Abstract
The current global health emergency in the form of the Coronavirus 2019 (COVID-19) pandemic has highlighted the need for fast, accurate, and efficient drug discovery pipelines. Traditional drug discovery projects relying on in vitro high-throughput screening (HTS) involve large investments and sophisticated experimental set-ups, affordable only to big biopharmaceutical companies. In this scenario, application of efficient state-of-the-art computational methods and modern artificial intelligence (AI)-based algorithms for rapid screening of repurposable chemical space [approved drugs and natural products (NPs) with proven pharmacokinetic profiles] to identify the initial leads is a powerful option to save resources and time. Structure-based drug repurposing is a popular in silico repurposing approach. In this review, we discuss traditional and modern AI-based computational methods and tools applied at various stages for structure-based drug discovery (SBDD) pipelines. Additionally, we highlight the role of generative models in generating molecules with scaffolds from repurposable chemical space.
Keywords: Drug repurposing, Machine learning, Force field, Quantum mechanics, Inverse design, Generative modeling
Introduction
Identifying small molecules that can lead to an alteration in biochemical mechanisms via interactions with specific biological targets has been the key aspect of modern rational drug discovery (DD). This idea revolutionized the DD pipeline, resulting in extensive development of combinatorial chemistry and HTS over the past few decades. However, these techniques involve very high costs and long assay development and standardization times, which are not affordable for all. In this scenario, a shift from traditional ways of synthesizing and screening huge chemical libraries to the concept of drug repositioning/repurposing/reprofiling (DR), in which drugs with known indications are repurposed for new indications is a safe and cost-effective alternative. This rapid drug development strategy involves evaluation of new disease pathways, identifying new targets and studying their structures, functions, and dynamics to rationally reposition suitable molecules from the known chemical space, rather than random screening.1, 2, 3 In silico DR has attracted the attention of the pharmaceutical industries and research communities worldwide during the current COVID-19 pandemic because the use of advanced computational algorithms can predict 3D structures of targets, detect binding pockets/interaction hotspots of new drug targets, and screen the known drug candidates against new target structures, dramatically reducing the time and cost required for DR.1
DR involves the identification of new applications for existing drugs at a lower cost and in a shorter time.2 There are different computational DR strategies. For example, computational DR approaches that have been applied to the COVID-19 pandemic can be broadly categorized into: (i) drug/target network-based models; (ii) structure-based approaches; and (iii) AI approaches.1 Network-based approaches are divided into two categories: network-based clustering approaches and network-based propagation approaches. Both network-based approaches enable the annotation of important patterns, the identification of proteins that are functionally associated with COVID-19, and the discovery of novel drug–disease or drug–target relationships useful for new therapies. Structure-based approaches enable the identification of small chemical compounds able to bind macromolecular targets to evaluate how a chemical compound can interact with its biological counterpart, to find new applications for existing drugs. AI-based networks currently appear less relevant because they need more data for their application.1 Rapidly emerging high-precision in silico techniques/algorithms and consistently increasing computational access to huge amounts of data regarding clinical research, pathways involved in diseases, gene expression profiles, drug target structures, pharmacophores, and so on, have supported the use of computational approaches to envisage new indications/placements for old drugs.2, 4 In silico DR pipelines involve a variety of approaches, such as genomics, systems biology, network biology, chemo/bioinformatics, and structural bioinformatics-based approaches to identify optimal ‘new target–known drug’ pairs. Among these in silico methods, structure-based drug repurposing (SBDR) is important in its own right, given that the 3D structure of the target is a prerequisite to screen the repurposable chemical space (RCS) and explore suitable ligand interactions with the target binding site through techniques including docking, pharmacophore modeling, and molecular dynamics (MD) simulations. Along with the approved drugs, the RCS can include: all the molecules that have passed preclinical in vitro/in vivo stages and have entered the clinical phase, as well as compounds from various NP databases, such as Ayurveda, IMPPAT Berdy’s Bioactive NP Database, Carotenoids Database, Chinese Traditional Medicinal Herbs database, FooDB, and TCMDB@Taiwan, the absorption, distribution, metabolism, and excretion (ADMET) and toxicity profiles of which are well established. Table 1 lists data sources of the RCS, drug targets, pathways, and drug–target complexes. Although traditionally SBDD mostly involves docking-based virtual screening (VS), computationally intensive methods, such as MD simulations to include flexibilities of the targets, binding free energy calculations, and quantum chemical (QM) calculations, can also be applied for accurate predictions when a considerably smaller chemical library, such as only approved drugs, is considered for a DR project. In addition, the rapidly emerging AI–machine learning (ML) methods have essential roles in overcoming the limitations of traditional methods and confer accurate predictions. In this review, we discuss traditional and the modern AI-based computational methods and tools applied at various stages of SBDR pipelines. Advanced ML techniques, such as generative modeling, are also discussed, which can be indirectly applied for SBDR. We also highlight recent successful applications of computational techniques for SBDR.
Table 1.
Data sources for repurposable chemical space, targets, pathways, and drug–target complexes.
| Database | URL | Content |
|---|---|---|
| Data sources for repurposable chemicals | ||
| DrugBank | https://go.drugbank.com/ | Detailed chemical, pharmacological, and pharmaceutical data of drugs and sequence, structure, and pathway information of drug targets |
| TCM | http://tcm.cmu.edu.tw/ | 170 000 traditional Chinese medicine compounds, which passed ADMET filters with 3D structures |
| e-Drug3D | https://chemoinfo.ipmc.cnrs.fr/MOLDB/index.php | 1822 compounds (maximum molecular weight: 2000), similar to the US Pharmacopeia of Small Drugs |
| SuperDRUG2 | http://cheminfo.charite.de/superdrug2/ | ∼ 4600 active pharmaceutical ingredients |
| DNP | http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml | The Natural Products subset of Dictionary of Organic Compounds |
| KEGG DRUG | www.genome.jp/kegg/drug/ | Drugs approved to be marketed in Europe, USA, and Japan, with information of their targets and other molecular interaction networks |
| Data sources to explore new targets/pathways/indications for the RCS | ||
| Therapeutic Target Database (TTD) | http://bidd.nus.edu.sg/group/cjttd/ | Studied and reported protein, RNA/DNA drug targets as well as pathways involved in targeted disease |
| STITCH | http://stitch.embl.de/ | Known and predicted interactions of chemicals and proteins |
| Small Molecule Pathway Database (SMPDB) | https://smpdb.ca/ | Information on ∼ 350 human small-molecule pathways |
| Transformer | https://bioinformatics.charite.de/transformer/ | Data on enzymatic/nonenzymatic transformation of various xenobiotics in humans; interactions and process of transport of drugs, prodrugs, traditional Chinese medicines etc. |
| Human Metabolome Database | https://hmdb.ca/ | Small-molecule metabolites in the human body |
| KEGG PATHWAY Database | www.genome.jp/kegg/pathway.html | Detailed information on targets, molecular interaction networks, and enzymes involved in metabolism of known drugs with references to several relevant databases and web-based tools |
| Data sources to train and test ML models for binding affinity prediction | ||
| Protein Data Bank (PDB) | www.rcsb.org/ | Experimental structures of biomacromolecules, such as proteins/nucleic acids, ribosomes etc. |
| PDBbind | www.pdbbind.org.cn/ | Experimentally measured IC50, Kd, Ki, and other binding affinity data of the PDB protein–ligand complexes |
| BindingDB | www.bindingdb.org/bind/index.jsp | Measured binding affinities of small, drug-like molecules and drugs with known drug targets |
| SCORPIO | http://scorpio.biophysics.ismb.lon.ac.uk/scorpio.html | Structurally resolved and thermodynamically characterised protein–ligand complexes |
| Ki Database | https://kidbdev.med.unc.edu/databases/kidb.php | Published and internally derived 55 472 Ki, or affinity values for a large number of drugs and drug candidates with GPCRs, ion channels, transporters, and enzymes |
| BAPPL complexes set | www.scfbio-iitd.res.in/software/drugdesign/proteinliganddataset.htm | 161 protein–ligand complexes with experimental and predicted free energies of binding |
| DNA Drug complex data set | www.scfbio-iitd.res.in/software/drugdesign/dnadrugdataset.jsp | DNA–drug complexes comprising 16 minimized crystal structures and 34 model-built structures, along with experimental affinities |
| DUD.E | http://dude.docking.org/ | Provides decoy molecules for testing docking and ML models; affinities of 22 886 active compounds against 102 different targets; includes 50 decoy molecules for each active molecule with similar physicochemical properties but dissimilar 2D topologies |
SBDR and AI/ML techniques in modern drug discovery
The fundamentals of SBDR are based on the abilities of the drug to bind to multiple protein-binding sites. Apart from their original therapeutic targets, the drugs show affinities for other proteins, so-called ‘off-targets’. These off-targets can be carrier proteins, transporters, plasma proteins, among others, to which the drugs bind to cause side effects, which are not always detrimental and open ways to explore new indications for the drugs. One of the earliest examples of such an off-target-based approach was repositioning of sildenafil, which was originally used to treat angina; observation of sildenafil interacting with a phosphodiesterase (PDE5) resulted in this drug being repurposed for the treatment of erectile dysfunction.5 SBDR methods depend on the availability of the receptor protein and ligand structures. These methods mostly comprise high-throughput VS6 of the RCS using molecular docking and/or pharmacophore models.7, 8
The past few years have witnessed a rapid increase in the area of data-driven ML applications in general, which are becoming a vital tool during early drug discovery efforts. Multiple factors, such as rapidly accumulating relevant experimental data (e.g., DrugBank, ChEMBL, PDB, PubChem, and PDBbind), development of modern ML methods, libraries, and affordable computational power, are fueling such a surge.
