Abstract
Clear cell renal cell carcinoma (ccRCC) is the most common and aggressive subtype of kidney cancer, with high mortality rates worldwide. The sonic hedgehog (SHH) molecular cascade is altered in various malignancies in tumorigenesis, and several SHH pathway inhibitors have been considered as potential anticancer drugs. The aim of the present study was to determine the expression profile of SHH signaling components and their target genes in ccRCC. Additionally, the present study examined the effects of SHH pathway inhibitory drugs (RU-SKI43, cyclopamine and GLI-antagonist 61) on cell viability, cell cycle progression, expression levels of SHH target genes and migration ability in 786-O, ACHN and HK2 cells. The study also included paired tumor and normal samples from 62 patients with ccRCC. The mRNA levels in clinical samples and cell lines were measured via reverse transcription-quantitative PCR. Cell viability was examined using a sulforhodamine B assay. Flow cytometry was used to investigate cell cycle progression and the migratory rate of cells was assessed using a wound healing assay. High mRNA levels of SHH, smoothened (SMO), glioma-associated zinc finger protein (GLI)1-3, BCL2 apoptosis regulator (BCL2), MYC proto-oncogene (MYC), vascular endothelial growth factor A (VEGFA) and cyclin D1 (CCND1) were observed in the tumor tissues, especially in early ccRCC, according to the TNM stage or World Health Organization/International Society of Urological Pathology (ISUP) grade. High expression levels of VEGFA, as well as low CCND1 mRNA expression, were associated with short overall survival, and increased VEGFA expression was an independent prognostic factor of a poor outcome in patients with advanced ISUP grade (Cox hazard ratio test). Cyclopamine treatment was found to arrest 786-O cells in the G2/M phase and decreased the expression levels of GLI1, BCL2, VEGFA and CCND1. RU-SKI43 inhibited cell migration and decreased the expression levels of BCL2, MYC and CCND1 in ACHN cells. Overall, the results of the present study suggested that SHH signaling may be involved in the early development of ccRCC, and the expression levels of CCND1 and VEGFA may serve as prognostic factors of this disease. Cyclopamine and RU-SKI43 appear to be potential anti-renal cell carcinoma drugs; however, this hypothesis requires verification by further in vivo studies.
Keywords: clear cell renal cell carcinoma, gene expression, quantitative PCR, sonic hedgehog, SHH, PTCH1, SMO, SUFU, GLI1-3, BCL2, MYC, VEGFA, CCND1, cyclin D1, prognostic factors, RU-SKI43, cyclopamine, GANT61, sunitinib
Introduction
The GLOBOCAN database reported 431,288 new kidney cancer cases worldwide in 2020 (1). Clear cell renal cell carcinoma (ccRCC) accounts for ~75% of renal cell carcinomas (RCCs) and is responsible for most RCC-related mortality (2,3). Although novel drugs have been implemented for ccRCC treatment since 2005, this cancer remains a therapeutic challenge, especially if diagnosed at the metastatic stage (3). Therefore, novel prognostic factors or therapeutic approaches for ccRCC based on molecular mechanisms involved in the pathogenesis of this cancer are required (4).
The Sonic hedgehog (SHH) pathway is a molecular signaling cascade, the activity of which is mainly limited to embryonic life (5). Through SHH/patched 1 (PTCH1)/smoothened (SMO)/glioma-associated zinc finger protein (GLI)2, 3 proteins, this signaling pathway stimulates the expression of the downstream activator GLI1 and PTCH1 repressor, followed by activation (via GLI1 protein) of several target genes, including MYC proto-oncogene (MYC), BCL2 apoptosis regulator (BCL2), vascular endothelial growth factor A (VEGFA) and cyclin D1 (CCND1) (Fig. 1) (6). Their molecular products are pro-carcinogenic proteins (7-10), since they stimulate cell proliferation, possess anti-apoptotic and pro-angiogenic properties, and activate the cell cycle. MYC and BCL2 gene amplifications, as well as higher mRNA levels of VEGFA and CCND1, are associated with shorter survival of patients with esophageal squamous cell carcinoma (11), diffuse large B-cell lymphoma (12), lung cancer (13) and gastric cancer (14), respectively. However, reports regarding the expression of the SHH pathway components and their effects on downstream genes in ccRCC remain limited and ambiguous (15-19).
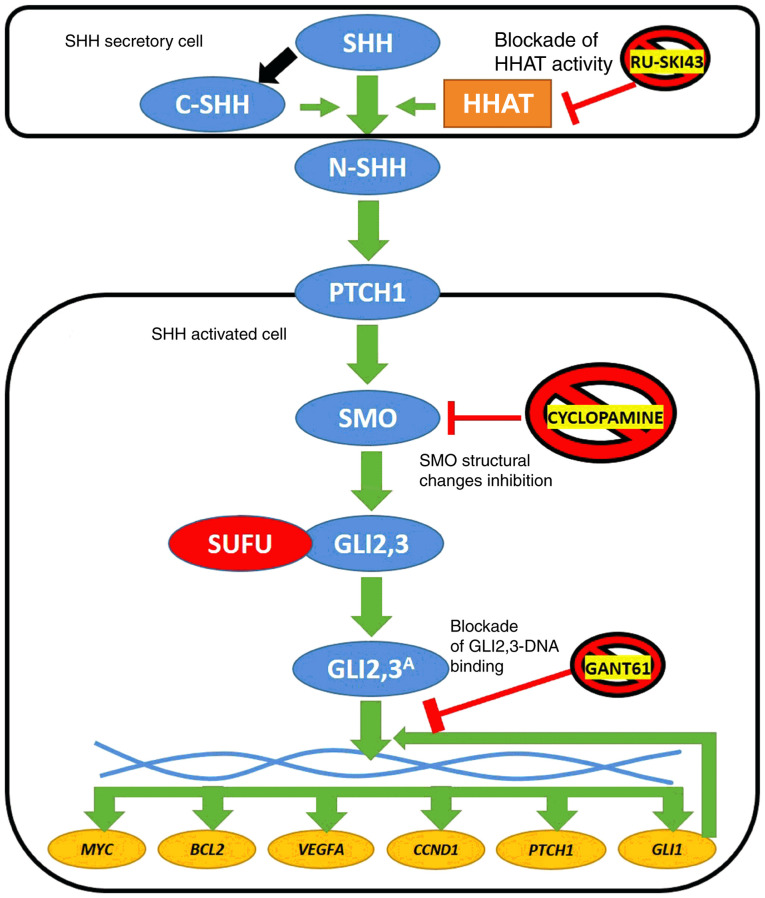
Figure 1.
Simplified diagram of SHH pathway activation in vertebrates. SHH ligand undergoes posttranslational cleavage into N- and C-terminal SHH. Structural modifications of N-SHH caused by C-SHH and HHAT lead to its binding with the PTCH1 receptor. This stimulates SMO protein to process GLI2 and GLI3, which dissociate from the SUFU protein and finally stimulate the expression of target genes (MYC, BCL2, VEGFA, CCND1, PTCH1 and GLI1). The latter encodes another transcriptional activator of SHH signaling (6). Three SHH pathway inhibitors are presented in the figure: RU-SKI43 blocks the activity of the HHAT enzyme (21), cyclopamine inhibits conformational changes of the SMO protein (23) and GANT61 modifies the structure of GLI1 and GLI2, preventing their interaction with double-stranded DNA (24). Blue circles, proteins of the SHH signaling pathway; red circle, negative SHH pathway regulator; yellow circles, SHH cascade target genes; and orange rectangle, enzyme. SHH, sonic hedgehog ligand; C-SHH, C-terminal part of the SHH ligand; N-SHH, N-terminal part of the SHH ligand; HHAT, hedgehog acyltransferase; PTCH1, patched 1; SMO, smoothened; SUFU, suppressor of fused homolog; GLI1,2,3, glioma-associated zinc finger protein 1, 2, 3; GLI2,3A, activated glioma-associated zinc finger protein 2, 3; VEGFA, vascular endothelial growth factor A; CCND1, cyclin D1; GANT61, GLI-antagonist 61.
Based on numerous reports on SHH pathway alterations in various cancer types, several inhibitory drugs have been developed (20). They may block signaling at different pathway stages. RU-SKI43 is a selective inhibitor of hedgehog acyltransferase (HHAT; Fig. 1) (21). HHAT provides chemical modifications the N-terminal part of the SHH ligand (N-SHH), thus providing upstream activation of the SHH pathway (22). Another SHH pathway inhibitor, cyclopamine, blocks conformational changes of the SMO protein, and thus prevents its activation by inhibiting GLI2-3 release (Fig. 1) (23). GLI-antagonist 61 (GANT61) is one of the most promising anticancer agents, since it can modify the structure of both GLI1 and GLI2, preventing them from DNA binding and action as transcription factors (TFs) (Fig. 1) (24,25). To the best of our knowledge, no comparative studies on the effect of the aforementioned inhibitors of the SHH pathway on RCC cells have been published.
Our previous research examining 37 ccRCC tissue samples revealed deregulation of the expression levels of SHH pathway genes (16). Therefore, in the present study, a more comprehensive analysis of the mRNA levels of SHH, PTCH1, SMO and suppressor of fused homolog (SUFU), TFs GLI1-3 and their target genes in ccRCC tissues and cell lines was performed. Furthermore, the study focused on comparative analysis concerning the effects of RU-SKI43, cyclopamine and GANT61 inhibitors on proliferation, cell cycle distribution, migration and gene expression in ccRCC cell lines and a normal kidney cell line.
Materials and methods
Patients and samples
ccRCC tumor tissues and morphologically unchanged kidney samples were obtained during radical nephrectomy from 62 patients who underwent surgery at the Department of Urology, Medical University of Gdańsk (Gdansk, Poland). The samples were collected over a 4-year period from 2017 to 2020. The study included 62 patients with ccRCC, including 21 women and 41 men (mean age ± SD, 63,84±11.25 years, age range: 33-86 years). The exclusion criteria included: other than ccRCC histological subtypes of RCC, multifocal and/or bilateral kidney tumors and Von Hippel-Lindau disease. The study was approved by the Independent Bioethics Committee for Scientific Research at Medical University of Gdańsk (decision nos. NKEBN/4/20 11 and NKBBN/370/2016). Written informed consent was acquired from each patient before surgery.
Material acquisition
Small (ca. 5×5×5 mm) pieces of ccRCC tumor tissues and control, morphologically unchanged kidney samples from the same patient, were placed into test tubes in the operating theater, no later than 20 min after kidney resection. Tissue samples for quantitative PCR (qPCR) analysis were placed in test tubes filled with 5 volumes of RNAlater (Ambion; Thermo Fisher Scientific, Inc.), and were placed at -80°C after 24 h incubation at 4°C. The other two samples of tumor tissues were fixed in 4% buffered formalin, embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) for histopathological assessment. The samples were included in the qPCR analysis if >60% cells in the respective histological sections in tumor samples presented characteristic features of ccRCC, while all cells of unchanged (control) samples presented normal morphology (26,27). If both conditions were not fulfilled, the material was excluded from the study.
Total RNA isolation
Total RNA from the collected samples was isolated using the ExtractMe Total RNA kit (Blirt, Inc.) according to the manufacturer's protocol. The collected samples were homogenized in 2-ml tubes with 300 µl lysis buffer and ceramic beads using the MagnaLyser apparatus (Roche Diagnostics) for 40 sec at 6,000 rpm at room temperature. The obtained RNA was dissolved in 70 µl nuclease-free water. The quantity and quality of RNA were measured using a spectrophotometer (NanoDrop ND 1000; Thermo Fisher Scientific, Inc.). RNA samples were stored at -80°C until further analysis. For first-strand cDNA synthesis, 1 µg RNA was reverse transcribed using 1 µl RevertAid reverse transcriptase (Fermentas; Thermo Fisher Scientific, Inc.) and 0.5 µg dT18 primers (Sigma-Aldrich; Merck KGaA) in a total volume of 20 µl. The reaction was performed according to the manufacturer's protocol (Thermo Fisher Scientific, Inc.). cDNA samples were stored at -20°C until further analysis.
Assessment of gene mRNA levels
mRNA assessment was performed using the qPCR technique. Primer sequences were designed using the Primer-BLAST software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Their concentrations, as well as experimentally established reaction conditions, are presented in Table I. The measurements were performed in duplicate using 1 µl 4X-diluted cDNA and AmplifyMe NoRox SybrGreen (Blirt, Inc.) chemistry in a total volume of 10 µl. The reaction was conducted on a separate PCR plate (4titude, Ltd.) for each gene with a negative control (water instead of cDNA) and 10X diluted pooled cDNA as a precision control. A StepOne Plus apparatus with StepOne Plus software ver. 2.3 (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for the amplification process and data analysis. Geometric mean of Ct (threshold cycle) values for each gene were normalized to the reference gene [glucuronidase β (GUSB)], according to our previous normalization study on ccRCC (28), using Livak's equation (29): x=2ΔCt, where x stands for expression of gene y and ΔCt=(Ct GUSB-Ct gene Y). Obtained raw expression data for each tumor sample were calibrated to average expression data of control samples (fold change; control sample=1).
Table I.
