Abstract
Many signal transductions resulting from ligand–receptor interactions occur at the cell surface. These signaling pathways play essential roles in cell polarization, membrane morphogenesis, and the modulation of membrane tension at the cell surface. However, due to the large number of membrane-binding proteins, including actin-membrane linkers, and transmembrane proteins present at the cell surface, the molecular mechanisms underlying the regulation at the cell surface are yet unclear. Here, we describe the molecular functions of one of the key players at the cell surface, ezrin/radixin/moesin (ERM) proteins from a biophysical point of view. We focus our discussion on biophysical properties of ERM proteins revealed by using biophysical tools in live cells and in vitro reconstitution systems. We first describe the structural properties of ERM proteins and then discuss the interactions of ERM proteins with PI(4,5)P2 and the actin cytoskeleton. These properties of ERM proteins revealed by using biophysical approaches have led to a better understanding of their physiological functions in cells and tissues.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12551-021-00928-0.
Keywords: Ezrin/radixin/moesin proteins; Phosphatidylinositol 4,5-bisphosphate; Actin cytoskeleton; Membrane tension; GUVs; Supported lipid bilayers
Introduction
The cell surface is composed of the plasma membrane, membrane proteins, and a thin layer of crosslinked actin networks underlying the membrane (Chalut and Paluch 2016; Chugh et al. 2017; Chugh and Paluch 2018; Svitkina 2020; Sitarska and Diz-Muñoz 2020). The cell surface acts as an interface between cells and the extracellular environment. Many outside-in/inside-out signals are transduced through the cell surface. Thus, biochemical reactions occurring at the cell surface need to be precisely regulated. Fine-tuned biochemical signals at the cell surface result in the formation of distinct compartments in the plasma membrane, which in turn define cell morphology and polarization (Wakayama et al. 2011; Valderrama et al. 2012; Hebert et al. 2012; Fröse et al. 2018). For example, microvilli at the apical surface of epithelial cells have distinct proteins and lipid compositions compared to the rest of the plasma membrane (Ikenouchi 2018). One of the key players for this compartmentalization in the plasma membrane are ezrin/radixin/moesin (ERM) proteins (Neisch and Fehon 2011; McClatchey 2014; Chugh and Paluch 2018; Senju and Lappalainen 2019). ERM proteins are evolutionarily conserved protein families (Fig. S1a) (Mu et al. 2018). ERM proteins have high sequence homology (75.9% amino acid sequence identity between ezrin and radixin, and 72.2% amino acid sequence identity between ezrin and moesin). At the N-terminus of ERM proteins, there is a FERM domain which has a high sequence identity (85.9% amino acid sequence identity between ezrin and radixin and 85.4% amino acid sequence identity between ezrin and moesin) among human ERM proteins (Fig. 1a) (Tsukita and Yonemura 1999). The FERM domain is composed of a cloverleaf-like-shaped structure with three subdomains (F1, F2, and F3). Recent in vivo experiments showed that the binding of the FERM domain to phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) is critical for the function of ERM proteins (Ramalho et al. 2020). PI(4,5)P2 is one of the phosphoinositides that is negatively charged and acts as a second messenger to play vital roles in signal transduction and protein localization at the membrane compartments in cells. At the C-terminus, ERM proteins have an actin-binding domain (C-ERMAD). Thus, ERM proteins play key roles in cell signaling, morphogenesis, motility, and metastasis partly by modulating the linkage between the actin cytoskeleton and the plasma membrane (Muriel et al. 2016; Schön et al. 2019).
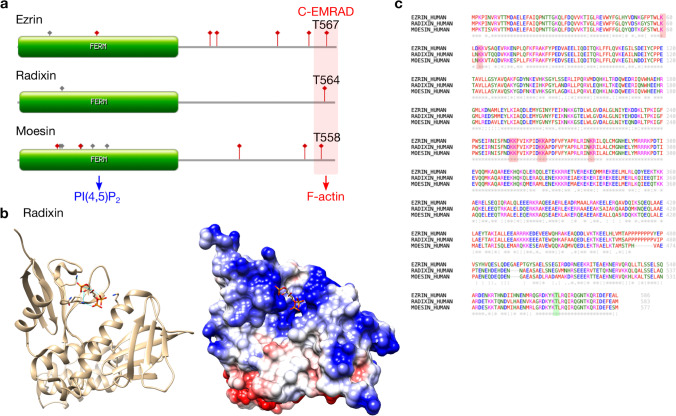
Fig. 1.
(a) Domain structure and post-translational modification of Homo sapiens ERM proteins are generated using PROSITE. Red (depicted with square) indicates the phosphorylation site, and gray indicates other post-translational modifications. The FERM domain interacts with PI(4,5)P2, and C-EMRAD interacts with the actin cytoskeleton. (b) Multiple sequence alignment of Homo sapiens ERM proteins is generated using Clustal Omega. Note that the phosphorylation sites of ROCK and other kinases (T567 for ezrin, T564 for radixin, and T558 for moesin, depicted with green box) and PI(4,5)P2-binding sites (depicted with red box) responsible for the activation of ERM proteins are well-conserved. (c) Ribbon and coulombic surface of radixin FERM domain-inositol-(1,4,5)-triphosphate (IP3) complex structure (PDB: 1GC6). IP3 (the head group of PI(4,5)P2) binds the negatively-charged surface (blue) of the radixin FERM domain. Hydrogen bonds between radixin FERM domain and three lysines (Lys60 and Lys63 from subdomain F1, and Lys278 from subdomain F3) and one asparagine (Asn62 from subdomain A) interacting with the three phosphate groups of IP3 are depicted by blue lines
ERM proteins can be scaffold proteins in the regulation of many fundamental physiological processes, such as the establishment of cell polarity (Wakayama et al. 2011; Valderrama et al. 2012; Hebert et al. 2012; Abeysundara et al. 2018; Fröse et al. 2018), microvilli formation (Zwaenepoel et al. 2012; Viswanatha et al. 2012, 2014; Dehapiot and Halet 2013), blebbing (Charras and Paluch 2008; Fritzsche et al. 2014; Ikenouchi and Aoki 2017; Hinojosa et al. 2017; Ikenouchi 2018), cell migration (Arpin et al. 2011; Parameswaran et al. 2011; Liu et al. 2012; Mak et al. 2012; Valderrama et al. 2012; Hsu et al. 2012; Saito et al. 2013; DeSouza et al. 2013; Parameswaran and Gupta 2013; Baeyens et al. 2013; García-Ortiz and Serrador 2020; Rahimi et al. 2021), cell division (Roubinet et al. 2011; Kunda et al. 2012; Hebert et al. 2012; Solinet et al. 2013; Sabino et al. 2015; Vilmos et al. 2016; Abeysundara et al. 2018; Yang et al. 2021; Rahimi et al. 2021), endocytosis (Li et al. 2017), exocytosis (Carmosino et al. 2012), phagocytosis (Mu et al. 2018; Roberts et al. 2020), podosome formation (Pan et al. 2013), transendothelial cell macroaperture (Stefani et al. 2017), cell–cell adhesions (Valderrama et al. 2012; Hsu et al. 2012; Amsellem et al. 2014), and epithelial-mesenchymal transition (Haynes et al. 2011; Wang et al. 2012; Fröse et al. 2018). Additionally, ERM proteins contribute to the modulation of some mechanical properties of the cell surface, such as tension, stiffness, and dynamics by regulating the actin cortex (Liu et al. 2012; Rouven Brückner et al. 2015; Stefani et al. 2017; Chugh and Paluch 2018; Roberts et al. 2020; Sitarska and Diz-Muñoz 2020). To accomplish the above-mentioned cellular processes, cells rely on the precise spatiotemporal regulations of ERM proteins via reversible activation cycles.
