Abstract
Objectives
Rheumatoid arthritis (RA) is an autoimmune disease strongly associated with the major histocompatibility complex (MHC) class II allele DRB1*04:01, which encodes a protein that binds self-peptides for presentation to T cells. This study characterises the autoantigen-presenting function of DRB1*04:01 (HLA-DRA*01:01/HLA-DRB1*04:01) at a molecular level for prototypic T-cell determinants, focusing on a post-translationally modified collagen type II (Col2)-derived peptide.
Methods
The crystal structures of DRB1*04:01 molecules in complex with the peptides HSP70289-306, citrullinated CILP982-996 and galactosylated Col2259-273 were determined on cocrystallisation. T cells specific for Col2259-273 were investigated in peripheral blood mononuclear cells from patients with DRB1*04:01-positive RA by cytofluorometric detection of the activation marker CD154 on peptide stimulation and binding of fluorescent DRB1*0401/Col2259-273 tetramer complexes. The cDNAs encoding the T-cell receptor (TCR) α-chains and β-chains were cloned from single-cell sorted tetramer-positive T cells and transferred via a lentiviral vector into TCR-deficient Jurkat 76 cells.
Results
The crystal structures identified peptide binding to DRB1*04:01 and potential side chain exposure to T cells. The main TCR recognition sites in Col2259-273 were lysine residues that can be galactosylated. RA T-cell responses to DRB1*04:01-presented Col2259-273 were dependent on peptide galactosylation at lysine 264. Dynamic molecular modelling of a functionally characterised Col2259-273-specific TCR complexed with DRB1*04:01/Col2259-273 provided evidence for differential allosteric T-cell recognition of glycosylated lysine 264.
Conclusions
The MHC-peptide-TCR interactions elucidated in our study provide new molecular insights into recognition of a post-translationally modified RA T-cell determinant with a known dominant role in arthritogenic and tolerogenic responses in murine Col2-induced arthritis.
Keywords: rheumatoid arthritis, autoimmunity, T-lymphocyte subsets
Key messages.
What is already known about this subject?
Rheumatoid arthritis (RA) is closely associated with HLA-DRB1*04:01-encoded major histocompatibility complex II molecules.
DRB1*04:01-restricted CD4+ T-cell responses to citrullinated autoantigens and post-translationally modified collagen II (Col2) have been described.
What does this study add?
This study contributes new crystal structure information that reveals the DRB1*04:01 function in presenting post-translationally modified antigenic determinants to T cells in patients with RA.
Through comparative analysis of RA T cells, T cell receptor (TCR) cloning and molecular modelling of a prototypic trimolecular complex, we gained insights into TCR recognition of unmodified and glycosylated Col2 in a DRB1*04:01 context.
How might this impact on clinical practice or future developments?
Our findings contribute to a better understanding of the role of posttranslational Col2 modification for TCR recognition in CD4+ T cells and has potential implications for induction of tolerance and onset of pathogenic autoimmunity.
Introduction
Rheumatoid arthritis (RA) is a chronic, inflammatory autoimmune disease targeting diarthrodial cartilaginous joints. The disease is believed to be initiated by the development of autoantibodies to various altered self-antigens, predominantly modified by citrullination for yet unknown reasons,1–4 followed years later by onset of joint inflammation and spreading of autoimmune responses to new structures, including cartilage proteins.5–7 The strong association of RA with major histocompatibility complex class II (MHCII) suggests activation of T cells and a maturation of an autoreactive B cell response, capable of orchestrating the immune attack on the joints.8 The origin of the T-cell activation is unknown but is likely to be dependent on presentation of antigenic peptides bound to the MHCII molecules. These peptides could have been derived from non-self-proteins, for example, from infectious agents giving help to autoreactive B cells, or could be modified self-peptides that have escaped tolerance selection.
The MHCII association with RA has been mapped to the DRB1 locus,9 10 which encodes a DR β-chain (DRB) and forms a peptide-binding receptor together with an invariant α-chain (DRA).11 According to a popular hypothesis, alleles of the highly polymorphic DRB1 locus associated with RA encode a shared peptide binding pocket for a selected set of self-peptides, thereby predisposing individuals to pathogenic T-cell activation.12 13 Most of the RA-associated DRB1 molecules have positively charged amino acids at position 71, favouring interactions with peptides that contain a negatively charged amino acid at the P4 position. Although a favoured binding of peptides with citrulline at this position (in contrast to a positively charged arginine) has been proposed,13 14 this hypothesis could not be confirmed in studies on larger sets of peptides.15 16
A potentially relevant self-antigen in arthritis pathogenesis is type II collagen (Col2) due to its abundance in cartilage and proven role as an arthritogenic immunogen in experimental arthritis.17 Moreover, antibodies to native and citrullinated Col218–22 and Col2-specific T cells18 are detectable in patients with RA. Interestingly, RA T-cell responses directed to the dominant Col2259-273 peptide were restricted by the RA-associated DR alleles 0401 and 0101, which were demonstrated to confer susceptibility to collagen-induced arthritis on transgenic expression in mice.23–25 However, it is crucial to consider that the major Col2259-273 peptide can be both hydroxylated and galactosylated at lysine residues and that RA T cells predominantly recognise the galactosylated form.18 Initial insight into the positioning of the galactosylated (gal) Col2259-273 peptide (galCol2259-273) in the binding pocket of the DRB1*04:01 molecule was provided by molecular modelling.26 27 The same target peptide and its post-translational modification (PTM) is also recognised by arthritogenic T cells in mice, provided that they express the natural murine antigen MHCII allele or transgenic human DRB1*04:01 or DRB1*01:01 molecules.18 23–25 Critical differences in central tolerance induction of T cells based on their specificity for unmodified (nCol2259-273) or galactosylated Col2259-273 (galCol2259-273)28 and in the tolerogenic potential of Col2 peptide vaccination against experimental arthritis have been observed in mice.29
To further elucidate the role of PTM in Col2 recognition by human T-cell receptors (TCR), we performed a comparative study of Col2259-273 recognition in either galactosylated or unmodified form using binding of tetramerised recombinant DRB1*04:01/Col2-peptide complexes and analysed Col2 peptide-induced T-cell activation in peripheral blood mononuclear cells (PBMCs) from patients with DRB1*04:01-positive RA. Moreover, we solved the X-ray crystallographic structure of DRB1*04:01(HLA-DRA1*01:01/HLA-DRB1*04:01) complexed with galCol2259-273 to gain the first molecular insights into the structural basis of PTM-dependent differential Col2 recognition. Together with sequence information obtained from a cloned human galCol2-specific TCR, these crystal structures allowed us to perform molecular modelling on the trimolecular MHCII/peptide/TCR complex.
