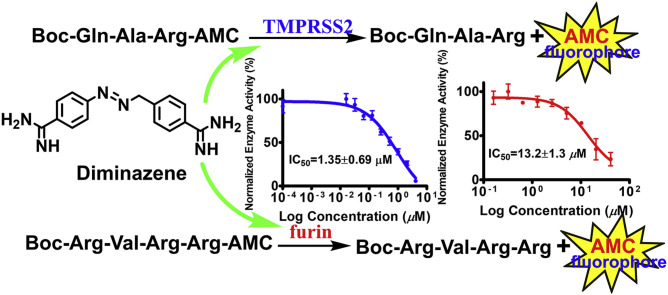
Abstract
The proteases TMPRSS2 (transmembrane protease serine 2) and furin are known to play important roles in viral infectivity including systematic COVID-19 infection through priming of the spike protein of SARS-CoV-2 and related viruses. To discover small-molecules capable of inhibiting these host proteases, we established convenient and cost-effective cell-based assays employing Vero cells overexpressing TMPRSS2 and furin. A cell-based proteolytic assay for broad-spectrum protease inhibitors was also established using human prostate cancer cell line LNCaP. Evaluation of camostat, nafamostat, and gabexate in these cell-based assays confirmed their known TMPRSS2 inhibitory activities. Diminazene, a veterinary medicinal agent and a known furin inhibitor was found to inhibit both TMPRSS2 and furin with IC50s of 1.35 and 13.2 μM, respectively. Establishment and the use of cell-based assays for evaluation TMPRSS2 and furin inhibitory activity and implications of dual activity of diminazene vs TMPRSS2 and furin are presented.
Keywords: SARS-CoV-2, TMPRSS2, Furin, Cell-based assays, Diminazene, Dual inhibitor
Graphical abstract
As of early June 2021, the pandemic COVID-19 caused by Severe Acute Respiratory Syndrome CoronaVirus-2 (SARS-CoV-2) has infected over 171 million individuals causing over 3.7 million deaths worldwide (World Health Organization Coronavirus), with over 33 million cases and 592,700 deaths reported in the U.S.(Centers for Disease Controls) Thanks to the recent development of vaccines, despite some concerns about their storage, distribution and vaccine-hesitant population, there is hope that the spread of the virus can be controlled. However, substantial genetic diversity, frequent recombination, and cumulative mutations of the RNA genome of SARS-CoV-2 have collectively resulted in a greater likelihood of the emergence of more transmissible and/or virulent mutants evading vaccine-derived immunity (Liu et al., 2020). In addition, there are a number of recent reports on serious adverse side-effects of COVID-19 vaccines including rare cases of myocarditis (inflammatory condition of the heart muscles) (Oster et al., 2022), neurological complications (Assiri et al., 2022), and rapidly progressive dementia (Chakrabarti et al., 2021). Thus, there is an urgent need for therapeutics to prevent/treat current and future outbreaks of Coronaviruses (CoVs). Therapeutic/preventative agents for CoVs can be targeted toward the virus, its proteins, or the host cells. Although drugs that directly target RNA viruses like CoVs have some advantages, drug resistance may develop rapidly after treatment, particularly when mutations occur frequently (Shyr et al., 2020). Conversely, those targeting host cells may slow the development of drug resistance as mutations in host cells are relatively rare, providing greater potential for prevention/treatment of SARS-CoV-2.
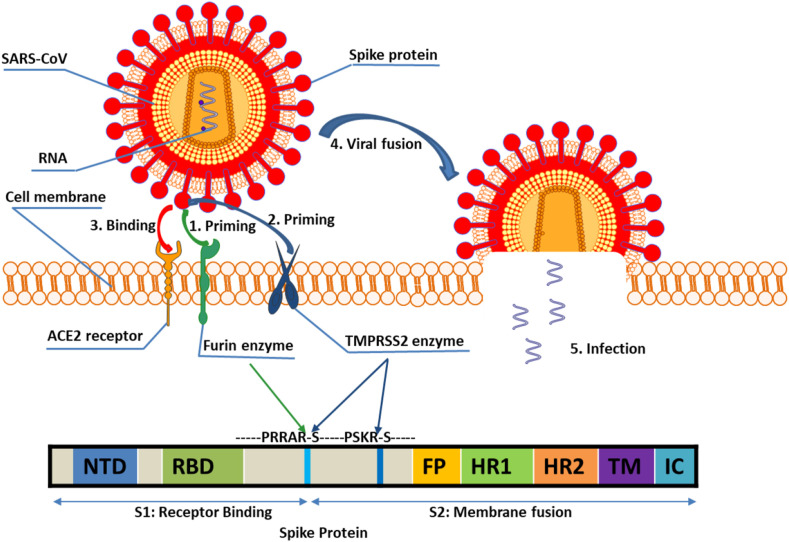
SARS-CoV-2 gains entry to the host cell through binding of the viral S protein to host ACE2 receptors on the surface of target cells (Shang et al., 2020). However, for the virus to enter host cells, double cleavage of the viral spike protein at the S1/S2 cleavage site and at the S2ʹ site is required (Fig. 1 ). Cleavage at S1/S2 site may be crucial for conformational changes required for receptor binding and/or subsequent exposure of S2ʹ site to host proteases for viral entry (Hoffmann et al., 2020a,b). Many proteases have been found to activate CoVs in vitro, including furin, cathepsin L, and trypsin-like serine proteases such as the transmembrane protease serine 2 (TMPRSS2), TMPRSS11A, and TMPRSS11D (Luan et al., 2020). Among these, TMPRSS2 and furin play major roles in proteolytic activation of a broad range of viruses (Bestle et al., 2020). TMPRSS2 is a type II transmembrane serine protease (TTSP) widely expressed in epithelial cells of the respiratory, gastrointestinal, and urogenital tract (Thunders and Delahunt, 2020). In addition, TMPRSS2 mutations have not been associated with any inherited pathologies in humans and TMPRSS2 knockout mice have no notable phenotype (Thunders and Delahunt, 2020). Importantly, functional inhibition of TMPRSS2 by camostat and nafamostat is known to reduce SARS-CoV-2 entry into cells (Qiao et al., 2021), and studies with TMPRSS2 transgenic knockout mice have shown that loss of TMPRSS2 reduces CoV replication in lungs resulting in a milder lung pathology (Iwata-Yoshikawa et al., 2019). Developmental regulation of the expression of the TMPRSS2 gene has been suggested to provide relative protection of infants and children from severe respiratory illness (Schuler et al., 2021), and epidemiological data has shown that the incidence and severity of diagnosed COVID-19 is higher in men than in women (Baughn et al., 2020). One possible explanation for this could be differences in levels of sex hormones, such as androgens, and the transcriptional signaling networks that subsequently occur in males vs females, including up-regulation of the TMPRSS2 host entry factor in males. This has led to the hypothesis that inhibition of androgen receptor (AR) activity and down-regulation of TMPRSS2 may prevent SARS-CoV-2 infection (Stopsack et al., 2020). A recent study to test this hypothesis has provided a strong rationale for the use of AR inhibitors such as enzalutamide [drug for castration-resistant prostate cancer (PC)] to decrease TMPRSS2 expression and in COVID-19 treatment (Qiao et al., 2021). Other studies have shown that enzalutamide treatment of mice dramatically decreased TMPRSS2 levels in the lung (Leach et al., 2020). It has recently been demonstrated that although enzalutamide effectively inhibited SARS-Cov-2 infection in human prostate cells, it had no protective role in treating COVID-19 through reducing TMPRSS2 expression in lung cells (Li et al., 2021). However, PC patients receiving androgen deprivation therapy appear to be partially protected from SARS-CoV-2 infection (Montopoli et al., 2020; Bahmad and Abou-Kheir, 2020; Mollica et al., 2020). Thus, targeting TMPRSS2 expression and/or activity could be a promising approach for potential intervention of corona viruses including COVID-19 (Kazuya et al., 2013).
