Abstract
Pexiganan, a 22-amino-acid antimicrobial peptide, is an analog of the magainin peptides isolated from the skin of the African clawed frog. Pexiganan exhibited in vitro broad-spectrum antibacterial activity when it was tested against 3,109 clinical isolates of gram-positive and gram-negative, anaerobic and aerobic bacteria. The pexiganan MIC at which 90% of isolates are inhibited (MIC90) was 32 μg/ml or less for Staphylococcus spp., Streptococcus spp., Enterococcus faecium, Corynebacterium spp., Pseudomonas spp., Acinetobacter spp., Stenotrophomonas spp., certain species of the family Enterobacteriaceae, Bacteroides spp., Peptostreptococcus spp., and Propionibacterium spp. Comparison of the MICs and minimum bactericidal concentrations (MBCs) of pexiganan for 143 isolates representing 32 species demonstrated that for 92% of the isolates tested, MBCs were the same or within 1 twofold difference of the MICs, consistent with a bactericidal mechanism of action. Killing curve analysis showed that pexiganan killed Pseudomonas aeruginosa rapidly, with 106 organisms/ml eliminated within 20 min of treatment with 16 μg of pexiganan per ml. No evidence of cross-resistance to a number of other antibiotic classes was observed, as determined by the equivalence of the MIC50s and the MIC90s of pexiganan for strains resistant to oxacillin, cefazolin, cefoxitin, imipenem, ofloxacin, ciprofloxacin, gentamicin, and clindamicin versus those for strains susceptible to these antimicrobial agents. Attempts to generate resistance in several bacterial species through repeated passage with subinhibitory concentrations of pexiganan were unsuccessful. In conclusion, pexiganan exhibits properties in vitro which make it an attractive candidate for development as a topical antimicrobial agent.
A recently emerged class of antibiotics with potential for use as human therapeutic agents is the antimicrobial peptides of animal origin (7). Over the past 15 years, more than 100 antimicrobial peptides, including magainins, cecropins, protegrins, and defensins, have been discovered in animals ranging from insects to humans (16, 20, 26). These peptides represent components of the system of host defense commonly called “innate immunity” and are used by animals to effectively deal with microbes in their environment (2, 9, 19, 21). Antimicrobial peptides selectively damage the membranes of bacteria through mechanisms which bacteria should theoretically find difficult to evade (2, 7, 16, 17). Among these animal-derived antibiotics are the magainins, discovered in the skin of the African clawed frog Xenopus laevis more than 12 years ago (8, 9, 12, 22, 24, 26). Through a series of amino acid substitutions and deletions, pexiganan (MSI-78) was constructed. Pexiganan exhibited an enhanced potency relative to that of magainin 2 against both gram-positive and gram-negative bacteria in several preliminary studies (3, 9). Pexiganan has been developed as a therapeutic antimicrobial agent for the topical treatment of infected diabetic foot ulcers (10).
In this paper, we report on the aspects of the in vitro activity of pexiganan relevant to its use as a topical anti-infective agent. A total of 3,109 clinical isolates including gram-positive and gram-negative microbes were screened for their susceptibilities to pexiganan. The results indicate that pexiganan is a broad-spectrum bactericidal peptide antibiotic.
MATERIALS AND METHODS
Organisms.
The microbial isolates tested were predominantly recent clinical isolates obtained from hospitals throughout the United States.
Agents.
The pexiganan (Gly-Ile-Gly-Lys-Phe-Leu-Lys-Lys-Ala-Lys-Lys-Phe-Gly-Lys-Ala-Phe-Val-Lys-Ile-Leu-Lys-Lys-NH2; molecular weight, 2478 [free peptide base]) used in the study was chemically synthesized by solid-phase procedures and was purified chromatographically by previously described procedures (26) at either Magainin Pharmaceuticals Inc. (MPI) or Bachem Bioscience (Torrance, Calif.). MSI-214, a peptide identical in sequence to pexiganan but comprising all d-amino acids, was synthesized and purified at MPI. Both pexiganan and MSI-214 were dissolved in deionized water before use. Imipenem was purchased from Merck Sharp & Dohme Research Laboratories (Rahway, N.J.), and ciprofloxacin was purchased from Miles, Inc. (West Haven, Conn.). Other antimicrobial agents were supplied by Sigma Chemical Co. (St. Louis, Mo.). Mueller-Hinton broth (MHB) and nutrient broth were supplied by Becton Dickinson Microbiology Systems (Cockeysville, Md.).
Antimicrobial testing.
The antimicrobial tests were carried out at three testing sites: MPI in Plymouth Meeting, Pa.; MRL Pharmaceutical Services Inc. in Franklin, Tenn. (the testing laboratory site is in Cypress, Calif.); and the Wadsworth Anaerobe Laboratory in Los Angeles, Calif. The method used to determine the MICs was the National Committee for Clinical Laboratory Standards (NCCLS) broth microdilution assay in microtiter plates (14, 15), with the exception that some isolates were initially tested (at the MPI site) in a total broth volume of 200 μl instead of 100 μl. In order to avoid any potential effect of cations in the test medium on the antimicrobial activity of pexiganan, the broth used for the testing of most aerobic bacteria with pexiganan was unsupplemented MHB. Anaerobe MIC broth (Wilkins-Chalgrin) was generally used in the MIC assays for anaerobic bacteria. If needed, 3% horse serum was added to the broth for MIC assays with anaerobic bacteria. The susceptibility and resistance breakpoints for the antibiotics other than pexiganan were based on NCCLS guidelines.
