ABSTRACT
The Lactobacillaceae are an intensively studied family of bacteria widely used in fermented food and probiotics, and many are native to the gut and vaginal microbiota of humans and other animals. Various studies have shown that specific Lactobacillaceae species produce metabolites that can inhibit the colonization of fungal and bacterial pathogens, but less is known about how Lactobacillaceae affect individual bacterial species in the endogenous animal microbiota. Here, we show that numerous Lactobacillaceae species inhibit the growth of the Lachnospiraceae family and the S24-7 group, two dominant clades of bacteria within the gut. We demonstrate that inhibitory activity is a property common to homofermentative Lactobacillaceae species, but not to species that use heterofermentative metabolism. We observe that homofermentative Lactobacillaceae species robustly acidify their environment, and that acidification alone is sufficient to inhibit growth of Lachnospiraceae and S24-7 growth, but not related species from the Clostridiales or Bacteroidales orders. This study represents one of the first in-depth explorations of the dynamic between Lactobacillaceae species and commensal intestinal bacteria, and contributes valuable insight toward deconvoluting their interactions within the gut microbial ecosystem.
KEYWORDS: Probiotics, lactobacilli, lactic acid bacteria, Bacteroidales, Clostridiales, microbiota, gut, acid stress, Lachnospiraceae, Muribaculaceae, S24-7
Introduction
Lactobacilli are an extensively studied clade of Gram-positive bacteria that are employed in a broad range of applications, including biotechnology, food fermentation, and probiotic formulations.1–4 Originally defined as a genus in 1901, this lactic acid-producing group has since expanded to comprise over 260 species based on morphology and fermentation products.5,6 However, recent 16S rRNA-based genotyping and genome sequencing efforts have shown that the lactobacilli are far more genetically diverse than most bacterial genera and even many bacterial families.7,8 As a result, in 2020 the Lactobacillus genus was formally reclassified into 25 genera under the umbrella of the Lactobacillaceae family.6 This reclassification was carried out using a polyphasic approach predicated on average nucleotide and amino acid identity and core genome phylogeny, while also taking physiology and ecology into consideration.6
Members of the newly defined Lactobacillaceae family cluster into two distinct clades depending on whether they utilize homofermentative or heterofermentative metabolism.9,10 Homofermentative species metabolize hexoses via the Embden-Meyerhof pathway, producing pyruvate as a key metabolic intermediate and lactate as an end product. Heterofermentative species metabolize hexoses via the phosphoketolase pathway, producing pyruvate and acetyl-phosphate as key intermediates with lactate and acetate or ethanol as end products. The split between the two types of metabolism appears to have occurred early in the evolution of the Lactobacillaceae, and their fermentation types correlate almost perfectly with phylogeny.10
However, beyond this binary metabolic delineation Lactobacillaceae species differ with respect to other aspects of their metabolism, physiology, and ecology – occupying niches ranging from free-living to strictly symbiotic.11 Importantly, many species from the Lactobacillaceae family are resident members of the gut and vaginal microbiota of humans and other animals. This fact, combined with the frequent use of Lactobacillaceae species in food and probiotics, has made studying representatives from this family an area of significant focus. Alongside investigating physiological and immunomodulatory effects on the host, over the past few years there has been increased interest in understanding how these species affect other members of the gut microbiota.12–14
Considerable research has explored the mechanisms by which specific Lactobacillaceae prevent growth of pathogenic bacteria or fungi. From nutrient or niche competition to production of antimicrobial compounds such as bacteriocins, lactic acid, and hydrogen peroxide, a bottom-up approach has been used to characterize the interactions between various Lactobacillaceae species and pathogens such as Listeria monocytogenes, Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, and Salmonella enterica.14–20 Fewer studies have examined how Lactobacillaceae species interact with commensal gut bacteria, and these have been almost exclusively carried out through a top-down approach by looking at changes in overall bacterial communities. Although this type of work has yielded valuable insights into the dynamics of the gut microbiota, variability in methodology across studies has led to inconsistent findings. For example, some studies have found that probiotic Lactobacillaceae species aid in restoring the composition of the gut microbiota after perturbation with antibiotics while others have found that they actively delay reestablishment of the native community.21,22 Consequently, elucidating the interactions between specific lactobacilli and the gut microbiota is integral to our understanding of these intricate microbial ecosystems and deciphering their effects on the host.
We recently established the Collection of Inflammation-Associated Mouse Intestinal Bacteria (CIAMIB), which contains isolates from four species of Lactobacillaceae (Wong et al., 2022; in press). In exploring the positive and negative interactions between individual isolates within this collection, we found that certain Lactobacillaceae species inhibit the growth of bacteria from the Lachnospiraceae family (order Clostridiales, phylum Firmicutes) and the S24-7 group (order Bacteroidales, phylum Bacteroidetes), while others do not. In the mammalian gut microbiota, the Firmicutes and Bacteroidetes phyla predominate, and Clostridiales and Bacteroidales are among the most abundant orders within these phyla. The relative abundance of the Lachnospiraceae family in the gut microbiota of mammals is generally above 10%, whereas the abundance of S24-7 bacteria ranges from approximately 2% in the human gut to over 20% in the gut of the laboratory mouse.23–27 Despite their prevalence, these taxa remain poorly characterized due to challenges in culturing representative isolates.25,26,28,29
Here, we characterized the inhibition of Lachnospiraceae and S24-7 isolates by Lactobacillaceae utilizing a metabolically diverse set of Lactobacillaceae isolates. We discovered that inhibitory activity was common to all homofermentative lactobacilli we tested. We observed that these homofermentative Lactobacillaceae species produce higher quantities of acid and allow the pH of the surrounding media to drop, whereas heterofermentative isolates maintain a more neutral environmental pH. Finally, we demonstrated that isolates from the S24-7 group and the Lachnospiraceae family are particularly susceptible to acidic conditions compared to related isolates from the Bacteroidales and Clostridiales orders. This work provides novel insight into the effect of species from the Lactobacillaceae family on commensal intestinal taxa and illustrates the importance of using bottom-up approaches to help deconvolute the complex network of interactions within the gut microbiota.
Results
Select species from the Lactobacillaceae family inhibit growth of commensal intestinal bacteria in a contact-independent manner
This study began with the exploration of synergistic or antagonistic microbe–microbe interactions using isolates from the CIAMIB. This collection comprises more than 40 strains isolated from the intestines of mice with inflammation and includes four species from the S24-7 group, including isolates from the Muribaculaceae family and a newly identified family. The collection also contains isolates of two novel species within the Lachnospiraceae family. Initial screening efforts involved spotting assays, where antagonistic interactions between isolates could be observed by the appearance of a zone of growth inhibition in the lawn surrounding the culture drop (Figure 1a). We consistently observed differences between Lactobacillaceae isolates in terms of their effect on the growth of isolates belonging to the S24-7 group or the Lachnospiraceae family (Table S1). Ligilactobacillus murinus (isolates NM26_J9 and NM28_3M-8), Lactobacillus johnsonii (isolate NM60_B2-8), and Lactobacillus intestinalis (isolate NM61_E11) inhibited the growth of all S24-7 and Lachnospiraceae isolates, whereas Limosilactobacillus reuteri (isolates NM11_1-41 and NM12_1-47) did not exhibit an inhibitory effect (Figure 1a; Figure S1).
Figure 1.
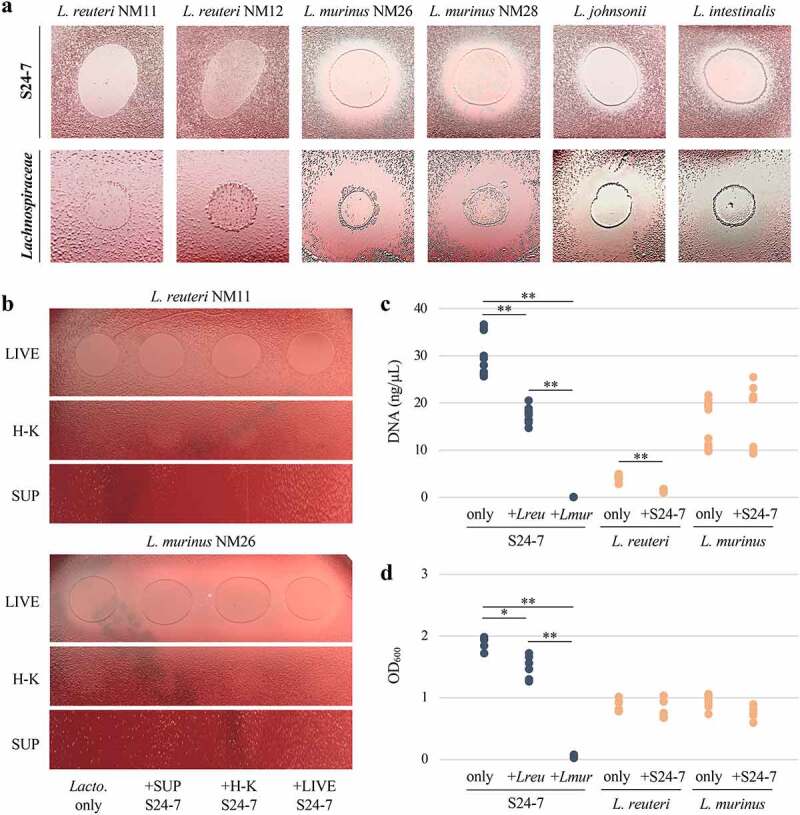
Specific Lactobacillaceae species inhibit the growth of representatives from the S24-7 group and Lachnospiraceae family in a contact-independent manner. For A) and B), representative images are shown (n = 3). A) Six Lactobacillaceae strains spotted onto lawns of NM74_B14 from the S24-7 group and NM01_1-7b from the Lachnospiraceae family. B) Live (LIVE), heat-killed (H-K), and supernatant (SUP) of uninhibitory L. reuteri NM11 and inhibitory L. murinus NM26 liquid cultures spotted onto NM74_B14. From left to right, spotting is Lactobacillaceae grown axenically (Lacto. only), with NM74_B14 supernatant (+SUP S24-7), heat-killed (+H-K S24-7), and live (+LIVE S24-7). C) Growth of NM74_B14 (S24-7), L. reuteri NM11, and L. murinus NM26 measured by qPCR based on DNA concentration. Data points are from four independent experiments. D) Growth measured by OD600 of NM74_B14 (S24-7), L. reuteri NM11, and L. murinus NM26 cultured in transwell plates alone or in the presence of another species. Data points are from three independent experiments. For C) and D), growth of NM74_B14 is shown in blue and growth of L. reuteri NM11 and L. murinus NM26 is shown in Orange. Welch’s t-test, * p < .05; ** p < .01.
