Abstract
BACKGROUND AND AIMS:
Hepatocytes play a central role in storage and utilization of fat by the liver. Selective breakdown of lipid droplets (LDs) by autophagy (also called lipophagy) is a key process utilized to catabolize these lipids as an energy source. How the autophagic machinery is selectively targeted to LDs, where it mediates membrane engulfment and subsequent degradation, is unclear. Recently, we have reported that two distinct GTPases, the mechanoenzyme, dynamin2 (Dyn2), and the small regulatory Rab GTPase, Rab10, work independently at distinct steps of lipophagy in hepatocytes.
APPROACH AND RESULTS:
In an attempt to understand how these proteins are regulated and recruited to autophagic organelles, we performed a nonbiased biochemical screen for Dyn2-binding partners and found that Dyn2 actually binds Rab10 directly through a defined effector domain of Rab10 and the middle domain of Dyn2. These two GTPases can be observed to interact transiently on membrane tubules in hepatoma cells and along LD-centric autophagic membranes. Most important, we found that a targeted disruption of this interaction leads to an inability of cells to trim tubulated cytoplasmic membranes, some of which extend from lipophagic organelles, resulting in LD accumulation.
CONCLUSIONS:
This study identifies a functional, and direct, interaction between Dyn2 and a regulatory Rab GTPase that may play an important role in hepatocellular metabolism.
Membrane trafficking plays an essential role in supporting many hepatocellular processes that include secretion of plasma proteins into the sinusoidal space, endocytic filtering of the blood, and secretion of bile into the apical canalicular space.(1) All of these processes are supported by a complex vesicular transport machinery that utilizes a cast of adaptors, molecular motors, and regulatory enzymes superimposed upon the cytoskeleton. In addition to hepatocellular secretory and endocytic functions, these membrane-trafficking pathways are also paramount to the liver’s ability to process and catabolize ingested lipids stored as lipid droplets (LDs). How the hepatocyte-trafficking machinery responds to a steady diet of cholesterol and saturated fats are of great interest because of the marked increase in western populations of alcoholic and nonalcoholic fatty liver as well as alcoholic and nonalcoholic steatohepatitis. Whereas LDs can be reduced by lipolysis by the action of cytoplasmic lipases,(2) it is now known that a modified autophagic pathway (lipophagy) plays a key role in LD catabolism in hepatocytes.(3–6) Thus, membrane trafficking is essential in mediating the targeting of LDs for lipophagic degradation, as well as the subsequent assembly, expansion, and maturation (lysosomal fusion) of the developing autophagosome.
All of the membrane-trafficking–dependent processes described above require the action of membrane-severing enzymes that vesiculate membranes from a donor compartment for subsequent transport to a desired location.
Key to this severing process is a well-conserved family of large GTPases termed the dynamins (Dyn1, −2, and −3) that convert guanosine triphosphate (GTP) hydrolysis into mechanical work to compress and break biological membranes. The conventional dynamins have been implicated to sever a variety of different membrane compartments from clathrin-coated pits(7,8) to caveolae(9) to autophagolysosomes.(10) In addition, dynamin2 (Dyn2), the ubiquitous form of dynamin that is expressed in all tissues,(11) has been implicated in the severing of autolysosomes in hepatocytes.(10) Importantly, inhibition of Dyn2 leads to the formation of exceptionally long autophagic lysosome reformation (ALR) tubules that are unable to generate nascent lysosomes used in the autophagic degradation of hepatocellular LDs resulting in fatty cells.(10,12)
How these vesiculated donor membrane compartments are transported and directed to the appropriate target membrane is a complex process and a topic of intense study given that mis-targeting of cargo can lead to a variety of disease conditions. One exceptionally large family of proteins that is known to regulate these key processes is the Rab family of small GTPases. There are well over 60 Rab proteins in the human genome that act as molecular switches to regulate various membrane-trafficking events. Most of these are expressed in the hepatocyte. Although the GTPase switch domains of these Rabs are highly conserved, they differ markedly at the “effector” and C-terminal domains that have been shown to interact with specific trafficking proteins that promote fusion, transport, and docking. For example, Rab7 is known to associate with late endocytic compartments, and in the GTP-bound state, Rab7 associates with Rab-interacting lysosomal protein that acts to connect this compartment to the cytoplasmic dynein motor for transport into the cell interior.(13) Rab7 has been shown to become activated in hepatocytes subjected to nutrient deprivation, stimulating its association with the LD surface and mediating recruitment of lysosomes to facilitate the lipophagic process.(14) More recently, a second Rab protein, Rab10, has also been shown to increase its association with LDs and has been implicated in recruitment of the autophagic machinery, such as light chain 3 (LC3) and autophagy-related 16 (Atg16), to hepatocyte LDs, where it mediates LD engulfment to facilitate lipophagy. Importantly, knockdown/knockout or inactivation of Rab10 attenuates lipophagy, resulting in steatotic hepatocytes.(15)
Thus, from the studies described above, it appears that two distinct families of large and small GTPases, Dyn2 and Rab10, may work synergistically to mediate the formation and maturation of a mature lipophagosome. In this study, we report that these GTPases act together in this process through a direct interaction to mediate LD catabolism in cultured human hepatoma and primary rat hepatocyte cells. In live cells coexpressing tagged Rab10 and Dyn2, we observed a dramatic, but transient, corecruitment of these proteins to membrane tubules and LDs. Biochemical analysis demonstrates a direct interaction of the Rab10 effector domain with the Dyn2 middle domain and a strong affinity for the Dyn2 middle domain with the LD surface. Importantly, knockdown of these proteins leads to membrane tubulation, deformed lipophagic organelles, and steatotic cells. These findings provide an example of an interaction between distinct large and small GTPases, and suggest that this binding plays a key function in hepatocellular lipophagy and lipid catabolism. The importance of the Rab10 family in regulating hepatocyte metabolism is discussed.
Materials and Methods
CELL CULTURE AND REAGENTS
The Hep3B2.1–7 (Hep3B) cell line was obtained from ATCC (HB-8064; ATCC, Manassas, VA); the HuH-7 cell line was from the laboratory of Dr. Greg Gores (Mayo Clinic, Rochester, MN). The Rab10 knockout fibroblasts were supplied by the laboratory of David James (University of Sydney, Australia). Cells were maintained in minimum essential medium (MEM) media containing 10% fetal bovine serum (FBS), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (Life Technologies, Carlsbad, CA), and grown at 37°C in 5% CO2.
The rabbit Rab10 antibody was from Sigma-Aldrich (R8906; St. Louis, MO). The Rab10(4E2) antibody was from GeneTex (GTX82800; Irvine, CA). Dyn2 antibodies were prepared as detailed.(9) Alexa Fluor secondary antibodies were used for immunofluorescence along with ProLong Antifade reagent (Thermo Fisher Scientific, Waltham, MA). Horseradish peroxidase (HRP) secondary antibodies (Invitrogen) for western blotting analysis, detected using SuperSignal West Pico substrate (Thermo Fisher Scientific). Diacylglycerol O-acyltransferase (DGAT) inhibitors, were purchased from Sigma-Aldrich.
BORTEZOMIB TREATMENT
Bortezomib (1 μM; ps-341) and (10 μM) MG-132 (Selleckchem, Munich, Germany) were applied to cells 18 hours before the green fluorescent protein (GFP)-Trap or glutathione-S-transferase (GST) pull-down assay.
CONSTRUCTS, SMALL INTERFERING RNA, AND TRANSFECTION
Generation of Rab10 constructs was described.(15) pGEX-4T-1 was obtained from GE Healthcare. The active human Q68L or dominant-negative human T23N forms of Rab10 were cloned into pEGFP-C1 and pCMV-HA. Constructs encoding the GFP-tagged middle domain of dynamin-2 were described.(16) The Dyn2-mCherry construct was generated by amplifying complementary DNA from a rat liver and cloning into the pmCherry-N1 vector (Clontech). Lipofectamine 2000 (Invitrogen) was used for transfection. The small interfering RNA (siRNA) oligos targeting Rab10 and Dyn2 were from Dharmacon/Horizon Discovery Group (Lafayette, CO) and cells transfected with RNAiMAX (Thermo Fisher Scientific).
FLUORESCENCE MICROSCOPY
Cells were processed for immunofluorescence as (17) Images were acquired by a Zeiss LSM 780 confocal microscope (Carl Zeiss, Oberkochen, Germany). For assessment of endogenous LD content, cells were processed for Oil Red O staining and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD) as described.(18)
TIME-LAPSE MICROSCOPY
Cells were plated onto glass-bottomed imaging dishes (Cell E&G LLC, San Diego, CA) following transfection. Live cells were imaged using a Zeiss LSM 780 confocal microscope (Carl Zeiss) and maintained at 37°C and 5% CO2. Visualization of LDs in live cells was achieved by labeling with BODIPY-C12 (558/568; Thermo Fisher Scientific) or with malic dehydrogenase (MDH) reagent (Abgent, WuXi App Tec, Shanghai, China).(19)
GST PULL-DOWN BINDING ASSAY
Cells were lysed in TEN100 lysis buffer (20 mM of Tris [pH 7.4], 0.1 mM of ethylenediaminetetraacetic acid [EDTA], and 100 mM of NaCl), containing complete protease inhibitors (Roche, Basel, Switzerland). Purified GST-fused proteins were incubated in cell lysate for 2 hours at 4°C. After washing the beads four times with NTEN300 buffer (20 mM of Tris [pH 7.4], 0.1 mM of EDTA, 300 mM of NaCl, 0.05% Nonidet P-40 (NP-40), and complete protease inhibitors; Roche), samples were subjected to sodium dodecyl sulfate (SDS)/polyacrylamide gel electrophoresis (PAGE). Desired proteins were detected by immunoblotting.
