Abstract
Neuropeptides are the most diverse messenger molecules in metazoans and are involved in regulation of daily physiology and a wide array of behaviors. Some neuropeptides and their cognate receptors are structurally and functionally well conserved over evolution in bilaterian animals. Among these are peptides related to gastrin and cholecystokinin (CCK). In mammals, CCK is produced by intestinal endocrine cells and brain neurons, and regulates gall bladder contractions, pancreatic enzyme secretion, gut functions, satiety and food intake. Additionally, CCK plays important roles in neuromodulation in several brain circuits that regulate reward, anxiety, aggression and sexual behavior. In invertebrates, CCK-type peptides (sulfakinins, SKs) are, with a few exceptions, produced by brain neurons only. Common among invertebrates is that SKs mediate satiety and regulate food ingestion by a variety of mechanisms. Also regulation of secretion of digestive enzymes has been reported. Studies of the genetically tractable fly Drosophila have advanced our understanding of SK signaling mechanisms in regulation of satiety and feeding, but also in gustatory sensitivity, locomotor activity, aggression and reproductive behavior. A set of eight SK-expressing brain neurons plays important roles in regulation of these competing behaviors. In males, they integrate internal state and external stimuli to diminish sex drive and increase aggression. The same neurons also diminish sugar gustation, induce satiety and reduce feeding. Although several functional roles of CCK/SK signaling appear conserved between Drosophila and mammals, available data suggest that the underlying mechanisms differ.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04214-4.
Keywords: Sulfakinin, Drosophila, Neuromodulation, Peptide hormone, Satiety, Behavior
Introduction
Neuropeptides are involved in the regulation of physiology and a wide array of vital behaviors of metazoans. They constitute secreted signals in neuronal circuits that are hierarchically arranged in the brain and partake in context-dependent orchestrating signaling by higher-order neurons, as well as in local executive modulation in specific circuits (see [1–5]). Thus, neuropeptides impart plasticity to the hardwired circuits of the central nervous system and in this review we highlight some specific roles of neuropeptides, especially cholecystokinin-like peptides, in regulation of competing behaviors such as feeding, mating and aggression.
Neuropeptides and peptide hormones constitute ancient signaling molecules that are genetically encoded on precursors that give rise to one or more bioactive peptides acting on different types of membrane receptors. Bioactive peptides are present already in organisms that lack a nervous system, such as for instance sponges [6] and the Placozoan Trichoplax adhaerens, [7]. The latter small marine animal utilizes a small number of peptides, derived from five precursors, to regulate simple behaviors associated with locomotion and food intake [8]. In organisms with simple nervous systems, such as cnidarians and ctenophores, there are more diverse sets on peptides produced by neurons, and thus referred to as neuropeptides [9–12]. The more evolved animals among the Bilateria produce numerous neuropeptides as well as peptide hormones that display a wide array of diverse functions in development, physiology and behavior [13–17]. Many of these peptidergic signaling pathways are evolutionarily conserved in the bilaterian phyla, including mammals [13, 14]. Among the conserved signaling pathways are those that utilize cholecystokinin (CCK)-related peptides and their receptors. It should be noted that in mammals CCK exists alongside a closely related (paralog) peptide named gastrin, which is encoded on a separate gene. We will discuss these two peptides in Sect. 2, but in the following we will refer to CCK signaling for simplicity. Interestingly, CCK signaling is functionally pleiotropic in most animals studied and regulates behavior and physiology associated with feeding, digestion, aggression and reproduction (reviewed in [16, 18–23]).
CCK signaling components have been identified in vertebrates and in several bilaterian invertebrate phyla [13–15, 24–31], as well as in deuterostome invertebrates such as echinoderms and invertebrate chordates (the latter also known as protochordates) [32, 33]. In mammals, CCK activity was first discovered in tissue already in 1906 [34] and CCK was isolated as a gut hormone in 1928 [35]. The peptide was initially identified as a factor released from the small intestine of cats and dogs that induced contractions of the gall bladder [35], and from pig intestine that triggered secretion of pancreatic enzyme [36]. Over the years, multiple additional functions have been discovered, including regulation of satiety and various neuromodulatory roles in the brain (see [18, 19, 37]).
The presence of CCK like peptides in insects and some other invertebrates was suggested early on [38–41], but was based solely on immunochemical detection with heterologous antisera. The first invertebrate CCK-type peptide was isolated in 1986 from the cockroach Leucophaea maderae (now Rhyparobia maderae) using head extract and monitoring myostimulatory action on the hindgut [24]. This peptide was named leucosulfakinin and closely related peptides, sulfakinins (SKs), have since been identified in multiple insects and other invertebrates, as well as in invertebrate chordates (see examples in Fig. 1). However, CCK-type peptides have not been found in non-bilaterians such as Porifera, Placozoa, Cnidaria, or Ctenophora (see [13–15]). After the identification of two SK receptors in Drosophila [42, 43], numerous invertebrate SK receptors are now known that display homologies to those of gastrin and CCK receptors in vertebrates (see [20, 32]). Signaling with CCK-type peptides has been assayed in multiple invertebrates and in this review we highlight the structure, distribution and functions of these peptides and their receptors, with some more detailed analysis in Drosophila and with comparisons to mammals. More specifically, we discuss the roles of SKs in satiety signaling, feeding, metabolism, reproductive behavior and aggression. Interestingly, it has been shown that DSK neurons are important in the regulation of competing behaviors. Thus, in male flies, DSK signaling diminishes sex drive, but increases aggression, in addition to its known role in inducing satiety and reduced feeding [23, 44, 45]. This peptidergic regulation of competing behaviors is the topic of one of the sections in this review. We also discuss how CCK-mediated satiety signaling in mammals differs mechanistically from SK signaling in Drosophila, although the outcome on food intake is similar. Finally, we outline some other neuromodulatory systems that use neuropeptides and monoamines to regulate feeding, reproductive behavior and aggression in association with SKs.
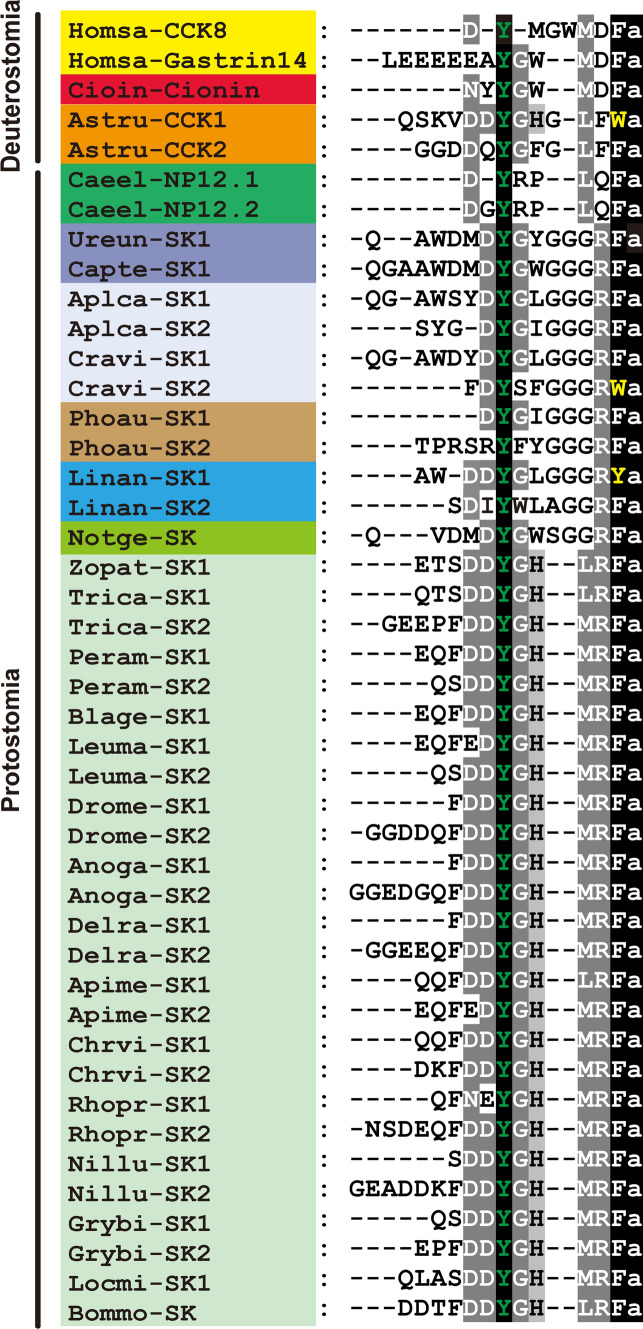
Fig. 1.
Sequence alignments of CCK, gastrin, sulfakinin and sulfakinin-like peptides from select species. Conserved residues are highlighted in black (identical) or gray (similar). Sulfated tyrosine was highlighted in green with black background. Species belonging to the same phyla have been highlighted with the same color. Note that pyroglutamate-blocked N-terminal residues are not indicated. Species names are as follows: Homsa (Homo sapiens), Cioin (Ciona intestinalis), Astru (Asterias rubens), Caeel (Caenorhabditis elegans), Ureun (Urechis unicinctus), Capte (Capitella teleta), Aplca (Aplysia californica), Cravi (Crassostrea virginica), Phoau (Phoronis australis), Linan (Lingula anatina), Notgen (Notospermus geniculatus), Zopat (Zophobas atratus), Trica (Tribolium castaneum), Peram (Periplaneta americana), Blage (Blattella germanica), Leuma (Leucophaea maderae), Drome (Drosophila melanogaster), Anoga (Anopheles gambiae), Delra (Delia radicum), Apime (Apis mellifera), Chrvi (Chrysis viridula), Rhopr (Rhodnius prolixus), Nillu (Nilaparvata lugens), Grybi (Gryllus bimaculatus), Locmi (Locusta migratoria), Bommo (Bombyx mori). Note that some insects only have one form of SK. The accession numbers of the sequences are listed in “Fig. 1
source data” in Supplementary data files
CCK signaling in mammals: a brief overview
Since there is a tremendous amount of published data on the distribution and functional roles of CCK and its receptors in mammals we will mainly focus on some of those features that relate to SK functions in invertebrates: satiety and feeding behavior, reproductive behavior, and aggression. For further details, several reviews are available that cover CCK signaling in mammals and other vertebrates [18, 19, 46–49].
CCK was one of the first hormones to be discovered in mammals, and was originally isolated from the small intestine as a factor that regulates gallbladder emptying and enzyme secretion from the pancreas [34, 35]. It can be noted that before sequencing of the factors acting on gall bladder and pancreas they were named CCK [35] and pancreozymin [36], respectively. After purification and sequencing in 1968, it turned out that the two activities were derived from a single peptide CCK/pancreozymin [50] and the name pancreozymin was abandoned. CCKs display strong sequence similarities to the peptide hormone gastrin, which was actually sequenced prior to CCK [51]. Gastrin was found in the duodenum and stomach and regulates gastric acid secretion [51]. Both CCK and gastrin exist in several forms that are N-terminal extensions of the same core peptides (CCK: 8, 33, 39, 58 or 83 amino acids; gastrin: 13, 17 or 34 amino acids) (see [18, 19, 52]). Note that CCK and gastrin are encoded on separate genes/precursors (see Fig. 2). The sequence of CCK8 in mammals is DYMGWMDFamide, where the tyrosine (Y) residue is commonly sulfated. The gastrin and CCK peptides share the C-terminal sequence GWMDFamide (see Fig. 1). It is interesting to note that, like several other neuropeptides, CCK-type peptides (e.g., cerulein) have also been identified in frog skin, where they are likely to be secreted as a deterrent against predators [53].
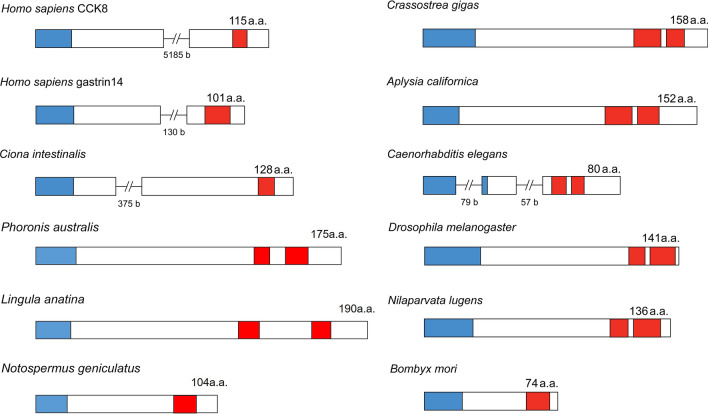
Fig. 2.
Schemes of CCK/SK precursors from representative species. Boxes and lines show exons and introns, respectively. Red boxes represent SK/CCK peptides and blue boxes indicate signal peptides. Note that DSK0 is not indicated in Drosophila. Crassostrea and Aplysia are mollusks, Ciona an invertebrate chordate, Caenorhabditis a nematode worm, Phoronis australis a phoronid, Lingula anatine a brachiopod, Notospermus geniculatus a nemertin and the others are insects. Introns have only been identified in mammals and C. elegans. The accession numbers of the sequences are listed in “Fig. 2
source data” in Supplementary data files
Mammalian CCK and gastrin act on the two GPCRs designated CCK1R and CCK2R, respectively (see [49]), the first of which was cloned in 1992 [54]. These receptors have different affinities for the gastrin/CCK peptide isoforms. Thus, CCK1R has a high affinity for sulfated CCK8-CCK58 and a 1000-fold lower affinity for gastrin, whereas CCK2R can be activated by both gastrin and CCK, even in their non-sulfated forms [19, 55]. The CCK1R is distributed in peripheral tissues such as the gastrointestinal tract, gallbladder, pancreas, and also in the anterior pituitary, nucleus accumbens, posterior hypothalamus, the brainstem (notably the nucleus of the solitary tract) and some areas of the midbrain, whereas the CCK2R is distributed predominantly in the brain, including amygdala, habenula, thalamus, cortex, hippocampus, and the olfactory bulb, and also in the pancreas and some parts of the gastrointestinal tract (reviewed in [18, 19]).
CCK, especially CCK8, is distributed widely in neurons in different parts of the brain, corresponding to the sites where the two CCK receptors have been found [18, 19, 56]. Importantly, CCK is also produced by a specific type of enteroendocrine cells (EECs), classified as type I cells, present in the duodenum and jejunum of the gastrointestinal tract (see [18, 46, 48, 57, 58]). It is CCK from these EECs that induces satiety.
Satiety in mammals is primarily induced by CCK signaling from the gut to the brain via the vagus nerve, and the mechanisms described next are based on several reviews [18, 46–48, 55]. Upon food intake and ensuing gastric distension, CCK is released from the EECs to act on afferent neurons in the adjacent vagal nerve (Fig. 3). These vagal afferent neurons (VANs) express CCKR1 and their axons connect to neurons in the nucleus of the solitary tract of the brainstem (hindbrain) (Fig. 3). CCK binding to the CCK1R in neurons of the VANs leads to activation of these neurons and thereby triggers a post-ingestive feedback to the hindbrain. The sensitivity of the VANs to CCK is modulated by the metabolic/energy state and CCK signaling regulates the expression of genes encoding other anorectic and orexigenic peptides and receptors in the VANs (e.g., receptors for leptin, urocortin, insulin, α-MSH, ghrelin and endocannabinoids). Thus, the VANs integrate several peptidergic signals and some sensory inputs to fine-tune food intake. It has been suggested that the VANs can adopt two states depending on the expression of these peptides and receptors, one associated with feeding (hunger) and another with inhibition of feeding (satiety) and that CCK acts as a gatekeeper that determines these states (see [47]). The CCK-induced signals in VANs propagate to the brainstem and result in efferent signals to the gastrointestinal tract that reduces food intake by controlling meal size. Mechanistically, these efferent satiety signals regulate (inhibit) gastric emptying, which in turn leads to reduced food ingestion (see [18, 47]). Furthermore, the VAN signals to the nucleus of solitary tract (NST) in the brainstem lead to activation of second-order neurons expressing for example neuropeptide Y, proopiomelanocortin (POMC) and dopamine. These NST neurons innervate several brain centers that regulate reward and food ingestion, such as the hypothalamus, mesolimbic system and nigro-striatal pathway. Thus, CCK-mediated satiety signaling originating in the intestine not only activates a direct feedback to the gastrointestinal tract, but also indirectly activates signaling within the brain that may mediate more long-lasting effects on behavior.
Fig. 3.
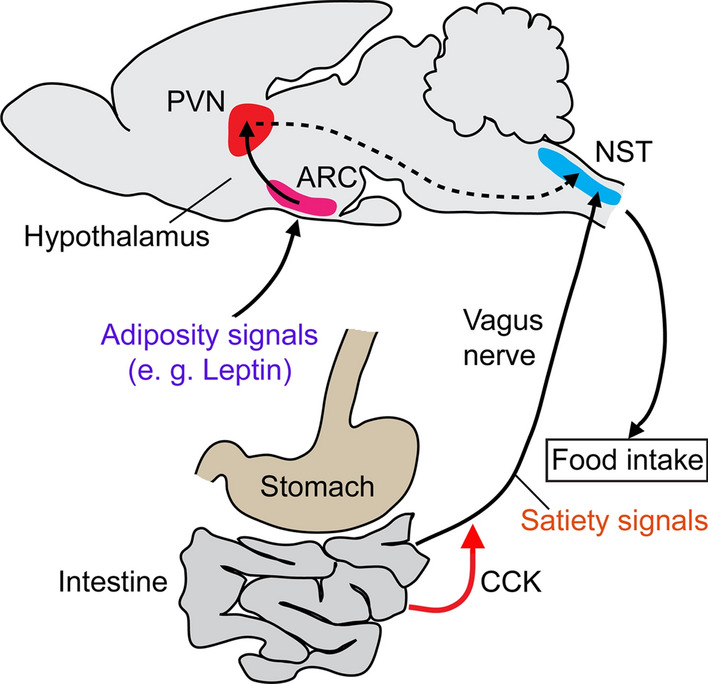
CCK and regulation of satiety in mammals. Upon stomach distension, CCK is released from enteroendocrine cells of the intestine and acts on CCK receptors on afferent neurons in the vagus nerve. These afferents signal to the nucleus of the solitary tract (NST) in the brainstem. Efferent neurons in the NST signal to regulate food intake by stomach emptying. Signals from adipose tissue (e.g., leptin) reach the neurons in the arcuate nucleus (ARC) in the hypothalamus, which in turn activate neurons in the paraventricular nucleus (PVN) and triggers signals to the NST that also regulate food ingestion. The vagus nerve afferents relay complex satiety signals that are gated by CCK (see text for further details). There is also CCK8-mediated signaling from nutrient-responsive CCK-producing neurons of the NST that innervate the PVN (not shown here). This figure is based on, but redrawn from, Morton et al. [286] and Dockray [47]
A direct central action of CCK in satiety has also been suggested. It was for instance shown that microinjection of CCK into the dorsomedial hypothalamus in rats leads to a reduced food intake and is mediated by the CCKR2 [59]. In mice, the neuronal substrate for this CCK8-mediated satiety signaling includes nutrient-responsive CCK-producing neurons of the NST that innervate the paraventricular nucleus of the hypothalamus [60]. Activation of these CCK neurons generates a long-term effect on appetite and reduction of body weight. In contrast, the CCK feedback action via the VAN/brainstem is associated with a short-term effect on food intake.