Details of the qPCR assays used in the study.
| Gene name | GeneBank transcript Acc. no. | Primer sequences | qPCR reaction conditions | qPCR reaction content |
|---|---|---|---|---|
| BCL2 | NM_000633.2 | 5′-GGATAACGGAGGCTGGGATG | 95°C, 2 min; 40 × (95°C, 5 sec; 59°C, 10 sec; 72°C, 12 sec; | 5 µl AmplifyMe |
| 5′-AGCCAGGAGAAATCAAACAGAG 300 nM each | 77°C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | NoRox SybrGreen (Blirt, Poland), Σ 10 µl | ||
| CCND1 | NM_053056.3 | 5′-TGTCCTACTTCAAATGTGTGCA | 95°C, 2 min; 40 × (95°C, 5 sec; 57°C, 10 sec; 72°C, 12 sec; | |
| 5′-ATTGGAAATGAACTTCACATCTGT 300 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| GLI1 | NM_005269.2 | 5′-CTACATCAACTCCGGCCAATAG | 95°C, 2 min; 40 × (95°C, 5 sec; 58°C, 10 sec; 72°C, 12 sec; | |
| 5′-TGACAGTATAGGCAGAGCTGAT 300 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| GLI2 | NM_005270.4 | 5′-CTCCAACGAGAAACCCTACATC | 95°C, 2 min; 40 × (95°C, 5 sec; 59°C, 10 sec; 72°C, 12 sec; | |
| 5′-CACTGTCCCCATTCTCTTTGAG 200 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| GLI3 | NM_000168.5 | 5′-CTCTGACCGATGGAGGTAGTAT | 95°C, 2 min; 40 × (95°C, 5 sec; 58°C, 12 sec; 72°C, 12 sec; | |
| 5′-TGTGTGCCATTTCCTATGAGAG 135 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| GUSB | NM_000181.4 | 5′-ATGCAGGTGATGGAAGAAGTGGTG | 95°C, 3 min; 40 × (95°C, 5 sec; 57°C, 10 sec; 72°C, 10 sec; | |
| 5′-AGAGTTGCTCACAAAGGTCACAGG 200 nM each | 75 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| MYC | NM_001354870.1 | 5′-AAACATCATCATCCAGGACTGT | 95°C, 2 min; 40 × (95°C, 5 sec; 58°C, 10 sec; 72°C, 30 sec; | |
| 5′-TCTTCTTGTTCCTCCTCAGAGT 300 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| PTCH1 | NM_000264.3 | 5′-GAATCCCTTTTGAGGACAGGAC | 95°C, 2 min; 40 × (95°C, 5 sec; 59°C, 10 sec; 72°C, 12 sec; | |
| 5′-GCATGGTAATCTGCGTTTCATG 200 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| SHH | NM_000193.3 | 5′-GCAAAGCAAAAAGACACTCGG | 95°C, 2 min; 40 × (95°C, 5 sec; 59°C, 10 sec; 72°C, 12 sec; | |
| 5′-ATTTAAGGCTCTTGAAGGTCCG 200 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| SMO | NM_005631.4 | 5′-ACTTCTTCAACCAGGCTGAGT | 95°C, 2 min; 40 × (95°C, 5 sec; 59°C, 10 sec; 72°C, 12 sec; | |
| 5′-TCATCTTGCTCTTCTTGATCCG 300 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| SUFU | NM_016169.3 | 5′-TTCTGTTGACCGAAGAGTTTGT | 95°C, 2 min; 40 × (95°C, 5 sec; 59°C, 10 sec; 72°C, 12 sec; | |
| 5′-TGGGAAGTTTGAATTCCTCTGG 200 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C | |||
| VEGFA | NM_001025366.2 | 5′-GTCTAATGCCCTGGAGCCT | 95°C, 2 min; 40 × (95°C, 5 sec; 59°C, 10 sec; 72°C, 30 sec; | |
| 5′-TTCGTTTAACTCAAGCTGCCTC 200 nM each | 77 °C, 10 sec-sample reading) Melting curve: 95°C, 15 sec; 60°C, 1 min; 60°C → 95°C reading every 0.3°C |
BCL2, BCL2 apoptosis regulator; CCND1, cyclin D1; GLI1,2,3, glioma-associated zinc finger protein 1, 2, 3; MYC, MYC proto-oncogene; PTCH1, patched 1; SHH, sonic hedgehog ligand; SMO, smoothened; SUFU, suppressor of fused homolog; VEGFA, vascular endothelial growth factor A.
Cell cultures and reagents
The 786-O (CRL1932™) and ACHN (CRL1611™) human RCC cell lines and the HK2 (CRL2190™) human proximal kidney tubule-derived cell line were obtained from the American Type Culture Collection (ATCC). The 786-O and ACHN cells were cultured in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA) and Eagle's Minimum Essential Medium (MEM; Sigma-Aldrich; Merck KGaA), respectively. Both media were supplemented with 10% FBS (Sigma-Aldrich; Merck KGaA) and 1% penicillin-streptomycin (Sigma-Aldrich; Merck KGaA). MEM was additionally supplemented with 2% L-glutamine (Sigma-Aldrich; Merck KGaA). HK-2 cells were cultured in keratinocyte serum-free medium (Gibco; Thermo Fisher Scientific, Inc.). The cells were cultured in a sterile incubator that was maintained at 37°C in an atmosphere of 5% CO2. RU-SKI43, cyclopamine, GANT61 and sunitinib were purchased from Sigma-Aldrich; Merck KGaA and dissolved in DMSO (Sigma-Aldrich; Merck KGaA) as stock solutions according to the manufacturer's recommendation. The final concentration of DMSO in the cell culture medium never exceeded 1%.
Cell viability assay
The cytotoxicity of RU-SKI43, cyclopamine, GANT61 and sunitinib was measured using a Sulforhodamine B (SRB) assay (30). Briefly, the cells were seeded into 96-well plates at the following density: 786-O, 5×103 cells/well; ACHN, 5×103 cells/well; and HK-2, 8×103 cells/well. Cells were incubated for 24 h in the appropriate medium. Different concentrations of each inhibitor (sunitinib, 0-20 µM; RU-SKI43, 0-60 µM; GANT61, 0-80 µM; and cyclopamine, 0-60 µM) were added to the wells. After incubation for 24, 48 or 72 h, the cells were fixed by adding trichloroacetic acid (TCA) to a final concentration of 10% TCA and incubated for 1 h at 4°C. Subsequently, the plates were washed with deionized water and thoroughly drained for ~30 min. SRB (100 µl), dissolved in 1% acetic acid, was added to each well and the cells were stained for 15 min at room temperature. Thereafter, the plates were washed with 1% acetic acid to remove the dye and then dried overnight. Next, 200 µl TRIS buffer (10 mM; pH 10.5) was added to each well and the plates were shaken gently to dissolve the stain. The absorbance was measured spectrophotometrically at 540 nm using a reference wavelength of 630 nm on an Epoch UV universal microplate reader (BioTek Instruments, Inc.).
Cell cycle analysis
786-O, ACHN and HK2 cells were seeded at a density of 2×105 cells/well and, after 24 h, the cells were treated with GANT61, RU-SKI43, cyclopamine and sunitinib at the half maximal effective concentration (EC50) and the 25% effective concentration (EC25; Table II). After 48 h, the cells were collected, washed with PBS (Sigma-Aldrich; Merck KGaA) and fixed in 70% ethyl alcohol for 48 h at 4°C. Directly before cytometric analysis, the cells were stained with a dye mixture containing RNase and propidium iodide (PI) (Sigma-Aldrich; Merck KGaA) for 30 min in the dark at 37°C. Fluorescence of the PI-stained cells was measured via flow cytometry (excitation, 536 nm; emission, 617 nm; FACSCalibur; Becton-Dickinson and Company). The results were analyzed using CellQuest Pro software (Becton, Dickinson and Company) and presented as a percentage of cells with DNA content corresponding to apoptotic/necrotic cells (subG1 fraction) or cells in the G0/G1, S and G2/M phases of the cell cycle.
Table II.
EC50 and EC25 concentrations of RU-SKI43, cyclopamine, GANT61, and sunitinib for the 786-O, ACHN, and HK2 cell lines.
| Conc. (µM) of each agent | Cell line
|
|||||
|---|---|---|---|---|---|---|
| 786-O
|
ACHN
|
HK2
|
||||
| EC50 | EC25 | EC50 | EC25 | EC50 | EC25 | |
| RU-SKI43 | 15.22 | 21.65 | 20.65 | 27.98 | 12.34 | 20.55 |
| Cyclopamine | 31.38 | 40.7 | 29.75 | 31.39 | 24.85 | 40.13 |
| GANT61 | 15.43 | 26.87 | 14.75 | 35.15 | 18.75 | 35.62 |
| Sunitinib | 4.36 | 8.72 | 6.43 | 10.41 | 3.71 | 8.60 |
EC50, half maximal effective concentration; EC25, 25% effective concentration.
qPCR analysis of mRNA levels in cultured cells
A total of 2×105 cells/well were seeded in 6-well plates and incubated for 24 h in the appropriate medium. Subsequently, the cells were treated for 48 h with inhibitors at the EC50 and EC25. Total RNA from the collected samples was isolated using the ExtractMe Total RNA kit (Blirt, Inc.). The obtained RNA was dissolved in 40 µl nuclease-free water. The quantity and quality of RNA were measured using a spectrophotometer (NanoDrop ND 1000; Thermo Fisher Scientific, Inc.). RNA samples were stored at -20°C until further analysis. Further analysis was performed as aforementioned for clinical samples.
Wound healing assay
The cells were seeded in 6-well plates at the following density: 786-O, 3×105 cells/well; and ACHN, 6×105 cells/well. Cells were incubated for 48 h in the appropriate medium: 786-O cells, RPMI1640; ACHN cells, Eagle's Minimum Essential Medium. Both media were supplemented with 10% FBS (31-34) and 1% penicillin-streptomycin. MEM was additionally supplemented with 2% L-glutamine. Confluent cell cultures were then scratched using a sterile tip and incubated with GANT61, RU-SKI43, cyclopamine and sunitinib at the EC25. The wound healing process was monitored (12 h for 786-O cells and 32 h for ACHN cells) and images of the scratches were captured at different points of time using an inverted microscope. Quantification was performed by measuring the number of pixels in each wound closure area using ImageJ software 1.53 (National Institutes of Health).
Statistical analysis
Statistical analysis was performed using GraphPad Prism ver. 6.07 (GraphPad Software, Inc.) and Statistic aver. 13.3 (StatSoft Ltd.) software. First, data normality was checked using the Shapiro-Wilk test. However, since most data for clinical samples did not pass it, the following non-parametric tests were applied: Mann-Whitney U for two groups, Wilcoxon signed-rank if samples were paired and Spearman's correlation. Outliers were identified using the ROUT test. The median values of mRNA levels for a particular gene in the control group were used as a threshold for the determination of upregulation and downregulation of a given gene in cancer tissues. In this way, 2×2 Fisher's exact test was performed. For outcome analysis of patients, Kaplan-Meier survival tests with log-rank (Mantel-Cox) tests were applied, with data acquired for overall survival (OS). Cox proportional hazard regression model with univariable (first step) and multivariable (second step) tests were applied. Survival associations were presented as hazard ratios (HRs) with their 95% confidence interval (CI) and P-values using Cox and Kaplan-Meier estimations (35,36). The results from cellular assays were analyzed using a unpaired Student's t-test, and for more than 2 groups-one or two-way ANOVA with post hoc Tukey's multiple comparison test. A two-sided P<0.05 was considered to indicate a statistically significant difference, with a 95% CI in all analyses.
Results
Clinicopathological characteristics of the patients
The clinicopathological features of the patients are presented in Table III. The study included 62 patients with ccRCC, including 21 women and 41 men (mean age ± SD, 63,84±11.25 years). According to the Union for International Cancer Control TNM 8th staging edition of RCC guidelines (4), 29 patients were classified as stage I (T1N0M0), 4 as stage II (T2N0M0), 27 as stage III (T1-2N1M0 or T3N0-2M0), and 2 as stage IV (T4N0-2M0 or T1-4N0-2M1). Local or distant metastases were diagnosed in 29 (47%) patients, at the time of nephrectomy. Histological Fuhrman/World Health Organization (WHO)/International Society of Urological Pathology (ISUP) grading assessment (37) revealed 4 ccRCC samples in grade 1, 22 samples in grade 2, 21 samples in grade 3, and 15 samples in grade 4. The mean follow-up period was 32 months (range, 3-72 months). The median OS rate was 24 months. All deaths were associated with ccRCC progression.
Table III.
Clinicopathological characteristics of the ccRCC patients and their association with SHH, PTCH1, SMO, SUFU, GLI1-3, BCL2, MYC, VEGFA, CCND1 mRNA levels.