To investigate the mechanism of this inhibition, we performed another set of spotting assays on the S24-7 isolate NM74_B14 and the Lachnospiraceae isolate NM01_1-7b, which were selected due to their relatively robust and consistent growth. We sought to determine if growth inhibition required live Lactobacillaceae cultures and if the inhibitory activity was stimulated by the presence of S24-7 or Lachnospiraceae. The lactobacilli were therefore grown axenically as well as with supernatant, heat-killed bacteria, or live bacteria from the inhibited NM74_B14 or NM01_1-7b isolates. These cultures were then spotted live, heat-killed, or filtered as supernatant onto lawns of NM74_B14 or NM01_1-7b. Only cultures with live L. murinus, L. johnsonii, and L. intestinalis retained their inhibitory activity. Neither heat-killed bacteria nor filtered Lactobacillaceae culture supernatants had an effect on growth (Figure 1b; Figure S2). This suggested that inhibition was either contact-dependent, mediated by a secreted factor that is unstable, or required continual production by living bacteria to reach a concentration necessary to exert its effect.
Using another approach to assess this interaction, we carried out a liquid competition assay where NM74_B14 was co-cultured with uninhibitory L. reuteri or inhibitory L. murinus and growth of each species was quantified using quantitative PCR (qPCR). Growth of NM74_B14 was completely inhibited in the presence of L. murinus, and halved relative to axenic growth when co-cultured with L. reuteri (Figure 1c). Notably, growth of L. reuteri was also significantly reduced when NM74_B14 was present, while L. murinus growth was unaffected by co-culturing. This reciprocal effect on growth of NM74_B14 and L. reuteri suggested either nutrient competition or possibly mutualistic behavior between these bacteria. This co-culture assay was not performed with Lachnospiraceae as both isolates from the CIAMIB grow poorly in liquid media.
To address whether inhibition was dependent on cell–cell contact, we conducted growth assays using transwell plates containing an insert with a membrane that is impermeable to bacterial cells but permeable to smaller molecules. In these plates NM74_B14 was cultured either alone, across the membrane from uninhibitory L. reuteri, or across the membrane from inhibitory L. murinus. Given that each species was grown in a separate compartment, turbidity was used to quantify growth. Similar to the qPCR assays, growth of NM74_B14 was decreased by L. reuteri and fully inhibited by L. murinus (Figure 1d). These results demonstrated that inhibition of S24-7 growth by L. murinus does not require cell-to-cell contact and is due to a secreted factor or metabolite.
Inhibitory and uninhibitory Lactobacillaceae species cluster according to phylogeny and metabolic properties
The considerable variation in gene content and low protein sequence homology in the genomes of L. murinus, L. johnsonii, L. intestinalis, and L. reuteri species (Figure S3) made it difficult to infer the cause of the observed growth inhibition from this small set of isolates. To test a larger set of species and catalog the distribution of inhibitory activity within the Lactobacillaceae family, we obtained a larger collection of Lactobacillaceae isolates selected to encompass a broad range of the genera from this family, amounting to 21 strains from 18 species, in addition to the 6 strains and 4 species from the CIAMIB (Table S2).
Spotting assays were repeated with the expanded panel of Lactobacillaceae isolates against NM74_B14 or NM01_1-7b. These assays showed that Limosilactobacillus vaginalis, Limosilactobacillus oris, Limosilactobacillus coleohominis, Levilactobacillus brevis, and Lentilactobacillus parafarraginis were uninhibitory (Table 1; Figure S4a). Conversely Ligilactobacillus ruminis, Lactobacillus gasseri, Lactobacillus crispatus, Lactobacillus jensenii, Lactobacillus iners, Lactobacillus psittaci, Lactobacillus delbrueckii, Lacticaseibacillus rhamnosus, Lactiplantibacillus plantarum, Loigolactobacillus coryniformis, Companilactobacillus farciminis, Companilactobacillus alimentarius, and Schleiferilactobacillus shenzhenensis all showed signs of inhibition (Table 1; Figure S4b). For the S24-7 isolate NM74_B14 the zones of clearance were smaller and difficult to detect for L. jensenii 269–3, L. iners, L. psittaci, and C. farciminis. This suggested that S24-7 species are slightly less susceptible to inhibition than Lachnospiraceae species. Notably, all isolates of the Ligilactobacillus, Lactobacillus, Lacticaseibacillus, Lactiplantibacillus, Loigolactobacillus, Companilactobacillus, and Schleiferilactobacillus genera were inhibitory to some extent. No isolates from the Limosilactobacillus, Levilactobacillus, and Lentilactobacillus genera showed an inhibitory effect on either S24-7 or Lachnospiraceae.
Table 1.
Inhibitory activity of a range of Lactobacillaceae species on growth of S24-7 and Lachnospiraceae representatives. Plus sign (+) indicates inhibition of NM74_B14 (S24-7) and NM01_1-7b (Lachnospiraceae). Minus sign (-) indicates no effect on growth of either isolate
| Genus | Species | Strain | Inhibitory activity |
|---|---|---|---|
| Limosilactobacillus | L. reuteri | NM11 | - |
| NM12 | - | ||
| L. vaginalis | EX336960VC11 | - | |
| L. oris | F0423 | - | |
| L. coleohominis | DSM14060 | - | |
| Levilactobacillus | L. brevis | DSM20054 | - |
| Lentilactobacillus | L. parafarraginis | DSM18390 | - |
| Ligilactobacillus | L. murinus | NM26 | + |
| NM28 | + | ||
| L. ruminis | DSM20403 | + | |
| Lactobacillus | L. johnsonii | NM60 | + |
| L. intestinalis | NM61 | + | |
| L. gasseri | JV-V03 | + | |
| EX336960VC01 | + | ||
| L. crispatus | PSS7772C | + | |
| EX849587VC03 | + | ||
| L. jensenii | 269–3 | + | |
| EX849587VC03 | + | ||
| L. iners | SPIN 2503V10-D | + | |
| L. psittaci | DSM15354 | + | |
| L. delbrueckii | N/A | + | |
| Lacticaseibacillus | L. rhamnosus | LMS2-1 | + |
| Lactiplantibacillus | L. plantarum | DSM20174 | + |
| Loigolactobacillus | L. coryniformis | DSM20001 | + |
| Companilactobacillus | C. farciminis | DSM20184 | + |
| C. alimentarius | DSM20249 | + | |
| Schleiferilactobacillus | S. shenzhenensis | DSM28193 | + |
Strikingly, the inhibitory genera clustered together both phylogenetically and according to their primary mode of fermentation (Figure 2). The inhibitory isolates all belonged to the homofermentative group of lactic acid bacteria that primarily use the Embden-Meyerhof pathway to metabolize hexoses, whereas the uninhibitory isolates were all heterofermentative bacteria that utilize the phosphoketolase pathway. The genomes of most isolates in our Lactobacillaceae collection were available and their putative metabolic capacities were further assessed using the KofamScan gene function annotation tool provided by the Kyoto Encyclopedia of Genes and Genomes (KEGG). This analysis revealed that all inhibitory strains encoded either 6-phosphofructokinase (pfkA, KEGG Orthology Number K00850) or 1-phosphofructokinase (fruK, K00882), while all uninhibitory strains lacked these genes (Table S3). Both phosphofructokinases generate fructose-1,6-bisphosphate, a central metabolic intermediate of the Embden-Meyerhof pathway, and their presence/absence is used to differentiate homofermentative from heterofermentative Lactobacillaceae.10,30 Additionally, two genes from the phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), fruA (K02770, transports fructose), and manX (K02794, transports mannose), were uniformly present in inhibitory strains and absent from uninhibitory strains. This coincides with the observation that heterofermentative Lactobacillaceae species harbor fewer PTS than homofermentative species, which is associated with a general loss of gene families related to carbohydrate transport and metabolism.10 The only KEGG functions identified that were common to uninhibitory strains and absent from all uninhibitory strains were 1,3-propanediol dehydrogenase (dhaT, K00086), which enables homofermentative lactobacilli to utilize glycerol as a hydrogen acceptor during the fermentation of glucose, and an uncharacterized putative glyoxalase (phnB, K04750).31,32
Figure 2.
