Abstract
Isogenic mutants derived from quinolone-susceptible isolate WT by introducing gyrA (S83L, D87G) and parC (S80I, E84K) mutations associated with quinolone resistance were characterized with respect to quinolone resistance, growth rate, and degree of global supercoiling. The latter was determined by use of a pair of reporter plasmids carrying supercoiling-dependent promoters pgyrA and ptopA, respectively, transcriptionally fused to the reporter gene bla coding for TEM-1 β-lactamase. The quotient (Qsc) of the β-lactamase specific activity determined for a mutant carrying either plasmid was taken as a measure of the degree of global supercoiling. These Qsc data were comparable to results obtained from the separation of topoisomers of plasmid pBR322 on chloroquine-containing agarose gels and indicate a reduced degree of negative supercoiling in resistant mutants relative to the parent, WT. The S83L mutation in gyrA had the strongest influence on quinolone resistance while leaving other parameters nearly unaffected. The gyrA double mutation (S83L plus D87G) had an effect on quinolone resistance similar to that of a single mutation. Phenotypic expression of the parC mutation (S80I) was dependent on the presence of at least one gyrA mutation. Expression of high-level fluoroquinolone resistance (ciprofloxacin MIC, >4 μg/ml) required a combination of the gyrA double mutation and one parC mutation (S80I or E84K). Such mutants showed considerable alterations of growth rate, global supercoiling, or both. Introduction of a parC mutation affected neither the doubling time nor the degree of supercoiling, while the presence of the gyrA D87G mutation was associated with a significant reduction in the degree of DNA supercoiling.
Since their introduction into clinical use in 1983, fluoroquinolones have played an essential role in the treatment of infectious diseases caused by enteric bacteria like Escherichia coli (25). The primary cellular target of fluoroquinolones in E. coli is a bacterial type II topoisomerase (DNA gyrase) consisting of two pairs of subunits, A and B (17). Gyrase is unique in catalyzing negative supercoiling of covalently closed circular double-stranded DNA in an ATP-consuming reaction and is therefore essential for maintenance of DNA topology. Recently, another type II topoisomerase, topoisomerase IV, which is responsible for decatenating the chromosomes before cell division, was identified as a secondary target of quinolones in E. coli (20, 33, 35). DNA gyrase and topoisomerase IV share extensive amino acid sequence homology, including highly conserved regions in both subunits A and B (31, 32).
In earlier reports, resistance to fluoroquinolones was rarely observed among clinical isolates of E. coli, and when it was observed, the isolates showed low-level resistance (3). However, in Germany, the prevalence of fluoroquinolone resistance among clinical isolates of E. coli increased from <1% to 5% between 1990 and 1995 (34).
Two basic mechanisms of resistance to fluoroquinolones have been identified: target alteration and reduced drug accumulation. A single mutation does not result in clinically relevant resistance (i.e., the MIC of ciprofloxacin [CIP] is <2 μg/ml) (20). Instead a combination of mutations is involved affecting the genes gyrA or gyrB (46, 62, 63) and parC or parE (6, 61), which code for subunits A and B of topoisomerases II and IV, respectively, and, as an example, the mar regulatory locus affecting both active drug efflux via the AcrAB-TolC complex (38, 59) and impaired access via reduced expression of outer membrane protein OmpF (7, 26).
Among clinical isolates, two types of mutants are predominantly found: low-level resistant isolates (CIP MIC, <2 μg/ml) most frequently carrying a single gyrA mutation altering serine 83 to leucine (S83L) (9, 15, 20, 55) and high-level resistant isolates (MIC, >4 μg/ml) carrying two gyrA mutations (S83L most commonly in combination with a mutation affecting aspartic acid 87 (D87) (22, 61) in addition to mutations affecting the analogous positions serine 80 (S80) and glutamic acid 84 (E84) in parC (20, 35, 61). Such isolates often show reduced drug accumulation (15, 21). However, the genetic basis of the latter mechanism remains obscure. Thus, while nothing is known about the sequence of the events following an initial gyrA mutation in clinical isolates (20) in mutants selected in vitro, mar-like mutations seem to occur as the second mutation preceding additional target mutations (23, 27, 52). Preliminary data indicate that highly resistant laboratory mutants have impaired viability, as demonstrated by significantly increased doubling times compared to that of the parent strain (WT) or a randomly chosen clinical isolate (205096) carrying similar gyrA and parC mutations (20, 23).
One possible explanation is that the accumulation of mutations in genes which code for essential enzymes involved in the control of DNA topology can affect the regulation of the degree of supercoiling and, thus, the expression of supercoiling-regulated genes in laboratory mutants. This might influence the growth rate by an unknown mechanism.
Therefore, this study aimed at (i) creating a set of isogenic mutants derived from isolate WT and carrying different combinations of known quinolone resistance mutations and (ii) investigating the impact of these mutations on the quinolone resistance, growth rate, and degree of negative supercoiling of the respective mutants.
MATERIALS AND METHODS
Bacterial strains.
Fluoroquinolone-resistant E. coli clinical isolate 205096 (22), quinolone-susceptible isolate WT, and its consecutive mutants MI, MII, MIII, MIVb, and R17 (selected in vitro) have been described previously (23).
Plasmids.
The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant markersa | Function(s)b | Source or reference |
|---|---|---|---|
| pBP507 | Mob+ Amir | Vector control | 22 |
| pBP517 | Mob+ AmirgyrAs | gyrA+ dominance test | 22 |
| pBP567 | Mob+ AmirparCs | parC+ dominance test | 20 |
| pMAK705 | Clmr Repts | Cloning | 19 |
| pMAK705gyrA3 | Clmr ReptsgyrA(S83L, D87G) | Mutagenesis | This study |
| pMAK705gyrA3.1 | Clmr ReptsgyrA(S83L) | Mutagenesis | This study |
| pMAK705gyrA3.2 | Clmr ReptsgyrA(D87G) | Mutagenesis | This study |
| pMAK705parC4 | Clmr ReptsparC(S80I) | Mutagenesis | This study |
| pBR322 | Ampr Tetr | Template for bla gene cloning, topoisomer distribution | 5 |
| pBR328 | Ampr Tetr | Control for Qsc determination | Boehringer |
| pBP523 | Mob+ Amir Ampr pgyrA-bla | Qsc determination | This study |
| pBP524 | Mob+ Amir Ampr ptopA-bla | Qsc determination | This study |
Relevant markers include determinants mediating resistance to amikacin (Amir), ampicillin (Ampr), chloramphenicol (Clmr), and tetracycline (Tetr); sequences required for mobilization transfer (Mob+) and thermosensitive replication (Repts); quinolone resistance mutations in genes gyrA and parC leading to amino acid exchanges of serine-83 to leucine and aspartate-87 to glycine (gyrAS83L and gyrD87G), as well as serine-80 to isoleucine (parCS80I), respectively; and reporter gene fusions of the bla gene coding for TEM-1 β-lactamase to promoters pgyrA and ptopA.
