Abstract
Human immunodeficiency virus (HIV) is the most extensively researched human pathogen. Despite this massive scientific endeavour, several fundamental viral processes remain enigmatic. One such critical process is uncoating—the event that releases the viral genome from the proteinaceous shell of the capsid during infection. While this process is conceptually simple, the molecular underpinnings, timing, regulation, and cellular location of uncoating remain contentious. This review describes the hurdles that have limited our understanding in this area and presents recently deployed in vitro and in cellulo techniques that have been developed expressly with the aim of directly visualising capsid uncoating at the single-particle level and understanding the mechanics behind this essential aspect of HIV infection.
Keywords: HIV, Capsid uncoating, TIRF microscopy
Introduction
Human immunodeficiency virus (HIV) is a lentivirus that infects the CD4 + cells of the immune system. Following fusion of viral and host cell membranes, the viral core, consisting of the genomic RNA and associated proteins surrounded by a proteinaceous capsid shell, is released into the cytoplasm (Campbell and Hope 2015). From this point, productive infection requires the coordinated execution of several processes: the virus must reverse transcribe its genome while traversing the cytoplasm, interact with the nuclear pore complex, enter the nucleus, and integrate the newly synthesised viral DNA into a host chromosome. The capsid mediates several of these processes and is essential during early stages of HIV infection; however, to release the viral DNA for integration, the capsid must open in a process known as uncoating. Despite its central role, the timing, cellular location, mechanism, and kinetics of uncoating remain elusive, and still less is known about its relationship with other aspects of the virus life cycle including reverse transcription and nuclear import.
The capsid plays a multi-faceted role in the HIV life cycle and has been implicated in maintenance of structural integrity, genome replication (Christensen et al. 2020; Xu et al. 2020), protection of the viral core from immune sensing (Sauter and Kirchhoff 2016; Sumner et al. 2020; Kumar et al. 2018), trafficking of the HIV core through the cytoplasm and into the nucleus, and determination of the integration site (Scoca and Di Nunzio 2021). Capsids have an average length of ~ 119 nm and width of ~ 61 nm (Briggs et al. 2003). Approximately 1,200–1,500 copies of capsid proteins (CA) assemble into roughly 200–250 hexamers and exactly 12 pentamers, with five pentamers distributed at the narrow end and seven at the wide end of the core to form the characteristic fullerene cone structure (Mattei et al. 2016; Ganser et al. 1999). CA is genetically fragile and exceptionally sensitive to mutation (Rihn et al. 2013), which reflects the need for the capsid to fulfil multiple functions and for its stability to be carefully balanced. The HIV-1 capsid retains an inherent plasticity and structural heterogeneity, which is attributed to variability in inter- and intra-hexamer interactions (Gres et al. 2015). Altering the distribution of pentamers contributes to variation in HIV-1 capsid morphology, as, when pentamers are distributed with six at each end of the capsid, a tubular capsid forms instead of the fullerene cone (Ganser et al. 1999; Briggs et al. 2003).
At least a large part of the capsid lattice needs to remain intact during the early post-entry stages to facilitate transport on microtubules in the cytoplasm, passage through the nuclear pore complex, and integration site targeting in the nucleus. What remains most debated is the place and time of when capsid uncoating is first initiated. There are three prominent models of uncoating (Fig. 1). The first proposes that uncoating initiates within the cytoplasm, after fusion of the viral membrane with the host cell and during transport of the core towards the nucleus; the second suggests that the capsid remains intact until it reaches the nuclear pore complex and that uncoating is directly linked to nuclear import; and the third identifies uncoating as occurring in the nucleus after passage through the nuclear pore complex.
Fig. 1.
Current main models of HIV uncoating and fluorescent markers used to provide evidence for each model. After fusion of the engineered HIV virion, the capsid is released into the cell. From left to right: loss of capsid integrity within the cytoplasm detected by using the iGFP system, with a portion of CA remaining associated with the genomic material (Mamede et al. 2017); uncoating at the nuclear pore complex detected using an indirect label for CA (Francis et al. 2016); and uncoating within the nucleus post-nuclear import using the iGFP system (Li et al. 2021), binding of the tagged OR3 protein to sequence engineered into the viral genome (Blanco-Rodriguez et al. 2020; Müller et al. 2021) and release of GFP-tagged CA from capsids (Burdick et al. 2020)
Historically, it has been difficult to observe the HIV capsid during uncoating. Initial insights into capsid stability were gained using a spectrum of indirect methods, none of which provided compelling evidence to distinguish between these three models. Biochemically, CA lattice formation can be determined using in vitro assembly of capsid-like cylinders, cones, or spheres from recombinant CA (Ganser-Pornillos et al. 2004; Ehrlich et al. 1992; Gross et al. 1997). Release of CA from capsids can be measured by separation of soluble dissociated CA from intact cores via ultracentrifugation and density gradients (Shah and Aiken 2011; Forshey et al. 2002; Wacharapornin et al. 2007), while the fate of capsid assay is a cell-based approach that expounds on a similar separation concept (Stremlau et al. 2006; Yang et al. 2014). More recently, the development of high-throughput assays such as Fast Assembly Inhibitor Test for HIV (FAITH) and Disassembly Inhibitor Test for HIV (DITH) (Hadravová et al. 2015; Dostálková et al. 2019) have helped elucidate the role of small molecules in lattice formation and capsid stability of mature particles (Dostálková et al. 2020).
Alongside these bulk biochemical assays, numerous cell-based assays provide insight into the timing and location of capsid uncoating. Examples include the cyclosporin A (CsA) washout assay, which measures the kinetics of the CsA-mediated rescue of HIV infection in cells expressing the capsid-targeting restriction factor Trim-CypA (Hulme et al. 2011); the Entry/Uncoating assay based on core-packaged RNA availability and Translation (EURT) assay, detecting translation of a reporter mRNA (which only becomes available upon uncoating) (Da Silva Santos et al. 2016); and inducible nuclear pore complex blockade to measure the nuclear import kinetics of capsids relative to other steps in the virus life cycle (Dharan et al. 2020).
The results from these studies have yielded differing conclusions with regard to the timing and regulation of capsid uncoating. The CsA washout assay identified the initiation of cytoplasmic uncoating within 1 h of viral fusion, whereby addition of reverse transcription inhibitors delayed uncoating (Ingram et al. 2020). In contrast, the EURT assay suggested that reverse transcription had no impact on capsid opening but led to a decreased sensitivity to the capsid inhibitor PF74, interpreted as a remodelling of the capsid (Da Silva Santos et al. 2016). Use of nuclear blockades demonstrated a relationship between nuclear import and the intact capsid (Dharan et al. 2020). Cores remained sensitive to PF74 for hours after they lost sensitivity to the nuclear pore blockade (Dharan et al. 2020), thus suggesting that uncoating occurs inside the nucleus.