COIMMUNOPRECIPITATION
Cells were collected into 1 mL of NP-40 lysis buffer containing complete protease inhibitors (Roche), sonicated, and centrifuged to remove nucleus and cell debris. Protein concentration was determined by bicinchoninic acid assay (Pierce/Thermo Fisher). Supernatants were incubated with Dyn2 antibodies for 2 hours before a 1-hour addition of Protein A sepharose (Sigma-Aldrich) beads. Samples were washed in NP-40 buffer containing 150 mM of NaCl three times and resolved by SDS-PAGE. For immunoprecipitations of GFP proteins, the protocol and reagents for GFP-Trap were used (Chromotek, Planegg-Martinsried, Germany).
ELECTRON MICROSCOPY
For conventional electron microscopy (EM), cells were processed and imaged as described.(9) For visualizing the late degradative compartments, cells were loaded with HRP for 90 minutes and chased for 2.5 hours before processing for transmission electron microscopy that included a peroxidase reaction step described.(10) Quantitative measurements of HRP tubule lengths were performed using Adobe Photoshop (Adobe Inc, San Jose, CA); full cell profiles of five cells were examined for each condition.
ISOLATION OF PRIMARY HEPATOCYTES
Hepatocytes were isolated from adult Sprague-Dawley rats as described.(20) Transfections were performed utilizing Lipofectamine 2000 (Invitrogen) and the RNAiMAX reagent (Thermo Fisher Scientific) for knockdown studies.
STATISTICAL ANALYSIS AND REPLICATION OF EXPERIMENTS
Statistical comparisons between two conditions were assessed by using the two-tailed paired Student t test, where numerical P values are reported as mean ± SD and represent data from a minimum of three independent experiments.
Results
A DIRECT INTERACTION OF GTPases ON MEMBRANOUS COMPARTMENTS IN HEPATOMA CELLS
To identify Dyn2-interacting factors that could mediate autophagosome (AP) membrane scission, we performed a screen from rat brain homogenate using the 220-amino-acid (aa) middle domain of Dyn2 immobilized to GST. Several prominent lower-molecular-weight bands were resolved that were not observed in the control samples of GST (Fig. 1A). These bands were sent for analysis by mass spectrometry and identified as the small GTPase, Rab10. To further confirm the association between Rab10 and Dyn2, immunoprecipitation of Dyn2 from Clone 9 cells also revealed an interaction with Rab10 (Fig. 1B). Unable to immunoprecipitate Rab10, we instead used a GST pull-down approach by combining bacterially expressed GST-Rab10 and His-Dyn2 proteins. Whereas GST alone did not bind His-Dyn2, the GST-Rab10 protein did pull down substantial Dyn2 protein, suggesting a direct interaction (Fig. 1C).
FIG. 1.
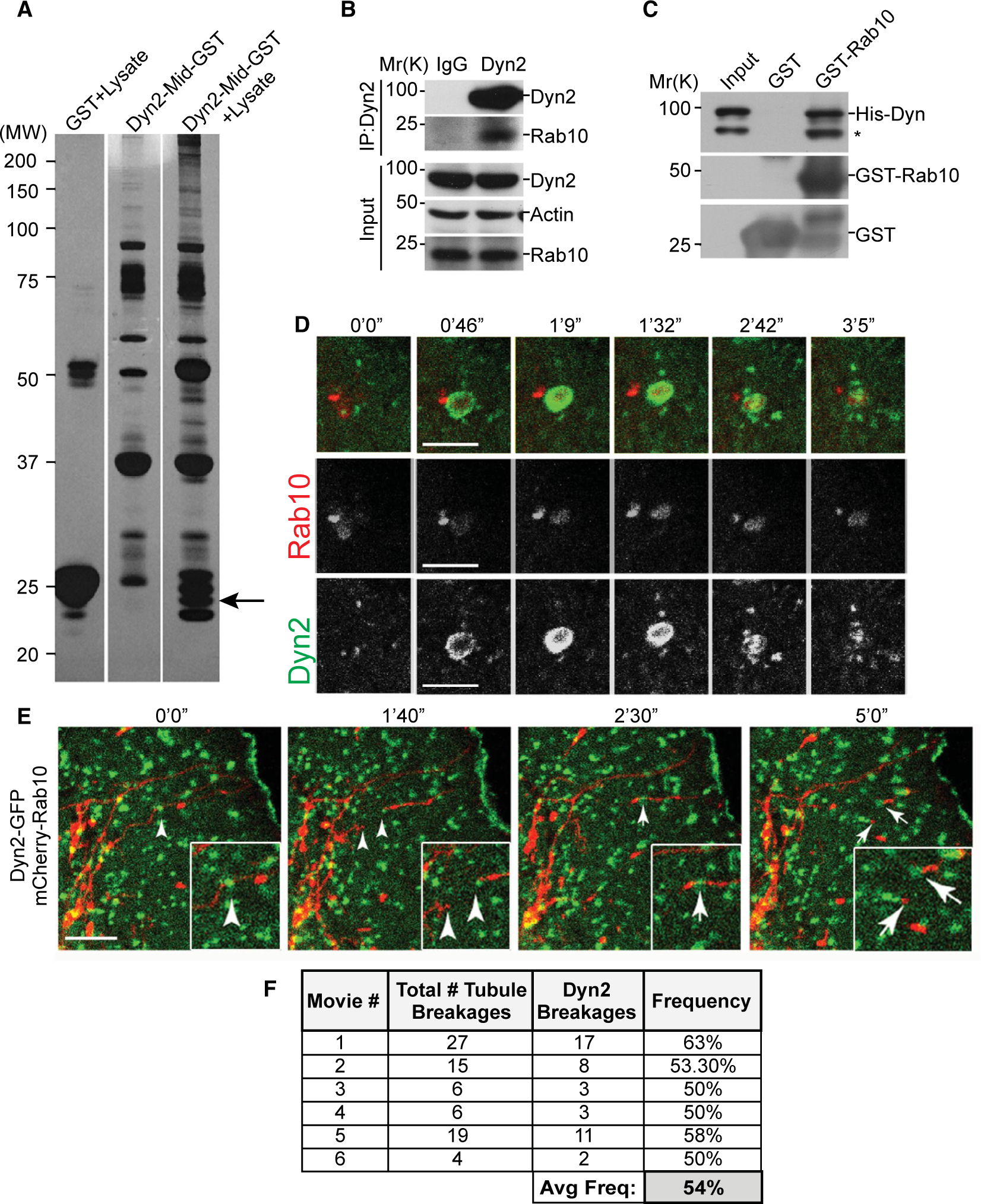
Rab10 and dynamin proteins interact in vitro and in living cells. (A) GST pull-down screen for proteins that bind to the middle domain of Dyn2 (Dyn2-Mid). Putative binding partners isolated from rat brain homogenate visualized by silver stain. (B) Immunoprecipitation of Rab10 by a Dyn2 antibody from Clone9 cell lysates. (C) Immunoblottings from the in vitro GST pull-down experiment using GST-Rab10 and Dyn2-His to examine direct binding. (D) Live-cell microscopy of 24-hour low-serum (0.1% FBS) starved Hep3B hepatoma cells cotransfected with Dyn2-GFP and mCherry-Rab10. (E) Time-lapse stills of a live Hep3B cell following 24-hour low-serum starvation, showing the recruitment of GFP-Dyn2 to a scission point of an mCherry-Rab10–coated membrane tubule. (F) Table displaying number and frequency of mCherry-Rab10 tubule scission events occurring with identifiable Dyn2-GFP puncta at a site of tubule breakage. Micron bars denote 5 μm. Abbreviations: Avg Freq, average frequency; His-Dyn, His-tagged dynamin; IgG, immunoglobulin G; IP, immunoprecipitation; MW, molecular weight.
With the biochemical confirmation of a Rab10-Dyn2 interaction, it was important to test whether these GTPases colocalize in live cells. Following expression of Dyn2-GFP and mCherry-Rab10, live cells were observed over 5- to 10-hour periods. Rab10-mCherry–labeled membranes were observed as crenated, vesicular structures that were observed to suddenly recruit a large intense spot of Dyn2-GFP represented as a yellow flash, demonstrating a transient, but deliberate association (Fig. 1D; Supporting Movie S1). This ephemeral overlap lasts just under 3 minutes and suggests a biological interaction.
In addition to the vesicular structures described above, Rab10 was also observed to coat long tubular membranes that extend randomly through the cytoplasm (Fig. 1E; Supporting Movie S2). Most interestingly, punctate spots of Dyn2-GFP could be observed to reside along these tubules at sites of ubule scission, supporting the premise of a functional interaction between Rab10 and Dyn2. Analysis revealed that over half of the scission events of these Rab10 tubules observed from multiple time-lapse series were coincident with the appearance of Dyn2-GFP puncta at the site of scission (Fig. 1F), strongly implying a role for Dyn2 in driving this event.
