ABSTRACT
Circulating miRNA may contribute to the development of adverse birth outcomes. However, few studies have investigated extracellular vesicle (EV) miRNA, which play important roles in intercellular communication, or compared miRNA at multiple time points in pregnancy. In the current study, 800 miRNA were profiled for EVs from maternal plasma collected in early (median: 12.5 weeks) and late (median: 31.8 weeks) pregnancy from 156 participants in the MADRES Study, a health disparity pregnancy cohort. Associations between miRNA and birth weight, birth weight for gestational age (GA), and GA at birth were examined using covariate-adjusted robust linear regression. Differences by infant sex and maternal BMI were also investigated. Late pregnancy measures of 13 miRNA were associated with GA at birth (PFDR<0.050). Negative associations were observed for eight miRNA (miR-4454+ miR-7975, miR-4516, let-7b-5p, miR-126-3p, miR-29b-3p, miR-15a-5p, miR-15b-5p, miR-19b-3p) and positive associations for five miRNA (miR-212-3p, miR-584-5p, miR-608, miR-210-3p, miR-188-5p). Predicted target genes were enriched (PFDR<0.050) in pathways involved in organogenesis and placental development. An additional miRNA (miR-107), measured in late pregnancy, was positively associated with GA at birth in infants born to obese women (PFDR for BMI interaction = 0.011). In primary analyses, the associations between early pregnancy miRNA and birth outcomes were not statistically significant (PFDR≥0.05). However, sex-specific associations were observed for early pregnancy measures of 37 miRNA and GA at birth (PFDR for interactions<0.050). None of the miRNA were associated with fetal growth measures (PFDR≥0.050). Our findings suggest that EV miRNA in both early and late pregnancy may influence gestational duration.
KEYWORDS: Mirna, extracellular vesicles, pregnancy, birth outcomes
Introduction
Fetal growth and gestational duration are important predictors of immediate and long-term health, influencing neonatal morbidity and mortality as well as risks for childhood obesity and metabolic syndrome, cognitive deficits, and cardiovascular disease in adulthood [1–6]. Preventing adverse birth outcomes therefore translates to widespread benefits for public health. Early biomarkers and a better understanding of the mechanisms contributing to the pathogenesis of adverse birth outcomes are needed to identify susceptible individuals and potential targets for intervention. However, early and minimally invasive biomarkers of birth outcomes are currently lacking. Furthermore, while some of the mechanisms contributing to reduced fetal growth and gestational duration have been elucidated, such as uteroplacental dysfunction, the underlying molecular mediators remain poorly understood [7].
Accumulating evidence suggests that miRNA play important roles in regulating normal fetal growth and development [8]. Distinct miRNA patterns have been reported in a variety of tissues, including maternal peripheral blood and cord blood cells, the placenta, and the cervix, in relation to both adverse birth outcomes and continuous measures of fetal growth and gestational duration [8–17]. In addition to being expressed in tissues, miRNA can be packaged into extracellular vesicles (EVs) and transported via circulation to other cells [18]. EV miRNA therefore have the ability to impact gene expression in distal target tissues and play critical roles in intercellular communication [18]. Prior studies have reported that EV levels are elevated in the circulation of pregnant women compared with non-pregnant women [19], and both EVs and EV miRNA increase in maternal circulation during gestation [19–21]. Given that a large fraction of these EVs are derived from the placenta [21,22], it has been hypothesized that EV miRNA play critical roles in maternal-placental-fetal crosstalk [23,24].
Maternal circulating EV miRNA can be measured in a minimally invasive manner and may therefore serve as promising early biomarkers of pregnancy complications and birth outcomes. In support of this, associations between maternal circulating EV miRNA and pregnancy complications, such as preeclampsia and gestational diabetes, have been observed [25–30]. A growing number of studies have also reported relationships between maternal circulating miRNA and birth outcomes, including fetal growth restriction and preterm birth [26,31–39]. However, few studies have specifically examined EV miRNA [26,31–33]. Furthermore, most studies have been small and focused on miRNA measures at a single time point in pregnancy [31,32,34–39]. The objectives of the current study were therefore to evaluate EV miRNA, measured in paired maternal plasma samples collected in early (median: 12.5-week gestation) and late (median: 31.8-week gestation) pregnancy, in relation to fetal growth and gestational duration. We investigated these relationships in 156 participants from the Maternal and Developmental Risks from Environmental and Social Stressors (MADRES) study, a health disparity cohort of predominantly lower-income Hispanic mother-infant pairs in Los Angeles [40].
Materials and methods
Study participants
The MADRES Study is an ongoing, prospective pregnancy cohort, which began enrolling participants in November 2015 and has been described previously [40]. Briefly, participants are recruited from four prenatal care providers in Los Angeles, California, most of which serve predominantly lower-income Hispanic populations. The participating clinics include two community health clinics, one county hospital prenatal clinic, and one private obstetrics and gynecology practice. Women are eligible to participate in MADRES if their pregnancy is at less than 30 weeks’ gestation at the time of recruitment, they are ≥18 years of age, and they can speak either English or Spanish fluently. Exclusion criteria include: 1) multiple gestation; 2) HIV positive status; 3) having a physical, mental, or cognitive disability that would prevent participation in the study or the ability to provide informed consent; and 4) current incarceration. Informed consent was obtained from each participant at study entry, and the study was approved by the University of Southern California’s Institutional Review Board. For the current study, we focused on 156 mother-infant pairs who 1) had reached the delivery period, 2) provided a blood sample in both early and late pregnancy, 3) had abstracted birth outcome data, and 4) had complete covariate information.
Maternal blood collection, processing, and EV RNA extraction
Blood samples were collected in EDTA vacutainers (BD Biosciences #366,643), transported on ice, and processed within 2 hours. Plasma aliquots were stored at −80°C until extraction. EV RNA was extracted from 500 µl of plasma using the Qiagen ExoRNeasy kit following the manufacturer’s instructions, with a second addition of 90 µL chloroform and an additional round of phase separation. Purified RNA aliquots were frozen at −80°C until quantification.
EV miRNA measures
Plasma EV miRNA were quantified by BioAnalyzer using a small RNA kit (Agilent Technologies, Inc. USA). If there was a sufficient quantity of miRNA (>100 pg/µl), 3 µl of the purified RNA were prepared for the NanoString NCounter Human v3 miRNA expression assay (Nanostring Technologies, Inc.) with a modified protocol; instead of using a 1:10 dilution after the addition of the hybridized probes, samples were diluted 1:2. Replicate samples (N = 6) were run across two chips to investigate possible technical variation, but were not found to differ significantly either in total or individual miRNA counts (P from Wilcoxon Rank Sum test≥0.050)
Birth outcomes
Birth weight (BW) information was abstracted from maternal medical records. If this information was missing (n = 2), proxy-reported information obtained from the mothers was used. Best estimates of gestational age (GA) at birth were ascertained using a hierarchy of methods, as described previously [41]. Briefly, a first trimester (<14 weeks GA) ultrasound measurement of crown-rump length was considered the most preferred and was used if available (n = 105). If unavailable, a second trimester (<28 weeks GA) ultrasound measurement of fetal biparietal diameter was used (n = 32). If measures from an early ultrasound were unavailable, GA at birth was determined based on the physician’s best clinical estimate, abstracted from the medical records (n = 19). BW for GA z-scores were calculated using a representative U.S. reference [42]. This reference was selected because it utilizes obstetric estimates of GA at birth and was updated recently, reflecting current trends in obesity and gestational diabetes, which can impact fetal growth [42].
Covariates
Questionnaires were administered to participants in either English or Spanish, depending on the participant’s preferred language. Maternal self-reported pre-pregnancy weight, ethnicity, birth country, prenatal vitamin use, and birth order were determined from a questionnaire that was administered during the first study visit. In this questionnaire, participants were also asked if they resided with a smoker during their pregnancy. During the first study visit, participants also completed the Pregnancy-Unique Quantification of Emesis/Nausea (PUQE) Index questionnaire to determine the severity of their nausea [43]. This variable was evaluated both continuously and as a binary variable (PUQE score >6 indicating moderate to severe nausea versus PUQE score ≤6 indicating mild to no nausea). Participants were asked about their highest attained education and if they had ever smoked during the pregnancy in questionnaires administered during their first, second, and third trimesters. Maternal standing height was measured twice by stadiometer (Perspectives Enterprises Model PE-AIM-101). Maternal pre-pregnancy BMI was calculated using the self-reported pre-pregnancy weight and measured height values (kg/m2). Each participant’s age was determined using the date of consent and her birth date. A combined variable indicating ethnicity by birthplace was created based on the participant’s self-reported ethnicity (Hispanic vs. non-Hispanic) and birth country (U.S. versus other). A combined variable was also created for any tobacco smoke exposure during the pregnancy, which was based on self-reported maternal smoking during the pregnancy or the participant sharing a residence with a smoker during the pregnancy. The season of birth was determined using the child’s birth date. Information on newborn sex was abstracted from medical records. If this information was missing from the maternal medical records, it was filled in using reports from a questionnaire administered to the mothers 7–14 days after birth. Maternal blood pressure measures and information on pregnancy-related hypertensive disorders were also abstracted from the maternal medical records. Women were classified as having preeclampsia or eclampsia if there was a physician diagnosis recorded in the medical records. Participants were classified as having gestational hypertension if they 1) had self-reported gestational hypertension, 2) had a physician diagnosis of gestational hypertension, or 3) met the American College of Obstetricians and Gynecologists criteria for gestational hypertension based on the abstracted blood pressures measures [44]. Information on physician-diagnosed diabetes prior to the pregnancy and gestational diabetes mellitus (GDM) during the pregnancy were abstracted from the maternal medical records. Results from glucose challenge tests (GCT) and oral glucose tolerance tests (OGTT) were also abstracted from the medical records. Participants were classified as having GDM if they 1) had a high positive GCT value or 2) a positive GCT value and either a physician diagnosis of GDM or at least two positive OGTT results. Participants were classified as having glucose intolerance during the pregnancy if they 1) had a positive GCT test and one positive OGTT result or 2) a positive GCT result but no follow up OGTT. Women were classified as having normal glucose during the pregnancy if their GCT results were negative or they had a positive GCT test result followed by a negative OGTT. GCTs were considered negative if they were below 120 mg/dL, positive if they were between 120 and 140 mg/dL, and high positive if they exceeded 140 mg/dL. OGTT cut-offs were as follows: fasting plasma ≥95 mg/dL, one-hour plasma ≥180 mg/dL, two-hour plasma ≥155 mg/dL, and three-hour plasma ≥140 mg/dL. For statistical analyses, GDM and glucose intolerance during the pregnancy were collapsed into one category.