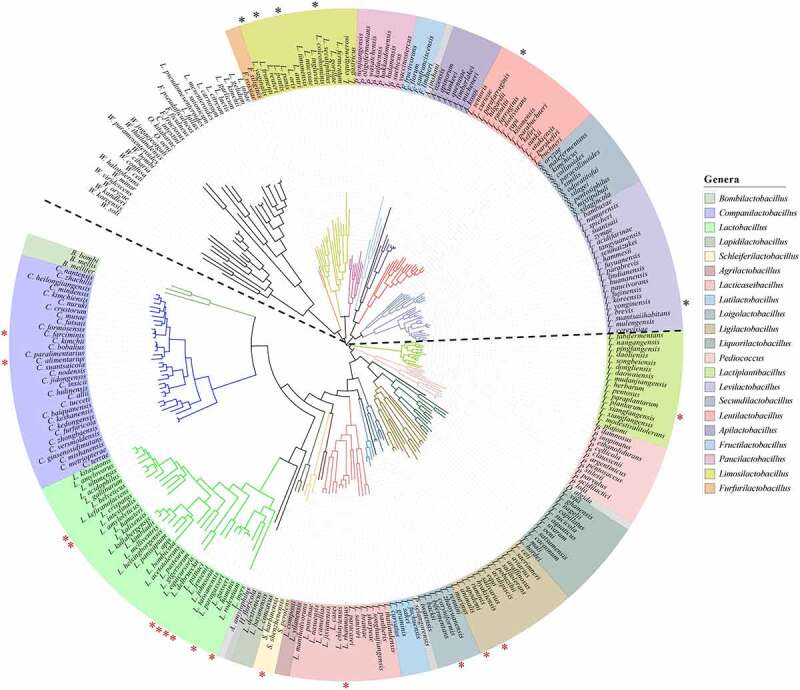
Inhibitory and uninhibitory Lactobacillaceae species cluster according to phylogeny. Phylogenomic analysis and tree constructed based on the concatenated alignment of protein sequences for 114 single-copy genes from type strains, containing 244 species from the Lactobacillaceae family (Zheng et al., 2020). Newly created genera (from what was previously classified as the Lactobacillus genus) are indicated by branch and label colors, and the legend on the right shows the name of each genus. Species labeled in gray are the sole representatives from their genus. Unlabeled species belong to the closely related Leuconostocaceae family (recently amalgamated into the Lactobacillaceae family). Species selected and tested against representatives from S24-7 (NM74_B14) and Lachnospiraceae (NM01_1-7b) are indicated by asterisks. Species that inhibited growth are below the dotted line, indicated by red asterisks. Species that were uninhibitory are above the dotted line, indicated by black asterisks.
Inhibitory Lactobacillaceae produce considerably higher quantities of lactate and pyruvate and lower the pH of the surrounding media
To define the metabolites produced by uninhibitory L. reuteri and L. brevis, and inhibitory L. murinus and L. plantarum, a quantitative compositional analysis of culture supernatants was performed using anion chromatography. Although the levels of several metabolites varied across each species, most did not correlate with the inhibitory and uninhibitory groups. However, the concentrations of lactate and formate were on average 1.5× and 2.0× higher, respectively, in inhibitory L. murinus and L. plantarum supernatants compared to uninhibitory L. reuteri and L. brevis supernatants (Table 2). The concentration of pyruvate was also elevated for inhibitory supernatants, with L. murinus containing 6.8 mg/L and L. plantarum containing 1.6 mg/L compared to 0.4 mg/L for L. reuteri and 0.1 mg/L for L. brevis. The higher concentrations of lactate and pyruvate in L. murinus and L. plantarum supernatant is consistent with their use of homofermentative metabolism, as lactate is the main end-product of this process and pyruvate is a key intermediate metabolite. Increased formate production by L. murinus and L. plantarum is likely due to the presence of formate C-acetyltransferase (pflB) in both genomes, a gene encoding an enzyme which catalyzes the conversion of pyruvate and coenzyme A (CoA) into formate and acetyl-CoA (Table S3). L. reuteri and L. brevis as well as other uninhibitory species lack this gene, however pflB is not found in all inhibitory strains of lactobacilli and therefore formate production is unlikely to be a central player in the inhibition of S24-7 and Lachnospiraceae.
Table 2.
Quantitative chromatographic analysis of anions present in culture supernatant of heterofermentative (inhibitory) and homofermentative (uninhibitory) Lactobacillaceae strains. Values are the average of three independent experiments. Standard deviation is shown for each value. n.d. = not detected
| |
Heterofermentative (uninhibitory) |
Homofermentative (inhibitory) |
||||
|---|---|---|---|---|---|---|
| Anions | Blank media (mg/L) | L. reuteri NM11 (mg/L) | L. brevis (mg/L) | L. murinus NM26 (mg/L) | L. plantarum (mg/L) | |
| Organic | Lactate | 1.113 ± 0.050 | 14.914 ± 0.105 | 14.479 ± 0.176 | 21.387 ± 0.472 | 23.135 ± 0.244 |
| Acetate | 2.661 ± 0.180 | 5.039 ± 0.051 | 5.726 ± 0.255 | 2.972 ± 0.276 | 5.973 ± 0.074 | |
| Propionate | 0.018 ± 0.015 | n.d. | n.d. | n.d. | n.d. | |
| Formate | 1.206 ± 0.077 | 1.110 ± 0.007 | 1.132 ± 0.014 | 2.124 ± 0.076 | 2.337 ± 0.031 | |
| Butyrate | 0.016 ± 0.003 | 0.072 ± 0.001 | 0.098 ± 0.001 | 0.042 ± 0.001 | 0.090 ± 0.015 | |
| Pyruvate | 9.362 ± 0.609 | 0.405 ± 0.048 | 0.118 ± 0.001 | 6.780 ± 0.224 | 1.595 ± 0.123 | |
| Succinate/Malate | 0.454 ± 0.031 | 0.427 ± 0.010 | 0.442 ± 0.003 | 0.442 ± 0.010 | 1.095 ± 0.013 | |
| Fumarate | 0.612 ± 0.081 | 0.444 ± 0.006 | 0.462 ± 0.014 | 0.699 ± 0.027 | 0.489 ± 0.076 | |
| Citrate | 0.812 ± 0.090 | 0.804 ± 0.004 | 0.809 ± 0.007 | 0.785 ± 0.022 | 0.023 ± 0.006 | |
| Inorganic | Chloride | 103.504 ± 6.783 | 98.583 ± 0.401 | 99.011 ± 0.972 | 99.002 ± 2.222 | 99.219 ± 1.254 |
| Nitrite | 0.002 ± 0.003 | n.d. | n.d. | n.d. | n.d. | |
| Sulfate | 0.075 ± 0.004 | 0.141 ± 0.004 | 0.079 ± 0.015 | 0.082 ± 0.004 | 0.115 ± 0.003 | |
| Nitrate | 0.825 ± 0.058 | 0.812 ± 0.005 | 0.796 ± 0.008 | 0.801 ± 0.020 | 0.820 ± 0.019 | |
| Phosphate | 16.718 ± 1.076 | 15.718 ± 0.033 | 15.794 ± 0.201 | 15.598 ± 0.375 | 15.284 ± 0.130 | |
An increase in acid production by homofermentative species might be expected to impact the pH of the culture media to a greater extent than for heterofermentative species. To determine whether homofermentative Lactobacillaceae have a pronounced effect on local pH, we assessed the effect of each strain on agar plates using the pH indicator bromocresol purple. Although all heterofermentative (uninhibitory) strains grew robustly on these plates, each one only produced a minimal shift in the pH of the surrounding media (Figure 3a). Conversely, all of the homofermentative (inhibitory) strains caused extensive acidification of the agar to below the pKa of 6.3 of bromocresol purple (Figure 3b). This strong effect on pH was notable even for the isolates that grew poorly on this media, such as L. gasseri JV-V03, L. coryniformis, and S. shenzhenensis. This demonstrated a clear correlation between media acidification and the ability of specific lactobacilli to inhibit the growth of S24-7 and Lachnospiraceae isolates.
Figure 3.
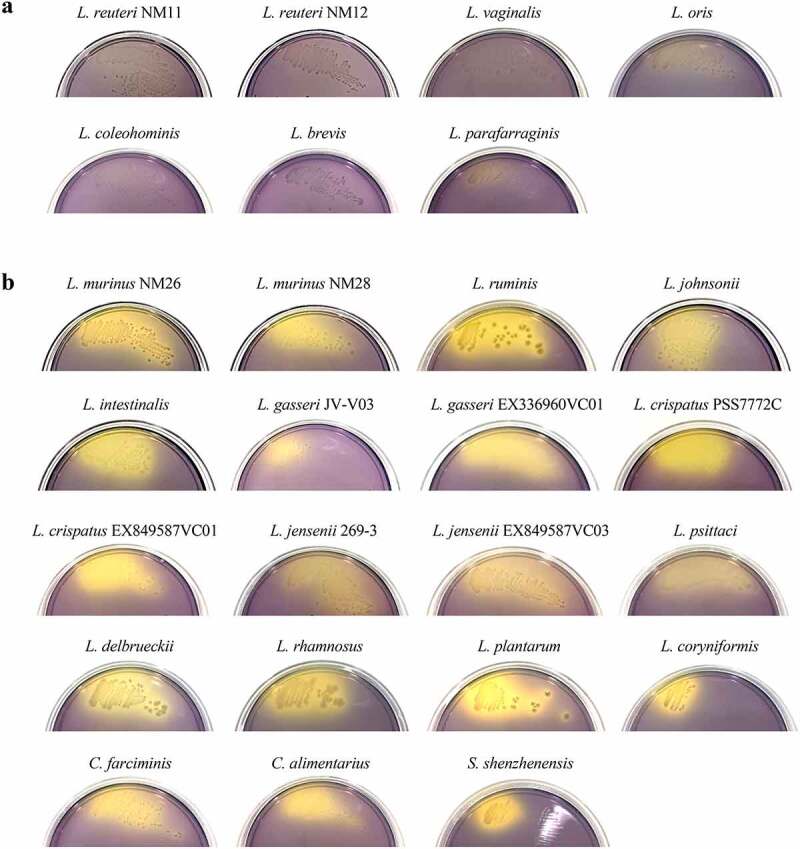
Inhibitory Lactobacillaceae species produce more acid than uninhibitory species. Each Lactobacillaceae strain was streaked out onto plates containing the pH indicator bromocresol purple (purple above pH 6.8, yellow below pH 5.2). Representative images are shown (n = 3). A) Strains that showed no signs of inhibition of NM74_B14 (S24-7) or NM01_1-7b (Lachnospiraceae). B) Strains that inhibited growth of both NM74_B14 and NM01_1-7b. L. iners was excluded as it did not grow on these plates.