To identify the regions of Rab10 and Dyn2 interaction, we pursued systematic deletion/mutation analysis of bacterially expressed proteins (Fig. 2). Rab GTPases are known to have an “effector” domain unique to each Rab that acts to recruit specific proteins key to their targeting and function (Fig. 2A). GST pulldowns of full-length or truncated Rab10 protein showed that only the Rab10-N domain, but not the Rab10-C domain, interacts with Dyn2 (Fig. 2B). Furthermore, a deletion of residues 38–46 (FISTIGIDFΔ9) within the Rab10-N region that included the effector domain markedly decreased its binding affinity with Dyn2. A more refined deletion of residues 38–40 (FISΔ3) exhibited an equally weak Dyn2-binding affinity (Fig. 2B). GFP-Trap–based immunoprecipitations of GFP tagged versions of wild-type (WT) and Rab10(Δ3) proteins that were expressed in primary rat hepatocytes confirmed a reduced association of this deletion mutant with endogenous Dyn2 protein (Supporting Fig. S1D).
FIG. 2.
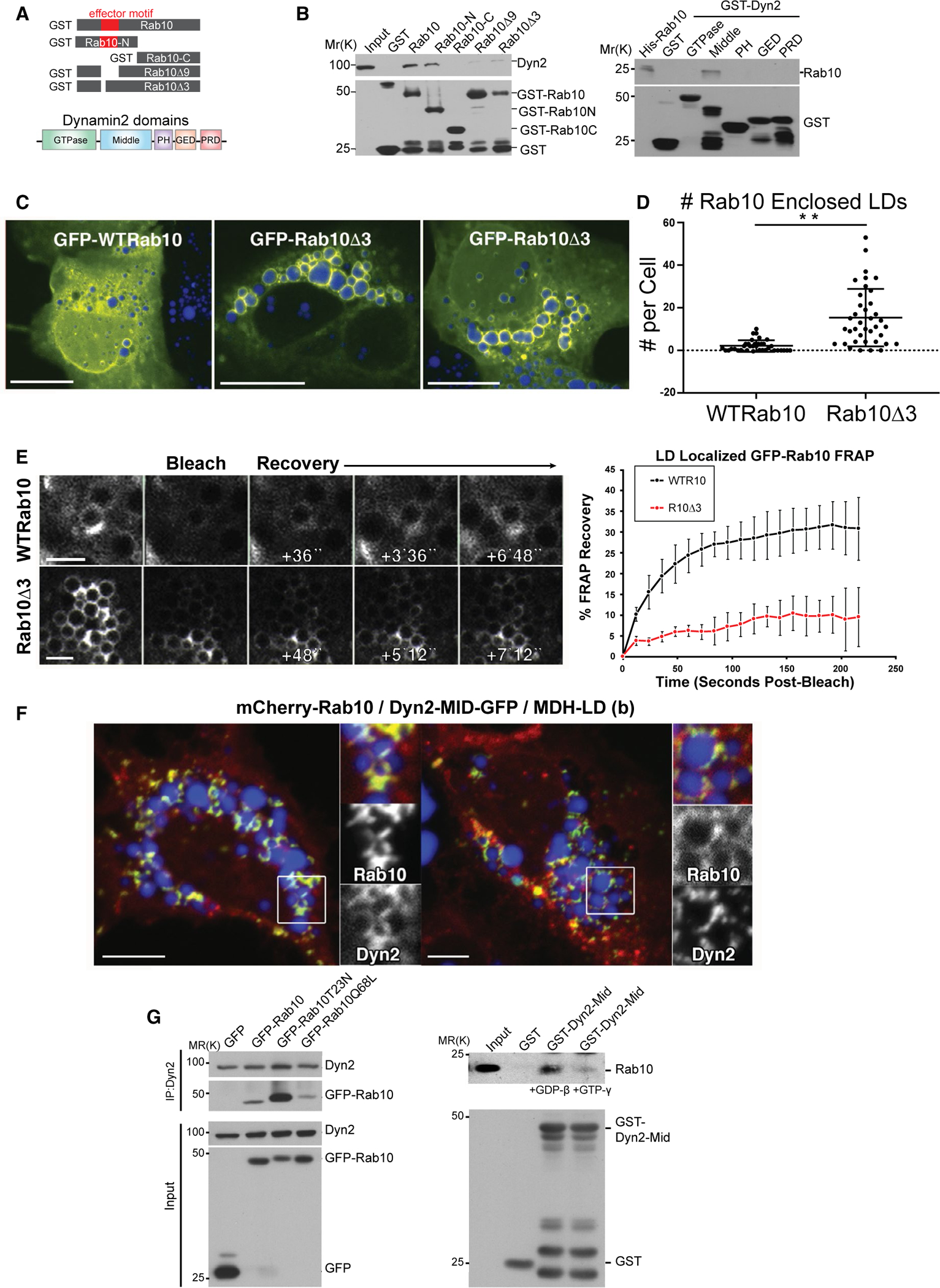
Mapping and characterization of direct interaction between Rab10 and Dyn2. (A) Schematic diagram of GST-tagged full-length Rab10, Rab10-N, Rab10- C, Rab10Δ9, and Δ3 deletion mutants as well as Dyn2 domains. (B) Immunoblotting analysis of a GST-Rab10–binding experiment in Clone9 cells. The second immunoblotting panel is the result of GST pull-down experiments in Clone9 cells using different Dyn2 domains tagged with GST. (C) Confocal images of Hep3B cells expressing either full-length GFP-Rab10 or the GFP-Rab10Δ3 mutant protein that does not bind Dyn2. (D) Graph displaying the average number of LDs encased in Rab10 that were observed in cells expressing either GFP-Rab10 or GFP-Rab10Δ3. Data from three independent experiments are represented. Error bars denote SE; **P < 0.01. (E) Images of Hep3B cells expressing wt or mutant GFP-Rab10 proteins that were exposed to FRAP analysis. Graph shows percent of fluorescence recovery over time (data from three trials, error bars represent SD). (F) Confocal fluorescence microscopy showing HuH7 cells coexpressing mCherry-Rab10 and the GFP-Dyn2 middle domain (Dyn2-MID), MDH-labeled LDs (blue). (G, left panel) Immunoblotting analysis of Dyn2 immunoprecipitations for GFP-tagged Rab10 from Hep3B cells expressing WT, inactive (T23N), and active (Q68L) forms of Rab10 and treated with bortezomib. A preferential binding of inactive Rab10 is observed. (G, right panel) Immunoblotting analysis of pulldowns with the GST-tagged mid domain of Dyn2 from Hep3B cell lysates preloaded with GDP-βS or GTP-γS demonstrate a stronger binding to endogenous Rab10 when GDP-bound. Micron bars: Panel C = 10 μm; panel D = 2 μm; panel E = 5 μm. Abbreviations: GED, GTPase effector domain; IP, immunoprecipitation; PH, pleckstrin homology.
In an attempt to define how Dyn2 might bind to Rab10, different GST-Dyn2 domains were expressed and tested for association with full-length Rab10 in vitro. We found that a 125-residue stretch (aa 746–870) of the Dyn2 middle domain exhibited the strongest affinity for Rab10 (Fig. 2B). This is the same region that was used to originally identify Rab10 as a putative binding partner in Fig. 1A. Given that this Rab10 (Δ3) mutant protein is predicted not to bind to Dyn2, we expressed GFP-Rab10(Δ3) protein in Hep3B cells to test whether it still might associate with LDs despite this reduced effector affinity. Surprisingly, a markedly increased affinity for LDs was observed compared to the GFP-Rab10 WT protein (Fig. 2C,D), suggesting that Rab10 may first associate with the LD before the recruitment of Dyn2. This increased affinity also suggested that the mutant protein undergoes less exchange from the LD surface. To test this premise, we performed fluorescence recovery after photobleaching (FRAP) in Hep3B cells expressing either the Rab10 WT or the Rab10(Δ3) mutant protein. GFP-Rab10 WT protein fluorescence recovered markedly faster than did the GFP-Rab10(Δ3) protein, indicating a minimal exchange rate of the mutant protein from the LD surface (Fig. 2E).
To further define the Rab10 interaction in living cells, this Dyn2 middle domain was GFP-tagged (MID-GFP), expressed in Hep3B cells coexpressing mCherry-Rab10, and the coassociation of these tagged proteins on LDs was scored. Many cells displayed a large number of MID-GFP “crescent” structures (Fig. 2F) polarized to one side of the LDs that mimicked the localization of Rab10 in these cells as published.(15) Significantly, whereas the MID-GFP domain was observed to associate with >20% of total LDs that were also mCherry-Rab10 coated, only 1% of non-Rab10-associated LDs were labeled with the MID-GFP protein (analysis from >20 cells), implying a Rab10-mediated recruitment of this tagged dynamin domain.
Given that binding and hydrolysis of GTP affects the capacity of Rab proteins to bind specific effector proteins, we next tested whether known GTPase mutants of Rab10 might alter the affinity for Dyn2. Accordingly, Hep3B cells were transfected to express Rab10-GFP–tagged WT, T23N (inactive, GTPase defective) and Q68L (constitutively active, GTP-bound) mutants while being incubated in the presence of the protesome inhibitor, bortezomib, for 18 hours before cell lysis to prevent cellular degradation of the mutant Rab10 proteins. Subsequently, cells were lysed and subjected to Dyn2 immunoprecipitation and western blotted for GFP-Rab10. We were surprised to find a marked increase in the association of Dyn2 with the inactive Rab10T23N form, compared to Rab10 WT or the active Rab10Q68L form (Fig. 2G). Consistent with this observation, we found that incubation of Hep3B cell lysates with 2 mM of guanosine 5′-diphosphate (GDP)-β, but not 0.2 mM of GTP-γ, increased Rab10-Dyn2 binding (Fig. 2G). Together, these data suggest that the state of GTP association with Rab10 alters its affinity for Dyn2.
