Abstract
Siderophores are iron-chelating molecules that solubilize Fe3+ for microbial utilization and facilitate colonization or infection of eukaryotes by liberating host iron for bacterial uptake. By fluorescently labeling membrane receptors and binding proteins, we created 20 sensors that detect, discriminate, and quantify apo- and ferric siderophores. The sensor proteins originated from TonB-dependent ligand-gated porins (LGPs) of Escherichia coli (Fiu, FepA, Cir, FhuA, IutA, BtuB), Klebsiella pneumoniae (IroN, FepA, FyuA), Acinetobacter baumannii (PiuA, FepA, PirA, BauA), Pseudomonas aeruginosa (FepA, FpvA), and Caulobacter crescentus (HutA) from a periplasmic E. coli binding protein (FepB) and from a human serum binding protein (siderocalin). They detected ferric catecholates (enterobactin, degraded enterobactin, glucosylated enterobactin, dihydroxybenzoate, dihydroxybenzoyl serine, cefidericol, MB-1), ferric hydroxamates (ferrichromes, aerobactin), mixed iron complexes (yersiniabactin, acinetobactin, pyoverdine), and porphyrins (hemin, vitamin B12). The sensors defined the specificities and corresponding affinities of the LGPs and binding proteins and monitored ferric siderophore and porphyrin transport by microbial pathogens. We also quantified, for the first time, broad recognition of diverse ferric complexes by some LGPs, as well as monospecificity for a single metal chelate by others. In addition to their primary ferric siderophore ligands, most LGPs bound the corresponding aposiderophore with ∼100-fold lower affinity. These sensors provide insights into ferric siderophore biosynthesis and uptake pathways in free-living, commensal, and pathogenic Gram-negative bacteria.
Keywords: siderophore, iron transport, fluorescent sensor, disulfide, site-directed mutagenesis, bacterial pathogenesis
Abbreviations: Abn, aerobactin; Acn, acinetobactin; Agn, anguibactin; B12, vitamin B12; Crn, corynebactin; DHB, dihydroxybenzoate; DHBS, dihydroxybenzoyl serine; Ent, enterobactin; Ent∗, degraded Ent; FBP, fluorescent binding protein; Fc, ferrichrome; FcA, ferrichrome A; FDC, cefidericol; FLGP, fluorescent ligand-gated porin; FM, fluorescein maleimide; FxB, ferrioxamine B; GEnt, glucosylated Ent; Hn, hemin; LGP, ligand-gated porin; OM, outer membrane; Pvd, pyoverdine; SCN, siderocalin; Ybt, yersiniabactin
Eukaryotic hosts and prokaryotic pathogens compete for iron (1), which influences the outcome of bacterial infections (2, 3). In aqueous solutions iron oxidizes and polymerizes as ferric oxyhydroxide (Fe(OH)n), the low solubility of which (KSP = 2.79 × 10−39 (4)) antagonizes its biological utilization. But, microbial siderophores (5) chelate Fe3+ with such high affinity (Ka = 1020–1052 (5)) that they release it from precipitates as hexacoordinate complexes that are available for assimilation (6). To the converse, humans and animals produce serum transferrin, lactoferrin, and cellular ferritin that bind, sequester, or store iron (7, 8). The vertebrate innate immune system (9) also actively inhibits siderophore-mediated iron acquisition by producing siderocalin (SCN; also called NGAL or lipocalin 2 (10, 11)). Nevertheless, aposiderophores can capture Fe3+ from the binding proteins [8,9,20–32,37], and Gram-negative bacteria internalize ferric siderophores [33–36], heme [27–29], and other porphyrins (12, 13) through TonB-dependent outer membrane (OM) receptor proteins. Consequently, siderophore biosynthesis and TonB-dependent iron uptake [9,12,14,84] promote colonization of the animal gut (14, 15, 16, 17, 18, 19, 20). Prokaryotic iron acquisition pathways are also a potential vulnerability, in that their disruption inhibits bacterial growth [1–7], reduces virulence, and may eliminate pathogenesis [2,8–15].
In Gram-negative cells, Fe3+ acquisition begins when cell surface receptors recognize and bind iron complexes. We refer to these OM proteins as ligand-gated porins (LGPs; (21)) because ligand binding triggers the TonB-dependent (22) opening of their normally closed, 22-stranded, transmembrane β-barrels (22, 23). The latter attribute categorizes them in the porin superfamily (24, 25). Unlike other porins (26), such as OmpF (27, 28) or LamB (29), LGP do not contain open transmembrane pores that act by diffusion. The C-terminal channels of LGP are occupied by an N-terminal, ∼150-residue globular domain that regulates ligand uptake by its interactions with the energy-dependent (30) cell envelope protein TonB (31, 32). As such, they are also called TonB-dependent transporters (TBDTs; (33)). All LGP-mediated Fe3+ transport requires the actions of TonB, which anchors in the inner membrane but spans the periplasm to associate with proteins in the OM (31, 32) (for additional information, see Fig. 1 in (1)).
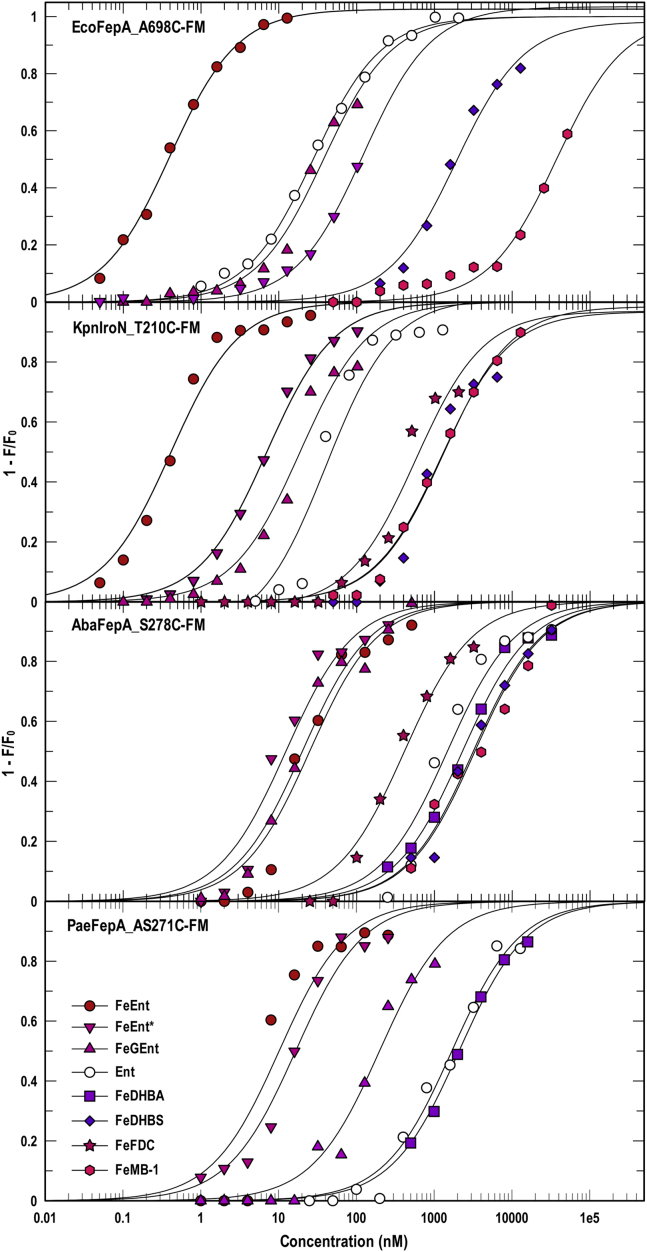
Figure 1.
Broad recognition of ferric catecholates by FepA orthologs.EcofepA, KpniroN, AbafepA, and PaefepA were cloned in pITS23, mutagenized to introduce single Cys residues, labeled with FM, and then assayed for binding of different ferric catecholate siderophores (see also Table 1). The comparisons revealed a hierarchy of affinities for mono- and tricatecholate iron complexes that was different among EcoFepA and its homologs in the three ESKAPE bacteria, which roughly mimicked their extents of sequence identity. For each data point we made three determinations of quenching in response to the addition of different concentrations of an iron complex. The curves represent nonlinear fits of the mean values to a single-site binding model using Grafit 6.0.12. The mean standard errors of the resulting KD values from the fitted curves were EcoFepA_A698C-FM, 17.3%; KpnIroNT210C-FM, 26.3%; AbaFepAS278C-FM, 21.1%; PaeFepA_S271C-FM, 22.4%. Statistics on the accuracy of each individual KD determination appear in Table 1.
Acquisition of certain ferric siderophores may promote pathogenesis and the invasion of specific host tissues. For example, the production of yersiniabactin (Ybt) by Yersinia pestis correlates with the development of bubonic or pneumonic plague (34, 35, 36). A variety of Gram-negative bacteria produce aerobactin (Abn) and utilize of ferric aerobactin (FeAbn), which enhances their virulence, invasiveness, and tissue tropism (37, 38, 39, 40, 41, 42). Similarly, elaboration of the catecholate enterobactin (Ent) and uptake of ferric enterobactin (FeEnt) facilitates bacterial colonization of eukaryotic hosts (14). Glycosylation of Ent (GEnt) (43, 44, 45) by the IroA system of Klebsiella pneumoniae and other pathogens (45, 46, 47, 48) reduces its neutralization by SCN (49, 50, 51), enhancing bacterial infection of specific tissues (for example, by K. pneumoniae (45, 52)). These relationships between specific siderophores and infectious diseases, as well as the patterns of siderophore expression and transport during colonization of host tissues, underscore the potential value of biochemical tools that may characterize or explicate such phenomena. Sensors of apo- and ferric siderophores (for additional information, see Fig. 1 in (53)) report kinetic and thermodynamic data on prokaryotic iron acquisition systems and may provide diagnostic information about the nature and virulence of bacterial infections. LGPs discriminate among >500 iron complexes (54), so their fluorescent derivatives (53) may sensitively inform about which siderophores are present in the microenvironment, or what their concentrations are in clinical samples, or how their utilization relates to pathogenicity. Toward those ends, the production of a fluorescent LGP (FLGP) in a transport-deficient, ΔtonB host creates a “decoy” sensor cell that detects and quantifies ligands in experimental suspensions or solutions. We used this strategy to produce 18 FLGP sensors and two fluorescent binding protein (FBP) sensors from siderocalin and EcoFepB. Collectively, they detected siderophore biosynthesis, measured siderophore concentrations, and monitored ferric siderophore uptake by cells of interest.
Results
Siderophore–sensor pairs
We engineered FLGP sensors of ferric catecholates (Ent, GEnt, dihydroxybenzoate [DHB], dihydroxybenzoyl serine [DHBS], cefidericol [FDC; (55, 56)], MB-1 (57, 58)), ferric hydroxamates (Abn, ferrichrome [Fc]), mixed chelators (Ybt, acinetobactin [Acn], anguibactin [Agn], pyoverdine [Pvd]), and porphyrins (hemin [Hn], vitamin B12 [B12]) (Table 1). In our experiments with these compounds we used the more readily available siderophore Agn as a biological equivalent of Acn, because the two chelators are nearly chemically identical (59, 60) and functionally equivalent with regard to recognition by Acinetobacter baumannii (61, 62). In total, the FLGP sensors originated from Escherichia coli (EcoFiu,† EcoFepA, EcoCir, EcoFhuA, EcoIutA, EcoBtuB), K. pneumoniae (KpnIroN, KpnFepA1658, KpnFepA4984, KpnFepA2380, KpnFyuA), A. baumannii (AbaFepA, AbaPirA, AbaPiuA, AbaBauA), Pseudomonas aeruginosa (PaeFepA, PaeFpvA), and Caulobacter crescentus (CcrHutA). We made additional FBP sensors from EcoFepB, a periplasmic binding protein that acts in ferric catecholate uptake, and human siderocalin (HsaSCN), which recognizes multiple siderophores. We cloned the LGP structural genes in the low-copy plasmid pHSG575 (63, 64, 65) and the binding protein genes in pET28a-c (+), that added a 6-His tag to their N termini. After verifying the clones by DNA sequencing, we changed a handful of residues in each protein to cysteine. Nearly all of these OM proteins are devoid of cysteine; the few exceptions contain Cys pairs that are unreactive unless subject to reduction by β-mercaptoethanol or other chemical agents (66). We chose the target residues for site-directed Cys mutagenesis from analysis of crystal structures (EcoFepA, EcoCir, EcoFiu, EcoFhuA, EcoBtuB, KpnFyuA, AbaPiuA, AbaPirA, AbaBauA, HsaSCN) or by their location in hypothetical structures predicted by the Modeller algorithm of CHIMERA (CcrHutA, EcoIutA, KpnFepA, KpnIroN, AbaFepA, PaeFepA). After construction and verification, we expressed the proteins in E. coli, subjected each Cys mutant LGP to modification by fluorescein maleimide (FM; 5 μM for 15 min at 37 °C; (67)), and then analyzed their expression, fluoresceination, and fluorescence quenching during binding of a metal complex. Relative to pathogenic host organisms, the production of FLGP in nonpathogenic, rough (rfaB) E. coli K-12 facilitated their optimum FM labeling in a cell surface environment that is unobscured by LPS O-antigen or capsule. We obtained high-level, usually stoichiometric modification of the engineered Cys sulfhydryls with extrinsic maleimide fluorophores (Figs. S2–S6). Certain Cys locations were better or worse for some LGP (e.g., AbaBauA, EcoFiu), but we always found accessible labeling sites in each OM protein. We then chose the most sensitive single mutant of each LGP and spectroscopically determined its specificities and affinities for the apo- and ferric siderophores in relevant chemical classes (Table 1). For production of HsaSCN and EcoFepB, we introduced Cys substitutions at positions near their binding sites, overexpressed the proteins, and purified them from cell lysates by metal ion chromatography (Fig. S6). For each LGP or soluble binding protein sensor, we studied three or more Cys substitutions to identify an effective site for attachment of an extrinsic fluorophore. To optimize detection and quantification of the individual metal complexes, we compared the fluorescence intensities of the different Cys mutant derivatives, as well as the extents of their quenching by their homologous and heterologous ligands.