Mitigating acidity alleviates the inhibitory effect of Lactobacillaceae species
To test if acidification of the surrounding media is the underlying cause of growth inhibition, L. reuteri (uninhibitory) and L. murinus (inhibitory) were spotted onto lawns of NM74_B14 or NM01_1-7b, either on standard media or media supplemented with MOPS buffer adjusted to pH 7. MOPS buffer had no effect on the growth of either NM74_B14 or NM01_1-7b when grown with L. reuteri, while the zones of growth inhibition for both isolates when grown with L. murinus were markedly reduced when the agar was supplemented with MOPS (Figure 4a).
Figure 4.
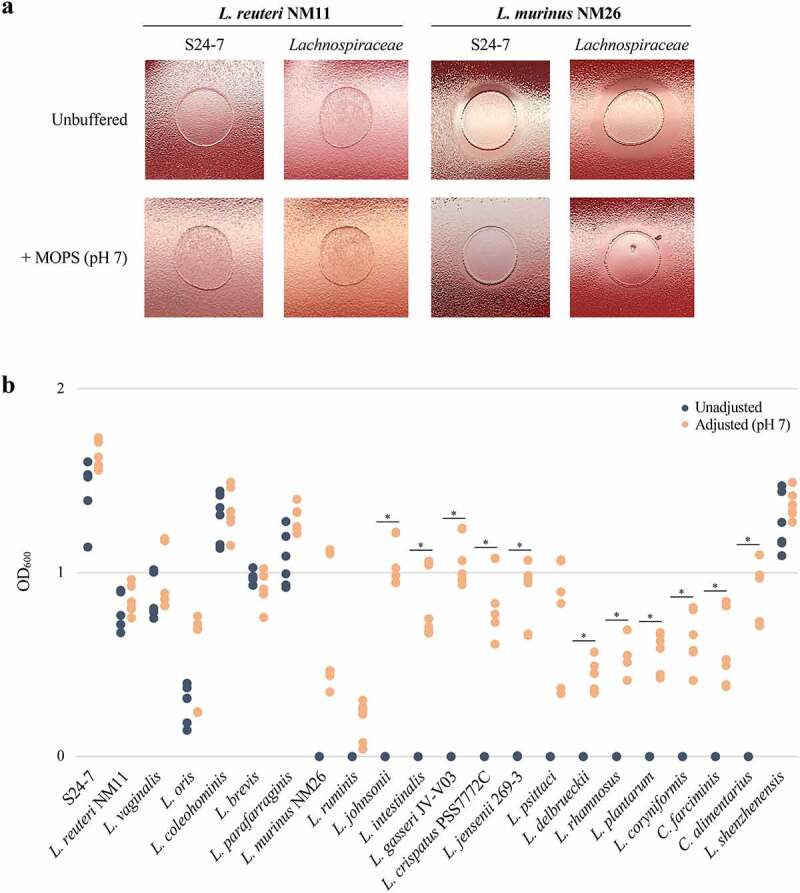
Mitigating acidity alleviates the inhibitory effect of Lactobacillaceae species against both S24-7 and Lachnospiraceae. A) Uninhibitory L. reuteri NM11 and inhibitory L. murinus NM26 spotted onto lawns of NM74_B14 (S24-7) and NM01_1-7b (Lachnospiraceae). The top row of plates is unbuffered, while the bottom row of plates are supplemented with MOPS buffer at pH 7. Representative images are shown (n = 3). B) Growth measured by OD600 of NM74_B14 in supernatant from liquid cultures of uninhibitory and inhibitory Lactobacillaceae species. For each Lactobacillaceae species, blue data points show growth of NM74_B14 in unadjusted supernatant while orange data points show growth in supernatant adjusted to pH 7 using NaOH (as indicated in the legend on the upper right). L. iners was excluded as it did not grow in this liquid media. ‘S24-7’ is a control of NM74_B14 grown in its own supernatant. Data points are from three independent experiments. Welch’s t-test, * = p < .05.
To examine the influence of pH more extensively across our entire collection of Lactobacillaceae species, we selected one isolate of each species to test the effects of their respective culture supernatants on the growth of the S24-7 strain NM74_B14 (with the exception of L. iners, which was unable to grow under these conditions). Supernatants were taken from cultures of each Lactobacillaceae species, either adjusted to pH 7 or left unadjusted, and subsequently inoculated with NM74_B14 to evaluate growth. Both turbidity and pH were measured for each Lactobacillaceae culture before taking supernatant, and turbidity measurements varied dramatically between Lactobacillaceae species independent of inhibitory capacity (Table S4). The pH of uninhibitory cultures ranged from pH 6.3–6.7 compared to pH 5.2–6.3 for inhibitory cultures, thereby corroborating the observations from the bromocresol purple assay. In the supernatants of uninhibitory Lactobacillaceae species NM74_B14 grew with and without pH adjustment, while for inhibitory species it only grew when the pH was adjusted to 7.0 (Figure 4b). The exception was S. shenzhenensis (inhibitory), which grew poorly in the liquid media we used and maintained a high culture pH relative to other inhibitory isolates. NM74_B14 grew in both unadjusted and adjusted S. shenzhenensis supernatants.
Due to the extremely poor growth of Lachnospiraceae isolates in liquid media, this assay could not be performed with NM01_1-7b. Instead, spotting assays were carried out on agar plates using MOPS to help buffer pH. In the presence of heterofermentative (uninhibitory) Lactobacillaceae, growth of NM01_1-7b was robust on both MOPS-supplemented and unbuffered media. In contrast, the zones of growth inhibition surrounding culture spots of homofermentative (inhibitory) Lactobacillaceae were substantially reduced on plates supplemented with MOPS buffer (Figure S5). Together, these findings indicate that media acidification is required for the inhibitory activity of homofermentative Lactobacillaceae against S24-7 and Lachnospiraceae.
S24s-7 and Lachnospiraceae species are more acid-sensitive than related species within the Bacteroidales and Clostridiales orders
Our initial screen examining the effect of lactobacilli on the growth of other members of the CIAMIB collection suggested that acid sensitivity might be characteristic of S24-7 and Lachnospiraceae, but not other Bacteroidales or Clostridiales bacteria. To investigate this, we selected species from the CIAMIB that belong to the Bacteroidales and Clostridiales orders most closely related to S24-7 and Lachnospiraceae species, respectively (Figure S6). Phocaeicola sartorii, Parabacteroides distasonis, Bacteroides faecichinchillae, and Bacteroides caecimuris were chosen as representatives from the Bacteroidaceae family within the Bacteroidales order, while Clostridium perfringens, Clostridium sartagoforme, and Clostridium chromiireducens were selected from the Clostridiaceae family within the Clostridiales order (Table S1).
The growth of S24-7 isolates M. intestinale, NM65_B17, NM74_B14, and NM86_A22 was compared against the four Bacteroidaceae species in liquid media adjusted to several pH levels (Figure 5). Despite variation in the extent of growth, at pH 7 both S24-7 and Bacteroidaceae species grew robustly. However, each decrease in pH considerably reduced growth of S24-7 species, and by pH 5.5 none showed any growth. Contrarily, P. sartorii, B. faecichinchillae, and B. caecimuris continued to grow even at pH 5.5. P. distasonis stopped growing at pH 6.0, but unlike S24-7 species its growth was not significantly reduced at pH 6.5 compared to pH 7.0. This increased sensitivity of P. distasonis at lower pH levels compared to other Bacteroidaceae species may reflect the phylogeny of these strains, as P. distasonis clusters more closely with the S24-7 species than the Bacteroidaceae species (Figure S6).
Figure 5.
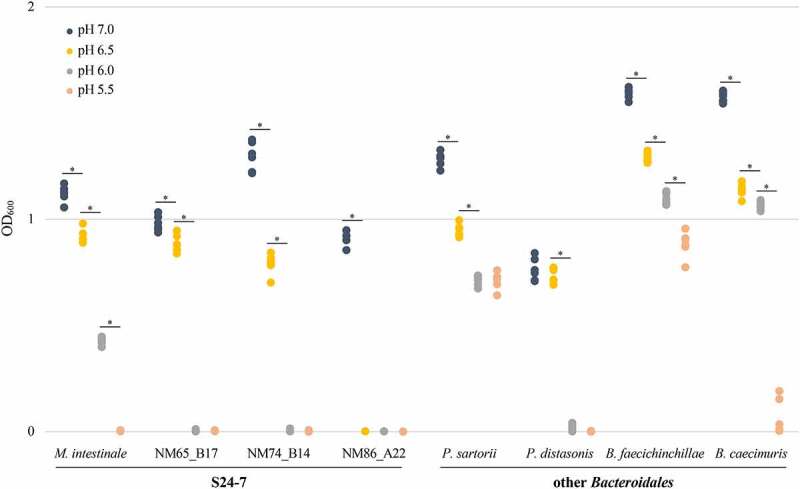
S24-7 species are more sensitive to acidity than other species within the Bacteroidales order. The four S24-7 isolates from the CIAMIB and four related species from within the Bacteroidales order were cultured in liquid media at pH 7.0, 6.5, 6.0, and 5.5, as indicated by the legend on the upper left. For each species at each pH, OD600 was measured 72 hours after inoculation as a proxy for growth. Data points are from three independent experiments. Welch’s t-test, * = p < .05.