Materials and methods
See online supplemental material.
annrheumdis-2021-220500supp001.pdf (749.4KB, pdf)
Results
The molecular complex of DRB1*04:01 with bound peptides
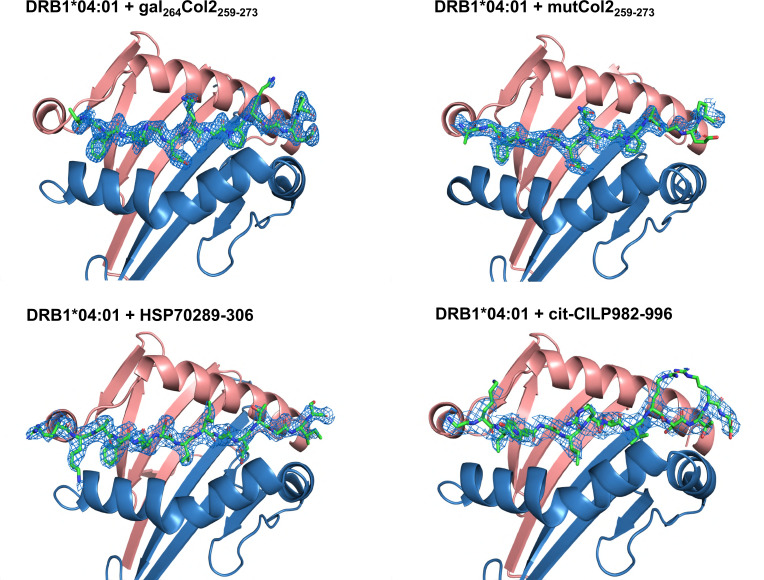
The investigated peptides nCol2259-273(GIAGFKGEQGPKGET),18 heat shock protein (HSP) HSP70289-306(TRKPFQSVIADTGISV) and citrullinated (cit) cartilage-intermediate protein (CILP) citCILP982-996 (GKLYGI[Cit]DV[Cit]STRDR) represent autoantigenic determinants recognised by T cells in RA.30 31 To establish the structural basis of their autoimmune recognition, we solved the crystal structures of DRB1*04:01 complexed with Col2259-273, mutated Col2259-273 containing alanine replacements at 264 (K264A, at P2) and 270 (K270A, at P8), HSP70289-306 and the CILP982-996 peptide citrullinated at positions 988 and 991 (figures 1 and 2, and online supplemental figure S1). All investigated T cell determinants, either unmodified or altered by citrullination or galactosylation, were bound in a conserved linear and extended conformation located in the classic binding groove of DRB1*04:01, thereby closely resembling previously solved DRB1*04:01 structures.32
Figure 1.
Top view of the DRAxDRB1*04:01 molecule binding peptides. The DRA (alpha chain) is shown in pink and the DRB (beta-chain) shown in blue. The 2Fo-Fc electron density map of each peptide is shown in blue mesh contoured at 1 σ.
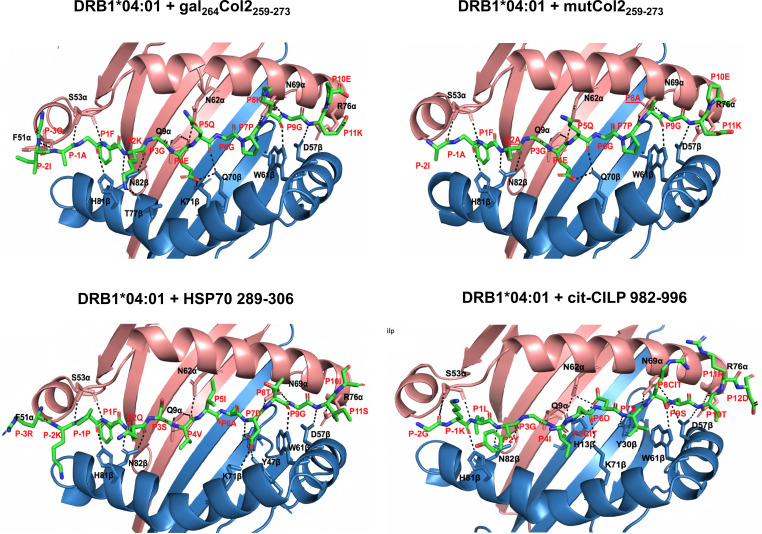
Figure 2.
Interaction of peptides with the DRAxDRB1*04:01 molecule. Hydrogen bonds are indicated by dashed black lines. Peptide residues are numbered in accordance with the numbering of the binding pockets. The residues from both α and β chains important for contacts with the peptide are represented as sticks.
The crystal structure of Col2259-273-bound DRB1*04:01 was determined at 1.9 Å resolution (online supplemental table S1). A weakly bound mutated human CLIP peptide (PVSKARMATGALAQA) occupying the DRB1*04:01 binding groove was exchanged with a synthetic glycosylated Col2259-273 (mono-[β-D-galactopyranosyl]-moiety at a lysine [K264] side chain) prior to protein crystallisation. However, we did not observe electron density for the galactose moiety electron density in the structure, most likely because it is mobile at its solvent-exposed position on the protein surface. As expected, the peptide occupied the P1, P4, P6, P7 and P9 pockets with P1-Phe, P4-Glu, P6-Gly, P7-Pro and P9-Gly, respectively, whereas the potential TCR contact residues are P2-Lys, P5-Gln and P8-Lys. We also determined DRB1*04:01 in complex with covalently attached mutated Col2259-273 (online supplemental table S1, figures 1 and 2, online supplemental figure S1 and figure S2). To investigate a possible influence on peptide binding by the two potential TCR contact residues P2-Lys and P8-Lys, we mutated them to Ala. We have previously found that the mutation of P2 from Lys to Ala slightly decreased the affinity of the peptide with DRB1*04:01, but also abolished T cell reactivity.25 As can be seen from the structure, the mutated peptide mimics the conformation and location of the wildtype peptide (online supplemental figure S2), but its P4-Glu did not engage with Lys71β, thus explaining the reduced affinity.
The HSP70289-306 peptide binds in a linear, extended manner with P1-Phe, P4-Val, P6-Ala and P9-Gly occupying the P1, P4, P6 and P9 pockets of DRB1*04:01, respectively, whereas P2-Gln, P5-Ile, P7-Asp, P8-Thr and P10-Ile represented potential TCR contact sites. The Gln70β within the shared epitope motif does not contact P6-Ala as seen in DRB1*04:01 where Gln70β hydrogen bonds to both P4E and P6G of Col2259-273. In contrast, P7-D is bound by both Lys71β and Tyr47β.
As expected, the citCILP982-996 peptide also binds in a linear, extended manner with P1-L, P4-I, P6-D and P9-S interacting with the DR molecules. Thus, the citrulline does not bind to the P4 pocket but instead is likely to face the TCR.
An overall comparison of the binding sites confirmed that the strongest DR binding site had a hydrophobic amino acid in the P1 position, whereas a considerable degree of flexibility was allowed at other positions. An important DR binding site is P4, in which an acidic side chain (glutamic acid) is favoured as it interacts with basic amino acids at position 71 in the beta chain, in line with the known association with RA. In contrast, the citrulline side chains of the CILP peptide did not bind within the P4 pocket. The TCR recognition sites for the Col2 peptide were the lysines at P2 and P10 as well as the glutamine at P5.
