Abstract
Single nucleotide polymorphisms (SNPs) are the most abundant type of genomic variation and the most accessible to genotype in large cohorts. However, they individually explain a small proportion of phenotypic differences between individuals. Ancestry, collective SNP effects, structural variants, somatic mutations or even differences in historic recombination can potentially explain a high percentage of genomic divergence. These genetic differences can be infrequent or laborious to characterize; however, many of them leave distinctive marks on the SNPs across the genome allowing their study in large population samples. Consequently, several methods have been developed over the last decade to detect and analyze different genomic structures using SNP arrays, to complement genome-wide association studies and determine the contribution of these structures to explain the phenotypic differences between individuals. We present an up-to-date collection of available bioinformatics tools that can be used to extract relevant genomic information from SNP array data including population structure and ancestry; polygenic risk scores; identity-by-descent fragments; linkage disequilibrium; heritability and structural variants such as inversions, copy number variants, genetic mosaicisms and recombination histories. From a systematic review of recently published applications of the methods, we describe the main characteristics of R packages, command-line tools and desktop applications, both free and commercial, to help make the most of a large amount of publicly available SNP data.
Keywords: structural variants, genomic structures, GWAS, bioinformatic methods, SNP arrays, software
Introduction
From a nucleotide change to a chromosome gain, genetic variation encompasses a huge spectrum that has been intensively studied for decades. Single nucleotide polymorphisms (SNPs) are the most abundant type of genetic variability. SNPs can be genotyped by many sequencing techniques from Sanger to Next Generation Sequencing (NGS). Microarrays and DNA chips are popular techniques that reduce cost by targeting a selected set of SNPs that are easily scalable to large population samples [1]. The Whitehead Institute and Affymetrix, Inc. developed the first SNP chip designed to genotype 1494 SNPs [2], giving birth to the first genome-wide association study (GWAS) that led to rapid growth of this technology. Now, there are hundreds of different types of chips, totally customizable depending on the research objectives, and some with more than a million markers to perform whole-genome SNP sequencing. Analysis and storage of these data have been largely made available in public repositories, such as Bioconductor and GitHub for methods, and dbGaP and EGA for data. As a result, today we have access to a great amount of freely SNP data from large cohorts, including several small studies and large consortia (Estonian Biobank, UK Biobank).
Before the GWAS explosion, it was soon realized that high-density SNP arrays could be used to detect structural variation, in addition to several other genomic features, that would be otherwise expensive and laborious to characterize in population samples. As a consequence, several research groups have been working on different methods to infer underlying genomic structures in SNP genotypes, and thus add important contributors to the genomic architecture of phenotypic traits. These added features of GWAS include the analysis of population structure and ancestry, the calculation of polygenic risk scores (PRS), the detection of identity-by-descent (IBD) fragments, the analysis of linkage disequilibrium (LD) and the estimation of the heritability of a trait. Algorithms have also been developed to detect genomic variation, including copy number variants (CNV), inversions, recombination patterns and genetic mosaicisms. SNP array genomic data in a large number of individuals provide an unmatched scenario to study the role of rare genomic variants in common phenotypes with enough statistical power. Researchers can then, for instance, enquire about the burden of rare variants to common diseases, study disease etiology by the identification of rare mutations in causal genes, or explore the role of these variants in the link between environmental exposures and phenotypes [3]. Recent reviews have described some of the biological processed that can be studied with high-density SNP data [4]. However, given the amount and diversity of methods to exploit this type of data, a comprehensive systematic review of the current methods in use is needed to help researchers in their choice.
We assessed a total of 105 different methods to infer underlying genomic structures in SNP data, including R-packages, command-line tools and desktop applications, both free and commercial. We selected the methods and tools with a systematic search of scientific literature and filtered those that were applied in at least one recent study of 88 selected publications between January 2020 and September 2021. We reviewed the available options to exploit SNP arrays, along with the methods and bioinformatics tools, that were used in those studies. Our aim is to facilitate the decision on choosing a current tool in use to extract relevant information from SNP array data, depending on individual needs, limitations and research interests.
References included in this review
We searched for relevant scientific literature following a strict protocol with specific MeSH terms that were chosen to obtain the most suitable articles for each section (Supplementary Table S1). In total, eight distinct queries were submitted to PubMed and, from each one, papers mentioning in their title the introduction of new software or method were selected. We filtered articles in which specific methods on SNP data were described [69]. Additional articles were included that appeared in the references of those from the initial selection and were considered relevant [36]. We then retrieved recent studies in humans applying at least one of the selected methods between January 2020 and September 2021 [88].
SNP genotypes from NGS data
Several programs that are presented in this review can be adapted to SNP genotypes obtained from NGS data, when appropriate formatting is provided [5, 6]. There are multiple algorithms for calling SNP genotypes from FASTQ files, either by applying heuristic (VarScan2, GSMapper, CLC Genomic Workbench, DNSTAR Lasergene) or by probabilistic methods (SAMtools, Beagle, GATK, Atlas-SNP2, SOAPsnp, SNVer), which follow both single and multi-sample approaches. The first step is usually to use an alignment tool that converts de FASTQ/FASTA files into SAM/BAM alignments, and then the variant calling tool to produce a VCF or text file with the polymorphic positions and genotypes. Different methods for analyzing SNP data can directly use these files. Alternatively, the VCF files can be further converted to other formats, including PLINK, or R data-structures such as GDS or snpMatrix objects, among others. While adaptation to NGS data is possible for numerous methods, tools that are based on BAF and/or LRR information (i.e. for CNVs and mosaicism detection) are only suitable for SNP arrays.
Quality control of SNP array data
Quality control (QC) of the SNP genotypes must be performed before their analysis. Some programs, as the Illumina BeadStudio, provide its own exhaustive QC steps for genotype calling [7, 8]. For those tools that do not include specific QC functions there are several specialized software options available, including the R packages GWASTools [9], QCGWAS [10] and SNPRelate [11]. The aim of QC functions is to filter out SNPs and samples with unreliable data. For filtering SNPs, the most common procedure is to discard those that have a high missing frequency, are not in Hardy–Weinberg Equilibrium (HWE) in the control group, or have a low minor allele frequency (MAF) (<1%, or 5%) [12–14]. Additionally, more specific controls can be applied depending on the available data and the research objectives, such as strand consistency [15]; detection of position mismatches, replicate errors or Mendelian inconsistencies; and the exclusion of A/T, G/C or non-HapMap SNPs [16–18]. Samples are usually filtered out with low call rates (i.e. individuals with several missing genotypes), high heterozygosity levels or a large Mendelian error rate. Those showing sex and race mismatch should be fixed or discarded. Moreover, population structure can also be controlled [19]. Additionally, if applicable, a correlation between samples is performed to detect and remove highly genetically related individuals [20].
Some programs require additional preprocessing of the SNP data such as phasing and imputation. For instance, programs that analyze population structure, detect IBD fragments or LD structures often start with phased haplotypes as input. In addition, some PRS tools need a previous imputation step, see the ‘Input Data’ column of the Tables for details. The phasing of the haplotypes, that is the separation between the maternally and paternally inherited copies of each chromosome, can be performed with tools like SHAPEIT 3, Eagle 2 and HAPI-UR [21, 22]. Genotype imputation, the estimation of missing genotypes from a genotype reference panel, can be performed with tools like Minimac 4 from the Michigan Imputation Server, Impute 5, Beagle 5.2 or PBWT from Sanger Imputation Server [23].
Enhance your gwas: the different ways to exploit SNP array data
The value of a GWAS can be substantially increased by studying additional genomic features that can be inferred from SNP array data, providing additional mechanisms, associations and insights into the genomic basis of phenotypic differences between individuals. We have identified the first group of methods that incorporate the joint effects of multiple SNPs and include those aimed at estimating population structure, ancestry, PRS and narrow-sense heritability, and the detection of regions with IBD and high LD. For each category, we have selected the top-five methods, according to the number of citations on PubMed between January 2020 and September 2021; see Tables 1–5. For those categories with more than five methods, all the tools are summarized in Supplementary Tables S2–S5.
Table 1.
Top-five tools (the most cited between January 2020 and September 2021 in PubMed) for the study of population structure and ancestry with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| STRUCTURE | Desktop App Command-line (C) | Free | Text file with genotypes and other optional fields | Bayesian with multiple tunning parameters | The first one Works with any type of multilocus genotype data | 2000 | [24] |
| EIGENSOFT | Command-line | Free | PLINK | Principal Components Analysis (PCA) | Combination of SMARTPCA and EIGENSTRAT Specific for case/control studies | 2006 | [30, 31] |
| ADMIXTOOLS | R Package Command-line (C) | Free | ‘ind’ file, ‘snp’ file and ‘geno’ file | Several methods | Infers proportion and dates of mixtures | 2012 | [33] |
| fastSTRUCTURE | Command-line (Python) | Free | Binary PLINK (BED/BIM/FAM) | Bayesian framework | Fast | 2014 | [25] |
| fineSTRUCTURE | Command-line | Free | Phased Haplotypes | ChromoPainter [38] (HMM-based) | Fine-scale population structure | 2012 | [38] |
Table 5.
Top-five tools (the most cited between January 2020 and September 2021 in PubMed) for the study of LD with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| Haploview | Desktop App Command-line (JAVA) | Free | Linkage format Phased Haplotypes HapMap Project Data Dumps PHASE PLINK | Two marker Expectation Maximization (EM) | Suite of tools | 2005 | [91] |
| Big-LD | R package gpart | Free | Text file with genotypes | Interval graph modeling of LD bins | Visualization options | 2018 | [93] |
| ALOHOMORA | Desktop App (Perl) | Free | Genotype data generated by GeneChip DNA Analysis Software (GDAS v3.0) from Affymetrix For other chips: MAP file, allele frequency file and genotype file in the Alohomora format | Several | Visualization options | 2005 | [90] |
| VarLD | Command-line (JAVA) | Free | Text file with genotypes | Quantification of the LD by the signed r2 metric | Performs inter-population comparisons | 2010 | [92] |
| LDExplorer | R Package | Free | Phased genotypes in VCF or HAPMAP2 format | MIG Algorithms | Deals with SNPs at any distance | 2014 | [94] |
Population structure and ancestry
Most of the GWASs performed to date are biased towards European populations, and tightly controlled for ancestry differences because uncontrolled admixture in a study is a confounding factor of single SNP associations. However, as most global populations are admixtures, researchers recognized that GWASs should be based on more representative samples and, therefore, the effect of ancestry on phenotypes needs to be better characterized. Tools assessing population structure and ancestry can be used to detect underlying admixed population structures, see Table 1 and Supplementary Table S2. One of the most popular and widely used tools for the estimation of ancestry is the program STRUCTURE [24], which assumes a model in which there are K populations (where K may be unknown), each of which is characterized by a set of allele frequencies at each locus. Then, it infers population structure and assigns individuals to subpopulations using a Bayesian method with multiple tuning parameters. STRUCTURE was remodeled in a command-line program named fastSTRUCTURE [25] that uses a variational Bayesian framework and is two orders of magnitude faster than its predecessor. ADMIXTURE [26] is another widely used software for the analysis of population structure that adopted the likelihood model embedded in STRUCTURE but is considerably faster. Other compiled programs that run from the command line are HaploPOP [27], which works by combining markers into haplotypes; RENT and RENT+ [28], which can be used to infer local genealogical trees from haplotypes with the presence of recombination; and POPSTR [29] that applies a Bayesian joint modeling framework that accepts both SNPs and CNVs. EIGENSOFT combines SMARTPCA [30] and the EIGENSTRAT [31] stratification correction method, which can be used on disease studies to explicitly model ancestry differences between cases and controls. Finally, SNP2pop [32] is a specific tool for tumoral samples that can classify individuals into 26 predefined population groups.
