Abstract
The rapidly evolving antigenic diversity of influenza A virus (IAV) genomes in swine makes it imperative to detect emerging novel strains and track their circulation. We analyzed in our review the sequencing technologies used for subtyping and characterizing swine IAV genomes. Google Scholar, PubMed, and International Nucleotide Sequence Database Collaboration (INSDC) database searches identified 216 studies that have utilized Sanger, second-, and third-generation sequencing techniques to subtype and characterize swine IAV genomes up to 31 March 2021. Sanger dideoxy sequencing was by far the most widely used sequencing technique for generating either full-length (43.0%) or partial (31.0%) IAV genomes in swine globally; however, in the last decade, other sequencing platforms such as Illumina have emerged as serious competitors for the generation of whole-genome sequences of swine IAVs. Although partial HA and NA gene sequences were sufficient to determine swine IAV subtypes, whole-genome sequences were critical for determining reassortments and identifying unusual or less frequently occurring IAV subtypes. The combination of Sanger and second-generation sequencing technologies also greatly improved swine IAV characterization. In addition, the rapidly evolving third-generation sequencing platform, MinION, appears promising for on-site, real-time sequencing of complete swine IAV genomes. With a higher raw read accuracy, the use of the MinION could enhance the scalability of swine IAV testing in the field and strengthen the swine IAV disease outbreak response.
Keywords: IAV subtyping; IAV surveillance; influenza A virus; MinION, next-generation sequencing; Sanger sequencing; swine IAV sequencing; whole-genome sequencing
Influenza A virus (IAV; Orthomyxoviridae, Alphainfluenzavirus) is the most prevalent of the 4 influenza virus types that have been reported in swine populations globally. 18 The tracheal receptors that efficiently bind human- and avian-origin IAVs make swine a favorable host or a mixing vessel for IAV inter-species transmission, reassortment, and evolution. 93 A broad range of antigenic diversity in hemagglutinin (HA) and neuraminidase (NA) genes exists in currently circulating swine IAV genomes,104,105 and forms the basis of IAV subtyping;3,110 however, mutations 118 and reassortments in internal gene segments are also critical for swine IAV evolution. The 2009 flu pandemic originated primarily from the interspecies transmission and reassortment of avian and human IAV strains within swine in Mexico, which triggered the emergence of a new IAV subtype termed the “A(H1N1)pdm09” virus. 65 In addition, by utilizing various sequencing technologies, numerous other novel and reassortant IAV subtypes have been reported in swine in recent years.97,100,103,106
Sanger dideoxy sequencing was the first sequencing platform that was introduced in 1977, 90 and, given the long (~800 bp) and high-quality reads produced, is considered the gold standard for DNA sequencing. 11 Sanger sequencing utilizes a chain-termination strategy that relies on a modified DNA polymerase enzyme, which incorporates dideoxynucleotides (ddNTPs) at a specific position in the DNA template. The initial studies for sequencing swine IAVs used 32 P-end–labeled oligonucleotide primers 95 and had to be manually read from the sequencing gel, making this method hazardous as well as labor- and time-intensive. The introduction of automated DNA sequencers, such as the ABI 3730 DNA sequencer 9 and the ABI 3130/3130xl genetic analyzer,66,69,70 as well as the replacement of radioactive-labeled primers with rhodamine-based fluorescent dyes, overcame these challenges and, as a result, this technology was used extensively to obtain up to 900 bases of IAV gene sequences per sequencing reaction.
The automation of Sanger sequencing along with the advent of next-generation sequencing (NGS) platforms, including the Roche 454 GS Junior in 2005, Illumina in 2007, and the Ion Torrent PGM in 2010, also facilitated the whole-genome sequencing (WGS) of IAV in swine.2,14,21 Although all next-generation technologies are capable of high-throughput sequencing, they use different sequencing strategies. Briefly, the Roche 454 GS Junior Titanium platform uses pyrosequencing by which it detects, in real-time, luminescence that results from the release of pyrophosphate at the incorporation of each nucleotide into the growing complementary DNA strand. 35 The Illumina platform utilizes a reversible chain termination strategy that uses fluorescently labeled reversible terminators (specially designed ddNTPs) that terminate primer extension during the sequencing reaction. In contrast, the Ion Torrent PGM works on the principle of proton detection by measuring the pH change that results from the release of a hydrogen ion upon incorporating a dNTP to the growing DNA template, using an ion semiconductor chip that offers a higher sequencing speed. 54
One of the significant limitations of Roche 454 pyrosequencing was the generation of false signals for homopolymers of adenine (A) in the sequencing reaction. 86 Similarly, one of the limitations of the Ion Torrent PGM lies in its low accuracy in recognizing homopolymers of >6 nucleotides in the DNA template. Illumina sequencing offers a highly specific “base-by-base” sequencing technology for eliminating homopolymer errors. Various Illumina sequencers, namely, Genome Analyzer IIx (GAIIx), NextSeq, MiSeq, and HiSeq, have been used to identify and characterize IAV genomes in swine populations. Notably, the MiSeq platform of Illumina, because of its user-friendly interface, is a popular second-generation sequencer for generating full-length IAV genomes in swine.
The advent of the Nanopore MinION, a third-generation sequencing platform, has further enhanced the existing capabilities of generating complete swine IAV genomes. 84 Given that high accuracy and low sequencing cost are critical for large-scale swine IAV testing, the continuous upgrading of the sequencing platforms, over a relatively short period of time, has made swine IAV WGS more affordable. The advancement and automation of sequencing technologies has therefore made it possible to conduct genomic surveillance of emerging subtypes, and monitoring of IAV evolution, in real-time. We present here a comprehensive overview of the use of various genome sequencing technologies and their applications, reliability, and cost-effectiveness in swine IAV testing, and their capacity for the characterization of novel and reassortant IAV subtypes in swine.
Review protocol and search criteria
We adopted a 2-tier approach for identifying the relevant records for IAV genome sequencing in swine. First, a comprehensive search of scientific databases, including NCBI-PubMed and Google Scholar, was conducted to identify full-text research articles that reported genome sequencing for identifying and characterizing IAV subtypes in swine populations globally. Search terms, including “influenza A virus outbreak in pigs”, “influenza A virus outbreak in swine”, “influenza A virus in swine”, “sequences of influenza A virus in swine”, and “influenza virus disease in swine”, were entered into the NCBI-PubMed and Google Scholar databases one by one. The title, abstract, methodology, and/or supplementary information associated with the research articles that emanated from the online database searches up to 31 March 2021 were screened for relevance for inclusion in our study. The supplementary data of publications were also used to assess the significance of articles for inclusion in our study. In a few cases, if the full-text research articles were not accessible online, those were requested from the authors through ResearchGate. The availability of full-length and partial IAV genomes and the information on sequencing methods were thoroughly verified through the NCBI-GenBank database using the accessions provided in each research article included in the analysis.