Qsc, measure of the relative degree of supercoiling (for a definition, see the text).
Oligonucleotides.
The oligonucleotides used in this study for amplification and sequence determination of fragments from genes gyrA and parC have been described previously (20, 22).
Antimicrobial agents.
Amikacin (Grünenthal, Aachen, Germany), nalidixic acid (Sterling-Winthrop, Guildford, United Kingdom), CIP, ampicillin (Bayer, Wuppertal, Germany), nitrocefin (Glaxo, Greenford, United Kingdom), and chloramphenicol (Bayer) were kindly supplied by the manufacturers. Novobiocin and tetracycline were obtained from Sigma (Deisenhofen, Germany).
Biochemicals.
All biochemicals, unless otherwise indicated, were from Boehringer (Mannheim, Germany). Taq DNA polymerase, restriction endonucleases, and buffers were purchased from Life Technologies (Eggenstein, Germany) and New England Biolabs (Schwalbach, Germany). DNA sequencing was performed by using the Silver Sequencing kit of Promega (Heidelberg, Germany) in accordance with the manufacturer’s instructions. Chloroquine was purchased from Sigma.
Chemicals and media.
All chemicals, unless otherwise stated, as well as standard broth no. I (NI agar and NI broth), were purchased from Merck (Darmstadt, Germany).
Mueller-Hinton broth was obtained from Difco (Detroit, Mich.). Luria-Bertani (LB) broth and agar were prepared by standard protocols as previously described (41).
Susceptibility testing.
MICs were determined by a broth microdilution method in accordance with the guidelines of the National Committee for Clinical Laboratory Standards (47) by using unsupplemented Mueller-Hinton broth. The drug susceptibilities of the plasmid-bearing strains used for detection of dominance were determined as single-cell MICs (24). Briefly, overnight cultures of a quinolone-resistant strain and the respective transconjugant grown in the presence of amikacin to maintain selective pressure against plasmid loss were serially diluted 10-fold. Three different inoculum sizes (10−7, 10−6, and 10−5) were spotted onto NI agar plates containing twofold serial dilutions of CIP to yield single CFU. By definition, the single-cell MIC was determined after 18 h of incubation as that concentration leading to a >10-fold reduction in the viable cell count compared with that of a drug-free control.
DNA isolation and transfer techniques.
Small amounts of plasmid DNA were prepared from transconjugant E. coli strains by alkaline lysis in accordance with a standard protocol (41). Large-scale preparation of plasmid DNA was performed either with the plasmid midi-kit (Quiagen) in accordance with the manufacturer’s guidelines or by cesium chloride buoyant density gradient centrifugation of cleared lysates (41).
Transformation of plasmid DNA was performed in accordance with standard protocols (41). Plasmids pBP507, pBP517, and pBP567, used for dominance tests, as well as pBP523 and pBP524, used for investigation of the degree of supercoiling, were transferred to the recipients by mobilization essentially as described previously (24), except for the use of E. coli K-12 strain S-17-1 (56) instead of C600SN(RP1H) as the donor.
DNA isolation for PCR.
Chromosomal DNA for the amplification of the promoter regions of gyrA and topA was isolated by resuspending three colonies in 100 μl of distilled water and incubating them for 15 min at 99°C. After brief centrifugation, 5 μl of the supernatant was used as the template for the subsequent PCR. Plasmid pBR322 was used as the source for the β-lactamase TEM-1 gene bla.
PCR and DNA sequencing.
Amplification of pgyrA and ptopA was performed with primers SB1 and SB2 and primers TOPB1 and TOPB2, respectively. Amplification of bla was performed with primers SB3 and SB4 for pBP523 and with TOPB3 and TOPB4 for pBP524. SB1 and TOPB1 contained restriction sites for EcoRI, whereas SB4 and TOPB4 contained restriction sites for XbaI. The primers used were SB1 (5′-GATTCAGGAACGAATT-3′), SB2 (5′-ACGGAAATGTTGAATACTCATCTAACCGCTAT-3′), SB3 (5′-GAGGGATAGCGGTTAGATGAGTATTCAACATTTC-3′), SB4 (5′-TAACTCTAGATCTGACGCTCAGTGGA-3′), TOPB1 (5′-GGCGAGCTCGAATTCGCGGTCGATGGGTTGTGT-3′), TOPB2 (5′-GAAATGTTGAATACTCATATTCACCTTACCTAATTT-3′), TOPB3 (5′-TTAGGTAAGGTGAATATGAGTATTCAACATTT-3′), and TOPB4 (5′-GCAGGTCGACTCTAGATCCTTTGATCTTT-3′). DNA fragments pgyrA, ptopA, and bla were amplified by using the following temperature profile: initial denaturation at 95°C for 5 min; 25 cycles of 40°C for 30 s, 72°C for x s, and 95°C for 30 s; and one final cycle of 40°C for 30 s and 72°C for 5 min (x = 30 for pgyrA, 60 for ptopA, and 90 for bla). The fragments were fused to a bla-containing DNA fragment by SOEing (splicing genes together by overlap extension) as previously described (43). The conditions for the fusion reactions were as follows: initial denaturation at 95°C for 5 min; 12 cycles of 72°C for 120 s and 95°C for 30 s; 20 cycles of 40°C for 30 s, 72°C for 90 s, and 95°C for 30 s; and one final cycle of 40°C for 30 s and 72°C for 5 min. This brought the bla gene under the control of the pgyrA (pBP523) and ptopA (pBP524) promoters, respectively.