DYN2 AND RAB10 INTERACT ON HEPATOCELLULAR LIPID DROPLETS
To better define the membrane organelles upon which the Dyn2 and Rab10 GTPases interact, we performed detailed time-lapse imaging of live cells. Rab10 associated with tubular vesicular structures (Fig. 1). We observe Rab10 tubules that have a pronounced affinity for LDs. Most interesting, these membrane tubules appeared to associate intimately with the LD surface, where they extend outward and are eventually severed (Fig. 3A; Supporting Movies S3–S5). Analysis of still frames from 40 live Hep3B cells revealed that an average of 2.6 of these Rab10 tubules were found in each cell, occurring on 2.7% of all LDs. From these images, it appears that Rab10 tubules are in a state of dynamic flux along each LD. This is consistent with our previous findings that Rab10 membranes regulate the subsequent recruitment of LC3 and other membrane-deforming components toward development of the nascent autophagolysosome used to engulf and digest smaller LDs during lipophagy.(15)
FIG. 3.
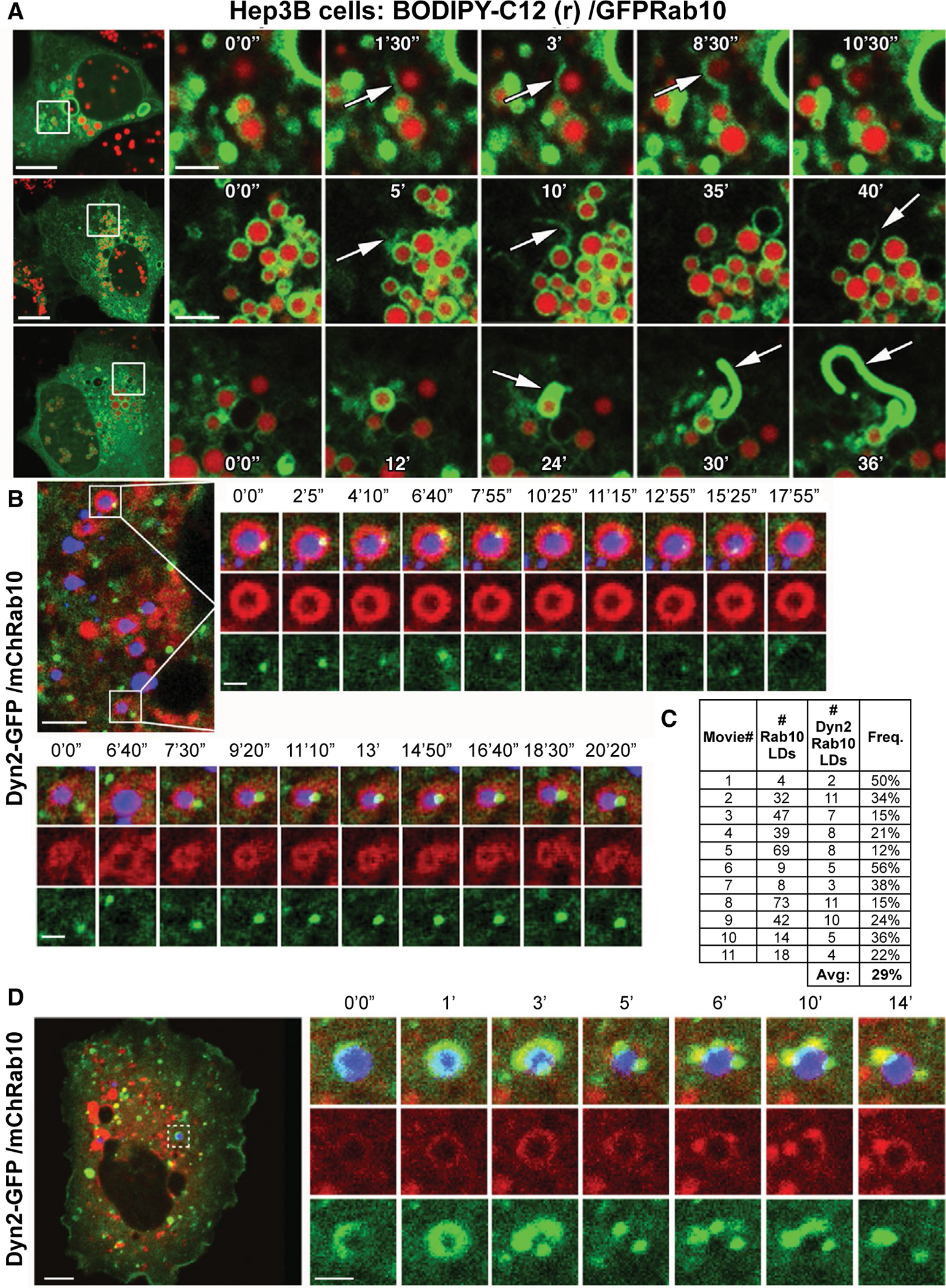
Dynamic remodeling of Rab10 tubules and interaction with Dyn2 on LDs. (A) Still images of live Hep3B cells showing dynamic extensions of GFP-Rab10–coated membrane tubules that are associated with LDs labeled with red fluorescent BODIPY-C12. (B,D) Live-cell confocal fluorescence microscopy time-lapse stills from Hep3B cells expressing GFP-Dyn2 and mCherry-Rab10. LDs are labeled by MDH (in blue). (C) Table displaying the frequency of colocalization between the mCherry-Rab10 and GFP-Dyn2 proteins on the perimeter of LDs within time-lapse series such as those displayed in (B) and (D). Micron bars: Panel A = 10 μm for low-magnification image, 2 μm for time-lapse image stills of boxed region; Panel B/C = 5 μm for low-magnification image, 1 μm for time-lapse image stills of boxed region. Abbreviations: Avg, average; Freq., frequency.
To test whether full-length Dyn2 might coassociate with these Rab10 centric membranes at the LD surface, mCherry-Rab10 and Dyn2-GFP were expressed in Hep3B cells and primary rat hepatocytes colabeled with MDH to indicate LDs. Interestingly, we observed that Rab10-coated LDs recruited Dyn2 puncta that actively moved along the LD perimeter (Fig. 3B,D; Supporting Movies S6 and S7). Similar examples of Dyn2 recruitment to LD-associated Rab10 membranes were observed in isolated primary hepatocytes, where an average of 12.5 LDs, 11.7% of total LDs analyzed from 21 fixed primary hepatocyte cells, exhibited coincidence of these two proteins (Supporting Fig. S1). Analysis of multiple time-lapse series of live Hep3B cells revealed that colocalization of Rab10 and Dyn2 proteins on the perimeter of LDs was frequent, occurring on average for 29% of all observed LDs (Fig. 3C). At times, these Dyn2 puncta would further vesiculate along with the Rab10-coated membranes (Fig. 3D), suggesting a severing event of these lipophagic competent compartments. Graphic examples of Dyn2-mediated scission of these Rab10 membranes emanating from an MDH-labeled LD can be seen in Supporting Fig. S2 and Supporting Movie S8. These observations are consistent with the concept of a Rab10-Dyn2 complex acting at the LD surface, perhaps to excise any excess membrane from developing degradative membranes/autophagosomes. Convergence of Rab10 and Dyn2 on LC3-GFP– or ATG16-GFP–positive autophagic membranes associated with LDs was observed in Hep3B cells (Supporting Fig. S3) and confirmed through independent labeling of Dyn2 and Rab10 in primary hepatocytes, where a surprisingly high average of 90.7% and 86.8% of LD-associated LC3-GFP spots were colabeled with Dyn2 and Rab10 proteins, respectively (analysis from 15 cells each; Supporting Fig. S4).
DYNAMIN SEVERS LD-ASSOCIATED RAB10 TUBULES
The intimate association of the Rab10 and Dyn2 GTPases on these dynamic cytoplasmic and LD-associated degradative structures characterized by tubules (Fig. 3) undergoing scission (Fig. 1E) implies a role for Dyn2 membrane pinchase activity in promoting fission of these Rab10-positive membranes. A depletion of Dyn2 would be expected to produce persistent Rab10 membrane tubules that are incapable of undergoing scission. This is indeed what we observed in cells that were depleted of Dyn2 through siRNA-mediated knockdown (Fig. 4A). Hep3B cells expressing GFP-tagged Rab10 often display transient and dynamic membrane tubules that emanate from multiple regions of the cell before disappearing (Figs. 1E and 3A; Supporting Movies S2–S4). Cells depleted for Dyn2, and expressing GFP-tagged Rab10, displayed an increased frequency, number, and overall length of Rab10 membrane tubules that remained following fixation (Fig. 4C). These findings suggest that the action of Dyn2 is required for driving the efficient scission of these Rab10-bearing membrane tubules, the normal action of which may be to support the editing of membrane compartments during Rab10-coordinated trafficking processes such as lipophagy.(15)
FIG. 4.