Statistical analyses
Statistical analyses were conducted in R (version 3.6.2) [45], and plots were created using ‘ggplot2’ and the ‘Enhanced Volcano’ packages [46,47]. Raw miRNA counts were normalized using the NanoStringNorm package (version 1.2.1) [48]. Sample-specific normalization was conducted based on total counts for the positive controls included on the nCounter platform and the geometric mean of the 75 miRNA with the highest counts. For primary analyses, we normalized miRNA counts separately by time point, given prior evidence that circulating miRNA may change during pregnancy [31,49]. In sensitivity analyses, we compared results after normalizing miRNA counts from both early and late pregnancy together. We retained miRNA with counts falling above sample-specific background levels for ≥60% of the participants, similar to previous studies [12,50]. Background levels were defined based on the mean + 1.5 SD of negative control values. Machine-read values were used for miRNA counts falling below these thresholds.
Major predictors of maternal circulating EV miRNA are largely unknown. To identify potential predictors of EV miRNA, we therefore ran principal component analyses (PCA) to reduce the dimensionality of the EV miRNA data using the ‘stats’ package in R [45]. Separate PCAs were conducted for miRNA measures from early versus late pregnancy. The top 10 principal components (PCs) explained approximately 80% of the variance in EV miRNA at each time point and were therefore retained (Figure S1). Variables that were correlated or associated with these miRNA PCs at either time point (P < 0.100) based on Spearman correlations (continuous variables), Wilcoxon rank sum tests (binary variables), or Kruskal Wallis tests (categorical variables) were considered possible predictors of EV miRNA in pregnancy. Variables investigated in relation to the miRNA PCs included maternal age, maternal pre-pregnancy BMI (continuous and categorical), GA at plasma collection, GA at birth (a potential precision variable for BW analyses), birth order, infant sex, maternal education, maternal ethnicity by birthplace, recruitment site, prenatal vitamin use, nausea severity in early pregnancy (continuous and categorical), in utero tobacco smoke exposure, birth season, and laboratory batch. For the late pregnancy miRNA PCs, we also investigated associations with pregnancy-related hypertensive disorders and GDM/glucose intolerance during the pregnancy. We did not include delivery type in these investigations, as it does not precede miRNA measures from either time point and therefore could not be a confounder. However, delivery type was investigated as a potential effect modifier in sensitivity analyses. Primary models were adjusted for recruitment site, maternal age, maternal ethnicity by birthplace, maternal pre-pregnancy BMI (continuous), infant sex, and laboratory batch, based on results from the PCAs (Figure S2, Table S1, Table S2), preliminary regression analyses, and a priori knowledge of each variable’s relationships with birth outcomes. Given that GA at birth may fall in the causal pathway from EV miRNA to fetal growth, BW models were compared with and without adjustment for this variable.
Because miRNA counts were largely right skewed with evidence of extreme outliers, we used robust linear regression, which is less sensitive to outliers compared with ordinary least squares regression [51]. Using this approach, we investigated associations between each miRNA and birth outcomes, adjusting for covariates. Separate models were run for each birth outcome. Given that early and late pregnancy measures for some of the miRNA were moderately to strongly correlated (median and maximum Spearman correlations: 0.39 and 0.70, respectively), miRNA measures from each time point were investigated in separate models. However, if a miRNA was significantly associated (PFDR<0.050) with a birth outcome in either early or late pregnancy, a secondary analysis was run in which the change in the miRNA from early to late pregnancy was modeled as a predictor of the relevant birth outcome.
Predicted target genes of miRNA associated (PFDR<0.050) with a given birth outcome were identified using mirDIP [52]. We selected mirDIP because it utilizes human miRNA-target predictions from 30 different resources and creates an integrated score to improve the accuracy of these predictions [52]. Predicted target genes were retained if they had a ‘very high’ mirDIP integrative confidence score. EnrichR was subsequently used to determine whether the predicted target genes identified by mirDIP were over- or under-represented (PFDR<0.050) in any PANTHER pathways [53,54]. Results from these pathway analyses were examined using bubble plots, which were created using code that we adapted from the ‘enrichment_chart’ function from the pathfindR package [55].
In secondary analyses, we investigated whether miRNA-birth outcome associations differed by infant sex or pre-pregnancy BMI (evaluated continuously) using cross-product terms in robust linear regression models. If a cross-product term was statistically significant (PFDR<0.050), we further examined the miRNA-birth outcome association in sex-stratified or BMI category-stratified analyses, respectively. In sensitivity analyses, we also investigated whether statistically significant (PFDR<0.050) results 1) were robust to the removal of extreme outliers (<Q1 – 3*IQR or >Q3 + 3*IQR), 2) were robust to the exclusion of pregnancy-related hypertensive disorders (preeclampsia, eclampsia, and gestational hypertension), 3) were robust to additional adjustment for the method of GA at birth ascertainment, and 4) differed by delivery type. Because there was a relatively small number of caesarean section deliveries (N = 34), scheduled versus unplanned caesarean sections were collapsed into one category for this last analysis and compared with all vaginal deliveries. We also evaluated whether results were sensitive to the exclusion of women who were diagnosed with diabetes prior to the pregnancy and whether late pregnancy miRNA results were sensitive to additional adjustment for GDM/glucose intolerance.
Given that approximately 20% of EVs in maternal circulation during pregnancy are estimated to originate in the placenta [22], we conducted a post hoc investigation to determine if any of the maternal circulating EV miRNA that were significantly associated (PFDR<0.050) with birth outcomes in primary analyses are expressed in the placenta. To investigate this, we used data previously generated for 230 participants in the Rhode Island Children’s Health Study (RICHS) [17]. Participant characteristics, placenta collection, and RNA isolation protocols for RICHS have been described previously [17]. Single-end RNA sequencing was used to profile placental miRNA expression. The methods used to generate and process these data have been described previously [17]. These methods are also summarized briefly in the supplement (Supplemental Methods). A miRNA was considered detectable if it had a normalized count >1 per million. For each miRNA, the percentage of placenta samples with a normalized count >1 per million was determined, and the median and range of the placental counts was also calculated. If a miRNA did not meet quality control standards, as described in the supplement (Supplemental Methods), it was excluded from this post-hoc investigation.
Results
Participant characteristics
Participant characteristics are shown in (Table 1). Women were between 18 and 45 years of age with a median age of 28. Prior to the pregnancy, nearly one-third of participants (32.1%) were overweight and more than a third (37.2%) were obese. Half of the participants were Hispanic and born outside the U.S., 35.3% were Hispanic and born in the U.S., and 14.7% were non-Hispanic. A quarter of the participants (25.6%) had not completed high school. More than a third of the participants (34.6%) developed GDM or glucose intolerance during the pregnancy, and 16.7% developed a pregnancy-related hypertensive disorder. There were more male (56.4%) compared with female (43.6%) infants in the study. Of the 800 miRNA profiled by the NanoString nCounter platform, 102 were detectable in maternal plasma EVs for at least 60% of the participants in early or late pregnancy and were therefore retained for statistical analyses.
Table 1.
