ABSTRACT
The gut microbiota was emerging as critical regulatory elements in shaping the outcome of cancer immunotherapy. However, the underlying mechanisms by which the gut commensal species enhance antitumor immunity remain largely unexplored. Here, we show that the gut microbiota from healthy individuals conferred considerable sensitivity to anti-PD-1 in the colorectal cancer (CRC) tumor-bearing mice, whereas gut microbiota from CRC patients failed to do so. By 16S rRNA gene sequencing, we identified Lactobacillus that was significantly increased in the mice with good response to anti-PD-1, and significantly correlated with anti-tumor immunity. After a series of screening, we isolated a novel Lacticaseibacillus strain, named L. paracasei sh2020. L. paracasei sh2020 showed the most notable anti-tumor immunity in the mice with gut dysbiosis. Mechanistically, the antitumor immune response elicited by L. paracasei sh2020 was dependent on CD8+ T cell. In vitro and in vivo studies revealed that L. paracasei sh2020 stimulation triggered the upregulated expression of CXCL10 in the tumors and subsequently enhanced CD8+ T cell recruitment. Meanwhile, the modulation of gut microbiota caused by L. paracasei sh2020 enhanced its antitumor effect and gut barrier function. Overall, our study offered novel insights into the mechanism by which gut microbiota shaped the outcome of cancer immunotherapy and, more importantly, the novel strain L. paracasei sh2020 might serve as an easy and effective way to promote anti-PD-1 effect in clinical practice.
KEYWORDS: Colorectal cancer, immunotherapy, anti-PD-1, gut microbiota, probiotics, tumor microenvironment
Introduction
Colorectal cancer (CRC) remains one of the common digestive tract tumors with increasing incidence and mortality worldwide. The prognosis of CRC remains dismal despite improved diagnostic and treatment strategies. In recent years, immunotherapy has become an emerging way in the treatment of solid tumors,1 which harness the immune system to produce anti-tumor effects. Anti-programmed cell death 1 (anti-PD-1) immunotherapy, as novel immunotherapy drug, has achieved favorable results in several advanced cancers.2,3 In contrast to other tumor types, CRC patients who benefit from anti-PD-1 are dismally limited.4,5 Therefore, how to make more CRC patients benefit from immunotherapy remains a hot topic in this field.
Recently, gut microbiota has fueled great enthusiasm in immunotherapy. Our recent publication has demonstrated that targeting gut microbiota using dietary fiber facilitated the anti-PD-1 efficacy in the gut humanized avatar mouse models.6 Moreover, the dietary fiber shifts the gut microbial community toward enrichment in Akkermansia, Lactobacillus and Bifidobacterium, which were significantly associated with good response to anti-PD-1 therapy.7–9 More recently, Mager L.F. et al isolated three bacterial species, including B. pseudolongum, L. johnsonii and Olsenella, and found that oral administration of these species could significantly enhance the therapeutic effect of immune checkpoint inhibitors in tumor models.10
Normal gut microbiota is a huge but relatively unexplored treasure trove.11 Although some specific bacteria have been associated with increased anti-tumor immunity, the research about single gut bacteria in immunotherapy is still far from experimentally clarified.12 Furthermore, the exact molecular mechanisms through which these gut microbes swayed immunotherapy has rarely been studied.13 In this study, we found that Lactobacillus was significantly associated with good response to anti-PD-1 in the tumor models of CRC. After a series of screening, we isolated and identified a novel Lactobacillus strain, named L. paracasei sh2020, which showed the most notable anti-tumor immunity in the gut humanized avatar mouse models. Subsequently, we put more efforts on the underlying mechanisms of the antitumor immunity induced by L. paracasei sh2020. Additionally, the favorable effects of L. paracasei sh2020 on gut homeostasis were confirmed in the context of anti-PD-1 therapy.
Results
Gut microbiota from healthy donors but not from CRC patients improved the efficacy of anti‑PD-1 in recipient mice
Five healthy donors (HD) and newly diagnosed CRC patients were collected from our institute. The detailed characteristics of all participants were listed in Table S1. Compared to HD, the CRC patients were characterized by decreased α-diversity (Figure S1a-b), distinct β-diversity (Figure S1c) as well as disturbed gut microbiota profiles (Figure S1d-f) consistent with recent studies.14,15 To determine whether and if so, to what extent, gut microbiota has influenced the response to anti-PD-1, we took advantage of avatar mouse models, which were gavaged with a broad-spectrum antibiotic cocktail (ATB) to depleted endogenous microbiota and subsequently humanized with feces from HD or CRC donors using fecal microbial transplantation (FMT). Such “humanized” mice were inoculated with syngeneic MC38 cells as described in our recent publication to investigate the role of gut microbiota in anti-PD-1 therapy (Figure 1a).6 As shown in Figure 1b, anti-PD-1 differently influenced tumor progression in an individual group of mice. Tumor growth was considerably inhibited in mouse recipients of healthy microbiota compared to mouse recipients of CRC microbiota. Specifically, in an individual group of mice, three individual groups (CRC1, CRC2, and CRC4) resulted in faster tumor growth, showing an impaired efficiency of anti-PD-1, while in the mice colonized with microbiota from HD, two individuals (HD1 and HD2) recipient mice almost completely abrogated the tumor progression (Figure 1c). Furthermore, increased CD4+ cells and CD8+ cells, and decreased Foxp3+ cells were observed in the tumors of the HD group than that of the CRC group, thereby fostering “hot” tumors (Figure 1d).
Figure 1.
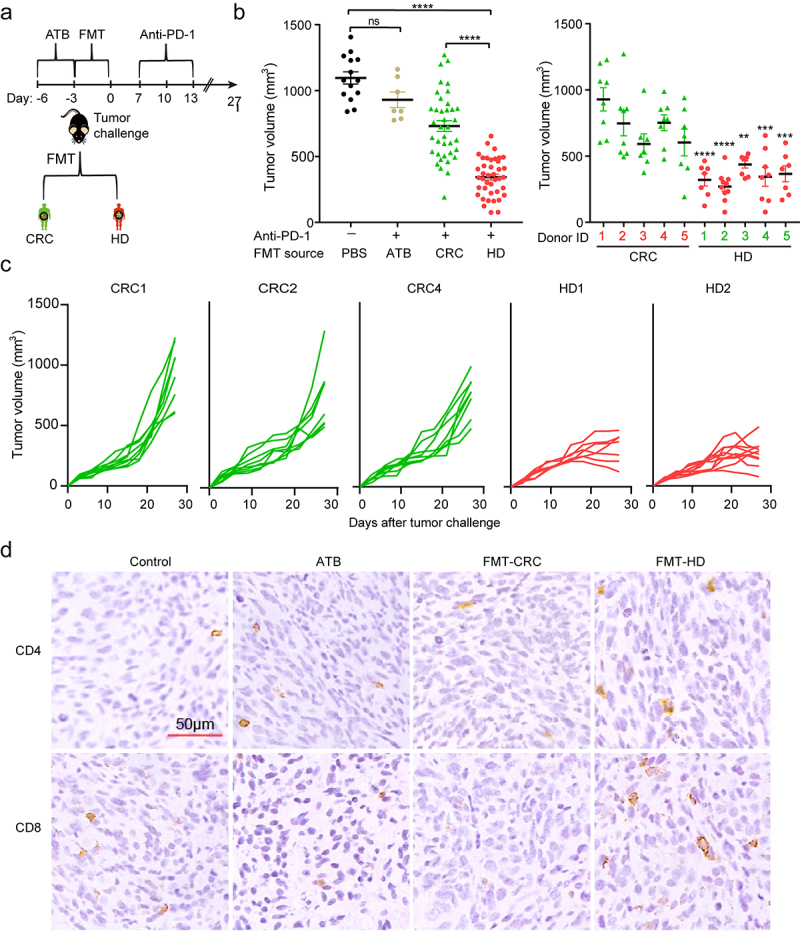
The gut microbiota from healthy donors, but not from CRC patients, improved therapeutic efficacy of anti‑PD‑1. (a) Experimental design: fecal microbiota transplantation (FMT) was performed after 3 days of ATB administration in mice. Five days later, MC38 cells were inoculated subcutaneously and anti-PD-1 was intraperitoneally administered every 3 days starting on day 7, in total three times. (b) The tumor volume of pooled groups of mice (left) and individual groups of mice (right) receiving gut microbiota from the CRC patients (CRC1-5) or healthy individuals (HD1-5) (n = 8–10). **P < .01, ***P < .001, ****P < .001 versus control (pooled data). (c) Individual tumor growth curves of the mice receiving gut microbiota from CRC patients (CRC1, CRC2, and CRC4) or healthy individuals (HD1, HD2) during treatment with anti-PD-1. (d) Representative images of IHC staining of CD4, and CD8 for tumors from each group.