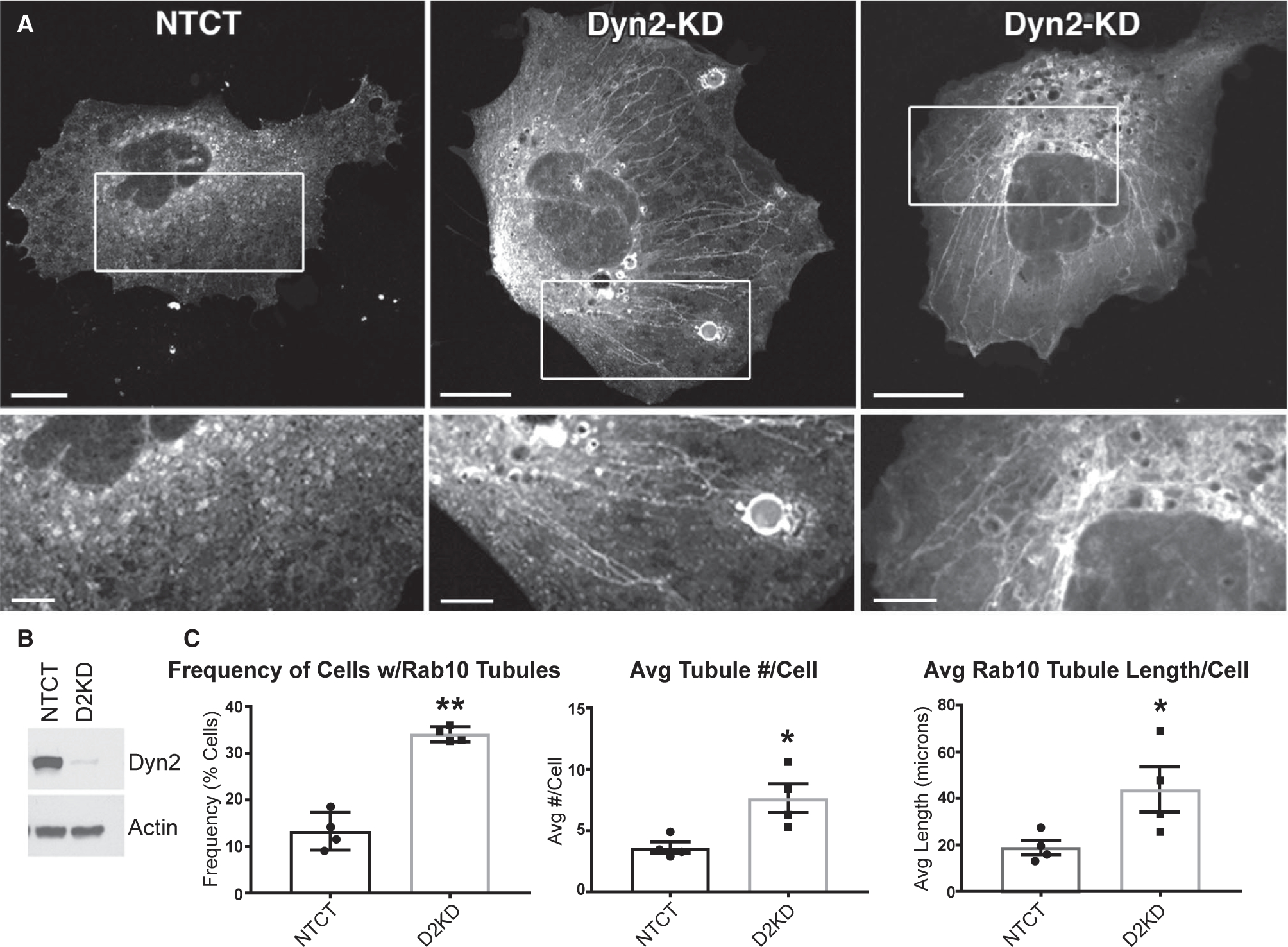
Persistence of Rab10 membrane tubules in Dyn2-depleted cells. (A) Images of fixed Hep3B cells expressing GFP-tagged Rab10 following transfection with nontargeting control siRNA (NTCT) or a duplex targeting Dyn2 show large numbers of persistent Rab10 membrane tubules in Dyn2 knockdown cells. (B) A representative immunoblotting for Dyn2 and actin proteins in lysates of Hep3B cells following a 72-hour knockdown period. (C) Graphs displaying the increased frequency, number, and average total Rab10 tubule lengths in cells following Dyn2 knockdown. *P < 0.05; **P < 0.01. Micron bars in A = 15 μm; inset = 5 μm. Abbreviations: Avg, average; D2KD, Dyn2 knockdown.
To provide ultrastructural information on the membranous tubules that accumulate on and around LDs, Hep3B cells were treated with siRNA to reduce Dyn2 levels, then loaded with HRP for 1.5 hours to label late endocytic membranes by EM (Figs. 1 and 3). Interestingly, numerous HRP-positive tubules could be viewed emanating from LDs encased in membranous organelles (Fig. 5). Most interesting are the long chains of tubules interconnecting very small LDs resulting in a “beads on a string” morphology. None of these LD-tubule structures were observed in nontargeting siRNA control-treated cells. This beaded-like morphology was highly prevalent throughout the cytoplasm in the Dyn2 knockdown cells compared to control cells, as displayed in a very-low-magnification tracing of sectioned Hep3B cells (Supporting Fig. S5). Analysis of five cell profiles for each condition yielded a 6.2-fold increase in average total HRP tubule length per cell, nontargeting control cells having 3.2 μm of tubules with Dyn2 knockdown cells having an average of 19.9 μm in total tubule length.
FIG. 5.
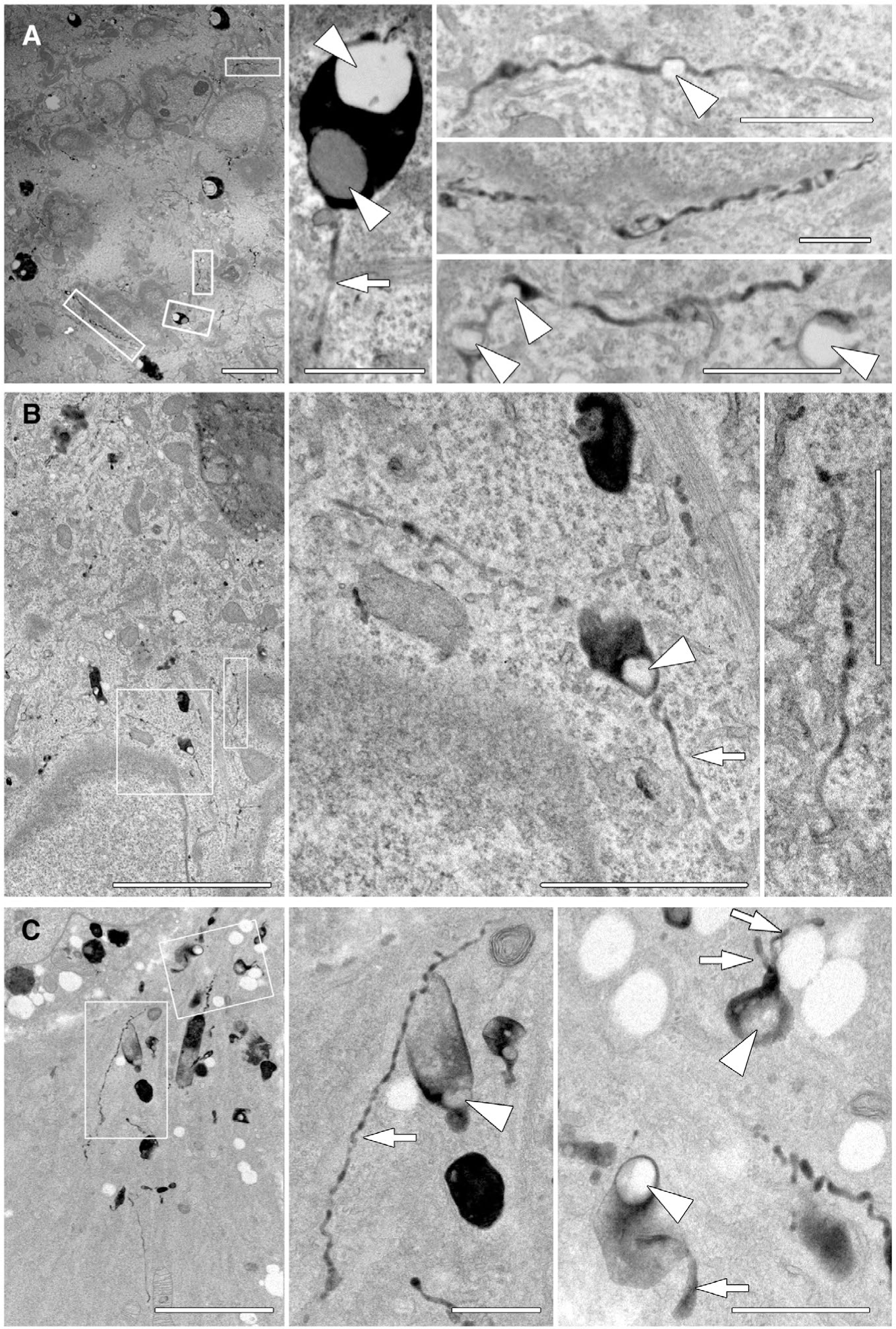
EM of thin HRP-positive tubules extending from lipid droplets in cells with reduced Dyn2 levels. Hep3B cells were treated with Dyn2 siRNA for 3 days, then serum starved for 24 hours; cells were then incubated with HRP for 90 minutes, chased 2 hours, and processed for EM and development of the HRP reaction product. (A-C) Hep3B cells that were treated with siRNA targeting endogenous Dyn2. Enlarged panels of boxed areas from low-magnification images show the presence of HRP-positive membranes that extend through the cytoplasm for significant distances. These tubules are not observed in control-treated cells and can encase exceptionally small LDs that would not be resolved by light microscopy. Arrows indicate HRP positive tubules and arrow heads indicate LDs. Micron bars in A-C = 5 μm; all increased magnifications of boxed regions = 1 μm.