The growth of Lachnospiraceae isolates NM01_1-7b and NM72_1-8 was compared against the three Clostridiaceae species by spotting onto plates adjusted to several pH levels (Figure 6). On plates where pH had not been adjusted (pH 7.4), both Lachnospiraceae and Clostridiaceae species grew robustly. On plates adjusted to pH 6.4 and 5.4, growth of both Lachnospiraceae isolates was completely inhibited whereas all Clostridiaceae species continued to grow well. For these assays we employed agar containing red blood cells and noticed that lysis of these cells was extensive on plates adjusted to pH 5.4. To control for the possibility that blood cell lysis released compounds that inhibit Lachnospiraceae independent of pH we examined the growth of Lachnospiraceae on chocolate agar plates, which are formulated using lysed red blood cells. All species grew on chocolate agar plates at neutral pH. Collectively, these solid and liquid growth assays established that species belonging to the S24-7 group and the Lachnospiraceae family are highly susceptible to acidic conditions, a feature they do not share with related taxa abundant in the gut microbiota.
Figure 6.
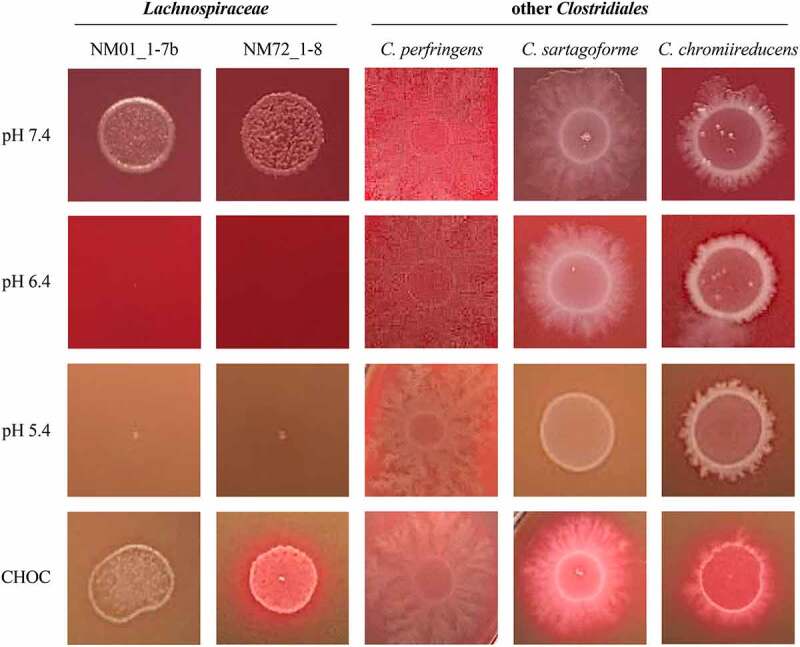
Lachnospiraceae species are more sensitive to acidity than other species within the Clostridiales order. The two Lachnospiraceae isolates from the CIAMIB and three related species from within the Clostridiales order were spotted onto agar plates adjusted to pH 7.4, 6.4, and 5.4, as well as chocolate agar (CHOC) plates. Representative images are shown (n = 3).
Discussion
Here, we report the inhibition of representatives from the prevalent Lachnospiraceae family and S24-7 group by a diverse set of Lactobacillaceae species. We find that this inhibitory activity is mediated by increased acidity and that it is congruent with the current phylogenetic and metabolic categorizations of the Lactobacillaceae family. This represents one of the first in-depth explorations into the effect of a broad range of Lactobacillaceae species on distinct taxa from the commensal gut microbiota. It is also the first report, to our knowledge, that Lachnospiraceae and S24-7 bacteria are highly sensitive to acid stress.
The benefits that Lactobacillaceae species have on human and animal health are frequently emphasized. However, many studies regarding the probiotic properties of lactobacilli do not address the physiological or ecological characteristics of the species or strain being studied and rarely is the impact these bacteria might have on the larger gut microbial ecosystem considered. The recent re-classification of fermentation types and division of this genus into multiple genera based on phylogeny, physiology, and ecology is a major step toward elucidating the hallmarks of each clade within the Lactobacillaceae family.6,10 These updated designations allowed us to determine that only homofermentative Lactobacillaceae species exert an inhibitory effect on Lachnospiraceae and S24-7 species.
Organic acid production and its effect on local pH levels has not been systematically explored as it relates to homofermentative and heterofermentative lactobacilli, nor have the ecological consequences of these differences. Prior work has demonstrated that homofermentative species produce high levels of lactic acid while heterofermentative species produce a mix of lactic acid and acetic acid.33–36 We found that homofermentative strains generated almost half again as much lactate as heterofermentative strains. Formate production was also considerably higher in homofermentative strains, although this is likely due to the presence of formate C-acetyltransferase in the genomes of both the L. murinus and L. plantarum isolates we selected to study. Conversely, acetate levels were comparable across groups.
The increased acidity that we observed in the media of homofermentative species may be a direct consequence of producing more organic acid, but the differences in total acid production were not as extreme as the differences in culture pH would indicate. It is possible that heterofermentative species buffer their surrounding environment through the production of ammonia or proton consumption via enzymes such as urease, arginine deiminase, and glutamate decarboxylase.37–41 However, the simplistic explanation that heterofermentative species actively neutralize their surrounding environment while homofermentative strains do not is complicated by the fact that many homofermentative species also encode deaminase and decarboxylase enzymes in their genomes.
The capacity of organic acids to inhibit bacterial growth is well established.14,18,19,42 One question is whether pH is the direct cause of growth inhibition as opposed to a specific organic acid such as lactate, but our experiments suggest that low pH is sufficient to inhibit Lachnospiraceae and S24-7 growth. We note that S24-7 was able to grow in Lactobacillaceae supernatants adjusted to a neutral pH, hence neutral lactate and acetate were not inhibitory. Additionally, neither S24-7 nor Lachnospiraceae were able to grow on media where the pH was lowered using inorganic acid – although the media was complex and may have contained low amounts of lactate and other acid salts.
The Lachnospiraceae and S24-7 species are poorly studied, largely due to a paucity of cultured isolates, and their sensitivity to stress has not been extensively examined. The apparent acid sensitivity of both taxa relative to other species from the Bacteroidales and Clostridiales orders warrants further investigation. The S24-7 group of bacteria has been shown to be highly intolerant to hyperosmotic conditions.43 Indeed, osmotic-induced diarrhea caused by the administration of polyethylene glycol (as routinely occurs prior to colonoscopy procedures) leads to an extinction of these bacteria from the gut. An inability to survive acid stress could render both S24-7 and Lachnospiraceae susceptible to elimination from the gut under circumstances that lower the pH of the colon including chronic inflammation or certain drugs, food additives, or xenobiotics.
It remains an open question whether interactions between lactobacilli and S24-7 or Lachnospiraceae occur in nature and are relevant in the context of the gut. Lachnospiraceae and S24-7 species are found in the colon (and cecum in mice) where the pH is typically close to neutral. Lactobacillaceae species, in contrast, are usually more abundant in the small intestine and stomach, where the pH is considerably more acidic.44–47 The host and other bacteria also modulate the composition and presence of organic acids through various mechanisms. A number of commensal gut bacteria from the Firmicutes phylum are able to convert lactate and acetate to short-chain fatty acids, particularly butyrate.48–50 In turn, the intestinal mucosa of the host can absorb lactate and short-chain fatty acids and control the pH of small intestinal contents through secretion of bicarbonate.51–54
There are many scenarios where inhibitory activity of specific lactobacilli on the gut microbiota could be important, including during the administration of lactobacilli as probiotics. Lactobacillaceae strains such as L. rhamnosus GG have been shown to at least transiently colonize the human colonic mucosa.55,56 Furthermore, the prevalence of both Lachnospiraceae and S24-7 species has been altered in numerous studies examining the effect of Lactobacillaceae species in food and probiotics on the gut microbiota.22,57–63 The effect that lactobacilli have on commensal bacteria like S24-7 and Lachnospiraceae may explain why administering some probiotics delays the reestablishment of the endogenous microbiota after antibiotic treatment. Regardless, this work highlights both the sensitivity of two abundant members of the gut microbiota to stress and the need to better characterize the effects that different species (and strains) of probiotic bacteria can have on the gut microbiota as a whole.
Methods
Bacterial growth media and conditions
All bacterial growth was carried out at 37°C in an anaerobic chamber (Anaerobe Systems, AS-580) using a gas mixture of 10% hydrogen, 10% carbon dioxide, and 80% nitrogen. Culture media was always transferred to anaerobic conditions at least 24 hours before use. Brucella Blood Agar (BRU) supplemented with hemin and vitamin K (Hardy Diagnostics, A30) was used to grow bacterial strains on solid media unless otherwise specified. For all assays, bacteria were streaked out onto BRU plates from stocks stored at −80°C in 10% glycerol and grown for 48–72 hours, then re-streaked onto fresh BRU plates and grown for another 48–72 hours. Brain Heart Infusion (BHI) Broth (Oxoid, CM1135) supplemented with 5 μg/mL hemin (Sigma, 51280), 1 μg/mL vitamin K1 (Sigma, 95271), and 0.5 mg/mL L-cysteine HCl after autoclaving was used to grow bacterial strains in liquid media. For liquid assays not requiring co-culturing or subsequent growth of NM74_B14 or NM01_1-7b, Lactobacillaceae species were cultured in de Man, Rogosa, and Sharpe (MRS) Broth (Sigma-Aldrich, 69966).