Despite the contributions of such assays, they rely on indirect readouts and ensemble averaging to monitor uncoating. This review will focus on recent cellular and in vitro approaches to directly visualise uncoating at the level of individual viral particles. It will summarise the evolution of techniques employed to study the process of HIV-1 uncoating, the contrasting models resulting from either a cytoplasm-based view of uncoating or a nuclear envelope and import-based view, and the remaining knowledge gaps.
Imaging uncoating in cells
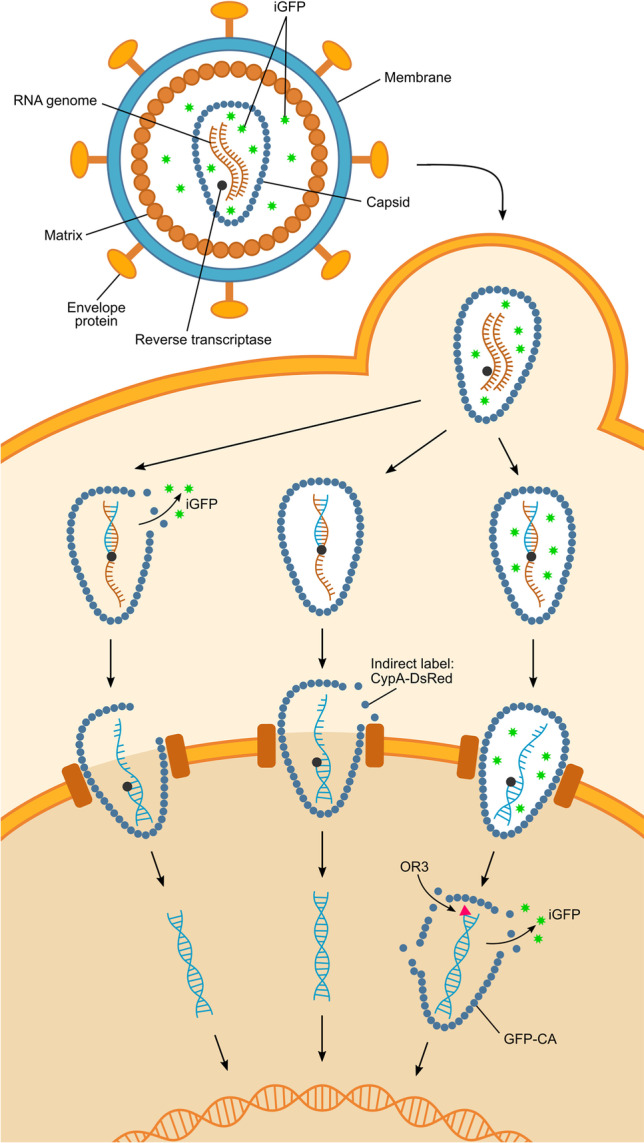
A variety of fluorescently tagged viral components (Fig. 2) have enabled the monitoring of virions through the HIV life cycle by fluorescence microscopy in live and fixed cells (Mukherjee et al. 2021). Fusing fluorescent proteins or self-labelling tags to viral proteins packaged into the capsid, such as integrase (IN), nucleocapsid, and the accessory protein Vpr, has been widely used to visualise the movement of viral cores within the cell; common designs include the fusion of fluorescent proteins to the N- or C-terminus of IN (Albanese et al. 2008; Ning et al. 2018). These markers can be partially lost from post-fusion cores before the onset of uncoating (e.g., as observed for tagged Vpr (Desai et al. 2015)) and/or remain associated with the viral genetic material after loss of CA from the core, allowing continued tracking of complexes post uncoating. Approaches to visualise capsid uncoating rely on (1) indirectly or directly tagging CA or (2) using fluorescent markers that are released from the capsid or gain access to bindings sites inside the capsid when it opens. All these methods need careful optimisation to maximise label incorporation for efficient detection in cells while not compromising viral behaviour and infectivity, as fusion of tags to viral components often leads to defects in capsid assembly or function (Mamede et al. 2021).
Fig. 2.
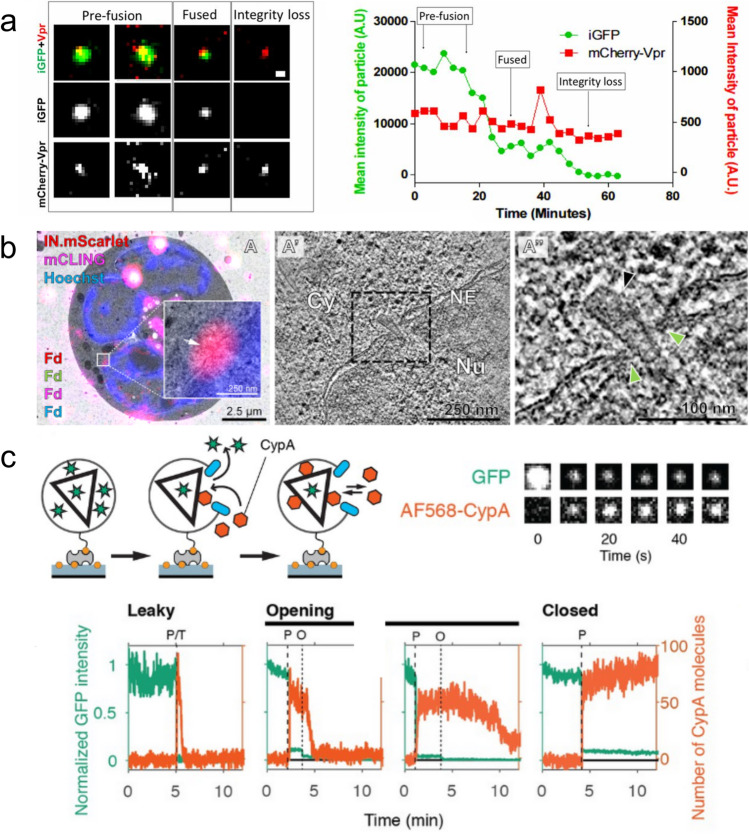
Selection of imaging methods for visualising uncoating at the level of individual cores. (a) Live-cell imaging of HIV iGFP/mCherry-Vpr particles. Two independent GFP signal drops are detected, with the first indicative of membrane fusion and the second indicative of capsid integrity loss. mCherry-Vpr is used to track the particles. The graph plots mean signal intensities of GFP (green) and mCherry-Vpr (red) over time. (Reproduced from Mamede et al. 2017.) (b) Correlative light and electron microscopy. (A) Overlay of a fluorescence image with the relevant electron micrograph, exhibiting an IN-mScarlet positive and mCLING-negative foci at the nuclear envelope. (A′, A″) A slice through a tomographic reconstruction of the position captured in (A), highlighting an intact capsid (indicated by the black arrowhead) deep inside the NPC (indicated by green arrowheads) with its narrow end pointed towards the nucleoplasm. (Reproduced from Zila et al. 2021.) (c) A schematic diagram of the TIRF assay, showing both GFP contained within the capsid to track capsid integrity and the binding of AF568-CypA to visualise capsid behaviour after membrane permeabilisation. Also included are snapshots of the GFP and AF568-CypA signal of a selected single viral particle at different time points. Example traces show leaky, opening, and closed traces, with green and orange used to indicate the GFP and CypA signals respectively. Capsid lifespan is indicated by loss of GFP signal and demarcated by the dotted lines. (Reproduced from Márquez et al. 2018)
Indirect labelling of CA: CypA-DsRed
CypA-DsRed, designed by Francis et al., is a fluorescently tagged oligomeric form of the CA-binding host protein cyclophilin A (CypA), and has high-avidity binding to the capsids (Francis et al. 2016). HIV particles efficiently incorporate CypA-DsRed when produced in cells expressing this protein. CypA-DsRed is bound sufficiently tightly to the capsid exterior, such that a decrease of its signal can be interpreted as loss of CA from the core as it undergoes uncoating in the target cell. The majority of cores (containing IN-GFP as a second label) lose the CypA-DsRed signal shortly after entry into the cytoplasm (Francis et al. 2016), exposing the complexes to proteasomal degradation (Francis and Melikyan 2018). Thus, premature uncoating did not lead to productive infection, whereby these cores may correspond to virions with improperly assembled capsids (Márquez et al. 2018). In contrast, virtually all cores docking at the nuclear membrane presented with a CypA-DsRed signal, marking CA as required for interaction with the nuclear envelope. Only cores that abruptly lost CypA-DsRed upon docking established infection, with cores that exhibited gradual or no loss failing to import (Francis and Melikyan 2018). Complexes inside the nucleus retained some level of CA and accumulated in nuclear speckles (Francis et al. 2020). Overall, infection was deemed to depend on the retention of CA until the nuclear membrane, upon which cores would undergo rapid uncoating prior to or during import. However, it is possible that the signal loss at the nuclear pore complex could be due to dissociation of CypA-DsRed from the capsid, e.g., as a result of binding competition from components of the nuclear pore complex or structural changes of the capsid during import.
