Abstract
Human leukocyte antigen (HLA)-G molecules are proposed to influence susceptibility to coronavirus disease 2019 (COVID-19). A case-control study was conducted on 209 patients with COVID-19 and198 controls to assess soluble HLA-G (sHLA-G) levels and HLA-G 14-bp insertion [Ins]/deletion [Del] polymorphism. Results revealed that median levels of sHLA-G were significantly higher in serum of COVID-19 patients than in controls (17.92 [interquartile range: 14.86–21.15] vs. 13.42 [9.95–17.38] ng/mL; probability <0.001). sHLA-G levels showed no significant differences between patients with moderate, severe or critical disease. Del allele was significantly associated with the risk of COVID-19 (odds ratio = 1.89; 95% confidence interval = 1.44–2.48; corrected probability = 0.001), while a higher risk was associated with Del/Del genotype (odds ratio = 2.39; 95% confidence interval = 1.25–4.58; corrected probability = 0.048). Allele and genotype frequencies of HLA-G 14-bp Ins/Del polymorphism stratified by gender or disease severity showed no significant differences in each stratum. Further, there was no significant impact of genotypes on sHLA-G levels. In conclusion, sHLA-G levels were up-regulated in COVID-19 patients regardless of disease severity. Further, it is suggested that HLA-G 14-bp Ins/Del polymorphism is associated with COVID-19 risk.
Keywords: COVID-19, SARS-CoV-2, HLA-G, 14-bp insertion/deletion polymorphism, 3′UTR
1. Introduction
For the second year, coronavirus disease 2019 (COVID-19) remains a challenge for scientists trying to understand the pathogenesis of the disease. COVID-19 is caused by the novel severe acute respiratory syndrome coronaviruse-2 (SARS-CoV-2) and is associated with high rates of morbidity and mortality [1]. Innate and adaptive immune responses are among the most important and indispensable host factors in controlling SARS-CoV-2 infection and the development of COVID-19 [2]. Studies revealed that COVID-19 patients are characterized by dysregulated immune responses manifested as hyper-inflammatory immunological humoral reactions with the hallmark of cytokine storm (cytokine release syndrome); a life-threatening systemic inflammatory syndrome [3], [4]. Besides, the cellular antiviral immune responses are impaired [5]. The molecular mechanisms behind dysregulated immune responses in COVID-19 patients are not well defined, and during viral infection, SARS-CoV-2 may also have developed various strategies to escape the host antiviral immune response and promote virus replication and disease progression [6].
During a viral infection, effective innate and adaptive immune cells are naturally developed. Natural killer (NK) cells are among the responsive innate immune cells that are involved in eliminating pathogens and virus-infected cells and suppressing viral spread. Activation of virus-specific CD8+ cytotoxic T cells is followed and outcomes in specific killing of virus-infected cells and activation of virus-specific CD4+ T cells. The activation of both NK cells and T cells is primarily controlled by human leukocyte antigens (HLA) [7]. HLA are highly polymorphic cell surface molecules organized into two classes; I and II, and are coded by a gene cluster in the short arm of human chromosome 6 [8]. HLA-class I molecules can be broadly divided into two subtypes; classical (HLA-A, B, and C) and non-classical (including HLA-E, F, G, and H) [9]. Classical HLA-class I are the most potent molecules in the immune response against viral infections. They are involved in presenting viral epitopes to T cells and inducing activation, differentiation and proliferation of these cells to become effective antiviral cells [10]. Whereas, non-classical HLA-class I molecules appear mostly to have immunosuppressive functions [11].
HLA-G is one of the non-classical HLA-class I molecules that was initially recognized to exhibit abundant and specific expression on extravillous trophoblasts under physiological conditions to protect the fetus from the maternal immune response [12]. Subsequently, it became clear that HLA-G molecules play a key role in enhancing and maintaining immune tolerance and have immunomodulatory properties [13]. Seven isoforms of HLA-G molecules have been identified, four of which are membrane-bound (HLA-G1, -G2, -G3 and -G4) and three are soluble (sHLA-G5, -G6 and -G7) [14]. Besides, other novel HLA-G isoforms have recently been described and included spliced isoforms with an extended 5′‐region and lacking transmembrane and α1 domains [15], [16]. Both membrane-bound and soluble HLA-G can exert several immunomodulatory effects by affecting CD4+ T, NK and CD8+ T cell functions, dendritic cell maturation, and B cell activation [17]. In addition, dysregulated expression of HLA-G molecules has been found in various pathological conditions including inflammatory, autoimmune and infectious diseases (including viral infections) [17], [18], [19], [20].
There is accumulating evidence that immunosuppressive mechanisms play a major role in promoting viral infection either by suppressing the ability of infected host cells to overcome viral infection or by preventing the elimination of virus-infected cells by immune cells. One common mechanism proposed for viruses to escape immune surveillance is the loss or down-regulation of classical HLA class Ia antigens and neoexpression of non-classical HLA class Ib antigens, such as HLA-E, -F and -G [21]. In COVID-19, it has been suggested that HLA-G may be considered as potent molecules that may suppress immune functions during SARS-CoV-2 infection and thus may enhance virus subversion and allow high rates of replication [22]. Another perspective also suggested that HLA-G molecules could cause profound immunosuppression, which favors SARS-CoV-2′s escape from immune attack [6]. In addition, significantly increased levels of sHLA-G have recently been demonstrated in serum of patients with severe COVID-19 [23].
The HLA-G locus exhibits limited polymorphism compared to classical HLA genes, and as of January 12, 2022, only 94 alleles have been identified at the molecular level (https://hla.alleles.org). HLA-G 14 base-pairs (14-bp) insertion (Ins)/deletion (Del) is another polymorphism of HLA-G gene located in the 3′ untranslated region (UTR) of exon eight and has been shown to affect mRNA stability and expression of HLA-G [24]. Besides, HLA-G 14 bp Ins/Del polymorphism has been associated with susceptibility to some viral infections; for instance, chronic hepatitis B, human cytomegalovirus and human immunodeficiency virus infections [25], [26], [27].
