Abstract
Shotgun cloning experiments with restriction enzyme-digested genomic DNA from Morganella morganii 1, which expresses high levels of cephalosporinase, into the pBKCMV cloning vector gave a recombinant plasmid, pPON-1, which encoded four entire genes: ampC, ampR, an hybF family gene, and orf-1 of unknown function. The deduced AmpC β-lactamase of pI 7.6 shared structural and functional homologies with AmpC from Citrobacter freundii, Escherichia coli, Yersinia enterocolitica, Enterobacter cloacae, and Serratia marcescens. The overlapping promoter organization of ampC and ampR, although much shorter in M. morganii than in the other enterobacterial species, suggested similar AmpR regulatory properties. The MICs of β-lactams for E. coli MC4100 (ampC mutant) harboring recombinant plasmid pACYC184 containing either ampC and ampR (pAC-1) or ampC (pAC-2) and induction experiments showed that the ampC gene of M. morganii 1 was repressed in the presence of ampR and was activated when a β-lactam inducer was added. Moreover, transformation of M. morganii 1 or of E. coli JRG582 (ΔampDE) harboring ampC and ampR with a recombinant plasmid containing ampD from E. cloacae resulted in a decrease in the β-lactam MICs and an inducible phenotype for M. morganii 1, thus underlining the role of an AmpD-like protein in the regulation of the M. morganii cephalosporinase. Fifteen other M. morganii clinical isolates with phenotypes of either low-level inducible cephalosporinase expression or high-level constitutive cephalosporinase expression harbored the same ampC-ampR organization, with the hybF and orf-1 genes surrounding them; the organization of these genes thus differed from those of ampC-ampR genes in C. freundii and E. cloacae, which are located downstream from the fumarate operon. Finally, an identical AmpC β-lactamase (DHA-1) was recently identified as being plasmid encoded in Salmonella enteritidis, and this is confirmatory evidence of a chromosomal origin of the plasmid-mediated cephalosporinases.
Species of the family Enterobacteriaceae including Citrobacter freundii, Enterobacter cloacae, Morganella morganii, Serratia marcescens, and Yersinia enterocolitica are naturally resistant to aminopenicillins and the early cephalosporins. This resistance phenotype is mediated by chromosomally encoded β-lactamases (AmpC) belonging to class C enzymes, also commonly named cephalosporinases (2, 5, 6). The inducible biosynthesis of these cephalosporinases has been reported phenotypically for these bacterial species. In C. freundii and E. cloacae, an ampR gene is located upstream of ampC and is divergently transcribed compared to ampC. Its deduced protein, a transcriptional regulator of the LysR family, acts as a repressor in the basal level of AmpC biosynthesis and favors its biosynthesis upon induction by several β-lactams (5). In these enterobacterial species, mutations in the promoter or the structural gene of ampD result in constitutive overproduction of cephalosporinase and explain the acquired resistance to expanded-spectrum cephalosporins (5, 11, 12, 17).
The mechanism of cephalosporinase expression has been studied in detail for E. cloacae and was found to be related to peptidoglycan components (13). Briefly, during normal growth in the absence of β-lactam as an inducer, the AmpR regulator is maintained in an inactive form by a peptidoglycan precursor, uridine pyrophosphoryl-N-acetyl muramyl-l-alanyl-d-glutamyl-meso-diaminopimelic acid-d-alanyl-d-alanine (UDP-MurNac-pentapeptide). This negative effect occurs by direct binding of UDP-MurNac-pentapeptide to the regulator. In this inactive form, AmpR binds to its operator site between the ampC and ampR structural genes, leading to repression of ampC expression. This inactivation of AmpR can be relieved by both knockout mutations in the ampD gene or the presence of β-lactams. Inactivation of ampD which encodes a cytosolic amidase specific for the recycling of muropeptides results in an increase in the concentration of its substrate, the 1,6-anhydro-N-acetyl-muramyl-l-alanyl-d-glutamyl-meso-diaminopimelic acid (anh-MurNac-tripeptide). The higher concentration of this muropeptide inside the cell is sufficient to displace the UDP-MurNac-pentapeptide from its AmpR binding site, thereby reactivating AmpR. In ampD+ cells, addition of β-lactams results in increased biosynthesis of cell wall degradation fragments and in a higher intracellular level of anh-MurNac-tripeptide by titrating the available AmpD activity. More recently, other muropeptides have been recognized as a signal for β-lactamase induction, such as anh-MurNac-pentapeptide (31).
As for other cephalosporinase-producing enterobacterial species, M. morganii strains may be grouped into two β-lactamase expression phenotypes, i.e., oxyimino-cephalosporin sensitive, with low-level and inducible cephalosporinase production on the one hand, and oxyimino-cephalosporin resistant, with high-level and constitutive cephalosporinase production on the other hand (32). While this work was in progress, the sequence of the cephalosporinase gene from an M. morganii isolate, strain SLM01, was reported along with the sequence of part of an ampR gene, but no evidence of a linkage between the presence of ampR and cephalosporinase regulation was reported (3). The purpose of our work was to identify ampR and ampC genes from an M. morganii isolate and correlate their presence with the regulatory properties of AmpR. The putative role of an AmpD-like protein was also investigated. A comparison with the plasmid-mediated ampC-ampR genes recently found in Salmonella enteritidis was also performed (4). Moreover, a comparison of ampR, ampC, and the surrounding sequences in M. morganii with those of other ampC-possessing enterobacterial species was undertaken.
MATERIALS AND METHODS
Bacterial strains and plasmid.