Spotting assays
For initial spotting assays using Lactobacillaceae species from the CIAMIB, S24-7 and Lachnospiraceae species were resuspended directly from BRU plates in BHI. S24-7 species were diluted to an OD600 of 0.1 and Lachnospiraceae species were diluted to an OD600 of 0.6 (due to their poor growth). For all subsequent spotting assays, a 1 μL inoculation loop full of NM74_B14 or NM01_1-7b from BRU plates was added to 4 mL of BHI to propagate enough bacteria to enable spotting of all Lactobacillaceae species. Cultures were incubated for 24 hours then diluted to an OD600 of 0.1 or 0.6, respectively. From this point onward, all spotting assays followed the same procedure. To create bacterial lawns, 600 μL of each dilution was spread onto individual BRU plates. Plates were tilted to evenly distribute bacteria, as using a spreader produced uneven swathes of S24-7 and Lachnospiraceae growth. Each Lactobacillaceae species was resuspended in 1X phosphate-buffered saline (PBS) containing 0.1% L-cysteine HCl and diluted to an OD600 of 1. After S24-7 or Lachnospiraceae lawns had dried, 5 μL of each Lactobacillaceae strain was spotted on top. Plates were incubated for 72 hours and imaged.
To test the effect of Lactobacillaceae supernatant and heat-killed bacteria compared to live bacteria, liquid cultures of NM74_B14 and NM01_1-7b (as described above) were grown for 24 hours, diluted to an OD600 of 1 and filtered using a 0.22 μM filter (to obtain supernatant), boiled for 30 minutes (to heat-kill bacteria), or left unmodified (live bacteria). For each Lactobacillaceae species tested, separate tubes containing 3.6 mL of MRS were supplemented with 400 μL of supernatant, heat-killed, or live NM74_B14 or NM01_1-7b. Each of these mixtures was inoculated with individual Lactobacillaceae species from BRU plates diluted as described above, alongside a set of tubes containing 4 mL MRS. After incubating for 24 hours, supernatant and heat-killed bacteria were obtained from each of these cultures by filtering and boiling. These combinations, as well as live bacteria, were then spotted onto lawns of NM74_B14 or NM01_1-7b in the same manner as described for previous spotting assays. Plates were incubated for 72 hours and imaged.
For buffered media assays, 1 M MOPS was prepared by dissolving 10.46 g MOPS (BioShop, MOP001) in distilled water (dH2O), adjusting to pH 7 with 10 N NaOH, filling to 50 mL, and filter-sterilizing. 200 μL of this 1 M MOPS buffered at pH 7 was spread onto BRU plates and allowed to dry completely before proceeding with spreading lawns of NM74_B14 and NM01_1-7b and spotting as described previously. Plates were incubated for 72 hours and imaged.
Co-culturing assay
NM74_B14 and Lactobacillaceae strains L. reuteri NM11 and L. murinus NM26 were each resuspended in BHI and diluted to an OD600 of 4. For axenic cultures, 5 μL of diluted NM74_B14, L. reuteri NM11, or L. murinus NM26 was added to 4 mL of BHI. For co-cultures, 5 μL of L. murinus NM11 or L. murinus NM26 was added along with 5 μL of NM74_B14 to 4 mL of BHI. Cultures were incubated for 72 hours, at which point OD600 was measured.
For quantitative PCR (qPCR), individual reactions were set up with 10 μL SsoFast EvaGreen Supermix (Bio-Rad, 1725203), 1 μL each of 10 μM forward and reverse primers, and 8 μL template DNA diluted 10X in nuclease-free water, to a total reaction volume of 20 μL. Primers for L. reuteri NM11 (forward: 5’-GGACTACCAGGGTATCTAA-3’; reverse: 5’-TCTCAACACCCGCCTTAATC-3’), L. murinus NM26 (forward: 5’-CCACATGCTAGTGAGCGTATC-3’; reverse: 5’-GTCCAGTTTCTTCTCGCTTCT-3’), and NM74_B14 (forward: 5’-GTGGAAACGAGAAGACTGTAGAA-3’; reverse: 5’-TTTCGTCTCTCAATCGGGAATAG-3’) were designed for this study. These primer sets targeted genes unique to each species (hypothetical proteins for L. reuteri and L. murinus, and a FMN adenylyltransferase/riboflavin kinase for NM74_B14) to ensure specificity, which was checked using PrimerBlast and confirmed experimentally. qPCR was carried out on an Eppendorf Mastercycler ep realplex in a 96-well format. Cycling conditions were 30 seconds at 95°C, 40 cycles of 5 seconds at 95°C and 10 seconds at 60°C, 15 seconds at 95°C followed by 15 seconds at 60°C, and a 20-minute ramp up to 95°C for 15 seconds. Five-point standard curves of 10X dilutions (20 ng to 0.002 ng or 10 ng to 0.001 ng) were set up in duplicate, while each sample was run in triplicate. Analysis of qPCR efficiency and accuracy was carried out using Eppendorf realplex software.
Transwell plate assay
NM74_B14 and Lactobacillaceae strains L. reuteri NM11 and L. murinus NM26 were resuspended from BRU plates in BHI. NM74_B14 was diluted to an OD600 of 0.6 and Lactobacillaceae strains were diluted to an OD600 of 0.1. 600 μL of BHI was aliquoted into the bottom compartment of the transwell plate (VWR, 10769–198) and 100 μL was aliquoted into the insert, which contains a 0.1 μM filter. Next, 5 μL of NM74_B14 was added to the bottom compartment and 5 μL of L. reuteri NM11 and L. murinus NM26 was added to separate inserts. Cultures were incubated for 72 hours, at which point OD600 was measured.
Phylogenetic tree construction
For the Lactobacillaceae family, phylogenomic analysis and tree were generated as described in Zheng et al., based on the concatenated alignment of protein sequences for 114 single-copy genes from type strains of all available Lactobacillaceae and Leuconostocaceae species retrieved August 2019 from GenBank.6 Visualization was performed with the Interactive Tree of Life.64 For clarity, both branches and bacterial names were highlighted based on the new Lactobacillaceae genera assignments, and a corresponding legend was added to allow easy referencing of updated groupings and genus names.
Phylogenetic trees of S24-7 and Bacteroidaceae species, as well as Lachnospiraceae and Clostridiaceae species, were constructed using 16S rRNA gene sequences. The full length (>1400 nucleotides) sequences of the 16S rRNA genes for each species were aligned with MUSCLE in MEGA7 using a 97% cutoff value.65,66 Psychroflexus gondwanensis (16S rRNA gene accession: JX986967.1) was used as an outgroup for the Bacteroidales tree. NM09_H32 from the CIAMIB (16S rRNA gene accession: MK929057.1) was used as an outgroup for the Clostridiales tree. This alignment was used to construct phylogenetic trees with the Neighbor-Joining method in MEGA7, with a bootstrap replication of 1000.66,67 Modifications to annotations were carried out in Inkscape 1.1.
Genomic analyses
Genomic comparisons between Lactobacillaceae species from the CIAMIB were conducted on the Pathosystems Resource Integration Center (PATRIC) through their Bacterial Bioinformatics Resource Center, using the genome assemblies for L. reuteri NM11, L. murinus NM26, L. johnsonii, and L. intestinalis (provided in Table S1).68 Bidirectional BLASTP was used to perform protein sequence-based genome comparisons in a pairwise manner between each species. The following default parameters were used: 30% minimum coverage of query and subject in BLAST; maximum E value of 1e-5; and 10% minimum identity of query and subject in BLAST.
For comparing KEGG Orthologs (KOs) between Lactobacillaceae species, coding sequences from the genome assemblies of all strains were downloaded from the National Center for Biotechnology Information (NCBI) RefSeq database in FASTA Nucleotide format. The Bio.SeqIO package from Biopython was used to translate each coding sequence to generate protein FASTA files for each genome.69 KofamScan version 1.3.0 was used to search each translated coding sequence against the KEGG database of Hidden Markov Models (HMMs), which represents a broad range of defined protein families.70 Significant hits were identified as those scoring above HMM-specific thresholds, and the KO identifier for the corresponding family was assigned to each translated coding sequence. All Lactobacillaceae strains in our collection were included in this analysis, with the exception of L. vaginalis, L. gasseri EX336960VC01, L. crispatus EX849587VC01, L. jensenii EX849587VC03, and L. delbrueckii as sequences for these strains were not available.
Anion chromatography
Each Lactobacillaceae strain was resuspended from BRU plates in BHI and diluted to an OD600 of 4, then 5 μL of this resuspension was transferred to 4 mL of BHI. Cultures were incubated for 24 hours, at which point supernatants were obtained for each strain by filtering cultures through 0.22 μM filters. Samples were frozen at −80°C until metabolic profiling was conducted. After thawing in preparation for chromatography, samples were diluted 20× and standards for anions of interest (acetate, chloride, citrate, citrate, formate, fumarate, lactate, nitrate, phosphate, pyruvate/oxaloacetate, and succinate/malate) were run at the following concentrations to generate standard curves: 0.5 mM, 0.2 mM, 0.05 mM, 0.01 mM, and 0.005 mM. Anion chromatography was carried out by the BioZone facility at the University of Toronto on a Thermo Scientific Dionex ICS-2100 ion chromatography system using an IonPac AS11 IC Column. For both standards and samples, the injection volume onto the column was 20 μL and the run time was 45 minutes. The flow rate was set to 1.0 mL/minute and the suppressor was set at 30 mM maximum [OH−] with a current of 75 mA. The eluent was a multi-step gradient of KOH that started at 0.5 mM. Data analysis was conducted on the Chromeleon Chromatography Data System software.
pH indicator plates
Indicator plates were created by supplementing 1 L worth of BHI with 15 g of agar and 0.02 g of bromocresol purple before autoclaving. After autoclaving, the agar was supplemented with 5 μg/mL hemin, 1 μg/mL vitamin K1, and 0.5 mg/mL L-cysteine HCl. Lactobacillaceae strains grown on BRU plates were resuspended in 1X PBS and diluted to an OD600 of 1. A 1 μL inoculation loop full of each strain was streaked out onto bromocresol purple agar plates. Plates were incubated for 72 hours, at which point images were taken.
