ABSTRACT
Neoantigens play a crucial role in cancer immunotherapy. However, the effectiveness and safety of neoantigen-based immunotherapies in patients with colorectal cancer (CRC), particularly in the Chinese population, have not been well studied. This study explored the feasibility and effectiveness of neoantigens in the treatment of CRC. Whole-exome sequencing (WES) and transcriptome sequencing were used to identify somatic mutations, RNA expression, and human leukocyte antigen (HLA) alleles. Neoantigen candidates were predicted, and immunogenicity was assessed. The neoantigens TSHZ3-L523P, RARA-R83H, TP53-R248W, EYA2-V333I, and NRAS-G12D from Patient 4 (PW4); TASP1-P161L, RAP1GAP-S215R, MOSPD1-V63I, and NAV2-D1973N from Patient 10 (PW10); and HAVCR2-F39V, SEC11A-R11L, SMPDL3B-T452M, LRFN3-R118Q, and ULK1-S248L from Patient 11 (HLA-A0201+PW11) induced a heightened neoantigen-reactive T cell (NRT) response as compared with the controls in peripheral blood lymphocytes (PBLs) isolated from patients with CRC. In addition, we identified neoantigen-containing peptides SEC11A-R11L and ULK1-S248L from HLA-A0201+PW11, which more effectively elicited specific CTL responses than the corresponding native peptides in PBLs isolated from HLA-A0201+PW11 as well as in HLA-A2.1/Kb transgenic mice. Importantly, adoptive transfer of NRTs induced by vaccination with two mutant peptides could effectively inhibit tumor growth in tumor-bearing mouse models. These data indicate that neoantigen-containing peptides with high immunogenicity represent promising candidates for peptide-mediated personalized therapy.
Abbreviations: CRC: colorectal cancer; DCs: dendritic cells; ELISPOT: enzyme-linked immunosorbent spot; E:T: effector:target; HLA: human leukocyte antigen; MHC: major histocompatibility complex; Mut: mutant type; NGS: next-generation sequencing; NRTs: neoantigen-reactive T cells; PBMCs: peripheral blood mononuclear cells; STR: short tandem repeat; PBLs: peripheral blood lymphocytes; PBS: phosphate-buffered saline; PD-1: programmed cell death protein 1; TILs: tumor-infiltrating lymphocytes; RNA-seq: RNA sequencing; Tg: transgenic; TMGs: tandem minigenes; WES: whole-exome sequencing; WT: wild-type.
KEYWORDS: Colorectal cancer, immunotherapy, neoantigens, tumor, cancer vaccine
Introduction
Colorectal cancer (CRC) is the third most common cause of cancer-related deaths worldwide.1 At present, CRC ranks fifth in incidence among systemic malignant tumors in China.2 Surgery is the primary treatment option for patients with CRC; however, adjuvant chemotherapy is used only for patients with stage II, stage III CRC, and stage II or III rectal cancer.3,4 With the introduction of screening programs and the development of new targeted drugs, survival has increased over the past 30 years. However, the 5-year relative survival rate of CRC is still only 68%. Notably, approximately one-third of patients undergoing curative surgery for CRC develop the recurrent disease because of incomplete tumor resection, and the standard therapies of surgery and adjuvant chemotherapy are often limited by side-effects and resistance to chemotherapy.5,6 Therefore, it is highly crucial to develop other strategies for treating CRC.
Tremendous advancements have been made in immunotherapy for cancer treatment.7,8 However, only less than 5% of patients with advanced CRC benefited from programmed cell death protein 1 (PD-1) checkpoint blocking immunotherapy, with limited efficacy in CRC.9 With the advent and availability of next-generation sequencing (NGS), the identification of patient-specific neoantigens for personalized vaccines has become feasible. Several clinical trials of melanoma, glioblastoma, and other types of cancers have reported neoantigen-based vaccines to be immunogenic and safe with promising clinical effects.10,11 In CRC, HLA-C*08:02-restricted tumor-infiltrating lymphocyte (TIL)-based immunotherapy targeting KRAS G12D oncogene mutation promoted tumor regression in one patient.12 However, the effectiveness and safety of neoantigen-based immunotherapy in CRC, particularly in the Chinese population, has not been well studied.
In this study, 13 patients with CRC were recruited to assess the feasibility and effectiveness of neoantigens and neoantigen-based immunotherapy. Tumor tissue samples and PBCs were collected to identify neoantigens, and the immunogenicity of neoantigens was determined. The anti-tumor effects of neoantigens on patients with CRC were determined using a neoantigen-reactive T cell (NRT)-induced in vitro cytotoxicity assay and by performing in vivo experiments in a tumor-bearing mice model. Our results indicated the possibility of the development of neoantigen-based personalized immunotherapies in CRC.
Materials and methods
Patient materials and cell lines
Tumor samples of patients were obtained from biopsy specimens. A part of each sample was fixed with formalin and embedded in paraffin. The remaining tumor tissue was immediately frozen in droplets and stored in gaseous liquid nitrogen. This study included adult patients with a histologically or cytologically confirmed CRC from the Second Affiliated Hospital of Wenzhou Medical University between February 2017 and February 2019. Written informed consent was collected from all donors for collecting tissue specimens. This study involving human samples was approved by the Medical Ethics Committee of the Second Affiliated Hospital, Wenzhou Medical University (MEC numbers: LCKY2018–67). All protocols were strictly performed according to the Helsinki Declaration and were performed after receiving patients’ consent.
Cell lines T2 (human TAP-deficient T2 cell line [human B and T cell hybrid expressing HLA-A2.1 molecules]) and the human colorectal adenocarcinoma SW480 (HLA-A2.1+) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and were cultured under standard conditions described by the ATCC. Peripheral blood mononuclear cells (PBMCs) were harvested for immunological monitoring or obtaining samples. These cells were isolated by density-gradient centrifugation using Ficoll–Paque solution (Amersham Biosciences, Bucks, UK) and collected from the buffy coat of healthy donors or from peripheral blood samples obtained from patients with CRC, whereas immature dendritic cells (DCs) were generated.13 SW480 cells were cultured under standard conditions described by the ATCC, and the cells were authenticated by short tandem repeat (STR) sequence analysis as described by the ATCC, whereas cell banks were generated and detected for Mycoplasma. All experiments were performed with Mycoplasma-free cells.
NGS
For RNA sequencing (RNA-seq), an RNeasy mini kit (Illumina, San Diego, CA, USA) was used to extract the RNA from fresh frozen tumor samples. RNA-seq libraries were constructed using the TruSeq Stranded mRNA Library Prep kit (Illumina) (for cell suspensions). Flow cytometry amplification and sequencing were performed using HiSeq 2500 according to the manufacturer’s instructions.