Participant characteristics (N = 156)
| Participant Characteristics | Mean ± SD or N (%) |
Median (Range) |
|---|---|---|
| Maternal Characteristic | ||
| Age, years | 28.6 ± 5.9 | 28.9 (18.3–45.5) |
| Pre-Pregnancy BMI, kg/m2 | 28.8 ± 6.6 | 27.8 (17.6–53.6) |
| Pre-Pregnancy BMI Category | ||
| Underweight/Normal Weight | 48 (30.8) | |
| Overweight | 50 (32.1) | |
| Obese | 58 (37.2) | |
| Ethnicity by Birth Place | ||
| Non-Hispanic | 23 (14.7) | |
| Hispanic Born in the U.S. | 55 (35.3) | |
| Hispanic Foreign-Born | 78 (50.0) | |
| Prenatal Vitamin Use | ||
| Ever | 141 (90.4) | |
| Never | 15 (9.6) | |
| Parity | ||
| Primiparous | 50 (32.1) | |
| Multiparous | 106 (67.9) | |
| Maternal Education | ||
| Less than High School | 40 (25.6) | |
| Completed High School | 52 (33.3) | |
| Completed Some College or Technical | 46 (29.5) | |
| Completed 4 Years of College | 18 (11.5) | |
| Pregnancy-Related Hypertension | ||
| None | 130 (83.3) | |
| Preeclampsia or Eclampsia | 14 (9.0) | |
| Gestational Hypertension | 12 (7.7) | |
| Diabetes | ||
| Diabetes Diagnosed Prior to Pregnancy | 4 (2.6) | |
| Gestational Diabetes Mellitus | 14 (9.0) | |
| Glucose Intolerance | 40 (25.6) | |
| Normal Glucose | 98 (62.8) | |
| GA at Early Pregnancy Plasma Collection, Weeks | 13.5 (4.0) | 12.5 (5.8–22.9) |
| GA at Late Pregnancy Plasma Collection, Weeks | 31.9 (1.9) | 31.8 (24.8–36.1) |
| Infant Characteristics | ||
| BW, grams | 3323 (447) | 3318 (2230–4560) |
| GA at Birth, weeks | 39.0 (1.5) | 39.3 (33.6–42.4) |
| BW for GA Z-Score, SD | 0.1 (1.0) | 0.0 (−2.9, 2.8) |
| Sex | ||
| Male | 88 (56.4) | |
| Female | 68 (43.6) | |
| Birth Season | ||
| Winter | 41 (26.3) | |
| Spring | 36 (23.1) | |
| Summer | 30 (19.2) | |
| Fall | 49 (31.4) |
Abbreviations Used: BW, birth weight; GA, gestational age
Robust linear regression results for early pregnancy miRNA
Only one miRNA (miR-340-5p) measured in early pregnancy was associated (P < 0.050) with BW in models adjusted for GA at birth (Table S3). In contrast, six miRNA were associated with BW (P < 0.050) when GA at birth was excluded from models (Table S3). 11 miRNA measured in early pregnancy were associated with GA at birth (P < 0.050) (Table S3). Associations between early pregnancy miRNA measures and birth outcomes were not statistically significant after multiple testing correction (PFDR≥0.050) (Table S3). The results were similar when raw miRNA counts from early and late pregnancy were normalized together rather than separately (Table S4) and were also similar after excluding participants who had been diagnosed with diabetes prior to the pregnancy (Table S5).
Robust linear regression results for late pregnancy miRNA
Two miRNA measured in late pregnancy were positively associated (P < 0.050) with BW in models adjusted for GA at birth (miR-148a-5p and miR-26b-5p) (Table S6). MiR-148a-5p remained positively associated (P < 0.050) with BW after removing GA at birth from the model, but miR-26b-5p did not (Table S6). An additional miRNA (miR-15a-5p) was associated (P < 0.050) with BW after removing GA at birth from the model (Table S6). Four miRNA in late pregnancy (miR-148a-3p, miR-26b-3p, miR-199a-5p, miR-937-3p) were associated with BW for GA (P < 0.050) (Table S6). Associations between late pregnancy miRNA and fetal growth measures (BW, BW for GA) did not remain statistically significant after multiple testing correction (PFDR≥0.050). The results were similar when miRNA counts from each time point were normalized together instead of separately, although some of the associations were no longer statistically significant (Table S7).
Thirty-three EV miRNA measured in late pregnancy were associated with GA at birth (P < 0.050) (Table 2). Of these, 13 remained statistically significant after correcting for multiple testing (PFDR<0.050) (Figure 1, Table 2). One probe, reflecting two miRNA (miR-4454+miR-7975), also remained statistically significant after a more conservative Bonferroni correction (PBonferroni<0.050). Results were similar when early and late pregnancy miRNA measures were normalized together rather than separately (Figure S3). Associations between these 13 miRNA and GA at birth were generally similar after removing extreme outliers (Table S8). Although the directions of the associations for these 13 miRNA were the same and remained statistically significant (P < 0.05) after excluding participants with pregnancy-related hypertensive disorders, the effect estimates were partially attenuated for many of the miRNA (miR-4516, miR-608, miR-29b-3p, miR-15a-5p, miR-210-3p, miR-15b-5p, miR-188-5p, miR-19b-3p) (Table S9). For 12 of the miRNA, associations with GA at birth were in consistent directions for infants delivered by caesarean section versus those delivered vaginally (Table S10). However, the inverse association between miR-4516 in late pregnancy and GA at birth was entirely driven by infants who were delivered vaginally (Table S10). We also examined the change in each of these 13 miRNA from early to late pregnancy as predictors of GA at birth; results were null for all but one probe (miR-4454+miR-7975) (Table S11). A doubling in miR-4454+miR-7975 from early to late pregnancy was associated with a −0.30 (95% CI: −0.57, −0.04) week shorter gestational duration (P = 0.024).
Table 2.
Associations between late pregnancy EV miRNA and GA at birtha for the 33 miRNA with a P-Value <0.050 (N = 156)
| miRNA | Effect Estimate (95% CI) | PUncorrected | PFDR | PBonferroni |
|---|---|---|---|---|
| miR-4454+miR-7975 | −0.51 (−0.72, −0.30) | 2.59 x 10−6 | 2.64 x 10−4 | 2.64 x 10−4 |
| miR-4516 | −0.46 (−0.72, −0.20) | 5.20 10–4 | 0.023 | 0.053 |
| miR-212-3p | 0.51 (0.22, 0.81) | 6.88 x 10−4 | 0.023 | 0.070 |
| miR-584-5p | 0.62 (0.25, 0.99) | 0.001 | 0.026 | 0.113 |
| let-7b-5p | −0.78 (−1.26, −0.30) | 0.001 | 0.026 | 0.134 |
| miR-608 | 0.47 (0.18, 0.76) | 0.002 | 0.026 | 0.156 |
| miR-126-3p | −0.46 (−0.75, −0.17) | 0.002 | 0.030 | 0.207 |
| miR-29b-3p | −0.52 (−0.85, −0.18) | 0.002 | 0.032 | 0.253 |
| miR-15a-5p | −0.41 (−0.68, −0.13) | 0.003 | 0.039 | 0.353 |
| miR-210–3p | 0.32 (0.10, 0.53) | 0.004 | 0.039 | 0.387 |
| miR-15b-5p | −0.45 (−0.77, −0.13) | 0.006 | 0.048 | 0.571 |
| miR-188-5p | 0.33 (0.10, 0.56) | 0.006 | 0.048 | 0.578 |
| miR-19b-3p | −0.48 (−0.82, −0.13) | 0.006 | 0.050 | 0.645 |
| miR-656-3p | 0.62 (0.17, 1.07) | 0.007 | 0.050 | 0.706 |
| miR-20a-5p+miR-20b-5p | −0.63 (−1.10, −0.16) | 0.009 | 0.058 | 0.868 |
| miR-199b-5p | 0.26 (0.06, 0.46) | 0.010 | 0.067 | 1 |
| miR-142-3p | −0.48 (−0.86, −0.11) | 0.012 | 0.070 | 1 |
| miR-1272 | 0.23 (0.04, 0.43) | 0.018 | 0.099 | 1 |
| let-7d-5p | −0.39 (−0.71, −0.06) | 0.020 | 0.099 | 1 |
| miR-21-5p | −0.42 (−0.77, −0.07) | 0.020 | 0.099 | 1 |
| miR-548y | 0.42 (0.06, 0.77) | 0.020 | 0.099 | 1 |
| miR-1296-3p | 0.42 (0.05, 0.78) | 0.025 | 0.116 | 1 |
| miR-16-5p | −0.34 (−0.64, −0.04) | 0.027 | 0.116 | 1 |
| miR-551b-3p | 0.41 (0.05, 0.78) | 0.027 | 0.116 | 1 |
| miR-937-3p | 0.29 (0.03, 0.55) | 0.029 | 0.118 | 1 |
| miR-150–5p | −0.39 (−0.75, −0.03) | 0.031 | 0.118 | 1 |
| miR-374a-5p | −0.54 (−1.04, −0.04) | 0.033 | 0.118 | 1 |
| miR-26b-5p | −0.63 (−1.22, −0.05) | 0.033 | 0.118 | 1 |
| miR-1258 | 0.38 (0.03, 0.74) | 0.034 | 0.118 | 1 |
| miR-221-3p | −0.58 (−1.12, −0.04) | 0.035 | 0.118 | 1 |
| miR-331-3p | 0.24 (0.02, 0.46) | 0.036 | 0.119 | 1 |
| miR-1827 | 0.44 (0.01, 0.86) | 0.044 | 0.139 | 1 |
| miR-320e | −0.24 (−0.47, 0.00) | 0.045 | 0.140 | 1 |
aResults are from robust linear regression models, which were adjusted for the GA at sample collection, maternal age, maternal pre-pregnancy BMI, maternal ethnicity by birthplace, recruitment site, infant sex, and laboratory batch. miRNA counts were log2-transformed. Effect estimates can therefore be interpreted as the difference in the specified outcome for a doubling in miRNA count.
Abbreviations Used: EV, extracellular vesicle; FDR, false discovery rate; GA, gestational age; miRNA, microrna
Figure 1.