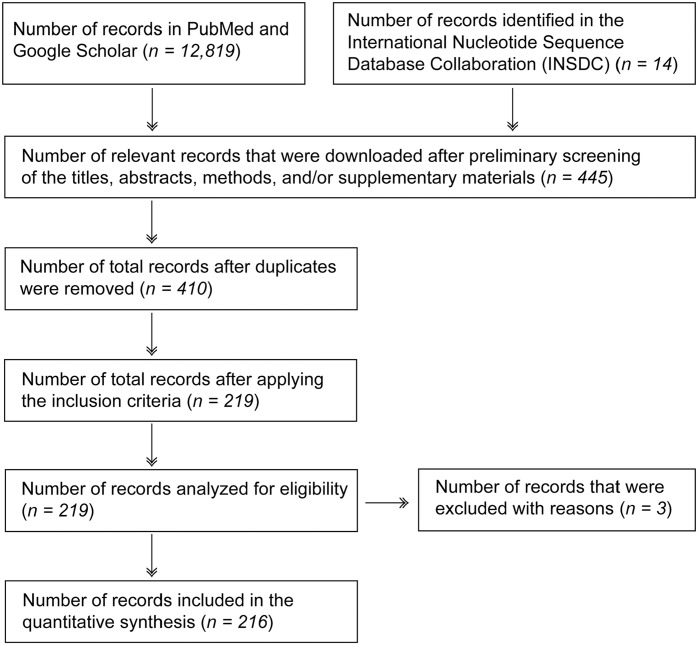
A further search of the International Nucleotide Sequence Database Collaboration (INSDC) database using the terms “influenza A virus in swine” and “influenza A virus genome sequencing” identified an additional 14 BioProjects reported from 9 countries that have attempted IAV genome sequencing in swine populations during 2017–2020 that were not available in the NCBI-PubMed and Google Scholar databases. The INSDC database is maintained by a collaboration of NCBI-GenBank, the European Bioinformatics Institute (EMBL-EBI), and the DNA Data Bank of Japan (DDBJ). INSDC publishes the technologies used for library preparation and sequencing. We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) 53 2009 flowchart to screen the literature and identify the relevant research articles because the PRISMA flowchart systematically explains each step for screening the literature and therefore helps to identify the relevant records (Fig. 1).
Figure 1.
The PRISMA chart illustrates the search strategy for relevant records that have utilized sequencing methods for subtyping and characterizing influenza A virus genomes from swine globally. We included 216 relevant records in our study, including 202 full-text research articles available in NCBI-PubMed and Google Scholar databases, and 14 BioProjects available in the INSDC database.
Inclusion and exclusion criteria
We included full-text original research articles and INSDC BioProjects that utilized genome sequencing to generate partial or complete IAV genomes in swine populations. Reviews, experimental studies, and serologic studies that did not attempt IAV genome sequencing in swine were not included in our analysis. Research articles in a language other than English were excluded from our analysis.
Records selected
As of 31 March 2021, we identified 216 records that have reported various methods for either partial or complete genome sequencing of IAV subtypes in swine populations globally (Fig. 1). The primary use of IAV genome sequencing was the subtyping and molecular characterization of IAV genomes to unravel the growing antigenic diversity of IAV in swine. Sanger dideoxy sequencing was the first method utilized for partial IAV genome sequencing in swine during the 1970s and 1980s in passive surveillance programs. 116 The full-length swine IAV genomes started appearing in the 1990s, still using Sanger technology. 78 The Illumina GAIIx was the first NGS platform to generate the complete genome of the A(H1N1)pdm09 virus from ill swine during the “swine-flu” pandemic in Mexico in 2009. 26 Although the Roche 454 GS Junior Titanium was among the earliest NGS platforms launched in 2005, it was not until 2010 that the Roche 454 GS Junior platform was utilized to sequence H1N1 and H3N2 viruses in swine in Spain. 62 The Ion Torrent PGM, another second-generation sequencing platform, generated a complete genome of a H1N2 subtype termed “A/swine/Denmark/10302-2/2012(H1N2)” during passive surveillance in pigs in Denmark in 2012. 14 Further technologic advancements in recent years enabled the launch of third-generation sequencing platforms. For example, Oxford Nanopore Technologies launched a real-time sequencing platform termed “MinION” for rapid WGS. To date, only one investigation has reported full-length swine IAV genomes using Nanopore MinION sequencing, 84 in which 13 full-length IAV genomes were sequenced and characterized from symptomatic swine on-site overnight at a swine exhibition in Iowa, USA. 84
Sanger sequencing
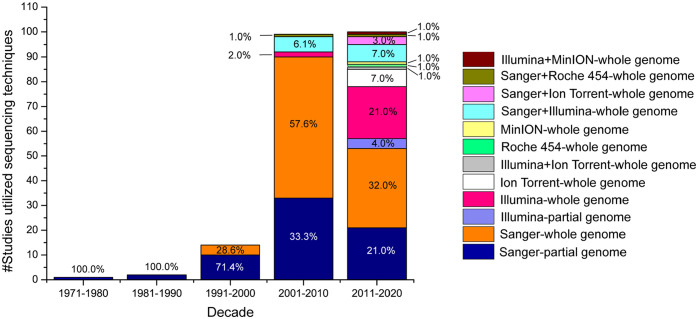
Sanger dideoxy sequencing was the most widely used sequencing method overall, with 93 (43.0%) studies using this technology to generate all 8 gene segments of IAV genomes; 67 (31.0%) studies amplified and sequenced only partial IAV genomes, including HA and NA genes for IAV subtyping. It was noteworthy that most of the novel and reassortant IAV subtypes were detected and characterized by Sanger dideoxy WGS (Table 1), which was also the most widely used sequencing method for whole genomes. Of the numerous studies that generated only partial IAV genomes in swine, there were also a few that identified novel or reassortant IAV subtypes.
Table 1.
Novel influenza A virus (IAV) subtypes in swine characterized by various sequencing approaches.