As an extension to these Dyn2 knockdown experiments, we asked whether a reduction in levels of the interacting Rab10 protein might lead to a similar phenotype. Mouse fibroblasts (MEFs) were utilized and isolated from Rab10 knockout neonatal mice given that the animals die shortly after birth, requiring the use of isolated MEFs as noted in our previous Rab10 study.(15) When comparing control and Rab10 knockout MEFs, prepared for HRP labeling and EM, we observed some significant differences. Whereas internalized electron-dense membranes appeared modest in number and were of a confined size in control MEFs (Supporting Fig. S6), endosomes in the Rab10 knockout cells were more prevalent in number and showed considerable deformities with anastomosing tubules and/or constricted shapes (Supporting Fig. S6). Furthermore, many of these dense organelles appeared to be full of small LDs, consistent with previous findings that these Rab10 knockout cells are steatotic and remain so even when placed under nutrient-deprived conditions.(15) Thus, these morphological studies, though correlative in nature, suggest that a knockdown/knockout of both Rab10 and Dyn2 lead to a common phenotype; mainly a disruption in a LD-associated compartment that appears to be defective in the ability to trim away excess membrane, an action consistent with the known function of the dynamin proteins.
Rab10-Dyn2 INTERACTIONS AND HEPATOCELLULAR LD CATABOLISM
The observations of aberrant membrane tubules associated with LDs in Dyn2 knockdown cells, described above, led us to predict that these manipulated cells might exhibit subsequent defects in LD catabolism. To test whether hepatoma cells might exhibit an increased association between Dyn2 and Rab10 when placed under nutrient deprivation, Huh7 cells expressing GFP, or GFP-Rab10, were shifted from normal media to reduced serum conditions (0.1%) for 24 hours before lysis and GFP-Trap–based immunoprecipitation. An increased association between the two GTPases occurs in cells during nutrient deprivation (Fig. 6A).
FIG. 6.
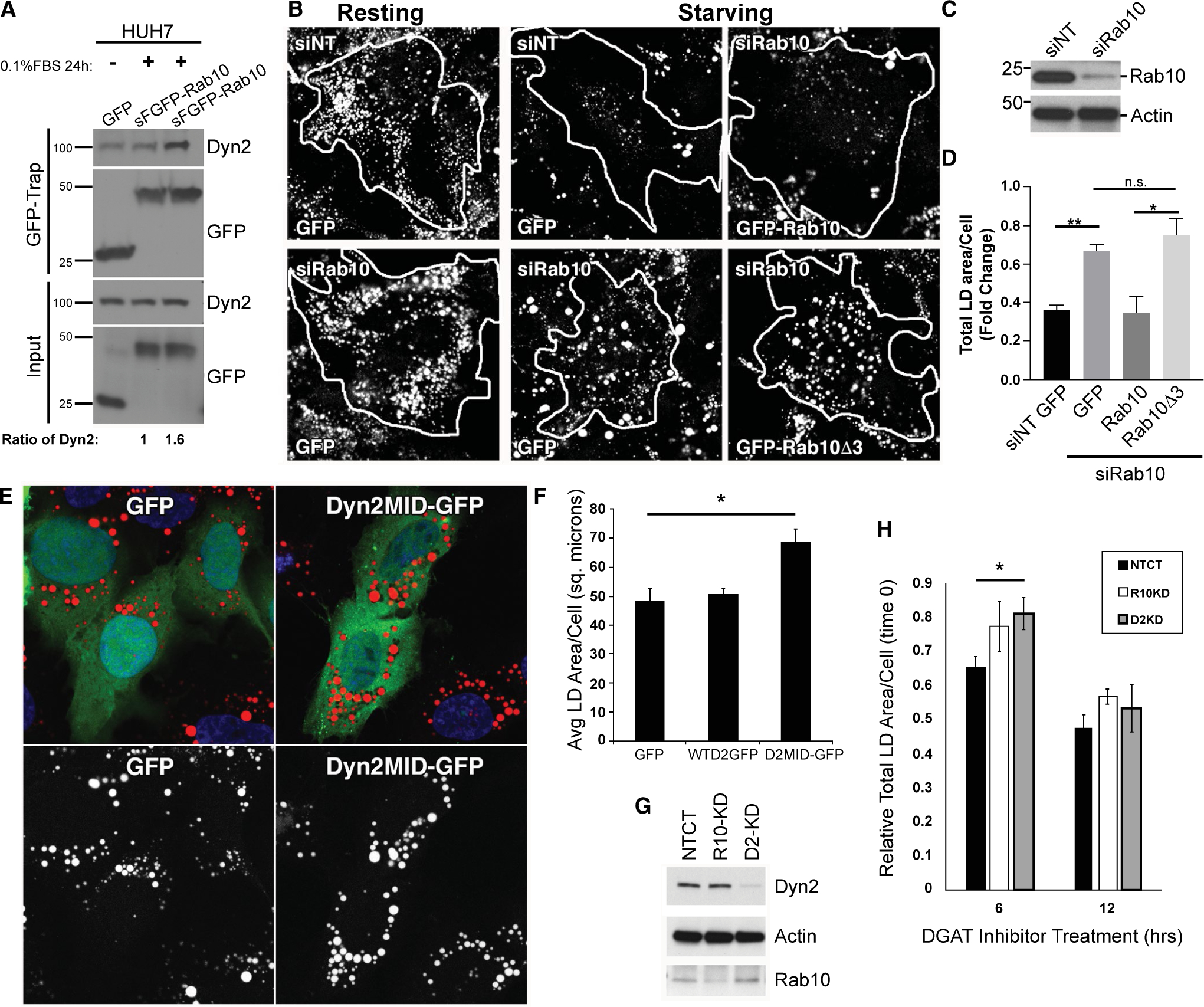
Disruption of Rab10 and Dyn2 interactions leads to cellular steatosis. (A) Interaction between Dyn2 and Rab10 under starvation conditions was analyzed by GFP-Trap immunoprecipitation. Lysates taken from HuH7 cells expressing GFP or GFP-Rab10 in MEM containing full or 0.1% FBS for 24 hours were subjected to immunoprecipitation. (B) Representative fluorescence images showing total Oil Red O–stained LD content in HuH7 cells following transfection with nontargeting control or Rab10-targeted siRNA (siNT and siRab10, respectively). Cells were treated with siRNA for 24 hours and then transfected with different GFP constructs. After another 24 hours, cells were cultured in either MEM with 10% or 0.1% FBS for an extra 48 hours. Tracings denote the perimeter of cells that were expressing GFP or GFP-tagged Rab10 proteins. (C) Representative immunoblottings showing the efficiency of the siRNA-mediated Rab10 knockdown in these HuH7 cells. (D) Quantification of total LD content per cell in starved HuH7 hepatoma cells from (B). All the total LD area data from the siNT-treated group were normalized to the data from siNT cells expressing GFP under resting conditions. Data from all the siRab10-treated group were normalized to data from siRab10 cells expressing GFP under resting conditions. (E) Hep3B cells transiently expressing GFP, GFP-tagged full-length Dyn2 protein (WTD2-GFP), or the Rab10-interacting middle domain of Dyn2 tagged with GFP (Dyn2MID) were loaded with 150 μM of oleate before a 24-hour period of starvation in 0.1% FBS media. Cells expressing the Dyn2MID domain had a reduced level of LD turnover indicated by the increase amount of Oil Red O–labeled lipid content after this starvation period. (F) Quantitative measures of average total area of Oil Red O–labeled LDs in Hep3B cells following the acute loading and starvation period. Values represent the average total area of LDs per cell from four separate experiments. (G) Representative immunoblottings showing the efficiency of siRNA-mediated knockdown of Rab10 and Dyn2 in isolated primary hepatocytes. (H) LD area measurements of primary hepatocytes following acute treatment with 10 μM of the DGAT1 and 2 inhibitors. Data are expressed relative to DMSO controls for each knockdown condition and represent average from five independent trials. Error bars denote SEM, with P values <0.05 denoted by asterisk (*) and <0.01 denoted by double asterisk (**). Abbreviations: Avg, average; DMSO, dimethyl sulfoxide; n.s., nonsignificant; sq. microns, square microns.