In a previous study by our group, up-regulated expression of sHLA-G was found in severe COVID-19 cases [23]. In the current study, our observation was extended to include patients with moderate and critical disease. In addition, the study sought to evaluate the association between HLA-G 14 bp Ins/Del polymorphism and COVID-19 in Iraqi patients. To the best of the researchers' knowledge, no study has been conducted in this context.
2. Materials and methods
2.1. Patients and controls
A case-control study was conducted on 209 patients with COVID-19 (mean age = 56.5 ± 14.1 years; age range = 18–85 years; 75.6% males) and 198 healthy controls (mean age = 42.3 ± 12.3 years; age range = 18–71 years; 76.3% males) to assess sHLA-G levels and HLA-G 14-bp Ins/Del polymorphism. The study protocol was approved by the Ethics Committee of the Iraqi Ministry of Health and Environment, and written consent was obtained from participants (Approval No. CSEC/0121/0014). Patients were admitted to three hospitals in Baghdad (Dar Al-Salam Field Hospital, Al-Karkh General Hospital and Al-Furat General Hospital) during September 2020–February 2021, and were included in the study 3–6 days after their admission (blood samples were collected from patients during these days). On the first day of admission, nasopharyngeal swabs were obtained from patients for the diagnosis of SARS-CoV-2 using the RealLine SARS-CoV-2 kit (Bioron Diagnostics GmbH), and the test result appeared within 24 h. In addition, a chest computerized tomography (CT) scan was performed to confirm diagnosis. Included patients were only those who showed a positive molecular test and were 18 years of age and older. Pregnant women were excluded. COVID-19 severity was determined according to the World Health Organization (WHO) Interim Guidance [28]. In light of this, 56 patients were in moderate condition (mean age = 52.4 ± 14.1 years; age range = 18–75 years; 80.4% males), 95 in severe condition (mean age = 56.3 ± 14.6 years; age range = 18–85 years; 76.8% males), and 58 in critical condition (mean age = 60.9 ± 12.0 years; age range = 32–84 years; 69.0% males). The control group included blood donors and health service personnel. They were healthy and had no respiratory diseases for the past 12 months. Their serum was negative for anti-SARS-CoV-2 IgM and IgG antibodies and C-reactive protein. Besides, their erythrocyte sedimentation rate (ESR) was below 20 mm/hour.
2.2. Immunoassay of sHLA-G
Serum level of sHLA-G was measured using a sandwich enzyme-linked immunosorbent assay (ELISA) kit designed for quantitative detection of total sHLA-G, and instructions of manufacturer were followed (Cat. No E0443Hu, Bioassay Technology Laboratory, China). The standard curve range of the kit was 0.3–90 ng/mL.
2.3. Detection of HLA-G 14-bp Ins/Del polymorphism
Genomic DNA was isolated using the ReliaPrep Blood gDNA Miniprep System kit (Promega, USA), following the supplied instructions of the manufacturer. A conventional polymerase chain reaction (PCR) assay was used to detect the 14-bp Ins/Del polymorphism using forward (5′-GTGATGGGCTGTTTAAAGTGTCACC-3′) and reverse (5′-GGAAGGAATGCAGTTCAGCATGA-3′) primers previously published [29]. The reaction mix consisted of 12.5 μL GoTaq Green Master Mix (Promega, USA), 5 μL DNA (50–60 ng/mL), 1 μL of each primer and 5.5 μL nuclease-free water (total volume = 25 μL). The thermal cycler (BioRad, USA) was programmed for the following optimized conditions: initial denaturation for 5 min at 94 °C, followed by 35 cycles of denaturation (30 s at 94 °C), annealing (30 s at 60 °C) and extension (30 s at 72 °C (30 s), and a final extension cycle for 5 min at 72 °C. Amplified PCR products were electrophoresed in a 3% agarose gel along with a 50 bp DNA ladder. A gel documentation system was used to visualize migrating bands, which were of two molecular sizes; 210 and 224 bp (corresponding to Del and Ins alleles, respectively).
2.4. Statistical analysis
Statistical analysis was performed using IBM SPSS Statistics 25.0 (Armonk, NY: IBM Corp.) and GraphPad Prism version 8.0.0 (San Diego, California USA). sHLA-G levels were expressed as median and interquartile range (IQR) because they did not show a normal distribution. Mann-Whitney U test (to compare two groups) or Kruskal-Wallis test (to compare more than two groups) was used to assess significant differences between medians. Allele and genotype frequencies of HLA-G 14-bp Ins/Del polymorphism were presented by number and percentage and significant differences were assessed using two-tailed Fisher exact test or Pearson Chi-square test. Pearson Chi-square goodness of fit test was used to assess the fit of genotype frequencies to Hardy–Weinberg equilibrium (HWE). Age- and gender-adjusted multinomial logistic regression analysis was performed to calculate odds ratio (OR) and 95% confidence interval (CI). The analysis was conducted under five genetic models; allele, recessive, dominant, overdominant and codominant. A probability (p) value ≤0.05 was considered statistically significant. The p-value was corrected (pc) for multiple comparisons using Bonferroni correction.