Further roles of CCK associated with feeding and metabolism include local actions in the intestine to decrease gastrointestinal motility, stimulate secretion of pepsinogen, inhibit gastric acid secretion, stimulate gallbladder contraction, and trigger secretion of hormones in the endocrine pancreas (reviewed in [18, 19, 61]). CCK peptides also stimulate secretion of calcitonin, insulin, and glucagon that are important regulators of metabolic homeostasis (see [19]).
Some evidence for a role of CCK in reproductive behavior is available. There is a sexual difference in neuronal CCK distribution in the bed nucleus of the stria terminalis and in the amygdala in rats [21]. This sexually dimorphic distribution of CCK neurons is in regions that are targets of steroid sex hormones and that are known to regulate aspects of reproduction [21]. This suggests that CCK is involved in a central integration of sensory input and steroid hormone action to modulate reproductive behavior [21]. Furthermore, it was reported that intraperitoneal administration of CCK-8 inhibits lordosis behavior (a mating response) in female rats, but not in males [62, 63].
A role of CCK in aggression in rats and mice has also been suggested. CCK-expressing neuronal projections have been identified within the limbic system, the brainstem, and the cerebral cortex, areas known to overlap with neuronal pathways that are involved in the modulation of fear, anxiety, and aggression (see [64]). It was shown that CCK signaling through the CCK1R in the brain induces hyperactivity and aggression [65]. Furthermore, overexpression of CCK2R in the mouse brain increases aggressive behavior, whereas mice lacking CCK2R display increased exploratory behavior and reduced anxiety [65, 66]. Other-CCK mediated regulatory functions in the brain have been discovered: in sleep, nociception, memory and learning processes, panic and anxiety (see [46, 67, 68]).
CCK/sulfakinin signaling components in invertebrates: a general overview
We now turn to signaling with CCK-type peptides, or SKs, in various invertebrate species. In this section, we describe general features of SKs, the structures of their precursors and SK receptors. We also point out structural similarities to vertebrate CCK and gastrin signaling components and evolutionary aspects of this signaling system. Finally, we discuss the distribution of peptides and receptors and their functional roles in invertebrates. A more detailed description of SK (DSK) signaling in Drosophila is given in Sect. 4, where we also highlight how DSK signaling serves to modulate competing behaviors.
Structure of invertebrate SK precursors and peptides
The first invertebrate CCK-type peptide was isolated from cockroach head extract and has the sequence EQFEDYGHMRFamide, which is highly similar to mammalian gastrin and CCK (see Fig. 1), including a sulfated tyrosine [24]. This peptide was designated leucosulfakinin (LSK). A second, pyroglutamate blocked, LSK (LSK-II, pQSDDYGHMRFamide) was identified soon after [69]. The first invertebrate gene encoding a SK precursor was cloned in Drosophila in 1988, and can give rise to the peptides DSK1, DSK2 and DSK0 [70] and later two DSK receptors (CCKLR1 and CCKLR2) were identified and characterized [42, 43].
Now CCK-type peptides have been identified in numerous invertebrate species of several bilaterian phyla, including the protostome phyla nematodes, mollusks, nemerteans, brachiopods, phoronids, annelids, tardigrades and arthropods [13–15, 24–31, 71–73], as well as in deuterostome invertebrates such as echinoderms and invertebrate chordates [32, 33]. Examples of SK sequences from different taxa are shown in Fig. 1. In all studied invertebrates, with the exception of spiders, there is only one gene encoding SK precursors [74]. In spiders, but not scorpions and mites, there are two genes encoding SK precursors [74, 75]. One of these spider precursors encodes two SK paracopies, and the other just a single SK. The organization of select CCK/SK precursors is shown in Fig. 2.
Interestingly, it seems that the presence of SKs is variable among species of certain taxa. For instance, among arthropods, SKs have not been identified in quite a number of studied beetles [76], and are lacking in the genomes of some parasite wasps [77, 78], in pea aphids [79, 80], in a stick insect, Carausius morosus [81], the Asian citrus phyllid Diaphorina citri (hemiptera) [82] and in a spider mite [83]. Interestingly, however, another mite, the house dust mite, does produce SKs [74]. It should be mentioned here that in cases where it has been investigated the lack of SK peptides is correlated with a lack of cognate CCK-type receptor. An exception to this is seen among the Xenacoelomorphs, a group that may have branched off from the other bilaterian phyla (Nephrozoa) early in evolution or constitute a sister group of Ambulacraria (see [84]). In this group SKs have not been identified, but two clades, Xenoturbella and Nemertodermatida, possess SK-type receptors, whereas the Acoela do not [29].
An important question to ask is whether other neuropeptides functionally replace SKs in those species that lack them, or whether the functional roles played by SKs are simply missing. Since SKs seem to play major roles in satiety signaling and regulation of feeding and digestion one might suspect that lack of SKs could reflect specific feeding habits. This seems not to be the case at least in beetles (coleopterans) where 31 species were analyzed of which 13 lack SKs [76]. When comparing the presence of SKs to life history parameters, including dietary behavior (herbivorous, mixed, predacious, saprophagous and xylophagous), no obvious correlation was found [76]. In the same study the authors also found no correlation between lack of SK (and some other neuropeptides) and other life history parameters or taxonomic relatedness of species. A few other neuropeptides are also lacking in several arthropod species, such as for example adipokinetic hormone/corazonin-related peptide (ACP), allatotropin, corazonin and leucokinin (see [16, 76, 82, 83, 85]). Thus, it remains to be understood why these losses have occurred and what the functional consequences are.
There are SK sequence entries for 116 insect species in the DINeR insect neuropeptide database (http://www.neurostresspep.eu/diner [86]). These entries reveal that insects commonly have two SKs, although several have only one form (e.g., Locusta migratoria, Bombyx mori and Manduca sexta), whereas the giant springtail Tetrodontophora bielanensis has three (see [87]). Also the shrimp Penaeus monodon has three bona fide SKs [88]. In Drosophila, melanogaster and other Drosophila species the precursor encodes a third shorter peptide (DSK0), whose sequence barely resembles a bona fide DSK ([70], DINeR database) and the presence of processed DSK0 in tissue has not been confirmed by mass spectrometry (see [89]). As mentioned, SKs are missing in some insect taxa, but since already the most basal apterygote insects possess SKs [87], it is suggestive that lack of SKs in various species is the result of a secondary loss.
Also in invertebrates in general, the precursors give rise to two SK paracopies (isoforms) in each species (see Fig. 2). Often, like in the starfish Asterias rubens [32] and several insect species such as for instance the cockroach P. americana [90] and bed bug Cimex lectularius [91], two SKs can be identified on the precursor, but mass spectrometry reveals that these exist both in sulfated and non-sulfated forms, suggesting the presence of four structural isoforms. A recent study of the locust Schistocerca gregaria found that the two paracopies SK1 (12 residues) and SK2 (27 residues) can be identified by mass spectrometry of dissected tissues in three forms each: SK1 as sulfated with N-terminal pQ (pyroglutamate), nonsulfated with pQ and sulfated without pQ, and SK2 as sulfated with pQ, nonsulfated with pQ and a truncated sulfated form (9 residues) [92]. In C. elegans the NPL-12a and 12b peptides only display weak sequence similarities to CCK (see Fig. 1), but activate two GPCRs related to CCK receptors (T23B3.4 and Y39A3B.5) [25, 93], suggesting the presence of CCK-type signaling in this worm.
In insects the SKs are C-terminally amidated and commonly between 9 and 17 amino acids long, and in many cases a species has one short and one longer form (see DINeR Database). However, with the exception of S. gregaria SK2 with 27 residues, there seems not to be any drastically extended SK forms in invertebrates, in contrast to mammalian CCK33, CCK58 and gastrin34. As noted above, some of the SKs are blocked with an N-terminal pyroglutamate (pQ), which adds a resistance to certain peptidases (e.g., in L. maderae, A. rubens, S. gregaria and Tribolium castaneum [32, 69, 92, 94]). The SKs have a conserved tyrosine (Y) residue that needs to be sulfated for full bioactivity of the peptide [90, 95, 96]. Curiously, in a bivalve mollusk, Crassostrea gigas, in the white shrimp Litopenaeus vannamei and the invertebrate chordate Ciona intestinalis, one of the SKs was identified with two sulfated tyrosines [33, 71, 97]. Several investigations of insects have found biological activity also for non-sulfated SKs (see e.g., [98–101]). It should be noted that this activity was detected in assays different from the ones used in canonical SK signaling in insects (see Table 1) and it is not known whether these SKs act on the bona fide SK receptors. In summary, the active core sequence in insect SKs for canonical bioactivity is Y(S03H)GHMRFamide [95, 96], which is well conserved also among other arthropods. As seen in Fig. 1, the sequences of other invertebrate SKs vary somewhat.
Table 1.
Functions of sulfakinins in invertebrates
| Species | Cells/neuronsa | Functionb | Reference | |
|---|---|---|---|---|
| Insects | ||||
| Drosophilac | IPCs or all DSK neurons | Decreases food intake, starvation resistance and dilp2 expression | [121] | |
| Drosophilac | IPCs or all DSK neurons | Decreases food intake, increases male aggression | [140] | |
| Drosophilac |
All DSK neurons CCKLR-17D1 |
Increases male aggression, promotes social dominance | [44] | |
| Drosophilac | IPCs or all DSK neurons | Increases male aggression (induced by social isolation) | [141] | |
| Drosophilac |
MP1/MP3 neurons CCKLR-17D3 |
Suppresses male sexual arousal | [23] | |
| Drosophilac | MP1/MP3 neurons | Suppresses sugar gustation and mediates satiety | [45] | |
| Drosophila | Peptide injections | Larval odor preference and locomotion | [98] | |
| Nilaparvata lugens |
Peptide injection SK knockdownd |
Suppresses sugar gustation and mediates satiety | [45] | |
| Nilaparvata lugens | Peptide injection | Reduces digestive enzyme activity | [142] | |
| Tribolium castaneum | Peptide injection and SKR2 knockdownd | Decreases food intake | [104, 105, 113, 143] | |
| Schistocerca gregaria | Peptide injection | Decreases food intake | [144] | |
| Gryllus bimaculatus | SK knockdownd | Decreases food intake | [145] | |
| Blattella germanica | Peptide injection | Decreases food intake | [146] | |
| Rhodnius prolixus | Peptide injection | Decreases food intake | [116] | |
| Rhodnius prolixus | SK and SKR knockdownd | Decreases food intake | [109] | |
| Phormia regina | Peptide injection | Decreases carbohydrate intake | [147] | |
| Periplaneta americana | Electrophysiology and SK application | Inhibits DUM neurons antago-nistically to AKHe | [118] | |
| Locusta migratoria | Peptide injection | Inhibits digestive enzyme secretion | [135] | |
| Rhynchophorus ferrugineus | Peptide injection | Stimulates release of α-amylase | [136] | |
| Leucophaea maderae | In vitro assay | Myostimulatory on hindgut | [24] | |
| Zophobas atratus | Peptide injection | Fatty acid composition and ILP levels in oenocytes, | [137] | |
| Tenebrio molitor | Peptide injection | Carbohydrate and ILP levels in hemolymph of larvae | [100] | |
| Other invertebrates | ||||
|
Homarus americanus (lobster) |
In vitro assay | Stimulates heart contractions | [148] | |
|
Asterias rubens (starfish) |
Peptide injection | Inhibits feeding and triggers stomach retraction | [32] | |
| Crassostrea gigas (oyster) | In vitro assay | Inhibits contractions in hindgutf | [71] | |
| Pecten maximus (scallop) | Peptide injection | Stimulates release of α-amylase | [136] | |
| C. elegans | Genetic manipulation | Modulates food-related behavior | [27] | |
aCells/neurons where SK signaling was manipulated. In other cases global action was assayed by various techniques
bFunction shows result of increased SK signaling
cFunction in Drosophila assessed by various genetic manipulations
dInjection of dsRNA
eInhibits octopaminergic dorsal unpaired median (DUM) neurons that regulate foraging activity antagonistically to the orexigenic peptide AKH
fAlso suggested that SK regulates feeding
Typically, only one gene encoding a CCK/gastrin type precursor has been found in each species of protostome invertebrates [20], as well as in the deuterostome invertebrates of the Ambulacraria (echinoderms and hemichordates) [32] and in invertebrate chordates, like the ascidian Ciona intestinalis [102]. Thus, a duplication of an ancestral CCK/gastrin gene that gave rise to separate genes encoding CCK and gastrin in the vertebrates seem to have occurred before the emergence of cartilaginous fish [52]. More specifically, synteny analysis has shown that this duplication arose through a whole genome duplication event (the second one known, 2R), probably before the emergence of cyclostomes [103].
Invertebrate SK receptors
Two DSK receptors (CCKLR1 and CCKLR2) were identified and characterized in Drosophila [42, 43] and later orthologs were characterized in Tribolium castaneum [104, 105]. These were followed by detection of numerous SK receptors (SKRs) in other invertebrates (see [20, 106]). Thus SKRs have been identified in several insects, as well as in for instance C. elegans, the water flea Daphnia pulex, a spiny lobster Panulirus argus, the sea squirt Ciona intestinalis, as well as the echinoderms Strongylocentrotus purpuratus and Asterias rubens (see [20, 25, 102, 107, 108]). However, cloning and characterization of SKRs has been performed only for a more limited number of species: Drosophila, T. castaneum, Rhodnius prolixus, A. rubens, C. elegans and C. intestinalis [20, 25, 32, 102, 105, 109]. Like in mammals and Drosophila, several insects and other invertebrates have two CCK-type receptors, or SKRs. However, in some only one SKR could be found in the genome: for instance brown planthopper Nilaparvata lugens, silkworm Bombyx mori and American cockroach P. americana, (see [20]). Also the starfish A. rubens seems to have only one SKR [32]. In cases that have been investigated, it appears that in insects that lack SK peptides also no SKR can be found in the genome, for example in pea aphid Acyrthosiphon pisum, the parasitic wasp Nasonia vitripennis and the parasitic nematode Meloidogyne incognita [20].
In Drosophila, sulfated DSK-1 and DSK-2 can both activate the two receptors CCKLR1 (CCKLR-17D3; CG32540) [42] and CCKLR2 (CCKLR-17D1; CG42301) [43] in different in vitro expression/assay systems. However, DSK-0, and the non-sulfated DSK-1 and DSK-2 cannot active the receptors at physiological concentrations. The signaling downstream of the CCKLRs in Drosophila has been analyzed to some extent. DSK/CCKLR2 regulation of larval neuromuscular junction growth was found to be mediated by the cyclic adenosine monophosphate (cAMP)—protein kinase A (PKA)—cAMP response element binding protein (CREB) pathway [110]. In the in vitro assays CCKLR1 coupled to both the Gq/G11 and Gi/Go signaling pathways [42] and the SK receptors of T. castaneum stimulated both the Ca2+ and cyclic AMP second messenger pathways [111]. Ligand-receptor interaction characteristics were modeled for the T. castaneum SKRs (TcSKR1 and TcSKR2) [112] and the structure–activity properties of different SKs were monitored in the cockroach hindgut contraction assay and it was found that the C-terminal heptapeptide DYGHMRFamide with sulfated tyrosine and amidation is critical for activity [95]. Using the flour beetle T. castaneum as a model, stable agonists and antagonists of SK were developed, injected and found effective on food intake [113].
As we shall see below, the two SKRs appear to be differentially distributed and contribute to distinct functions in invertebrates (in those few species that have been studied).
Distribution of SK peptides and receptors in invertebrates
Peptide distribution
In general, the cellular SK peptide expression in the nervous system is conserved among studied insects, but the distribution in other arthropods has not been described in detail. Thus, it is hard to make wide comparisons even among the arthropod taxa. Also in the other invertebrates, data on the cellular SK distribution is scarce.
In insects SK expression is primarily seen in a small number of neurons and neurosecretory cells with cell bodies in the brain [114–118]. Generally, SK has not been found in neuronal cell bodies of the ventral nerve cord (VNC), or in enteroendocrine cells (EECs) of the intestine. One exception is the mosquito Aedes aegypti where SK immunoreactivity was detected in EECs of the midgut [119]. In the insect brain, the number of SK-producing neurons is small: for example, about 26 neurons have been detected in R. prolixus [116], 20–24 in Drosophila (Fig. 4) [23, 44, 114, 120, 121], and about 30 neurons in P. americana [117, 118]. In all these species the numbers of neurons include small sets of neurosecretory cells in the pars intercerebralis, with axon terminations in the corpora cardiaca and neurohemal areas on the anterior aorta, crop and foregut, suggesting roles of SKs as circulating hormones (or local modulators of endocrine cells or muscles). In Drosophila, it was shown that the DSK-expressing pars intercerebralis neurons are a subpopulation of the 14 insulin-producing cells (IPCs) [121, 122]. It cannot be excluded that the axon terminations of the DSK/DILP-containing IPCs on the crop release peptide that acts on crop muscle, as was shown for the peptide myosuppressin released from similar neurosecretory cells [123, 124]. All studied insect species appear to have one or two pairs of large SK-expressing neurons with cell bodies and extensive processes in the brain and axons descending throughout the VNC (see [23, 114–116, 118, 125]).
Fig. 4.
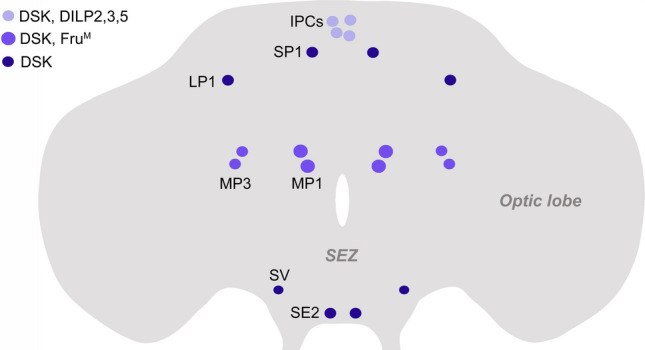
Distribution of cell bodies of neurons expressing DSK in the Drosophila brain. Four of the insulin-producing cells (IPCs) co-express DSK (the other IPCs are not shown). The MP1 and MP3 neurons express the male splice form of the transcription factor fruitless (FruM). The designations of the DSK neurons are based on [114, 287, 288]
The available data on the cellular distribution of SK in insects suggests a rather unique pattern compared to many other brain neuropeptides (see [16]). In addition to being excluded from gut EECs, SK is expressed in a few neurons with wide arborizations that invade neuropils interspersed between the so-called structured neuropils (also known as glomerular or columnar/layered neuropils). One exception is the DSK processes invading the lobula of the optic lobe in Drosophila, which is a columnar neuropil. Thus, there are no reports of SK-expressing neuronal branches in the central complex, the mushroom bodies, the antennal lobes or the rest of the optic lobe of insects (see Fig. 5). Also, there are no reports on clock neurons expressing SK. The arborizations in the Drosophila lobula are diffuse and irregular and derived from the two MP1a neurons (each neuron with branches bilaterally in both lobulae) (Fig. 5c, d). In Drosophila and other insects, two pairs of brain neurons have axons that descend throughout the VNC [23, 44, 114, 115, 125]. In Drosophila, these are the MP1a and MP1b neurons [44].