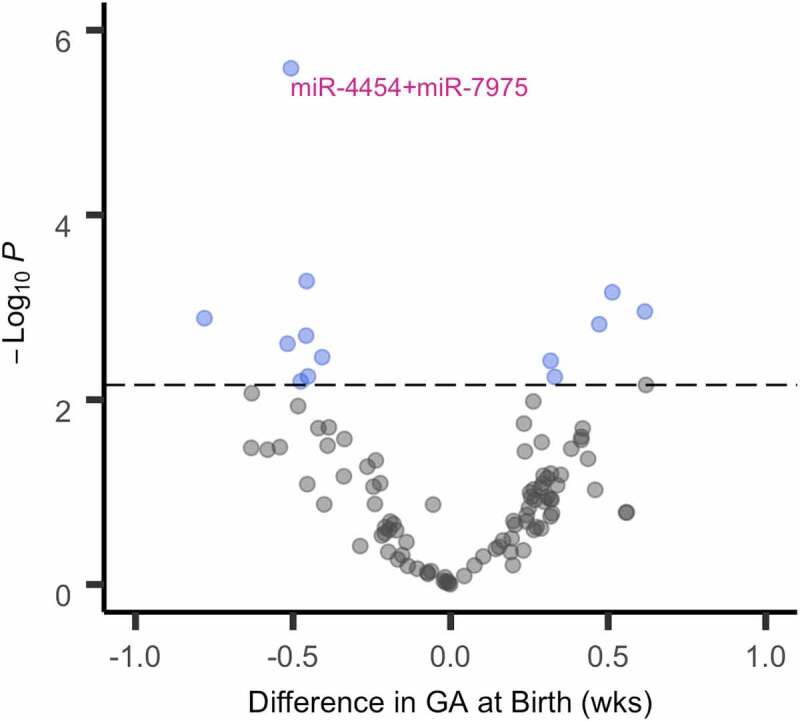
Volcano plot for late pregnancy EV miRNA and GA at birth (N = 156). The x-axis shows the difference in GA at birth for a doubling in the miRNA count. The y-axis represents the -log10(p-value). Each dot represents a miRNA that was detectable in at least 60% of MADRES participants in either early or late pregnancy. The dashed horizontal line shows the threshold for statistical significance based on a PFDR<0.050. miRNA that were statistically significant after a more conservative Bonferroni correction (PBonferroni<0.050) are annotated. Results are from robust linear regression models, which were adjusted for the GA at sample collection, maternal age, maternal pre-pregnancy BMI, maternal ethnicity by birthplace, recruitment site, infant sex, and laboratory batch. miRNA counts were log2-transformed. Effect estimates can therefore be interpreted as the difference in the specified outcome for a doubling in miRNA count. Abbreviations Used: EV, extracellular vesicle; FDR, false discovery rate; GA, gestational age; MADRES, Maternal and Developmental Risks from Environmental and Social Stressors study; miRNA, microRNA.
Results for late pregnancy EV miRNA and birth outcomes were similar after excluding participants diagnosed with diabetes prior to the pregnancy and after additional adjustment for GDM/glucose intolerance during the pregnancy (Tables S12 and S13 and Figures S4 and S5). Associations between late pregnancy EV miRNA and GA at birth were also similar after additional adjustment for the method that was used to ascertain GA (Table S14).
Interactions with infant sex and maternal pre-pregnancy BMI
Sex-specific associations were identified for early pregnancy measures of 37 miRNA (PFDR for interaction<0.050) and GA at birth (Table 3). Twenty-eight miRNA in early pregnancy were significantly associated with GA at birth among male infants only, while four miRNA in early pregnancy were significantly associated with GA at birth among female infants only. Additionally, five miRNA in early pregnancy were significantly associated with GA at birth in both males and females, but in opposite directions (Table 3). The most pronounced difference between males and females was observed for let-7g-5p (PFDR for interaction = 9.63 x 10−4). A doubling in let-7g-5p in early pregnancy was associated with a 1.52 (95% CI: 0.72, 2.31) week longer gestational duration among female infants only (Table 3). Additionally, we observed that early pregnancy maternal plasma EV levels of let-7g-5p were significantly higher in women carrying female (median count: 120) compared with male fetuses (median count: 113) (PWilcoxon<2.2 x 10−16). Sex-specific associations between early pregnancy miRNA counts and GA at birth were also observed for two other members of the let-7 family (let-7a-5p and let-7d-5p). Several members of the miR-548 family (miR-548ar-3p, miR-548e-5p, and miR-548y) and the C14MC cluster (miR-299, miR-1197, and miR-656) were also associated with GA at birth in a sex-dependent manner. Sex differences were not identified for late pregnancy miRNA measures and GA at birth and were not identified for either early or late pregnancy miRNA measures and fetal growth measures (BW, BW for GA) (PFDR for interaction ≥0.050).
Table 3.
Sex-Specific associations between early pregnancy EV miRNA and GA at birtha (PFDR for sex interaction<0.050)
| miRNA | Chromosome | Males (N = 88) | Females (N = 68) | Pinteraction | ||||
|---|---|---|---|---|---|---|---|---|
| Effect Estimate (95% CI) | P | Effect Estimate (95% CI) | P | PUncorrected | PFDR | PBonferroni | ||
| let-7g-5p | Chr3 | −0.51 (−1.14, 0.12) |
0.114 |
1.52 (0.72, 2.31) |
1.99 x 10−4 | 9.63 x 10−4 | 0.027 | 0.098 |
| miR-656-3p | Chr14 |
0.82 (0.40, 1.24) |
1.45 x 10−4 | −0.52 (−1.40, 0.36) |
0.250 | 0.001 | 0.027 | 0.111 |
| miR-548y | Chr14 |
0.86 (0.31, 1.41) |
0.002 |
−0.55 (−1.00, −0.10) |
0.017 | 0.001 | 0.027 | 0.119 |
| miR-374a-5p | ChrX |
−1.02 (−1.55, −0.49) |
1.46 x 10−4 | 0.67 (−0.17, 1.52) |
0.119 | 0.001 | 0.027 | 0.132 |
| miR-142-3p | Chr17 |
−0.87 (−1.26, −0.48) |
1.44 x 10−5 | 0.31 (−0.21, 0.82) |
0.248 | 0.001 | 0.027 | 0.148 |
| miR-887-5p | Chr5 |
0.54 (0.17, 0.91) |
0.004 |
−0.47 (−0.82, −0.12) |
0.009 | 0.002 | 0.027 | 0.173 |
| miR-34a-5p | Chr1 |
0.69 (0.27, 1.10) |
0.001 | −0.47 (−0.96, 0.03) |
0.064 | 0.002 | 0.027 | 0.192 |
| miR-1285-5p | Chr7 |
0.58 (0.10, 1.07) |
0.019 |
−0.70 (−1.28, −0.12) |
0.018 | 0.003 | 0.028 | 0.256 |
| miR-584-5p | Chr5 |
0.67 (0.32, 1.02) |
1.70 x 10−4 | −0.55 (−1.18, 0.09) |
0.094 | 0.003 | 0.028 | 0.256 |
| miR-15a-5p | Chr13 |
−0.61 (−1.06, −0.16) |
0.008 | 0.50 (−0.03, 1.03) |
0.067 | 0.004 | 0.035 | 0.384 |
| miR-1296-3p | Chr10 |
0.54 (0.19, 0.89) |
0.002 | −0.35 (−0.80, 0.09) |
0.121 | 0.004 | 0.035 | 0.418 |
| miR-223-3p | ChrX |
−0.47 (−0.83, −0.11) |
0.011 | 0.49 (−0.12, 1.11) |
0.118 | 0.005 | 0.035 | 0.490 |
| miR-1290 | Chr1 |
0.61 (0.09, 1.12) |
0.021 | −0.53 (−1.18, 0.11) |
0.106 | 0.005 | 0.035 | 0.502 |
| miR-301a-5p | Chr17 |
0.62 (0.15, 1.08) |
0.010 | −0.41 (−0.83, 0.01) |
0.055 | 0.005 | 0.035 | 0.513 |
| miR-607 | Chr10 |
0.55 (0.07, 1.03) |
0.026 |
−0.53 (−1.05, −0.02) |
0.042 | 0.005 | 0.035 | 0.558 |
| miR-299-5p | Chr14 |
0.82 (0.48, 1.16) |
2.24 x 10−6 | −0.46 (−1.05, 0.13) |
0.129 | 0.005 | 0.035 | 0.558 |
| miR-1268b | Chr17 |
0.67 (0.27, 1.07) |
0.001 | −0.54 (−1.20, 0.12) |
0.111 | 0.006 | 0.035 | 0.616 |
| miR-23b-3p | Chr9 | 0.42 (−0.08, 0.91) |
0.098 |
−0.68 (−1.15, −0.21) |
0.005 | 0.006 | 0.035 | 0.639 |
| miR-188-5p | ChrX |
0.47 (0.24, 0.70) |
7.71 x 10−5 | −0.14 (−0.45, 0.16) |
0.360 | 0.006 | 0.035 | 0.661 |
| let-7d-5p | Chr9 |
−0.82 (−1.43, −0.21) |
0.009 | 0.52 (−0.08, 1.12 |
0.089 | 0.007 | 0.036 | 0.714 |
| miR-937-3p | Chr8 |
0.55 (0.28, 0.82) |
6.14 x 10−5 | −0.22 (−0.61, 0.16) |
0.257 | 0.008 | 0.036 | 0.791 |
| miR-302b-3p | Chr4 |
0.70 (0.45, 0.95) |
4.74 x 10−8 | −0.22 (−0.65, 0.22) |
0.331 | 0.008 | 0.036 | 0.795 |
| miR-183-5p | Chr7 |
0.52 (0.01, 1.03) |
0.045 | −0.37 (−0.93, 0.19) |
0.191 | 0.009 | 0.039 | 0.906 |
| miR-551b-3p | Chr3 |
0.60 (0.13, 1.07) |
0.012 |
−0.53 (−1.02, −0.03) |
0.038 | 0.009 | 0.039 | 0.935 |
| miR-525-5p | Chr19 |
0.60 (0.22, 0.98) |
0.002 | −0.45 (−1.04, 0.14) |
0.137 | 0.010 | 0.041 | 1 |
| miR-1236-3p | Chr6 |
0.60 (0.11, 1.10) |
0.016 | −0.35 (−0.82, 0.13) |
0.150 | 0.011 | 0.041 | 1 |
| miR-584-3p | Chr5 |
0.45 (0.07, 0.84) |
0.020 | −0.45 (−0.95, 0.05) |
0.081 | 0.011 | 0.041 | 1 |
| miR-15b-5p | Chr3 |
−0.75 (−1.21, −0.30) |
0.001 | 0.29 (−0.33, 0.91) |
0.361 | 0.011 | 0.041 | 1 |
| miR-1197 | Chr14 |
0.63 (0.15, 1.11) |
0.010 | −0.41 (−0.97, 0.15) |
0.153 | 0.012 | 0.044 | 1 |
| miR-888-5p | ChrX |
0.46 (0.07, 0.86) |
0.021 | −0.53 (−1.07, 0.01) |
0.054 | 0.014 | 0.047 | 1 |
| miR-106a-5p + miR-17-5p |
ChrX + Chr13 | −0.39 (−0.85, 0.07) |
0.094 |
0.69 (0.08, 1.30) |
0.028 | 0.015 | 0.047 | 1 |
| let-7a-5p | Chr9 |
−0.15 (−0.25, −0.05) |
0.003 | 0.10 (−0.08, 0.28) |
0.270 | 0.015 | 0.047 | 1 |
| miR-1972 | Chr16 |
0.59 (0.15, 1.02) |
0.008 | −0.46 (−1.22, 0.29) |
0.229 | 0.016 | 0.049 | 1 |
| miR-939-5p | Chr8 |
0.52 (0.01, 1.03) |
0.046 | −0.43 (−0.89, 0.04) |
0.072 | 0.017 | 0.049 | 1 |
| miR-548e-5p | Chr10 |
0.56 (0.19, 0.93) |
0.003 | −0.41 (−1.10, 0.29) |
0.251 | 0.018 | 0.049 | 1 |
| miR-548ar-3p | Chr13 | 0.25 (−0.23, 0.73) |
0.307 |
−0.56 (−1.08, −0.03) |
0.038 | 0.018 | 0.049 | 1 |
| miR-1827 | Chr12 |
0.51 (0.13, 0.88) |
0.008 | −0.35 (−0.95, 0.25) |
0.256 | 0.018 | 0.049 | 1 |
aResults are from robust linear regression models, which were adjusted for the gestational age at sample collection, maternal age, maternal pre-pregnancy BMI, maternal ethnicity by birthplace, recruitment site, infant sex, and batch. miRNA counts were log2-transformed. Effect estimates can therefore be interpreted as the difference in the specified outcome for a doubling in miRNA count. Significant (P < 0.050) associations are bolded.