| Technique/Novel IAV subtypes reported | Virus strains sequenced | Year | PMID | Citation |
|---|---|---|---|---|
| Sanger dideoxy WGS | ||||
| Reassortant H9N2 | A/Swine/Hong Kong/9/98(H9N2) | 1998 | 11559800 | Ref. [78] |
| H4N6 | A/Swine/Ontario/01911-1/99 | 1999 | 10982381 | Ref. [40] |
| Avian H3N3 | A/Swine/Ontario/42729A/01 | 2001 | 15365042 | Ref. [41] |
| A/Swine/Ontario/K01477/01 | 2001 | |||
| Reassortant H7N2 | A/swine/KU/16/2001 | 2001 | 21741185 | Ref. [45] |
| Avian H1N1 | A/Swine/Saskatchewan/18789/02 | 2002 | 15365042 | Ref. [41] |
| Reassortant H3N1 | A/Swine/Minnesota/00395/2004 | 2004 | 16641303 | Ref. [59] |
| Equine H3N8 | A/swine/Chibi/01/2005(H3N8) | 2005 | 19396578 | Ref. [102] |
| A/swine/Anhui/01/2006(H3N8) | 2006 | |||
| H5N1 | A/swine/Banten/UT2071/2005(H5N1) | 2005 | 20875275 | Ref. [76] |
| A/swine/Banten/UT3063/2005(H5N1) | 2005 | |||
| A/swine/Banten/UT6001/2006(H5N1) | 2006 | |||
| Reassortant H2N3 | A/swine/Missouri/4296424/2006(H2N3) | 2006 | 18093945 | Ref. [60] |
| A/swine/Missouri/2124514/2006(H2N3) | 2006 | |||
| Reassortant avian-origin H9N2 | A/Swine/Guangxi/7/07(H9N2) | 2007 | 18403137 | Ref. [112] |
| Reassortant H5N2 | A/Swine/Korea/C12/08 | 2008 | 19359528 | Ref. [49] |
| A/Swine/Korea/C13/08 | 2008 | |||
| Avian H10N5 | A/swine/Hubei/10/2008/H10N5 | 2008 | 23166264 | Ref. [106] |
| H5N1 | A/swine/Jiangsu/1/2008 | 2008 | 23836394 | Ref. [36] |
| A/swine/Jiangsu/2/2009 | 2009 | |||
| Novel H4N1 | A/Swine/HuBei/06/2009(H4N1) | 2009 | 23166273 | Ref. [38] |
| Novel reassortant H3N2 | A/Swine/Guangxi/NS2783/10(H3N2) | 2010 | 25008935 | Ref. [52] |
| Eurasian avian-like H1N1 genotype 1 | A/swine/Henan/201/2011 | 2011 | 32601207 | Ref. [99] |
| Reassortant H3N1 | A/swine/Chachoengsao/NIAH105583-062-46/2012 | 2012 | 26115167 | Ref. [1] |
| Eurasian avian-like H1N1 genotype 1 | A/swine/Hebei/156/2012 | 2012 | 32601207 | Ref. [99] |
| A/swine/Jilin/625/2013 | 2013 | |||
| Eurasian avian-like triple reassortant H1N1 genotype 5 | A/swine/Shandong/S113/2014 | 2014 | 32601207 | Ref. [99] |
| Eurasian avian-like triple reassortant H1N1 genotype 6 | A/swine/Anhui/1227/2015 | 2015 | 32601207 | Ref. [99] |
| Eurasian avian-like triple reassortant H1N1 genotype 4 | A/swine/Shandong/16/2016 | 2016 | 32601207 | Ref. [99] |
| A/swine/Hebei/0113/2017 | 2017 | |||
| A/swine/Henan/SN10/2018 | 2018 | |||
| Novel reassortant H1N1 | A/swine/China/Qingdao/2018(H1N1) | 2018 | 31535780 | Ref. [113] |
| Sanger dideoxy (partial) genome sequencing | ||||
| Reassortant H3N2 | A/swine/Potsdam/35/1982(H3N2) | 1982 | 32868846 | Ref. [116] |
| Reassortant H1N7 | A/Swine/England/191973/92 | 1992 | 9191869 | Ref. [15] |
| H9N2 | A/swine/Henan/2/2004(H9N2) | 2004 | 18401696 | Ref. [21] |
| A/swine/Henan/3/2004(H9N2) | 2004 | |||
| H5N1 | A/swine/Egypt/165/2015(H5N1) | 2015 | 29075888 | Ref. [29] |
| H9N2 | A/swine/Egypt/151/2015(H9N2) | 2015 | ||
| Avian-origin H5N1 | A/swine/Nigeria/49/2016(H5N1) | 2016 | 29651056 | Ref. [66] |
| Roche 454 GS Junior WGS | ||||
| H1N1, H1N2 | A/swine/Ontario/13-1/2012(H1N1) | 2012 | 26030614 | Ref. [32] |
| A/swine/Ontario/68/2012(H1N2) | 2012 | |||
| Illumina WGS | ||||
| Novel reassortant H3N2 | A/swine/Rietberg/19732/2014(H3N2) | 2014 | 32868846 | Ref. [116] |
| Avian-origin H4N6 | A/swine/Missouri/A01727926/2015(H4N6) | 2015 | 28841443 | Ref. [2] |
| Unusual reassortant H1N1 | A/swine/Siberia/1sw/2016(H1N1) | 2016 | 28883131 | Ref. [98] |
| H1N2 variants | A/swine/Denmark/18-6662-26_PB2/2018 (H1N2) | 2018 | 32927910 | Ref. [12] |
| Ion Torrent PGM WGS | ||||
| H5N2 | A/swine/Estado de Mexico/EdoMexDMZC03/2015(H5N2) | 2015 | 30126057 | Ref. [88] |
| Avian H5N2 | Feral swine/Campeche/DMZC-DEFSAL-UIFMVZ19-12 (H5N2) | 2019 | 32403268 | Ref. [63] |
| Oxford Nanopore MinION WGS | ||||
| H1N1 | A/swine/Iowa/18Tosu0505/2018(H1N1) | 2018 | 32024713 | Ref. [84] |
| H1N2 | A/swine/Iowa/18TOSU0374/2018(H1N2) | 2018 | ||
| H3N2 | A/swine/Iowa/18Tosu0394/2018(H3N2) | 2018 | ||
WGS = whole-genome sequencing.
Second-generation sequencing
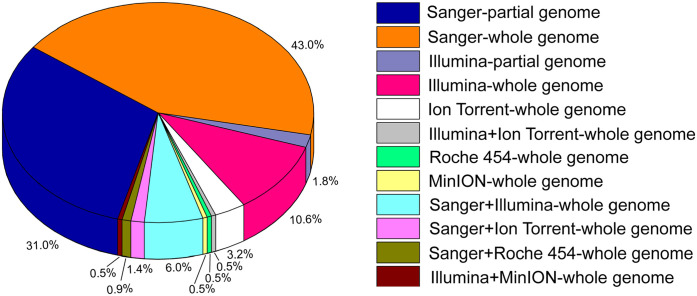
The introduction of various NGS platforms facilitated the detection and characterization of swine IAV WGS in active and passive surveillance undertaken in several countries. A total of 23 (10.6%) studies used only Illumina sequencing, which was the most used NGS platform for generating complete swine IAV genomes. In addition, 3 (1.8%) studies generated partial IAV genomes using Illumina sequencing alone. Ion Torrent PGM was used in 7 (3.2%) studies to generate complete IAV genomes; 1 (0.5%) study utilized a combination of Illumina and Ion Torrent PGM for generating full-length swine IAV genomes. Only one (0.5%) study utilized the Roche 454 GS Junior Titanium platform for IAV WGS.
Even though second-generation sequencers offered much promise for generating full-length IAV genomes, there were certain limitations, one of which was the generation of short reads.5,31 The alignment of short reads into a contiguous sequence (contig) is challenging, especially in cases in which there are repetitions in the genome sequences, often resulting in gaps in the genome assembly (e.g., where genome repeats are longer than the individual read lengths or where there is insufficient coverage). 81 Although mapping the assembled genome with a reference genome may resolve these gaps, in the case of de novo assembly, filling these gaps becomes challenging and usually requires a bioinformatics pipeline with a sufficiently trained workforce.
Sanger sequencing and NGS in combination
A total of 13 (6.0%) studies used a combination of Illumina and Sanger dideoxy sequencing to generate complete swine IAV genomes. Most of these studies were large-scale and spanned a long period of time, with the incorporation of the Illumina platform once it became available.114-116 In addition, 3 (1.4%) studies combined Ion Torrent PGM and Sanger dideoxy sequencing for generating full-length IAV genomes, whereas the Roche 454 GS Junior Titanium and Sanger dideoxy sequencing were used for full-length sequencing in 2 (0.9%) studies.