Although several cell types and tissues express more than one ERM protein, the expression levels of each ERM protein vary in cells and tissues according to their distinct functions (Fig. S1b) (Wang et al. 2012). For example, ezrin is highly expressed in intestinal epithelial cells, radixin in hepatocytes, and moesin in vascular endothelial cells (Fehon et al. 2010). Thus, although ERM proteins share high homology sequences, individual ERM proteins may have specific and unique physiological functions in different tissues.
In this review, we discuss several biophysical characterizations of ERM proteins, focusing on their activation upon PI(4,5)P2-binding and phosphorylation, and the regulation of the dynamics of the actin cytoskeleton revealed by using biophysical tools in live cells and in vitro reconstitution systems.
ERM protein regulation in cells
The regulation (activation and inactivation) of ERM proteins is reversible and fine-tuned at the cell surface to achieve their physiological functions (Tachibana et al. 2015). The inactivated ERM proteins have a closed configuration as cytosolic monomers or dimers due to a head-to-tail intra or intermolecular interaction between the FERM domain and the C-ERMAD; in this closed configuration, the actin-binding site of ERM proteins is masked. “Opening up” ERM proteins requires FERM domain-PI(4,5)P2 binding and post-translational modifications (Bosk et al. 2011; Jayasundar et al. 2012; Maniti et al. 2012; Shabardina et al. 2016; Lubart et al. 2018). So far, the precise mechanism of ERM protein regulation remains unclear.
The X-ray crystal structure of radixin has revealed that its FERM domain binds to the IP3 (head group of PI(4,5)P2) (Fig. 1b) (Hamada et al. 2000). The interaction is based on two major binding sites: the “pocket” and the “patch” (two pairs of lysine residues). The Lys60, Lys63 and Lys278, and clustered patches Lys253-Lys254 and Lys262-Lys263 of subdomain F3 are responsible for PI(4,5)P2 binding. These residues are well-conserved among the FERM domains of ERM proteins, indicating a similar binding mode to PI(4,5)P2 (Fig. 1c). FERM domains have a certain structural orientation against the membrane, as determined by their PI(4,5)P2 interactions. The patch is proposed to be more accessible than the pocket in autoinhibited moesin (Ben-Aissa et al. 2012). Thus, PI(4,5)P2 may bind to the more accessible patch first and then to the pocket (Ben-Aissa et al. 2012).
Several post-translational modifications have been reported for ERM proteins. Of these, the phosphorylation of T567 for ezrin (Homo sapiens), T564 for radixin (Homo sapiens), and T558 for moesin (Homo sapiens) are well-conserved and critical for the activation of ERM proteins (Fig. 1c) (Pelaseyed et al. 2017; Lubart et al. 2018; Ramalho et al. 2020; 2017). In the closed conformation of ERM proteins, these phosphorylation sites are buried at the interface between the FERM domain and the C-ERMAD. The binding of the FERM domain to PI(4,5)P2 changes the conformational structure of the ERM proteins to render the actin-binding site of the C-ERMAD more accessible for threonine phosphorylation (Bosk et al. 2011; Ben-Aissa et al. 2012; Braunger et al. 2014; Shabardina et al. 2016). Phosphorylation at the C-terminus of ERM proteins then “opens up” the proteins by the repulsive interaction between the FERM domain and the C-ERMAD due to the negative charge of the additional phosphate group. Thus, binding to PI(4,5)P2 followed by the phosphorylation of threonine in the C-ERMAD cooperatively promotes the full activation of ERM proteins (Fig. 2) (Pelaseyed et al. 2017).
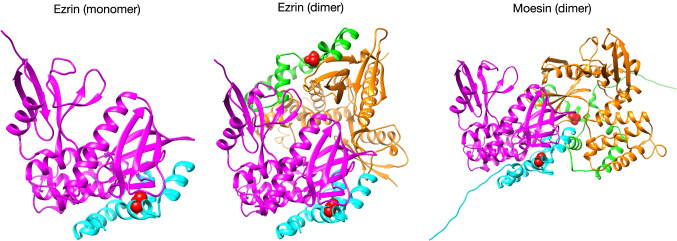
Fig. 2.
Crystal structures of auto-inhibition with head-to-tail interaction of ERM proteins (PDB: 4RM9 for ezrin monomer, PDB: 4RM8 for ezrin dimer, PDB: 1EF1 for moesin dimer). The phosphorylation sites buried at the interface between the FERM domain and the C-ERMAD are depicted with red sphere
Phosphorylation of ERM proteins is specifically mediated by ROCK (Belkina et al. 2009). PKC isoforms (Wald et al. 2008; Hong et al. 2011), LOK/STK10 (Belkina et al. 2009; Viswanatha et al. 2012; Pelaseyed et al. 2017), SLK (Viswanatha et al. 2012), and JNK (Pan et al. 2013) can also phosphorylate the above-mentioned sites of ERM proteins; however, their distinct mechanisms of physiological regulation remain elusive. Of note, LOK and SLK both localize at the apical membrane of epithelial cells, where PI(4,5)P2 is present, and specifically phosphorylate ezrin at the cell surface (Viswanatha et al. 2012; Pelaseyed et al. 2017).
Phosphorylated ERMs are dephosphorylated by several phosphatases, such as MLCP (Kovacs-Kasa et al. 2016) and protein phosphatase 1 (PP1) (Yang et al. 2012; Kunda et al. 2012; Canals et al. 2012). These phosphatases inactivate ERM proteins to form a closed conformation in the cytoplasm. In the closed conformation, the FERM domain of ERM proteins interact with C-ERMAD in a head-to-tail manner by masking the membrane and actin-binding surfaces (Ben-Aissa et al. 2012; Jayasundar et al. 2012).