Fig. 1.
Binding of TMPRSS2 and furin priming Spike protein to ACE2 receptor before cellular entry and viral membrane fusion enabled by TMPRSS2 priming. NTD: N-terminal domain; RBD: receptor-binding domain; FP: fusion peptide; HR: heptad repeats; TM: transmembrane domain; CT: cytoplasmic tail.
Furin is a member of the proprotein convertases (PCs) family containing a catalytic serine protease domain of the subtilisin type which exists in both membrane-bound and secreted forms (van de Ven et al., 1990). Furin is involved in numerous normal physiological and pathogenic processes including viral propagation, bacterial toxin activation, dementia, cancer, and metastasis (Thomas, 2002). It has recently been proposed that the SARS-CoV-2 spike glycoprotein (S) contains a furin cleavage complex (FCC) and that FCC contributes to its infectivity and pathogenicity in multiple ways (Coutard et al., 2020). Furin efficiently cleaves S1/S2 with the sequence RRAR↓S in vitro, which is an important step for the spike protein of SARS-Cov-2 to bind to host ACE receptor and gain entry into the host cell (Hoffmann et al., 2020a,b). SARS-Cov-2 also uses endogenous furin to cleave the S protein in the trans-Golgi network soon after virion assembly. This latter mechanism separates furin from other virally hijacked proteases such as TMPRSS2 potently increasing the pathogenicity of SARS-CoV-2 (Fitzgerald, 2020). Furin's cleavage site requirements have been used to produce potent peptide- and protein-based inhibitors capable of blocking in vitro and in vivo furin activity (Becker et al., 2010). Furthermore, replication of SARS-Cov-2 in HEK293 cells was strongly inhibited by the furin inhibitor MI-1851(Hoffmann et al., 2020a,b).
To date, most of the TMPRSS2 and furin inhibitory assays developed for SARS-CoV-2 drug discovery involved the use of purified recombinant human TMPRSS2 and furin proteins expressed in various organisms. Using TMPRSS2 enzyme expressed in yeast, Shrimp et al. confirmed the TMPRSS2 inhibitory activities of three small-molecule drugs camostat, nafamostat, and gabexate approved in Japan to treat pancreatitis (Shrimp et al., 2020). However, the FDA-approved mucolytic cough suppressant bromhexine that was previously reported to be an inhibitor of TMPRSS2 using recombinant human TMPRSS2 expressed in the fungus, Pichia pastoris (Maggio and Corsini, 2020), was found to be inactive in this assay. Using an assay involving recombinant furin enzyme expressed in cells of the insect, Spodoptera frugiperda, Peng et al. found that the natural product luteolin inhibited furin activity and restricted dengue virus replication (Peng et al., 2017). By using the human recombinant furin expressed in human embryonic kidney (HEK293) cells, a recent report confirmed that diminazene was able to inhibit this enzyme (Wu et al., 2020).
Although the use of enzyme-based biochemical assays for drug discovery has certain advantages, the effect of a drug on an organism is complex and involves interactions at multiple levels that cannot be predicted using biochemical assays. Contrary to biochemical assays, cell-based assays are more biologically relevant surrogates to predict the response of the organism. Furthermore, at some point in the drug discovery process predicting cellular toxicity is important and eukaryotic cell culture is accepted as the model system of choice to obtain a first approximation of toxicity. TMPRSS2 enzyme exists mostly as a transmembrane protein. However, some of it is excreted to the extracellular environment near membrane to activate other proteins on the membrane (Böttcher-Friebertshäuser, 2018). In contrast, furin is an intracellular enzyme enriched in Golgi apparatus, but some of it is present as a membrane protein that is excreted to the extracellular environment (Vidricaire et al., 1993). The first cell-based TMPRSS2 assay was reported using porcine small intestinal epithelial cells (IPEC-J2) that led to the discovery of the small-molecule synthetic inhibitor I-432(Pászti-Gere et al., 2016). However, the abundance of matriptase in IPEC-J2 cells with proteolytic activity similar to TMPRSS2 limited the use of this assay for TMPRSS2 selective inhibitors (Pászti-Gere et al., 2015). A number of stably transfected cell lines overexpressing TMPRSS2 have been developed especially for the purpose of understanding the viral infection of host cells. These include human lung carcinoma cells A549/hACE2/TMPRSS2 (Tran et al., 2021), human colon epithelial cells Caco-2/TMPRSS2/TMPRSS4 (Bertram et al., 2010), human embryonic kidney cells HEK-293T/TMPRSS2 (Azouz et al., 2020), and African green monkey kidney epithelial cells VeroE6/TMPRSS2 (Schuler et al., 2021). Of these, HEK-293T cells transfected with PLX-TMPRSS2 plasmid has been used to screen for TMPRSS2 inhibitors that led to the discovery of alpha 1 antitrypsin (A1AT), a protein belonging to the serpin superfamily (Azouz et al., 2020). Stably transfected furin-overexpressing cell lines have been produced and used to investigate the role of furin protease in hepatocellular carcinoma, heart development, and viral infection of host cells. These include human hepatoma cells Huh7/furin (Huang et al., 2012), human head and neck squamous cell carcinoma (HNSCC) cells SCC12/furin and SCC15/furin (Bassi et al., 2003), Chinese hamster ovary cells CHO-GRAP/furin (Coppola et al., 2007; Ramos-Molina et al., 2015), CHO-FD11/furin (Susan-Resiga et al., 2011), and African green monkey kidney epithelial cells Vero/furin (Mukherjee et al., 2016). Of these, the CHO-GRAP/furin cell line contains a recombinant reporter consisting of a Golgi retention (GR) signal and a protease cleavage sequence fused to alkaline phosphatase (AP) (Coppola et al., 2007; Ramos-Molina et al., 2015), and the method focused on the measurement of protease activity on Golgi-resident protease.
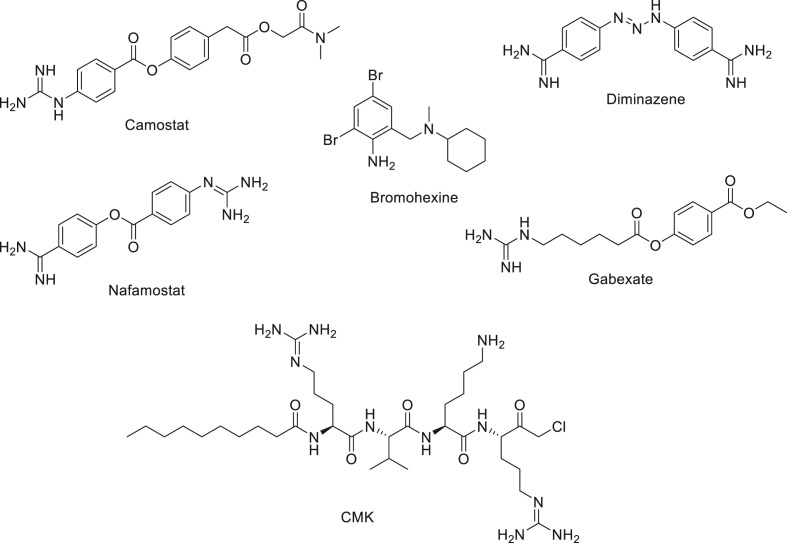
In this study, we established and used cell-based assays for inhibition of TMPRSS2 and furin employing VeroE6/TMPRSS2 cells overexpressing TMPRSS2 (Schuler et al., 2021), and Vero/furin cells overexpressing furin (Mukherjee et al., 2016). The human prostate cancer (PC) cell line LNCaP known to overexpress TMPRSS2 enzyme (Lucas et al., 2014; Chen et al., 2010) was also used to further evaluate compounds active in our VeroE6/TMPRSS2 cell-based assay. Herein, we report the establishment of these cell-based assays and the application of these assays to evaluate known TMPRSS2 inhibitors including camostat, nafamostat, gabexate (Shrimp et al., 2020), and the controversial bromhexine (Shrimp et al., 2020; Maggio and Corsini, 2020) and furin inhibitor Decanoyl-Arg-Val-Lys-Arg-CMK (CMK) (Fig. 2 ) (Angliker et al., 1993) Our cell-based assays also led to the discovery of diminazene (Fig. 2), a veterinary drug used as a treatment of trypanosomiasis and babesiosis (Qaradakhi et al., 2020), as a dual inhibitor of TMPRSS2 and furin.