The minimum bactericidal concentrations (MBCs) were determined according to NCCLS guidelines (13). The killing curve assay was performed on the basis of a previously published standard protocol (11). The experiment was done in duplicate. Cells from the logarithmic phase of growth were collected and were incubated at 37°C with different concentrations of pexiganan in a total volume of 5 ml of cation-adjusted MHB (105 to 106 organisms/ml). At different time points, 0.1 ml of the culture was collected and was mixed with 25 ml of molten agar for the preparation of agar pour plates. Since the drug had been diluted at least 250-fold in the plates, the antibiotic carryover effect was minimal. In addition, to obtain appropriate numbers of CFU in an individual plate (fewer than 150 colonies/plate) to ensure accurate colony counting, 0.2 ml of the culture was taken from different time points, and a series of 10-fold dilutions (10−1 to 10−7) was prepared. Then, 0.1 ml of these diluted cells was used to prepare the plates as described above. The plates were incubated overnight at 37°C.
In vitro resistance study.
In brief, the in vitro passage study involved the incorporation of pexiganan into Mueller-Hinton agarose at a concentration of one-half the previously established MIC determined by a broth microdilution assay. Agarose was used to replace agar as the support matrix in the in vitro passage study since our preliminary experimental results indicated that alginic acid, a major anionic component in agar, binds to pexiganan and significantly inhibits its antimicrobial activity (data not shown). The organisms were cultured in duplicate on the agarose plates (both antibiotic-containing and control plates) for 7 to 14 sequential passages. For each organism, the MIC was determined prior to the initiation of the study, after the 4th passage, after the 7th passage, and in some cases, after the 11th and 14th passages. If the MIC after the fourth passage was greater than 1 twofold dilution higher than the original MIC, then for passes 5, 6, and 7, the amount of pexiganan in the agarose was increased to one-half of that new MIC. A similar approach was also used in the instances in which 14 passages were conducted.
RESULTS AND DISCUSSION
In vitro antibacterial activity.
A total of 3,108 clinical isolates were tested, including 2,692 aerobes and 416 anaerobes. The MICs at which 50 and 90% of isolates are inhibited (MIC50s and MIC90s) and the distributions of MICs for these organisms are summarized in Table 1. Pexiganan demonstrated potency against gram-positive and gram-negative aerobes and anaerobes. Among all the gram-positive aerobic microbes tested except Streptococcus sanguis and Enterococcus faecalis, 90% of staphylococci, streptococci, vancomycin-resistant Enterococcus faecium (VREF), Micrococcus spp., and Corynebacterium spp. were inhibited by pexiganan at a concentration of 16 μg/ml or less. Pexiganan exhibited potency against the majority of gram-negative aerobic bacteria tested, such as members of the family Enterobacteriaceae, Pseudomonas spp., Acinetobacter baumannii, and Stenotrophomonas maltophilia, and 90% of clinical isolates of these species were inhibited by pexiganan at 64 μg/ml or less. Three species (Alcaligenes faecalis, S. sanguis, and E. faecalis) were less sensitive to pexiganan (MIC90s, ≥256 μg/ml), but the MIC50s of pexiganan for these three species were 16 μg/ml (A. faecalis), 64 μg/ml (S. sanguis), and 128 μg/ml (E. faecalis). The anaerobes tested, including Clostridium spp., Peptostreptococcus spp., Bacteroides spp., Prevotella spp., Fusobacterium nucleatum, and Propionibacterium acnes, were generally more sensitive to pexiganan than aerobic bacteria, and 90% of these isolates were inhibited by pexiganan at a concentration of 64 μg/ml or less. Pexiganan was most active against Bacteroides spp. and P. acnes, with an MIC90 of ≤8 μg/ml.
TABLE 1.