Detection of T lymphocytes specific for Col2259-273
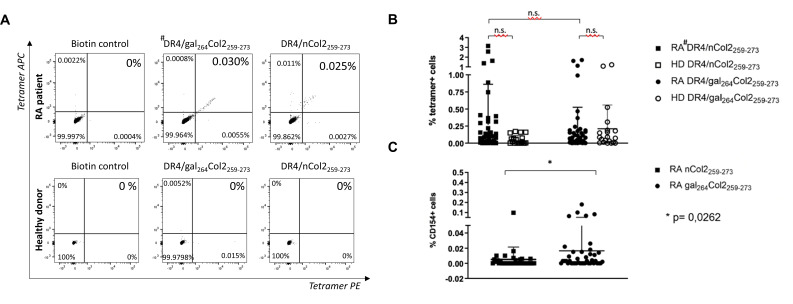
DRB1*04:01 allele carriers (patients with RA and healthy donors) were investigated for the presence of Col2259-273 peptide (nCol2 or gal264Col2)-specific CD4+ T cells in peripheral blood by flow cytometric analysis of tetramer-stained PBMCs. A representative result in figure 3A shows the detection of Col2 epitope-specific cells that stain double-positive for two identical but differently fluorescence-labelled DRB1*04:01/Col2 tetramer complexes. The frequency of double-positive cells was 0.030% for the gal264Col2259-273-peptide and 0.025% for the nCol2259-273-peptide containing tetramers in the total CD4+ T-lymphocyte population. In a concomitantly analysed blood sample from a healthy donor, no DRB1*04:01/Col2-specific T cells could be identified.
Figure 3.
Detection of human antigen-specific T cells in the peripheral blood of HLA-DRB1*04:01 carriers by flow cytometry using #DR4 (HLADRB1*04:01)-tetramers with specificity for the gal264Col2259-273 or nCol2259-273 peptide. (A) Representative dot blots: the CD4+ enriched T cells from PBMCs were stained with a dead/live marker and DRB1*04:01/Col2 peptide tetramers conjugated with two different fluorophores (PE and APC). Subsequent flowcytometric analysis reveals the Col2-specific cells in the live double positive stained subpopulation. Representative dot blots of specific DR4/gal264Col2 and DR4/nCol2 tetramer binding to T helper cells in samples from a patient with RA and a HD are shown. Biotin-streptavidin complexes without a specific peptide conjugate served as a negative control. (B) Specific tetramer binding of T helper cells from patients with RA (n=55) and HD (n=20) using #DR4 (DRB1*04:01)-tetramers with different peptide specificity (gal264Col2 vs nCol2). In PBMCs of patients with RA, the frequencies of DR4/nCol2 and DR4/gal264Col2 staining CD4+T cells do not differ significantly (n.s.). Depicted values represent processed data in which for each tetramer staining datapoint the respective biotin background has already been subtracted from the raw value. (C) Detection of antigen-specific T cells in PBMCs of patients with RA (HLA-DRB1*04:01) on in vitro stimulation by synthetic Col2 peptides using flow cytometry. PBMCs from patients with RA were stimulated with a Col2 peptide (gal264Col2, nCol2) and anti-CD40 for 7 hours. Subsequently, peptide-induced upregulation of the activation marker CD154 on the surface of the live CD4+T cell population was detected by flow cytometry. The frequency of CD154-positive CD4-positive T helper cells in PBMCs was significantly elevated in response to vitro challenge by the galCol2 peptide (n=41) compared with unmodified nCol2 (n=35, p=0.0262). Statistical significance was determined using the Mann-Whitney test. APC, allophycocyanin; HD, healthy donors; PBMC, peripheral blood mononuclear cells; PE, phycoerythrin; RA, rheumatoid arthritis; TCR, T-cell receptor.
The studies on our entire sample size using DRB1*04:01 tetramers containing either unmodified nCol2 or gal264Col2259-273 peptide confirmed detectability of antigen-specific T cells in PBMCs from patients with RA at the expected low precursor frequencies. The frequencies of DRB1*04:01/nCol2-staining and DRB1*04:01/gal264Col2-staining CD4+ T cells staining CD4+ T cells were identical: 27.27%. Staining positivity was defined by a value exceeding the threshold set by the mean of negative biotin control +3 SD (see Methods section). Interestingly, CD4+ T cells staining with the DRB1*04:01/nCol2-tetramers as well the DRB1*04:01/gal264Col2-tetramers were also detectable at a percentage of 5% and 30%, respectively, in the small cohort of healthy DRB1*04:01 carriers (figure 3B). No significant differences were detectable between the RA and healthy donor groups (figure 3B). In this respect, the small sample size constitutes a certain constraint of our study mainly due to limited access to biomaterial from HLA-typed healthy blood donors.
Interestingly, the DRB1*04:01/Col2-tetramer-positive T-cell population detectable in the peripheral blood of patients with RA exhibited a difference in responsiveness to in vitro stimulation of PBMCs with synthetic Col2 peptides. Stimulation with the gal264Col2 peptide resulted in an elevated frequency of antigen-activated CD4+ T lymphocytes compared with nCol2 as determined by peptide-induced upregulation of the activation marker CD154 detected by flow cytometry (figure 3C). Taken together, the results demonstrate a functional impact of the Col2 peptide structure presented in the context of a DRB1*04:01-encoded MHCII molecule on autoimmune recognition by CD4+ T cells in the peripheral blood of patients with RA.
Analysis of a cloned TCR derived from a single sorted Col2-reactive T lymphocyte
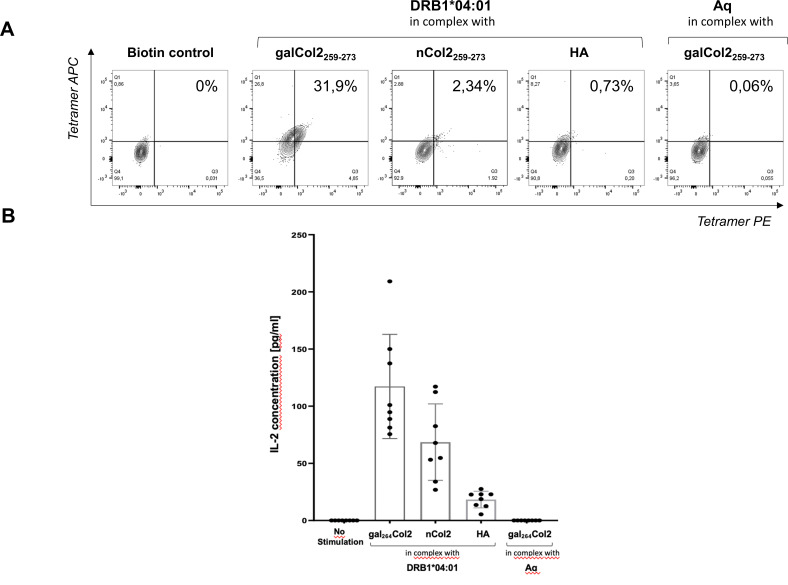
We next aimed to characterise TCRs with DRB1*04:01-restricted recognition of Col2259-273. Single cells of in vitro expanded gal264Col2259-273 peptide-reactive CD4+ T lymphocytes from patients with DRB1*04:01-positive RA were sorted according to staining with DRB1*04:01/gal264Col2259-273 tetramers and used for Vα-TCR and Vβ-TCR gene amplification by PCR. A prototypic TCR (TCR#16), for which we obtained the complete cDNA sequence of the paired α-chain and β-chain (see online supplemental figures S3 and S4), was further characterised by recombinant expression in the TCR-deficient Jurkat 76 cell line on lentiviral gene transfer (see online supplemental figure S5 for studies on two additionally transduced human TCRs). As shown in figure 4A, TCR-transduced Jurkat cells exhibited a specific positive staining with the DRB1*04:01/gal264 Col2259-273 tetramers and at a clearly reduced level with DRB1*04:01/nCol2259-273 tetramers, whereas control constructs consisting either of MHCII complexes in which DRB1*04:01 is replaced by the murine analogue Aq (Aq/gal264Col2259-273) or DRB1*04:01 complexed with the influenza hemagglutinin (HA) peptide (HA306-318: PKYVKQNTLKLAT) (DRB1*0401/HA-peptide) remained negative. In addition, Jurkat cells transduced with a human HA-specific TCR (HA1.7)32 stained positive with DRB1*04:01/HA306-318 while remaining negative when stained with the DRB1*04:01/gal264Col2259-273 tetramer (data not shown).