Fig. 2.
Some TMPRSS2 and furin inhibitors.
1. Results and discussion
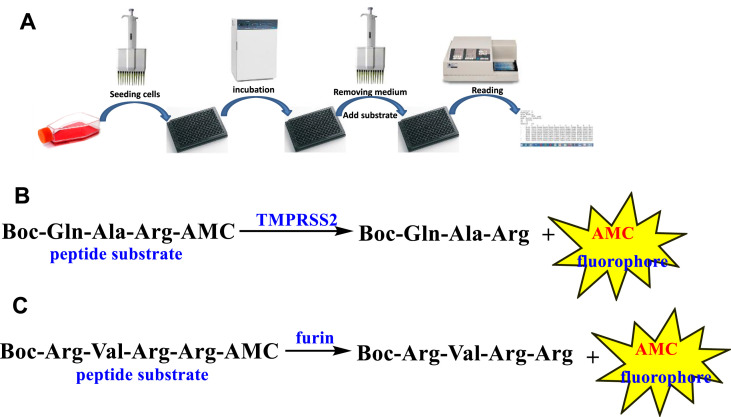
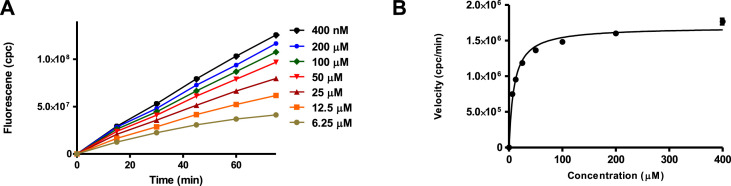
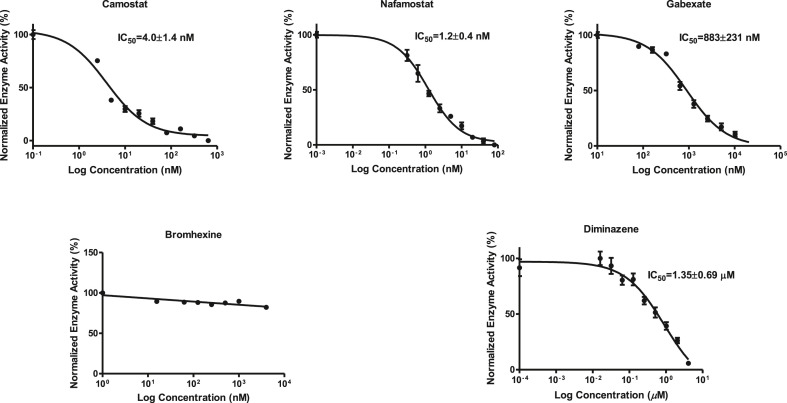
The commercially available fluorogenic peptides, Boc-Gln-Ala-Arg-AMC and Boc-Arg-Val-Arg-Arg-AMC, previously shown to be good substrates for TMPRSS2 (Shrimp et al., 2020) and furin (Wu et al., 2020), respectively were selected for optimizing the experimental conditions of the cell-based enzymatic assays. The assay scheme is illustrated in Fig. 3 . The VeroE6/TMPRSS2 overexpressing cells produced by Matsuyama et al. (2020) were obtained from Japanese Collection of Research Bioresources (JCBR) cell bank. Cells were cultured in DMEM supplemented with 10% heat inactivated fetal bovine serum containing 100 unit/mL of penicillin, 100 μg/mL of streptomycin, and 1.0 mg/mL of geneticin in a T-flask by following the instructions of JCBR cell bank. Furin overexpressing Vero/furin cells produced by Mukherjee et al. (2016) were obtained from the U.S. National Institute of Allergy and Infectious Diseases. Vero/furin cells were cultured in DMEM with GlutaMAX medium supplemented with 7% FBS, 100 unit/mL of penicillin, 100 μg/mL of streptomycin, and 5 μg/mL of blasticidin (Mukherjee et al., 2016). When 90–100% confluence was achieved, the cells were seeded to a 96-well black and clear bottom microplate. Based on cell seeding number test (Supplementary Figures S1 and S7), a seeding density of 1 × 104 cells/well was selected for both TMPRSS2 and furin assays resulting in consistency in cell growth rate and health. Further experimentation suggested that the use of a detergent for furin assay is necessary for increasing permeability of the cell membrane since most of the enzyme resides intracellularly (Supplementary Figure S4). The substrate concentration used for the bioassay is an additional important factor which is known to affect the cost and sensitivity of the assay. Through the titration experiments of the substrate (Fig. 4 A), the K m of the TMPRSS2 assay was determined to be 9.5 μM (Fig. 4B) and the K m of the furin assay was determined to be 44.6 μM (Supplementary Figure S6). It is known that the substrate concentration should be less than K m in assays involving purified enzymes. However, cell-based enzyme assays involve heterogeneous reactions which means the substrate concentration around the enzyme will be much lower compared to assays using purified enzymes suggesting that a substrate concentration higher than K m will be required for cell-based enzyme assays. Considering detection noise due to the background fluorescence of the substrate and the plates, together with the sensitivity of the instrument, 50 or 100 μM concentrations of the substrate with 30 or 60 min pre-incubation before reading fluorescence intensity were found to satisfy the assay requirements for sensitivity and consistency within the period of initial reaction rate. The final protocols used for the cell-based TMPRSS2 and furin inhibitory assays are presented in Table 1, Table 2 , respectively. The known TMPRSS2 inhibitors camostat, nafamostat, and gabexate were then evaluated using the above cell-based TMPRSS2 assay resulting in IC50s of 4.0 ± 0.7 nM for camostat, 1.2 ± 0.2 nM for nafamostat, and 883 ± 231 nM for gabexate (Fig. 5 ). Bromhexine was found to be inactive up to a concentration of 4.0 μM. Diminazene, a veterinary drug structurally related to camostat and nafamostat, showed a moderate TMPRSS2 inhibitory activity with an IC50 of 1.35 ± 0.69 μM in our cell-based assay.
Fig. 3.
(A) Cell-based enzyme inhibition assay workflow. (B) Enzymatic hydrolysis of the peptide substrate Boc-Gln-Ala-Arg-AMC by TMPRSS2. (C) Enzymatic hydrolysis of the peptide substrate Boc-Arg-Val-Arg-Arg-AMC by furin.
Fig. 4.
(A) TMPRSS2 substrate titration curves. (B) Kinetic data from Figure A were plotted to estimate the Km by curve fitting of the Michaelis–Menten equation (Km = 9.5 μM). Curve fitting was carried out using GraphPad Prism. Error bars indicate the standard error from the mean.
Table 1.
