Abstract
Background and Aims
Regenerating islet-derived protein type 3 [Reg3] lectins are antimicrobial peptides at mucosal surfaces of the gut, whose expression is regulated by pathogenic gut microbes via interleukin-22- or Toll-like receptor signalling. In addition to antimicrobial effects, tissue protection is hypothesized, but has been poorly investigated in the gut.
Methods
We applied antibiotic-induced microbiota perturbations, gnotobiotic approaches and a dextran-sodium sulfate [DSS] colitis model to assess microbial Reg3 regulation in the intestines and its role in colitis. We also used an intestinal organoid model to investigate this axis in vitro.
Results
First, we studied whether gut commensals are involved in Reg3 expression in mice, and found that antibiotic-mediated reduction of Clostridia downregulated intestinal Reg3B. A loss in Clostridia was accompanied by a significant reduction of short-chain fatty acids [SCFAs], and knock-out [KO] mice for SCFA receptors GPR43 and GPR109 expressed less intestinal Reg3B/-G. Propionate was found to induce Reg3 in intestinal organoids and in gnotobiotic mice colonized with a defined, SCFA-producing microbiota. Investigating the role of Reg3B as a protective factor in colitis, we found that Reg3B-KO mice display increased inflammation and less crypt proliferation in the DSS colitis model. Propionate decreased colitis and increased proliferation. Treatment of organoids exposed to DSS with Reg3B or propionate reversed the chemical injury with a loss of expression of the stem-cell marker Lgr5 and Olfm4.
Conclusions
Our results suggest that Clostridia can regulate Reg3-associated epithelial homeostasis through propionate signalling. We also provide evidence that the Reg3–propionate axis may be an important mediator of gut epithelial regeneration in colitis.
Keywords: Microbiome, Reg3, colitis, propionate
1. Introduction
Multifaceted microbiota–host interactions are essential for physiological homeostasis, and subtle changes of this complex microbial milieu can have short- and long-term pathological effects on the host. In the gastrointestinal [GI] tract, intestinal epithelial cells [IECs] are pivotal in maintaining homeostasis between the microbiota and the host by mucus barrier construction, priming of humoral immune responses or secretion of antimicrobial peptides [AMPs].1 C-type lectins of the regenerating islet-derived protein type 3 [Reg3] family are among the most important AMPs produced within the gut mucosa. For instance, Reg3gamma [Reg3G], the murine orthologue to human Reg3A, has bactericidal activity against Gram-positive pathogens,2 whereas Reg3beta [Reg3B], a closely related isoform, targets Gram-negative bacteria and protects against Salmonella enteritidis or Yersinia pseudotuberculosis infections.3–5 In addition, Reg3B restricts the mucosa-associated gut microbiota and prevents translocation of commensal bacteria after GI tissue injury.6,7 A genetic knock-out [KO] of Reg3B, in turn, leads to dysbiosis-like microbiota changes.6
It has been demonstrated that colonization of the gut of germ-free mice with commensal bacteria induces the intestinal expression of Reg3G2 and Reg3B,8 and treatment with broad-spectrum antibiotics reduces the expression of both Reg3 proteins in mice,9 suggesting a general role of the gut commensal microbiota in Reg3 regulation. Toll-like receptor [TLR]-dependent signalling has been suggested as one important inducer of Reg3 upon intestinal pathogen exposure.10–12 In addition, interleukin-22 [IL-22] signalling also drives Reg3 expression, primarily in inflammatory conditions of the gut.13 Beyond that, the roles of commensals and their metabolites in the regulation of Reg3 and Reg3-associated epithelial homeostasis are less well understood.
Here, we hypothesized that certain members of the commensal gut flora can regulate intestinal Reg3 expression under steady-state conditions via metabolite-driven microbe–host interactions. To study this, we partially depleted the gut microbiota with antibiotics, and additionally colonized germ-free mice with a restricted gut flora. Furthermore, the intestinal organoid model was used to investigate the effects of microbial metabolites on Reg3 expression.
Reg3 lectins are highly upregulated in inflammatory bowel disease or acute graft-versus-host disease,14,15 both inflammatory conditions associated with injuries of the commensal gut microbiota. These injuries, in turn, are associated with a depletion of beneficial microbial metabolites such as short-chain fatty acids [SCFAs].16 For this reason, we finally assessed the role of Reg3B and SCFAs in modulating gut inflammatory responses and regeneration using the dextran sulfate sodium [DSS] model of chemical colitis in mice and in the organoid model.
2. Methods
2.1. Animals
Male C57BL/6J mice were obtained from Envigo or the Jackson Laboratories. Male MyD88-KO mice were obtained from Dr Thorsten Buch, University of Zurich, and bred at the animal facility of the Institute for Medical Microbiology, Techn. Univ. Munich; GPR43- and GPR109-KO mice were bred at the animal facility of Memorial Sloan-Kettering Cancer Center [MSKCC], and Reg3B-KO mice were bred and maintained at the animal facility of UC San Diego.17 Mice used for experiments were 7–12 weeks old and were housed in groups of 3–5 mice per cage under specific pathogen-free [SPF] conditions with unlimited access to water and food. Wild-type [WT] littermates were used as controls. Germ-free C56BL/6J mice were bred at the gnotobiotic facility of MSKCC and housed in Sentry SPP isocages for gnotobiotic animals [Allentown Inc.]. All experiments were performed according to the permission and guidelines of the local ethical committees and state veterinary office.
2.2. Antibiotic treatment, microbial colonization and DSS colitis
2.2.1. Antibiotics
Mice received either a broad-spectrum antimicrobial treatment according to a standard protocol18 with ampicillin [1 g/L], vancomycin [500 mg/L], neomycin [1 g/L], metronidazole [500 mg/L] and fluconazole [1 g/L; to prevent outgrowth of Candida 19] administered via drinking water. Another batch of mice received a cocktail of amipicillin [500 mg/L] plus enrofloxacin [250 mgl/L] which we recently found to also induce gut decontamination in mice.20
Alternatively, mice were treated with rifaximin [150 mg/kg, p.o. gavage once daily; dose according to Xu et al. 21]; controls received PBS. Antibiotic administration was done for 10 days unless otherwise stated in the text.
2.2.2. ASF colonization
Germ-free mice were colonized with an altered Schaedler flora [ASF] obtained from Taconic Bioscience. Fresh ASF containing faecal pellets were dissolved in sterile PBS under anaerobic conditions [Bactron anaerobic chamber, Shel Lab] and mice received p.o. gavages of this suspension. Experiments were performed 2–3 weeks after colonization.
2.2.3. DSS colitis
Animals were treated with 2, 3 or 4% of DSS [MP Biomedical] via drinking water for 5 days and then DSS was switched to tap water for another 3 days as indicated for each experiment. Animal weights were monitored daily and intestinal tissue was harvested at the end of the experiments to assess colitis and regeneration.
2.3. Tissue harvest and processing
Mice were killed by cervical dislocation. Ileum, colon and caecum, and caecal contents were harvested and immediately frozen at −80°C for RNA extraction. Colon or ileum were also immersed in different tissue fixation solutions for histological analyses.