Table 1.
Affinities (KD values; nM or uM) of LGP sensors for apo and ferric siderophores and porphyrins
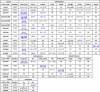 |
1LGP acronyms abbreviate the genus and species of their origin: E. coli FepA, EcoFepA; K. pneumoniae FepA locus 1658, KpnFepA1658; Yersinia pestis FyuA, YpeFyuA; Homo sapien SCN, HsaSCN, etc.
2Location of the Cys substitution in the mature protein sequence.
3The table lists KD values (blue text: nM; black text: uM) from analysis of fluorescence quenching with ferric or apo (parenthetic values) siderophores or porphyrins (see Figure 1, Figure 2, Figure 3). We performed each quenching titration 2 or 3 times; the tabulated KD values result from a single representative experiment and are listed with their associated standard errors from a nonlinear curve fit to a single-site binding model, using Grafit 6.012 (Erithacus Ltd, Middlesex, UK). The KD values of preferred natural ligands are underlined. FeEnt∗, partially degraded FeEnt; NB, no binding; ND, no data.
LGP binding promiscuity
The high (nanomolar) affinity of certain LGPs for a particular ligand suggested potential exclusivity in their binding reactions. EcoFepA, for example, tightly adsorbs FeEnt (KD = 0.1–0.4 nM (53, 68, 69, 70)), which allows it to scavenge subnanomolar concentrations of FeEnt. However, we found that EcoFepA does not have exclusive specificity for FeEnt. Like PaeFpvA (71, 72), the EcoFepA-FM sensor had considerable affinity for the aposiderophore, Ent (KD = 27 nM). As we surveyed other FLGP in our panel this result became a general trend: 11 other FLGPs showed ∼100-fold lower binding affinity for their corresponding aposiderophores. Thus, FeEnt was the primary ligand of EcoFepA, KpnFepA1658 and 4984, AbaFepA, PaeFepA, KpnIroN, and EcoFepB, but they all also adsorbed Ent with 70- to 200-fold lower affinity (Table 1). Receptors for nonphenolate iron complexes showed the same duality: EcoFhuA, EcoIutA, and KpnFyuA bound apoFc, Abn, and Ybt with 60- to 200-fold lower affinity than seen for their ferric complexes. In addition to aposiderophores, EcoFepA also recognized other chemically similar iron complexes. Several different forms of catecholate chelators are relevant to Gram-negative bacterial iron acquisition. Besides glucosylation, the three catecholate groups of FeEnt may oxidize to quinones, and the lactone ring that conjoins them may hydrolyze to yield linear tri-, di-, and monocatecholates. These degradation products (FeEnt∗) arose even when FeEnt was stored on ice. Although we typically removed them by chromatography on Sephadex LH20 (73), we also studied their interactions with the FLGP. EcoFiu and EcoCir were previously linked to the uptake of ferric monocatecholates (74), but unexpectedly, decoy assays showed that EcoFepA-FM adsorbed FeEnt∗ (KD = 38 nM) and had weak affinity for FeGEnt (KD = 122 nM) and for FeDHBS (KD = 2000 nM). This was another trend: besides highest affinity for a primary ligand (EcoFepA: FeEnt; EcoFiu: FeEnt∗; EcoCir: FeEnt∗, EcoFhuA: Fc), many LGP sensors manifested secondary, lower affinities for related metal chelates (Fig. 1 and Table 1).
The specificities of orthologous LGP in related organisms were unpredictable and complicated, as illustrated by the ferric catecholate uptake systems of Gram-negative ESKAPE pathogens. First, the EcoFepA orthologs KpnFepA1658 (82% identity) and PaePfeA (61% identity) had the same ligand preferences as EcoFepA (FeEnt > FeEnt∗ > FeGEnt). However, AbaFepA (46% identity) differed, in that it bound FeEnt∗ slightly better (KD = 12 nM) than FeEnt and FeGEnt (KD ≈ 25 nM). Second, other orthologous ferric catecholate receptors displayed both recognition correspondences and differences. EcoCir and AbaPirA were a pair (37% identity), with considerably lower (micromolar) affinities for iron complexes. Both bound FeEnt∗ (KD values of 7 and 13 μM, respectively), but both also recognized other iron complexes, with preferences (Table 1):
EcoCir: FeFDC > FeEnt∗ ≈ FeDHBS > Fe-MB-1 > FeEnt;
AbaPirA: FeFDC > FeEnt > FeDHB > FeEnt∗ > FeMB-1 > FeDHBS.
Similarly, EcoFiu and AbaPiuA (31% identity) both manifested lower (generally micromolar) binding affinities for ferric catecholates (Table 1), which produced different priorities:
EcoFiu: FeDHBS > FeGEnt > FeEnt∗ > FeEnt ≈ FeFDC.
AbaPiuA: FeEnt∗ > FeEnt > FeDHB > FeMB-1 > FeFDC.
EcoFiu did not bind FeDHB; AbaPiuA did not bind FeGEnt or FeDHBS. So, in both cases, despite their annotation as orthologs, EcoCir and AbaPirA, and EcoFiu and AbaPiu were quite different in their binding specificities.
Lastly, KpnIroN grouped with EcoFiu (21% identity), EcoFepA (51% identity), and AbaFepA (49.4% identity), in that all four proteins recognized FeEnt, FeEnt∗, and FeGEnt, but with different priorities:
EcoFepA: FeEnt > FeEnt∗ > FeGEnt;
EcoFiu: FeGEnt > FeEnt∗ > FeEnt;
KpnIroN: FeEnt > FeGEnt > FeEnt∗;
AbaFepA: FeEnt∗ > FeGEnt ≈ FeEnt. All the FepA orthologs also recognized some combination of ferric monocatecholates, with lower affinity.
Besides their divergent specificities, some LGP in the catecholate receptor group had much lower binding and transport affinities for their ligands. Overall, the ferric catecholate receptors spanned a 10,000-fold range of affinity, from subnanomolar to 10 to 15 μM KD values, suggesting that certain receptors (AbaPiuA, EcoCir, AbaPirA, AbaBauA) are only functionally relevant when their ferric siderophore ligands are present at comparatively high (i.e., μM) concentrations (Fig. 1 and Table 1). Overall, the catecholate-specific FLGP revealed that Gram-negative bacteria produce assortments of OM proteins that discriminate among the ferric siderophores they encounter in nature or in the host environment. The different selectivities and affinities of these receptors allow the bacteria to adapt to a variety of siderophore compositions and concentrations.
LGP binding selectivity
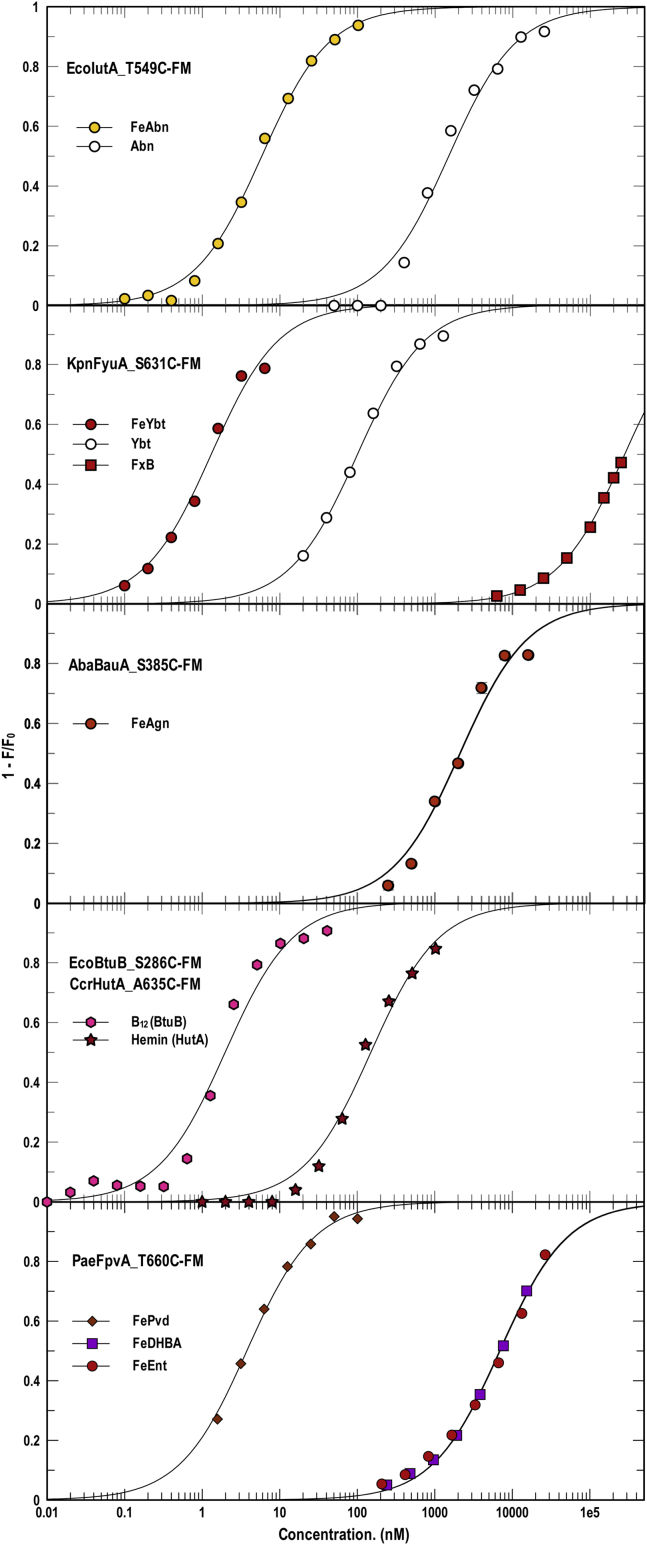
Although the ferric catecholate LGP sensors displayed broad recognition specificity (Fig. 1), other LGP sensors showed singularity for one metal complex (Fig. 2). EcoIutA, EcoBtuB, CcrHutA, KpnFyuA, AbaBauA, and PaeFpvA were nearly or fully exclusive in their binding of FeAbn, B12, hemin, FeYbt, FeAgn, and FePvd, respectively. Among those, only KpnFyuA (that weakly bound FxB) and PaeFpvA (that weakly bound FeEnt, FeEnt∗, and FeDHB) had any affinity for other ferric siderophores besides their primary ligands. IutA, for example, had high affinity for FeAbn (KD = 6 nM) and did not measurably bind any other ferric hydroxamates (Fc, FcA, FxB). Each of these more specific receptors was aptly suited to its biochemical role. Abn biosynthesis and FeAbn uptake are often encoded together on transferable plasmids; bacteria that harbor such plasmids express a complete system for siderophore production and ferric siderophore acquisition, which confers invasiveness in the host (75). Hence, EcoIutA's selectivity for FeAbn, and its blindness to other ferric complexes, well complement Abn production by virulent strains. Likewise, KpnFyuA and AbaBauA exclusively bound ferric siderophores associated with the pathogenicity of their producers. CcrHutA and EcoBtuB were also specific for iron and cobalt porphyrins, respectively (Fig. 2).
Figure 2.
Specific recognition of ferric siderophores by FLGP. Binding of FeAbn, FeYbt, FeAgn, B12, hemin, and FePvD by EcoIutA, KpnFyuA, AbaBauA, EcoBtuB, CcrHutA, and PaeFpvA, respectively. Structural genes of each of the LGPs were cloned in pITS23, mutagenized to introduce single Cys residues, labeled with FM, and then assayed for binding of different hydroxamate and mixed-chelation siderophores (see also Table 1). For each data point we made three determinations of quenching in response to the addition of different concentrations of an iron complex. The curves represent nonlinear fits of the mean values to a single-site binding model using Grafit 6.0.12. The mean standard errors of the resulting KD values from the fitted curves were EcoIutA_T549C-FM, 13%; KpnFyuA_S631C-FM, 12%; AbaBauA_S385C-FM, 19%; EcoBtuB_S286C-FM, 16%; CcrHutA_A635C-FM, 18%; PaeFpvA_T660C-FM, 9.7%. Statistics on the accuracy of each individual KD determination appear in Table 1. The experiment revealed nearly exclusive binding of all the metal complexes by their respective LGP receptor proteins. KpnFyuA also weakly bound FxB, and PaeFpvA weakly bound FeEnt and FeDHB.