Population stratification is also accessible in a number of R packages and MATLAB tools. There are several options, which include ADMIXTOOLS [33], AWclust [34], PC-AiR [35], IPCAPS [36]. The former is a suite of methods that can infer proportions and dates of the mixture between populations, while the second one uses Ward’s minimum variance to estimate sub-clusters and does not require the markers to be unlinked. Additionally, it can work with an unknown number of populations. PC-AiR can do robust population structure inference in the presence of known or cryptic relatedness, first identifying a subset of unrelated individuals that is representative of all ancestries in the sample, and then performing a PCA and predicting components of variation for all remaining individuals based on genetic similarities. IPCAPS resolves fine-scale population structure assigning individuals to genetically similar subgroups. Finally, KIND [37] is based on MATLAB and utilizes the spatial distribution of minor-allele SNP variants, to construct a vector for each individual and calculate a pair-wise kinship coefficient. More recently, fineSTRUCTURE [38] has emerged as a command-line and R option to detect more subtle changes in population structure from haplotypes. fineSTRUCTURE runs ChromoPainter, a tool used by other algorithms to analyze admixture events, such as GLOBETROTTER [39] and GTMix [40].
Specific ancestry, rather than unknown substructure, can be inferred with several options including tsinfer [41], PCAdmix [42], LAMP [43], HAPMIX [44], and RFMix [45], the R packages ELAI [46], Summix [47], FastPop [48] and FamANC [49] and the program MI-MAAP [50]. tsinfer, a Python-based software, is used to infer whole-genome histories through the succinct tree sequence, PCAdmix is PCA-based and LAMP works with Hidden Markov Models (HMM). LAMP and HAPMIX perform well in recently admixed populations. RFMix and ELAI are specialized in local ancestry. Other R packages include Summix that estimates ancestry proportions from summary data, FastPop that can infer ancestries on data involving two or more intercontinental origins and FamANC that can be used for local ancestry in large pedigrees. MI-MAAP [50] is a web-based bioinformatics tool designed to prioritize informative markers although it can also classify multi-ancestry admixed populations.
Identity by descent
Recent shared ancestry between pairs of individuals can be estimated by the identification of shared chromosomal segments, i.e. IBD genomic fragments. Some methods have been implemented in specific software tools, almost all of them run from the command-line, see Table 2 and Supplementary Table S3. They can be divided into two distinct groups: those that need phased data and those that do not. In the first group there is GERMLINE [51] that deals with errors in the genotypes and iLASH [52], RaPID [53], hap-IBD [54], FastSMC [55] and fastIBD [56], all of them reporting improved speed. Also in this group, there is RefinedIBD [57], which does not allow for genotype errors but uses the GERMLINE algorithm for the identification of shared haplotypes exceeding a threshold length. Both Refined IBD and fastIBD are implemented in the BEAGLE software. Methods that allow unphased datasets include: Parente2 [58] that applies an embedded log-likelihood ratio method; IBIS [59] that finds identical-by-descent segments via identical-by-state method; and TRUFFLE [60] that admits genotyping errors and can be applied to raw variant calls from VCF files. In addition, IBDLD [61] and IBD_Haplo [62] (included in the MORGAN software and in the R package IBDhaploRtools) can analyze both phased and unphased data. Another popular program is RELATE [63], that accounts for genotyping errors, missing data and LD without pruning away SNPs and is available both as a C++ command-line software or R package. After IBD fragments are identified, programs such as ibd-ends [64] give the probability distribution for each endpoint; also, IBDkin [65] and SNPRelate [11] can estimate kinship coefficients and relatedness from IBD data.
Table 2.
Top-five tools (the most cited between January 2020 and September 2021 in PubMed) for the study of identity by descent fragments with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| RefinedIBD | Command-line (JAVA) BEAGLE Software | Free | Phased Data VCF file with genotypes | GERMLINE Algorithm + probabilistic approach | Does not allow genotype errors | 2013 | [57] |
| GERMLINE | Command-line (C++) | Free | Phased Data PLINK haplotype data | Dynamic Programming | Allows genotype errors | 2009 | [51] |
| fastIBD | Command-line (JAVA) BEAGLE Software | Free | Phased Data Text file with genotypes | Estimation of frequencies of shared haplotypes | Fast | 2011 | [56] |
| Hap-IBD | Command-line (JAVA) | Free | Phased Data VCF file with genotypes PLINK text files (PED/MAP) | Positional Burrows-Wheeler transform PBWT | Fast and simple | 2020 | [54] |
| RaPID | Command-line (Python) | Free | Phased Data VCF file with haplotypes | Random Projection (based on the positional Burrows-Wheeler transform, PBWT) | Fast Configurable parameters | 2019 | [53] |
Heritability
Phenotypic variance can be explained by many factors, the percentage that is due to genetic factors is called heritability, which can be defined in a narrow-sense (h2) or in a broad-sense (H2). While the first one refers only to additive genetic variation, the last also includes interaction effects such as dominance and epistasis. Single SNP associations are part of the narrow-sense heritability. One individual SNP is unlikely to explain a sizable part of heritability, as the magnitudes of single SNP associations are usually small. However, the addition of several SNP effects across the genome can explain an important part of the narrow-sense heritability, other contributors being rare structural variants. When considering a complex trait in a GWAS, it is interesting to estimate the contribution to its h2 given by the adding effects of all the SNPs in the study. There are many bioinformatics tools that estimate h2, see Table 3. Implemented in R, HERRA [66] is a heritability estimator that works with machine-learning methods and can handle continuous or dichotomous outcomes. It runs from the command line. LD SCore (LDSC) [67, 68] computes heritability, LD scores and genetic correlations and RHE-mc [69] estimates the variation in a complex trait that can be attributed to additive and dominance effects. Alternatively, LDAK [70] works with a modified kinship matrix in which SNPs are weighted according to local LD, reducing the bias and increasing the precision of narrow-sense heritability estimates. The implementation includes methods to calculate heritability from either summary statistics or individual-level data.
Table 3.
Tools for the study of heritability with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| LD Score (LDSC) | Command-line (Python) | Free | GWAS summary statistics | Linkage Disequilibrium Score Regression | Suite of tools | 2015 | [67, 68] |
| LDAK | Command-line (Compiled) | Free | PLINK | Modified kinship matrix Restricted maximum likelihood (REML) Haseman Elston (HE) regression Phenotype-correlation, genotype-correlation (PCGC) regression | Suite of tools | 2012 | [70] |
| HERRA | R Code | Free | Matrix with genotypes, disease status and covariates | Machine learning | Continuous or Dichotomous outcomes | 2017 | [66] |
| RHE-mc | Command-line (C++) | Free | PLINK | Randomized algorithm Method-of-moments (MoM) estimator | Estimates the variation that can be attributed to additive and dominance deviation | 2021 | [69] |
Polygenic risk scores
Estimation of heritability informs on the overall genetic architecture of phenotypic traits. However, it does not provide, as single SNP associations, an estimate of individual patient risk. Numerous SNPs with the highest associations can be combined into a PRS to substantially increase their independent risks into a collective one. Most of the algorithms that can calculate PRS have been developed during the last 5 years, see Table 4 and Supplementary Table S4. Although PRS has been implemented in PLINK [71] for some time, the first dedicated PRS software, PRSice [72] was published in 2015 and upgraded in 2019 (PRSice-2 [73]). PRSice runs from the command line and includes some automated steps from PLINK together with some additional steps in quality control. These two methods apply the most straightforward approach, named Pruning and Thresholding (P + T) [74].
Table 4.
Top-five (the most cited between January 2020 and September 2021 in PubMed) tools for the study of PRS with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| PRSice | Command-line (C++, Compiled, R for plotting) | Free | Binary PLINK BED/BIM/FAM) or imputed (Oxford .bgen) | Pruning and Thresholding (P + T) | Visualization options with R | 2015 | [72, 73] |
| PRS-CS | Command-line (Python) | Free | GWAS summary statistics External LD reference panel | Continuous shrinkage (CS) on SNP effect sizes + High-dimensional Bayesian regression framework | External LD reference panel | 2019 | [84] |
| SBLUP/BLUP GCTA | Command-line (C++, Compiled) | Free | Binary PLINK BED/BIM/FAM) or imputed (Oxford .bgen v1.2) | Linear mixed-effects model | Analyses individual chromosomes | 2020 v1.93.2beta | [75, 76] |
| SBayesR GCTB | Command-line (C++, Compiled) | Free | Binary PLINK BED/BIM/FAM) | Bayesian mixture model | Uses low computational resources | 2019 | [77] |
| lassosum | R Package bigstatsr | Free | Binary PLINK BED/BIM/FAM) | Regularized regression model | External LD reference panel Pseudovalidation | 2017 | [81] |
In addition, the Program in Complex Trait Genomics of the IMB (University of Queensland) developed GCTA (tool for Genome-wide Complex Trait Analysis) and GCTB (tool for Genome-wide Complex Trait Bayesian analysis) that run from the command line and include software for the analysis of PRS. While GCTA provides SBLUP [75, 76], which works with a linear mixed-effects model, GCTB includes SBayesR [77] that applies a Bayesian mixture model. In addition to those, there is also PRSoS [78] that works with a P + T approach and XPA [79], which is specialized in non-European populations and works within a cross-population analysis framework.
Some R packages are also able to calculate PRSs from genotype data: bigsnpr [80], lassosum [81] and EBPRS [82]. The first one is based on the package bigstatsr, a tool for scalable statistical analysis. It provides LDPred2 [83] to calculate PRS using both a Bayesian mixture model and P + T, and has been recently updated with the inclusion of lassosum2 that applies a regularized regression model. With this fusion, both methods can be used in the same package with input files in bed format. As for EBPRS, one of its main advantages is the independence of tuning parameters or external information. There are other interesting tools that can construct PRS from the summary statistics of a GWAS. These are PRS-CS [84], RSS [85], R2BGLiMS [86], penRegSum [87], ggmix [88], XPASS [79] and NPS [89].
Linkage disequilibrium
In GWASs of complex traits, neighboring and significant SNPs are likely in LD with the causal variant. Thus, the understanding of LD patterns and their block-like structures can guide the interpretation of association studies, see Table 5 and Supplementary Table S5. ALOHOMORA [90] is an open-source desktop app devoted to the linkage analysis of Affymetrix GeneChip® Human Mapping 10 K SNP array. Another desktop app performing the same function is Haploview [91], a very useful tool for the computation of LD statistics and population haplotype patterns. The quantification of LD variation between two populations can be done with varLD [92], which is a JAVA program that allows genome-wide assessment of LD variation as well as targeted analysis of a specific genomic region.
The Big-LD [93] algorithm of the gpart R package is a block partition method that uses interval graph modeling of LD bins, clustering strong pairwise LD SNPs that are not necessarily physically consecutive. LDExplorer [94] is another R package that includes the MIG algorithm, which computes the LD between SNPs at any distance, without maximal block length restrictions. Finally, also written in R, MATILDE [95] is an MCMC algorithm that can be used as a dimension reduction tool to identify blocks of LD for clustering contiguous SNPs.
Go beyond: genotyping structural variants
SNP data can also be used to extract information about changes in the structure of the genome. In other words, there are algorithms capable of genotyping structural variants from SNP array data, including chromosomal inversions, CNVs, mosaicism and recombination patterns. For each category we have selected the top-five methods, according to the number of citations on PubMed between January 2020 and September 2021; see Tables 6–8. All the tools for the genotyping of CNV and mosaicisms are summarized in Supplementary Tables S6 and S7.
Table 6.
Tools for the study of inversions with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| invClust | R package | Free | PLINK | Mixture model, uses all the SNPs in the inverted segment | Detects 20 human inversions from the invFest database, experimentally validated and greater than 0.2 Mb Allows including ancestry information | 2015 | [97] |
| PFIDO | R package | Free | PLINK | Pairwise identity-by-state distance matrix transformed by MDS. Model-based approach with 18 parameterized Gaussian mixture models | Detects 8p23 inversion-type Does not rely on any specific SNP | 2012 | [99] |
| inveRsion | R package | Free | Text files with 0/1/2 coded genotypes | Sliding window scan, uses linkage between groups of SNPs | Detects inversions directly from genotypes Can detect new possible inversion regions Optimal for homogenous samples and old inversions. | 2012 | [96] |
| scoreInvHap | R package | Free | PLINK or VCF files | Comparison with reference haplotype-genotypes | Detects 20 human inversions from the invFest database, experimentally validated and greater than 0.2 Mb | 2019 | [98] |
| RecombClust | R package | Free | Phased VCF files | LDmixture model | Detects chromosomal subpopulations with distinct recombination histories | 2020 | [102] |
Table 8.