| Patients/genes
|
Subgroups |
SHH
|
SMO
|
SUFU
|
GLI2
|
GLI3
|
GLI1
|
PTCH1
|
BCL2
|
CCND1
|
MYC
|
VEGFA
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
qPCR results (%)
|
||||||||||||||||||||||||
| n=62 | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P 1 | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | ↓ | ↑ | P 1 | ↓ | ↑ | P-valuea | ↓ | ↑ | P-valuea | |
| Age (years) | ≤64 | 14 | 18 | 0.799 | 13 | 19 | 0.610 | 11 | 21 | 0.125 | 8 | 24 | 0.579 | 11 | 21 | 0.303 | 8 | 24 | 0.579 | 11 | 21 | 0.021 | 12 | 20 | 1.000 | 11 | 21 | 1.000 | 2 | 30 | 0.418 | 9 | 23 | 0.189 |
| Mean ± SD: | n=32 | (22.6) | (29.0) | (21.0) | (30.6) | (17.7) | (33.9) | (12.9) | (38.7) | (17.7) | (33.9) | (12.9) | (38.7) | (17.7) | (33.9) | (19.4) | (32.3) | (17.7) | (33.9) | (3.2) | (48.4) | (14.5) | (37.1) | |||||||||||
| 63,84±11.25 | >64 | 15 | 15 | 15 | 15 | 17 | 13 | 10 | 20 | 15 | 15 | 10 | 20 | 20 | 10 | 12 | 18 | 11 | 19 | 4 | 26 | 14 | 16 | |||||||||||
| Range: 33-86 | n=30 | (24.2) | (24.2) | (24.2) | (24.2) | (27.4) | (21.0) | (16.1) | (32.3) | (24.2) | (24.2) | (16.1) | (32.3) | (32.3) | (16.1) | (19.4) | (29.0) | (17.7) | (30.6) | (6.5) | (41.9) | (22.6) | (25.8) | |||||||||||
| Sex | Female | 12 | 9 | 0.289 | 10 | 11 | 0.794 | 11 | 10 | 0.434 | 9 | 12 | 0.138 | 11 | 10 | 0.283 | 5 | 16 | 0.570 | 9 | 12 | 0.592 | 8 | 13 | 1.000 | 6 | 15 | 0.576 | 3 | 18 | 0.398 | 9 | 12 | 0.583 |
| n=21 | (19.4) | (14.5) | (16.1) | (17.7) | (17.7) | (16.1) | (14.5) | (19.4) | (17.7) | (16.1) | (8.1) | (25.8) | (14.5) | (19.4) | (12.9) | (21.0) | (9.7) | (24.2) | (4.8) | (29.0) | (14.5) | (19.4) | ||||||||||||
| Male | 17 | 24 | 18 | 23 | 17 | 24 | 9 | 32 | 15 | 26 | 13 | 28 | 22 | 19 | 16 | 25 | 16 | 25 | 3 | 38 | 14 | 27 | ||||||||||||
| n=41 | (27.4) | (38.7) | (29.0) | (37.1) | (27.4) | (38.7) | (14.5) | (51.6) | (24.2) | (41.9) | (21.0) | (45.2) | (35.5) | (30.6) | (25.8) | (40.3) | (25.8) | (40.3) | (4.8) | (61.3) | (22.6) | (43.5) | ||||||||||||
| Tumor size (cm) | ≤7 cm | 19 | 20 | 0.794 | 17 | 22 | 0.796 | 17 | 22 | 0.796 | 11 | 28 | 1.000 | 14 | 25 | 0.288 | 8 | 31 | 0.082 | 18 | 21 | 0.600 | 12 | 27 | 0.112 | 10 | 29 | 0.054 | 3 | 36 | 0.662 | 13 | 26 | 0.587 |
| n=39 | (30.6) | (32.3) | (27.4) | (35.5) | (27.4) | (35.5) | (17.7) | (45.2) | (22.6) | (40.3) | (12.9) | (50.0) | (29.0) | (33.9) | (19.4) | (43.5) | (16.1) | (46.8) | (4.8) | (58.1) | (21.0) | (41.9) | ||||||||||||
| >7 cm | 10 | 13 | 11 | 12 | 11 | 12 | 7 | 16 | 12 | 11 | 10 | 13 | 13 | 10 | 12 | 11 | 12 | 11 | 3 | 20 | 10 | 13 | ||||||||||||
| n=23 | (16.1) | (21.0) | (17.7) | (19.4) | (17.7) | (19.4) | (11.3) | (16.1) | (19.4) | (17.7) | (16.1) | (21.0) | (21.0) | (16.1) | (19.4) | (17.7) | (19.4) | (17.7) | (4.8) | (32.3) | (16.1) | (21.0) | ||||||||||||
| ISUP | 1 + 2 | 12 | 14 | 1.000 | 12 | 14 | 1.000 | 11 | 15 | 0.798 | 8 | 18 | 1.000 | 9 | 17 | 0.435 | 6 | 20 | 0.412 | 10 | 16 | 0.198 | 6 | 20 | 0.038 | 5 | 21 | 0.032 | 4 | 22 | 0.227 | 8 | 18 | 0.434 |
| Histologicalgrade | n=26 | (19.4) | (22.6) | (19.4) | (22.6) | (17.7) | (24.2) | (12.9) | (29.0) | (14.5) | (27.4) | (9.7) | (32.3) | (16.1) | (25.8) | (9.7) | (32.3) | (8.1) | (33.9) | (6.5) | (35.5) | (12.9) | (29.0) | |||||||||||
| 3 + 4 | 17 | 19 | 16 | 20 | 17 | 19 | 10 | 26 | 17 | 19 | 12 | 24 | 21 | 15 | 18 | 18 | 17 | 19 | 2 | 34 | 15 | 21 | ||||||||||||
| n=36 | (27.4) | (30.6) | (25.8) | (32.3) | (27.4) | (30.6) | (16.1) | (41.9) | (27.4) | (30.6) | (19.4) | (38.7) | (33.9) | (24.2) | (29.0) | (29.0) | (27.4) | (30.6) | (3.2) | (54.8) | (24.2) | (33.9) | ||||||||||||
| TNM stage | Non-metastatic | 13 | 20 | 0.308 | 13 | 20 | 0.444 | 12 | 21 | 0.201 | 10 | 23 | 1.000 | 12 | 21 | 0.441 | 8 | 25 | 0.413 | 11 | 22 | 0.010 | 10 | 23 | 0.194 | 7 | 26 | 0.017 | 2 | 31 | 0.405 | 9 | 24 | 0.116 |
| n=33 | (21.0) | (32.3) | (21.0) | (32.3) | (19.4) | (33.9) | (16.1) | (37.1) | (19.4) | (33.9) | (12.9) | (40.3) | (17.7) | (35.5) | (16.1) | (37.1) | (11.3) | (41.9) | (3.2) | (50.0) | (14.5) | (38.7) | ||||||||||||
| Metastatic | 16 | 13 | 15 | 14 | 16 | 13 | 8 | 21 | 14 | 15 | 10 | 19 | 20 | 9 | 14 | 15 | 15 | 14 | 4 | 25 | 14 | 15 | ||||||||||||
| n=29 | (25.8) | (21.0) | (24.2) | (22.6) | (25.8) | (21.0) | (12.9) | (33.9) | (22.6) | (24.2) | (16.1) | (30.6) | (32.3) | (14.5) | (22.6) | (24.2) | (24.2) | (22.6) | (6.5) | (40.3) | (22.6) | (24.2) | ||||||||||||
High (↑) and low (↓) expression groups are based on the median values in normal biopsies. aP-values were calculated by Fisher's 2×2 test. Cut off values: SHH=1.023, SMO=0.228, SUFU=0.096, GLI2=0.0009, GLI3=0.016, GLI1=0.027, PTCH1=0.214, BCL2=0.414, MYC=0.077, VEGFA=0.001, CCND1=0.251. ccRCC, clear cell renal cell carcinoma; qPCR, quantitative polymerase chain reaction; SHH-sonic hedgehog; PTCH1, patched 1; SMO, smoothened; SUFU, suppressor of fused homolog; GLI1, 2, 3, glioma-associated zinc finger protein 1, 2, 3; BCL2, BCL2 apoptosis regulator; MYC, MYC proto-oncogene; BHLH, transcription factor; VEGFA, vascular endothelial growth factor A; CCND1, cyclin D1.
The SHH pathway genes and their targets are overexpressed in ccRCC tissues
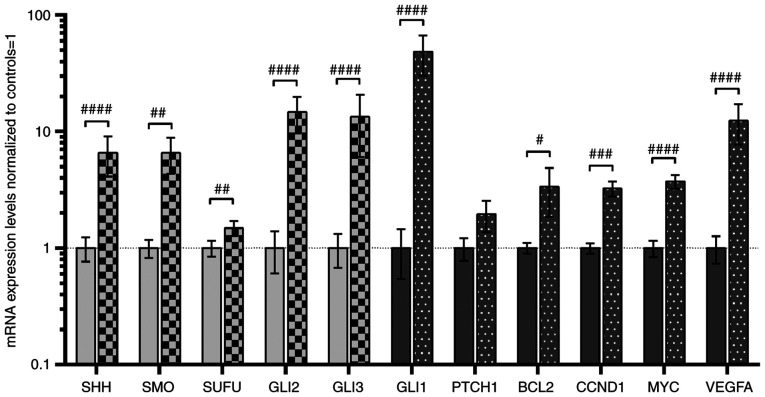
A total of two assessment methods were applied (16,35,36): Direct quantitative comparisons between tumor and normal kidney tissues, are shown in Fig. 2, and comparisons of mRNA levels of genes in tumor samples divided into high (↑) and low (↓) expression groups based on the median values in normal biopsies, are shown in Table III. Similar to our previous study (16), increased expression of all analyzed SHH pathway genes was observed in tumor samples (light grey bars in Fig. 2), with the highest levels of signaling TFs, GLI2 and GLI3, with ~14-fold higher mRNA levels in tumors (Fig. 2). Furthermore, higher mRNA level of GLI2 and GLI3 was observed in 44 and 36 patients with ccRCC, respectively (Table III). High (~48-fold upregulation) GLI1 gene expression was observed in tumor tissues of 44 patients with ccRCC (Table III), and this also acts as positive feedback in the SHH signaling pathway (6). Although the mRNA levels of PTCH1, the second gene involved in negative feedback regulation of the SHH signaling pathway (6), were not quantitatively changed in direct comparison with normal kidney samples, increased levels were observed in tumor ccRCC tissues of younger patients (<64 years of age; Table III), which is in line with our previous research (16). The expression levels of putative SHH pathway-upregulated genes were also increased (dark-grey bars in Fig. 2), with high expression levels of VEGFA (~12-fold upregulation) identified in 39 patients with ccRCC (Table III). Subsequently, the present study examined whether GLI1-3 expression was associated with mRNA levels of their upstream regulator SHH and their target genes. There was a strong association between SHH expression and all studied GLI TFs (Table IV; GLI1, r=0.680, P<0.0001; GLI2, r=0.600, P<0.0001; GLI3, r=0.644, P<0.0001). The association between the expression of SHH and GLI1 was also observed in our previous study (16). Furthermore, in the cancer tissues, GLI1 was strongly upregulated by GLI2 and GLI3 (r=0.767 and r=0.894, respectively; P<0.0001; Table IV). The expression levels of other genes activated by GLIs presented weaker correlations, with the lowest r values observed for PTCH1. However, a weak positive association was observed between CCND1 gene expression and GLI2 gene expression. Interestingly, the expression levels of VEGFA exhibited a strong (r>0.75) correlation with the expression levels of GLI1-3 (Table IV).
Figure 2.
Analysis of the expression of SHH pathway genes (SHH, SMO, SUFU, GLI2 and GLI3; light grey bars) and SHH pathway target genes (GLI1, PTCH1, BCL2, CCND1, MYC and VEGFA; dark grey bars). Comparison between ccRCC and normal kidney samples. Gene expression was assessed as described in the Materials and methods section. Bars and whiskers represent the mean ± SD normalized to control kidney samples. #P<0.05, ##P<0.01, ###P<0.001, ####P<0.0001 (Wilcoxon signed-rank). SHH, sonic hedgehog ligand; SMO, smoothened; SUFU, suppressor of fused homolog; GLI1,2,3, glioma-associated zinc finger protein 1, 2, 3; PTCH1, patched 1; CCND1, cyclin D1; VEGFA, vascular endothelial growth factor A; ccRCC, clear cell renal cell carcinoma; qPCR, quantitative PCR.
Table IV.
Correlation between the SHH, GLI TFS, and their targeted genes at the mRNA levelsa.
| Genes
|
GLI2
|
GLI3
|
GLI1
|
|||
|---|---|---|---|---|---|---|
| Correlation results | rb | P-valueb | rb | P-valueb | rb | P-valueb |
| SHH | 0.600 | <0.0001 | 0.644 | <0.0001 | 0.680 | <0.0001 |
| GLI1 | 0.767 | <0.0001 | 0.894 | <0.0001 | - | - |
| PTCH1 | 0.560 | 0.0002 | 0.548 | 0.0003 | 0.577 | 0.0002 |
| BCL2 | 0.649 | <0.0001 | 0.517 | 0.0003 | 0.628 | <0.0001 |
| MYC | 0.696 | <0.0001 | 0.600 | <0.0001 | 0.526 | 0.0002 |
| VEGFA | 0.752 | <0.0001 | 0.771 | <0.0001 | 0.755 | <0.0001 |
| CCND1 | 0.333 | 0.038 | 0.188 | 0.239 | 0.164 | 0.306 |
GLI TFs are placed horizontally; SHH and the affected genes are vertical.
r and P-values were calculated by Spearman's test: results with statistically significant values are indicated in bold print. SHH, sonic hedgehog; PTCH1, patched 1; GLI1,2,3, glioma-associated zinc finger protein 1, 2, 3; BCL2, BCL2 apoptosis regulator; MYC, MYC proto-oncogene; TFs, transcription factorS; VEGFA, vascular endothelial growth factor A; CCND1, cyclin D1.