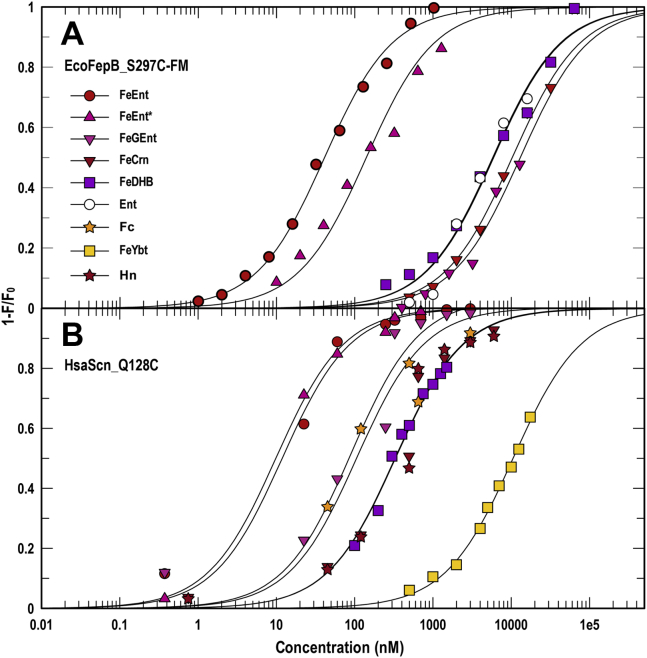
Specificities of EcoFepB and HsaSCN
The periplasmic protein FepB recognizes and binds FeEnt after its internalization through the OM, and the innate immune component SCN opposes the iron scavenging of bacterial pathogens by adsorbing apo- and ferric siderophores, thereby reducing their concentrations in tissues, body fluids, blood, and serum (76, 77, 78). After cloning EcofepB and the cDNA of the human gene HsaSCN in pET28, we genetically engineered the substitutions T297C and Q128C, respectively, purified the overexpressed His-tagged mutant binding proteins by metal affinity chromatography, and labeled them with FM. As expected, the periplasmic binding protein EcoFepB-FM preferentially recognized FeEnt (KD = 43 μM), about the same as reported for the native protein (79), and with affinity that was about 100-fold less than EcoFepA. EcoFepB also bound Ent, FeEnt∗, FeGEnt, FeCrn, and FeDHB, with a range of affinities. In that sense, it resembled HsaSCN. The serum FBP sensor preferred FeEnt and FeEnt∗ (KD = 10–12 nM) among the apo and ferric siderophores we tested. HsaSCN-FM also showed submicromolar affinity for other ferric catecholates, including FeGEnt (KD = 0.09 μM), FeDHB (KD = 0.33 μM), and FeCrn (KD = 0.33 μM). It similarly adsorbed ferric hydroxamates and mixed ferric chelates, usually with submicromolar affinity: Fc (KD = 0.11 μM), FxB (KD = 0.18 μM), FeAgn (KD = 0.16 μM), FeYbt (KD = 11 μM). Overall, HsaSCN adsorbed many iron chelates, more than any of the LGPs, with affinities over 3 logs of concentration (10 nM–11 μM; Fig. 3).
Figure 3.
Binding of ferric siderophores by EcoFepB and HsaSCN. After expression and purification of EcoFepB_T297C-FM (A) and the HsaSCN _Q128C-FM (B), we evaluated the affinities of the two binding proteins for a spectrum of apo and ferric siderophores. For each data point we made three determinations of quenching in response to the addition of different concentrations of an iron complex. The curves represent nonlinear fits of the mean values to a single-site binding model using Grafit 6.0.12. The mean standard errors of the resulting KD values from the fitted curves were EcoFepB_T297C-FM, 23%; HsaScn_Q128C-FM, 21%. Statistics on the accuracy of each individual KD determination appear in Table 1.
Measurement of siderophores in complex samples
Decoy sensors that possess high affinity for a single ligand can unambiguously detect, identify, and quantify that compound in experimental samples. To illustrate this capability we grew laboratory (MG1655 (80)), probiotic (Nissle 1917 (81, 82, 83)), and pathogenic (CP9 (84)) strains of E. coli and two hypervirulent strains of K. pneumoniae (HvKp1, HvKp2 (39)) in iron-deficient MOPS (85) and M9 (86) minimal media, removed the cells by centrifugation, and analyzed the supernatants with decoy sensors. Under these conditions the bacteria maximize siderophore production, in some cases so much that ferration of the spent media resulted in deeply red- or orange-colored solutions. The genomic and biosynthetic pathways of these strains (53, 68, 69, 70) implied the production of Ent, GEnt, Abn, and Ybt. Therefore, we interrogated their ferrrated supernatants with EcoFepA-FM, EcoIutA-FM, KpnIroN-FM, and KpnFyuA-FM (Table 2 and Fig. 4). The sensors found only three of the four siderophores in the CP9 and HvKpn1 supernatants, and they were different between the two: in the former case, FeEnt, FeGEnt, and FeYbt; in the latter case, FeEnt, FeGEnt, and FeAbn. HvKpn2, on the other hand, produced all four siderophores, which enhances its potential virulence relative to the other strains.
Table 2.
Concentrations (μM) of Ent, GEnt, Abn, and Ybt in spent bacterial media
| Conditions |
Strain |
|||||||
|---|---|---|---|---|---|---|---|---|
| FLGP | Ligand | Mediuma | BN1071 | MG1655 | Nissle | CP9 | HvKp1 | HvKp2 |
| EcoFepA | FeEnt | MOPS | 0 | 32.5 ± 0.13 | 4.6 ± 0.01 | 8.4 ± 0.7 | 2.1 ± 0.07 | 1.4 ± 0.07 |
| “ | “ | M9 | 0 | 3.2 ± 0.1 | 0.04 ± 0.01 | 0.2 ± 0.07 | 0.43 ± 0.03 | 0.267 ± 0.02 |
| KpnIroN | FeGEnt | MOPS | 0 | 11.1 ± 1.5 | 71.3 ± 0.2 | 84 ± 0.7 | 6.9 ± 0.07 | 14.9 ± 0.07 |
| “ | “ | M9 | 0 | 1.6 ± 0.2 | 0.3 ± 0.01 | 5.1 ± 0.05 | 2.8 ± 0.03 | 0.9 ± 0.1 |
| EcoIutA | FeAbn | MOPS | 0 | 0 | 180 ± 14 | 0 | 444 ± 91 | 626 ± 90 |
| “ | “ | M9 | 0 | 0 | 26.5 ± 0.9 | 0 | 210 ± 11 | 186 ± 17 |
| KpnFyuA | FeYbt | MOPS | 0 | 0.01 ± 0.001 | 6.3 ± 0.12 | 19 ± 0.08 | 0.02 ± 0.001 | 1.3 ± 0.23 |
| “ | “ | M9 | 0 | 0.02 ± 0.001 | 0.8 ± 0.004 | 1.2 ± 0.02 | 0.03 ± 0.001 | 8 ± 0.07 |
After growth of E. coli and K. pneumoniae strains in either MOPS or M9 media, we added FeCl3 to 1 mM in the culture supernatants and used fluoresceinated EcoFepA, KpnIroN, EcoIutA, and KpnFyuA to determine the concentrations of FeEnt, FeGEnt, FeAbn, and FeYbt, respectively. See Figure 4 and Experimental procedures for additional information. We repeated the experiment four times; the tabulated data are from a single, representative experiment, in which each culture supernatant was assayed with each sensor in triplicate. The results are listed with the standard deviations of those three determinations.
In this experiment the cell densities of the six strains (tabulated left to right above), after 24 h of growth in iron-restricted M9, were 0.1, 0.18, 0.19, 0.2, 0.44, and 0.43 × 109 cells/ml, respectively. The densities of the six strains after 24 h of growth in iron-restricted MOPS were 1.25, 1.52, 1.9, 1.56, 1.42, and 1.83 × 109 cells/ml, respectively.
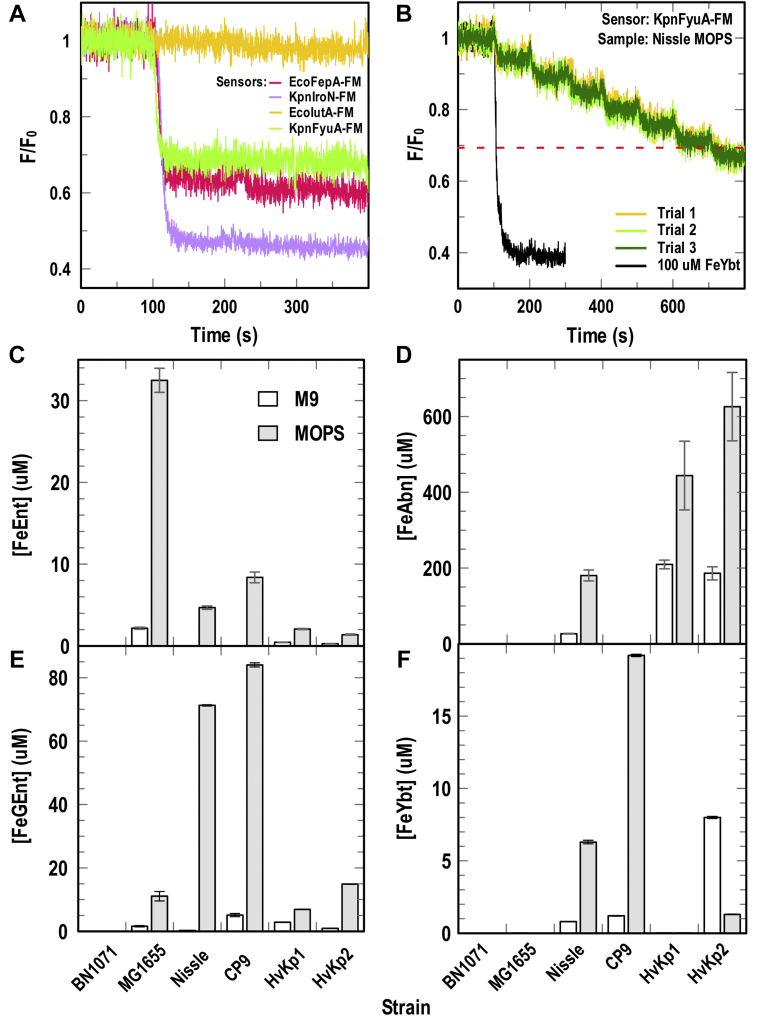
Figure 4.
Decoy sensor assays of siderophores in culture supernatants. After bacterial growth in iron-deficient media, we removed the cells by centrifugation, ferrated the culture supernatants and used four fluoresceinated sensor proteins (EcoFepA, KpnIroN, EcoIutA, and KpFyuA) to assay for the presence of FeEnt, FeGEnt, FeAero, and FeYbt, respectively, in media from E. coli strains B1071, MG1655, Nissle 1917, and CP9 and K. pneumoniae strains HvKp1 and HvKp2. A, detection of ferric siderophores. Undiluted culture supernatant, in this case from CP9, immediately revealed the presence or absence of the compounds of interest. The quenching patterns from the four FLGP sensors, when an aliquot of supernatant was added at 100 s, showed that CP9 made Ent, GEnt, and Ybt, but not Abn. B, determination of ferric siderophore concentrations in culture supernatants. For several FD sensors (e.g., KpnFyuA, shown here), the quenching from sequential additions of spent ferrated media (in this case, 3 μl aliquots of a 50-fold dilution of the Nissle 1917 (139) supernatant), added at 100 s intervals, revealed the concentration of ferric siderophore. In each such experiment, we made three individual trials, averaged the spectra, and calculated the variance between the trials, which was minimal. At a level of 50% quenching (dashed red line), [FeYbt] = KD: 1 nM, so we calculated [FeYbt] in the original supernatant from the dilution factor. C–F, production of siderophores by E. coli and K. pneumoniae strains in iron-limited M9 and MOPS media. Using the same sequential titration approach, we interrogated the spent media of several bacterial strains. BN1071 (entA) is a control E. coli laboratory strain (137) that does not make any siderophores; MG1655 is a prototypic (80) laboratory E. coli strain, whereas NIssle 1917, CP9, and the K. pneumoniae strains HvKp1 and HvKp2 are wild isolates. Titrations of supernatant aliquots from cultures of these strains divulged the presence and concentrations of Ent (C), Abn (D), GEnt (E), and Ybt (F). The numerical data underlying (C–F), and their statistical validation, are listed in Table 2.