Figure 4.
Binding of DRB1*04:01 tetramers to TCR-deficient Jurkat cells and induction of IL-2. (A) Flow cytometric analysis of binding of DRB1*04:01/gal264Col2259-273 tetramers to TCR-deficient Jurkat 76 cells after gene transfer of the cloned α and β chains of TCR #16. Transduction of a TCR-deficient Jurkat 76 cell line was performed with a lentiviral vector encoding the α and β chains of TCR #16 cloned from a single-cell sorted CD4+ gal264Col2-specific T cell. Transduced Jurkat cells were stained with a dead/live marker and DRB1*04:01/peptide tetramers conjugated with two different fluorophores (PE and APC). Flow cytometric analysis revealed tetramer-specific cells in the live double-positive stained subpopulation. A biotin-streptavidin complex without a specific peptide conjugate served as negative control. (B) Induction of specific IL-2 responses in transduced Jurkat 76 cells expressing the human TCR #16 receptor by stimulation with DRB1*04:01/Col2259-273 peptide complexes. Transduced Jurkat cells were incubated with soluble DRB1*04:01/peptide complexes for 24 hours. Specific activation of cells via the TCR was measured by induced IL-2 release specific capture ELISA. Unstimulated cells served as a negative control. The MHCII restriction of TCR was performed by stimulation with the murine Aq/gal264Col2 peptide complex. Bars indicate mean values, lines indicate SD and dots represent separate experiments. APC, allophycocyanin; Aq, murine MHCII allele; HA, influenza hemagglutinin 306–318; IL, interleukin; PE, phycoerythrin; TCR, T-cell receptor.
Subsequent functional studies using lentiviral gene transfer from a single sorted gal264Col2259-273-specific T-cell of a patient with HLA-DRB1*04:01-positive RA demonstrated the selective capability of recombinant monomeric DRB1*04:01/Col2-peptide complexes to induce IL-2 production in TCR-reconstituted Jurkat 76 cells (figure 4B). The challenge with monomeric DRB1*04:01/gal264Col2259-273 induced the strongest IL-2 response. Stimulation with DRB1*04:01 complexes containing the nCol2 peptide resulted in a considerably reduced IL-2 release that nevertheless clearly exceeded the levels induced by DRB1*04:01/HA306-318 or Aq/gal264Col2259-273 control complexes (figure 4B). To confirm this result, we tested antigen-presenting cells (APCs) homozygously expressing DR*0401 obtained from DRB1*04:01 knock-in mice as well as APCs from the peripheral blood of DR*0401individuals (online supplemental figure S6). The presentation of gal264Col2259-273 in a DRB1*04:01 context on the surface of either murine or human fixed APC after preloading with peptides was specifically recognised by TCR#16 mRNA transfected nuclear factor of activated T cells (NFAT) luciferase Jurkat reporter cells and associated with stronger NFAT activation compared with the stimulatory effect of the nCOL2 peptide under identical conditions. The CLIP control peptide did not lead to any activation of the TCR mRNA transfected Jurkat reporter cells. Accordingly, these results are in agreement with the studies on the specificity of tetramer-induced IL-2 responses via the recombinantly expressed TCR #16 in lentivirally transduced Jurkat 76 cells lacking an endogenous TCR.
Modelling of molecular interactions in the trimolecular complex of the DRB1*04:01, Col2259-273 peptide and TCR
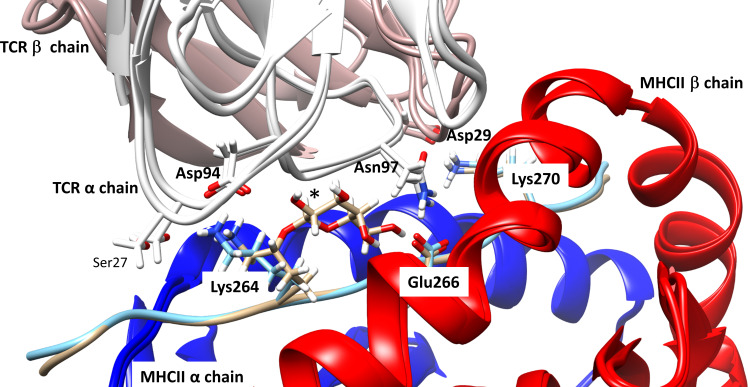
Based on the identified sequence of the human Col2-specific TCR#16 and the solved crystal structure of the DRB1*04:01/Col2259-273 complex, molecular modelling was performed using the template of a published TCR cocrystallised with an influenza peptide-containing DRB1*04:01 molecule (HA1.7).32 The overview of the entire modelled 3D structure of the multicomponent system consisting of the DRB1*04:01/Col2 peptide/TCR complex is shown in online supplemental figure S7. More detailed insights into critical amino acid residues involved in interactions between the unmodified or gal264Col2 peptides and TCR #16 variable regions in the DRB1*04:01 complex are provided in figure 5, which depicts the superimposition of the minimised starting geometries for the two trimolecular complexes. Molecular interaction of the Col2 peptide with the TCR occurs via three side chain bonds irrespective of Col2259-273 galactosylation. Two interactions involve the CDR3 region of the TCRα-chain (Asp94—Lys264 [Col2259-273] and Asn97—Glu266 [Col2259-273]) and an additional salt bridge involves the CDR1 region of the TCRβ-chain (Asp29—Lys270 [Col2259-273]). The galactose residue at Lys264 is in close contact with the TCRα-CDR3 backbone but is not involved in side chain interactions.
Figure 5.
Molecular model of the three-dimensional structure of the multicomponent DRB1*04:01/Col2 peptide/TCR complex. Superimposition of minimised starting geometries for the trimolecular complexes of TCR#16 with DRB1*04:01 containing either the unmodified (K264) or galactosylated (gal264) version of the Col2259-273 peptide. The image depicts three side chain bonds irrespective of Col2259-273 galactosylation: two involving the CDR3 region of the TCRα-chain, Asp94 - Lys264 (Col2259-273) and Asn97—Glu266 (Col2259-273) and one in the CDR1 region of the TCRβ-chain, Asp29—Lys270 (Col2259-273). The galactose (*) residue at Lys264 is not involved in side chain interactions with the TCR. TCR, T-cell receptor.