2.3.1. RNA extraction, reverse transcription and qPCR
Total RNA was isolated from homogenized tissue using the RNeasy Mini Kit [Qiagen] according to the manufacturer’s instructions including on-column DNase digestion. RNA was converted to cDNA using a combination of random hexamer primers and M-MLV reverse transcriptase [Promega]. Subsequently, cDNAs were amplified using a CFX384 qPCR cycler [Bio-Rad] or a QuantStudio 7 Flex RT PCR system [Applied Biosystems]. The sequences of the gene-specific primers are listed in Supplementary Table 1. GAPDH was used as an endogenous control to normalize the target gene expression. For analysis, the fold change for the target gene was calculated using the 2−ΔΔCT method after normalization to controls.
2.3.2. Fixation
For Muc2-immunohistochemistry, a solution containing 60% methanol, 30% chloroform and 10% glacial acetic acid was used as fixation solution as described previously.22 A 4% formalin solution was used for H&E histology and Reg3B immunohistochemisty. The tissue was fixated for up to 24 h at 4°C. Afterwards, a standard dehydration procedure was performed followed by paraffin embedding. Subsequently, intestinal tissue was sectioned into 4-µm thin sections except for intestinal organoid immunofluorescence [see below].
2.4. Immunohistochemistry and histology
2.4.1. Muc2, Reg3B and Ki67 immunohistochemistry
Immunostaining was performed according to previously described protocols.22,23 For mucus visualization, sections were incubated with an anti-Muc2 rabbit polyclonal antibody [H300, Santa Cruz, 1:100 in blocking solution] at 4°C overnight. Following incubation with primary antibody, tissues were washed in PBS and incubated with goat anti-rabbit Alexa 488 antibody [1:100 in PBS] for 2 h and counterstained with DAPI [1:1000]. For Ki67 staining, paraffin-embedded tissue sections were deparaffinized, boiled in sodium citrate buffer [pH 6.0, 10 min] for antigen retrieval, incubated with 3% hydrogen peroxide [10 min] at room temperature, and finally blocked with 5% normal goat serum. Anti-Ki67 rabbit monoclonal antibody [1:400; Cell Signaling Technology] was incubated overnight at 4°C, and following PBS washing a horseradish peroxidase [HRP]-conjugated anti-rabbit secondary antibody [1:200; Promega] was added for 1 h at room temperature. Ki67 detection was performed with 3,3′-diaminobenzidine staining [SignalStain DAB Substrate Kit, Cell Signaling Technology], and tissue was counterstained with haematoxylin.
For Reg3B immunostaining, 4% paraformaldehyde [PFA]-fixed organoids were incubated with a polyclonal sheep anti-mouse Reg3B antibody [R&D Systems, 1:100 in blocking buffer], and as secondary antibody an Alexa Fluor 594 donkey anti-sheep antibody [dilution 1:500 in blocking buffer] was used. Ki67 staining was done by co-incubating with anti-Ki67 rabbit monoclonal antibody [1:400; Cell Signaling Technology], and as secondary antibody Alexa 488 goat anti-rabbit [dilution 1:500 in blocking buffer]. Sections were cover-slipped with Vectashield mounting media [H-1000].
Haematoxylin and eosin [HE] staining for conventional histology was performed according to a standard protocol. All images were acquired with a Leica DMRBE microscope and the AxioCamMRm and Axio vision imaging software 4.8.
2.4.2.Analyses
The width of the inner mucus layer was determined from the average of four measurements per field with four fields measured per section. Thickness measurements were done using the NIH ImageJ software. Histological scoring of DSS colitis severity was done by assessing, first, the infiltration with lymphocytes, second, the amount of oedema and, third, the area of ulceration according to Knoop et al.24. The scores were summed to a cumulative value between 0 and 9. Analysis of Ki67-stained sections was performed according to a previously published protocol.25 Briefly, tissue sections were assessed for crypts with an open, straight lumen and a clearly visible base. All of the epithelial cells in the crypt were counted and the percentage of cells that showed nuclear staining for Ki67 in both basal and non-basal regions was recorded. For each mouse, an average of ten crypts was assessed. Scoring of inflammation and Ki67 staining was performed in a blinded fashion.
2.5. Intestinal organoids
2.5.1. Crypt isolation
Isolation of crypts from small intestines of mice was done as previously described.26,27 Briefly, after the mice were killed and intestines were harvested, the gut was opened longitudinally and washed with PBS. The luminal side was scraped using a glass slide to remove intestinal contents and villous structures. To dissociate the crypts, the intestine was cut into pieces, and they were incubated in EDTA [20 mM] for 20 min at 4°C with gentle vortexing. After washing with PBS the crypts were released by manual shaking of the suspension for 3 min. The supernatant was collected and passed through a 70-µm strainer. The remaining tissue pieces were resuspended in PBS, again manually shacked, and passed through a 70-µm strainer. The crypt-containing solution was centrifuged at 800 r.p.m. for 5 min at 4°C. The pellet was resuspended in Advanced DMEM/F12 [Invitrogen] and centrifuged at 600 r.p.m. for 5 min at 4°C to remove debris and single cells.
2.5.2. Organoid culture
The freshly isolated crypts were embedded directly in growth-factor-reduced, phenol red-free Matrigel [BD Biosciences]. Droplets of 20 µL were cast at the bottom of 24-well plates and polymerized for 15 min at 37°C. A crypt medium containing complete EGF/Noggin/R-spondin-1 [ENR]-medium of advanced DMEM/F12 [Sigma Aldrich], 2 mM Glutamax [Invitrogen], 10 mM HEPES [Sigma Aldrich], 100 U/mL penicillin/100 μg/mL streptomycin [Sigma Aldrich], 1 mM N-acetyl cysteine [Sigma Aldrich], B27 supplement [Invitrogen], N2 supplement [Invitrogen], 50 ng/mL mEGF [Peprotech], 100 ng/mL mNoggin [Peprotech] and 100 ng/mL mR-spondin [Peprotech] was added to small intestine crypt cultures.26 Medium was replaced every 2–3 days; for passaging of organoids after 5–7 days of culture, organoids were mechanically disrupted with a seropipette and cold media to depolymerize the Matrigel and generate organoid fragments. After washing away the old Matrigel by spinning down at 600 r.p.m., organoid fragments were re-plated in liquid Matrigel and new crypt medium was added. Organoids were treated with SCFAs by adding sodium butyrate, sodium propionate and sodium acetate [all Sigma Aldrich] to the crypt medium at concentrations of 1 and 5 mM for 10 h. The concentrations of SCFAs used in the present study were largely derived from previous reports on the effects of SCFAs in intestinal organoids and our own preliminary dose–response experiments.28 In separate experiments, chemical organoid injury was carried out by adding 0.01% DSS to the organoid media for 10 h. After these treatments organoids were harvested in TRIzol reagent [Invitrogen] to analyse mRNA expression by qPCR.
2.6. 16S rRNA analyses of faecal microbiota
2.6.1. DNA extraction
DNA was extracted from mouse stool samples by using QIAamp DNA Stool Mini Kit [Quiagen] according to the manufacturer’s manual and stored at −20°C until further use.
2.6.2. qPCR target sequencing of 16S rRNA
Faecal DNA [diluted to 10 ng/µL] was analysed for relative abundances of selected 16S rRNA targets applying a previously published real-time PCR-based Gut Low-Density Array [GULDA].29,30 The target genes represent major taxonomic groups within the five predominant bacterial phyla of the gut, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia and Euryarchaeota: Firmicutes, Lactobacillus spp., Clostridia clusters IV and XIVa, Enterococcus spp., Bacteroidetes, Prevotella spp., Alistipes spp., Bifidobacterium spp., Enterobacteriaceae, Akkermansia muciniphila, Methanobrevibacter smithii and Desulfobrivio spp. Primers are listed in Supplementary Table 1. qPCR efficiencies for each amplicon were calculated by LINREGPCR software, and the efficiencies for each amplicon group were then used to determine the initial concentration N 0 of the DNA target [i.e. 16S rRNA gene]. Finally, the relative abundance of the each specific amplicon group was obtained by normalization to the N 0-value obtained for the universal bacterial amplicon group determined in the same array.29 For the comparison of total 16S rRNA amounts, N 0-values from antibiotic-treated mice were normalized to non-treated controls.