The detection of noncatecholate ferric siderophores (FeAbn, FeYbt) was relatively straightforward because their FLGP sensors (EcoIutA-FM and KpnFyuA-FM, respectively) were virtually monospecific and unimpaired by interference from other ferric complexes (Tables 1 and 2; Fig. 2). The quantification of ferric catecholates was more complex, because the best sensors for FeEnt (EcoFepA-FM) and FeGEnt (KpnIroN-FM) also bound (albeit with lower affinity) other catecholate iron chelates (Table 1 and Fig. 1). This dual recognition of FeEnt and FeGEnt by EcoFepA-FM and KpnIroN-FM complicated the quantification of GEnt. However, because of its ∼300-fold higher affinity for FeEnt than for FeGEnt (Table 1), EcoFepA-FM accurately measured [FeEnt], even if FeGEnt was present at >100-fold higher levels (Table 2 and Fig. 4). Once the [FeEnt] in a solution was known, the extent of quenching of KpnIroN (which binds FeGEnt; KD = 7 nM) by the same solution provided information about the presence of FeGEnt (Experimental procedures).
These sensor measurements revealed other things about the strains and their growth in the two media. First, all strains grew more in MOPS media, achieving cell densities of 3 to 5 × 109/ml, relative to 0.5 to 1 × 109/ml in M9 (Table 2). The sensors showed that siderophore biosynthesis was correspondingly greater in MOPS, usually 5- to 10-fold higher, or more, than in M9 (Table 2 and Fig. 4). Second, despite these general trends, the strains were individually unique with regard to siderophore production in iron-deficient conditions. MG1655 only produced Ent, whereas the wild isolates Nissle, CP9, HvKp1, and HvKp2 produced combinations of the siderophores that we assayed: Nissle and HvKp2 produced all four, and as noted above; CP9 did not make Abn; and HvKp1 did not make Ybt (Fig. 4). Third, after 24 h growth in iron-deficient MOPS media, the total siderophore concentrations approached millimolar levels. This abundant growth and copious siderophore production illustrated the suitability of MOPS minimal medium for imposition of iron deficiency. Lastly, correlations between siderophore biosynthesis and bacterial virulence were not only discernable but also cryptic. The ExPEC strain CP9 excreted a profuse amount of GEnt but no Abn. Conversely, the defining characteristic of both hypervirulent K. pneumoniae strains was massive secretion of Abn, ∼50-fold more than their level of GEnt production.
Measurements of metal transport
Besides their recognition and discrimination of metal complexes, decoy sensors monitored the uptake of their ligands by cells of interest ((53), Fig. 5). In ΔtonB hosts LGP can bind but cannot transport their metal complexes. When FeEnt binds to OKN13(ΔtonB)/pEcoFepA-FM it quenches fluorescence, but the strain's TonB deficiency prevents FeEnt uptake. As a result, the fluorescence of EcoFepA-FM inversely relates to [FeEnt] in its environment (53). When such decoy sensor cells cohabit an environment with transport-active bacteria they reveal their ligand's concentration and, therefore, also its uptake by the cohabitant. EcoFepA avidly binds FeEnt (KD = 0.1–0.4 nM (69, 70)); 40 nM FeEnt quenches EcoFepA-FM ∼99%. Such high-affinity receptors, with nanomolar KD values, are most effective decoy sensors, because they rapidly detect the transport (depletion) of nanomolar amounts of ferric siderophores or porphyrins by the bacteria. Iron-deficient Gram-negative bacteria, with maximally expressed chromosomal FepA, transport FeEnt at ∼50 pmol/109 cells/min (53, 87), so in FLGP assays with ∼2.5 × 107 cells/ml in a cuvette, such strains deplete 40 nM FeEnt in <40 min. Lower-affinity FLGP with higher KD values (e.g., AbaFepA, KD = 17 nM [Table 1]) require higher concentrations of ligand (1.7 μM FeEnt) to comparably quench the sensor. Consequently, it takes longer for test strains to deplete this 100-fold higher concentration of FeEnt, and assays with an AbaFepA-FM sensor show slower fluorescence recovery.
Figure 5.
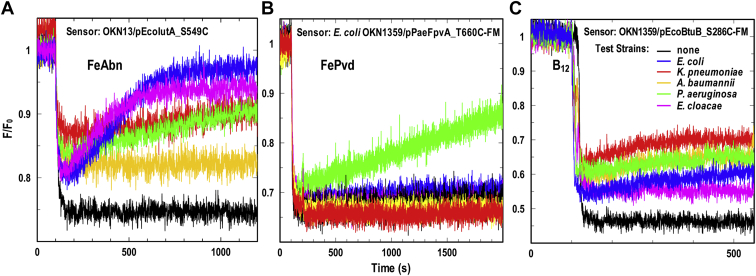
Decoy sensor measurements of FeAbn, FePvd, and B12uptake by CRE/ESKAPE pathogens. We expressed three of the FGLP sensors in Table 1 in TonB-deficient E. coli hosts and incubated the cells at 37 °C with bacteria that were grown in iron-deficient MOPS minimal media: E. coli MG1655 (blue); K. pneumoniae Kp52.145 (red); A. baumannii 17978 (yellow); P. aeruginosa PAO1 (green); E. cloacae (magenta); sensor only (black). The assays observed uptake of FeAbn by all the strains except A. baumannii (A), exclusive transport of FePvd by P. aeruginosa (B), and uptake of B12 by all the strains except E. cloacae (C).
Besides EcoFepA-FM–FeEnt (53), we tested the abilities of three other decoy sensor–ligand pairs (EcoIutA-FM–FeAbn; PaeFpvA-FM–FePvd; EcoBtuB-FM–B12) (Fig. 5) to monitor metal uptake by Gram-negative CRE/ESKAPE bacteria. The sensors revealed complex patterns of utilization. Almost all of the strains acquired FeAbn (except A. baumannii), but at different rates: iron-restricted E. coli and Escherichia cloacae transported FeAbn roughly 2-fold faster than P. aeruginosa or K. pneumoniae. Similarly, all the test strains but one (E. cloacae) transported B12, but at a slower rate than the uptake of ferric siderophores by the test strains. Lastly, only P. aeruginosa transported FePvd, and it did so at a considerably slower rate than E. coli transported FeEnt (53) or FeAbn. Thus, the FLGP quantitatively defined the transport of the noted compounds by the test strains, demonstrating the generality of the decoy sensor approach to uptake measurements.
Discussion
The conversion of binding proteins into decoy sensors is an evolution of Cys scanning mutagenesis, which Kaback and colleagues used to comprehensively analyze and manipulate the membrane transporter LacY (88, 89, 90, 91, 92, 93). After exhaustive mutagenesis they concluded that neither the elimination of native Cys residues nor the introduction and chemical modification of genetically engineered Cys side chains were strongly detrimental to the function of the lactose permease (94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110). Similar research on EcoFepA and other LGPs reiterated the benefits of site-directed chemical modifications of OM proteins (23, 53, 66, 68, 69, 111, 112, 113, 114, 115, 116, 117). One of the noteworthy attributes of FLGP and FBP sensors was their facile determination of the breadth of their own specificity. The relative affinities of their binding reactions defined each receptor's ligand preference. In a broader sense, these data reveal which nutrients, vitamins, and metals a bacterium acquires from its environment. The orthologs and paralogs of EcoFepA illustrated the insight that FLGP sensors provide. Despite its annotation, AbaFepA had little similarity to EcoFepA in regard to its affinities and specificities for catecholate iron complexes. EcoFepA, KpnFepA1658, and PaeFepA, on the other hand, were consistent in their primary affinity for FeEnt. EcoFiu and EcoCir were previously implicated in the uptake of monocatecholates (74), but besides compounds like FeDHBS, FeFDC, and FeMB-1 they bound FeEnt, FeEnt∗, and FeGEnt (EcoFiu only). These findings highlighted a general promiscuity in the catecholate receptors and showed the utility of FLGP in unraveling the complexities of their own specificities. In addition, the tactic of site-directed fluorescence modification of receptors and transporters creates a biochemical path to the verification of genomic sequence annotations.
The differences in fluoresceination levels of the different proteins in our study derives from two main factors. Primarily, the production and labeling of foreign proteins in E. coli may be considerably reduced by codon usage differences from the host species. Second, even when guided by crystallographic data, the selection of suitable sites in LGP surface loops for covalent modification is unpredictable and requires experimental evaluation. Their dynamic motion in vivo (67, 118, 119) underlies this uncertainty: certain sites may be sterically hindered by the motion or configuration of contiguous amino acid side chains. Together these factors led to lower fluorescence labeling of certain LGPs (e.g., KpnFyuA, Fig. S4; AbaBauA, Fig. S5).
The sensor assays described hierarchies of receptor preferences for different forms of catecholates compounds, including the siderophore antibiotics MB-1 (57, 58) and FDC (55, 120). Such studies are valuable in early phase discovery of new antibacterial drugs. A candidate compound’s minimum inhibitory concentration against target pathogens depends on its accumulation in the correct bacterial compartment (the periplasm for β-lactams), which is difficult to measure. The influx rate for siderophore–antibacterials depends on TonB-dependent intake through one or more LGPs (1). Without knowing which LGPs are involved, it is a blind empirical discovery process, which has led to the costly failure of many siderophore–β-lactams and the success of only one compound, FDC (121, 122). Information from FLGP sensors will inform and de-risk the drug discovery process, by discerning the spectrum of pathogens (or limitations) for candidate compounds, as well as risks from “adaptation-based” or mutational resistance. It is problematical that the identities and affinities of the LGP involved in FDC uptake are still unknown in all the species for which it is approved for clinical use. Just as it is important to know the target of an antibacterial drug, it is also important to know the route of its entry into each pathogen. In this sense, it was of interest that the first licensed Trojan Horse antibiotic, FDC (FeTroja, Shionogi Inc), was recognized by several FLGPs. Its clinical efficacy was already demonstrated against A. baumannii (121), which concurs with the observation that, besides EcoFiu, EcoCir, and KpnIroN, it bound to AbaPiuA, AbaFepA, and AbaPirA.
The extension of this technology to the analysis of clinical samples from human or animal hosts may reveal physiological functions that confer pathogenicity or infectivity. The production of particular siderophores connects to the colonization, tropism, invasiveness, or lethality of particular bacteria. Some microbes produce virulence-enhancing siderophores, as illustrated by A. baumannii's synthesis of acinetobactin, baumoferrins, and fimsbactins. At present, the utilization of iron complexes of these three siderophores appears species specific and unique to the pathogenicity of A. baumannii (123). Other siderophores, or groups of siderophores, more broadly correlate with virulence. The association of Abn with invasiveness was the first example of this relationship (42), but the simultaneous elaboration of Abn, Ybt, and GEnt by isolates of E. coli, K. pneumoniae (17, 52), Salmonella typhimurium (43), and other bacteria reaffirmed this phenomenon. Each of the siderophores in this mixture promotes the infection of specific organs, increasing the virulence of organisms like hypervirulent K. pneumoniae (40). Certain siderophores also affect the overall outcome of encounters between bacteria and humans, by influencing the fine line between colonization and pathogenesis. The production of Ent and utilization of FeEnt facilitate colonization of the mammalian gut (14), whereas glucosylation of Ent to form GEnt, and utilization of FeGEnt, promotes pathogenesis (45, 47, 124, 125, 126). The distinction between the naked or glucosylated siderophore is that SCN adsorbs Ent/FeEnt and removes them from circulation, but its lower affinity for GEnt/FeGEnt means that it does not similarly eliminate them from fluids and tissues. This evasion of SCN allows the accumulation of potentially nutritive amounts of GEnt in blood, serum, and lymph, which may enhance bacterial proliferation. HsaSCN-FM bound a spectrum of apo and ferric siderophores, which concurs with its innate immune function against pathogenic microbes (54). Nevertheless, besides GEnt/FeGEnt, neither Abn/FeAbn nor Ybt/FeYbt nor Agn/FeAgn adsorbed to HsaSCN: these siderophores consistently link to Gram-negative bacterial pathogenicity. The binding specificities of HsaSCN were already largely known (11, 50, 127, 128, 129, 130), but the FBP sensor determinations ranked its ligands in the overall hierarchy of its biological activity.
Siderophore production contributes to the infectivity and tissue tropism of K. pneumoniae (39, 40, 45, 52), and we found that hypervirulent K. pneumoniae (HvKpn2) made significant amounts of Ent, GEnt, Ybt, and Abn. However, its massive production of Abn (0.6 mM in MOPS) was ∼40-fold more than that of any of the other siderophores and ∼97% of the total amount of secreted siderophores, supporting the known role of aerobactin in promoting bacterial invasiveness and pathogenicity (37, 39, 131, 132, 133, 134, 135). To the contrary, E. coli Nissle 1917, a nonpathogenic probiotic strain (83), made the same four siderophores in comparable but different amounts. It also produced more aerobactin (0.2 mM) than any other siderophore. Hence, elaboration of siderophores is not a stand-alone virulence determinant of the Enterobacteraceae. Other attributes of K. pneumoniae (as, for example, its hypermucoviscosity (136)), synergize with siderophore biosynthetic and transport systems to maximize the infectivity of certain pathogenic strains.