Detailed protocol for cell-based TMPRSS2 enzyme inhibitory assay using VeroE6/TMPRSS2 cells.
| Step no. | Process | Notes |
|---|---|---|
| 1 | Cells plated at a density of 10,000 viable cells/well in triplicate in 96-well microplate with 100 μL/well culture medium. | DMEM (e.g., Corning cat.# 10-013-CV), supplemented with 10% heat inactivated fetal bovine serum (e.g., Millipore-Sigma cat.# 12306C-100 ML), 100 unit/mL penicillin and streptomycin, and 1.0 mg/mL geneticin used as culture medium. Black and clear bottom 96-well plate (e.g., Corning Corsta 3603). |
| 2 | The plate incubated at 37 °C and 5% CO2 for 24 h | |
| 3 | Take out the plate from the incubator, remove the culture medium completely. Wash wells with 100 μL/well of DPBS (1x) and completely remove the DPBS. | The monolayer of cells in the wells should stay at the bottom of the wells when washing. The DPBS (1x) buffer should be without calcium, magnesium, and phenol red (e.g., Corning cat# 21-031-cv) |
| 4 | Add 50 μL/well of inhibitor solution in DPBS (1x) and stand at RT for 5 min. | The inhibitor stock solution prepared by dissolving the inhibitor in DMSO or H2O. Stock solutions diluted with DPBS (1x). Final DMSO concentration should be less than 0.2%. |
| 5 | Add 50 μL/well of substrate solution in DPBS (1x). | |
| 6 | Read fluorescence intensity after 30 min at excitation 360 nm/emission 460 nm. | Instrument parameter settings is depend on the microplate reader used. For LJL Biosystems Analyst AD, a default parameters, including Z height: middle of the well; integration time: 100 ms; Attenuator: off; PMT mode: SmartRead with sensitivity 2; Plate shaking time: 10 s; Optics: bottom with 50/50 dichroic mirror. |
Table 2.
Detailed protocol for cell-based furin enzyme inhibitory assay using Vero/furin cells.
| Step no. | Process | Notes |
|---|---|---|
| 1 | Cells plated at a density of 10,000 viable cells/well in triplicate in 96-well microplate with 100 μL/well culture medium. | DMEM with glutaMAX (e.g., Gibco cat.# 10566-016), supplemented with 7% fetal bovine serum (e.g., Hyclone cat.# SH30396.02), 100 unit/mL penicillin and streptomycin, and 5 μg/mL of blasticidin used as culture medium. Black and clear bottom 96-well plate (e.g., Corning Corsta 3603). |
| 2 | The plate incubated in 37 °C and 5% CO2 for 24 h | |
| 3 | Take out the plate from incubator and remove culture medium completely. Wash wells with 100 μL/well of DPBS (1x) and completely remove DPBS. | Monolayer of cells in the wells should stay at the bottom of the wells when washing. DPBS (1x) buffer should be without calcium, magnesium, and phenol red (e.g. Corning cat# 21-031-cv) |
| 4 | Add 50 μL/well of inhibitor solution in DPBS (1x) containing 0.2% Tween 20 and stand at RT for 30 min. | Inhibitor stock solution prepared by dissolving the inhibitor in DMSO or H2O. Stock solution diluted with DPBS (1x) containing 0.2% Tween 20. Final DMSO concentration should be less than 0.2%. |
| 5 | Add 50 μL/well of substrate solution in DPBS (1x). | |
| 6 | Read fluorescence intensity after 30 min at excitation 360 nm/emission 460 nm. | The instrument parameter setting is dependent on the microplate reader used. For LJL Biosystems Analyst AD, a default parameters, including Z Height: middle of the well; Integration time: 100 ms; Attenuator: off; PMT mode: SmartRead with sensitivity 2; Plate shaking time: 10 Sec.; Optics: bottom with 50/50 Dichroic mirror. |
Fig. 5.
Inhibitory activities of camostat, nafamostat, gabexate, and bromhexine against TMPRSS2 in VeroE6/TMPRSS2 cell-based assay.
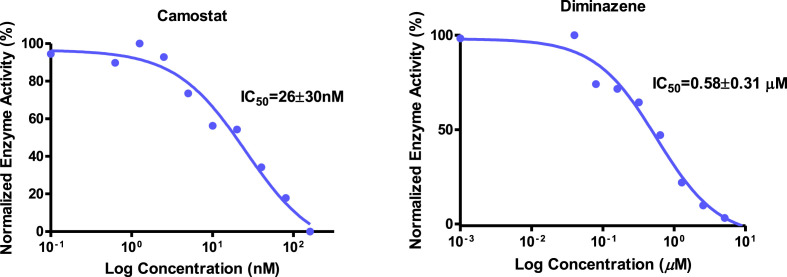
It is known that besides overexpressing TMPRSS2, LNCaP cells express matriptase, a type II transmembrane serine protease with a similar proteolytic activity (Ko et al., 2015). Matriptase has been confirmed to activate influenza virus infection (Beaulieu et al., 2013), and was considered to be one of the major membrane-bound serine proteases priming the spike protein of SARS-CoV-2 (Fuentes-Prior, 2020). These findings suggested that an LNCaP cell-based assay may be useful for evaluating compounds for their ability to inhibit a wide range of proteases. The cell-based assay with LNCaP cells was installed by replacing VeroE6/TMPRSS2 cells by LNCaP cells in the above cell-based TMPRSS2 enzyme assay (Supplementary Figure S2). Evaluation of camostat and diminazene using LNCaP cell-based proteolytic enzyme inhibitory assay suggested their IC50s to be 26 ± 30 nM and 0.58 ± 0.31 μM, respectively (Fig. 6 ). Interestingly, camostat was found to be over six times less active in LNCaP cell-based assay compared to VeroE6/TMPRSS2 cell-based assay, but the inhibitory activity of diminazene was over two times higher in LNCaP cell-based assay than in VeroE6/TMPRSS2 cell-based assay. These results suggested that camostat is a more selective TMPRSS2 enzyme inhibitor, while diminazene in addition to TMPRSS2, inhibits a wide range of proteolytic enzymes. This finding was further supported by the ability of diminazene to inhibit furin in our Vero/furin cell-based assay as described below.
Fig. 6.
Inhibitory activities of camostat and diminazene in TMPRSS2 in LNCaP cell-based assay.
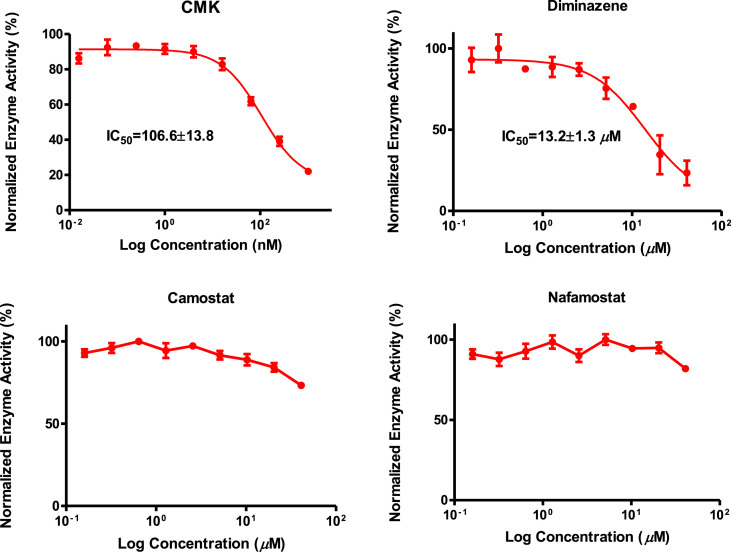
A known furin enzyme inhibitor, furin inhibitor I (Decanoyl-Arg-Val-Lys-Arg-CMK, abbreviated as CMK) and three TMPRSS2 inhibitors, camostat, nafamostat, and diminazene, were tested for their ability to inhibit furin using our Vero/furin cell-based assay. The results revealed that CMK exhibited strong furin inhibitory activity with an IC50 of 107 ± 14 nM and diminazene showed moderate activity with an IC50 of 13.2 ± 1.3 μM (Fig. 7 ). Interestingly, camostat and nafamostat were found to be inactive in Vero/furin cell-based assay up to a concentration of 40.0 μM. Diminazene has been shown to have furin inhibitory activity in an assay using the purified enzyme (Wu et al., 2020). Our data supports the previous finding (Shrimp et al., 2020) that camostat and nafamostat are selective TMPRSS2 inhibitors and that diminazene is a dual inhibitor of the proteolytic enzymes TMPRSS2 and furin.