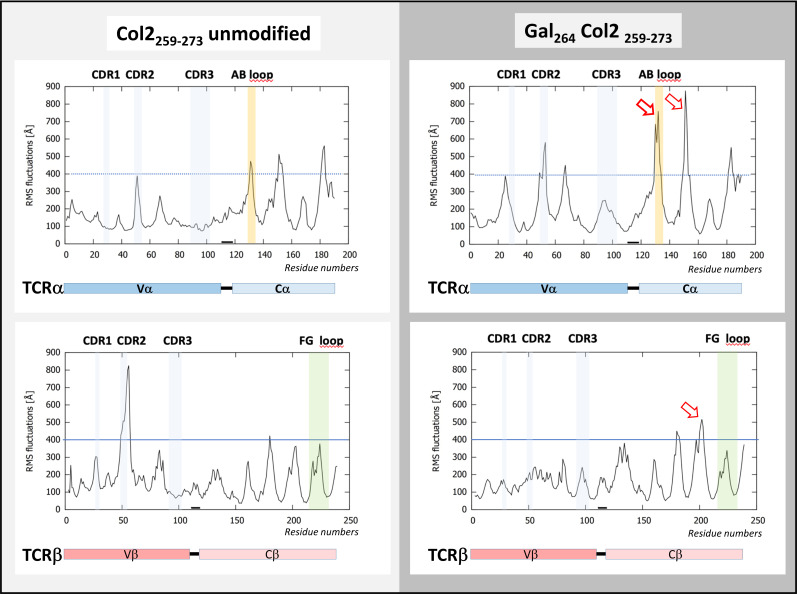
Additional insight was provided by comparative molecular dynamic simulations of both trimolecular complexes (unmodified or galactosylated Col2 peptide). Snapshots at 950 ns of dynamic modelling revealed critical differences imposed by the galactosyl residue at lysine 264 in the Col2 peptide (figure 6). TCR#16 interaction with the complex containing nCol2 at the initial salt bridge Lys264—Asp94 (TCRα-CDR3) caused the complex to open up to allow neoformation of a bond to Asn97 (TCRα-CDR3). Consequently, Glu266 of Col2259-273 formed a new salt bridge with Lys71 in the β-chain of DRB1*04:01 at expense of the initial bonding to Asn97 (TCRα-CDR3) (figure 6A). By contrast, the presence of a galactose residue at position 264 allowed formation of a hydrogen bond to the TCRα-CDR3 backbone, also resulting in the stabilisation of both side chain bonds likely due to limitation in lysine 264 mobility (figure 6B). The stabilising allosteric effect is depicted in figure 7, which shows a comparative overview of all intermolecular bonds formed in the trimolecular complexes consisting of TCR#16 and DRB1*04:01 associated with either nCol2 or gal264Col2. The graphic illustrates that the galactose is the only moiety bonded to all three molecules (the Col2 peptide, the DRB1*04:01 β-chain and the TCR). The molecular dynamics simulations for a 1000 ns period exhibited a rather low degree of molecular fluctuation in the TCR V-regions contacting the DRB1*04:01/Col2 peptide complex (figure 8). An exception was a peak of molecular mobility detectable in a solvent-exposed loop with reduced protein contacts carboxyterminal of TCRβ-CDR2 in the complex with nCol2-bound DRB1*04:01 (figure 8). Even more remarkable was the increase in molecular fluctuations in the constant region of TCR#16 affecting a Cβ domain just proximal to the so-called FG-loop and a Cα region that included the AB-loop.33 The molecular flexibility of these functional domains, which localise near ectodomains of the signal-transducing CD3 membrane complex,33 was clearly more pronounced in the trimolecular complex containing gal264Col2 (figure 8).
Figure 6.
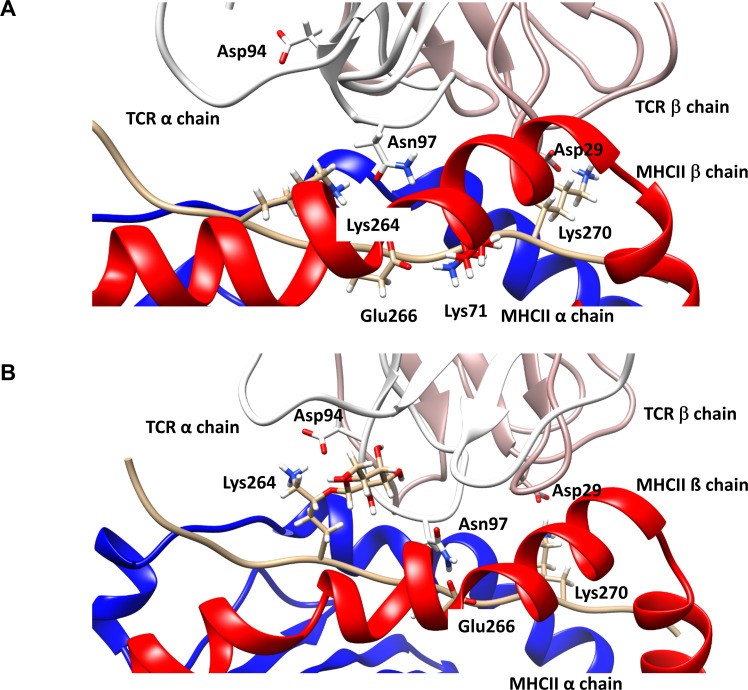
Geometry of the trimolecular complexes after 950 ns of molecular dynamics simulation. (A) Trimolecular complex consisting of TCR#16 and DRB1*04:01 complexed with the unmodified Col2259-273 peptide. The initial salt bridge Lys264 - Asp94 (TCRα-CDR3) breaks to allow for neoformation of a bond between Lys264 of Col2259-273 and Asn97 (TCRα-CDR3). Glu266 of Col2259-273, which initially interacted with Asn97 in the TCRα (figure 5), now forms a salt bridge with Lys71 in the β-chain of DRB1*04:01, whereas the initial bond between Lys270 (Col2259-273)-Asp29 (TCRβ-CDR1) remains preserved. (B) Trimolecular complex consisting of TCR#16 and DRB1*04:01 complexed with the gal264Col2259-273 peptide. The initial salt bridge Lys264 - Asp94 (TCRα-CDR3) remains intact, likely due to the stabilising impact of the sugar ring by decreasing the mobility of Lys264. The galactose ring in close contact to the CDR3 backbone of TCRα forms a hydrogen bond to the backbone. The two other side chain bonds of the starting geometry (figure 5), Glu266 (gal264Col2259-273)-Asn97 (TCRα-CDR3) and Lys270 (gal264Col2259-273)-Asp29 (TCRβ-CDR1), remain detectable. CDR, complementarity determining region; MHC, major histocompatibility complex; TCR, T-cell receptor.
Figure 7.
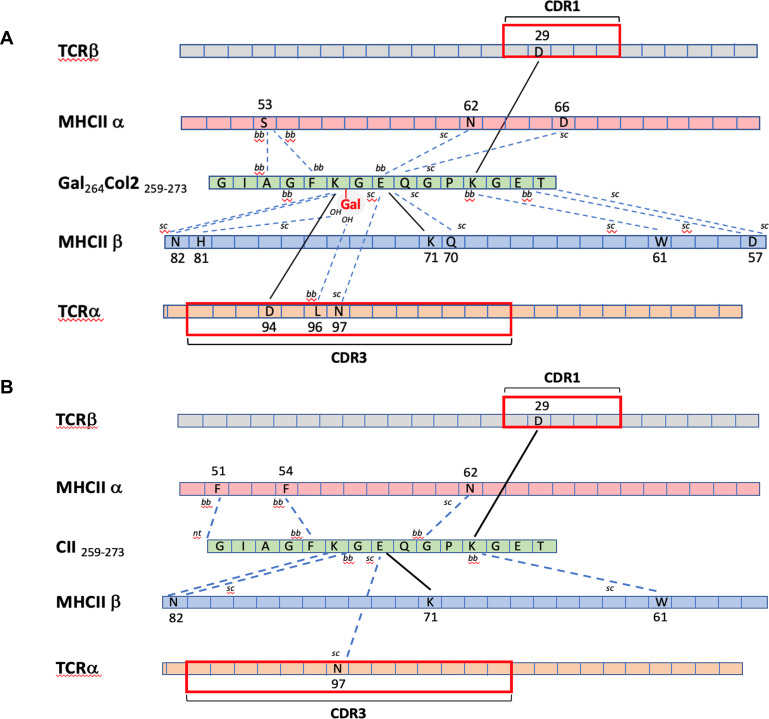
Comparative overview of intermolecular bonds formed in the trimolecular complexes of TCR#16 and DRB1*04:01 (MHCII) associated with either (A) gal264Col2 or (B) nCol2. Amino acids are shown in one letter code and their positions in the respective protein sequences indicated by number. Bold lines indicate salt bridges and dashed lines indicate hydrogen bonds. ai, aromatic interaction; bb, backbone; CDR, complementarity determining region; MHC, major histocompatibility complex; sc, side chain, TCR, T-cell receptor.