In addition to filamentous actin (F-actin) and microtubules (see below), ERM proteins directly or indirectly bind to several other proteins (Fig. S1c), for example, scaffold proteins (IQGAP1, NHERF-1, NHERF-2), actin-regulatory proteins (ELMO, EPS8, RhoGAP conundrum), transporter proteins (anoctamin-1, aquaporin-2, NHE-3, NKCC2), receptors (CD44, thrombomodulin), metal-binding proteins (PDZD8, S100-A4), and adhesion molecules (ICAM-2, podocalyxin, TMIGD1) Henning et al. 2011; Carmosino et al. 2012; Hsu et al. 2012; Perez-Cornejo et al. 2012; Zwaenepoel et al. 2012; Boratkó and Csortos 2013; Neisch et al. 2013; Amsellem et al. 2014; Liu et al. 2014; Singh et al. 2014; Viswanatha et al. 2014; Chen et al. 2015; Epting et al. 2015; Nammalwar et al. 2015; Biri-Kovács et al. 2017; Li et al. 2017; Fröse et al. 2018; Rahimi et al. 2021). Upon PI(4,5)P2 binding, some of the interaction partners of ERM proteins bind selectively with either phosphorylated or non-phosphorylated forms of ERM proteins (Viswanatha et al. 2013; Biri-Kovács et al. 2017). Although validations of this interactome are ongoing, the activation of ERM proteins involves several selective protein–protein interactions which are spatiotemporally regulated at specific subcellular compartments.
ERM proteins link the cytoskeleton to the plasma membrane
Activated ERM proteins can link the actin cytoskeleton to the plasma membrane or integral membrane proteins (CD43/44, CFTR, ICAM-1/2, and NHE3) (Tsukita and Yonemura 1999; Bretscher et al. 2002; Fehon et al. 2010; Neisch and Fehon 2011; Braunger et al. 2014). This interaction is achieved directly, or indirectly with scaffold proteins, for example, NHERF (Terawaki et al. 2006; Kawaguchi et al. 2017). The interaction between ERM proteins and F-actin with or without their interaction partners is essential for regulating cortical actin assembly for instance in microvilli of epithelial cells and filopodia of migrating cells (Sauvanet et al. 2015). Thus, ERM proteins regulate spatiotemporal turnover of actin assembly and disassembly in specific subcellular structures (Muriel et al. 2016). It was shown that moesin promotes F-actin network formation on early endosomes. Besides actin, moesin has also been demonstrated to interact with microtubules (Solinet et al. 2013; Lubart et al. 2018). It was shown that moesin directly binds to microtubules upon phosphorylation in vitro and stabilizes microtubules at the cell cortex in vivo. This ERM–microtubule interaction is required for regulating spindle organization during metaphase and cell shape transformation after anaphase onset; however, the detailed molecular mechanisms of ERM–microtubule interaction and the corresponding physiological roles remain elusive.
ERM protein dynamics on the membrane and with the actin cytoskeleton
The complexity of studying ERM–membrane interactions in cells can be circumvented by using purified ERM proteins and model membranes (Maniti et al. 2013; Sarkis and Vié 2020). The methodological advances have enabled the generation of model membranes containing PI(4,5)P2 (Carvalho et al. 2008; Maniti et al. 2013; Drücker et al. 2014; Beber et al. 2019; Schäfer et al. 2020). In the past decades, rich and insightful information on how ERM proteins are activated and how they bind to PI(4,5)P2 and F-actin have been provided by in vitro reconstitution systems composed of purified ERM proteins and model membranes. Typical model membranes are supported lipid bilayers (SLBs), multilamellar vesicles (MLVs, typical diameters larger than 500 nm), small unilamellar vesicles (SUVs, typical diameters smaller than 100 nm), large unilamellar vesicles (LUVs, typical diameters of 100–1000 nm), and giant unilamellar vesicles (GUVs, typical diameters larger than 1 μm) (Lin et al. 2010; Sezgin and Schwille 2012; Dimova and Marques 2019).
MLVs, LUVs, and SUVs have been intensively used to study protein–membrane interactions quantitatively, for example, by using EM microscopy, co-sedimentation assays, and spectroscopic techniques (Blin et al. 2008; Maniti et al. 2012; Senju and Zhao 2021; Senju et al. 2021). These vesicles have a relatively smaller size compared to cells. It is noteworthy that when using SUVs, their membrane curvature could contribute to how proteins interact with the membranes. This is because the diameter of SUVs, which is usually on the order of 100 nm, is only a few times larger than protein sizes; for instance, Bin/amphiphysin/Rvs (BAR) domains have a length of around 20 nm (Carman and Dominguez 2018). Therefore, when binding on SUVs, proteins may be bent or may have a certain configuration that would allow binding to the curved SUV surfaces. Besides, the curvature of SUVs could be quite different from that of the membranes of cellular organelles, and hence this should be considered when comparing lipid–protein interactions on SUVs with those in cell membranes. Furthermore, as the membrane tension of these three vesicle types is generally high compared with that of GUVs, protein-driven membrane deformation, if any, is not readily assessable.
Using PI(4,5)P2-containing MLVs, LUVs, or SUVs combined with co-sedimentation assays or fluorescence correlation spectroscopy (FCS), the binding affinities and modes of ERM proteins with PI(4,5)P2 have been obtained (Blin et al. 2008; Maniti et al. 2013; Senju et al. 2017). The apparent dissociation constant (Kd) is around 5 μM for ezrin and moesin to PI(4,5)P2-containing liposomes whose lipid compositions are similar to that of the plasma membrane. Cooperative binding of PI(4,5)P2 with ERM proteins has also been observed (Jayasundar et al. 2012; Lubart et al. 2018). Thus, the initial binding of one PI(4,5)P2 molecule to ERM proteins may promote additional PI(4,5)P2 binding, thereby inducing PI(4,5)P2 clustering.
GUVs have been well-recognized as an important model system to study protein–membrane interactions given that GUV membranes resemble many, if not all, properties of cellular membranes (Litschel et al. 2021). Some key advantages of using GUVs are (1) micron-size allowing readily observation by conventional microscopy; (2) deformable, free-standing membranes; and (3) readiness for optical, mechanical and chemical manipulations, for instance to change membrane tension or GUV shapes.