Experimental procedures
Bacteria and plasmids
E. coli laboratory strains descended from BN1071 (entA; (137)): OKN1 (ΔtonB), OKN3 (ΔfepA), OKN7 (ΔfhuA), OKN13 (ΔtonB, ΔfepA) (114), and MG1655 (80). E. coli strains Nissle 1917 (83, 138, 139) and CP9 (84) are probiotic and ExPEC wild isolates, respectively. ESKAPE strains and other wild isolates included K. pneumoniae Kp52.145 ((140); courtesy of Regis Tournebiz, Institut Pasteur), A. baumannii ATCC 17978, and P. aeruginosa PA01 (courtesy of Stephen Lory, Harvard University). K. pneumoniae Kp52.145 was the source of four annotated kpnfepA structural genes: chromosomal loci 1658, 2380, 4984, and plasmid (pII) locus 0027. We obtained the HsaSCN structural gene in plasmid GST-h SCN_pGEX-4t-1 from Roland K. Strong.
Media, siderophores, and other metal complexes
We grew bacteria in LB (141) and subcultured at 0.5% to 1% into iron-deficient MOPS (85) or M9 (86) minimal media for 6 to 24 h to achieve iron limitation. We prepared apo and iron complexes from a collection (142) of ∼40 purified catecholate, hydroxamate, and mixed chelation siderophores or purified the compounds of interest from bacterial culture supernatants (Fig. S1). In each case, we measured and adjusted their concentrations from their UV-visible absorption spectra. For determinations with acinetobactin (Acn), we substituted anguibactin (Agn). The two mixed chelation siderophores are nearly chemically identical, but the iron complex of the latter is more readily available and known to bind to the BauA receptor of A. baumannii (61, 143). Other compounds were from commercial sources or colleagues (DHBS, KeyOrganics; FDC, Venatorx, H. Ron Kaback; MB-1, Venatorx).
Selection of Cys substitution mutant targets
For each LGP of interest we chose multiple candidate residues, located in the surface loops of their outer vestibules, for Cys substitution and fluorescent modification. In most cases, crystal structures guided the selections (EcoFepA [1FEP; (144)], EcoBtuB [1NQF; (145)], EcoFhuA [1BY5; (146)], EcoFiu [6BPM; (147)], EcoCir [2HDF; (148)], AbaBauA [6H7F; (143)], AbaPirA [5FR8; (149)], AbaPiuA [5FP1; (149)], YpeFyuA [4EPA; (150)]). In other cases (CcrHutA, EcoIutA, KpnFepA, KpnIroN, PaeFepA) we used CLUSTALΩ (151) to identify the closest structurally solved phylogenetic ortholog or paralog of the unsolved LGP, on the basis of percent sequence identity, and then predicted its structure with the MODELLER function of CHIMERA (UCSF). After ensuing construction of multiple mutants for each receptor protein, we evaluated their accessibility to chemical modification and their sensitivity to fluorescence quenching to determine the best construct for each of the siderophores under investigation (Figs. S2–S6).
Site-directed Cys mutagenesis
As a general approach, we used PCR to clone the LGP of interest (Table 1) from the chromosomes of selected bacterial species. Because we ultimately expressed all the LGP-derived sensors in the E. coli OM, we precisely cloned the nucleotide sequences encoding their mature proteins and inserted them downstream from the native promoter and signal peptide of EcofepA in pITS23 (64), a derivative of the low-copy pHSG575 (65). We used QuikChange (Stratagene) for single Cys substitutions in the LGP of interest, with complementary oligonucleotides flanking the mutation, followed by digestion of the wild-type vector by DpnI. We confirmed the mutations by sequencing (Genewiz) of purified plasmids.
Cys mutant protein expression and fluorescent labeling
The vector for LGP production, pITS23 (152), carries wild-type EcofepA under control of its native, Fur-regulated promoter. For expression of each of the E. coli LGP (Fiu, FhuA, IutA, Cir, BtuB) we replaced EcofepA in pITS23 with the alternate LGP structural gene, with its own signal sequence, such that the iron-regulated EcofepA promoter controlled biosynthesis. For LGPs from other species (KpnFepA, KpnIroN, KpnFyuA, AbaBauA, PaeFepA, PaeFpvA, CcrHutA), we replaced the sequence encoding mature EcoFepA with the sequence encoding the foreign mature LGP, downstream from the EcoFepA signal sequence, and regulated by the EcofepA promoter. This strategy allowed the EcoFepA signal sequence to direct secretion and assembly of the foreign OM protein in the E. coli OM. We transformed the constructs into entA, tonB+ (OKN3, OKN359) or entA, ΔtonB (OKN13, OKN1359) E. coli hosts, such that growth in iron-deficient MOPS media caused overexpression of the sensor proteins. We fluoresceinated the LGP of E. coli/pITS23 constructs in situ, in living cells. For the soluble binding proteins, we purified the 6His-tagged Cys mutant proteins from cell lysates by metal affinity chromatography, modified them with 5 μM FM at pH 6.77 for 15 min, and repurified the fluorescently labeled sensor by gel filtration chromatography. We evaluated the expression and labeling of each of the cloned LGP by growing the appropriate E. coli host strain (OKN3, OKN359, OKN13, or OKN1359), harboring the plasmid constructs of interest, in iron-deficient MOPS medium to late log phase (A600nm = 2.5–3.5). We subsequently analyzed SDS-PAGE–resolved samples of bacterial OM, or purified protein fractions, for fluorescence emissions at 520 nM before staining with Coomassie blue R (Figs. S1–S3). In some cases, we used mouse monoclonal (α-EcoFepA) antibodies or rabbit polyclonal (α-FepB) antisera to determine the expression levels and stoichiometry of FM labeling, which allowed quantification of expression. In each case, we obtained comparable, but sometimes less, iron-regulated expression of the heterologous LGP under control of the EcofepA promoter.
Siderophore nutrition tests
To qualitatively evaluate the FeEnt uptake abilities encoded by the mutant fepA alleles, we performed siderophore nutrition tests (68, 153). We expressed the results as the diameter of bacterial growth around the paper disk and compared mutant halos to the growth halo conferred by a strain harboring pITS23 (fepA+). These experiments confirmed the transport functionality of the cloned proteins.
Preparation of cell envelope fractions
After growth in LB broth overnight, we subcultured FepA mutant derivatives on pITS23 in OKN3, OKN13, OKN359, or OKN1359 at 1% into 20 ml of iron-deficient MOPS media and incubated the cultures with shaking for 5.5 to 6 h at 37 °C, to an A600 nm = 1 to 1.2 (5–6 × 108 cells/ml). In some cases, we subjected the cells to fluorescence modification at this stage, as discussed below. After washing with 50 mM NaHPO4, pH 6.7, we collected the cells by centrifugation at 7500g, resuspended the pellets in 2 ml of PBS containing trace amounts of DNase and RNase, and passed the cell suspensions (3×) through a French pressure cell at 14,000 psi (154). After spinning the lysates at 3000g for 10 min to remove unbroken cells and debris, we transferred the supernatants to microcentrifuge tubes and pelleted the cell envelopes by centrifugation at 13,000g for 45 min. We resuspended the cell envelopes in 400 μl of 50 mM Tris-Cl, pH 7.4 and analyzed expression and the extent of fluorescence labeling by SDS-PAGE and UV illumination.
Fluorescence modifications
For modifications with maleimide fluorophores, we inoculated E. coli strains OKN3, OKN13, OKN359, or OKN1359 harboring plasmids that encoded a Cys mutant LGP from frozen stocks into LB, grew them overnight, and subcultured at 1% into iron-deficient MOPS minimal media, with shaking (200 rpm) at 37 °C for 10 to 12 h, until late exponential phase. After collecting the cells by centrifugation at 7500g for 15 min, and washing with 50 mM NaHPO4, pH 6.7, we labeled the Cys-mutant LGP with 5 μM FM in 50 mM NaHPO4, pH 6.7 for 15 min at 37 °C FM. We terminated the labeling reactions with 100 μM β-mercaptoethanol. After collecting the fluoresceinated cells by centrifugation at 7500g for 15 min, and washing with 50 mM NaHPO4, pH 6.7, we resuspended them in PBS plus 0.2% glucose. We immediately used the labeled cells in spectroscopic experiments or rested them on ice (up to 24 h), or added glycerol to 15% and stored them (indefinitely) as 1-ml aliquots at −70 °C. In the latter case, after thawing the labeled cells we pelleted them by centrifugation in a microfuge, washed them once with and resuspended them in PBS containing 0.2% glucose. Lastly, for evaluation of protein expression or the extent of FM labeling we solubilized aliquots of the cell suspensions with sample buffer and subjected them to SDS-PAGE. After electrophoresis, we first visualized the fluorescence labeling on a Typhoon 8600 Biomolecular Scanner (GE/Amersham) and then stained the gels with Coomassie blue R (155).
SDS-PAGE and Western immunoblots
We electrophoretically analyzed cell envelope fractions of the mutants. For SDS-PAGE (156, 157) we suspended 30 μg of cell envelope protein (calculated from absorbance at 280 nm) in sample buffer containing 1% SDS and 3% βME, boiled the sample for 5 min, and resolved the samples on 11% acrylamide/0.3% bis acrylamide slabs (156) at 30 mA. To enhance the separation of proteins in the 80-kDa range, we continued electrophoresis at 30 mA for an additional 45 min after the tracking dye left the gels. [125I]-protein A immunoblots provided precise data on the concentration of EcoFepA, and the stained SDS-PAGE gels revealed the concentrations of the other LGP in the OM, relative to EcoFepA.
Fluorescence spectroscopic binding determinations
We observed fluorophore-labeled cells in an SLM AMINCO 8100 fluorescence spectrometer, upgraded with an OLIS operating system and software (OLIS SpectralWorks, OLIS Inc), to control its shutters, polarizers, and data collection. We also utilized an OLIS Clarity fluorescence spectrometer for fluorescence assays. For binding determinations we deposited 2.5 × 107 labeled cells in a quartz cuvette (final volume, 2 ml) with stirring at 37 °C, measured the initial fluorescence (F0), and then added increasing concentrations of an iron complex while monitoring the quenching of fluorescence emissions (F) at 520 nm. We performed each measurement in triplicate and calculated the mean value of F/F0 at each ligand concentration. We plotted 1-F/F0 versus [ligand] and analyzed the data by the 1-site binding model of Grafit 6.0.12 (Erithacus Ltd), which fits data to a single site saturation curve, where the amount of ligand bound is plotted as a function of the amount free:
These plots yielded KD values for each of the receptor-ligand interactions, with associated standard errors.
Measurement of siderophore concentrations in spent media
For analysis of culture supernatants, we inoculated 10 to 25 ml volumes of iron-deficient M9 or MOPS minimal media with strains of E. coli or K. pneumoniae, shook the cultures at 37 °C for 24 h, removed the cells by centrifugation at 7000g for 15 min, and added FeCl3 to 1 mM. After incubating the ferrated supernatants for at least an hour at room temperature, we stored them on ice until they were analyzed.
For concentration measurements using FLGP sensors, we first expressed and fluoresceinated (53) the appropriate LGP, expressed in either OKN13 (ΔtonB, ΔfepA) or OKN1359 (ΔtonB, ΔfepA, Δcir, Δfiu). We next ascertained the initial fluorescence intensity of 2.5 × 107 sensor cells in 2 ml of PBS in a quartz cuvette (λex = 492 nm; λem = 520 nm) and their maximal quenching by excess ligand. To measure the supernatant concentrations of FeEnt (with EcoFepA-FM), FeAbn (EcoIutA-FM), and FeYbt (KpnFyuA-FM), we removed any precipitated or aggregated material by microcentrifugation at 13,000 rpm for 2 min and added sequential aliquots of the clarified, ferrated supernatant to the sensor cell suspension, until 50% quenching occurred (i.e., [ferric siderophore] = KD). The dilution factors to 50% quenching divulged the [siderophore] in the original culture supernatant.
Because Ent is the precursor of GEnt, both catecholates are usually present in the media of GEnt producers (Fig. 4). This fact complicates measurement of [FeGEnt]. EcoFepA and KpnIroN bind FeEnt with the same affinity (KD = 0.4 nM), but the latter protein also binds FeGEnt (KD = 6.3 nM) (53). As a result, the comparative quenching of EcoFepA-FM and KpnIroN-FM by spent media allowed estimation of its [FeGEnt], as follows. After finding the volume of supernatant required to achieve 50% quenching of 2.5 × 107 cells of OKN1359/pEcoFepA-FM, we measured the extent of quenching of 2.5 × 107 cells of OKN1359/pKpnIroN-FM by the same volume of the same supernatant. From the relationship
that expresses the fraction of bound (FrBd) sensor (KpnIroN) to its saturation by both FeEnt and FeGEnt. From experimentally determined F/F0 and [FeEnt] (=0.4 nM at 50% quenching), we solved for [FeGEnt].