For whole-exome sequencing (WES), DNA was extracted from fresh-frozen tumor samples or cultured tumor cells using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA). Genomic DNA was fragmented, end-repaired, and simultaneously ligated to the bar-coded sequencing adapters (Illumina), amplified, and size-selected. Whole-exome capture was performed using Agilent SureSelect Human All Exon 44-Mb version 2.0 bait set (Agilent Technologies, Santa Clara, CA, USA).14 The resulting libraries were quantified using a quantitative polymerase chain reaction (qPCR), and 76 base-paired end reads were generated and sequenced using HiSeq 2000 or 2500 sequencers (Illumina).
Bioinformatics and mutation discovery
All mutations in a single patient were analyzed using the Python programming language. DNA libraries required at least 150 × 106 paired-end 50 nucleotide reads, whereas RNA libraries required at least 75 × 106 paired-end 50 nucleotide reads. For mutation detection, DNA reads were compared with the reference genome hg19 using BWA.15 Duplicate exomes from tumor and matched normal samples were analyzed for single-nucleotide variants. The sites were identified and screened for homozygous genotypes in normal samples, thus maintaining a high degree of specificity in single-nucleotide variation. The remaining sites were further examined to determine the presumed homozygous or heterozygous mutation event. The suspicious sites were screened to exclude potential false positives, check the sum of duplicates, and merge the duplicates. We compared the genomic coordinates of the identified variants with the detailed known gene transcription coordinates in the UCSC and further determined the relationship between the mutations and genes, transcription, expression values derived from RNA-seq, and the changes in potential amino acid sequences. For RNA-seq, RNA reads were compared with the hg19 reference genome and transcriptome using Bowtie.16 We determined gene expression by comparing known gene transcripts with detailed exon coordinates in UCSC and then normalized to the RPKM units.17
Human leukocyte antigen typing
The HLA alleles of each patient were identified from the WES data using OptiType (CeGaT GmbH, Tubingen, Germany)18 with the default settings after filtering the reads aligning to the HLA region using RazerS version 3.4.0 (GitHub Inc., San Francisco, USA).19
Neoantigen candidate identification
The non-synonymous somatic mutations, including single-nucleotide variants, insertions, deletions, and frameshifts, were used to predict neoantigens by MuPeXI (DTU Health Tech, Denmark),20 which is a pipeline for neoantigen identification. First, 8 to 11 and 12 to 15 amino acid long mutant peptides were generated for major histocompatibility complex (MHC) I- and MHC II-restricted neoantigen prediction, respectively. The expression of mutant genes was determined from RNA-seq data in transcripts per million. The mutant allele frequency was determined with the variant caller Mutect2.21 Each peptide was provided a priority score based on HLA binding affinity, expression, similarity to self-peptides, and mutant allele frequency. Peptides with a priority score greater than 0 were selected as neoantigen candidates.
Synthesis of long peptides and final vaccine preparation
The non-synonymous mutations were analyzed in the context of the respective 27-mer amino acid epitope with the mutated amino acid in the center (position 14). The peptides with purity greater than 95% were analyzed by mass spectrometry and synthesized by GL Biochem (Shanghai, China) using fluorenylmethoxycarbonyl chemistry and purified using reversed-phase high-performance liquid chromatography. The lyophilized peptides were dissolved in dimethyl sulfoxide, diluted to a concentration of 10 mM in a phosphate buffer brine (pH 7.4), and stored as isocyanates at −80°C.
Recombinant minigenes-pcDNA3.1 plasmids
Tandem minigenes (TMGs) can be constructed, as described previously.22 Minigenes were constructed for mutations; they encoded the mutated amino acid and surrounding upstream and downstream native amino acids for a total length of 27 amino acids. Multiple minigenes were arranged in tandem without additional linker sequences and synthesized (Sangon Biotech Co., Ltd., Shanghai). Each tandem minigene construct was inserted into a pcDNA3.1 vector using available EcoRI, and BamHI cut sites, namely minigene-pcDNA3.1 plasmids. The aforementioned procedure was performed strictly following the manufacturer’s instructions.
Establishment of stable transfectants of SW480
SW480 cells in the logarithmic growth phase were stably transfected with minigene-pcDNA3.1 plasmids using Lipofectamine and Plus reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s instructions and as previously described.23 G418 (Invitrogen, San Diego, CA, USA) was used to select positively transfected cells.24 The efficiency of transfection for minigenes was confirmed by RT-PCR.
Induction of NRTs by co-culturing with CRC patient-derived peripheral blood lymphocytes in vitro
PBMCs from one HLA-A2.1+patient with CRC and human peripheral blood monocyte-derived DCs were generated as described previously.13,25 NRTs were generated as described previously with minor modifications.26 After seven days of co-culturing with peptide-pulsed autologous DCs, lymphocytes were restimulated with peptide-pulsed autologous DCs in a medium containing 10 ng/mL IL-7 and subsequently supplemented with 50 IU/mL rhIL-2 (ThermoFisher) 72 h later. Lymphocytes were restimulated at an interval of seven days in the same manner. Half of the medium was replaced every three days with a fresh medium containing rhIL-2 (50 IU/mL) and expanded as necessary. On day 7, after the third stimulation, the number of cytokine-producing T cells was examined by enzyme-linked immunosorbent spot (ELISPOT) assays, and the cells were tested for cytotoxicity using CCK-8 assays.
Generation of neoantigen-specific T cells in HLA-A2.1/Kb transgenic mice
All experimental procedures and animal protocols were approved by the AAALAC-accredited Animal Studies Committee of Wenzhou Medical University (AEC number: WYDW2019–0924) and were in compliance with all relevant ethical regulations. All procedures were designed in accordance with the ethical standards of the National Research Council to minimize the suffering and number of animals. All animals used in this study were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). The mice were kept at 20 ± 2°C, humidity of 55% ± 10%, lighting of 350 lux (at bench level), and 12/12-hour light/dark cycle. All mice were fed the standard rodent diet with free access to water. The health of the animals was monitored twice daily during the feeding period. No adverse events were observed. This study follows the ARRIVE Guidelines for animal studies.27
For the in vivo efficacy evaluation studies, we used eight 6- to 8-week-old female HLA-A2.1/Kb transgenic mice (mean body weight: 20 ± 2 g), which were raised under the standard conditions and cared for as per the animal care facility guidelines.28 To avoid the interferences of sex in the experiments, only female mice were used in this study. Bone marrow-derived DCs were prepared from Tg mice, and HLA-A2.1/Kb transgenic mice were inoculated with peptide-pulsed DCs.29 The spleen cells of the mice without red blood cells were suspended in the RPMI 1640 medium containing 10% FCS and subsequently counted and placed in 6-well plates. Mouse spleen cells were restimulated by the addition of the corresponding peptides (20 µM) in vitro. After seven days, the numbers of T cell-producing IFN-γ were determined using ELISPOT, and specific killing activity was assessed using CCK8 assay.