Figure 8.
Molecular dynamics simulations of trimolecular complexes consisting of TCR#16 and DRB1*04:01 associated with either the galactosylated (gal264) (left) or unmodified Col2259-273 peptide (right). Depicted are the root mean squares of fluctuation values for the variable and constant region residues of TCRα and TCRβ within the trimolecular complex over a simulation period of 1000 ns. In complex with the unmodified Col2 peptide, a peak of molecular mobility is detectable carboxyterminal of the TCRβ-CDR2 domain and to a minor degree in TCRα-CDR2 in complex with the gal264Col2 peptide. Most notable is the increase of molecular fluctuations in the constant region of TCR#16 (arrows), especially in the α-chain, also affecting the CαAB loop and clearly much more pronounced in complex with gal264 compared with the unmodified Col2 peptide. CDR, complementarity-determining region; RMS, root mean square; TCR, T-cell receptor.
Discussion
Our studies characterised molecular details of antigen presentation by the DRB1*04:01 molecule, an allelic variant strongly associated with RA in Caucasian patients, by structure determinations following co-crystallisation with several peptides known to trigger autoreactive RA T cells. A crucial residue for DR molecules associated with RA is position 71 in the β-chain. This residue, which is a lysine in DRB1*04:01, critically interacts with glutamic acid in position 266 of the Col2 peptide. However, there is some freedom in this interaction as other peptides derived from HSP and CILP contain different amino acids in position 266. The residues of valine 11 (V11b) and histidine 13 (H13b) of the β chain in HLA-DRB1*04:01 have been shown previously to be associated with susceptibility of seropositive RA in genome-wide association studies.11 Our crystal structure analysis reveals that both residues likely contribute to the stability of the MHCII molecule as they are located where α and β chains pair in the beta-plated sheet. Moreover, a likely contribution to antigen presentation of the CILP982-996 peptide is provided by the residue H13β that forms a hydrogen bond with the carboxyl group of the aspartic acid residue in the P6 pocket (P6D) of the CILP peptide. This hydrogen bond is missing in the other three complexed MHCII/peptide crystals, as the P6A (HSP70289-306) and P6G (Col259-273) lack the corresponding carboxyl group. In addition, the side chains of A74β orientate toward the P4 pocket, likely influencing the binding specificity of the P4 residue.
Similar to its interaction with murine Aq,34 the DRB1*04:01 bound Col2259-273 peptide exposes two lysine residues that are physiologically modified by hydroxylation and glycosylation, and these modified variants can be recognised by T cells.18 35 Whereas in the mouse Col2 immunisation activates Col2259-273 -specific T cells, thereby inducing a severe form of erosive arthritis, it is not clear to what extent MHCII-restricted Col2-specific T cells play a regulatory role in humans. In this context, there has been a long-standing question concerning why these T cells are not deleted from the repertoire by central tolerance. A possible explanation could be that TCR affinity for PTM variants of Col2 peptides promotes escape from thymic selection; this hypothesis is supported by our previous finding that non-modified Col2, but not glycosylated Col2-determinants, could be expressed by mouse or human thymic epithelium.29 Notably, earlier studies have detected glycopeptide-reactive T cells in patients with DRB1*04:01-positive and DRB1*01:01-positive RA,18 36 and the present investigation provides new evidence that peripheral T cells of healthy individuals also express TCRs with binding affinity for DRB1*01:01/ gal264Col2 complexes in respective MHCII allele carriers.
The present investigation of human PBMCs from RA and healthy DRB1*04:01 carriers, which used tetramer staining as well as parallel peptide-induced T cell activation assays, provides new insight into the impact of PTM on Col2 T-cell recognition. In contrast to a comparable prevalence of CD4+ T cells that recognise nCol2 or gal264Col2 in DRB1*04:01 tetramer complexes, T-cell responsiveness was increased on peptide challenge with the galactosylated variant. The mechanism for this glycosylation-dependent impact on T-cell activation is not entirely clear and might be multifactorial, but one attractive hypothesis is that an altered TCR interaction with the DRB1*04:01-bound galactosylated Col2-peptide affects TCR signal transmission. Accordingly, our functional and structural characterisation of a prototypic human TCR and its interaction with nCol2 or gal264Col2 in the context of DRB1*04:01 presentation provide first experimental support for this hypothesis. The comparative dynamic modelling of the respective trimolecular complexes containing either the galactosylated or unmodified Col2 variant revealed a major difference in the propagation of TCR dynamics from the V-regions to key allosteric sites in the Cα and Cβ region pertaining particularly to the so-called CαAB loop.33 This domain was previously described for its role in the allosteric regulation of TCR signalling as evidenced by fluorescence-based conformational changes on MHCII/peptide perception and signalling impairment by mutational analysis.37 Another region exhibiting reinforced molecular fluctuations on TCR recognition of the DRB1*04:01/gal264Col2 in our studies is in immediate proximity to the CβFG loop that has been critically incriminated in T-cell activation and thymic selection.38 39
Thus, our dynamic modelling studies provide evidence for allosteric changes initiated by CDR3α-region interaction with a single galactose residue in the DRB1*04:01-bound gal264Col2 peptide, which propagates to result in increased conformational flexibility at distant sites in the constant TCR regions contacting the CD3 signalling complex. Whereas it remains enigmatic how these allosteric changes are transmitted across the cell membrane, our complementary functional T-cell studies provide experimental support for translation into a reinforced TCR signal in response to challenge by gal264Col2-bound versus nCol2-bound DRB1*04:01complexes. Moreover, the data obtained by analysis of RA T cells and the prototypic TCR #16 might reflect mechanisms of central tolerance by thymic medullary epithelial cells that do not express galactosylated Col2 and accordingly execute clonal deletion exclusively via presentation of unmodified Col2 determinants.29 Thus, T cells rescued from thymic selection due to weak TCR recognition of DRB1*04:01/nCol2 complexes and the resulting low TCR signal intensities could subsequently become activated on MHCII presentation of physiologically galactosylated Col2 in peripheral tissues39 via reinforced signalling dynamics initiated by the recognition of posttranslational Col2 peptide modifications. However, the outcome of the activation, which can result in either arthritogenic or tolerogenic T cell responses, would remain context dependent. An obvious objection to our findings on the lack of a contribution of citrulline residues to MHCII binding in the two crystallised control complexes containing citrullinated epitopes of RA T cell responses and our focus on TCR recognition of Col2 peptides is how our proposed concept could be linked to citrulline-specific immunity and its strong association with DRB1*04:01 in RA. However, it has been shown that Col2 can be citrullinated in vivo, both in mice and in humans with RA.22 Moreover, the autoantibody response to citrullinated Col2 is prominent in B cell recognition of Col2. Accordingly, it is easily conceivable that B cells specific for citrullinated Col2 can present the non-citrullinated 259–273 peptide to T cells and vice versa.40 Such T cells could help to activate B cells specific for citrullinated epitopes, potentially breaking tolerance and allowing pathogenic epitope spreading of the anticitrullinated protein antibody response target, joint cartilage.40
Although we have provided indirect evidence for the proposed scenario, further studies are clearly needed to provide additional experimental support and to answer the question of whether this hypothesis might also apply to citrullinated antigens, such as the CILP-derived peptide cocrystallised with DRB1*04:01 in this study.