Collectively, gut microbiota from CRC tended to suppress the efficacy of anti-PD-1 treatment, which might partly explain the low response to anti-PD-1 among CRC patients. On the other hand, gut microbiota from HD conferred sensitivity to anti-PD-1, providing the possibility that there existed effector bacteria in the gut, which might play a dominant role in facilitating anti-PD-1 therapy.
Distinct gut microbiota development in recipient mice colonized with fecal microbiota from CRC and healthy donors
We conducted 16S rRNA sequencing to analyze the fecal samples collected from mice at day 10 and 30, respectively. The number of observed OTUs was significantly higher in the FMT-HD group at days 30 (Figure 2a-b), which reflected a higher community richness of FMT-HD. Upon analyzing β-diversity across groups, the overall community at day 10 revealed a distinct clustering pattern between the two groups (Figure 2c) and was sustained at 30 days (Figure 2d). We then assessed the landscape of gut microbiota overtime in two groups. No significant change was observed among the most abundant phyla, including Firmicutes and Bacteroidetes (Figure 2e). However, the Firmicutes/Bacteroidetes ratio in FMT-HD group was significantly higher than that in the FMT-CRC group, suggesting a better capacity to maintain normal gut homeostasis (Figure 2f). 16,17 At the lower taxonomic level, FMT-HD group showed restoration toward gut normobiosis. For instance, several well-known commensals used in probiotic preparations,18,19 including Lactobacillaceae, Bifidobacteriaceae, Erysipelotrichaceae, and Ruminococcaceae, which are immunomodulatory SCFA-producing taxa, and associated with healthy gut ecosystems,20 increased upon FMT from healthy individuals at day 10. At day 30, the relative abundance of Lactobacillaceae, Bifidobacteriaceae, and Ruminococcaceae increased progressively in the FMT-HD group (Figure 2g). These findings demonstrated the distinct gut microbiota development between the recipient mice colonized with fecal from CRC and HD.
Figure 2.

The gut microbiota from healthy donors restored normobiotic ecology of gut microbiota. (a-b) Changes in the α-diversity between the FMT-CRC and FMT-HD group was determined using the OUT numbers (a), and Chao index (b) (n = 7). (c-d) PCoA of β-diversity using the Bray-Curtis dissimilarity metric among samples of individual groups of mice in each group at day 10 (c), and day 30 (d). (e) Component proportion of bacterial phylum in each group. (f) Firmicutes-to-Bacteroidetes ratio in individual groups of mice in each group at days 30. (g) Comparison of relative abundance of bacterial class level between FMT-CRC (inner rings) and FMT-HD group (outer ring) at days 10 (left), and 30 (right). *P < .05.
Gut microbial signature potentially associated with anti-PD-1 efficacy
To identify the microbial taxa that might be potentially associated with the improved efficacy of anti-PD-1, we performed a differential taxa analysis between the two groups. In total, 10 taxa were differentially abundant at the genus level. Notably, FMT-HD group consistently exhibited increased relative abundance of Lactobacillus (Figure 3a). Furthermore, Lactobacillus was negatively correlated with almost all other genera, with different relative abundances between the two groups (Figure 3b). A microbial signature with high accuracy in predicting the outcomes of anti-PD-1 therapy was identified using selbal.21 This signature consists of the two genera, Lactobacillus and Phascolarctobacterium as a numerator, and Akkermansia and Bacteroides as a denominator (Figure 3c). More importantly, the signature produced an area under the receiver operating characteristic curve (AUC) of 1.00 (Figure 3d). A positive balance value in the FMT-HD group suggested that the two genera in the numerator had a much higher abundance than that in the denominator (Akkermansia and Bacteroides). Lactobacillus, the abundance of which increased most in the FMT-HD group, were also ranking as the top three most important taxon to the microbiota community (Figure 3e).
Figure 3.
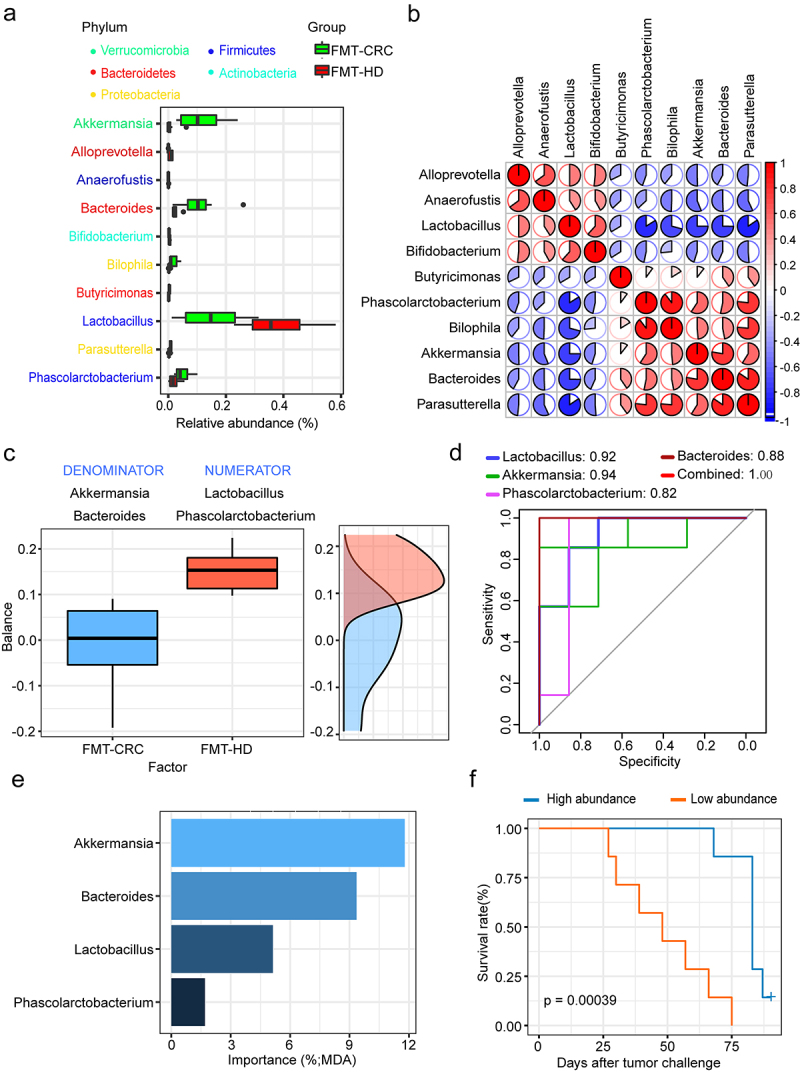
Lactobacillus was positively associated with antitumor immune response. (a) Differential relative abundance of taxa at the genus level between the FMT-CRC and FMT-HD group (n = 7). (b) Relationship of genera with different abundances. Lactobacillus was negatively correlated with almost all other genera with different relative abundances. (c) Gut microbial signature associated with anti-PD-1 outcomes in mice. (d) ROC analysis of the power of four genera as predictive of anti-PD-1 outcomes. (e) The ranking of four genera by their importance to the gut microbial community. (f) Kaplan-Meier survival analysis based on the abundance of Lactobacillus in the tumor-bearing mice (n = 14). MDA, mean decrease in accuracy.