Top-five tools (the most cited between January 2020 and September 2021 in PubMed) for the study of mosaicism with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| GISTIC | Command-line (MATLAB) | Free | Segmented Data | Ziggurat Deconstruction (ZD) | Specific for cancer samples | 2007 | [156] |
| MoChA | Command-line (C) (bcfools extension) R for graphical outputs | Free | VCF files with LRR and BAF values (raw Affymetrix or Illumina files if using a complementary pipeline) | Hidden Markov Model (HMM) | Detects LOH Visualization options | 2020 | [143] |
| PICNIC | Command-line (MATLAB) | Free (under license) | Affymetrix CEL files | Hidden Markov Model (HMM) | Specific for cancer samples Predicts absolute copy number Visualization options | 2010 | [151] |
| BAFSegmentation | Command-line (perl, R) | Free | Preprocessed files with BAF and LRR | Segmentation-based | Specific for cancer samples Provides percentage Detects LOH Visualization options | 2008 | [147] |
| hapLOH | Command-line (Python, Perl) | Free | BAF file and phased genotypes | Hidden Markov Model (HMM) | Supports low aberrant cell proportions | 2013 | [157] |
Chromosomal inversions
Chromosomal inversions are chromosomic rearrangements that appear when two breaks occur in the same chromosome and the cleaved fragment rotates before re-joining, see Figure 1. Although inversions can be detected by many methods from FISH to NGS, using SNP data is a promising cost-effective option, as it allows a rapid analysis of thousands of samples at zero cost, see Table 6. Inversion genotyping from SNPs depends on their LD within and across the inverted region. The first genotyping approach relies on the detection of the high LD that is associated with SNPs flanking the inversion and SNPs within the inversion but contiguous to the breakpoints. The other approach is based on the detection of distinct clusters in genomic divergence created by the lack of recombination that occurs between the non-collinear regions of individuals heterozygous for the inversion. When the two fragments evolve separately and accumulate mutations, they represent two distinct lineages that can be detected by a clustering tool of the SNPs within the inversion, as if they were from different populations.
Figure 1.
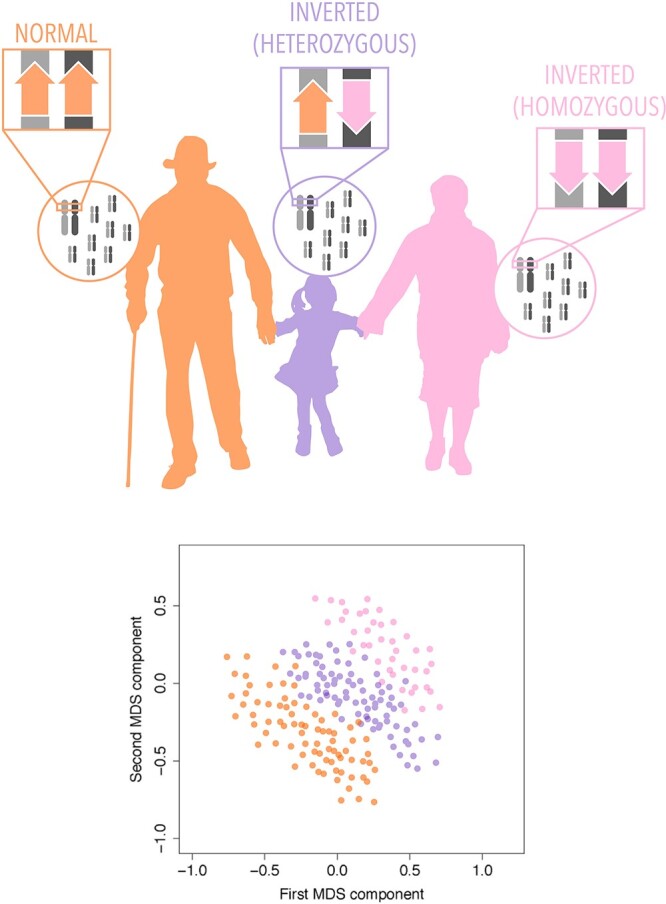
Chromosomic inversions appear when two breaks occur in the same chromosome and the cleaved fragment rotates before re-joining. They can be found in heterozygosis (center) or homozygosis (right). One of the methods for inversion detection is the clustering detection performed by invClust, which classifies the inversion genotypes into clusters of similar haplotype origin.
There are three software tools capable of genotyping several inversions from SNPs: inveRsion [96], invClust [97] and scoreInvHap [98]. inveRsion was the first to be developed and uses linkage differences in SNP groups across inversion alleles. It first employs a sliding window scan that phases and pairs haplotype blocks around potential breakpoints to identify regions likely to have an inversion. Then, it applies a mixture model to identify candidate inverted regions and to determine the inversion genotypes of the individuals. The second tool, invClust, tests the existence of extended haplotypes by exploiting all the SNPs inside the inverted segment, instead of only using those at the breakpoints. The method classifies the inversion genotypes into clusters of similar haplotype origin, accounting for differences in ancestries (Figure 1). The clustering detection is then performed with a mixture model that includes specific constraints based on a previous observation of the data, which helps to reduce the degrees of freedom and improve inversion genotyping. invClust allows ancestry information to be included in the mixture model. Finally, the last of the methods, scoreInvHap, also relies on the haplotype structures generated by the inversions. The method uses reference haplotype-genotypes, previously linked to reported experimental inversion-genotypes, and compares the SNPs of a new individual with those in the reference. The algorithm works under stringent conditions of SNP coverage and sample sizes. Another feature of scoreInvHap is its capacity to confidently call inversions with multiple haplotypes, as such, it can genotype inversions that other methods cannot.
In addition to the three previous methods, there is an algorithm specialized in the genotyping of the 8p23 human inversion named Phase Free Inversion Detection Operator (PFIDO) [99]. The method is based on the clustering of multidimensional scaling axes on a pairwise identity-by-state distance matrix across individuals. It identifies the axis displaying most sub-structure and clusters individuals with a18 parameter Gaussian mixture modelling. The most parsimonious model is selected and the conditional probability of an individual belonging to each cluster is calculated using a z-score.
There are two other pipelines to genotype inversions that have been developed by different laboratories [100, 101]. However, they are difficult to apply given the lack of support software. Finally, inversions can also be detected by their specific recombination patterns. The role of recombination as a source of genetic variability for adaptation and evolution is widely known. Moreover, it has been recently reported that recombination patterns can define distinct chromosomal subpopulations that may influence phenotypic traits. RecombClust [102] is a tool that is able to detect chromosomal subpopulations based on recombination histories using SNP array data. RecombClust can be used to detect inversions, regions under selection or where recombination is under regulation in a subgroup of individuals.
Copy number variants
In addition to the duplication of genetic material in homologous chromosomes, occasionally, there are chromosomal segments with a varying number of repeats between individuals known as CNV. CNVs can represent gains of material when an individual has more than the two expected copies, and losses when an individual has less than two copies, see Figure 2. CNVs can also lead to a copy number (CN) neutral loss of heterozygosity (CNN LOH) of the segment when a loss in one chromosome is repaired by making a copy of the remaining allele, which results in having two identical copies (LOH) but with a normal CN (CNN). It is also possible to characterize global homozygosity and heterozygosity of individuals, using standard tools such as PLINK and BCFTools.
Figure 2.
Representation of a CNV region in a normal state, gain of genetic material, loss of genetic material and CNN LOH.
Although the possibility of genotyping CNVs from SNP arrays with specialized tools has been available for some time, bioinformaticians are still challenged to develop reliable algorithms capable of detecting CNVs with high accuracy. Ideally, these tools should achieve good specificity and sensitivity, and low false positive and negative rates, besides being applicable to different types of arrays. To date, the development of many different methods aiming to fit all these requirements has been scrutinized in different articles that aim to determine the methods with the best performances [103–116]. The algorithms are, however, difficult to compare, not only due to their different requirements in terms of input data but also because most of them have different tuning parameters. Therefore, adjusting and finding the right parameters plays a determinant role. Some authors argue that algorithms that are specifically developed for a certain SNP array tend to perform better than platform-independent tools or algorithms that have been readapted [106]. Another relevant issue is that deletions are easier to detect than duplications, given that the first represents a 50% decrease in signal intensity while the latter implies a 33% increase. Even when using the same type of array, some programs will perform better than others depending on the characteristics of the samples. Because there is not yet a gold standard for CNV genotyping, the consensus is that no single algorithm is sufficiently powered and results should be accepted only if obtained by two or more different tools.
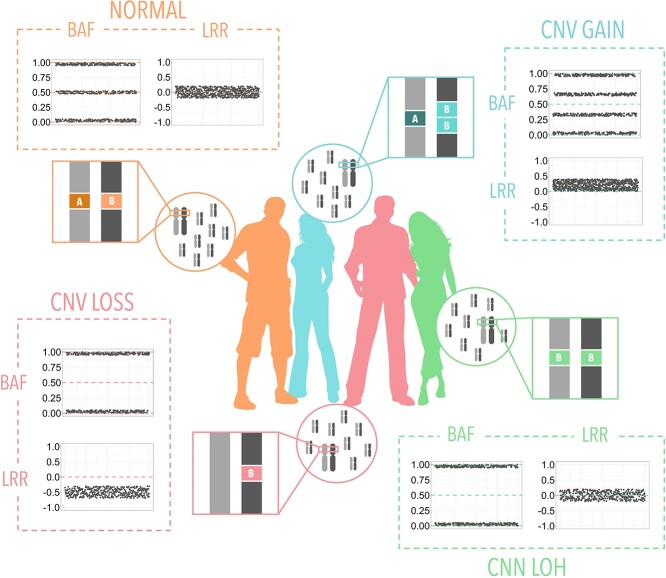
The genotyping of CNVs from SNP arrays is based on the analysis of the B allele frequency (BAF) and the logR Ratio (LRR) (Figure 3), with two basic steps involving the normalization of signals, which is used to clean the data; and the detection of the region and its CN, which can be done by different approaches. Depending on the detection method, we can roughly divide the algorithms into those based on Hidden Markov Models (HMM), segmentation or a combination of both. More practically, we can divide the algorithms into groups regarding the genotyping platform for which they are most suitable, see Table 7 and Supplementary Table S6. The first set includes those methods for Illumina arrays, the second group for Affymetrix, and the third for both or other platforms. In general, CNV genotyping tools provide their own normalization step, but for those that do not implement it, there are tools like ITALICS [117] or affy2sv [118].
Figure 3.
Changes in the BAF and LRR within CNVs of different types. (Orange) Normal state where the BAF (a measure of heterozygosity) is on average 0 or 1 for homozygous probes and 0.5 for heterozygous probes and the LLR (a normalized measure of DNA content) is on average 0. (Blue) CN gain is represented by a split of the BAF signal at 1/3 and 2/3 and a gain in LRR. (Red) CN loss is represented by a loss BAF for heterozygous probes (0.5) and a loss in LRR signal. (Green) Loss of heterozygosis by CNV is represented by a loss BAF for heterozygous probes and no change in the LRR signal.
Table 7.