M. morganii 1 was isolated at the Hôpital Antoine Béclère (Clamart, France) in 1997 from a clinical specimen (dermatous ulcer) and was identified with the API 20E system (bioMérieux, Marcy l’Etoile). The other unrelated M. morganii clinical isolates (strains 2 to 16) were identified from patient specimens in 1997 at the Hôpital de Bicêtre (Le Kremlin-Bicêtre, France). Escherichia coli DH10B (Life Technologies, Eragny, France) was used as the host strain for cloning experiments. β-Lactamase expression was studied by using E. coli MC4100 lacking an ampC gene or E. coli JRG582 from which its ampDE genes were deleted (12). The cloning vectors were either the pBKCMV phagemid (Stratagene, La Jolla, Calif.), which confers kanamycin resistance, or pACYC184, which confers chloramphenicol and tetracycline resistance (7). Plasmid pNH5 containing an HpaI fragment with the ampD gene from E. cloacae into pBGS18 conferred kanamycin resistance (12).
Antimicrobial agents and MIC determinations.
The agents and their sources were as follows: amoxicillin, clavulanic acid, and ticarcillin, SmithKline French-Beecham (Nanterre, France); aztreonam and cefepime, Bristol-Myers Squibb (Paris, France); ceftazidime, Glaxo (Paris, France); cephalothin, cefamandole, and moxalactam, Eli Lilly (Saint-Cloud, France); piperacillin and tazobactam, Lederle (Oullins, France); cefotaxime and cefpirome, Hoechst-Roussel (Paris, France); cefoxitin and imipenem, Merck Sharp & Dohme-Chibret (Paris, France); ceftriaxone (Roche, Neuilly, France); and kanamycin and chloramphenicol, Sigma (Saint-Quentin Falavier, France).
Antibiogram for the M. morganii clinical isolates and the recombinant E. coli strains were first done by a routine agar disk diffusion assay with Mueller-Hinton agar plates and antibiotic-containing disks (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France). The MICs were then determined by an agar dilution technique on Mueller-Hinton agar plates with a Steers multiple inoculator and an inoculum of 104 CFU per spot (26). All plates were incubated at 37°C for 18 h.
Genetic techniques.
Genomic DNAs of M. morganii 1 to 16 were extracted as described previously (24). Fragments of genomic DNA from M. morganii 1 partially digested with Sau3AI (Pharmacia Biotech, Orsay, France) were ligated into the BamHI site of the pBKCMV phagemid. Ligation was performed at a 1:1 vector-insert ratio at a final concentration of 200 ng of DNA in a ligation mixture containing 1 U of T4 DNA ligase (Boehringer, Meylan, France) at 4°C for 18 h. Recombinant plasmids were transformed by electroporation (gene pulser II; Bio-Rad, Ivry-sur-Seine, France) into electrocompetent E. coli DH10B cells (Bio-Rad). Antibiotic-resistant colonies were selected on Trypticase soy agar plates containing 50 μg of amoxicillin per ml and 30 μg of kanamycin per ml.
Recombinant plasmid DNA was obtained from 100-ml Trypticase soy broth cultures grown with amoxicillin (100 μg/ml) overnight at 37°C. The plasmid DNA was prepared with Qiagen columns (Qiagen, Courtaboeuf, France). Fragments sizes were estimated according to those on 1-kb DNA ladder (Pharmacia), which was used as a molecular size standard. A 5.9-kb cloned DNA fragment from pPON-1 (see Results section) was sequenced from both strands by using laboratory-designed successive primers and an Applied Biosystems sequencer (ABI 311). The nucleotide sequence and the deduced protein sequence were analyzed with Pedro’s biomolecular tools (29). Multiple sequence alignment of deduced peptide sequences was carried out with the GCG program Pileup, which uses a simplification of the progressive alignment method of Feng and Doolittle (9). The deduced AmpC and AmpR proteins were compared with the corresponding proteins of the enterobacterial species possessing known ampC and/or ampR genes: E. coli (14), C. freundii (16), E. cloacae (11), S. marcescens (25), and Y. enterocolitica (28).
DNA-DNA hybridizations were performed as described by Maniatis et al. (19). Genomic DNA from M. morganii 1 was digested with either EcoRI, EcoRV, or PstI. The products of these digestions were separated on a 0.8% agarose gel prior to a Southern transfer onto an N+ Hybond nylon membrane (Amersham) followed by UV light cross-linking. The membranes were incubated for 1 h at 42°C in a prehybridization solution made of 100 μg of salmon sperm DNA per ml, 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate, 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 30% formamide. The DNA probe used consisted of the 385-bp SacI-BssHII fragment from recombinant plasmid pPON-1 containing an internal part of the ampC gene. The probe was radiolabelled with [α-32P]dATP with a random-primer DNA labelling kit (Boehringer). Hybridization was revealed by autoradiography with Kodak films after exposure at −80°C for 18 h.
Comparison of the ampC-ampR-coding regions and of the surrounding sequences in the different expression phenotypes.
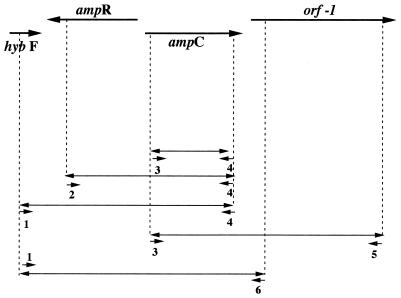
By using the DNA sequences obtained from the recombinant plasmid pPON-1 (Fig. 1), sets of primers were desigted to PCR amplify from M. morganii isolates of both inducible and noninducible phenotypes either the hybF, ampR, and ampC genes (primer 1 [5′-TGAGTGCGGCGGACATTATC-3′] and primer 4 [5′-GGCTTTGACTCTTTCGGTATTC-3′]), the ampR and ampC genes (primer 2 [5′-GTTTCCGTACGGGACTGTAAC-3′], and primer 4), the ampC gene alone (primer 3 [5′-TTCTGCCGCTGATAATGTCGC-3′] and primer 4), or the ampC gene and orf-1 (primer 3 and primer 5 [5′-ACCACCACAAAGCGCGAGTC-3′]) (Fig. 2). Recombinant plasmids pAC-1 and pAC-2 were obtained by cloning PCR-amplified fragments (primers 1 and 6 and primers 2 and 6, respectively [primer 6, 5′-CCATAAAACAGCCCATAAAGC-3′]) into the EcoRV site of pACYC184. pAC-1 contained the ampR and ampC genes, and pAC-2 contained only the ampC gene (Fig. 1 and 2). E. coli MC4100 was transformed with pAC-1 and pAC-2, E. coli JRG582 was transformed with pAC-1 with or without pNH5, and M. morganii 1 was transformed with pNH5 by the calcium chloride method as described elsewhere (19).