Expression levels of SHH pathway genes and their targets are upregulated in early ccRCC
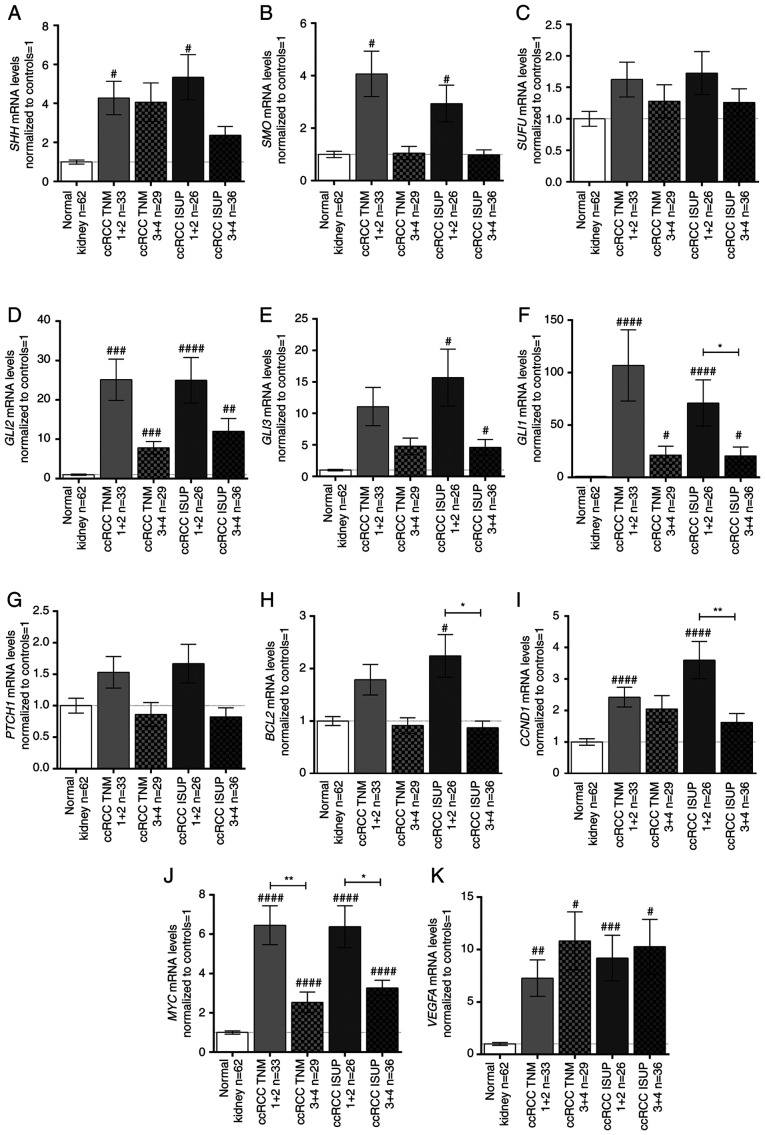
With the exception of PTCH1, the expression levels of the analyzed genes were not associated with age, sex or tumor size (Table III; plots not shown); however, differences were observed when cancer tissues were classified by TNM stage and ISUP grade (Fig. 3). Generally, the analyzed SHH signaling pathway genes were upregulated in early TNM (1+2) stages and ISUP (1+2) grades (Fig. 3A, B, D, E) with the exception of SUFU (Fig. 3C). GLI2 was strongly upregulated in all TNM stages/ISUP grades with ~25-fold higher levels in early stage/grade tissues compared with in paired normal kidney tissues (Fig. 3D). On the other hand, GLI3 was upregulated ~15- and ~5-fold in early and advanced ISUP stages, respectively, with no relation to TNM stage (Fig. 3E). The mRNA expression levels of GLI1 exhibited a different pattern: GLI1 mRNA expression was ~107- and ~71-fold higher in early TNM stages and ISUP grades, respectively; however, in advanced stage/grade tissues, GLI1 expression was still at least ~20 times higher than in control tissues (Fig. 3F). PTCH1 exhibited an opposite pattern of expression according to TNM stages (Table III); however, no significant difference, as assessed using the Mann-Whitney U test, was noted (Fig. 3G). The BCL2 and CCND1 genes showed a similar expression pattern to the SHH pathway genes: BCL2 was upregulated in ISUP grades 1+2 (Fig. 3H; Table III), while tumors in both early TNM and ISUP stages/grades were characterized by higher mRNA expression levels of CCND1 (Fig. 3I; Table III). MYC expression in ccRCC was ~6- and 3-fold higher in early and advanced stages/grades, respectively, than in control tissues (Fig. 3J). Furthermore, there was a marked difference in MYC mRNA expression between the cancer tissues assessed as stage/grade 1+2 and 3+4 according to both TNM and ISUP classifications (Fig. 3J). Interestingly, all tumor samples exhibited increased VEGFA mRNA expression compared with control tissues independent of ccRCC stage/grade (Fig. 3K).
Figure 3.
mRNA expression levels of the (A) SHH, (B) SMO, (C) SUFU, (D) GLI2, (E) GLI3, (F) GLI1, (G) PTCH1, (H) BCL2, (I) CCND1, (J) MYC, and (K) VEGFA genes in tissue samples of patients with ccRCC, related to TNM stage and ISUP grade. Gene expression was measured by qPCR. Bars and whiskers represent the mean ± SEM normalized to control kidney samples. P-values between groups (Mann-Whitney U test) are noted: #P<0.05, ##P<0.01, ###P<0.001, ####P<0.0001 to compare significance between control and ccRCC samples; *P<0.05, **P<0.01, to compare significance between ccRCC TNM 1+2 and TNM 3+4 samples as well as ccRCC ISUP 1+2 and ISUP 3+4 samples. SHH, sonic hedgehog ligand; SMO, smoothened; SUFU, suppressor of fused homolog; GLI1,2,3, glioma-associated zinc finger protein 1, 2, 3; PTCH1, patched 1; CCND1, cyclin D1; VEGFA, vascular endothelial growth factor A; ccRCC, clear cell renal cell carcinoma; qPCR, quantitative PCR; ISUP, International Society of Urological Pathology.
Low CCND1 and high VEGFA expression levels in ccRCC tissues are associated with shorter OS
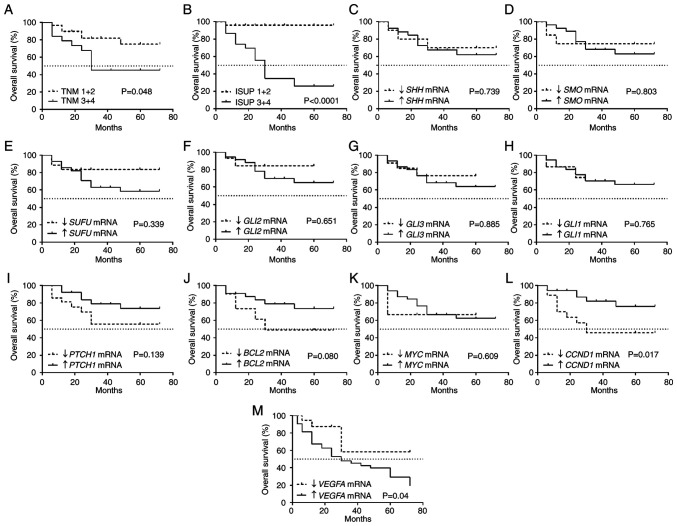
It was observed that tumors characterized by advanced TNM stages and ISUP grades were associated with shorter OS (Fig. 4A and B) with a 50% survival rate of 30 months. The mRNA levels of SHH, SMO, SUFU, GLI2, GLI3, GLI1, PTCH1, BCL2 and MYC (Fig. 4D-K) were not associated with OS. Shorter OS was associated with lower CCND1 expression (Fig. 4L) as well as high VEGFA expression (Fig. 4M).
Figure 4.
Kaplan-Meier overall survival analysis for patients with ccRCC related to clinicopathological and molecular data. (A) TNM classification. (B) ISUP grade. (C) SHH mRNA expression. (D) SMO mRNA expression. (E) SUFU mRNA expression. (F) GLI2 mRNA expression. (G) GLI3 mRNA expression. (H) GLI1 mRNA expression. (I) PTCH1 mRNA expression. (J) BCL2 mRNA expression. (K) MYC mRNA expression. (L) CCND1 mRNA expression. (M) VEGFA mRNA expression. Cut-off values for increased and decreased expression were determined using the median expression value of each gene in the control samples. The log-rank (Mantel-Cox) test was applied. SHH, sonic hedgehog ligand; SMO, smoothened; SUFU, suppressor of fused homolog; GLI1,2,3, glioma-associated zinc finger protein 1, 2, 3; PTCH1, patched 1; CCND1, cyclin D1; VEGFA, vascular endothelial growth factor A; ccRCC, clear cell renal cell carcinoma; qPCR, quantitative PCR; ISUP, International Society of Urological Pathology.
Cox proportional hazards test with univariable regression analysis revealed that patients with high ISUP grade, high VEGFA expression and low CCND1 expression were at risk of death due to ccRCC. Multivariable regression analysis indicated that increased VEGFA mRNA expression was an independent prognostic factor of a worse outcome in patients with advanced ISUP histological grade. Patients with ccRCC with advanced (3+4) histological grade had a high risk of death (HR=12.55) if they also exhibited increased VEGFA mRNA expression in tumor tissues (HR=3.72; Table V).
Table V.
Univariable and multivariable Cox regression analysis of the overall survival rate of the ccRCC patients.
| Parameters | Univariable analysis
|
Multivariable analysis
|
||||
|---|---|---|---|---|---|---|
| χ2 | P-value | HR (95 CI) | χ2 | P-value | HR (95 CI) | |
| Sex | ||||||
| Female vs. Male | 0.48 | 0.486 | 0.52 (0.08-3.21) | |||
| Age (years) | ||||||
| >62 vs. ≤62 | 1.99 | 0.157 | 0.32 (0.06-1.54) | |||
| Tumor size (cm) | ||||||
| >7 vs. ≤7 | 1.08 | 0.297 | 2.62 (0.42-16.06) | |||
| Tumor stage | ||||||
| T3+4 vs. T1+2 | 0.04 | 0.824 | 1.30 (2.64-585.07) | |||
| Histological ISUP grade | ||||||
| 3+4 vs. 1+2 | 7.11 | 0.007 | 39.37 (0.12-13.51) | 10.94 | <0.001 | 12.55 (2.80-56.18) |
| SHH mRNA levels | ||||||
| ↑ vs. ↓ | 2.84 | 0.091 | 23.76 (0.59-945.48) | |||
| SMO mRNA levels | ||||||
| ↑ vs. ↓ | 2.58 | 0.107 | 0.07 (0.01-1.76) | |||
| SUFU mRNA levels | ||||||
| ↑ vs. ↓ | 0.10 | 0.747 | 2.04 (0.026-159.89) | |||
| GLI2 mRNA levels | ||||||
| ↑ vs. ↓ | 0.15 | 0.698 | 0.47 (0.01-20.22) | |||
| GLI3 mRNA levels | ||||||
| ↑ vs. ↓ | 0.17 | 0.675 | 0.49 (0.02-13.27) | |||
| GLI1 mRNA levels | ||||||
| ↑ vs. ↓ | 1.24 | 0.265 | 8.10 (0.20-321.45) | |||
| PTCH1 mRNA levels | ||||||
| ↑ vs. ↓ | 3.72 | 0.053 | 0.18 (0.03-1.02) | |||
| BCL2 mRNA levels | ||||||
| ↑ vs. ↓ | 0.64 | 0.420 | 0.44 (0.06-3.16) | |||
| CCND1 mRNA levels | ||||||
| ↓ vs. ↑ | 3.93 | 0.047 | 12.63 (1.03-154.95) | 0.07 | 0.781 | 1.14 (0.43-3.01) |
| MYC mRNA levels | ||||||
| ↑ vs. ↓ | 0.01 | 0.901 | 1.29 (0.02-78.98) | |||
| VEGFA mRNA levels | ||||||
| ↑ vs. ↓ | 4.12 | 0.042 | 10.09 (1.08-93.97) | 4.17 | 0.041 | 3.72 (1.05-13.17) |
HR, hazard ratio; CI, confidence interval; ccRCC, clear cell renal cell carcinoma; SHH, sonic hedgehog; SMO, smoothened; SUFU, suppressor of fused homolog; GLI1, 2, 3, glioma-associated zinc finger protein 1, 2, 3; PTCH1, patched 1; BCL2, BCL2 apoptosis regulator; CCND1, cyclin D1; MYC, MYC proto-oncogene; VEGFA, vascular endothelial growth factor A.
Effects of SHH pathway inhibitors on proliferation and the cell cycle of 786-O, ACHN and HK2 cells SHH pathway inhibitors decrease proliferation of the 786-O, ACHN and HK2 cells
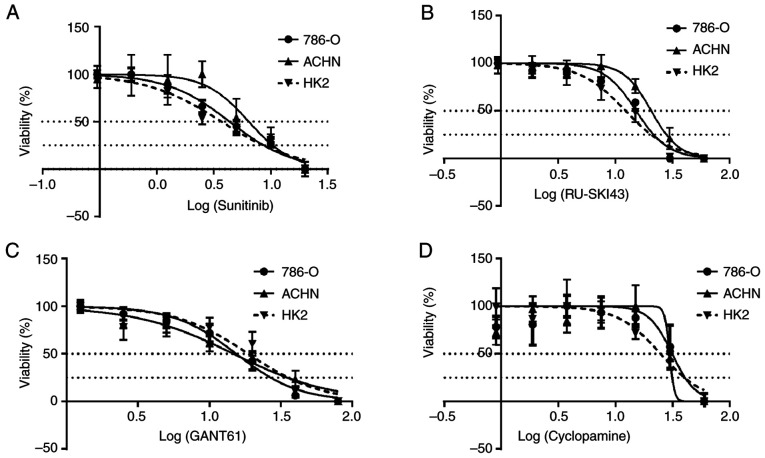
The SRB cell viability assay revealed that sunitinib, RU-SKI43, GANT61 and cyclopamine markedly inhibited the proliferation of 786-O and ACHN kidney cancer cells and HK2 normal kidney cells after 48 h (Fig. 5A-D). Data for incubation times of 24 and 72 h are not shown, since cell proliferation did not change after 24 h and/or all cells died after 72 h of drug treatment. The calculated EC50 and EC25 values for individual compounds for a particular cell line are presented in Table II (also shown as dotted lines in Fig. 5A-D) and were used in further analyses.
Figure 5.
Effects of (A) sunitinib, (B) RU-SKI43, (C) GANT61 and (D) cyclopamine on the viability of 786-O, ACHN and HK2 cells. The cells were treated with serial dilutions of sunitinib (0-20 µM), RU-SKI (0-60 µM), GANT61 (0-80 µM) and cyclopamine (0-60 µM). Data are presented as the mean of three independent experiments ± SD on a logarithmic scale. Statistical significance was estimated using one-way ANOVA.