We then stratified all gut humanized mice based on the median relative abundance of Lactobacillus. Kaplan–Meier analysis showed significantly longer survival that was observed in the Lactobacillus high group (Figure 3f). Indeed, recent studies demonstrate that some strains of Lactobacillus can modulate the immune response against tumors in murine model.22,23 Collectively, we thus hypothesized that members of this genus might represent a major component underlying the improved antitumor immune effects in FMT-HD group.
Lactobacillus cocktail improved response to anti-PD-1 in tumor-bearing mice
To determine causality for at least one Lactobacillus member, all stools of the healthy donors were cultured with Man Rogosa Sharpe medium, which is a selective cultural medium designed to favor the Lactobacillus growth. As a result, three bacterial species, including L. reuteri, L. plantarum, and L. paracasei, were able to be cultured and identified by 16S rRNA. We thus created a live cocktail with the three species and administered it via oral gavage to gut humanized mice using the fecal from CRC1 donor (Figure 4a). Notably, this cocktail significantly reversed the impaired efficacy of anti-PD-1 observed in the recipient mice (Figure 4b-d).
Figure 4.
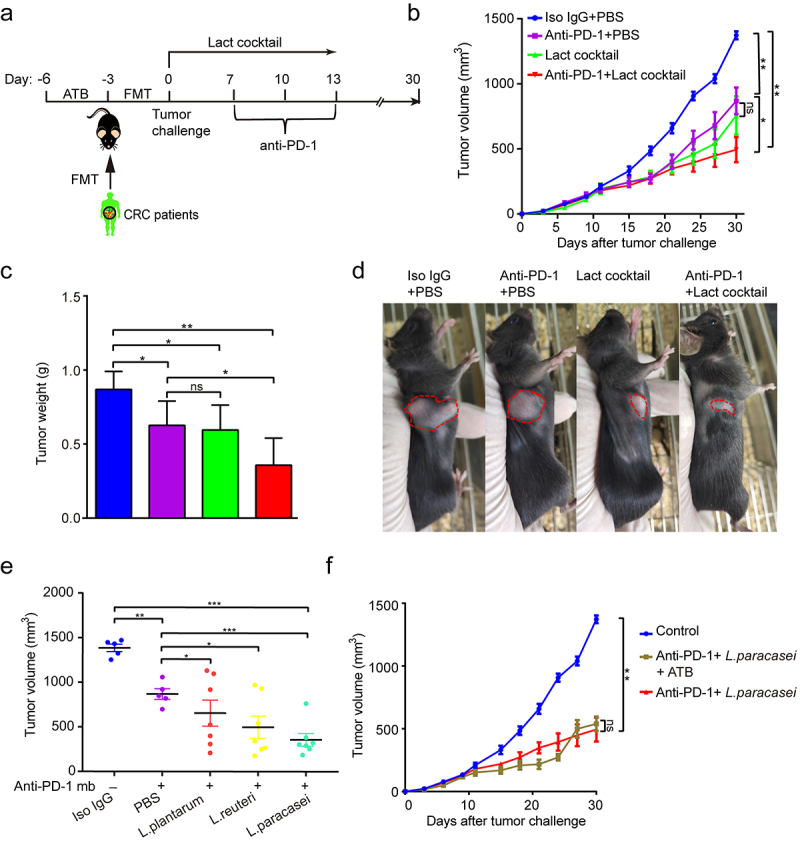
Lactobacillus cocktail improved response to anti-PD-1 in the mice with gut dysbiosis. (a) Experimental design: FMT from C1 patient was conducted after 3 days of ATB treatment in SPF mice. After the FMT complement, the tumor-bearing mice were orally administrated with Lactobacillus cocktail once a day for 10 days after tumor challenge. (b) Tumor growth from the mice after treatment with isotype IgG, or anti-PD-1 alone or Lactobacillus cocktail alone, anti-PD-1 combined with Lactobacillus cocktail (n = 6–7). (c) Tumor weight of the tumor-bearing mice in each group (n = 6–7). (d) Representative photograph of tumors on day 20 after tumor challenge. (e) Tumor volumes from the tumor-bearing mice receiving different strains. (f) Tumor growth in the tumor-bearing mice pre-treated with antibiotic cocktail (n = 6–7). ns, no significant difference, *P < .05, **P < .01, ***P < .001.
Among this cocktail, we further mined the functional strain that improved the efficacy of anti-PD-1. As shown in Figure 4e, the isolated L. plantarum had no apparent effect on the tumor growth when combined with anti-PD-1. The isolated L. reuteri showed mild anti-tumor effects. In contrast, the isolated L. paracasei elicited the most notable anti-tumor ability, demonstrating that L. paracasei was likely the key functional strain responsible for the enhanced anti-tumor immunity. Interestingly, the isolated L. paracasei retained its favorable effect in controlling the tumor growth in the mice basically depleted of gut microbiota (Figure 4f). In addition, neither morphological nor pathological damage was found in major organs (Figures S2), indicating that the isolated L. paracasei did not exert a significant adverse influence in normal tissues. Then, the whole genome of the isolated L. paracasei was sequenced and assembled using Oxford Nanopore combined with Illumina high throughput next-generation sequencing technologies.24 The genome comparative analyses among the isolated L. paracasei and other 26 L.casei strains available in the NCBI database were presented in Figures S3 and Table S2, suggesting that the isolated L. paracasei was a novel strain, and thus we named it L. paracasei sh2020.
Transcriptome analysis revealed upregulation of T cell trafficking-associated genes within tumors following L. paracasei sh2020
To explore the potential mechanisms of L. paracasei sh2020, we performed RNA-seq analysis of tumor tissues harvested from the mice treated with vehicle, or L. paracasei sh2020. First, GSEA analysis identified high-level categories, such as regulation of T cell mediated immunity, leukocyte migration, positive regulation of adaptive immune response, were enriched in the L. paracasei sh2020-treated tumors (Figure 5a). Notably, this group promoted the expression of T cell chemoattractant chemokines (Figure 5b-c), such as CXCL9, CXCL10, and CXCL11, previously described to be involved in modulating the tumor immune environment.25 Next, the RNA-seq data were then analyzed to estimate the tumor-infiltrating immune cell. We also found that there was a greater proportion of CD8+ T cells in the L. paracasei sh2020-treated tumors (Figure S4). These results suggested that L. paracasei sh2020 might elicit chemokines expression in the tumors, which ultimately promoted CD8+ T cell infiltration into the tumor beds.
Figure 5.

Transcriptome analysis revealed the increased T cell trafficking-associated genes following L. paracasei sh2020. (a) Enriched gene sets in L. paracasei sh2020-treated tumors identified by GSEA (n = 5). (b-c) GSEA enrichment plots of positive regulation of leukocyte chemotaxis and chemokine-mediated signaling pathway that were enriched in the L. paracasei sh2020-treated tumors were shown (n = 5).
CD8+ T cell mediated the anti-tumor immunity of L. paracasei sh2020
Next, flow cytometry was used to examine the immune cell infiltrates of control and L. paracasei sh2020-treated tumors, which showed a prominent expansion of CD8+ T cells in L. paracasei sh2020-treated tumors, and no significant changes were found in other immune subsets (Figure 6a). Furthermore, IFN-γ+CD8+ T cells were significantly increased after L. paracasei sh2020 administration (Figure 6b-c). Moreover, these CD8+T cells had improved activation status following L. paracasei sh2020, since ICOS was upregulated on them (Figures S5). To identify which T cell subsets were required for the efficacy of L. paracasei sh2020, the CD4+ or CD8+ T cells were depleted using in vivo neutralizing antibodies against CD8, CD4 (Figure 6d). 26 Depletion of CD8+ T cells completely abrogated the effect of L. paracasei sh2020, suggesting that CD8+ T cells were indispensable for the anti-tumor immunity of L. paracasei sh2020. In contrast, depletion of CD4+ T cells did not influence the anti-tumor effect of L. paracasei sh2020 (Figure 6e). Collectively, L. paracasei sh2020 promoted CD8+ T cell infiltration and induced a CD8+ T cell-dependent anti-tumor immunity.
Figure 6.