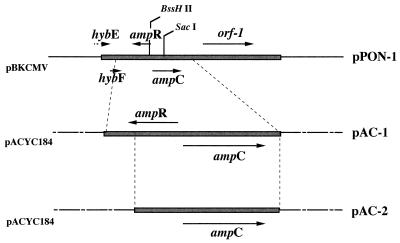
FIG. 1.
Restriction endonuclease maps of the inserts from recombinant plasmid pPON-1, which codes for the ampR and ampC genes from M. morganii 1, and of pAC-1 (ampR, ampC) and pAC-2 (ampC) pACYC184 derivatives. The thick lines represent the cloned inserts from M. morganii 1, the thin lines indicate vector pBKCMV, and the dotted lines represent vector pACYC184. The five ORFs found in the 5,914-bp sequenced are indicated, as are their translation directions.
FIG. 2.
Map of pPON-1 insert from M. morganii 1 and the primers used to PCR amplify the indicated genes from 15 M. morganii clinical isolates: primers 1 and 4 for hybF, ampR, and ampC; primers 2 and 4 for ampR and ampC; primers 3 and 4 for ampC; and primers 3 and 5 for ampC and orf-1. Cloning of ampR-ampC genes or of the ampC gene into pACYC184, giving recombinant plasmids pAC-1 or pAC-2, respectively, used similarly PCR-amplified fragments.
β-Lactamase assays.
Cultures of E. coli harboring the pPON-1 recombinant plasmid and M. morganii 1 were grown overnight at 37°C in 200 ml of Trypticase soy broth with amoxicillin at 100 μg/ml. After low-speed centrifugation, the bacterial pellet was resuspended prior to sonification (two times for 30 s each time at 20 Hz; phospholyser Vibra Cell 300; Bioblock, Illkirch, France) and subsequent high-speed centrifugation (twice, 30 min, 48,000 × g, 4°C). The residual nucleic acids in the supernatant were precipitated by treatment with 7% (vol/vol) spermin (0.2 M; Sigma) for 2 h at 4°C. This suspension was ultracentrifuged at 100,000 × g for 1 h at 4°C. The supernatants containing the crude enzyme extracts were subjected to isoelectric focusing.
The β-lactamase activity of E. coli DH10B cultures harboring pPON-1 was assayed by UV spectrophotometry (spectrophotometer Ultrospec 2000; Pharmacia Biotech, Orsay, France) at 30°C in 100 mM phosphate buffer (pH 7.0). The antibiotic wavelengths were chosen as described previously by Matagne et al. (20). Antibiotic solutions were freshly prepared in 100 mM phosphate buffer (pH 7.0). Kinetic parameters were derived from the initial velocity obtained with four to six substrate concentrations. Km values were determined according to the Eadie-Hofstee representation [V = f(Vi/S), where Vi is the initial velocity and S is the substrate concentration]. Vmax values were expressed relative to that of benzylpenicillin, which was set equal to 100. Enzyme inhibition was studied with cephalothin (100 μM) as the substrate. Clavulanic acid and tazobactam, at various concentrations, were preincubated with enzyme for 3 min at 30°C before addition of the substrate.
Analytical isoelectric focusing was performed with a mini IEF 111 (Bio-Rad) with a pH gradient of 3.5 to 9.5 ampholine polyacrylamide gel according to the manufacturer’s instructions. The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Oxoid, Paris, France) in distilled water. The pI values were determined and compared to those from molecular standards and to those of known β-lactamases (26).
β-Lactamase basal level determination and induction assays were performed as suggested previously (32) with M. morganii 1 to 16 and E. coli MC4100 harboring either recombinant plasmid pAC-1 or pAC-2 (see Results section). Briefly, overnight cultures were diluted 1:10 and were grown for 1 h and 30 min in a preincubated Trypticase soy broth on a rotating shaker at 37°C. Then, the cultures were grown for an additional 2 h in the presence of an inducer. Imipenem (0.5 μg/ml) was used as the inducer since this β-lactam was reported to be the most powerful cephalosporinase inducer for M. morganii (32), and cefoxitin (4 μg/ml) was used for the induction of E. coli MC4100 with recombinant plasmids. One unit of enzyme activity was defined as the activity which hydrolyzed 1 μmol of cephalothin per min. The total protein content was measured with bovine albumin as the standard (Bio-Rad DC Protein assay kit).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the Genbank/EMBL nucleotide databases under the accession no. AF055067.
RESULTS AND DISCUSSION
Cloning of the cephalosporinase gene and susceptibility testing.
Cloning from genomic DNA of M. morganii 1 gave several recombinant clones containing 5.9- to 12-kb inserts. One of them, which harbored recombinant plasmid pPON-1 with a 5.9-kb insert, was retained for further analysis (Fig. 1). MIC determinations showed that M. morganii 1 was resistant to amoxicillin, cephalothin, ceftazidime, and cefotaxime, of intermediate susceptibility to ticarcillin and cefoxitin, and fully susceptible to moxalactam and imipenem (Table 1). This antibiotic resistance pattern corresponded to that of an M. morganii strain producing high levels of cephalosporinase. As reported previously (32) and as opposed to other enterobacterial species which possess a cephalosporinase, such a strain was, surprisingly, of intermediate susceptibility to ticarcillin and cefoxitin, despite being resistant to expanded-spectrum cephalosporins. This strain remained susceptible to cefepime and cefpirome, likely due to the poor affinity of the M. morganii cephalosporinase for these β-lactams and/or better intracellular penetration (27).