Fig. 7.
Inhibitory activities of CMK, diminazene, camostat, and nafamostat in Vero/furin cell-based assay.
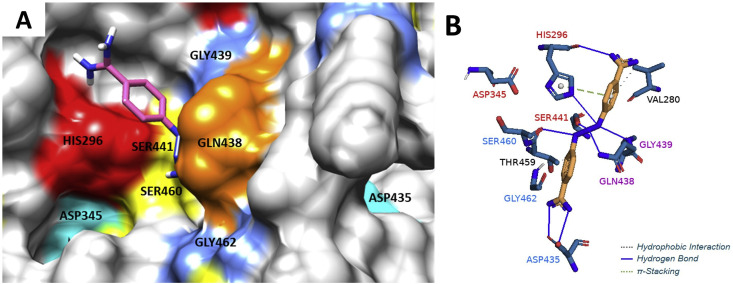
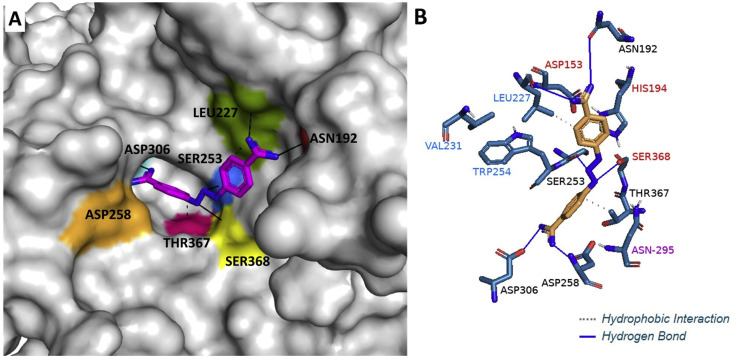
In order to explain the ability of diminazene to inhibit TMPRSS2 and furin and to compare its binding with that of camostat and nafamostat to these targets, molecular docking studies were performed using several docking programs. Molecular docking of diminazene by using Autodock Vina (Trott and Olson, 2010) revealed that it can bind to TMPRSS2 catalytic binding site consisting of the triad residues HIS296, ASP345, and SER441 (Fig. 8 , Supplementary Table S8-1) similar to camostat and nafamostat (Zhu et al., 2020), suggesting they may share the same inhibitory mechanism. Diminazene can also bind to the furin catalytic/binding site consisting of the residues ASN192, LEU227, SER253, ASP258, ASP306, THR367, and SER368 (Fig. 9 ). Diminazene showed similar binding pose observed for the furin inhibitor MI-52 [m-guanidinomethyl-phenylacetyl-Arg-Val-Arg-(4-amidomethyl)-benzamidine] crystallized with the protein (PDB code 5JXH) (Dahms et al., 2016) (Supplementary Figure S8-3). For further understanding the activity difference between diminazene and camostat/nafamostat against furin, a series of docking experiments were performed by using Autodock Vina (Trott and Olson, 2010), SwissDock (Grosdidier et al., 2011) and DockThor (Santos et al., 2020) which uses different algorithms. The results obtained by DockThor, a free web-based docking software considering the protein flexibility implicitly, showed the lowest binding energy for diminazene and higher binding energy for camostat and nafamostat (Supplementary Table S8-2). Further examination of the docking results from the above three docking software found nafamostat and camostat also favor binding to the non-active sites of furin (Supplementary Figures S8-2B and S8-2C) and to present different binding pose depending on the algorithm used by the software while diminazene binds to the same active pocket even when blind docking was used (Supplementary Figure S8-2A). These results suggested that diminazene has more affinity for the catalytic binding site of furin, which agrees with the results from our cell-based assay for inhibition of furin.
Fig. 8.
(A) Binding mode of diminazene (pink) in the TMPRSS2 catalytic site containing the triad residues HIS296 (red), SER441 (yellow) and ASP345 (cyan), and the binding site containing the residues SER460 (yellow), GLY462 (blue), and ASP435 (cyan). Picture was made using UCSF Chimera. (B) 3D representation of hydrogen bonds, hydrophobic interactions and π-Stacking of diminazene within TMPRSS2 biding site. Picture was made using Pymol. Diminazene forms favorable interactions with residues found at the catalytic site (residue names are shown in red), at the substrate binding site (residue names are shown in blue), and with the conserved residues forming the oxyanium hole (residue names are shown in purple). Molecular docking was carried out using AutoDock Vina 1.1.2 and the grid box parameters were set to cover the catalytic and binding sites of TMPRSS2 (homologue model, UniProtKB id = O15393). Preparation of protein and receptor was made using the software MarvinSketch, AutoDock tolls 1.5.6, Mopac 2016, and UCSF Chimera 1.15. Binding energy of −6.7 kcal/mol.
Fig. 9.
(A) Binding mode of diminazene (pink) in the furin catalytic/binding site containing the ASN192 (red), LEU227 (green), SER253 (blue), ASP258 (orange), ASP306 (cyan), SER368 (yellow) and THR367 (pink). Molecular docking was carried out using DockThor and picture was made using Pymol. (B) 3D representation of hydrogen bonds and hydrophobic interactions of diminazene within furin biding site. Picture was made using Pymol. Diminazene forms favorable interactions with residues found at the catalytic site (residue names are shown in red) and the substrate binding site (residue names are shown in blue). Binding energy of −18.3 kcal/mol.
2. Conclusion
In summary, we established and validated three convenient and cost-effective cell-based proteolytic enzyme inhibitory assays employing the cell lines known to overexpress TMPRSS2 (VeroE6/TMPRSS2 and LNCaP) and furin (Vero/furin). Combined use of these assays should lead to the discovery of selective and/or broad-spectrum protease inhibitors with potential to block host infection of SARS-CoV-2 and other viruses. Evaluation of compounds known to inhibit either TMPRSS2 or furin led to the discovery of diminazene as a dual inhibitor of TMPRSS2 and furin. This is significant as it is known that several membrane-associated serine proteinases contribute to activation of the SARS-CoV-2 spike protein individually or in synergy with each other (Fuentes-Prior, 2020), and that a combination of TMPRSS2 inhibitors aprotinin or MI-432 with furin inhibitor MI-1851 has recently been shown to have an enhanced antiviral activity vs SARS-CoV-2 in human airway cells at lower doses compared to treatment with each inhibitor alone (Bestle et al., 2020). SARS-CoV-2 is also known to target angiotensin-converting enzyme 2 (ACE2) causing lowering of ACE2 levels resulting in augmented pulmonary injury and worsened outcomes in patients with pre-existing cardiovascular comorbidities (Verdecchia et al., 2020; Zaman et al., 2020). As diminazene is known to be an ACE2 activator (Qaradakhi et al., 2020), it has recently been proposed that restoring ACE2 functions through administration of diminazene may promote cardiovascular and pulmonary protection vs SARS-CoV-2 and may improve COVID-19 patients (with and without pre-existing comorbidities) outcomes (Gadanec et al., 2021). Further studies to evaluate the potential of diminazene when used in combination with other therapeutics to block SARS-CoV-2 infection and/or treat patients experiencing organ dysfunction caused by loss of ACE2 function are warranted.