Distribution of MICs of pexiganan for aerobic bacteria
| Microorganism | No. of isolates | No. of strains for which the MIC (μg/ml) is as follows:
|
MIC (μg/ml)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | >256 | 50% | 90% | ||
| Aerobes | |||||||||||||||
| Gram-positive aerobes | |||||||||||||||
| Staphylococcus aureus | 512 | 6 | 53 | 332 | 93 | 24 | 4 | 8 | 16 | ||||||
| Staphylococcus epidermidis | 137 | 1 | 54 | 45 | 28 | 8 | 1 | 4 | 8 | ||||||
| Staphylococcus haemolyticus | 50 | 10 | 24 | 12 | 4 | 4 | 8 | ||||||||
| Other coagulase-negative staphylococcia | 117 | 14 | 43 | 54 | 3 | 1 | 2 | 8 | 8 | ||||||
| Streptococcus agalactiae | 105 | 48 | 35 | 13 | 9 | 8 | 16 | ||||||||
| Streptococcus pyogenes | 253 | 118 | 95 | 28 | 8 | 1 | 1 | 2 | 4 | 8 | |||||
| Streptococcus sanguis | 30 | 1 | 1 | 4 | 2 | 1 | 6 | 4 | 3 | 8 | 64 | >256 | |||
| Other streptococcic | 83 | 1 | 16 | 32 | 26 | 5 | 2 | 1 | 8 | 16 | |||||
| Enterococcus faecalis | 101 | 1 | 1 | 6 | 13 | 37 | 32 | 11 | 128 | >256 | |||||
| Enterococcus faeciumb | 10 | 9 | 1 | 4 | 4 | ||||||||||
| Micrococcus spp. | 37 | 10 | 8 | 15 | 4 | 4 | 8 | ||||||||
| Corynebacterium spp.d | 49 | 2 | 6 | 31 | 6 | 3 | 1 | 4 | 8 | ||||||
| Gram-negative aerobes | |||||||||||||||
| Acinetobacter baumanii | 113 | 13 | 49 | 38 | 13 | 2 | 8 | ||||||||
| Alcaligenes faecalis | 24 | 2 | 1 | 5 | 5 | 2 | 2 | 4 | 1 | 2 | 16 | 256 | |||
| Citrobacter diversus | 78 | 7 | 53 | 16 | 1 | 1 | 8 | 16 | |||||||
| Citrobacter freundii | 105 | 4 | 72 | 29 | 8 | 16 | |||||||||
| Enterobacter aerogenes | 102 | 23 | 63 | 13 | 3 | 16 | 32 | ||||||||
| Enterobacter cloacae | 117 | 3 | 37 | 48 | 13 | 7 | 5 | 2 | 2 | 16 | 64 | ||||
| Escherichia coli | 137 | 2 | 6 | 13 | 37 | 54 | 20 | 5 | 16 | 32 | |||||
| Klebsiella oxytoca | 100 | 3 | 46 | 44 | 5 | 1 | 1 | 16 | 16 | ||||||
| Klebsiella pneumoniae | 123 | 4 | 47 | 55 | 15 | 1 | 1 | 8 | 16 | ||||||
| Pseudomonas aeruginosa | 150 | 1 | 20 | 45 | 58 | 18 | 6 | 2 | 16 | 32 | |||||
| Other Pseudomonas spp.e | 35 | 9 | 16 | 8 | 2 | 8 | 16 | ||||||||
| Stenotrophomonas maltophilia | 124 | 10 | 31 | 56 | 22 | 5 | 8 | 16 | |||||||
| Anaerobes | |||||||||||||||
| Gram-positive anaerobes | |||||||||||||||
| Clostridium innocuum | 26 | 1 | 5 | 11 | 6 | 1 | 1 | 1 | 8 | 16 | |||||
| Clostridium perfringens | 31 | 7 | 4 | 3 | 7 | 5 | 2 | 3 | 16 | 64 | |||||
| Clostridium ramosum | 24 | 1 | 11 | 9 | 3 | 4 | 16 | ||||||||
| Clostridium sporogenes | 20 | 6 | 9 | 5 | 8 | 16 | |||||||||
| Peptostreptococcus anaerobius | 23 | 1 | 5 | 2 | 7 | 4 | 2 | 1 | 1 | 8 | 32 | ||||
| Peptostreptococcus magnus | 29 | 2 | 1 | 3 | 8 | 9 | 3 | 1 | 1 | 1 | 2 | 8 | |||
| Peptostreptococcus prevotii | 25 | 2 | 8 | 7 | 6 | 2 | 2 | 4 | |||||||
| Propionibacterium acnes | 33 | 3 | 6 | 18 | 4 | 2 | 4 | 8 | |||||||
| Gram-negative anaerobes | |||||||||||||||
| Bacteroides distasonis | 30 | 1 | 2 | 8 | 17 | 2 | 4 | 4 | |||||||
| Bacteroides fragilis | 34 | 1 | 1 | 11 | 19 | 1 | 1 | 4 | 4 | ||||||
| Bacteroides ovatus | 20 | 17 | 2 | 1 | 4 | 8 | |||||||||
| Bacteroides thetaiotaomicron | 35 | 1 | 1 | 12 | 13 | 5 | 1 | 1 | 1 | 4 | 8 | ||||
| Fusobacterium nucleatum | 29 | 3 | 9 | 6 | 6 | 3 | 2 | 8 | 64 | ||||||
| Prevotella bivia | 35 | 1 | 1 | 17 | 3 | 4 | 7 | 2 | 4 | 32 | |||||
| Prevotella melaninogenica | 22 | 1 | 4 | 9 | 2 | 3 | 1 | 2 | 4 | 64 | |||||
| Total | 3,108 | 14 | 3 | 44 | 353 | 711 | 1,073 | 563 | 150 | 62 | 61 | 44 | 30 | ||
Includes 32 Staphylococcus hominis, 30 Staphylococcus simulans, 27 Staphylococcus warneri, and 27 Staphylococcus xylosus isolates and 1 Staphylococcus intermedius isolate.
All isolates were vancomycin resistant (MIC, >32 μg/ml).
Includes 38 Streptococcus bovis (group D), 11 Streptococcus group C, and 29 Streptococcus group G isolates, 1 Streptococcus salivarius isolate, 3 Streptococcus equisimilis isolates, and 1 Streptococcus mitis isolate.
Includes 24 Corynebacterium xerosis, 10 C. minutissimum, 7 C. striatum, 3 Corynebacterium group ANF, 2 Corynebacterium group B, and 2 Corynebacterium group G2 isolates and 1 Corynebacterium group I isolate.