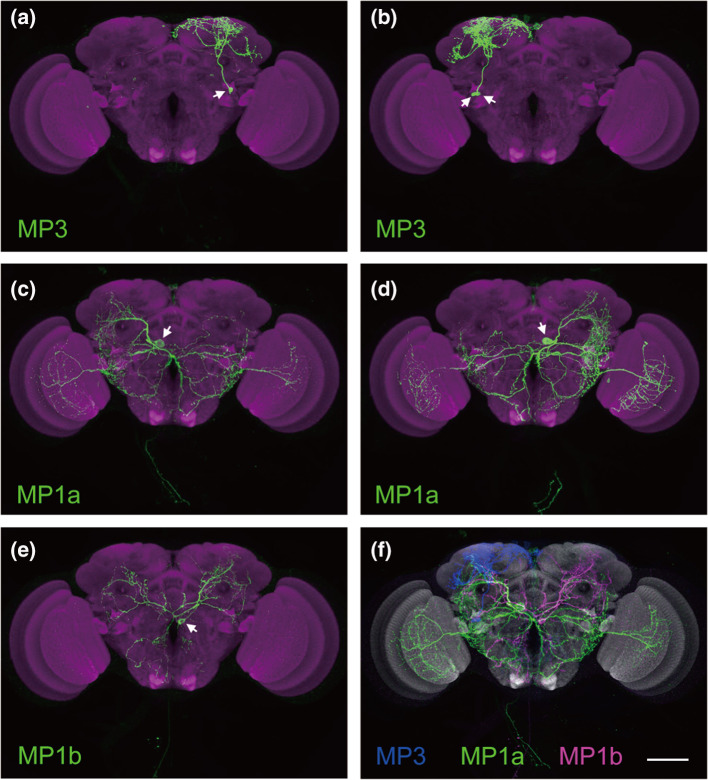
Fig. 5.
Single-neuron labeling of DSK-producing MP neurons. a–f. Stochastic labeling of single MP3 (a), two MP3 (b), single MP1a (c and d) and single MP1b (e) neurons. Two MP3, one MP1a and one MP1b neurons are registered in a standard brain (f). Scale bars, 50 μm. Note that the MP1/MP3 neurons do not innervate central complex, antennal lobes or mushroom bodies. Figure from Wu et al. [23], with permission
There are other neuropeptides/peptide hormones in Drosophila that can be seen only in neurons with processes in non-structured neuropils. These are CAPA-PK, CCAP, DH44, GPB5, hugin-PK, DILPs, ITP, and LK (see [16], and furthermore they are not expressed in gut EECs [126]. Three of these peptides, DH44, hugin-PK, ITP, are however, also present in clock neurons [127, 128]. Interestingly the peptides listed above are known to regulate metabolism, feeding and/or water and ion homeostasis in adult flies (see [16]. In summary, it appears that in insects SK is not a brain–gut peptide, in contrast to many other peptides (see [1, 129, 130]). However, SK is likely to function both as a local neuromodulator and a circulating hormone. We will describe the detailed anatomy of the Drosophila DSK neurons in relation to their functions in a later section (Sect. 4).
In crustaceans, there is scarce information about cellular distribution of SK. In the shrimp Penaeus monodon, SK-immunolabeled neuronal cell bodies were seen only in the brain [88]. One pair of large cell bodies give rise to descending axons running throughout the VNC, similar to those in insects, and there are six to eight additional smaller brain neurons [88]. SK was detected in axon terminations of the neurohemal organ in the eyestalks designated the sinus gland [131] and in the neurohemal pericardial organ [132] of the crab Cancer borealis. CCK-type peptide was furthermore identified in neuronal processes in the stomatogastric nervous system (STN) of this crab [133, 134]. The STN is known to house neuronal circuits that regulate rhythmic movements of elements in the internal feeding apparatus such as the gastric mill (for chewing) and pylorus (for pumping and filtering of chewed food). The distribution of CCK-type peptide suggests that it may act both as a neuromodulator in local STN neurons, and as a circulating hormone to regulate rhythm-generating networks utilized during feeding and food processing [133, 134]. However, although several neuropeptides/peptide hormones have been investigated for their modulatory actions in the circuits of the STN, the function of CCK-type peptide remains to be tested.
In C. elegans the CCK-type neuropeptide NPL-12 is localized in one tail neuron with its cell body in the dorso-rectal ganglion, and was identified as the ring interneuron DVA [25]. This neuron connects to the SMB motoneuron that regulates head movements during feeding and ventral cord motoneurons controlling body wall muscles and thereby locomotion (see [27, 93]). The ventral cord motoneurons express the CCK receptor CKR2 and in presence of food the worms dwell and feed, whereas in absence of food they disperse under control of this circuit, which also includes sensory cells, as well as dopaminergic (PDE) and other peptidergic (AVK) neurons [27].
In the starfish A. rubens (Echinodermata) the CNS is organized in a radial fashion with a circumoral nerve ring and radial nerve cords. SK peptide was detected by in situ hybridization and immunohistochemistry to neuronal cell bodies and axonal processes in the so-called ectoneural and hyponeural regions of the CNS [32]. The neurons in the ectoneural region are predominantly sensory neurons and interneurons, whereas the hyponeural region houses motoneurons [32]. Furthermore these authors found SK neurons in the digestive system (esophagus, peristomial membrane, cardiac stomach, pyloric stomach, pyloric duct, pyloric caeca, intestine, and rectal caeca), tube feet and body wall. SK peptides were shown to trigger stomach retraction and inhibition of feeding in the starfish [32].
SK receptor distribution
The general distribution of SK receptors in major tissues has been assayed in some insects by PCR [100, 104, 105, 109]. There are no data for the two DSK receptors in tissues of Drosophila in FlyAtlas, probably due to low expression levels. The cellular distribution of SK receptors has to our knowledge only been investigated in Drosophila [23, 44, 45], the cockroach P. americana [118], and in the starfish A. rubens [32], but not in other invertebrates.
In Drosophila, different knock-in GAL4s into the gene loci of CCKLR-17D3 and CCKLR-17D1 were utilized for displaying the distribution of the DSK receptors [23, 44, 45]. The two receptors appear to be largely differentially expressed, but detailed analysis was not performed to screen for any colocalization or for detailed neuronal expression (but see below). Both receptors display widespread distributions in numerous neurons in the brain and VNC [23, 44, 45]. CCKLR-17D1 is expressed in neurons of the optic lobe, fan-shaped body of the central complex, SEZ and numerous protocerebral neurons [44]. CCKLR-17D3 is expressed in fan-shaped body of the central complex, mushroom bodies and in a smaller number of neurons scattered in the protocerebrum and SEZ [23, 44]. Furthermore, it was found that CCKLR-17D3 is expressed in subsets of gustatory receptor neurons (GRNs) in the proboscis and proleg tarsi that house the sweet sensing gustatory receptors Gr64f [45]. Recent work showed that CCKLR-17D1 is needed for modulation of aggression [44], whereas CCKLR-17D3 in suppressing sexual arousal and in sweet sensing [23, 45].
In the cockroach P. americana SK receptor protein was detected in octopaminergic dorsal unpaired median (DUM) neurons of the VNC and it was found that as a satiety signal SK depresses activity in these DUM neurons antagonistically to the orexigenic peptide AKH [118].
In the starfish A. rubens, the distribution of the single SK receptor was localized by immunocytochemistry to neurons in the CNS, tube feet, apical muscle, and digestive system [32]. Close apposition between SK peptide and SK receptor was seen in the ectoneural neuropil in the circumoral nerve ring and radial nerve cords. These authors furthermore found that some SK producing neurons also expressed the SK receptor.
Functional roles of SK signaling in invertebrates
As seen in Table 1, SK signaling has been investigated in several species of insects, as well as in a few other invertebrates. Since Drosophila and the brown planthopper Nilaparvata lugens have been studied in some more detail we will describe DSK functions in these insects separately in the next section.
In several insect species injection of SK peptide and double stranded RNA for RNA-interference (RNAi) to knock down SK and SKR have been utilized to show that SK signaling decreases food intake (references in Table 1). DSK injections furthermore decrease secretion of digestive enzymes in a locust [135], but increased α-amylase secretion in the palm weevil Rhynchophorus ferrugineus [136]. Other effects of SK determined after injections or by in vitro assays are stimulation of gut muscle contractions [24], effects on fatty acid-, carbohydrate- and insulin-like peptide levels in beetles [100, 137]. In the ascidian Styela clava mammalian CCK8 peptide acts to stimulate gastric enzyme secretion [138] and in another ascidian C. intestinalis the distribution of SK receptor in the siphon and ovary suggests a role for CCK-type signaling in feeding and reproduction [102].
Could loss of SK signaling in some insects be correlated with altered physiology and/or behavior? Some papers suggest so. For instance, SK/SKR could not be found in the genomes of the peach aphid Myzus persicae and pea aphid Acyrthosiphon pisum [139]. Since SK/SKR plays an important role in feeding behavior and aphids excrete honeydew, which results in a substantial loss of energy, it was proposed that loss of the SK signaling system might lead to increased food intake to compensate for the energy loss [79]. Another suggestion is that a specific insect lifestyle could result in loss of SK signaling since parasitic wasps lack SK/SKR, whereas the social honeybee A. mellifera does not [78]. However, as mentioned above in an earlier section, lack of SK signaling could not be correlated with differences in feeding behaviors and other life history parameters or taxonomic relatedness of species in a study of 31 species of beetles 13 of which lack SKs [76]. Thus, it remains to be clarified whether a loss of SK signaling reflects behavior and physiology or if other peptidergic systems take over the role of SKs.
Multiple functional roles of CCK-type signaling in Drosophila
Since DSK signaling in Drosophila has been quite well investigated at the cellular level, we provide a more detailed description here. We first outline the anatomy of the DSK neurons and the circuits they are part of. These circuits modulate aggression, courtship behavior, taste, foraging and feeding and seem to integrate multiple inputs from external and internal sensors. The studies reviewed here pertain to adult Drosophila males if nothing else is stated.
DSK signaling in Drosophila
In the Drosophila brain there are 20 distinct DSK-expressing neurons (Fig. 4) and a small number of additional neurons that are less consistently seen with immunolabeling and Gal4-expression [23, 44, 45, 114]. No DSK-producing neuronal cell bodies were detected in the ventral nerve cord or intestine, although brain-derived axonal processes can be seen in the ventral nerve cord [44, 114]. The major types of DSK neurons that we will discuss here are a small subset of the IPCs in the pars intercerebralis [121, 122], and four pairs of median posterior neurons designated MP1 and MP3 [23, 44, 114]. The functions of the other DSK neurons are not yet known.
It is not clear how many of the 14 DILP-producing IPCs that co-express DSK, since the DSK expression is variable, but at least four cells can consistently be seen in adults [121]. The IPCs send axons to the retrocerebral complex (corpora cardiaca–corpora allata and hypocerebral ganglion) as well as the anterior aorta, foregut and crop and additionally have branches (presumed dendrites and/or peptide release sites) in pars intercerebralis, tritocerebrum and SEZ (see [149, 150]). DSK-immunolabeling can be seen in these brain processes, but the peptide distribution in the other sites has unfortunately not been examined in Drosophila. However, in the American cockroach, the corresponding neurons have SK expressing axon terminations in the corpora cardiaca–corpora allata [117].
The MP1 and MP3 neurons are of three distinct types shown in Fig. 5 [23, 44]. Each of the two pairs of MP3 neurons has wide ipsilateral arborizations dorsally in one brain hemisphere (Fig. 5a, b). The MP1a neurons are bilateral with branches in the lobula and ventrolateral brain neuropils (including subesophageal zone, SEZ) in both hemispheres (Fig. 5c, d), as well as axons descending into the VNC where they innervate the accessory mesothoracic neuropil. MP1b neurons are bilaterally supplying branches to the midbrain and SEZ (Fig. 5e) and have axons to the VNC. Together these DSK neurons innervate a substantial volume of the brain (Fig. 5f), but avoid the prominent centers such as the mushroom bodies, central complex and antennal lobes. Also the lamina, medulla and lobula plate of the optic lobe are devoid of DSK processes.
As seen in Fig. 6, the DSK-producing IPCs and the MP1/MP3 neurons are in male flies part of circuits regulating sugar sensing, feeding, daily activity, aggression and courtship behavior (females have not been specifically studied). Furthermore, this figure shows that the DSK expressing neurons are of different functional types (and molecular set-up). Thus, the IPCs express the octopamine receptor OAMB and are under influence of octopaminergic neurons [140, 151, 152], whereas the MP1/MP3 neurons express the male splice form of the transcription factor Fruitless (FruM) and are connected reciprocally to the P1 neuron cluster [23, 44]. Both of these types of DSK neurons receive direct or indirect signals reporting nutritional status and the MP1/MP3 neurons are additionally influenced by other internal and external factors (age and housing conditions) [23, 45, 153]. It can be noted that the IPCs are furthermore regulated by a number of other neurotransmitters, neuropeptides and systemic inputs that will be discussed in a later section. The other DSK neuron types have not been specifically investigated. We shall get back to this figure in the next section in the context of functional DSK circuits.
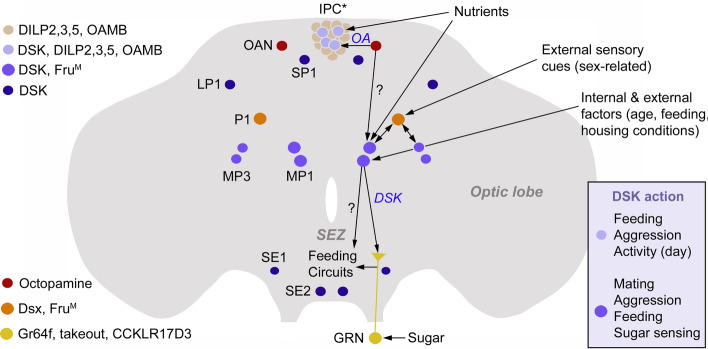
Fig. 6.
DSK-expressing neurons in the Drosophila brain and their interactions with other neurons in regulation of behavior and physiology. Of the DSK expressing neurons shown, only the MP1, MP3 and a subpopulation of insulin-producing cells (IPCs) in the pars intercerebralis have been specifically implicated in regulation of feeding/metabolism, aggression and mating behavior. Their functions are shown in the box with DSK action. The functional roles of the remaining (dark blue) neurons are not yet known. Other neurons shown are a pair of octopaminergic neurons (OAN), a pair (only a pair from a larger neuron cluster is shown) of fruitlessM (FruM) and doublesex (Dsx) expressing neurons (P1) and a sweet-sensing gustatory receptor neuron (GRN) that expresses the gustatory receptor Gr64f, takeout and the DSK receptor CCKLR-17D3 (CCKLR). The OANs regulate activity in IPCs, probably including the ones expressing DSK, thereby regulating satiety (feeding), aggression and daily activity [140, 151]. It is not known if the OANs also interact with the MP1/MP3 neurons (indicated by ?). The MP1/MP3 neurons act on the P1 neurons to suppress male sexual behavior [23]. The P1 neurons also regulate activity in MP1/MP3 neurons to modulate male aggression, and MP1/MP3 neurons are postsynaptic to P1 neurons as shown by trans-tango [44]. Furthermore, the MP1/MP3 neurons receive inputs mediating internal and external cues about age, metabolic status, and housing conditions and thereby regulate mating, feeding and sugar sensing [23, 45]. After food intake, the MP1/MP3 neurons suppress activity in the Gr64f-expressing GRNs to diminish sugar sensitivity and thereby food search and feeding [45]. The GRNs signal to circuits that regulate motivation to feed (Feeding Circuits)
As mentioned earlier, the distribution of the two DSK receptors CCKLR-17D1 and CCKLR-17D3 has been mapped in the Drosophila CNS by Gal4-driven fluorescence [23, 44]. The distribution has not been described at the cellular level though, except that a subset of the gustatory receptor neurons (GRNs) in the proboscis and proleg tarsi that also express the sugar receptor Gr64f have co-localized CCKLR-17D3 [45]. In male flies a subset of the P1 neurons also express CCKLR-17D3 [23]. Generally, the distribution of the two DSK receptors appear to match that of DSK producing neurons processes, but they can also be seen in processes in the fan-shaped body of the central complex (17D1 and 17D3), in the optic lobe (17D1 and 17D3), and in the mushroom body lobes (17D3) [23, 44, 45]. This reflects the Gal4-driven GFP expression in the receptor expressing neurons and not necessarily the distribution of receptor protein; the polarity of these neurons in terms of dendrites and axon terminations/output sites needs to be resolved.
Aggression and courtship behavior
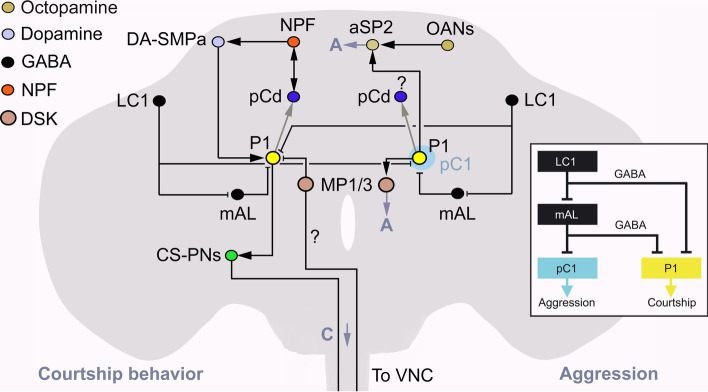
In male flies, the MP1 and MP3 neurons express FruM and receive inputs from the male-specific P1 neurons that also express FruM [23, 44] (Fig. 6). These DSK neurons are thus part of a circuit that regulates male-specific behavior. The P1 neurons constitute a heterologous set of about 20 neurons that express FruM and are known to integrate chemosensory and other cues from potential mates and control sexual arousal, or cues from other males and regulate aggression [154–160]. Thus, the P1 neurons weigh sensory inputs to control two opposing behaviors. Interestingly, the MP1/MP3 neurons play important roles in this circuitry by being postsynaptic to a subset of the P1 neurons and acting on FruM positive neurons that express the DSK receptor CCKL-17D3 for sexual arousal and CCKL-17D1 for aggression [23, 44]. In Fig. 7, we show a simplified scheme of the circuitry that regulates aggression and courtship behavior, including the MP1/MP3 neurons. These circuits are interconnected by GABAergic neurons that form a switch between courtship and aggression [157]. In Fig. 6, we show that P1 neurons and some DSK neurons receive inputs from the external and internal environment that provide cues for activation of specific circuits.
Fig. 7.