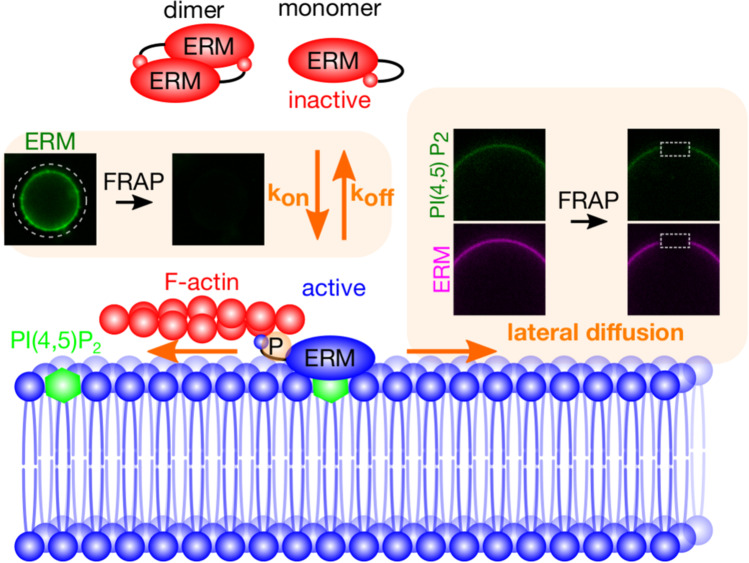
Fluorescence recovery after photobleaching (FRAP) experiments have revealed that ezrin and moesin are stably associated to PI(4,5)P2-containing GUV membranes, whose lipid composition is similar to that of the plasma membrane (Fig. 3) (Senju et al. 2017). Previous FRAP studies in epithelial cell microvilli and melanoma cell blebs have indicated the presence of more than two pools of ezrin fluorescence recovery with different timescales (Coscoy et al. 2002; Fritzsche et al. 2014). The fast turnover pool of ezrin in cell blebs reflects its binding and unbinding to F-actin (Fritzsche et al. 2014). Interestingly, in microvilli, ezrin was found to be “immobile” when binding to F-actin. These different observations indicate the plausible contribution of membrane shape (the rather flat membrane in blebs and the highly curved membrane in microvilli) in the physiological function of ERM proteins. In addition to the change in the binding/unbinding constant owing to the differences in membrane curvatures, it could also be difficult for proteins to diffuse in or out of the microvilli because of their small diameter. In cellular structures and on GUVs, the slow ezrin turnover indicates that ezrin binds to PI(4,5)P2-containing membranes with high affinity and slow dissociation dynamics. Furthermore, the FERM domain of ezrin was found to regulate PI(4,5)P2 lateral diffusion by slowing it down (Senju et al. 2017). This slow diffusion is postulated to be the result of protein crowding or the oligomerization of the ezrin on the membrane. PI(4,5)P2 is known to be clustered in specific microdomains in cells (Wen et al. 2021). Thus, ERM proteins would compensate for the negative charge of the PI(4,5)P2 head group by binding with their positively charged residues on the membrane. Recently, using a newly developed tool, bioluminescence resonance energy transfer (BRET)-based conformational biosensors, a pool of closed inactive but membrane-associated ezrin was found in cells. This pool of ezrin was rapidly activated before the further recruitment of other closed inactive ezrin from the cytosol to the plasma membrane. This recent work provides new insights into the long-standing question of the mechanism of ERM protein regulation.
Fig. 3.
A working model for the dynamics of ERM proteins on PI(4,5)P2-containing membranes. The association/dissociation of ERM proteins with membranes, and the lateral diffusion of ERM proteins and PI(4,5)P2 can be analyzed by using giant unilamellar vesicles (GUVs) and fluorescent recovery after photo bleaching (FRAP)
SLBs are lipid bilayers formed on solid substrates such as glass and mica. Thanks to the solid support, one can change buffers, and protein type and concentration readily, for instance by using microfluidic devices. Additionally, since the membrane is sitting on top of the substrate, one can easily implement total internal reflection fluorescence (TIRF) microscopy to achieve an outstanding signal-to-noise ratio, as well as several microscopy techniques such as FCS (Haustein and Schwille 2007) and fluorescence resonance energy transfer (FRET), and super-resolution microscopies such as direct stochastic optical reconstruction microscopy (dSTORM) and stimulated emission depletion (STED) microscopy to reveal protein clustering and assembly at a nanometer resolution (Mashaghi et al. 2014; Sezgin 2017; Migliorini et al. 2018; Barbotin et al. 2020). A series of studies using SLBs and purified ezrin, for instance the mutant ezrin T567D that mimics the phosphorylation of the threonine residue, has been carried out (Bosk et al. 2011; Shabardina et al. 2016). They showed that the phosphorylation of ezrin T567 enhances its conformational change to the active state upon PI(4,5)P2-binding, which is capable of binding to F-actin. Recently, the roles of actin crosslinkers, fascin and α-actinin, in the architecture of the ezrin-mediated actin cortex assembled on PI(4,5)P2-SLBs were investigated (Schön et al. 2019). The addition of these crosslinkers was found to influence the architecture of the ezrin-mediated actin network. Moreover, ezrin can be recruited to negatively curved membrane tubes via a direct interaction with the I-BAR domain of IRSp53, which is reminiscent of cellular filopodia (Tsai et al. 2018). The molecular details provided in the above-mentioned studies contribute to our understanding of the vital roles of PI(4,5)P2 and the actin cytoskeleton in ERM protein association and signaling in cells.
Conclusions and future perspectives
ERM proteins have many vital physiological functions in cell polarity, morphogenesis, and the modulation of membrane tension, partly via their actin–membrane linking ability. To carry out this wide range of functions, ERM proteins orchestrate the assembly of protein complexes at the cell surface.
This review highlights the need for further investigation of the interactions of PI(4,5)P2 molecules with the FERM domain of ERM proteins, for example, their cooperative or non-cooperative binding. The physiological role of moesin and microtubule interaction, and moesin-mediated interplay between the actin cytoskeleton and microtubule need to be investigated. Additionally, it remains unclear how PI(4,5)P2 hydrolysis may contribute to the inactivation of ERM proteins and induce their subsequent dissociation from the plasma membrane in vivo. A better understanding of the physiological roles of post-translational modifications, other than the well-characterized threonine phosphorylation at the C-ERMAD of ERM proteins, is also needed. To gain further insights into the regulation of ERM proteins, the clarification of whether ERM proteins function as monomers, dimers with a head-to-tail orientation (Phang et al. 2016; Lubart et al. 2018), or oligomers at the cell surface (Lubart et al. 2018) is needed. The generally accepted knowledge in cells is that the monomeric form of ERM proteins is active and localized at the plasma membrane, whereas the dimeric form is inactive and mostly found in the cytoplasm. The recently found pool of inactive ERM proteins that are stably associated with the plasma membrane calls for future studies to answer this question in cell biology. Little is known regarding how ERM proteins are spatiotemporally regulated in vivo. Biophysical approaches using the well-defined systems that we have introduced here will certainly provide new insights into these fundamental questions.
Supplementary Information
Below is the link to the electronic supplementary material.
Fig. S1 (EPS 1805 kb)
(a) A simple phylogenetic tree of ERM proteins.
(b) The microarray gene expression profiles of Homo sapiens ERM proteins in various cell lines and tissues generated by BioGPS. Note that the expression levels of ezrin/radoxin/moesin are different in cell lines and tissues.