For DNA cycle sequencing with the Silver Sequencing and Staining kit (Promega), 100 fmol of a PCR fragment was used in accordance with the manufacturer’s instructions.
Allelic exchange of chromosomal genes.
To introduce point mutations associated with quinolone resistance into the chromosomal gyrA and parC genes, the allelic exchange technique (19) was applied with the following modifications. Briefly, PCR fragments carrying the respective mutant allele flanked by suitable restriction sites were cloned into the polylinker region of plasmid pMAK705: gyrA genes were isolated as BamHI/XbaI fragments from plasmid pBP7614 (22) and inserted into the BamHI/XbaI sites of plasmid pMAK705. The parC gene of E. coli MIII was isolated from plasmid pBP567-4 (20) as an NsiI/SacI fragment and inserted into the compatible PstI/SacI sites of pMAK705. The presence of the respective mutant allele in recombinant plasmids was confirmed by DNA sequencing of the quinolone resistance-determining region (QRDR). For allelic exchange, the respective plasmid was introduced into the strain to be mutagenized by electroporation using a Gene Pulser (BioRad, Munich, Germany) by following the manufacturer’s instructions. About 30 colonies of recombinant cells growing at 30°C on LB agar containing chloramphenicol (30 μg/ml for JM83, WT, and MI or 60 μg/ml for MII) were used to inoculate 100 ml of LB broth.
Gel electrophoretic separation of topoisomers.
DNA of plasmid pBR322 was transferred to strain WT and different mutants by the CaCl2 transformation method, and the supercoiled topoisomers were isolated by cesium chloride buoyant density gradient centrifugation (41). The purified plasmid DNA was separated on 1.8% agarose gels in 1× TAE buffer (40 mM Tris-HCl [pH 8.3], 25 mM sodium acetate, 1 mM EDTA). Gels containing different concentrations of chloroquine (2, 5, and 10 μg/ml in 1% acetic acid) were run for 16 h at 2.5 V/cm. Three washing steps of 30 min each were performed in 10 mM MgSO4, in 1× TAE buffer, and in distilled water. Gels were stained with SYBR Green (Biozym, Hessisch Oldendorf, Germany) diluted 10,000-fold in TAE buffer for 15 min.
Determination of doubling time.
For determining the doubling time, freshly grown cells were diluted 1:1,000 in prewarmed LB broth and incubated at 37°C under agitation (250 rpm). Every 15 min, samples were taken for determining the viable cell count. Doubling times during log phase were determined from the linear part of a semilogarithmic plot of the number of CFU per milliliter against time.
Determination of β-lactamase specific activity.
Cells were grown to mid-log phase (optical density at 546 nm, 0.5). Twenty milliliters was harvested by centrifugation (8,000 × g), washed in 5 ml of ice-cold phosphate buffer (0.1 M, pH 7.0), and resuspended in 1 ml of phosphate buffer. Cells were lysed by two 10-s ultrasonification steps (Branson Sonifier B-12) with a 1-min cooling interval. After centrifugation (8,000 × g, 4°C), the supernatant was used for determination of β-lactamase specific activity using nitrocefin as a chromogenic substrate (48). Protein content was measured by the method of Lowry et al. (37).
RESULTS
Isolation and genotypic characterization of topoisomerase mutants.
A series of isogenic derivatives of isolate WT carrying various combinations of mutations in genes gyrA and parC were obtained either by introducing a point mutation via the allelic-exchange technique or by a one-step selection procedure. The resulting strains are included in Table 2. Since the genetic background of the source strain, WT, which is a randomly chosen quinolone-susceptible isolate, is not defined, E. coli K-12 strain JM83 was used as a control for some first- and second-step mutants (Table 2).
TABLE 2.
Results of genetic and phenotypic characterization of quinolone-resistant mutants
| E. coli strain | Mutation(s) in QRDR of:
|
MIC (μg/ml)b
|
Doubling time (min) | Change in doubling time (%)c | Qsc | ΔQscd (%) | ςe | ||
|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | CIP | CLM | ||||||
| K-12 JM83 | —g | — | 0.015 | 16 | 24 | 0 | 1.57 | 0 | 0.05 |
| JM83/3.1 | S83L | — | 1 | 16 | 32 | 33 | 1.23 | 22 | 0 |
| JM83/3.2 | D87G | — | 0.5 | 16 | 32 | 33 | 0.92 | 41 | 0.05 |
| JM83/3 | S83L, D87G | — | 2 | 16 | 49 | 104 | 1.29 | 18 | 0.01 |
| WT | — | — | 0.015 | 4 | 25 | 0 | 1.60 | 0 | 0.05 |
| MI | S83L | — | 0.5 | 4 | 26 | 4 | 1.37 | 14 | 0.02 |
| MIIa | S83L | — | 4 | 8 | 27 | 8 | 1.38 | 13 | 0.05 |
| WT-3.2 | D87G | — | 0.25 | 8 | 27 | 8 | 1.17 | 27 | 0.12 |
| WT-4 | — | S80I | 0.03 | NDh | ND | ND | 1.56 | 2 | 0.07 |
| WT-3 | S83L, D87G | — | 1 | 8 | 36 | 44 | 1.49 | 6 | 0.06 |
| MI-4 | S83L | S80I | 2 | ND | ND | ND | 1.33 | 17 | 0.05 |
| MII-3.2a | D87G | — | 2 | 16 | 29 | 16 | 1.13 | 29 | 0.03 |
| MII-3a | S83L, D87G | — | 8–16 | 16 | 75 | 200 | 0.50 | 32 | 0.02 |
| WT-3-4M4 | S83L, D87G | S80I | 32 | ND | ND | ND | 1.56 | 3 | 0.03 |
| WT-3-4M21 | S83L, D87G | E84K | 32 | ND | ND | ND | 1.62 | −2 | 0.00 |
| MIIIa | S83L, D87G | S80I | 64 | 32 | 35 | 40 | 1.00 | 36 | 0.06 |
| MIVba | S83L, D87G | S80I | 256 | 64 | 127 | 408 | 0.36 | 77 | 0.06 |
| R17 | S83L, D87G | S80I | 256 | 32 | 37 | 48 | 0.44 | 72 | 0.05 |
| 205096f | S83L, D87G | E84K | 64 | 32 | 21 | −16 | 1.47 | 6 | 0.06 |
MII and its derivatives carry a mutation reducing CIP accumulation (23).