3. Results
3.1. sHLA-G level
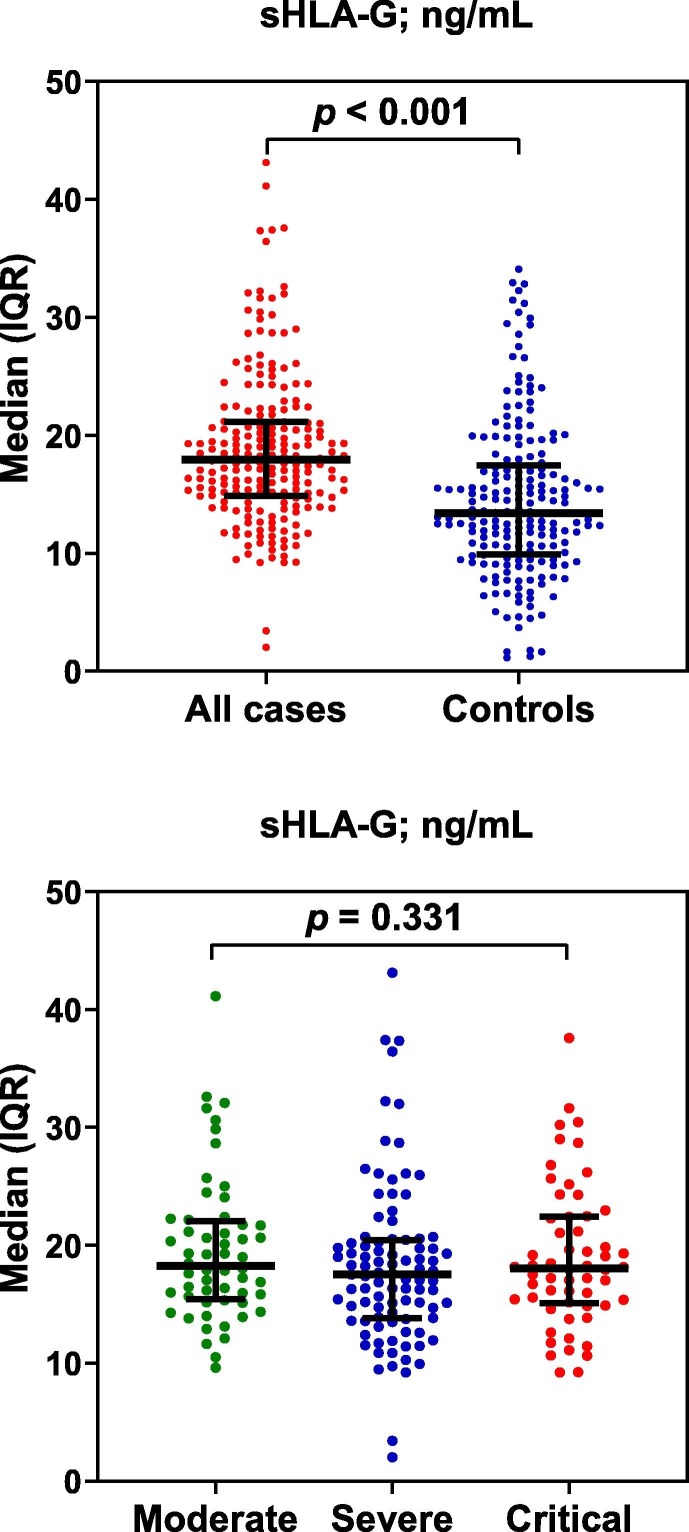
Median levels of sHLA-G were significantly higher in serum of COVID-19 patients than in controls (17.92 [IQR: 14.86–21.15] vs. 13.42 [IQR: 9.95–17.38] ng/mL; p < 0.001). In the case of disease severity, sHLA-G levels did not show significant differences between patients with moderate, severe or severe illness (p = 0.331) (Fig. 1 ).
Fig. 1.
Scatter dot plots of sHLA-G in all cases of COVID-19 and controls, and in cases distributed according to severity of disease. Horizontal lines indicate medians, while vertical lines indicate interquartile range (IQR). Significant differences were assessed with Mann-Whitney U test (to compare two groups) or Kruskal-Wallis test (to compare three groups).
3.2. HLA-G 14-bp Ins/Del polymorphism
Genotype frequencies of HLA-G 14-bp Ins/Del polymorphism were compatible with HWE in COVID-19 patients and controls (p = 0.738 and 0.371, respectively). Under allele model, logistic regression analysis demonstrated that Del allele was significantly associated with the risk of COVID-19 in patients compared to controls (56.7 vs. 44.4%; OR = 1.89; 95% CI = 1.44–2.48; p < 0.001; pc = 0.001). A higher risk was associated with Del/Del genotype under codominant model (31.6 vs. 18.2%; OR = 2.39; 95% CI = 1.25–4.58; p = 0.008; pc = 0.048). In the recessive and dominant models, no significant differences were maintained after applying Bonferroni correction, while no significant difference was found in the overdominant model (Table 1 ).
Table 1.
Logistic regression and Hardy-Weinberg analyses of HLA-G 14-bp Insertion/Deletion polymorphism in COVID-19 cases compared to controls.
| Model† | Allele/genotype | Cases (N = 209) |
Controls (N = 198) |
OR | 95% CI | p-value (pc) | ||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | |||||
| Allele | Ins | 181 | 43.3 | 220 | 55.6 | Reference | ||
| Del | 237 | 56.7 | 176 | 44.4 | 1.89 | 1.44–2.48 | <0.001 (0.001) | |
| Recessive | Ins/Ins + Ins/Del | 143 | 68.4 | 162 | 81.8 | Reference | ||
| Del/Del | 66 | 31.6 | 36 | 18.2 | 1.74 | 1.03–2.93 | 0.039 (0.234) | |
| Dominant | Ins/Ins | 38 | 18.2 | 58 | 29.3 | Reference | ||
| Ins/Del + Del/Del | 171 | 81.8 | 140 | 70.7 | 1.82 | 1.08–3.09 | 0.026 (0.156) | |
| Overdominant | Ins/Ins + Del/Del | 104 | 49.8 | 94 | 47.5 | Reference | ||
| Ins/Del | 105 | 50.2 | 104 | 52.5 | 1.03 | 0.66–1.61 | 0.889 (1.0) | |
| Codominant | Ins/Ins | 38 | 18.2 | 58 | 29.3 | Reference | ||
| Ins/Del | 105 | 50.2 | 104 | 52.5 | 1.60 | 0.92–2.79 | 0.097 (0.582) | |
| Del/Del | 66 | 31.6 | 36 | 18.2 | 2.39 | 1.25–4.58 | 0.008 (0.048) | |
| HWE-p-value | 0.738 | 0.371 | ||||||
†: The analysis was adjusted for age and gender; HWE: Hardy-Weinberg equilibrium; Ins: Insertion; Del: Deletion; OR: Odds ratio; CI: Confidence interval; p: Two-tailed Fisher exact probability; pc: Bonferroni correction probability. Significant p-value is indicated in bold.