Direct labelling of CA: fusion with fluorescent proteins or self-labelling tags
Fusion of a fluorescent protein to the N- or C-terminus of CA is challenging, severely compromising virion production and infectivity (Bönisch et al. 2020; Pereira et al. 2011), presumably due to effects on capsid assembly and function. For example, tags at the N-terminus would be expected to prevent formation of the N-terminal β-hairpin, a structural feature essential for capsid maturation and proposed to gate access of polyanions to the central pore in CA hexamers and thus regulate dNTP import (Jacques et al. 2016) and IP6-mediated CA lattice stabilisation (Mallery et al. 2018). While infectivity can be rescued by mixing tagged CA with an excess of untagged CA, this approach can result in insufficient levels of fluorescence for faithfully detecting particles and measuring uncoating kinetics, as observed, for example, with CA tagged at the C-terminus with GFP (CA-GFP) (Bönisch et al. 2020). In a recent advance, Burdick et al. produced HIV from a mixture of CA labelled at its N-terminus with GFP (GFP-CA) and an excess of untagged CA, which retained approximately 50% of their infectivity in several cell types, including macrophages, and contained efficiently labelled cores with normal morphology (Burdick et al. 2020). By using spinning disk confocal microscopy, they captured viral complexes entering the nucleus without detecting a drop in the GFP-CA signal, suggesting that intact or nearly intact capsids traverse the nuclear pore complex. Uncoating was later detected as the complete and sudden loss of the GFP signal of particles within ~ 1.5 μm of their chromosomal integration sites. Their results suggest that nuclear import precedes the rapid disassembly of the capsid. In contrast, capsids containing amino acid substitutions that prevent nuclear entry disassemble while remaining stuck at the nuclear envelope, whereby the timing of disassembly remained unaffected, suggesting that uncoating is triggered by virion-intrinsic processes, such as completion of reverse transcription, rather than by cellular location.
Alternative strategies using self-labelling tags have focused on the insertion of tetracysteine (TC) tags, either at the N- or C-terminus or within the CA sequence (Bönisch et al. 2020; Pereira et al. 2011). These small, six to twelve amino acid motifs are bound with high affinity by the fluorescein arsenical helix binder (FlAsH), a dye that has been used to label TC-tagged matrix for tracking HIV production (Turville et al. 2008) and to label TC-tagged IN for super-resolution imaging of HIV in infected cells (Lelek et al. 2012). However, the potential for non-specific labelling and rapid photobleaching of these dyes represents hurdles for imaging uncoating kinetics. As observed for GFP fusions, insertion of the tag at either the N-terminus or the C-terminus of the CA protein abolished infectivity (Pereira et al. 2011), suggesting that the small tag still compromises the stability of the CA lattice.
Detecting capsid integrity loss: iGFP as a solution phase marker
HIV with internal GFP (iGFP) is produced by insertion of a GFP sequence into the Gag polyprotein (Hübner et al. 2007). Flanking protease cleavage sites ensure release of GFP as a solution phase marker upon proteolysis during virion maturation, with ~ 10–20% of the released GFP being taken up into the capsid and the remainder being retained within the membrane but exterior to the core. This ratio of GFP inside versus outside the capsid is expected for the relative volumes of these two compartments, confirming that GFP is a solution-phase marker. Using live-cell imaging, Mamede et al. showed that HIV iGFP infection is characterised by two distinct drops in the GFP signal (Fig. 2a) (Mamede et al. 2017). Most of the GFP signal is lost from the particle when the GFP outside the capsid is released upon fusion of the viral particle with the plasma membrane. The GFP-loaded core can then be tracked as a dimmer diffraction-limited spot on its journey from the cellular periphery to the nucleus, whereby the sudden loss of the residual GFP signal pinpoints the precise moment when the capsid loses its integrity. In contrast to labelled CA, the iGFP system is exquisitely sensitive to detecting the appearance of the first defect in the capsid that is sufficiently large to allow passage of GFP, but the subsequent disassembly process of capsid post-opening cannot be monitored.