2.6.3. 16S rRNA amplicon sequencing
In a validation experiment for targeted 16S rRNA sequencing, faecal 16S rRNA DNA was amplified by PCR [30 cycles; AccuPrime II Taq DNA polymerase, Invitrogen] using primers directed against the V3 region of the 16S rRNA. Amplicon sequencing was performed on an Ion PGM platform [Life Technologies] applying 400-bp sequencing kits and 314v2 chips according to the manufacturer’s instructions. Base calling and run demultiplexing were performed by using TorrentServer software, version 3.6.2, with default parameters for the general sequencing application. Sequences were analysed using the MetaGenome Rapid Annotation using Subsystem Technology [MG-RAST] pipeline.31 Sequences with distance-based similarity of at least 97% were assigned the same OTU [operational taxonomic unit] and taxonomy assignments were done using the RDP reference database.
2.7. Analysis of short-chain fatty acids
SCFA levels from caecal contents and faeces were determined by 1H-NMR spectroscopy using a UNITY INOVA 600 NMR spectrometer [Varian] as previously described.32 Spectra were analysed using the Chenomx NMR software.
2.8. Statistical analyses
Data are presented as mean ± standard error of the mean [SEM]. Data were analysed by one-sample t-tests for gene expression results and by unpaired t-tests for 16S rRNA results and histological scores. Two-factorial analyses of variance were performed for weight data of the DSS experiments. Analyses were performed using GraphPad Prism 7.0 [GraphPad] and R software [The R Project for Statistical Computing]. Statistical significance was accepted at p < 0.05.
3. Results
3.1. Antibiotic-induced microbiota injuries reduce Reg3B expression
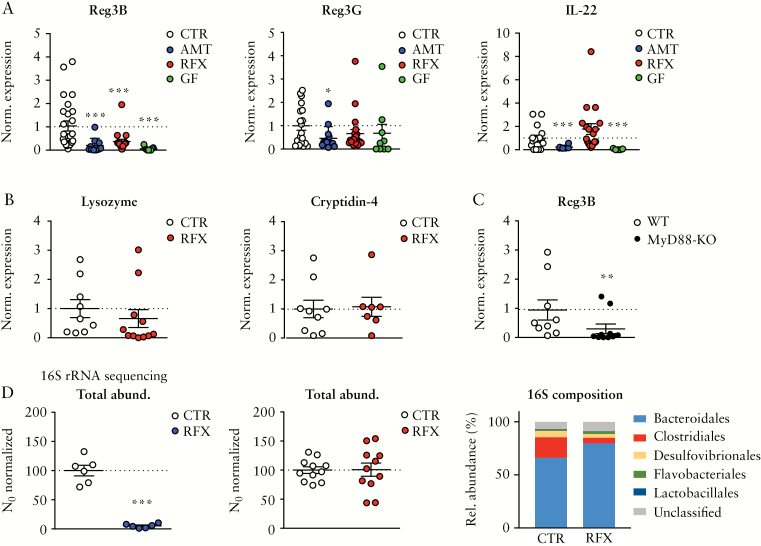
To investigate whether the gut microbiota is involved in intestinal Reg3B expression, C57BL/6 mice were treated either with a broad-spectrum antibiotic cocktail [AMT; ampicillin, vancomycin, neomycin, metronidazole] or with the non-absorbable, narrow-spectrum antibiotic rifaximin [RFX]33 to induce varying degrees of microbial dysbiosis. We observed that AMT and RFX treatments significantly reduced Reg3B expression in the mouse caecum almost to a level of germ-free [GF] mice that served as a reference [Figure 1A]. In contrast, the expression of the Reg3G lectin showed only a trend towards lower levels after RFX treatment and in GF mice [Figure 1A and Supplementary Figure 1B], but was significantly reduced in AMT mice. The expression of IL-22, an upstream regulator of Reg3B, was not significantly changed after RFX treatment (Figure 1A; IL-22 protein levels in ileal homogenates were not different between RFX- and PBS-treated mice [data not shown]). The reduction in Reg3B expression after RFX administration was also observed in the small intestine and the colon [Supplementary Figure 1A for small intestine and colon]. Notably, intraperitoneal administration of recombinant murine IL-22 [rmIL-22] rescued the RFX-induced downregulation of Reg3B [Supplementary Figure 1C]. Expression levels of lysozyme or cryptidin-4, representing other classes of mouse defensins, were not changed after RFX treatment [Figure 1B]. We further investigated Reg3B expression in mice deficient for MyD88, a major component of microbial stimulated TLR signalling. We found a significantly lower expression of Reg3B in MyD88 knock-out [KO] mice compared to littermate controls at steady state in the caecum [Figure 1C], confirming previous observations on the effect of TLR signalling in the regulation of Reg3 lectins.7
Figure 1.
Antibiotic-induced Reg3 modulation and microbiota changes. [A] mRNA levels of Reg3B [left], Reg3G [middle], IL-22 [right] in the caecum after treatment with an antibiotic cocktail [AMT, ampicillin, neomycin, vancomycin, metronidazole], rifaximin [RFX] or in germ-free [GF] mice compared to untreated SPF mice [CTR]. [B] mRNA levels of lysozyme and cryptidin-4 after RFX treatment. [C] Expression of Reg3B in MyD88-KO mice at steady-state compared to littermate WT mice. [D] Total bacterial load assessed by 16S rRNA qPCRs after AMT [left], or RFX treatment [middle]; right, OTU binning on order level of faecal microbiota in RFX vs CTR mice. Data are means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; n = 6–20 mice per group.
16S rRNA analyses from faeces of antibiotic-treated mice was performed using a qPCR approach with primers selected for specific taxa [GULDA29,30]. AMT treatment depleted the gut microbiota as shown by a significant reduction in universal bacterial amplicons, whereas RFX administration did not change the total bacterial load in mice [Figure 1D]. Analysing selected bacterial taxa by GULDA, we found that RFX treatment significantly reduced the relative abundance of Firmicutes, Clostridia Clusters IV and XIVa, Akkermansia muciniphila and Alistipes spp., but it significantly increased Bifidobacterium spp., Prevotella spp. and Enterobacteriaceae with a trend towards a higher relative abundance of Bacteroidetes [Supplementary Figure 1D]. To validate the results of the 16S rRNA qPCRs, we performed 16S rRNA amplicon sequencing on an Ion-PGM sequencer, and again found a reduction of Clostridiales and an increase in Bacteroidales [Figure 1D].