Top-five tools for the study of CNVs with their characteristics
| Tool | Type | Availability | Input data | Algorithm | Characteristics | Year | Reference |
|---|---|---|---|---|---|---|---|
| PennCNV | Command-line (Perl) | Free | Processed Intensity files with LRR and BAF + PFB (Population frequency of B allele) files (supplied with the package for several Affymetrix/Illumina arrays) | Hidden Markov Model (HMM) | Visualization options | 2007 | [127] |
| QuantiSNP | Command-line (MATLAB) | Free | Illumina Infinium I/II or Affymetrix 500 K and SNP 6.0 processed intensity files with LRR and BAF | Objective Bayes Hidden Markov Model (OB-HMM) | Visualization options Detects Loss of Heterozygosity | 2007 | [128] |
| Birdsuite | Command-line (Bash, needs R, JAVA, Matlab, Python) | Free | Affymetrix CEL files (Genome-Wide Human SNP Array 6.0) Illumina 610 (beta version) | Birdseye - Hidden Markov Model (HMM) Canary - One-dimensional Gaussian mixture model (GMM) | Linux only PLINK conversion pipeline | 2008 | [124] |
| SCIMMkit | Command-line (Perl, R) | Free | Final call report from Illumina BeadStudio (Infinium II and GoldenGate BeadXpress chips) | SCIMM (SNP-Conditional Mixture Modeling) - Mixture-likelihood based clustering SCOUT (SNP-Conditional Outlier detection) - Scoring function. | Visualization options (scatterplots) | 2008 | [121] |
| GLAD | R package | Free | Preprocessed files with LRR values | Segmentation based on Adaptive Weights Smoothing (AWS) | Specific for cancer samples | 2004 | [133] |
CNV algorithms for Illumina data
Illumina provides its own algorithm named CNVPartition that uses bivariate Gaussian distributions for CNV genotyping. This tool is part of the GenomeStudio platform, which can be freely downloaded from their website. It is fast and easy for having a general overview of the data [103, 108], it also has high specificity [105] and positive predicted rate both with deletions and duplications [113] but performs poorly in terms of sensitivity [103, 105, 113]. CamCNV [119] is an alternative R-package capable of detecting rare CNVs with at least three probes. The command-line tool, PlatinumCNV [120] is also specific for Illumina data. This algorithm applies a Gaussian mixture model with a cross-sample approach that is able to detect allele-specific patterns. It provides some additional R functions to plot the results but its main disadvantage is the requirement of large sample sizes (of several thousand) for sufficient power to detect genome-wide CNVs. SCIMMkit [121, 122] is another command-line software that includes three tools for CN detection: SCIMM that genotypes deletions, SCIMMSearch that generates probe sets to be used by SCIMM, and SCOUT that detects rare deletion and duplication variants. The modelling algorithm, that requires Perl and R to run, needs statistical knowledge for correct use [103] but provides high detection rates [108]. Finally, Trityper [123] applies a cross-sample approach but, although it can interpret single and tri-allelic SNPs, it is only able to genotype deletions and not duplications [103, 108].
CNV algorithms for Affymetrix data
Affymetrix also provides their own tools for the analysis of CNVs. They are implemented on a desktop application called Affymetrix Genotyping Console (GTC), which is also available in the command-line through the Affymetrix Array Power Tools (APT). For Human Mapping 100 and 500 K arrays, they provide the Affymetrix GeneChip Chromosome Copy Number Analysis Tool (CNAT) version CN4 and for Genome-Wide Human SNP 6.0 Array they offer the CNAT version CN5 and the Canary algorithm, which is part of Birdsuite [124]. Birdsuite is a set of four tools developed by the Broad Institute including (i) Canary, that genotypes common CNVs, (ii) Birdseed, that genotypes biallelic SNPs, (iii) Birdseye, that detects rare and de novo CNVs and (iv) Fawkes, that integrates CNV information to produce consistent SNP genotypes even for non-biallelic cases. While Birdsuite has low reproducibility of its own results [109, 111], it has a high success rate on validated CNVs with more than 20 markers [115]. Moreover, it also performs well in the detection of rare CNVs. Birdsuite was trained in HapMap samples and that raises the possibility of a biased outcome, which has been confirmed by some authors [109] and rejected by others [106].
Finally, COKGEN [125] is an R-package ready to perform different steps including normalization and a two-stage CNV detection based on optimization [103], with adjustable parameters to facilitate the detection of rare CNVs. It retrieves fewer events than the others but a high percentage of them can be validated, which results in a high concordance rate but low sensitivity [126].
CNV algorithms for multiple platforms
The last group of algorithms includes those that can deal with more than one type of data, they include command-line programs, a few R packages and three commercial desktop applications.
PennCNV [127] is one of the most used command-line tools and thus, and one of the most reviewed. In general, it stands out for its easy use with well-supported instructions and detailed guidelines with quality control metrics to deal with problematic data [103, 108, 109, 112, 116] as well as a good performance in terms of specificity, reliability, reproducibility and bias, both in detecting the CNVs and assessing the number of copies. It can be used to detect varying degrees of genetic relatedness [110], and it is reported as the most suitable tool by the majority of performance studies. The weakness of PennCNV, as reported by in some articles [105, 126], is a low sensitivity that increases using pedigree information [111]. This is probably related to the detection of small CNVs [112], since sensitivity increases in large CNVs [107, 111, 115].
Another widely used command-line tool is QuantiSNP [128], which provides easily modifiable parameters and has been reported to perform well in datasets with diverse characteristics, detecting a high number of CNVs [112]. As many detections may not be validated with other tools [115], it has medium sensitivity and specificity together with high false positive and negative rates [105, 113, 126]. Although originally developed for earlier versions of the Affymetrix chips, dChipSNP [129] is another tool that can now deal with both Illumina and Affymetrix platforms. The software automatically determines the optimal parameters, which cannot be accessed nor modified by the user [114] and seem to be best fitted for Affymetrix data [108]. According to some authors, dChipSNP is biased towards the detection of duplications over deletions [109, 114], and this can be explained by the fact that this software was originally developed to detect LOH regions and clustering in cancer samples [129]. The last command-line tool, cnvHap, [130] works with Illumina, Affymetrix and Agilent data. Performing haplotype-based detection, the algorithm is especially successful with small CNVs of less than 10 probes. According to some authors [126], cnvHap finds approximately from 5 to 10 times more CNVs than other tools depending on the dataset, which results in lower concordance rates but higher sensitivity than others.
Regarding R packages, R-GADA [131] provides a complete and flexible pipeline, being able to genotype CNVs, graphically display the results and perform association analysis for Affymetrix, Illumina or aCGH arrays [103]. This package uses pre-computed LRR values and applies a segmentation algorithm based on Genome Alteration Detection Analysis (GADA) which gives a clear advantage in processing speed [108]. Another two R packages that also implement a segmentation-based algorithm to pre-computed LRR values are VEGA [132] and GLAD [133]. The first one is based on the Mumford and Shah model, while the second was initially developed for aCGH arrays but it can be applied to SNP arrays applying an Adaptive Weights Smoothing algorithm [115]. In general, segmentation-based algorithms tend to find more CNV segments than other tools [112, 115].
Finally, Partek Genomics Suite (PGS), SNP & Variation Suite (SVS) and Nexus Copy Number are three commercial tools developed by Partek, Golden Helix and BioDiscovery, respectively. They can be purchased and downloaded from their own websites and all of them provide a user-friendly graphical interface with data viewers and support microarray data from several platforms. In terms of performance, PGS detects fewer events than the others but with a high validation percentage, and it tends to perform well with frequent and large CNVs. It also shows a low sensitivity that increases with quantile normalization [110, 111]. On the contrary, SVS is one of the algorithms that find more CNVs, which are not validated with other tools [110]. Finally, Nexus Copy Number includes two algorithms named Rank and SNPRank that perform well but are affected by high false-positive rates and high sample-to-sample variation. For lowering false-positive findings DeepCNV [134] and SeeCiTe [135] are two tools to perform an automatic validation of the obtained CNV calls. Both programs can be used after running any of the mentioned programs, keeping in mind that the last one works with trios’ data.
Genomic mosaicism
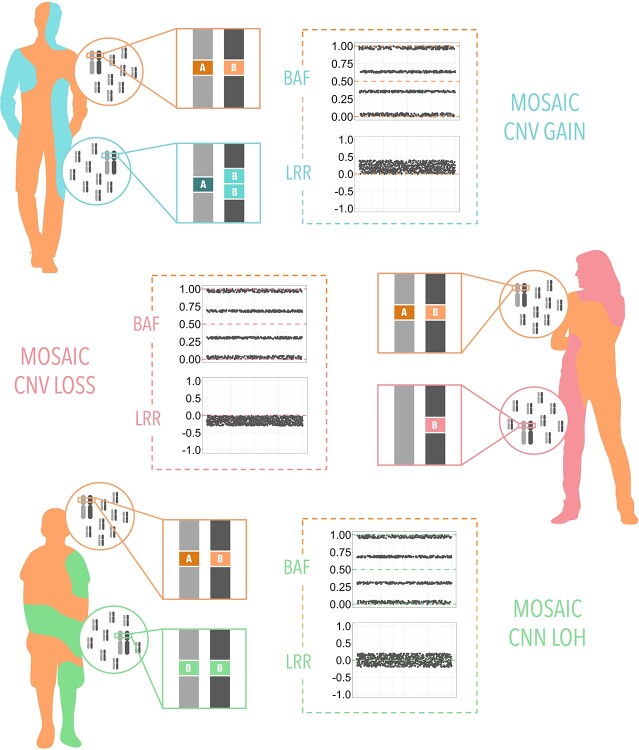
Mutations occurring during stages of continuous cell divisions can lead to the coexistence of groups of cells with genotypic differences within the same organism, a phenomenon called mosaicism. Mutations that imply duplications, deletions or reorganization of small DNA fragments are called CNV mosaicisms, which are the existence of a CNV but only in a percentage of the organism’s cells. The detection of mosaic events is also derived from the BAF and LRR signals but with different patterns from those seen for constitutional CNVs (Figure 4). It is important to note that most of the algorithms that have been developed to detect CNVs cannot deal with mosaicisms, which are usually ignored or misclassified. For this reason, it has been necessary to create new specific tools. We have listed algorithms that have been designed to detect mosaic events in SNP array data, see Table 8 and Supplementary Table S7. We make the distinction between methods applied in non-cancer and cancer studies.
Figure 4.
Changes in the BAF and LRR depending on the type of mosaicism. (Top) A mosaic CN gain is represented by a split of the BAF signal between in values between 1/3 and 2/3 and a gain in LRR. (Middle) A mosaic CN loss is represented by a BAF split between 0 and 1 and a loss of LRR. (Bottom) A mosaic loss of heterozygosity is represented by a BAF split between 0 and 1 and a normal LRR.
Non-cancer samples
Mosaic Alteration Detection (MAD) [136] is probably one of the most used tools. First, it applies the GADA algorithm [131] to perform segmentation on the deviation of the BAF signal from its expected value. Then, it classifies the events depending on the type of mosaicism found using the LRR signal. It can detect deletions, duplications and copy neutral changes as well as regions of homozygosity due to IBD. Its high sensitivity and specificity allow the capture of both previously described alterations and new variants, even if they are small or affect a low percentage of the cells in the sample. The MAD algorithm has been implemented in an R package [131] to facilitate the analyses. Aging-related mosaic loss of chromosome Y (mLOY) is the most commonly acquired mutation in males’ genome. LOY is a neutral mutation [137] that is associated with several human diseases including cancer [138, 139], Alzheimer’s disease [140] and cardiovascular disease [141]. The specific tool MADLoy [142] for mLOY detection provides a robust and efficient calling for large studies.
There is an extension of bcftools software named MoChA [143] that runs on the command line but needs phased VCF files with either BAF and LRR or allelic depth (AD) values. A recently developed pipeline that converts raw data from both Affymetrix and Illumina platforms to VCF files has made the tool more accessible. MoChA detects losses and gains of DNA as well as LOH regions. Finally, parent–child trios data can be analyzed with triPOD [144]. This method, which is a Perl script that uses R for graphical visualization, has a command-line version. The method is also accessible from a web application. The tool can detect deletions, amplifications, and uniparental disomies, heterodisomies and isodisomies, and has the peculiarity that it can tell if the aberration is inherited from the father or mother, or if there exists a paternal or maternal contribution. The calling method, namely Parent-of-Origin-based Detection (POD), is different than other programs. POD is based on the identification of SNPs which are informative for abnormal parental contribution, i.e. when the comparison of progeny and parental genotypes potentially reveals abnormal parental contribution in the region.