Abbreviations Used: FDR, false discovery rate; EV, extracellular vesicle; GA, gestational age; miRNA, microRNA
A statistically significant interaction was identified for one miRNA (miR-107), measured in late pregnancy and pre-pregnancy BMI (PFDR for interaction = 0.011). Among infants born to obese women, a doubling in miR-107 in late pregnancy was associated with a 1.05 (95% CI: 0.34, 1.76) week longer gestational duration (P = 0.004). Associations were null for infants born to overweight and normal/underweight women. No other significant interactions were identified for pre-pregnancy BMI (PFDR for interaction≥0.050).
Predicted target genes and pathway analyses
MirDIP identified 6,012 unique high-confidence predicted target genes for the 13 miRNA which, in late pregnancy, were associated with GA at birth (Table S15). These predicted target genes were overrepresented (PFDR<0.050) in 57 PANTHER pathways (Table S16). The top 10 pathways included the Wnt signaling pathway, angiogenesis, the EGF receptor signalling pathway, Cadherin signalling, apoptosis signaling, integrin signalling, the CCKR signaling map ST pathway, the FGF signaling pathway, the PDGF signaling pathway, and the Ras pathway (Figure 2).
Figure 2.
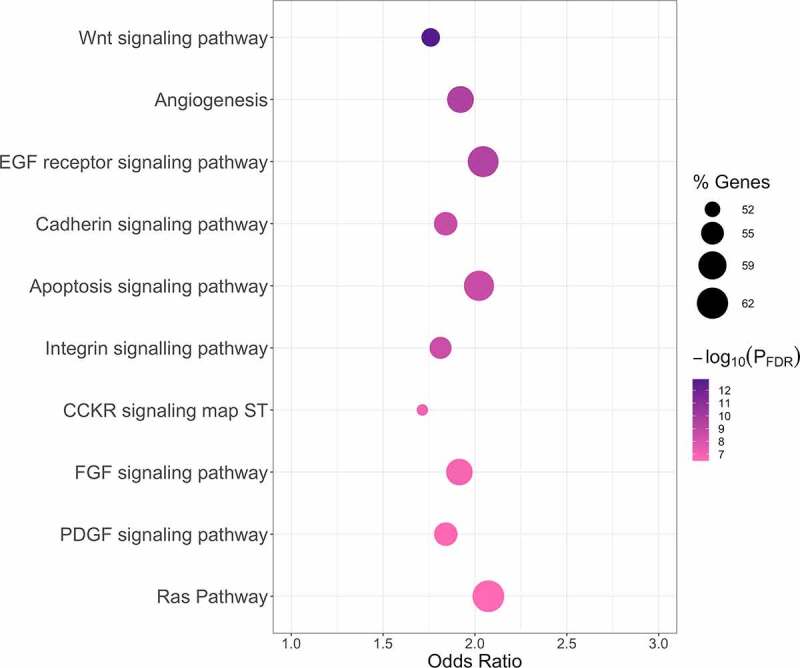
Top 10 PANTHER pathways associated with predicted target genes identified for late pregnancy EV miRNA associated with GA at birth. PANTHER pathway enrichment analyses were conducted using enrichR [53]. Results are presented in a bubble plot. The dot color represents the -log10(PFDR) from Fisher’s Exact tests (dark purple represents smaller p-values and light pink represents larger p-values). The dot size indicates the percentage of pathway members that are represented by the predicted target genes identified for late pregnancy miRNA associated with GA at birth (larger dots represent larger percentages). The top 10 PANTHER pathways based on statistical significance are shown on the y-axis and are ordered from smallest to largest PFDR. Odds ratios from Fisher’s Exact tests are shown on the x-axis. Abbreviations Used: EV, extracellular vesicle; FDR, false discovery rate; GA, gestational age; miRNA, microRNA.
A total of 7,779 unique high-confidence target genes were identified by mirDIP for the 33 miRNA which, in early pregnancy, were associated with GA at birth in male infants (Table S17). These genes were overrepresented (PFDR<0.050) in 66 PANTHER pathways (Table S18). The top 10 pathways included the CCKR signaling map ST pathway, the EGF Receptor signaling pathway, the integrin signaling pathway, angiogenesis, the FGF signaling pathway, the p53 pathway, the Ras pathway, the TGF-beta signaling pathway, the PDGF signaling pathway, and the Wnt signaling pathway (Figure S6). Additionally, 5,638 unique high-confidence target genes were identified by mirDIP for the nine miRNA which, in early pregnancy, were associated with GA at birth in female infants (Table S19). These genes were overrepresented (PFDR<0.050) in 31 PANTHER pathways (Table S20), all of which were nested within the 66 PANTHER pathways identified for males. The top 10 PANTHER pathways for females included the CCKR signaling map ST pathway, the PDGF signaling pathway, the TGF-beta signaling pathway, the Wnt signaling pathway, angiogenesis, the EGF receptor signaling pathway, the integrin signaling pathway, the apoptosis signaling pathway, the PI3 kinase pathway, and the Beta1 adrenergic receptor signaling pathway (Figure S7).
Post-hoc investigation of placental miRNA expression in RICHS
Of the 33 miRNA that, in late pregnancy, were associated with GA at birth in MADRES prior to multiple testing correction (P < 0.050), 28 met the RICHS quality control criteria and were therefore included in the post-hoc investigation (Table 4). All 28 miRNA were expressed in at least 23% of the RICHS placenta samples, and 24 of the miRNA were expressed in 100% of the placenta samples (Table 4). A subset of five miRNA were highly expressed in the placenta, with normalized counts per million ranging in the thousands (miR-16-5p, miR-221-3p) or tens of thousands (miR-126-3p, miR-21-5p, miR-26b-5p) (Table 4).
Table 4.