3.2. RFX treatment reduces luminal short-chain fatty acids
Clostridia clusters IV and XIV contain the genera Clostridium, Faecalibacterium, Ruminococcus, Lachnospira and Eubacterium.34 Clostridia deploy several enzymes capable of degrading polysaccharides, either endogenous or dietary in origin, into SCFAs with acetate, propionate and butyrate as the most abundant SFCAs.35 These compounds can activate host cells via signalling through G-protein coupled receptors, such as GPR43 with high affinities to acetate and propionate and GPR109 with high affinity to butyrate.36,37
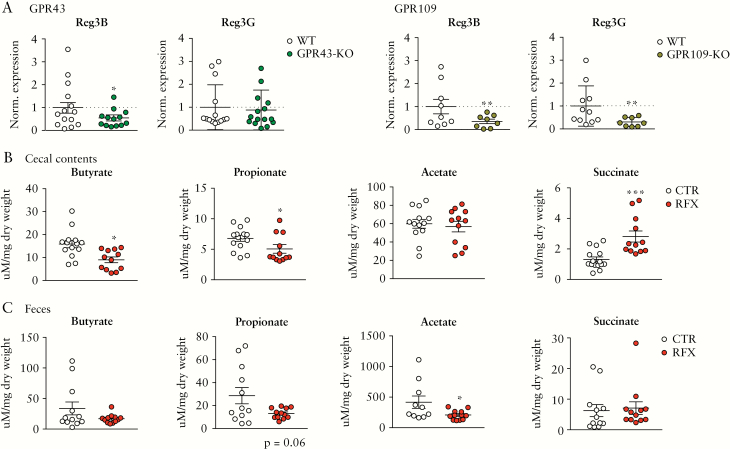
Given that RFX treatment leads to a depletion of intestinal Clostridia and also reduces Reg3B expression, we hypothesized that SCFA signalling is involved in the regulation of gut mucosal Reg3 lectins. In a first approach, we analysed Reg3 mRNA expression in mice lacking GPR43 or GPR109 and observed significantly lower levels of Reg3B in GPR43- and GPR109-KO mice and significantly less expression of Reg3G in GPR109-KO mice compared to WT animals [Figure 2A]. Next, we investigated gut luminal SCFA levels after RFX treatment. Intriguingly, the RFX-induced depletion in Clostridia XIV bacteria was accompanied by a significant reduction of butyrate and propionate in caecal contents and less acetate in the faeces when compared to controls [Figure 2B, C]. In contrast, succinate was increased in caecal contents after RFX treatment [Figure 2B]. We next measured the concentrations of SCFAs in the caecal contents of WT and GRP43-KO mice and observed no genotype-related differences at steady state [when receiving PBS]. RFX treatment reduced propionate, and to a lesser extent butyrate, in WT mice, and the same trend was observed in GPR43-KO mice [Supplementary Figure 3A], which is also reflected by a similar composition of SCFA-producing bacteria in the faeces of both WT and GPR43-KO mice, as described previously.38
Figure 2.
Reg3 expression in GPR43 and GPR109-KO mice and luminal SCFA reduction after antibiotic treatment. [A] mRNA levels of Reg3B or -G expression in the caecum of GPR43- [left] and GPR109-KO mice [right] mice compared to WT controls. [B] Caecal luminal contents of butyrate, propionate, acetate and succinate after RFX treatment measured by 1H-NMR. [C] Levels of butyrate, propionate, acetate and succinate analysed in faeces after RFX treatment. Data are means ± SEM. *p < 0.05, ***p < 0.001; n = 8–15 mice per group.
SCFAs have been reported to increase mucus production and secretion.39,40 We therefore tested whether RFX treatment reduced the thickness of the inner mucus layer. We could not detect any changes in its thickness, suggesting that several additional factors other than SCFAs contribute to gut mucus layer formation [Supplementary Figure 1E; AMT served as a positive control].
3.3. Propionate induces Reg3 expression via SCFA receptor signalling
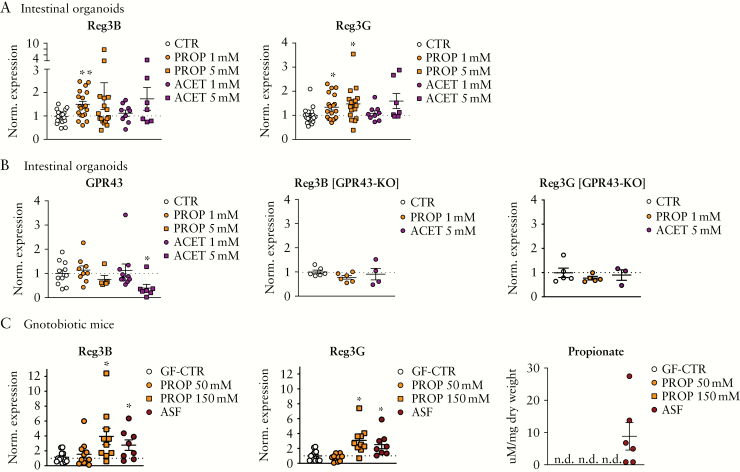
We next studied whether SCFAs directly regulate Reg3B expression on IECs using intestinal organoids, a reductionist in vitro system to investigate gene expression regulation on the level of IECs. The reason for using organoids derived from small intestinal epithelium as a model system to investigate the regulation of Reg3s is that we have previously established small intestinal organoids as a model for testing SCFA effects.16 Furthermore, the levels of expression of Reg3B or -G and also for the SCFA receptor GPR43 are comparable across different parts of the intestine in SPF mice,41,42 which allows us to use either small or large intestinal organoids to study the interaction of both. We chose butyrate, propionate and acetate as all were found reduced in faeces and/or caecal contents after RFX treatment. We administered sodium butyrate to the organoid media at doses reported to be effective in organoid growth.16 We did not find any changes in Reg3B expression after 1 mM butyrate treatment. Higher doses of sodium butyrate [5 mM] even reduced the expression of Reg3B and Reg3G [Supplementary Figure 4A]. This could be due to toxic effects as the cell cultures showed apoptotic cells, a phenomenon already reported for butyrate.35 In contrast, treatment of intestinal organoids with sodium propionate significantly upregulated Reg3B and Reg3G [Figure 3A; significant effect for Reg3B with 1 mM and for Reg3G with 1 and 5 mM propionate]. Sodium acetate incubation did not show a significant effect on Reg3 expression. In a proof-of-concept experiment, we stained Reg3B protein in intestinal organoids after propionate incubation [1 mM, 12 h] using immunofluorescence-based confocal imaging. Propionate treatment, and rmIL-22 stimulation as a reference, induced a brighter cytoplasmic Reg3B signal in organoid enterocytes with several high Reg3B-positive cells compared to non-treated, naïve mouse organoids [Supplementary Figure 2]. In unstimulated cells, Reg3B staining was mainly observed at the basal compartment, reflecting the Reg3B staining pattern in the small intestines of mice at steady state.43 As propionate and acetate mainly signal through GPR43, we also investigated the expression levels of GPR43 itself in the intestinal organoids after SCFA stimulation and found no changes after SCFA treatment. Only the high dose of acetate [5 mM] had an effect, indicating an internalization/feedback process [Figure 3B].
Figure 3.
Reg3 regulation by SCFAs. [A] Expression of Reg3B [left] and Reg3G [right] in small intestinal organoids after adding sodium propionate or sodium acetate to culture media. [B] Left, mRNA levels of GPR43 in small intestinal organoids after sodium propionate or sodium acetate treatment; right, Reg3B and Reg3G expression levels after SCFA treatment in small intestine organoids derived from GPR43-KO mice. [C] Caecum mRNA expression levels of Reg3B [left] and Reg3G [middle] in GF mice after oral treatment with propionate or in ASF gnotobiotic mice compared to GF controls [GF-CTR]; left, 1H-NMR analyses of propionate in caecal contents after propionate treatment or ASF colonization [n.d.: not detected/below level of detection]. Data are means ± SEM. *p < 0.05; SI organoids: n = 4–18 biological replicates per group; gnotobiotic study: n = 8–13 mice per group.