3. Experimental section
3.1. General methods
VeroE6/TMPRSS2 cells (# JCRB 1819) were obtained from Japanese Collection of Research Bioresources (JCRB) Cell Bank and Sekisui XenoTech, LLC. Vero/furin overexpressing cells were kindly provided by Dr. Ted Pierson of the U.S. National Institute of Allergy and Infectious Diseases. LNCaP and VeroE6 cells were obtained from ATCC. Diminazene and camostat were purchased from MedChemExpress. Nafamostat, gabexate, and bromhexine were obtained from Selleck Chemicals. The furin inhibitor I (Decanoyl-Arg-Val-Lys-Arg-CMK) was purchased from EMD Millipore, TMPRSS2 substrate Boc-Gln-Ala-Arg-AMC from Peptides International Inc., and furin substrate Boc-Arg-Val-Arg-Arg-AMC from Bachem Americas, Inc. Dulbecco's Modification of Eagle's Medium (DMEM, # 10-013-CV), RPMI 1640 medium (# 10-040-CV), black clear bottom 96 wells plates (Corning Corstar 3603) were obtained from Corning. DMEM with GlutaMAX (Cat# 10564-011), blasticin (Cat# 15070-063), and 2-mercaptoethanol (1000x, # 21985-023) were obtained from Gibco Inc. Heat inactivated fetal bovine serum (# 2306C-100 ML), 100 mM sodium pyruvate (#S8636-100 mL), and non-essential amino acids (100X, #M7145-100 mL) were purchased from Sigma. Hyclone fetal bovine serum (# 30396.02) was obtained from Cytiva. Geneticin (G-418, VWR# 45000-630) was obtained from VWR. Penicillin-streptomycin (100x, # SV30010) and 10 mM HEPES (# 15630056) was obtained from Fisher Scientific. The LJL Biosystems Analyst AD 96/384 plate reader set to excitation/emission 360 nm/460 nm fluorescence intensity method was used for the assay with default parameters, including Z Height: middle of the well; Integration time: 100 ms; Attenuator: off; PMT mode: SmartRead with sensitivity 2; Plate shaking time: 10 Sec.; Optics: bottom with 50/50 Dichroic mirror, was used for fluorescence intensity measurement. The data were processed by GraphPad Prism 5.03. DMEM medium, supplemented with 10% FBS, 100 unit/mL of penicillin, 100 μg/mL of streptomycin, and 1.0 mg/mL of geneticin (G418) was used for culturing VeroE6/TMPRSS2 overexpressing cells. DMEM medium supplemented with 10% FBS used for culturing VeroE6 cells. DMEM with glutaMAX medium, supplemented with 7% FBS, 100 unit/mL of penicillin, 100 μg/mL of streptomycin, and 5 μg/mL of blasticidin, was used for culturing Vero/furin overexpressing cells. RPMI medium supplemented with 5% FBS, 0.1 mM HEPES, 1x non-essential amino acids, 50 μM 2-mercaptoethanol, 1.0 mM sodium pyruvate, and 100 unit/mL of penicillin, and 100 μg/mL of streptomycin were used for culturing LNCaP cells.
3.2. Determination of TMPRSS2 enzyme activity
The cells (VeroE6/TMPRSS2 overexpressing cells or LNCaP cells) were seeded in a black, clear and flat bottom, 96-well plate at 10,000 cells/well density with 100 μL/well corresponding medium. After incubation at 37 °C and 5% CO2 for 24 h, 50 μL/well medium was transferred to another black, bottom clear, 96-well plate. The remainder of the medium was completely removed, and the cell layer was washed with 100 μL/well DPBS (1x). The proteolytic activity of the medium and cells were measured separately by adding the fluorogenic substrate Boc-Gln-Ala-Arg-AMC to a final concentration of 100 μM. The fluorescence intensity was read every 15 min at room temperature using LJL Biosystem Analyst AD with excitation/emission set to 360 nm/460 nm.
3.3. TMPRSS2 enzyme inhibition assay
The cells (VeroE6/TMPRSS2 overexpressing cells or LNCaP cells) were seeded in a black, clear and flat bottom, 96-well plate at 10,000 cells/well density with 100 μL/well of the corresponding medium. After incubation at 37 °C and 5% CO2 for 24 h, the medium was completely removed, and the cell layer was washed with 100 μL/well DPBS (1x). The inhibitor solution (50 μL/well), prepared by diluting the DMSO stock solution with DPBS (1x), was added to the plate. After allowing the plate to stand at room temperature for 30 min, the fluorogenic substrate Boc-Gln-Ala-Arg-AMC (50 μL/well) was added to a final concentration of 100 μM. The fluorescence intensity was determined after 30 min incubation at RT by using LJL Biosystem Analyst AD with excitation/emission set to 360 nm/460 nm.
3.4. Determination of furin enzyme activity
The cells (Vero/furin overexpressing cells) were seeded in a black, clear and flat bottom, 96-well plate at 10,000 cells/well density with 100 μL/well of the corresponding medium. After incubation at 37 °C and 5% CO2 for 24 h, 50 μL/well medium was transferred to another black, clear and flat bottom, 96-well plate, and the remaining medium was completely removed. The cell layer was washed with 100 μL/well of DPBS (1x) after which 50 μL/well of DPBS (1x) containing 0.2% Tween 20 was added and the plate was incubated at room temperature for 30 min. The proteolytic activity in the medium and cells were measured separately by adding the fluorogenic substrate Boc-Arg-Val-Arg-Arg-AMC to a final concentration of 50 μM. The fluorescence intensity was read every 30 min at room temperature using LJL Biosystem Analyst AD with excitation/emission set to 360 nm/460 nm.
3.5. Furin enzyme inhibition assay
The cells (Vero/furin overexpressing cells) were seeded in a black, clear and flat bottom 96-well plate at 10,000 cells/well density with 100 μL/well of the corresponding medium. After incubation at 37 °C and 5% CO2 for 24 h, the medium was completely removed, and the cell layer was washed with 100 μL/well of DPBS (1x). A solution of the inhibitor (50 μL/well), prepared by diluting the DMSO stock solution with DPBS (1x) containing 0.2% Tween 20 was added to the plate. After incubation at room temperature for 30 min, the fluorogenic substrate Boc-Arg-Val-Arg-Arg-AMC (50 μL/well) was added to a final concentration of 50 μM. The fluorescence intensity was read after additional 60 min incubation at room temperature by using LJL Biosystem Analyst AD with excitation/emission set to 360 nm/460 nm.