Fluorescence spectroscopic uptake measurements
We employed decoy sensors (53) to monitor TonB-dependent uptake of various siderophores by bacterial pathogens. In the fluorescence spectroscopic uptake studies E. coli OKN13 (ΔtonB, ΔfepA) or OKN1359 (ΔtonB, Δfiu, ΔfepA, Δcir) were host strains for pITS23 derivatives carrying single Cys mutants of an LGP of interest. The FLGPs were decoy sensors that monitored ferric siderophore uptake by test strains. For each assay we used 107 sensor cells (e.g., OKN13/pFepA-FM (53)) in a 2-ml quartz cuvette containing PBS + 0.2% glucose at 37 °C. After recording fluorescence intensity for 100 s we added an iron complex (e.g., FeEnt) at a final concentration of ∼10 nM. After incubating the sample for 100 s, during which time fluorescence was quenched (e.g., ∼60% quenching for EcoFepA_A698C-FM), we added 0.5 to 2 × 107 cells of the test strain (E. coli MG1655, K. pneumoniae Kp52.145, A. baumannii I7978, P. aeruginosa PA01, E. cloacae) and monitored the time course of fluorescence emissions at 520 nm for 15 to 40 min, with stirring. Transport of the iron complex by the test strain resulted in an increase in fluorescence intensity as the cells depleted it from solution.
Data availability
All data are contained within the article and the Supplemental Information file.
Supporting information
This article contains supporting information.
Conflict of interest
D. A. S. is employed and compensated by Venatorx and may own stock or stock options in Venatorx as part of his remuneration for employment.
Acknowledgments
The authors thank Stephen Lory, Keith Poole, and Regis Tournebiz for providing bacterial strains, Roland Strong and Peter Rupert for providing GST-hSCN_pGEX-4t-1, the source of the HsaSCN structural gene, and Ron Kaback for a sample of FDC.
Author contributions
A. K., T. Y., S. C., A. M., B. L. N., D. A. S., M. B. L., L. A. A., M. M., T. A. R., S. M. N., and P. E. K. conceptualization; A. K., T. Y., S. C., A. M., B. L. N., S. M. N., and P. E. K. methodology; A. K., T. Y., S. C., A. M., B. L. N., S. M. N., and P. E. K. validation; A. K., T. Y., S. C., A. M., B. L. N., S. L. P., N. M. d. S., M. B. L., L. A. A., M. M., T. A. R., S. M. N., and P. E. K. investigation; P. E. K. writing – original draft; A. K., D. A. S., M. B. L., L. A. A., T. A. R., and P. E. K. writing – review & editing; D. A. S. and T. A. R. supervision; S. M. N. and P. E. K. project administration.
Funding and additional information
T. A. R. was supported by the Veterans Administration Western New York Healthcare System, Buffalo, New York, USA, the Department of Veterans Affairs VA Merit Review 1I01BX004677-01A1, and NIH 1R21AI141826-01A1. D. A. S. was supported by the Defense Threat Reduction Agency (DTRA) under Contract No. HDTRA117C0070 and NIAID, National Institutes of Health under Contract No. 75N93020C00016. M. B. L. was supported by NIH grants R21AI135225 and R01AI148241 and S. L. P. by NIH F31AI147404. N. M. d. S. was supported by the São Paulo Research Foundation (FAPESP 2019/08514-0), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Brasil 307974/2017-0), FAPESP (2017/02127-9), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES 88887.470428/2019-00). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Chris Whitfield
Footnotes
For the designation of LGP from different bacterial species, we created acronyms that emulate the designation of restriction enzymes: abbreviation of their genus and species of origin, followed by the enzyme's name (e.g., EcoRI for E. coli restriction enzyme I). Hence, EcoFepA for E. coli FepA; KpnIroN for K. pneumoniae IroN; AbaBauA for A. baumannii BauA, etc.
Supporting information
References
- 1.Klebba P.E., Newton S.M.C., Six D.A., Kumar A., Yang T., Nairn B.L., Munger C., Chakravorty S. Iron acquisition systems of gram-negative bacterial pathogens define TonB-dependent pathways to novel antibiotics. Chem. Rev. 2021;121:5193–5239. doi: 10.1021/acs.chemrev.0c01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassat J.E., Skaar E.P. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganz T. Iron and infection. Int. J. Hematol. 2018;107:7–15. doi: 10.1007/s12185-017-2366-2. [DOI] [PubMed] [Google Scholar]
- 4.Neilands J.B. In: Microbial Iron Metabolism - A Comprehensive Treatise. Neilands J.B., editor. Academic Press; New York, NY: 1974. Iron and its role in microbial physiology; pp. 4–34. [Google Scholar]
- 5.Neilands J.B. Microbial iron compounds. Annu. Rev. Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 6.Noinaj N., Guillier M., Barnard T.J., Buchanan S.K. TonB-dependent transporters: Regulation, structure, and function. Annu. Rev. Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson G.J., Frazer D.M. Current understanding of iron homeostasis. Am. J. Clin. Nutr. 2017;106:1559s–1566s. doi: 10.3945/ajcn.117.155804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wish J.B. Assessing iron status: Beyond serum ferritin and transferrin saturation. Clin. J. Am. Soc. Nephrol. 2006;1 Suppl 1:S4–S8. doi: 10.2215/CJN.01490506. [DOI] [PubMed] [Google Scholar]
- 9.Coffey R., Ganz T. Iron homeostasis: An anthropocentric perspective. J. Biol. Chem. 2017;292:12727–12734. doi: 10.1074/jbc.R117.781823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fluckinger M., Haas H., Merschak P., Glasgow B.J., Redl B. Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob. Agents Chemother. 2004;48:3367–3372. doi: 10.1128/AAC.48.9.3367-3372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes M.A., Paulsene W., Jide X., Ratledge C., Strong R.K. Siderocalin (Lcn 2) also binds carboxymycobactins, potentially defending against mycobacterial infections through iron sequestration. Structure. 2005;13:29–41. doi: 10.1016/j.str.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Bradbeer C. The proton motive force drives the outer membrane transport of cobalamin in Escherichia coli. J. Bacteriol. 1993;175:3146–3150. doi: 10.1128/jb.175.10.3146-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradbeer C., Kenley J.S., Di Masi D.R., Leighton M. Transport of vitamin B12 in Escherichia coli. Corrinoid specificities of the periplasmic B12-binding protein and of energy-dependent B12 transport. J. Biol. Chem. 1978;253:1347–1352. [PubMed] [Google Scholar]
- 14.Pi H., Jones S.A., Mercer L.E., Meador J.P., Caughron J.E., Jordan L., Newton S.M., Conway T., Klebba P.E. Role of catecholate siderophores in gram-negative bacterial colonization of the mouse gut. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdelhamed H., Lu J., Lawrence M.L., Karsi A. Ferric hydroxamate uptake system contributes to Edwardsiella ictaluri virulence. Microb. Pathog. 2016;100:195–200. doi: 10.1016/j.micpath.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Holden V.I., Bachman M.A. Diverging roles of bacterial siderophores during infection. Metallomics. 2015;7:986–995. doi: 10.1039/c4mt00333k. [DOI] [PubMed] [Google Scholar]
- 17.Holden V.I., Breen P., Houle S., Dozois C.M., Bachman M.A. Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. mBio. 2016;7 doi: 10.1128/mBio.01397-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramakrishnan G. Iron and virulence in Francisella tularensis. Front. Cell. Infect. Microbiol. 2017;7:107. doi: 10.3389/fcimb.2017.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saha P., Xiao X., Yeoh B.S., Chen Q., Katkere B., Kirimanjeswara G.S., Vijay-Kumar M. The bacterial siderophore enterobactin confers survival advantage to Salmonella in macrophages. Gut Microbes. 2019;10:412–423. doi: 10.1080/19490976.2018.1546519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy T.A., Moreland S.M., Andrews-Polymenis H., Detweiler C.S. The ferric enterobactin transporter Fep is required for persistent Salmonella enterica serovar typhimurium infection. Infect. Immun. 2013;81:4063–4070. doi: 10.1128/IAI.00412-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutz J.M., Liu J., Lyons J.A., Goranson J., Armstrong S.K., McIntosh M.A., Feix J.B., Klebba P.E. Formation of a gated channel by a ligand-specific transport protein in the bacterial outer membrane. Science. 1992;258:471–475. doi: 10.1126/science.1411544. [DOI] [PubMed] [Google Scholar]
- 22.Newton S.M., Trinh V., Pi H., Klebba P.E. Direct measurements of the outer membrane stage of ferric enterobactin transport: Postuptake binding. J. Biol. Chem. 2010;285:17488–17497. doi: 10.1074/jbc.M109.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang X., Payne M.A., Cao Z., Foster S.B., Feix J.B., Newton S.M., Klebba P.E. Ligand-specific opening of a gated-porin channel in the outer membrane of living bacteria. Science. 1997;276:1261–1264. doi: 10.1126/science.276.5316.1261. [DOI] [PubMed] [Google Scholar]
- 24.Saier M.H., Jr. Families of proteins forming transmembrane channels. J. Membr. Biol. 2000;175:165–180. doi: 10.1007/s00232001065. [DOI] [PubMed] [Google Scholar]
- 25.Yen M.R., Peabody C.R., Partovi S.M., Zhai Y., Tseng Y.H., Saier M.H. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta. 2002;1562:6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- 26.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J. Biol. Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 27.Nikaido H., Rosenberg E.Y. Porin channels in Escherichia coli: Studies with liposomes reconstituted from purified proteins. J. Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenbusch J.P. Structural and functional properties of porin channels in E. coli outer membranes. Experientia. 1990;46:167–173. [PubMed] [Google Scholar]
- 29.Luckey M., Nikaido H. Specificity of diffusion channels produced by lambda phage receptor protein of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1980;77:167–171. doi: 10.1073/pnas.77.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan L.D., Zhou Y., Smallwood C.R., Lill Y., Ritchie K., Yip W.T., Newton S.M., Klebba P.E. Energy-dependent motion of TonB in the gram-negative bacterial inner membrane. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11553–11558. doi: 10.1073/pnas.1304243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pawelek P.D., Croteau N., Ng-Thow-Hing C., Khursigara C.M., Moiseeva N., Allaire M., Coulton J.W. Structure of TonB in complex with FhuA, E. coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 32.Shultis D.D., Purdy M.D., Banchs C.N., Wiener M.C. Outer membrane active transport: Structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 33.Schauer K., Rodionov D.A., de Reuse H. New substrates for TonB-dependent transport: Do we only see the 'tip of the iceberg'? Trends Biochem. Sci. 2008;6:6. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 34.Fetherston J.D., Kirillina O., Bobrov A.G., Paulley J.T., Perry R.D. The yersiniabactin transport system is critical for the pathogenesis of bubonic and pneumonic plague. Infect. Immun. 2010;78:2045–2052. doi: 10.1128/IAI.01236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehring A.M., DeMoll E., Fetherston J.D., Mori I., Mayhew G.F., Blattner F.R., Walsh C.T., Perry R.D. Iron acquisition in plague: Modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 36.Perry R.D., Fetherston J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011;13:808–817. doi: 10.1016/j.micinf.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brock J.H., Williams P.H., Liceaga J., Wooldridge K.G. Relative availability of transferrin-bound iron and cell-derived iron to aerobactin-producing and enterochelin-producing strains of Escherichia coli and to other microorganisms. Infect. Immun. 1991;59:3185–3190. doi: 10.1128/iai.59.9.3185-3190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLauchlin J. Human listeriosis in Britain, 1967-85, a summary of 722 cases. 1. Listeriosis during pregnancy and in the newborn. Epidemiol. Infect. 1990;104:181–189. doi: 10.1017/s0950268800059343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russo T.A., Olson R., MacDonald U., Beanan J., Davidson B.A. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 2015;83:3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russo T.A., Shon A.S., Beanan J.M., Olson R., MacDonald U., Pomakov A.O., Visitacion M.P. Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams P., Smith M.A., Stevenson P., Griffiths E., Tomas J.M. Novel aerobactin receptor in Klebsiella pneumoniae. J. Gen. Microbiol. 1989;135:3173–3181. doi: 10.1099/00221287-135-12-3173. [DOI] [PubMed] [Google Scholar]
- 42.Williams P.H. Novel iron uptake system specified by ColV plasmids: An important component in the virulence of invasive strains of Escherichia coli. Infect. Immun. 1979;26:925–932. doi: 10.1128/iai.26.3.925-932.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bister B., Bischoff D., Nicholson G.J., Valdebenito M., Schneider K., Winkelmann G., Hantke K., Sussmuth R.D. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. Biometals. 2004;17:471–481. doi: 10.1023/b:biom.0000029432.69418.6a. [DOI] [PubMed] [Google Scholar]