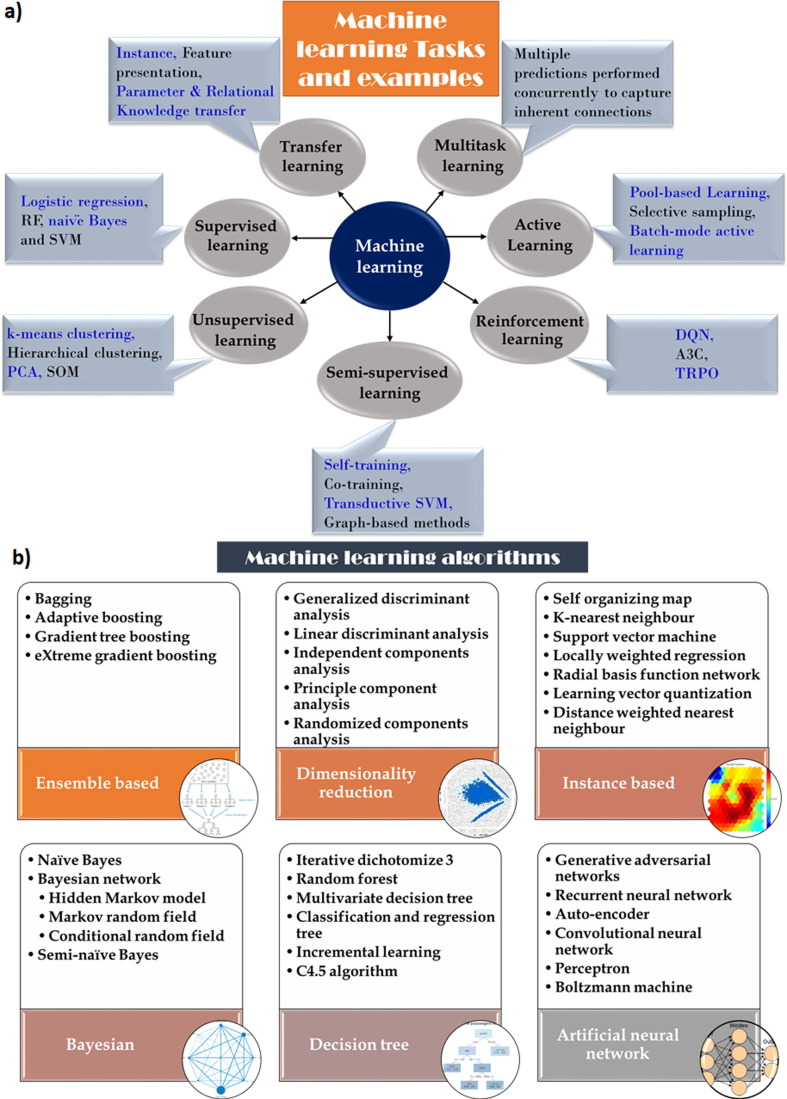
ML algorithms have relevant and potential applications at almost all steps of the SBDR pipeline and beyond, such as drug screening, target screening, target structure/binding site prediction, lead optimization, prediction of drug–drug interactions, and ADMET property prediction.9 ML methods aim to learn from existing data and predict properties instead of using physics-based understanding to explicitly compute properties.10 These methods can broadly be classified as supervised learning, unsupervised learning, and reinforcement learning. In supervised learning, markers or labels of new samples are predicted through ML models that are trained from samples with known markers. Unsupervised learning, in which the training samples without any labels are used to develop a model, is used to recognize complex patterns and to transform data to a lower dimension in general. Reinforcement learning attempts to perform reward-driven learning, in which an agent attempts to find an ideal set of actions to endorse some outcome through analysis of the environment combined with performing actions to alter that environment. Fig. 1 shows various categories of ML tasks and algorithms that are commonly used in drug design exercises. Naive Bayesian (NB), support vector machine (SVM), decision trees, random forest (RF), and artificial neural networks (ANNs) are the most popular classical ML algorithms, whereas deep Boltzmann machine (DBM), deep belief networks (DBNs), generative adversarial networks (GANs), variational autoencoders (VAEs), and adversarial autoencoders (AAEs) are some of the modern ML methods for discriminative, regression, clustering, regularization, dimensionality reduction, and generative tasks.
Figure 1.
Title. (a) Classification of machine-learning (ML) tasks based on principle of learning; (b) different types of ML algorithm. For definitions of abbreviations, please see the main text.
Despite issues with their use in other research areas, AI/ML methods have been used continuously in drug design efforts over the past 25 years or so. Earlier applications in drug design activities were dominated by classical ML methods. NB algorithms, a supervised learning method, have been successful in processing massive amounts of information and in predictive modeling, while having a unique tolerance of data noise. For example, NB models in combination with extended-connectivity fingerprints (ECFPs) were used by Pang et al. to classify active and inactive molecules and predict their biological activity as estrogen receptor antagonists.11 Similarly, Wei et al. developed multiple quantitative structure activity relationship (QSAR) models using NB as a classifier in combination with SVM to identify HIV and hepatitis C inhibitors.12
RF comprises an ensemble of multiple uncorrelated decision trees, where, for a given task, each tree independently performs one prediction and the one with the maximum votes is selected as the best fit. Training of several decision trees minimizes individual errors and maximizes the efficiency because the final prediction is the best out of several independent predictions, unlike other algorithms. Cano et al. applied RF methods to predict protein–ligand binding affinities in a VS project, in which they trained the algorithm with a data set comprising kinases, nuclear hormone receptors, and their ligands.13 Rahman et al. predicted drug response confidence level for a particular genome by using multivariate RF, in which the input data were genetic and epigenetic attributes.14
SVMs are popular in computer-aided drug design (CADD) on account of their ability to differentiate between actives and inactives through binary class prediction or to train regression models that predict the activities and ranking compounds. SVM are trained to separate nonlinearly separable low-dimensional input data in a higher-dimensional latent space through feature mapping.15 SVM models that are specifically designed to predict drug–receptor interactions take into account protein-binding site as well as protein–ligand interaction features as important components for predictive modelling. Wang et al. developed and trained SVM models with diverse features, such as chemical structural features, pharmacological or therapeutic effects, and genomics data of the proteins, to predict drug–target interactions.16 Kawaii et al. used SVM models in which the drug molecules were allowed to match with numerous targets from different pathways to predict their bioactivities against multiple pathways.17
ANNs, analogous to nerve cells or neurons, obtain frequent input signals, calculate the weighted sum of the inputs via a nonlinear activation function, and produce an initiation response. The resulting connected neurons then receive the output signals passed on from preceding neurons. A typical ANN comprises three components: (i) an input layer; (ii) a hidden layer; and (iii) an output layer.18 The middle hidden layer comprises fully or partially connected processing nodes, which receive the input variables from the input nodes and transform them into the output nodes, which ultimately compute the output signal. ANN algorithms are iteratively trained via back propagation. The performance of ANN methods might be inferior to that of RF and SVM, especially when the data set is small, resulting in problems such as overfitting. However, with availability of big data, ANNs have re-emerged as deep learning (DL) algorithms,19 which are based on the feed-forward NNs of ANN with several hidden layers. These hidden layers account for the learning abilities of the computational models from multidimensional data. DL algorithms are at the development front-line in most scientific and technological fields. DL-based methods have brought about a paradigm shift in the field of CADD, from QSAR, target identification, VS to lead molecule design and optimization because they are able to recognize, interpret as well as generate complex data. Deep NNs (DNNs), recurrent NNs (RNNs), and convolutional NNs (CNNs) are the major NNs that are used in DD projects. These can be used for both prediction of molecular properties and generating molecular structures with requisite properties.19
Traditional and AI/ML-aided methods at different stages of SBDR pipelines
SBDR methods depend on the availability of receptor protein and ligand structures. Fig. 2 provides examples of approaches used in SBDR projects. The first step of most SBDR pipelines is to obtain high-quality 3D structures of the new targets. If a structure is not solved experimentally, one can model it computationally. Once a good-quality target structure is available, identifying and characterizing the ligand-binding sites in the receptor is the next step so that the RCS can be screened against them. This is followed by high-throughput VS6 of the RCS using molecular docking and/or pharmacophore models7 , 8 to obtain initial repurposing candidates. These candidates are further ranked, screened, or optimized using computationally intensive MD simulations, MM-GB(PB)SA and QM-based binding energy estimations. Once a NP or an existing drug has been found to have significant affinity for a given target, it can be used as a lead for further development to improve the binding affinity. In other words, by preserving the overall structural skeleton/scaffold of the molecule, one can attempt to change the functional groups around the structure until the desired property is achieved. Here, we highlight how classical and modern ML methods along with traditional computational methods are used at all the above-discussed stages of SBDR and the rapidly evolving generative models for generating small molecules containing privileged scaffolds (from NPs or existing drugs).