TABLE 1.
MICs of β-lactams for clinical isolates M. morganii 1 and 5, E. coli DH10B harboring recombinant plasmid pPON-1, and the E. coli DH10B reference strain
| β-Lactam | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| M. morganii 1 | E. coli DH10B (pPON-1) | E. coli DH10B | M. morganii 5 | |
| Amoxicillin | >512 | >512 | 2 | 512 |
| Amoxicillin + Claa | 512 | >512 | 2 | 512 |
| Amoxicillin + Tazb | 32 | 32 | 2 | 32 |
| Ticarcillin | 8 | 64 | 2 | 0.12 |
| Ticarcillin + Cla | 8 | 64 | 1 | 0.25 |
| Ticarcillin + Taz | 2 | 32 | 1 | 0.25 |
| Piperacillin | 32 | 32 | 1 | 0.5 |
| Cephalothin | >512 | >512 | 4 | >512 |
| Cefamandole | 64 | 32 | 1 | >512 |
| Cefoxitin | 16 | 128 | 8 | 16 |
| Cefepime | 0.06 | 0.06 | 0.03 | <0.06 |
| Cefpirome | 0.5 | 0.06 | 0.03 | 1 |
| Ceftazidime | 4 | 16 | 0.12 | <0.06 |
| Ceftazidime + Clav | 4 | 32 | 0.12 | <0.06 |
| Ceftazidime + Taz | 0.06 | 2 | 0.12 | <0.06 |
| Cefotaxime | 4 | 4 | 0.12 | 0.12 |
| Ceftriaxone | 2 | 1 | 0.06 | <0.06 |
| Imipenem | 1 | 0.12 | 0.06 | 1 |
| Aztreonam | 1 | 1 | 0.12 | <0.06 |
| Moxalactam | 0.125 | 0.5 | 0.25 | <0.06 |
Cla, clavulanic acid at a fixed concentration of 2 μg/ml.
Taz, tazobactam at a fixed concentration of 4 μg/ml.
E. coli harboring pPON-1 showed a resistance profile similar to that of M. morganii 1 except for resistance to cefoxitin (Table 1). The addition of clavulanic acid did not modify the resistance pattern. However, as described for other M. morganii cephalosporinases (1), the addition of tazobactam decreased the MICs of ticarcillin and ceftazidime, which is an uncommon property for cephalosporinases (Table 1).
Identification of the cephalosporinase.
The entire 5.9-kb insert from recombinant plasmid pPON-1 was sequenced on both strands (Fig. 1). Analysis of this insert for coding regions revealed an open reading frame (ORF) of 1,137 bp encoding a 379-amino-acid protein. This ORF was preceeded by −35 and a −10 regions consistent with a putative enterobacterial promoter (Fig. 3). The overall GC content of this ORF was 51%, which is within the expected range of the GC contents of enterobacterial genes. Within the deduced amino acid sequence of the protein, a serine-valine-phenylalanine-lysine tetrad (S-V-F-K) at positions 67 to 70 was found (Fig. 3); it included the conserved serine and lysine amino acid residues characteristic of β-lactamases possessing a serine active site (15). Three structural elements characteristic of class C β-lactamases (cephalosporinases) were found (21): YAN at positions 150 to 152, DAES at positions 217 to 220, and KTG at positions 314 to 316 (Fig. 4). This enzyme shared 56, 55, 54, 53, and 39% homologies with AmpC from E. coli K-12, C. freundii OS60, E. cloacae MHN-1, Y. enterocolitica IP97, and S. marcescens SR50, respectively.
FIG. 3.
Nucleotide sequence of the 2,126-bp fragment of pPON-1 containing the ampC- and ampR-coding regions. The deduced amino acid sequences are designated in single-letter code. The putative promoter sequences are represented by −35 and −10 regions (boxed). The start and stop codons of these genes are underlined.
FIG. 4.
Comparison of amino acid sequences of AmpC from M. morganii SLM01 and DHA-1 from M. morganii 1. Dotted lines indicate identical amino acids. The boldface amino acids are characteristic either of serine β-lactamases (S-V-S-K) or of class C β-lactamases.
Kinetic parameters of the identified β-lactamase (called DHA-1) revealed that it had strong activity against benzylpenicillin and cephalothin but poorly hydrolyzed ceftazidime cefotaxime, and ticarcillin (Table 2). Its activity was strongly inhibited by tazobactam and was poorly inhibited by clavulanic acid. Their 50% inhibitory concentrations were 0.6 and >100 μM, respectively (28% inhibition for 300 μM clavulanic acid). These hydrolysis parameters correlated to those obtained with another M. morganii isolate described by Yang and Livermore (32). M. morganii 1 produced the same β-lactamase of pI 7.6 as the E. coli DH10B strain harboring recombinant plasmid pPON-1 (data not shown).
TABLE 2.
Kinetic parameters of various β-lactam antibiotics for AmpC β-lactamase (DHA-1) from M. morganii 1a
| Substrate | Vmaxb | Km (μM) | Vmax/Kmb |
|---|---|---|---|
| Ampicillin | 3.5 | 29 | 3.4 |
| Aztreonam | <0.1 | —c | — |
| Benzylpenicillin | 100 | 28 | 100 |
| Cefotaxime | <0.1 | — | — |
| Cefepime | <0.1 | — | — |
| Ceftazidime | 0.5 | 27 | 0.5 |
| Cephalothin | 246 | 53 | 120 |
| Cefoxitin | <0.1 | — | — |
| Imipenem | <0.1 | — | — |
| Piperacillin | 2.4 | 22 | 3 |
| Ticarcillin | <0.1 | — | — |
The β-lactamase extracts were from E. coli DH10B cultures harboring recombinant plasmid pPON-1.
Vmax and Vmax/Km are expressed relative to that of benzylpenicillin, which was set equal to 100.
In these cases, the hydrolysis parameters could not be determined (Vmax was too low or Km was too high).