Includes 17 Pseudomonas fluorescens, 10 Pseudomonas putida, 2 Pseudomonas stutzeri, 2 Pseudomonas pickettii, and 2 Pseudomonas species isolates and 1 isolate each of Pseudomonas paucimobilis and Pseudomonas vesicularis.
In order to establish reference MICs for pexiganan, we tested a number of strains from the American Type Culture Collection (ATCC) for their sensitivities to pexiganan. As indicated in Table 2, the MICs for the ATCC strains were generally consistent with those for the clinical isolates tested (Table 1 and 2). Of these ATCC strains, Haemophilus influenzae, Helicobacter pylori, Streptococcus pneumoniae, and Veillonella parvula, clinical strains of which had not been tested in this study, were also inhibited by pexiganan (MICs, ≤32 μg/ml). The MIC of pexiganan was >256 μg/ml for Proteus mirabilis and Serratia marcescens, as noted previously for magainins (26).
TABLE 2.
MICs of pexiganan and all-d-amino-acid pexiganan (MSI-214) for ATCC reference strains
| ATCC strain | MIC (μg/ml)
|
|
|---|---|---|
| Pexiganan | MSI-214 | |
| Staphylococcus aureus ATCC 29213 | 8–16 | 16 |
| Staphylococcus aureus ATCC 33591 (methicillin resistant) | 16–32 | NDa |
| Staphylococcus epidermidis ATCC 12228 | 2–4 | ND |
| Enterococcus faecium ATCC 51559 (vancomycin resistant) | 8 | 4 |
| Enterococcus faecalis ATCC 29212 | 64 | 16 |
| Group A Streptococcus ATCC 49399 | 4 | ND |
| Streptococcus pneumoniae ATCC 49619 | 32 | ND |
| Streptococcus bovis ATCC 49133 | 4 | 4 |
| Bacillus cereus ATCC 11778 | 8 | ND |
| Micrococcus luteus ATCC 4698 | 2 | 2–4 |
| Corynebacterium group A ATCC 49676 | 2–4 | ND |
| Escherichia coli ATCC 25922 | 8–16 | 8–16 |
| Enterobacter cloacae ATCC 35030 | 8 | ND |
| Proteus mirabilis ATCC 29245 | >256 | >256 |
| Serratia marcescens ATCC 8100 | >256 | >256 |
| Helicobacter pylori ATCC 43504 | 2 | ND |
| Haemophilus influenzae ATCC 49247 | 8 | ND |
| Pseudomonas aeruginosa ATCC 27853 | 8–16 | 16–32 |
| Bacteroides fragilis ATCC 90028 | 2–4 | ND |
| Bacteroides thetaiotaomicron ATCC 29741 | 2–4 | ND |
| Clostridium difficile ATCC 43255 | 4 | ND |
| Propionibacterium acnes ATCC 29399 | 8 | ND |
| Veillonella parvula ATCC 10790 | 8 | ND |
ND, the MIC was not determined.
The broad antibacterial spectrum of pexiganan is consistent with the antibacterial spectrum previously observed for antimicrobial peptides of animal origin (2, 7, 9) and confirms a recently published preliminary study of the activity of pexiganan (3). The antimicrobial spectrum of pexiganan is broader than those of the current commercially available peptide antibiotics, such as polymyxin B or polymyxin E. Although the antimicrobial activities of polymyxin and pexiganan against many species of gram-negative bacteria are comparable, polymyxin exhibits no significant activity against gram-positive bacteria, such as staphylococci (11).
Use of pexiganan composed of all d-amino acids (MSI-214) to elucidate mechanisms of resistance.
MSI-214 is an analog of pexiganan. Its amino acid sequence is identical to that of pexiganan, but it is composed of all d-amino acids and it is resistant to the actions of known natural peptidases (1, 23). To understand whether proteolytic degradation plays a role in the resistance of certain bacterial species to pexiganan, we compared the MIC of pexiganan with that of MSI-214 for nine species. Included were comparisons of both more sensitive organisms (i.e., VREF and Micrococcus luteus) and less sensitive organisms (i.e., P. mirabilis and S. marcescens). As shown in Table 2, the differences in MICs between the l and the d forms of pexiganan were less than 2 twofold dilutions for the same species of bacterium. MSI-214 showed a slightly enhanced activity only against E. faecalis. In addition, intact pexiganan peptide could be recovered from the culture medium following overnight growth of P. mirabilis and S. marcescens (data not shown). The data suggest that elaboration of proteolytic enzymes is not a primary mechanism for the generation of bacterial resistance to pexiganan. Analogous to other antimicrobial peptides of animal origin, the natural resistance to pexiganan in some species may be explained by a failure of the peptide to interact with either the inner or the outer membrane or by the action of a peptide-transporting efflux pump (4–6, 18).
Potency against antibiotic-resistant strains.