Figure 2.
Possible strategies for structure-based drug repurposing (SBDR) for screening of molecules from repurposable chemical space.
Target structure prediction
The first step in SBDR is identification of the relevant target/s of interest and the availability of their 3D structure. VS of RCS using structure-based methods (traditional or ML) requires that the 3D structure of the target is available. experimental 3D structures from X-ray crystallography, NMR spectroscopy or cryo-electron microscopy can be obtained from the Protein Data Bank (PDB), which contains more than 150 000 bimolecular structures. Given that there is a large gap between the number of potential targets and the number of available experimental 3D structures, there is a tremendous interest in developing computational methods that can predict protein structures reliably. In silico methods, such as threading, ab initio techniques, and homology modeling have essential roles in predicting the structure of the desired targets.20 Homology modeling is the most popular structure prediction method,21 in which the structure of the target protein is modeled based on the experimental structure of a homologous template protein. In the absence of a homologous template structure, the fold recognition or threading method is used, in which each residue of the target is aligned to a position in the template and a template is selected based on the best alignment. If a target sequence does not have a suitable template either through homology or threading, the structures are modeled from scratch by optimizing the enthalpic and entropic parameters to generate the thermodynamically most-stable 3D conformation of the target protein.22 I-TASSER is a widely used structure prediction tool, which uses a combination of ab initio modeling, threading, and atomistic energy refinement to generate the 3D structure of a protein from its sequence.23
Although comparative modeling, ab initio modeling, and threading methods have had successes, they have major limitations. Over the past few years, ML methods have been helping to push the predictive capabilities of protein structures from sequences toward experimental accuracy.24 ML methods are capable of learning the relationship between primary sequences of proteins and known 3D structures, to develop predictive models. In CASP13, a DL-based ab initio protein structure prediction method named AlphaFold25 showed the best performance. AlphaFold comprises a core distance map predictor, which is implemented as a deep residue-NN with 220 residue slabs handling a depiction of dimensionality, analogous to input features calculated from two 64-amino acid fragments. The NN predictions include backbone torsion angles and pairwise distances between residues. Each residue slab has three layers containing a dilated convolutional layer and the blocks phase through dilation of values 1, 2, 4, and 8. The DL model has 21 million parameters, including 1D and 2D parameters, their combinations, and the evolutionary/coevolutionary profiles, of a training set of ∼ 29 000 proteins curated from various sources. Along with a distance map, AlphaFold predicts the φ and ψ angles to generate an initial predicted structure. Recently, AlphaFold 2.0 was proposed in CASP14 to outperform all the methods known so far, to the extent that the authors claim this to be the ‘solution to a 50-year-old grand challenge in biology’. The recently developed DL-based RoseTTAFold tool has shown promise in fast, correct protein structure and interaction predictions using a three-track network incorporating sequence (1D), topological distance map 2D, and spatial position (3D) information.26
Binding site prediction
The logic that proteins with similar structures might have affinities for similar ligands and seem to be involved in similar functions forms the basis of SBDR. Studies reported that similar ligands could bind to multiple targets with similar local binding sites despite the low global sequence similarity, demonstrating the importance of binding site/binding pocket detection and comparison in DR. Binding sites for ligands are mostly concave surfaces characterized by specific amino acid residues in a specific geometric orientation suitable for molecular recognition and molecular function of the protein. Conventional pocket detection algorithms can be broadly classified as sequence-based, geometry-based, and energy-based methods.27 Geometry-based methods were the first binding site prediction methods, and use 3D structural information to explore the pockets/clefts/cavities on the protein surface. These methods are efficient but do not consider the flexibilities of the protein surface. Surfnet,28 proposed by Laskowski, Fpocket algorithm,29 LIGSITEcsc,30 and PASS31 are examples of geometry-based methods.
Energy-based methods predict the most suitable binding site on the protein surface based on estimation of interaction energies of flexible probe molecules throughout the surface. One of the first methods was developed by Goodford,27 who calculated H-bond, electrostatic, and van der Waals components of interaction energies for different grid points on the protein surface and predicted the binding sites according to these interaction energies. Q-SiteFinder32 and PocketFinder are examples of energy-based methods. COACH33 is a combination of FINDSITE34 and ConCavity,35 which performed better than either method alone. FunFOLD,36 CHED, and HemeBIND37 also generate prediction models using a combination of different methods. Recently, ML-based methods, such as DeepSite,38 DeeplyTough,39 DeepDrug3D,40 and BionoiNet,41 were shown to be extremely efficient, achieving experimental accuracy for the prediction of binding sites.
RCS screening and lead optimization
Structure-based VS represents a highly efficient methodology for repositioning of known drug molecules to bind to potential new targets. Structure-based VS is mostly molecular docking based.20 Docking finds the suitable binding poses of molecules in the target binding site using a scoring function and the best-scored compounds from a large chemical library for a biomolecular target are further ranked based on the protein–ligand interactions.42 The RCS constitutes various classes of privileged structure43 with proven bioavailability and compatibility, reducing the probability of the best hits obtained via VS failing downstream in vitro/in vivo or ADMET tests. Molecular docking can be a single-target approach, in which only interactions between the known drugs and an individual target are identified, or it can be an ‘inverse docking’ approach, in which binding interactions of a molecule with multiple known targets are explored5, 44 to estimate its target selectivity. The molecular docking method typically comprises three key steps: modeling and predocking preparation of target and ligand structures; generation and sampling of the ligand conformers in the binding pocket of the receptor; and evaluation of the docking score reflecting the binding energy of the ligand–target complexes.45
To address the issue of ligand flexibility, several methods are commonly used, with stochastic methods being popular. Monte Carlo (MC) and/or genetic algorithms (GA) are two such examples. The MC algorithm stochastically alters a single parameter each time to produce new conformations that are allowed or disallowed based on Boltzmann distributions.4 A sufficiently high temperature is assigned at the start of modeling to ensure a high chance of the next sampled conformation being accepted. Then, the temperature is gradually lowered during docking, during which a low-energy protein–ligand complex is captured as a result of the lower conformational flexibility. Conversely, GA adopts a methodology inspired by Darwin’s evolution theory, which is initialized by an arbitrary population of conformations modeled as a set of chromosomes that can randomly crossover and mutate to produce a new set of conformations. The compound conformations with the lowest binding energies with the target are considered the ‘fittest’ and are accepted as start points to yield a new generation. This sequence is iteratively repeated until the target–ligand complex reaches a local energy minimum.4
There are three broad classes of traditional scoring function: (i) empirical; (ii) knowledge based; and (iii) force-field based.46 In the first class, different types of polar and nonpolar intermolecular interactions are extracted from a training set comprising the reported experimental structures, and parameters equivalent to each type of interactions are standardized with a certain weightage. The coefficients of these parameters are optimized through multiple linear regression models, using the reported binding affinity values of the training set molecules as the independent variable. Force-field-based scoring functions compute the potential energy of the entire ligand–target complex by adding up contributions from van der Waals or electrostatic interaction energies between the atoms of the ligand and those of the receptor. In knowledge-based scoring, the reported receptor–ligand complexes are analyzed to obtain structural information, which is further used to develop atomic interaction potentials that refer to the interactions between the ligand and receptor atoms.47 Fig. 3 depicts the popular computational tools/software available for tasks at different stages of SBDR.
Figure 3.
In silico tools for structure-based drug repurposing (SBDR).
Consistent efforts are being made to improve the performance of existing scoring functions by including additional terms for precise assessment of the ligands or entropy changes during receptor binding.48 Consensus scoring (i.e., using several scoring functions in parallel) has been developed for superior estimation of the binding affinity and to minimize false positive results. The computationally demanding, yet more accurate, QM techniques are being used to improve accuracies of the scoring functions, as discussed below. Finally, multiple scoring functions can be used in concert for so-called ‘consensus scoring’.
