-7 |
| Pepstatin A | MP Biomedicals | Cat#195368; CAS: 26305-03-3 |
| Bafilomycin A1 | Alfa Aesar | Cat#J61835; CAS: 88899-55-2 |
| Leu-Leu methyl ester hydrobromide (LLOMe) | Sigma Aldrich | Cat#L7393; CAS: 16689-14-8 |
| Torin1 | Tocris Bioscience | Cat#4247; CAS: 1222998-36-8 |
| Rapamycin | Calbiochem | Cat#553210; CAS: 53123-88-9 |
| Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) | Sigma Aldrich | Cat#C2759; CAS: 555-60-2 |
| LysoTracker Red DND-99 | ThermoFisher | Cat#L7528; CAS: 231946-72-8 |
| LysoLive GalGreen fluorogenic substrate | MarkerGene Technologies | Cat#M2776 |
| OptiPrep Density Gradient Medium | Sigma Aldrich | Cat#D1556; CAS: 92339-11-2 |
| Methyl-ß-cyclodextrin (MCD) | Sigma Aldrich | Cat#C4555; CAS: 128446-36-6 |
| Cholesterol | Sigma Aldrich | Cat#C3045; CAS: 57-88-5 |
| Human low-density lipoprotein (LDL) | Alfa Aesar | Cat#J65309 |
| Normal Donkey Serum | Jackson ImmunoResearch | Cat#017-000-121; RRID: AB_2337258 |
| 15:0-18:1 phosphocholine | Avanti Polar Lipids | Cat#330716; CAS: 253127-55-8 |
| Tert-butyl methyl ether (TBME) | Sigma Aldrich | Cat#306975; CAS: 1634-04-4 |
| OSW-1 | Shair Laboratory, Harvard University | Burgett, et al., 2011 |
| Filipin | Sigma Aldrich | Cat#F9765; CAS: 11078-21-0 |
| U18666A | Tocris Bioscience | Cat#1638; CAS: 3039-71-2 |
| Deposited Data | ||
| Lyso-IP Proteomics | Zoncu Laboratory | This study (table S1) |
| Lyso-IP Lipidomics | Zoncu Laboratory | This study (table S2) |
| Experimental Models: Cell Lines | ||
| HEK293T sgNT | Zoncu Laboratory | Castellano, et al. 2017 |
| HEK293T sgNPC1 | Zoncu Laboratory | Castellano, et al. 2017 |
| HEK293T sgNT TMEM192-RFP-3xHA | Zoncu Laboratory | This study |
| HEK293T sgNPC1 TMEM192-RFP-3xHA | Zoncu Laboratory | This study |
| HEK293T sgNT TMEM192-FLAG | Zoncu Laboratory | This study |
| HEK293T sgNPC1 NPC1(WT)-FLAG | Zoncu Laboratory | This study |
| HEK293T sgNPC1 NPC1(P202/3A)-FLAG | Zoncu Laboratory | This study |
| HEK293T sgNPC1 NPC1(F503/4A)-FLAG | Zoncu Laboratory | This study |
| HEK293T sgNPC1 NPC1(P691S)-FLAG | Zoncu Laboratory | This study |
| HEK293T sgNPC1 NPC1(D786N)-FLAG | Zoncu Laboratory | This study |
| MEF WT (NPC1+/+) | (Loftus et al.,1997) | |
| MEF NPC1−/− | (Loftus et al., 1997) | |
| MEF WT TMEM192-RFP-3xHA | Zoncu Laboratory | This study |
| MEF NPC1−/− TMEM192-RFP-3xHA | Zoncu Laboratory | This study |
| hIPSC Control (NPC1+/+) | Goldstein Laboratory | This study |
| hIPSC NPC1−/− | Goldstein Laboratory | This study |
| Recombinant DNA | ||
| pLJM1-TMEM192-RFP-3xHA | Zoncu Laboratory | Lim, et al. 2019 |
| pLJM1-TMEM192-FLAG | Zoncu Laboratory | This study |
| pLVX-NPC1(WT)-FLAG | Zoncu Laboratory | This study |
| pLVX-NPC1(P202/3A)-FLAG | Zoncu Laboratory | This study |
| pLVX-NPC1(F503/4A)-FLAG | Zoncu Laboratory | This study |
| pLVX-NPC1(P691S)-FLAG | Zoncu Laboratory | This study |
| pLVX-NPC1(D786N)-FLAG | Zoncu Laboratory | This study |
| pLKO.1-shLuciferase | The RNAi Consortium, Broad Institute | https://portals.broadinstitute.org/gpp/public/ |
| pLKO.1-shLAMTOR5 | The RNAi Consortium, Broad Institute | https://portals.broadinstitute.org/gpp/public/ |
| Software and Algorithms | ||
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | BD Biosciences | https://www.flowjo.com/ |
| FIJI v.2.0.0-rc-69/1.52i | NIH | https://imagej.net/Fiji |
| Other | ||
| Pierce Anti-HA Magnetic Beads | ThermoFisher | Cat#88836 |
Highlights:
Proteomic profiling of NPC lysosomes reveal both proteolytic and structural defects
Loss of cholesterol transport activity by NPC1 causes aberrant mTORC1 signaling
mTORC1 inhibition restores lysosomal and mitochondrial function in NPC cells
Acknowledgements
We thank all members of the Zoncu Lab for helpful insights. This work was supported by NIH R01GM127763 and R01GM130995, a Shirl and Kay Curci Foundation Grant and a University of Notre Dame/APMRF grant to R.Z., a University of Notre Dame/APMRF 20174028 grant to M.P.O. and a 2019 AACR-Amgen Fellowship in Clinical/Translational Cancer Research (19–40-11-SHIN) to H.R.S.
Footnotes
Declaration of Interests
