Abstract
Objective
To date, there are no predictive biomarkers to guide selection of patients with gastric cancer (GC) who benefit from paclitaxel. Stomach cancer Adjuvant Multi-Institutional group Trial (SAMIT) was a 2×2 factorial randomised phase III study in which patients with GC were randomised to Pac-S-1 (paclitaxel +S-1), Pac-UFT (paclitaxel +UFT), S-1 alone or UFT alone after curative surgery.
Design
The primary objective of this study was to identify a gene signature that predicts survival benefit from paclitaxel chemotherapy in GC patients. SAMIT GC samples were profiled using a customised 476 gene NanoString panel. A random forest machine-learning model was applied on the NanoString profiles to develop a gene signature. An independent cohort of metastatic patients with GC treated with paclitaxel and ramucirumab (Pac-Ram) served as an external validation cohort.
Results
From the SAMIT trial 499 samples were analysed in this study. From the Pac-S-1 training cohort, the random forest model generated a 19-gene signature assigning patients to two groups: Pac-Sensitive and Pac-Resistant. In the Pac-UFT validation cohort, Pac-Sensitive patients exhibited a significant improvement in disease free survival (DFS): 3-year DFS 66% vs 40% (HR 0.44, p=0.0029). There was no survival difference between Pac-Sensitive and Pac-Resistant in the UFT or S-1 alone arms, test of interaction p<0.001. In the external Pac-Ram validation cohort, the signature predicted benefit for Pac-Sensitive (median PFS 147 days vs 112 days, HR 0.48, p=0.022).
Conclusion
Using machine-learning techniques on one of the largest GC trials (SAMIT), we identify a gene signature representing the first predictive biomarker for paclitaxel benefit.
Trial registration number
UMIN Clinical Trials Registry: C000000082 (SAMIT); ClinicalTrials.gov identifier, 02628951 (South Korean trial)
Keywords: gastric cancer, chemotherapy, adjuvant treatment
Significance of this study.
What is already known on this subject?
Paclitaxel is an active chemotherapeutic agent in the treatment of gastric cancer (GC).
Stomach cancer Adjuvant Multi-Institutional group Trial (SAMIT) was a randomised 2×2 factorial phase III trial, studying the role of adjuvant paclitaxel in patients with GC.
There are currently no biomarkers for selecting patients with GC who benefit from paclitaxel
Machine-learning modelling using genomic data is an emerging and novel strategy to identify gene signature based predictive biomarkers
What are the new findings?
NanoString transcriptomic profiling was performed on SAMIT surgical resection samples.
Using the trial’s 2×2 factorial study design, a gene signature predicting paclitaxel survival benefit was developed with a random forest machine-learning method.
The model was validated internally within SAMIT samples, and independently in an external validation cohort.
How might it impact on clinical practice in the foreseeable future?
These findings represent the first predictive biomarker for paclitaxel benefit in GC.
Given the rising utilisation of taxanes in GC in the adjuvant and perioperative setting, this biomarker may guide in identifying patients who benefit from taxane based therapy, following further validation in prospective trials.
Introduction
The identification of predictive biomarkers to personalise treatment of patients with gastric cancer (GC) is challenging due to the complex genomic and molecular landscape of this malignancy.1 Adding chemotherapy to surgery has improved patient with GC survival, when administered in either the perioperative or postoperative setting.2 Besides 5-fluorouracil (5FU) and platinum, taxanes have demonstrated activity in patients with GC, with docetaxel and paclitaxel both showing a survival benefit in first-line and second-line treatment of patients with metastatic GC, respectively.3 4 Recently, the use of docetaxel has shown survival improvements when combined with 5FU and platinum-based therapy in the perioperative setting (FLOT4).5 The triplet regimen of 5FU/platinum/docetaxel is considered a standard of care in several Western nations.4 Similarly, in Japan, addition of docetaxel to S-1 adjuvant chemotherapy has demonstrated a survival benefit.6 However, other GC trials investigating the role of taxanes in the adjuvant and metastatic setting were unable to demonstrate survival improvement.7 8 These contradictory results underpin the urgent clinical need for predictive biomarkers to identify patients who will benefit from taxane chemotherapy. While there are data for biomarkers predicting 5FU and platinum benefit, there are currently no predictive biomarkers for taxane therapy in GC described in the literature.9–13
Stomach cancer Adjuvant Multi-Institutional group Trial (SAMIT) was a phase III randomised trial conducted in Japan, to assess superiority of adjuvant sequential treatment (paclitaxel followed by UFT or paclitaxel followed by S-1) compared with monotherapy (UFT or S-1 alone), and to assess the non-inferiority of UFT compared with S-1 after curative surgery.14 UFT is an oral combination of uracil (an inhibitor of dihydropyrimidine dehydrogenase (DPD)) and tegafur (a 5FU prodrug).15 S-1 is an oral combination of tegafur, gimeracil (a DPD inhibitor) and oteracil (a compound that localises preferentially to the gut and inhibits orotate phosphoribosyl-transferase, reducing activation of 5FU and gastrointestinal toxicity).16 The clinical results of SAMIT, reported in 2014, showed that sequential paclitaxel did not improve disease-free survival (DFS) and UFT was not non-inferior to S-1.14
Taxanes are thought to exert their anti-cancer effects through aberrant stabilisation of microtubules, leading to defects in chromosome segregation, mitotic arrest and activation of the spindle assembly checkpoint, where prolonged activation results in cell death. Studies in patients with breast cancer suggest that altered expression of genes involved in the spindle assembly checkpoint may affect cellular sensitivity to paclitaxel,17 18 and immunogenic cell death after chemotherapy.19 20 We hypothesised that in GC the expression of genes involved in chromosomal stability and/or immunogenic cell death may predict benefit from paclitaxel. We aimed to test this hypothesis using samples and clinical data from the SAMIT trial. Findings from the SAMIT study were validated in an independent phase 2 GC trial cohort from South Korea.
Methods
SAMIT trial
SAMIT trial was a 2×2 factorial design phase III study conducted in Japan.14 In total, 1495 patients were randomised to either single agent UFT (UFT arm), single agent S-1 (S-1 arm) or sequential paclitaxel-S-1(Pac-S-1 arm) or sequential paclitaxel-UFT (Pac-UFT arm).
Surgical sample processing and RNA extraction for biomarker cohort
Formalin-fixed, paraffin-embedded (FFPE) blocks or unstained cut sections from gastrectomy specimens were collected from Japanese sites participating in the SAMIT study.RNA was extracted from the primary tumour and prepared for NanoString profiling (online supplemental methods).
gutjnl-2021-324060supp001.pdf (159.1KB, pdf)
NanoString analysis
A custom designed NanoString (NanoString Technologies, USA) panel of 476 genes was used (online supplemental table 1). Gene ontologies were broadly categorised into
gutjnl-2021-324060supp004.xlsx (1.6MB, xlsx)
Genes involved in the spindle assembly checkpoint.
Genes with therapeutic implications in GC.
Genes involved in the GC tumour microenvironment.
Genes relevant to oncogenic signalling pathways.
Genes with frequent genomic alterations in GC.
Immune-related genes in GC.
DNA damage repair genes.
Genes with reported predictive benefit in GC chemotherapy from the literature.