Acknowledgments
We thank Dr Anna Linusson, University of Umeå, Sweden, for the kind provision of parameters for molecular modelling of the non-standard residues 5-hydroxylysine and galactosylated 5-hydroxylysine and Sharon L. Cross, Ph.D., Mission Viejo, CA for editorial assistance supported by Fraunhofer Institute for Translational Medicine and Pharmacology. We are grateful to Luna Ribaric, Division of Rheumatology, University Hospital Frankfurt, Goethe University, Frankfurt am Main, for excellent technical support in the performance of human T cell assays and PD Dr Christof Geisen, Institute of Transfusion Medicine and Immunohematology, Goethe University Hospital, Frankfurt, Germany for providing anonymous blood samples from healthy carriers of the HLA-DRB1*04:01 allele. HL gratefully acknowledges the computer resources and support provided by the Erlangen Regional Computing Centre (RRZE) and by the Erlangen National High Performance Computing Centre at Friedrich-Alexander-University Erlangen-Nürnberg.
Footnotes
Handling editor: Josef S Smolen
CG and SW contributed equally.
Contributors: CG, SW, BX, RH and HB contributed to the study planning and design. CG, SW, BX, DD, JV, JK, N-ND, NS, HL, RH and HB contributed to the acquisition, analysis or interpretation of data and critical revision of the manuscript. GC, SW, N-ND, HL, RH and HB contributed to the acquisition of data and drafting of the manuscript. RH and HB had access to all data and act as guarantors for the study. All the authors have read and approved the final version of the manuscript.
Funding: HB received support from the German Federal Ministry of Education and Research (GO-Bio-project aidCURE; 031A385), the Federal State of Hesse (LOEWE-project 13, IME Fraunhofer Project Group TMP at Goethe University) and the Fraunhofer Cluster of Excellence for Immune-Mediated Diseases CIMD.RH and CG were supported by grants from the Knut and Alice Wallenberg Foundation (KAW 2015.0063), the Swedish Association against Rheumatism (R-757331), the Swedish Foundation for Strategic Research (RB13-0156), the Swedish Research Council (2015-02662), the Erling Persson Foundation and the European Union Innovative Medicine Initiative project BeTheCure (115142). JK and JV were supported by the Swedish Research Council (2017-06104) and the Erling Persson Foundation.
Competing interests: NS, BX, SW, RH and HB are listed as inventors on Patent EP2020072287 (https://www.onscope.com/ipowner/en/ip/ptwo/EP2020072287.html). SW, N-ND, RH and HB are lasted as inventors on Patent EP2020072280 (https://www.onscope.com/ipowner/en/ip/ptwo/EP2020072280.html). The owner of both patents is Fraunhofer-Gesellschaft zur Förderung der Angewandten Forschung E.V. (Germany). SW is listed on these patents under her maiden name of Sylvia Cienciala.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as supplementary information. The crystallographic coordinates and structure factors elucidated in this study have been deposited in the Protein Data Bank with the accession codes listed in online supplemental table S1 (7NZE, 7NZF, 7NZH, 7O00).41–44
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All blood sample donors gave prior written consent for study inclusion. The current study has been approved by the ethical approval committee of the University Hospital Frankfurt, Germany.
References
- 1. Aho K, Palosuo T, Raunio V, et al. When does rheumatoid disease start? Arthritis Rheum 1985;28:485–9. 10.1002/art.1780280503 [DOI] [PubMed] [Google Scholar]
- 2. Scherer HU, Huizinga TWJ, Krönke G, et al. The B cell response to citrullinated antigens in the development of rheumatoid arthritis. Nat Rev Rheumatol 2018;14:157–69. 10.1038/nrrheum.2018.10 [DOI] [PubMed] [Google Scholar]
- 3. Kurki P, Aho K, Palosuo T, et al. Immunopathology of rheumatoid arthritis. Antikeratin antibodies precede the clinical disease. Arthritis Rheum 1992;35:914–7. 10.1002/art.1780350810 [DOI] [PubMed] [Google Scholar]
- 4. Rantapää-Dahlqvist S, de Jong BAW, Berglin E, et al. Antibodies against cyclic citrullinated peptide and IgA rheumatoid factor predict the development of rheumatoid arthritis. Arthritis Rheum 2003;48:2741–9. 10.1002/art.11223 [DOI] [PubMed] [Google Scholar]
- 5. Brink M, Hansson M, Mathsson L, et al. Multiplex analyses of antibodies against citrullinated peptides in individuals prior to development of rheumatoid arthritis. Arthritis Rheum 2013;65:899–910. 10.1002/art.37835 [DOI] [PubMed] [Google Scholar]
- 6. Pereira RS, Black CM, Duance VC, et al. Disappearing collagen antibodies in rheumatoid arthritis. Lancet 1985;2:501–2. 10.1016/S0140-6736(85)90436-2 [DOI] [PubMed] [Google Scholar]
- 7. Clague RB, Firth SA, Holt PJ, et al. Serum antibodies to type II collagen in rheumatoid arthritis: comparison of 6 immunological methods and clinical features. Ann Rheum Dis 1983;42:537–44. 10.1136/ard.42.5.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ge C, Holmdahl R. The structure, specificity and function of anti-citrullinated protein antibodies. Nat Rev Rheumatol 2019;15:503–8. 10.1038/s41584-019-0244-4 [DOI] [PubMed] [Google Scholar]
- 9. Stastny P. Mixed lymphocyte cultures in rheumatoid arthritis. J Clin Invest 1976;57:1148–57. 10.1172/JCI108382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014;506:376–81. 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raychaudhuri S, Sandor C, Stahl EA, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 2012;44:291–6. 10.1038/ng.1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 1987;30:1205–13. 10.1002/art.1780301102 [DOI] [PubMed] [Google Scholar]
- 13. Hill JA, Southwood S, Sette A, et al. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J Immunol 2003;171:538–41. 10.4049/jimmunol.171.2.538 [DOI] [PubMed] [Google Scholar]
- 14. Ting YT, Petersen J, Ramarathinam SH, et al. The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis. J Biol Chem 2018;293:3236–51. 10.1074/jbc.RA117.001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Auger I, Sebbag M, Vincent C, et al. Influence of HLA-DR genes on the production of rheumatoid arthritis-specific autoantibodies to citrullinated fibrinogen. Arthritis Rheum 2005;52:3424–32. 10.1002/art.21391 [DOI] [PubMed] [Google Scholar]
- 16. Sidney J, Becart S, Zhou M, et al. Citrullination only infrequently impacts peptide binding to HLA class II MHC. PLoS One 2017;12:e0177140. 10.1371/journal.pone.0177140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holmdahl R, Andersson M, Goldschmidt TJ, et al. Type II collagen autoimmunity in animals and provocations leading to arthritis. Immunol Rev 1990;118:193–232. 10.1111/j.1600-065X.1990.tb00817.x [DOI] [PubMed] [Google Scholar]