Along with pexiganan, several other classes of antibiotics were also tested against the isolates, including cephalosporins, carbapenems, fluoroquinolones, lincosamides, and aminoglycosides. These data were used to determine whether strains that are resistant to other classes of antibiotics and strains that are susceptible to other classes of antibiotics have differences in sensitivity to pexiganan. Among the isolates tested, a significant number of resistant bacteria were included: oxacillin-resistant staphylococci (175 isolates), ofloxacin-resistant staphylococci (70 isolates), cefazolin-resistant staphylococci (19 isolates), imipenem-resistant staphylococci (18 isolates), imipenem-resistant Pseudomonas spp. (24 isolates), ciprofloxacin-resistant Pseudomonas spp. (22 isolates), gentamicin-resistant Pseudomonas spp. (17 isolates), ciprofloxacin-resistant Acinetobacter baumannii (34 isolates), clindamycin-resistant Clostridium spp. (35 isolates), and cefoxitin-resistant Bacteroides spp. (11 isolates). There were no significant differences in the MIC50s and MIC90s of pexiganan for those strains resistant to oxacillin, cefazolin, cefoxitin, imipenem, ofloxacin, ciprofloxacin, gentamicin, and clindamicin and the MIC50s and MIC90s of pexiganan for those strains susceptible to these antimicrobial agents. With respect to the peptide antibiotics polymyxin B and polymyxin E, only more limited studies were conducted. For three clinical isolates of P. aeruginosa that were resistant to polymyxin B (MICs 32 to 128 μg/ml) and polymyxin E (MICs, 256 to >256 μg/ml), pexiganan MICs were identical to those for nonresistant strains of Pseudomonas aeruginosa. Non-cross-resistance between polymyxin B and a magainin peptide has been observed elsewhere (25), suggesting subtle differences in the mechanisms of action of different classes of peptide antimicrobial agents. Taken together, the results indicate that no evidence of cross-resistance between pexiganan and the other commonly used antimicrobial agents was observed.
Effect of culture conditions on antimicrobial activity of pexiganan.
Since the initial interaction between a cationic peptide and the membrane of the target cell is electrostatic in nature, the antibiotic activity of pexiganan as a function of the pH of the medium was explored. In an assay conducted in nutrient broth, 30 clinical isolates of P. aeruginosa (9 isolates), Staphylococcus aureus (11 isolates), and Staphylococcus epidermidis (10 isolates) were tested at pHs ranging from 5.0 to 8.0. No notable difference in potency was observed, although a modest increase in MICs, found at pH 5.5 with S. aureus, was noted (Table 3). These results indicate that the fundamental antimicrobial activity of pexiganan does not notably decrease with changes in pH.
TABLE 3.
Effect of pH on MIC of pexiganan when tested in nutrient broth
| Organism | No. of clinical isolates | Mean MIC (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|---|
| pH 5.0 | pH 5.5 | pH 6.0 | pH 6.5 | pH 7.0 | pH 7.5 | pH 8.0 | ||
| Pseudomonas aeruginosa | 9 | 1.9 | 3.4 | 5.2 | 5.1 | 4.9 | 6.4 | 6.8 |
| Staphylococcus aureus | 11 | 7.7 | 14.7 | 10.7 | 6.9 | 5.4 | 4.3 | 3.6 |
| Staphylococcus epidermidis | 10 | NDb | 3.0 | 3.5 | 3.5 | 3.7 | 3.0 | 3.5 |
The geometric mean MICs for all isolates tested.
ND, the MIC was not determined.
One possible concern with the use of a cationic antimicrobial peptide such as pexiganan is its possible inactivation by serum, either through the action of proteases or through interactions with proteins or lipids. To determine the effect of serum on the antimicrobial activity of pexiganan, the MIC of pexiganan was evaluated in the presence of sera from various animals (Table 4). Against the three species of organisms tested, human serum had a minimal effect on the activity of pexiganan, with the changes in the MIC being less than twofold. In contrast, mouse serum completely eliminated the activity of pexiganan, while sera from other animals inhibited the activity of pexiganan at various levels. In a separate experiment, we compared heat-inactivated mouse serum with fresh mouse serum. The heat-treated (55°C for 30 min) serum exhibited markedly reduced inhibitory activity, with the MICs falling from >256 μg/ml (unheated serum) to 16 μg/ml (heat-treated serum) for S. aureus and from 256 μg/ml (unheated serum) to 32 to 64 μg/ml (heat-treated serum) for E. coli. The original MICs in MHB for the species tested were 2 μg/ml (S. aureus) and 4 μg/ml (E. coli). A heat-sensitive component in mouse serum appears to be, at least in part, responsible for inactivation of the antimicrobial activity of pexiganan. These observations are relevant to in vivo studies with this and related peptides, since the infected mouse is a commonly used animal model of antibiotic efficacy.
TABLE 4.
Effect of sera from different animals on antibacterial activity of pexiganana
| Organism | MIC (μg/ml) in 50% serum from the following animal + 50% MHB:
|
MIC (μg/ml) in 100% MHB | |||||
|---|---|---|---|---|---|---|---|
| Mouse | Rat | Rabbit | Swine | Bovine | Human | ||
| Staphylococcus aureus ATCC 25923 | >256 | 16 | 16 | 8 | 128 | 8 | 4 |
| Escherichia coli ATCC 25922 | 128 | 256 | 16 | 32 | 128 | 16 | 8 |
| Pseudomonas aeruginosa ATCC 27853 | >256 | >256 | 128 | 32 | 32 | 32 | 16 |
The protocol used for evaluation of the inhibition of antimicrobial activity of pexiganan by serum was a variation of the NCCLS broth microdilution assay (15) with a standard (100 μl) volume containing 50 μl of serum and 50 μl of unsupplemented MHB. The results were compared to those for growth controls without serum. The serum samples used were not heat inactivated.