ELISPOT assay
IFN-γ ELISPOT kit (Shenzhen Dakewei Biotechnology Co. Ltd, Anshan, China) was used to determine the number of cytokine-producing T cells after overnight activation with peptide.30 We used several culture protocols to analyze T cell-mediated immune responses. Peptide-stimulated PBMCs were added to two wells for approximately 18 h, or DCs pulsed with a peptide were added and co-cultured with T cells. The plate was first washed, and a diluted test antibody (1:100 dilution) was added. Afterward, the plate was incubated at 37°C for 1 h. After washing the plate again, streptavidin–horseradish peroxidase diluted to the above-mentioned year-on-year dilution was added, and the plate was incubated at the same temperature for 1 h. Moreover, a 3-amino-9-ethylcarbazole solution was prepared according to the manufacturers’ instructions and added to each well. The plate was next incubated at room temperature for approximately 15 to 20 min in the dark, after which deionized water was added to stop the development. The plates were scanned using an ELISPOT CTL Reader (Cellular Technology Ltd., Shaker Heights, OH, USA) and analyzed using an ELISPOT software (AIDELISPOT, Strassberg, Germany). A response was considered as positive if a minimum of five spots per 1 × 105 cells were detected and were at least twice over those in the peptide-free (medium only) control.
In vitro T cell cytotoxicity assay
The cytotoxic activity of neoantigen- and tumor-specific cytotoxic T cells can be assessed using the CCK8 assay.31 The formula for determining the percentage specific lysis was as follows: cell cytotoxicity = (1 − (Ae − Ab)/(Ac − Ab)) × 100%, where Ae is the absorbance of the experimental group, Ac is the absorbance of the control group, and Ab is the absorbance of a blank well. Mutated protein-pulsed T2 cells (T2 cells were incubated overnight at 37°C with 20 μM of the corresponding peptide in serum-free RPMI-1640 medium supplemented with 100 ng/mL β2-microglobulin) and minimally nucleated SW480 cells (HLA-A2.1+, minigene expression) were used as peptide-specific targets.
Adoptive immunotherapy in tumor-bearing nude mice
For the adoptive transfer model, we used 6- to 8-week-old female C57BL/6nu/nu mice (mean body weight: 20 ± 2 g). To avoid the interferences of sex in the experiments, only female mice were used in this study. SW480-minigene tumor cells (5 × 106) were injected into the C57BL/6nu/nu mice breast fat pads, forming homogeneous tumors. After three days, the mice were injected intravenously with splenocytes (1 × 108 cells per mouse) from vaccinated HLA-A2.1/Kb transgenic mice in each group that were stimulated with 20 µM HAVCR2-F39V, SEC11A-R11L, and ULK1-S248L for seven days. This adoptive infusion was performed twice at one week of intervals, and intraperitoneal injection of 2000 U of hIL-2 was administered every two days. The control mice accepted splenocytes from transgenic mice by immunization with non-pulsed DCs or injecting only IL-2. The size of the tumor (two-dimensional) was measured thrice per week, and the mice were sacrificed on day 80 after tumor inoculation.
Statistical analysis
A two-tailed Student’s t-test was used to determine the statistical significance of the differences between the means. Tumor sizes were compared between the groups using the Mann–Whitney U test. Survival analyses were plotted using Kaplan–Meier curves. A p-value of < 0.05 was considered statistically significant. All statistical tests were performed using SPSS manager software (v21; IBM Inc., Beijing, China).
Results
Somatic mutation identification and neoantigen prediction for patients with CRC
We recruited 13 patients with CRC (Table 1) and collected tumor tissue and peripheral blood cell samples from each patient for WES and RNA-seq. The tumor tissue sample of patient PW1 was not qualified for sequencing. The sequencing data of PW3 and PW5 did not meet the requirements for neoantigen analysis; therefore, sequencing results for only ten patients were obtained. HLA alleles of these ten patients were identified using OptiType using whole-exome samples, and the results are provided in Supplementary Table 1. A median of 54 (range, 29–89) non-synonymous somatic mutations were identified (Figure 1a) and selected to predict neoantigens that bind to MHC I and II molecules. The total number of mutations in each patient’s exon region is shown. A median of 36 (range, 16–61) and 15 (range, 9–28) MHC I- and MHC II-restricted neoantigens were predicted using MuPeXI (priority score for both >0), respectively (Supplementary Table 2). Among all predicted neoantigens, a median of 9 (range, 5–18) and 46 (range, 22–72) neoantigens were strong and weak binders with a percentage rank less than 0.5 and greater than 0.5, respectively (Figure 1b).
Table 1.
Clinical characteristics of 13 patients receiving personalized immunotherapy
| Patient ID | Age | sex | Primary tumor |
Pathology | Grade | TNM (Stage) |
Metastatic Sites | Prior Therapy | PS* |
|---|---|---|---|---|---|---|---|---|---|
| PW1 | 78 | F | colon | Adenocarcinoma | G3 | T4bN2bM1b (IVB) |
Liver, Peritumor and mesenteric lymph nodes | Surgery | 1 |
| PW2 | 59 | M | colon | Adenocarcinoma | G2 | T3N2bM0 (IIIC) |
Peritumor lymph node | Surgery, Oxaliplatin+capecitabine | 0 |
| PW3 | 33 | F | colon | Adenocarcinoma | G2 | T3N0M1 (IV) |
Liver, Lung, ovary, brain, bone | Oxaliplatin+tegafur, Bevacizumab, Oxaliplatin+capecitabine, Surgery, Irinotecan+5-Fu+calcium folinatc, Apixaban, Oxaliplatin+raltitrexed | 4 |
| PW4 | 52 | F | rectum | Adenocarcinoma | G2 | T3N1aM1 (IV) |
Lung, Peritumor lymph node | Surgery, Bevacizumab+oxaliplatin +capecitabine |
1 |
| PW5 | 72 | F | colon | Adenocarcinoma | G2 | T3N0M1 (IV) |
Liver | Surgery, Oxaliplatin+cetuximab +capecitabine |
1 |
| PW6 | 71 | M | colon | Adenocarcinoma | G2 | T4bN0M1 (IV) |
Liver | Surgery | 1 |
| PW7 | 74 | M | rectum | Adenocarcinoma | G2 | T3N1bM1 (IV) |
Liver, Mesenteric and tumor distal lymph nodes | Surgery, Oxaliplatin+capecitabine | 1 |
| PW8 | 59 | F | colon | Adenocarcinoma | G1-2 | T4aN0M1 (IV) |
Liver | Surgery, Oxaliplatin+cetuximab +capecitabine, Irinotecan+5-Fu +calcium folinatc+cetuximab |
1 |
| PW9 | 49 | F | rectum | Adenocarcinoma | G2 | T3N2M1 (IV) |
Left adnexa, tumor distal lymph nodes | Surgery, Oxaliplatin+capecitabine | 0 |
| PW10 | 66 | F | rectum | Adenocarcinoma | G2 | T4aN1bM1 (IV) |
Uterus, Right fallopian tube, Peritumor lymph nodes | Surgery, Oxaliplatin+capecitabine | 1 |
| PW11 | 57 | M | rectum | Adenocarcinoma | G3 | T4N2M1 (IV) |
Bladder, Peritumor and left internal iliac artery lymph nodes, Napes and omentum majus nodules | Surgery, Oxaliplatin+capecitabine, Bevacizumab, Irinotecan+capecitabine, Bevacizumab+irinotecan+capecitabine, Bevacizumab+raltitrexed, 50 Gy/25 F radiotherapy | 3 |
| PW12 | 89 | M | colon | Adenocarcinoma | G2 | T3N1bM1 (IV) |
Liver, Lung, Mesenteric lymph nodes | Surgery | 2 |
| PW13 | 68 | M | colon | Adenocarcinoma | G3 | T4aN2M1 (IV) |
Peritumor, mesenteric and retroperitoneal lymph nodes | Surgery, Oxaliplatin+capecitabine, Bevacizumab+oxaliplatin +capecitabine, Bevacizumab, Bevacizumab+capecitabine |
1 |
*PS: Performance status: ECOG, Eastern Cooperative Oncology Group.