Quality control (QC) analyses between triplicates and corrections for batch effects are described in the supplement (online supplemental methods and figure S1). Normalised NanoString gene expression data and correlative clinicopathological characteristics are provided in online supplemental table 1.
gutjnl-2021-324060supp003.pdf (2MB, pdf)
Gene signature development using machine-learning models
The random forest method uses an ensemble of classification trees of several variables.21 Metrics for measuring the predictive performance included accuracy, precision, recall, F-measure and area under curve (AUC). The F-measure has been shown to handle class imbalances in the dataset better than positive predictive value and accuracy.22 23
Random forests return a prediction from a collection of classification trees (in our analyses we used ntree=3000) (online supplemental methods). Each tree is grown by using a bootstrap sample of the data set and, only a random subset of the original variables is examined at each node. Multiple iterations of the random forest algorithm were run, performing a variable importance analysis and eliminating variables that did not contribute to the classification resulting in improved predictive performance.24 The model was trained based on a DFS< or ≥2 years classifier. As the random forest algorithm works best when trained on equally sized groups, this clinically relevant cut-off of DFS was also selected to create relatively equal groups.5 A sensitivity and variable importance analysis for F-measure was performed to identify the optimal number of genes to be included in the gene signature. The classes identified by the gene signature derived from the random forest algorithm were labelled ‘Pac-Sensitive’ (paclitaxel-sensitive, ie, those patients that derive a survival benefit from paclitaxel) and ‘Pac-Resistant’ (paclitaxel-resistant, ie, those patients that lack a survival benefit from paclitaxel). The randomForest R package was used.
Creation of training and validation cohorts for machine learning
The aim was to train the random forest model on a training cohort to generate a classifier, which would then be applied on a validation cohort to confirm accuracy, F-measure and AUC of the classifier. The classifier would then be tested on an ‘external independent validation cohort’ (described in the next section). AUC was calculated using a time-dependent receiver operating characteristic (ROC) curve for 2-year DFS, using the risksetROC package.
We took advantage of the unique 2×2 factorial design of the SAMIT trial. Essentially, each arm of the study could function as individual cohort of patients for training and validation. As SAMIT was a randomised phase III trial, the cohorts (arms) would be well balanced with respect to confounding factors. Thus, two paclitaxel treated (Pac-S-1 and Pac-UFT) and two paclitaxel untreated (S-1 and UFT) cohorts were formed. Tumour samples from one paclitaxel treated arm would be used to train the model, while the other would be used to validate the classifier. Since S-1 currently represents the standard of care for GC chemotherapy in Japan, we elected to use the Pac-S-1 arm (n=128) as the training cohort and the Pac-UFT arm (n=123) as the validation cohort. We also tested the interaction between the classifier and treatment with paclitaxel by applying the classifier on the non-paclitaxel containing arms (S-1 and UFT).
External independent validation South Korean paclitaxel-ramucirumab trial cohort (Pac-Ram)
Data for this validation cohort were derived from a single-arm non-randomised phase II trial of patients with GC with metastatic disease treated with paclitaxel and ramucirumab (n=47).25 Patients with metastatic GC who had progressed on at least 1 line of chemotherapy which included platinum/fluoropyrimidine were enrolled in a phase II trial at the Samsung Medical Center, South Korea. All patients provided written informed consent before enrolment. A tumour biopsy was obtained between day −42 and day 1 prior to initiation of study treatment. Intravenous paclitaxel and ramucirumab were administered until documented disease progression, unacceptable toxicity or patients’ refusal was reported. RNA-Seq was performed and data were aligned to GENCODE V.19 transcript annotation using STAR. Transcripts per million abundance measure were generated using RSEM. RNA-seq transcripts mapping to genes profiled using the NanoString panel were extracted. Random forest classifiers trained on the SAMIT study were applied to the normalised RNA-seq data to predict benefit from paclitaxel therapy.
The Cancer Genome Atlas analysis
Gene expression data and clinical data for the The Cancer Genome Atlas (TCGA) stomach adenocarcinoma (STAD) cohort were downloaded from Firebrowse.26 Illumina HiSeq RNA-SeqV2 RSEM normalised gene values were used and applied through a similar pipeline as the Pac-Ram cohort RNA-Seq samples for generation of the random forest gene signature. HER2 status of TCGA STAD samples were derived from the HER2 index of Li et al.27 The HER2 index is an expression-based classifier reflecting the HER2-enriched transcriptional pattern for tumours harbouring HER2 aberrations. A cut-off 0.75 was used to classify samples as positive.
Statistical analyses
The primary objective of this translational study was to identify a gene signature generated by a machine-learning model that predicts survival benefit from paclitaxel chemotherapy in patients from the SAMIT trial, and to validate these findings in an independent GC patient cohort.
All analyses were done using R (V.3.6.1) with statistical significance set at two-tailed p<0.05. Fisher’s exact test was used to evaluate associations with categorical variables. Kaplan-Meier curves with log-rank statistics were used to compare overall survival (OS), DFS in the SAMIT cohort and progression-free survival (PFS) in the Korean cohort. OS was calculated from the date of randomisation to the date of death by any cause, DFS was calculated from the date of randomisation to the date of first event (relapse of stomach cancer, death from any cause or occurrence of a second cancer), PFS was calculated from the date of first dose of paclitaxel/ramucirumab to the date of disease progression or death from any cause). The assumption of proportional hazards was checked using the scaled Schoenfeld residuals method and was supported. Cox regression was performed to present HRs and 95% confidence intervals (CI). Univariate and multivariate analysis for DFS was performed.
Results
SAMIT biomarker cohort characteristics
RNA was extracted from 552 GC resection samples (37% of the whole SAMIT trial population) and profiled by a custom-made NanoString assay. 53 samples (10%) failed QC and were excluded from further analyses leaving a total of 499 GCs for final analyses (figure 1A). There were no statistically significant differences in patient characteristics between the original SAMIT trial population and subset of patients in the biomarker cohort (online supplemental table S2). Patient characteristics were well balanced between the arms in the biomarker cohort (table 1). Median age was 65 years (range 29–80 years) and 69% (n=343) were male. The median follow-up time was 60.5 months (IQR: 47.5–78.3 months), and there were no survival differences between the biomarker cohort and original SAMIT trial population (online supplemental figure S2). There were also no differences in the clinicopathologic characteristics of the samples that failed QC and the ones that did not (online supplemental table S3).