(c) Protein-protein interaction (PPI) network analysis. Some known and predicted PPIs of ERM proteins are analyzed using stringApp in Cytoscape.
Acknowledgements
We thank Prof. Pekka Lappalainen (HiLIFE - Institute of Biotechnology, University of Helsinki, Finland), Prof. Patricia Bassereau (Institut Curie, France), Prof. Emmanuel Lemichez (Institut Pasteur, France), and Prof. Ilpo Vattulainen (Department of Physics, University of Helsinki, Finland) for the insightful discussions.
Author contribution
Yosuke Senju and Feng-Ching Tsai had the idea for the article. Yosuke Senju and Feng-Ching Tsai performed the literature search. Yosuke Senju and Feng-Ching Tsai drafted and critically revised the work.
Funding
This study was supported by the FY 2015 Researcher Exchange Program between the Japan Society for the Promotion of Science (JSPS) and Academy of Finland (AF), Astellas Foundation for Research on Metabolic Disorders, Scandinavia-Japan Sasakawa Foundation, Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care, The Association for Fordays Self-Reliance Support in Japan, Okayama Foundation for Science and Technology, ONO Medical Research Foundation, Takeda Science Foundation, The Naito Foundation, The Company of Biologists, European Biophysical Societies Association (EBSA), and Institut Curie, Centre National de la Recherche Scientifique (CNRS).
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yosuke Senju, Email: yosuke.senju@okayama-u.ac.jp.
Feng-Ching Tsai, Email: feng-ching.tsai@curie.fr.
References
- Abeysundara N, Simmonds AJ, Hughes SC. Moesin is involved in polarity maintenance and cortical remodeling during asymmetric cell division. Mol Biol Cell. 2018;29:419–434. doi: 10.1091/mbc.E17-05-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsellem V, Dryden NH, Martinelli R, et al. ICAM-2 regulates vascular permeability and N-cadherin localization through ezrin-radixin-moesin (ERM) proteins and Rac-1 signalling. Cell Commun Signal. 2014;12:12. doi: 10.1186/1478-811X-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpin M, Chirivino D, Naba A, Zwaenepoel I. Emerging role for ERM proteins in cell adhesion and migration. Cell Adh Migr. 2011;5:199–206. doi: 10.4161/cam.5.2.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens N, Latrache I, Yerna X, et al. Redundant control of migration and adhesion by ERM proteins in vascular smooth muscle cells. Biochem Biophys Res Commun. 2013;441:579–585. doi: 10.1016/j.bbrc.2013.10.118. [DOI] [PubMed] [Google Scholar]
- Barbotin A, Urbančič I, Galiani S, et al. z-STED imaging and spectroscopy to investigate nanoscale membrane structure and dynamics. Biophys J. 2020;118:2448–2457. doi: 10.1016/j.bpj.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beber A, Alqabandi M, Prévost C, et al. Septin-based readout of PI(4,5)P2 incorporation into membranes of giant unilamellar vesicles. Cytoskeleton (hoboken) 2019;76:92–103. doi: 10.1002/cm.21480. [DOI] [PubMed] [Google Scholar]
- Belkina NV, Liu Y, Hao J-J, et al. LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc Natl Acad Sci U S A. 2009;106:4707–4712. doi: 10.1073/pnas.0805963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aissa K, Patino-Lopez G, Belkina NV, et al. Activation of moesin, a protein that links actin cytoskeleton to the plasma membrane, occurs by phosphatidylinositol 4,5-bisphosphate (PIP2) binding sequentially to two sites and releasing an autoinhibitory linker. J Biol Chem. 2012;287:16311–16323. doi: 10.1074/jbc.M111.304881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biri-Kovács B, Kiss B, Vadászi H, et al. Ezrin interacts with S100A4 via both its N- and C-terminal domains. PLoS ONE. 2017;12:e0177489. doi: 10.1371/journal.pone.0177489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin G, Margeat E, Carvalho K, et al. Quantitative analysis of the binding of ezrin to large unilamellar vesicles containing phosphatidylinositol 4,5 bisphosphate. Biophys J. 2008;94:1021–1033. doi: 10.1529/biophysj.107.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratkó A, Csortos C. NHERF2 is crucial in ERM phosphorylation in pulmonary endothelial cells. Cell Commun Signal. 2013;11:99. doi: 10.1186/1478-811X-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosk S, Braunger JA, Gerke V, Steinem C. Activation of F-actin binding capacity of ezrin: synergism of PIP2 interaction and phosphorylation. Biophys J. 2011;100:1708–1717. doi: 10.1016/j.bpj.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunger JA, Brückner BR, Nehls S, et al. Phosphatidylinositol 4,5-bisphosphate alters the number of attachment sites between ezrin and actin filaments: a colloidal probe study. J Biol Chem. 2014;289:9833–9843. doi: 10.1074/jbc.M113.530659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Canals D, Roddy P, Hannun YA. Protein phosphatase 1α mediates ceramide-induced ERM protein dephosphorylation: a novel mechanism independent of phosphatidylinositol 4, 5-biphosphate (PIP2) and myosin/ERM phosphatase. J Biol Chem. 2012;287:10145–10155. doi: 10.1074/jbc.M111.306456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman PJ, Dominguez R. BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys Rev. 2018;10:1587–1604. doi: 10.1007/s12551-018-0467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmosino M, Rizzo F, Procino G, et al. Identification of moesin as NKCC2-interacting protein and analysis of its functional role in the NKCC2 apical trafficking. Biol Cell. 2012;104:658–676. doi: 10.1111/boc.201100074. [DOI] [PubMed] [Google Scholar]
- Carvalho K, Ramos L, Roy C, Picart C. Giant unilamellar vesicles containing phosphatidylinositol(4,5)bisphosphate: characterization and functionality. Biophys J. 2008;95:4348–4360. doi: 10.1529/biophysj.107.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalut KJ, Paluch EK. The actin cortex: a bridge between cell shape and function. Dev Cell. 2016;38:571–573. doi: 10.1016/j.devcel.2016.09.011. [DOI] [PubMed] [Google Scholar]
- Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- Chen X, Khajeh JA, Ju JH, et al. Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-ezrin heterocomplex, as revealed by small angle neutron scattering. J Biol Chem. 2015;290:6639–6652. doi: 10.1074/jbc.M114.589523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Clark AG, Smith MB, et al. Actin cortex architecture regulates cell surface tension. Nat Cell Biol. 2017;19:689–697. doi: 10.1038/ncb3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Paluch EK (2018) The actin cortex at a glance. J Cell Sci 131:jcs186254. 10.1242/jcs.186254 [DOI] [PMC free article] [PubMed]