To explore whether the increased interaction between these two proteins induced by acute starvation might play a role in LD catabolism, Huh7 cells were treated with control nontargeting siRNA or siRab10 and placed in normal media or low-serum conditions for 48 hours. These cells, under “resting” fed conditions, accumulated a modest number of LDs upon Rab10 knockdown and significantly maintained LD numbers when placed in low-serum conditions during the 48 hours. In comparison, control nontargeting siRNA–treated cells catabolized most of their LDs (Fig. 6B) upon nutrient deprivation.(15) To extend this observation, we tested whether cells with reduced Rab10 levels could be “rescued” to catabolize LDs by the expression of Rab10-GFP WT or the Rab10(Δ3) mutant protein that fails to bind to Dyn2 (Fig. 2) As predicted, siRab10-depleted cells were able to catabolize LDs upon reexpression of the Rab10-GFP WT protein. In contrast, cells transfected to reexpress the Rab10Δ3-GFP mutant were defective in LD catabolism and exhibited a high LD content (Fig. 6B–D). The importance of Rab10-Dyn2 interactions supporting LD catabolism are further implicated by the inhibitory effects observed following expression of the Dyn2 middle domain. Hep3B cells expressing either GFP-tagged Dyn2 WT or GFP alone had lower LD content following a 24-hour starvation period than cells where a GFP-tagged version of the Dyn2 middle domain was expressed, likely serving as a dominant negative in binding to LD localized Rab10 (as seen in Fig. 2E) and disrupting the recruitment of full-length Dyn2 in support of lipophagy-mediated LD breakdown (Fig. 6E,F). Acute LD catabolism was also imposed upon isolated primary hepatocytes over a span of hours following treatment with TAG synthesis inhibitors. Using this experimental approach, we observed attenuated LD loss from these cells that were first treated with DGAT1 and −2 inhibitors for 6 hours following Rab10 and Dyn2 knockdown to prevent the synthesis of new LDs (Fig. 6G,H). These findings are consistent with the premise that Rab10-Dyn2 interactions are important for normal LD catabolism.
Discussion
This study has provided findings on the membrane-trafficking machinery utilized by the hepatocyte to mediate formation and maturation of the developing AP and other degradative vesicles as they envelope LDs for catabolic breakdown. We focused on the action of a specific small GTPase, Rab10, that has been shown by several groups to associate intimately with the LD surface(21) and play an important role in the formation of a functional interface between the AP and LD, what we have termed the “lipophagic synapse.” Thus, these findings suggest that Rab10, situated at both of these organelles, is key in the recruitment of the lipophagic machinery, such as LC3 and autophagy-related proteins, as well as membrane-deforming
ATPases, such as EH domain-binding protein 1/EH domain protein 2, that aid in the envelopment of the LD by the growing AP.(15) In a separate study, we have reported that a large mechano-GTPase, Dyn2, participates in the process of ALRs to sever tubular lysosomes and thereby generate nascent lysosomes for utilization by the developing AP.(10) Thus, although both GTPases are known to function in related, but distinct, cellular lipophagic processes, we were interested to find, through an initial blinded biochemical assay, that these enzymes interact directly (Fig. 1).
Dyn2 has been demonstrated to bind perhaps a dozen or more different cytoskeletal or membrane-trafficking effectors,(22) but, to our knowledge, none of these effectors represent Rab proteins. Furthermore, it was particularly surprising that the Dyn2 middle domain or “stalk,” not the proline-rich domain (PRD), was the region responsible for Rab10 interaction. This domain is generally thought to mediate self-assembly of the Dyn2 polymer.(23) In the polymerized state, this region may not be accessible for interaction with other proteins, such as Rab10, suggesting that it may be recruited to a Rab10-enriched membrane as it assembles. Indeed, most models of dynamin assembly implicate a coassembly-recruitment process such as that observed with in vitro assays using isolated membranes and dynamin proteins.(24,25) The ephemeral recruitment of Dyn2 puncta at Rab10-positive membranes (Fig. 1D and Supporting Figs. S1 and S3; Supporting Movies S1, S6, and S8) and extended Rab10 tubules (Fig. 1E) we observed in Hep3B cells is consistent with the premise of a rapid recruitment, assembly, severing, and disassembly that we and others have observed during the internalization of clathrin-coated pits at the plasma membrane.(26,27)
The middle domain of Dyn2 has been shown to bind to another protein, such as gamma tubulin, that is situated at the centrosome(16) and at the midbody of dividing cells.(28,29) Central to the Rab10 interaction is the “effector domain” present in all Rab proteins that bind to key functional proteins when in the GTP-bound state.(30) We were surprised to find that the Rab10T23N mutant defective in binding GTP exhibited a higher biochemical affinity for Dyn2 than did the active Rab10Q68L mutant. This suggests that Dyn2, while acting as a binding partner, may not represent a true Rab10 “effector” protein. Although rare, there is precedence for a GDP-bound Rab10 family member (Rab8) exhibiting increased association at the LD-LD interface to mediate lipid and protein exchange between these organelles.(31)
An interesting characteristic of Rab10 is the presence of the specific effector sequence of 38-FISTIGIDF-46 that appears unique to the Rab10 subfamily that includes Rab8, Rab10, and Rab13. These other Rabs also appear to mediate Golgi, endosomal membrane traffic,(32–35) as well as transport of components into primary cilia(36) and are associated with LDs,(21,37,38) although to a lesser extent than Rab10. Removal of this 9-aa sequence prevents binding to Dyn2 (Fig. 2), and substitution of this sequence with the effector domain of the nonrelated Rab6 also prevents binding (data not shown).
Because Rab proteins have multiple effector proteins, it is equally interesting and challenging to define which effector is recruited and functional in a cellular context at any given time. Our goal with the substantial live cell quantiation was to provide some insights into these interactions. Toward this end, this study provides observations that, first, Rab10 and Dyn2 exhibit an intimate, and transient, colocalization on both membrane tubules (Fig. 1; Supporting Movies S2 and S8) and LDs (Fig. 3; Supporting Movies S6–S8). Second, as mentioned above, mutation of the effector domain prevents binding to the Dyn2 mid domain (Fig. 2). Third, the attenuated interacting domains of these proteins exhibit increased association with the LD surface (Fig. 2), and, fourth, knockdown of Dyn2 affects the function and morphology of Rab10 tubules (Fig. 4) whereas knockdown/knockout of Dyn2 and Rab10, respectively, independently leads to deformed and tubulated lipophagic organelles (Fig. 5 and Supporting Fig. S6). Finally, manipulation of both proteins through knockdown or expression of binding mutant proteins(10) (Fig. 6) leads to similar defects in LD catabolism through lipophagy, while having no apparent effect on LD synthesis or cytosolic lipase recruitment to the LD surface (data not shown). Taken together, all of these findings are consistent with the premise that these two GTPases interact at the LD surface and on cytoplasmic tubules to mediate Rab10 tubule scission (Fig. 7).
FIG. 7.
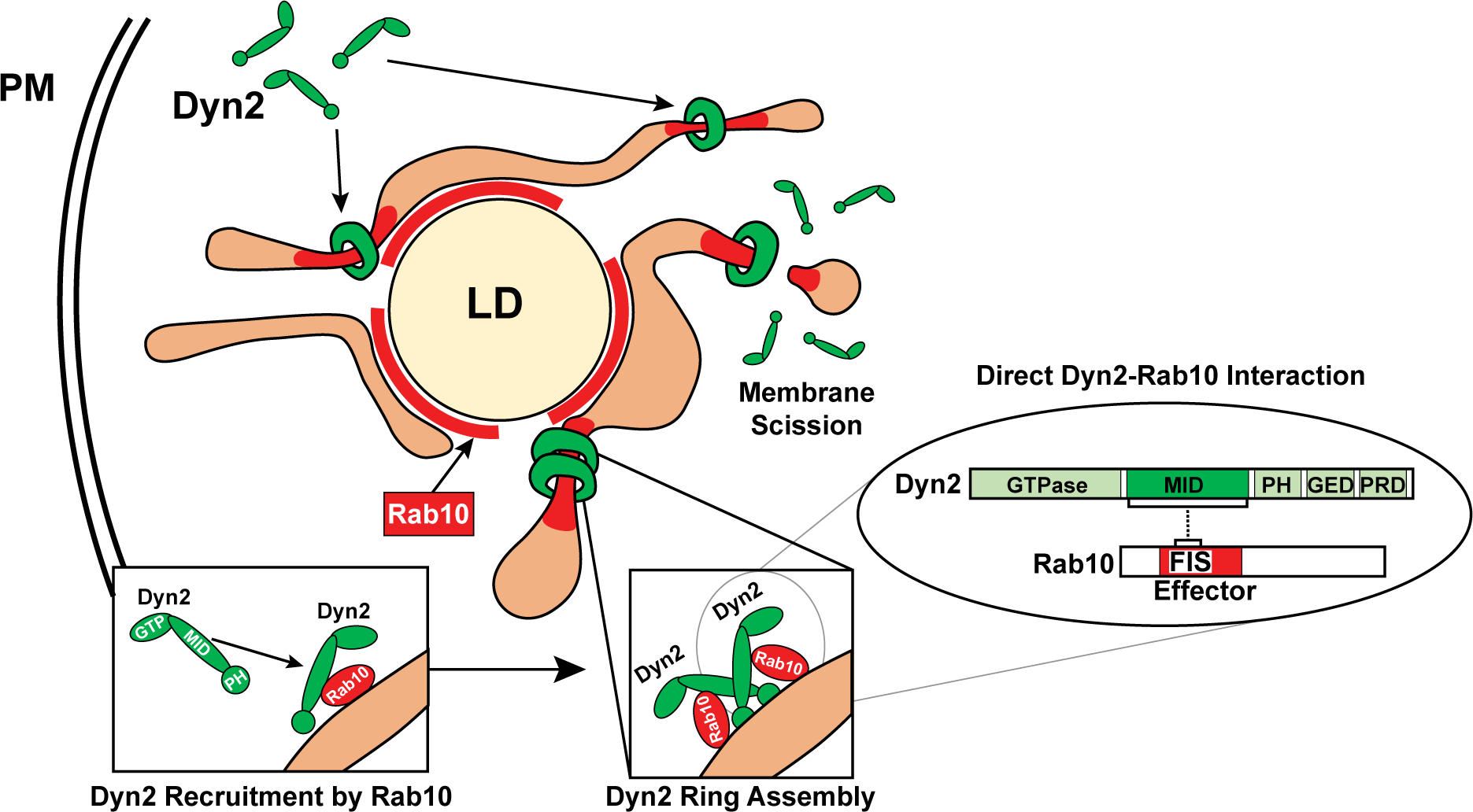
Maturation of lipophagic organelles in hepatocytes is dependent upon a Rab10-Dyn2 complex. A model depicting direct interaction of the Rab10 and Dyn2 GTPases by effector and middle domains, respectively. Rab10 may coordinate the recruitment of Dyn2 dimers to LD-associated membranes, leading to Dyn2 contractile polymer assembly and eventual tubulation/scission of these membrane targets. The coordinated efforts of Rab10 and Dyn2 are required for functional maturation of these lipophagic compartments in regulating hepatic lipid stores. Abbreviations: GED, GTPase effector domain; MID, middle; PH, pleckstrin homology; PM, plasma membrane; PRD, proline-rich domain.