Funding
We thank the College of Agricultural and Life Sciences of the University of Arizona for award of a COVID-19 Pilot Project grant. A.A.L.G. has disclosed financial interests in Regulonix, LLC. USA.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crchbi.2022.100023.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Angliker H., Wikstrom P., Shaw E., Brenner C., Fuller R.S. The synthesis of inhibitors for processing proteinases and their action on the Kex2 proteinase of yeast. Biochem. J. 1993;293(1):75–81. doi: 10.1042/bj2930075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri S.A., Althaqafi R.M.M., Alswat K., Alghamdi A.A., Alomairi N.E., Nemenqani D.M., Ibrahim Z.S., Elkady A. Post COVID-19 vaccination-associated neurological complications. Neuropsychiatric Dis. Treat. 2022;18:137–154. doi: 10.2147/NDT.S343438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azouz N.P., Klingler A.M., Callahan V., Akhrymuk I.V., Elez K., Raich L., Henry B.M., Benoit J.L., Benoit S.W., Noé F., Kehn-Hall K., Rothenberg M.E. Alpha 1 antitrypsin is an inhibitor of the SARS-CoV-2–priming protease TMPRSS2. bioRxiv. 2020 doi: 10.1101/2020.05.04.077826. 2020.05.04.077826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmad H.F., Abou-Kheir W. Crosstalk between COVID-19 and prostate cancer. Prostate Cancer Prostatic Dis. 2020;23(4):561–563. doi: 10.1038/s41391-020-0262-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi D.E., Mahloogi H., Lopez De Cicco R., Klein-Szanto A. Increased furin activity enhances the malignant phenotype of human head and neck cancer cells. Am. J. Pathol. 2003;162(2):439–447. doi: 10.1016/s0002-9440(10)63838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughn L.B., Sharma N., Elhaik E., Sekulic A., Bryce A.H., Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 infection. Mayo Clin. Proc. 2020;95(9):1989–1999. doi: 10.1016/j.mayocp.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu A., Gravel E., Cloutier A., Marois I., Colombo É., Désilets A., Verreault C., Leduc R., Marsault É., Richter M.V. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J. Virol. 2013;87(8):4237–4251. doi: 10.1128/JVI.03005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker G.L., Sielaff F., Than M.E., Lindberg I., Routhier S., Day R., Lu Y., Garten W., Steinmetzer T. Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics. J. Med. Chem. 2010;53(3):1067–1075. doi: 10.1021/jm9012455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Blazejewska P., Soilleux E., Allen P., Danisch S., Steffen I., Choi S.Y., Park Y., Schneider H., Schughart K., Pöhlmann S. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010;84(19):10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Van Lam van T., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C., Klenk H.-D., Garten W., Steinmetzer T., Böttcher-Friebertshäuser E. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3(9) doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshäuser E. Membrane-anchored serine proteases: host cell factors in proteolytic activation of viral glycoproteins. Activ. Virus. Host Proteas. 2018:153–203. [Google Scholar]
- Centers for disease control COVID data tracker. https://covid.cdc.gov/covid-data-tracker/#datatracker-home accessed June 04, 2021)
- Chakrabarti S.S., Tiwari A., Jaiswal S., Kaur U., Kumar I., Mittal A., Singh A., Chakrabarti S. Rapidly progressive dementia with asymmetric rigidity following ChAdOx1 nCoV-19 vaccination. Aging Dis. 2021 doi: 10.14336/ad.2021.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Lee M.S., Lucht A., Chou F.P., Huang W., Havighurst T.C., Kim K., Wang J.K., Antalis T.M., Johnson M.D., Lin C.Y. TMPRSS2, a serine protease expressed in the prostate on the apical surface of luminal epithelial cells and released into semen in prostasomes, is misregulated in prostate cancer cells. Am. J. Pathol. 2010;176(6):2986–2996. doi: 10.2353/ajpath.2010.090665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola J.M., Hamilton C.A., Bhojani M.S., Larsen M.J., Ross B.D., Rehemtulla A. Identification of inhibitors using a cell-based assay for monitoring Golgi-resident protease activity. Anal. Biochem. 2007;364(1):19–29. doi: 10.1016/j.ab.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahms S.O., Arciniega M., Steinmetzer T., Huber R., Than M.E. Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. Proc. Natl. Acad. Sci. U.S.A. 2016;113:11196–11201. doi: 10.1073/pnas.1613630113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K. Furin protease: from SARS Cov-2 to anthrax, diabetes, and hypertension. Perm. J. 2020;24(4):148–149. doi: 10.7812/TPP/20.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P. Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J. Biol. Chem. 2020;296:100135. doi: 10.1074/jbc.REV120.015980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadanec L.K., Qaradakhi T., McSweeney R.K., Ali A.B., Zulli A., Apostolopoulos V. Dual targeting of Toll-like receptor 4 and angiotensin-converting enzyme 2: a proposed approach to SARS-CoV-2 treatment. Future Microbiol. 2021;16:205–209. doi: 10.2217/fmb-2021-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier A., Zoete V., Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr366. Web Server issue), W270–W277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease receptor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.H., Lin K.H., Liao C.H., Lai M.W., Tseng Y.H., Yeh C.T. Furin overexpression suppresses tumor growth and predicts a better postoperative disease-free survival in hepatocellular carcinoma. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0040738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93(6):e01815–e01818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazuya S., Miyuki K., Shutoku M. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87(23):12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko C.J., Huang C.C., Lin H.Y., Juan C.P., Lan S.W., Shyu H.Y., Wu S.R., Hsiao P.W., Huang H.P., Shun C.T., Lee M.S. Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Res. 2015;75(14):2949–2960. doi: 10.1158/0008-5472.CAN-14-3297. [DOI] [PubMed] [Google Scholar]
- Leach D.A., Isac A.-M., Bevan C.L., Brooke G.N. TMPRSS2, required for SARS-CoV-2 entry, is downregulated in lung cells by enzalutamide, a prostate cancer therapeutic. Res. Square. 2020 doi: 10.21203/rs.3.rs-34503/v1. [DOI] [Google Scholar]
- Li F., Han M., Dai P., Xu W., He J., Tao X., Wu Y., Tong X., Xia X., Guo W., Zhou Y., Li Y., Zhu Y., Zhang X., Liu Z., Aji R., Cai X., Li Y., Qu D., Chen Y., Jiang S., Wang Q., Ji H., Xie Y., Sun Y., Lu L., Gao D. Distinct mechanisms for TMPRSS2 expression explain organ-specific inhibition of SARS-CoV-2 infection by enzalutamide. Nat. Commun. 2021;12(1):866. doi: 10.1038/s41467-021-21171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Guan W.-J., Zhong N.-S. Strategies and advances in combating COVID-19 in China. Engineering. 2020;6(10):1076–1084. doi: 10.1016/j.eng.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan B., Huynh T., Cheng X., Lan G., Wang H.-R. Targeting proteases for treating COVID-19. J. Proteome Res. 2020;19:4316–4326. doi: 10.1021/acs.jproteome.0c00430. [DOI] [PubMed] [Google Scholar]