Binding energy estimations using traditional computational methods
In force field-based MD simulations, the systems comprise atoms and ions and the electrons are not considered explicitly. MD simulations allow us to keep track of the positions and momenta of these fundamental particles as a function of time. The atoms located in different molecular centers interact with each other through van der Waals and electrostatic interactions. Usually, the former is described using the Lennard–Jones-like potential energy function, which has –rij –6 and rij –12 dependence on the distance between the atoms, whereas the latter has inverse distance dependence. The dynamics of the system can be followed by solving Newton’s equation of motion. The time step usually used is 1–2 fs for modeling the biological systems in ambient conditions (300 K and 1 atm pressure). Once trajectories of sufficient timescale are established, thermodynamic properties can be computed using the positions and momenta of all the particles. To study the kinetics of association and dissociation of protein–ligand complexes, one needs to carry out long timescale simulations, which is usually computationally demanding. However, this can be handled with the use of steered MD or simulations with enhanced sampling techniques along selected reaction coordinates. In some implementations, one has to define the egression (unbinding) pathway explicitly, whereas, in some recent implementations (such as random acceleration MD), by setting the acceleration threshold for the ligand (to help the ligand to identify the pathway for release) alone helps the algorithm finds the regression pathway. In umbrella sampling simulations, the reaction coordinate for the dissociation is defined and the free energies for the unbinding are computed from the potential mean force. These methods have the advantage of traditional MD and provide free energy changes along the protein–ligand association or dissociation pathway. In certain targets, the residence time (RT) of the ligand within a target dictates the pharmacological activity rather than its binding affinity itself49 and, in these cases, enhanced sampling MD simulations can provide direct information about the RT, which is inversely proportional to koff. Targets, such as G-protein-coupled receptors (GPCR), HIV protease inhibitors, kinase inhibitors, and translocator proteins (TSPOs) are those targets for which RT is a key parameter for optimizing the potent ligands. In the case of TSPO targets, the sampling MD simulations were able to explain different koff for a specific ligand compared with the remaining two compounds, even though all three ligands had comparable binding affinity.50 The interaction of its naphthyl group with the LP1 loop along the egression pathway has been attributed to its increased residence time.50
Binding free energy calculations using MM-GB(PB)SA
Molecular docking approaches have been in use for more than three decades but their success rate in predicting the lead drug compounds from a chemical library is low, limiting their application.51 Binding free energies and docking poses from molecular docking approaches were found to be inaccurate in many cases. Nevertheless, they are the workhorses when compounds from larger chemical libraries needed to be screened. As the entries in certain chemical spaces are expected to grow exponentially, there will be no end to the use of molecular docking approaches.52 In addition, for obtaining potential lead compounds, one can use these approaches for prescreening, with the most promising compounds then being screened using a more reliable scoring function. This approach has been shown to be promising in ranking various protein–ligand complexes.53, 54
MM-GB(PB)SA-based binding free energies are widely used scoring functions for ranking protein–ligand complexes next to those used in molecular docking approaches. In both approaches, the binding free energies are obtained as the sum of van der Waals, electrostatic interactions, polar and nonpolar solvation free energies. Both MM-GBSA and MM-PBSA approaches differ with respect to the solvation-free energies, with the former two terms remain the same. In the MM-GBSA approach, the polar contribution solvation-free energies are obtained by solving the electrostatics of the complex in an aqueous solvent environment using the Generalized Born approach, whereas in the case of MM-PBSA, they are obtained using the Poisson–Boltzmann equation. The nonpolar contributions to solvation-free energies in MM-GBSA approach are obtained from the solvent accessible surface area. The binding free energy in these approaches is generally obtained as the difference in the free energies of the end products. In other words, the free energies are computed for the reactants (i.e., the protein and ligands in unbound state in an aqueous solvent environment) and products (protein–ligand complex in an aqueous solvent environment); the free energy difference of these two states is referred to as the binding free energy. The binding free energies are computed in two different ways, referred to as 1A-MM-GB(PB)SA or 3A-MM-GB(PB)SA depending upon whether the binding free energies were computed using a trajectory of the complex alone or using trajectories of subsystems (i.e., protein and ligand) and the complex.55 The former approach is computationally less demanding because a single MD simulation is carried out for the complex and the binding free energies for the three systems (complex, protein, and ligand) are obtained by using the coordinates of the system of interest and by stripping out the rest of the system coordinates. Another advantage of using a single trajectory for computing the binding free energies is that the change in internal energies associated with the complexation process is zero. Even though it is expensive, one can compute the entropic contributions from a normal mode analysis. In most instances, the entropic contributions are not computed because it is assumed that they do not have a major role in estimating the relative binding free energy differences of different ligands.
The binding free energies computed using the MM-GBSA and MM-PBSA approaches are not explicitly treating the effect of nonbonded interactions between the solvent (hydrogen bonds in particular) with the protein and ligands. In certain cases, in which the protein binding sites are occupied by ‘crystalline water’, these implicit models might not perform well and contributions from such water molecules need to be added in addition to the contributions obtained from implicit solvent models. The binding free energies are generally reported as an average over various configurations from the MD simulations and so these approaches account for the conformational flexibility of proteins and ligands, which is one of the merits of these approaches.
These approaches have shown success in ranking various protein–ligand complexes and there are reports of them outperforming the molecular docking-based ranking. For example, Rastelli et al. compared the performance of MM-GBSA and MM-PBSA with AutoDock in identifying active compounds from decoys against Plasmodium falciparum DHFR; the former two methods were able to rank the compounds in excellent agreement with experimental binding affinities.56
On the negative side, there were also many benchmark studies that showed larger fluctuations in binding free energies computed using a longer timescale. Instead, it was suggested that the binding free energies should be computed from many independent simulations of shorter timescales. In the case of avdin complexed with biotin analogs, it was shown that the average binding free energies over 5–50 independent MD simulations were needed to get an accuracy of 1 kJ/mol.57 Other studies also reported that the longer timescale MD simulations were not beneficial but that timescales limited to 5 ns yielded better accuracy in binding free energies.58
Binding free energy calculations from QM-based approaches
The binding free energies obtained using force-field approaches suffer from the use of fixed charges for the ligands in aqueous and protein environments. Naturally, the electronic structure, atomic charges, and molecular dipole moments depend on the nature of the environment and force-field methods do not account for such effects. To describe the electrostatics in solvent and protein environments, we need to use electronic structure theory-based approaches. However, these are computationally very demanding and memory intensive. The expense of electronic structure theory calculations is in the order of N 3–N 7, where N is the number of one electron wavefunctions of the system; thus, the size of the system that can be handled is limited to 100–200 atoms. Here, we are interested in the interaction energies of protein–ligand complexes, which are many times larger than this. Thus, approximate methods were developed that facilitate the use of QM theory for large-scale systems, such as protein–ligand complexes: (i) QM cluster models; (ii) hybrid QM/MM models; (iii) QM fragmentation approaches; and (iv) fragment molecular orbitals.
QM cluster models are based on the approximation that the binding site residues make larger contributions to the protein–ligand binding free energies. One can obtain the model for the protein–ligand cluster by using a cut-off, and the binding site residues within this distance from the center of mass of the ligand are included. It is essential to add suitable capping atoms in which the peptide bonds are cut. Given that, in many cases, the structure of the binding site is stabilized by the rest of the residues in the protein, the free optimization of the cluster can lead to changes in the binding mode/pose of the ligand within the binding site. Therefore, the terminal atoms of amino acids are fixed and partial optimizations are carried out to estimate the interaction energies. The interaction energies are given as the difference between the energy of the cluster to the sum of energies of the ligand and amino acids.
Hybrid QM/MM models use an effective Hamiltonian to describe the interaction between the protein–ligand subsystems, in which these systems are described using molecular mechanics and QM, respectively. The polarization of the ligand by the environment is correctly captured by the model, but the effect resulting from back polarization (i.e., polarization of the protein environments by the ligand) is not accounted for. Since we are mainly interested in the energetics of the ligands, this approach is reliable and also computationally less demanding. The whole protein and solvents can be included in the MM region without any difficulty and their polarization effect on the ligands can be modeled correctly using this approach. However, this approximation has issues when there is significant charge transfer between the binding site residues or solvents to ligand or when the QM subsystem is covalently bonded to MM region (as in the irreversible inhibitors), which is nicely described in QM cluster models. The charge transfer effect can be accounted for by describing the whole system involved in the charge transfer as a QM system and the rest as the MM system. This requires the treatment of the bonded region connecting the QM and MM subsystems using the hydrogen capping method and, in certain cases, overpolarization of the QM region connected through the MM region by covalent bonds has to be screened using a damping function.