Third-generation sequencing
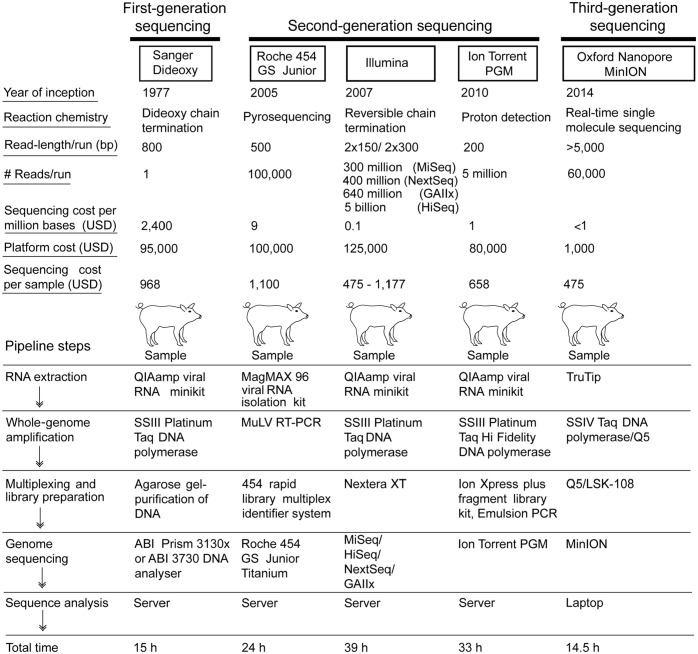
Oxford Nanopore Technologies developed a portable sequencing device called the “MinION” to overcome the challenges with NGS technologies, and to offer single-molecule real-time sequencing. 39 To date, one (0.5%) study utilized only the MinION sequencing platform for generating complete IAV genomes in swine; one study (0.5%) utilized the combination of Illumina and MinION sequencing to generate swine IAV whole genomes (Fig. 2). One of the significant advantages of the MinION is the generation of long reads (~5,000 bases). The portable Nanopore MinION device is also available at an affordable price of ~USD 1,000. The Nanopore MinION device detects the sequences using an applied electric current as the DNA template passes through the biological nanopore. 39 Other salient features of the MinION include its portability, miniature size, rapid results, and laptop-based sequence analysis. These features make the MinION an attractive option as a scalable sequencing technique for large-scale real-time IAV surveillance in swine. Interestingly, the MinION can generate swine IAV sequences much faster than the other NGS platforms. For example, the pipeline steps from RNA extraction until sequencing using the MinION may take 14 h 43 or 14.5 h 84 compared to Illumina (39 h), 84 Roche 454 GS Junior (24 h), 54 Ion Torrent PGM (33 h), 43 and Sanger sequencing (15 h). The advent of third-generation sequencing technology appears promising for real-time and large-scale IAV surveillance in swine populations.
Figure 2.
Illustration of sequencing approaches for partial and full-length genome sequencing of influenza A virus (IAV) subtypes from swine globally. Various sequencing techniques were used in 216 studies to generate IAV genomes in swine up to 31 March 2021. Sanger dideoxy sequencing76,99 was the sequencing technique used most widely, followed by second-generation Illumina sequencing. 2 Since 2014, a third-generation sequencing method, MinION, has also been used for generating IAV genomes from swine. 84
Sequencing approaches used by NGS studies
Overall, 5 different sequencing platforms have been used to generate IAV genomes in swine populations based on various reaction chemistries (Fig. 3; Table 2). Although amplicon sequencing was the more popular sequencing approach in NGS studies (70.4%), a few studies (27.3%) used random primers for cDNA synthesis for library preparation; the one remaining (2.3%) NGS study did not mention the sequencing approach utilized.
Figure 3.
Comparison of Sanger, second-, and third-generation sequencing technologies that have been used for influenza A virus (IAV) sequencing in swine populations. The sequencing cost varied among different sequencing platforms, especially between Sanger and second-generation sequencing platforms. The Oxford Nanopore MinION sequencing generated IAV genomes from swine samples within 14.5 h, the shortest reaction time among all next-generation sequencing platforms.
Table 2.
An overview of sequencing approaches to generate influenza A virus (IAV) genome sequences in swine.
| Sequencing technology | IAV subtypes reported | Strategy for IAV sequencing | Citation |
|---|---|---|---|
| First-generation sequencing | |||
| Sanger dideoxy WGS | H1N1, H1N2, H3N2, A(H1N1)pdm09, H2N3, H3N1, H3N3, H3N8, H4N1, H4N6, H5N1, H5N2, H7N2, H9N2, H10N5 | All 8 segments were fully amplified using IAV gene-specific/universal primers. The amplicons were sequenced. | Refs. [1,4,9,24,36–38,40,41,45,49,52,59,60,75,76,78,94,99,102,106,108,117] |
| Sanger dideoxy sequencing (partial genome) | H3N2, H9N2 | All 8 segments were partially amplified. The amplicons were sequenced. | Refs. [21,95] |
| H1N1, H1N2, H3N2, A(H1N1)pdm09, H5N1 | The HA and NA genes were amplified using gene-specific primers. The amplicons were sequenced. | Refs. [34,66,80,96,101] | |
| Avian H5, H9 | The HA gene was amplified using gene-specific primers. The amplicons were sequenced. | Ref. [29] | |
| Second-generation sequencing | |||
| Illumina MiSeq/HiSeq 2000/GAIIx WGS | H1N1, H1N2, A(H1N1)pdm09, reassortant H1N1, reassortant H3N2, reassortant A(H1N1)pdm09 | Reverse transcription used random hexamers. Double-stranded cDNA was used for library preparation. | Refs. [20,47,98,114–116] |
| Illumina MiSeq | H3N2, A(H1N1)pdm09 | Reverse transcription used IAV universal primers. Double stranded cDNA was used for library preparation. | Ref. [67] |
| Illumina MiSeq (partial genome) | H1, H3 | Reverse transcription used random hexamers. Only HA gene was amplified using gene-specific primers. | Ref. [46] |
| H1 | Only HA gene was amplified using gene-specific primers. The amplicons were used for library preparation. | Ref. [100] | |
| Illumina MiSeq/HiSeq/NextSeq 500/GAIIx WGS | H1N1, H1N2, reassortant H1N2, H3N2, H4N6, A(H1N1)pdm09 | All 8 segments were fully amplified using IAV gene-specific primers. The amplicons were used for library preparation. | Refs. [2,12,16,19,22,25,64,65,68,71,73,77,87,91,92,109] |
| Ion Torrent PGM | H1N1, H1N2, H3N2, A(H1N1)pdm09, H5N2 | All 8 segments were fully amplified using IAV gene-specific primers. The amplicons were used for library preparation. | Refs. [7,14,44,58,63,72,88,89] |
| Roche 454 GS Junior | H1N1, H1N2, H3N2 | Reverse transcription used random primers. Double-stranded cDNA was used for library preparation. | Refs. [32,33,62] |
| Third-generation sequencing | |||
| Oxford Nanopore MinION | H1N1, H1N2, H3N2 | All 8 segments were fully amplified using IAV gene-specific primers. The amplicons were used for library preparation. | Ref. [84] |
WGS = whole-genome sequencing.