Figure 1.
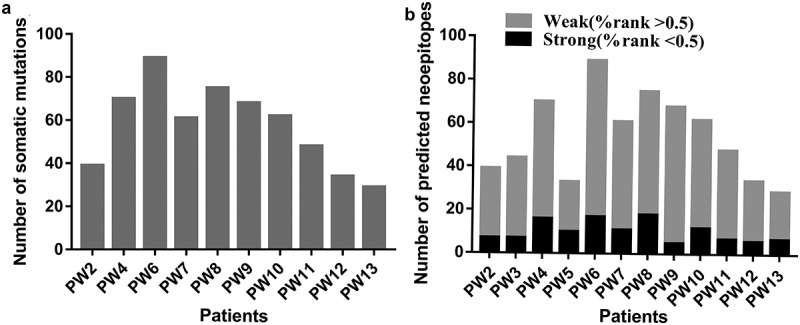
Number of somatic mutations and predicted neoantigens in 10 patients with CRC (a) WES and RNA-seq were performed in 10 patients with CRC. Tumor-specific non-synonymous somatic mutations were identified. The number of somatic mutations of each patient is shown. (b) Neoantigens were predicted for each patient. The number of neoantigens as well as strong (%rank <0.5) and weak binders (0.5< %rank <2) of each patient is shown.
Neoantigen induces enhanced NRT responses from patients with CRC in vitro
PBMCs of patients PW4, PW10, and PW11 were used to evaluate the immunogenicity of neoantigen candidates because they had sufficient PBMCs. Ten hypothesized mutation-related long peptides (27 aa) with strong HLA affinity were selected from PW4 and PW10 for chemical synthesis, respectively. In addition, to further assess the immunogenicity of the polypeptides in HLA-A2.1/Kb transgenic mice, nine HLA-A0201-restricted neoantigen candidates from PW11 were synthesized (Supplementary Table 3). A simple and rapid method was used to determine the peptide immunogenicity in cancer patients26 with a slight modification. The immunogenicity of each synthesized polypeptide was assessed in PBMCs from the corresponding patient. ELISPOT analysis showed that compared with the medium (no peptide) or the unrelated peptide VSV-NP43–69 (STKVALNDLRAYVYQGIKSGNPSILHI), five neoantigen candidates from PW4 (TSHZ3-L523P, RARA-R83H, TP53-R248W, EYA2-V333I, and NRAS-G12D), four from PW10 (TASP1-P161L, RAP1GAP-S215R, MOSPD1-V63I, and NAV2-D1973N), and five from PW11 (HAVCR2-F39V, SEC11A-R11L, SMPDL3B-T452M, LRFN3-R118Q, and ULK1-S248L) induced peptide-specific T cell responses. In particular, RARA-R83H, TP53-R248W, and NRAS-G12D from PW4; TASP1-P161L, RAP1GAP-S215R, and NAV2-D1973N from PW10; and HAVCR2-F39V, SEC11A-R11L, and ULK1-S248L from PW11 induced antigen-specific T cell responses Figure 2(a-c).
Figure 2.
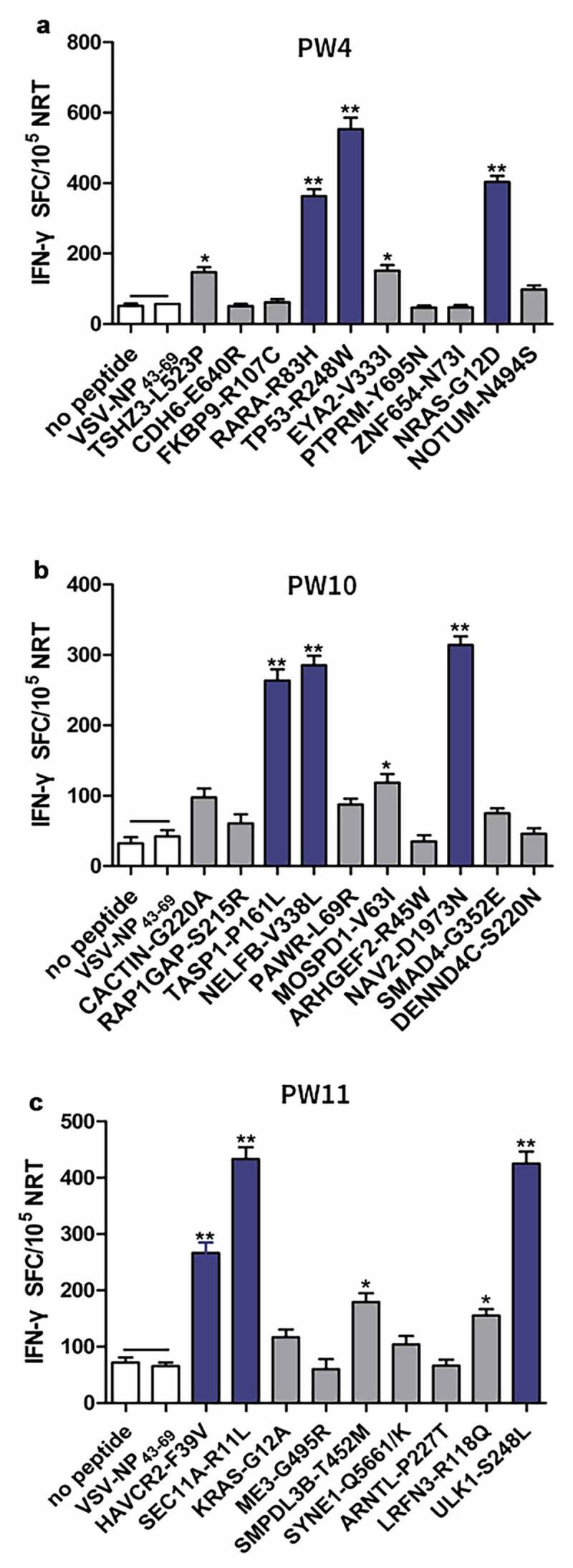
Immunogenicity testing of neoantigens from patients with CRC. Autologous PBMCs were stimulated with candidate mutated peptides every 3 days in the presence of IL-2. On day 10, T-cell responses to each antigen were measured by an IFN-γ ELISPOT assay. The PBMCs in a, b, and c were obtained from PW4, PW10, and PW11 with CRC, respectively. No peptide (medium alone) or VSV-NP43-69 (STKVALNDLRAYVYQGIKSGNPSILHI) stimulation was used as a control. Data are presented as mean ± S.D. of three independent experiments. **p< .01 and *p< .05 compared with IFN-γ production by PBMCs stimulated without peptide or with VSV-NP43-69. NRTs, neoantigen-reactive T cells; SFC, spot-forming cell; VSV-NP, nucleoprotein of vesicular stomatitis virus; S.D., standard deviation.