- Lucas J.M., Heinlein C., Kim T., Hernandez S.A., Malik M.S., True L.D., Morrissey C., Corey E., Montgomery B., Mostaghel E., Clegg N., Coleman I., Brown C.M., Schneider E.L., Craik C., Simon J.A., Bedalov A., Nelson P.S. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4(11):1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio R., Corsini G.U. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS-CoV-2 infection. Pharmacol. Res. 2020;157:104837. doi: 10.1016/j.phrs.2020.104837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U.S.A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollica V., Rizzo A., Massari F. The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer. Future Oncol. 2020;16(27):2029–2033. doi: 10.2217/fon-2020-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montopoli M., Zumerle S., Vettor R., Rugge M., Zorzi M., Catapano C.V., Carbone G.M., Cavalli A., Pagano F., Ragazzi E., Prayer-Galetti T., Alimonti A. Androgen-deprivation therapies for prostate cancer and risk of infection by SARS-CoV-2: a population-based study (N 4532) Ann. Oncol. 2020;31(8):1040–1045. doi: 10.1016/j.annonc.2020.04.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Sirohi D., Dowd K.A., Chen Z., Diamond M.S., Kuhn R.J., Pierson T.C. Enhancing dengue virus maturation using a stable furin overexpressing cell line. Virology. 2016;497:33–40. doi: 10.1016/j.virol.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster M.E., Shay D.K., Su J.R., Gee J., Creech C.B., Broder K.R., Edwards K., Soslow J.H., Dendy J.M., Schlaudecker E., Lang S.M., Barnett E.D., Ruberg F.L., Smith M.J., Campbell M.J., Lopes R.D., Sperling L.S., Baumblatt J.A., Thompson D.L., Marquez P.L., Strid P., Woo J., Pugsley R., Reagan-Steiner S., DeStefano F., Shimabukuro T.T. Myocarditis cases reported after mRNA-based COVID-19 vaccination in the US from December 2020 to August 2021. JAMA. 2022;327(4):331–340. doi: 10.1001/jama.2021.24110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pászti-Gere E., McManus S., Meggyesházi N., Balla P., Gálfi P., Steinmetzer T. Inhibition of matriptase activity results in decreased intestinal epithelial monolayer integrity in vitro. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0141077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pászti-Gere E., Czimmermann E., Ujhelyi G., Balla P., Maiwald A., Steinmetzer T. In vitro characterization of TMPRSS2 inhibition in IPEC-J2 cells. J. Enzym. Inhib. Med. Chem. 2016;31(Suppl. 2):123–129. doi: 10.1080/14756366.2016.1193732. [DOI] [PubMed] [Google Scholar]
- Peng M., Watanabe S., Chan K., He Q., Zhao Y., Zhang Z., Lai X., Luo D., Vasudevan S.G., Li G. Luteolin restricts dengue virus replication through inhibition of the proprotein convertase furin. Antivir. Res. 2017;143:176–185. doi: 10.1016/j.antiviral.2017.03.026. [DOI] [PubMed] [Google Scholar]
- Qaradakhi T., Gadanec L.K., McSweeney K.R., Tacey A., Apostolopoulos V., Levinger I., Rimarova K., Egom E.E., Rodrigo L., Kruzliak P., Kubatka P., Zulli A. The potential actions of angiotensin-converting enzyme II (ACE2) activator diminazene aceturate (DIZE) in various diseases. Clin. Exp. Pharmacol. Physiol. 2020;47(5):751–758. doi: 10.1111/1440-1681.13251. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Wang X.-M., Mannan R., Pitchiaya S., Zhang Y., Wotring J.W., Xiao L., Robinson D.R., Wu Y.-M., Tien J.C.-Y., Cao X., Simko S.A., Apel I.J., Bawa P., Kregel S., Narayanan S.P., Raskind G., Ellison S.J., Parolia A., Zelenka-Wang S., McMurry L., Su F., Wang R., Cheng Y., Delekta A.D., Mei Z., Pretto C.D., Wang S., Mehra R., Sexton J.Z., Chinnaiyan A.M. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Nat. Acad. Sci. USA. 2021;118(1) doi: 10.1073/pnas.2021450118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Molina B., Lick A.N., Nasrolahi Shirazi A., Oh D., Tiwari R., El-Sayed N.S., Parang K., Lindberg I. Cationic cell-penetrating peptides are potent furin inhibitors. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos K.B., Guedes I.A., Karl A.L.M., Dardenne L.E. Highly flexible ligand docking: benchmarking of the DockThor program on the LEADS-PEP protein-peptide data set. J. Chem. Inf. Model. 2020;60(2):667–683. doi: 10.1021/acs.jcim.9b00905. [DOI] [PubMed] [Google Scholar]
- Schuler B.A., Habermann A.C., Plosa E.J., Taylor C.J., Jetter C., Negretti N.M., Kapp M.E., Benjamin J.T., Gulleman P., Nichols D.S., Braunstein L.Z., Hackett A., Koval M., Guttentag S.H., Blackwell T.S., Webber S.A., Banovich N.E., Kropski J.A., Sucre J.M. Age-determined expression of priming protease TMPRSS2 and localization of SARS-CoV-2 in lung epithelium. J. Clin. Invest. 2021;131(1) doi: 10.1172/JCI140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimp J.H., Kales S.C., Sanderson P.E., Simeonov A., Shen M., Hall M.D. An enzymatic TMPRSS2 assay for assessment of clinical candidates and discovery of inhibitors as potential treatment of COVID-19. ACS Pharmacol. Transl. Sci. 2020;3(5):997–1007. doi: 10.1021/acsptsci.0c00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyr Z.A., Gorshkov K., Chen C.Z., Zheng W. Drug discovery strategies for SARS-CoV-2. J. Pharmacol. Exp. Therapeut. 2020;375(1):127–138. doi: 10.1124/jpet.120.000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopsack K.H., Mucci L.A., Antonarakis E.S., Nelson P.S., Kantoff P.W. TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov. 2020;10(6):779–782. doi: 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susan-Resiga D., Essalmani R., Hamelin J., Asselin M.C., Benjannet S., Chamberland A., Day R., Szumska D., Constam D., Bhattacharya S., Prat A., Seidah N.G. Furin is the major processing enzyme of the cardiac-specific growth factor bone morphogenetic protein 10. J. Biol. Chem. 2011;286(26):22785–22794. doi: 10.1074/jbc.M111.233577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thunders M., Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2) J. Clin. Pathol. 2020;73(12):773–776. doi: 10.1136/jclinpath-2020-206987. [DOI] [PubMed] [Google Scholar]
- Tran H.T.T., Le N.P.K., Gigl M., Dawid C., Lamy E. Common dandelion (Taraxacum officinale) efficiently blocks the interaction between ACE2 cell surface receptor and SARS-CoV-2 spike protein D614, mutants D614G, N501Y, K417N and E484K in vitro. bioRxiv. 2021 doi: 10.1101/2021.03.19.435959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven W.J., Voorberg J., Fontijn R., Pannekoek H., van den Ouweland A.M., van Duijnhoven H.L., Roebroek A.J., Siezen R.J. Furin is a subtilisin-like proprotein processing enzyme in higher eukaryotes. Mol. Biol. Rep. 1990;14:265–275. doi: 10.1007/BF00429896. [DOI] [PubMed] [Google Scholar]
- Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidricaire G., Denault J.B., Leduc R. Characterization of a secreted Form of human furin endoprotease. Biochem. Biophys. Res. Commun. 1993;195(2):1011–1018. doi: 10.1006/bbrc.1993.2145. [DOI] [PubMed] [Google Scholar]
- World Health Organization coronavirus (COVID-19) dashboard. https://covid19.who.int/ accessed June 04, 2021)
- Wu C., Zheng M., Yang Y., Gu X., Yang K., Li M., Liu Y., Zhang Q., Zhang P., Wang Y., Wang Q., Xu Y., Zhou Y., Zhang Y., Chen L., Li H. Furin: a potential therapeutic target for COVID-19. iScience. 2020;23(10):101642. doi: 10.1016/j.isci.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Maclsaac A.I., Jennings G.L.R., Schlaich M.P., Inglis S.C., Arnold R., Kumar S., Thomas L., Wahi S., Lo S., Naismith C., Duffy S.J., Nicholls S.J., Newcomb A., Almeida A.A., Wong S., Lund M., Chew D.P., Kritharides L., Chow C.K., Bhindi R. Cardiovascular disease and COVID-19: Australian and New Zealand consensus statement. Med. J. Aust. 2020;213(4):182–187. doi: 10.5694/mja2.50714. [DOI] [PubMed] [Google Scholar]
- Zhu H., Du W., Song M., Liu Q., Herrmann A., Huang Q. Spontaneous binding of potential COVID-19 drugs (camostat and nafamostat) to human serine protease TMPRSS2. Comput. Struct. Biotechnol. J. 2020;19:467–476. doi: 10.1016/j.csbj.2020.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.