Mamede et al. found that most capsids released GFP approximately 30 min after fusion, while a small proportion of particles retained GFP for longer than 2 h (Mamede et al. 2017). However, only virions that lost their GFP signal early (within 45 min) led to infection. Quantification of CA by immunofluorescence further showed that GFP loss in the cytoplasm was associated with shedding of most CA subunits from the capsid. Thus, the authors proposed that infectivity depends on early uncoating in the cytoplasm. In contrast, Li et al. used the iGFP system to show that GFP-positive cores lost their signal inside the nucleus prior to integration, leading to productive infection (Li et al. 2021). The signal of a fluorescence-labelled protein binding to CA hexamers (i.e. recognising assembled CA) disappeared within 1–3 min after GFP release. Based on these observations, the authors suggested that the intact capsid enters the nucleus and opens only shortly before complete uncoating and the release of the pre-integration complex.
It is not clear what contributed to these conflicting results. Using live-cell imaging to pinpoint where and when capsids lose their integrity for the subset of particles that lead to productive infection is challenging, because it represents a short event in a process that takes several hours from entry into the cytoplasm to integration into a chromosome. Phototoxicity and photobleaching typically put constraints on the imaging protocol with respect to temporal resolution and observation time such that individual studies necessarily focus on a particular stage or subcellular compartment. Ideally, cellular imaging would entail unbiased tracking of all viral particles throughout the entire life cycle from fusion at the plasma membrane to transcription of viral RNA from integrated proviruses, an endeavour that could capitalise on recent advances in fluorescence microscopy such as lattice light sheet imaging and automated particle tracking (Aguet et al. 2016; Willy et al. 2021). It is possible that all types of behaviours can be observed in the cell, whereby the challenge is to identify which pathways predominate and lead to infection versus those that lead to degradation.
Detecting capsid integrity loss: ANCHOR system for labelling viral DNA
The HIV-1 ANCHOR system, first adapted by Blanco-Rodriguez et al., is designed to detect viral DNA that is no longer shielded inside an intact capsid in live or fixed cells (Blanco-Rodriguez et al. 2020). The bacterial-derived ANCH sequence is introduced into the viral genome and, following reverse transcription and opening of the capsid lattice, allows for the GFP-tagged OR3 protein to access and specifically bind to this sequence. The authors detected viral DNA in association with CA primarily inside the nucleus but also at the nuclear pore complex and suggested a model whereby the capsid is remodelled during passage through the nuclear pore complex. Müller et al. showed that HIV cores (labelled with an integrase fusion protein) in the cytoplasm and the nucleus were associated with CA (detected by immunofluorescence), whereby viral DNA became detectable with OR3-GFP only after nuclear entry and the OR3-GFP signal subsequently separated from the CA signal (Müller et al. 2021). Correlative light and electron microscopy (CLEM) in combination with tomography revealed that nuclear particles lacking OR3-GFP represented cone-shaped structures containing electron-dense material, consistent with the viral nucleoprotein complex. Tomograms at later time points revealed structures resembling broken capsids or capsid-like remnants that lacked internal density. Thus, uncoating and release of the viral DNA for integration were proposed to occur within the nucleus, representing a rupture of the capsid, but not a complete dissociation of the lattice.
Identifying fused virions for CLEM using mCLING
Approaches to distinguish between cores that have entered the cytoplasm following a productive fusion event and virions that were merely endocytosed rely on detecting the separation of a core marker (such as labelled Vpr or IN) from a membrane marker. The membrane marker can be incorporated into the viral membrane during particle assembly in producer cells as demonstrated using the fusion protein S15-mCherry (Campbell et al. 2007). Alternatively, the fluorescent endocytic probe mCLING (Revelo et al. 2014) has been used to stain the plasma membrane of target cells before entry of HIV labelled with IN-mScarlet (Zila et al. 2021, 2019). This approach has been used as part of a ground-breaking correlative light and electron microscopy (CLEM) study to characterise intact capsids and uncoating intermediates in cells by cryoelectron tomography (cryoET), whereby post-fusion complexes were identified as IN-positive foci in the cytoplasm that lacked mCLING signal. Tomograms of these areas revealed the presence of conical structures in the cytosol next to nuclear pores and deep within the nuclear pore complex, containing highly dense material presumed to be the viral genome (Fig. 2b). In contrast, structures identified inside the nucleus appeared to be open and empty. These images support the same model as above, whereby essentially intact capsids traverse the nuclear pore complex and break open in the nucleus to release the viral DNA for integration.
Imaging uncoating in vitro
Although the cellular context is lost, in vitro reconstitution of capsid uncoating for imaging offers greater system control, superior imaging conditions, and, in some cases, increased statistical power to dissect the intrinsic properties of the capsids and the effect of specific perturbation such as co-factor binding or reverse transcription. Sample preparation involves isolation of viral cores by stripping the viral membrane with detergent followed by density gradient centrifugation, or permeabilisation of virions in situ to gain access to the capsid. Imaging modalities include fluorescence microscopy to visualise with high temporal resolution and single-molecule sensitivity the kinetics of capsid uncoating utilising the same labelling strategies as employed for cellular imaging; atomic force microscopy (AFM) to probe changes in mechanical properties leading to uncoating; and cryoelectron tomography (cryoET) to visualise the structure of uncoating intermediates.
Imaging uncoating kinetics by fluorescence microscopy
In pioneering work, Francis et al. adhered HIV co-labelled with IN-GFP and CypA-DsRed on coverslips, and permeabilised particles with the detergent saponin before fixing and immunostaining for CA at different times after permeabilisation (Francis et al. 2016). While the number of IN-GFP spots (each corresponding to an adhered viral particle) remained constant over time, the CypA-DsRed and CA spots disappeared with a half-life of ~ 10 min, providing a measure for the intrinsic uncoating kinetics of the capsid. Mutations in CA known to affect capsid stability and the addition of cell lysate (containing stabilising factors) resulted in the expected corresponding changes in uncoating kinetics.