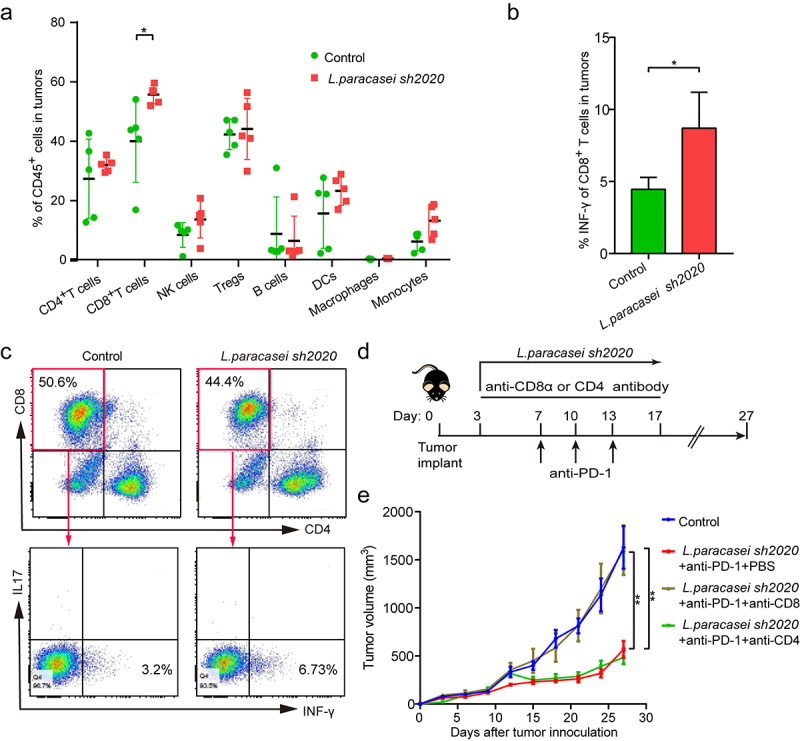
CD8+ T cells were required for anti-tumor effect of L. paracasei sh2020. (a) The percentage of tumor-infiltrating immune cell in individual mice in the control and L. paracasei sh2020-treated group (n = 5). (b-c) The expression of INF-γ on the CD8+ T cells within tumors from mice in the control and L. paracasei sh2020-treated group at the end of the experiment, representative flow sample and statistics were showed in c and d, respectively (n = 4–5). (d) Experimental design: C57BL/6 mice were implanted subcutaneously with 5.0 × 105 MC38 cells and treated with anti-PD-1 + L. paracasei sh2020. The anti-mouse CD8α or CD4 antibody was initiated 1 day before anti-PD-1 + L. paracasei sh2020 treatment and continued twice a week for two weeks. (e) Tumor growth in the tumor-bearing mice treated with L. paracasei sh2020 and depleted for CD4+ or CD8+ T cells (n = 5–6). *P < .05, **P < .01.
L. paracasei sh2020 increased the expression and release of CXCL10 in the tumors
Our next goal was to explore the underlying mechanisms by which L. paracasei sh2020 facilitate the CD8+ T cell infiltration in the tumors. Analysis of RNA-seq data showed that specific chemokines that promoted T cell recruitment were upregulated after L. paracasei sh2020 administration, suggesting the pivotal role of L. paracasei sh2020 in boosting chemokine expression and release in tumor microenvironment. Among them, CXCL10 showed the most increase compared with other chemokines (Figure 5b-c). qRT-PCR showed that L. paracasei sh2020 made a drastic increase in the level of CXCL10 in tumor tissue, whereas no changes were found in CXCL9, and CXCL11 (Figure 7a). This observation was also confirmed by IHC of CXCL10 in the tumors (Figure 7b-c).
Figure 7.

L. paracasei sh2020 promoted the expression and secretion of CXCL10 in vivo and in vitro. (a) The expression of CXCL9, CXCL10 and CXCL11 in tumor tissues from control and L. paracasei sh2020-treated mice was detected by qRT-PCR. (b-c) Representative images (b) and quantification (c) of IHC staining of CXCL10 in the tumor tissues from control and L. paracasei sh2020-treated tumors. (d) Tumor growth in each group. (e) The levels of CXCL10 in the conditioned medium. (f-g) Tumor growth in the tumor-bearing mice with intratumoral injection of L. paracasei sh2020 (n = 5–6). (h-i) Representative images (h) and quantification (i) of IHC staining of CXCL10 and CD8 in each group (n = 4–5). (j) The serum levels of CXCL10 were examined by ELISA. ns, no significant difference, *P < .05, **P < .01, ***P < .001.
Then, L. paracasei sh2020 was pasteurized and the dead L. paracasei sh2020 abrogated the tumor control observed in the tumor-bearing mice (Figure 7d), indicating the live bacteria but not the bacterial debris, induced antitumor immunity. Hence, live L. paracasei sh2020 was cultured and used to investigate its role in the expression and release of CXCL10 in vitro. The tumor cells were incubated with L. paracasei sh2020 for 4 h, 8 h, 16 h, 24 h, and 36 h, respectively, secretion of CXCL10 protein was measured using ELISA assays. L. paracasei sh2020 promoted CXCL10 secretion after 8 h, 16 h, 24 h, and 36 h, and highest level of CXCL10 protein was achieved after 24 h of incubation (Figure 7e). To further confirm this result in vivo, intratumoral injection of live bacteria was performed according to the recent studies.27,28 We found that intratumoral injection of live L. paracasei sh2020 also considerably prevented tumor growth (Figure 7f-g), induced CXCL10 expression, and CD8+ T cell infiltration in the tumor beds (Figure 7h-i). In addition, increased CXCL10 levels were observed in the serum of these mice compared to those of the controls (Figure 7j). Together, L. paracasei sh2020 promoted CXCL10 secretion in vitro and in vivo, which was known to facilitate recruitment of cytotoxic T cells into tumors.29,30
CXCL10 controlled CD8+ T cell infiltration mediated by L. paracasei sh2020
Furthermore, the CXCL10 and CD8 expressions were detected in the control and L. paracasei sh2020-treated tumors using IHC (Figure 8a). It was found that L. paracasei sh2020-treated tumors showed an increase in CD8 expression, as well as CXCL10 staining. In contrast, control tumors with a low score of CXCL10 showed lower levels of CD8 stain (Figure 8b). Collectively, a negative correlation between CXCL10 and CD8+ T cell infiltration was observed in the tumor tissues (Figure 8c). To further investigate the role of CXCL10 on the effect of L. paracasei sh2020 in vivo, we blocked CXCL10 using anti-CXCL10-neutralizing antibody in the L. paracasei sh2020-treated mice, showing that neutralizing CXCL10 in vivo completely dampened the effect of L. paracasei sh2020 (Figure 8d-e). This rescue of the tumor growth was associated with significantly decreased CD8+ T cell infiltration (Figure 8f, and Figure S6), suggesting that CXCL10-mediated CD8+ T cell infiltration played a crucial role in L. paracasei sh2020-induced tumor inhibition.
Figure 8.
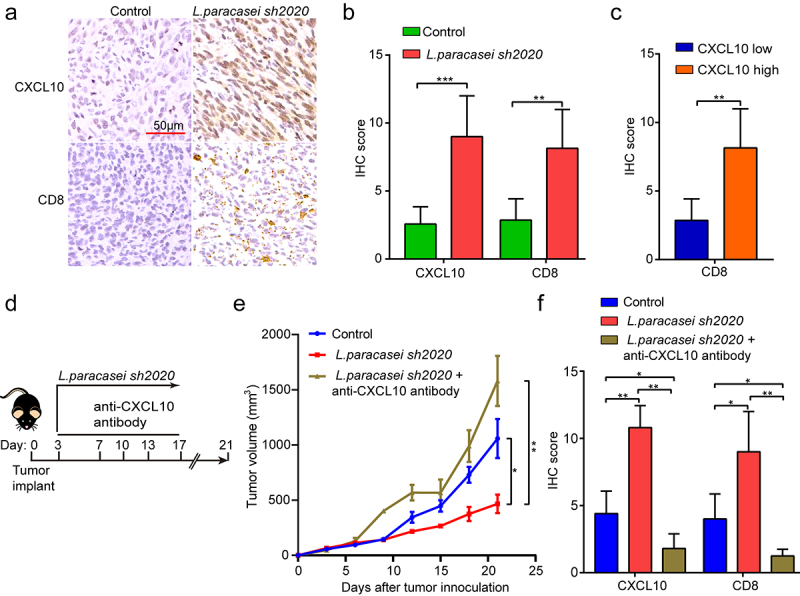
CXCL10 controlled CD8+ T cell migration and the effect of L. paracasei sh2020 in vivo. (a-b) Representative images of IHC staining of CD8 and CXCL10 (a), and quantification (b) for the control and L. paracasei sh2020-treated tumors (n = 6–7). (c) IHC analysis of CD8 in tumors, which were divided into two groups according to CXCL10 high and low expression. (d) Experimental design: C57BL/6 mice were implanted subcutaneously with 5.0 × 105 MC38 cells and was treated with control vehicle or anti-CXCL10 antibody by intraperitoneal injection, every 3 days starting on D3, in total three times. The mice were given L. paracasei sh2020 with a dose of 1.0 × 109 CFU by gavage starting from D0 to D13. (e) Tumor growth in tumor-bearing mice in d. (f) Quantification of IHC staining of CXCL10 and CD8 in the tumors after neutralizing CXCL10 in vivo (n = 4–5). ns, no significant difference, *P < .05, **P < .01, ***P < .001.