The QM fragmentation scheme allows one to estimate the interaction of protein–ligand complexes using electronic structure theory. As the whole protein can not be treated using QM theory, the protein is fragmented into individual amino acids and the contributions from each fragment to the interaction energy with the ligand are computed and added together to obtain the total interaction energy. In other words, the total protein–ligand interactions are computed as the sum over the individual amino acid–ligand interactions. Usually, the bonds are cut along peptide bonds and capped with hydrogens or certain capping groups, such as acetyl or N-methyl amino groups. However, when we use such capping groups, their interaction energy contributions to the total protein–ligand interaction energy should be removed at the end. Since each amino acid and ligand intermolecular complex is handled separately, even the interaction energies can be obtained using highly correlated methods, such as MP2 and coupled-cluster theory. In general, dispersion corrected DFT or Minnesota functionals (namely MO6-2X) can be adopted to best describe the interaction between the individual amino acid fragments and ligand. In QM-based approaches, the binding enthalpies are approximated for binding free energies because the interaction energies are computed from the optimized structure for protein–ligand complexes. With the use of dispersion-corrected DFT (B3LYP/6-31G* -D), the performance of a QM fragmentation scheme referred to as EE-GMFCC-CPCM was tested on biotin and biotin analogs bound to avidin; the correlation between the experimental and predicted binding affinities was ∼ 0.88. The study was based on protein–ligand configurations obtained from MD; by averaging over more configurations, the correlation was shown to improve.59
AI/ML-based scoring functions and binding affinity prediction
One of the major efforts in VS is to be able to calculate binding affinities accurately. Whereas MD-based free energy methods can yield accurate values, they are slow; by contrast, scoring functions are fast but are less accurate. ML methods are thought of as having the potential to be fast/efficient and simultaneously significantly better than traditional scoring functions.60, 61 An SVM model was trained by coupling distinct docking-energy terms with the experimentally reported binding affinity of the training set of PDE inhibitors, to identify direct inhibitors of Mycobacterium tuberculosis, which was one of the first applications of the ML technique in the context of drug repositioning. Recently, the element-specific persistent homology (ESPH) method was used in association with CNN by Wei and coworkers to develop TopologyNet,62 a multichannel topological NN, in which the topological features represented biomacromolecular geometry diminishing the dimensionality of the complex 3D data. The gradient boosting decision tree (GBDT) regression was combined with the ESPH method to develop T-Bind. Here, element-specific topological fingerprints generated the features represented as binned barcodes and the models were fed by these features. The 3D voxel representation of both ligands and receptors were generated applying 3D CNN to devise KDEEP.63 Ashtawy and Mahapatra established two new scoring functions, BgN-Score and BsN-Score, based on bagging and boosting ensembles of NN models, respectively, using features that were combinations of the terms from X-Score, AffiScore, GOLD, and RF-Score.64 Later, Pande and coworkers proposed a scoring function known as PotentialNet65 based on staged graph CNN (GCN), which encompassed steps such as covalent-only, dual noncovalent–covalent propagations, and ligand-based graph using atom types, bonds, and interatomic distances as input descriptors; the authors emphasized the fact that the whole data set as well as the methods used for splitting the data, affect the relative performance of scoring functions. Twelve ML-based scoring functions were proposed and evaluated by Khamis and Gomaa on the PDBbind (v2013) core sets. They performed principal component analysis (PCA) to decrease the dimensionality of the huge set of input features to seven principal components using RF, kNN, NN, and SVM, which initially featured 108 terms from RF-Score, BALL, X-Score, and SLIDE.66 Li et al. developed the first XGBoost-based scoring function XGB-Score, implementing GBDT for amplified accuracy and speed.67 Su et al. also reported similar observations from their systematic study including six ML algorithms, namely Bayesian Ridge Regression (BRR), K-Nearest Neighbors (KNN), Decision Trees (DTs), Linear Support Vector Regression (L-SVR), Multilayer Perceptron (MLP), and RF.68 Yang et al. emphasized the importance of large, diverse, unbiased data sets for training AI/ML-based models, where they found overperformance (Pearson R2 = 0.73) of atomic CNN models trained on the PDBbind data set and recognized the property and topology biases in the DUD-E data set leading to artificially increased enrichment.69 Morrone et al. developed modular graph-based CNN models trained on structural data from protein − ligand complexes generated by molecular docking, to predict activity and binding mode.70 The algorithm presents a dual-graph architecture with separate subnetworks for the receptor–ligand contact maps and the ligand bond connectivities. Moro and coworkers used a combination of convolutional and fully connected NNs to develop a model to predict the performance of different common docking protocols from a protein structure and a small ligand molecule.71 Deep Docking is a new platform based on DL, which is able to dock billions of compounds with optimized speed and accuracy. This approach predicts the docking scores using deep QSAR models that learn from docking scores of a training set compound library.72 OnionNet73 is a DNN model to accurately predict the protein–ligand binding affinities based on rotation-free element pair-specific contacts between ligands and protein atoms. The efficiency of the model was assessed and compared with the contemporary scoring functions using the CASF-2013 benchmark and PDBbind database (v2016 core set). Sirimulla and colleagues established a DNN-based scoring function trained by 384 molecular descriptors, such as electrostatic interactions and H-bonds, calculated from the binding pockets of the PDBbind v2016 data set using BINANA software.74 Several other DL-based scoring functions have recently been developed to achieve speed and accuracy to predict target–receptor binding affinity, as discussed in recent reviews.75, 76, 77, 78
Generative modeling
Once a NP or an existing drug has been found to have significant affinity toward a given target, it can be taken as a lead for further development to improve its binding affinity. In other words, preserving the overall structural skeleton/scaffold of the molecule, one attempts to change the functional groups around the structure until the desired property is achieved. Over the last 2 to 3 years, modern DL method-enabled generative modeling has been shown to be effective for such purposes. Molecular design typically involves the measurement or prediction of a given property of interest for guess molecules using experiments or computational methods. This is followed by understanding of the structure–property relationship; upon multiple iterations between the two steps, molecules with desired properties are obtained. In other words, traditionally, one goes from the chemical space to the property space. However, generative models allow us to go from the property space to the chemical space. In other words, these methods are capable of generating molecular structures with the desired physicochemical and other pharmacodynamic/pharmacokinetic properties. The two major tasks of a generative model is to propose valid chemical structures, and to condition the generation toward certain biases. Four main methods have been successful in this aspect: (i) RNNs; (ii) Reinforcement Learning (RL); (iii) GANs; and (iv) VAEs. In the context of molecular design in the DD process, the chemical space is essentially infinite and, hence, such generative modeling approaches are useful for exploring this space to identify molecules that exhibit the desired properties. For optimization in the context of improving the binding affinity or other pharmacokinetic properties of NPs or existing drugs, generative models can be conditioned with multiple objectives such as the presence of a given scaffold and exhibition of desired properties.
Recurrent neural networks
RNN-based models are considered powerful generative models in the natural language-processing domain. These models are trained on the string representation of molecules, such as simplified molecular input line entry systems (SMILES),79 and learn the semantics of the representation,80, 81, 82, 83 helping to generate new molecules without explicitly defining the rules for molecule design.
Variational autoencoders
DL models based on VAEs comprise an encoder and a decoder. Generally, molecules are mapped to a latent space using an encoder, and a decoder is used to map latent vector representation back to the molecule.84, 85, 86 The latent space is often combined with optimization techniques to generate new molecules with the desired properties.
Generative adversarial networks
GANs comprise two ML models, the generator and discriminator, which are trained simultaneously to compete with each other. The generator generates a molecule and the discriminator performs a binary classification if that molecule belongs to the data set or is synthetic.87, 88 The generator helps to sample new molecules from the learned distribution.
Reinforcement learning
RL methods aid generative models with the objective of maximizing the reward of the generated molecules. RL techniques have been combined with SMILES-based models to generate new molecules but have low chemical validity.90, 91, 92, 93 To overcome this problem, a graph convolutional policy network (GCPN)94 was proposed achieving 100% validity of generated molecules. Fig. 4 shows a schematic of different generative models using different modern ML methods.
Figure 4.
Schematics of simple generative models using different modern machine-learning (ML) methods; (a) recurrent neural network (RNN); (b) variational auto encoder (VAE); (c) generative adversarial network (GAN); and (d) reinforcement learning (RL).