SCFAs can modulate target cell function beyond GPR43 signalling, for example by chromatin remodelling.16 To test whether GPR43 is mediating SCFA effects on Reg3B expression, we incubated intestinal organoids from GPR43-KO mice with propionate or acetate and found no induction of Reg3B or Reg3G expression [Figure 3B]. In the organoid system, both Reg3 proteins were expressed at similar levels between WT and GPR43-KO mice at steady state, and growth measured as organoid area was similar [Supplementary Figure 3B].
In another approach, we investigated whether propionate can also induce intestinal Reg3B expression in vivo. GF mice received either sterile sodium propionate via drinking water [doses according to Smith et al.44] or were colonized with an ASF that contains acetate- and propionate-producing strains but almost no butyrate producers.45 We observed a significant induction of Reg3B and Reg3G in caecal tissue of gnotobiotic ASF mice compared to GF mice [Figure 3C]. Propionate treatment induced a significant increase in caecal Reg3G and a trend towards higher Reg3B expression compared to GF mice [Figure 3C]. ASF strains produced propionate and acetate as measured in caecal contents of colonized mice [Figure 3C; Supplementary Figure 4B], whereas the levels of these SCFAs and butyrate were below detection limits in caecal contents from mice treated with sodium propionate or GF controls.
3.4. Reg3B and propionate protect against DSS-induced colitis and promote regeneration
Beyond a function as an antimicrobial protein, Reg3 proteins have been demonstrated to promote regeneration after tissue injury.46 Given that gut microbiota and microbial metabolites influence IECs to maintain homeostasis in inflammatory states,18 we hypothesized that Reg3B and propionate as the major SCFA regulating Reg3 expression may also play an important role in colitis.
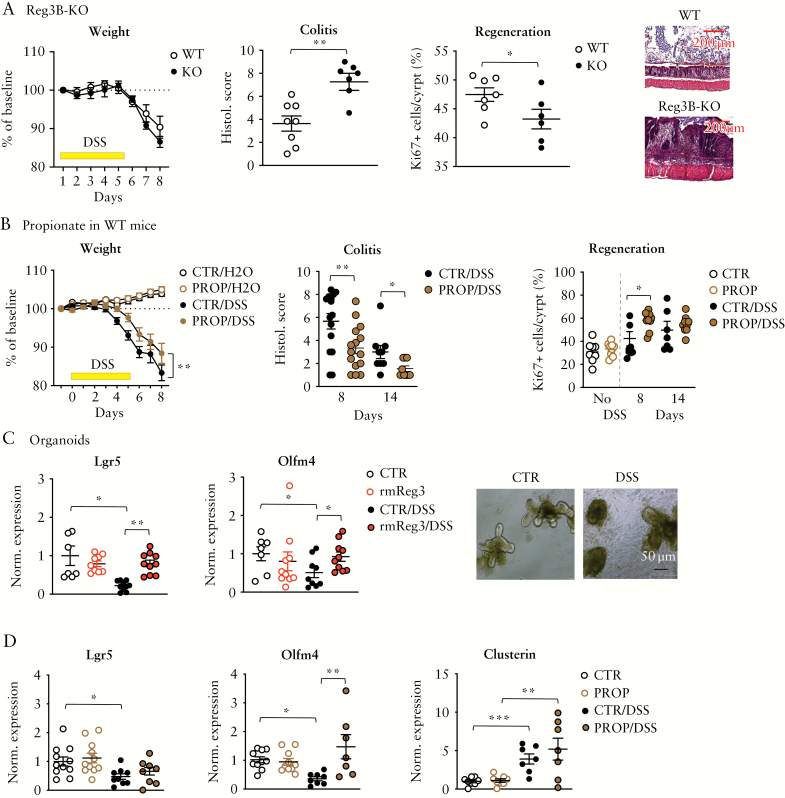
In a first experiment, Reg3B-KO mice and WT littermate controls received 2% DSS to induce an acute, chemical colitis. As shown in Figure 4A, Reg3B-KO mice showed a more severe DSS colitis compared to WT littermates, as observed by significantly higher histological colitis scores, although the weight loss was not significantly different between the two genotypes. As Reg3B is considered to contribute to tissue regeneration, we performed a Ki67 immunohistochemistry staining of colon sections and found a significantly reduced proportion of proliferating, Ki67-positive cells in crypts of DSS-treated Reg3B-KO mice. Supplementary Figure 5A shows the faecal microbiota composition of WT and Reg3B-KO mice at steady state.
Figure 4.
Reg3B modulates DSS colitis. [A] Body weight changes [left] and histological scores [middle] of inflamed gut epithelium during/after a 2% DSS treatment of Reg3B-KO mice vs controls; right, per cent Ki-67+ cells per total cells per colon crypt in WT vs Reg3B-KO mice. Representative DSS day-8 images of colon sections stained with haematoxylin and eosin. [B] Body weight changes [left], histological scores [middle] and Ki-67 staining of colon crypts [right] in WT mice treated with propionate [50 mM] vs control and 3% DSS for another 5 days; propionate was administered 2 days before DSS until tissue harvest on day 8 or 14. [C] Expression of Lgr5 [left] and Olfm4 [right] in small intestinal organoids after adding recombinant murine Reg3B [100 µM] and/or 0.01% DSS to culture media; photomicrographs of non-treated [CTR] and DSS-treated [DSS] organoids. [D] mRNA expression in organoids of Lgr5, Olfm4 or clusterin after adding propionate [PROP, 1 mM] and/or 0.01% DSS to the culture media. Data are means ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. SI organoids: n = 7–10 biological replicates per group; mouse experiments: n = 7–12 mice per group.
Next, we investigated whether propionate, a major inducer of intestinal Reg3, modifies the DSS colitis phenotype. Propionate treatment [50 mM, via drinking water] significantly attenuated acute colitis-related weight loss and tissue injury, and enhanced crypt proliferation as seen by an increased number of Ki67-positive cells [mainly on day 8 after start of DSS], a surrogate for tissue regeneration [Figure 4B].
We followed an additional batch of WT mice until day 14 and observed that propionate-treated mice recovered slightly faster after DSS injury [Supplementary Figure 5B] Propionate treatment upregulated Reg3B and -G expression in the colon of DSS-treated mice [Supplementary Figure 5C]. RFX treatment had only minor effects on DSS-induced colitis. Mice were pretreated with RFX for 7 days and received DSS afterwards. We observed a trend towards more weight loss (ANOVA [interaction treatment × time]: F 6,60 = 1.53, p = 0.18], and a higher colitis score [p = 0.11] in RFX-treated mice [Supplementary Figure 5D]. We further treated mice with ampicillin plus enrofloxacin [AE],20 which we reported to deplete SCFAs such as propionate in the gut.47 To investigate whether propionate depletion affects proliferation and crypt regeneration after injury, we administered AE to DSS-treated mice and observed significantly reduced Ki67-positive cells in the crypts of antibiotic-treated mice compared to non-AE DSS-treated mice [Supplementary Figure 5E].