Neuronal circuits that regulate courtship behavior and aggression in Drosophila. In this simplified scheme, single neurons are shown even when multiple neurons are involved (e.g., P1, pC1, MP1/3 neurons). Circuits to the left regulate courtship behavior and those to the right aggression. Arrows show activation and stop bars inhibition. The gray arrows indicate indirect action via other neurons. The double arrows indicate a stimulatory recurrent circuit that involves pCD and NPF neurons that is known to sustain courtship motivation [167]. The neurotransmitters and neuropeptides utilized by some of the neurons are shown in the upper left corner. This scheme does not include the sensory inputs to P1 neurons and other neurons that mediate various signals from conspecific male and female flies. The P1 neurons are a subpopulation of the double-sex-expressing pC1 neurons (light blue circle), as shown to the right [157]. Approximately 20 P1 neurons (FruM-expressing) are central in initiating courtship, whereas an unspecified number of P1/pC1 neurons trigger aggression [157, 161–163]. The DSK-expressing MP1/3 neurons are functionally associated with P1 neurons in both behaviors [23, 44]. Importantly, the Fru-expressing LC1 and mAL neurons use GABA to switch P1 (or P1/pC1) neuron activity between courtship and aggression [157]. Thus, LC1 neurons are shown here schematically as interconnecting P1 and P1/pC1 circuits reciprocally in the two behavior circuits. In the real fly, this circuit is present in each hemisphere as shown in the inset. There is an inhibitory recurrent signal from copulation-reporting neurons in the abdominal ganglion to brain NPF neurons (not shown here). In the VNC, another set of abdominal neurons utilize GABA, glutamate and corazonin to regulate copulation under modulation by dopamine. In aggression P1 neurons and octopaminergic neurons (OANs) converge on aSP2 neurons to promote aggression. Like in the courtship circuitry, pCd neurons ensure sustained aggression (A), but it is not known whether also here the NPF neurons are involved (indicated by ?). For further details, see the text. This figure is based on a figure in Lee and Wu [289] and is redrawn and updated with MP1/MP3 circuits
The P1 neurons are a subpopulation of the doublesex-expressing pC1 neurons [157]. About 20 P1 neurons receive various sensory cues from female flies and are central in initiating courtship, whereas an unspecified number of P1/pC1 neurons trigger aggression based on sensory cues from other males or non-conspecifics [157, 161–163]. The DSK-expressing MP1/MP3 neurons are functionally associated with P1 neurons in both behaviors [23, 44]. We shall start by describing the courtship circuitry (Fig. 7, left side). In courtship, the P1 neurons are triggered by sensory inputs and are modulated by dopamine from DA-SMPa neurons [164, 165], inhibited by DSK from MP1/3 neurons [23] and indirectly, via neuropeptide F receptor (NPFR) expressing DA-SMPa neurons, by NPF [166]. In support of the action of DSK from MP1/MP3 on P1 neurons, it was shown that P1 neurons express the receptor CCKLR-17D3 [23]. A stimulatory recurrent circuit including pCD and NPF neurons is known to sustain courtship motivation [167]. P1 neurons indirectly act on pCd neurons to extend duration of courtship singing [168]. A set of Fru-expressing LC1 and mAL neurons use GABA to switch P1 (or P1/pC1) neuron activity between courtship and aggression based on sensory cues from flies of either sex [157]. The P1 neurons act on descending neurons (known as P2b/plP10 neurons) to activate courtship singing via pacemaker circuits in the thoracic neuromeres of the VNC [169]. Via various inputs the MP1/MP3 neurons monitor internal and external signals such as metabolic status, housing conditions and ageing (Fig. 6) and based on the valence of these inputs they can inhibit the P1 neurons and thereby suppress courtship behavior [23]. The MP1/MP3 neurons have axons descending into the VNC [44, 114], but whether they are involved in regulating circuits controlling courtship in these ganglia is not known. After copulation there is an inhibitory recurrent signal from copulation reporting neurons in the abdominal ganglion to brain NPF neurons that ensure that the activation of P1 neurons cease [167]. Local circuits in the VNC comprise a set of abdominal neurons that utilize GABA, glutamate and corazonin to regulate copulation motor behavior under modulation by dopamine [170]. The MP1/MP3 neurons not only inhibit sexual arousal, they also suppress wakefulness and spontaneous walking, suggesting that DSK release has a general inhibitory effect on activity [23].
In virgin female flies, DSK also inhibits reproductive behavior via the CCKLR-17D3 receptor, by diminishing receptivity to courting males [23]. Female flies lack P1 neurons, but have doublesex-expressing neurons (e.g., PC1 and pCd) that are critical for female receptivity [171]. Labeling experiments suggest that the DSK neurons are presynaptic to the sex-specific doublesex neurons. Thus, both in males and females DSK neurons interact with the sex-dimporphic doublesex neurons to suppress reproductive behavior [23]. A recent study (preprint) also reported a role of DSK in female reproductive behavior [172]. These authors found that the DSK expressing MP1 neurons act on the CCKLR-17D3 receptor to regulate virgin female receptivity to mating males, as measured by copulation rate and copulation latency. Thus, the MP3 neurons appear to play no role in this behavior [172].
In male flies, P1 neurons and octopaminergic neurons (OANs) converge on aSP2 neurons to promote aggressive behavior [173] (Fig. 7). Like in the courtship circuitry, pCd neurons ensure sustained aggression [167], but it is not known whether also here the NPF neurons are involved in the circuitry, although NPF is involved in regulation of aggression [174]. As mentioned for courtship, the LC1-mAL circuit acts to switch P1 (or pC1) neuron activity between courtship and aggression. The MP1/MP3 neurons are postsynaptic to the P1 neurons and upon activation they promote aggression by acting on neurons expressing the DSK receptor CCKL-17D1 [44]. There are additional neurons that regulate aspects of aggressive behavior that utilize NPF, tachykinin and serotonin [174–176], but their relation to the MP1/MP3 circuits are not known (and they are not shown in Fig. 7).
It can be noted that also in mammals there is an association between circuits regulating aggression, reproduction and feeding, although not necessarily involving CCK. Such circuits are found in the hypothalamus and the neuroendocrine system, and it has been shown that neurons that are activated during aggression are inhibited during mating [177]. To some extent the functional analog of the hypothalamus in insects is the pars intercerebralis [178–180]. In Drosophila, this region houses several types of peptidergic neurosecretory cells, including the 14 IPCs that produce DILPs and DSK [121, 149, 180–182]. There is some evidence that the IPCs are involved in the regulation of male aggression [140, 141, 151, 183], and there seems to be to be a role of the co-localized DSK [140, 141], but the role of DSK in IPCs in courtship has not been tested.
Satiety and feeding in Drosophila and brown planthopper
Of the DSK expressing neurons, only the MP1, MP3 and a subpopulation of IPCs in the pars intercerebralis have been specifically implicated in regulation of satiety, feeding and metabolism [45, 121, 184] (Fig. 6). Knockdown of DSK in the IPCs or all DSK neurons, or inactivation of these neurons via expression of a hyperpolarizing ion channel, leads to flies that ingest more food [121]. Interestingly, it suffices to manipulate DSK in the IPCs alone to affect feeding. The same manipulations also affect the flies’ choice of food so that they are less discriminative against bitter food [121]. It was also found that knockdown of DSK in IPCs or all DSK neurons led to an increase in dilp2, 3, and 5 transcript levels in fed, but not in starved flies. Reversely, knockdown of dilp2, 3, and 5, using a triple mutant, diminished Dsk levels in fed flies [121]. Thus, there seems to be an endocrine or autocrine feedback regulation of the two sets of neuropeptides. Another study also demonstrated that DSK knockdown in IPCs increased total amount of food ingested and number of feeding bouts [184]. That study found that octopamine increases feeding, but also Dsk transcription, suggesting a negative feedback between octopamine and DSK in regulation of feeding. Neither of these studies addressed the mechanisms by which DSK induces satiety and decreased food ingestion.
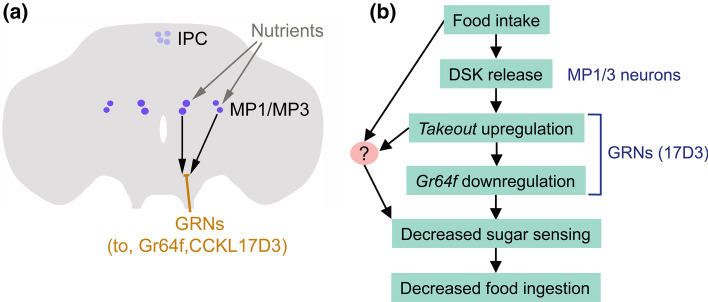
More recently, experiments on Drosophila and the brown planthopper N. lugens dissected the role of DSK signaling in satiety in some more detail [45]. This study focused on the MP1/MP3 neurons and their interactions with other neurons (see Fig. 8). Feeding upregulates Dsk transcription in the brain and more specifically induces elevated DSK immunolabeling in MP1/MP3 neurons, as well as increased spontaneous activity and calcium signaling in these neurons [45]. Thus, the MP1/MP3 neurons receive inputs from nutrient sensing neurons. Further analysis was guided by experiments in the planthopper. In this insect, an RNA-seq transcriptome analysis was performed after genetic downregulation of SK. Out of multiple genes with altered expression, a few genes of interest were found upregulated, namely those encoding sweet sensing gustatory receptor neurons (GRNs) and the takeout gene [45]. This is interesting since gustation is necessary for probing the palatability of food sources [185, 186] and takeout is important in feeding behavior of flies [187, 188]. Further experiments in Drosophila [45] showed that food ingestion downregulates the sweet gustatory receptor Gr64f (and starvation increases it). Optogenetic activation of GRNs that express Gr64f increases the flies’ motivation to feed. Furthermore, knockdown of dsk leads to an upregulation of Gr64f transcription and optogenetic activation of the DSK expressing MP1/MP3 neurons decreases the sugar sensitivity of gustatory neurons [45]. The DSK receptor CCKLR-17D3 could be found in a subpopulation of the Gr64f-expressing GRNs in proleg tarsi, proboscis and maxillary palps, and feeding dowregulates expression of this receptor in the appendages [45]. It was also found that silencing of the Dsk gene negatively regulates takeout expression and intriguingly knockdown of takeout leads to an upregulation of Gr64f expression. Thus, in summary, food intake leads to DSK release from MP1/MP3 neurons which upregulates takeout and downregulates Gr64f in CCKLR-17D3-expressing GRNs. This decreases the sugar sensing and thereby food ingestion is reduced (Fig. 8). In the planthopper similar mechanisms were revealed [45]. Thus, DSK signaling nutrient-dependently modulates the sensitivity of sweet-sensing GRNs both in Drosophila and the planthopper, suggesting a conserved peptidergic signaling pathway in these distantly related insects.
Fig. 8.
DSK regulates sugar sensitivity and feeding in Drosophila. (a) The MP1/MP3 neurons receive nutrient signals of unknown origin. Upon feeding DSK is released and inhibits responsiveness of afferent gustatory receptor neurons (GRNs) that express the DSK receptor CCKL17D3, as well as the sugar receptor Gr64f and the gene takeout (to). (b) Scheme depicting the action of MP1/MP3 neurons after food intake. It is likely that a parallel pathway acts on unknown neurons (indicated by ?) to decrease sugar sensing. Panel b is based on Guo et al. [45]
In Drosophila, it appears that the DSK-producing IPCs are not necessary for the interactions with sugar gustation and ensuing decreased feeding. This suggests that satiety signaling induced by DSK from the IPCs [121] is mechanistically distinct and further studies are required to determine whether it is by hormonal activity or maybe action on contractions of the crop, which is innervated by the IPCs. It is also important to note that the DSK/SK satiety signaling described by Guo et al. [45] is mechanistically very different from that shown for CCK in the mammalian gut—vagus nerve—hindbrain axis shown in Fig. 3. However, it might be that more similarities will be discovered in insects when compared to circuits in the brainstem and hypothalamus (see Sect. 2).
Since a subset of the IPCs produce DSK [121] it is possible that the mechanisms that are known to regulate production and release of DILPs affect the colocalized DSK in a similar way. This remains to be investigated. However, it was shown that Dilp2, 3, 5 mutant flies display decreased DSK transcript levels and also that DSK-knockdown in DSK-neurons or in IPCs leads to increased dilp transcripts [121], suggesting feedbacks between these peptidergic systems. Thus, it is possible that the intrinsic nutrient sensitivity of IPCs and the neuropeptides and neurotransmitters that regulate IPC activity also affect DSK production and release. These signaling substances are listed in Table 2 and would be of interest to investigate in relation to DSK signaling.
Table 2.
Neuropeptides, peptide hormones and neurotransmitters that regulate IPCs in adult Drosophila, some of which produce DSK
| Peptide/SMN | Source | Receptor in IPCs | References |
|---|---|---|---|
| Allatostatin A | DAR2 | [189] | |
| CCHamide2 | EECs | CCHa2-R | [190, 191] |
| DILP2,3,5 | Autocrine feedback | dInR | [192] |
| DILP6 | Fat body | dInR | [193] |
| DILP8 | Tumors | Lgr3 | [194] |
| Dopamine | Interneurons | Dopamine R1 | [195] |
| DSK | Interneurons or autocrine | Not shown | [121] |
| GABA | Interneurons | GABABR2 | [196] |
| Leucokinin | Interneurons | LKR | [197, 198] |
| Limostatin | Corpora cardiaca | Pyrokinin receptor 1 | [199] |
| NPF | EECs | NPFR | [200] |
| Octopamine | Interneurons | OAMB receptor | [151] |
| Pigment-dispersing factor | Clock neurons | PDFR | [201] |
| Serotonin | Interneurons | 5-HT1A receptor | [202] |
| sNPF | Interneurons/LNCs | sNPFR1 | [203] |
| Tachykinin | Interneurons | TkR99D (DTKR)a | [204] |
Does DSK interact with other peptidergic and aminergic systems in Drosophila?
There are several other neuropeptides involved in the regulation of feeding, aggression and courtship in Drosophila. They are listed in Tables 3, 4 and 5, and the peptide distribution in neurons involved in feeding is shown in Fig. 9. These peptides act at different levels of relevant circuits either as neuromodulators or as circulating hormones and their distribution in the brain is widespread with cell bodies in many different areas (Fig. 9). The details of the circuits involving these neurons are beyond the scope of this review and the reader is referred to the papers listed in the tables for further information.
Table 3.
Neuropeptides that regulate aggression
Table 4.
Neuropeptides that regulate mating and copulation
Table 5.
Neuropeptides that regulate food search/feeding
| Peptide | Neurons | References |
|---|---|---|
| AKH | Corpora cardiaca cells | [238, 239] |
| AstA | Interneurons, gut EECs | [189, 240–242] |
| CCHa2 | Gut EECs, fat body | [190, 191] |
| CRZ | LNCs | [243] |
| DH44 | MNCs and VNC neurons | [244–246] |
| DILPs | IPCs | [247–250] |
| DSK | IPCs, MP1/MP3 | [45, 121] |
| Hugin | SEZ neurons | [251] |
| ITP | LNCs | [252] |
| LK | Brain neurons | [197, 253] |
| MIP | Brain neurons | [254, 255] |
| NPF | Interneurons | [256–262] |
| Sex peptide | Via sperm act in females | [263–266] |
| SIFamide | Interneurons | [267] |
| sNPF | OSNs-PNs, Interneurons MB circuits | [250, 257, 261, 268, 269] |
| TK | OSNs-PNs | [270] |
Fig. 9.
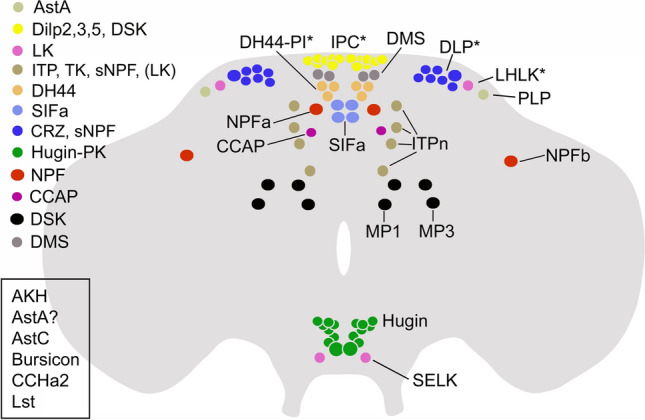
Peptidergic neuroendocrine systems in the Drosophila brain that regulate feeding and associated behaviors in addition to the DSK neurons. The figure shows the distribution of cell bodies of peptidergic neurons that have been implicated in feeding-related behavior. These are neurosecretory cells in MNC (IPC and DH44-PI) and LNC groups (ITPn and DLP) and interneurons located in distinct brain regions (MP1 and MP3, CCAP, LHLK, PLP, NPF, SIFa, Hugin and SELK); a few of the Hugin cells are neurosecretory cells. The neuron groups indicated with asterisks are cell autonomously nutrient sensing (only a subset of the DLPs), the MP1 and MP3 receive nutrient inputs, and the Hugin cells in the subesophageal zone receive gustatory inputs. The peptides released from these cells are shown in the legend in the upper left part of the figure. Note that also circuits associated with the mushroom bodies are linked to some of the peptidergic systems shown and are involved in regulation of food seeking and feeding [261]. There are also peptides derived from the intestine or corpora cardiaca that act on brain circuits to regulate feeding directly or indirectly (listed in the box in the bottom left of the figure). See text for further literature references. This figure was updated from Nässel and Zandawala [180]
Also monoamines like dopamine, octopamine, tyramine and serotonin play important roles in these behaviors. Octopaminergic neurons regulate courtship [207] or choice between courtship and aggression [208, 209], aggression [140, 210], appetite [211], and feeding behavior [184, 212, 213]. Tyramine is a satiety signal that in males supports courtship rather than feeding [214]. Dopaminergic neurons modulate courtship [164, 215], aggression [216], food search [217] and feeding behavior [218]. Serotonin modulates courtship [219, 220], aggression [176], and feeding behavior [221, 222]. In circuits that regulate aggression and courtship behavior, the MP1/MP2 neurons cooperate with neurons utilizing NPF, dopamine and octopamine as shown in Figs. 6, and 7, but in other cases it is not known whether the monoamines and other neuropeptides interact with DSK neurons and DSK signaling.
Conclusions and future perspectives
In this review, we have discussed the evolutionary conservation of the structures of CCK-type peptides and their receptors and some of their functions in bilaterian invertebrates and vertebrates. The relatedness between invertebrate and mammalian CCK-type signaling components was suggested already many years ago [24, 26, 33, 42, 106], but now we can see that also several of the functional roles of CCK signaling appear to be evolutionarily conserved (see [32, 106, 271]). These include roles in satiety signaling and regulation of feeding, digestion, aggression and courtship behavior [18, 19, 23, 44–47, 62, 121, 135, 140, 272]. As will be discussed below, it is important to note that the “conserved functions” are only superficially similar and at a mechanistic level they differ in details and complexity between taxons. In contrast to the situation for mammals, there is not yet enough data in Drosophila or other invertebrates to reveal the complete circuits and pathways underlying DSK signaling in feeding, digestion, aggression and courtship behavior. In Drosophila some of this behavior regulation is by paracrine signaling in circuits within the CNS, and probably another part is by hormonal action via brain neurosecretory cells. In mammals, there are further layers of CCK signaling served by peripheral neurons (e.g., CCKR-expressing vagus nerve neurons, VANs) and gut EECs [18, 19, 46, 47]. These layers seem to be lacking in insects, but may be present in a simpler form in echinoderms [32]. It is, however, possible that DSK released from IPCs in Drosophila acts directly on the crop, which is supplied by axon terminations of the IPCs. This might regulate crop contractions and thus provide a satiety-induced effect similar to the gastric emptying triggered by the CCK-induced activity in the circuits of the vagus nerve afferents—brainstem efferents [18, 19, 46, 47].