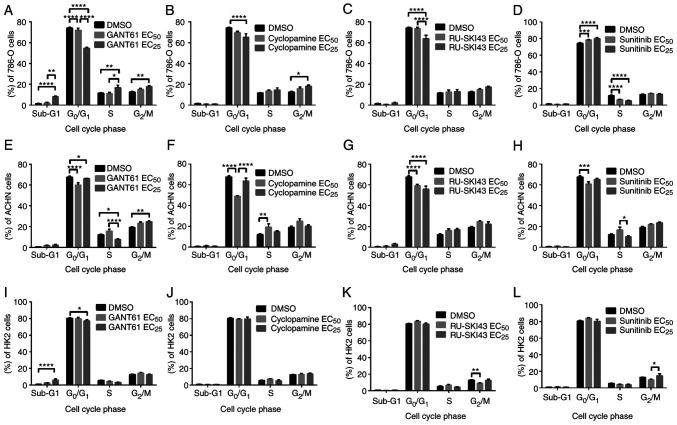
SHH pathway inhibitors decrease the number of 786-O and ACHN cells in the G0/G1 phase
Flow cytometry analysis revealed that the treatment of 786-O cells with GANT61 at the EC25 led to a significant dose-dependent decrease in the number of cells in the G0/G1 phase and a significant increase in the number of cells in sub-G1 (indicative of apoptosis induction), S and G2/M phases (Fig. 6A). Similar to GANT61, cyclopamine at the EC25 significantly reduced the number of 786-O cells in the G0/G1 phase and significantly increased the number of these cells in the G2/M phase (Fig. 6B). The treatment of 786-O cells with RU-SKI43 at the EC25 significantly decreased the number of cells in the G0/G1 phase without any significant effect on the other cell cycle phases (Fig. 6C). Different effects of sunitinib (at the EC25) on 786-O cells were observed, including a dose-dependent increase in the number of cells in the G0/G1 phase and a decreased number of cells in the S phase of the cell cycle (Fig. 6D). Treatment with GANT61 significantly reduced the number of ACHN cells in the G0/G1 phase at both the EC50 and EC25, and in S phase at the EC25. Furthermore, GANT61 at the EC25 significantly increased the number of ACHN cells in the G2/M phase (Fig. 6E). Cyclopamine at the EC50 significantly decreased the number of ACHN cells in the G0/G1 phase and significantly increased the number of cells in the S phase (Fig. 6F). After incubation of ACHN cells with RU-SKI43 at both the EC50 and EC25, and with sunitinib at the EC50, a reduced number of cells in the G0/G1 phase was observed, without any significant changes in the other cell cycle phases (Fig. 6G and H, respectively).
Figure 6.
Effects of (A, E and I) GANT61, (B, F and J) cyclopamine, (C, G and K) RU-SKI43 and (D, H and L) sunitinib on the cell cycle distribution of 786-O, ACHN and HK2 cells after 48 h of incubation with each compound at the EC50 or EC25. Cells were harvested, stained with PI and analyzed by flow cytometry. Statistical significance was estimated using two-way ANOVA. Data are presented as the mean ± SEM of three independent experiments carried out in triplicate. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. control. EC25, 25% effective concentration; EC50, half maximal effective concentration.
GANT61 increases the number of HK2 cells in the sub-G1 phase
Incubation of HK2 cells with GANT61 at the EC25 increased the number of cells in the sub-G1 phase. Furthermore, treatment with GANT61 at the EC25 reduced the number of cells in the G0/G1 phase (Fig. 6I). Cyclopamine at both concentrations had no significant effect on the cell cycle of HK2 cells (Fig. 6J). RU-SKI43 at the EC50 decreased the number of HK2 cells in the G2/M phase, without any effect on the other cell cycle phases (Fig. 6K). Sunitinib at the EC25 increased the number of cells in the G2/M phase compared with HK2 cells incubated with a lower (EC50) sunitinib concentration (Fig. 6L). In parallel control experiments, the solvent, DMSO, had no effect on the cell cycle of 786-O, ACHN and HK2 cells.
Effects of SHH pathway inhibitors on the expression levels of SHH pathway target genes in 786-O, ACHN and HK2 cells
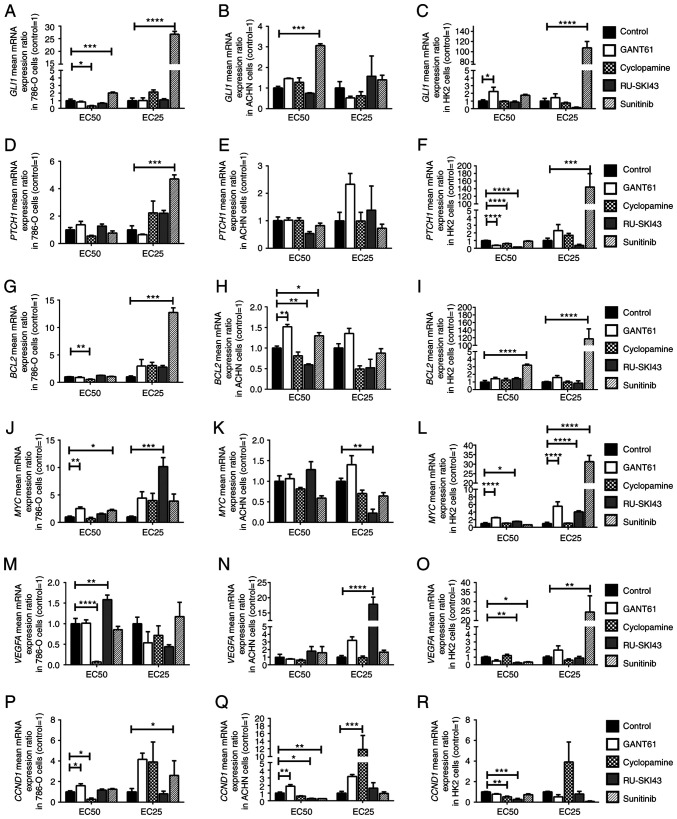
The results concerning the expression levels of SHH target genes in 786-O, ACHN, and HK2 cells are presented in Fig. 7: GLI1 (A-C), PTCH1 (D-F), BCL2 (G-I), MYC (J-L), VEGFA (M-O) and CCND1 (P-R).
Figure 7.
Effects of GANT61, cyclopamine, RU-SKI43 and sunitinib on (A-C) GLI1, (D-F) PTCH1, (G-I) BCL2, (J-L) MYC, (M-O) VEGFA and (P-R) CCND1 mRNA expression in 786-O, ACHN and HK2 cells. The cells were treated with each compound at the EC50 and EC25 for 48 h. cDNA was obtained from cells by RNA isolation and further reverse transcription. Gene expression was measured by qPCR. Statistical significance was estimated using one-way ANOVA. Data are presented as the mean ± SEM of three independent experiments carried out in duplicate. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 vs. control. GLI1, glioma-associated zinc finger protein 1; PTCH1, patched 1; CCND1, cyclin D1; VEGFA, vascular endothelial growth factor A; qPCR, quantitative PCR; EC25, 25% effective concentration; EC50, half maximal effective concentration.
GANT61 increases the expression level of SHH pathway target genes in 786-O, ACHN and HK2 cells
The incubation of the studied cell lines with GANT61 at the EC50 led to the significant upregulation of MYC and CCND1 mRNA expression in 786-O cells (Fig. 7J and P), and also significantly increased the mRNA expression levels of BCL2 and CCND1 in ACHN cells (Fig. 7H and Q). In HK2 cells, GANT61 at the EC50 significantly increased GLI1 gene expression (Fig. 7C). Furthermore, GANT61 at the EC50 and EC25 increased the expression levels of MYC (Fig. 7L), while it decreased PTCH1 mRNA expression at the EC50 (Fig. 7F).
Cyclopamine decreases the expression level of SHH pathway target genes in 786-O cells
The incubation of 786-O cells with cyclopamine at the EC50 decreased the expression levels of GLI1, BCL2, VEGFA and CCND1 (Fig. 7A, G, M and P). Cyclopamine at the EC25 stimulated the expression of the CCND1 gene in ACHN cells (Fig. 7Q). In HK2 cells, cyclopamine at the EC50 significantly decreased the mRNA expression levels of PTCH1 and CCND1 (Fig. 7F and R).
RU-SKI43 decreases the expression level of SHH pathway target genes in ACHN cells
Significantly increased expression levels of MYC and VEGFA were observed after incubation of 786-O cells with RU-SKI43, at the EC25 and EC50, respectively (Fig. 7J and M). RU-SKI43 at the EC50 significantly decreased the mRNA expression levels of BCL2 and CCND1 in ACHN cells (Fig. 7H and Q), and RU-SKI43 at the EC25 significantly decreased the mRNA expression levels of MYC (Fig. 7K). In ACHN cells incubated with RU-SKI43 at the EC25 the expression levels of VEGFA were upregulated (Fig. 7N). In HK2 cells, RU-SKI43 at both the EC50 and EC25 significantly increased the expression levels of MYC (Fig. 7L); however, at the EC50, RU-SKI43 significantly reduced the expression levels of the PTCH1, VEGFA and CCND1 genes (Fig. 7F, O and R).
Sunitinib increases the expression level of SHH pathway target genes in 786-O, ACHN and HK2 cells
The incubation of 786-O cells with sunitinib at the EC50 and EC25 was associated with the upregulation of GLI1 mRNA expression (Fig. 7A). Furthermore, upregulation of the expression levels of PTCH1, BCL2 and CCND1 genes in 786-O cells cultured with sunitinib at the EC25 (Fig. 7D, G and P), and higher MYC mRNA expression in 786-O cells incubated with sunitinib at the EC50 were revealed (Fig. 7J). Following the incubation of ACHN cells with sunitinib at the EC50, increased expression levels of GLI1 and BCL2 (Fig. 7B and H), and reduced CCND1 mRNA expression were revealed (Fig. 7Q). The treatment of HK2 cells with sunitinib at the EC25 led to the significant upregulation of the expression levels of GLI1 (Fig. 7C), PTCH1 (Fig. 7F), MYC (Fig. 7L) and VEGFA (Fig. 7O). Sunitinb at both the EC50 and EC25 significantly increased the expression levels of BCL2 in HK2 cells (Fig. 7I). In HK2 normal kidney cells, sunitinib at the EC50 significantly decreased the mRNA expression levels of VEGFA (Fig. 7O).
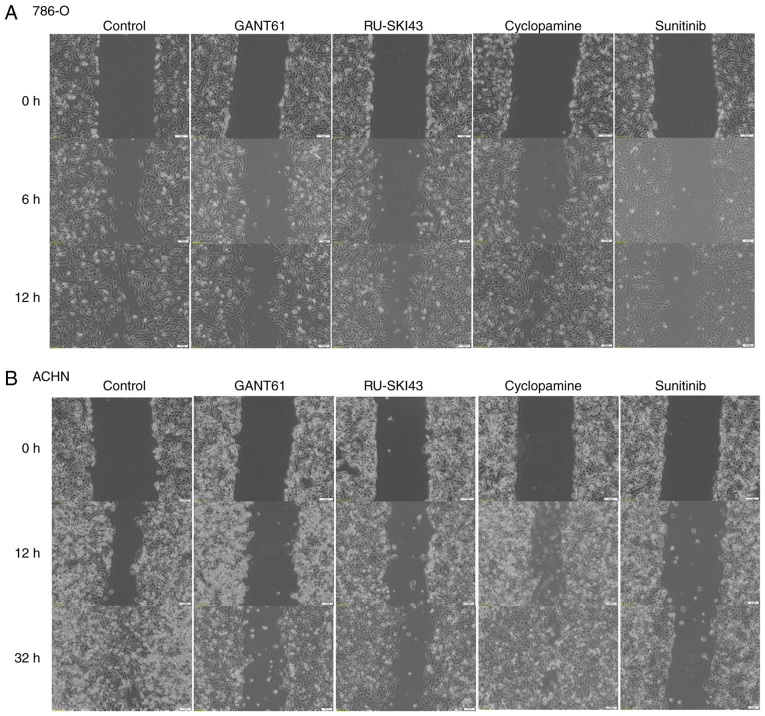
Effects of SHH pathway inhibitors on 786-O, ACHN and HK2 cell migration GANT61 and RU-SKI43 inhibit the migration of renal cancer cell lines
The observation times for 786-O and ACHN cells were first adjusted, as these cell lines exhibited different migration rates. The final time for the complete gap closure for 786-O cells was 12 h (Figs. 8A and 9A), while ACHN cells required at least 32 h to completely close the gap (Figs. 8B and 9B). Therefore, at 12 h after scratching, it was observed that inhibition of 786-O cell migration was caused by sunitinib (41.83±13% wound closure) and GANT61 (51.83±7.46% wound closure). Additionally, RU-SKI43 and sunitinib inhibited the migration of ACHN cells at 32 h after scratching (40.02±10.55% and 49.76±15.31% wound closure, respectively), while GANT61 and cyclopamine did not alter the rate of ACHN cell migration. The bar graph illustrating percentage wound closure, as well as representative images documenting the cell migration process at the indicated time points during the scratch wound assay, are presented in Figs. 8 and 9, respectively.
Figure 8.
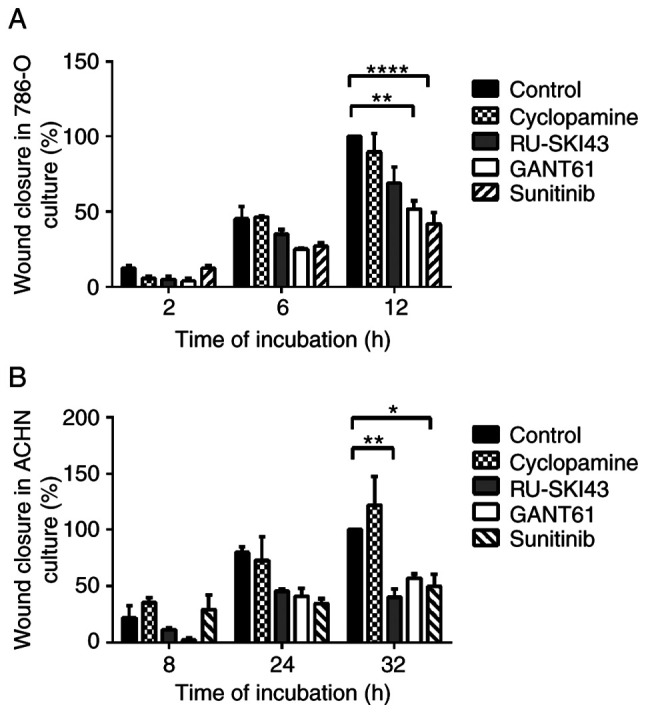
Summary bar graph illustrating percentage wound closure at indicated time points during the scratch wound assay for (A) 786-O and (B) ACHN cell cultures incubated with GANT61, RU-SKI43, cyclopamine and sunitinib. Data are presented as the mean ± SEM of three independent experiments carried out in duplicate. *P<0.05, **P<0.01, ****P<0.0001 vs. control.
Figure 9.