Oral intake of L. paracasei sh2020 maintained gut microbiota homeostasis in the context of anti-PD-1 therapy
ral intake of live probiotics can be expected to influence the gut homeostasis.31 Finally, we performed 16S rRNA sequencing to characterize the effects of L. paracasei sh2020 oral administration on gut microbiota profiles. Oral intake of L. paracasei sh2020 increased bacterial community diversity in the context of anti-PD-1 therapy (Figure 9a-b). 3D-PCoA analysis suggested that the overall gut bacterial community of the anti-PD-1 + L. paracasei sh2020 group had gradually deviated from the control or anti-PD-1 group (Figure 9c). And the abundance of gut bacteria was considerably changed by L. paracasei sh2020 in context of anti-PD-1 therapy (Figure 9c). L. paracasei sh2020 increased the relative abundance of Lactobacillus in the anti-PD-1-treated mice. Furthermore, the correlations between Lactobacillus and the level of tumor-infiltration immune cells were also testified, confirming the strongly positive correlation between Lactobacillus and antitumor immune response (Figure 9e). In addition, H&E staining showed that L. paracasei sh2020 might help to maintain gut homeostasis (Figure 9f), thereby providing an intact gut barrier against immune checkpoint inhibitor-associated diarrhea and colitis.32
Figure 9.
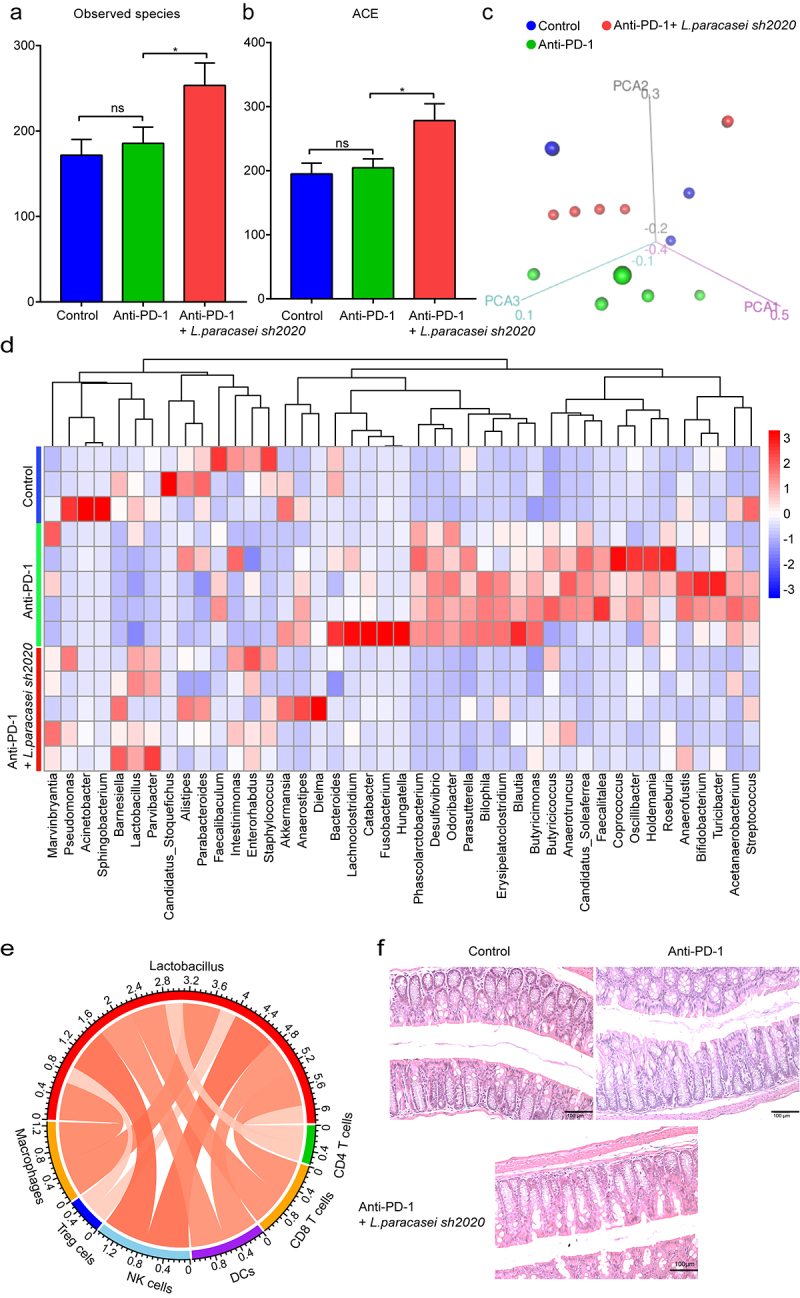
Oral administration of L. paracasei sh2020 improved gut microbiota homeostasis in the context of anti-PD-1 therapy. (a-b) The α-diversity differences between the groups were estimated by the observed species (a), and ACE (b) (n = 3–5). (c) 3D-PCoA analysis of fecal samples based on the Bray-Curtis dissimilarity metric. (d) Community heatmap analysis at the genus level among different treatment groups (n = 3–5). (e) Correlation networks of Lactobacillus and tumor-infiltrating immune cells. (f) Representative images of H&E staining of the colon tissues in each group. ns, no significant difference, *P < .05.
Discussion
Gut microbiota is heavily influenced by both genetic and environmental factors including age, gender, genotype, diet, and environmental exposures.33 It also emerged from several recent studies that different research institutions or commercial vendors usually harbor considerable variations in microbiological environment,15,34 leading to differences in gut microbiota structure and composition. Since the microbiome deficit of one individual may not necessarily mirror that of another individual, it is not surprising that patients respond differently to FMT. In this context, humanized microbiome mice model is a valuable donor-recipient matching approach for identifying the “super-donor”, whose stool confers significantly more successful FMT outcomes than the stool of other donors. In our study, we performed FMT using individual stool from five healthy donors, giving a personalized cocktail of organisms that were fed to the mice. One strength of our study could be that all human donors were young, and all of them have led a healthy lifestyle, including healthy balanced eating, regular physical activities, proper sleep, quit smoke and alcohol, and staying happy and positive. Studies have shown that a healthy functional microbial ecosystem is heavily shaped by these healthy lifestyles, which is fundamental to instruct the immune system toward homeostasis. Hence, it is of significance to screen the “super-donor” among this population, and thus guarantee FMT success in CRC patients underwent immunotherapy. Interestingly, even in the young and healthy individuals with healthy lifestyle, their gut microbiota differently influenced tumor progression in the presence of anti-PD-1 mb. The observed inter-individual variation among these donors was accompanied by specific gut microbiota profiles as well as T cell phenotypes in the TME. However, the precise mechanisms underlying these variations in recipient mice need to be investigated in future detail, focusing on individual microbiotas and specific pathways. Additionally, the number of healthy individuals in our study was small, and future studies with larger cohorts are required.