Susceptibilities were determined as MICs of CIP and chloramphenicol (CLM) by broth microdilution assay.
The change in the doubling time of a mutant compared to that of its parent (strain WT or JM83) is given.
Differences in Qsc values were calculated in comparison to the respective isogenic parent (ΔQsc = 0).
ς, standard deviation.
The data determined for isolate 205096 are calculated in relation to those for strain WT (100%).
—, no mutation detected.
ND, not determined.
Quinolone resistance phenotype of mutants.
Independently of the genetic background in WT, MII, or JM83, a single gyrA S83L mutation (MI, MII, or JM83-3.1, respectively) conferred a higher level of quinolone resistance (by one serial dilution step) than a single gyrA D87G mutation (WT-3.2, MII-3.2, or JM83-3.2). Introducing a gyrA double mutation (S83L and D87G) into WT and JM83 (yielding WT-3 and JM83-3, respectively) resulted in an increase in the CIP MIC of one serial dilution step and an increase of two dilution steps for MII-3 (MII genetic background) compared to S83L (MI, JM83-3.1; Table 2). In contrast, a single parC mutation (S80I) in strain WT did not affect the quinolone susceptibilities of the resulting derivative, WT-4, while it significantly affected those of the gyrA double mutant WT-3 (WT-3-4M4), as did the parC mutation E84K (WT-3-4M21) (Table 2). Again, the combination of three mutations (gyrA S83L, D87G, and parC S80I) was associated with higher quinolone MICs (by one serial dilution step) in the background of MII (MIII) compared to WT-3-4M4. MIII resembles clinical isolate 205096 by carrying a similar combination of topoisomerase mutations and showing an identical MIC of CIP (64 μg/ml).
Reductions in the MICs of CIP and nalidixic acid after transfer into the different mutants of plasmid-coded alleles of gyrA+ (pBP517) or parC+ (pBP567), respectively, indicated that both types of mutations contribute to the expression of quinolone resistance (data not shown).
Doubling times of topoisomerase mutants.
E. coli K-12 JM83-3.1, carrying the single S83L mutation in gyrA, as well as JM83-3.2 (D87G), showed a doubling time increase of 33%. The doubling times of respective mutants MI and WT3.2 derived from strain WT were less affected by a single mutation in gyrA (4 to 8%). For all of the mutants investigated which carry the double mutation in gyrA—including consecutive mutant MIII—the doubling time is increased by at least 40%. In contrast, the doubling time of clinical isolate 205096, with mutations in gyrA and parC identical to those in mutant MIII, is even slightly reduced compared with that of strain WT (21 versus 25 min; Table 2).
Relative degrees of DNA supercoiling.
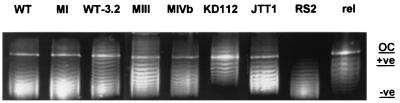
Supercoiling alterations were investigated by isolating plasmid pBR322 from the different strains and separating topoisomers on chloroquine-containing agarose gels. Depending on the concentration of the intercalating dye, the agarose concentration, and the plasmid used, the electrophoretic mobilities of the different topoisomers vary. The relative position of the topoisomer with the average linking number gives an estimate of the mean degree of supercoiling of the DNA. The results are shown in Fig. 1. At the chloroquine concentration used in the assays (5 μg/ml), more relaxed topoisomers migrated as positive supercoils while negatively supercoiled forms migrated as negative supercoils. In accordance, compared to the mobility of the mean topoisomer band(s) of reference strain JTT1, that of strain RS2 (increased negative supercoiling) is higher while that of strain KD112 (reduced negative supercoiling) is lower, irrespective of the chloroquine concentration used (2 to 10 μg/ml; Fig. 1, lanes 6 to 8, and data not shown). Comparably, in the presence of 5-μg/ml chloroquine, strains WT and MI show similar topoisomer distribution patterns, while those of mutants WT-3.2, MIII, and MIVb are shifted to lower mobilities (Fig. 1). The slight difference between WT and MIVb observed at a chloroquine concentration of 5 μg/ml is increased in the presence of chloroquine at 2 and 10 μg/ml (data not shown).
FIG. 1.
Electrophoretic separation of plasmid pBR322. Supercoiled DNA of plasmid pBR322 was isolated from different E. coli strains as described in Materials and Methods. Topoisomers were separated in a 1.8% agarose gel containing chloroquine at 5 μg/ml in TAE buffer. Samples were run at 2.5 V/cm for 16 h. Under these conditions relaxed control DNA of pBR322 (rel), which was obtained by treatment of supercoiled DNA with calf thymus topoisomerase I (Gibco-BRL) in accordance with the manufacturer’s recommendations, runs as positively supercoiled (+ve) while negatively supercoiled topoisomers still run as negatively supercoiled (−ve). OC, open circular DNA.
As an alternative approach, the expression of the reporter gene bla coding for TEM-1 β-lactamase was transcriptionally fused to the pgyrA and ptopA promoters, and cloned into plasmid pBP507 to yield plasmids pBP523 (pgyrA-bla) and pBP524 (ptopA-bla), respectively. These plasmids were introduced separately into bacterial strains by transformation for determining the expression of β-lactamase. The quotient Qsc was calculated by dividing the β-lactamase specific activity of cells carrying plasmid pBP524 by that of cells carrying plasmid pBP523. Qsc was determined in at least three separate experiments for each strain and was taken as a measure of the relative degree of DNA supercoiling.
To verify that the reporter gene assay responds to alterations in the relative degree of supercoiling, β-lactamase specific activities of cells containing either reporter plasmid in comparison to that of cell carrying pBR328 containing the native TEM-1 promoter were determined in the presence of increasing concentrations of novobiocin. Novobiocin, a competitive inhibitor of ATP binding to gyrase subunit B, is known to relax supercoiled DNA. Table 3 summarizes the results. Irrespective of the novobiocin concentration used (0 to 75 μg/ml), the expression of β-lactamase from plasmid pBP523 was always higher and that of plasmid pBP524 was always lower than the intermediate expression level from plasmid pBR328. As shown, decreasing Qsc values reflected the relaxation of DNA by novobiocin.