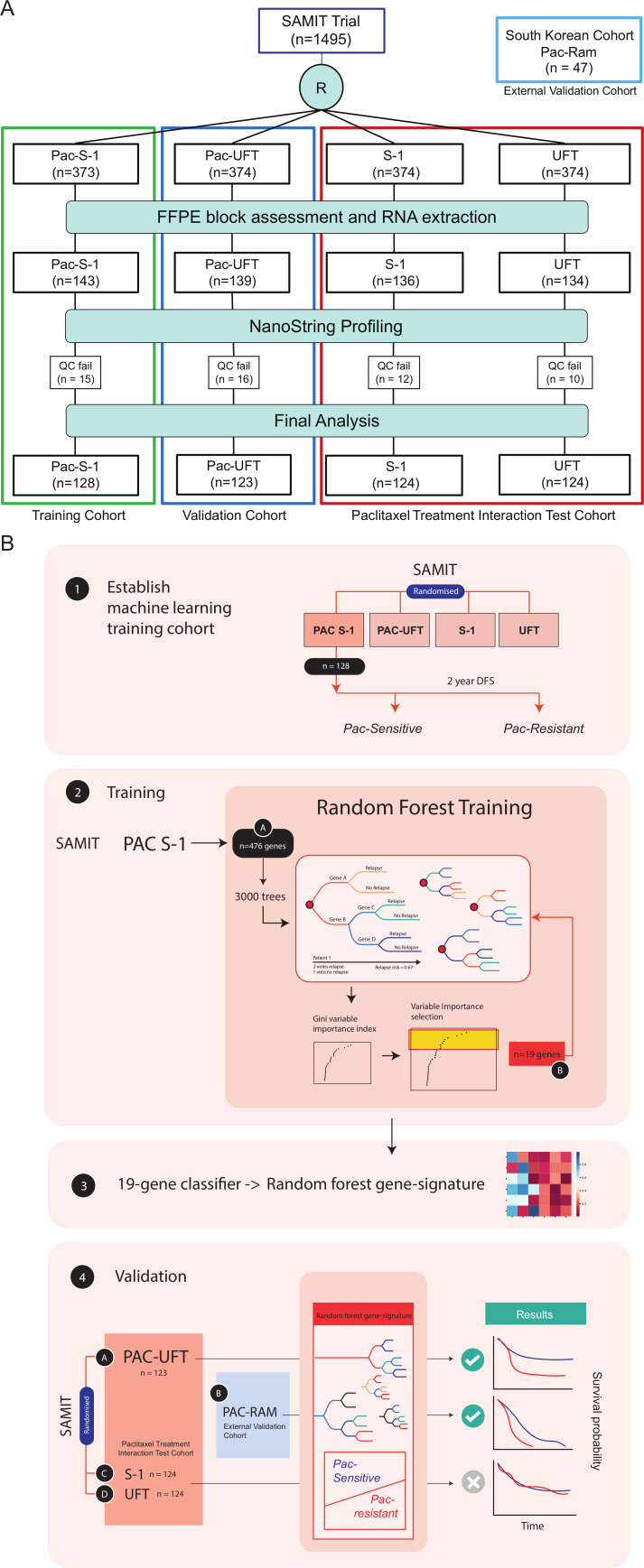
Figure 1.
Flow chart of sample analysis from SAMIT trial A: In total, 1495 patients were randomised after surgery in the SAMIT trial to four arms (UFT alone, S-1 alone, Pac-UFT and Pac-S-1). After assessment of formalin-fixed paraffin embedded blocks (FFPE) for tumour content, RNA was extracted and available for 552 samples which were profiled using the NanoString platform. After quality control postprofiling, 499 samples were included in the final analysis. (B) Samples from the Pac-S1 arm were selected to train the random forest machine-learning model. Samples were trained using a 2-year DFS to define Pac-Sensitive and Pac-Resistant groups. Using 476 genes in the custom-made NanoString panel, a variable importance analysis was performed, and the top 19 genes selected. The final model was retrained with these 19 genes to generate a random forest gene signature. This signature was then applied on the Pac-UFT arm as an internal validation cohort and the S-1 and UFT arms to test for interaction with paclitaxel treatment. The Pac-Ram samples were tested as an external validation cohort. SAMIT, Stomach cancer Adjuvant Multi-Institutional group Trial.
Table 1.
Patient characteristics of biomarker cohort of SAMIT study
| Factors | UFT (n=124) |
S-1 (n=124) |
Pac-UFT (n=123) |
Pac-S-1 (n=128) |
P value | |||||
| n | % | n | % | n | % | n | % | |||
| Age | Median (range) | 65 (29–80) | 65 (38–80) | 66 (30–80) | 67(37–80) | 0.78 | ||||
| Gender | Male | 87 | 70 | 77 | 62 | 86 | 70 | 93 | 73 | 0.31 |
| Female | 37 | 30 | 47 | 38 | 37 | 30 | 35 | 27 | ||
| Performance status | 0 | 107 | 86 | 103 | 83 | 101 | 82 | 105 | 82 | 0.77 |
| 1 | 17 | 14 | 21 | 17 | 22 | 18 | 23 | 18 | ||
| Histological type | Differentiated | 45 | 36 | 52 | 42 | 49 | 40 | 55 | 43 | 0.73 |
| Undifferentiated | 77 | 63 | 70 | 56 | 71 | 58 | 72 | 56 | ||
| Other | 1 | 1 | 2 | 2 | 2 | 2 | 1 | 1 | ||
| T stage | pT1 | 2 | 2 | 4 | 3 | 0 | 0 | 0 | 0 | 0.69 |
| pT2 | 40 | 32 | 38 | 30 | 32 | 26 | 42 | 33 | ||
| pT3 | 77 | 62 | 80 | 65 | 83 | 67 | 81 | 64 | ||
| pT4 | 5 | 4 | 2 | 2 | 7 | 6 | 4 | 3 | ||
| N stage | pN0 | 17 | 14 | 25 | 20 | 34 | 28 | 28 | 22 | 0.19 |
| pN1 | 46 | 37 | 51 | 41 | 41 | 33 | 39 | 31 | ||
| pN2 | 59 | 47 | 46 | 37 | 46 | 38 | 55 | 43 | ||
| pN3 | 2 | 2 | 2 | 2 | 1 | 1 | 5 | 4 | ||
American Joint Committee on Cancer/Union for International Cancer Control (AJCC/UICC) sixth edition.
gutjnl-2021-324060supp002.pdf (132.9KB, pdf)
Development of a predictive gene signature of paclitaxel benefit using random forest analysis
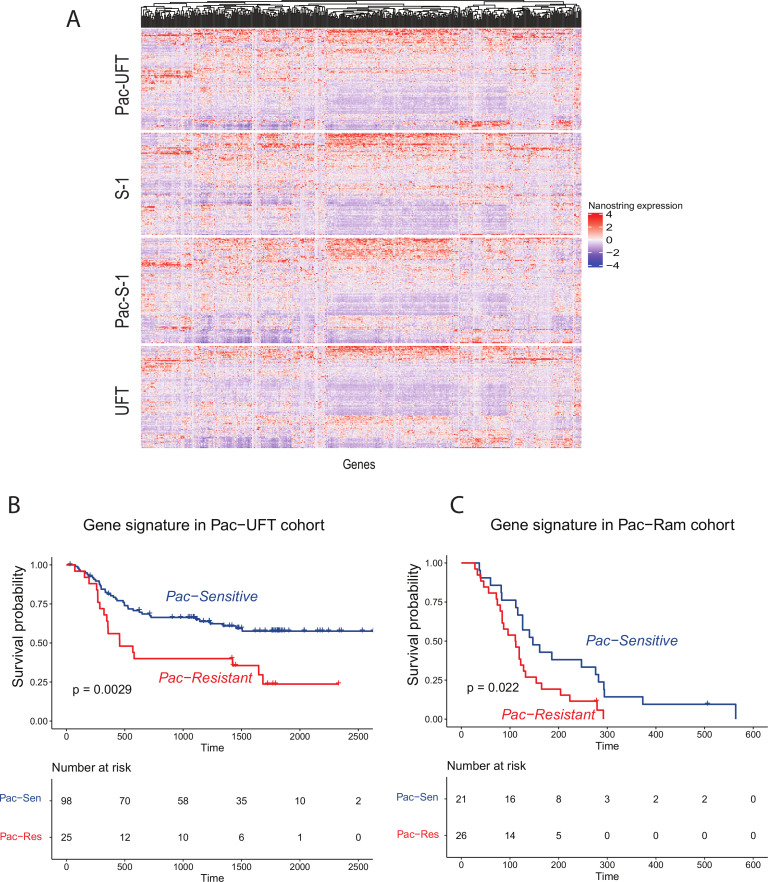
NanoString profiles did not reveal any significant differences in gene expression between treatment arms, and none of the single gene expression levels were associated with DFS by Cox univariate regression after correction for multiplicity using the false discovery rate method (online supplemental table 1 and figure 2A).