Rab10 AND Dynamin2 AS IMPORTANT HEPATOCELLULAR METABOLIC REGULATORS
This study has led to a testable model, suggesting that Rab10-associated tubules participate in the maturation of LD-associated degradative autophagic/lysosomal membranes and require Dyn2 for severing. Tubules with enriched domains of Rab10 in the GDP-bound state may favor recruitment of Dyn2 dimers through an effector/mid-domain interaction, leading to polymer assembly, tubule constriction, and membrane scission over a period of just 5–10 minutes (Fig. 1; Supporting Movies S2–S4). These intimate scission sites could remove and trim away any unneeded excessive tubules from the LD, facilitating membrane recycling and lipophagosome maturation. Prevention of these latter steps leads to a break in cyclical utilization of Rab10 membranes, as well as ALR utilization, and subsequently LD accumulation. We view the long chains of LDs connected by delicate membrane tubules in the Dyn2 knockdown cells displayed in Fig. 5 and Supporting Fig. S5 as particularly graphic and may represent a form of cellular microsteatosis that is unresolvable by light microscopy or histology and requires EM.
Whereas we have previously demonstrated that Rab10 is associated with lipophagosomes in primary hepatocytes,(15) this current study has utilized human hepatoma cells as biological models to study the synergistic function of Rab10 and Dyn2. Rab10 has recently been reported to be highly expressed in hepatocellular carcinoma and is required for tumor cell growth and metastasis while correlating with tumor aggressiveness and poor patient outcome.(39) Similarly, we have reported that an increased expression of Dyn2 potentiates metastatic migration of pancreatic ductular adenocarcinoma.(40) Expression of these GTPases may thus facilitate the efficient catabolic degradation of stored lipids to drive the high energy demands of continued cell growth and active metastatic invasion. Thus, because of the relevance to both hepatic steatosis and tumor progression, we believe that further defining the expression, interaction, effectors, and regulation of these dual GTPases is particularly important.
Supplementary Material
Acknowledgments
Supported by NIH grants 5R37DK044650 NIDDK (to M.A.M.) and 5RO1AA020735 NIAAA (to M.A.M. and C.A.C.), Mayo Clinic Center for Cell Signaling in Gastroenterology (NIDDK P30DK084567), and 17PRE33660888 (to K.D.M.).
Abbreviations:
- aa
amino acid(s)
- ALR
autophagic lysosome reformation
- AP
autophagosome
- DGAT
diacylglycerol O-acyltransferase
- Dyn2
dynamin 2 GTPase
- EM
electron microscopy
- FBS
fetal bovine serum
- FRAP
fluorescence recovery after photobleaching
- GDP
guanosine 5′-diphosphate
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- HRP
horseradish peroxidase
- LC3
light chain 3
- LD
lipid droplet
- MDH
malic dehydrogenase
- MEFs
mouse embryonic fibroblasts
- MEM
minimum essential medium
- MID-GFP
GFP-tagged middle domain
- NP-40
Nonidet P-40
- PRD
proline-rich domain
- Rab10
Rab10 GTPase
- siRNA
small interfering RNA
- WT
wild type
Footnotes
Potential conflict of interest: Nothing to report.
Supporting Information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.31059/suppinfo.
REFERENCES
- 1).Treyer A, Musch A. Hepatocyte polarity. Compr Physiol 2013;3:243–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Zechner R, Madeo F, Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol 2017;18:671–684. [DOI] [PubMed] [Google Scholar]
- 3).Schulze RJ, Drizyte K, Casey CA, McNiven MA. Hepatic lipophagy: new insights into autophagic catabolism of lipid droplets in the liver. Hepatol Commun 2017;1:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Martinez-Lopez N, Singh R. Autophagy and lipid droplets in the liver. Annu Rev Nutr 2015;35:215–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol 2012;2012:282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature 2009;458:1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Kaksonen M, Roux A. Mechanisms of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 2018;19:313–326. [DOI] [PubMed] [Google Scholar]
- 8).McNiven MA, Cao H, Pitts KR, Yoon Y. The dynamin family of mechanoenzymes: pinching in new places. Trends Biochem Sci 2000;25:115–120. [DOI] [PubMed] [Google Scholar]
- 9).Henley JR, Krueger EW, Oswald BJ, McNiven MA. Dynamin-mediated internalization of caveolae. J Cell Biol 1998;141:85–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Schulze RJ, Weller SG, Schroeder B, Krueger EW, Chi S, Casey CA, et al. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol 2013;203:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Cook TA, Urrutia R, McNiven MA. Identification of dynamin 2, an isoform ubiquitously expressed in rat tissues. Proc Natl Acad Sci U S A 1994;91:644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Schulze RJ, McNiven MA. A well-oiled machine: DNM2/dynamin 2 helps keep hepatocyte lipophagy running smoothly. Autophagy 2014;10:388–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, et al. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol 2001;11:1680–1685. [DOI] [PubMed] [Google Scholar]
- 14).Schroeder B, Schulze RJ, Weller SG, Sletten AC, Casey CA, McNiven MA. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 2015;61:1896–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Li Z, Schulze RJ, Weller SG, Krueger EW, Schott MB, Zhang X, et al. A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci Adv 2016;2:e1601470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Thompson HM, Cao H, Chen J, Euteneuer U, McNiven MA. Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat Cell Biol 2004;6:335–342. [DOI] [PubMed] [Google Scholar]
- 17).Cao H, Garcia F, McNiven MA. Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell 1998;9:2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Schott MB, Rasineni K, Weller SG, Schulze RJ, Sletten AC, Casey CA, et al. beta-Adrenergic induction of lipolysis in hepatocytes is inhibited by ethanol exposure. J Biol Chem 2017;292:11815–11828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Yang HJ, Hsu CL, Yang JY, Yang WY. Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS One 2012;7:e32693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Schott MB, Weller SG, Schulze RJ, Krueger EW, Drizyte-Miller K, Casey CA, et al. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J Cell Biol 2019;218:3320–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, et al. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev Cell 2018;44:97–112.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol 2012;13:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Chen YJ, Zhang P, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nat Struct Mol Biol 2004;11:574–575. [DOI] [PubMed] [Google Scholar]
- 24).Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 1995;374:190–192. [DOI] [PubMed] [Google Scholar]
- 25).Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol 1999;1:33–39. [DOI] [PubMed] [Google Scholar]
- 26).Cao H, Krueger EW, McNiven MA. Hepatocytes internalize trophic receptors at large endocytic “hot spots”. HEPATOLOGY 2011;54:1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol 2002;4:691–698. [DOI] [PubMed] [Google Scholar]
- 28).Thompson HM, Skop AR, Euteneuer U, Meyer BJ, McNiven MA. The large GTPase dynamin associates with the spindle mid-zone and is required for cytokinesis. Curr Biol 2002;12:2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Konopka CA, Schleede JB, Skop AR, Bednarek SY. Dynamin and cytokinesis. Traffic 2006;7:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 2011;91:119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Wu L, Xu D, Zhou L, Xie B, Yu L, Yang H, et al. Rab8a-AS160-MSS4 regulatory circuit controls lipid droplet fusion and growth. Dev Cell 2014;30:378–393. [DOI] [PubMed] [Google Scholar]
- 32).Huber LA, Pimplikar S, Parton RG, Virta H, Zerial M, Simons K. Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J Cell Biol 1993;123:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Marzesco AM, Dunia I, Pandjaitan R, Recouvreur M, Dauzonne D, Benedetti EL, et al. The small GTPase Rab13 regulates assembly of functional tight junctions in epithelial cells. Mol Biol Cell 2002;13:1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Yamamura R, Nishimura N, Nakatsuji H, Arase S, Sasaki T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol Biol Cell 2008;19:971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Ang AL, Taguchi T, Francis S, Folsch H, Murrells LJ, Pypaert M, et al. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol 2004;167:531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol 2007;178:363–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res 2007;6:3256–3265. [DOI] [PubMed] [Google Scholar]
- 38).Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem 2004;279:3787–3792. [DOI] [PubMed] [Google Scholar]
- 39).Wang W, Jia WD, Hu B, Pan YY. RAB10 overexpression promotes tumor growth and indicates poor prognosis of hepatocellular carcinoma. Oncotarget 2017;8:26434–26447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Eppinga RD, Krueger EW, Weller SG, Zhang L, Cao H, McNiven MA. Increased expression of the large GTPase dynamin 2 potentiates metastatic migration and invasion of pancreatic ductal carcinoma. Oncogene 2012;31:1228–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