Allele and genotype frequencies of HLA-G 14-bp Ins/Del polymorphism stratified by gender and disease severity showed no significant differences in each stratum (Table 2 ).
Table 2.
Allele and genotype frequencies of HLA-G 14-bp insertion/deletion polymorphism in COVID-19 cases distributed according to gender and disease severity.
| Characteristic |
Ins |
Del |
p-value | Ins/Ins |
Ins/Del |
Del/Del |
p-value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | N | % | |||
| Gender | ||||||||||||
| Male | 138 | 43.7 | 178 | 56.3 | 0.819 | 30 | 19.0 | 78 | 49.4 | 50 | 31.6 | 0.848 |
| Female | 43 | 42.2 | 59 | 57.8 | 8 | 15.7 | 27 | 52.9 | 16 | 31.4 | ||
| Disease severity | ||||||||||||
| Moderate | 52 | 46.4 | 60 | 53.6 | 0.721 | 12 | 21.4 | 28 | 50.0 | 16 | 28.6 | 0.474 |
| Severe | 81 | 42.6 | 109 | 57.4 | 19 | 20.0 | 43 | 45.3 | 33 | 34.7 | ||
| Critical | 48 | 41.4 | 68 | 58.6 | 7 | 12.1 | 34 | 58.6 | 17 | 29.3 | ||
Ins: Insertion; Del: Deletion; p: Pearson Chi-square test probability (significant p-value is indicated in bold).
3.3. Impact 14-bp Ins/Del polymorphism on sHLA-G levels
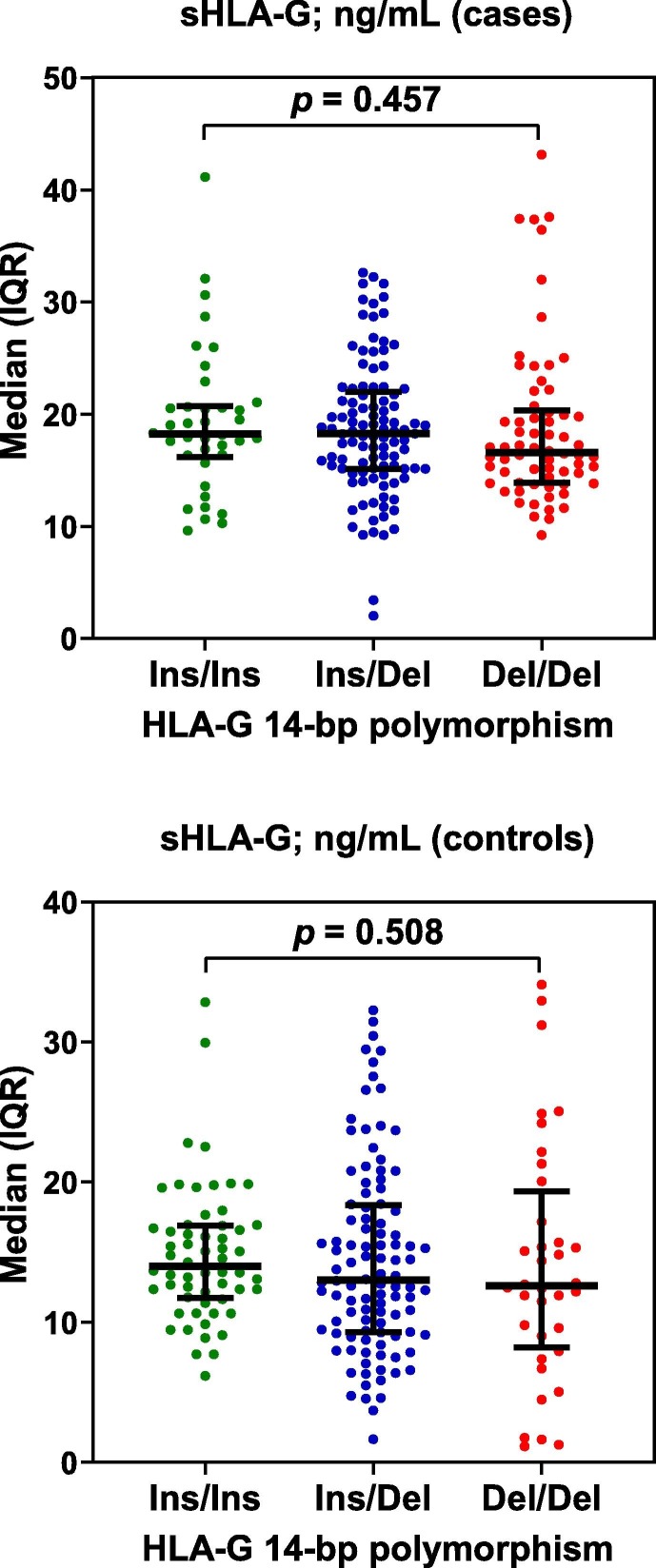
Although Del/Del genotype tended to show lower levels of sHLA-G compared to Ins/Ins and Ins/Del genotypes in patients and controls, the differences were not significant (p = 0.457 and 0.508, respectively) (Fig. 2 ).
Fig. 2.
Scatter dot plots of sHLA-G in COVID-19 cases and controls distributed according to genotypes of HLA-G 14-bp insertion (Ins)/deletion (Del) polymorphism. Horizontal lines indicate medians, while vertical lines indicate interquartile range (IQR). Significant differences were assessed with Kruskal-Wallis test.