Figure 2.
Performance of random forest gene signature. (A) Heatmap of NanoString expression profiles from SAMIT samples by treatment arm. NanoString gene expression is represented in columns, scaled. Blue to red denotes transcript expression, with blue indicating low gene expression and red indicating high gene expression. There is no global difference in gene expression profiles between the four arms. (B) Kaplan-Meier curve of disease free survival (DFS) of patients classified by the random forest gene signature as either Pac-Sensitive (blue) or Pac-Resistant (red) in the validation Pac-UFT arm. A 3-year DFS 66% (Pac-Sensitive) vs 40% (Pac-Resistant) (HR 0.44, 95% CI 0.25 to 0.76, logrank p=0.0029) (C) Kaplan-Meier curve of progression-free survival (PFS) of patients classified by the random forest gene signature as either Pac-Sensitive (blue) or Pac-Resistant (red) in the external validation Pac-Ram cohort. Median PFS 147 days vs 112 days, HR 0.48, 95% CI 0.25 to 0.91, logrank p=0.022. SAMIT, Stomach cancer Adjuvant Multi-Institutional group Trial.
To identify gene signatures for paclitaxel-benefit, we found that application of the machine-learning approach by randomly dividing SAMIT samples into training and validation cohort failed to deliver a gene signature that could be validated in the Pac-Ram external cohort (online supplemental methods). We, therefore, decided to take advantage of the unique 2×2 factorial design of the SAMIT trial, with each arm of the study representing an individual cohort. The cohorts were well balanced with respect to clinicopathological factors (table 1). Using the Pac-S-1 arm (n=128) as the training cohort, we labelled patients as Pac-Sensitive if they derived a survival benefit from paclitaxel and as Pac-Resistant if they did not have a survival benefit from paclitaxel (DFS≥ or <2 years) (figure 1B).
From the Pac-S-1 cohort, random forest and variable importance analysis identified the top genes contributing to the model, on which the classifier was trained. A sensitivity analysis using different numbers of genes within the signature was performed, using F-measure and accuracy (online supplemental figure S3), to decide on the optimal number of genes within the signature. A 19-gene signature (random forest gene signature) trained on the Pac-S-1 cohort and tested on the Pac-UFT validation cohort (n=123) had the highest accuracy and F-measure, and was selected for further analysis. In the Pac-UFT arm, patients classified as Pac-Sensitive by the random forest gene signature had a significantly longer DFS compared with those classified as Pac-Resistant: 3-year DFS 66% (Pac-Sensitive) vs 40% (Pac-Resistant) (HR 0.44, 95% CI 0.25 to 0.76, logrank p=0.0029) (figure 2B). Accuracy of the random forest gene signature in the Pac-UFT validation cohort was 0.61, F-measure was 0.71 and AUC was 0.75 (95% CI 0.50 to 0.99).
Clinicopathological features of the Pac-Resistant patients in Pac-UFT revealed no significant differences when compared with Pac-Sensitive patients (online supplemental table S4). When clinical features of the Pac-UFT cohort were considered, in both univariate and multivariate survival (DFS) analysis, depth of invasion (pT), lymph node status (pN) and random forest gene signature were significant predictors of survival (table 2). After adjusting the survival model for depth of invasion and lymph node status, the random forest gene signature remained significantly associated with survival (HR 0.45, 95% CI 0.26 to 0.80, p=0.006).
Table 2.
Univariate and multivariate survival analysis in Pac-UFT
| Factors | Univariate | Multivariate | ||
| N=123 | HR (95% CI) | P value | HR (95% CI) | P value |
|
Random forest gene signature
Pac-Sensitive versus Pac-Resistant |
0.44 (0.25 to 0.76) | 0.0038 | 0.42 (0.24 to 0.76) | 0.0037 |
| Age | 1.00 (0.97 to 1.03) | 0.79 | 0.99 (0.96 to 1.03) | 0.70 |
| Gender: Female versus male | 0.70 (0.38 to 1.28) | 0.25 | 0.57 (0.30 to 1.09) | 0.09 |
| ECOG Performance Status: PS 1 versus PS 0 | 1∙27 (0.66 to 2.47) | 0.47 | 1.43 (0.70 to 2.91) | 0.33 |
|
Histology
Undifferentiated/others versus differentiated |
1.43 (0.81 to 2.51) | 0.22 | 1.55 (0.85 to 2.81) | 0.15 |
| Depth of invasion pT3/4 vs pT1/2 | 2.48 (1.17 to 5.26) | 0.018 | 2∙43 (1.12 to 5.26) | 0.024 |
| Lymph node status pN2/3 vs pN0/1 | 2.89 (1.69 to 4.94) | <0.001 | 2∙56 (1.47 to 4.48) | <0.001 |
AJCC/UICC sixth edition.
Significant p values (< 0.05) in bold.
Validation of the random forest gene signature in an independent external cohort
After successful internal validation of the random forest gene signature, we tested the signature in an independent external cohort (Pac-Ram cohort). In the Pac-Ram cohort, the random forest gene signature predicted that patients with Pac-Sensitive GC have longer PFS compared with patients with Pac-Resistant GC (median PFS 147 days vs 112 days, HR 0.48, 95% CI 0.25 to 0.91, logrank p=0.022) (figure 2C). In the Pac-Ram cohort, F-measure was 0.62, accuracy 0.64 and AUC was 0.88 (95% CI 0.68 to 1.0). This suggests that the random forest model trained on the Pac-S1 cohort identified a robust classifier which was able to predict paclitaxel benefit in both, the internal and the external validation cohorts. There was no association between the random forest gene signature and radiologically measured objective response rates in the Pac-Ram cohort.
To study the interaction of the random forest gene signature with paclitaxel treatment, we ran the model on the UFT alone and S-1 alone arms of the SAMIT study and compared results to the Pac-UFT arm. There was no survival difference between patients with Pac-Sensitive and Pac-Resistant GC in the UFT or S1 arms (HR 0.99, 95% CI 0.55 to 1.83, p=0.99)(supplementary figure S4). The test of interaction for the paclitaxel containing regimens with random forest gene signature was significant (p<0.001). The test for heterogeneity to validate the interaction of paclitaxel on DFS was significant (p<0.001). These findings suggest that the random-forest gene signature is predictive for survival benefit with paclitaxel treatment and is not merely a prognostic biomarker.
Transcriptomic characteristics of GC classified by the random forest gene signature
To understand the expression profiles of individual genes within the random forest gene signature, the 19 selected genes were studied in greater detail in the entire SAMIT dataset. The 19 genes in the signature include: CD209, TOP3B, BCL2, MCM2, RAD9A, IL10, HLA-DMB, MCM10, PTPRC, FANCM, ADORA2A, MS4A1, FANCG, CD3G, DSCR6, TBX21, ZWILCH, IL17A and FCGR3A. Heatmaps of genes split by Pac-Sensitive and Pac-Resistant GC revealed some differences in gene-expression profiles, although these are unlikely to have led to separate clusters using traditional unsupervised hierarchical clustering techniques (online supplemental figure 5A). Pairwise comparisons of genes revealed that some genes were significantly higher expressed in Pac-Sensitive GC compared with Pac-Resistant GC (or vice-versa), while others had similar median expression levels between the two groups (online supplemental figure 5B). These findings highlight the importance of considering multigene interactions for the generation of a signature, which may be achieved through machine-learning pattern recognition.