We further translated the DSS-induced gut injury into the in vitro organoid model by adding 0.01% DSS for 10 h to the organoid culture medium before tissue harvest. This chemical injury induces disintegration of the 3D culture and apoptosis of enterocytes [Figure 4C], and is associated with a significant reduction in the expression of leucine-rich repeat-containing G-protein coupled receptor 5 [Lgr5] and olfactomedin 4 [Olfm4] [Figure 4C]. Lgr5 is expressed in intestinal stem cells of the crypt base and Olfm4 is another marker for Lgr5+ stem cells in the intestines48,49; the expression of these markers is considered a surrogate for adult stem cells in the gut.50 Adding rmReg3B to the culture media during the DSS challenge reversed the DSS-induced reduction of Lgr5 and Olfm4 expression in mouse organoids [Figure 4C]. Similarly, treating the organoids with propionate [1 mM] also reversed the DSS-accompanied reduction of Olfm4 reduction in organoids, but had little effect on Lgr5 expression [Figure 4D]. We also assessed the expression of clusterin, a multifunctional protein involved in cell proliferation, differentiation and survival of cells.51 It is also enriched in Lgr5+ intestinal stem cells52 and is specifically upregulated in these stem cells after tissue damage.53 Here, we observed that the expression of clusterin is also upregulated in DSS-treated intestinal organoids, and administration of propionate further augments its expression [Figure 4D, right].
4. Discussion
In the present study, we observed that broad- and narrow-spectrum antibiotic treatment in mice reduced the intestinal expression of the antimicrobial Reg3 peptides. As a putative mechanism, antibiotic-induced reduction of Clostridia strains and the consecutive depletion of SCFAs, specifically propionic acid, are postulated to underlie Reg3B modulation on IECs.
Both AMT as a broad-spectrum antimicrobial treatment and RFX, a more selective antibiotic drug used for traveller’s diarrhoea or to treat hepatic encephalopathy,54,55 downregulated Reg3B expression in several parts of the intestinal tract. AMT significantly depleted the gut flora as observed by 16S rRNA qPCR and downregulated Reg3B almost to a level of GF mice, which are reported to have low expression levels of Reg3 AMPs.2,56 In contrast, RFX did not reduce the total faecal microbial load, which has been observed previously in a rat model and in clinical studies.33,57,58 In addition, it also spared an intact mucus layer. Overall, RFX induced shifts in the composition of the microbiota with a reduction of Firmicutes, especially bacteria from Clostridia Clusters IV and XIV, and an increase in Bidiobacterium spp., Prevotella spp. and Enterobacteriaceae, reflecting a dysbiotic state of the gut microbiota. These composition changes in the gut’s microbial ecosystem and a modest effect of RFX on bacterial richness have been observed in other RFX treatment studies59 and contrast with its potent effects on aerobic and anaerobic bacteria in vitro.60 Given that Enterobacteriaceae are significantly enriched after RFX treatment, a bacterial family of potent lipopolysaccharide producers, the TLR pathway does not appear to be involved in the reduced Reg3B expression after RFX treatment. An alternative mechanism regulating Reg3B expression is the production of SCFAs by certain microbial species. As RFX treatment reduced Clostridiales, which are major SCFA producers, we investigated their luminal levels after treatment and observed significantly less butyrate, propionate and acetate, mainly in the caecum. We also attempted to measure SCFAs in ileal contents, but butyrate and propionate as major SCFAs affected by RFX could not be reliably detected. Intriguingly, a clinical study also observed less butyrate after RFX treatment in patients with irritable bowel syndrome.61 Along these lines, we observed that mice deficient in GPR43, a major receptor for SCFA signalling, also showed lower levels in Reg3B compared to controls, thereby confirming another recent study that reported lower expression of Reg3G and other beta-defensins in GPR43-KO mice.62 In their study, the authors also found that butyrate administration in mice or in vitro to IECs is able to upregulate Reg3G expression in a STAT3-dependent manner.
Using the intestinal organoid system, we also investigated the effects of different SCFAs on in vitro Reg3 expression. Here, we did not find butyrate to be able to induce Reg3B or Reg3G, but rather an opposite effect, which might be caused by toxic effects of butyrate to IECs in high concentrations. Instead, propionate, and to a lesser extent acetate, were found as potent inductors of Reg3B and Reg3G expression in vitro. Treating intestinal organoids from GPR43-KO mice with SCFAs did not change Reg3 expression, indicating the propionate-induced Reg3 expression depends on GPR43 signalling. To confirm our findings in vivo, we colonized GF mice with ASF, a synthetic commensal gut flora harbouring propionate- and acetate-producing strains, and found a significant induction of intestinal Reg3B and Reg3G expression. Oral treatment with propionate had a modest, dose-dependent effect on lectin expression, perhaps due to potential absorption in proximal parts of the GI tract and SCFA amounts below the detection limit in the caecum. Another line of evidence for an important role of SCFA modulation of Reg3B expression on the level of IECs comes from the observation that RFX treatment did not change IL-22 expression. IL-22 is produced by different types of immune cells located in the lamina propria of the intestines63 and is considered a major upstream signal for AMP expression on enterocytes via STAT3 signalling.64,65
Finally, we investigated whether Reg3 and propionate play a role in maintaining mucosal homeostasis during colitis by analysing the effects of DSS as a colitogenic agent in our animal models. First, using a genetic KO approach, mice deficient in Reg3B showed a significantly more severe colitis phenotype and less crypt regeneration as determined by Ki67 compared to their WT littermates. As we and others have shown, Reg3B KO mice have a slightly different faecal microbiota composition compared to WT mice at steady state.6 To exclude that this colitis phenotype is caused by gut microbiota dysbiosis or enhanced bacterial translocation, faecal transfer of KO flora into GF mice could give more insights, but may be biased because of an induction of Reg3 antimicrobial peptides.2 On the other hand, propionate treatment in WT mice attenuated DSS-induced colitis and promoted crypt regeneration in vivo and in the organoid model. Although propionate can induce the expression of Reg3 in the epithelium, it has several other actions on the mucosa as well,35 which can explain the increase in proliferation. Reg3B itself, however, can stimulate intestinal stem cells upon tissue injury as observed in vitro in the gut organoid model.
We further investigated whether RFX pretreatment and a consecutive reduction of SCFAs and Reg3B increases the severity of DSS colitis as SCFA–GPR43 signalling has been shown to protect the host against chemical colitis66 or a T-cell transfer model of colitis.44 RFX treatment increased DSS colitis severity compared to controls, but without statistical significance. At this point we cannot exclude that DSS plus RFX led to additional microbiota changes that may have counteracted the loss of Clostridiales that we observed at steady-state levels. In addition, there is evidence that RFX exerts beneficial effects on host physiology as a drug itself that occur independent of microbiota changes. For instance, it has been shown that it mitigates pro-inflammatory processes in vitro.67–69 Of note, evidence for a microbiota-dependent effect of Reg3 in colitis comes from a recent report showing that transgenic hepatic overexpression of Reg3A being secreted into the gut lumen via bile preserves gut microbiota from oxidative stress and thereby prevents inflammation.70 Alternatively, a loss of Reg3 could also aggravate inflammatory lesions by impaired regeneration, as this lectin is hypothesized to drive tissue repair after injury in the skin,46 the pancreas71 or the liver.72 In addition, it could affect macrophage trafficking to control immune cell infiltrations—an effect described in the heart during experimental myocardial ischaemia.73
In conclusion, our results suggest a novel mechanism by which specific members of the gut commensal microbiota can regulate epithelial homeostasis and defence in the gut. It also suggests a novel role of Reg3 in the gut acting beyond a simple antimicrobial peptide.