There is growing evidence that NRTs can achieve tumor regression in patients receiving adoptive cell therapy.17,32 In addition, to evaluate the ability of NRTs to respond to Mut and WT peptides, we used HLA-A0201+autologous DCs from PW11 loaded with HAVCR2-F39V, SEC11A-R11L, or ULK1-S248L to stimulate PBMCs for generating NRTs in vitro. Mutated peptide-specific NRTs were produced in vitro, and their immune responses were compared with WT peptide-induced NRTs. The ELISPOT analysis revealed that NRTs secreted higher levels of IFN-γ in response to Mut peptide stimulation than that the corresponding WT peptide stimulation (Figure 3a).
Figure 3.
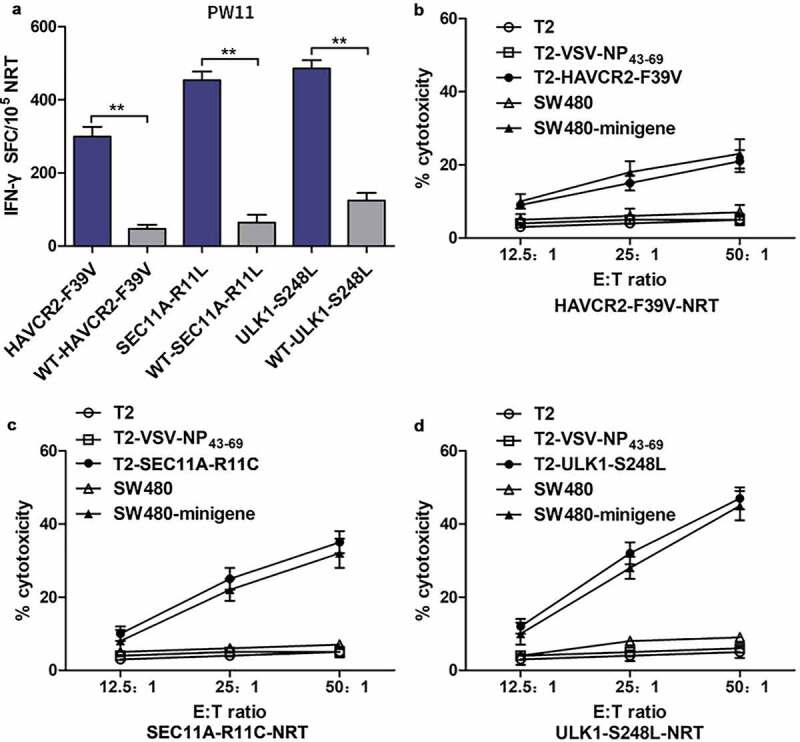
Cytotoxicity of NRTs raised by in vitro stimulation of PBLs of HLA-A0201+PW11. NRTs were induced with autologous HAVCR2-F39V, SEC11A-R11L, or ULK1-S248L-pulsed DCs derived from the PBLs of PW11. On day 7, after the third stimulation, the NRTs were harvested for analysis. (a) IFN-γ secretion by neoantigen-reactive T-cell lines in response to mutated and WT peptides. WT-HAVCR2-F39V, WT-SEC11A-R11L, and WT-ULK1-S248L represent wild type peptides, and HAVCR2-F39V, SEC11A-R11L, and ULK1-S248L represent mutant peptides. IFN-γ-positive SFCs/105 NRTs were detected by cytokine-specific ELISPOT (b, c, and d). Cytotoxicity measured by a CCK8 kit at the specified E:T ratio. Peptide-specific targets were mutated protein-pulsed T2 cells and minimally nucleated SW480 cells (HLA-A2.1+, minigene expression), whereas VSV-NP43-69-pulsed T2 cells, T2 cells alone, and SW480 cells alone were used as controls. Data are expressed as the mean ± standard error of the mean (SEM) of three independent experiments. **p< .01. NRTs, neoantigen-reactive T cells; WT, wild-type; E:T, effector: target; PBL, peripheral blood lymphocyte; SFC, spot-forming cells; SEM, standard error of the mean.
To further examine the cytotoxicity of NRTs, HLA-A0201+T2 cells loaded with a mutant polypeptide and HLA-A0201+SW480 cells transfected with the corresponding mutant polypeptide minigene were used as target cells. The results are shown in Figure 3(b–d). At different E:T ratios, bulk NRTs against HAVCR2-F39V, SEC11A-R11L, and ULK1-S248L, particularly NRTs against SEC11A-R11L and ULK1-S248L, efficiently killed the target cells expressing the corresponding antigen. NRTs showed no significant cytotoxic effect against T2 cells that were not loaded with peptide or loaded with the irrelevant peptide VSV-NP43–69 or with SW480 cells that did not express the mutant peptide.
HLA-A2.1-restricted neoantigens can induce NRTs in HLA-A2.1/Kb transgenic mice in vivo
To evaluate the immunogenicity of candidate polypeptides in vivo, we selected the mutant polypeptides HAVCR2-F39V, SEC11A-R11L, and ULK1-S248L from HLA-A0201+PW11, which were confirmed to be highly immunogenic in vitro, to immunize eight transgenic mice. On days 0 and 7, HAVCR2-F39V, SEC11A-R11L, and ULK1-S248L (100 µg/per peptide) and 50 µg of poly (I:C) were mixed, and subcutaneously injected into transgenic mice for immunization. Splenocytes were harvested seven days after the last immunization, and some splenocytes were evaluated using IFN-γ ELISPOT. The remaining splenocytes were cultured for seven days under stimulation with the corresponding peptide, and T cells were collected for cytotoxicity analysis. The ELISPOT results showed that the NRTs against SEC11A-R11L and ULK1-S248L from HLA-A2.1/Kb mice produced a stronger IFN-γ-secretory response to stimulation with the corresponding peptide than the WT peptide. Stimulation with medium (no peptide) and the unrelated peptide VSV-NP43–69 produced only a background-level secretion response. Moreover, NRTs against HAVCR2-F39V produced only background-level secretion responses to stimulation with various peptides (Figure 4a). In a cytotoxicity assay, the NRTs, especially induced by SEC11A-R11L and ULK1-S248L, expressed significant cytotoxicity to T2 cells and SW480-minigene cells loading or expressing the corresponding mutant peptides. NRTs showed no significant cytotoxic effect against T2 cells that were not loaded with the peptide or loaded with the irrelevant peptide VSV-NP43–69 or SW480 cells that did not express the mutant peptide Figure 4(c, d). NRTs did not exhibit a significant cytotoxic effect against HAVCR2-F39V (Figure 4b). No adverse reactions were observed in mice throughout the experiment.