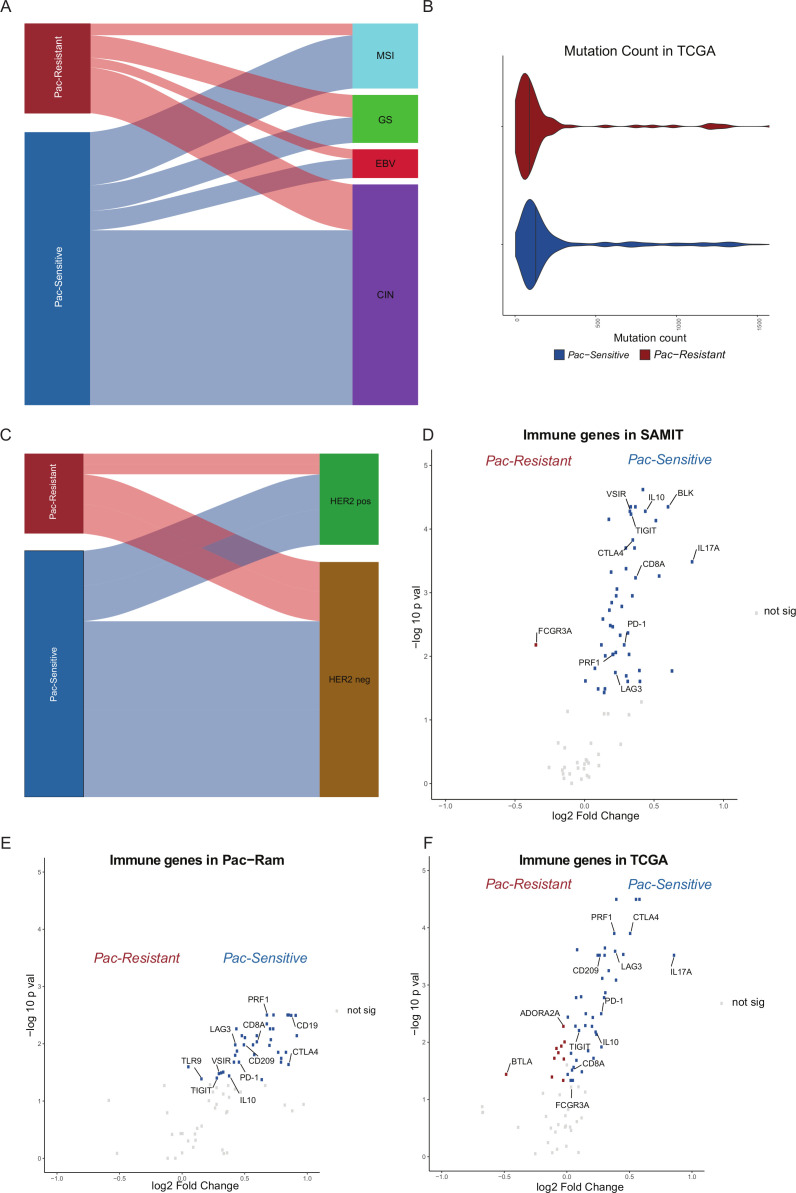
We tested the random forest gene signature on TCGA STAD samples (n=375), and 76% were classified as Pac-Sensitive. Of the four TCGA subtypes: chromosomal instable (CIN), genome stable (GS), microsatellite instable and Epstein-Barr virus associated,28 GS GC were more frequently classified as Pac-Resistant than Pac-Sensitive (25% vs 9%), while CIN GC were more frequently classified as Pac-Sensitive (64% vs 51%; Fisher’s exact p=0.0019) (figure 3A). Mutation counts were higher in the Pac-Sensitive group (Wilcoxon p=0.00031) (figure 3B). There were no differences in the HER2 status between Pac-Sensitive and Pac-Resistant GC (Fisher’s exact p=0.68) (figure 3C).
Figure 3.
Transcriptomic characteristics of random forest gene signature. (A) Alluvial plot of TCGA STAD samples by gastric cancer (GC) subtype. TCGA samples (n=375) were divided into Pac-Sensitive and Pac-Resistant by the random forest gene signature. These groups were correlated with TCGA GC subtypes chromosomal instable (CIN), genome stable (GS), microsatellite instable (MSI) and Epstein-Barr virus associated (EBV). There was a statistically higher proportion of GS patients in the Pac-Resistant group (Fisher’s exact p=0.0019). (B) Violin plot of mutation count between Pac-Sensitive and Pac-Resistant TCGA samples. Mutation count was higher in the Pac-Sensitive group (Wilcoxon p=0.00031). (C) Alluvial plot of TCGA STAD samples by HER2 status. Samples were correlated with HER2 status, with no significant difference. (D) Volcano plot comparing immune-related gene expression between Pac-Sensitive and Pac-Resistant in SAMIT. X-axis: log2 fold change (log2FC) of gene expression between Pac-Sensitive and Pac-Resistant. Y-axis: log10 adjusted p values after false discovery rate correction. Genes of interest have been annotated within the plot. Grey dots represent genes with similar expression in Pac-Sensitive and Pac-Resistant GC. Blue dots represent genes which are overexpressed in Pac-Sensitive GC. Red dots represent genes which are overexpressed in Pac-Resistant GC. (E) Volcano plot of immune-related genes comparing gene expression level between Pac-Sensitive and Pac-Resistant in Pac-Ram cohort. (F) Volcano plot of immune-related genes comparing gene expression level between Pac-Sensitive and Pac-Resistant in TCGA STAD samples. SAMIT, Stomach cancer Adjuvant Multi-Institutional group Trial; STAD, stomach adenocarcinoma; TCGA, The Cancer Genome Atlas.
Finally, the association between the Pac-Resistant/Pac-Sensitive groups and immune-related gene expression profiles was explored. The customised NanoString panel used in this study included 79 immune-related genes. In the SAMIT samples, Pac-Sensitive GC had a higher expression of markers of cytolytic T-cells such as CD8A and PRF1 (figure 3D).29 The expression of immune checkpoints genes such as CTLA-4, PD-1 and LAG3 was also higher in Pac-Sensitive GC. Similar associations with the expression of immune-related genes were also observed in the Pac-Ram and TCGA datasets (figure 3E, F).
Discussion
The 2×2 factorial trial design of the SAMIT study and the relatively large number of collected tumours provided a good experimental setting to search for biomarkers of paclitaxel benefit in patients with resectable GC. The SAMIT trial design allowed the creation of training and validation cohorts treated in a uniform manner with balanced patient and tumour characteristics between treatment arms. The presence of paclitaxel and non-paclitaxel containing treatment arms allowed us to distinguish between the predictive and prognostic value of potential biomarkers, and to identify treatment interactions specific to paclitaxel. Given the limited amount of FFPE tissue collected from the trial patients, we chose to create a customised NanoString panel to investigate the expression of a relatively large number of genes to identify biomarkers of paclitaxel benefit. The NanoString platform has proven to provide reliable results when performed on RNA extracted from FFPE tissue. Previous studies have shown good correlation between NanoString and RNASeq results in GC.30 31 Based on these data, we used RNA-seq data from a separate trial of GC patients treated with paclitaxel (and ramucirumab) as external validation.