While this work was in progress, the sequence of an ampC gene from an M. morganii isolate (isolate SLM01) was published (3). It shares 98.9% amino acid identity with DHA-1 (Fig. 4). The cephalosporinases of M. morganii SLM01 and DHA-1 have pI values of 7.4 and 7.6, respectively. Their sequences differ at four positions for SLM01 and DHA-1: position 48, isoleucine versus valine, respectively; position 84, alanine versus threonine, respectively; position 90, alanine versus glutamic acid, respectively; and position 133, asparagine versus histidine, respectively (Fig. 4). The first difference at position 48 is neutral. None of these amino acid changes are located within the putative catalytic site, as deduced from the closely related cephalosporinase from E. cloacae (18). These changes did not extend the hydrolysis profile of DHA-1 compared to that of a previously published cephalosporinase for another M. morganii isolate (3).
Hybridization experiments were then performed with genomic DNA from M. morganii 1 restricted with either EcoRI, EcoRV, or PstI. These restriction enzymes did not cut within the blaDHA-1 internal probe consisting of the 385-bp SacI-BssHII fragment from pPON-1 (Fig. 1). Single hybridization bands of various sizes (7, 3.2, and 9 kb, respectively) were obtained, thus indicating that blaDHA-1 is unique in M. morganii 1 chromosomal DNA, excluding any ampC duplication.
Genetic environment of the cephalosporinase gene.
Downstream from the ampC gene, another ORF (orf-1) of unknown function was found. An identical orf-1 was found downstream from ampC in M. morganii SLM01 (3), thus suggesting a conserved position of ampC in M. morganii species. Upstream from the ampC gene, three ORFs were also found in pPON-1 (Fig. 1). Two of them coded for HybF and HybE (partial sequence); the two proteins shared sequence homologies with hydrogenase subunits of E. coli, and HybF shared 66% homology with HybF from E. coli (23). Their functions in M. morganii have not yet been studied. Immediately upstream from the ampC gene, an ampR gene was found, and its deduced protein was 99% homologous to the partial sequence of AmpR reported from M. morganii SLM01. The ampR gene had an overlapping and divergently oriented promoter, as described for other ampC-ampR regulatory systems (Fig. 5) (5). Analysis of the intercistronic region revealed close similarity with those of the other ampC-ampR systems of C. freundii, E. cloacae, S. marcescens, and Y. enterocolitica (Fig. 5) (5, 11, 17, 28). Upstream from the −35 and −10 consensus sequences for the ampC promoter, a deletion has occurred in M. morganii species compared to those of the other enterobacterial species. The significance of this result remains unknown; it is, however, unlikely that it may significantly influence the effect of AmpR binding, but it may affect AmpC expression. The deduced AmpR protein of M. morganii 1 shared homology with AmpR proteins of other species: 81, 62, 60, and 48% for E. cloacae MHN-1, C. freundii OS60, Y. enterocolitica IP97, and S. marcescens S50, respectively (25). This homology was mainly within the N-terminal sequence, where a helix-turn-helix motif is required for binding to the intercistronic regions between ampR and ampC (Fig. 6). The homologies among the AmpR proteins were different from those found for AmpC proteins, although AmpR and AmpC from S. marcescens SR50 shared the least homology with both AmpR and AmpC from M. morganii 1.
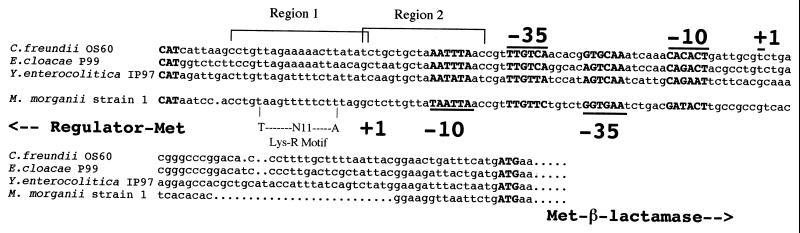
FIG. 5.
Alignment of the intercistronic region of ampC-ampR from M. morganii, C. freundii, Y. enterocolitica, S. marcescens, and E. cloacae. The start codons and the −35 and −10 regions of the promoters are shown below the DNA sequences for ampR and above for the DNA sequences for ampC. The +1 sign indicates the putative mRNA transcription start site. The sequences marked Region 1 and Region 2 correspond to those conserved among ampC-ampR intercistronic regions. Region 1 and region 2 are the two components of the putative AmpR binding site. Only region 1 contains a LysR binding motif (T-N11-A).
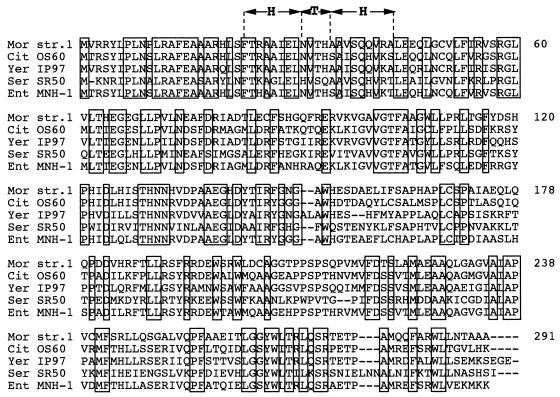
FIG. 6.
Multiple-sequence alignment of amino acid sequences of AmpR regulating cephalosporinase expression. The origins of AmpR are as follows: M. morganii 1 (Mor str. 1), C. freundii OS60 (Cit OS60), Y. enterocolitica IP97 (Yer IP97), S. marcescens SR50 (Ser SR50), and E. cloacae MHN-1 (Ent MHN-1). Identical amino acids are boxed. The predicted helix-turn-helix DNA-binding motif of the LysR family is shown (HTH).
Regulation of cephalosporinase expression.