Just as a side water and soil raises side people, different environments and customs influence the gut microbiota structure and composition. Since we and other researchers have demonstrated that anti-PD-1 outcomes are heavily influenced by gut microbiota,35,36 it is not surprising that patients from different countries and regions respond differently to anti-PD-1 therapy. Moreover, the effector bacteria that are associated with good response to anti-PD-1 remain as high variation among different research institutions or commercial vendors. For example, A.muciniphila was reported to facilitate anti-PD-1 in metastatic melanoma patients.37 However, A.muciniphila abundance was not significantly changed after FMT from healthy donors in our study, indicating that the favorable antitumor effect of healthy gut microbiota did not follow the A. muciniphila way. In our study, we cultured and isolated a novel L. paracasei from the feces samples of healthy donors, which was named L. paracasei sh2020. We found that L. paracasei sh2020 was effective to reduce tumor growth when combined with anti-PD-1. Surprisingly, single L. paracasei sh2020 even produced a stronger antitumor effect than the anti-PD-1 therapy alone.
Tumor-infiltrating lymphocytes (TILs) represented the major component of the tumor microenvironment. Recent studies have divided solid tumors into two subtypes: “hot” tumor and “cold” tumor.38,39 The latter, with less TILs, usually represented poor response to immunotherapy,40 which was a major obstacle to effective treatment with anti-PD-1 among CRC patients.41,42 And researchers tried to develop strategies to turn “cold” tumors into “hot” tumors these days. Furthermore, improving T cell infiltration in the tumors via manipulating gut microbiota is an area of active research, which still stays at its infant stage. In our study, selective depletion of T cell subsets highlighted the indispensable role of CD8+ T cells in the L. paracasei sh2020 treatment, which was also confirmed in the patients treated with anti-PD-1. Thus, we concluded that the effect of L. paracasei sh2020 was largely dependent on CD8+ T cell in the tumors. Thus, increased T cell infiltration induced by L. paracasei sh2020 was a promising strategy to improve poorly infiltrated tumor microenvironment, transforming “cold” tumors to “hot” ones.
Furthermore, we strived to clarify the precise mechanisms for responding to L. paracasei sh2020. We compared gene expression profiles in the L. paracasei sh2020-treated tumors with that in the control tumors, and GSEA of these genes highlighted Leukocyte migration-associated pathways as the key influenced pathway mediated by L. paracasei sh2020. Most noteworthy was that chemokines involved in these pathways also showed significance in a comparison of gene expression profiles between the control and L. paracasei sh2020-treated tumors, indicating that L. paracasei sh2020 might promote T cell infiltration through reversing the expression of these genes. In fact, chemokines play crucial roles in the lymphocyte recruitment within the tumor microenvironment.43 More importantly, we identified that CXCL10 was the as the most significantly influenced chemokines by L. paracasei sh2020. CXCL10 was a T helper 1 type chemokine and governed the trafficking of the main antitumor immune cells, including CD8+ T cells, into the tumor beds. Considering this, we confirmed that L. paracasei sh2020 treatment confers high levels of CXCL10 than the control tumor in both vitro and vivo. In the in vitro tumor cell culture model, stimulation by L. paracasei sh2020 could result in the increased production of CXCL10, which was further confirmed by the L. paracasei sh2020-treated tumors in vivo. Hence, L. paracasei sh2020 in antitumor immunity or as immunomodulator to facilitate immunotherapy.
In a review of the oral administration route for L. paracasei sh2020, we consequently outlined gut bacterial community in the mice using 16S rRNA gene sequencing. Oral administration of L. paracasei sh2020 significantly altered the microbial community and increased the microbial community diversity in the context of anti-PD-1 therapy, which had favorable roles in the improved antitumor immunity. A healthy gut environment, shaped by the balanced gut microbiota, is fundamental to the presence of normal gut barrier.20 However, anti-PD-1 can cause immune-related adverse effects, especially in the patients with the dysbiosis of gut microbiota.44 One of the most common toxicities is anti-PD-1-associated colitis. Here, histopathological analysis of colons confirmed that L. paracasei sh2020 administration might help to maintain intestinal homeostasis by modulating gut microbiota, which was a novel and promising strategy to mitigate the immune checkpoint inhibitor-associated colitis.32 In all, L. paracasei sh2020 could ameliorate gut microbiota changes induced by anti-PD-1 therapy, and thus restore the homeostasis of gut microecology.
In conclusion, our study identified a novel commensal L. paracasei sh2020 that was able to trigger anti-tumor immunity and prevent tumor growth in the mice. anti-PD-1 combined with L. casei sh2020 markedly trig tumor suppression, making it superior to anti-PD-1 or L. paracasei sh2020 monotherapy. Notably, the modulation of gut microbiota caused by L. paracasei sh2020 might play a synergic role in enhancing immunotherapy.
Materials and methods
Cell line and culture
Mouse colon adenocarcinoma cell-line MC38 was obtained from Type Culture Collection of Chinese Academy of Sciences, China (Shanghai). The cell was cultured in the RPMI-1640 medium (Gibco, USA) with 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (Hyclone, USA) in a humidified atmosphere containing 5% CO2 at 37°C.
Mice
All animal experiments were approved by the Ethics Committee of Minhang Hospital, Fudan University. The female C57BL/6 mice (6 to 8-weeks old) were obtained from Charles River Laboratories, China. All mice were housed under specific pathogen-free conditions and allowed to acclimate for one week before performing experiments. The ethics of animal research has been reviewed and approved by the Ethics Committee of Fudan-Minhang academic health system (Approval number: 2020 Minhang Hospital JS-006).
Fecal microbiota transplantation (FMT) experiment
Five patients newly diagnosed with CRC and healthy volunteers were enrolled in the study, according to the criteria described previously.34 All Subjects information was provided in Table S1. Fresh stools from each subject were immediately suspended in an equal volume of PBS containing 20% glycerol, snap-frozen in liquid nitrogen, and stored in −80°C refrigerator until use. FMT was performed as follows: we gave antibiotics to adult mice (6–8 weeks of age) by oral gavage with 200 mg/kg of ampicillin, metronidazole, and neomycin, and 100 mg/kg of vancomycin daily for 3 days. Then, stool samples for humans were thawed and suspended in an equal volume of PBS, vortexed, and the supernatant was centrifuged. 100 µL of the fecal suspension was administered to mice by oral gavage, and another 100 µL was applied on the fur of each animal for 3 days. The study protocol has been reviewed and approved by the Ethics Committee of Fudan-Minhang academic health system and informed consent was obtained from all participants.
Bacteria stains isolation, and identification
Fecal sample was homogenized in sterile physiological saline in a 15 ml sterile centrifuge tube. After centrifugation (1000 g, 1 min), the supernatant was incubated in a modified MRS medium for 18–24 h. After which, 10 µL of the enrichment cultures were streaked on the Rogosa agar, a highly selective media for the isolation of Lactobacillus spp,45 and incubated at 37°C for 24–48 h under aerobic conditions. After incubation, a sterile inoculation needle was used to pick up single colonies and streak them on clean plates to obtain pure cultures. Genome DNA was isolated for 16s rRNA PCR to verify the purified strain type. As for whole-genome sequencing, the bacterial genomic DNA was extracted for Oxford Nanopore and Illumina sequencing. Sequence blasts were carried out in the National Center for Biotechnology Information (NCBI) database. The high-quality reads were assembled by Miniasm.24 The genomes data for other Lactobacillus strains available in the NCBI database were downloaded for the comparative analysis.
Mouse tumor models and treatments
When MC38 cells grew reaching about 80% confluence, they were harvested and washed three times in phosphate-buffered saline (PBS). 5.0 × 105 cells were inoculated subcutaneously into the right flank of each mouse. Tumor volume and mouse weights were monitored every 3 days. Tumor volume was calculated as length × width2 × 0.5. The survival status of these mice was checked and recorded every day. For anti-PD-1 treatment, mice were intraperitoneally injected with anti-PD-1 (J43, BE0033-2, BioXcell) at a dose of 200 μg/mouse every 3 days for three times.