RICHS placenta expression levels (N = 230) of miRNA associated with GA at birth (P < 0.050) in MADRESa.
| miRNA | N (%) Detectable | Median Count | Count Range |
|---|---|---|---|
| let-7b-5p* | 230 (100.0) | 317 | 132–1,800 |
| let-7d-5p | 230 (100.0) | 93 | 34–1095 |
| miR-126-3p* | 230 (100.0) | 16,085 | 8,629–30,756 |
| miR-142-3p | 230 (100.0) | 101 | 26–503 |
| miR-150–5p | 230 (100.0) | 30 | 11–227 |
| miR-15a-5p* | 230 (100.0) | 330 | 103–994 |
| miR-15b-5p* | 230 (100.0) | 590 | 228–4505 |
| miR-16-5p | 230 (100.0) | 4,016 | 2,187–18,293 |
| miR-188-5p* | 230 (100.0) | 7 | 1–22 |
| miR-199b-5p | 230 (100.0) | 693 | 326–1,279 |
| miR-19b-3p* | 230 (100.0) | 251 | 56–940 |
| miR-20a-5p | 230 (100.0) | 451 | 132–898 |
| miR-20b-5p | 230 (100.0) | 21 | 8–121 |
| miR-21-5p | 230 (100.0) | 74,655 | 39,337–165,849 |
| miR-210-3p* | 230 (100.0) | 29 | 5–114 |
| miR-212-3p* | 122 (53.0) | 1 | 0–18 |
| miR-221-3p | 230 (100.0) | 3,499 | 1,770–5,658 |
| miR-26b-5p | 230 (100.0) | 20,053 | 7,981–31,070 |
| miR-29b-3p* | 230 (100.0) | 393 | 117–1,721 |
| miR-320e | 149 (64.8) | 1 | 0–5 |
| miR-331–3p | 230 (100.0) | 73 | 34–120 |
| miR-374a-5p | 230 (100.0) | 971 | 281–4,855 |
| miR-4454* | 230 (100.0) | 6 | 2–27 |
| miR-548y | 174 (75.7) | 2 | 0–5 |
| miR-551b-3p | 230 (100.0) | 42 | 8–161 |
| miR-584-5p* | 230 (100.0) | 373 | 170–741 |
| miR-656-3p | 230 (100.0) | 69 | 19–177 |
| miR-937-3p | 22.6 | 1 | 0–3 |
aThe percentage of RICHS placenta samples that had detectable levels (normalized count per million>1) of the specified miRNA is shown with the median and range placental count. If a miRNA did not meet the quality control standards for RICHS, as described in the Supplement, it was excluded from the post hoc investigation. *miRNA was significantly associated with GA at birth in MADRES after multiple testing correction (PFDR<0.050).
Abbreviations Used: FDR, false discovery rate; GA, gestational age; MADRES, Maternal and Developmental Risks from Environmental and Social Stressors Study; miRNA, microRNA; RICHS, Rhode Island Children’s Health Study
Discussion
In a cohort of predominantly low-income Hispanic mother-infant pairs in urban Los Angeles, late pregnancy measures of 13 miRNA in maternal circulating EVs were associated with GA at birth. Five of these miRNA were positively associated with GA at birth, while eight were negatively associated with GA at birth, and none of the associations differed by infant sex. In BMI-stratified analyses, we identified an additional miRNA (miR-107) which, in late pregnancy, was positively associated with GA at birth among infants born to obese women. Although early pregnancy measures of EV miRNA were not associated with birth outcomes in primary analyses, sex-dependent associations were identified for early pregnancy measures of 37 miRNA and GA at birth. Neither early nor late pregnancy miRNA were associated with fetal growth measures (BW or BW for GA) after correcting for multiple testing.
Many of the miRNA associated with GA at birth in MADRES have been implicated in adverse birth outcomes or related pregnancy disorders in prior studies. For example, miR-19b-3p, miR-212-3p, miR-126, miR-210, miR-584, miR-188-5p, and miR-29b have previously been associated with preterm birth or preeclampsia [10,28,29,33,36,38,56–61]. Several of these miRNA (e.g. miR-126, miR-212, miR-29b) also play important roles in angiogenesis [60,62,63], a process that is critical for normal placental development and function, which in turn affects gestational duration [64–66]. Consistent with this, we observed that the predicted target genes of the 13 miRNA associated with GA at birth were overrepresented in pathways involved in placental development, including angiogenesis, and pathways important for embryogenesis, such as the Wnt and EGF receptor pathways [67,68].
Few studies have specifically examined EV miRNA in relation to birth outcomes. To our knowledge, the largest to study to widely profile EV miRNA in maternal circulation and investigate relationships with birth outcomes was conducted by Rodosthenous et al. [33]. They profiled >700 miRNA in EVs from second trimester serum samples of 100 women in the PROGRESS cohort and identified 14 that were associated with BW for GA [33]. However, none of the associations remained statistically significant after correcting for multiple testing, and associations with GA at birth were not investigated [33]. A relatively large case-control study by Hromadnikova et al. (N = 64 cases, N = 102 controls) also examined EV miRNA in relation to fetal growth restriction [26]. Using a candidate gene approach, they profiled six miRNA that are abundantly expressed in the placenta in first trimester maternal plasma samples [26]. One of these miRNA (miR-525-5p) was measured and detectable in MADRES, but was not associated with fetal growth. Two very small case-control studies have also profiled EV miRNA using RNA-sequencing and examined relationships with preterm birth [31,32]. One study (N = 11 cases, N = 11 controls) measured EV miRNA in first trimester maternal plasma and reported significant associations for 164 miRNA, but did not account for multiple testing [32]. None of these miRNA were significantly associated with GA at birth in MADRES after multiple testing corrections. The second study by Menon et al. 2019 (N = 10 preterm, N = 20 controls) measured EV miRNA in maternal plasma samples collected at each trimester of pregnancy [31]. After multiple testing corrections, they identified 173 miRNA that changed differentially across gestation in preterm versus term pregnancies. Seven of these miRNA were measured and detectable in MADRES, and several (miR-223-3p, miR-107, miR-4516) were associated with GA at birth either in primary or secondary analyses. This included one miRNA (miR-4516) that in late pregnancy was inversely associated with GA at birth in MADRES and also in a small study (N = 53) that profiled cervical miRNA in mid-pregnancy [12]. This miRNA may therefore play an important role in regulating gestational length.
Although we did not observe significant associations between early pregnancy miRNA measures and birth outcomes when evaluating male and female infants together, sex-dependent associations were identified for early pregnancy measures of 37 miRNA and GA at birth. Twenty-eight of these miRNA were associated with GA at birth in males only, while four were associated with GA at birth in females only. Five miRNA were associated with GA at birth in both males and females but in opposite directions. While many of the miRNA associated with GA at birth differed for males and females, the predicted target genes of these miRNA were enriched in similar PANTHER pathways, which suggests overlapping functions. To our knowledge, only one previous study (by Rodosthenous et al.) has investigated sex differences in miRNA-birth outcome associations [33]. However, this study focused exclusively on fetal growth, thus potential sex differences in associations between miRNA and GA at birth were not examined [33].
In MADRES, the most pronounced sex difference was identified for let-7g-5p (Pinteraction = 9.63 x 10−4), which was positively associated with GA at birth in female infants only. Plasma EV levels of this miRNA were also higher on average in women carrying female, compared with male, fetuses. Interestingly, studies in adults have similarly observed higher total plasma levels of let-7g-5p in women compared with men [69], and sex-dependent associations between serum let-7g-5p and metabolic syndrome have been reported [70]. We also observed sex-specific associations for two additional members of the let-7 family (let-7a-5p and let-7d-5p), which is sensitive to estrogen [71]. Notably, four of the miRNA associated with GA at birth in males only were located on the X chromosome (miR-374a-5p, miR-223-3p, miR-188-5p, miR-888-5p). One of these miRNA (miR-223-3p) has been associated with preterm birth or shorter gestational duration in several prior studies, which measured miR-223-3p in diverse biospecimens, including plasma and peripheral blood mononuclear cells, as well as cervical and amniotic tissues [11,12,35,72,73]. This miRNA is highly expressed in the uterus and has been hypothesized to play important roles in labor induction [74]. In fact, two previous studies have reported higher miR-223 levels in uterine tissue from women undergoing spontaneous term labor deliveries compared with women undergoing term non-labor deliveries [73,74]. MiR-223-3p is also sensitive to oxytocin, a hormone that induces myometrial contractility in both term and preterm labors, and may therefore be important for regulating parturition [75]. We also identified three miRNA (miR-299, miR-1197, and miR-656) associated with GA at birth among males only that are located on the C14MC cluster, which is only found in placental mammals and plays critical roles in placentation and fetal growth [76]. In humans, this miRNA cluster is primarily expressed in the placenta, with higher expression levels reported in early pregnancy [76,77]. Although it is currently unclear why we observed sex differences for early, but not late, pregnancy EV miRNA, one explanation may be that placentation occurs in early pregnancy and is highly dependent on maternal-fetal crosstalk, which differs by fetal sex [78,79].
Generally, we did not observe differences in miRNA-birth outcome associations according to maternal pre-pregnancy BMI, although we did identify one miRNA (miR-107) that was positively associated with GA at birth in infants born to obese women only. One previous study similarly observed an association between miR-107 and preterm birth, but they did not investigate differences by maternal BMI [31]. Although the mechanism by which miR-107 may differentially impact GA at birth according to maternal obesity status is unknown, Toll-like receptors may play a role, as they are known to repress miR-107 expression and are upregulated in obesity [80]. Prior studies have also reported that miR-107 is dysregulated in rodent models of obesity and diabetes and that this miRNA regulates insulin sensitivity and lipid storage in human adipocytes [80,81]. In a sheep model, maternal obesity has also been associated with increased hepatic miR-107 expression in the offspring [82]. There is therefore growing evidence that miR-107 is influenced by obesity status and may also be involved in the development of obesity.
In a post hoc investigation, we observed that the majority of miRNA associated with GA at birth in MADRES were detectable in placenta samples from the RICHS cohort. Five of these miRNA (miR-126-3p, miR-16-5p, miR-21-5p, miR-221-3p, miR-26b-5p) were highly expressed in the placenta, with normalized counts per million ranging in the thousands and tens of thousands. A study in Australia has similarly reported high placental expression levels of these miRNA [83]. It is therefore possible that some of the EV miRNA associated with gestational duration in MADRES originated in the placenta. This would be consistent with previous studies, which have estimated that approximately 20% of EVs in maternal circulation during late pregnancy are derived from the placenta [22], possibly to facilitate maternal-placental-fetal crosstalk [23,24].