A comprehensive literature review was performed to select the probes for the NanoString panel, covering a range of mechanisms of action of paclitaxel and cell death, GC-specific oncogenesis and other previously proposed predictive markers of taxane benefit. We chose a machine-learning model to identify a gene signature that was potentially predictive of paclitaxel benefit. The random forest method has been used by other groups as a reliable machine-learning algorithm for gene classifier creation.32 33 One advantage of the random forest method is the ability to incorporate a large number of variables (in this situation, genes) in the model. Few algorithms can process data with a variable size which is much larger than the number of samples in the training dataset and continue to demonstrate significant predictive performance.24
Recently, a four gene classifier was described to predict benefit of adjuvant chemotherapy in GC.34 This classifier was the first to suggest that GC patients classified as ‘immune high’ and low-risk derive no benefit from adjuvant cytotoxic chemotherapy after curative D2 gastrectomy. It is interesting to note that the gene signature identified in our study includes immune related genes such as CD209, a dendritic-cell marker, T-cell surface glycoprotein gamma chain CD3G and interleukin genes IL10 and IL17A.35 36 The Pac-Resistant and Pac-Sensitive GC also appeared to have different expression of other immune-related genes including immune checkpoints and markers of T-cell cytolytic activity.29 31 Other studies have suggested that patients with immune cell-rich colorectal cancers do not benefit from chemotherapy.37 Our random forest gene signature included cell-cycle checkpoint gene RAD9A and BCL2, a regulator of apoptosis.38 39 RAD9A is a component of the 9-1-1 DNA clamp, which has multiple roles in DNA repair. As Pac-Sensitive tumours had higher levels of RAD9A, it could be possible that paclitaxel induced DNA damage in the setting of high RAD9A expression increases mitotic cell death. RAD9A has also been reported to bind and neutralise BCL2, thereby inducing a proapoptotic state.40 Targeting the BCL2 pathway has been shown to reverse paclitaxel resistance in oesophageal cancer cell lines.41 TOP3B, a DNA topoisomerase enzyme included in our gene signature, was found to be prognostic when overexpressed in ovarian cancer.42 It is likely that there are interactions, yet to be fully defined, between several genes in these important pathways which subsequently lead to sensitivity or resistance to paclitaxel.
An important point of consideration is the similarities and differences between the SAMIT trial patients and the external Pac-Ram cohort. Both cohorts included GC patients treated with paclitaxel. However, in SAMIT, paclitaxel was combined with 5FU based chemotherapy, while in the Pac-Ram cohort paclitaxel was combined with ramucirumab, an anti-angiogenic monoclonal antibody. SAMIT was a study of patients treated in the adjuvant setting including earlier stage tumours, while Pac-Ram comprised patients with late stage GC treated in the metastatic setting after progression on first-line platinum-based chemotherapy. While DFS was used to measure survival in the SAMIT study, PFS was measured in the Pac-Ram clinical trial. Despite these differences, the random forest gene signature was able to predict paclitaxel benefit in both cohorts. The supporting data from the S-1/UFT arms suggests that the signature is not a prognostic biomarker. These findings suggest that this signature is relatively specific for predicting benefit from paclitaxel chemotherapy in GC. Our ability to analyse RNA from only a subset of the original trial population is a limitation of this study. However, clinicopathological variables and survival were similar between our biomarker study cohort and the original cohort. As the paclitaxel-containing cohorts in our study were combined with various other agents (S-1, UFT and ramucirumab), the effects of these agents on the performance of the random forest gene signature could not be fully determined due to the lack of a GC patient cohort treated with paclitaxel alone. The accuracy and F-measure in our internal validation cohort was moderate (0.61 and 0.71, respectively). The signature, however, was validated in an external cohort, and further study is required to improve these metrics. As a precedent, similar measurements have been seen in more robust and validated tests such as Oncotype Dx in breast cancer, a tool to predict for lack of benefit for chemotherapy in hormone receptor positive breast cancer.43 In the initial oncotype Dx report performed on National Surgical Adjuvant Breast and Bowel Project B-14 and B-20 studies, accuracy was 72% and F-measure was 76% (calculated from data provided in manuscript), which is similar to our study. Another limitation of this study is the post hoc nature of this translational analysis. A prospective clinical trial would be required to confirm the clinical utility of this gene signature.
In conclusion, this study is the first and largest performed to identify a gene signature predictive of paclitaxel benefit in GC. Given the increasing use of taxanes in GC in the adjuvant and perioperative setting, this biomarker has significant potential to guide clinicians in identifying patients with GC who might benefit from taxane based therapy. Validation of the gene signature in prospective trials is warranted.
Footnotes
Correction notice: This article has been corrected since it published Online First. Online supplementary table 1 has been added.
Collaborators: We are deeply grateful to Kazuaki Tanabe (Hiroshima University), Michiya Kobayashi (Kochi University), Shigehumi Yoshino (Yamaguchi University), Masazumi Takahashi (Yokohama Citizens Hospital), Nobuhiro Takiguchi (Chiba Cancer Center), Norio Mitsumori (Tokyo Jikeikai University), Kazumasa Fujitani (Osaka Prefectural General Medical Center), Ryoji Fukushima (Teikyo University), Isao Noguchi (Shikoku Cancer Center), Yoshihiro Kakechi (Kobe University), Naoki Hirabayashi (Hiroshima City Asa Citizens Hospital), Yukihiko Tokunaga(Osaka Kita Teishin Hospital), Akinori Takagane (Hakodate Goryokaku Hospital), and Kazuhiro Nishikawa (Osaka Medical Center) for providing clinical samples from the SAMIT trial.
Contributors: Conceptualisation: RS, NBK, MMH, ADJ, AT, PT. Data curation: RS, TO, KY, TY, YM, YR, MM, JG, JS, ST, AL-KT, CCYN, HG, JL and AT. Formal analysis: RS, NBK, YHC, JG, ST and MDS. Funding acquisition: RS, TO, AT, JL and PT. Methodology: RS, NBK, YHC, ADJ, JG, ST and PT. Project administration: RS, AL-KT, PT. Resources: TO, AT, JL and PT. Supervision: TO, HG, AT and PT. Visualisation: RS and NBK. Writing-original draft : RS, NBK. Writing-review and editing: YHC, JG, ST, HG and PT. Approval of final version of manuscript: all authors.
Funding: This work was supported by the Epidemiological & Clinical Research Information Network (ECRIN) and Kanagawa Standard Anti-Cancer Therapy Support System (KSATSS), which are non-profit organizations, JSPS KAKENHI Grant Numbers 842038 and 26461984, the Project Promoting Clinical Trials for Development of New Drugs (18lk0201061t0003 and 20lk0201061t0005) from the Japan Agency for Medical Research and Development (AMED), and a Grant-in-Aid for Scientific Research in Singapore. RS is supported by a National Medical Research Council (NMRC) Fellowship (NMRC/Fellowship/0059/2018), Singapore. PT is supported by Duke-NUS Medical School and the Genome Institute of Singapore, Agency for Science, Technology and Research. PT was also supported by the Cancer Science Institute of Singapore, NUS, under the National Research Foundation Singapore and the Singapore Ministry of Education under its Research Centres of Excellence initiative. This research was supported by the Singapore Ministry of Health’s National Medical Research Council under its Open Fund-Large Collaborative Grant (“OF-LCG”) (MOH-OFLCG18May-0003). This work was also supported by National Medical Research Council grants NR13NMR111OM, and NMRC/STaR/0026/2015.