Supplementary Material
Acknowledgments
The authors would like to thank Raphaela Semper for help with qPCR experiments, Martina Grandl with intestinal organoids, and Prof. Anne Krug for help with antibiotics and DSS experiments. Thanks to Jonathan U. Peled for comments on the manuscript.
Contributor Information
Danica Bajic, Klinik für Innere Medizin II, Klinikum rechts der Isar, Techn. Univ. Munich, Munich, Germany.
Adrian Niemann, Klinik für Innere Medizin II, Klinikum rechts der Isar, Techn. Univ. Munich, Munich, Germany.
Anna-Katharina Hillmer, Klinik für Innere Medizin II, Klinikum rechts der Isar, Techn. Univ. Munich, Munich, Germany.
Raquel Mejias-Luque, Institute for Medical Microbiology, Immunology and Hygiene, Techn. Univ. Munich, Munich, Germany; University Hospital Zurich, Division of Gastroenterology and Hepatology, Zurich, Switzerland.
Sena Bluemel, UC San Diego School of Medicine, Division of Gastroenterology, San Diego, USA; University Hospital Zurich, Division of Gastroenterology and Hepatology, Zurich, Switzerland.
Melissa Docampo, Memorial Sloan-Kettering Cancer Center, Immunology Program, New York, USA.
Maja C Funk, Division Signaling and Functional Genomics, German Cancer Research Center (DKFZ) and Heidelberg University, Heidelberg, Germany.
Elena Tonin, Division Signaling and Functional Genomics, German Cancer Research Center (DKFZ) and Heidelberg University, Heidelberg, Germany.
Michael Boutros, Division Signaling and Functional Genomics, German Cancer Research Center (DKFZ) and Heidelberg University, Heidelberg, Germany.
Bernd Schnabl, UC San Diego School of Medicine, Division of Gastroenterology, San Diego, USA.
Dirk H Busch, Institute for Medical Microbiology, Immunology and Hygiene, Techn. Univ. Munich, Munich, Germany; German Center for Infection Research (DZIF), Partner Site Munich, Munich, Germany.
Tsuyoshi Miki, Department of Microbiology, School of Pharmacy, Kitasato University, Japan.
Roland M Schmid, Klinik für Innere Medizin II, Klinikum rechts der Isar, Techn. Univ. Munich, Munich, Germany.
Marcel R M van den Brink, Memorial Sloan-Kettering Cancer Center, Immunology Program, New York, USA.
Markus Gerhard, Institute for Medical Microbiology, Immunology and Hygiene, Techn. Univ. Munich, Munich, Germany; German Center for Infection Research (DZIF), Partner Site Munich, Munich, Germany.
Christoph K Stein-Thoeringer, Klinik für Innere Medizin II, Klinikum rechts der Isar, Techn. Univ. Munich, Munich, Germany; Division Microbiome and Cancer, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Funding
S.B. was supported by grants from the Swiss National Science Foundation [P2SKP3 158649, P3400PB 171581]. C.S.T. was supported by a clinical leave scholarship by DZIF and the Else-Kroener-Fresenius Forschungskolleg. B.S. was supported by the NIDDK-funded San Diego Digestive Diseases Research Center (P30 DK120515). M.F. was funded by the Helmholtz Alliance Aging and Metabolic Programming AMPro.
Conflict of Interest
The authors declare no conflicts of interest.
Author Contributions
D.B., A.N., K.H., S.B., M.D., M.C.F., E.T. and C.S.T. performed experiments; D.B., R.M.L. and C.S.T. performed data analysis; R.M.S., D.B., M.B., B.S., M.V.D.B. and M.G. contributed to concept and interpretation; D.B., R.M.L. and C.S.T. wrote the manuscript.
References
- 1. Okumura R, Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med 2017;49:e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006;313:1126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stelter C, Käppeli R, König C, et al. Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLoS One 2011;6:e20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Ampting MT, Loonen LM, Schonewille AJ, et al. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect Immun 2012;80:1115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dessein R, Gironella M, Vignal C, et al. Toll-like receptor 2 is critical for induction of reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut 2009;58:771–6. [DOI] [PubMed] [Google Scholar]
- 6. Wang L, Fouts DE, Stärkel P, et al. Intestinal reg3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe 2016;19:227–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin Regiiigamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011;334:255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atarashi K, Tanoue T, Ando M, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015;163:367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu YY, Hsu CM, Chen PH, Fung CP, Chen LW. Toll-like receptor stimulation induces nondefensin protein expression and reverses antibiotic-induced gut defense impairment. Infect Immun 2014;82:1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis 2010;201:534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stockinger S, Duerr CU, Fulde M, et al. TRIF signaling drives homeostatic intestinal epithelial antimicrobial peptide expression. J Immunol 2014;193:4223–34. [DOI] [PubMed] [Google Scholar]
- 12. Pedicord VA, Lockhart AAK, Rangan KJ, et al. Exploiting a host-commensal interaction to promote intestinal barrier function and enteric pathogen tolerance. Sci Immunol 2016;1:eaai7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aden K, Rehman A, Falk-Paulsen M, et al. Epithelial IL-23R signaling licenses protective Il-22 responses in intestinal inflammation. Cell Rep 2016;16:2208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ogawa H, Fukushima K, Naito H, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis 2003;9:162–70. [DOI] [PubMed] [Google Scholar]
- 15. Ferrara JL, Harris AC, Greenson JK, et al. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood 2011;118:6702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mathewson ND, Jenq R, Mathew AV, et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol 2016;17:505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bluemel S, Wang L, Martino C, et al. The role of intestinal C-type regenerating islet derived-3 lectins for nonalcoholic steatohepatitis. Hepatol Commun 2018;2:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004;118:229–41. [DOI] [PubMed] [Google Scholar]
- 19. Holzscheiter M, Layland LE, Loffredo-Verde E, et al. Lack of host gut microbiota alters immune responses and intestinal granuloma formation during schistosomiasis. Clin Exp Immunol 2014;175:246–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Staffas A, Burgos da Silva M, Slingerland AE, et al. Nutritional support from the intestinal microbiota improves hematopoietic reconstitution after bone marrow transplantation in mice. Cell Host Microbe 2018;23:447–457.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu D, Gao J, Gillilland M 3rd, et al. Rifaximin alters intestinal bacteria and prevents stress-induced gut inflammation and visceral hyperalgesia in rats. Gastroenterology 2014;146:484–96.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johansson ME, Phillipson M, Petersson J, et al. The inner of the two muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 2008;105:15064–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O’Rourke KP., Dow LE, Lowe SW. Immunofluorescent staining of mouse intestinal stem cells. Bio-protocol. 2016;6:e1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knoop KA, McDonald KG, Kulkarni DH, Newberry RD. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 2016;65:1100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Montrose DC, Zhou XK, McNally EM, et al. Celecoxib alters the intestinal microbiota and metabolome in association with reducing Polyp burden. Cancer Prev Res (Phila) 2016;9:721–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindemans CA, Calafiore M, Mertelsmann AM, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 2015;528:560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gjorevski N, Sachs N, Manfrin A, et al. Designer matrices for intestinal stem cell and organoid culture. Nature 2016;539:560–4. [DOI] [PubMed] [Google Scholar]