Our study had many notable strengths. To our knowledge, this is the largest study to examine maternal circulating miRNA in relation to birth outcomes, which allowed us to investigate important modifiers of these relationships, including infant sex and maternal pre-pregnancy BMI, which have rarely been investigated in this context. This is also one of few studies to specifically focus on EV miRNA, which play important roles in intercellular, and potentially maternal-fetal, communication [23,24]. We also used the NanoString nCounter platform to profile 800 different miRNA in maternal circulating EVs. This platform covers 100% of the high-confidence miRNA annotated by miRBase, allowing us to comprehensively investigate EV miRNA in relation to birth outcomes. A novel aspect of our study was the comparison of EV miRNA in both early and late pregnancy to identify potential windows of importance. Our focus on a predominantly low-income Hispanic pregnancy cohort is also a major strength of the study, as epigenetic research in more diverse populations is greatly needed. However, this may limit the generalizability of our findings. It is also important to acknowledge that we were underpowered to examine preterm birth, small for gestational age, or large for gestational age as outcomes due to the small number of participants in each of these categories. Furthermore, while we were able to investigate possible differences in the miRNA-GA at birth associations based on delivery type (i.e. vaginal versus caesarean section deliveries), we did not have sufficient statistical power to investigate differences by delivery sub-types, such as planned versus unplanned caesarean section deliveries, which merits future investigation.
In conclusion, we identified 13 miRNA in maternal circulating EVs, measured in late pregnancy, that were associated with GA at birth in a health disparity pregnancy cohort. In a post hoc investigation using data from the RICHS cohort, we observed that the majority of these miRNA are expressed in the placenta. In secondary analyses, we identified a miRNA (miR-107) which, in late pregnancy, was associated with GA at birth only among infants born to obese women. We also identified 37 miRNA which, in early pregnancy, were associated with GA at birth in a sex-dependent manner, including several miRNA that are predominantly expressed in the placenta. Our findings suggest that A) EV miRNA in both early and late pregnancy contribute to gestational duration, B) important sex differences may exist in early pregnancy, and C) a subset of these miRNA may originate in the placenta. Additional studies in diverse populations are needed to determine if these miRNA may serve as early and minimally invasive biomarkers of gestational duration and to elucidate the mechanisms by which these miRNA may influence gestational length.
Supplementary Material
Acknowledgments
We would like to thank the MADRES participants, the study staff, and our community clinic partners for their many contributions. The work described in this paper was supported by NIH grants R01 MD011698, R00 ES030400, P50 ES026086, P50 MD015705, 4UH3 OD023287-03, P30 ES007048, R01 ES025145, and EPA grant 83615801-0. The content is solely the responsibility of the authors and does not necessarily represent the official views of these funding organizations.
Funding Statement
This work was supported by the NIH Office of the Director [4UH3OD023287-03]; National Institute of Environmental Health Sciences [P30 ES007048]; National Institute of Environmental Health Sciences [R00 ES030400]; National Institute of Environmental Health Sciences [P50 ES026086]; National Institute of Environmental Health Sciences [R01 ES025145]; National Institute on Minority Health and Health Disparities [R01 MD011698]; National Institute on Minority Health and Health Disparities [P50 MD015705]; U.S. Environmental Protection Agency [83615801-0].
Disclosure statement
The authors declare no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Boyle EM, Poulsen G, Field DJ, et al. Effects of gestational age at birth on health outcomes at 3 and 5 years of age: population-based cohort study. Bmj. 2012;344:e896. mar01 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Beauregard JL, Drews-Botsch C, Sales JM, et al. Preterm birth, poverty, and cognitive development. Pediatrics. 2018;141(1). 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Markopoulou P, Papanikolaou E, Analytis A, et al. Preterm birth as a risk factor for metabolic syndrome and cardiovascular disease in adult life: a systematic review and meta-analysis. J Pediatr. 2019;210:69–80. e5. [DOI] [PubMed] [Google Scholar]
- [4].McCormick MC. The contribution of low birth weight to infant mortality and childhood morbidity. N Engl J Med. 1985;312(2):82–90. [DOI] [PubMed] [Google Scholar]
- [5].Oudgenoeg-Paz O, Mulder H, Jongmans MJ, et al., Van der Stigchel S. The link between motor and cognitive development in children born preterm and/or with low birth weight: a review of current evidence. Neurosci Biobehav Rev. 2017;80:382–393. [DOI] [PubMed] [Google Scholar]
- [6].Visentin S, Grumolato F, Nardelli GB, et al. Early origins of adult disease: low birth weight and vascular remodeling. Atherosclerosis. 2014;237(2):391–399. [DOI] [PubMed] [Google Scholar]
- [7].Ilekis JV, Tsilou E, Fisher S, et al. Placental origins of adverse pregnancy outcomes: potential molecular targets: an executive workshop summary of the Eunice Kennedy Shriver national institute of child health and human development. Am J Obstet Gynecol. 2016;215(1):S1–S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cai M, Kolluru GK, Ahmed A. Small molecule, big prospects: microRNA in pregnancy and its complications. J Pregnancy. 2017;2017. 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Elovitz MA, Brown AG, Anton L, et al. Distinct cervical microRNA profiles are present in women destined to have a preterm birth. Am J Obstet Gynecol. 2014;210(3):221.e1-. e11. [DOI] [PubMed] [Google Scholar]
- [10].Gródecka-Szwajkiewicz D, Ulańczyk Z, Zagrodnik E, et al. Differential secretion of angiopoietic factors and expression of microRNA in umbilical cord blood from healthy Appropriate-For-Gestational-Age preterm and term Newborns—in search of biomarkers of Angiogenesis-Related processes in preterm birth. Int J Mol Sci. 2020;21(4):1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Winger EE, Reed JL, Ji X, et al. MicroRNAs isolated from peripheral blood in the first trimester predict spontaneous preterm birth. PloS One. 2020;15(8):e0236805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sanders AP, Burris HH, Just AC, et al. microRNA expression in the cervix during pregnancy is associated with length of gestation. Epigenetics. 2015;10(3):221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jiang H, Wu W, Zhang M, et al. Aberrant upregulation of miR-21 in placental tissues of macrosomia. J Perinatol. 2014;34(9):658–663. [DOI] [PubMed] [Google Scholar]
- [14].Zhang JT, Cai QY, Ji SS, et al. Decreased miR-143 and increased miR-21 placental expression levels are associated with macrosomia. Mol Med Rep. 2016;13(4):3273–3280. [DOI] [PubMed] [Google Scholar]
- [15].Li J, Chen L, Tang Q, et al. The role, mechanism and potentially novel biomarker of microRNA-17-92 cluster in macrosomia. Sci Rep. 2015;5:17212. 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rodil-Garcia P, Edc A-L, Montoya-Contreras A, et al. Analysis of microRNA expression in newborns with differential birth weight using newborn screening cards. Int J Mol Sci. 2017;18(12):2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kennedy EM, Hermetz K, Burt A, et al. Placental microRNA expression associates with birthweight through control of adipokines: results from two independent cohorts. Epigenetics. 2020:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Salomon C, Torres MJ, Kobayashi M, et al. A gestational profile of placental exosomes in maternal plasma and their effects on endothelial cell migration. PloS One. 2014;9(6):e98667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sarker S, Scholz-Romero K, Perez A, et al. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J Transl Med. 2014;12(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mitchell MD, Peiris HN, Kobayashi M, et al. Placental exosomes in normal and complicated pregnancy. Am J Obstet Gynecol. 2015;213(4):S173–S81. [DOI] [PubMed] [Google Scholar]
- [22].Elfeky O, Longo S, Lai A, et al. Influence of maternal BMI on the exosomal profile during gestation and their role on maternal systemic inflammation. Placenta. 2017;50:60–69. [DOI] [PubMed] [Google Scholar]
- [23].Buca D, Bologna G, D’Amico A, et al. Extracellular vesicles in Feto–Maternal crosstalk and pregnancy disorders. Int J Mol Sci. 2020;21(6):2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bowman CE, Arany Z, Wolfgang MJ. Regulation of maternal–fetal metabolic communication. Cellular and molecular life sciences. 2020:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Salomon C, Guanzon D, Scholz-Romero K, et al. Placental exosomes as early biomarker of preeclampsia: potential role of exosomal microRNAs across gestation. J Clin Endocrinol Metab. 2017;102(9):3182–3194. [DOI] [PubMed] [Google Scholar]
- [26].Hromadnikova I, Dvorakova L, Kotlabova K, et al. The prediction of gestational hypertension, preeclampsia and fetal growth restriction via the first trimester screening of plasma exosomal C19MC microRNAs. Int J Mol Sci. 2019;20(12):2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Devor E, Santillan D, Scroggins S, et al. Trimester-specific plasma exosome microRNA expression profiles in preeclampsia. J Matern Fetal Neonatal Med. 2020;33(18):3116–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Biró O, Alasztics B, Molvarec A, et al. Various levels of circulating exosomal total-miRNA and miR-210 hypoxamiR in different forms of pregnancy hypertension. Pregnancy Hypertens. 2017;10:207–212. [DOI] [PubMed] [Google Scholar]
- [29].Sandrim V, Luizon M, Palei A, et al. Circulating micro RNA expression profiles in pre‐eclampsia: evidence of increased miR‐885‐5p levels. BJOG. 2016;123(13):2120–2128. [DOI] [PubMed] [Google Scholar]