Disclaimer: The funders of the study had no role in study design, data collection, analysis, interpretation, or writing of the manuscript. PT, TO and AT had full access to all data in the study and as corresponding authors had final responsibility for the decision to submit for publication.
Competing interests: RS: Advisory board: BMS, Merck, Eisai, Bayer, Taiho; honoraria for talks: MSD, Eli Lilly, BMS, Roche, Taiho; Travel funding: Roche, Astra Zeneca, Taiho, Eisai; Research funding: Paxman Coolers, MSD. These are outside the submitted work.TO: Research Funding: Taiho pharmaceutical, Chugai pharmaceutical, Ono pharmaceutical, Daiitisankyo pharmaceutical, Nippon Kayaku and Eli Lilly Japan K. K. Lecture fees: Nippon Kayaku, Ono pharmaceutical and Bristol-Myers Squibb K. K. Speaker Bureau: Taiho pharmaceutical, Chugai pharmaceutical, Ono pharmaceutical, Bristol-Myers Squibb K. K and Eli Lilly Japan K. K. These are outside the submitted work. TY: Lecture fees from: MSD, ONO, BMS, Taiho, Chugai, Daiichi-Sankyo, Lilly, Johnson & Johnson, Covidien and Olympus. Personal grant from Lilly. These are outside the submitted work. KY: Personal fees from Taiho Pharm and Bristol-Myers Squibb, during the conduct of the study; grants and personal fees from Asahi Kasei Pharma, Chugai Pharm., Covidien Japan, Daiichi Sankyo, Eisai, Eli Lilly Japan, Johnson & Johnson, MerkSerono, MSD, Nippon Kayaku, Novartis, Ono Pharm., Otsuka Pharm., Sanofi, Tsumura, Yakult Honsha, Takeda Pharm., grants from Abbott, Abbvie, Astellas, Biogen Japan, Celgene, GlaxoSmithKline, KCI, Kyowa Kirin, Meiji Seika Pharma, Toray Medical, Koninklijke Philips, personal fees from AstraZeneka, Denka, EA Pharma, Olympus, Pfizer, Sanwa Kagaku Kenkyusho, SBI Pharma, Teijin Phamra, TERUMO. These are outside the submitted work. YR: Speaker Bureau from; Daiichi-Sankyo, Johnson & Johnson, Otsuka, Lilly, Taiho pharmaceutical, Bristol-Myers Squibb. Research Funding: Taiho pharmaceutical, Abbott, Asahi Kasei, Daiichi-Sankyo, Tsumura & Co., Covidien, Zeria pharmaceutical, Otsuka, EA Pharma, Johnson & Johnson. These are outside the submitted work. YM: Lecture fees from AstraZeneca, Taiho, Chugai, and Daiichi-Sankyo. Consigned research fund from Toso company, Japan. These are outside the submitted work. MM: Research Funding from Chugai pharmaceutical, Teijin pharmaceutical, Daiitisankyo pharmaceutical, Takeda pharmaceutical, Terumo, Japan Lifelin, Senkod. These are outside the submitted work. ST: Lecture fee: Bayer Yakuhin, Amgen Astellas BioPharma K.K. Consultation fee: Boehringer Ingelheim. These are outside the submitted work. ADJ: honoraria from AstraZeneca, Janssen and MSD, travel funding from Perkin Elmer, and research funding from Janssen. These are outside the submitted work. HG: honoraria for participation in an expert meeting from MSD. These are outside the submitted work. AL-KT: Lecture fees Chugai Pharmaceutical. These are outside the submitted work. PT: Travel: Illumina, Research funding: Thermo Fisher, Kyowa Hakko Kirin. These are outside the submitted work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information. Contact Corresponding Author: PT: gmstanp@duke-nus.edu.sg for further data provision if required.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
All available samples were used in this translational substudy, which was approved by the individual local Institutional Review Boards. The translational study analysis was approved by the Domain Specific Review Board (DSRB), Singapore (Ethics approval Ref: 2019/00429). The South Korean trial protocol was approved by the Institutional Review Board of Samsung Medical Center (Seoul, Korea).
References
- 1. Sundar R, Tan P. Genomic analyses and precision oncology in gastroesophageal cancer: forwards or backwards? Cancer Discov 2018;8:14–16. 10.1158/2159-8290.CD-17-1295 [DOI] [PubMed] [Google Scholar]
- 2. Kang Y-K, Cho H. Perioperative FLOT: new standard for gastric cancer? Lancet 2019;393:1914–6. 10.1016/S0140-6736(18)33189-1 [DOI] [PubMed] [Google Scholar]
- 3. Kanagavel D, Fedyanin M, Tryakin A, et al. Second-line treatment of metastatic gastric cancer: current options and future directions. World J Gastroenterol 2015;21:11621–35. 10.3748/wjg.v21.i41.11621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Cutsem E, Moiseyenko VM, Tjulandin S, et al. Phase III study of docetaxel and cisplatin plus fluorouracil compared with cisplatin and fluorouracil as first-line therapy for advanced gastric cancer: a report of the V325 Study Group. J Clin Oncol 2006;24:4991–7. 10.1200/JCO.2006.06.8429 [DOI] [PubMed] [Google Scholar]
- 5. Al-Batran S-E, Homann N, Pauligk C, et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet 2019;393:1948–57. 10.1016/S0140-6736(18)32557-1 [DOI] [PubMed] [Google Scholar]
- 6. Kodera Y, Yoshida K, Kochi M, et al. A randomized phase III study comparing S-1 plus docetaxel with S-1 alone as a postoperative adjuvant chemotherapy for curatively resected stage III gastric cancer (JACCRO GC-07 trial). Journal of Clinical Oncology 2018;36:4007. 10.1200/JCO.2018.36.15_suppl.4007 [DOI] [Google Scholar]
- 7. Bajetta E, Floriani I, Di Bartolomeo M, et al. Randomized trial on adjuvant treatment with FOLFIRI followed by docetaxel and cisplatin versus 5-fluorouracil and folinic acid for radically resected gastric cancer. Ann Oncol 2014;25:1373–8. 10.1093/annonc/mdu146 [DOI] [PubMed] [Google Scholar]
- 8. Yamada Y, Boku N, Mizusawa J, et al. Phase III study comparing triplet chemotherapy with S-1 and cisplatin plus docetaxel versus doublet chemotherapy with S-1 and cisplatin for advanced gastric cancer (JCOG1013). J Clin Oncol 2018;36:4009. 10.1200/JCO.2018.36.15_suppl.4009 [DOI] [Google Scholar]
- 9. Tan IB, Ivanova T, Lim KH, et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology 2011;141:476–85. 85.e1-11. 10.1053/j.gastro.2011.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yong WP, Rha SY, Tan IB-H, et al. Real-Time Tumor Gene Expression Profiling to Direct Gastric Cancer Chemotherapy: Proof-of-Concept "3G" Trial. Clin Cancer Res 2018;24:5272–81. 10.1158/1078-0432.CCR-18-0193 [DOI] [PubMed] [Google Scholar]
- 11. Sundar R, Ng A, Zouridis H, et al. Dna methylation signature predictive of benefit from neoadjuvant chemotherapy in esophageal adenocarcinoma: results from the MRC OEO2 phase III trial. JCO 2019;37:43. 10.1200/JCO.2019.37.4_suppl.43 [DOI] [PubMed] [Google Scholar]