None of the tools can provide the percentage of cells that carry the chromosomic aberration. For this purpose, there is a tool named Distribution Analysis by Fitting Integrated Probabilities (DANFIP) [145]: it provides the frequency of big aberrations—that have to be previously known—with a precision of at least 0.1% and an accuracy of at least 4%. The method uses an inverse continuous distribution function that can assess the degree of mosaicism from the BAF signal. It performs well for simple deletion monosomy, partial monosomy, simple trisomy, partial trisomy and uniparental disomy with trisomy mosaicism. There are two other algorithms for smaller unknown CNVs in mosaic that give the percentage of affected cells. The first one, MONTAGE [146], written in Perl and Bash languages, uses BAF and LRR values in a sliding window approach that looks for allelic imbalances and classifies them as a deletion or duplication in relation to normal diploid CN. Also, it detects LOH regions. An interesting characteristic is that MONTAGE results can be directly analyzed for phenotype associations with ParseCNV. HaplotypeCN [126] is a command-line tool with two important strengths: first, it can detect parent-specific CN change on either chromosome and provides haplotype-specific results; second, the CN are provided as fractional numbers, so it can detect somatic mutations in heterogeneous cell populations. HaplotypeCN finds a low number of CNVs high concordance rate but low sensitivity [126].
Cancer samples
Mosaicism appears as a consequence of a mutation during a stage of continuous cell divisions. Tumorigenesis is, therefore, a prolific scenario for mosaicisms, being one of the characteristics of cancer cells. For this reason, there are many CNV mosaicism detection tools that are specialized in the detection of genetic aberrations in cancer samples. These tools are particularly designed to deal with a heterogeneous sample. In cancer, the presence of normal and cancerous cells sums up the inherent intratumor heterogeneity. The tools can also detect aneuploidies.
BAFSegmentation [147] is one of the most complete tools, as it detects LOH and allelic imbalances including gains, loses and hemizygous loses and is able to provide an estimation of the percentage of cancerous cells. This last functionality is as well a characteristic of ASCAT [148], also designed for tumor samples. CNAG [149] and TAFFYS [150] have been specifically developed for the analysis of CN alterations and LOH in cancer cells and are freely available. In addition, the Copy Number Analyser for Affymetrix GeneChip arrays (CNAG) is a software from the university of Tokio that, as reported in Baross et al. [114], detects a higher proportion of duplications and a lower proportion of deletions than other programs. Its HMM parameters are optimized to detect full CN changes in mostly diploid samples, so it is recommended to adjust them for detection of mosaic CNVs, among others. Finally, TAFFYS runs in MATLAB and can work with diluted tumor samples with a minimum of 30% of cancer cells. After suppressing and modelling the signal noise, it applies an HMM for CN inference with visualization capacity. Other methods for CNV in cancer are PICNIC [151], that predicts absolute CN; PennCNV-tumor [152], which is able to quantify the intratumor heterogeneity; TAPS [153], that has high sensitivity; OncoSNP [154]; genoCN [155]; GISTIC [156] and hapLOH [157], which can work with a very low proportion of aberrant cells, either in tumor-normal mixture samples or with clonal aberrations in non-malignant tissues.
Most of the algorithms that detect CNVs, both in mosaicism and not, report them as a list of genome regions. Some of them provide their own algorithms to perform associations once the calls have been obtained (see Tables 7 and 8, Supplementary Tables S6 and S7). For those not offering the option, this step can be done externally with any association analysis tool able to work with genomic regions, for example, the R packages regioneR [158] and CNVassoc [159].
Recent studies applying inference methods of SNP data
Our selection of methods for this review was based on the methods’ reported application between January 2020 and September 2021 in studies with relevant results. Here, we describe the context of use of the most popular ones.
Software devoted to the analysis of population structure and ancestry was not only used to infer population patterns [40, 160–163] but also to support studies of diseases and traits in which population admixture and ancestry play determinant roles [164–173]. The same could be observed for IBD detection, with some articles centered on population structure [55, 174–176] and others on practical applications [177, 178]. In ancestry studies, more than one algorithm was usually applied, and we did not observe a clear trend towards any of the tools, as almost all of them had been referenced during the analyzed period. By contrast, in IBD calculations, one run of the tool seemed to be sufficient for estimations. fastIBD (BEAGLE) along with GERMLINE were the most recurrently employed.
Regarding heritability, LD Score (LDSC) was, for instance, applied to study face and brain shape [179] as well as tissue-specific gene sets [180], while LDAK helped to reach a better understanding of heritability and variance of the phenotypic traits [181, 182]. For the calculation of PRS, PRSice and PRS-CS were the first choices, followed by LDPred, lassosum and SBayesR. While PRS tools have been mostly used on neurological studies involving Alzheimer’s [183–185], depression [186–188], self-harm [189, 190] or substance use [191–193], they have also been applied on other diseases and conditions [194–199]. Most studies applying LD tools were also on neurological conditions [200–203]. Including these and other phenotypes, Haploview was the most popular tool [204–206].
Regarding CNVs, PennCNV and QuantiSNP were used to detect several rare CNV associations in over 100 000 European ancestry subjects with autoimmune, cardio-metabolic, oncologic and neurological/psychiatric diseases [207]. Studies involving CNVs related to neurological outcomes were the most frequent [208–215], all of them applied PennCNV. In particular, CNV genotyping was crucial in studies involving diseases such as schizophrenia [216, 217], bipolar disorder [216], Parkinson’s disease [218], Autism Spectrum Disorder [219] and attention deficit hyperactivity disorder [220]. Other CNVs studies included different outcomes such as HBV [221, 222], autophagy [223], gallstones disease [224], vesicoureteral reflux [225], esotropia [226], cheap screenings for primary immunodeficiency [227] and COVID-19 potential targets [228]. For these studies, PennCNV was the most popular, followed by Nexus Copy Number, Birdsuite and QuantiSNP. In the context of cancer, the same algorithms were used for the research on cervical cancer [229], multiple myeloma [230], germline PTEN mutations [231] and tumor evolution and response to drugs [232] and thyroid carcinoma [233].
Studies on mosaicism detection in cancer mainly used BAFSegmentation, TAPS, PICNIC, GISTIC and CNAG relating acute myeloid leukemia [234], hyperdiploid childhood acute lymphoblastic leukemia [235], ossification of fibromyxoid tumors [236], gastric adenocarcinoma [237], hepatoblastoma [238], lymphoblastic leukemia [239] and non-Hodgkin B-cell lymphoma [240]. Also, mosaicism genotyping was performed with MoChA, MAD, BAFSegmentation and PICNICfor mastocytosis [241] and drug resistance. Relevant findings of mosaic detection include their contribution to Autism Spectrum Disorder risk [242], predisposition to infections [243] and clonal hematopoiesis [143]. Recent applications on the detection of inversions, included the genotyping of 20 inversions with scoreInvHap to study their role in obesity-related diseases [244], while a similar study in admixed population in Brazil used invClust [245].
Discussion
We have reviewed the current landscape of available options to exploit SNP array data. However, new tools are being developed every year. In addition to SNP arrays, NGS is another important tool to detect multiple genomic substructures and, particularly those with low frequency, such as rare variants and point mutations. Nevertheless, NGS is limited by two important aspects. First, the storage and analysis of large datasets demand large computational resources. Second, its high cost drives most studies to opt for alternative sequencing techniques, which translates into a fewer number of publicly available NGS datasets. Consequently, SNP arrays still remain an important option for large population studies.
Given the high availability of microarray SNP data and methods, extraction of underlying genomic structure is becoming a mainstream addition to GWASs. In addition, huge amounts of SNP data are continuously generated from GWAS studies and made available in public repositories. These datasets can still be exploited and relevant information extracted from them. The genotyping of structural variants is one of the most promising ones. Among the distinct algorithms that have been specially developed to this end, the ones focused on CNV are the most abundant, followed by CNV mosaicism in cancer samples. By contrast, there are only three tools that can genotype human inversions. Whereas for inversion genotyping it is a good option to choose one single method, for CNV genotyping it is recommended to use at least two of them. First, because most of the tools do not cover the entire spectrum of CNV size. Second, because the performance of each algorithm depends on the characteristics of the dataset. Regarding the tools for population structure and ancestry as well as the analysis of LD and IBD, the best option is choosing a program that fits the characteristics of the population. If those are unknown, using more than one tool is recommended. In addition, we encourage to run more than one software to achieve a more accurate calculation of PRSs and the estimation of the heritability of complex traits.
Key Points
Large consortia and public databases have generated large amounts of freely available SNP array data that are underexploited.
SNP array data can be used to extract information on population structure and ancestry, polygenic risk scores, identity-by-descent fragments, linkage disequilibrium, heritability and to genotype structural variants, such as inversions, copy number variants, mosaicisms and recombination histories.
Choosing a tool to make the most from SNP data is challenged by the amount of methods and their diversity.
This review presents software summary tables of many R packages, command-line tools and desktop applications, describing their advantages and disadvantages, input requirements and context of use.
We review current tools in use in recent published literature.
Supplementary Material
The authors work in the Bioinformatic Research Group in Epidemiology, a research group devoted to the development of statistical methods and software for genomics, transcriptomics and exposomics to be applied in environmental epidemiology.
Laura Balagué-Dobón is a PhD student in the Bioinformatics Research Group in Epidemiology of ISGlobal.
Alejandro Cáceres is a principal bioinformatician in the Bioinformatics Research Group in Epidemiology of ISGlobal where he as worked on omics data analysis and methods development since 2008.
Juan R. González leads the Bioinformatics Research Group in Epidemiology of ISGlobal where develops new statistical methods and bioinformatic tools to exploit omic data.
Contributor Information
Laura Balagué-Dobón, Bioinformatics Research Group in Epidemiology of ISGlobal.
Alejandro Cáceres, Bioinformatics Research Group in Epidemiology of ISGlobal.
Juan R González, Bioinformatics Research Group in Epidemiology of ISGlobal.
Funding
This work was supported by the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 874583, ATHLETE); the Ministerio de Ciencia, Innovación y Universidades (MICIU), Agencia Estatal de Investigación (AEI) and Fondo Europeo de Desarrollo Regional, UE (RTI2018-100789-B-I00) also through the Centro de Excelencia Severo Ochoa 2019-2023 Program (CEX2018-000806-S); the Catalan Government through the CERCA Program; and the CIBERESP (ESPG38/2022).