Bactericidal activity.
The difference between the MICs and the MBCs has been established as an index of the bactericidal activity of an antibiotic (11). A total of 143 isolates representing 32 aerobic species were tested to determine both the MICs and the MBCs of pexiganan. As indicated in Table 5, of 143 isolates tested, MBCs were the same or within 1 doubling dilution higher of the MICs for 132 (92%) isolates. For only two isolates of S. sanguis were MBCs increased more than 4 twofold dilutions. These findings are consistent with a bactericidal mechanism of action for pexiganan.
TABLE 5.
Comparison of the difference between MIC and MBC of pexiganan for aerobic bacteria
| Organism | No. of isolates | No. of isolates for which the MIC was different from the MBC by the following no. of doubling dilutionsa:
|
||||
|---|---|---|---|---|---|---|
| 0 | +1 | +2 | +3 | ≥+4 | ||
| Staphylococcus aureus | 5 | 3 | 1 | 1 | ||
| Coagulase-negative staphylococcib | 25 | 22 | 3 | |||
| Streptococcus agalactiae | 5 | 5 | ||||
| Streptococcus pyogenes | 5 | 3 | 1 | 1 | ||
| Other streptococcic | 23 | 14 | 5 | 1 | 1 | 2 |
| Enterococcus faecalis | 4 | 2 | 1 | 1 | ||
| Micrococcus spp. | 4 | 4 | ||||
| Corynebacterium spp.d | 9 | 9 | ||||
| Enterobacteriaceaee | 35 | 25 | 9 | 1 | ||
| Pseudomonas spp.f | 10 | 6 | 4 | |||
| Acinetobacter baumannii | 5 | 5 | ||||
| Stenotrophomonas maltophilia | 5 | 5 | ||||
| Alcaligenes faecalis | 8 | 3 | 3 | 2 | ||
| Total | 143 | 106 | 26 | 6 | 3 | 2 |
| Cumulative % | 74 | 92 | 97 | 99 | 100 | |
The difference between the MIC and the MBC was present as follows: 0, the MBC equals the MIC; +1, the MBC is 1 doubling dilution higher than the MIC; +2, the MBC is 2 doubling dilutions higher than the MIC; +3, the MBC is 3 doubling dilutions higher than the MIC; ≥+4, the MBC is 4 doubling dilutions or more than four doubling dilutions higher than the MIC.
Includes five Staphylococcus epidermidis, five Staphylococcus haemolyticus, three Staphylococcus hominis, four Staphylococcus simulans, four Staphylococcus warneri, and four Staphylococcus xylosus isolates.
Includes three Streptococcus bovis, five Streptococcus group C, three Streptococcus group G, seven Streptococcus intermedius, and five Streptococcus sanguis isolates.
Includes five Corynebacterium minutissimum and three Corynebacterium striatum isolates and one Corynebacterium xerosis isolate.
Includes five Citrobacter freundii, four Citrobacter koseri, five Enterobacter aerogenes, six Enterobacter cloacae, five Escherichia coli, five Klebsiella oxytoca, and five Klebsiella pneumoniae isolates.
Includes five Pseudomonas aeruginosa and five Pseudomonas fluorescens isolates.
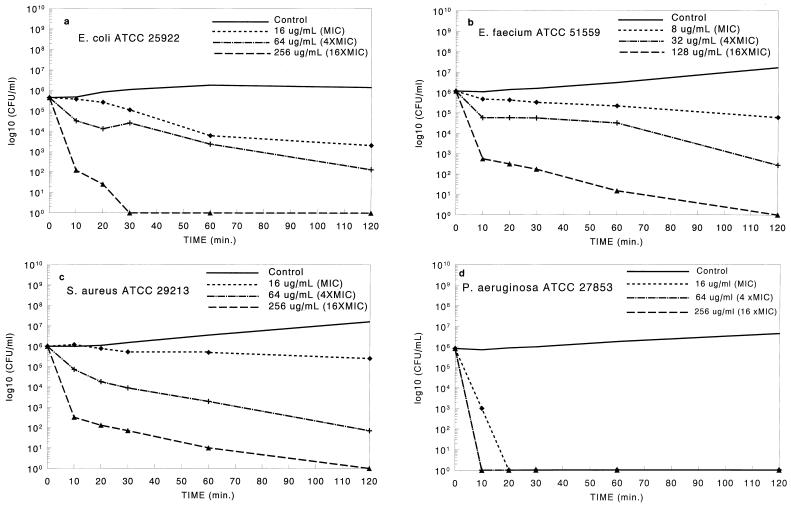
The bactericidal action of antibiotic peptides such as pexiganan is thought to result from irreversible membrane-disruptive damage (2, 7, 9). On the basis of the mechanism of action of magainin, pexiganan would be expected to exert its antimicrobial action rapidly, as reported previously for related molecules (26). The kinetics of bacterial killing were evaluated against four species (E. coli, S. aureus, P. aeruginosa, and VREF) at three concentrations of pexiganan (1×, 4×, and 16× the MIC). Cell viability within the first 2 h was measured. As shown in Fig. 1, for all four species, the rate of bacterial killing was dependent on the concentration of pexiganan, with greater than 105 organisms/ml being eradicated (reductions to <5 CFU/ml) within 2 h with the highest concentration studied. Bacterial cultures were monitored for up to 24 h, and no regrowth was observed; for all four species tested, no colony was found after 0.2 ml of the 24-h cultures was plated. The best activity was found against P. aeruginosa: more than 106 P. aeruginosa organisms/ml were eradicated within 20 min with 16 μg of pexiganan per ml (1× the MIC). The results indicate that at appropriate concentrations, pexiganan is a rapidly acting bactericidal agent. Bactericidal action occurs, as assessed with S. aureus, E. coli, P. aeruginosa, and VREF, and was clearly dependent on the concentration of pexiganan to which the organisms were exposed.