By utilising total internal reflection fluorescence (TIRF) microscopy, Márquez et al. followed the real-time uncoating kinetics of permeabilised HIV iGFP particles (lacking the envelope protein) captured onto glass coverslips using surface chemistry and microfluidics (Márquez et al. 2018, 2019). Permeabilisation was achieved using a pore-forming protein (Fig. 2c) that assembles into large (~ 30–40 nm) ring-shaped membrane pores that permit the passage of proteins while retaining the viral core. As discussed earlier, release of the GFP content marker in two steps allowed pinpointing the time of membrane permeabilisation followed by capsid opening. To image the disassembly kinetics after capsid opening (GFP release), fluorescently labelled CypA was used to ‘paint’ the CA lattice, generating a signal proportional to the number of CA proteins without interfering with the uncoating kinetics. Analysis of hundreds of cores revealed different degrees of capsid stability, mirroring the structural heterogeneity of the capsid (Mattei et al. 2016). The majority of capsids are leaky (incompletely or improperly assembled) and uncoat immediately, similar to the subset of non-infectious cores observed by live-cell imaging leaky (Francis et al. 2016; Francis and Melikyan 2018). Metastable capsids retained GFP for varying times (half-life of ~ 10 min), whereby uncoating was a catastrophic process, with the capsid rapidly collapsing after the initial opening event. Some capsids remained closed throughout the observation period, possibly due to higher stability.
The TIRF uncoating assay has been instrumental in resolving the differential effects of capsid-binding host molecules and drugs on capsid uncoating (Márquez et al. 2018; Mallery et al. 2018). The cellular polyanion IP6 is selectively packaged into virions and promotes assembly of the conical capsid by neutralising the positive charge of the arginine-lined pore in the centre of CA hexamers (Mallery et al. 2018; Dick et al. 2018). IP6 prevents capsid opening (increase in half-life from minutes to hours) as required for transport of a closed capsid to the nucleus but does not prevent CA lattice disassembly once the capsid has become defective (Mallery et al. 2018). Interfering with IP6 packaging or binding by mutation results in improper capsid assembly and premature uncoating (Renner et al. 2021; Mallery et al. 2021, 2019). In contrast, the capsid-binding drug PF74 induces rapid rupture of the capsid but prevents disassembly of the lattice thereafter (Márquez et al. 2018). It is tempting to speculate that binding of host cofactors to the same pocket as occupied by PF74 could induce uncoating by rupturing the capsid, possibly leading to the broken but stable capsid remnants observed by cryoET in the nucleus (see above).
Imaging mechanical properties of capsids by atomic force microscopy
Atomic force microscopy studies of isolated HIV-1 cores adhered to solid substrates have expanded our understanding of the mechanisms that initiate uncoating by measuring the mechanical properties and morphology of the capsid (Mallery et al. 2018; Ramalho et al. 2016; Rankovic et al. 2021, 2017). After addition of dNTPs to initiate reverse transcription in IP6-stabilised cores, Rankovic et al. recently observed characteristic spikes in capsid stiffness that corresponded to transient morphological changes (apparent as a conversion from a conical to a spherical structure) (Rankovic et al. 2021). While the underlying molecular processes for these transient states remain to be identified, the authors speculated that stiffness spikes correspond to specific stages of reverse transcription. Interestingly, loss of part of the capsid (predominantly at the narrow end of the cone) was observed after the third stiffness spike, ultimately leading to disappearance of the capsid. All of these transformations depended on reverse transcription as shown by inhibiting reverse transcriptase activity using drugs or mutation. These AFM measurements are difficult to perform, limiting the temporal resolution and number of cores that can be analysed, but provide unique insight into the interplay between reverse transcription and capsid mechanics at the level of individual particles, providing evidence that uncoating is triggered by mechanical stresses resulting from conversion of the viral RNA into stiffer double-stranded viral DNA.
In vitro reconstitution of reverse transcription and cryoET of uncoating intermediates
The encapsidated reverse transcription (ERT) assay provides PCR-based quantification of the level of early or late reverse transcription products formed in isolated cores and thus provides a complementary ensemble method to study the link between reverse transcription and uncoating in vitro, for example by showing the role of IP6-mediated capsid stabilisation for enhancing ERT (Jennings et al. 2020; Sowd et al. 2021). Christensen et al. optimised reconstitution of reverse transcription in viral particles permeabilised with the detergent mellitin and in the presence of IP6 and cell lysate, allowing the authors to capture disassembly intermediates via cryoET (Christensen et al. 2020). While most capsids remained largely intact, some were seen with defects ranging from small ruptures to loss of large patches of hexamers, indicating that uncoating is not an all-or-none process but that the capsid is instead lost in a more piecewise fashion. The authors observed nucleic acids looping out of these rupture points, suggesting that increasing pressure from the double-stranded DNA can trigger capsid breakage and subsequent uncoating. These DNA loops were detected primarily at late time points, but it remains unclear how far reverse transcription had progressed in these examples.
Taken together, the different imaging approaches discussed above helped establish the pivotal role of IP6 for promoting capsid stability and reverse transcription. While TIRF analysis suggests that removal of the first CA protein(s) from the lattice leads to a rapid collapse of the entire lattice, AFM and cryoET show broken capsids with missing lattice pieces, consistent with uncoating occurring via capsid rupture and gradual release of lattice pieces. The reason for such contradictory results is not clear but may be related to differences in experimental conditions; for example, TIRF measurements have so far been conducted only in the absence of reverse transcription. It is tempting to speculate that the mode of uncoating differs before and after reverse transcription, switching from a catastrophic process that propagates from a lattice void due to spontaneous CA dissociation to a rupture of the capsid due to build-up of pressure, whereby the broken capsid may be stabilised by compressive strain in the lattice (Yu et al. 2021). It is also worth noting the technical differences between these techniques. The TIRF uncoating assay enables simultaneous imaging of many single-particle uncoating reactions at high temporal resolution from the moment of membrane permeabilisation, allowing the identification of capsids with different intrinsic stability levels (including defective capsids), whereby it remains unclear which stability type can support infection. Short-lived uncoating intermediates are presumably not detectable by the lower temporal resolution in AFM (which also requires a core isolation protocol that enriches for stable capsids) or by cryoET, which captures a single snapshot in time.