Since pBKCMV is a multicopy vector (200 to 300 copies), expression of ampC was studied after its cloning into a plasmid, pACYC184, of lower copy number (20 to 30 copies). pAC-1 and pAC-2 were obtained by cloning the PCR-amplified ampC and ampR genes and the ampC gene, respectively. E. coli MC4100(pAC-1) had a cephalosporinase inducible phenotype in the presence of cefoxitin (6.7-fold increase), while E. coli MC4100(pAC-2), which lacked the ampR gene, showed an increase in its level of basal cephalosporinase expression (5.8-fold), together with a loss of inducibility (Table 3). Similarly, the MICs of the β-lactams were higher for E. coli MC4100(pAC-2) than for E. coli MC4100(pAC-1), and this was most noticeable for cefotaxime (Table 4). Similar results were obtained with the C. freundii and E. cloacae cephalosporinases, for which ampR deletion results in an increase in β-lactamase expression (2.4-fold) and a lack of induction (5, 17). Thus, AmpR seems to act in M. morganii like it does in other enterobacterial species: as a negative regulator of cephalosporinase expression in the absence of a β-lactam inducer and as an activator in the presence of an inducer.
TABLE 3.
β-Lactamase activity of M. morganii 1 with or without pNH5 (ampD of E. cloacae) and E. coli MC4100 harboring either recombinant plasmid pAC-1 (ampR and ampC genes) or pAC-2 (ampC gene)
| Strain | β-Lactamase activity (mU/mg of protein)a
|
|
|---|---|---|
| Basal level | Induced | |
| M. morganii 1b | 1,430 | 1,350 |
| M. morganii 1(pNH5)b | 8 | 400 |
| E. coli MC4100(pAC-1)c | 200 | 1,350 |
| E. coli MC4100(pAC-2)c | 1,160 | 1,100 |
One unit of β-lactamase is defined as 1 μmol of cephalothin hydrolyzed per min. The β-lactamase activities are geometric mean determinations for three independent cultures. The standard deviations were within 10%.
Imipenem (0.5 μg/ml) was used as the inducer.
Cefoxitin (4 μg/ml) was used as the inducer.
TABLE 4.
MICs of β-lactams for E. coli strains and M. morganii 1 with different genotypes
| Strain | Relevant genotype | MIC (μg/ml)
|
|||
|---|---|---|---|---|---|
| Amoxicillin | Piperacillin | Ceftazidime | Cefotaxime | ||
| E. coli | |||||
| MC4100 | ampD+ ampE+ ampC | 4 | 0.5 | 0.5 | <0.06 |
| MC4100(pAC-1) | ampD+ ampE+ ampC+ ampR+ | 128 | 8 | 2 | 0.5 |
| MC4100(pAC-2) | ampD+ ampE+ ampC+ ampR | 512 | 32 | 16 | 2 |
| JRG582 | Δ(ampDE) | 1 | <0.5 | 0.5 | <0.06 |
| JRG582(pAC-1) | Δ(ampDE) ampC+ ampR+ | 256 | 8 | 8 | 1 |
| JRG582(pAC-1)(pNH5) | Δ(ampDE) ampC+ ampR+ ampD+ | 128 | 8 | 4 | 0.25 |
| M. morganii | |||||
| 1 | >512 | 32 | 4 | 4 | |
| 1(pNH5) | ampD+ | 16 | 2 | 0.5 | 0.12 |
The level of cephalosporinase from M. morganii 1 indicated high-level and constitutive expression of this enzyme, as inferred by negative results of induction assays (Table 3). M. morganii 1 might possess mutations either in the promoter of an ampD-like gene or within the structural gene. To test this hypothesis, M. morganii 1 was transformed with plasmid pNH5 containing an ampD gene from E. cloacae. A decrease in the β-lactam MICs was obtained (Table 4). Similarly, the β-lactam MICs for E. coli JRG582 (ΔampDE) harboring the ampC and ampR genes from M. morganii 1 with or without the same ampD gene were lower in the presence of ampD (Table 4), and E. coli JRG582 had an inducible cephalosporinase expression phenotype. Additionally, M. morganii 1 transformed with pNH5 (ampD gene) recovered inducible cephalosporinase expression (50-fold increase) (Table 3). Moreover, these results indicated that an AmpD from E. cloacae may act in trans as a regulatory protein for expression of M. morganii AmpC in a similar manner as for the AmpC of E. cloacae.
Induction experiments and basal cephalosporinase levels divided the 15 other M. morganii isolates into two groups: those with low-level (although variable) and inducible expression of cephalosporinase and those with high-level and constitutive expression of cephalosporinase (Table 5). Detailed MICs of the β-lactams for M. morganii 5 (low-level and inducible cephalosporinase expression) confirmed these results (Table 1). Both groups correlated with strains with oxyiminocephalosporin-susceptible and -resistant phenotypes, respectively. The only exception was M. morganii 16, which had a relatively high level of cephalosporinase expression, but it remained inducible (Table 5). Therefore, it is likely that M. morganii strains with high-level and constitutive cephalosporinase expression, like M. morganii 1, corresponded to those possessing a nonfunctional AmpD-like protein.
TABLE 5.
β-Lactamase expression phenotypes and MICs at which 90% of strains are inhibited for selected β-lactams for 16 M. morganii clinical strains
| Phenotype | Isolate | β-Lactamase activity (mU/mg of protein)a
|
MIC90 (μg/ml)b
|
|||
|---|---|---|---|---|---|---|
| Basal | Inducedc | Ceftazidime | Cefotaxime | Piperacillin | ||
| High level, constitutive | 1 | 1,430 | 1,350 | 4 | 4 | 32 |
| 2 | 5,031 | 10,487 | 256 | 32 | 64 | |
| 3 | 8,761 | 6,113 | 8 | 4 | 16 | |
| 4 | 960 | 1,990 | 32 | 8 | 64 | |
| Low level, inducible | 5 | 3 | 41 | 0.06 | 0.12 | 0.25 |
| 6 | 3 | 98 | <0.06 | <0.06 | 0.25 | |
| 7 | 3 | 229 | 0.06 | <0.06 | 0.25 | |
| 8 | 3 | 242 | <0.06 | <0.06 | 0.25 | |
| 9 | 4 | 50 | <0.06 | <0.06 | 0.25 | |
| 10 | 4 | 93 | <0.06 | <0.06 | 0.25 | |
| 11 | 5 | 16 | 0.06 | 0.06 | 0.12 | |
| 12 | 5 | 3,644 | <0.06 | <0.06 | 0.50 | |
| 13 | 10 | 430 | <0.06 | <0.06 | 0.50 | |
| 14 | 35 | 5,532 | <0.06 | <0.06 | 0.50 | |
| 15 | 70 | 3,165 | <0.06 | <0.06 | 0.50 | |
| 16 | 365 | 2,774 | 1 | 1 | 8 | |
One unit of β-lactamase is defined as 1 μmol of cephalothin hydrolyzed per min.