Advantages of WGS over partial IAV genome sequencing
Although partial genome sequences (specifically HA and NA genes) are sufficient to determine IAV subtypes and their origin, in recent decades, more studies have focused on generating complete swine IAV genomes because it provides crucial information about swine IAV evolution (Fig. 4). An example of this is a recent whole genome study on swine in China in which they found reassortant HA and NA genes of Eurasian-avian origin, the PA gene of avian-origin, NS gene of the triple-reassortant lineage, and other internal genes of the A(H1N1)pdm09 virus lineage. These sequences, therefore, provided information on how the swine viruses had evolved to facilitate human infection. 99 Another example is a 2017 study in the United States, in which whole-genome phylogenetic analyses of swine IAVs identified numerous swine genotypes with triple-reassortant internal genes (TRIGs). 83 In addition, whole-genome sequences have been used to identify potential mammalian adaptation markers in IAVs isolated from swine.61,82 These markers indicate that circulating human 82 as well as avian IAVs17,61 are adapting to swine. Other advantages of sequencing whole IAV genomes include the use of this information for reverse genetics. An example illustrating this use is a study in which reverse genetics experiments showed that the 2009 pandemic emerged not only as a result of IAV gene reassortments, but also mutations in the genome. 118
Figure 4.
A graphical illustration of the trend of reporting sequencing approaches for either partial or complete genome sequencing of influenza A virus (IAV) from swine globally. Partial swine IAV gene sequencing started during the 1970s. Although Sanger dideoxy sequencing continues to be the sequencing method used most widely for generating IAV genomes from swine, the emergence of next-generation sequencing platforms has challenged the dominance of Sanger dideoxy sequencing since ~2010. Various second- and third-generation sequencing methods have generated full-length IAV genomes from swine samples in recent decades. A few long-term, large-scale studies utilized a combination of Sanger and second-generation sequencing techniques to generate IAV genomes in swine.
Applications of swine IAV WGS
WGS has also indirectly assisted with IAV vaccine development.57,105 For example, genetic information from a swine-like triple-reassortant H3N2 virus allowed scientists to develop a live attenuated vaccine against the A(H1N1)pdm09 virus 79 through modifications of the PB1 and PB2 polymerase genes of the swine virus. Similarly, genetic information from IAV genomes enabled the expression of a truncated NS1 protein from swine H3N2 virus for a modified-live virus vaccine. 85 Sequences from circulating strains have also provided information about amino acid substitutions that affect vaccine efficacy. 105 For this type of application, Sanger sequencing and NGS technologies have an advantage because they can efficiently measure the occurrence of variants of concern (resistant or capable of immune escape) in a complex IAV population. 57 In addition, swine IAV, like several other RNA viruses, may form quasi-species given their error-prone polymerase enzyme. 23 NGS deep-sequencing platforms, such as Illumina, have efficiently detected minority variants in a diverse IAV population,10,56 and have shown their strength in analyzing antigenic drift 50 and identifying existing antigenic diversity in swine IAVs, which are helpful with vaccine-related decisions.13,51,104
Most NGS studies using amplicon sequencing for generating all 8 swine IAV genome segments 28 have facilitated the identification of complex IAV populations. For example, identification of avian-origin H4N6 virus in swine in Canada used amplicon sequencing to generate the entire H4N6 virus genome. 2 Furthermore, generating whole-genome swine IAV sequences using NGS has enabled the analysis of antigenic shift (reassortments) 74 in the genomes, giving insights into swine IAV evolution. 8
Advantages of NGS over RT-PCR
Although commonly occurring IAV subtypes, such as H1 and H3, as well as N1 and N2, can be determined using reverse-transcription PCR (RT-PCR) assays targeting the HA and NA genes, the existing broad genetic diversity of IAVs in swine could make subtyping challenging for some of these variants as well as other less-frequently occurring IAV subtypes,especially in events of spillover from other host species to swine. Genetic information gained from sequencing, therefore, has an advantage. For example, one nasal swab sample obtained from a pig with clinical signs of influenza-like illness was found to be RT-PCR positive during the IAV screening assay. The virus was isolated on Madin–Darby canine kidney (MDCK) cells, but it could not be subtyped using H1 and H3 as well as N1 and N2 subtyping assays; however, IAV screening RT-PCR remained positive. The virus isolate was sequenced on the Illumina MiSeq, which successfully generated the complete genome of an avian-origin H4N6 virus. 2 In addition, nonspecific amplification during PCR might result in a false IAV positive. For example, 4 swine oropharyngeal swabs that had been identified incorrectly as IAV positive by RT-PCR were subsequently identified as non-target contigs and not actual viral RNA contigs, using the MiSeq platform. 55 NGS has also shown potential in IAV testing with full-length IAV genomes generated from RT-PCR–positive swine samples with Ct values <30.27,107 However, real-time RT-PCR–positive samples with Ct values >35 failed to generate IAV sequences because of the very low abundance of virus sequences in those samples with high Ct values. 27
Cost-effectiveness of various sequencing platforms
Although Sanger sequencing is still cost-effective for partial swine IAV sequencing for subtype identification, NGS platforms have reduced the sequencing cost per million bases compared to Sanger dideoxy sequencing. For example, Sanger sequencing cost ~USD 2,400 per megabase (Mb), compared to the Roche 454 GS Junior platform cost of USD 9. The Ion Torrent PGM further reduced this cost to USD 1; Illumina sequencing costs only USD 0.1 per Mb,54,111 and the Nanopore MinION costs <USD 1 per Mb. A 2021 study analyzed the cost-effectiveness of Sanger and NGS platforms for influenza virus WGS in the reference laboratories in Europe and determined that Sanger WGS at the Friedrich-Loeffler-Institut (FLI; Germany) costs € 836 (USD 968) per sample. 6 In comparison, the Ion Torrent PGM costs € 568 (USD 658) per sample at the FLI. 6 The Illumina MiSeq WGS at the Animal and Plant Health Agency (APHA, UK) costs € 1,017 (USD 1,177). 6 It was reported that the NGS cost varies between the laboratories because of several factors, including batch size for sample preparation as well as supplier related costs of equipment and consumables. 6
In our experience, the Illumina MiSeq at the Agricultural Research Council (ARC), Onderstepoort, Pretoria, South Africa costs ~USD 475 per sample; Illumina MiSeq at the Michigan State University, USA starts from USD 524 per sample, depending on the genome coverage (https://rtsf.natsci.msu.edu/genomics/pricing/). Although a specific cost for IAV sequencing on the Roche 454 GS Junior platform (which is no longer supported) could not be obtained, general sequencing costs are reported to be USD 1,100 per sample (http://www.personalizedgenes.com/). As a result, the competitive cost of the NGS platforms have facilitated the use of large-scale whole genome studies for IAV surveillance in swine. The competitive running cost of the Nanopore MinION at ~USD 475 per sample has the potential to establish it as a preferred sequencing application for swine IAV detection in research laboratories.