Cell migration (scratch wound healing assay). (A) 786-O and (B) ACHN cells were incubated with GANT61, RU-SKI43, cyclopamine and sunitinib for 12 and 32 h, respectively. Representative images from three independent experiments are shown.
Discussion
Sonic hedgehog (SHH) signaling is involved in the human embryogenesis of epithelial tissue (38) and the respiratory system (39), and it is critical in the development of the nervous system (40-42) and limbs (43,44). Although some authors have suggested that this molecular cascade also remains active in postnatal life in organs such as the brain (45) or lungs (46), the signaling becomes silenced in almost all other tissues (47). Physiological reactivation of the SHH signaling pathway has been observed during the process of wound healing (48,49). However, upregulation of the expression levels of SHH pathway genes, either at the mRNA or protein level, has been observed in various malignancies, including gastric cancer (50), retinoblastoma (51), glioma (52) and chronic myeloid leukemia (53). Therefore, SHH signaling appears to be involved in the molecular mechanism of carcinogenesis and may provide a promising anticancer target.
The present study included 62 clear cell renal cell carcinoma (ccRCC) patients. The clinicopathological features of the patients, including age, female/male ratio, and stage, grade and tumor size distribution, were similar to those of patients in our previous research (16,35,36). The age and sex distribution complied with the ccRCC group presented by Hsieh et al (3). Similar to our previous study (16), the present study revealed markedly higher mRNA expression levels of PTCH1 in the group of younger patients, which, to the best of our knowledge, is a novel observation.
One of the aims of the present study was to examine the molecular pattern of SHH pathway components at the mRNA level in ccRCC tumor tissues compared with paired morphologically unchanged kidney tissue samples. The data obtained via qPCR demonstrated that the expression levels of almost all analyzed genes were upregulated in cancer tissues. The largest increase in expression in tumor samples (~100-fold upregulation) was reported for GLI1. Similar to the current results, the upregulation of the expression levels of GLI1 and GLI2 in ccRCC was noted by Zhou et al (17) at the mRNA (qPCR) and protein levels [immunohistochemical staining (IHC)] in 58 (qPCR) or 17 (IHC) ccRCC cases. However, contrary to the present findings, they reported lower mRNA levels of SHH and PTCH1 in tumor tissues and did not observe a difference in the expression levels of GLI3 between ccRCC and control samples (17). Therefore, while the upregulation of the expression levels of the GLI1 and GLI2 transcription factors (TFs) in ccRCC appears to be confirmed, further studies are required to investigate the expression of the upstream components of the SHH signaling pathway in kidney tumor tissues. The association between the expression of SHH pathway genes and tumor grade has also been analyzed in colorectal carcinoma (54). The study revealed higher SHH immunoexpression in well-differentiated tumors than in poorly differentiated tumors, which is a similar result to the current findings for SHH mRNA expression and ccRCC grade (54). However, in breast cancer tissues, the immunoreactivity of SHH, GLI1 and GLI2 was found to be positively associated with histological grade (55), which may suggest different tendencies in the expression of SHH signaling genes depending on cancer type and histological grade.
The crosstalk of SHH signaling and other cellular pathways in malignancies has recently been featured in our previous review (6). The associations between mRNA levels of GLI1-3 and their molecular target genes in ccRCC were determined since there are no related data in the available literature. First, it was revealed that mRNA levels of GLI1 were associated with GLI2-3, which confirmed their canonical relationship in the analyzed pathway. The mRNA expression levels of the SHH gene were positively associated with those of the GLI1-3 TFs. Furthermore, the mRNA expression levels of PTCH1, BCL2, MYC and VEGFA were related to the mRNA expression levels of GLI1-3. Therefore, the present data suggested that the SHH cascade was involved in the stimulation of GLI1, PTCH1, BCL2, MYC and VEGFA gene activity in ccRCC, providing novel information. A similar association between SHH pathway components was also observed by Saze et al (50) in gastric cancer. They found positive associations between the mRNA expression levels of SHH and PTCH1, as well as among the expression levels of PTCH1, SMO and GLI1 in gastric cancer tissues (50). Zhou et al (17) suggested that GLI1/2 expression in ccRCC is regulated by PI3K/AKT signaling, due to the association between GLI1/2 mRNA expression and the content of total as well as the phosphorylated form of AKT protein. Although the CCND1 gene encoding cyclin D1, an important cell cycle regulator which forms a complex with Cdk4 and Cdk6 during the G1 phase (56), is listed as one of the target genes of the SHH pathway (57), the present results revealed weak effects of this molecular cascade on CCND1 expression in ccRCC.
The present study analyzed the association of the expression levels of the genes involved in the SHH pathway and its target genes with tumor stage (TNM 8th staging edition of RCC) (4) and histological grade (WHO/ISUP grading system) (58). The mRNA levels of almost all studied genes were increased in terms of both TNM staging and histological grading, predominantly in the group of less advanced cancers (TNM 1+2; ISUP 1+2). A clear tendency to downregulation of gene expression in ccRCC tumors assessed as TNM 3+4 and ISUP 3+4 was revealed, which is a novel observation in this type of cancer. These data suggest that the expression levels of genes of the SHH pathway and associated target genes may be a promising molecular marker of early kidney cancer development; however, additional research investigating this hypothesis must be performed. The observed decrease in GLI1, BCL2, MYC and CCND1 mRNA levels in higher grades of ccRCC (ISUP 3+4) compared with the levels in lower grades (ISUP 1+2) indicated the prognostic potential of these parameters. The aforementioned results were confirmed by using a 2×2 Fisher's test for BCL2 and CCND1 expression. Jäger et al (59) determined the prognostic significance of the expression of SHH pathway proteins in 140 samples with ccRCC via IHC. They revealed higher immunoreactivity of GLI3, PTCH1 and SHH in ISUP 3+4 (G3/G4) tumors than in ISUP 1+2 (G1/G2) ccRCC tumors (59). The disparities between their results and the present results may be caused by the differences in the applied research techniques, IHC vs. qPCR, respectively, with the first being considered to be qualitative or at most semi-quantitative. Furthermore, post-translational modifications of SHH and GLI proteins, such as structural cleavage (22), phosphorylation or ubiquitination (60) have been reported, and such changes can influence the immunoreactivity.
BCL2 belongs to the BCL2 family of proteins, with anti-apoptotic properties (8). It is involved in the intrinsic apoptotic pathway, where it blocks activation of BCL2-antagonist/killer and BAX proteins in/during the absence of pro-apoptotic signals (61). The negative association between BCL2 expression and ccRCC grade reported in the present study is consistent with the results of a study by Itoi et al (62). They assessed 101 RCC (including 76 ccRCC cases) specimens via IHC and western blotting and observed higher expression levels of BCL2 in the early TNM stages (pT1-2) and ISUP grades 1+2 than in advanced RCCs (62). Furthermore, patients with BCL-2-positive RCC also exhibit a longer survival period than patients with BCL-2-negative RCC, and thus, the expression/presence of BCL-2 could be a favorable prognostic factor in RCC (62). The same conclusion may apply to the expression levels of the CCND1 gene, since the present results were consistent with the results of studies by Yang et al (63) and Wang et al (64) which analyzed the mRNA levels of CCND1 in a large number of ccRCC samples from Gene Expression Omnibus datasets and revealed a negative association between CCND1 gene expression and tumor grade.
The MYC proto-oncogene acts as a TF for a broad range of target genes encoding proteins involved in the progression of the cell cycle and cell proliferation, differentiation, metabolism and apoptosis (7,65). The present study confirmed the oncogenic role of MYC due to its upregulation in ccRCC, which was consistent with the results of a study by Tang et al (66). They also demonstrated increased MYC expression at the mRNA and protein levels in 44 ccRCC tumors, as well as in 786-O, 769-P, A498, ACHN and Caki-1 RCC cells (66). MYC mRNA expression was associated with the expression levels of BCL2, VEGFA and CCND1, indicating that those genes may be targets for MYC oncoprotein in ccRCC (66). These data suggest that MYC protein may serve an important role in ccRCC development.
The analysis of the survival period revealed a worse prognosis for patients with TNM stages 3+4 and ISUP grades 3+4, which was consistent with our previous studies on ccRCC (16,35,36). Although higher overall CCND1 expression was observed in ccRCC tissues, its decreased expression was associated with worse patient outcomes. Generally, involvement in the progression of the cell cycle makes CCND1 a potential proto-oncogene (67). An association between CCND1 upregulation and poor prognosis has been observed in head and neck cancer (68) and gastric cancer (69). However, studies investigating ccRCC (63,64) and breast cancer (70-72) have revealed contrary data. Yang et al (63) performed a comprehensive analysis of the online databases containing data on gene expression profiles in ccRCC. Similar to the present results, they revealed an association between a longer survival period and higher mRNA levels of CCND1 in cancer tissues. Furthermore, both univariate and multivariate Cox regression analyses demonstrated an association between CCND1 expression and poor OS (63), which was consistent with the present results of univariable analysis of CCND1 mRNA expression in ccRCC. A study by Wang et al (64) revealed higher immunoreactivity of cyclin D1 in ccRCC samples compared with normal kidney tissues, which was in line with the mRNA expression results of the present study. Furthermore, statistical analysis of the database of patients with ccRCC indicated that decreased mRNA levels of CCND1 were related to tumor recurrence (64), which is in agreement with the present observations. Therefore, the CCND1 expression profile may have a prognostic significance in ccRCC; however, more studies that also focus on cyclin D1 protein expression in ccRCC tissues are required.
The results concerning the association between increased VEGFA expression and worse outcomes were consistent with other studies (73,74) and our previous research (36). ccRCC development is closely related to mutations in the VHL tumor suppressor gene and accumulation of hypoxia-inducible factors (HIFs), which stimulate the activation of vascular endothelial growth factors (75). A high concentration of VEGFs induces neoangiogenesis, which promotes tumor growth and facilitates hematogenous metastases (76). Therefore, the compliance of the obtained results with the widely-accepted pattern of kidney cancer molecular development (77) suggests that VEGFA gene expression may be a promising prognostic factor in ccRCC (73,74,77). This observation was strengthened by the results of Cox multivariable analysis, where patients with advanced histological ISUP grade were at high risk of death, but only with coexisting increased VEGFA mRNA expression levels in the tumors. This observation is in accord with our previous report that patients with ccRCC with advanced ISUP grades and high levels of VEGFA (at the mRNA or protein level) are at higher risk of death or ccRCC recurrence (36).
The current observation concerning upregulation of the expression levels of the SHH pathway components and their target genes in ccRCC prompted the examination of the influence of the SHH pathway inhibitors on renal cancer cells in vitro. The 786-O cell line is derived from human renal cell adenocarcinoma and possesses features of ccRCC such as mutated VHL with altered HIF and VEGF signaling pathways (78). ACHN is a metastatic cell line with uncertain RCC histotype; however, with the molecular characterization of papillary RCC (78). The present study also used epithelial HK2 cells derived from kidney proximal tubules to determine the effects of inhibitors on normal kidney cells (78).
The present study examined inhibitors of the SHH pathway acting on three different targets. RU-SKI43 blocks HHAT, an enzyme which catalyzes posttranslational modifications of SHH, thus inhibiting secretion of N-SHH (21). Treatment of AsPC-1 and Panc-1 pancreatic cancer cells with RU-SKI43 was found to decrease their proliferation rate but also reduced the expression levels of GLI1 in AsPC-1 cells (79). These results are associated with the inhibition of HHAT activity, since C-2, a compound structurally related to RU-SKI43 with no HHAT blocking ability, has no effect on pancreatic cell lines (79). Furthermore, RU-SKI43 was found to decrease the proliferation of T47D breast cancer cells, which exhibit high expression levels of HHAT (80). These promising results regarding RU-SKI43 acting on pancreatic and breast cancer cell lines prompted the selection of this SHH ligand inhibitor for assessment in the present study, which, for the first time, assessed its effects on renal cancer cells. Cell cycle analysis revealed a dose-dependent decrease in the fraction of 786-O and ACHN cells in the G0/G1 phase after RU-SKI43 treatment. The mRNA levels of some SHH target genes were downregulated in ACHN cells; however, in 786-O cells, RU-SKI43 increased the expression levels of the MYC and VEGFA oncogenes. The latter finding may be explained by low activity of the HHAT enzyme in these cells, as this compound does not inhibit the proliferation of HHAT-negative pancreatic cell lines (79); however, further experiments should be performed to confirm this thesis.
Among numerous SHH inhibitors, cyclopamine, a compound blocking SMO protein activity, has been widely tested in different types of cancer cells and tissues (81-83). At present, three functional analogs of cyclopamine, vismodegib, sonidegib (used for the treatment of metastatic basal cell carcinoma) and glasdegib (treatment of acute myeloid leukemia), have been approved by the Food and Drug Administration (FDA) (84). The inhibition of cell division indicates the possible usefulness of cyclopamine as a promising anticancer agent in ccRCC. Furthermore, the present results revealed that cyclopamine blocked the expression of SHH target genes to a higher degree than the other analyzed SHH pathway inhibitors. However, wound healing analysis did not reveal any significant effect of cyclopamine on the migration of renal cancer cells. The analysis performed by Dormoy et al (85) demonstrated anti-proliferative and pro-apoptotic properties of cyclopamine in relation to 786-O cells. Furthermore, cyclopamine treatment decreased the mRNA levels of almost all SHH pathway genes, including GLI1, which is consistent with previous results (85). In vivo cyclopamine studies have demonstrated tumor regression in nude mice bearing human ccRCC tumors (85). The present results were also consistent with those obtained by D'Amato et al (86) who analyzed the effects of sonidegib on 786-O cells. This SHH inhibitor, when used alone, does not have a significant effect on cell viability and migration; however, it decreases the levels of GLI1 protein measured by western blotting (86). A decrease in 786-O cell survival and migration rates is only observed when sonidegib is used in combination with everolimus (inhibitor of mTOR signaling) or sunitinib (multi-targeted RTK inhibitor) and similar results have been obtained for sunitinib-resistant 786-O SuR cells (86). Therefore, it was hypothesized that blockade of the SHH pathway at the SMO protein level by binding with cyclopamine or its derivatives appears to have the best anticancer properties in ccRCC among other analyzed inhibitors, especially in combination with other drugs targeting other cellular processes and signaling pathways, such as everolimus or sunitinib.