4. Discussion
The current study demonstrated that sHLA-G levels were significantly elevated in COVID-19 patients regardless of disease severity. These findings were consistent with our previous observation that sHLA-G levels were up-regulated in serum of patients with severe disease [23]. These data confirm the recently addressed hypothetical views implicating HLA-G in immunopathogenesis of COVID-19 and disease severity [6], [22]. Under normal physiological conditions, HLA-G can function as an inhibitor of T and NK cell cytotoxicity and an enhancer of T regulatory cells [30]. Therefore, up-regulated levels of HLA-G are associated with the potential for immunosuppression, through which SARS-CoV-2 can evade the host's immune response, and thus COVID-19 can progress [6]. Interestingly, in vitro evidence showed that a large number of genes related to HLA-class I (including HLA-G), HLA-class II, or antigen presentation were found to be up-regulated 36 h post-infection of a human lung epithelial cell line with SARS-CoV [31].
In viral infections, two hypotheses have been put forward to explain the role of HLA-G in the immunopathogenesis of viruses. HLA-G may enhance immune escape mechanisms that favor viral persistence, and this is sustained by the immunosuppressive properties of HLA-G. Otherwise, HLA-G expression and/or secretion may reflect a balanced response to the inflammatory responses that occur during viral infection [19]. Up-regulation of HLA-G has been associated with various viral infections such as human immunodeficiency virus, human cytomegalovirus, influenza A virus, hepatitis B and C viruses, and accordingly, it has been speculated that up-regulation of HLA-G expression can be considered as a potential virus strategy to avoid immune responses [32], [33], [34]. Zidi also suggested that up-regulated levels of sHLA-G in patients infected with SARS-CoV-2 may be due to delocalization of surface-bound HLA-G into the plasma after its cleavage by matrix metalloproteinases produced by connective tissue and pro-inflammatory cells such as activated neutrophils [22]. In addition, it has been indicated that HLA-G expression can be induced by some cytokines and hormones such as interleukin (IL)-10 and cortisol [35]. IL-10 is an anti-inflammatory cytokine that can be produced by monocytes, macrophages and lymphocytes to counteract exacerbated inflammation [36]. Cortisol is a stress-related steroid hormone synthesized from cholesterol [37]. It is interesting to note that IL-10 and cortisol show up-regulated levels in serum of COVID-19 patients and are associated with disease severity and increased mortality [38], [39].
Up-regulated expression of HLA-G is also a prominent feature of cancerous cells associated with immune evasion and is strongly linked to disease progression and prognosis in patients with various malignancies [40]. Thus, cancerous cells and cells infected with SARS-CoV-2 may share similar mechanisms of the HLA-G signaling pathway in evading the immune system. Indeed, evidence in patients with malignancies or COVID-19 indicates that HLA-G molecules mediate the effects of immune tolerance by binding to their specific receptors, immunoglobulin-like transcripts (ILTs), which are expressed in most immune cells. Further, HLA-G can interact with CD8 and the killer inhibitory receptor 2DL4 and thus provoke T cell apoptosis and NK cell senescence [6], [41]. In addition, HLA-G 14-bp Ins/Del polymorphism has been associated with the risk of various cancers, particularly breast cancer and esophageal cancer, and the Ins/Ins genotype and Ins allele versus Del/Del genotype and Del allele, respectively have shown protective effects [42]. In the present study, it was also demonstrated that HLA-G 14-bp Ins/Del polymorphism may influence susceptibility to COVID-19. It was found that individuals with Del allele and Del/Del genotype were more likely to develop CΟVΙD-19 compared to Ins allele and Ins/Ins genotype, respectively (OR = 1.89 and 2.39, respectively). Perhaps, the current study has been the first that investigated HLΑ-G 14-bp Ins/Del polymorphism in CΟVΙD-19. The only available evidence is a genome-wide association study conducted on critically-ill patients with CΟVΙD-19 from the United Kingdom and identified that the HLA-G variant rs9380142 was associated with susceptibility to disease [43]. Of note, this variant is located in exon eight of the HLA-G gene in the vicinity of the 14-bp Ins/Del polymorphism. This observation may suggest that genetic variants of HLA-G are associated with susceptibility to CΟVΙD-19, and therefore, it is warranted to guide research involving HLA-G variants in the disease susceptibility.
When other viral infections were considered, there has been inconsistent observations regarding the association of HLΑ-G 14-bp Ins/Del polymorphism with the susceptibility to viral infections. Some investigator groups have linked this polymorphism with human cytomegalovirus and human T-lymphotropic virus type-1 (HTLV-1) infections [27], [44], while others indicated no clear association with viral infections [25], [45]. However, a meta-analysis study of 10 studies revealed that the overall analysis favored no association between different viral infections and the 14-bp Ins/Del polymorphism, but subgroup analysis demonstrated a significant association with HTLV-1 infection [46]. An additional study has demonstrated that HLΑ-G 14-bp Ins/Del polymorphism was significantly linked to chronic hepatitis B virus in Brazilians [26]. These findings, together with findings of this study, may highlight the significance of HLA-G 14-bp Ins/Del polymorphism in etiopathogenesis of viral infections including CΟVΙD-19, but further investigations are warranted.
The HLA-G 14-bp Ins/Del polymorphism has been shown to affect sHLA-G expression in plasma of a normal Chinese population. Individuals with Ins/Ins genotype showed a significantly lower level of sHLA-G compared to individuals with Del/Del genotype [24]. In this study, an opposite observation was made and the Ins/Ins genotype appeared to increase sHLA-G expression, while the Del/Del genotype showed decreased sHLA-G expression in serum of COVID-19 patients or controls, but the differences were not significant. No specific cause may explain these adverse results, but the sample evaluated may have an effect. The Chinese study used plasma while the current study used serum. It has been indicated that plasma shows higher levels of sHLA-G than the serum of the same individuals. It has been suggested that during clot formation an amount of sHLA-G is trapped and/or consumed [47]. In addition, there are other causes that can affect sHLA-G level in relation to HLA-G 14-bp Ins/Del polymorphism including therapies and co-morbidities (for instance diabetes and cardiovascular disease), which were not identified in the current study, as it was observed that sHLA-G level may be affected by them [11], [14], [17], [23], [48].