R.Z. is co-founder, stockholder and scientific advisor for Frontier Medicines Corp.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, and Sabatini DM (2017). Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandapadamanaban M, Masson GR, Perisic O, Berndt A, Kaufman J, Johnson CM, Santhanam B, Rogala KB, Sabatini DM, and Williams RL (2019). Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrzejewska Z, Nevo N, Thomas L, Chhuon C, Bailleux A, Chauvet V, Courtoy PJ, Chol M, Guerrera IC, and Antignac C. (2016). Cystinosin is a Component of the Vacuolar H+-ATPase-Ragulator-Rag Complex Controlling Mammalian Target of Rapamycin Complex 1 Signaling. J. Am. Soc. Nephrol. 27, 1678–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo MEG, Naschberger A, Fürnrohr BG, Stasyk T, Dunzendorfer-Matt T, Lechner S, Welti S, Kremser L, Shivalingaiah G, Offterdinger M, et al. (2017). Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science 358, 377–381. [DOI] [PubMed] [Google Scholar]
- Ballabio A, and Gieselmann V. (2009). Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta 1793, 684–696. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, and Sabatini DM (2012). Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomeo R, Cinque L, De Leonibus C, Forrester A, Salzano AC, Monfregola J, De Gennaro E, Nusco E, Azario I, Lanzara C, et al. (2017). mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J. Clin. Invest. 127, 3717–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, and Manning BD (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloëtta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. (2008). Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8, 411–424. [DOI] [PubMed] [Google Scholar]
- Brown RA, Voit A, Srikanth MP, Thayer JA, Kingsbury TJ, Jacobson MA, Lipinski MM, Feldman RA, and Awad O. (2019). mTOR hyperactivity mediates lysosomal dysfunction in Gaucher’s disease iPSC-neuronal cells. Dis Model Mech 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgett AWG, Poulsen TB, Wangkanont K, Anderson DR, Kikuchi C, Shimada K, Okubo S, Fortner KC, Mimaki Y, Kuroda M, et al. (2011). Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nat. Chem. Biol. 7, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, and Mootha VK (2016). MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle REN, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, et al. (2017). Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355, 1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, and Roux A. (2015). Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 163, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C, Puthanveetil P, Ory DS, and Lieberman AP (2016). Genetic and pharmacological evidence implicates cathepsins in Niemann-Pick C cerebellar degeneration. Hum. Mol. Genet. 25, 1434–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY-S, Shin HR, Berdan CA, Ford B, Ward CC, Olzmann JA, Zoncu R, and Nomura DK (2019). Covalent targeting of the vacuolar H+-ATPase activates autophagy via mTORC1 inhibition. Nat. Chem. Biol. 15, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Fendt S-M, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, et al. (2013). The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, and Puigserver P. (2007). mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740. [DOI] [PubMed] [Google Scholar]
- Danyukova T, Ariunbat K, Thelen M, Brocke-Ahmadinejad N, Mole SE, and Storch S. (2018). Loss of CLN7 results in depletion of soluble lysosomal proteins and impaired mTOR reactivation. Hum. Mol. Genet. 27, 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, and De Camilli P. (2016). Endosome-ER Contacts Control Actin Nucleation and Retromer Function through VAP-Dependent Regulation of PI4P. Cell 166, 408–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. (2010). Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Saffari A, Wahlster L, Di Nardo A, Turner D, Lewis TL, Conrad C, Rothberg JM, Lipton JO, Kölker S, et al. (2016). Impaired Mitochondrial Dynamics and Mitophagy in Neuronal Models of Tuberous Sclerosis Complex. Cell Rep 17, 1053–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrick MJ, Yu T, Chung C, and Lieberman AP (2012). Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum. Mol. Genet. 