MIC90, MIC at which 90% of isolates are inhibited.
Imipenem (0.5 μg/ml) was used as the inducer.
Comparison of the ampC- and ampR-coding regions and surrounding sequences in M. morganii isolates with different β-lactamase expression phenotypes.
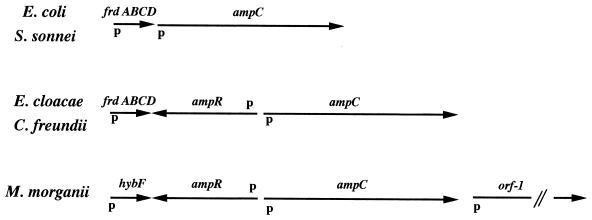
Using different pairs of primers designed from the pPON-1 sequence (Fig. 1), we amplified hybF-ampR, ampC-ampR, ampC, and ampC–orf-1 DNA fragments from the genomic DNAs of 15 M. morganii isolates, strains 2 to 16. In each case, the same fragments expected from the pPON-1 sequence were obtained, i.e., 2,418 bp for the hybF-ampR-ampC fragment, 2,043 bp for the ampR-ampC fragment, 1,047 bp for the ampC fragment, and 3,073 bp for the ampC-hybF fragment. In all M. morganii strains studied, ampR, orf-1, and hybF were found at the same locations at which they were found in M. morganii 1, whatever the β-lactamase expression phenotype was. Compared to other enterobacterial species in which an ampC gene is found (8). M. morganii species did not possess a fumarate operon upstream from the ampC gene (Fig. 7). This result may infer that M. morganii species are more distantly related to E. cloacae species and C. freundii species than the latter two species are to one another. Surprisingly, HybF shared 53% homology with an ORF, ORFB, of unknown function, with its gene being located just downstream from the fumarate operon in Proteus vulgaris, an ampC-negative enterobacterial species. It remained intriguing that HybF is a protein involved in dihydrogen uptake in E. coli, with its gene being a member of an operon which can be differentially induced to high levels when cells are grown in medium containing dihydrogen as an electron donor and fumarate as an electron acceptor (23).
FIG. 7.
Organization of the sequences surrounding ampC ampC in various enterobacterial species. The positions and directions of the fumarate operon (frdABCD), hybF, hybE, orf-1, ampC, and ampR genes are indicated with arrows. Also indicated are the locations of the putative promoters.
Finally, Barnaud et al. (4) reported on an inducible plasmid-mediated cephalosporinase gene along with a functional ampR gene from Salmonella enteritidis (4). Sequence analysis revealed that this plasmid-mediated cephalosporinase was 100% homologous to DHA-1 (the reason why the same name was retained), with their genes differing by only seven base-pair substitutions (data not shown). The AmpR sequences were 98% identical at the DNA level and 99% identical at the protein level. The investigators reported an unusual integron carrying aadA2, sulI along with blaDHA-1, and ampR (30). In this integron, no 59-bp element typical of an integron recombination site was located closed to blaDHA-1 or ampR (30). aadA2 and sulI were not chromosomally located around the ampR-ampC genes in our M. morganii strains. In contrast, the three chromosomally located ORFs found around ampR-ampC genes in M. morganii strains were not found to be located on an integron in S. enteritidis (30). Analysis of the DNA sequence located on each side of the ampC-ampR sequence in M. morganii strains and in the ampC-ampR-possessing S. enteritidis strain did not locate any hot-spot recombination site, therefore giving no explanation for the presence of ampC-ampR within this integron.
Taken together, the results indicating a plasmid location of blaDHA-1 in S. enteritidis and the results of our work indicate that almost identical ampR-ampC operons may be located in either the chromosome or the plasmid for different enterobacterial species. The encoded AmpC sequences were 100% identical, in contrast to other reported plasmid-mediated cephalosporinases, which share at most 96% homology with any chromosomal cephalosporinases (10). This is therefore confirmatory evidence of the chromosomal origin of the plasmid-located cephalosporinases which are now spread worldwide (22).
ACKNOWLEDGMENTS
We thank S. T. Cole and N. Honoré for the gift of E. coli MC4100, E. coli JRG582, and plasmid pNH5 and G. Arlet for sharing unpublished sequence data. We are grateful to S. Bellais for help in enzymatic determinations.
This work was funded by the Ministère de l’Education Nationale et de la Recherche (grant JE2227) and a grant-in-aid from Institut SmithKline-Beecham (La-Défense) of France.