It has been shown that CCK/SK act as local neuromodulators in different circuits of the CNS in both insects and mammals, but detailed comparisons of the circuitry remains to be performed [19, 23, 44, 46, 273, 274]. In mammals, CCK is co-expressed in various brain neurons with dopamine (DA), serotonin, GABA and other neuropeptides such as substance P, enkephalin, oxytocin and corticotropin-releasing hormone [46, 275, 276]; see also [277] for further co-expression patterns suggested from single cell transcriptomics data. Thus, CCK may function as a co-transmitter of dopamine, serotonin and GABA and as a co-modulator with other neuropeptides. Among invertebrates, we so far only know of the colocalization of DSK with DILPs in the IPCs of the Drosophila brain [121, 122], but systematic analysis has not been performed (however, see [278, 279]).
As noted above, the mechanisms by which CCK/SK induce satiety differ between insects and mammals. In mammals, gastric distension leads to CCK release from EECs of the gastrointestinal tract that trigger CCKR expressing afferent neurons (VANs) of the vagus nerve to signal to neurons in the brainstem, leading to a reduction in food intake [18, 46–48]. Concomitantly, second-order neurons in the nucleus of the solitary tract in the brainstem signal to several brain centers (including hypothalamus) that regulate feeding, reward and ingestion. In insects the CCK-mediated satiety signaling known so far is by means of circuits in the brain that affect carbohydrate sensing by gustatory neurons and thereby food intake, although DSK from the brain neurosecretory cells (IPCs) also seem to contribute to other satiety mechanisms [45, 121, 140]. In C. elegans, the CCK signaling acts to alter locomotion associated with feeding, rather than direct reduction of food intake [27] and in starfish CCK-type peptide acts on muscle to retract the stomach and thereby stop food intake [32]. There is a need to further investigate the insect circuits involving SK and feeding since we do not know how the metabolic state is conveyed to the SK neurons or the neuronal mechanisms for the reduced food ingestion upon satiety. The regulation of feeding is complex in mammals, and both the peripheral and central mechanisms involve numerous neurotransmitters and neuropeptides [18, 280]. Also in Drosophila, multiple brain circuits and peptidergic systems, as well as a few peptides released from EECs, have also been identified in regulation of feeding and metabolism (see Fig. 9 and Table 2). Due to this complexity, comparisons between mammals and insects regarding the complete circuitry are beyond the scope of this review. Also the DSK-associated circuits that regulate aggression and courtship behavior in Drosophila [23, 44] need further analysis and especially how the balance between the competing behaviors is accomplished.
Overall, the complexity of CCK/SK signaling varies depending on phylogenetic position of the organism and, thus, in mammals the range of pleiotropic actions is much wider than in insects. Yet we can show here that a relatively small number of DSK expressing neurons in the Drosophila brain have important roles in regulation of taste, satiety, feeding, activity, aggression and courtship. Most of these are interneurons that utilize DSKs in paracrine signaling in neuronal circuits within the CNS [23, 44, 45, 141], but a small set are neurosecretory cells (IPCs) that probably release DSKs and insulin-like peptides into the circulation for action on close or distant target tissues/organs, such as the crop, intestine and others [121]. In insects the hormonal action of SKs has not been explicitly verified by determination of peptides in the circulation. Some data, however, suggest hormonal action. Injection of SK regulates digestive enzyme activity and motility in the gut of insects, but the main part of the intestine is not innervated by SK producing neurons and no SK expressing EECs have been found [135, 136, 142, 147]. Thus, the endogenous source of SK acting on the intestine is likely to be the brain neurosecretory cells. To confirm this, SK release needs to be detected in the hemolymph and the distribution of DSK receptors in the periphery mapped to detect sites of action of SKs. Receptor expression data indicates low amounts of SKR1 and 2 transcript in heart and reproductive organs of R. prolixus [109], but data are lacking for other insects, including Drosophila.
One important finding is that DSK and specific small sets of DSK-producing neurons in Drosophila play a role in several behaviors, some of which are conflicting. These are the FruM-expressing MP1/MP3 neurons that inhibit male courtship behavior, but stimulate aggression [23, 44]. The same neurons are also part of a satiety-inducing circuit that down-regulates sugar sensitivity and appetite in fed flies [45]. The circuits involved in regulation of aggression, courtship and satiety/feeding are each composed of multiple neurons that utilize a multitude of neurotransmitters and neuropeptides (Tables 3, 4 and 5). Nevertheless, the MP1/MP3 neurons appear to be involved in the switching between conflicting behaviors, probably by integrating different types of inputs that relay information about the external and internal environment. Other similar peptidergic brain systems are constituted for example by the four widely arborizing SIFamide-producing neurons that integrate sexual behavior, feeding and sleep by interactions with multiple brain and VNC circuits [237, 267, 281, 282] and Hugin neurons that integrate homeostatic sleep signals and the circadian clock, and relays locomotor activity output in adults [283, 284]. In larvae, Hugin neurons receive gustatory inputs and form a hub between feeding and locomotion [251, 285], a function that is not yet explored in adult flies.
In conclusion, we still have a long way to go to fully understand the fascinating roles of CCK/SK and their receptors in physiology and behavior of invertebrates. This peptidergic system is also interesting from the evolutionary point of view, since it illustrates how CCK-type signaling can induce specific states such as satiety in mechanistically distinct ways in insects and mammals.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Meet Zandawala for comments on an earlier version of this paper.
Abbreviations
- AKH
Adipokinetic hormone
- AstA
Allatostatin A
- CAPA-PK
Capability pyrokinin
- CCAP
Crustacean cardioactive peptide
- CCHa2
CCHamide-2
- CCK
Cholecystokinin
- CRZ
Corazonin
- DH44
Diuretic hormone 44
- DILP
Drosophila Insulin-like peptide
- DSK
Drosophila Sulfakinin
- GABA
Gamma aminobutyric acid
- GPB5
Glycoprotein B5
- Hugin-PK
Hugin-derived pyrokinin
- ILP
Insulin-like peptide
- ITP
Ion transport peptide
- LK
Leucokinin
- MIP
Myoinhibitory peptide
- NPF
Neuropeptide F
Pigment-dispersing factor
- SIFa
SIFamide
- SK
Sulfakinin
- sNPF
Short neuropeptide F
- TK
Tachykinin
Authors' contributions
DRN prepared the first draft of the manuscript. SFW assisted with subsequent drafts and both authors approved the final version. The figures were prepared by DRN and SFW.
Funding
Open access funding provided by Stockholm University. The research was financed by the Swedish Research Council (Vetenskapsrådet), Grant number 2015–04626 (to DRN) and National Natural Science Foundation of China, Grant number 32022011 (to SFW).
Availability of data and material
This article is a literature review and does not contain any new data. Source data files for Figs. 1 and 2 are in Supplementary material.
Declarations
Conflict of interests
The authors declare that there is no conflict of interest.
Ethical approval and consent to participate
This article is a literature review and does not contain any animal experiments or studies with human participants performed by any of the authors.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nässel DR, Pauls D, Huetteroth W. Neuropeptides in modulation of Drosophila behavior: how to get a grip on their pleiotropic actions. Curr Opin Insect Sci. 2019;36:1–8. doi: 10.1016/j.cois.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan HS, Salazar Thula O, Khoss N, Zimmer M. Nested neuronal dynamics orchestrate a behavioral hierarchy across timescales. Neuron. 2020;105(3):562–76.e9. doi: 10.1016/j.neuron.2019.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson DJ. Circuit modules linking internal states and social behaviour in flies and mice. Nat Rev Neurosci. 2016;17(11):692–704. doi: 10.1038/nrn.2016.125. [DOI] [PubMed] [Google Scholar]
- 4.Brezina V. Beyond the wiring diagram: signalling through complex neuromodulator networks. Philos Trans R Soc Lond B Biol Sci. 2010;365(1551):2363–2374. doi: 10.1098/rstb.2010.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SM, Su CY, Wang JW. Neuromodulation of innate behaviors in Drosophila. Annu Rev Neurosci. 2017;40:327–348. doi: 10.1146/annurev-neuro-072116-031558. [DOI] [PubMed] [Google Scholar]
- 6.Luis Alfonso Y-G, Daniel T, Gáspár J. Pre-metazoan origin of neuropeptide signalling. BioRxiv. 2021 doi: 10.1101/2021.11.19.469228. [DOI] [Google Scholar]
- 7.Nikitin M. Bioinformatic prediction of Trichoplax adhaerens regulatory peptides. Gen Comp Endocr. 2015;212:145–155. doi: 10.1016/j.ygcen.2014.03.049. [DOI] [PubMed] [Google Scholar]
- 8.Senatore A, Reese TS, Smith CL. Neuropeptidergic integration of behavior in Trichoplax adhaerens, an animal without synapses. J Exp Biol. 2017;220(18):3381. doi: 10.1242/jeb.162396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen SKD, Koch TL, Hauser F, Garm A, Grimmelikhuijzen CJP. De novo transcriptome assembly of the cubomedusa Tripedalia cystophora, including the analysis of a set of genes involved in peptidergic neurotransmission. BMC Genomics. 2019;20(1):175. doi: 10.1186/s12864-019-5514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch TL, Grimmelikhuijzen CJP. Global neuropeptide annotations from the genomes and transcriptomes of Cubozoa, Scyphozoa, Staurozoa (Cnidaria: Medusozoa), and Octocorallia (Cnidaria: Anthozoa) Front Endocrinol (Lausanne). 2019 doi: 10.3389/fendo.2019.00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510(7503):109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachkova MY, Nordmann E-L, Soto-Àngel JJ, Meeda Y, Górski B, Naumann B, et al. Neuropeptide repertoire and 3D anatomy of the ctenophore nervous system. Curr Biol. 2021;31(23):5274–85.e6. doi: 10.1016/j.cub.2021.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Jekely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci U S A. 2013;110(21):8702–8707. doi: 10.1073/pnas.1221833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirabeau O, Joly JS. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci U S A. 2013;110(22):E2028–E2037. doi: 10.1073/pnas.1219956110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elphick MR, Mirabeau O, Larhammar D. Evolution of neuropeptide signalling systems. J Exp Biol. 2018;221(3):151092. doi: 10.1242/jeb.151092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nässel DR, Zandawala M. Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Progr Neurobiol. 2019;179:101607. doi: 10.1016/j.pneurobio.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Jékely G, Melzer S, Beets I, Kadow ICG, Koene J, Haddad S, et al. The long and the short of it—a perspective on peptidergic regulation of circuits and behaviour. J Exp Biol. 2018;221(3):166710. doi: 10.1242/jeb.166710. [DOI] [PubMed] [Google Scholar]
- 18.Cawthon CR, de La Serre CB. The critical role of CCK in the regulation of food intake and diet-induced obesity. Peptides. 2021;138:170492. doi: 10.1016/j.peptides.2020.170492. [DOI] [PubMed] [Google Scholar]
- 19.Rehfeld JF. Cholecystokinin-from local gut hormone to ubiquitous messenger. Front Endocrinol (Lausanne) 2017;8:47. doi: 10.3389/fendo.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu N, Smagghe G. CCK(-like) and receptors: structure and phylogeny in a comparative perspective. Gen Comp Endocr. 2014;209:74–81. doi: 10.1016/j.ygcen.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Micevych P, Akesson T, Elde R. Distribution of cholecystokinin-immunoreactive cell bodies in the male and female rat: II. Bed nucleus of the stria terminalis and amygdala. J Comp Neurol. 1988;269(3):381–391. doi: 10.1002/cne.902690306. [DOI] [PubMed] [Google Scholar]
- 22.Markowski VP, Hull EM. Cholecystokinin modulates mesolimbic dopaminergic influences on male rat copulatory behavior. Brain Res. 1995;699(2):266–274. doi: 10.1016/0006-8993(95)00918-g. [DOI] [PubMed] [Google Scholar]
- 23.Wu S, Guo C, Zhao H, Sun M, Chen J, Han C, et al. Drosulfakinin signaling in fruitless circuitry antagonizes P1 neurons to regulate sexual arousal in Drosophila. Nat Commun. 2019;10(1):4770. doi: 10.1038/s41467-019-12758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nachman RJ, Holman GM, Haddon WF, Ling N. Leucosulfakinin, a sulfated insect neuropeptide with homology to gastrin and cholecystokinin. Science. 1986;234(4772):71–73. doi: 10.1126/science.3749893. [DOI] [PubMed] [Google Scholar]
- 25.Janssen T, Meelkop E, Lindemans M, Verstraelen K, Husson SJ, Temmerman L, et al. Discovery of a cholecystokinin-gastrin-like signaling system in nematodes. Endocrinology. 2008;149(6):2826–2839. doi: 10.1210/en.2007-1772. [DOI] [PubMed] [Google Scholar]
- 26.Johnsen AH, Rehfeld JF. LymnaDFamides, a new family of neuropeptides from the pond snail, Lymnaea stagnalis Clue to cholecystokinin immunoreactivity in invertebrates? Eur J Biochem. 1993;213(2):875–879. doi: 10.1111/j.1432-1033.1993.tb17831.x. [DOI] [PubMed] [Google Scholar]
- 27.Oranth A, Schultheis C, Tolstenkov O, Erbguth K, Nagpal J, Hain D, et al. Food sensation modulates locomotion by dopamine and neuropeptide signaling in a distributed neuronal network. Neuron. 2018;100(6):1414–1428. doi: 10.1016/j.neuron.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Koziol U. Precursors of neuropeptides and peptide hormones in the genomes of tardigrades. Gen Comp Endocr. 2018;267:116–127. doi: 10.1016/j.ygcen.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Thiel D, Franz-Wachtel M, Aguilera F, Hejnol A. Xenacoelomorph neuropeptidomes reveal a major expansion of neuropeptide systems during early bilaterian evolution. Mol Biol Evol. 2018;35(10):2528–2543. doi: 10.1093/molbev/msy160. [DOI] [Google Scholar]
- 30.Thiel D, Yañez Guerra LA, Franz-Wachtel M, Hejnol A, Jékely G. Nemertean, brachiopod and phoronid neuropeptidomics reveals ancestral spiralian signalling systems. BioRxiv. 2021 doi: 10.1101/2021.03.03.433790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Oliveira AL, Calcino A, Wanninger A. Extensive conservation of the proneuropeptide and peptide prohormone complement in mollusks. Sci Rep. 2019;9(1):4846. doi: 10.1038/s41598-019-40949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tinoco AB, Barreiro-Iglesias A, Yañez Guerra LA, Delroisse J, Zhang Y, Gunner EF, et al. Ancient role of sulfakinin/cholecystokinin-type signalling in inhibitory regulation of feeding processes revealed in an echinoderm. Elife. 2021;10:e65667. doi: 10.7554/eLife.65667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnsen AH, Rehfeld JF. Cionin: a disulfotyrosyl hybrid of cholecystokinin and gastrin from the neural ganglion of the protochordate Ciona intestinalis. J Biol Chem. 1990;265(6):3054–3058. doi: 10.1016/S0021-9258(19)39732-7. [DOI] [PubMed] [Google Scholar]
- 34.Edkins JS. The chemical mechanism of gastric secretion. J Physiol. 1906;34(1–2):133–144. doi: 10.1113/jphysiol.1906.sp001146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivy AC, Oldberg E. A hormone mechanism for gallbladder contraction and evacuation. Am J Physiol. 1928;86:559–613. doi: 10.1152/ajplegacy.1928.86.3.599. [DOI] [Google Scholar]
- 36.Harper AA, Raper HS. Pancreozymin, a stimulant of the secretion of pancreatic enzymes in extracts of the small intestine. J Physiol. 1943;102(1):115–125. doi: 10.1113/jphysiol.1943.sp004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Q, Ou L, Wang W, Guo D-Y. Gastrin, cholecystokinin, signaling, and biological activities in cellular processes. Front Endocrinol (Lausanne). 2020 doi: 10.3389/fendo.2020.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer KJ, Speirs RD, Childs CN. Immunochemical evidence for a gastrin-like peptide in insect neuroendocrine system. Gen Comp Endocr. 1977;32(4):423–426. doi: 10.1016/0016-6480(77)90224-6. [DOI] [PubMed] [Google Scholar]
- 39.Larson BA, Vigna SR. Species and tissue distribution of gholecystokinin/gastrin-like substances in some invertebrates. Gen Comp Endocr. 1983;50(3):469–475. doi: 10.1016/0016-6480(83)90268-X. [DOI] [PubMed] [Google Scholar]
- 40.El-Salhy M, Abou-El-Ela R, Falkmer S, Grimelius L, Wilander E. Immunohistochemical evidence of gastro-entero-pancreatic neurohormonal peptides of vertebrate type in the nervous system of the larva of a dipteran insect, the hoverfly Eristalis aeneus. Regul Peptides. 1980;1(3):187–204. doi: 10.1016/0167-0115(80)90271-2. [DOI] [PubMed] [Google Scholar]
- 41.Duve H, Thorpe A. Gastrin/cholecystokinin (CCK)-like immunoreactive neurones in the brain of the blowfly, Calliphora erythrocephala (Diptera) Gen Comp Endocr. 1981;43(3):381–391. doi: 10.1016/0016-6480(81)90298-7. [DOI] [PubMed] [Google Scholar]
- 42.Kubiak TM, Larsen MJ, Burton KJ, Bannow CA, Martin RA, Zantello MR, et al. Cloning and functional expression of the first Drosophila melanogaster sulfakinin receptor DSK-R1. Biochem Biophys Res Commun. 2002;291(2):313–320. doi: 10.1006/bbrc.2002.6459. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Peterson J, Nachman R, Ganetzky B. Drosulfakinin activates CCKLR-17D1 and promotes larval locomotion and escape response in Drosophila. Fly. 2012;6:4. doi: 10.4161/fly.21534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu F, Deng B, Xiao N, Wang T, Li Y, Wang R, et al. A neuropeptide regulates fighting behavior in Drosophila melanogaster. Elife. 2020;9:e54229. doi: 10.7554/eLife.54229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo D, Zhang Y-J, Zhang S, Li J, Guo C, Pan Y-F, et al. Cholecystokinin-like peptide mediates satiety by inhibiting sugar attraction. PLoS Genet. 2021;17(8):e1009724. doi: 10.1371/journal.pgen.1009724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawley JN, Corwin RL. Biological actions of cholecystokinin. Peptides. 1994;15(4):731–755. doi: 10.1016/0196-9781(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 47.Dockray GJ. Cholecystokinin and gut–brain signalling. Regul Peptides. 2009;155(1):6–10. doi: 10.1016/j.regpep.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 48.Moran TH. Cholecystokinin and satiety: current perspectives. Nutrition. 2000;16(10):858–865. doi: 10.1016/s0899-9007(00)00419-6. [DOI] [PubMed] [Google Scholar]