Recent examples of SBDR
Drug repurposing was considered the most efficient route to develop therapeutics for COVID-19-like virus-associated infections. A review article published in 2019 showed that from 2012 to 2017, 172 drugs were repurposed, with 70% in different stages of clinical development.89 Aspirin, bevacizumab, canakinumab, difluprednate, dimethyl fumarate, sildenafil, bupropion, and thalidomide are some of the drugs from repurposable chemical space that have since been approved for treating different diseases.89, 90 A bibliometric review of drug repurposing showed that > 60% of the 35 000 drugs or drug candidates have been tested against more than one disease, whereas 189 chemicals have been tested against > 300 diseases.91 Drugs, such as prednisolone, dexamethasone, prednisone, and methylprednisolone, have been repurposed for treating > 1000 diseases.91 Such promising results have also attracted researchers working toward the development of therapeutics for various virus-associated infections, such as Ebola virus, Middle East respiratory syndrome-coronavirus (MERS-CoV), and severe acute respiratory syndrome SARS-CoV-1 over the past decade. During the recent emergence of SARS-CoV-2-associated COVID-19, drug repurposing based on computational approaches has been used to identify potential drug compounds.91 The chemical library of approved antipolymerase drugs,92 the DrugBank database52, 93, 94 and chemical libraries of natural products were used. 3CLpro, PLpro, envelope (E) protein, spike protein, RNA dependent RNA polymerase (RdRp) and methyltransferase proteins were considered as potential targets from the virus,91 whereas, in humans, those that mediate the interaction with the viral spike protein, such as ACE-2, TMPRSS2, and Cathepsin-L, were also considered potential targets.91 For example, Yadav et al. recently performed docking and MD simulations to explore the repurposing of two approved bile salts, chenodeoxycholate and ursodeoxycholate, to bind to the SARS-CoV-2 envelope protein95 (Fig. 5 ). A sequential approach involving molecular docking and binding free energy calculations using MM-GBSA was used to repurpose compounds from the DrugBank database for COVID-19 therapeutics.96 Fig. 6 shows the binding mode of lead compounds from the DrugBank database within the four viral targets.
Figure 5.
Molecular dynamic (MD) simulation studies reveal a high influx of water molecules into the transmembrane channel of the severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) envelope protein (a) when bound to the approved drug chenodeoxycholate (b), which is a natural bile salt.
Figure 6.
Binding mode of lead compounds from the DrugBank database within the four viral targets from severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2): (a) 3CLPro; (b) PLPro; (c) RdRp; and (d) Spike protein.
Concluding remarks and prospects
Fully exploring the chemical space with currently available experimental and computational approaches is not possible. The upper limit for the number of entries in chemical space is reported to be 10180 and the number of possible small organic molecules is suggested to be 1060. Even if we had access to exascale computing facilities that could screen a compound per second, we still need the lifetime of the universe to scan all the compounds. Then, even if we were able to identify top compounds with superior binding affinity, there is no assurance that these compounds would have favorable pharmacodynamic and pharmacokinetic properties (i.e., ADMET, solubility and bioavailability). Thus, in situations such as the current COVID-19 pandemic and rapidly emerging SARS-CoV-2 variants, where one has to urgently find a scalable solution, repurposing existing drugs and screening of existing NPs with experimentally annotated pharmacokinetic profiles are appropriate approaches to identify potential compounds toward any therapeutic target associated with a disease of interest within a reasonable timeline.
The limited size of the repurposable chemical space can be handled easily with currently available SBDD approaches. Here, we have summarized traditional methods applied at each stage of SBDR as well as recently developed AI algorithms, which can be used either instead of, or in association with, traditional methods to achieve accurate predictions. Computationally intensive MD simulations and QM-based methods that can be used conveniently for small RCS for efficient binding energy estimation have also been discussed. Whereas traditional methods, such as docking-based VS, are extremely quick to screen a few thousand molecules of RCS against new targets, the accuracy of the calculated molecular properties, such as binding affinity, is low because of the severe approximations used. Alternatively, free energy calculations using MD simulations and QM methods are capable of providing accurate values. In recent years, modern ML methods have been seen as potential methods that will make every task throughout the DD process more efficient. Although classical ML methods are still valuable in situations where the data set size is limited, modern ML methods are proving to be disruptive and are changing the way that different tasks in DD processes are being undertaken. Recent studies have shown that ML methods can help in identifying targets, predicting 3D structures of target proteins from the sequence, helping to screen large numbers of small druglike molecules, performing generative tasks to suggest new ligands, providing retrosynthetic pathways for synthesis, controlling robotic systems to physically synthesize compounds, processing the signal corresponding to molecule characterization based on spectra, and predicting outcomes of clinical trials. For VS applications, NN-based methods have been shown to be useful for developing ML-based scoring functions that are accurate and computationally tractable. Additionally, generative methods are capable of suggesting molecules that have scaffolds identified from NPs and existing drugs. Hence, careful combination of traditional methods and data-driven methods is expected to speed up the whole DD process in general and drug repurposing in particular.
Acknowledgments
C.C. thanks DST, India for financial support in the form of an INSPIRE Faculty award. U.D.P. thanks DST-SERB (Grant No.: CVD/2020/000343), and IHub-Data, IIIT Hyderabad for financial assistance.
References
- 1.Dotolo S., Marabotti A., Facchiano A., Tagliaferri R. A review on drug repurposing applicable to COVID-19. Briefings in Bioinformatics. 2021;22:726–741. doi: 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanhaelen Q., editor. Computational Methods for Drug Repurposing. Springer; New York: 2019. [Google Scholar]
- 3.Issa N.T., Kruger J., Byers S.W., Dakshanamurthy S. Drug repurposing a reality: from computers to the clinic. Expert Review of Clinical Pharmacology. 2013;6(2):95–97. doi: 10.1586/ecp.12.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badrinarayan P., Choudhury C., Sastry G.N. In: Systems and Synthetic Biology. Singh V., Dhar P.K., editors. Amsterdam; Springer; 2015. Molecular modeling; pp. 93–128. [Google Scholar]
- 5.Jarada T.N., Rokne J.G., Alhajj R. A review of computational drug repositioning: strategies, approaches, opportunities, challenges, and directions. J Cheminform. 2020;12(1):46. doi: 10.1186/s13321-020-00450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma D.L., Chan D.S.H., Leung C.H. Drug repositioning by structure-based virtual screening. Chem Soc Rev. 2013;42(5):2130. doi: 10.1039/c2cs35357a. [DOI] [PubMed] [Google Scholar]
- 7.Choudhury C., Priyakumar U.D., Sastry G.N. Dynamic ligand-based pharmacophore modeling and virtual screening to identify mycobacterial cyclopropane synthase inhibitors. J Chem Sci. 2016;128(5):719–732. doi: 10.1021/ci500737b. [DOI] [PubMed] [Google Scholar]
- 8.Choudhury C., Priyakumar U.D., Sastry G.N. Dynamics based pharmacophore models for screening potential inhibitors of mycobacterial cyclopropane synthase. J Chem Inf Model. 2015;55(4):848–860. doi: 10.1021/ci500737b. [DOI] [PubMed] [Google Scholar]
- 9.Yang X., Wang Y., Byrne R., Schneider G., Yang S. Concepts of artificial intelligence for computer-assisted drug discovery. Chem Rev. 2019;119(18):10520–10594. doi: 10.1021/acs.chemrev.8b00728. [DOI] [PubMed] [Google Scholar]
- 10.Ballester P.J. Machine learning for molecular modelling in drug design. Biomolecules. 2019;9(6):216. doi: 10.3390/biom9060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang X., Fu W., Wang J., Kang D., Xu L., Zhao Y., et al. Identification of estrogen receptor α antagonists from natural products via in vitro and in silico approaches. Oxidative Medicine and Cellular Longevity. 2018;2018:1–11. doi: 10.1155/2018/6040149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Y., Li W., Du T., Hong Z., Lin J. Targeting HIV/HCV coinfection using a machine learning-based multiple quantitative structure-activity relationships (multiple QSAR) method. IJMS. 2019;20(14):3572. doi: 10.3390/ijms20143572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cano G., Garcia-Rodriguez J., Garcia-Garcia A., Perez-Sanchez H., Benediktsson J.A., Thapa A., et al. Automatic selection of molecular descriptors using random forest: application to drug discovery. Expert Systems with Applications. 2017;72:151–159. [Google Scholar]