REFERENCES
- 1.Akova M, Yang Y, Livermore D M. Interactions of tazobactam and clavulanate with inducibility- and constitutively-expressed class I β-lactamases. J Antimicrob Chemother. 1990;25:199–208. doi: 10.1093/jac/25.2.199. [DOI] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Barnaud G, Arlet G, Danglot C, Philippon A. Cloning and sequencing of the gene encoding the AmpC β-lactamase of Morganella morganii. FEMS Microbiol Lett. 1997;148:15–20. doi: 10.1111/j.1574-6968.1997.tb10260.x. [DOI] [PubMed] [Google Scholar]
- 4.Barnaud G, Arlet G, Verdet C, Gaillot O, Lagrange P H, Philippon A. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob Agents Chemother. 1998;42:2352–2358. doi: 10.1128/aac.42.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett P M, Chopra I. Molecular basis of beta-lactamase induction in bacteria. Antimicrob Agents Chemother. 1993;37:153–158. doi: 10.1128/aac.37.2.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole S T. Nucleotide sequence and comparative analysis of the frd operon encoding the fumarate reductase of Proteus vulgaris. Eur J Biochem. 1987;167:481–488. doi: 10.1111/j.1432-1033.1987.tb13362.x. [DOI] [PubMed] [Google Scholar]
- 9.Feng D F, Doolittle R F. Progressive alignment and phylogenetic tree construction of protein sequences. Meth Enzymol. 1990;183:375–387. doi: 10.1016/0076-6879(90)83025-5. [DOI] [PubMed] [Google Scholar]
- 10.Gazouli M, Tzouvelekis L S, Prinarakis E, Miriagou V, Tzelepi E. Transferable cefoxitin resistance in enterobacteria from Greek hospitals and characterization of a plasmid-mediated group 1 β-lactamase (LAT-2) Antimicrob Agents Chemother. 1996;40:1736–1740. doi: 10.1128/aac.40.7.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honoré N, Nicolas M H, Cole S T. Inducible cephalosporinase production in clinical isolates of Enterobacter cloacae is controlled by a regulatory gene that has been deleted from Escherichia coli. EMBO J. 1986;5:3709–3714. doi: 10.1002/j.1460-2075.1986.tb04704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honoré N, Nicolas M H, Cole S T. Regulation of enterobacterial cephalosporinase production: the role of a membrane-bound sensory transducer. Mol Microbiol. 1989;3:1121–1130. doi: 10.1111/j.1365-2958.1989.tb00262.x. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs C, Frère J M, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 14.Jaurin B, Grunström T. ampC cephalosporinase of Escherichia coli K-12 has a different evolutionary origin from that of β-lactamases of the penicillinase type. Proc Natl Acad Sci USA. 1981;78:4897–4901. doi: 10.1073/pnas.78.8.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joris B, Ghuysen J M, Dive G, Renard A, Dideberg O, Charlier P, Frère J M, Kelly J A, Boyington J C, Moews P C, Knox J R. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 dd-peptidase family. Biochem J. 1988;250:313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindberg F, Normark S. Sequence of the Citrobacter freundii OS60 chromosomal ampC beta-lactamase gene. Eur J Biochem. 1986;156:441–445. doi: 10.1111/j.1432-1033.1986.tb09601.x. [DOI] [PubMed] [Google Scholar]
- 17.Lindberg F, Westman L, Normark S. Regulatory components in Citrobacter freundii ampC β-lactamase induction. Proc Natl Acad Sci USA. 1987;82:4620–4624. doi: 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobkovsky E, Moews P C, Liu Zhao H, Frère J M, Knox J R. Evolution of an enzyme activity: crystallographic structure at 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc Natl Acad Sci USA. 1993;90:11257–11261. doi: 10.1073/pnas.90.23.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 20.Matagne A, Misselyn-Baudouin A M, Joris B, Erpicum B, Granier B, Frère J M. The diversity of the catalytic properties of class A β-lactamases. Biochem J. 1990;265:131–146. doi: 10.1042/bj2650131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumara N, Minami S, Mitsuhashi S. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob Agents Chemother. 1998;42:176–179. doi: 10.1128/aac.42.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medeiros A A. Evolution and dissemination of β-lactamases accelerated by generation of β-lactam antibiotics. Clin Infect Dis. 1997;24:S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 23.Messon K N, Chatelus C Y, Dervartanian M, Wendt J C, Shanmugen K T, Peck H D, Jr, Przybula A E. Cloning, sequencing and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase. J Bacteriol. 1994;176:4416–4423. doi: 10.1128/jb.176.14.4416-4423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naas T, Nordmann P. Analysis of a carbapenem-hydrolysing class A β-lactamase from Enterobacter cloacae and of its Lys-R type regulatory protein. Proc Natl Acad Sci USA. 1994;91:7693–7696. doi: 10.1073/pnas.91.16.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Normura K, Yoshida T. Nucleotide sequence of the Serratia marcescens SR50 chromosomal ampC beta-lactamase gene. FEMS Lett. 1990;58:295–299. doi: 10.1111/j.1574-6968.1990.tb13992.x. [DOI] [PubMed] [Google Scholar]
- 26.Philippon L N, Naas T, Bouthors A T, Barakett V, Nordmann P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philips D J, Carlton D D, Farrell C A, Kessler R E. Affinity of cephalosporinases for β-lactamases as a factor in antibacterial efficacy. Antimicrob Agents Chemother. 1986;29:845–848. doi: 10.1128/aac.29.5.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seoane A, Francia M V, Garcia L J. Nucleotide sequence of the ampC-ampR region from the chromosome of Yersinia enterocolitica. Antimicrob Agents Chemother. 1992;36:1049–1056. doi: 10.1128/aac.36.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swofford D L. PAUP (version 3.0): phylogenetic analysis using parsimony. Champaign: Illinois Natural History Survey; 1989. [Google Scholar]
- 30.Verdet C, Arlet G, Barnaud G, Lagrange P H, Philippon A. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Novel integron carrying ampC and ampR genes on a plasmid from Salmonella enteritidis, abstr. C-125; p. 68. [Google Scholar]
- 31.Wiedemann B, Dietz H, Pfeifle D. Induction of β-lactamase in Enterobacter cloacae. Clin Infect Dis. 1998;27:S42–S47. doi: 10.1086/514921. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, Livermore D M. Chromosomal β-lactamase expression and resistance of β-lactam antibiotics in Proteus vulgaris and Morganella morganii. Antimicrob Agents Chemother. 1988;32:1385–1391. doi: 10.1128/aac.32.9.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]