- 49.Dufresne M, Seva C, Fourmy D. Cholecystokinin and Gastrin Receptors. Physiol Rev. 2006;86(3):805–847. doi: 10.1152/physrev.00014.2005. [DOI] [PubMed] [Google Scholar]
- 50.Mutt V, Jorpes JE. Structure of porcine cholecystokinin-pancreozymin. 1. Cleavage with thrombin and with trypsin. Eur J Biochem. 1968;6(1):156–162. doi: 10.1111/j.1432-1033.1968.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 51.Gregory RA, Tracy HJ. The constitution and properties of two gastrins extracted from hog antral mucosa. Gut. 1964;5(2):103–114. doi: 10.1136/gut.5.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnsen AH. Phylogeny of the cholecystokinin/gastrin family. Front Neuroendocrinol. 1998;19(2):73–99. doi: 10.1006/frne.1997.0163. [DOI] [PubMed] [Google Scholar]
- 53.Lazarus LH, Attila M. The toad, ugly and venomous, wears yet a precious jewel in his skin. Prog Neurobiol. 1993;41(4):473–507. doi: 10.1016/0301-0082(93)90027-P. [DOI] [PubMed] [Google Scholar]
- 54.Kopin AS, Lee YM, McBride EW, Miller LJ, Lu M, Lin HY, et al. Expression cloning and characterization of the canine parietal cell gastrin receptor. Proc Natl Acad Sci. 1992;89(8):3605. doi: 10.1073/pnas.89.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller LJ, Desai AJ. Metabolic actions of the type 1 cholecystokinin receptor: its potential as a therapeutic target. Trends Endocrinol Metab. 2016;27(9):609–619. doi: 10.1016/j.tem.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rehfeld JF, Friis-Hansen L, Goetze JP, Hansen TV. The biology of cholecystokinin and gastrin peptides. Curr Top Med Chem. 2007;7(12):1154–1165. doi: 10.2174/156802607780960483. [DOI] [PubMed] [Google Scholar]
- 57.Buchan AM, Polak JM, Solcia E, Capella C, Hudson D, Pearse AG. Electron immunohistochemical evidence for the human intestinal I cell as the source of CCK. Gut. 1978;19(5):403–407. doi: 10.1136/gut.19.5.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polak JM, Bloom SR, Rayford PL, Pearse AG, Buchan AM, Thompson JC. Identification of cholecystokinin-secreting cells. Lancet (London, England) 1975;2(7943):1016–1018. doi: 10.1016/s0140-6736(75)90297-4. [DOI] [PubMed] [Google Scholar]
- 59.Rust VA, Crosby KM. Cholecystokinin acts in the dorsomedial hypothalamus of young male rats to suppress appetite in a nitric oxide-dependent manner. Neurosci Lett. 2021;764:136295. doi: 10.1016/j.neulet.2021.136295. [DOI] [PubMed] [Google Scholar]
- 60.D'Agostino G, Lyons DJ, Cristiano C, Burke LK, Madara JC, Campbell JN, et al. Appetite controlled by a cholecystokinin nucleus of the solitary tract to hypothalamus neurocircuit. Elife. 2016;5:e12225. doi: 10.7554/eLife.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen RT. Involvement of cholecystokinin/gastrin-related peptides and their receptors in clinical gastrointestinal disorders. Pharmacol Toxicol. 2002;91(6):333–350. doi: 10.1034/j.1600-0773.2002.910611.x. [DOI] [PubMed] [Google Scholar]
- 62.Bloch GJ, Babcock AM, Gorski RA, Micevych PE. Effects of cholecystokinin on male copulatory behavior and lordosis behavior in male rats. Physiol Behav. 1988;43(3):351–357. doi: 10.1016/0031-9384(88)90198-9. [DOI] [PubMed] [Google Scholar]
- 63.Bloch GJ, Babcock AM, Gorski RA, Micevych PE. Cholecystokinin stimulates and inhibits lordosis behavior in female rats. Physiol Behav. 1987;39(2):217–224. doi: 10.1016/0031-9384(87)90012-6. [DOI] [PubMed] [Google Scholar]
- 64.Zwanzger P, Domschke K, Bradwejn J. Neuronal network of panic disorder: the role of the neuropeptide cholecystokinin. Depress Anxiety. 2012;29(9):762–774. doi: 10.1002/da.21919. [DOI] [PubMed] [Google Scholar]
- 65.Li Q, Deng X, Singh P. Significant increase in the aggressive behavior of transgenic mice overexpressing peripheral progastrin peptides: associated changes in CCK2 and serotonin receptors in the CNS. Neuropsychopharmacology. 2007;32(8):1813–1821. doi: 10.1038/sj.npp.1301304. [DOI] [PubMed] [Google Scholar]
- 66.Raud S, Innos J, Abramov U, Reimets A, Kõks S, Soosaar A, et al. Targeted invalidation of CCK2 receptor gene induces anxiolytic-like action in light-dark exploration, but not in fear conditioning test. Psychopharmacology. 2005;181(2):347–357. doi: 10.1007/s00213-005-2255-x. [DOI] [PubMed] [Google Scholar]
- 67.Moran TH, Schwartz GJ. Neurobiology of cholecystokinin. Crit Rev Neurobiol. 1994;9(1):1–28. [PubMed] [Google Scholar]
- 68.Kennedy JL, Bradwejn J, Koszycki D, King N, Crowe R, Vincent J, et al. Investigation of cholecystokinin system genes in panic disorder. Mol Psychiatry. 1999;4(3):284–285. doi: 10.1038/sj.mp.4000507. [DOI] [PubMed] [Google Scholar]
- 69.Nachman RJ, Holman GM, Cook BJ, Haddon WF, Ling N. Leucosulfakinin-II, a blocked sulfated insect neuropeptide with homology to cholecystokinin and gastrin. Biochem Biophys Res Commun. 1986;140(1):357–364. doi: 10.1016/0006-291x(86)91098-3. [DOI] [PubMed] [Google Scholar]
- 70.Nichols R, Schneuwly SA, Dixon JE. Identification and characterization of a Drosophila homologue to the vertebrate neuropeptide cholecystokinin. J Biol Chem. 1988;263(25):12167–12170. doi: 10.1016/S0021-9258(18)37731-7. [DOI] [PubMed] [Google Scholar]
- 71.Schwartz J, Dubos MP, Pasquier J, Zatylny-Gaudin C, Favrel P. Emergence of a cholecystokinin/sulfakinin signalling system in Lophotrochozoa. Sci Rep. 2018;8(1):16424. doi: 10.1038/s41598-018-34700-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conzelmann M, Williams EA, Krug K, Franz-Wachtel M, Macek B, Jékely G. The neuropeptide complement of the marine annelid Platynereis dumerilii. BMC Genom. 2013;14:906. doi: 10.1186/1471-2164-14-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang M, Wang Y, Li Y, Li W, Li R, Xie X, et al. Identification and characterization of neuropeptides by transcriptome and proteome analyses in a bivalve mollusc Patinopecten yessoensis. Front Genet. 2018;9:197. doi: 10.3389/fgene.2018.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Veenstra JA. Neuropeptide evolution: chelicerate neurohormone and neuropeptide genes may reflect one or more whole genome duplications. Gen Comp Endocrinol. 2016;229:41–55. doi: 10.1016/j.ygcen.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 75.Christie AE. In silico characterization of the neuropeptidome of the Western black widow spider Latrodectus hesperus. Gen Comp Endocr. 2015;210:63–80. doi: 10.1016/j.ygcen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 76.Pandit AA, Davies S-A, Smagghe G, Dow JAT. Evolutionary trends of neuropeptide signaling in beetles—A comparative analysis of Coleopteran transcriptomic and genomic data. Insect Biochem Molec. 2019;114:103227. doi: 10.1016/j.ibmb.2019.103227. [DOI] [PubMed] [Google Scholar]
- 77.Chang J, Zhao J, Tian X. In silico prediction of neuropeptides in Hymenoptera parasitoid wasps. PLoS ONE. 2018;13(2):e0193561. doi: 10.1371/journal.pone.0193561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hauser F, Neupert S, Williamson M, Predel R, Tanaka Y, Grimmelikhuijzen CJP. Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. J Proteome Res. 2010;9(10):5296–5310. doi: 10.1021/pr100570j. [DOI] [PubMed] [Google Scholar]
- 79.Li C, Yun X, Hu X, Zhang Y, Sang M, Liu X, et al. Identification of G protein-coupled receptors in the pea aphid Acyrthosiphon pisum. Genomics. 2013;102(4):345–354. doi: 10.1016/j.ygeno.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 80.Huybrechts J, Bonhomme J, Minoli S, Prunier-Leterme N, Dombrovsky A, Abdel-Latief M, et al. Neuropeptide and neurohormone precursors in the pea aphid Acyrthosiphon pisum. Insect Mol Biol. 2010;19(Suppl 2):87–95. doi: 10.1111/j.1365-2583.2009.00951.x. [DOI] [PubMed] [Google Scholar]
- 81.Liessem S, Ragionieri L, Neupert S, Büschges A, Predel R. Transcriptomic and neuropeptidomic analysis of the stick insect Carausius morosus. J Proteome Res. 2018;17(6):2192–2204. doi: 10.1021/acs.jproteome.8b00155. [DOI] [PubMed] [Google Scholar]
- 82.Wang Z, Zhou W, Hameed MS, Liu J, Zeng X. Characterization and expression profiling of neuropeptides and G-protein-coupled receptors (GPCRs) for neuropeptides in the asian citrus psyllid, Diaphorina citri (Hemiptera: Psyllidae) Int J Mol Sci. 2018 doi: 10.3390/ijms19123912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Veenstra JA, Rombauts S, Grbic M. In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem Mol. 2012;42(4):277–295. doi: 10.1016/j.ibmb.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 84.Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, et al. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 2011;470(7333):255–258. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nässel DR, Wu S-F. Leucokinins: multifunctional neuropeptides and hormones in insects and other invertebrates. Int J Mol Sci. 2021 doi: 10.3390/ijms22041531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeoh JGC, Pandit AA, Zandawala M, Nässel DR, Davies SA, Dow JAT. DINeR: database for Insect Neuropeptide Research. Insect Biochem Mol Biol. 2017;86:9–19. doi: 10.1016/j.ibmb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 87.Derst C, Dircksen H, Meusemann K, Zhou X, Liu S, Predel R. Evolution of neuropeptides in non-pterygote hexapods. BMC Evol Biol. 2016;16(1):51. doi: 10.1186/s12862-016-0621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnsen AH, Duve H, Davey M, Hall M, Thorpe A. Sulfakinin neuropeptides in a crustacean. Eur J Biochem. 2000;267(4):1153–1160. doi: 10.1046/j.1432-1327.2000.01113.x. [DOI] [PubMed] [Google Scholar]
- 89.Wegener C, Gorbashov A. Molecular evolution of neuropeptides in the genus Drosophila. Genome Biol. 2008;9(8):R131. doi: 10.1186/gb-2008-9-8-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Predel R, Brandt W, Kellner R, Rapus J, Nachman RJ, Gäde G. Post-translational modifications of the insect sulfakinins: sulfation, pyroglutamate-formation and O-methylation of glutamic acid. Eur J Biochem. 1999;263(2):552–560. doi: 10.1046/j.1432-1327.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- 91.Predel R, Neupert S, Derst C, Reinhardt K, Wegener C. Neuropeptidomics of the bed bug cimex lectularius. J Proteome Res. 2018;17(1):440–454. doi: 10.1021/acs.jproteome.7b00630. [DOI] [PubMed] [Google Scholar]
- 92.Ragionieri L, Verdonck R, Verlinden H, Marchal E, Vanden Broeck J, Predel R. Schistocerca neuropeptides—an update. J Insect Physiol. 2021 doi: 10.1016/j.jinsphys.2021.104326. [DOI] [PubMed] [Google Scholar]
- 93.Ramachandran S, Banerjee N, Bhattacharya R, Lemons ML, Florman J, Lambert CM, et al. A conserved neuropeptide system links head and body motor circuits to enable adaptive behavior. BioRxiv. 2021 doi: 10.1101/2020.04.27.064550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li B, Predel R, Neupert S, Hauser F, Tanaka Y, Cazzamali G, et al. Genomics, transcriptomics, and peptidomics of neuropeptides and protein hormones in the red flour beetle Tribolium castaneum. Genome Res. 2008;18(1):113–122. doi: 10.1101/gr.6714008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nachman RJ, Holman GM, Haddon WF. Structural aspects of gastrin/CCK-like insect leucosulfakinins and FMRF-amide. Peptides. 1988;9:137–143. doi: 10.1016/0196-9781(88)90237-9. [DOI] [PubMed] [Google Scholar]
- 96.Schoofs L, Nachman RJ. CHAPTER 28—Sulfakinins. In: Kastin AJ, editor. Handbook of biologically active peptides. Burlington: Academic Press; 2006. pp. 183–187. [Google Scholar]
- 97.Torfs P, Baggerman G, Meeusen T, Nieto J, Nachman RJ, Calderon J, et al. Isolation, identification, and synthesis of a disulfated sulfakinin from the central nervous system of an arthropods the white shrimp Litopenaeus vannamei. Biochem Biophys Res Commun. 2002;299(2):312–320. doi: 10.1016/s0006-291x(02)02624-4. [DOI] [PubMed] [Google Scholar]
- 98.Nichols R, Egle JP, Langan NR, Palmer GC. The different effects of structurally related sulfakinins on Drosophila melanogaster odor preference and locomotion suggest involvement of distinct mechanisms. Peptides. 2008;29(12):2128–2135. doi: 10.1016/j.peptides.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nichols R. The first nonsulfated sulfakinin activity reported suggests nsDSK acts in gut biology. Peptides. 2007;28(4):767–773. doi: 10.1016/j.peptides.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 100.Słocińska M, Chowański S, Marciniak P. Identification of sulfakinin receptors (SKR) in Tenebrio molitor beetle and the influence of sulfakinins on carbohydrates metabolism. J Comp Physiol B. 2020;190(5):669–679. doi: 10.1007/s00360-020-01300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Slocinska M, Antos-Krzeminska N, Rosinski G, Jarmuszkiewicz W. Nonsulfated sulfakinin changes metabolic parameters of insect fat body mitochondria. Arch Insect Biochem Physiol. 2016;93(4):177–189. doi: 10.1002/arch.21350. [DOI] [PubMed] [Google Scholar]
- 102.Sekiguchi T, Ogasawara M, Satake H. Molecular and functional characterization of cionin receptors in the ascidian, Ciona intestinalis: the evolutionary origin of the vertebrate cholecystokinin/gastrin family. J Endocrinol. 2012;213(1):99–106. doi: 10.1530/joe-11-0410. [DOI] [PubMed] [Google Scholar]
- 103.Dupré D, Tostivint H. Evolution of the gastrin-cholecystokinin gene family revealed by synteny analysis. Gen Comp Endocrinol. 2014;195:164–173. doi: 10.1016/j.ygcen.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 104.Yu N, Nachman RJ, Smagghe G. Characterization of sulfakinin and sulfakinin receptor and their roles in food intake in the red flour beetle Tribolium castaneum. Gen Comp Endocr. 2013;188:196–203. doi: 10.1016/j.ygcen.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 105.Yu N, Smagghe G. Characterization of sulfakinin receptor 2 and its role in food intake in the red flour beetle Tribolium castaneum. Peptides. 2014;53:232–237. doi: 10.1016/j.peptides.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 106.Staljanssens D, Azari EK, Christiaens O, Beaufays J, Lins L, Van Camp J, et al. The CCK(-like) receptor in the animal kingdom: Functions, evolution and structures. Peptides. 2011;32(3):607–619. doi: 10.1016/j.peptides.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 107.Christie AE. Identification of putative neuropeptidergic signaling systems in the spiny lobster, Panulirus argus. Invert Neurosci. 2020;20(1):2. doi: 10.1007/s10158-020-0235-9. [DOI] [PubMed] [Google Scholar]
- 108.Tanaka Y, Suetsugu Y, Yamamoto K, Noda H, Shinoda T. Transcriptome analysis of neuropeptides and G-protein coupled receptors (GPCRs) for neuropeptides in the brown planthopper Nilaparvata lugens. Peptides. 2014;53:125–133. doi: 10.1016/j.peptides.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 109.Bloom M, Lange AB, Orchard I. Identification, functional characterization, and pharmacological analysis of two sulfakinin receptors in the medically-important insect Rhodnius prolixus. Sci Rep. 2019;9(1):13437. doi: 10.1038/s41598-019-49790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen X, Ganetzky B. A neuropeptide signaling pathway regulates synaptic growth in Drosophila. J Cell Biol. 2012;196(4):529–543. doi: 10.1083/jcb.201109044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zels S, Verlinden H, Dillen S, Vleugels R, Nachman RJ, Broeck JV. Signaling properties and pharmacological analysis of two sulfakinin receptors from the red flour beetle, Tribolium castaneum. PLoS ONE. 2014;9(4):e94502. doi: 10.1371/journal.pone.0094502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu N, Zotti MJ, Scheys F, Braz ASK, Penna PHC, Nachman RJ, et al. Flexibility and extracellular opening determine the interaction between ligands and insect sulfakinin receptors. Sci Rep. 2015;5(1):12627. doi: 10.1038/srep12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu N, Benzi V, Zotti MJ, Staljanssens D, Kaczmarek K, Zabrocki J, et al. Analogs of sulfakinin-related peptides demonstrate reduction in food intake in the red flour beetle, Tribolium castaneum, while putative antagonists increase consumption. Peptides. 2013;41:107–112. doi: 10.1016/j.peptides.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 114.Nichols R, Lim IA. Spatial and temporal immunocytochemical analysis of drosulfakinin (Dsk) gene products in the Drosophila melanogaster central nervous system. Cell Tissue Res. 1996;283(1):107–116. doi: 10.1007/s004410050518. [DOI] [PubMed] [Google Scholar]
- 115.Agricola HJ, Bräunig P. Comparative aspects of peptidergic signaling pathways in the nervous systems of arthropods. In: Breidbach O, Kutsch W, editors. The nervous systems of invertebrates: an evolutionary and comparative approach. Basel: Birkhäuser Verlag; 1994. pp. 303–327. [Google Scholar]
- 116.Al-Alkawi H, Lange AB, Orchard I. Cloning, localization, and physiological effects of sulfakinin in the kissing bug Rhodnius prolixus. Peptides. 2017;98:15–22. doi: 10.1016/j.peptides.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 117.East PD, Hales DF, Cooper PD. Distribution of sulfakinin-like peptides in the central and sympathetic nervous system of the American cockroach, Periplaneta americana (L.) and the field cricket, Teleogryllus commodus (Walker) Tissue Cell. 1997;29(3):347–354. doi: 10.1016/S0040-8166(97)80010-9. [DOI] [PubMed] [Google Scholar]
- 118.Wicher D, Derst C, Gautier H, Lapied B, Heinemann SH, Agricola HJ. The satiety signaling neuropeptide perisulfakinin inhibits the activity of central neurons promoting general activity. Front Cell Neurosci. 2007;1:3. doi: 10.3389/neuro.03.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Veenstra JA, Lau GW, Agricola H-J, Petzel DH. Immunohistological localization of regulatory peptides in the midgut of the female mosquito Aedes aegypti. Histochem Cell Biol. 1995;104(5):337–347. doi: 10.1007/BF01458127. [DOI] [PubMed] [Google Scholar]
- 120.Nichols R, McCormick J, Lim I. Dromyosuppressin and drosulfakinin, two structurally related Drosophila neuropeptides, are uniquely expressed in the adult central nervous system. Ann N Y Acad Sci. 1997;814:315–318. doi: 10.1111/j.1749-6632.1997.tb46173.x. [DOI] [PubMed] [Google Scholar]
- 121.Söderberg JA, Carlsson MA, Nässel DR. Insulin-producing cells in the Drosophila brain also express satiety-inducing cholecystokinin-like peptide. Drosulfakinin Front Endocrinol. 2012;3:109. doi: 10.3389/fendo.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Park D, Veenstra JA, Park JH, Taghert PH. Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS ONE. 2008;3(3):e1896. doi: 10.1371/journal.pone.0001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hadjieconomou D, King G, Gaspar P, Mineo A, Blackie L, Ameku T, et al. Enteric neurons increase maternal food intake during reproduction. Nature. 2020;587(7834):455–459. doi: 10.1038/s41586-020-2866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gough CS, Fairlamb GM, Bell P, Nachman RJ, Audsley N, Isaac RE. Peptidergic control in a fruit crop pest: the spotted-wing drosophila, Drosophila suzukii. PLoS ONE. 2017;12(11):e0188021. doi: 10.1371/journal.pone.0188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Duve H, Rehfeld JF, East P, Thorpe A. Localisation of sulfakinin neuronal pathways in the blowfly Calliphora vomitoria. Cell Tissue Res. 1994;275(1):177–186. doi: 10.1007/BF00305385. [DOI] [PubMed] [Google Scholar]