References
- 1. Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet 2011;12:363–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang DG, Fan JB, Siao CJ, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science (80-) 1998;280:1077–82. [DOI] [PubMed] [Google Scholar]
- 3. Peters A, Nawrot TS, Baccarelli AA. Hallmarks of environmental insults. Cell 2021;184:1455–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Samuels DC, Below JE, Ness S, et al. Alternative applications of genotyping array data using multivariant methods. Trends Genet 2020;36:857–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mielczarek M, Szyda J. Review of alignment and SNP calling algorithms for next-generation sequencing data. J Appl Genet 2016;57:71–9. [DOI] [PubMed] [Google Scholar]
- 6. Nielsen R, Paul S, et al. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet 2011;12:443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo Y, He J, Zhao S, et al. Illumina human exome genotyping array clustering and quality control. Nat Protoc 2014;9:2643–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao S, Jing W, Samuels DC, et al. Strategies for processing and quality control of Illumina genotyping arrays. Brief Bioinform 2018;19:765–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gogarten SM, Bhangale T, Conomos MP, et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics 2012;28:3329–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Der Most PJ, Vaez A, Prins BP, et al. QCGWAS: a flexible R package for automated quality control of genome-wide association results. Bioinformatics 2014;30:1185–6. [DOI] [PubMed] [Google Scholar]
- 11. Zheng X, Levine D, Shen J, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 2012;28:3326–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teo YY. Common statistical issues in genome-wide association studies: a review on power, data quality control, genotype calling and population structure. Curr Opin Lipidol 2008;19:133–43. [DOI] [PubMed] [Google Scholar]
- 13. Anderson CA, Pettersson FH, Clarke GM, et al. Data quality control in genetic case-control association studies. Nat Protoc 2010;5:1564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Turner S, Armstrong LL, Bradford Y, et al. Quality control procedures for genome wide association studies NIH public access. Curr Protoc Hum Genet 2011:68:1.19.1–1.19.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J, Samuels DC, Shyr Y, et al. StrandScript: evaluation of Illumina genotyping array design and strand correction. Bioinformatics 2017;33:2399–401. [DOI] [PubMed] [Google Scholar]
- 16. Laurie CC, Doheny KF, Mirel DB, et al. Quality control and quality assurance in genotypic data for genome-wide association studies NIH public access author manuscript. Genet Epidemiol 2010;34:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hunter-Zinck H, Shi Y, Li M, et al. Genotyping array design and data quality control in the million veteran program. Am J Hum Genet 2020;106:535–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Psaty BM, O’Donnell CJ, Gudnason V, et al. Methods in genetics and clinical interpretation cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009;2:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clayton DG, Walker NM, Smyth DJ, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet 2005;37:1243–6. [DOI] [PubMed] [Google Scholar]
- 20. Gondro C, Lee SH, Lee HK, et al. Quality control for genome-wide association studies. Methods Mol Biol 2013;1019:129–47. [DOI] [PubMed] [Google Scholar]
- 21. Al Bkhetan Z, Chana G, Ramamohanarao K, et al. Evaluation of consensus strategies for haplotype phasing. Brief Bioinform 2021;22:bbaa280. [DOI] [PubMed] [Google Scholar]
- 22. Marino A De, Mahmoud AA, Bose M, et al. A comparative analysis of current phasing and imputation software. bioRxiv 2021; 2021.11.04.467340 https://www.biorxiv.org/content/10.1101/2021.11.04.467340v1.full.pdf+html. [DOI] [PMC free article] [PubMed]
- 23. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics 2000;155:945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raj A, Stephens M, Pritchard JK. FastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 2014;197:573–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alexander DH, Lange K. Enhancements to the admixture algorithm for individual ancestry estimation. BMC Bioinformatics 2011;12:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duforet-Frebourg N, Gattepaille LM, Blum MGB, et al. HaploPOP: a software that improves population assignment by combining markers into haplotypes. BMC Bioinformatics 2015;16:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mirzaei S, Wu Y. RENT+: an improved method for inferring local genealogical trees from haplotypes with recombination. Bioinformatics 2017;33:1021–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahn J, Conkright B, Boca SM, et al. POPSTR: inference of admixed population structure based on single-nucleotide polymorphisms and copy number variations. J Comput Biol 2018;25:417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Patterson N, Price AL, Reich D. Population structure and Eigenanalysis. PLoS Genet 2006;2:2074–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–9. [DOI] [PubMed] [Google Scholar]
- 32. Huang Q, Baudis M. Enabling population assignment from cancer genomes with SNP2pop. Sci Rep 2020;10:4846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patterson N, Moorjani P, Luo Y, et al. Ancient admixture in human history. Genetics 2012;192:1065–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao X, Starmer JD. AWclust: point-and-click software for non-parametric population structure analysis. BMC Bioinformatics 2008;9:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol 2015;39:276–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chaichoompu K, Abegaz F, Tongsima S, et al. IPCAPS: an R package for iterative pruning to capture population structure. Source Code Biol Med 2019;14:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee H, Chen L. Inference of kinship using spatial distributions of SNPs for genome-wide association studies. BMC Genomics 2016;17:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lawson DJ, Hellenthal G, Myers S, et al. Inference of population structure using dense haplotype data. PLoS Genet 2012;8:e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hellenthal G, Busby GBJ, Band G, et al. A genetic atlas of human admixture history. Science 2014;343:747–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Y. Inference of population admixture network from local gene genealogies: a coalescent-based maximum likelihood approach. Bioinformatics 2020;36:i326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelleher J, Wong Y, Wohns AW, et al. Inferring whole-genome histories in large population datasets. Nat Genet 2019;51:1330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brisbin A, Bryc K, Byrnes J, et al. Pcadmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol 2012;84:343–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baran Y, Pasaniuc B, Sankararaman S, et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics 2012;28:1359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Price AL, Tandon A, Patterson N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet 2009;5:e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maples BK, Gravel S, Kenny EE, et al. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet 2013;93:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guan Y. Detecting structure of haplotypes and local ancestry. Genetics 2014;196:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arriaga-MacKenzie IS, Matesi G, Chen S, et al. Summix: a method for detecting and adjusting for population structure in genetic summary data. Am J Hum Genet 2021;108:1270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y, Byun J, Cai G, et al. FastPop: a rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinformatics 2016;17:122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang H, Sofer T, Zhang X, et al. Local ancestry inference in large pedigrees. Sci Rep 2020;10:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen S, Ghandikota S, Gautam Y, et al. MI-MAAP: marker informativeness for multi-ancestry admixed populations. BMC Bioinformatics 2020;21:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gusev A, Lowe JK, Stoffel M, et al. Whole population, genome-wide mapping of hidden relatedness. Genome Res 2009;19:318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shemirani R, Belbin GM, Avery CL, et al. Rapid detection of identity-by-descent tracts for mega-scale datasets. Nat Commun 2021;12:3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Naseri A, Liu X, Tang K, et al. RaPID: ultra-fast, powerful, and accurate detection of segments identical by descent (IBD) in biobank-scale cohorts. Genome Biol 2019;20:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou Y, Browning SR, Browning BL. A fast and simple method for detecting identity-by-descent segments in large-scale data. Am J Hum Genet 2020;106:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saada JN, Kalantzis G, Shyr D, et al. Identity-by-descent detection across 487,409 British samples reveals fine scale population structure and ultra-rare variant associations. Nat Commun 2020;11:6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. Am J Hum Genet 2011;88:173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Browning BL, Browning SR. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 2013;194:459–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rodriguez JM, Bercovici S, Huang L, et al. Parente2: a fast and accurate method for detecting identity by descent. Genome Res 2015;25:280–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seidman DN, Shenoy SA, Kim M, et al. Rapid, phase-free detection of long identity-by-descent segments enables effective relationship classification. Am J Hum Genet 2020;106:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dimitromanolakis A, Paterson AD, Sun L. Fast and accurate shared segment detection and relatedness estimation in un-phased genetic data via truffle. Am J Hum Genet 2019;105:78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Han L, Abney M. Identity by descent estimation with dense genome-wide genotype data. Genet Epidemiol 2011;35:557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brown MD, Glazner CG, Zheng C, et al. Inferring coancestry in population samples in the presence of linkage disequilibrium. Genetics 2012;190:1447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Albrechtsen A, Korneliussen TS, Moltke I, et al. Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet Epidemiol 2009;33:266–74. [DOI] [PubMed] [Google Scholar]
- 64. Browning SR, Browning BL. Probabilistic estimation of identity by descent segment endpoints and detection of recent selection. Am J Hum Genet 2020;107:895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou Y, Browning SR, Browning BL. IBDkin: fast estimation of kinship coefficients from identity by descent segments. Bioinformatics 2020;36:4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gorfine M, Berndt SI, Chang-Claude J, et al. Heritability estimation using a regularized regression approach (HERRA): applicable to continuous, dichotomous or age-at-onset outcome. PLoS One 2017;12:e0181269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Finucane HK, Bulik-Sullivan B, Gusev A, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015;47:1228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pazokitoroudi A, Chiu AM, Burch KS, et al. Quantifying the contribution of dominance deviation effects to complex trait variation in biobank-scale data. Am J Hum Genet 2021;108:799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Speed D, Hemani G, Johnson MR, et al. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet 2012;91:1011–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Euesden J, Lewis CM, O’Reilly PF. PRSice: polygenic risk score software. Bioinformatics 2015;31:1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Choi SW, O'Reilly PF. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience 2019;8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Igo RP, Kinzy TG, Cooke Bailey JN. Genetic risk scores. Curr Protoc Hum Genet 2019;104:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang J, Lee SH, Goddard ME, et al. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Robinson MR, Kleinman A, Graff M, et al. Genetic evidence of assortative mating in humans the life lines cohort study †, Genetic Investigation of Anthropometric Traits (GIANT) consortium. Nat Hum Behav 2017;1:16. [Google Scholar]
- 77. Lloyd-Jones LR, Zeng J, Sidorenko J, et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nat Commun 2019;10:5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen LM, Yao N, Garg E, et al. PRS-on-spark (PRSoS): a novel, efficient and flexible approach for generating polygenic risk scores. BMC Bioinformatics 2018;19:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cai M, Xiao J, Zhang S, et al. A unified framework for cross-population trait prediction by leveraging the genetic correlation of polygenic traits. Am J Hum Genet 2021;108:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Prive F, Aschard H, Ziyatdinov A, et al. Efficient analysis of large-scale genome-wide data with two R packages: Bigstatsr and bigsnpr. Bioinformatics 2018;34:2781–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mak TSH, Porsch RM, Choi SW, et al. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol 2017;41:469–80. [DOI] [PubMed] [Google Scholar]
- 82. Song S, Jiang W, Hou L, et al. Leveraging effect size distributions to improve polygenic risk scores derived from summary statistics of genome-wide association studies. PLoS Comput Biol 2020;16:e1007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Privé F, Arbel J, Vilhjálmsson BJ. LDpred2: better, faster, stronger. Bioinformatics 2021;36:5424–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ge T, Chen CY, Ni Y, et al. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun 2019;10:1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhu X, Stephens M. Bayesian large-scale multiple regression with summary statistics from genome-wide association studies. Ann Appl Stat 2017;11:1561–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Newcombe PJ, Nelson CP, Samani NJ, et al. A flexible and parallelizable approach to genome-wide polygenic risk scores. Genet Epidemiol 2019;43:730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pattee J, Pan W. Penalized regression and model selection methods for polygenic scores on summary statistics. PLoS Comput Biol 2020;16:e1008271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bhatnagar SR, Yang Y, Lu T, et al. Simultaneous SNP selection and adjustment for population structure in high dimensional prediction models. PLoS Genet 2020;16:e1008766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chun S, Imakaev M, Hui D, et al. Non-parametric polygenic risk prediction via partitioned GWAS summary statistics. Am J Hum Genet 2020;107:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rüschendorf F, Nürnberg P. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics 2005;21:2123–5. [DOI] [PubMed] [Google Scholar]
- 91. Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- 92. Ong Twee-Hee R, Teo Y-Y. varLD: a program for quantifying variation in linkage disequilibrium patterns between populations. Bioinformatics 2010;26:1269–70. [DOI] [PubMed] [Google Scholar]