Figure 4.
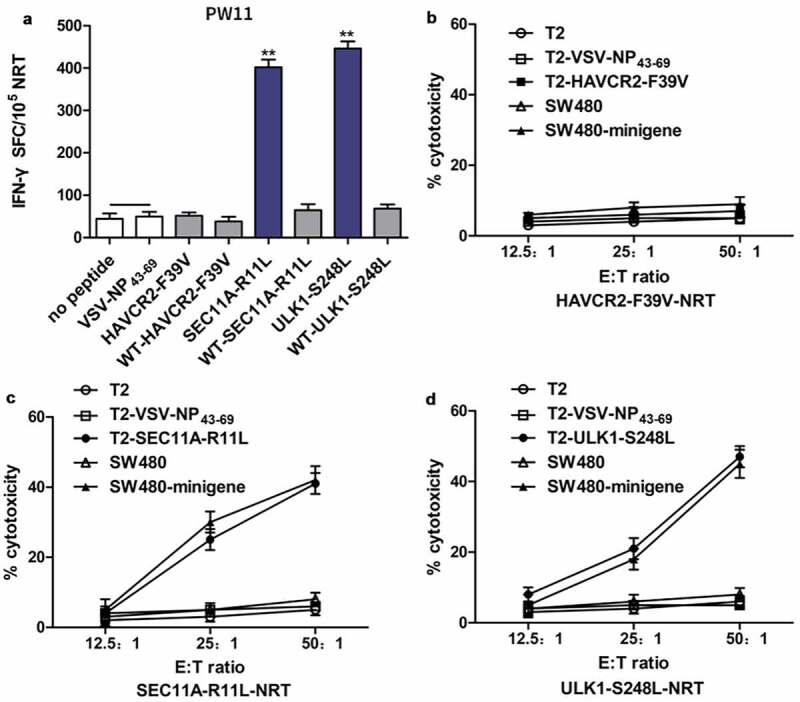
SEC11A-R11L and ULK1-S248L induce more efficient NRT responses than WT epitopes in HLA-A2.1/Kb-Tg mice. (a) Splenocytes of mice (n= 5) vaccinated with mutated peptides were tested by ELISPOT for the recognition of mutated peptides compared to the recognition of the corresponding WT sequences. Data are presented as mean ± standard deviation (SD) of three independent experiments. **p< .01 compared with IFN-γ production by splenocytes stimulated without peptide or with VSV-NP43-69 (b, c, and d). Splenocytes from HLA-A2.1/Kb-Tg mice immunized with mutated peptides were restimulated in vitro with the same mutated peptide for 7 days. Ex vivo cytotoxicity against corresponding mutated peptide-pulsed T2 cells and minigene-nucleofected SW480 cells were examined by CCK8 kit assays at the indicated E:T ratio. VSV-NP43-69-pulsed T2 cells, T2 cells alone, and SW480 cells alone were used as controls. Data are presented as the mean ± S.E.M. of three independent experiments. **p< .01. E:T, effector:target; SFC, spot-forming cell.
Adoptive NRT immunotherapy of C57BL/6nu/nu mice bearing human CRC tumors
To determine whether SEC11A-R11L and ULK1-S248L peptides could be used as a potent vaccine to limit tumor growth in vivo, we obtained HLA-A2.1/Kb transgenic mouse-derived NRTs, which were adoptively transferred into SW480-minigene human CRC-bearing C57BL/6nu/nu mice. Therefore, we developed an adoptive transfer therapy model in nude mice. As shown in Figure 5a, mice subjected to adoptive transfer of SEC11A-R11L- and ULK1-S248L-stimulated NRTs showed significantly delayed tumor growth, whereas NRTs induced by irrelevant peptide could not prevent tumor growth. In addition, 6/10 of the SEC11A-R11-vaccinated mice and 7/10 of the ULK1-S248L-vaccinated mice had a significant long-term survival after tumor inoculation (over 80 days) (Figure 5b). All mice in the control group died on days 20 to 40. Therefore, these results indicated that immunization with SEC11A-R11L and ULK1-S248L peptides induced an effective anti-tumor response in vivo. No adverse reactions were observed in the experimental and control mice.
Figure 5.
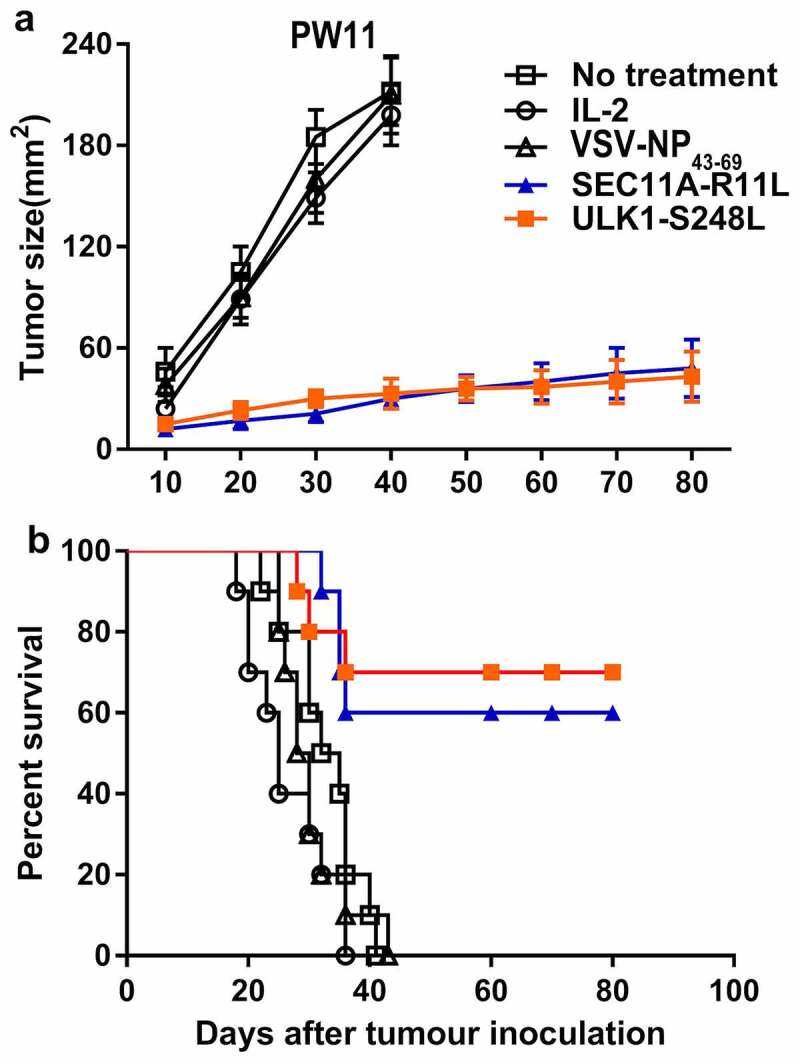
Adoptive immunotherapy of minigene-nucleofected SW480 tumor-bearing nude mice. Small gene-nuclear transfected SW480 tumor cells (5 × 106 cells/mouse) were injected into the flanks of C57BL/6nu/nu mice, and 3 days later, HLA-matched immunization with SEC11A-R11L and ULK1-S248L was performed. Splenocytes (1 × 108 cells/mouse) from HLA-A2.1/Kb-Tg mice were prepared as described in the materials and methods. The control group received only IL-2 or did not receive any treatment. (a) Tumor growth was observed every 3 days and recorded as the mean tumor size (mm2). (b) Mouse survival after tumor inoculation was monitored (n= 10 mice per group).