- Coscoy S, Waharte F, Gautreau A, et al. Molecular analysis of microscopic ezrin dynamics by two-photon FRAP. Proc Natl Acad Sci U S A. 2002;99:12813–12818. doi: 10.1073/pnas.192084599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehapiot B, Halet G. Ran GTPase promotes oocyte polarization by regulating ERM (ezrin/radixin/moesin) inactivation. Cell Cycle. 2013;12:1672–1678. doi: 10.4161/cc.24901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSouza LV, Matta A, Karim Z, et al. Role of moesin in hyaluronan induced cell migration in glioblastoma multiforme. Mol Cancer. 2013;12:74. doi: 10.1186/1476-4598-12-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova R, Marques CM, editors. The giant vesicle book. Boca Raton: CRC Press; 2019. [Google Scholar]
- Drücker P, Grill D, Gerke V, Galla H-J. Formation and characterization of supported lipid bilayers containing phosphatidylinositol-4,5-bisphosphate and cholesterol as functional surfaces. Langmuir. 2014;30:14877–14886. doi: 10.1021/la503203a. [DOI] [PubMed] [Google Scholar]
- Epting D, Slanchev K, Boehlke C, et al. The Rac1 regulator ELMO controls basal body migration and docking in multiciliated cells through interaction with ezrin. Development. 2015;142:174–184. doi: 10.1242/dev.112250. [DOI] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche M, Thorogate R, Charras G. Quantitative analysis of ezrin turnover dynamics in the actin cortex. Biophys J. 2014;106:343–353. doi: 10.1016/j.bpj.2013.11.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröse J, Chen MB, Hebron KE, et al. Epithelial-mesenchymal transition induces podocalyxin to promote extravasation via ezrin signaling. Cell Rep. 2018;24:962–972. doi: 10.1016/j.celrep.2018.06.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ortiz A, Serrador JM. ERM proteins at the crossroad of leukocyte polarization, migration and intercellular adhesion. Int J Mol Sci. 2020;21:E1502. doi: 10.3390/ijms21041502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, Shimizu T, Matsui T, et al. Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 2000;19:4449–4462. doi: 10.1093/emboj/19.17.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haustein E, Schwille P. Fluorescence correlation spectroscopy: novel variations of an established technique. Annu Rev Biophys Biomol Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- Haynes J, Srivastava J, Madson N, et al. Dynamic actin remodeling during epithelial–mesenchymal transition depends on increased moesin expression. Mol Biol Cell. 2011;22:4750–4764. doi: 10.1091/mbc.E11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AM, DuBoff B, Casaletto JB, et al. Merlin/ERM proteins establish cortical asymmetry and centrosome position. Genes Dev. 2012;26:2709–2723. doi: 10.1101/gad.194027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning MS, Stiedl P, Barry DS, et al. PDZD8 is a novel moesin-interacting cytoskeletal regulatory protein that suppresses infection by herpes simplex virus type 1. Virology. 2011;415:114–121. doi: 10.1016/j.virol.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Hinojosa LS, Holst M, Baarlink C, Grosse R. MRTF transcription and ezrin-dependent plasma membrane blebbing are required for entotic invasion. J Cell Biol. 2017;216:3087–3095. doi: 10.1083/jcb.201702010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-H, Osborne T, Ren L, et al. Protein kinase C regulates ezrin-radixin-moesin phosphorylation in canine osteosarcoma cells. Vet Comp Oncol. 2011;9:207–218. doi: 10.1111/j.1476-5829.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-Y, Shi G-Y, Kuo C-H, et al. Thrombomodulin is an ezrin-interacting protein that controls epithelial morphology and promotes collective cell migration. FASEB J. 2012;26:3440–3452. doi: 10.1096/fj.12-204917. [DOI] [PubMed] [Google Scholar]
- Ikenouchi J. Roles of membrane lipids in the organization of epithelial cells: old and new problems. Tissue Barriers. 2018;6:1–8. doi: 10.1080/21688370.2018.1502531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenouchi J, Aoki K. Membrane bleb: a seesaw game of two small GTPases. Small GTPases. 2017;8:85–89. doi: 10.1080/21541248.2016.1199266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasundar JJ, Ju JH, He L, et al. Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J Biol Chem. 2012;287:37119–37133. doi: 10.1074/jbc.M112.380972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Yoshida S, Hatano R, Asano S. Pathophysiological roles of ezrin/radixin/moesin proteins. Biol Pharm Bull. 2017;40:381–390. doi: 10.1248/bpb.b16-01011. [DOI] [PubMed] [Google Scholar]
- Kovacs-Kasa A, Gorshkov BA, Kim K-M, et al. The protective role of MLCP-mediated ERM dephosphorylation in endotoxin-induced lung injury in vitro and in vivo. Sci Rep. 2016;6:39018. doi: 10.1038/srep39018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Rodrigues NTL, Moeendarbary E, et al. PP1-mediated moesin dephosphorylation couples polar relaxation to mitotic exit. Curr Biol. 2012;22:231–236. doi: 10.1016/j.cub.2011.12.016. [DOI] [PubMed] [Google Scholar]
- Li W, Jin WW, Tsuji K, et al. Ezrin directly interacts with AQP2 and promotes its endocytosis. J Cell Sci. 2017;130:2914–2925. doi: 10.1242/jcs.204842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-C, Yu C-H, Triffo S, Groves JT. Supported membrane formation, characterization, functionalization, and patterning for application in biological science and technology. Curr Protoc Chem Biol. 2010;2:235–269. doi: 10.1002/9780470559277.ch100131. [DOI] [PubMed] [Google Scholar]
- Litschel T, Kelley CF, Holz D, et al. Reconstitution of contractile actomyosin rings in vesicles. Nat Commun. 2021;12:2254. doi: 10.1038/s41467-021-22422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Guidry JJ, Worthylake DK. The conserved sequence repeats of IQGAP1 mediate binding to ezrin. J Proteome Res. 2014;13:1156–1166. doi: 10.1021/pr400787p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Belkina NV, Park C, et al. Constitutively active ezrin increases membrane tension, slows migration, and impedes endothelial transmigration of lymphocytes in vivo in mice. Blood. 2012;119:445–453. doi: 10.1182/blood-2011-07-368860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubart Q, Vitet H, Dalonneau F, et al. Role of phosphorylation in moesin interactions with PIP2-containing biomimetic membranes. Biophys J. 2018;114:98–112. doi: 10.1016/j.bpj.2017.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak H, Naba A, Varma S, et al. Ezrin phosphorylation on tyrosine 477 regulates invasion and metastasis of breast cancer cells. BMC Cancer. 2012;12:82. doi: 10.1186/1471-2407-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniti O, Carvalho K, Picart C. Model membranes to shed light on the biochemical and physical properties of ezrin/radixin/moesin. Biochimie. 2013;95:3–11. doi: 10.1016/j.biochi.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniti O, Khalifat N, Goggia K, et al. Binding of moesin and ezrin to membranes containing phosphatidylinositol (4,5) bisphosphate: a comparative study of the affinity constants and conformational changes. Biochim Biophys Acta. 2012;1818:2839–2849. doi: 10.1016/j.bbamem.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashaghi A, Mashaghi S, Reviakine I, et al. Label-free characterization of biomembranes: from structure to dynamics. Chem Soc Rev. 2014;43:887–900. doi: 10.1039/c3cs60243e. [DOI] [PubMed] [Google Scholar]