Sequencing challenges
Just as Sanger sequencing offered high accuracy of raw reads (99.9%), the second-generation sequencing platforms (e.g., Roche 454 GS Junior [99.9%] and Illumina [98%]), offered comparable accuracy when aligned to reference genomes. 54 However, one challenge that the Roche 454 GS Junior sequencer faced was its inability to correctly identify homopolymers of ≥6 nucleotides. 54 The Ion Torrent PGM was reported to be 1.5 times less accurate than the Illumina MiSeq for sequencing IAV genomes as a result of insertions and deletions that occurred mainly in the homopolymer regions. In contrast, the errors that occurred on the Illumina MiSeq platform were mostly nucleotide substitutions. 103
The accuracy of MinION sequencing, when aligned to a reference genome, has been reported to be much lower than the other NGS platforms (up to 71.5%) 48 ; however, de novo assembly using the MinION offered improved assembly compared to Illumina. 30 Intriguingly, the recent advances in algorithms in MinION are reported to improve its raw read accuracy up to 98.3% (as of May 2021; Oxford Nanopore Technologies), with further improvements ongoing to gain a higher raw read accuracy. Given that we are under constant threat of the emergence of another influenza pandemic, it is imperative for a sequencing technology such as the MinION to become deployable for a scalable outbreak response in the field.
Prospects of RNA sequencing for IAV detection
Direct RNA sequencing of IAV using the Nanopore MinION generates longer reads of the IAV genome in a shorter time by eliminating the requirements for cDNA synthesis and PCR amplification. 42 The complete coding region of an IAV genome has been sequenced directly from RNA using a reverse genetically constructed “rA/Puerto Rico/8/1934” virus, a candidate vaccine virus, and a standard laboratory strain. 42 Using a custom-designed adaptor to target the negative-sense RNA into a protein nanopore on the MinION platform, 100% nucleotide coverage was generated successfully, with 99% of reads mapped to the IAV genome 42 ; the study should pave the way for performing direct RNA sequencing for other RNA viruses and may also be applied to swine IAV clinical samples.
Conclusion
The ongoing reports of novel IAV subtypes and genotypes in swine populations exemplify the need for active IAV genomic surveillance. Sanger dideoxy sequencing has contributed significantly to unravelling the existing genetic and antigenic diversity of IAV genomes in swine. More recently, the applications of second-generation sequencing, especially the Illumina MiSeq, have facilitated large-scale surveillance for investigating IAV disease burden in swine populations. Interestingly, the on-site real-time sequencing ability of the emerging third-generation Nanopore MinION technology, given the low cost, portability, and laptop-based analysis, makes it a serious competitor to the existing second-generation sequencing platforms. Optimization of the MinION for direct RNA sequencing may transform the swine IAV genome sequencing landscapes in upcoming years.
Acknowledgments
We thank Dr. Vivienne Russell for reviewing the manuscript. We thank the College of Health Sciences of the University of KwaZulu-Natal, Durban, for a 3-y CHS Research Scholarship to Ravendra P. Chauhan. We acknowledge the reviewers and the editor for providing valuable suggestions, which were critical to improving the manuscript. We sincerely thank the editor-in-chief for the critical review of the manuscript and the useful comments, which further improved the presentation of the manuscript.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Ravendra P. Chauhan  https://orcid.org/0000-0002-4674-8255
https://orcid.org/0000-0002-4674-8255
Michelle L. Gordon  https://orcid.org/0000-0001-9945-1394
https://orcid.org/0000-0001-9945-1394
References
- 1. Abe H, et al. Co-infection of influenza A viruses of swine contributes to effective shuffling of gene segments in a naturally reared pig. Virology 2015;484:203–212. [DOI] [PubMed] [Google Scholar]
- 2. Abente EJ, et al. Detection and characterization of an H4N6 avian-lineage influenza A virus in pigs in the Midwestern United States. Virology 2017;511:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Nat Acad Sci U S A 1981;78:7639–7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ali A, et al. Identification of swine H1N2/pandemic H1N1 reassortant influenza virus in pigs, United States. Vet Microbiol 2012;158:60–68. [DOI] [PubMed] [Google Scholar]
- 5. Alkan C, et al. Limitations of next-generation genome sequence assembly. Nat Methods 2011;8:61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alleweldt F, et al. Economic evaluation of whole genome sequencing for pathogen identification and surveillance—results of case studies in Europe and the Americas 2016 to 2019. Euro Surveill 2021;26:1900606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson BD, et al. Prospective surveillance for influenza A virus in Chinese swine farms. Emerg Microbes Infect 2018;7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anderson TK, et al. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med 2021;11:a038737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baratelli M, et al. Genetic characterization of influenza A viruses circulating in pigs and isolated in north-east Spain during the period 2006–2007. Res Vet Sci 2014;96:380–388. [DOI] [PubMed] [Google Scholar]
- 10. Barbezange C, et al. Seasonal genetic drift of human influenza A virus quasispecies revealed by deep sequencing. Front Microbiol 2018;9:2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Besser J, et al. Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 2018;24:335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhatta TR, et al. Infection dynamics of swine influenza virus in a Danish pig herd reveals recurrent infections with different variants of the H1N2 swine influenza A virus subtype. Viruses 2020;12:1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bolton MJ, et al. Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016. Influenza Other Respir Viruses 2019;13:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Breum S, et al. Influenza A virus with a human-like n2 gene is circulating in pigs. Genome Announc 2013;1:e00712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown IH, et al. Genetic characterization of an influenza A virus of unusual subtype (H1N7) isolated from pigs in England. Arch Virol 1997;142:1045–1050. [DOI] [PubMed] [Google Scholar]
- 16. Chastagner A, et al. Bidirectional human-swine transmission of seasonal influenza A(H1N1)pdm09 virus in pig herd, France, 2018. Emerg Infect Dis 2019;25:1940–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chauhan RP, Gordon ML. Deciphering transmission dynamics and spillover of avian influenza viruses from avian species to swine populations globally. Virus Genes 2021;57:541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chauhan RP, Gordon ML. A systematic review analyzing the prevalence and circulation of influenza viruses in swine population worldwide. Pathogens 2020;9:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiapponi C, et al. Temporal insight into the natural generation of a new reassortant porcine influenza virus in a swine holding. Vet Microbiol 2014;174:9–15. [DOI] [PubMed] [Google Scholar]
- 20. Clavijo A, et al. Identification and analysis of the first 2009 pandemic H1N1 influenza virus from U.S. feral swine. Zoonoses Public Health 2013;60:327–335. [DOI] [PubMed] [Google Scholar]
- 21. Cong YL, et al. Swine infection with H9N2 influenza viruses in China in 2004. Virus Genes 2008;36:461–469. [DOI] [PubMed] [Google Scholar]
- 22. Diaz A, et al. Complete genome sequencing of influenza A viruses within swine farrow-to-wean farms reveals the emergence, persistence, and subsidence of diverse viral genotypes. J Virol 2017;91:e00745-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Domingo E, et al. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis 1998;4:521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ducatez MF, et al. Influenza A(H1N1)pdm09 virus in pigs, Togo, 2013. Vet Microbiol 2015;177:201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ducatez MF, et al. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis 2011;17:1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Escalera-Zamudio M, et al. Characterization of an influenza A virus in Mexican swine that is related to the A/H1N1/2009 pandemic clade. Virology 2012;433:176–182. [DOI] [PubMed] [Google Scholar]