- 44.Fischbach M.A., Lin H., Liu D.R., Walsh C.T. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 45.Bachman M.A., Lenio S., Schmidt L., Oyler J.E., Weiser J.N. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. mBio. 2012;3 doi: 10.1128/mBio.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischbach M.A., Lin H., Zhou L., Yu Y., Abergel R.J., Liu D.R., Raymond K.N., Wanner B.L., Strong R.K., Walsh C.T., Aderem A., Smith K.D. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U. S. A. 2006;103:16502–16507. doi: 10.1073/pnas.0604636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hantke K., Nicholson G., Rabsch W., Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith K.D. Iron metabolism at the host pathogen interface: Lipocalin 2 and the pathogen-associated iroA gene cluster. Int. J. Biochem. Cell Biol. 2007;39:1776–1780. doi: 10.1016/j.biocel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan Y.R., Liu J.S., Pociask D.A., Zheng M., Mietzner T.A., Berger T., Mak T.W., Clifton M.C., Strong R.K., Ray P., Kolls J.K. Lipocalin 2 is required for pulmonary host defense against Klebsiella infection. J. Immunol. 2009;182:4947–4956. doi: 10.4049/jimmunol.0803282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Correnti C., Strong R.K. Mammalian siderophores, siderophore-binding lipocalins, and the labile iron pool. J. Biol. Chem. 2012;287:13524–13531. doi: 10.1074/jbc.R111.311829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sia A.K., Allred B.E., Raymond K.N. Siderocalins: Siderophore binding proteins evolved for primary pathogen host defense. Curr. Opin. Chem. Biol. 2013;17:150–157. doi: 10.1016/j.cbpa.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachman M.A., Oyler J.E., Burns S.H., Caza M., Lepine F., Dozois C.M., Weiser J.N. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 2011;79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakravorty S., Shipelskiy Y., Kumar A., Majumdar A., Yang T., Nairn B.L., Newton S.M., Klebba P.E. Universal fluorescent sensors of high-affinity iron transport, applied to ESKAPE pathogens. J. Biol. Chem. 2019;294:4682–4692. doi: 10.1074/jbc.RA118.006921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hider R.C., Kong X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 55.Ito A., Nishikawa T., Matsumoto S., Yoshizawa H., Sato T., Nakamura R., Tsuji M., Yamano Y. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016;60:7396–7401. doi: 10.1128/AAC.01405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohira N., West J., Ito A., Ito-Horiyama T., Nakamura R., Sato T., Rittenhouse S., Tsuji M., Yamano Y. In vitro antimicrobial activity of a siderophore cephalosporin, S-649266, against Enterobacteriaceae clinical isolates, including carbapenem-resistant strains. Antimicrob. Agents Chemother. 2016;60:729–734. doi: 10.1128/AAC.01695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomaras A.P., Crandon J.L., McPherson C.J., Banevicius M.A., Finegan S.M., Irvine R.L., Brown M.F., O'Donnell J.P., Nicolau D.P. Adaptation-based resistance to siderophore-conjugated antibacterial agents by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013;57:4197–4207. doi: 10.1128/AAC.00629-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomaras A.P., Crandon J.L., McPherson C.J., Nicolau D.P. Potentiation of antibacterial activity of the MB-1 siderophore-monobactam conjugate using an efflux pump inhibitor. Antimicrob. Agents Chemother. 2015;59:2439–2442. doi: 10.1128/AAC.04172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Actis L.A., Fish W., Crosa J.H., Kellerman K., Ellenberger S.R., Hauser F.M., Sanders-Loehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1) J. Bacteriol. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S., Okujo N., Sakakibara Y. Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch. Microbiol. 1994;162:249–254. doi: 10.1007/BF00301846. [DOI] [PubMed] [Google Scholar]
- 61.Dorsey C.W., Tomaras A.P., Connerly P.L., Tolmasky M.E., Crosa J.H., Actis L.A. The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology. 2004;150:3657–3667. doi: 10.1099/mic.0.27371-0. [DOI] [PubMed] [Google Scholar]
- 62.Song W.Y., Jeong D., Kim J., Lee M.W., Oh M.H., Kim H.J. Key structural elements for cellular uptake of acinetobactin, a major siderophore of Acinetobacter baumannii. Org. Lett. 2017;19:500–503. doi: 10.1021/acs.orglett.6b03671. [DOI] [PubMed] [Google Scholar]
- 63.Hashimoto-Gotoh T., Franklin F.C., Nordheim A., Timmis K.N. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981;16:227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- 64.Scott D.C., Cao Z., Qi Z., Bauler M., Igo J.D., Newton S.M., Klebba P.E. Exchangeability of N termini in the ligand-gated porins of Escherichia coli. J. Biol. Chem. 2001;276:13025–13033. doi: 10.1074/jbc.M011282200. [DOI] [PubMed] [Google Scholar]
- 65.Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 66.Liu J., Rutz J.M., Klebba P.E., Feix J.B. A site-directed spin-labeling study of ligand-induced conformational change in the ferric enterobactin receptor, FepA. Biochemistry. 1994;33:13274–13283. doi: 10.1021/bi00249a014. [DOI] [PubMed] [Google Scholar]
- 67.Smallwood C.R., Jordan L., Trinh V., Schuerch D.W., Gala A., Hanson M., Shipelskiy Y., Majumdar A., Newton S.M., Klebba P.E. Concerted loop motion triggers induced fit of FepA to ferric enterobactin. J. Gen. Physiol. 2014;144:71–80. doi: 10.1085/jgp.201311159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao Z., Qi Z., Sprencel C., Newton S.M., Klebba P.E. Aromatic components of two ferric enterobactin binding sites in Escherichia coli fepA. Mol. Microbiol. 2000;37:1306–1317. doi: 10.1046/j.1365-2958.2000.02093.x. [DOI] [PubMed] [Google Scholar]
- 69.Cao Z., Warfel P., Newton S.M., Klebba P.E. Spectroscopic observations of ferric enterobactin transport. J. Biol. Chem. 2003;278:1022–1028. doi: 10.1074/jbc.M210360200. [DOI] [PubMed] [Google Scholar]
- 70.Newton S.M., Igo J.D., Scott D.C., Klebba P.E. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol. Microbiol. 1999;32:1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 71.Schalk I.J., Kyslik P., Prome D., van Dorsselaer A., Poole K., Abdallah M.A., Pattus F. Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron-free ligand: Implications for siderophore-mediated iron transport. Biochemistry. 1999;38:9357–9365. doi: 10.1021/bi990421x. [DOI] [PubMed] [Google Scholar]
- 72.Schalk I.J., Hennard C., Dugave C., Poole K., Abdallah M.A., Pattus F. Iron-free pyoverdin binds to its outer membrane receptor FpvA in Pseudomonas aeruginosa: A new mechanism for membrane iron transport. Mol. Microbiol. 2001;39:351–360. doi: 10.1046/j.1365-2958.2001.02207.x. [DOI] [PubMed] [Google Scholar]
- 73.Wayne R., Neilands J.B. Evidence for common binding sites for ferrichrome compounds and bacteriophage phi 80 in the cell envelope of Escherichia coli. J. Bacteriol. 1975;121:497–503. doi: 10.1128/jb.121.2.497-503.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nikaido H., Rosenberg E.Y. Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: Study with beta-lactam antibiotics containing catechol and analogous groups. J. Bacteriol. 1990;172:1361–1367. doi: 10.1128/jb.172.3.1361-1367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Warner P.J., Williams P.H., Bindereif A., Neilands J.B. ColV plasmid-specific aerobactin synthesis by invasive strains of Escherichia coli. Infect. Immun. 1981;33:540–545. doi: 10.1128/iai.33.2.540-545.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clifton M.C., Corrent C., Strong R.K. Siderocalins: Siderophore-binding proteins of the innate immune system. Biometals. 2009;22:557–564. doi: 10.1007/s10534-009-9207-6. [DOI] [PubMed] [Google Scholar]
- 77.Flo T.H., Smith K.D., Sato S., Rodriguez D.J., Holmes M.A., Strong R.K., Akira S., Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 78.Goetz D.H., Holmes M.A., Borregaard N., Bluhm M.E., Raymond K.N., Strong R.K. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 79.Sprencel C., Cao Z., Qi Z., Scott D.C., Montague M.A., Ivanoff N., Xu J., Raymond K.M., Newton S.M., Klebba P.E. Binding of ferric enterobactin by the Escherichia coli periplasmic protein fepB. J. Bacteriol. 2000;182:5359–5364. doi: 10.1128/jb.182.19.5359-5364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blattner F.R., Plunkett G., 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Gregor J., Davis N.W., Kirkpatrick H.A., Goeden M.A., Rose D.J., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 81.Deriu E., Liu J.Z., Pezeshki M., Edwards R.A., Ochoa R.J., Contreras H., Libby S.J., Fang F.C., Raffatellu M. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kokesová A., Frolová L., Kverka M., Sokol D., Rossmann P., Bártová J., Tlaskalová-Hogenová H. Oral administration of probiotic bacteria (E. coli Nissle, E. coli O83, Lactobacillus casei) influences the severity of dextran sodium sulfate-induced colitis in BALB/c mice. Folia Microbiol. (Praha) 2006;51:478–484. doi: 10.1007/BF02931595. [DOI] [PubMed] [Google Scholar]
- 83.Pradhan S., Weiss A.A. Probiotic properties of Escherichia coli Nissle in human intestinal organoids. mBio. 2020;11 doi: 10.1128/mBio.01470-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nazareth H., Genagon S.A., Russo T.A. Extraintestinal pathogenic Escherichia coli survives within neutrophils. Infect. Immun. 2007;75:2776–2785. doi: 10.1128/IAI.01095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Neidhardt F.C., Bloch P.L., Smith D.F. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roberts R.B., Abelson P.H., Cowie D.B., Bolton E.T., Britten R.J. Vol 607. Carnegie Institution; Washington, DC: 1957. p. 5. (Studies of Biosynthesis in Escherichia coli). [Google Scholar]
- 87.Rutz J.M., Abdullah T., Singh S.P., Kalve V.I., Klebba P.E. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J. Bacteriol. 1991;173:5964–5974. doi: 10.1128/jb.173.19.5964-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frillingos S., Gonzalez A., Kaback H.R. Cysteine-scanning mutagenesis of helix IV and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. off. Biochemistry. 1997;36:14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 89.Frillingos S., Kaback H.R. Cysteine-scanning mutagenesis of helix VI and the flanking hydrophilic domains on the lactose permease of Escherichia coli. Biochemistry. 1996;35:5333–5338. doi: 10.1021/bi953068d. [DOI] [PubMed] [Google Scholar]
- 90.Frillingos S., Sun J., Gonzalez A., Kaback H.R. Cysteine-scanning mutagenesis of helix II and flanking hydrophilic domains in the lactose permease of Escherichia coli. Biochemistry. 1997;36:269–273. doi: 10.1021/bi9618629. [DOI] [PubMed] [Google Scholar]
- 91.He M.M., Sun J., Kaback H.R. Cysteine-scanning mutagenesis of transmembrane domain XII and the flanking periplasmic loop in the lactose permease of Escherichia coli. Biochemistry. 1996;35:12909–12914. doi: 10.1021/bi960876b. [DOI] [PubMed] [Google Scholar]
- 92.Sahin-Toth M., Kaback H.R. Cysteine scanning mutagenesis of putative transmembrane helices IX and X in the lactose permease of Escherichia coli. Protein Sci. 1993;2:1024–1033. doi: 10.1002/pro.5560020615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weitzman C., Kaback H.R. Cysteine scanning mutagenesis of helix V in the lactose permease of Escherichia coli. Biochemistry. 1995;34:9374–9379. doi: 10.1021/bi00029a013. [DOI] [PubMed] [Google Scholar]
- 94.Ermolova N., Madhvani R.V., Kaback H.R. Site-directed alkylation of cysteine replacements in the lactose permease of Escherichia coli: Helices I, III, VI, and XI. Biochemistry. 2006;45:4182–4189. doi: 10.1021/bi052631h. [DOI] [PubMed] [Google Scholar]
- 95.Frillingos S., Kaback H.R. Probing the conformation of the lactose permease of Escherichia coli by in situ site-directed sulfhydryl modification. Biochemistry. 1996;35:3950–3956. doi: 10.1021/bi952601m. [DOI] [PubMed] [Google Scholar]
- 96.Frillingos S., Kaback H.R. The role of helix VIII in the lactose permease of Escherichia coli: II. Site-directed sulfhydryl modification. Protein Sci. 1997;6:438–443. doi: 10.1002/pro.5560060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guan L., Kaback H.R. Site-directed alkylation of cysteine to test solvent accessibility of membrane proteins. Nat. Protoc. 2007;2:2012–2017. doi: 10.1038/nprot.2007.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang X., Nie Y., Kaback H.R. Site-directed alkylation studies with LacY provide evidence for the alternating access model of transport. Biochemistry. 2011;50:1634–1640. doi: 10.1021/bi101988s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jung K., Jung H., Kaback H.R. Dynamics of lactose permease of Escherichia coli determined by site-directed fluorescence labeling. Biochemistry. 1994;33:3980–3985. doi: 10.1021/bi00179a026. [DOI] [PubMed] [Google Scholar]
- 100.Kaback H.R., Dunten R., Frillingos S., Venkatesan P., Kwaw I., Zhang W., Ermolova N. Site-directed alkylation and the alternating access model for LacY. Proc. Natl. Acad. Sci. U. S. A. 2007;104:491–494. doi: 10.1073/pnas.0609968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwaw I., Zen K.C., Hu Y., Kaback H.R. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: Helices IV and V that contain the major determinants for substrate binding. Biochemistry. 2001;40:10491–10499. doi: 10.1021/bi010866x. [DOI] [PubMed] [Google Scholar]