- McClatchey AI. ERM proteins at a glance. J Cell Sci. 2014;127:3199–3204. doi: 10.1242/jcs.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini E, Weidenhaupt M, Picart C. Practical guide to characterize biomolecule adsorption on solid surfaces (review) Biointerphases. 2018;13:06D303. doi: 10.1116/1.5045122. [DOI] [PubMed] [Google Scholar]
- Mu L, Tu Z, Miao L, et al. A phosphatidylinositol 4,5-bisphosphate redistribution-based sensing mechanism initiates a phagocytosis programing. Nat Commun. 2018;9:4259. doi: 10.1038/s41467-018-06744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel O, Tomas A, Scott CC, Gruenberg J. Moesin and cortactin control actin-dependent multivesicular endosome biogenesis. Mol Biol Cell. 2016;27:3305–3316. doi: 10.1091/mbc.E15-12-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nammalwar RC, Heil A, Gerke V. Ezrin interacts with the scaffold protein IQGAP1 and affects its cortical localization. Biochim Biophys Acta. 2015;1853:2086–2094. doi: 10.1016/j.bbamcr.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Neisch AL, Fehon RG. Ezrin, radixin and moesin: key regulators of membrane-cortex interactions and signaling. Curr Opin Cell Biol. 2011;23:377–382. doi: 10.1016/j.ceb.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch AL, Formstecher E, Fehon RG. Conundrum, an ARHGAP18 orthologue, regulates RhoA and proliferation through interactions with moesin. Mol Biol Cell. 2013;24:1420–1433. doi: 10.1091/mbc.E12-11-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y-R, Tseng W-S, Chang P-W, Chen H-C. Phosphorylation of moesin by Jun N-terminal kinase is important for podosome rosette formation in Src-transformed fibroblasts. J Cell Sci. 2013;126:5670–5680. doi: 10.1242/jcs.134361. [DOI] [PubMed] [Google Scholar]
- Parameswaran N, Gupta N. Re-defining ERM function in lymphocyte activation and migration. Immunol Rev. 2013;256:63–79. doi: 10.1111/imr.12104. [DOI] [PubMed] [Google Scholar]
- Parameswaran N, Matsui K, Gupta N. Conformational switching in ezrin regulates morphological and cytoskeletal changes required for B cell chemotaxis. J Immunol. 2011;186:4088–4097. doi: 10.4049/jimmunol.1001139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaseyed T, Viswanatha R, Sauvanet C et al (2017) Ezrin activation by LOK phosphorylation involves a PIP2-dependent wedge mechanism. eLife 6:e22759. 10.7554/eLife.22759 [DOI] [PMC free article] [PubMed]
- Perez-Cornejo P, Gokhale A, Duran C, et al. Anoctamin 1 (Tmem16A) Ca2+-activated chloride channel stoichiometrically interacts with an ezrin-radixin-moesin network. Proc Natl Acad Sci U S A. 2012;109:10376–10381. doi: 10.1073/pnas.1200174109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang JM, Harrop SJ, Duff AP, et al. Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin. Biochem J. 2016;473:2763–2782. doi: 10.1042/BCJ20160541. [DOI] [PubMed] [Google Scholar]
- Rahimi N, Ho RXY, Chandler KB, et al. The cell adhesion molecule TMIGD1 binds to moesin and regulates tubulin acetylation and cell migration. J Biomed Sci. 2021;28:61. doi: 10.1186/s12929-021-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho JJ, Sepers JJ, Nicolle O, et al (2020) C-terminal phosphorylation modulates ERM-1 localization and dynamics to control cortical actin organization and support lumen formation during Caenorhabditis elegans development. Development 147:dev188011. 10.1242/dev.188011 [DOI] [PMC free article] [PubMed]
- Roberts RE, Dewitt S, Hallett MB. Membrane tension and the role of ezrin during phagocytosis. Adv Exp Med Biol. 2020;1246:83–102. doi: 10.1007/978-3-030-40406-2_6. [DOI] [PubMed] [Google Scholar]
- Roubinet C, Decelle B, Chicanne G, et al. Molecular networks linked by moesin drive remodeling of the cell cortex during mitosis. J Cell Biol. 2011;195:99–112. doi: 10.1083/jcb.201106048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouven Brückner B, Pietuch A, Nehls S, et al. Ezrin is a major regulator of membrane tension in epithelial cells. Sci Rep. 2015;5:14700. doi: 10.1038/srep14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino D, Gogendeau D, Gambarotto D, et al. Moesin is a major regulator of centrosome behavior in epithelial cells with extra centrosomes. Curr Biol. 2015;25:879–889. doi: 10.1016/j.cub.2015.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Yamamoto H, Mukaisho K, et al. Mechanisms underlying cancer progression caused by ezrin overexpression in tongue squamous cell carcinoma. PLoS ONE. 2013;8:e54881. doi: 10.1371/journal.pone.0054881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis J, Vié V. Biomimetic models to investigate membrane biophysics affecting lipid-protein interaction. Front Bioeng Biotechnol. 2020;8:270. doi: 10.3389/fbioe.2020.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvanet C, Wayt J, Pelaseyed T, Bretscher A. Structure, regulation, and functional diversity of microvilli on the apical domain of epithelial cells. Annu Rev Cell Dev Biol. 2015;31:593–621. doi: 10.1146/annurev-cellbio-100814-125234. [DOI] [PubMed] [Google Scholar]
- Schäfer J, Nehls J, Schön M, et al. Leaflet-dependent distribution of PtdIns[4,5]P2 in supported model membranes. Langmuir. 2020;36:1320–1328. doi: 10.1021/acs.langmuir.9b03793. [DOI] [PubMed] [Google Scholar]
- Schön M, Mey I, Steinem C. Influence of cross-linkers on ezrin-bound minimal actin cortices. Prog Biophys Mol Biol. 2019;144:91–101. doi: 10.1016/j.pbiomolbio.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Senju Y, Kalimeri M, Koskela EV, et al. Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc Natl Acad Sci USA. 2017;114:E8977–E8986. doi: 10.1073/pnas.1705032114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senju Y, Lappalainen P. Regulation of actin dynamics by PI(4,5)P2 in cell migration and endocytosis. Curr Opin Cell Biol. 2019;56:7–13. doi: 10.1016/j.ceb.2018.08.003. [DOI] [PubMed] [Google Scholar]