- 126.Veenstra JA. Peptidergic paracrine and endocrine cells in the midgut of the fruit fly maggot. Cell Tissue Res. 2009 doi: 10.1007/s00441-009-0769-y. [DOI] [PubMed] [Google Scholar]
- 127.Johard HA, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Förster C, et al. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. 2009;516(1):59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- 128.Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, et al. RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet. 2017;13(2):e1006613. doi: 10.1371/journal.pgen.1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reiher W, Shirras C, Kahnt J, Baumeister S, Isaac RE, Wegener C. Peptidomics and peptide hormone processing in the Drosophila midgut. J Proteome Res. 2011;10(4):1881–1892. doi: 10.1021/pr101116g. [DOI] [PubMed] [Google Scholar]
- 130.Wegener C, Veenstra JA. Chemical identity, function and regulation of enteroendocrine peptides in insects. Curr Opin Insect Sci. 2015;11:8–13. doi: 10.1016/j.cois.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 131.DeLaney K, Hu M, Hellenbrand T, Dickinson PS, Nusbaum MP, Li L. Mass spectrometry quantification, localization, and discovery of feeding-related neuropeptides in Cancer borealis. ACS Chem Neurosci. 2021;12(4):782–798. doi: 10.1021/acschemneuro.1c00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Christie AE. Crustacean neuroendocrine systems and their signaling agents. Cell Tissue Res. 2011;345(1):41–67. doi: 10.1007/s00441-011-1183-9. [DOI] [PubMed] [Google Scholar]
- 133.Marder E, Bucher D, Schulz DJ, Taylor AL. Invertebrate central pattern generation moves along. Curr Biol. 2005;15(17):R685–R699. doi: 10.1016/j.cub.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 134.Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- 135.Zels S, Dillen S, Crabbe K, Spit J, Nachman RJ, Vanden BJ. Sulfakinin is an important regulator of digestive processes in the migratory locust Locusta migratoria. Insect Biochem Mol Biol. 2015;61:8–16. doi: 10.1016/j.ibmb.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 136.Nachman RJ, Giard W, Favrel P, Suresh Y, Sreekumar S, Holman GM. Insect myosuppressins and sulfakinins stimulate release of the digestive enzyme a-amylase in two invertebrates: the scallop Pecten maximus and insect Rhynchophorus ferrugineus. Ann N Y Acad Sci. 1997;814:335–338. doi: 10.1111/j.1749-6632.1997.tb46178.x. [DOI] [Google Scholar]
- 137.Szymczak-Cendlak M, Gołębiowski M, Chowański S, Pacholska-Bogalska J, Marciniak P, Rosiński G, et al. Sulfakinins influence lipid composition and insulin-like peptides level in oenocytes of Zophobas atratus beetles. J Comp Physiol B. 2021 doi: 10.1007/s00360-021-01398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thorndyke MC, Bevis PJR. Comparative studies on the effects of cholecystokinins, caerulein, bombesin 6–14 nonapeptide, and physalaemin on gastric secretion in the ascidian Styela clava. Gen Comp Endocr. 1984;55(2):251–259. doi: 10.1016/0016-6480(84)90109-6. [DOI] [PubMed] [Google Scholar]
- 139.The International Aphid Genomics C Genome sequence of the pea aphid Acyrthosiphon pisum. PLOS Biol. 2010;8(2):e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Williams MJ, Goergen P, Rajendran J, Klockars A, Kasagiannis A, Fredriksson R, et al. Regulation of aggression by obesity-linked genes TfAP-2 and Twz through octopamine signaling in drosophila. Genetics. 2014;196(1):349–362. doi: 10.1534/genetics.113.158402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Agrawal P, Kao D, Chung P, Looger LL. The neuropeptide Drosulfakinin regulates social isolation-induced aggression in Drosophila. J Exp Biol. 2020 doi: 10.1242/jeb.207407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Guo D, Zhang S, Zhang Y-J, Ma J-Y, Gao C-F, Wu S-F. Sulfakinin inhibits activity of digestive enzymes in the brown planthopper, Nilaparvata lugens. J Asia-Pacific Entomol. 2020;23(4):1073–1082. doi: 10.1016/j.aspen.2020.09.004. [DOI] [Google Scholar]
- 143.Lin X, Yu N, Smagghe G. Insulin receptor regulates food intake through sulfakinin signaling in the red flour beetle Tribolium castaneum. Peptides. 2016;80:89–95. doi: 10.1016/j.peptides.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 144.Wei Z, Baggerman G, Goldsworthy G, Verhaert P, De Loof A, et al. Sulfakinins reduce food intake in the desert locust Schistocerca gregaria. J Insect Physiol. 2000;46(9):1259–1265. doi: 10.1016/S0022-1910(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 145.Meyering-Vos M, Muller A. RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J Insect Physiol. 2007;53(8):840–848. doi: 10.1016/j.jinsphys.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 146.Maestro JL, Aguilar R, Pascual N, Valero ML, Piulachs MD, Andreu D, et al. Screening of antifeedant activity in brain extracts led to the identification of sulfakinin as a satiety promoter in the German cockroach. Are arthropod sulfakinins homologous to vertebrate gastrins-cholecystokinins? Eur J Biochem. 2001;268(22):5824–5830. doi: 10.1046/j.0014-2956.2001.02527.x. [DOI] [PubMed] [Google Scholar]
- 147.Downer KE, Haselton AT, Nachman RJ, Stoffolano JG., Jr Insect satiety: sulfakinin localization and the effect of drosulfakinin on protein and carbohydrate ingestion in the blow fly, Phormia regina (Diptera: Calliphoridae) J Insect Physiol. 2007;53(1):106–112. doi: 10.1016/j.jinsphys.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 148.Dickinson PS, Stevens JS, Rus S, Brennan HR, Goiney CC, Smith CM, et al. Identification and cardiotropic actions of sulfakinin peptides in the American lobster Homarus americanus. J Exp Biol. 2007;210(Pt 13):2278–2289. doi: 10.1242/jeb.004770. [DOI] [PubMed] [Google Scholar]
- 149.Cao C, Brown MR. Localization of an insulin-like peptide in brains of two flies. Cell Tissue Res. 2001;304(2):317–321. doi: 10.1007/s004410100367. [DOI] [PubMed] [Google Scholar]
- 150.Nässel DR, Vanden BJ. Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides. Cell Mol Life Sci. 2016;73(2):271–290. doi: 10.1007/s00018-015-2063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Luo J, Lushchak OV, Goergen P, Williams MJ, Nässel DR. Drosophila insulin-producing cells are differentially modulated by serotonin and octopamine receptors and affect social behavior. PLoS ONE. 2014;9(6):e99732. doi: 10.1371/journal.pone.0099732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Crocker A, Shahidullah M, Levitan IB, Sehgal A. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron. 2010;65(5):670–681. doi: 10.1016/j.neuron.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Park S, Alfa RW, Topper SM, Kim GE, Kockel L, Kim SK. A genetic strategy to measure circulating Drosophila insulin reveals genes regulating insulin production and secretion. PLoS Genet. 2014;10(8):e1004555. doi: 10.1371/journal.pgen.1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pan Y, Meissner GW, Baker BS. Joint control of Drosophila male courtship behavior by motion cues and activation of male-specific P1 neurons. Proc Natl Acad Sci U S A. 2012;109(25):10065–10070. doi: 10.1073/pnas.1207107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Clowney EJ, Iguchi S, Bussell JJ, Scheer E, Ruta V. Multimodal chemosensory circuits controlling male courtship in drosophila. Neuron. 2015;87(5):1036–1049. doi: 10.1016/j.neuron.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kallman BR, Kim H, Scott K. Excitation and inhibition onto central courtship neurons biases Drosophila mate choice. Elife. 2015;4:e11188. doi: 10.7554/eLife.11188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Koganezawa M, Kimura K-I, Yamamoto D. The neural circuitry that functions as a switch for courtship versus aggression in drosophila males. Curr Biol. 2016;26(11):1395–1403. doi: 10.1016/j.cub.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 158.Asahina K. Sex differences in Drosophila behavior: qualitative and quantitative dimorphism. Curr Opin Physio. 2018;6:35–45. doi: 10.1016/j.cophys.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yamamoto D. Brain sex differences and function of the fruitless gene in Drosophila. J Neurogenet. 2008 doi: 10.1080/01677060802298491. [DOI] [PubMed] [Google Scholar]
- 160.Yamamoto D, Koganezawa M. Genes and circuits of courtship behaviour in Drosophila males. Nat Rev Neurosci. 2013;14(10):681–692. doi: 10.1038/nrn3567. [DOI] [PubMed] [Google Scholar]
- 161.Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121(5):785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 162.Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, Baker BS. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436(7049):395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 163.Vrontou E, Nilsen SP, Demir E, Kravitz EA, Dickson BJ. fruitless regulates aggression and dominance in Drosophila. Nat Neurosci. 2006;9(12):1469–1471. doi: 10.1038/nn1809. [DOI] [PubMed] [Google Scholar]
- 164.Zhang Stephen X, Rogulja D, Crickmore MA. Dopaminergic circuitry underlying mating drive. Neuron. 2016;91(1):168–181. doi: 10.1016/j.neuron.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 165.Zhang SX, Miner LE, Boutros CL, Rogulja D, Crickmore MA. Motivation, perception, and chance converge to make a binary decision. Neuron. 2018;99(2):376–88.e6. doi: 10.1016/j.neuron.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu W, Ganguly A, Huang J, Wang Y, Ni JD, Gurav AS, et al. Neuropeptide F regulates courtship in Drosophila through a male-specific neuronal circuit. Elife. 2019;8:e49574. doi: 10.7554/eLife.49574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang SX, Rogulja D, Crickmore MA. Recurrent circuitry sustains drosophila courtship drive while priming itself for satiety. Curr Biol. 2019;29(19):3216–28.e9. doi: 10.1016/j.cub.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jung Y, Kennedy A, Chiu H, Mohammad F, Claridge-Chang A, Anderson DJ. Neurons that function within an integrator to promote a persistent behavioral state in Drosophila. Neuron. 2020;105(2):322–33.e5. doi: 10.1016/j.neuron.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kohatsu S, Koganezawa M, Yamamoto D. Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron. 2011;69(3):498–508. doi: 10.1016/j.neuron.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 170.Pavlou HJ, Lin AC, Neville MC, Nojima T, Diao F, Chen BE, et al. Neural circuitry coordinating male copulation. Elife. 2016;5:e20713. doi: 10.7554/eLife.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhou C, Pan Y, Robinett CC, Meissner GW, Baker BS. Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron. 2014;83(1):149–163. doi: 10.1016/j.neuron.2014.05.038. [DOI] [PubMed] [Google Scholar]
- 172.Zhou C, Wang T, Jing B, Deng B, Shi K, Li J, et al. Drosulfakinin signaling modulates female sexual receptivity in Drosophila. BioRxiv. 2021 doi: 10.1101/2021.12.09.471924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Watanabe K, Chiu H, Pfeiffer BD, Wong AM, Hoopfer ED, Rubin GM, et al. A circuit node that integrates convergent input from neuromodulatory and social behavior-promoting neurons to control aggression in Drosophila. Neuron. 2017;95(5):1112–28.e7. doi: 10.1016/j.neuron.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dierick HA, Greenspan RJ. Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat Genet. 2007;39(5):678–682. doi: 10.1038/ng2029. [DOI] [PubMed] [Google Scholar]
- 175.Asahina K, Watanabe K, Duistermars BJ, Hoopfer E, Gonzalez CR, Eyjolfsdottir EA, et al. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156(1–2):221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alekseyenko OV, Lee C, Kravitz EA. Targeted manipulation of serotonergic neurotransmission affects the escalation of aggression in adult male Drosophila melanogaster. PLoS ONE. 2010;5(5):e10806. doi: 10.1371/journal.pone.0010806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lin D, Boyle MP, Dollar P, Lee H, Lein ES, Perona P, et al. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470(7333):221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hartenstein V. The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J Endocrinol. 2006;190(3):555–570. doi: 10.1677/joe.1.06964. [DOI] [PubMed] [Google Scholar]
- 179.Scharrer B. Insects as models in neuroendocrine research. Annu Rev Entomol. 1987;32:1–16. doi: 10.1146/annurev.en.32.010187.000245. [DOI] [PubMed] [Google Scholar]
- 180.Nässel DR, Zandawala M. Hormonal axes in Drosophila: regulation of hormone release and multiplicity of actions. Cell Tissue Res. 2020;382(2):233–266. doi: 10.1007/s00441-020-03264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296(5570):1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 182.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11(4):213–221. doi: 10.1016/S0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 183.Davis SM, Thomas AL, Nomie KJ, Huang L, Dierick HA. Tailless and atrophin control drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat Commun. 2014 doi: 10.1038/ncomms4177. [DOI] [PubMed] [Google Scholar]
- 184.Williams MJ, Goergen P, Rajendran J, Zheleznyakova G, Hägglund MG, Perland E, et al. Obesity-linked homologues TfAP-2 and Twz establish meal frequency in Drosophila melanogaster. PLoS Genet. 2014;10(9):e1004499. doi: 10.1371/journal.pgen.1004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117(7):981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 186.Dethier VG. The hungry fly: a physiological study of the behavior associated with feeding. Oxford: Harvard Press; 1976. p. 489. [Google Scholar]
- 187.Meunier N, Belgacem YH, Martin JR. Regulation of feeding behaviour and locomotor activity by takeout in Drosophila. J Exp Biol. 2007;210(Pt 8):1424–1434. doi: 10.1242/jeb.02755. [DOI] [PubMed] [Google Scholar]
- 188.Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101(6):647–656. doi: 10.1016/S0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 189.Hentze JL, Carlsson MA, Kondo S, Nässel DR, Rewitz KF. The neuropeptide allatostatin A regulates metabolism and feeding decisions in Drosophila. Sci Rep. 2015;5:11680. doi: 10.1038/srep11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ren GR, Hauser F, Rewitz KF, Kondo S, Engelbrecht AF, Didriksen AK, et al. CCHamide-2 is an orexigenic brain-gut peptide in Drosophila. PLoS ONE. 2015;10(7):e0133017. doi: 10.1371/journal.pone.0133017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sano H, Nakamura A, Texada MJ, Truman JW, Ishimoto H, Kamikouchi A, et al. The nutrient-responsive hormone CCHamide-2 controls growth by regulating insulin-like peptides in the brain of Drosophila melanogaster. PLoS Genet. 2015;11(5):e1005209. doi: 10.1371/journal.pgen.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6(2):e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bai H, Kang P, Tatar M. Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell. 2012;11(6):978–985. doi: 10.1111/acel.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yeom E, Shin H, Yoo W, Jun E, Kim S, Hong SH, et al. Tumour-derived Dilp8/INSL3 induces cancer anorexia by regulating feeding neuropeptides via Lgr3/8 in the brain. Nat Cell Biol. 2021;23(2):172–183. doi: 10.1038/s41556-020-00628-z. [DOI] [PubMed] [Google Scholar]
- 195.Andreatta G, Kyriacou CP, Flatt T, Costa R. Aminergic signaling controls ovarian dormancy in drosophila. Sci Rep. 2018;8(1):2030. doi: 10.1038/s41598-018-20407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Enell LE, Kapan N, Söderberg JA, Kahsai L, Nässel DR. Insulin signaling, lifespan and stress resistance are modulated by metabotropic GABA receptors on insulin producing cells in the brain of Drosophila. PLoS ONE. 2010;5(12):e15780. doi: 10.1371/journal.pone.0015780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zandawala M, Yurgel ME, Liao S, Texada MJ, Rewitz KF, Keene AC, et al. Modulation of Drosophila post-feeding physiology and behavior by the neuropeptide leucokinin. PLoS Genet. 2018;14(11):e1007767. doi: 10.1371/journal.pgen.1007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yurgel ME, Kakad P, Zandawala M, Nässel DR, Godenschwege TA, Keene AC. A single pair of leucokinin neurons are modulated by feeding state and regulate sleep-metabolism interactions. PLoS Biol. 2019;17(2):e2006409. doi: 10.1371/journal.pbio.2006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Alfa RW, Park S, Skelly KR, Poffenberger G, Jain N, Gu X, et al. Suppression of insulin production and secretion by a decretin hormone. Cell Metab. 2015;21(2):323–333. doi: 10.1016/j.cmet.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yoshinari Y, Kosakamoto H, Kamiyama T, Hoshino R, Matsuoka R, Kondo S, et al. The sugar-responsive enteroendocrine neuropeptide F regulates lipid metabolism through glucagon-like and insulin-like hormones in Drosophila melanogaster. Nat Commun. 2021;12(1):4818. doi: 10.1038/s41467-021-25146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nagy D, Cusumano P, Andreatta G. Peptidergic signaling from clock neurons regulates reproductive dormancy in Drosophila melanogaster. PLos Genet. 2019;15(6):e1008158. doi: 10.1371/journal.pgen.1008158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Luo J, Becnel J, Nichols CD, Nässel DR. Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5-HT(1A) receptor. Cell Mol Life Sci. 2012;69(3):471–484. doi: 10.1007/s00018-011-0789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kapan N, Lushchak OV, Luo J, Nässel DR. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell Mol Life Sci. 2012;69:4051–4066. doi: 10.1007/s00018-012-1097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Birse RT, Söderberg JA, Luo J, Winther ÅM, Nässel DR. Regulation of insulin-producing cells in the adult Drosophila brain via the tachykinin peptide receptor DTKR. J Exp Biol. 2011;214:4201–4208. doi: 10.1242/jeb.062091. [DOI] [PubMed] [Google Scholar]