TABLE 3.
β-Lactamase specific activities and Qscs of novobiocin-treated strain WT, reference strain JTT1, and its topoisomerase mutants
| Strain | Reference | Δsc (%)a | Novobiocin concn (μg/ml)b | β-Lactamase sp act (U/mg)c for strain carrying plasmid:
|
Qsc | ΔQsc (%)d | ||
|---|---|---|---|---|---|---|---|---|
| pBR328 | pBP523 | pBP524 | ||||||
| WT | 23 | 10.8 | 7.5 | 12.1 | 1.61 | 0 | ||
| WT | 25 | 15.1 | 13.6 | 13.8 | 1.01 | −37 | ||
| WT | 50 | 21.3 | 26.1 | 19.6 | 0.75 | −53 | ||
| WT | 75 | 24.6 | 29.4 | 13.2 | 0.45 | −72 | ||
| JTT1 | 58 | 0 | NDe | 4.1 | 7.7 | 1.87 | 0 | |
| KD112 | 57 | −17 | ND | 5.8 | 8.3 | 1.44 | −23 | |
| RS 2 | 58 | +8 | ND | 1.2 | 6.8 | 5.54 | +296 | |
The differences in the degree of supercoiling (Δ sc) relative to that of the wild-type parent (JTT1; Δsc = 0) are from reference 58. A negative value indicates a reduction in the degree of negative supercoiling, and a positive value indicates an increase.
Cells were grown in the presence of subinhibitory concentrations of novobiocin as indicated.
The bla gene was transcriptionally fused to promoter pbla (plasmid pBR328), pgyrA (plasmid pBP523), or ptopA (plasmid pBP524).
Differences in Qsc values (ΔQsc) were calculated in comparison to the respective isogenic parent (ΔQsc = 0).
ND, not determined.
Furthermore, the functionality of the reporter gene assay was verified by determining the Qscs of reference strain E. coli K-12 JTT1, showing a normal relative degree of supercoiling, and its derivatives RS2 (increased negative supercoiling due to a topA deletion) and KD112 (reduced negative supercoiling due to the gyrB226 mutation) (58). The different Qscs reflected the increased degree of supercoiling of RS2, as well as the reduced degree of supercoiling of KD112 (Table 3).
The results of the Qsc determination for the various mutants are summarized in Table 2. The introduction of the mutation S83L into gyrA of strain WT had no detectable impact on the distribution of DNA topoisomers (Fig. 1) and caused a slight (14%) decrease in Qsc. For JM83-3.1, the Qsc value decreased by 22% compared to that of its parent, JM83. Introduction of the mutation D87G into gyrA resulted in greater alterations of the Qsc values compared to that of the respective parent strain (41% for JM83-3.2 and 27% for WT-3.2). However, mutant MII carrying the S83L gyrA mutation in a mar background shows nearly the same Qsc value as the immediate parent MI, while the Qsc of mutant MII-3.2 (D87G mutation in the mar background) is decreased by 18% in relation to mutant MII (Table 2).
Compared to strains carrying a single S83L mutation in gyrA (MI, MII, and MI-4), the gyrA double mutation (S83L and D87G) in mutant WT-3 yielded a similar decrease in the Qsc value (6% versus 14, 13, and 17%). A remarkable decrease in the Qsc of 32% compared to that of WT was detected for mutant MII-3 carrying the gyrA double mutation in the mar background (Table 2). Similarly, decreases in Qsc values of 36 to 77% were detected in gyrA double mutants MIII, MIVb, and R17 selected in vitro.
The S80I mutation of parC had no influence on the degree of supercoiling, whether introduced alone (WT-4) or in combination with S83L (MI-4). In agreement with this finding, the Qscs of mutants WT-3-4M4 and WT-3-4M21, which carry the S80I and E84K mutations in parC, respectively, show Qsc values comparable to that of parent strain WT-3.
DISCUSSION
According to the currently accepted alternating-target model, high-level fluoroquinolone resistance in E. coli develops by stepwise acquisition of target mutations (20, 33, 35). This idea is supported by data showing that the two isolated target enzymes, DNA gyrase and topoisomerase IV, are differently sensitive to quinolones (28). However, clinical isolates which carry single mutations in gyrA and parC, like E. coli 3204917 (20), and which are intermediately resistant to CIP are found with a very low prevalence (15). This raises the question of whether the development of high-level fluoroquinolone resistance, besides known resistance mutations, requires the acquisition of additional stabilizing mutations that do not necessarily confer a higher level of resistance. Since the genetic background of clinical isolates is generally poorly defined, in the present study, mutants carrying defined quinolone resistance mutations in the identical genetic background of quinolone-susceptible isolate WT were generated. These mutants were investigated for their levels of quinolone resistance, their relative degrees of DNA supercoiling, and their growth rates in order to obtain a set of comparable data.
Mutants MI, MII, and MIII, which had been preferentially selected in vitro, show a higher level of quinolone resistance than in vitro-generated mutants belonging to the same fictive selection step. Nevertheless, the latter mutants are viable even in the absence of a stabilizing mutation(s) but have reduced quinolone susceptibilities and, in most cases, bear other disadvantages, like an increased doubling time, a decreased degree of supercoiling, or both. Thus, during the selection of quinolone-resistant mutants in vitro, the level of quinolone resistance conferred by a respective mutation seems to be the major factor determining the sequence of mutations. Besides these general findings, a certain mutation with an observed phenotype is not generally obvious. (i) Mutant MI not only shows a higher MIC of CIP than WT-4 and WT-3.2 but also is slightly less affected in its growth rate than WT-3.2, which carries a D87G mutation frequently found secondary to an S83L gyrA mutation (Table 2) (15, 20, 35, 36, 61). Thus, the data presented here provide a plausible explanation for the high prevalence of the gyrA S83L mutation in clinical isolates. The finding that a parC mutation alone does not alter the quinolone susceptibility of a gyrA+ strain (WT-4; Table 2) supports the view that gyrase is the primary target of CIP in E. coli.