Conclusion
The continued evolution of imaging techniques to follow the state of capsids at the level of single virions has provided the most detailed direct visualisation of uncoating to date and how it coordinates with the other elements of the HIV life cycle. In vitro reconstitution can define the intrinsic properties of the capsid, resolve the dynamic interplay with (host) molecules affecting uncoating kinetics, and thus identify the minimal components to control capsid-associated processes. Complementary observations from cellular systems place these observations into the physiologically relevant context, identify engagement with different cellular components and processes, and map events to subcellular compartments. The recent evidence suggests that capsids can traverse the nuclear pore complex and that the formation of double-stranded DNA, a process associated with the capsid itself, may trigger uncoating. Thus, the timing and location of uncoating may depend on the kinetics of transport processes relative to the kinetics of reverse transcription. Control of these processes may in turn be specific for different cell types, e.g., by availability of host cofactors. A scenario whereby a capsid enters the nucleus before it uncoats might be particularly relevant in cell types that trigger an innate immune response upon exposure of viral DNA in the cytoplasm (Towers and Noursadeghi 2014; Sumner et al. 2020).
This review highlights the recent progress that has been made towards understanding the process of uncoating. However, several questions remain unaddressed: (1) Does reverse transcription require the closed capsid and what controls its kinetics? For example, reverse transcription kinetics could be controlled by the opening and closing of the import channels for dNTPs (Jacques et al. 2016). (2) How does the capsid traverse the nuclear pore complex? Capsids may function as a karyopherin that can diffuse into the selective barrier formed by components of the nuclear pore complex. (3) What are the mechanisms and triggers of uncoating? While reverse transcription is a likely trigger, how the capsid breaks and whether other components are involved remain unclear. Recent evidence suggests that capsids associate with distinct cellular proteins in different compartments (cytoplasm-nuclear pore complex-nucleus) (Rebensburg et al. 2021) that utilise the same binding sites on the capsid but may have differential effects on capsid-associated processes. It has also been observed that capsids partition into phase-separated compartments in the nucleus (Rensen et al. 2021; Francis et al. 2020), which may further alter the intrinsic uncoating behaviour. Further advances in imaging methods and uncoating markers, such as minimally invasive labelling of CA using non-natural amino acids and click chemistry (Schifferdecker et al. 2021), have the potential to resolve the seemingly contradictory observations outlined in this review and further refine our understanding of the multiple roles of the HIV capsid in infection.
Acknowledgements
The authors acknowledge the Bedegal people who are the Traditional Custodians of the Land on which work included in this review was performed, and to Elders past and present.
Funding
JHS is supported by NHMRC grant APP1182212.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aguet F, Upadhyayula S, Gaudin R, Chou Y-Y, Cocucci E, He K, Chen B-C, Mosaliganti K, Pasham M, Skillern W, Legant WR, Liu T-L, Findlay G, Marino E, Danuser G, Megason S, Betzig E, Kirchhausen T (2016) Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Mol Biol Cell 27(22):3418–3435. 10.1091/mbc.E16-03-0164 [DOI] [PMC free article] [PubMed]
- Albanese A, Arosio D, Terreni M, Cereseto A (2008) HIV-1 pre-integration complexes selectively target decondensed chromatin in the nuclear periphery. PLoS ONE 3(6):e2413. 10.1371/journal.pone.0002413 [DOI] [PMC free article] [PubMed]
- Blanco-Rodriguez G, Gazi A, Monel B, Frabetti S, Scoca V, Mueller F, Schwartz O, Krijnse-Locker J, Charneau P, Di Nunzio F. Remodeling of the core leads HIV-1 preintegration complex into the nucleus of human lymphocytes. J Virol. 2020;94(11):e00135–e120. doi: 10.1128/JVI.00135-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönisch IZ, Dirix L, Lemmens V, Borrenberghs D, Wit FD, Vernaillen F, Rocha S, Christ F, Hendrix J, Hofkens J, Debyser Z, Kirchhoff F (2020) Capsid-labelled HIV to investigate the role of capsid during nuclear import and integration. J Virol 94 (7):e01024-01019. 10.1128/JVI.01024-19 [DOI] [PMC free article] [PubMed]
- Briggs JAG, Wilk T, Welker R, Kräusslich H-G, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22(7):1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick RC, Li C, Munshi M, Rawson JMO, Nagashima K, Hu W-S, Pathak VK. HIV-1 uncoats in the nucleus near sites of integration. Proc Natl Acad Sci U S A. 2020;117(10):5486–5493. doi: 10.1073/pnas.1920631117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Hope TJ. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol. 2015;13(8):471–483. doi: 10.1038/nrmicro3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Perez O, Melar M, Hope TJ. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360(2):286–293. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Ganser-Pornillos BK, Johnson JS, Pornillos O, Sundquist WI. Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science. 2020;370(6513):eabc8420. doi: 10.1126/science.abc8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva SC, Tartour K, Cimarelli A. A novel entry/uncoating assay reveals the presence of at least two species of viral capsids during synchronized HIV-1 infection. PLoS Pathog. 2016;12(9):e1005897. doi: 10.1371/journal.ppat.1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai TM, Marin M, Sood C, Shi J, Nawaz F, Aiken C, Melikyan GB. Fluorescent protein-tagged Vpr dissociates from HIV-1 core after viral fusion and rapidly enters the cell nucleus. Retrovirology. 2015;12:88. doi: 10.1186/s12977-015-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharan A, Bachmann N, Talley S, Zwikelmaier V, Campbell EM. Nuclear pore blockade reveals that HIV-1 completes reverse transcription and uncoating in the nucleus. Nat Microbiol. 2020;5(9):1088–1095. doi: 10.1038/s41564-020-0735-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick RA, Zadrozny KK, Xu C, Schur FKM, Lyddon TD, Ricana CL, Wagner JM, Perilla JR, Ganser-Pornillos BK, Johnson MC, Pornillos O, Vogt VM. Inositol phosphates are assembly co-factors for HIV-1. Nature. 2018;560(7719):509–512. doi: 10.1038/s41586-018-0396-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostálková A, Hadravová R, Kaufman F, Křížová I, Škach K, Flegel M, Hrabal R, Ruml T, Rumlová M. A simple, high-throughput stabilization assay to test HIV-1 uncoating inhibitors. Sci Rep. 2019;9(1):17076. doi: 10.1038/s41598-019-53483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostálková A, Kaufman F, Křížová I, Vokatá B, Ruml T, Rumlová M (2020) In vitro quantification of the effects of IP6 and other small polyanions on immature HIV-1 particle assembly and core stability. J Virol 94 (20). 10.1128/jvi.00991-20 [DOI] [PMC free article] [PubMed]