- 93. Kim SA, Cho CS, Kim SR, et al. A new haplotype block detection method for dense genome sequencing data based on interval graph modeling of clusters of highly correlated SNPs. Bioinformatics 2018;34:388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Taliun D, Gamper J, Pattaro C. Efficient haplotype block recognition of very long and dense genetic sequences. BMC Bioinformatics 2014;15:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Pattaro C, Ruczinski I, Fallin DM, et al. Haplotype block partitioning as a tool for dimensionality reduction in SNP association studies. BMC Genomics 2008;9:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cáceres A, Sindi SS, Raphael BJ, et al. Identification of polymorphic inversions from genotypes. BMC Bioinformatics 2012;13:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Cáceres A, González JR. Following the footprints of polymorphic inversions on SNP data: from detection to association tests. Nucleic Acids Res 2015;43:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ruiz-Arenas C, Cáceres A, López-Sánchez M, et al. scoreInvHap: inversion genotyping for genome-wide association studies. PLoS Genet 2019;15:e1008203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Salm MPA, Horswell SD, Hutchison CE, et al. The origin, global distribution, and functional impact of the human 8p23 inversion polymorphism. Genome Res 2012;22:1144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bansal V, Bashir A, Bafna V. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res 2007;17:219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Boettger LM, Handsaker RE, Zody MC, et al. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet 2012;44:881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ruiz-Arenas C, Cáceres A, López M, et al. Identifying chromosomal subpopulations based on their recombination histories advances the study of the genetic basis of phenotypic traits. Genome Res 2020;31:1802–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Winchester L, Ragoussis J. Algorithm implementation for cnv discovery using Affymetrix and Illumina snp array data. Methods Mol Biol 2012;838:291–310. [DOI] [PubMed] [Google Scholar]
- 104. Li W, Olivier M. Current analysis platforms and methods for detecting copy number variation. Physiol Genomics 2013;45:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Marenne G, Rodríguez-Santiago B, Closas MG, et al. Assessment of copy number variation using the Illumina Infinium 1M SNP-array: a comparison of methodological approaches in the Spanish bladder cancer/EPICURO study. Hum Mutat 2011;32:240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pinto D, Darvishi K, Shi X, et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat Biotechnol 2011;29:512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kim S-Y, Kim J-H, Chung Y-J. Effect of combining multiple CNV defining algorithms on the reliability of CNV calls from SNP genotyping data. Genomics Inform 2012;10:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Winchester L, Yau C, Ragoussis J. Comparing CNV detection methods for SNP arrays. Brief Funct Genomic Proteomic 2009;8:353–66. [DOI] [PubMed] [Google Scholar]
- 109. Zhang X, Du R, Li S, et al. Evaluation of copy number variation detection for a SNP array platform. BMC Bioinformatics 2014;15:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Castellani CA, Melka MG, Wishart AE, et al. Biological relevance of CNV calling methods using familial relatedness including monozygotic twins. BMC Bioinformatics 2014;15:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zhang D, Qian Y, Akula N, et al. Accuracy of CNV detection from GWAS data. PLoS One 2011;6:e14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Dellinger AE, Saw SM, Goh LK, et al. Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Res 2010;38:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lin P, Hartz SM, Wang JC, et al. Copy number variation accuracy in genome-wide association studies. Hum Hered 2011;71:141–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Baross Á, Delaney AD, Li HI, et al. Assessment of algorithms for high throughput detection of genomic copy number variation in oligonucleotide microarray data. BMC Bioinformatics 2007;8:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nutsua ME, Fischer A, Nebel A, et al. Family-based benchmarking of copy number variation detection software. PLoS One 2015;10:e0133465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Eckel-Passow JE, Atkinson EJ, Maharjan S, et al. Software comparison for evaluating genomic copy number variation for Affymetrix 6.0 SNP array platform. BMC Bioinformatics 2011;12:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Rigaill G, Hupé P, Almeida A, et al. ITALICS: an algorithm for normalization and DNA copy number calling for Affymetrix SNP arrays. Bioinformatics 2008;24:768–74. [DOI] [PubMed] [Google Scholar]
- 118. Hernandez-Ferrer C, Quintela Garcia I, Danielski K, et al. affy2sv: an R package to pre-process Affymetrix CytoScan HD and 750K arrays for SNP, CNV, inversion and mosaicism calling. BMC Bioinformatics 2015;16:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dennis J, Walker L, Tyrer J, et al. Detecting rare copy number variants from Illumina genotyping arrays with the CamCNV pipeline: segmentation of z-scores improves detection and reliability. Genet Epidemiol 2021;45:237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kumasaka N, Fujisawa H, Hosono N, et al. PlatinumCNV: a Bayesian Gaussian mixture model for genotyping copy number polymorphisms using SNP array signal intensity data. Genet Epidemiol 2011;35:831–44. [DOI] [PubMed] [Google Scholar]
- 121. Zerr T, Cooper GM, Eichler EE, et al. Targeted interrogation of copy number variation using SCIMMkit. Bioinformatics 2009;26:120–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cooper GM, Zerr T, Kidd JM, et al. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet 2008;40:1199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Franke L, de Kovel CGF, Aulchenko YS, et al. Detection, imputation, and association analysis of small deletions and null alleles on oligonucleotide arrays. Am J Hum Genet 2008;82:1316–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008;40:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yavaş G, Koyutürk M, Ozsoyoğlu M, et al. COKGEN: a software for the identification of rare copy number variation from SNP microarrays. Pac Symp Biocomput 2010;15:371–82. [PubMed] [Google Scholar]
- 126. Lin YJ, Chen YT, Hsu SN, et al. HaplotypeCN: copy number haplotype inference with hidden markov model and localized haplotype clustering. PLoS One 2014;9:e96841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 2007;17:1665–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Colella S, Yau C, Taylor JM, et al. QuantiSNP: an objective bayes hidden-Markov model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res 2007;35:2013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lin M, Wie LJ, Sellers WR, et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics 2004;20:1233–40. [DOI] [PubMed] [Google Scholar]
- 130. Coin LJM, Asher JE, Walters RG, et al. CnvHap: an integrative population and haplotype-based multiplatform model of SNPs and CNVs. Nat Methods 2010;7:541–6. [DOI] [PubMed] [Google Scholar]
- 131. Pique-Regi R, Cáceres A, González JR. R-Gada: a fast and flexible pipeline for copy number analysis in association studies. BMC Bioinformatics 2010;11:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Morganella S, Cerulo L, Viglietto G, et al. VEGA: variational segmentation for copy number detection. Bioinformatics 2010;26:3020–7. [DOI] [PubMed] [Google Scholar]
- 133. Hupé P, Stransky N, Thiery JP, et al. Analysis of array CGH data: from signal ratio to gain and loss of DNA regions. Bioinformatics 2004;20:3413–22. [DOI] [PubMed] [Google Scholar]
- 134. Glessner JT, Hou X, Zhong C, et al. DeepCNV: a deep learning approach for authenticating copy number variations. Brief Bioinform 2021;22:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lavrichenko K, Helgeland Ø, Njølstad PR, et al. SeeCiTe: a method to assess CNV calls from SNP arrays using trio data. Bioinformatics 2021;37:1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. González JR, Rodríguez-Santiago B, Cáceres A, et al. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinformatics 2011;12:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Guo X, Dai X, Zhou T, et al. Mosaic loss of human Y chromosome: what, how and why. Hum Genet 2020;139:421–46. [DOI] [PubMed] [Google Scholar]
- 138. Forsberg LA, Rasi C, Malmqvist N, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet 2014;46:624–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Noveski P, Madjunkova S, Stefanovska ES, et al. Loss of Y chromosome in peripheral blood of colorectal and prostate cancer patients. PLoS One 2016;11:e0146264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Dumanski JP, Lambert JC, Rasi C, et al. Mosaic loss of chromosome Y in blood is associated with Alzheimer disease. Am J Hum Genet 2016;98:1208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Haitjema S, Kofink D, Van Setten J, et al. Loss of y chromosome in blood is associated with major cardiovascular events during follow-up in men after carotid endarterectomy. Circ Cardiovasc Genet 2017;10:e001544. [DOI] [PubMed] [Google Scholar]
- 142. González JR, López-Sánchez M, Cáceres A, et al. MADloy: robust detection of mosaic loss of chromosome Y from genotype-array-intensity data. BMC Bioinformatics 2020;21:533–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Loh P-R, Genovese G, McCarroll SA. Monogenic and polygenic inheritance become instruments for clonal selection. Nature 2020;584:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Baugher JD, Baugher BD, Shirley MD, et al. Sensitive and specific detection of mosaic chromosomal abnormalities using the parent-of-origin-based detection (POD) method. BMC Genomics 2013;14:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Markello TC, Carlson-Donohoe H, Sincan M, et al. Sensitive quantification of mosaicism using high density SNP arrays and the cumulative distribution function. Mol Genet Metab 2012;105:665–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Glessner JT, Chang X, Liu Y, et al. MONTAGE: a new tool for high-throughput detection of mosaic copy number variation. BMC Genomics 2021;22:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Staaf J, Lindgren D, Vallon-Christersson J, et al. Segmentation-based detection of allelic imbalance and loss-of-heterozygosity in cancer cells using whole genome SNP arrays. Genome Biol 2008;9:R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Van LP, Nilsen G, Nordgard SH, et al. Analyzing cancer samples with SNP arrays. Methods Mol Biol 2012;802:57–72. [DOI] [PubMed] [Google Scholar]
- 149. Nannya Y, Sanada M, Nakazaki K, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res 2005;65:6071–9. [DOI] [PubMed] [Google Scholar]
- 150. Liu Y, Li A, Feng H, et al. TAFFYS: an integrated tool for comprehensive analysis of genomic aberrations in tumor samples. PLoS One 2015;10:e0129835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Greenman CD, Bignell G, Butler A, et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 2010;11:164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Chen GK, Chang X, Curtis C, et al. Precise inference of copy number alterations in tumor samples from SNP arrays. Bioinformatics 2013;29:2964–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Rasmussen M, Sundström M, Göransson Kultima H, et al. Allele-specific copy number analysis of tumor samples with aneuploidy and tumor heterogeneity. Genome Biol 2011;12:R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Yau C. OncoSNP-SEQ: a statistical approach for the identification of somatic copy number alterations from next-generation sequencing of cancer genomes. Bioinformatics 2013;29:2482–4. [DOI] [PubMed] [Google Scholar]
- 155. Sun W, Wright FA, Tang Z, et al. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res 2009;37:5365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Vattathil S, Scheet P. Haplotype-based profiling of subtle allelic imbalance with SNP arrays. Genome Res 2013;23:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Gel B, Díez-Villanueva A, Serra E, et al. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 2016;32:289–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Subirana I, Diaz-Uriarte R, Lucas G, et al. CNVassoc: association analysis of CNV data using R. BMC Med Genomics 2011;4:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kutanan W, Liu D, Kampuansai J, et al. Reconstructing the human genetic history of mainland Southeast Asia: insights from genome-wide data from Thailand and Laos. Mol Biol Evol 2021;38:3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Chaichoompu K, Abegaz F, Cavadas B, et al. A different view on fine-scale population structure in Western African populations. Hum Genet 2020;139:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yang X-Y, Rakha A, Chen W, et al. Tracing the genetic legacy of the Tibetan empire in the Balti. Mol Biol Evol 2021;38:1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kerminen S, Cerioli N, Pacauskas D, et al. Changes in the fine-scale genetic structure of Finland through the 20th century. PLoS Genet 2021;17:e1009347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Fachal L, Aschard H, Beesley J, et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat Genet 2020;52:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Pairo-Castineira E, Clohisey S, Klaric L, et al. Genetic mechanisms of critical illness in COVID-19. Nat 2020;591:92–8. [DOI] [PubMed] [Google Scholar]