21, 4876–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltes M, Gale SE, Moores S, Ory DS, and Schaffer JE (2020). Monitoring the itinerary of lysosomal cholesterol in Niemann-Pick Type C1-deficient cells after cyclodextrin treatment. J. Lipid Res. 61, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhou Y, Goldstein JL, Brown MS, and Radhakrishnan A. (2017). Cholesterol-induced conformational changes in the sterol-sensing domain of the Scap protein suggest feedback mechanism to control cholesterol synthesis. J. Biol. Chem. 292, 8729–8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Qian H, Zhou X, Wu J, Wan T, Cao P, Huang W, Zhao X, Wang X, Wang P, et al. (2016). Structural Insights into the Niemann-Pick C1 (NPC1)-Mediated Cholesterol Transfer and Ebola Infection. Cell 165, 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämälistö S, Stahl JL, Favaro E, Yang Q, Liu B, Christoffersen L, Loos B, Guasch Boldú C, Joyce JA, Reinheckel T, et al. (2020). Spatially and temporally defined lysosomal leakage facilitates mitotic chromosome segregation. Nat Commun 11, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante RE, and Radhakrishnan A. (2017). Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, and Goldstein JL (2008). NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15287–15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, et al. (2012). Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature 482, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova EA, van den Heuvel LP, Elmonem MA, De Smedt H, Missiaen L, Pastore A, Mekahli D, Bultynck G, and Levtchenko EN (2016). Altered mTOR signalling in nephropathic cystinosis. J. Inherit. Metab. Dis. 39, 457–464. [DOI] [PubMed] [Google Scholar]
- Jia J, Claude-Taupin A, Gu Y, Choi SW, Peters R, Bissa B, Mudd MH, Allers L, Pallikkuth S, Lidke KA, et al. (2020). Galectin-3 Coordinates a Cellular System for Lysosomal Repair and Removal. Dev. Cell 52, 69–87.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, and Youle RJ (2010). Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, and Youle RJ (2014). PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, and Muqit MMK (2014). Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BE, Madreiter CT, Vishnu N, Malli R, Graier WF, and Karten B. (2014). Adaptations of energy metabolism associated with increased levels of mitochondrial cholesterol in Niemann-Pick type C1-deficient cells. J. Biol. Chem. 289, 16278–16289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Behl C, and Dikic I. (2016). Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol. 26, 6–16. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, and Guan K-L (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Beuchat MH, Lindsay M, Frias S, Palmiter RD, Sakuraba H, Parton RG, and Gruenberg J. (1999). Late endosomal membranes rich in lysobisphosphatidic acid regulate cholesterol transport. Nat. Cell Biol. 1, 113–118. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, and Infante RE (2009). Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. (2012). Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RE, and Zoncu R. (2019). The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, and Youle RJ (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Brown MS, and Goldstein JL (2010). Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 3441–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Saha P, Li J, Blobel G, and Pfeffer SR (2016). Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proc. Natl. Acad. Sci. U.S.A. 113, 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C-Y, Davis OB, Shin HR, Zhang J, Berdan CA, Jiang X, Counihan JL, Ory DS, Nomura DK, and Zoncu R. (2019). ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat. Cell Biol. 21, 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY, and Sabatini DM (2020). mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu EA, Schultz ML, Mochida C, Chung C, Paulson HL, and Lieberman AP (2020). Fbxo2 mediates clearance of damaged lysosomes and modifies neurodegeneration in the Niemann-Pick C brain. JCI Insight. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld MA, Tagle DA, et al. (1997). Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277, 232–235. [DOI] [PubMed] [Google Scholar]
- Lu F, Liang Q, Abi-Mosleh L, Das A, De Brabander JK, Goldstein JL, and Brown MS (2015). Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. (2013). Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, and Fairn GD (2015). Complementary probes reveal that phosphatidylserine is required for the proper transbilayer distribution of cholesterol. J. Cell. Sci. 128, 1422–1433. [DOI] [PubMed] [Google Scholar]