- 12. Ivanova T, Zouridis H, Wu Y, et al. Integrated epigenomics identifies BMP4 as a modulator of cisplatin sensitivity in gastric cancer. Gut 2013;62:22–33. 10.1136/gutjnl-2011-301113 [DOI] [PubMed] [Google Scholar]
- 13. Smyth E, Zhang S, Cunningham D, et al. Pharmacogenetic analysis of the UK MRC (medical Research Council) magic trial: association of polymorphisms with toxicity and survival in patients treated with perioperative epirubicin, cisplatin, and 5-fluorouracil (ECF) chemotherapy. Clin Cancer Res 2017;23:7543–9. 10.1158/1078-0432.CCR-16-3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsuburaya A, Yoshida K, Kobayashi M, et al. Sequential paclitaxel followed by tegafur and uracil (UFT) or S-1 versus UFT or S-1 monotherapy as adjuvant chemotherapy for T4a/b gastric cancer (SAMIT): a phase 3 factorial randomised controlled trial. Lancet Oncol 2014;15:886–93. 10.1016/S1470-2045(14)70025-7 [DOI] [PubMed] [Google Scholar]
- 15. Köhne CH, Peters GJ. Uft: mechanism of drug action. Oncology 2000;14:13–18. [PubMed] [Google Scholar]
- 16. Maehara Y. S-1 in gastric cancer: a comprehensive review. Gastric Cancer 2003;6(Suppl 1):2–8. 10.1007/s10120-003-0232-9 [DOI] [PubMed] [Google Scholar]
- 17. Weaver BA. How Taxol/paclitaxel kills cancer cells. Mol Biol Cell 2014;25:2677–81. 10.1091/mbc.e14-04-0916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Swanton C, Marani M, Pardo O, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell 2007;11:498–512. 10.1016/j.ccr.2007.04.011 [DOI] [PubMed] [Google Scholar]
- 19. Hodge JW, Garnett CT, Farsaci B, et al. Chemotherapy-induced immunogenic modulation of tumor cells enhances killing by cytotoxic T lymphocytes and is distinct from immunogenic cell death. Int J Cancer 2013;133:624–36. 10.1002/ijc.28070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kroemer G, Galluzzi L, Kepp O, et al. Immunogenic cell death in cancer therapy. Annu Rev Immunol 2013;31:51–72. 10.1146/annurev-immunol-032712-100008 [DOI] [PubMed] [Google Scholar]
- 21. Breiman L. Random forests. Mach Learn 2001;45:5–32. 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 22. Rijsbergen CJV. Information retrieval: Butterworth-Heinemann 1979.
- 23. Forman G, Scholz M. Apples-to-apples in cross-validation studies: pitfalls in classifier performance measurement. SIGKDD Explor Newsl 2010;12:49–57. [Google Scholar]
- 24. Alvarez S, Diaz-Uriarte R, Osorio A. A predictor based on the somatic genomic changes of the BRCA1/BRCA2 breast cancer tumors identifies the non-BRCA1/BRCA2 tumors with BRCA1 promoter hypermethylation. Clin Cancer Res 2005;11:1146–53. [PubMed] [Google Scholar]
- 25. Kim ST, Sa JK, Oh SY, et al. Comprehensive molecular characterization of gastric cancer patients from phase II second-line ramucirumab plus paclitaxel therapy trial. Genome Med 2021;13:11. 10.1186/s13073-021-00826-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deng M, Brägelmann J, Kryukov I, et al. FirebrowseR: an R client to the broad Institute's Firehose pipeline. Database 2017;2017. 10.1093/database/baw160. [Epub ahead of print: 06 Jan 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Z, Chen S, Feng W, et al. A pan-cancer analysis of HER2 index revealed transcriptional pattern for precise selection of HER2-targeted therapy. EBioMedicine 2020;62:103074. 10.1016/j.ebiom.2020.103074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qamra A, Xing M, Padmanabhan N, et al. Epigenomic promoter alterations amplify gene isoform and immunogenic diversity in gastric adenocarcinoma. Cancer Discov 2017;7:630–51. 10.1158/2159-8290.CD-16-1022 [DOI] [PubMed] [Google Scholar]
- 31. Sundar R, Huang KK, Qamra A, et al. Epigenomic promoter alterations predict for benefit from immune checkpoint inhibition in metastatic gastric cancer. Ann Oncol 2019;30:424–30. 10.1093/annonc/mdy550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kursa MB. Robustness of random Forest-based gene selection methods. BMC Bioinformatics 2014;15:8. 10.1186/1471-2105-15-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kong Y, Yu T. A deep neural network model using random forest to extract feature representation for gene expression data classification. Sci Rep 2018;8:16477. 10.1038/s41598-018-34833-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheong J-H, Yang H-K, Kim H, et al. Predictive test for chemotherapy response in resectable gastric cancer: a multi-cohort, retrospective analysis. Lancet Oncol 2018;19:629–38. 10.1016/S1470-2045(18)30108-6 [DOI] [PubMed] [Google Scholar]
- 35. Liu L, Chen Z, Shi W, et al. Breast cancer survival prediction using seven prognostic biomarker genes. Oncol Lett 2019;18:2907–16. 10.3892/ol.2019.10635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wirth JR, Molano I, Ruiz P, et al. Tlr7 agonism accelerates disease and causes a fatal myeloproliferative disorder in NZM 2410 lupus mice. Front Immunol 2019;10:3054. 10.3389/fimmu.2019.03054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Angell HK, Bruni D, Barrett JC, et al. The immunoscore: colon cancer and beyond. Clin Cancer Res 2020;26:332–9. 10.1158/1078-0432.CCR-18-1851 [DOI] [PubMed] [Google Scholar]
- 38. Broustas CG, Hopkins KM, Panigrahi SK, et al. RAD9A promotes metastatic phenotypes through transcriptional regulation of anterior gradient 2 (AGR2). Carcinogenesis 2019;40:164–72. 10.1093/carcin/bgy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch Toxicol 2015;89:289–317. 10.1007/s00204-014-1448-7 [DOI] [PubMed] [Google Scholar]
- 40. Komatsu K, Miyashita T, Hang H, et al. Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat Cell Biol 2000;2:1–6. 10.1038/71316 [DOI] [PubMed] [Google Scholar]
- 41. Shi X, Dou Y, Zhou K, et al. Targeting the Bcl-2 family and P-glycoprotein reverses paclitaxel resistance in human esophageal carcinoma cell line. Biomed Pharmacother 2017;90:897–905. 10.1016/j.biopha.2017.04.043 [DOI] [PubMed] [Google Scholar]
- 42. Bai Y, Li L-D, Li J, et al. Targeting of topoisomerases for prognosis and drug resistance in ovarian cancer. J Ovarian Res 2016;9:35. 10.1186/s13048-016-0244-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–26. 10.1056/NEJMoa041588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2021-324060supp001.pdf (159.1KB, pdf)
gutjnl-2021-324060supp004.xlsx (1.6MB, xlsx)
gutjnl-2021-324060supp003.pdf (2MB, pdf)
gutjnl-2021-324060supp002.pdf (132.9KB, pdf)
Data Availability Statement
Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information. Contact Corresponding Author: PT: gmstanp@duke-nus.edu.sg for further data provision if required.