Although the current study showed that the HLΑ-G 14-bp Ins/Del polymorphism may influence susceptibility to COVID-19 and expression levels of HLA-G, it was revealed by a recent meta-analysis study of several human diseases (autoimmune, inflammatory and infectious diseases) that this polymorphism should not be used as a single genetic marker of disease susceptibility and the full 3′UTR fragment must be analyzed in this context [49]. It has been found that other variants in the 3′UTR of HLA-G gene are involved in the posttranscriptional control of HLA-G expression; for instance, +3010 G/C (rs1710), +3027 C/A (rs17179101) and +3035 C/T (rs17179108), and at least two additional SNPs have been functionally studied and associated with the regulation of HLA-G expression levels (+3142 C/G [rs1063320] and +3187A/G [rs9380142]) [35]. Moreover, an extreme linkage disequilibrium (LD) can be observed along the entire 3′UTR and HLA-G gene [50]. Then, it is paramount to consider this LD once different 3′UTR haplotype combinations harbored each HLΑ-G 14-bp Ins/Del polymorphism. For instance, the 14-bp (insertion) may appear in UTR-2, UTR-5, or UTR-7, and each of these haplotypes is more frequently associated with a specific promoter and coding region haplotypes, producing different HLA-G proteins [51]. Finally, it is important to make clear that the associations reported in the literature between HLΑ-G 14-bp Ins/Del polymorphism and diseases are inconsistent [49].
The study faced the limitation of a relatively low sample size of patients with moderate or critical disease. Besides, other SNPs in exon 8 in the 3′UTR region of the HLA-G gene were not included in the study. Therefore, it would be useful to sequence this region to determine related SNPs.
In conclusion, sHLA-G levels were up-regulated in serum of COVID-19 patients regardless of disease severity. Further, Del allele and Del/Del genotype of HLA-G 14-bp Ins/Del polymorphism were proposed to be associated with increased susceptibility to COVID-19. Genotypes of this polymorphism did not influence sHLA-G serum levels in patients or controls.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The kind assistance and cooperation of the medical staff at Dar Al-Salam Field Hospital, Al-Karkh General Hospital and Al-Furat General Hospital (Baghdad) are appreciated by authors.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
The authors declare that there were no conflicts of interest.
References
- 1.Wang C., Wang Z., Wang G., Lau J.Y.N., Zhang K., Li W. COVID-19 in early current status and looking forward. Signal Transduct. Target. Ther. 2021;6(2021):1–14. doi: 10.1038/s41392-021-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hosseini A., Hashemi V., Shomali N., Asghari F., Gharibi T., Akbari M., Gholizadeh S., Jafari A. Innate and adaptive immune responses against coronavirus. Biomed. Pharmacother. 2020;132 doi: 10.1016/j.biopha.2020.110859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahaghoghi-Hajghorbani S., Zafari P., Masoumi E., Rajabinejad M., Jafari-Shakib R., Hasani B., Rafiei A. The role of dysregulated immune responses in COVID-19 pathogenesis. Virus Res. 2020;290 doi: 10.1016/j.virusres.2020.198197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L., Li R., Song G., Scholes G.D., She Z.-S. Impairment of T cells’ antiviral and anti-inflammation immunities may be critical to death from COVID-19. R. Soc. Open Sci. 2021;8 doi: 10.1098/rsos.211606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin A., Yan W.-H. Perspective of HLA-G induced immunosuppression in SARS-CoV-2 infection. Front. Immunol. 2021;12:5192. doi: 10.3389/fimmu.2021.788769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wit J., Borghans J.A.M., Kesmir C., van Baarle D. Editorial: role of HLA and KIR in viral infections. Front. Immunol. 2016;7:286. doi: 10.3389/fimmu.2016.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trowsdale J., Knight J.C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet. 2013;14:301–323. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieczorek M., Abualrous E.T., Sticht J., Álvaro-Benito M., Stolzenberg S., Noé F., Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imrie A., McCarthy S. HLA and immunodominance in viral infection: T-cell responses in protection and immunopathogenesis. Microbiol. Aust. 2021;42:84–86. doi: 10.1071/MA21020. [DOI] [Google Scholar]