(ii) Mutant MII carries a mar mutation, reducing drug accumulation (30), in addition to the gyrA S83L mutation. All other in vitro mutants carrying two mutations (WT-3, MII-3.2, and MI-4) are less resistant and more affected in growth rate, supercoiling, or both (Table 2). The finding that the increase in the MIC of CIP for gyrA double mutant WT-3 is only one serial dilution step compared to gyrA single mutant MI and, thus, is less than that for MI-4 (gyrA S83L, parC S80I) provides direct evidence that a gyrA double mutation is not sufficient to confer high-level fluoroquinolone resistance. However, in practice, a mar mutation and not, as postulated, a parC mutation has been identified secondary to a gyrA mutation in the course of in vitro selection.
(iii) In the third selection step, mutant MIII was obtained, which is the first high-level fluoroquinolone-resistant mutant in this series of consecutive mutants selected in vitro. Curiously, this mutant had acquired two mutations (gyrA D87G and parC S80I) in a single selection step (20, 23). Moreover, despite the simultaneous acquisition of an increased doubling time and a reduced relative degree of supercoiling, this statistically unlikely event has been reproducibly confirmed by two independent in vitro selection experiments using unrelated susceptible isolates as parents (2). Using mutant WT-3 instead of MII, high-level fluoroquinolone-resistant mutants can be obtained easily with a natural mutation frequency of about 2 × 10−8. Two resulting mutants from a single experiment, WT-3-4M4 and WT-3-4M21, have acquired one additional parC mutation (S80I and E84K, respectively) and show a degree of global supercoiling comparable to that of their immediate parent, WT-3, and nearly identical to that of the progenitor strain, WT (Table 2). Mutant MII-3, which is a possible candidate for an intermediate mutant in the selection from MII to MIII, shows slightly increased MICs of CIP in comparison with MII. However, the CIP MIC remains below that for MIII. Additionally, the increased growth rate of MII-3 and its reduced relative degree of supercoiling indicate reduced viability.
Taken together, the data provide direct evidence that three target mutations—two in gyrA and one in parC—are the minimal requirements for high-level fluoroquinolone resistance in E. coli, i.e., a MIC of CIP of >8 μg/ml. This is in agreement with the current view that both targets, DNA gyrase and topoisomerase IV, are involved in the expression of fluoroquinolone resistance. However, considering the sequence of events following an initial gyrA S83L mutation, results obtained from laboratory mutants are different from observations made with clinical isolates. While a mar mutation seems to be an important early step in vitro (8, 23, 27, 52), the prevalence of this mutation is less than 15% among fluoroquinolone-resistant clinical isolates of E. coli, as a recent study revealed (40). Thus, mar mutations do not seem to be the predominant mutations affecting quinolone accumulation in clinical isolates (15, 40). Nevertheless, reduced quinolone accumulation has been detected in several resistant clinical isolates (15, 21, 49), but the genetic basis remains obscure.
Since the primary target, DNA gyrase, is an essential enzyme involved in the regulation of global DNA supercoiling, it is tempting to speculate that the accumulation of mutations in the QRDR, a region highly conserved even among distantly related species, affects not only the intrinsic activities of quinolones but also the enzymatic activity of gyrase.
The intracellular level of DNA supercoiling is maintained by the opposing enzymatic activities of DNA gyrase (negative supercoiling) and topoisomerase I (relaxing), and by homeostatic control of the corresponding genes, gyrA/gyrB and topA, respectively (17, 44).
A standard technique for determination of the relative degree of supercoiling of cellular DNA is gel electrophoretic separation of reporter plasmids on agarose gel containing an intercalating dye like ethidium bromide or chloroquine. These compounds alter the pitch of the DNA double helix and thereby reduce the superhelical tension of a covalently closed circular DNA double strand. For the discrimination of positive and negative supercoiling, two-dimensional separation of topoisomers can be used (for a summary, see reference 14).
As an alternative approach, supercoiling-affected promoters instead of supercoiling-dependent structural alterations have successfully been used to determine supercoiling-dependent gene expression. The results were consistent for supercoiling-regulated promoters pgyrA, pproU, and plac, irrespective of their location on the bacterial chromosome, and indicated a homogeneous level of DNA supercoiling of the different chromosomal supercoiling domains (45, 51).
We have developed a similar system for determination of the supercoiling-dependent expression of the bla gene coding for TEM-1 β-lactamase in E. coli wild-type strains and their fluoroquinolone-resistant derivatives by using promoters pgyrA and ptopA, respectively. These promoters respond reciprocally to alterations of the degree of supercoiling: pgyrA activity is increased over that of ptopA at a low degree of negative supercoiling, while ptopA activity is increased over that of pgyrA at a high degree of supercoiling (44). At the transcriptional level, this contributes to homeostatic control, i.e., the maintenance of a constant degree of chromosomal DNA supercoiling. In the present study, to sense alterations of the level of supercoiling, two broad-host-range plasmids have been developed which carry the TEM-1 β-lactamase gene bla as a reporter gene transcriptionally fused to promoters ptopA and pgyrA, respectively. Thus, alterations of the level of global supercoiling affect β-lactamase expression differently with the different promoters and can be expressed as the Qsc quotients of the respective β-lactamase activities (Tables 2 and 3).
The functionality of this method for the determination of the relative degree of supercoiling is demonstrated in several ways. (i) Compared to the parent strain, E. coli K-12 JTT1, topA mutant RS2, known to have a greater degree of negative supercoiling, yields higher Qsc values, while gyrB mutant KD112 yields a lesser degree of negative supercoiling (Table 3). (ii) Selective inhibition of gyrase by quinolones or coumarins, which results in the relaxation of DNA (53), yields Qsc values significantly lower than that of the untreated control (Table 3 and data not shown) (18, 42, 54). (iii) Repeated determinations with different strains yielded reproducible results with standard deviations of <10% (Table 2). (iv) Parent strain WT and some selected mutants showed qualitatively comparable results in assays of both Qsc and topoisomer distribution, indicating a reduction in the relative degree of negative supercoiling. However, no quantitative correlations between the two methods were detectable.