Supernatant assay
Lactobacillaceae strains and NM74_B14 were individually resuspended from BRU plates in BHI and diluted to an OD600 of 4, then 13.75 μL of this resuspension was transferred to 11 mL of BHI. Cultures were incubated for 24 hours, then filtered with 0.22 μM filters to obtain 10 mL of supernatant. To adjust pH, a 2 mL aliquot of each supernatant was measured and adjusted to pH 7 using 10 N NaOH. The remaining 8 mL of supernatant was split into four 2 mL aliquots, and two of these aliquots were adjusted to pH 7 using the same quantity of 10 N NaOH (without measuring pH, to maintain sterility). Next, NM74_B14 was resuspended from BRU plates into BHI and diluted to an OD600 of 2. 5 μL of this dilution was added to each Lactobacillaceae supernatant and the NM74_B14 supernatant as a control. Cultures were incubated for 72 hours, at which point OD600 was measured.
Testing acid sensitivity
For liquid assays with S24-7 and other Bacteroidales species, BHI was adjusted to pH 7.0, 6.5, 6.0, and 5.5 using 6 N HCl. After adjusting pH, media was filter-sterilized through a 0.22 μM filter and added in 2 mL aliquots to individual wells in 24-well plates. Each Bacteroidales species was resuspended in BHI from BRU plates and diluted to an OD600 of 2, then 5 μL of each species was added to aliquots of media adjusted to the four different pH levels. Cultures were incubated for 72 hours, at which point OD600 was measured.
For plate assays with Lachnospiraceae and other Clostridiales species, customized plates were created using Brucella Broth (BD, B11088) with 15 g of agar per 1 L of dH2O. After autoclaving, media was supplemented with 50 mL of sheep blood (Cedarlane, CL2581-100D). At this point, media was left either unmodified (pH 7.4), or adjusted to pH 6.4 or 5.4 by adding 6 N HCl. To make chocolate agar (CHOC), sheep blood was warmed in a water bath to 55°C for two hours before adding to media. Cultures of Clostridiales species were set up by taking a 1 μL inoculation loop full of each species and inoculating 4 mL of BHI. After incubating for 24 hours, cultures were diluted to an OD600 of 0.6 and 10 μL of each species was spotted onto pH-adjusted and CHOC plates. Plates were incubated for 72 hours, at which point images were taken.
Supplementary Material
Acknowledgments
We thank Dr. Jinshui Zheng (Huazhong Agricultural University, China) for the use of the Lactobacillaceae phylogenetic tree from Zheng et al., 2020. We thank Line Lomheim from the laboratory of Dr. Elizabeth Edwards (University of Toronto, Canada) for performing and analyzing the anion chromatography data. We thank Dr. Dana Philpott (University of Toronto, Canada) for the use of their anaerobic chamber and their L. delbrueckii isolate. We also thank Jordan Lin for his feedback on the manuscript. This work was supported by a Team Grant from the Canadian Institutes of Health Research (#144628) and a Weston Family Microbiome Initiative Grant from the W. Garfield Weston Foundation.
Funding Statement
This work was supported by the Amgen Foundation [Scholars Program]; Canadian Institutes of Health Research [144628]; W. Garfield Weston Foundation.
Authors’ contributions
EJEB and WWN conceived the study. EJEB, DC, and WWN designed the study and interpreted data. EJEB and DC carried out experimental work. EOYW generated phylogenetic trees and cultivated/sequenced the CIAMIB isolates used in this study. DK and WWN performed the computational analyses. EJEB wrote the manuscript. All authors read, edited, and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data Availability
The data that support the findings of this study are available from the corresponding author (WWN) upon reasonable request. Genomic sequence data from previously unpublished strains that appear in this manuscript can be obtained from NCBI Bioproject #474907. Strains used in this study are available from WWN.
Supplementary material
Supplemental data for this article can be accessed here.
References
- 1.Bermúdez-Humarán LG, Aubry C, Motta JP, Deraison C, Steidler L, Vergnolle N, Chatel JM, Langella P.. Engineering lactococci and lactobacilli for human health. Curr Opin Microbiol. 2013;16(3):278–19. doi: 10.1016/j.mib.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Bourdichon F, Casaregola S, Farrokh C, Frisvad JC, Gerds ML, Hammes WP, Harnett J, Huys G, Laulund S, Ouwehand A, et al. Food fermentations microorganisms with technological beneficial use. Int J Food Microbiol. 2012;154(3):87–97. doi: 10.1016/j.ijfoodmicro.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 3.Saxelin M. Probiotic formulations and applications, the current probiotics market, and changes in the marketplace: a European perspective. Clin Infect Dis. 2008;46(Suppl 2):S76–79. doi: 10.1086/523337. [DOI] [PubMed] [Google Scholar]
- 4.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document: the International scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 5.Kandler O. Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek. 1983;49(3):209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- 6.Zheng J, Wittouck S, Salvetti E, Franz CMAP, Harris HMB, Mattarelli P, O’Toole PW, Pot B, Vandamme P, Walter J, et al. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst and Evol Microbiol. 2020;70(4):2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 7.Sun Z, Harris HMB, McCann A, Guo C, Argimón S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun. 2015;6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salvetti E, Torriani S, Felis GE. The Genus Lactobacillus: a taxonomic update. Probiotics Antimicrob Proteins. 2012;4(4):217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- 9.Gänzle MG. Lactic metabolism revisited: metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr Opin Food Sci. 2015;2:106–117. doi: 10.1016/j.cofs.2015.03.001. [DOI] [Google Scholar]
- 10.Zheng J, Ruan L, Sun M, Gänzle M, Björkroth J. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol. 2015;81(20):7233–7243. doi: 10.1128/AEM.02116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev. 2017;41(Supp 1):S27–48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 12.Thomas CM, Versalovic J. Probiotics-host communication: modulation of signaling pathways in the intestine. Gut Microbes. 2010;1(30):148–163. doi: 10.4161/gmic.1.3.11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgado S, Sánchez B, Margolles A, Ruas-Madiedo P, Ruiz L. Molecules produced by probiotics and intestinal microorganisms with immunomodulatory activity. Nutrients. 2020;12(2):391. doi: 10.3390/nu12020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebeer S, Vanderleyden J, De Keersmaecker SCJ. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 2008;72(4):728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corr SC, Li Y, Riedel CU, Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A. 2007;104(18):7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CC, Lai CC, Huang HL, Huang WY, Toh HS, Weng TC, Chuang YC, Lu YC, Tang HJ. Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front Microbiol. 2019;10:789. doi: 10.3389/fmicb.2019.00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe. 2008;14(3):166–171. doi: 10.1016/j.anaerobe.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28(4):405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Turpin W, Humblot C, Thomas M, Guyot JP. Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int J Food Microbiol. 2010;143(3):87–102. doi: 10.1016/j.ijfoodmicro.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 20.Li J, Wang W, Xu SX, Magarvey NA, McCormick JK. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc Natl Acad Sci U S A. 2011;108(8):3360–3365. doi: 10.1073/pnas.1017431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korpela K, Salonen A, Vepsäläinen O, Suomalainen M, Kolmeder C, Varjosalo M, Miettinen S, Kukkonen K, Savilahti E, Kuitunen M, et al. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome. 2018;6(1):182. doi: 10.1186/s40168-018-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, Zur M, Regev-Lehavi D, Brik RBZ, Federici S, et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174(6):1406–1423. doi: 10.1016/j.cell.2018.08.047. [DOI] [PubMed] [Google Scholar]
- 23.Salzman NH, de Jong H, Paterson Y, Harmsen HJM, Welling GW, Bos NA. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology (Reading). 2002;148:3651–3660. doi: 10.1099/00221287-148-11-3651. [DOI] [PubMed] [Google Scholar]
- 24.Ormerod KL, Wood DLA, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal’Molin CGO, Palfreyman RW, Nielsen LK, Cooper MA, et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4(1):36. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagkouvardos I, Lesker TR, Hitch TCA, Gálvez EJC, Smit N, Neuhaus K, Wang J, Baines JF, Abt B, Stecher B, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. 2019;7(1):28. doi: 10.1186/s40168-019-0637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6(3):703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung YW, Gwak HJ, Moon S, Rho M, Ryu JH, Dong Q. Functional dynamics of bacterial species in the mouse gut microbiome revealed by metagenomic and metatranscriptomic analyses. PLoS One. 2020;15(1):e0227886. doi: 10.1371/journal.pone.0227886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533(7604):543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyake S, Ding Y, Soh M, Low A, Seedorf H. Muribaculum gordoncarteri sp. nov., an anaerobic bacterium from the faeces of C57BL/6J mice. Int J Syst Evol Microbiol. 2020;70(8):4725–4729. doi: 10.1099/ijsem.0.004338. [DOI] [PubMed] [Google Scholar]