Discussion
Cancer immunotherapy is rapidly evolving and has been effectively used in clinical practice. Neoantigens-based immunotherapy is a new type of tumor immunity;33 however, studies on the use of neoantigens in CRC, particularly in the Chinese population, are rare. To study the effectiveness of neoantigens in CRC, a workflow was suggested to predict and assess the neoantigens in patients with CRC. Neoantigens were identified for ten patients with CRC, and the immunogenicity of neoantigens was assessed for three patients. Approximately half of the predicted neoantigens for all three patients stimulated T cell responses in PBMCs of patients. In addition, the predicted neoantigens from PW11 (HLA-A0201) showed promising anti-tumor efficacy in HLA-A2.1/Kb transgenic mice and tumor-bearing mouse models.
In a previous study on T cell transfer therapy for CRC, Tran et al. screened cultures of TILs from patients for reactivity against all identified mutant neoepitopes and identified one neoepitope KRAS G12D that stimulated CD8+T cell response.12 Identifying neoantigens was laborious and time-consuming. Thus, we used computational tools to identify neoantigens from hundreds of mutations. However, one of the most crucial problems of in silico neoantigen identification is the low immunogenicity of predicted neoantigen candidates. The state-of-art tools that are primarily based on the binding affinity between peptide and MHC molecules, such as NetMHCpan and NetMHCIIpan, have a low validation rate of approximately 20% to 30%.34 Therefore, MuPeXI was used, which is a computational tool that integrates binding affinity, RNA expression, mutant allele frequency, and similarity to self-peptides, to determine the probability of neoantigen candidates in activating immune responses.20 Our results showed that 50% of neoantigens measured in vitro could induce reactive T cells.
Chen et al. found that indel-related neoantigens covered a large proportion of MSI-H CRC patients.35 About 10 to 20 neoantigen peptides are usually synthesized simultaneously to prepare vaccines or other immunotherapy products.10 Jitske et al. investigated the presence of neoantigen-specific T cell responses in TIL and PBL of seven MMR-p CRC patients.36 Another research group demonstrated the existence of NRTs in several types of metastasis of MMR-p gastrointestinal tumors, including CRC.37 These data show that neoantigen-specific T cells reside in both primary tumors and metastasis of CRC.
Increasing evidence shows that personalized peptide vaccines, in combination with NRT adoptive therapy, have successfully been used to treat several human solid tumors,17 thus confirming the use of NRT-based personalized tumor therapy. We used HLA-A2.1-restricted neoantigens to induce NRTs in HLA-A2.1/Kb transgenic mice and transferred the transgenic mouse-derived lymphocytes into human CRC tumor-bearing C57BL/6nu/nu mice. The CRC mice model showed a significant tumor regression, indicating the effectiveness of NRT cell therapy in CRC.
This study confirmed the presence of numerous neoantigens in CRC. In addition, NRTs induced by neoantigens in CRC can also produce anti-tumor effects.
Limitations: The limitations of this study are as follows: First, the sample size of the experiment was small. Although ten patients were tested for neoantigens, blood samples collected from only three patients were sufficient for vaccine evaluation. In addition, the NRT anti-tumor experiment only from one patient with HLA-A0201 was performed. In further studies, a larger sample size is required to verify the efficacy of neoantigens in anti-tumor immunity. Second, these results are only from in vitro studies or mouse model. Clinical trials are required to evaluate the anti-tumor effects of neoantigens. We are currently conducting clinical trials of neoantigens-based vaccines for CRC. The registration number of our clinical trials is ChiCTR1900022372.
Supplementary Material
Acknowledgments
The authors thank the members of our laboratories for their contributions and helpful discussions. The authors also thank for the analysis of neoantigens by P&PMed (www.pnp-med.com).
Funding Statement
The authors disclosed the receipt of the following financial support for the research, authorship, and/or publication of this manuscript. This work was supported by the National Natural Science Foundation of China (Grant no. 3197080252); Qingdao Science and Technology Project (for benefiting the people, 19–6–1–27-nsh); Qingdao Outstanding Health Professional Development Fund; the Wenzhou Major Science and Technology Special Project (2018ZY004) from Wenzhou Science and Technology Bureau; the Public Benefit Technology Research Project of Zhejiang Science and Technology Department (LGF20H160019); and the Research Foundation of Academician Helin Workstation (item no. 19331103).
Availability of data and materials
The dataset(s) supporting the findings of this study are included in the manuscript.
Consent for publication
Written informed consent for publication was obtained from all participants.
Code availability
Not applicable.
Cell line authentication
Cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA).
Data availability Statement
This article title (DOI: 10.21203/rs.3.rs-22032/v1) is present as a pre-print on the Research Square website and can be accessed on https://www.researchsquare.com/article/rs-22032/v1.
Disclosure statement
The authors declare that they have no conflicts of interest.
Ethical statement
In this study, all investigations and experiments have obtained patients’ consent and been approved by the Medical Ethics Committee of the Second Affiliated Hospital, Wenzhou Medical University (MEC numbers: LCKY2018–67). This study was conducted in accordance with the Declaration of Helsinki. Informed consent was obtained from all participants included in the study.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2021.1891814.