FIG. 1.
Killing curve study. The killing activity of pexiganan against four ATCC strains was monitored for the first 2 h. (a) Escherichia coli ATCC 25922; (b) Enterococcus faecium ATCC 51559 (vancomycin resistant); (c) Staphylococcus aureus ATCC 29213; (d) Pseudomonas aeruginosa ATCC 27853. For P. aeruginosa, the killing curves were identical (overlapping in the figure) for 4× the MIC and 16× the MIC. Under the assay conditions used in this study, the lowest detectable level was 5 CFU/ml.
In vitro development-of-resistance studies.
To explore the development of resistance to pexiganan in vitro, two selection procedures were conducted. In the first procedure, several organisms were passed repeatedly in the presence of concentrations of pexiganan insufficient to effect complete killing, a process which selects for rare resistant mutants that may exist or develop in a population. For a total of 27 clinical isolates representing eight bacterial species (Table 6), after seven sequential passages in the presence of subinhibitory concentrations of pexiganan, the change in average MICs for individual isolates and species was less than twofold. In addition, the in vitro development of resistance to pexiganan was compared to that to two other topical antibiotics, mupirocin and fusidic acid, for several species. After seven passages in vitro in the presence of subinhibitory concentrations of mupirocin, there were increases in the MICs for two S. aureus isolates (a 64-fold increase for the mupirocin-susceptible strain and an 8-fold increase for the mupirocin-resistant strain). In contrast, no increase in the MICs was observed for the same isolates treated with pexiganan. A similar pattern was also seen with fusidic acid. After 14 in vitro passages in the presence of subinhibitory concentrations of fusidic acid with two isolates (one S. aureus isolate and one S. epidermidis isolate), resistance to fusidic acid developed in both isolates: the MICs rose from 0.06 to 64 to 128 μg/ml (S. aureus) and from 0.06 to 128 μg/ml (S. epidermidis). In contrast, no change in the MIC was seen for isolates exposed to pexiganan. The results of these experiments, which were limited in terms of the scope of isolates and species tested, indicate that in vitro resistance acquired by the selection of mutations within the population of a given bacterial isolate may occur with mupirocin and fusidic acid but, for the same isolate, not with pexiganan. Consistent with this concept, antimicrobial peptides of animal origin have previously been suggested to possess a low potential for the induction of bacterial resistance (7).
TABLE 6.
Changes in MIC after sequential in vitro passages with a subinhibitory concentration
| Organism | No. of isolates | Mean MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|---|
| Pexiganan, initial | After four passesb
|
After seven passesb
|
||||
| Pexiganan | Control | Pexiganan | Control | |||
| Staphylococcus aureus | 3 | 5.8 | 5.8 | 4.6 | 8.3 | 6.6 |
| Coagulase-negative staphylococci | 3 | 1.4 | 2.3 | 2 | 2 | 2 |
| Enterobacter cloacae | 4 | 6.8 | 8.3 | 9.6 | 10.2 | 8.1 |
| Klebsiella pneumoniae | 4 | 13.6 | 9.6 | 9.2 | 14.3 | 12.6 |
| Pseudomonas aeruginosa | 5 | 8.2 | 11.3 | 9.8 | 19.7 | 10.4 |
| Acinetobacter baumannii | 4 | 7.7 | 12.1 | 12.4 | 10.7 | 8.8 |
| Stenotrophomonas maltophilia | 4 | 5.3 | 9.1 | 9.6 | 15.5 | 13.6 |
The geometric mean of MICs for all isolates tested.
The pexiganan-treated groups were passed on plates containing pexiganan at 50% of its MIC, while the controls were passed under the same conditions on plates without pexiganan.
In a second selection procedure, we intentionally introduced genomic mutations in vitro by exposure of S. aureus and P. aeruginosa to either a chemical mutagen or UV light. No pexiganan-resistant mutants were produced by either chemical or UV mutagenesis (data not shown).
Thus, the development of increased resistance of an isolate to pexiganan, through multiple-passage studies or mutagenesis, has not been observed, as noted both in this study and in the course of many years of experimentation (27). In addition, no evidence exists, either through our own observations or within the published literature, of the transfer of a resistance phenotype between an intrinsically resistant and susceptible isolate through either chromosomal or plasmid-mediated mechanisms.
In conclusion, pexiganan, an analog of the animal-derived antibiotic magainin, has been shown to exhibit broad-spectrum microbicidal activity and acts with a bactericidal mechanism against which the likelihood of the development of resistance may be low. In addition, no evidence of cross-resistance to a number of other antibiotic classes was observed, as determined by the equivalence of the MIC50s and MIC90s of pexiganan for those strains resistant to oxacillin, cefazolin, cefoxitin, imipenem, ofloxacin, ciprofloxacin, gentamicin, and clindamicin and the MIC50s and MIC90s of pexiganan for those strains susceptible to these antimicrobial agents. These in vitro properties have made pexiganan an attractive candidate for development as a topical antimicrobial agent (7, 9, 10).