- Mahoney SJ, Narayan S, Molz L, Berstler LA, Kang SA, Vlasuk GP, and Saiah E. (2018). A small molecule inhibitor of Rheb selectively targets mTORC1 signaling. Nat Commun 9, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, and Puertollano R. (2012). MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, and Manning BD (2014). Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, and Antonny B. (2013). A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155, 830–843. [DOI] [PubMed] [Google Scholar]
- Millard EE, Gale SE, Dudley N, Zhang J, Schaffer JE, and Ory DS (2005). The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J. Biol. Chem. 280, 28581–28590. [DOI] [PubMed] [Google Scholar]
- Morita M, Gravel S-P, Chénard V, Sikström K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. (2013). mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711. [DOI] [PubMed] [Google Scholar]
- Morita M, Prudent J, Basu K, Goyon V, Katsumura S, Hulea L, Pearl D, Siddiqui N, Strack S, McGuirk S, et al. (2017). mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol. Cell 67, 922–935.e5. [DOI] [PubMed] [Google Scholar]
- Nguyen HC, Talledge N, McCullough J, Sharma A, Moss FR, Iwasa JH, Vershinin MD, Sundquist WI, and Frost A. (2020). Membrane constriction and thinning by sequential ESCRT-III polymerization. Nat. Struct. Mol. Biol. 27, 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez MP (2012). Defective mitophagy in human Niemann-Pick Type C1 neurons is due to abnormal autophagy activation. Autophagy 8, 1157–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez MP, Roberts EA, Kidwell CU, Yuan SH, Plaisted WC, and Goldstein LSB (2012). Disruption and therapeutic rescue of autophagy in a human neuronal model of Niemann Pick type C1. Hum. Mol. Genet. 21, 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong S-E, Walford GA, Sugiana C, Boneh A, Chen WK, et al. (2008). A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RM, and Zoncu R. (2016). The Lysosome as a Regulatory Hub. Annu. Rev. Cell Dev. Biol. 32, 223–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. (2011). mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR (2019). NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J. Biol. Chem. 294, 1706–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt FM, d’Azzo A, Davidson BL, Neufeld EF, and Tifft CJ (2018). Lysosomal storage diseases. Nat Rev Dis Primers 4, 27. [DOI] [PubMed] [Google Scholar]
- Puente C, Hendrickson RC, and Jiang X. (2016). Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy. J. Biol. Chem. 291, 6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic M, Schink KO, Wenzel EM, Nähse V, Bongiovanni A, Lafont F, and Stenmark H. (2018). ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repnik U, Česen MH, and Turk B. (2016). The Use of Lysosomotropic Dyes to Exclude Lysosomal Membrane Permeabilization. Cold Spring Harb Protoc 2016. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, and Ferguson SM (2012). The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, de Stanchina E, et al. (2016). Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 534, 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogala KB, Gu X, Kedir JF, Abu-Remaileh M, Bianchi LF, Bottino AMS, Dueholm R, Niehaus A, Overwijn D, Fils A-CP, et al. (2019). Structural basis for the docking of mTORC1 on the lysosomal surface. Science 366, 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P, and Klumperman J. (2009). Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10, 623–635. [DOI] [PubMed] [Google Scholar]
- Sakamachi Y, Morioka S, Mihaly SR, Takaesu G, Foley JF, Fessler MB, and Ninomiya-Tsuji J. (2017). TAK1 regulates resident macrophages by protecting lysosomal integrity. Cell Death Dis 8, e2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, and Sabatini DM (2010). Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. (2009). A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Carroll B, Buganim Y, Maetzel D, Ng AHM, Cassady JP, Cohen MA, Chakraborty S, Wang H, Spooner E, et al. (2013). Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Rep 5, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedin S, Sindelar PJ, Pentchev P, Brunk U, and Dallner G. (1997). Peroxisomal impairment in Niemann-Pick type C disease. J. Biol. Chem. 272, 6245–6251. [DOI] [PubMed] [Google Scholar]