- 14.Rahman R., Otridge J., Pal R. IntegratedMRF: random forest-based framework for integrating prediction from different data types. Bioinformatics. 2017;33(9):1407–1410. doi: 10.1093/bioinformatics/btw765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maltarollo V.G., Kronenberger T., Espinoza G.Z., Oliveira P.R., Honorio K.M. Advances with support vector machines for novel drug discovery. Expert Opinion on Drug Discovery. 2019;14(1):23–33. doi: 10.1080/17460441.2019.1549033. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y.C., Zhang C.H., Deng N.Y., Wang Y. Kernel-based data fusion improves the drug–protein interaction prediction. Computational Biology and Chemistry. 2011;35(6):353–362. doi: 10.1016/j.compbiolchem.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Kawai K., Fujishima S., Takahashi Y. Predictive activity profiling of drugs by topological-fragment-spectra-based support vector machines. J Chem Inf Model. 2008;48(6):1152–1160. doi: 10.1021/ci7004753. [DOI] [PubMed] [Google Scholar]
- 18.Baskin I.I., Winkler D., Tetko I.V. A renaissance of neural networks in drug discovery. Expert Opinion on Drug Discovery. 2016;11(8):785–795. doi: 10.1080/17460441.2016.1201262. [DOI] [PubMed] [Google Scholar]
- 19.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 20.Batool M., Ahmad B., Choi S. A structure-based drug discovery paradigm. IJMS. 2019;20(11):2783. doi: 10.3390/ijms20112783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ginalski K. Comparative modeling for protein structure prediction. Current Opinion in Structural Biology. 2006;16(2):172–177. doi: 10.1016/j.sbi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Floudas C.A., Fung H.K., McAllister S.R., Mönnigmann M., Rajgaria R. Advances in protein structure prediction and de novo protein design: a review. Chemical Engineering Science. 2006;61(3):966–988. [Google Scholar]
- 23.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torrisi M., Pollastri G., Le Q. Deep learning methods in protein structure prediction. Computational and Structural Biotechnology Journal. 2020;18:1301–1310. doi: 10.1016/j.csbj.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577(7792):706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 26.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373(6557):871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche D., Brackenridge D., McGuffin L. Proteins and their interacting partners: an introduction to protein–ligand binding site prediction methods. IJMS. 2015;16(12):29829–29842. doi: 10.3390/ijms161226202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski R.A. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. Journal of Molecular Graphics. 1995;13(5):323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 29.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinformatics. 2009;10(1):168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang B., Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6(1):19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brady G.P., Jr., Stouten P.F.W. Fast prediction and visualization of protein binding pockets with PASS. Journal of Computer-Aided Molecular Design. 2000;14(4):383–401. doi: 10.1023/a:1008124202956. [DOI] [PubMed] [Google Scholar]
- 32.Laurie A.T.R., Jackson R.M. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21(9):1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 33.Wu Q., Peng Z., Zhang Y., Yang J. COACH-D: improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Research. 2018;46(W1):W438–W442. doi: 10.1093/nar/gky439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brylinski M., Skolnick J. FINDSITE-metal: Integrating evolutionary information and machine learning for structure-based metal-binding site prediction at the proteome level: metal-binding site prediction by FINDSITE-Metal. Proteins. 2011;79(3):735–751. doi: 10.1002/prot.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacCallum R.M., Martin A.C.R., Thornton J.M. Antibody-antigen Interactions: contact analysis and binding site topography. Journal of Molecular Biology. 1996;262(5):732–745. doi: 10.1006/jmbi.1996.0548. [DOI] [PubMed] [Google Scholar]
- 36.Roche D.B., Tetchner S.J., McGuffin L.J. FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics. 2011;12(1):160. doi: 10.1186/1471-2105-12-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y.F., Cheng C.W., Shih C.S., Hwang J.K., Yu C.S., Lu C.H. MIB: metal ion-binding site prediction and docking server. J Chem Inf Model. 2016;56(12):2287–2291. doi: 10.1021/acs.jcim.6b00407. [DOI] [PubMed] [Google Scholar]
- 38.Jiménez J., Doerr S., Martínez-Rosell G., Rose A.S., De Fabritiis G. DeepSite: protein-binding site predictor using 3D-convolutional neural networks. Bioinformatics. 2017;33(19):3036–3042. doi: 10.1093/bioinformatics/btx350. [DOI] [PubMed] [Google Scholar]
- 39.Simonovsky M., Meyers J. DeeplyTough: learning structural comparison of protein binding sites. J Chem Inf Model. 2020;60(4):2356–2366. doi: 10.1021/acs.jcim.9b00554. [DOI] [PubMed] [Google Scholar]
- 40.Pu L., Govindaraj R.G., Lemoine J.M., Wu H.C., Brylinski M. DeepDrug3D: classification of ligand-binding pockets in proteins with a convolutional neural network. PLoS Comput Biol. 2019;15(2) doi: 10.1371/journal.pcbi.1006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi W., Lemoine J.M., Shawky A.A., Singha M., Pu L., Yang S., et al. BionoiNet: ligand-binding site classification with off-the-shelf deep neural network. Bioinformatics. 2020;36(10):3077–3083. doi: 10.1093/bioinformatics/btaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurumurthy C., Sambasiva Rao P., Veeraswamy B., Santhosh Kumar G., Shanthan Rao P., Kotamraju S., et al. A facile and single pot strategy for the synthesis of novel naphthyridine derivatives under microwave irradiation conditions using ZnCl2 as catalyst, evaluation of AChE inhibitory activity, and molecular modeling studies. Med Chem Res. 2012;21(8):1785–1795. [Google Scholar]
- 43.Choudhury C., Deva Priyakumar U., Narahari S.G. Structural and functional diversities of the hexadecahydro-1H-cyclopentaphenanthrene framework, a ubiquitous scaffold in steroidal hormones. Mol Inf. 2016;35(3–4):145–157. doi: 10.1002/minf.201600005. [DOI] [PubMed] [Google Scholar]
- 44.Vanhaelen Q., Mamoshina P., Aliper A.M., Artemov A., Lezhnina K., Ozerov I., et al. Design of efficient computational workflows for in silico drug repurposing. Drug Discovery Today. 2017;22(2):210–222. doi: 10.1016/j.drudis.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Kumar S. Molecular docking: a structure-based approach for drug repurposing. In: XXXX eds. In Silico Drug Design. Amsterdam, Elsevier; 2019: 161-189.
- 46.Huang S.Y., Grinter S.Z., Zou X. Scoring functions and their evaluation methods for protein–ligand docking: recent advances and future directions. Phys Chem Chem Phys. 2010;12(40):12899. doi: 10.1039/c0cp00151a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J., Fu A., Zhang L. An overview of scoring functions used for protein–ligand interactions in molecular docking. Interdiscip Sci Comput Life Sci. 2019;11(2):320–328. doi: 10.1007/s12539-019-00327-w. [DOI] [PubMed] [Google Scholar]
- 48.March-Vila E., Pinzi L., Sturm N., Tinivella A., Engkvist O., Chen H., et al. On the integration of in silico drug design methods for drug repurposing. Front Pharmacol. 2017;8:298. doi: 10.3389/fphar.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixon T., Lotz S.D., Dickson A. Predicting ligand binding affinity using on- and off-rates for the SAMPL6 SAMPLing challenge. J Comput Aided Mol Des. 2018;32(10):1001–1012. doi: 10.1007/s10822-018-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruno A., Barresi E., Simola N., Da Pozzo E., Costa B., Novellino E., et al. Unbinding of translocator protein 18 kDa (TSPO) ligands: from in vitro residence time to in vivo efficacy via in silico simulations. ACS Chem Neurosci. 2019;10(8):3805–3814. doi: 10.1021/acschemneuro.9b00300. [DOI] [PubMed] [Google Scholar]
- 51.Wang G., Zhu W. Molecular docking for drug discovery and development: a widely used approach but far from perfect. Future Med Chem. 2016;8(14):1707–1710. doi: 10.4155/fmc-2016-0143. [DOI] [PubMed] [Google Scholar]
- 52.Arús-Pous J., Blaschke T., Ulander S., Reymond J.L., Chen H., Engkvist O. Exploring the GDB-13 chemical space using deep generative models. J Cheminform. 2019;11(1):20. doi: 10.1186/s13321-019-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem. 2011;32(5):866–877. doi: 10.1002/jcc.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun H., Li Y., Shen M., Tian S., Xu L., Pan P., et al. Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys Chem Chem Phys. 2014;16(40):22035–22045. doi: 10.1039/c4cp03179b. [DOI] [PubMed] [Google Scholar]