- 30.Buron-Moles G, Chailyan A, Dolejs I, Forster J, Mikš MH. Uncovering carbohydrate metabolism through a genotype-phenotype association study of 56 lactic acid bacteria genomes. Appl Microbiol Biotechnol. 2019;103(7):3135–3152. doi: 10.1007/s00253-019-09701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talarico TL, Axelsson LT, Novotny J, Fiuzat M, Dobrogosz WJ. Utilization of glycerol as a hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD oxidoreductase. Appl Environ Microbiol. 1990;56(4):943–948. doi: 10.1128/aem.56.4.943-948.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veiga da Cunha M, Foster MA. Sugar-glycerol cofermentations in lactobacilli: the fate of lactate. J Bacteriol. 1992;174(3):1013–1019. doi: 10.1128/jb.174.3.1013-1019.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Annuk H, Shchepetova J, Kullisaar T, Songisepp E, Zilmer M, Mikelsaar M. Characterization of intestinal lactobacilli as putative probiotic candidates. J Appl Microbiol. 2003;94(3):403–412. doi: 10.1046/j.1365-2672.2003.01847.x. [DOI] [PubMed] [Google Scholar]
- 34.Burgé G, Saulou-Bérion C, Moussa M, Allais F, Athes V, Spinnler HE. Relationships between the use of Embden Meyerhof pathway (EMP) or Phosphoketolase pathway (PKP) and lactate production capabilities of diverse Lactobacillus reuteri strains. J Microbiol. 2015;53(10):702–710. doi: 10.1007/s12275-015-5056-x. [DOI] [PubMed] [Google Scholar]
- 35.Lee JJ, Choi YJ, Lee MJ, Park SJ, Oh SJ, Yun YR, Min SG, Seo HY, Park SH, Lee MA. Effects of combining two lactic acid bacteria as a starter culture on model kimchi fermentation. Food Res Int. 2020;136:109591. doi: 10.1016/j.foodres.2020.109591. [DOI] [PubMed] [Google Scholar]
- 36.Muck RE, Nadeau EMG, McAllister TA, Contreras-Govea FE, Santos MC, Kung L. Silage review: recent advances and future uses of silage additives. J Dairy Sci. 2018;101(5):3980–4000. doi: 10.3168/jds.2017-13839. [DOI] [PubMed] [Google Scholar]
- 37.Krumbeck JA, Marsteller NL, Frese SA, Peterson DA, Ramer-Tait AE, Hutkins RW, Walter J. Characterization of the ecological role of genes mediating acid resistance in Lactobacillus reuteri during colonization of the gastrointestinal tract. Environ Microbiol. 2016;18(7):2172–2184. doi: 10.1111/1462-2920.13108. [DOI] [PubMed] [Google Scholar]
- 38.Teixeira JS, Seeras A, Sanchez-Maldonado AF, Zhang C, Su MSW, Gänzle MG. Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol. 2014;42:172–180. doi: 10.1016/j.fm.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 39.De Angelis M, Mariotti L, Rossi J, Servili M, Fox PF, Rollán G, Gobbetti M. Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl Environ Microbiol. 2002;68(12):6193–6201. doi: 10.1128/AEM.68.12.6193-6201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotter PD, Hill C. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol Mol Biol Rev. 2003;67(3):429–453. doi: 10.1128/MMBR.67.3.429-453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lyu C, Zhao W, Peng C, Hu S, Fang H, Hua Y, Yao S, Huang J, Mei L. Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production. Microb Cell Fact. 2018;17(1):180. doi: 10.1186/s12934-018-1029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Keersmaecker SCJ, Verhoeven TLA, Desair J, Marchal K, Vanderleyden J, Nagy I. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol Lett. 2006;259(1):89–96. doi: 10.1111/j.1574-6968.2006.00250.x. [DOI] [PubMed] [Google Scholar]
- 43.Tropini C, Moss EL, Merrill BD, Ng KM, Higginbottom SK, Casavant EP, Gonzalez CG, Fremin B, Bouley DM, Elias JE, et al. Transient osmotic perturbation causes long-term alteration to the gut microbiota. Cell. 2018;173(7):1742–1754. doi: 10.1016/j.cell.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheithauer TPM, Dallinga-Thie GM, de Vos WM, Nieuwdorp M, van Raalte DH. Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol Metab. 2016;5(9):759–770. doi: 10.1016/j.molmet.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nardone G, Compare D, Rocco A. A microbiota-centric view of diseases of the upper gastrointestinal tract. Lancet Gastroenterol Hepatol. 2017;2(4):298–312. doi: 10.1016/S2468-1253(16)30108-X. [DOI] [PubMed] [Google Scholar]
- 46.Araújo JR, Tomas J, Brenner C, Sansonetti PJ. Impact of high-fat diet on the intestinal microbiota and small intestinal physiology before and after the onset of obesity. Biochimie. 2017;141:97–106. doi: 10.1016/j.biochi.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 47.Gu S, Chen D, Zhang JN, Lv X, Wang K, Duan LP, Nie Y, Wu XL, Dale C. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One. 2013;8(10):e74957. doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66(4):1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourriaud C, Robins RJ, Martin L, Kozlowski F, Tenailleau E, Cherbut C, Michel C. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J Appl Microbiol. 2005;99(1):201–212. doi: 10.1111/j.1365-2672.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- 50.Muñoz-Tamayo R, Laroche B, É W, Doré J, Duncan SH, Flint HJ, Leclerc M. Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol Ecol. 2011;76(3):615–624. doi: 10.1111/j.1574-6941.2011.01085.x. [DOI] [PubMed] [Google Scholar]
- 51.Ding Z, Xu Y. Lactic acid is absorbed from the small intestine of sheep. J Exp Zool A Comp Exp Biol. 2003;295(1):29–36. doi: 10.1002/jez.a.10212. [DOI] [PubMed] [Google Scholar]
- 52.Ewaschuk JB, Naylor JM, Zello GA. D-lactate in human and ruminant metabolism. J Nutr. 2005;135(7):1619–1625. doi: 10.1093/jn/135.7.1619. [DOI] [PubMed] [Google Scholar]
- 53.Wong JMW, de Souza R, Kendall CWC, Emam A, Jenkins DJA. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 54.Hogan DL, Ainsworth MA, Isenberg JI. Review article: gastroduodenal bicarbonate secretion. Aliment Pharmacol Ther. 1994;8(5):475–488. doi: 10.1111/j.1365-2036.1994.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 55.Alander M, Satokari R, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl Environ Microbiol. 1999;65(1):351–354. doi: 10.1128/AEM.65.1.351-354.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarem-Damerdji L, Sarem F, Marchal L, Nicolas JP. In vitro colonization ability of human colon mucosa by exogenous Lactobacillus strains. FEMS Microbiol Lett. 1995;131(2):133–137. doi: 10.1111/j.1574-6968.1995.tb07767.x. [DOI] [PubMed] [Google Scholar]
- 57.Usui Y, Kimura Y, Satoh T, Takemura N, Ouchi Y, Ohmiya H, Kobayashi K, Suzuki H, Koyama S, Hagiwara S, et al. Effects of long-term intake of a yogurt fermented with Lactobacillus delbrueckii subsp. bulgaricus 2038 and Streptococcus thermophilus 1131 on mice. Int Immunol. 2018;30(7):319–331. doi: 10.1093/intimm/dxy035. [DOI] [PubMed] [Google Scholar]
- 58.Park SE, Kwon SJ, Cho KM, Seo SH, Kim EJ, Unno T, Bok SH, Park DH, Son HS. Intervention with kimchi microbial community ameliorates obesity by regulating gut microbiota. J Microbiol. 2020;58(10):859–867. doi: 10.1007/s12275-020-0266-2. [DOI] [PubMed] [Google Scholar]
- 59.Beck BR, Park GS, Jeong DY, Lee YH, Im S, Song WH, Kang J. Multidisciplinary and comparative investigations of potential psychobiotic effects of Lactobacillus strains isolated from newborns and their impact on gut microbiota and ileal transcriptome in a healthy murine model. Front Cell Infect Microbiol. 2019;9:269. doi: 10.3389/fcimb.2019.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yin X, Yan Y, Kim EB, Lee B, Marco ML. Short communication: effect of milk and milk containing Lactobacillus casei on the intestinal microbiota of mice. J Dairy Sci. 2014;97(4):2049–2055. doi: 10.3168/jds.2013-7477. [DOI] [PubMed] [Google Scholar]
- 61.Aktas B, de Wolfe TJ, Tandee K, Safdar N, Darien BJ, Steele JL. The effect of Lactobacillus casei 32G on the mouse cecum microbiota and innate immune response is dose and time dependent. PLoS One. 2015;10(12):e0145784. doi: 10.1371/journal.pone.0145784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Cesare A, Sirri F, Manfreda G, Moniaci P, Giardini A, Zampiga M, Meluzzi A. Effect of dietary supplementation with Lactobacillus acidophilus D2/CSL (CECT 4529) on caecum microbioma and productive performance in broiler chickens. PLoS One. 2017;12(5):e0176309. doi: 10.1371/journal.pone.0176309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xing Z, Tang W, Yang Y, Geng W, Rehman RU, Wang Y. Colonization and gut flora modulation of Lactobacillus kefiranofaciens ZW3 in the intestinal tract of mice. Probiotics Antimicrob Proteins. 2018;10(2):374–382. doi: 10.1007/s12602-017-9288-4. [DOI] [PubMed] [Google Scholar]
- 64.Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:256–259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 68.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, et al. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:606–612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36(7):2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (WWN) upon reasonable request. Genomic sequence data from previously unpublished strains that appear in this manuscript can be obtained from NCBI Bioproject #474907. Strains used in this study are available from WWN.