- 27. Fischer N, et al. Evaluation of unbiased next-generation sequencing of RNA (RNA-seq) as a diagnostic method in influenza virus-positive respiratory samples. J Clin Microbiol 2015;53:2238–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fitzpatrick AH, et al. High throughput sequencing for the detection and characterization of RNA viruses. Front Microbiol 2021;12:621719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gomaa MR, et al. Evidence of infection with avian, human, and swine influenza viruses in pigs in Cairo, Egypt. Arch Virol 2018;163:359–364. [DOI] [PubMed] [Google Scholar]
- 30. Goodwin S, et al. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res 2015;25:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goodwin S, et al. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grgić H, et al. Genetic characterization of H1N1 and H1N2 influenza A viruses circulating in Ontario pigs in 2012. PLoS One 2015;10:e0127840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grgić H, et al. Molecular characterization of H3N2 influenza A viruses isolated from Ontario swine in 2011 and 2012. Virol J 2014;11:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haach V, et al. Evaluation of two multiplex RT-PCR assays for detection and subtype differentiation of Brazilian swine influenza viruses. Braz J Microbiol 2020;51:1447–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harrington CT, et al. Fundamentals of pyrosequencing. Arch Pathol Lab Med 2013;137:1296–1303. [DOI] [PubMed] [Google Scholar]
- 36. He L, et al. Isolation and characterization of two H5N1 influenza viruses from swine in Jiangsu Province of China. Arch Virol 2013;158:2531–2541. [DOI] [PubMed] [Google Scholar]
- 37. Hoffmann E, et al. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 2001;146:2275–2289. [DOI] [PubMed] [Google Scholar]
- 38. Hu Y, et al. Complete genome sequence of a novel H4N1 influenza virus isolated from a pig in central China. J Virol 2012;86:13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jain M, et al. Improved data analysis for the MinION nanopore sequencer. Nat Methods 2015;12:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karasin AI, et al. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol 2000;74:9322–9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karasin AI, et al. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol 2004;42:4349–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Keller MW, et al. Direct RNA sequencing of the coding complete influenza A virus genome. Sci Rep 2018;8:14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. King J, et al. Rapid multiplex MinION nanopore sequencing workflow for influenza A viruses. BMC Infect Dis 2020;20:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krog JS, et al. Triple-reassortant influenza A virus with H3 of human seasonal origin, NA of swine origin, and internal A(H1N1) pandemic 2009 genes is established in Danish pigs. Influenza Other Respir Viruses 2017;11:298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kwon TY, et al. Genetic characterization of H7N2 influenza virus isolated from pigs. Vet Microbiol 2011;153:393–397. [DOI] [PubMed] [Google Scholar]
- 46. Kyriakis CS, et al. Molecular epidemiology of swine influenza A viruses in the Southeastern United States, highlights regional differences in circulating strains. Vet Microbiol 2017;211:174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lange J, et al. Reassortants of the pandemic (H1N1) 2009 virus and establishment of a novel porcine H1N2 influenza virus, lineage in Germany. Vet Microbiol 2013;167:345–356. [DOI] [PubMed] [Google Scholar]
- 48. Laver T, et al. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol Detect Quantif 2015;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee JH, et al. Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J Virol 2009;83:4205–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lewis NS, et al. Substitutions near the hemagglutinin receptor-binding site determine the antigenic evolution of influenza A H3N2 viruses in U.S. swine. J Virol 2014;88:4752–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis NS, et al. The global antigenic diversity of swine influenza A viruses. Elife 2016;5:e12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liang H, et al. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J Virol 2014;88:10864–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liberati A, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 2009;339:b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu L, et al. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012;2012:251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lu H, et al. Next-generation sequencing confirmation of real-time RT-PCR false positive influenza-A virus detection in waterfowl and swine swab samples. J Next Gener Seq Appl 2016;3:134. [Google Scholar]
- 56. Lu IN, et al. Applying next-generation sequencing to unravel the mutational landscape in viral quasispecies. Virus Res 2020;283:197963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Luciani F, et al. Next generation deep sequencing and vaccine design: today and tomorrow. Trends Biotechnol 2012;30:443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ma M-J, et al. Evidence for cross-species influenza A virus transmission within swine farms, China: a One Health, prospective cohort study. Clin Infect Dis 2018;66:533–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ma W, et al. Isolation and genetic characterization of new reassortant H3N1 swine influenza virus from pigs in the midwestern United States. J Virol 2006;80:5092–5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ma W, et al. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci U S A 2007;104:20949–20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mancera Gracia JC, et al. Effect of serial pig passages on the adaptation of an avian H9N2 influenza virus to swine. PLoS One 2017;12:e0175267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Martín-Valls GE, et al. Phylogeny of Spanish swine influenza viruses isolated from respiratory disease outbreaks and evolution of swine influenza virus within an endemically infected farm. Vet Microbiol 2014;170:266–277. [DOI] [PubMed] [Google Scholar]
- 63. Maya-Badillo BA, et al. Eco-epidemiological evidence of the transmission of avian and human influenza A viruses in wild pigs in Campeche, Mexico. Viruses 2020;12:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McBride DS, et al. Tracing the source of influenza A virus zoonoses in interconnected circuits of swine exhibitions. J Infect Dis 2021;224:458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mena I, et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 2016;5:e16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meseko C, et al. Evidence of exposure of domestic pigs to highly pathogenic avian influenza H5N1 in Nigeria. Sci Rep 2018;8:5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mine J, et al. Genetic and antigenic dynamics of influenza A viruses of swine on pig farms in Thailand. Arch Virol 2019;164:457–472. [DOI] [PubMed] [Google Scholar]
- 68. Mine J, et al. Genetic characterization of influenza A viruses in Japanese swine in 2015 to 2019. J Virol 2020;94:e02169-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moreno A, et al. First pandemic H1N1 outbreak from a pig farm in Italy. Open Virol J 2010;4:52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nagarajan K, et al. Influenza A H1N1 virus in Indian pigs & its genetic relatedness with pandemic human influenza A 2009 H1N1. Indian J Med Res 2010;132:160–167. [PubMed] [Google Scholar]