- Ehrlich LS, Agresta BE, Carter CA. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66(8):4874–4883. doi: 10.1128/JVI.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76(11):5667–5677. doi: 10.1128/jvi.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis AC, Melikyan GB. Single HIV-1 imaging reveals progression of infection through CA-dependent steps of docking at the nuclear pore, uncoating, and nuclear transport. Cell Host Microbe. 2018;23(4):536–548.e536. doi: 10.1016/j.chom.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis AC, Marin M, Shi J, Aiken C, Melikyan GB. Time-resolved imaging of single HIV-1 uncoating in vitro and in living cells. PLoS Pathog. 2016;12(6):e1005709. doi: 10.1371/journal.ppat.1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis AC, Marin M, Singh PK, Achuthan V, Prellberg MJ, Palermino-Rowland K, Lan S, Tedbury PR, Sarafianos SG, Engelman AN, Melikyan GB. HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Nat Commun. 2020;11(1):3505. doi: 10.1038/s41467-020-17256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283(5398):80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78(5):2545–2552. doi: 10.1128/jvi.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, Sarafianos SG (2015) Structural virology. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science (New York, NY) 349 (6243):99–103. 10.1126/science.aaa5936 [DOI] [PMC free article] [PubMed]
- Gross I, Hohenberg H, Kräusslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249(2):592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- Hadravová R, Rumlová M, Ruml T. FAITH - Fast Assembly Inhibitor Test for HIV. Virology. 2015;486:78–87. doi: 10.1016/j.virol.2015.08.029. [DOI] [PubMed] [Google Scholar]
- Hübner W, Chen P, Del Portillo A, Liu Y, Gordon RE, Chen BK. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J Virol. 2007;81(22):12596–12607. doi: 10.1128/jvi.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A. 2011;108(24):9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram Z, Taylor M, Okland G, Martin R, Hulme AE. Characterization of HIV-1 uncoating in human microglial cell lines. Virol J. 2020;17(1):31–31. doi: 10.1186/s12985-020-01301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques DA, McEwan WA, Hilditch L, Price AJ, Towers GJ, James LC. HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature. 2016;536(7616):349–353. doi: 10.1038/nature19098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings J, Shi J, Varadarajan J, Jamieson PJ, Aiken C (2020) The host cell metabolite inositol hexakisphosphate promotes efficient endogenous HIV-1 Reverse Transcription by stabilizing the viral capsid. mBio 11 (6). 10.1128/mBio.02820-20 [DOI] [PMC free article] [PubMed]
- Kumar S, Morrison JH, Dingli D, Poeschla E. HIV-1 Activation of innate immunity depends strongly on the intracellular level of TREX1 and sensing of incomplete reverse transcription products. J Virol. 2018;92(16):e00001–00018. doi: 10.1128/JVI.00001-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelek M, Di Nunzio F, Henriques R, Charneau P, Arhel N, Zimmer C. Superresolution imaging of HIV in infected cells with FlAsH-PALM. Proc Natl Acad Sci U S A. 2012;109(22):8564–8569. doi: 10.1073/pnas.1013267109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Burdick RC, Nagashima K, Hu WS, Pathak VK (2021) HIV-1 cores retain their integrity until minutes before uncoating in the nucleus. Proc Natl Acad Sci U S A 118 (10). 10.1073/pnas.2019467118 [DOI] [PMC free article] [PubMed]
- Mallery DL, Márquez CL, McEwan WA, Dickson CF, Jacques DA, Anandapadamanaban M, Bichel K, Towers GJ, Saiardi A, Böcking T, James LC. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. Elife. 2018;7:e35335. doi: 10.7554/eLife.35335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery DL, Faysal KMR, Kleinpeter A, Wilson MSC, Vaysburd M, Fletcher AJ, Novikova M, Böcking T, Freed EO, Saiardi A, James LC. Cellular IP(6) levels limit HIV Production while viruses that cannot efficiently package IP(6) are attenuated for infection and replication. Cell Rep. 2019;29(12):3983–3996.e3984. doi: 10.1016/j.celrep.2019.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery DL, Kleinpeter AB, Renner N, Faysal KMR, Novikova M, Kiss L, Wilson MSC, Ahsan B, Ke Z, Briggs JAG, Saiardi A, Böcking T, Freed EO, James LC (2021) A stable immature lattice packages IP(6) for HIV capsid maturation. Sci Adv 7 (11). 10.1126/sciadv.abe4716 [DOI] [PMC free article] [PubMed]
- Mamede JI, Cianci GC, Anderson MR, Hope TJ. Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc Natl Acad Sci U S A. 2017;114(34):E7169–E7178. doi: 10.1073/pnas.1706245114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamede JI, Griffin J, Gambut S, Hope TJ (2021) A new generation of functional tagged proteins for HIV fluorescence imaging. Viruses 13 (3). 10.3390/v13030386 [DOI] [PMC free article] [PubMed]
- Márquez CL, Lau D, Walsh J, Shah V, McGuinness C, Wong A, Aggarwal A, Parker MW, Jacques DA, Turville S, Böcking T. Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. Elife. 2018;7:e34772. doi: 10.7554/eLife.34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez CL, Lau D, Walsh J, Rifat Faysal KM, Parker MW, Turville SG, Böcking T. Fluorescence microscopy assay to measure HIV-1 capsid uncoating kinetics in vitro. Bio-Protoc. 2019;9(13):e3297. doi: 10.21769/BioProtoc.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei S, Glass B, Hagen WJH, Kräusslich H-G, Briggs JAG. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science. 2016;354(6318):1434–1437. doi: 10.1126/science.aah4972. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Boutant E, Réal E, Mély Y, Anton H (2021) Imaging viral infection by fluorescence microscopy: focus on HIV-1 early stage. Viruses 13 (2). 10.3390/v13020213 [DOI] [PMC free article] [PubMed]