The bacterial culture and administration
The isolated bacteria were resuspended and cultured in MRS broth at 37°C for 16–24 hours. Supernatants from the media were harvested, passed through a 0.45 µm filter, and either stored at −80°C or used directly for coculture with tumor cells. The strains were then centrifuged at 8000 g for 5 minutes and the pellets were diluted in the PBS. Each mouse was treated once-daily with 1.0 × 109 CFU (L. plantarum, L. paracasei, and L. reuteri) cocktail for 15 days by oral gavage.
Depletion of T cell subsets and CXCL10 in vivo
In the in vivo T cell subsets depletion study, InVivoMAb anti-mouse CD4 (YTS191, BE0119, BioXCell), CD8α (YTS169.4, BE0117 BioXcell) antibodies were intraperitoneally injected at a dose of 200 μg per mouse. To neutralize Cxcl10 in vivo, 50 µg of anti-mouse Cxcl10 (134013, MAB466, R&D systems) was intraperitoneally injected into mice every 3 days for 2 weeks.
16S rRNA sequencing analysis
Total bacterial DNA was extracted from fecal samples using the DNA extraction kit (TIANGEN, China). For analysis of the taxonomic composition of gut microbiota, the hypervariable regions V3-V4 region of 16S rRNA genes was pooled and sequenced at the Illumina MiSeq 2500 platform (Illumina MiSeq, USA). Reads were trimmed and classified using QIIME (V 1.8.0).46 After quality filtering and chimera removal, these sequences were assigned to operational taxonomic units (OTUs) with ≥ 97% similarity using UPARSE47 and RDP classifier48 was used to classify OTUs at a given taxonomic rank. The gut microbial signature was identified using selbal,49 a greedy stepwise algorithm for balance selection, and unsupervised RandomForest classification analysis.
Next generation RNA sequencing (RNA-seq) analysis
Total RNA was extracted from tumors that were treated with or without L. casei sh2020. RNA-seq was performed using Illumina HiSeq (Novogene Bioinformatics Technology Co., Ltd.). After quality control, RNA-seq reads were mapped to the mouse reference haploid genome sequence (GRCm38.p3 C57BL/6, NCBI). Quantified transcripts were collapsed into gene counts, which were subsequently transformed to normalized FPKM values for all downstream analyses. Differentially expressed genes were identified using “limma” R package.
To explore signaling pathways enrichment, Gene Set Enrichment Analysis (GSEA) was performed using GSEA software version 4.0.0. The CIBERSORT algorithm50 was used to estimate the relative fractions of 25 distinct immune cell types within a complex mixture of RNA-seq data using deconvolution algorithm.
Immunohistochemistry (IHC)
Formalin-fixed, paraffin-embedded tissues were sectioned at 5 μm thickness. And then the sections were dewaxed and repaired. The primary antibodies included rat anti-mouse CD8α (1:400, eBioscience,14–0808), rabbit anti-mouse CD4 (1:400, Abcam, ab183685), rabbit anti-Foxp3 (1:400, Abcam, ab54501), and rabbit anti-CXCL10 (1:800, absin, 135937). The staining intensity was scored from 0 to 3 (no staining, 0; weak staining, 1; moderate staining, 2; strong staining, 3). The staining extent was scored from 0 to 4 according to the positive proportion of cells (0%, 0; 1–25%, 1; 26–50%, 2; 51–75%, 3; >76%, 4). The final score was calculated as multiplying the staining intensity and extent score.
Flow cytometry
Fresh tumor tissues were minced into small pieces and digested in an RPMI medium containing collagenase D (1 mg/ml; Roche) and DNase1 (150 UI/ml; Sigma) for 30 minutes at 37°C and then washed and filtered twice using 100 µm cell strainers (Falcon®). Fixable Viability Stain 620 (BD Biosciences) was used to discriminate live and dead cells. The cells were then blocked with Fc-block (BD Biosciences) and stained with anti-mouse antibodies. The following antibodies were used: PE/Cyanine7 anti-human/mouse CD45R (B220) (60–0452-U025, TONBO), Brilliant Violet 711TM anti-mouse CD45 (103147, BioLegend), APC Anti-mouse NK1.1y (CD161) Antibody (20–5941-U025, TONBO), FITC Anti-mouse MHC Class II (I-A/I-E) Antibody (35–5321-U025, TONBO), PE Anti-mouse CD11c Antibody (N418) (50–0114-U025, TONBO), 710 Anti-mouse CD11b Antibody (M1/70) (80–0112-U025, TONBO), 450 Anti-mouse F4/80 (BM8.1) (75–4801-U025, TONBO), PE Anti-mouse/rat Foxp3 (FJK-16s) (12–5773-82, eBioscience), APC Anti-human/mouse/rat CD278 Antibody (ICOS) (APC ICOS 313510, Biolegend), eFluor 450 anti-mouse CD8a Antibody (48–0081-82, eBioscience), FITC anti-mouse CD4 Antibody (GK1.5) (11–0041-82, eBioscience), and APC anti-mouse INF gamma Antibody (XMG1.2) (17–7311-82, eBioscience).
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total mRNA was extracted using TRIzol reagent (Life Technologies). cDNA was synthesized by a PrimeScriptTM RT kit (Takara). RNA concentration purity was measured on a Nanodrop1000 spectrophotometer (Agilent). Quantitative real-time PCR was performed on cDNA using gene-specific primers in the presence of SYBR Green (Applied BioSystems). The results were analyzed the 2−ΔΔCT method. Primer pairs used in this study were shown as follows:
| Genes | Forward (5’ – 3’) | Reverse (5’ – 3’) |
|---|---|---|
| CXCL9 | GAAGTCCGCTGTTCTTTTCC | TTGACTTCCGTTCTTCAGTGT |
| CXCL10 | GCTGCAACTGCATCCATATC | AGGAGCCCTTTTAGACCTTT ATGTAC |
| CXCL11 | CTTATGTTCAAACAGGGGCG | TGCATTATGAGGCGAGCTT |
| β-actin | CGCAAAGACCTGTATGCCAAT | GGGCTGTGATCTCCTTCTGC |
Enzyme linked immunosorbent assay (ELISA)
For the quantitative determination of CXCL10 secreted protein levels in the tumor cell supernatants, as well as in the serum of mice, we used the Mouse CXCL10/IP-10 ELISA Kit (MULTISCIENCES, EK268/2), according to the manufacturer’s instructions.
Statistical analysis
All data were presented as the mean ± SEM. Differences between groups were determined by two-tailed unpaired Student’s t-test using GraphPad Prism 7.0 software or as otherwise stated in the figure legend.
Supplementary Material
Funding Statement
The present study was supported by grants from National Natural Science Foundation of China (81872245 and 81803601), and grants from Shanghai Sailing Program (17YF1416700).
Contributors
SLZ and LSW designed and finalized the study. SLZ conducted the bioinformatics analysis. SLZ, YQM, and ZYZ performed the experiments. SLZ, CYK, HLC, PRC, YW, and BH helped on samples collection. SLZ, HB, and YQM drew the charts and wrote the paper. LSW and GQC supervised the project. All the authors approved the final version submitted.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethics approval
The study protocol has been reviewed and approved by the Ethics Committee of Fudan-Minhang academic health system (Approval number: 2020 Minhang Hospital JS-006).
Provenance and peer review
Not commissioned; externally peer reviewed.