ACKNOWLEDGMENTS
We thank D. York for technical assistance with the antimicrobial assays and R. Waldron for assistance in review of the manuscript.
REFERENCES
- 1.Bessalle R, Kapitkovshy A, Gorea A, Shalit I, Fridkin M. All d-magainin: chirality antimicrobial activity, and proteolytic resistance. FEBS Lett. 1990;274:151–155. doi: 10.1016/0014-5793(90)81351-n. [DOI] [PubMed] [Google Scholar]
- 2.Boman H G. Peptide antibiotics and their role in innate immunity. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs P C, Barry A L, Brown S D. In vitro antimicrobial activity of MSI-78, a magainin analog. Antimicrob Agents Chemother. 1998;42:1213–1216. doi: 10.1128/aac.42.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groisman E A. How bacteria resist killing by host-defense peptides. Trends Microbiol. 1994;2:444–449. doi: 10.1016/0966-842x(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 5.Groisman E A, Aspedon A. The genetic basis of microbial resistance to antimicrobial peptides. Methods Mol Biol. 1995;78:205–215. doi: 10.1385/0-89603-408-9:205. [DOI] [PubMed] [Google Scholar]
- 6.Gunn J S, Lim K B, Krueger J, Kim K, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 7.Hancock R E W. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 8.Iwahori A, Hirota Y, Sampe P, Miyano S, Takahashi N, Sasatsu M, Kondo I, Numao N. On the antibacterial activity of normal and reversed magainin 2 analogs against Helicobacter pylori. Biol Pharm Bull. 1997;20:805–808. doi: 10.1248/bpb.20.805. [DOI] [PubMed] [Google Scholar]
- 9.Jacob L, Zasloff M. Potential therapeutic application of magainins and other antimicrobial agents for animal origin. Ciba Found Symp. 1994;186:197–216. doi: 10.1002/9780470514658.ch12. [DOI] [PubMed] [Google Scholar]
- 10.Lipsky B A, Litka P A, Zasloff M, Nelson K the MSI-78-303 and 304 Study Group. Program and abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Microbial eradication and clinical resolution of infected diabetic foot ulcers treated with topical MSI-78 vs. oral ofloxacin, abstr. LM-57; p. 374. [Google Scholar]
- 11.Lorian V. Antibiotics in laboratory medicine. Baltimore, Md: The Williams & Wilkins Co.; 1996. [Google Scholar]
- 12.Ludtke S J, Heller W, Harroun T A, Yang L, Huang H W. Membrane pores induced by magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for determining bactericidal activity of antimicrobial agents. Proposed guideline (M26-P). Approved standard (M11-A3). Wayne, Pa: National Committee for Clinical Laboratory Standards; 1987. [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 3rd ed. Approved standard (M11-A3). Wayne, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard (M7-A4). Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 16.Nicolas P, Mor A. Peptides as weapons against microorganisms in the chemical defense system of vertebrates. Annu Rev Microbiol. 1995;49:277–304. doi: 10.1146/annurev.mi.49.100195.001425. [DOI] [PubMed] [Google Scholar]
- 17.Nissen-Meyer J, Nes I F. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol. 1997;167:67–77. [PubMed] [Google Scholar]
- 18.Peterson A A, Fesik S W, McGroarty E J. Decreased binding of antibiotics to lipopolysaccharides from polymyxin-resistant strains of Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1987;31:230–237. doi: 10.1128/aac.31.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schonwetter B S, Stolzenberg E D, Zasloff M A. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 20.Steinberg D A, Hurst M A, Fujii C A, Kung H C, Ho J F, Cheng F C, Loury D J, Fiddes J C. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob Agents Chemother. 1997;41:1738–1742. doi: 10.1128/aac.41.8.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stolzenberg E D, Anderson G M, Ackermann M R, Whitlock R H, Zasloff M. Epithelial antibiotic induced in states of disease. Proc Natl Acad Sci USA. 1997;94:8686–8690. doi: 10.1073/pnas.94.16.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tytler E M, Anantharamaiah G M, Walker D E, Mishra V K, Palgunachari M N, Segrest J P. Molecular basis for prokaryotic specificity of magainin-induced lysis. Biochemistry. 1995;34:4393–4401. doi: 10.1021/bi00013a031. [DOI] [PubMed] [Google Scholar]
- 23.Wade D, Boman A, Wahlin B, Drain C M, Andreu D, Boman H G, Merrifield R B. All-d amino acid-containing channel-forming antibiotic peptides. Proc Natl Acad Sci USA. 1990;87:4761–4765. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wenk M R, Seelig J. Magainin 2 amide interaction with lipid membranes: calorimetric detection of peptide binding and pore formation. Biochemistry. 1998;37:3909–3916. doi: 10.1021/bi972615n. [DOI] [PubMed] [Google Scholar]
- 25.Wright, S. Personal communication.
- 26.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zasloff, M., D. MacDonald, and Y. Ge. Unpublished data.