- Schöneberg J, Lee I-H, Iwasa JH, and Hurley JH (2017). Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 18, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber KH, Arriola Apelo SI, Yu D, Brinkman JA, Velarde MC, Syed FA, Liao C-Y, Baar EL, Carbajal KA, Sherman DS, et al. (2019). A novel rapamycin analog is highly selective for mTORC1 in vivo. Nat Commun 10, 3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz ML, Krus KL, Kaushik S, Dang D, Chopra R, Qi L, Shakkottai VG, Cuervo AM, and Lieberman AP (2018). Coordinate regulation of mutant NPC1 degradation by selective ER autophagy and MARCH6-dependent ERAD. Nat Commun 9, 3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. (2012). A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra ML, Schlesinger PH, Naismith TV, and Hanson PI (2018). Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat DE, Sun P, Wiseman JA, Huang L, El-Banna M, Zheng H, Moore DF, and Lobel P. (2013). Extending the mannose 6-phosphate glycoproteome by high resolution/accuracy mass spectrometry analysis of control and acid phosphatase 5-deficient mice. Mol. Cell Proteomics 12, 1806–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M-Y, Morris KL, Kim DJ, Fu Y, Lawrence R, Stjepanovic G, Zoncu R, and Hurley JH (2017). Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex. Mol. Cell 68, 835–846.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, and Gray NS (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, and Prinz WA (2013). Direct imaging reveals stable, micrometer-scale lipid domains that segregate proteins in live cells. J. Cell Biol. 202, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Fujimoto M, Tatematsu T, Cheng J, Orii M, Takatori S, and Fujimoto T. (2017). Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley SU, and Suzuki K. (2004). Consequences of NPC1 and NPC2 loss of function in mammalian neurons. Biochim. Biophys. Acta 1685, 48–62. [DOI] [PubMed] [Google Scholar]
- Wang S, Tsun Z-Y, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, et al. (2015). Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Simicek M, Schubert A, and Komander D. (2015). Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RA, Yen FS, Nicholson SPV, Alwaseem H, Bayraktar EC, Alam M, Timson RC, La K, Abu-Remaileh M, Molina H, et al. (2020). Maintaining Iron Homeostasis Is the Key Role of Lysosomal Acidity for Cell Proliferation. Mol. Cell 77, 645–655.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler MBL, Kidmose RT, Szomek M, Thaysen K, Rawson S, Muench SP, Wüstner D, and Pedersen BP (2019). Structural Insight into Eukaryotic Sterol Transport through Niemann-Pick Type C Proteins. Cell 179, 485–497.e18. [DOI] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, and Sabatini DM (2017). mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell 171, 642–654.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe D, Xia Z-P, Adams CM, and Rawson RB (2002). Three mutations in sterol-sensing domain of SCAP block interaction with insig and render SREBP cleavage insensitive to sterols. Proc. Natl. Acad. Sci. U.S.A. 99, 16672–16677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yambire KF, Fernandez-Mosquera L, Steinfeld R, Mühle C, Ikonen E, Milosevic I, and Raimundo N. (2019a). Mitochondrial biogenesis is transcriptionally repressed in lysosomal lipid storage diseases. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yambire KF, Rostosky C, Watanabe T, Pacheu-Grau D, Torres-Odio S, Sanchez-Guerrero A, Senderovich O, Meyron-Holtz EG, Milosevic I, Frahm J, et al. (2019b). Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gong J-S, Ko M, Garver WS, Yanagisawa K, and Michikawa M. (2005). Altered cholesterol metabolism in Niemann-Pick type C1 mouse brains affects mitochondrial function. J. Biol. Chem. 280, 11731–11739. [DOI] [PubMed] [Google Scholar]
- Yuan SH, Martin J, Elia J, Flippin J, Paramban RI, Hefferan MP, Vidal JG, Mu Y, Killian RL, Israel MA, et al. (2011). Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS ONE 6, e17540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, and Sabatini DM (2011). mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of proteins and spectral abundances identified from Lyso-IP proteomics. Related to Figures 1, 2, and 6.
Table S2. List of LC-MS peak intensities for lipid species extracted from purified lysosomes. Related to Figure 4.
Data Availability Statement
The supplemental tables accompanying the published article includes all datasets generated in this study.