- Müller TG, Zila V, Peters K, Schifferdecker S, Stanic M, Lucic B, Laketa V, Lusic M, Müller B, Kräusslich HG (2021) HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. Elife 10. 10.7554/eLife.64776 [DOI] [PMC free article] [PubMed]
- Ning J, Zhong Z, Fischer DK, Harris G, Watkins SC, Ambrose Z, Zhang P (2018) Truncated CPSF6 forms higher-order complexes that bind and disrupt HIV-1 capsid. J Virol 92 (13). 10.1128/jvi.00368-18 [DOI] [PMC free article] [PubMed]
- Pereira CF, Ellenberg PC, Jones KL, Fernandez TL, Smyth RP, Hawkes DJ, Hijnen M, Vivet-Boudou V, Marquet R, Johnson I, Mak J. Labeling of multiple HIV-1 proteins with the biarsenical-tetracysteine system. PLoS ONE. 2011;6(2):e17016. doi: 10.1371/journal.pone.0017016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho R, Rankovic S, Zhou J, Aiken C, Rousso I. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology. 2016;13:17. doi: 10.1186/s12977-016-0250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankovic S, Varadarajan J, Ramalho R, Aiken C, Rousso I. Reverse transcription mechanically initiates HIV-1 capsid disassembly. J Virol. 2017;91(12):e00289–e217. doi: 10.1128/JVI.00289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankovic S, Deshpande A, Harel S, Aiken C, Rousso I (2021) HIV-1 uncoating occurs via a series of rapid biomechanical changes in the core related to individual stages of reverse transcription. J Virol 95 (10). 10.1128/jvi.00166-21 [DOI] [PMC free article] [PubMed]
- Rebensburg SV, Wei G, Larue RC, Lindenberger J, Francis AC, Annamalai AS, Morrison J, Shkriabai N, Huang SW, KewalRamani V, Poeschla EM, Melikyan GB, Kvaratskhelia M. Sec24C is an HIV-1 host dependency factor crucial for virus replication. Nat Microbiol. 2021;6(4):435–444. doi: 10.1038/s41564-021-00868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner N, Mallery DL, Faysal KMR, Peng W, Jacques DA, Böcking T, James LC. A lysine ring in HIV capsid pores coordinates IP6 to drive mature capsid assembly. PLoS Pathog. 2021;17(2):e1009164. doi: 10.1371/journal.ppat.1009164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensen E, Mueller F, Scoca V, Parmar JJ, Souque P, Zimmer C, Di Nunzio F. Clustering and reverse transcription of HIV-1 genomes in nuclear niches of macrophages. EMBO J. 2021;40(1):e105247. doi: 10.15252/embj.2020105247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelo NH, Kamin D, Truckenbrodt S, Wong AB, Reuter-Jessen K, Reisinger E, Moser T, Rizzoli SO. A new probe for super-resolution imaging of membranes elucidates trafficking pathways. J Cell Biol. 2014;205(4):591–606. doi: 10.1083/jcb.201402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD. Extreme genetic fragility of the HIV-1 capsid. PLoS Pathog. 2013;9(6):e1003461–e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Kirchhoff F. HIV replication: a game of hide and sense. Curr Opin HIV AIDS. 2016;11(2):173–181. doi: 10.1097/coh.0000000000000233. [DOI] [PubMed] [Google Scholar]
- Schifferdecker S, Zila V, Mueller TG, Sakin V, Anders-Oesswein M, Laketa V, Kraeusslich H-G, Mueller B (2021) Direct capsid labeling of infectious HIV-1 by genetic code expansion allows detection of largely complete nuclear capsids and suggests nuclear entry of HIV-1 complexes via common routes. bioRxiv. 10.1101/2021.09.14.460218 [DOI] [PMC free article] [PubMed]
- Scoca V, Di Nunzio F. The HIV-1 capsid: from structural component to key factor for host nuclear invasion. Viruses. 2021;13(2):273. doi: 10.3390/v13020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah VB, Aiken C (2011) In vitro uncoating of HIV-1 cores. J Vis Exp (57). 10.3791/3384 [DOI] [PMC free article] [PubMed]
- Sowd GA, Shi J, Aiken C (2021) HIV-1 CA inhibitors are antagonized by inositol phosphate stabilization of the viral capsid in cells. J Virol:Jvi0144521. 10.1128/jvi.01445-21 [DOI] [PMC free article] [PubMed]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner RP, Harrison L, Touizer E, Peacock TP, Spencer M, Zuliani-Alvarez L, Towers GJ. Disrupting HIV-1 capsid formation causes cGAS sensing of viral DNA. EMBO J. 2020;39(20):e103958–e103958. doi: 10.15252/embj.2019103958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers GJ, Noursadeghi M. Interactions between HIV-1 and the cell-autonomous innate immune system. Cell Host Microbe. 2014;16(1):10–18. doi: 10.1016/j.chom.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turville SG, Aravantinou M, Stössel H, Romani N, Robbiani M. Resolution of de novo HIV production and trafficking in immature dendritic cells. Nat Methods. 2008;5(1):75–85. doi: 10.1038/nmeth1137. [DOI] [PubMed] [Google Scholar]
- Wacharapornin P, Lauhakirti D, Auewarakul P. The effect of capsid mutations on HIV-1 uncoating. Virology. 2007;358(1):48–54. doi: 10.1016/j.virol.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Willy NM, Colombo F, Huber S, Smith AC, Norton EG, Kural C, Cocucci E (2021) CALM supports clathrin-coated vesicle completion upon membrane tension increase. Proc Natl Acad Sci U S A 118(25):e2010438118. 10.1073/pnas.2010438118 [DOI] [PMC free article] [PubMed]
- Xu C, Fischer DK, Rankovic S, Li W, Dick RA, Runge B, Zadorozhnyi R, Ahn J, Aiken C, Polenova T, Engelman AN, Ambrose Z, Rousso I, Perilla JR. Permeability of the HIV-1 capsid to metabolites modulates viral DNA synthesis. PLoS Biol. 2020;18(12):e3001015–e3001015. doi: 10.1371/journal.pbio.3001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Luban J, Diaz-Griffero F. The fate of HIV-1 capsid: a biochemical assay for HIV-1 uncoating. Methods Mol Biol. 2014;1087:29–36. doi: 10.1007/978-1-62703-670-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A, Lee EM, Briggs JA, Ganser-Pornillos BK, Pornillos O, Voth GA (2021) Strain and crack propagation of HIV-1 capsids during uncoating. bioRxiv. 10.1101/2021.09.30.462583 [DOI] [PMC free article] [PubMed]
- Zila V, Margiotta E, Turoňová B, Müller TG, Zimmerli CE, Mattei S, Allegretti M, Börner K, Rada J, Müller B, Lusic M, Kräusslich H-G, Beck M. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell. 2021;184(4):1032–1046.e1018. doi: 10.1016/j.cell.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zila V, Müller TG, Laketa V, Müller B, Kräusslich H-G, Lingappa JR (2019) Analysis of CA content and CPSF6 dependence of early HIV-1 replication complexes in SupT1-R5 cells. mBio 10 (6):e02501–02519. 10.1128/mBio.02501-19 [DOI] [PMC free article] [PubMed]