- 102.Majumdar D.S., Smirnova I., Kasho V., Nir E., Kong X., Weiss S., Kaback H.R. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nie Y., Ermolova N., Kaback H.R. Site-directed alkylation of LacY: Effect of the proton electrochemical gradient. J. Mol. Biol. 2007;374:356–364. doi: 10.1016/j.jmb.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Venkatesan P., Hu Y., Kaback H.R. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: Helix X. Biochemistry. 2000;39:10656–10661. doi: 10.1021/bi0004403. [DOI] [PubMed] [Google Scholar]
- 105.Venkatesan P., Kwaw I., Hu Y., Kaback H.R. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: Helix VII. Biochemistry. 2000;39:10641–10648. doi: 10.1021/bi000438b. [DOI] [PubMed] [Google Scholar]
- 106.Venkatesan P., Liu Z., Hu Y., Kaback H.R. Site-directed sulfhydryl labeling of the lactose permease of Escherichia coli: N-ethylmaleimide-sensitive face of helix II. Biochemistry. 2000;39:10649–10655. doi: 10.1021/bi0004394. [DOI] [PubMed] [Google Scholar]
- 107.Voss J., He M.M., Hubbell W.L., Kaback H.R. Site-directed spin labeling demonstrates that transmembrane domain XII in the lactose permease of Escherichia coli is an alpha-helix. Biochemistry. 1996;35:12915–12918. doi: 10.1021/bi9608774. [DOI] [PubMed] [Google Scholar]
- 108.Voss J., Hubbell W.L., Hernandez-Borrell J., Kaback H.R. Site-directed spin-labeling of transmembrane domain VII and the 4B1 antibody epitope in the lactose permease of Escherichia coli. Biochemistry. 1997;36:15055–15061. doi: 10.1021/bi971726j. [DOI] [PubMed] [Google Scholar]
- 109.Wu J., Voss J., Hubbell W.L., Kaback H.R. Site-directed spin labeling and chemical crosslinking demonstrate that helix V is close to helices VII and VIII in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10123–10127. doi: 10.1073/pnas.93.19.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang W., Hu Y., Kaback H.R. Site-directed sulfhydryl labeling of helix IX in the lactose permease of Escherichia coli. Biochemistry. 2003;42:4904–4908. doi: 10.1021/bi034078e. [DOI] [PubMed] [Google Scholar]
- 111.Annamalai R., Jin B., Cao Z., Newton S.M., Klebba P.E. Recognition of ferric catecholates by FepA. J. Bacteriol. 2004;186:3578–3589. doi: 10.1128/JB.186.11.3578-3589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hanson M., Jordan L.D., Shipelskiy Y., Newton S.M., Klebba P.E. High-throughput screening assay for inhibitors of TonB-dependent iron transport. J. Biomol. Screen. 2016;21:316–322. doi: 10.1177/1087057115613788. [DOI] [PubMed] [Google Scholar]
- 113.Klug C.S., Su W., Liu J., Klebba P.E., Feix J.B. Denaturant unfolding of the ferric enterobactin receptor and ligand- induced stabilization studied by site-directed spin labeling. Biochemistry. 1995;34:14230–14236. doi: 10.1021/bi00043a030. [DOI] [PubMed] [Google Scholar]
- 114.Ma L., Kaserer W., Annamalai R., Scott D.C., Jin B., Jiang X., Xiao Q., Maymani H., Massis L.M., Ferreira L.C., Newton S.M., Klebba P.E. Evidence of ball-and-chain transport of ferric enterobactin through FepA. J. Biol. Chem. 2007;282:397–406. doi: 10.1074/jbc.M605333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Majumdar A., Trinh V., Moore K.J., Smallwood C.R., Kumar A., Yang T., Scott D.C., Long N.J., Newton S.M., Klebba P.E. Conformational rearrangements in the N-domain of Escherichia coli FepA during ferric enterobactin transport. J. Biol. Chem. 2020;295:4974–4984. doi: 10.1074/jbc.RA119.011850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nairn B.L., Eliasson O.S., Hyder D.R., Long N.J., Majumdar A., Chakravorty S., McDonald P., Roy A., Newton S.M., Klebba P.E. Fluorescence high-throughput screening for inhibitors of TonB action. J. Bacteriol. 2017;199 doi: 10.1128/JB.00889-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Payne M.A., Igo J.D., Cao Z., Foster S.B., Newton S.M., Klebba P.E. Biphasic binding kinetics between FepA and its ligands. J. Biol. Chem. 1997;272:21950–21955. doi: 10.1074/jbc.272.35.21950. [DOI] [PubMed] [Google Scholar]
- 118.Ferguson A.D., Chakraborty R., Smith B.S., Esser L., van der Helm D., Deisenhofer J. Structural basis of gating by the outer membrane transporter FecA. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- 119.Scott D.C., Newton S.M., Klebba P.E. Surface loop motion in FepA. J. Bacteriol. 2002;184:4906–4911. doi: 10.1128/JB.184.17.4906-4911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wu J.Y., Srinivas P., Pogue J.M. Cefiderocol: A novel agent for the management of multidrug-resistant gram-negative organisms. Infect. Dis. Ther. 2020;9:17–40. doi: 10.1007/s40121-020-00286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mabayoje D.A., NicFhogartaigh C., Cherian B.P., Tan M.G.M., Wareham D.W. Compassionate use of cefiderocol for carbapenem-resistant Acinetobacter baumannii prosthetic joint infection. JAC Antimicrob. Resist. 2021;3:i21–i24. doi: 10.1093/jacamr/dlab055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wright H., Bonomo R.A., Paterson D.L. New agents for the treatment of infections with gram-negative bacteria: Restoring the miracle or false dawn? Clin. Microbiol. Infect. 2017;23:704–712. doi: 10.1016/j.cmi.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 123.Sheldon J.R., Skaar E.P. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Caza M., Lepine F., Milot S., Dozois C.M. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect. Immun. 2008;76:3539–3549. doi: 10.1128/IAI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee I.R., Molton J.S., Wyres K.L., Gorrie C., Wong J., Hoh C.H., Teo J., Kalimuddin S., Lye D.C., Archuleta S., Holt K.E., Gan Y.H. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci. Rep. 2016;6:29316. doi: 10.1038/srep29316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Williams P.H., Rabsch W., Methner U., Voigt W., Tschape H., Reissbrodt R. Catecholate receptor proteins in Salmonella enterica: Role in virulence and implications for vaccine development. Vaccine. 2006;24:3840–3844. doi: 10.1016/j.vaccine.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 127.Abergel R.J., Moore E.G., Strong R.K., Raymond K.N. Microbial evasion of the immune system: Structural modifications of enterobactin impair siderocalin recognition. J. Am. Chem. Soc. 2006;128:10998–10999. doi: 10.1021/ja062476+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Allred B.E., Correnti C., Clifton M.C., Strong R.K., Raymond K.N. Siderocalin outwits the coordination chemistry of vibriobactin, a siderophore of Vibrio cholerae. ACS Chem. Biol. 2013;8:1882–1887. doi: 10.1021/cb4002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoette T.M., Abergel R.J., Xu J., Strong R.K., Raymond K.N. The role of electrostatics in siderophore recognition by the immunoprotein Siderocalin. J. Am. Chem. Soc. 2008;130:17584–17592. doi: 10.1021/ja8074665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang J., Goetz D., Li J.Y., Wang W., Mori K., Setlik D., Du T., Erdjument-Bromage H., Tempst P., Strong R., Barasch J. An iron delivery pathway mediated by a lipocalin. Mol. Cell. 2002;10:1045–1056. doi: 10.1016/s1097-2765(02)00710-4. [DOI] [PubMed] [Google Scholar]
- 131.Der Vartanian M., Jaffeux B., Contrepois M., Chavarot M., Girardeau J.P., Bertin Y., Martin C. Role of aerobactin in systemic spread of an opportunistic strain of Escherichia coli from the intestinal tract of gnotobiotic lambs. Infect. Immun. 1992;60:2800–2807. doi: 10.1128/iai.60.7.2800-2807.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Faundez G., Figueroa G., Troncoso M., Cabello F.C. Characterization of enteroinvasive Escherichia coli strains isolated from children with diarrhea in Chile. J. Clin. Microbiol. 1988;26:928–932. doi: 10.1128/jcm.26.5.928-932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Garcia E.C., Brumbaugh A.R., Mobley H.L. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect. Immun. 2011;79:1225–1235. doi: 10.1128/IAI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Montgomerie J.Z., Bindereif A., Neilands J.B., Kalmanson G.M., Guze L.B. Association of hydroxamate siderophore (aerobactin) with Escherichia coli isolated from patients with bacteremia. Infect. Immun. 1984;46:835–838. doi: 10.1128/iai.46.3.835-838.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Torres A.G., Redford P., Welch R.A., Payne S.M. TonB-dependent systems of uropathogenic Escherichia coli: Aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 2001;69:6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Russo T.A., Marr C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019;32 doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Klebba P.E., McIntosh M.A., Neilands J.B. Kinetics of biosynthesis of iron-regulated membrane proteins in Escherichia coli. J. Bacteriol. 1982;149:880–888. doi: 10.1128/jb.149.3.880-888.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Massip C., Branchu P., Bossuet-Greif N., Chagneau C.V., Gaillard D., Martin P., Boury M., Sécher T., Dubois D., Nougayrède J.P., Oswald E. Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sonnenborn U. Escherichia coli strain Nissle 1917-from bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 2016;363 doi: 10.1093/femsle/fnw212. [DOI] [PubMed] [Google Scholar]
- 140.Lery L.M., Frangeul L., Tomas A., Passet V., Almeida A.S., Bialek-Davenet S., Barbe V., Bengoechea J.A., Sansonetti P., Brisse S., Tournebize R. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 2014;12:41. doi: 10.1186/1741-7007-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Groman N.B. Factors in lysis and lysis inhibition by lambda bacteriophage. J. Bacteriol. 1965;90:1563–1568. doi: 10.1128/jb.90.6.1563-1568.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Balhesteros H., Shipelskiy Y., Long N.J., Majumdar A., Katz B.B., Santos N.M., Leaden L., Newton S.M., Marques M.V., Klebba P.E. TonB-dependent heme/hemoglobin utilization by Caulobacter crescentus HutA. J. Bacteriol. 2017;199 doi: 10.1128/JB.00723-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moynié L., Serra I., Scorciapino M.A., Oueis E., Page M.G., Ceccarelli M., Naismith J.H. Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogen Acinetobacter baumannii. Elife. 2018;7 doi: 10.7554/eLife.42270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Buchanan S.K., Smith B.S., Venkatramani L., Xia D., Esser L., Palnitkar M., Chakraborty R., van der Helm D., Deisenhofer J. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 145.Chimento D.P., Mohanty A.K., Kadner R.J., Wiener M.C. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 146.Locher K.P., Rees B., Koebnik R., Mitschler A., Moulinier L., Rosenbusch J.P., Moras D. Transmembrane signaling across the ligand-gated FhuA receptor: Crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 147.Grinter R., Lithgow T. The structure of the bacterial iron-catecholate transporter Fiu suggests that it imports substrates via a two-step mechanism. J. Biol. Chem. 2019;294:19523–19534. doi: 10.1074/jbc.RA119.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Buchanan S.K., Lukacik P., Grizot S., Ghirlando R., Ali M.M., Barnard T.J., Jakes K.S., Kienker P.K., Esser L. Structure of colicin I receptor bound to the R-domain of colicin ia: Implications for protein import. EMBO J. 2007;26:2594–2604. doi: 10.1038/sj.emboj.7601693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Moynié L., Luscher A., Rolo D., Pletzer D., Tortajada A., Weingart H., Braun Y., Page M.G., Naismith J.H., Köhler T. Structure and function of the PiuA and PirA siderophore-drug receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02531-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lukacik P., Barnard T.J., Keller P.W., Chaturvedi K.S., Seddiki N., Fairman J.W., Noinaj N., Kirby T.L., Henderson J.P., Steven A.C., Hinnebusch B.J., Buchanan S.K. Structural engineering of a phage lysin that targets gram-negative pathogens. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9857–9862. doi: 10.1073/pnas.1203472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 152.Scott D.C. University of Oklahoma; 2000. Mechanism of Ferric Enterobactin Transport through Escherichia coli FepA: Evolution of a Bacterial Venus Fly trap. Ph. D. thesis. [Google Scholar]
- 153.Wayne R., Frick K., Neilands J.B. Siderophore protection against colicins M, B, V, and Ia in Escherichia coli. J. Bacteriol. 1976;126:7–12. doi: 10.1128/jb.126.1.7-12.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Smit J., Kamio Y., Nikaido H. Outer membrane of Salmonella typhimurium: Chemical analysis and freeze- fracture studies with lipopolysaccharide mutants. J. Bacteriol. 1975;124:942–958. doi: 10.1128/jb.124.2.942-958.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ames G.F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J. Biol. Chem. 1974;249:634–644. [PubMed] [Google Scholar]
- 156.Hancock R.E., Braun V. The colicin I receptor of Escherichia coli K-12 has a role in enterochelin-mediated iron transport. FEBS Lett. 1976;65:208–210. doi: 10.1016/0014-5793(76)80481-4. [DOI] [PubMed] [Google Scholar]
- 157.Lugtenberg B., Meijers J., Peters R., van der Hoek P., van Alphen L. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and the Supplemental Information file.