References
- 1.Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Biondi A, Vacante M, Ambrosino I, Cristaldi E, Pietrapertosa G, Basile F. Role of surgery for colorectal cancer in the elderly. World J Gastrointest Surg. 2016;8(9):606–13. doi: 10.4240/wjgs.v8.i9.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biondi A, Grosso G, Mistretta A, Marventano S, Toscano C, Drago F, Gangi S, Basile F. Laparoscopic vs. open approach for colorectal cancer: evolution over time of minimal invasive surgery. BMC Surg. 2013;13 Suppl 2(Suppl2):S12. doi: 10.1186/1471-2482-13-s2-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Loon K, Venook AP. Curable patient with metastatic colorectal cancer: balancing effective therapies and toxicities. J Clin Oncol. 2014;32(10):991–96. doi: 10.1200/jco.2013.53.5195. [DOI] [PubMed] [Google Scholar]
- 6.Sharif S, O’Connell MJ, Yothers G, Lopa S, Wolmark N. FOLFOX and FLOX regimens for the adjuvant treatment of resected stage II and III colon cancer. Cancer Invest. 2008;26(9):956–63. doi: 10.1080/07357900802132550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardoll D, Allison J. Cancer immunotherapy: breaking the barriers to harvest the crop. Nat Med. 2004;10(9):887–92. doi: 10.1038/nm0904-887. [DOI] [PubMed] [Google Scholar]
- 8.Lesterhuis WJ, Haanen JB, Punt CJ. Cancer immunotherapy–revisited. Nat Rev Drug Discov. 2011;10(8):591–600. doi: 10.1038/nrd3500. [DOI] [PubMed] [Google Scholar]
- 9.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi: 10.1158/2159-8290.cd-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565(7738):234–39. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–21. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. T-Cell transfer therapy targeting mutant KRAS in Cancer. N Engl J Med. 2016;375(23):2255–62. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Wan T, Zhou X, Wang B, Yang F, Li N, Chen G, Dai S, Liu S, Zhang M, et al. Hsp70-like protein 1 fusion protein enhances induction of carcinoembryonic antigen-specific CD8+ CTL response by dendritic cell vaccine. Cancer Res. 2005;65(11):4947–54. doi: 10.1158/0008-5472.can-04-3912. [DOI] [PubMed] [Google Scholar]
- 14.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27(2):182–89. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh S, Chan CK. Analysis of RNA-seq data using tophat and cufflinks. Methods Mol Biol. 2016;1374:339–61. doi: 10.1007/978-1-4939-3167-5_18. [DOI] [PubMed] [Google Scholar]
- 17.Chen F, Zou Z, Du J, Su S, Shao J, Meng F, Yang J, Xu Q, Ding N, Yang Y, et al. Neoantigen identification strategies enable personalized immunotherapy in refractory solid tumors. J Clin Invest. 2019;129(5):2056–70. doi: 10.1172/jci99538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30(23):3310–16. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weese D, Holtgrewe M, Reinert K, Razer S. 3: faster, fully sensitive read mapping. Bioinformatics. 2012;28(20):2592–99. doi: 10.1093/bioinformatics/bts505. [DOI] [PubMed] [Google Scholar]
- 20.Bjerregaard AM, Nielsen M, Hadrup SR, Szallasi Z, Eklund AC. MuPeXI: prediction of neo-epitopes from tumor sequencing data. Cancer Immunol Immunother. 2017;66(9):1123–30. doi: 10.1007/s00262-017-2001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim B, Won D, Jang M, Kim H, Choi JR, Kim TI, Lee ST. Next-generation sequencing with comprehensive bioinformatics analysis facilitates somatic mosaic APC gene mutation detection in patients with familial adenomatous polyposis. BMC Med Genomics. 2019;12(1):103. doi: 10.1186/s12920-019-0553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344(6184):641–45. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He JD, Luo HL, Li J, Feng WT, Chen LB. Influences of the interferon induced transmembrane protein 1 on the proliferation, invasion, and metastasis of the colorectal cancer SW480 cell lines. Chin Med J (Engl). 2012;125:517–22. [PubMed] [Google Scholar]
- 24.Ogura M, Kakuda T, Takarada T, Nakamichi N, Fukumori R, Kim YH, Hinoi E, Yoneda Y. Promotion of both proliferation and neuronal differentiation in pluripotent P19 cells with stable overexpression of the glutamine transporter slc38a1. PLoS One. 2012;7(10):e48270. doi: 10.1371/journal.pone.0048270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Yu Y, Zhang M, Wang W, Cao X. The involvement of TNF-alpha-related apoptosis-inducing ligand in the enhanced cytotoxicity of IFN-beta-stimulated human dendritic cells to tumor cells. J Immunol. 2001;166:5407–15. doi: 10.4049/jimmunol.166.9.5407. [DOI] [PubMed] [Google Scholar]
- 26.Sun W, Wei X, Niu A, Ma X, Li JJ, Gao D. Enhanced anti-colon cancer immune responses with modified eEF2-derived peptides. Cancer Lett. 2015;369:112–23. doi: 10.1016/j.canlet. [DOI] [PubMed] [Google Scholar]
- 27.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. Osteoarthritis Cartilage. 2012;20:256–60. doi: 10.1016/j.joca.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Slater AM, Cao LA. Protocol for housing mice in an enriched environment. J Visualized Exp. 2015;(100)e52874. doi: 10.3791/52874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S, Yi L, Ling M, Jiang J, Song L, Liu J, Cao X. HSP70L1-mediated intracellular priming of dendritic cell vaccination induces more potent CTL response against cancer. Cell Mol Immunol. 2018;15(2):135–45. doi: 10.1038/cmi.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su S, Hu B, Shao J, Shen B, Du J, Du Y, Zhou J, Yu L, Zhang L, Chen F, et al. CRISPR-Cas9 mediated efficient PD-1 disruption on human primary T cells from cancer patients. Sci Rep. 2016;6:20070. doi: 10.1038/srep20070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu C, Zheng Y, Tang J, Wang D, Ma Z, Li S, Chang X. Stimulation of DC-CIK with PADI4 protein can significantly elevate the therapeutic efficiency in esophageal cancer. J Immunol Res. 2019;2019:6587570. doi: 10.1155/2019/6587570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med. 2018;24(6):724–30. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jinushi M, Hodi FS, Dranoff G. Enhancing the clinical activity of granulocyte-macrophage colony-stimulating factor-secreting tumor cell vaccines. Immunol Rev. 2008;222:287–98. doi: 10.1111/j.1600-065X.2008.00618.x. [DOI] [PubMed] [Google Scholar]
- 34.Bjerregaard AM, Nielsen M, Jurtz V, Barra CM, Hadrup SR, Szallasi Z, Eklund AC. An analysis of natural T cell responses to predicted tumor neoepitopes. Front Immunol. 2017;8:1566. doi: 10.3389/fimmu.2017.01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Liu S, Qu R, Li B. Recurrent neoantigens in colorectal cancer as potential immunotherapy targets. Biomed Res Int. 2020;2020:2861240. doi: 10.1155/2020/2861240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Bulk J, Verdegaal EME, Ruano D, Ijsselsteijn ME, Visser M, van der Breggen R, Duhen T, van der Ploeg M, de Vries NL, Oosting J, et al. Neoantigen-specific immunity in low mutation burden colorectal cancers of the consensus molecular subtype 4. Genome Med. 2019;11:87. doi: 10.1186/s13073-019-0697-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parkhurst MR, Robbins PF, Tran E, Prickett TD, Gartner JJ, Jia L, Ivey G, Li YF, El-Gamil M, Lalani A, et al. Unique neoantigens arise from somatic mutations in patients with gastrointestinal cancers. Cancer Discov. 2019;9:1022–35. doi: 10.1158/2159-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset(s) supporting the findings of this study are included in the manuscript.
This article title (DOI: 10.21203/rs.3.rs-22032/v1) is present as a pre-print on the Research Square website and can be accessed on https://www.researchsquare.com/article/rs-22032/v1.