- 11.Wyatt R.C., Lanzoni G., Russell M.A., Gerling I., Richardson S.J. What the HLA-I!—Classical and non-classical HLA class I and their potential roles in type 1 diabetes. Curr. Diab. Rep. 2019;19:1–11. doi: 10.1007/s11892-019-1245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nilsson L.L., Djurisic S., Hviid T.V.F. Controlling the immunological crosstalk during conception and pregnancy: HLA-G in reproduction. Front. Immunol. 2014;5:198. doi: 10.3389/fimmu.2014.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carosella E.D., Rouas-Freiss N., Le Roux D.T., Moreau P., LeMaoult J. HLA-g. An immune checkpoint molecule. Adv. Immunol., Academic Press. 2015:33–144. doi: 10.1016/bs.ai.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Arnaiz-Villena A., Juarez I., Suarez-Trujillo F., López-Nares A., Vaquero C., Palacio-Gruber J., Martin-Villa J.M. HLA-G: Function, polymorphisms and pathology. Int. J. Immunogenet. 2021;48:172–192. doi: 10.1111/iji.12513. [DOI] [PubMed] [Google Scholar]
- 15.Lin A., Zhang X., Zhang R.L., Zhang J.G., Zhou W.J., Yan W.H. Clinical significance of potential unidentified HLA-G isoforms without a1 domain but containing intron 4 in colorectal cancer patients. Front. Oncol. 2018;8:361. doi: 10.3389/fonc.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tronik-Le Roux D., Renard J., Vérine J., Renault V., Tubacher E., LeMaoult J., Rouas-Freiss N., Deleuze J.F., Desgrandschamps F., Carosella E.D. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol. Oncol. 2017;11:1561–1578. doi: 10.1002/1878-0261.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contini P., Murdaca G., Puppo F., Negrini S. HLA-G expressing immune cells in immune mediated diseases. Front. Immunol. 2020;11:1613. doi: 10.3389/fimmu.2020.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laaribi A.B., Bortolotti D., Hannachi N., Mehri A., Hazgui O., Ben Yahia H., Babay W., Belhadj M., Chaouech H., Yacoub S., Letaief A., Ouzari H.I., Boudabous A., Di Luca D., Boukadida J., Rizzo R., Zidi I. Increased levels of soluble HLA-G molecules in Tunisian patients with chronic hepatitis B infection. J. Viral Hepat. 2017;24:1016–1022. doi: 10.1111/jvh.12718. [DOI] [PubMed] [Google Scholar]
- 19.Amiot L., Vu N., Samson M. Immunomodulatory properties of HLA-G in infectious diseases. J. Immunol. Res. 2014;2014 doi: 10.1155/2014/298569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albayati Z., Alyami A., Alomar S., Middleton D., Bonnett L., Aleem S., Flanagan B.F., Christmas S.E. The influence of cytomegalovirus on expression of HLA-G and its ligand KIR2DL4 by human peripheral blood leucocyte subsets. Scand. J. Immunol. 2017;86:396–407. doi: 10.1111/sji.12594. [DOI] [PubMed] [Google Scholar]
- 21.Jasinski-Bergner S., Schmiedel D., Mandelboim O., Seliger B. Role of HLA-G in viral infections. Front. Immunol. 2022;13:342. doi: 10.3389/fimmu.2022.826074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zidi I. Puzzling out the COVID-19: therapy targeting HLA-G and HLA-E. Hum. Immunol. 2020;81:697–701. doi: 10.1016/j.humimm.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Bayatee N.T., Ad’hiah A.H. Soluble HLA-G is upregulated in serum of patients with severe COVID-19. Hum. Immunol. 2021;82:726–732. doi: 10.1016/j.humimm.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X.Y., Yan W.H., Lin A., Xu H.H., Zhang J.G., Wang X.X. The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens. 2008;72:335–341. doi: 10.1111/j.1399-0039.2008.01107.x. [DOI] [PubMed] [Google Scholar]
- 25.da Silva G.K., Vianna P., Veit T.D., Crovella S., Catamo E., Cordero E.A.A., Mattevi V.S., Lazzaretti R.K., Sprinz E., Kuhmmer R., Chies J.A.B. Influence of HLA-G polymorphisms in human immunodeficiency virus infection and hepatitis C virus co-infection in Brazilian and Italian individuals. Infect. Genet. Evol. 2014;21:418–423. doi: 10.1016/j.meegid.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 26.J. Michel Wolf, V.R. Zingalli Bueno Pereira, P.A. Zanetti Ballardin Roncato, A. Castagna Wortmann, G.Z. Stumm, F. Oliveira da Silva, V.R. Lunge, D. Simon, The HLA-G 14-bp insertion/deletion polymorphism is associated with chronic hepatitis B in Southern Brazil: A case-control study, Hum. Immunol. 81 (2020) 79–84. https://doi.org/10.1016/j.humimm.2020.01.003. [DOI] [PubMed]
- 27.Zheng X.Q., Zhu F., Shi W.W., Lin A., Yan W.H. The HLA-G 14 bp insertion/deletion polymorphism is a putative susceptible factor for active human cytomegalovirus infection in children. Tissue Antigens. 2009;74:317–321. doi: 10.1111/j.1399-0039.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization, Clinical management of severe acute respiratory infection when novel coronavirus (2019-nCoV) infection is suspected: interim guidance 28 January 2020, Who. (2020) 10. WHO/2019-nCoV/clinical/2020.5%0ACC BY-NC-SA 3.0 IGO%0AWHO/2019-nCoV/clinical/2020.5%0ACC BY-NC-SA 3.0 IGO%0Ahttps://apps.who.int/iris/handle/10665/330893e.