- 18. Bäcklund J, Carlsen S, Höger T, et al. Predominant selection of T cells specific for the glycosylated collagen type II epitope (263-270) in humanized transgenic mice and in rheumatoid arthritis. Proc Natl Acad Sci U S A 2002;99:9960–5. 10.1073/pnas.132254199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burkhardt H, Sehnert B, Bockermann R, et al. Humoral immune response to citrullinated collagen type II determinants in early rheumatoid arthritis. Eur J Immunol 2005;35:1643–52. 10.1002/eji.200526000 [DOI] [PubMed] [Google Scholar]
- 20. Lindh I, Snir O, Lönnblom E, et al. Type II collagen antibody response is enriched in the synovial fluid of rheumatoid joints and directed to the same major epitopes as in collagen induced arthritis in primates and mice. Arthritis Res Ther 2014;16:R143. 10.1186/ar4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang B, Ge C, Lönnblom E, et al. The autoantibody response to cyclic citrullinated collagen type II peptides in rheumatoid arthritis. Rheumatology 2019;58:1623–33. 10.1093/rheumatology/kez073 [DOI] [PubMed] [Google Scholar]
- 22. Haag S, Schneider N, Mason DE, et al. Identification of new citrulline-specific autoantibodies, which bind to human arthritic cartilage, by mass spectrometric analysis of citrullinated type II collagen. Arthritis Rheumatol 2014;66:1440–9. 10.1002/art.38383 [DOI] [PubMed] [Google Scholar]
- 23. Rosloniec EF, Brand DD, Myers LK, et al. An HLA-DR1 transgene confers susceptibility to collagen-induced arthritis elicited with human type II collagen. J Exp Med 1997;185:1113–22. 10.1084/jem.185.6.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rosloniec EF, Brand DD, Myers LK, et al. Induction of autoimmune arthritis in HLA-DR4 (DRB1*0401) transgenic mice by immunization with human and bovine type II collagen. J Immunol 1998;160:2573–8. [PubMed] [Google Scholar]
- 25. Andersson EC, Hansen BE, Jacobsen H, et al. Definition of MHC and T cell receptor contacts in the HLA-DR4restricted immunodominant epitope in type II collagen and characterization of collagen-induced arthritis in HLA-DR4 and human CD4 transgenic mice. Proc Natl Acad Sci U S A 1998;95:7574–9. 10.1073/pnas.95.13.7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindgren C, Tyagi M, Viljanen J, et al. Dynamics determine signaling in a multicomponent system associated with rheumatoid arthritis. J Med Chem 2018;61:4774–90. 10.1021/acs.jmedchem.7b01880 [DOI] [PubMed] [Google Scholar]
- 27. Broddefalk J, Bäcklund J, Almqvist F, et al. T cells recognize a glycopeptide derived from type II collagen in a model for rheumatoid arthritis. J Am Chem Soc 1998;120:7676–83. 10.1021/ja980489k [DOI] [Google Scholar]
- 28. Raposo B, Merky P, Lundqvist C, et al. T cells specific for post-translational modifications escape intrathymic tolerance induction. Nat Commun 2018;9:353. 10.1038/s41467-017-02763-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dzhambazov B, Nandakumar KS, Kihlberg J, et al. Therapeutic vaccination of active arthritis with a glycosylated collagen type II peptide in complex with MHC class II molecules. J Immunol 2006;176:1525–33. 10.4049/jimmunol.176.3.1525 [DOI] [PubMed] [Google Scholar]
- 30. Shoda H, Hanata N, Sumitomo S, et al. Immune responses to mycobacterial heat shock protein 70 accompany self-reactivity to human BiP in rheumatoid arthritis. Sci Rep 2016;6:22486. 10.1038/srep22486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. James EA, Rieck M, Pieper J, et al. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol 2014;66:1712–22. 10.1002/art.38637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hennecke J, Wiley DC. Structure of a complex of the human alpha/beta T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA*0101 and DRB1*0401): insight into TCR cross-restriction and alloreactivity. J Exp Med 2002;195:571–81. 10.1084/jem.20011194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mariuzza RA, Agnihotri P, Orban J. The structural basis of T-cell receptor (TCR) activation: an enduring enigma. J Biol Chem 2020;295:914–25. 10.1016/S0021-9258(17)49904-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kjellén P, Brunsberg U, Broddefalk J, et al. The structural basis of MHC control of collagen-induced arthritis; binding of the immunodominant type II collagen 256-270 glycopeptide to H-2Aq and H-2Ap molecules. Eur J Immunol 1998;28:755–66. [DOI] [PubMed] [Google Scholar]
- 35. Michaëlsson E, Malmström V, Reis S, et al. T cell recognition of carbohydrates on type II collagen. J Exp Med 1994;180:745–9. 10.1084/jem.180.2.745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Snir O, Bäcklund J, Boström J, et al. Multifunctional T cell reactivity with native and glycosylated type II collagen in rheumatoid arthritis. Arthritis Rheum 2012;64:2482–8. 10.1002/art.34459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beddoe T, Chen Z, Clements CS, et al. Antigen ligation triggers a conformational change within the constant domain of the alphabeta T cell receptor. Immunity 2009;30:777–88. 10.1016/j.immuni.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 38. Rangarajan S, He Y, Chen Y, et al. Peptide-Mhc (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites. J Biol Chem 2018;293:15991–6005. 10.1074/jbc.RA118.003832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sasada T, Touma M, Chang H-C, et al. Involvement of the TCR Cbeta FG loop in thymic selection and T cell function. J Exp Med 2002;195:1419–31. 10.1084/jem.20020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Uysal H, Nandakumar KS, Kessel C, et al. Antibodies to citrullinated proteins: molecular interactions and arthritogenicity. Immunol Rev 2010;233:9–33. 10.1111/j.0105-2896.2009.00853.x [DOI] [PubMed] [Google Scholar]
- 41. Ge C, Holmdahl R. Data from: X-ray diffraction. protein Databank in Europe, entry awaiting author approval [dataset]. Available: https://dxdoiorg/102210/pdb7nze/pdb
- 42. Ge C, Dobritzsch D, Holmdahl R. Data from: crystal structure of HLA-DR4 in complex with a mutated human collagen type II peptide. protein Databank in Europe, entry awaiting author approval [dataset]. Available: https://dxdoiorg/102210/pdb7nzf/pdb
- 43. Ge C, Holmdahl R. Data from: crystal structure of HLA-DR4 in complex with a citrullinated CILP peptide. protein Databank in Europe, processed, waiting for author review and approval [dataset]. Available: http://dxdoiorg/102210/pdb7nzh/pdb
- 44. Ge C, Holmdahl R. Data from: crystal structure of HLA-DR4 in complex with a Hsp70 peptide. protein Databank in Europe, entry withheld until publication [dataset]. Available: https://dxdoiorg/102210/pdb7o00/pdb
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2021-220500supp001.pdf (749.4KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. All data relevant to the study are included in the article or uploaded as supplementary information. The crystallographic coordinates and structure factors elucidated in this study have been deposited in the Protein Data Bank with the accession codes listed in online supplemental table S1 (7NZE, 7NZF, 7NZH, 7O00).41–44