- 71. Nasamran C, et al. Persistence of pdm2009-H1N1 internal genes of swine influenza in pigs, Thailand. Sci Rep 2020;10:19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nelson MI, et al. The emergence and evolution of influenza A (H1α) viruses in swine in Canada and the United States. J Gen Virol 2017;98:2663–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nelson MI, et al. Influenza A viruses of human origin in swine, Brazil. Emerg Infect Dis 2015;21:1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nelson MI, et al. Human-origin influenza A(H3N2) reassortant viruses in swine, Southeast Mexico. Emerg Infect Dis 2019;25:691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ngo LT, et al. Isolation of novel triple-reassortant swine H3N2 influenza viruses possessing the hemagglutinin and neuraminidase genes of a seasonal influenza virus in Vietnam in 2010. Influenza Other Resp Viruses 2012;6:6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nidom CA, et al. Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis 2010;16:1515–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nirmala J, et al. Genetic diversity of influenza A viruses circulating in pigs between winter and summer in a Minnesota live animal market. Zoonoses Public Health 2020;67:243–250. [DOI] [PubMed] [Google Scholar]
- 78. Peiris JS, et al. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol 2001;75:9679–9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Pena L, et al. Modifications in the polymerase genes of a swine-like triple-reassortant influenza virus to generate live attenuated vaccines against 2009 pandemic H1N1 viruses. J Virol 2011;85:456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pippig J, et al. Influenza A viruses detected in swine in southern Germany after the H1N1 Pandemic in 2009. Zoonoses Public Health 2016;63:555–568. [DOI] [PubMed] [Google Scholar]
- 81. Radford AD, et al. Application of next-generation sequencing technologies in virology. J Gen Virol 2012;93:1853–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rajao DS, et al. Adaptation of human influenza viruses to swine. Front Vet Sci 2019;5:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rajão DS, et al. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol 2017;91:e01763-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rambo-Martin BL, et al. Influenza A virus field surveillance at a swine-human interface. mSphere 2020;5:e00822-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Richt JA, et al. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol 2006;80:11009–11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ronaghi M, et al. Pyrosequencing for discovery and analysis of DNA sequence variations. Pharmacogenomics 2007;8:1437–1441. [DOI] [PubMed] [Google Scholar]
- 87. Ryt-Hansen P, et al. Substantial antigenic drift in the Hemagglutinin protein of swine influenza A viruses. Viruses 2020;12:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Saavedra-Montañez M, et al. Identification and genomic characterization of influenza viruses with different origin in Mexican pigs. Transbound Emerg Dis 2019;66:186–194. [DOI] [PubMed] [Google Scholar]
- 89. Sánchez-Betancourt JI, et al. Complete genome sequence of a novel influenza A H1N2 virus circulating in swine from Central Bajio region, Mexico. Transbound Emerg Dis 2017;64:2083–2092. [DOI] [PubMed] [Google Scholar]
- 90. Sanger F, et al. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 1977;74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Schaefer R, et al. A human-like H1N2 influenza virus detected during an outbreak of acute respiratory disease in swine in Brazil. Arch Virol 2015;160:29–38. [DOI] [PubMed] [Google Scholar]
- 92. Schmidt C, et al. Full-genome sequence of a reassortant H1N2 influenza A virus isolated from pigs in Brazil. Genome Announc 2014;2:e01319-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Scholtissek C. Pigs as ‘mixing vessels’ for the creation of new pandemic influenza A viruses. Med Princ Pract 1990–91;2:65–71. [Google Scholar]
- 94. Shin JY, et al. Isolation and characterization of novel H3N1 swine influenza viruses from pigs with respiratory diseases in Korea. J Clin Microbiol 2006;44:3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shu LL, et al. Evidence for interspecies transmission and reassortment of influenza A viruses in pigs in southern China. Virology 1994;202:825–833. [DOI] [PubMed] [Google Scholar]
- 96. Smith DW, et al. Respiratory illness in a piggery associated with the first identified outbreak of swine influenza in Australia: assessing the risk to human health and zoonotic potential. Trop Med Infect Dis 2019;4:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Smith GJD, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009;459:1122–1125. [DOI] [PubMed] [Google Scholar]
- 98. Sobolev I, et al. Genome sequence of an unusual reassortant H1N1 swine influenza virus isolated from a pig in Russia, 2016. Genome Announc 2017;5:e00747-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sun H, et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci U S A 2020;117:17204–17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tapia R, et al. Antigenic characterization of novel H1 influenza A viruses in swine. Sci Rep 2020;10:4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Tinoco YO, et al. Transmission dynamics of pandemic influenza A(H1N1)pdm09 virus in humans and swine in backyard farms in Tumbes, Peru. Influenza Other Respir Viruses 2016;10:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tu J, et al. Isolation and molecular characterization of equine H3N8 influenza viruses from pigs in China. Arch Virol 2009;154:887–890. [DOI] [PubMed] [Google Scholar]
- 103. Van den Hoecke S, et al. Analysis of the genetic diversity of influenza A viruses using next-generation DNA sequencing. BMC Genomics 2015;16:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Vincent AL, et al. A brief introduction to influenza A virus in swine. Methods Mol Biol 2014;1161:243–258. [DOI] [PubMed] [Google Scholar]
- 105. Vincent AL, et al. Influenza A virus vaccines for swine. Vet Microbiol 2017;206:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang N, et al. Complete genome sequence of an H10N5 avian influenza virus isolated from pigs in central China. J Virol 2012;86:13865–13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. World Health Organization (WHO). Next-generation sequencing of influenza viruses. WHO, October 2019. https://www.who.int/influenza/gisrs_laboratory/national_influenza_centres/NGS_guidance_for_NICs.pdf [Google Scholar]
- 108. World Health Organization (WHO). Sequencing primers and protocol. WHO, 12 May 2009. https://www.who.int/csr/resources/publications/swineflu/GenomePrimers_20090512.pdf [Google Scholar]
- 109. Wong FYK, et al. Divergent human-origin influenza viruses detected in Australian swine populations. J Virol 2018;92:e00316-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wright KE, et al. Typing and subtyping of influenza viruses in clinical samples by PCR. J Clin Microbiol 1995;33:1180–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Young C, Argáez C. CADTH rapid response reports. In: Rapid Genome-Wide Testing: A Review of Clinical Utility, Cost-Effectiveness, and Guidelines. Ottawa, ON: Canadian Agency for Drugs and Technologies in Health, 2019. https://www.ncbi.nlm.nih.gov/books/NBK549546/ [PubMed] [Google Scholar]
- 112. Yu H, et al. Isolation and genetic characterization of avian origin H9N2 influenza viruses from pigs in China. Vet Microbiol 2008;131:82–92. [DOI] [PubMed] [Google Scholar]
- 113. Yu Z, et al. A novel reassortant influenza A (H1N1) virus infection in swine in Shandong Province, eastern China. Transbound Emerg Dis 2020;67:450–454. [DOI] [PubMed] [Google Scholar]
- 114. Zell R, et al. Cocirculation of swine H1N1 influenza A virus lineages in Germany. Viruses 2020;12:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zell R, et al. Displacement of the Gent/1999 human-like swine H1N2 influenza A virus lineage by novel H1N2 reassortants in Germany. Arch Virol 2020;165:55–67. [DOI] [PubMed] [Google Scholar]
- 116. Zell R, et al. Novel reassortant swine H3N2 influenza A viruses in Germany. Sci Rep 2020;10:14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhao G, et al. Isolation and phylogenetic analysis of pandemic H1N1/09 influenza virus from swine in Jiangsu province of China. Res Vet Sci 2012;93:125–132. [DOI] [PubMed] [Google Scholar]
- 118. Zhao X, et al. Characterization of an artificial swine-origin influenza virus with the same gene combination as H1N1/2009 virus: a genesis clue of pandemic strain. PLoS One 2011;6:e22091. [DOI] [PMC free article] [PubMed] [Google Scholar]