- 29.Abdul-Hussein S.S., Ali E.N., Alkhalidi N.M.F., Zaki N.H., Ad’hiah A.H. Susceptibility role of soluble HLA-G and HLA-G 14-bp insertion/deletion polymorphism in inflammatory bowel disease. Egypt. J. Med Hum. Genet. 2020;21:1–12. doi: 10.1186/s43042-020-00104-1. [DOI] [Google Scholar]
- 30.Xu X., Zhou Y., Wei H. Roles of HLA-G in the maternal-fetal immune microenvironment. Front. Immunol. 2020;11:2767. doi: 10.3389/fimmu.2020.592010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., Yount B.L., Graham R.L., Baric R.S., Katzea M.G. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4 doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dias F.C., Castelli E.C., Collares C.V.A., Moreau P., Donadi E.A. The role of HLA-G molecule and HLA-G gene polymorphisms in tumors, viral hepatitis, and parasitic diseases. Front. Immunol. 2015;6:9. doi: 10.3389/fimmu.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan W.H., Lin A., Chen B.G., Chen S.Y. Induction of both membrane-bound and soluble HLA-G expression in active human cytomegalovirus infection. J. Infect. Dis. 2009;200:820–826. doi: 10.1086/604733. [DOI] [PubMed] [Google Scholar]
- 34.C. Li, I. Toth, J. Schulze zur Wiesch, F. Pereyra, J. Rychert, E.S. Rosenberg, J. van Lunzen, M. Lichterfeld, X.G. Yu, Functional Characterization of HLA-G+ Regulatory T Cells in HIV-1 Infection, PLoS Pathog. 9 (2013) e1003140. https://doi.org/10.1371/journal.ppat.1003140. [DOI] [PMC free article] [PubMed]
- 35.Castelli E.C., Veiga-Castelli L.C., Yaghi L., Moreau P., Donadi E.A. Transcriptional and posttranscriptional regulations of the HLA-G gene. J. Immunol. Res. 2014;2014 doi: 10.1155/2014/734068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer S.S., Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit. Rev. Immunol. 2012;32:23–63. doi: 10.1615/critrevimmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L. Thau, S. Sharma, Physiology, Cortisol, StatPearls Publishing, 2019. https://www.ncbi.nlm.nih.gov/books/NBK538239/ (accessed January 30, 2022). [PubMed]
- 38.Lu L., Zhang H., Dauphars D.J., He Y.W. A potential role of interleukin 10 in COVID-19 pathogenesis. Trends Immunol. 2021;42:3–5. doi: 10.1016/j.it.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan T., Khoo B., Mills E.G., Phylactou M., Patel B., Eng P.C., Thurston L., Muzi B., Meeran K., Prevost A.T., Comninos A.N., Abbara A., Dhillo W.S. Association between high serum total cortisol concentrations and mortality from COVID-19. Lancet Diabetes Endocrinol. 2020;8:659–660. doi: 10.1016/S2213-8587(20)30216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin A., Yan W.H. Heterogeneity of HLA-G expression in cancers: facing the challenges. Front. Immunol. 2018;9:2164. doi: 10.3389/fimmu.2018.02164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loustau M., Anna F., Dréan R., Lecomte M., Langlade-Demoyen P., Caumartin J. HLA-G neo-expression on tumors. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y., Lu J., Wu Y.E., Zhao X., Li L. Genetic variation in the HLA-G 3UTR 14–bp insertion/deletion and the associated cancer risk: Evidence from 25 case–control studies. Biosci. Rep. 2019;39 doi: 10.1042/BSR20181991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., Furniss J., Richmond A., Gountouna E., Wrobel N., Harrison D., Wang B., Wu Y., Meynert A., Griffiths F., Oosthuyzen W., Kousathanas A., Moutsianas L., Yang Z., Zhai R., Zheng C., Grimes G., Beale R., Millar J., Shih B., Keating S., Zechner M., Haley C., Porteous D.J., Hayward C., Yang J., Knight J., Summers C., Shankar-Hari M., Klenerman P., Turtle L., Ho A., Moore S.C., Hinds C., Horby P., Nichol A., Maslove D., Ling L., McAuley D., Montgomery H., Walsh T., Pereira A.C., Renieri A., Shen X., Ponting C.P., Fawkes A., Tenesa A., Caulfield M., Scott R., Rowan K., Murphy L., Openshaw P.J.M., Semple M.G., Law A., Vitart V., Wilson J.F., Baillie J.K. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 44.Cilião Alves D.C., Haddad R., Rocha-Júnior M.C., De Deus Wagatsuma V.M., Martelli-Palomino G., Marques A.A., Takayanagui O.M., Covas D.T., Kashima S., Donadi E.A. HLA-G 3′-untranslated region polymorphisms are associated with HTLV-1 infection, proviral load and HTLV-associated myelopathy/tropical spastic paraparesis development. J. Gen. Virol. 2016;97:2742–2752. doi: 10.1099/jgv.0.000559. [DOI] [PubMed] [Google Scholar]
- 45.S. da C. Ferreira, S.G.F. Chachá, F.F. Souza, A.C. Teixeira, R. de C. Santana, N.H.S. Deghaide, S. Rodrigues, L.A. Marano, C.T. Mendes-Junior, L.N.Z. Ramalho, S. Zucoloto, E.A. Donadi, A. de L.C. Martinelli, The HLA-G 14-base pair deletion allele and the deletion/deletion genotype are associated with persistent HBe antigenemia in chronic hepatis B infection, Hum. Immunol. 78 (2017) 166–171. https://doi.org/10.1016/j.humimm.2016.12.011. [DOI] [PubMed]
- 46.Lv H., Lv H., Lin Z., Chen L., Zhu M., Hong D. Meta-analysis of correlationship between HLA-G 3′UTR 14-bp Ins/Del polymorphism and virus susceptibility. Med. (United States) 2018;97 doi: 10.1097/MD.0000000000012262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudstein-Svetlicky N., Loewenthal R., Horejsi V., Gazit E. HLA-G levels in serum and plasma. Tissue Antigens. 2006;67:111–116. doi: 10.1111/j.1399-0039.2006.00540.x. [DOI] [PubMed] [Google Scholar]
- 48.Agnihotri V., Gupta A., Kumar L., Dey S. Serum sHLA-G: significant diagnostic biomarker with respect to therapy and immunosuppressive mediators in Head and Neck Squamous Cell Carcinoma. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-60811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Almeida B.S., Muniz Y.C.N., Prompt A.H., Castelli E.C., Mendes-Junior C.T., Donadi E.A. Genetic association between HLA-G 14-bp polymorphism and diseases: A systematic review and meta-analysis. Hum. Immunol. 2018;79:724–735. doi: 10.1016/j.humimm.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Drabbels J.J.M., Welleweerd R., van Rooy I., Johnsen G.M., Staff A.C., Haasnoot G.W., Westerink N., Claas F.H.J., Rozemuller E., Eikmans M. HLA-G whole gene amplification reveals linkage disequilibrium between the HLA-G 3′UTR and coding sequence. HLA. 2020;96:179–185. doi: 10.1111/tan.13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poras I., Yaghi L., Martelli-Palomino G., Mendes C.T., Muniz Y.C.N., Cagnin N.F., De Almeida B.S., Castelli E.C., Carosella E.D., Donadi E.A., Moreau P. Haplotypes of the HLA-G 3′ untranslated region respond to endogenous factors of HLA-G+ and HLA-G- cell lines differentially. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169032. [DOI] [PMC free article] [PubMed] [Google Scholar]