This was not unexpected, since there is not necessarily a linear correlation between alteration of the activity of a promoter and the distribution of topoisomers. In addition, the impact of an alteration of the degree of supercoiling on various promoters is different. Nevertheless, the data presented in this study provide evidence of the applicability of an approach using reporter plasmids to qualitatively sense alterations in the degree of supercoiling. This view is supported by data presented by others (45, 51).
In contrast to earlier studies showing that gyrB mutations which confer resistance to coumarin lead to different levels of reduction in gyrase activity while gyrA mutations conferring quinolone resistance usually do not affect enzyme activity (1), in the present study, different gyrA mutations showed graded effects on DNA supercoiling. This might be due to a higher sensitivity of the enzymatic reporter gene assay used. A single gyrA mutation S83L (in mutants JM83-3.1 and MI), alone or in combination with a second mutation (WT-3, MI-4, or MII), results in a less dramatic reduction in the degree of DNA supercoiling than the gyrA mutation D87G, either alone (WT-3.2 or JM83-3.2) or in combination with a mar mutation (MII-3.2): the Qsc values are reduced by up to 30% (Table 2). This is indicative of topologically underwound DNA in these mutants (Fig. 1). Whereas the D87G mutation had the strongest influence on global supercoiling, its impact on quinolone resistance was lower than that of the S83L mutation (Table 2). These findings may provide a reasonable explanation for the high prevalence in clinical isolates of mutation S83L as the first gyrA mutation instead of D87G.
As the transcription of many genes is known to be dependent on the degree of supercoiling, bacteria with alterations in DNA superhelicity show different patterns of protein expression, even if the alterations are small (57). The increases in the doubling times of some mutants may be due to changes in DNA supercoiling. In vitro mutants MIII to MIVb and R17, showing high-level resistance to fluoroquinolones, reveal considerable decreases in Qsc (36 to 77%; Table 2). In addition, the doubling time is increased at least by 40%. In contrast, mutants WT-3.2 and MII-3.2, obtained by in vitro mutagenesis, have Qsc reductions between 27 and 29%, while the doubling times are only slightly increased (8 to 16%). Moreover, mutant WT-3, with an increase in doubling time of 44%, reveals only a minimal reduction in the degree of supercoiling of 6%. These data indicate that for a given mutant, alterations in doubling time and Qsc are not necessarily interrelated. A reasonable explanation could be that the process of gene transcription, like other gyrase-involving reactions, e.g., recombination, might be more sensitive to local changes in the degree of supercoiling than to global alterations (12).
The finding that changes in the degree of supercoiling occur in vitro during the development of quinolone resistance makes it tempting to speculate that they even play a role in vivo. The alterations in the degree of supercoiling, globally as well as locally, may occur transiently and may be compensated for by additional mutations that restore the viability of the cell. Mutations which restore the degree of supercoiling to a level like that of the wild type have been detected in genes coding for type I and II topoisomerases (1, 10, 11, 50), as well as in genes coding for histone-like DNA-binding protein H-NS or HU (13, 29). HU is thought to play a physiological role in chromosomal DNA topology, probably by facilitating the action of gyrase (39). Mutations in genes coding for proteins that are involved in the process of transcription, like integration host factor (IHF), could compensate for mutations in gyrA that affect local disturbance during gene expression. Due to the involvement of IHF in affecting the activity of a large number of operons in E. coli, mutations in IHF could render transcriptional regulation to levels like that of the wild type (16). Under in vivo conditions, i.e., at the site of an infection, variations in the global gene expression pattern might also cause alterations in the response to environmental stimuli and, thus, affect the type of mutant preferentially selected. As a consequence, mutations selected in vitro would differ from those selected in vivo, as has been observed in the present study.
Further analysis of the in vitro-selected mutants, as well as clinical isolates, is necessary to elucidate the interrelationship among quinolone resistance, DNA supercoiling, and compensatory mutations.
ACKNOWLEDGMENTS
The expert technical assistance of Daniela Olsoczki is gratefully acknowledged.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to P.H. (He1864/1-3). V.H. was supported by the Pinguin Stiftung, Düsseldorf, Germany.
REFERENCES
- 1.Aleixandre V, Urios A, Herrera G, Blanco M. New Escherichia coli gyrA and gyrB mutations which have a graded effect on DNA supercoiling. Mol Gen Genet. 1989;219:306–312. doi: 10.1007/BF00261192. [DOI] [PubMed] [Google Scholar]
- 2.Bagel, S. 1998. Personal communication.