GANT61 is a downstream SHH pathway inhibitor, with the ability to change the conformation of GLI1 and GLI2, thus preventing the expression of their target genes (24,25). The reports of the resistance to FDA-approved cancer therapies targeting SHH signaling (87), and the involvement of other pathways, such as PI3K/AKT, in the stimulation of GLI-mediated transcription (17) suggest that direct inhibition of GLIs may be more effective than targeting the upstream SHH pathway components. Although the promising effects of using GANT61 have been observed in experiments on ovarian (88) or glioblastoma cells (89), this compound has not yet been examined in clinical trials. The present analysis revealed that GANT61 arrested ACHN and 786-O kidney cancer cells in the G2/M phase and inhibited the migration of 786-O cells. However, GANT61 at a high dose (EC25) increased the number of HK2 cells in the sub-G1 phase of the cell cycle, which may suggest that this SHH pathway inhibitor stimulates apoptosis of normal kidney cells. This observation suggests that GANT61 does not act specifically on cancer cells, thus its clinical usefulness seems to be weak. Moreover, an unexpected increased expression of SHH target genes after GANT61 treatment, was observed. The latter finding suggests that the transcriptional activity of GLIs in kidney cancer cells can be stimulated by pathways other than SHH, as suggested by Zhou et al (17). They demonstrated that dual blockade of GLI TFs by GANT61 and perifosine (an AKT inhibitor) provided improved anti-proliferative and pro-apoptotic effects on 786-O and 769-P RCC cells compared with the separate treatment with these two drugs (17). Furthermore, in vivo treatment of cancer-bearing mice with both GANT61 and perifosine was found to be associated with higher renal cancer suppression than treatment with either of these agents alone, and this is reflected by the reduction in tumor volume and increased immunoreactivity of an apoptotic marker (cleaved-caspase 3) in cancer tissues (17). It has been demonstrated that activation of the MAPK/ERK signaling pathway stimulates the transcriptional activity of GLIs in gastric cancer (6). Therefore, due to the possible involvement of other signaling pathways in the regulation of transcriptional activity of GLIs in kidney cancer, the antitumor properties of GANT61 appear to be weak.
To compare the effects of SHH inhibitors with the drug widely used for RCC therapy we included in the study sunitinib-the first-line chemotherapeutic applied for RCC treatment. The present analysis revealed a stronger effect of sunitinib on SHH-activated genes in kidney cancer cell lines than that of GANT61. This novel finding of the increased expression levels of GLI1 and VEGFA in RCC cell lines after sunitinib treatment may correspond to the widely observed acquisition of sunitinib-resistance in advanced ccRCC cases (90). It was hypothesized that upregulation of GLI1 and VEGFA expression may lead to enhanced viability and aggressiveness of tumor cells even if cell motility after treatment of cells with sunitinib was widely reduced. It was also suggested that, in RCC tumors, some cancer cells secrete high doses of VEGFA in response to sunitinib, enhancing the survival of cancer by increasing neoangiogenesis and enriching the tumor microenvironment. This could explain the results of our previous study, which documented a strong relationship between patient survival and occurrence of metastasis (high HRs), treatment with sunitinib and high expression of VEGFA (at the mRNA and protein level) in ccRCC tissues (36). However, this hypothesis requires further analysis in in vitro and in vivo models.
In conclusion, the present study indicated that, similarly to some other cancer types, the SHH pathway is active in ccRCC; however, the expression levels of its components are mainly upregulated in tumors at the early stages and grades of the disease. The SHH effectors GLI1-3 were strongly associated with the upregulation of MYC, BCL2 and VEGFA oncogenes in ccRCC tissues; however, the expression of those genes in RCC cell lines was not inhibited by SHH pathway-targeting drugs, with the exception of cyclopamine. The ambiguous results of the effects of GANT61 on SHH target expression may exclude this drug from further studies in ccRCC. Therefore, it was suggested that RU-SKI43 and cyclopamine and its derivatives should be further investigated, especially in early and intermediate stages of ccRCC in in vitro and in vivo models. Despite the apparent inhibition of RCC cell migration by sunitinib, the observed upregulation of GLI1, BCL2, MYC and VEGFA oncogenes is yet another challenge to overcome in the first-line therapy of advanced ccRCC with sunitinib (36). Another novelty of the present study was the observation of worse outcome in patients with ccRCC with low expression levels of CCND1. Although cyclin D has been acknowledged as an oncoprotein, the role of its decreased expression in ccRCC compared with other malignancies should be further investigated. Furthermore, the relationship between high VEGFA expression and low CCND1 expression and patient outcomes should be taken into consideration in the identification of potential prognostic factors in ccRCC.
Acknowledgments
Not applicable.
Funding Statement
This study was supported by the ST-12 and ST-02-0117/07 statutory funds of the Medical University of Gdańsk, as well as a grant from the Polish Ministry of Science and Higher Education (contract grant no. MB 664/280/63/73-3309).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
AKC and AR performed the molecular analyses and statistical tests and prepared the manuscript. JK collected the tissue samples and acquired the data. MM collected the tissue samples, acquired the data, and provided funding. ZK revised the manuscript in light of the analyzed data, give the final approval of the version to be published, and provided funding. PMW designed and supervised the study and revised the manuscript. All authors have read and approved the final manuscript for publication. AKC and PMW confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The present study received the approval of the Independent Bioethics Commission at the Medical University of Gdańsk (decision nos. NKEBN/4/2011 and NKBBN/370/2016; Gdansk, Poland), and written informed consent was obtained before surgery from each patient. All experimental procedures were performed according to the regulations and internal biosafety and bioethics guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A, Vakiti A, Rawla P, Barsouk A. Epidemiology of renal cell carcinoma. World J Oncol. 2020;11:79–87. doi: 10.14740/wjon1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Escudier B, Porta C, Schmidinger M, Rioux-Leclercq N, Bex A, Khoo V, Grünwald V, Gillessen S, clinicalguidelines@esmo.org Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30:706–720. doi: 10.1093/annonc/mdz056. [DOI] [PubMed] [Google Scholar]
- 5.Carballo GB, Honorato JR, de Lopes GPF, Spohr TCLSE. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018;16:11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kotulak-Chrząszcz A, Kmieć Z, Wierzbicki P. Sonic Hedgehog signaling pathway in gynecological and genitourinary cancer (Review) Int J Mol Med. 2021;47:106. doi: 10.3892/ijmm.2021.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Sig Transduct Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warren CFA, Wong-Brown MW, Bowden NA. BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis. 2019;10:177. doi: 10.1038/s41419-019-1407-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273:114–127. doi: 10.1111/joim.12019. [DOI] [PubMed] [Google Scholar]
- 10.Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med (Berl) 2016;94:1313–1326. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Jiang D, Zhu T, Wang Y, Wang H, Wang Q, Tan L, Zhu H, Yao J. Prognostic significance of c-MYC amplification in esophageal squamous cell carcinoma. Ann Thorac Surg. 2019;107:436–443. doi: 10.1016/j.athoracsur.2018.07.077. [DOI] [PubMed] [Google Scholar]
- 12.Huang S, Nong L, Wang W, Liang L, Zheng Y, Liu J, Li D, Li X, Zhang B, Li T. Prognostic impact of diffuse large B-cell lymphoma with extra copies of MYC, BCL2 and/or BCL6: Comparison with double/triple hit lymphoma and double expressor lymphoma. Diagn Pathol. 2019;14:81. doi: 10.1186/s13000-019-0856-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang SD, Mccrudden CM, Kwok HF. Prognostic significance of combining VEGFA, FLT1 and KDR mRNA expression in lung cancer. Oncol Lett. 2015;10:1893–1901. doi: 10.3892/ol.2015.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shan YS, Hsu HP, Lai MD, Hung YH, Wang CY, Yen MC, Chen YL. Cyclin D1 overexpression correlates with poor tumor differentiation and prognosis in gastric cancer. Oncol Lett. 2017;14:4517–4526. doi: 10.3892/ol.2017.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behnsawy HM, Shigemura K, Meligy FY, Yamamichi F, Yamashita M, Haung WC, Li X, Miyake H, Tanaka K, Kawabata M, et al. Possible role of sonic hedgehog and epithelial-mesenchymal transition in renal cell cancer progression. Korean J Urol. 2013;54:547. doi: 10.4111/kju.2013.54.8.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotulak-Chrzaszcz A, Klacz J, Matuszewski M, Kmiec Z, Wierzbicki P. Expression of the Sonic Hedgehog pathway components in clear cell renal cell carcinoma. Oncol Lett. 2019;18:5801–5810. doi: 10.3892/ol.2019.10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Zhu G, Huang J, Li L, Du Y, Gao Y, Wu D, Wang X, Hsieh JT, He D, Wu K. Non-canonical GLI1/2 activation by PI3K/AKT signaling in renal cell carcinoma: A novel potential therapeutic target. Cancer Lett. 2016;370:313–323. doi: 10.1016/j.canlet.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa J, Miyake H, Fujisawa M. GLI2 expression levels in radical nephrectomy specimens as a predictor of disease progression in patients with metastatic clear cell renal cell carcinoma following treatment with sunitinib. Mol Clin Oncol. 2016;5:186–192. doi: 10.3892/mco.2016.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Wu K, Gao D, Zhu G, Wu D, Wang X, Chen Y, Du Y, Song W, Ma Z, et al. Reciprocal regulation of hypoxia-inducible factor 2α and GLI1 expression associated with the radioresistance of renal cell carcinoma. Int J Radiat Oncol Biol Phys. 2014;90:942–951. doi: 10.1016/j.ijrobp.2014.06.065. [DOI] [PubMed] [Google Scholar]
- 20.Xin M, Ji X, De La Cruz LK, Thareja S, Wang B. Strategies to target the Hedgehog signaling pathway for cancer therapy. Med Res Rev. 2018;38:870–913. doi: 10.1002/med.21482. [DOI] [PubMed] [Google Scholar]
- 21.Lanyon-Hogg T, Masumoto N, Bodakh G, Konitsiotis AD, Thinon E, Rodgers UR, Owens RJ, Magee AI, Tate EW. Synthesis and characterisation of 5-acyl-6,7-dihydrothieno[3,2-c]pyridine inhibitors of Hedgehog acyltransferase. Data Brief. 2016;7:257–281. doi: 10.1016/j.dib.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei J, Yan N. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science. 2018;361:eaas8935. doi: 10.1126/science.aas8935. [DOI] [PubMed] [Google Scholar]
- 23.Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the Sonic Hedgehog signaling pathway: Review of smoothened and GLI inhibitors. Cancers (Basel) 2016;8:22. doi: 10.3390/cancers8020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonnissen A, Isebaert S, Haustermans K. Targeting the Hedgehog signaling pathway in cancer: Beyond smoothened. Oncotarget. 2015;6:13899–13913. doi: 10.18632/oncotarget.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eble JN, Sauter G, Epstein JI, Sesterhenn IA, editors. World Health Organization Classification of Tumours. IARC Press; Lyon: 2004. Pathology and genetics of tumours of the urinary system and male genital organs. https://www.patologi.com/WHO%20kidney%20testis.pdf . [Google Scholar]
- 27.The Cancer Genome, Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wierzbicki PM, Klacz J, Rybarczyk A, Slebioda T, Stanislawowski M, Wronska A, Kowalczyk A, Matuszewski M, Kmiec Z. Identification of a suitable qPCR reference gene in metastatic clear cell renal cell carcinoma. Tumor Biol. 2014;35:12473–12487. doi: 10.1007/s13277-014-2566-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 31.Kang CW, Han YE, Kim J, Oh JH, Cho YH, Lee EJ. 4-Hydroxybenzaldehyde accelerates acute wound healing through activation of focal adhesion signalling in keratinocytes. Sci Rep. 2017;7:14192. doi: 10.1038/s41598-017-14368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan YJ, Wei LL, Wu XJ, Huo FC, Mou J, Pei DS. MiR-106a-5p inhibits the cell migration and invasion of renal cell carcinoma through targeting PAK5. Cell Death Dis. 2017;8:e3155. doi: 10.1038/cddis.2017.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinto BI, Cruz ND, Lujan OR, Propper CR, Kellar RS. In vitro scratch assay to demonstrate effects of arsenic on skin cell migration. J Vis Exp. 2019;3791:58838. doi: 10.3791/58838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y. MiR-630 functions as a tumor oncogene in renal cell carcinoma. Arch Med Sci. 2016;12:473–478. doi: 10.5114/aoms.2016.59918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rybarczyk A, Klacz J, Wronska A, Matuszewski M, Kmiec Z, Wierzbicki PM. Overexpression of the YAP1 oncogene in clear cell renal cell carcinoma is associated with poor outcome. Oncol Rep. 2017;38:427–439. doi: 10.3892/or.2017.5642. [DOI] [PubMed] [Google Scholar]