- 166. Hamid I, Korunes KL, Beleza S, et al. Rapid adaptation to malaria facilitated by admixture in the human population of Cabo Verde. Elife 2021;10:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Ostrom QT, Egan KM, Nabors LB, et al. Glioma risk associated with extent of estimated European genetic ancestry in African-Americans and Hispanics. Int J Cancer 2020;146:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Jorgenson E, Choquet H, Yin J, et al. Genetic ancestry, skin pigmentation, and the risk of cutaneous squamous cell carcinoma in Hispanic/Latino and non-Hispanic white populations. Commun Biol 2020;3:765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Huynh-Le M-P, Fan CC, Karunamuni R, et al. Polygenic hazard score is associated with prostate cancer in multi-ethnic populations. Nat Commun 2021;12:1236–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Cheng H, Sewda A, Marquez-Luna C, et al. Genetic architecture of cardiometabolic risks in people living with HIV. BMC Med 2020;18:288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Shan MA, Meyer OS, Refn M, et al. Analysis of skin pigmentation and genetic ancestry in three subpopulations from Pakistan: Punjabi, Pashtun, and Baloch. Genes (Basel) 2021;12:733–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Zhang C, Ostrom QT, Hansen HM, et al. European genetic ancestry associated with risk of childhood ependymoma. Neuro Oncol 2020;22:1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kebede T, Bech N, Allienne J-F, et al. Genetic evidence for the role of non-human primates as reservoir hosts for human schistosomiasis. PLoS Negl Trop Dis 2020;14:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Finke K, Kourakos M, Brown G, et al. Ancestral haplotype reconstruction in endogamous populations using identity-by-descent. PLoS Comput Biol 2021;17:e1008638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Naseri A, Tang K, Geng X, et al. Personalized genealogical history of UK individuals inferred from biobank-scale IBD segments. BMC Biol 2021;19:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Tagore D, Aghakhanian F, Naidu R, et al. Insights into the demographic history of Asia from common ancestry and admixture in the genomic landscape of present-day Austroasiatic speakers. BMC Biol 2021;19:61–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Bae S, Won S, Kim H. Selection and evaluation of bi-allelic autosomal SNP markers for paternity testing in Koreans. Int J Leg Med 2021;135:1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Asgari S, Luo Y, Akbari A, et al. A positively selected FBN1 missense variant reduces height in Peruvians. Nature 2020;582:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Naqvi S, Sleyp Y, Hoskens H, et al. Shared heritability of human face and brain shape. Nat Genet 2021;53:830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Luo Y, Li X, Wang X, et al. Estimating heritability and its enrichment in tissue-specific gene sets in admixed populations. Hum Mol Genet 2021;30:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Athanasiadis G, Speed D, Andersen MK, et al. Estimating narrow-sense heritability using family data from admixed populations. Heredity (Edinb) 2020;124:751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Pazokitoroudi A, Wu Y, Burch KS, et al. Efficient variance components analysis across millions of genomes. Nat Commun 2020;11:4020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Wu HM, Goate AM, O’Reilly PF. Heterogeneous effects of genetic risk for Alzheimer’s disease on the phenome. Transl Psychiatry 2021;11:406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Leonenko G, Baker E, Stevenson-Hoare J, et al. Identifying individuals with high risk of Alzheimer’s disease using polygenic risk scores. Nat Commun 2021;12:4506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Zhang Q, Sidorenko J, Couvy-Duchesne B, et al. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. Nat Commun 2020;11:4799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Cao Z, Yang H, Ye Y, et al. Polygenic risk score, healthy lifestyles, and risk of incident depression. Transl Psychiatry 2021;11:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kang H-J, Park Y, Yoo K-H, et al. Sex differences in the genetic architecture of depression. Sci Rep 2020;10:9927–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Lobo JJ, McLean SA, Tungate AS, et al. Polygenic risk scoring to assess genetic overlap and protective factors influencing posttraumatic stress, depression, and chronic pain after motor vehicle collision trauma. Transl Psychiatry 2021;11:359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Warrier V, Baron-Cohen S. Childhood trauma, life-time self-harm, and suicidal behaviour and ideation are associated with polygenic scores for autism. Mol Psychiatry 2021;26:1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Campos AI, Verweij KJH, Statham DJ, et al. Genetic aetiology of self-harm ideation and behaviour. Sci Rep 2020;10:9713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Park H, Forthman KL, Kuplicki R, et al. Polygenic risk for neuroticism moderates response to gains and losses in amygdala and caudate: evidence from a clinical cohort. J Affect Disord 2021;293:124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Barr PB, Ksinan A, Su J, et al. Using polygenic scores for identifying individuals at increased risk of substance use disorders in clinical and population samples. Transl Psychiatry 2020;10:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Sanchez-Roige S, Cox NJ, Johnson EO, et al. Alcohol and cigarette smoking consumption as genetic proxies for alcohol misuse and nicotine dependence. Drug Alcohol Depend 2021;221:108612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Mars N, Widén E, Kerminen S, et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat Commun 2020;11:6383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Vaura F, Kauko A, Suvila K, et al. Polygenic risk scores predict hypertension onset and cardiovascular risk. Hypertension 2021;77:1119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Actkins KV, Singh K, Hucks D, et al. Characterizing the clinical and genetic spectrum of polycystic ovary syndrome in electronic health records. J Clin Endocrinol Metab 2021;106:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Lanca C, Kassam I, Patasova K, et al. New polygenic risk score to predict high myopia in Singapore Chinese children. Transl Vis Sci Technol 2021;10(8):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Wang Y-F, Zhang Y, Lin Z, et al. Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity between ancestral groups. Nat Commun 2021;12:772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Batra A, Chen LM, Wang Z, et al. Early life adversity and polygenic risk for high fasting insulin are associated with childhood impulsivity. Front Neurosci 2021;15:704785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Polushina T, Banerjee N, Giddaluru S, et al. Identification of pleiotropy at the gene level between psychiatric disorders and related traits. Transl Psychiatry 2021;11:410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Novikova G, Kapoor M, Tcw J, et al. Integration of Alzheimer’s disease genetics and myeloid genomics identifies disease risk regulatory elements and genes. Nat Commun 2021;12:1610–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Alves-Ferreira M, Quintas M, Sequeiros J, et al. A genetic interaction of NRXN2 with GABRE, SYT1 and CASK in migraine patients: a case-control study. J Headache Pain 2021;22:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Qadeer MI, Amar A, Huang Y-Y, et al. Association of serotonin system-related genes with homicidal behavior and criminal aggression in a prison population of Pakistani origin. Sci Rep 2021;11:1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Haddad D, John SE, Mohammad A, et al. SARS-CoV-2: possible recombination and emergence of potentially more virulent strains. PLoS One 2021;16:e0251368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Harper AR, Goel A, Grace C, et al. Common genetic variants and modifiable risk factors underpin hypertrophic cardiomyopathy susceptibility and expressivity. Nat Genet 2021;53:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Meyer OS, Salvo NM, Kjærbye A, et al. Prediction of eye colour in Scandinavians using the EyeColour 11 (EC11) SNP set. Genes (Basel) 2021;12:821–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Li YR, Glessner JT, Coe BP, et al. Rare copy number variants in over 100,000 European ancestry subjects reveal multiple disease associations. Nat Commun 2020;11:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Smajlagić D, Lavrichenko K, Berland S, et al. Population prevalence and inheritance pattern of recurrent CNVs associated with neurodevelopmental disorders in 12,252 newborns and their parents. Eur J Hum Genet 2021;29:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Sønderby IE, van der Meer D, Moreau C, et al. 1q21.1 distal copy number variants are associated with cerebral and cognitive alterations in humans. Transl Psychiatry 2021;11:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Yamasaki M, Makino T, Khor S-S, et al. Sensitivity to gene dosage and gene expression affects genes with copy number variants observed among neuropsychiatric diseases. BMC Med Genomics 2020;13:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Sønderby IE, Gústafsson Ó, Doan NT, et al. Dose response of the 16p11.2 distal copy number variant on intracranial volume and basal ganglia. Mol Psychiatry 2020;25:584–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Giner-Delgado C, Villatoro S, Lerga-Jaso J, et al. Evolutionary and functional impact of common polymorphic inversions in the human genome. Nat Commun 2019;10:4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Group WC, , et al. Association of copy number variation of the 15q11.2 BP1-BP2 region with cortical and subcortical morphology and cognition. JAMA. Psychiatry 2020;77:420–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Bliskunova T, Genis-Mendoza A-D, Martínez-Magaña JJ, et al. Association of MGAT4C with major neurocognitive disorder in the Mexican population. Gene 2021;778:145484. [DOI] [PubMed] [Google Scholar]
- 215. Niestroj L-M, Perez-Palma E, Howrigan DP, et al. Epilepsy subtype-specific copy number burden observed in a genome-wide study of 17 458 subjects. Brain 2020;143:2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Vega-Sevey JG, Martínez-Magaña JJ, Genis-Mendoza AD, et al. Copy number variants in siblings of Mexican origin concordant for schizophrenia or bipolar disorder. Psychiatry Res 2020;291:113018. [DOI] [PubMed] [Google Scholar]
- 217. Warland A, Kendall KM, Rees E, et al. Schizophrenia-associated genomic copy number variants and subcortical brain volumes in the UK biobank. Mol Psychiatry 2020;25:854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Sarihan EI, Pérez-Palma E, Niestroj L-M, et al. Genome-wide analysis of copy number variation in Latin American Parkinson’s disease patients. Mov Disord 2021;36:434–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Sakamoto Y, Shimoyama S, Furukawa T, et al. Copy number variations in Japanese children with autism spectrum disorder. Psychiatr Genet 2021;31:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Martin J, Hosking G, Wadon M, et al. A brief report: de novo copy number variants in children with attention deficit hyperactivity disorder. Transl Psychiatry 2020;10:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Sun F, Tan W, Dan Y, et al. Copy number gain of pro-inflammatory genes in patients with HBV-related acute-on-chronic liver failure. BMC Med Genomics 2020;13:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Kikuchi M, Kobayashi K, Nishida N, et al. Genome-wide copy number variation analysis of hepatitis B infection in a Japanese population. Hum genome Var 2021;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Petukhova L, Patel AV, Rigo RK, et al. Integrative analysis of rare copy number variants and gene expression data in alopecia areata implicates an aetiological role for autophagy. Exp Dermatol 2020;29:243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Pérez-Palma E, Bustos BI, Lal D, et al. Copy number variants in lipid metabolism genes are associated with gallstones disease in men. Eur J Hum Genet 2020;28:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Verbitsky M, Krithivasan P, Batourina E, et al. Copy number variant analysis and genome-wide association study identify loci with large effect for vesicoureteral reflux. J Am Soc Nephrol 2021;32:805–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Whitman MC, Di Gioia SA, Chan W-M, et al. Recurrent rare copy number variants increase risk for Esotropia. Invest Ophthalmol Vis Sci 2020;61:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Suratannon N, van Wijck RTA, Broer L, et al. Rapid low-cost microarray-based genotyping for genetic screening in primary immunodeficiency. Front Immunol 2020;11:614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Zarubin A, Stepanov V, Markov A, et al. Structural variability, expression profile, and pharmacogenetic properties of TMPRSS2 gene as a potential target for COVID-19 therapy. Genes (Basel) 2021;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Dai J, Wang L, Li L, et al. Interplay of microRNAs to genetic, epigenetic, copy number variations of cervical cancer related genes. J Reprod Immunol 2020;142:103184. [DOI] [PubMed] [Google Scholar]
- 230. Lee N, Kim S-M, Lee Y, et al. Prognostic value of integrated cytogenetic, somatic variation, and copy number variation analyses in Korean patients with newly diagnosed multiple myeloma. PLoS One 2021;16:e0246322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Yehia L, Seyfi M, Niestroj L-M, et al. Copy number variation and clinical outcomes in patients with germline PTEN mutations. JAMA Netw Open 2020;3:e1920415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Shukla A, Nguyen THM, Moka SB, et al. Chromosome arm aneuploidies shape tumour evolution and drug response. Nat Commun 2020;11:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Araujo AN, Camacho CP, Mendes TB, et al. Comprehensive assessment of copy number alterations uncovers recurrent AIFM3 and DLK1 copy gain in medullary thyroid carcinoma. Cancers (Basel) 2021;13:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Wang Y, Zhou W, McReynolds LJ, et al. Prognostic impact of pre-transplant chromosomal aberrations in peripheral blood of patients undergoing unrelated donor hematopoietic cell transplant for acute myeloid leukemia. Sci Rep 2021;11:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Moura-Castro LH, Peña-Martínez P, Castor A, et al. Sister chromatid cohesion defects are associated with chromosomal copy number heterogeneity in high hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 2021;60:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Hofvander J, Jo VY, Fletcher CDM, et al. PHF1 fusions cause distinct gene expression and chromatin accessibility profiles in ossifying fibromyxoid tumors and mesenchymal cells. Mod Pathol 2020;33:1331–40. [DOI] [PubMed] [Google Scholar]
- 237. Peille A-L, Vuaroqueaux V, Wong S-S, et al. Evaluation of molecular subtypes and clonal selection during establishment of patient-derived tumor xenografts from gastric adenocarcinoma. Commun Biol 2020;3:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Sekiguchi M, Seki M, Kawai T, et al. Integrated multiomics analysis of hepatoblastoma unravels its heterogeneity and provides novel druggable targets. NPJ Precis Oncol 2020;4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Yang M, Safavi S, Woodward EL, et al. 13q12.2 deletions in acute lymphoblastic leukemia lead to upregulation of FLT3 through enhancer hijacking. Blood 2020;136(946). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Matsumoto Y, Chinen Y, Shimura Y, et al. Recurrent intragenic exon rearrangements of SOBP and AUTS2 in non-Hodgkin B-cell lymphoma. Int J Hematol 2020;111:75–83. [DOI] [PubMed] [Google Scholar]
- 241. Galatà G, García-Montero A, Kristensen T, et al. Genome-wide association study identifies novel susceptibility loci for KIT D816V positive mastocytosis. Am J Hum Genet 2021;108:284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Sherman MA, Rodin RE, Genovese G, et al. Large mosaic copy number variations confer autism risk. Nat Neurosci 2021;24:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Zekavat SM, Lin S-H, Bick AG, et al. Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection. Nat Med 2021;27:1012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. González JR, Ruiz-Arenas C, Cáceres A, et al. Polymorphic inversions underlie the shared genetic susceptibility of obesity-related diseases. Am J Hum Genet 2020;106:846–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Secolin R, Gonsales MC, Rocha CS, et al. Exploring a region on chromosome 8p23.1 displaying positive selection signals in Brazilian admixed populations: additional insights into predisposition to obesity and related disorders. Front Genet 2021;12:636542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.