- 28. Lukovac S, Belzer C, Pellis L, et al. Differential modulation by akkermansia muciniphila and faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio 2014;5:e01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergström A, Licht TR, Wilcks A, et al. Introducing gut low-density array (GULDA): a validated approach for qPCR-based intestinal microbial community analysis. FEMS Microbiol Lett 2012;337:38–47. [DOI] [PubMed] [Google Scholar]
- 30. Ellekilde M, Selfjord E, Larsen CS, et al. Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Sci Rep 2014;4:5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer F, Paarmann D, D’Souza M, et al. The metagenomics rast server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 2008;9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shono Y, Docampo MD, Peled JU, et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci Transl Med 2016;8:339ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim MS, Morales W, Hani AA, et al. The effect of rifaximin on gut flora and Staphylococcus resistance. Dig Dis Sci 2013;58:1676–82. [DOI] [PubMed] [Google Scholar]
- 34. Collins MD, Lawson PA, Willems A, et al. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 1994;44:812–26. [DOI] [PubMed] [Google Scholar]
- 35. Gill PA, van Zelm MC, Muir JG, Gibson PR. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther 2018;48:15–34. [DOI] [PubMed] [Google Scholar]
- 36. Brown AJ, Goldsworthy SM, Barnes AA, et al. The orphan g protein-coupled receptors gpr41 and gpr43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 2003;278:11312–9. [DOI] [PubMed] [Google Scholar]
- 37. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 2016;165:1332–45. [DOI] [PubMed] [Google Scholar]
- 38. Fujiwara H, Docampo MD, Riwes M, et al. Microbial metabolite sensor gpr43 controls severity of experimental gvhd. Nat Commun 2018;9:3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Finnie IA, Dwarakanath AD, Taylor BA, Rhodes JM. Colonic mucin synthesis is increased by sodium butyrate. Gut 1995;36:93–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barcelo A, Claustre J, Moro F, et al. Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut 2000;46:218–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larsson E, Tremaroli V, Lee YS, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through myd88. Gut 2012;61:1124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sivaprakasam S, Gurav A, Paschall AV, et al. An essential role of ffar2 (gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogenesis 2016;5:e238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Earle KA, Billings G, Sigal M, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 2015;18:478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Biggs MB, Medlock GL, Moutinho TJ, et al. Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J 2017;11:426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lai Y, Li D, Li C, et al. The antimicrobial protein reg3a regulates keratinocyte proliferation and differentiation after skin injury. Immunity 2012;37:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sorbara MT, Dubin K, Littmann ER, et al. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J Exp Med 2019;216:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 2009;137:15–7. [DOI] [PubMed] [Google Scholar]
- 49. Stange DE, Clevers H. Concise review: the yin and yang of intestinal [cancer] stem cells and their progenitors. Stem Cells 2013;31:2287–95. [DOI] [PubMed] [Google Scholar]
- 50. Kim HS, Lee C, Kim WH, Maeng YH, Jang BG. Expression profile of intestinal stem cell markers in colitis-associated carcinogenesis. Sci Rep 2017;7:6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peix L, Evans IC, Pearce DR, et al. Diverse functions of clusterin promote and protect against the development of pulmonary fibrosis. Sci Rep 2018;8:1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shapiro B, Tocci P, Haase G, Gavert N, Ben-Ze’ev A. Clusterin, a gene enriched in intestinal stem cells, is required for L1-mediated colon cancer metastasis. Oncotarget 2015;6:34389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ayyaz A, Kumar S, Sangiorgi B, et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 2019;569:121–5. [DOI] [PubMed] [Google Scholar]
- 54. DuPont HL. Review article: the antimicrobial effects of rifaximin on the gut microbiota. Aliment Pharmacol Ther 2016;43 Suppl 1:3–10. [DOI] [PubMed] [Google Scholar]
- 55. Bajaj JS. Review article: potential mechanisms of action of rifaximin in the management of hepatic encephalopathy and other complications of cirrhosis. Aliment Pharmacol Ther 2016;43 Suppl 1:11–26. [DOI] [PubMed] [Google Scholar]
- 56. Reikvam DH, Erofeev A, Sandvik A, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 2011;6:e17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maccaferri S, Vitali B, Klinder A, et al. Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: an in vitro approach using a continuous culture colonic model system. J Antimicrob Chemother 2010;65:2556–65. [DOI] [PubMed] [Google Scholar]
- 58. Kimer N, Pedersen JS, Tavenier J, et al. Rifaximin has minor effects on bacterial composition, inflammation, and bacterial translocation in cirrhosis: a randomized trial. J Gastroenterol Hepatol 2018;33:307–14. [DOI] [PubMed] [Google Scholar]
- 59. Ponziani FR, Zocco MA, D’Aversa F, Pompili M, Gasbarrini A. Eubiotic properties of rifaximin: disruption of the traditional concepts in gut microbiota modulation. World J Gastroenterol 2017;23:4491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pistiki A, Galani I, Pyleris E, et al. In vitro activity of rifaximin against isolates from patients with small intestinal bacterial overgrowth. Int J Antimicrob Agents 2014;43:236–41. [DOI] [PubMed] [Google Scholar]
- 61. Acosta A, Camilleri M, Shin A, et al. Effects of rifaximin on transit, permeability, fecal microbiome, and organic acid excretion in irritable bowel syndrome. Clin Transl Gastroenterol 2016;7:e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhao Y, Chen F, Wu W, et al. Gpr43 mediates microbiota metabolite scfa regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mtor and stat3. Mucosal Immunol 2018;11:752–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Parks OB, Pociask DA, Hodzic Z, Kolls JK, Good M. Interleukin-22 signaling in the regulation of intestinal health and disease. Front Cell Dev Biol 2015;3:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zheng Y, Valdez PA, Danilenko DM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med 2008;14:282–9. [DOI] [PubMed] [Google Scholar]
- 65. Pickert G, Neufert C, Leppkes M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med 2009;206:1465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009;461:1282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mencarelli A, Migliorati M, Barbanti M, et al. Pregnane-X-receptor mediates the anti-inflammatory activities of rifaximin on detoxification pathways in intestinal epithelial cells. Biochem Pharmacol 2010;80:1700–7. [DOI] [PubMed] [Google Scholar]
- 68. Kang DJ, Kakiyama G, Betrapally NS, et al. Rifaximin exerts beneficial effects independent of its ability to alter microbiota composition. Clin Transl Gastroenterol 2016;7:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sartor RB. Review article: the potential mechanisms of action of rifaximin in the management of inflammatory bowel diseases. Aliment Pharmacol Ther 2016;43 Suppl 1:27–36. [DOI] [PubMed] [Google Scholar]
- 70. Darnaud M, Dos Santos A, Gonzalez P, et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology 2018;154:1009–23.e14. [DOI] [PubMed] [Google Scholar]
- 71. Malka D, Vasseur S, Bödeker H, et al. Tumor necrosis factor alpha triggers antiapoptotic mechanisms in rat pancreatic cells through pancreatitis-associated protein I activation. Gastroenterology 2000;119:816–28. [DOI] [PubMed] [Google Scholar]
- 72. Lieu HT, Batteux F, Simon MT, et al. HIP/PAP accelerates liver regeneration and protects against acetaminophen injury in mice. Hepatology 2005;42:618–26. [DOI] [PubMed] [Google Scholar]
- 73. Lörchner H, Pöling J, Gajawada P, et al. Myocardial healing requires Reg3β-dependent accumulation of macrophages in the ischemic heart. Nat Med 2015;21:353–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.