- 36.Wierzbicki PM, Klacz J, Kotulak-Chrzaszcz A, Wronska A, Stanislawowski M, Rybarczyk A, Ludziejewska A, Kmiec Z, Matuszewski M. Prognostic significance of VHL, HIF1A, HIF2A, VEGFA and p53 expression in patients with clear-cell renal cell carcinoma treated with sunitinib as first-line treatment. Int J Oncol. 2019;55:371–390. doi: 10.3892/ijo.2019.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM. The 2016 WHO classification of tumours of the urinary system and male genital organs-part A: Renal, penile, and testicular tumours. Eur Urol. 2016;70:93–105. doi: 10.1016/j.eururo.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 38.Zheng L, Rui C, Zhang H, Chen J, Jia X, Xiao Y. Sonic hedgehog signaling in epithelial tissue development. Regen Med Res. 2019;7:3. doi: 10.1051/rmr/190004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandes-Silva H, Correia-Pinto J, Moura R. Canonical Sonic Hedgehog signaling in early lung development. J Dev Biol. 2017;5:3. doi: 10.3390/jdb5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Memi F, Zecevic N, Radonjić N. Multiple roles of Sonic Hedgehog in the developing human cortex are suggested by its widespread distribution. Brain Struct Funct. 2018;223:2361–2375. doi: 10.1007/s00429-018-1621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SM, Jang HJ, Lee JH. Roles of primary cilia in the developing brain. Front Cell Neurosci. 2019;13:218. doi: 10.3389/fncel.2019.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Chavez E, Scheerer C, Wizenmann A, Blaess S. The zinc finger transcription factor GLI3 is a regulator of precerebellar neuronal migration. Development. 2018;145:dev166033. doi: 10.1242/dev.166033. [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Rios J. The many lives of SHH in limb development and evolution. Semin Cell Dev Biol. 2016;49:116–124. doi: 10.1016/j.semcdb.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 44.Tickle C, Towers M. Sonic Hedgehog signaling in limb development. Front Cell Dev Biol. 2017;5:14. doi: 10.3389/fcell.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivell A, Petralia RS, Wang YX, Clawson E, Moehl K, Mattson MP, Yao PJ. Sonic hedgehog expression in the postnatal brain. Biol Open. 2019;8:bio.040592. doi: 10.1242/bio.040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Cassandras M, Peng T. The role of Hedgehog signaling in adult lung regeneration and maintenance. J Dev Biol. 2019;7:14. doi: 10.3390/jdb7030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fattahi S, Pilehchian Langroudi M, Akhavan-Niaki H. Hedgehog signaling pathway: Epigenetic regulation and role in disease and cancer development. J Cell Physiol. 2018;233:5726–5735. doi: 10.1002/jcp.26506. [DOI] [PubMed] [Google Scholar]
- 48.Le H, Kleinerman R, Lerman OZ, Brown D, Galiano R, Gurtner GC, Warren SM, Levine JP, Saadeh PB. Hedgehog signaling is essential for normal wound healing: Hedgehog signaling is essential for normal wound healing. Wound Repair Regen. 2008;16:768–773. doi: 10.1111/j.1524-475X.2008.00430.x. [DOI] [PubMed] [Google Scholar]
- 49.Takebe H, Shalehin N, Hosoya A, Shimo T, Irie K. Sonic Hedgehog regulates bone fracture healing. Int J Mol Sci. 2020;21:677. doi: 10.3390/ijms21020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saze Z, Terashima M, Kogure M, Ohsuka F, Suzuki H, Gotoh M. Activation of the Sonic Hedgehog pathway and its prognostic impact in patients with gastric cancer. Dig Surg. 2012;29:115–123. doi: 10.1159/000336949. [DOI] [PubMed] [Google Scholar]
- 51.Choe JY, Yun JY, Jeon YK, Kim SH, Choung HK, Oh S, Park M, Kim JE. Sonic hedgehog signalling proteins are frequently expressed in retinoblastoma and are associated with aggressive clinicopathological features. J Clin Pathol. 2015;68:6–11. doi: 10.1136/jclinpath-2014-202434. [DOI] [PubMed] [Google Scholar]
- 52.Li Q, Zhang Y, Zhan H, Yuan Z, Lu P, Zhan L, Xu W. The Hedgehog signalling pathway and its prognostic impact in human gliomas: Role of Hedgehog pathway in gliomas. ANZ J Surg. 2011;81:440–445. doi: 10.1111/j.1445-2197.2010.05585.x. [DOI] [PubMed] [Google Scholar]
- 53.Dierks C, Beigi R, Guo GR, Zirlik K, Stegert MR, Manley P, Trussell C, Schmitt-Graeff A, Landwerlin K, Veelken H, Warmuth M. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14:238–249. doi: 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 54.Al Ghamdi D, Gomaa W, Abulaban A, Al-Ahwal M, Buhmeida A, Al-Qahtani M, Al-Maghrabi J. The significance of sonic hedgehog immunohistochemical expression in colorectal carcinoma. J Microsc Ultrastruct. 2015;3:169–174. doi: 10.1016/j.jmau.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurebayashi J, Kanomata N, Koike Y, Ohta Y, Saitoh W, Kishino E. Comprehensive immunohistochemical analyses on expression levels of Hedgehog signaling molecules in breast cancers. Breast Cancer. 2018;25:759–767. doi: 10.1007/s12282-018-0884-2. [DOI] [PubMed] [Google Scholar]
- 56.Pestell RG. New roles of Cyclin D1. Am J Pathol. 2013;183:3–9. doi: 10.1016/j.ajpath.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skoda AM, Simovic D, Karin V, Kardum V, Vranic S, Serman L. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn J Basic Med Sci. 2018;18:8–20. doi: 10.17305/bjbms.2018.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delahunt B, Cheville JC, Martignoni G, Humphrey PA, Magi-Galluzzi C, McKenney J, Egevad L, Algaba F, Moch H, Grignon DJ, et al. The International society of urological pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am J Surg Pathol. 2013;37:1490–1504. doi: 10.1097/PAS.0b013e318299f0fb. [DOI] [PubMed] [Google Scholar]
- 59.Jäger W, Thomas C, Fazli L, Hurtado-Coll A, Li E, Janssen C, Gust KM, So AI, Hainz M, Schmidtmann I, et al. DHH is an independent prognosticator of oncologic outcome of clear cell renal cell carcinoma. J Urol. 2014;192:1842–1848. doi: 10.1016/j.juro.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 60.Niewiadomski P, Niedziółka SM, Markiewicz Ł, Uśpieński T, Baran B, Chojnowska K. Gli Proteins: Regulation in development and cancer. Cells. 2019;8:147. doi: 10.3390/cells8020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radha G, Raghavan SC. BCL2: A promising cancer therapeutic target. Biochim Biophys Acta Rev Cancer. 2017;1868:309–314. doi: 10.1016/j.bbcan.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Itoi T, Yamana K, Bilim V, Takahashi K, Tomita F. Impact of frequent Bcl-2 expression on better prognosis in renal cell carcinoma patients. Br J Cancer. 2004;90:200–205. doi: 10.1038/sj.bjc.6601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang JF, Shi SN, Xu WH, Qiu YH, Zheng JZ, Yu K, Song XY, Li F, Wang Y, Wang R, et al. Screening, identification and validation of CCND1 and PECAM1/CD31 for predicting prognosis in renal cell carcinoma patients. Aging (Albany NY) 2019;11:12057–12079. doi: 10.18632/aging.102540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang QS, Li F, Liao ZQ, Li K, Yang XL, Lin YY, Zhao YL, Weng SY, Xia Y, Ye Y, et al. Low level of Cyclin-D1 correlates with worse prognosis of clear cell renal cell carcinoma patients. Cancer Med. 2019;8:4100–4109. doi: 10.1002/cam4.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang XN, Su XX, Cheng SQ, Sun ZY, Huang ZS, Ou TM. MYC modulators in cancer: A patent review. Expert Opin Ther Pat. 2019;29:353–367. doi: 10.1080/13543776.2019.1612878. [DOI] [PubMed] [Google Scholar]
- 66.Tang SW, Chang WH, Su YC, Chen YC, Lai YH, Wu PT, Hsu CI, Lin WC, Lai MK, Lin JY. MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Lett. 2009;273:35–43. doi: 10.1016/j.canlet.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 67.Tchakarska G, Sola B. The double dealing of cyclin D1. Cell Cycle. 2020;19:163–178. doi: 10.1080/15384101.2019.1706903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moradi Binabaj M, Bahrami A, Khazaei M, Ryzhikov M, Ferns GA, Avan A, Mahdi Hassanian S. The prognostic value of cyclin D1 expression in the survival of cancer patients: A meta-analysis. Gene. 2020;728:144283. doi: 10.1016/j.gene.2019.144283. [DOI] [PubMed] [Google Scholar]
- 69.Kumari S, Puneet, Prasad SB, Yadav SS, Kumar M, Khanna A, Dixit VK, Nath G, Singh S, Narayan G. Cyclin D1 and cyclin E2 are differentially expressed in gastric cancer. Med Oncol. 2016;33:40. doi: 10.1007/s12032-016-0754-8. [DOI] [PubMed] [Google Scholar]
- 70.Lehn S, Tobin NP, Berglund P, Nilsson K, Sims AH, Jirström K, Härkönen P, Lamb R, Landberg G. Down-regulation of the oncogene cyclin D1 increases migratory capacity in breast cancer and is linked to unfavorable prognostic features. Am J Pathol. 2010;177:2886–2897. doi: 10.2353/ajpath.2010.100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mylona E, Tzelepis K, Theohari I, Giannopoulou I, Papadimitriou C, Nakopoulou L. Cyclin D1 in invasive breast carcinoma: Favourable prognostic significance in unselected patients and within subgroups with an aggressive phenotype. Histopathology. 2013;62:472–480. doi: 10.1111/his.12013. [DOI] [PubMed] [Google Scholar]
- 72.Ortiz AB, Garcia D, Vicente Y, Palka M, Bellas C, Martin P. Prognostic significance of cyclin D1 protein expression and gene amplification in invasive breast carcinoma. PLoS One. 2017;12:e0188068. doi: 10.1371/journal.pone.0188068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebru T, Fulya OP, Hakan A, Vuslat YC, Necdet S, Nuray C, Filiz O. Analysis of various potential prognostic markers and survival data in clear cell renal cell carcinoma. Int Braz J Urol. 2017;43:440–454. doi: 10.1590/s1677-5538.ibju.2015.0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X, Zhang J, Wang Y, Tu M, Wang Y, Shi G. Upregulated VEGFA and DLL4 act as potential prognostic genes for clear cell renal cell carcinoma. Onco Targets Ther. 2018;11:1697–1706. doi: 10.2147/OTT.S150565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Zhang Q. VHL and hypoxia signaling: Beyond HIF in cancer. Biomedicines. 2018;6:35. doi: 10.3390/biomedicines6010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aziz SA, Sznol J, Adeniran A, Colberg JW, Camp RL, Kluger HM. Vascularity of primary and metastatic renal cell carcinoma specimens. J Transl Med. 2013;11:15. doi: 10.1186/1479-5876-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turner KJ, Moore JW, Jones A, Taylor CF, Cuthbert-Heavens D, Han C, Leek RD, Gatter KC, Maxwell PH, Ratcliffe PJ, et al. Expression of hypoxia-inducible factors in human renal cancer: Relationship to angiogenesis and to the von Hippel-Lindau gene mutation. Cancer Res. 2002;62:2957–2961. [PubMed] [Google Scholar]
- 78.Brodaczewska KK, Szczylik C, Fiedorowicz M, Porta C, Czarnecka AM. Choosing the right cell line for renal cell cancer research. Mol Cancer. 2016;15:83. doi: 10.1186/s12943-016-0565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petrova E, Matevossian A, Resh MD. Hedgehog acyltransferase as a target in pancreatic ductal adenocarcinoma. Oncogene. 2015;34:263–268. doi: 10.1038/onc.2013.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matevossian A, Resh MD. Hedgehog Acyltransferase as a target in estrogen receptor positive, HER2 amplified, and tamoxifen resistant breast cancer cells. Mol Cancer. 2015;14:72. doi: 10.1186/s12943-015-0345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu HY, Dong Z. Gli3 silencing enhances cyclopamine suppressive effects on ovarian cancer. Onco Targets Ther. 2014;7:2007–2011. doi: 10.2147/OTT.S57346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mukherjee S, Frolova N, Sadlonova A, Novak Z, Steg A, Page GP, Welch DR, Lobo-Ruppert SM, Ruppert JM, Johnson MR, Frost AR. Hedgehog signaling and response to cyclopamine differ in epithelial and stromal cells in benign breast and breast cancer. Cancer Biol Ther. 2006;5:674–683. doi: 10.4161/cbt.5.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Steg A, Amm HM, Novak Z, Frost AR, Johnson MR. Gli3 mediates cell survival and sensitivity to cyclopamine in pancreatic cancer. Cancer Biol Ther. 2010;10:893–902. doi: 10.4161/cbt.10.9.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peer E, Tesanovic S, Aberger F. Next-generation Hedgehog/GLI pathway inhibitors for cancer therapy. Cancers (Basel) 2019;11:538. doi: 10.3390/cancers11040538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dormoy V, Danilin S, Lindner V, Thomas L, Rothhut S, Coquard C, Helwig JJ, Jacqmin D, Lang H, Massfelder T. The sonic hedgehog signaling pathway is reactivated in human renal cell carcinoma and plays orchestral role in tumor growth. Mol Cancer. 2009;8:123. doi: 10.1186/1476-4598-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.D'Amato C, Rosa R, Marciano R, D'Amato V, Formisano L, Nappi L, Raimondo L, Di Mauro C, Servetto A, Fulciniti F, et al. Inhibition of Hedgehog signalling by NVP-LDE225 (Erismodegib) interferes with growth and invasion of human renal cell carcinoma cells. Br J Cancer. 2014;111:1168–1179. doi: 10.1038/bjc.2014.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carpenter RL, Ray H. Safety and tolerability of Sonic Hedgehog pathway inhibitors in Cancer. Drug Saf. 2019;42:263–279. doi: 10.1007/s40264-018-0777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang Y, Chen H, Duan J, Wu W, Le F, Mou F. The inhibitory effect and safety of GANT61 on HeLa cells in nude mice. Exp Mol Pathol. 2020;113:104352. doi: 10.1016/j.yexmp.2019.104352. [DOI] [PubMed] [Google Scholar]
- 89.Carballo GB, Ribeiro JH, Lopes GPF, Ferrer VP, Dezonne RS, Pereira CM, Spohr TCLS. GANT-61 induces autophagy and apoptosis in glioblastoma cells despite their heterogeneity. Cell Mol Neurobiol. 2021;41:1227–1244. doi: 10.1007/s10571-020-00891-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown LC, Desai K, Zhang T, Ornstein MC. The immunotherapy landscape in renal cell carcinoma. BioDrugs. 2020;34:733–748. doi: 10.1007/s40259-020-00449-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.