- Senju Y, Lappalainen P, Zhao H. Liposome co-sedimentation and co-flotation assays to study lipid-protein interactions. Methods Mol Biol. 2021;2251:195–204. doi: 10.1007/978-1-0716-1142-5_14. [DOI] [PubMed] [Google Scholar]
- Senju Y, Zhao H. Fluorescence assays to study membrane penetration of proteins. Methods Mol Biol. 2021;2251:215–223. doi: 10.1007/978-1-0716-1142-5_16. [DOI] [PubMed] [Google Scholar]
- Sezgin E. Super-resolution optical microscopy for studying membrane structure and dynamics. J Phys Condens Matter. 2017;29:273001. doi: 10.1088/1361-648X/aa7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Schwille P. Model membrane platforms to study protein-membrane interactions. Mol Membr Biol. 2012;29:144–154. doi: 10.3109/09687688.2012.700490. [DOI] [PubMed] [Google Scholar]
- Shabardina V, Kramer C, Gerdes B, et al. Mode of ezrin-membrane interaction as a function of PIP2 binding and pseudophosphorylation. Biophys J. 2016;110:2710–2719. doi: 10.1016/j.bpj.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Lin R, Yang J, et al. AKT and GSK-3 are necessary for direct ezrin binding to NHE3 as part of a C-terminal stimulatory complex: role of a novel Ser-rich NHE3 C-terminal motif in NHE3 activity and trafficking. J Biol Chem. 2014;289:5449–5461. doi: 10.1074/jbc.M113.521336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarska E, Diz-Muñoz A. Pay attention to membrane tension: mechanobiology of the cell surface. Curr Opin Cell Biol. 2020;66:11–18. doi: 10.1016/j.ceb.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinet S, Mahmud K, Stewman SF, et al. The actin-binding ERM protein moesin binds to and stabilizes microtubules at the cell cortex. J Cell Biol. 2013;202:251–260. doi: 10.1083/jcb.201304052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani C, Gonzalez-Rodriguez D, Senju Y, et al. Ezrin enhances line tension along transcellular tunnel edges via NMIIa driven actomyosin cable formation. Nat Commun. 2017;8:15839. doi: 10.1038/ncomms15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM. Actin cell cortex: structure and molecular organization. Trends Cell Biol. 2020;30:556–565. doi: 10.1016/j.tcb.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K, Haghparast SMA, Miyake J. Inhibition of cell adhesion by phosphorylated ezrin/radixin/moesin. Cell Adh Migr. 2015;9:502–512. doi: 10.1080/19336918.2015.1113366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terawaki S, Maesaki R, Hakoshima T. Structural basis for NHERF recognition by ERM proteins. Structure. 2006;14:777–789. doi: 10.1016/j.str.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Tsai F-C, Bertin A, Bousquet H, et al. Ezrin enrichment on curved membranes requires a specific conformation or interaction with a curvature-sensitive partner. Elife. 2018;7:e37262. doi: 10.7554/eLife.37262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Yonemura S. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J Biol Chem. 1999;274:34507–34510. doi: 10.1074/jbc.274.49.34507. [DOI] [PubMed] [Google Scholar]
- Valderrama F, Thevapala S, Ridley AJ. Radixin regulates cell migration and cell–cell adhesion through Rac1. J Cell Sci. 2012;125:3310–3319. doi: 10.1242/jcs.094383. [DOI] [PubMed] [Google Scholar]
- Vilmos P, Kristó I, Szikora S, et al. The actin-binding ERM protein moesin directly regulates spindle assembly and function during mitosis. Cell Biol Int. 2016;40:696–707. doi: 10.1002/cbin.10607. [DOI] [PubMed] [Google Scholar]
- Viswanatha R, Bretscher A, Garbett D. Dynamics of ezrin and EBP50 in regulating microvilli on the apical aspect of epithelial cells. Biochem Soc Trans. 2014;42:189–194. doi: 10.1042/BST20130263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R, Ohouo PY, Smolka MB, Bretscher A. Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J Cell Biol. 2012;199:969–984. doi: 10.1083/jcb.201207047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R, Wayt J, Ohouo PY, et al. Interactome Analysis Reveals Ezrin Can Adopt Multiple Conformational States. J Biol Chem. 2013;288:35437–35451. doi: 10.1074/jbc.M113.505669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakayama Y, Miura K, Sabe H, Mochizuki N. EphrinA1-EphA2 signal induces compaction and polarization of Madin-Darby canine kidney cells by inactivating Ezrin through negative regulation of RhoA. J Biol Chem. 2011;286:44243–44253. doi: 10.1074/jbc.M111.267047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald FA, Oriolo AS, Mashukova A, et al. Atypical protein kinase C (iota) activates ezrin in the apical domain of intestinal epithelial cells. J Cell Sci. 2008;121:644–654. doi: 10.1242/jcs.016246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-C, Liau J-Y, Lu Y-S, et al. Differential expression of moesin in breast cancers and its implication in epithelial-mesenchymal transition. Histopathology. 2012;61:78–87. doi: 10.1111/j.1365-2559.2012.04204.x. [DOI] [PubMed] [Google Scholar]
- Wen Y, Vogt VM, Feigenson GW. PI(4,5)P2 Clustering and Its Impact on Biological Functions. Annu Rev Biochem. 2021;90:681–707. doi: 10.1146/annurev-biochem-070920-094827. [DOI] [PubMed] [Google Scholar]
- Yang G, Hiruma S, Kitamura A, et al. Molecular basis of functional exchangeability between ezrin and other actin-membrane associated proteins during cytokinesis. Exp Cell Res. 2021;403:112600. doi: 10.1016/j.yexcr.2021.112600. [DOI] [PubMed] [Google Scholar]
- Yang Y, Primrose DA, Leung AC, et al. The PP1 phosphatase flapwing regulates the activity of Merlin and Moesin in Drosophila. Dev Biol. 2012;361:412–426. doi: 10.1016/j.ydbio.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel I, Naba A, Cunha MLD, M, , et al. Ezrin regulates microvillus morphogenesis by promoting distinct activities of Eps8 proteins. Mol Biol Cell. 2012;23:1080–1095. doi: 10.1091/mbc.E11-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 (EPS 1805 kb)
(a) A simple phylogenetic tree of ERM proteins.
(b) The microarray gene expression profiles of Homo sapiens ERM proteins in various cell lines and tissues generated by BioGPS. Note that the expression levels of ezrin/radoxin/moesin are different in cell lines and tissues.
(c) Protein-protein interaction (PPI) network analysis. Some known and predicted PPIs of ERM proteins are analyzed using stringApp in Cytoscape.
Data Availability Statement
Not applicable.
Not applicable.