- [30].Gillet V, Ouellet A, Stepanov Y, et al. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J Clin Endocrinol Metab. 2019;104(11):5157–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Menon R, Debnath C, Lai A, et al. Circulating exosomal miRNA profile during term and preterm birth pregnancies: a longitudinal study. Endocrinology. 2019;160(2):249–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fallen S, Baxter D, Wu X, et al. Extracellular vesicle RNA s reflect placenta dysfunction and are a biomarker source for preterm labour. J Cell Mol Med. 2018;22(5):2760–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rodosthenous RS, Burris HH, Sanders AP, et al. Second trimester extracellular microRNAs in maternal blood and fetal growth: an exploratory study. Epigenetics. 2017;12(9):804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Higashijima A, Miura K, Mishima H, et al. Characterization of placenta‐specific microRNAs in fetal growth restriction pregnancy. Prenat Diagn. 2013;33(3):214–222. [DOI] [PubMed] [Google Scholar]
- [35].Gray C, McCowan LM, Patel R, et al. Maternal plasma miRNAs as biomarkers during mid-pregnancy to predict later spontaneous preterm birth: a pilot study. Sci Rep. 2017;7(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cook J, Bennett PR, Kim SH, et al. First trimester circulating microRNA biomarkers predictive of subsequent preterm delivery and cervical shortening. Sci Rep. 2019;9(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mouillet J-F, Chu T, Hubel CA, et al. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31(9):781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Whitehead CL, Teh WT, Walker SP, et al. Circulating MicroRNAs in maternal blood as potential biomarkers for fetal hypoxia in-utero. PLoS One. 2013;8(11):e78487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Elovitz MA, Anton L, Bastek J, et al. Can microRNA profiling in maternal blood identify women at risk for preterm birth? Am J Obstet Gynecol. 2015;212(6):782.e1-. e5. [DOI] [PubMed] [Google Scholar]
- [40].Bastain TM, Chavez T, Habre R, et al. Study design, protocol and profile of the Maternal And Developmental Risks from Environmental and Social Stressors (MADRES) pregnancy cohort: a prospective cohort study in predominantly low-Income Hispanic women in urban Los Angeles. BMC Pregnancy Childbirth. 2019;19(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Howe CG, Claus Henn B, Eckel SP, et al. Prenatal metal mixtures and birth weight for gestational age in a predominantly lower-Income Hispanic pregnancy cohort in Los Angeles. Environ Health Perspect. 2020;128(11):117001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aris IM, Kleinman KP, Belfort MB, et al. 2017 US reference for singleton birth weight percentiles using obstetric estimates of gestation. Pediatrics. 2019;144(1):e20190076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Koren G, Boskovic R, Hard M, et al. (pregnancy-unique quantification of emesis and nausea) scoring system for nausea and vomiting of pregnancy. Am J Obstet Gynecol. 2002;186(5):S228–S31. [DOI] [PubMed] [Google Scholar]
- [44].ACo O, Gynecologists. gestational hypertension and preeclampsia: ACOG practice bulletin, number 222. Obstetrics and gynecology. 2020;135(6):e237–e60. [DOI] [PubMed] [Google Scholar]
- [45].Team RCR: A language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria. 2020 [Google Scholar]
- [46].Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- [47].Blighe K, Rana S, Lewis M. EnhancedVolcano:Publication-ready volcano plots with enhanced colouring and labeling. R Package Version. 2019;1. [Google Scholar]
- [48].Waggott D, Chu K, Yin S, et al. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28(11):1546–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Miura K, Miura S, Yamasaki K, et al. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem. 2010;56(11):1767–1771. [DOI] [PubMed] [Google Scholar]
- [50].Sanders AP, Burris HH, Just AC, et al. Altered miRNA expression in the cervix during pregnancy associated with lead and mercury exposure. Epigenomics. 2015;7(6):885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Venables WNR. Modern Applied Statistics with S. Springer, New York: 2002. [Google Scholar]
- [52].Tokar T, Pastrello C, Rossos AE, et al. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46(D1):D360–D70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mi H, Poudel S, Muruganujan A, et al. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44(D1):D336–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ulgen E, Ozisik O, Sezerman OU. pathfindR: an R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front Genet. 2019;10:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tang Y, Ji H, Liu H, et al. Pro-inflammatory cytokine-induced microRNA-212-3p expression promotes myocyte contraction via methyl-CpG-binding protein 2: a novel mechanism for infection-related preterm parturition. Mol Hum Reprod. 2019;25(5):274–282. [DOI] [PubMed] [Google Scholar]
- [57].Ortiz-Dosal A, Del Carmen Arellanes-licea E, Rodil-García P, et al. Circulating microRNAs overexpressed in macrosomia: an experimental and bioinformatic approach. J Dev Orig Health Dis. 2020:1–9. [DOI] [PubMed] [Google Scholar]
- [58].Tang Y, Ji H, Liu H, et al. Identification and functional analysis of microRNA in myometrium tissue from spontaneous preterm labor. Int J Clin Exp Pathol. 2015;8(10):12811. [PMC free article] [PubMed] [Google Scholar]
- [59].Yang S, Li H, Ge Q, et al. Deregulated microRNA species in the plasma and placenta of patients with preeclampsia. Mol Med Rep. 2015;12(1):527–534. [DOI] [PubMed] [Google Scholar]
- [60].Li P, Guo W, Du L, et al. microRNA-29b contributes to pre-eclampsia through its effects on apoptosis, invasion and angiogenesis of trophoblast cells. Clin Sci. 2013;124(1):27–40. [DOI] [PubMed] [Google Scholar]
- [61].Goldenberg RL, Culhane JF, Iams JD, et al. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kumarswamy R, Volkmann I, Beermann J, et al. Vascular importance of the miR-212/132 cluster. Eur Heart J. 2014;35(45):3224–3231. [DOI] [PubMed] [Google Scholar]
- [63].Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Huppertz B, Peeters LL. Vascular biology in implantation and placentation. Angiogenesis. 2005;8(2):157–167. [DOI] [PubMed] [Google Scholar]
- [65].Salafia CM, Maas E, Thorp JM, et al. Measures of placental growth in relation to birth weight and gestational age. Am J Epidemiol. 2005;162(10):991–998. [DOI] [PubMed] [Google Scholar]
- [66].Morgan TK. Role of the placenta in preterm birth: a review. Am J Perinatol. 2016;33(3):258–266. [DOI] [PubMed] [Google Scholar]
- [67].Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018;145(11):dev146589. [DOI] [PubMed] [Google Scholar]
- [68].Dackor J, Caron KM, Threadgill DW. Placental and embryonic growth restriction in mice with reduced function epidermal growth factor receptor alleles. Genetics. 2009;183(1):207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wu J, Cai H, Xiang Y-B, et al. Intra-individual variation of miRNA expression levels in human plasma samples. Biomarkers. 2018;23(4):339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang Y-T, Tsai P-C, Liao Y-C, et al. Circulating microRNAs have a sex-specific association with metabolic syndrome. J Biomed Sci. 2013;20(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Klinge CM miRNAs and estrogen action. Trends Endocrinol Metab. 2012;23(5):223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Enquobahrie DA, Hensley M, Qiu C, et al. Candidate gene and microRNA expression in fetal membranes and preterm delivery risk. Reprod Sci. 2016;23(6):731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hassan SS, Romero R, Pineles B, et al. MicroRNA expression profiling of the human uterine cervix after term labor and delivery. Am J Obstet Gynecol. 2010;202(1):80.e1-. e8. [DOI] [PubMed] [Google Scholar]
- [74].Ackerman IVWE, Buhimschi IA, Brubaker D, et al. Integrated microRNA and mRNA network analysis of the human myometrial transcriptome in the transition from quiescence to labor. Biol Reprod. 2018;98(6):834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cook JR, MacIntyre DA, Samara E, et al. Exogenous oxytocin modulates human myometrial microRNAs. Am J Obstet Gynecol. 2015;213(1):65.e1-. e9. [DOI] [PubMed] [Google Scholar]
- [76].Malnou EC, Umlauf D, Mouysset M, et al. Imprinted MicroRNA gene clusters in the evolution, development, and functions of mammalian placenta. Front Genet. 2019;9:706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Morales-Prieto D, Chaiwangyen W, Ospina-Prieto S, et al. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33(9):725–734. [DOI] [PubMed] [Google Scholar]
- [78].Sun T, Gonzalez TL, Deng N, et al. Sexually dimorphic crosstalk at the maternal-fetal interface. J Clin Endocrinol Metab. 2020;105(12):dgaa503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gonzalez TL, Sun T, Koeppel AF, et al. Sex differences in the late first trimester human placenta transcriptome. Biol Sex Differ. 2018;9(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Foley NH, OˈNeill LA. miR‐107: a Toll‐like receptor‐regulated miRNA dysregulated in obesity and type II diabetes. J Leukoc Biol. 2012;92(3):521–527. [DOI] [PubMed] [Google Scholar]
- [81].Ahonen MA, Haridas PN, Mysore R, et al. miR-107 inhibits CDK6 expression, differentiation, and lipid storage in human adipocytes. Mol Cell Endocrinol. 2019;479:110–116. [DOI] [PubMed] [Google Scholar]
- [82].Nicholas LM, Rattanatray L, MacLaughlin SM, et al. Differential effects of maternal obesity and weight loss in the periconceptional period on the epigenetic regulation of hepatic insulin‐signaling pathways in the offspring. Faseb J. 2013;27(9):3786–3796. [DOI] [PubMed] [Google Scholar]
- [83].Smith MD, Pillman K, Jankovic-Karasoulos T, et al. Large-scale transcriptome-wide profiling of microRNAs in human placenta and maternal plasma at early to mid gestation. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