Data availability statement
All data relevant to the study were included in the article or uploaded as supplementary information.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Hoos A. Development of immuno-oncology drugs - from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov. 2016;15(4):235–19. doi: 10.1038/nrd.2015.35. [DOI] [PubMed] [Google Scholar]
- 2.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48(3):434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi G, Yang Q, Zhang Y, Jiang Q, Lin Y, Yang S, Wang H, Cheng L, Zhang X, Li Y, et al. Modulating the tumor microenvironment via oncolytic viruses and CSF-1R inhibition synergistically enhances anti-PD-1 Immunotherapy. Mol Ther. 2019;27(1):244–260. doi: 10.1016/j.ymthe.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, Diaz LA Jr. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol. 2019;16(6):361–375. doi: 10.1038/s41575-019-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang SL, Mao YQ, Zhang ZY, Li ZM, Kong CY, Chen HL, Cai PR, Han B, Ye T, Wang LS. Pectin supplement significantly enhanced the anti-PD-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics. 2021;11(9):4155–4170. doi: 10.7150/thno.54476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salgia NJ, Bergerot PG, Maia MC, Dizman N, Hsu J, Gillece JD, Folkerts M, Reining L, Trent J, Highlander SK, et al. Stool microbiome profiling of patients with metastatic renal cell carcinoma receiving anti-PD-1 immune checkpoint inhibitors. Eur Urol. 2020;78(4):498–502. doi: 10.1016/j.eururo.2020.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli E, Segata N, Desnoyer A, Pietrantonio F, Ferrere G, et al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol. 2020;78(2):195–206. doi: 10.1016/j.eururo.2020.04.044. [DOI] [PubMed] [Google Scholar]
- 9.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 10.Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, Paik S, Stagg J, Groves RA, Gallo M, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 2020;369(6510):1481–1489. doi: 10.1126/science.abc3421. [DOI] [PubMed] [Google Scholar]
- 11.Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 12.Allen-Vercoe E, Coburn B. A microbiota-derived metabolite augments cancer immunotherapy responses in mice. Cancer Cell. 2020;38(4):452–453. doi: 10.1016/j.ccell.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer. 2019;7(1):108. doi: 10.1186/s40425-019-0574-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goedert JJ, Gong Y, Hua X, Zhong H, He Y, Peng P, Yu G, Wang W, Ravel J, Shi J, et al. Fecal microbiota characteristics of patients with colorectal adenoma detected by screening: a population-based study. EBioMed. 2015;2(6):597–603. doi: 10.1016/j.ebiom.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Misra BB, Liang L, Bi D, Weng W, Wu W, Cai S, Qin H, Goel A, Li X, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics. 2019;9(14):4101–4114. doi: 10.7150/thno.35186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TM, Comelli EM. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes. 2014;4e121. doi: 10.1038/nutd.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 18.Bowen JM, Stringer AM, Gibson RJ, Yeoh AS, Hannam S, Keefe DM. VSL#3 probiotic treatment reduces chemotherapy-induced diarrhea and weight loss. Cancer Biol Ther. 2007;6(9):1449–1454. doi: 10.4161/cbt.6.9.4622. [DOI] [PubMed] [Google Scholar]
- 19.Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgard L, Wettergren Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017;4(1):e000145. doi: 10.1136/bmjgast-2017-000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Lopez G, Troisi J, Colucci A, Guglietta S, Carloni S, Guglielmetti S, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018;9(1):5184. doi: 10.1038/s41467-018-07359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivera-Pinto J, Egozcue JJ, Pawlowsky-Glahn V, Paredes R, Noguera-Julian M, Calle ML, Lozupone C. Balances: a new perspective for microbiome analysis. mSystems. 2018;3(4). doi: 10.1128/mSystems.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacouton E, Michel M-L, Torres-Maravilla E, Chain F, Langella P, Bermúdez-Humarán LG. Elucidating the immune-related mechanisms by which probiotic strain lactobacillus casei BL23 displays anti-tumoral properties. Front Microbiol. 2019;9(3281). doi: 10.3389/fmicb.2018.03281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacouton E, Michel ML, Torres-Maravilla E, Chain F, Langella P, Bermudez-Humaran LG. Elucidating the immune-related mechanisms by which probiotic strain lactobacillus casei BL23 displays anti-tumoral properties. Front Microbiol. 2018;93281. doi: 10.3389/fmicb.2018.03281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis KE, Selby MJ, Masters G, Valle J, Dito G, Curtis WR, Garcia R, Mink KA, Waggie KS, Holdren MS, et al. Interleukin-21 combined with PD-1 or CTLA-4 blockade enhances antitumor immunity in mouse tumor models. Oncoimmunology. 2017;7(1):e1377873. doi: 10.1080/2162402x.2017.1377873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, Lin W, Chang EB, Weichselbaum RR, Fu YX. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. 2020;217(5). doi: 10.1084/jem.20192282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, Vitale I, Goubar A, Baracco EE, Remédios C, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. 2014;20(11):1301–1309. doi: 10.1038/nm.3708. [DOI] [PubMed] [Google Scholar]
- 30.Chow MT, Ozga AJ, Servis RL, Frederick DT, Lo JA, Fisher DE, Freeman GJ, Boland GM, Luster AD. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity. 2019;50(6):1498–1512.e5. doi: 10.1016/j.immuni.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Si W, Liang H, Bugno J, Xu Q, Ding X, Yang K, Fu Y, Weichselbaum RR, Zhao X, Wang L. Lactobacillus rhamnosus GG induces cGAS/STING-dependent type I interferon and improves response to immune checkpoint blockade. Gut. 2021;71(3):521–533. doi: 10.1136/gutjnl-2020-323426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, Jiang ZD, Abu-Sbeih H, Sanchez CA, Chang CC, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. 2018;24(12):1804–1808. doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costea PI, Hildebrand F, Arumugam M, Bäckhed F, Blaser MJ, Bushman FD, de Vos WM, Ehrlich SD, Fraser CM, Hattori M, et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2018;3(1):8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, Xiao X, Kwong TNY, Tsoi H, Wkk W, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. 2017;153(6):1621–1633.e6. doi: 10.1053/j.gastro.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Sun H, Li B, Zhang X, Cui J, Yun J, Yang Y, Zhang L, Meng Q, Wu S, et al. probiotics ameliorate colon epithelial injury induced by ambient ultrafine particles exposure. Adv Sci (Weinh). 2019;6(18):1900972. doi: 10.1002/advs.201900972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Sung CY, Lee N, Ni Y, Pihlajamäki J, Panagiotou G, El-Nezami H. Probiotics modulated gut microbiota suppresses hepatocellular carcinoma growth in mice. Proc Natl Acad Sci U S A. 2016;113(9):E1306–15. doi: 10.1073/pnas.1518189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, Gajewski TF. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Byrne KT, Yan F, Yamazoe T, Chen Z, Baslan T, Richman LP, Lin JH, Sun YH, Rech AJ, et al. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49(1):178–193.e7. doi: 10.1016/j.immuni.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362(6411). doi: 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, Olszanski AJ, Malvehy J, Cebon J, Fernandez E, et al. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell. 2017;170(6):1109–1119.e10. doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garris CS, Arlauckas SP, Kohler RH, Trefny MP, Garren S, Piot C, Engblom C, Pfirschke C, Siwicki M, Gungabeesoon J, et al. Successful anti-PD-1 cancer immunotherapy requires T cell-dendritic cell crosstalk involving the cytokines IFN-gamma and IL-12. Immunity. 2018;49(6):1148–1161.e7. doi: 10.1016/j.immuni.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pérez-Ruiz E, Jiménez-Castro J, Berciano-Guerrero MA, Valdivia J, Estalella-Mendoza S, Toscano F, Rodriguez de la Borbolla Artacho M, Garrido-Siles M, Martínez-Bautista MJ, Villatoro Roldan R, et al. Impact of intestinal dysbiosis-related drugs on the efficacy of immune checkpoint inhibitors in clinical practice. Clin Transl Oncol. 2020;22(10):1778–1785. doi: 10.1007/s12094-020-02315-9. [DOI] [PubMed] [Google Scholar]
- 45.Volokhov DV, Amselle M, Beck BJ, Popham DL, Whittaker P, Wang H, Kerrigan E, Chizhikov VE. Lactobacillus brantae sp. nov., isolated from faeces of Canada geese (Branta canadensis). Int J Syst Evol Microbiol. 2012;62(9):2068–2076. doi: 10.1099/ijs.0.033852-0. [DOI] [PubMed] [Google Scholar]
- 46.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/aem.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 48.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(Database issue):D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivera-Pinto E, Pawlowsky-Glahn P, Pawlowsky-Glahn V, Paredes R, Noguera-Julian M, Calle ML. Balances: a new perspective for microbiome analysis. mSystems. 2018. 17;3(4):e00053–18. doi: 10.1128/mSystems.00053-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to the study were included in the article or uploaded as supplementary information.