- 55.Genheden S., Ryde U. Comparison of end-point continuum-solvation methods for the calculation of protein-ligand binding free energies. Proteins. 2012;80(5):1326–1342. doi: 10.1002/prot.24029. [DOI] [PubMed] [Google Scholar]
- 56.Rastelli G., Rio A.D., Degliesposti G., Sgobba M. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem. 2009;31(4):797–810. doi: 10.1002/jcc.21372. [DOI] [PubMed] [Google Scholar]
- 57.Genheden S., Ryde U. How to obtain statistically converged MM/GBSA results. J Comput Chem. 2010;31(4):837–846. doi: 10.1002/jcc.21366. [DOI] [PubMed] [Google Scholar]
- 58.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu J., Wang X., Zhang J.Z.H., He X. Calculation of protein–ligand binding affinities based on a fragment quantum mechanical method. RSC Adv. 2015;5(129):107020–107030. [Google Scholar]
- 60.Khamis M.A., Gomaa W., Ahmed W.F. Machine learning in computational docking. Artificial Intelligence in Medicine. 2015;63(3):135–152. doi: 10.1016/j.artmed.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 61.Melville J., Burke E., Hirst J. Machine learning in virtual screening. CCHTS. 2009;12(4):332–343. doi: 10.2174/138620709788167980. [DOI] [PubMed] [Google Scholar]
- 62.Cang Z., Wei G.W. TopologyNet: topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLoS Comput Biol. 2017;13(7) doi: 10.1371/journal.pcbi.1005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiménez J., Škalič M., Martínez-Rosell G., De Fabritiis G. KDEEP : protein–ligand absolute binding affinity prediction via 3D-convolutional neural networks. J Chem Inf Model. 2018;58(2):287–296. doi: 10.1021/acs.jcim.7b00650. [DOI] [PubMed] [Google Scholar]
- 64.Ashtawy H.M., Mahapatra N.R. BgN-Score and BsN-Score: bagging and boosting based ensemble neural networks scoring functions for accurate binding affinity prediction of protein-ligand complexes. BMC Bioinformatics. 2015;16(S4):S8. doi: 10.1186/1471-2105-16-S4-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feinberg E.N., Sur D., Wu Z., Husic B.E., Mai H., Li Y., et al. PotentialNet for molecular property prediction. ACS Cent Sci. 2018;4(11):1520–1530. doi: 10.1021/acscentsci.8b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khamis M.A., Gomaa W. Comparative assessment of machine-learning scoring functions on PDBbind 2013. Engineering Applications of Artificial Intelligence. 2015;45:136–151. [Google Scholar]
- 67.Li H., Peng J., Sidorov P., Leung Y., Leung K.S., Wong M.H., et al. Classical scoring functions for docking are unable to exploit large volumes of structural and interaction data. Bioinformatics. 2019;35(20):3989–3995. doi: 10.1093/bioinformatics/btz183. [DOI] [PubMed] [Google Scholar]
- 68.Su M., Feng G., Liu Z., Li Y., Wang R. Tapping on the black box: how is the scoring power of a machine-learning scoring function dependent on the training set? J Chem Inf Model. 2020;60(3):1122–1136. doi: 10.1021/acs.jcim.9b00714. [DOI] [PubMed] [Google Scholar]
- 69.Yang J., Shen C., Huang N. Predicting or pretending: artificial intelligence for protein-ligand interactions lack of sufficiently large and unbiased datasets. Front Pharmacol. 2020;11:69. doi: 10.3389/fphar.2020.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morrone J.A., Weber J.K., Huynh T., Luo H., Cornell W.D. Combining docking pose rank and structure with deep learning improves protein–ligand binding mode prediction over a baseline docking approach. J Chem Inf Model. 2020;60(9):4170–4179. doi: 10.1021/acs.jcim.9b00927. [DOI] [PubMed] [Google Scholar]
- 71.Jiménez-Luna J., Cuzzolin A., Bolcato G., Sturlese M., Moro S. A deep-learning approach toward rational molecular docking protocol selection. Molecules. 2020;25(11):2487. doi: 10.3390/molecules25112487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gentile F., Agrawal V., Hsing M., Ton A.T., Ban F., Norinder U., et al. Deep docking: a deep learning platform for augmentation of structure based drug discovery. ACS Cent Sci. 2020;6(6):939–949. doi: 10.1021/acscentsci.0c00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng L., Fan J., Mu Y. OnionNet: a multiple-layer intermolecular-contact-based convolutional neural network for protein–ligand binding affinity prediction. ACS Omega. 2019;4(14):15956–15965. doi: 10.1021/acsomega.9b01997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hassan M, Mogollon DC, Fuentes O, Suman S. DLSCORE: a deep learning model for predicting protein-ligand binding affinities. ChemRxiv Published online April 20, 2018. http://dx.doi.org/10.26434/chemrxiv.6159143.v1.
- 75.Ballester P.J. Selecting machine-learning scoring functions for structure-based virtual screening. Drug Discovery Today: Technologies. 2019;32–33:81–87. doi: 10.1016/j.ddtec.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 76.Shen C, Hu Y, Wang Z, Zhang X, Zhong H, Wang G, et al. Can machine learning consistently improve the scoring power of classical scoring functions? Insights into the role of machine learning in scoring functions. Briefings in Bioinformatics. Published online January 25, 2020: bbz173. [DOI] [PubMed]
- 77.Li H., Sze K., Lu G., Ballester P.J. Machine-learning scoring functions for structure-based drug lead optimization. WIREs Comput Mol Sci. 2020;10(5) [Google Scholar]
- 78.Shen C., Ding J., Wang Z., Cao D., Ding X., Hou T. From machine learning to deep learning: advances in scoring functions for protein–ligand docking. WIREs Comput Mol Sci. 2020;10(1) [Google Scholar]
- 79.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Model. 1988; 28(1): 31-36.
- 80.Segler M.H.S., Kogej T., Tyrchan C., Waller M.P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent Sci. 2018;4(1):120–131. doi: 10.1021/acscentsci.7b00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ertl P, Lewis R, Martin E, Polyakov V. In silico generation of novel, drug-like chemical matter using the LSTM neural network. arXiv. Published online January 8, 2018. https://doi.org/10.48550/arXiv.1712.07449.
- 82.Gupta A., Müller A.T., Huisman B.J.H., Fuchs J.A., Schneider P., Schneider G. Generative recurrent networks for de novo drug design. Mol Inf. 2018;37(1–2):1700111. doi: 10.1002/minf.201700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bjerrum EJ, Threlfall R. Molecular generation with recurrent neural networks (RNNs). arXiv. Published online May 17, 2017. https://doi.org/10.48550/arXiv.1705.04612.
- 84.Gómez-Bombarelli R., Wei J.N., Duvenaud D., et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent Sci. 2018;4(2):268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffiths R.R., Hernández-Lobato J.M. Constrained Bayesian optimization for automatic chemical design using variational autoencoders. Chem Sci. 2020;11(2):577–586. doi: 10.1039/c9sc04026a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dai H, Tian Y, Dai B, Skiena S, Song L. Syntax-directed variational autoencoder for structured data. arXiv. Published online February 23, 2018. 10.48550/arXiv.1802.08786. [DOI]
- 87.Blaschke T., Olivecrona M., Engkvist O., Bajorath J., Chen H. Application of generative autoencoder in de novo molecular design. Mol Inf. 2018;37(1–2):1700123. doi: 10.1002/minf.201700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Cao N, Kipf T. MolGAN: An implicit generative model for small molecular graphs. arXiv: 180511973 . Published online May 30, 2018. Accessed January 31, 2021. http://arxiv.org/abs/1805.11973.
- 89.Polamreddy P., Gattu N. The drug repurposing landscape from 2012 to 2017: evolution, challenges, and possible solutions. Drug Discov Today. 2019;24(3):789–795. doi: 10.1016/j.drudis.2018.11.022. [DOI] [PubMed] [Google Scholar]
- 90.Baker N.C., Ekins S., Williams A.J., Tropsha A. A bibliometric review of drug repurposing. Drug Discov Today. 2018;23(3):661–672. doi: 10.1016/j.drudis.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Y.W., Xie Y., Tang L.S., Pu D., Zhu Y.J., Liu J.Y., et al. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies. Sig Transduct Target Ther. 2021;6(1):317. doi: 10.1038/s41392-021-00733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368(6493):829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- 93.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khelfaoui H., Harkati D., Saleh B.A. Molecular docking, molecular dynamics simulations and reactivity, studies on approved drugs library targeting ACE2 and SARS-CoV-2 binding with ACE2. Journal of Biomolecular Structure and Dynamics. 2021;39(18):7246–7262. doi: 10.1080/07391102.2020.1803967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yadav R, Choudhury C, Kumar Y, Bhatia A. Virtual repurposing of ursodeoxycholate and chenodeoxycholate as lead candidates against SARS-Cov2-envelope protein: a molecular dynamics investigation. Journal of Biomolecular Structure and Dynamics. Published online December 31, 2020. 10.1080/07391102.2020.1868339. [DOI] [PMC free article] [PubMed]
- 96.Murugan N.A., Kumar S., Jeyakanthan J., Srivastava V. Searching for target-specific and multi-targeting organics for Covid-19 in the Drugbank database with a double scoring approach. Sci Rep. 2020;10(1):19125. doi: 10.1038/s41598-020-75762-7. [DOI] [PMC free article] [PubMed] [Google Scholar]