- 205.Qi W, Wang G, Wang L. A novel satiety sensor detects circulating glucose and suppresses food consumption via insulin-producing cells in Drosophila. Cell Res. 2021;31(5):580–588. doi: 10.1038/s41422-020-00449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Meschi E, Léopold P, Delanoue R. An EGF-responsive neural circuit couples insulin secretion with nutrition in Drosophila. Dev Cell. 2019;48(1):76–86.e5. doi: 10.1016/j.devcel.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 207.Zhou C, Huang H, Kim SM, Lin H, Meng X, Han KA, et al. Molecular genetic analysis of sexual rejection: roles of octopamine and its receptor OAMB in Drosophila courtship conditioning. J Neurosci. 2012;32(41):14281–14287. doi: 10.1523/jneurosci.0517-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Certel SJ, Leung A, Lin CY, Perez P, Chiang AS, Kravitz EA. Octopamine neuromodulatory effects on a social behavior decision-making network in Drosophila males. PLoS ONE. 2010;5(10):e13248. doi: 10.1371/journal.pone.0013248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Certel SJ, Savella MG, Schlegel DCF, Kravitz EA. Modulation of Drosophila male behavioral choice. Proc Natl Acad Sci. 2007;104(11):4706. doi: 10.1073/pnas.0700328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hoyer SC, Eckart A, Herrel A, Zars T, Fischer SA, Hardie SL, et al. Octopamine in male aggression of Drosophila. Curr Biol. 2008;18(3):159–167. doi: 10.1016/j.cub.2007.12.052. [DOI] [PubMed] [Google Scholar]
- 211.Zhang T, Branch A, Shen P. Octopamine-mediated circuit mechanism underlying controlled appetite for palatable food in <em>Drosophila</em>. Proc Natl Acad Sci. 2013;110(38):15431. doi: 10.1073/pnas.1308816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Selcho M, Pauls D. Linking physiological processes and feeding behaviors by octopamine. Curr Opin Insect Sci. 2019;36:125–130. doi: 10.1016/j.cois.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 213.Youn H, Kirkhart C, Chia J, Scott K. A subset of octopaminergic neurons that promotes feeding initiation in Drosophila melanogaster. PLoS ONE. 2018;13(6):e0198362. doi: 10.1371/journal.pone.0198362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cheriyamkunnel SJ, Rose S, Jacob PF, Blackburn LA, Glasgow S, Moorse J, et al. A neuronal mechanism controlling the choice between feeding and sexual behaviors in Drosophila. Curr Biol. 2021;31(19):4231–45.e4. doi: 10.1016/j.cub.2021.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ishimoto H, Kamikouchi A. A feedforward circuit regulates action selection of pre-mating courtship behavior in female <em>Drosophila</em>. Curr Biol. 2020;30(3):396–407.e4. doi: 10.1016/j.cub.2019.11.065. [DOI] [PubMed] [Google Scholar]
- 216.Alekseyenko OV, Chan Y-B, Li R, Kravitz EA. Single dopaminergic neurons that modulate aggression in Drosophila. Proc Natl Acad Sci. 2013;110(15):6151. doi: 10.1073/pnas.1303446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Landayan D, Feldman DS, Wolf FW. Satiation state-dependent dopaminergic control of foraging in Drosophila. Sci Rep. 2018;8(1):5777. doi: 10.1038/s41598-018-24217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Liu Q, Tabuchi M, Liu S, Kodama L, Horiuchi W, Daniels J, et al. Branch-specific plasticity of a bifunctional dopamine circuit encodes protein hunger. Science. 2017;356(6337):534–539. doi: 10.1126/science.aal3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pooryasin A, Fiala A. Identified serotonin-releasing neurons induce behavioral quiescence and suppress mating in Drosophila. J Neurosci. 2015;35(37):12792–12812. doi: 10.1523/jneurosci.1638-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Becnel J, Johnson O, Luo J, Nässel DR, Nichols CD. The serotonin 5-HT7Dro receptor is expressed in the brain of Drosophila, and is essential for normal courtship and mating. PLoS ONE. 2011;6(6):e20800. doi: 10.1371/journal.pone.0020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Albin SD, Kaun KR, Knapp JM, Chung P, Heberlein U, Simpson JH. A subset of serotonergic neurons evokes hunger in adult Drosophila. Curr Biol. 2015;25(18):2435–2440. doi: 10.1016/j.cub.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 222.Gasque G, Conway S, Huang J, Rao Y, Vosshall LB. Small molecule drug screening in Drosophila identifies the 5HT2A receptor as a feeding modulation target. Sci Rep. 2013;3:2120. doi: 10.1038/srep02120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kim YK, Saver M, Simon J, Kent CF, Shao LS, Eddison M, et al. Repetitive aggressive encounters generate a long-lasting internal state in Drosophila melanogaster males. Proc Natl Acad Sci U S A. 2018;115(5):1099–1104. doi: 10.1073/pnas.1716612115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jiang H, Lkhagva A, Daubnerova I, Chae HS, Simo L, Jung SH, et al. Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc Natl Acad Sci U S A. 2013;110(37):E3526–E3534. doi: 10.1073/pnas.1310676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Tayler TD, Pacheco DA, Hergarden AC, Murthy M, Anderson DJ. A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proc Natl Acad Sci U S A. 2012;109(50):20697–20702. doi: 10.1073/pnas.1218246109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zer-Krispil S, Zak H, Shao L, Ben-Shaanan S, Tordjman L, Bentzur A, et al. Ejaculation induced by the activation of Crz neurons is rewarding to drosophila males. Curr Biol. 2018;28(9):1445–1452. doi: 10.1016/j.cub.2018.03.039. [DOI] [PubMed] [Google Scholar]
- 227.Lee KM, Daubnerova I, Isaac RE, Zhang C, Choi S, Chung J, et al. A neuronal pathway that controls sperm ejection and storage in female Drosophila. Curr Biol. 2015;25(6):790–797. doi: 10.1016/j.cub.2015.01.050. [DOI] [PubMed] [Google Scholar]
- 228.Jang YH, Chae HS, Kim YJ. Female-specific myoinhibitory peptide neurons regulate mating receptivity in Drosophila melanogaster. Nat Commun. 2017;8(1):1630. doi: 10.1038/s41467-017-01794-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Kim WJ, Jan LY, Jan YN. A PDF/NPF neuropeptide signaling circuitry of male Drosophila melanogaster controls rival-induced prolonged mating. Neuron. 2013;80(5):1190–1205. doi: 10.1016/j.neuron.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shohat-Ophir G, Kaun KR, Azanchi R, Heberlein U. Sexual deprivation increases ethanol intake in Drosophila. Science. 2012;335(6074):1351–1355. doi: 10.1126/science.1215932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gendron CM, Kuo TH, Harvanek ZM, Chung BY, Yew JY, Dierick HA, et al. Drosophila life span and physiology are modulated by sexual perception and reward. Science. 2014;343(6170):544–548. doi: 10.1126/science.1243339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Krupp JJ, Billeter JC, Wong A, Choi C, Nitabach MN, Levine JD. Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in Drosophila. Neuron. 2013;79(1):54–68. doi: 10.1016/j.neuron.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Yang CH, Rumpf S, Xiang Y, Gordon MD, Song W, Jan LY, et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61(4):519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chen PS, Stumm-Zollinger E, Aigaki T, Balmer J, Bienz M, Bohlen P. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell. 1988;54(3):291–298. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 235.Wolfner MF. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity (Edinb) 2002;88(2):85–93. doi: 10.1038/sj.hdy.6800017. [DOI] [PubMed] [Google Scholar]
- 236.Terhzaz S, Rosay P, Goodwin SF, Veenstra JA. The neuropeptide SIFamide modulates sexual behavior in Drosophila. Biochem Biophys Res Commun. 2007;352(2):305–310. doi: 10.1016/j.bbrc.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 237.Sellami A, Veenstra JA. SIFamide acts on fruitless neurons to modulate sexual behavior in Drosophila melanogaster. Peptides. 2015;74:50–56. doi: 10.1016/j.peptides.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 238.Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167(1):311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bharucha KN, Tarr P, Zipursky SL. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J Exp Biol. 2008;211(Pt 19):3103–3110. doi: 10.1242/jeb.016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Hergarden AC, Tayler TD, Anderson DJ. Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc Natl Acad Sci U S A. 2012;109(10):3967–3972. doi: 10.1073/pnas.1200778109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yu Y, Huang R, Ye J, Zhang V, Wu C, Cheng G, et al. Regulation of starvation-induced hyperactivity by insulin and glucagon signaling in adult Drosophila. Elife. 2016;5:e15693. doi: 10.7554/eLife.15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Chen J, Reiher W, Hermann-Luibl C, Sellami A, Cognigni P, Kondo S, et al. Allatostatin a signalling in drosophila regulates feeding and sleep and is modulated by PDF. PLoS Genet. 2016;12(9):e1006346. doi: 10.1371/journal.pgen.1006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kubrak OI, Lushchak OV, Zandawala M, Nässel DR. Systemic corazonin signalling modulates stress responses and metabolism in Drosophila. Open Biol. 2016 doi: 10.1098/rsob.160152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zandawala M, Marley R, Davies SA, Nässel DR. Characterization of a set of abdominal neuroendocrine cells that regulate stress physiology using colocalized diuretic peptides in Drosophila. Cell Mol Life Sci CMLS. 2018;75(6):1099–1115. doi: 10.1007/s00018-017-2682-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yang Z, Huang R, Fu X, Wang G, Qi W, Mao D, et al. A post-ingestive amino acid sensor promotes food consumption in Drosophila. Cell Res. 2018;28(10):1013–1025. doi: 10.1038/s41422-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Dus M, Lai JSY, Gunapala KM, Min S, Tayler TD, Hergarden AC, et al. Nutrient sensor in the brain directs the action of the brain-gut axis in drosophila. Neuron. 2015;87(1):139–151. doi: 10.1016/j.neuron.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhan YP, Liu L, Zhu Y. Taotie neurons regulate appetite in Drosophila. Nat Commun. 2016;7:13633. doi: 10.1038/ncomms13633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Pool AH, Scott K. Feeding regulation in Drosophila. Curr Opin Neurobiol. 2014;29:57–63. doi: 10.1016/j.conb.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lin S, Senapati B, Tsao C-H. Neural basis of hunger-driven behaviour in Drosophila. Open Biol. 2019;9(3):180259. doi: 10.1098/rsob.180259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell. 2011;145(1):133–144. doi: 10.1016/j.cell.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 2005;3(9):e305. doi: 10.1371/journal.pbio.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Galikova M, Dircksen H, Nässel DR. The thirsty fly: Ion transport peptide (ITP) is a novel endocrine regulator of water homeostasis in Drosophila. PLoS Genet. 2018;14(8):e1007618. doi: 10.1371/journal.pgen.1007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Al-Anzi B, Armand E, Nagamei P, Olszewski M, Sapin V, Waters C, et al. The leucokinin pathway and its neurons regulate meal size in Drosophila. Curr Biol. 2010;20(11):969–978. doi: 10.1016/j.cub.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 2008;7(2):199–206. doi: 10.1111/j.1474-9726.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yang T, Yuan Z, Liu C, Liu T, Zhang W. A neural circuit integrates pharyngeal sensation to control feeding. Cell Rep. 2021 doi: 10.1016/j.celrep.2021.109983. [DOI] [PubMed] [Google Scholar]
- 256.Chung BY, Ro J, Hutter SA, Miller KM, Guduguntla LS, Kondo S, et al. Drosophila neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 2017;19(12):2441–2450. doi: 10.1016/j.celrep.2017.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Shen P, Cai HN. Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J Neurobiol. 2001;47(1):16–25. doi: 10.1002/neu.1012. [DOI] [PubMed] [Google Scholar]
- 258.Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci U S A. 2005;102(37):13289–13294. doi: 10.1073/pnas.0501914102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. 2005;8(10):1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
- 260.Pu Y, Zhang Y, Zhang Y, Shen P. Two drosophila neuropeptide Y-like neurons define a reward module for transforming appetitive odor representations to motivation. Sci Rep. 2018;8(1):11658. doi: 10.1038/s41598-018-30113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Tsao C-H, Chen C-C, Lin C-H, Yang H-Y, Lin S. Drosophila mushroom bodies integrate hunger and satiety signals to control innate food-seeking behavior. Elife. 2018;7:e35264. doi: 10.7554/eLife.35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang Y, Pu Y, Shen P. Neuropeptide-gated perception of appetitive olfactory inputs in Drosophila larvae. Cell Rep. 2013;3(3):820–830. doi: 10.1016/j.celrep.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 263.Carvalho GB, Kapahi P, Anderson DJ, Benzer S. Allocrine modulation of feeding behavior by the sex peptide of Drosophila. Curr Biol. 2006;16(7):692–696. doi: 10.1016/j.cub.2006.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Barnes AI, Wigby S, Boone JM, Partridge L, Chapman T. Feeding, fecundity and lifespan in female Drosophila melanogaster. Proc Biol Sci. 2008;275(1643):1675–1683. doi: 10.1098/rspb.2008.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kubli E. Sex-peptides: seminal peptides of the Drosophila male. Cell Mol Life Sci. 2003;60(8):1689–1704. doi: 10.1007/s00018-003-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Chapman T, Bangham J, Vinti G, Seifried B, Lung O, Wolfner MF, et al. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci U S A. 2003;100(17):9923–9928. doi: 10.1073/pnas.1631635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Martelli C, Pech U, Kobbenbring S, Pauls D, Bahl B, Sommer MV, et al. SIFamide translates hunger signals into appetitive and feeding behavior in Drosophila. Cell Rep. 2017;20(2):464–478. doi: 10.1016/j.celrep.2017.06.043. [DOI] [PubMed] [Google Scholar]
- 268.Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem. 2004;279(49):50781–50789. doi: 10.1074/jbc.M407842200. [DOI] [PubMed] [Google Scholar]
- 269.Hong SH, Lee KS, Kwak SJ, Kim AK, Bai H, Jung MS, et al. Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in drosophila and mammals. PLoS Genet. 2012;8(8):e1002857. doi: 10.1371/journal.pgen.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ko KI, Root CM, Lindsay SA, Zaninovich OA, Shepherd AK, Wasserman SA, et al. Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. Elife. 2015 doi: 10.7554/eLife.08298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Nässel DR, Williams MJ. Cholecystokinin-like peptide (DSK) in Drosophila, not only for satiety signaling. Front Endocrinol (Lausanne) 2014;5:219. doi: 10.3389/fendo.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Panksepp J, Burgdorf J, Beinfeld MC, Kroes RA, Moskal JR. Regional brain cholecystokinin changes as a function of friendly and aggressive social interactions in rats. Brain Res. 2004;1025(1):75–84. doi: 10.1016/j.brainres.2004.07.076. [DOI] [PubMed] [Google Scholar]
- 273.Crawley JN. Comparative distribution of cholecystokinin and other neuropeptides. Ann N Y Acad Sci. 1985;448(1):1–8. doi: 10.1111/j.1749-6632.1985.tb29900.x. [DOI] [PubMed] [Google Scholar]
- 274.Lee SY, Soltesz I. Cholecystokinin: a multi-functional molecular switch of neuronal circuits. Dev Neurobiol. 2011;71(1):83–91. doi: 10.1002/dneu.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hökfelt T, Rehfeld JF, Skirboll L, Ivemark B, Goldstein M, Markey K. Evidence for coexistence of dopamine and CCK in meso-limbic neurones. Nature. 1980;285(5765):476–478. doi: 10.1038/285476a0. [DOI] [PubMed] [Google Scholar]
- 276.Hökfelt T, Millhorn D, Seroogy K, Tsuruo Y, Ceccatelli S, Lindh B, et al. Coexistence of peptides with classical neurotransmitters. Experientia. 1987;43(7):768–780. doi: 10.1007/BF01945354. [DOI] [PubMed] [Google Scholar]
- 277.Smith SJ, Sümbül U, Graybuck LT, Collman F, Seshamani S, Gala R, et al. Single-cell transcriptomic evidence for dense intracortical neuropeptide networks. Elife. 2019;8:e47889. doi: 10.7554/eLife.47889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Croset V, Treiber CD, Waddell S. Cellular diversity in the Drosophila midbrain revealed by single-cell transcriptomics. BioRxiv. 2017 doi: 10.1101/237818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Davie K, Janssens J, Koldere D, De Waegeneer M, Pech U, Kreft L, et al. A single-cell transcriptome atlas of the aging Drosophila brain. Cell. 2018;174:982–998. doi: 10.1016/j.cell.2018.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zeltser LM. Feeding circuit development and early-life influences on future feeding behaviour. Nat Rev Neurosci. 2018;19(5):302–316. doi: 10.1038/nrn.2018.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Huang H, Possidente DR, Vecsey CG. Optogenetic activation of SIFamide (SIFa) neurons induces a complex sleep-promoting effect in the fruit fly Drosophila melanogaster. Physiol Behav. 2021;239:113507. doi: 10.1016/j.physbeh.2021.113507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wong K, Schweizer J, Nguyen K-NH, Atieh S, Kim WJ. Neuropeptide relay between SIFa signaling controls the experience-dependent mating duration of male Drosophila. BioRxiv. 2019 doi: 10.1101/819045. [DOI] [Google Scholar]
- 283.Schwarz JE, King AN, Hsu CT, Barber AF, Sehgal A. Hugin neurons link the sleep homeostat to circadian clock neurons. BioRxiv. 2021 doi: 10.1101/2020.04.29.068627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.King AN, Barber AF, Smith AE, Dreyer AP, Sitaraman D, Nitabach MN, et al. A peptidergic circuit links the circadian clock to locomotor activity. Curr Biol. 2017;27(13):1915–1927. doi: 10.1016/j.cub.2017.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schlegel P, Texada MJ, Miroschnikow A, Schoofs A, Huckesfeld S, Peters M, et al. Synaptic transmission parallels neuromodulation in a central food-intake circuit. Elife. 2016 doi: 10.7554/eLife.16799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Morton GJ, Blevins JE, Williams DL, Niswender KD, Gelling RW, Rhodes CJ, et al. Leptin action in the forebrain regulates the hindbrain response to satiety signals. J Clin Investig. 2005;115(3):703–710. doi: 10.1172/JCI200522081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.White K, Hurteau T, Punsal P. Neuropeptide-FMRFamide-like immunoreactivity in Drosophila: development and distribution. J Comp Neurol. 1986;247(4):430–438. doi: 10.1002/cne.902470403. [DOI] [PubMed] [Google Scholar]
- 288.Schneider LE, Obrien MA, Taghert PH. In situ hybridization analysis of the FMRFamide neuropeptide gene in Drosophila. I. Restricted expression in embryonic and larval stages. J Comp Neurol. 1991;304(4):608–622. doi: 10.1002/cne.903040408. [DOI] [PubMed] [Google Scholar]
- 289.Lee SS, Wu MN. Neural circuit mechanisms encoding motivational states in Drosophila. Curr Opin Neurobiol. 2020;64:135–142. doi: 10.1016/j.conb.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article is a literature review and does not contain any new data. Source data files for Figs. 1 and 2 are in Supplementary material.