- 3.Barry A L, Fuchs P C, Pfaller M A, Allen S D, Gerlach E H. Prevalence of fluoroquinolone resistant bacterial isolates in four medical centers during the first quarter of 1990. Eur J Clin Microbiol Infect Dis. 1990;9:906–908. doi: 10.1007/BF01967511. [DOI] [PubMed] [Google Scholar]
- 4.Blanche F, Cameron B, Bernard F X, Maton L, Manse B, Ferrero L, Ratet N, Lecoq C, Goniot A, Bisch D, Crouzet J. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob Agents Chemother. 1996;40:2714–2720. doi: 10.1128/aac.40.12.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 6.Breines D M, Ouabdesselam S, Ng E Y, Tankovic J, Shah S, Soussy C J, Hooper D C. Quinolone resistance locus nfxD of Escherichia coli is a mutant allele of the parE gene encoding a subunit of topoisomerase IV. Antimicrob Agents Chemother. 1997;41:175–179. doi: 10.1128/aac.41.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen S L, McMurry L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen S P, Hooper D C, Wolfson J S, Souza K S, McMurry L M, Levy S B. Endogenous active efflux of norfloxacin in susceptible Escherichia coli. Antimicrob Agents Chemother. 1988;32:1187–1191. doi: 10.1128/aac.32.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen M E, Wyke A W, Kuroda R, Fisher L M. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother. 1989;33:886–894. doi: 10.1128/aac.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 11.Dorman C J, Lynch A S, Bhriain N N, Higgins C F. DNA supercoiling in Escherichia coli: topA mutations can be suppressed by DNA amplifications involving the tolC locus. Mol Microbiol. 1989;3:531–540. doi: 10.1111/j.1365-2958.1989.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 12.Dove S L, Dorman C J. The site-specific recombination system regulating expression of the type 1 fimbrial subunit gene of Escherichia coli is sensitive to changes in DNA supercoiling. Mol Microbiol. 1994;14:975–988. doi: 10.1111/j.1365-2958.1994.tb01332.x. [DOI] [PubMed] [Google Scholar]
- 13.Dri A M, Moreau P L, Rouviere-Yaniv J. Role of the histone-like proteins OsmZ and HU in homologous recombination. Gene. 1992;120:11–16. doi: 10.1016/0378-1119(92)90003-8. [DOI] [PubMed] [Google Scholar]
- 14.Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 15.Everett M J, Jin Y F, Ricci V, Piddock L J. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:2380–2386. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freundlich M, Ramani N, Mathew E, Sirko A, Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol Microbiol. 1992;6:2557–2563. doi: 10.1111/j.1365-2958.1992.tb01432.x. [DOI] [PubMed] [Google Scholar]
- 17.Gellert M, Mizuuchi K, O’Dea M H, Nash H A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gellert M, O’Dea M H, Itoh T, Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc Natl Acad Sci USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heisig P. Mechanisms and epidemiology of fluoroquinolone resistance in Enterobacteriaceae. In: Hakenbeck R, editor. Biospektrum special edition 1997: antibiotic resistance—recent development and future perspectives. Heidelberg, Germany: Spektrum Akademischer Verlag; 1997. pp. 42–46. [Google Scholar]
- 22.Heisig P, Schedletzky H, Falkenstein-Paul H. Mutations in the gyrA gene of a highly fluoroquinolone-resistant clinical isolate of Escherichia coli. Antimicrob Agents Chemother. 1993;37:696–701. doi: 10.1128/aac.37.4.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother. 1994;38:1284–1291. doi: 10.1128/aac.38.6.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heisig P, Wiedemann B. Use of a broad-host-range gyrA plasmid for genetic characterization of fluoroquinolone-resistant gram-negative bacteria. Antimicrob Agents Chemother. 1991;35:2031–2036. doi: 10.1128/aac.35.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooper D C, Wolfson J S. Mechanisms of bacterial resistance to quinolones. Washington, D.C: American Society for Microbiology; 1992. pp. 97–118. [Google Scholar]
- 26.Hooper D C, Wolfson J S, Bozza M A, Ng E Y. Genetics and regulation of outer membrane protein expression by quinolone resistance loci nfxB, nfxC, and cfxB. Antimicrob Agents Chemother. 1992;36:1151–1154. doi: 10.1128/aac.36.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooper D C, Wolfson J S, Ng E Y, Swartz M N. Mechanisms of action of and resistance to ciprofloxacin. Am J Med. 1987;82:12–20. [PubMed] [Google Scholar]
- 28.Hoshino K, Kitamura A, Morrissey I, Sato K, Kato J-I, Ikeda H. Comparison of inhibition of Escherichia coli topoisomerase IV by quinolones with DNA gyrase inhibition. Antimicrob Agents Chemother. 1994;38:2623–2627. doi: 10.1128/aac.38.11.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulton C S, Seirafi A, Hinton J C, Sidebotham J M, Waddell L, Pavitt G D, Owen-Hughes T, Spassky A, Buc H, Higgins C F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 30.Hüllen V, Heisig P, Wiedemann B. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Role of marR mutations in the development of clinical resistance of Escherichia coli against fluoroquinolones, abstr. C-64; p. 57. [Google Scholar]
- 31.Kato J-I, Nishimura Y, Imamura Y, Niki H, Hiraga S, Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 32.Kato J-I, Nishimura Y, Yamada M, Suzuki H, Hirota Y. Gene organization in the region containing a new gene involved in chromosome partition in Escherichia coli. J Bacteriol. 1988;170:3967–3977. doi: 10.1128/jb.170.9.3967-3977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kresken M, Hafner D. Prävalenz der Antibiotikaresistenz bei klinisch wichtigen Infektionserregern in Mitteleuropa. Chemother J. 1996;5:225–230. [Google Scholar]
- 35.Kumagai Y, Kato J I, Hoshino K, Akasaka T, Sato K, Ikeda H. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob Agents Chemother. 1996;40:710–714. doi: 10.1128/aac.40.3.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehn N, Stower-Hoffmann J, Kott T, Strassner C, Wagner H, Kronke M, Schneider-Brachert W. Characterization of clinical isolates of Escherichia coli showing high levels of fluoroquinolone resistance. J Clin Microbiol. 1996;34:597–602. doi: 10.1128/jcm.34.3.597-602.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 38.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 39.Malik M, Bensaid A, Rouviere-Yaniv J, Drlica K. Histone-like protein HU and bacterial DNA topology: suppression of an HU deficiency by gyrase mutations. J Mol Biol. 1996;256:66–76. doi: 10.1006/jmbi.1996.0068. [DOI] [PubMed] [Google Scholar]
- 40.Maneewannakul K, Levy S B. Identification of mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:1695–1698. doi: 10.1128/aac.40.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 42.Maxwell A. The interaction between coumarin drugs and DNA gyrase. Mol Microbiol. 1993;9:681–686. doi: 10.1111/j.1365-2958.1993.tb01728.x. [DOI] [PubMed] [Google Scholar]
- 43.McArn Horton R, Pease L R. SOEing. In: McPherson M J, editor. Directed mutagenesis. Oxford, England: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 44.Menzel R, Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983;34:105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- 45.Miller W G, Simons R W. Chromosomal supercoiling in Escherichia coli. Mol Microbiol. 1993;10:675–684. doi: 10.1111/j.1365-2958.1993.tb00939.x. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura S, Nakamura M, Kojima T, Yoshida H. gyrA and gyrB mutations in quinolone-resistant strains of Escherichia coli. Antimicrob Agents Chemother. 1989;33:254–255. doi: 10.1128/aac.33.2.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-T. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1996. [Google Scholar]
- 48.O’Callaghan C H, Muggleton P W, Ross G W. Effects of β-lactamase from gram-negative organisms on cephalosporins and penicillins. 1968. pp. 57–63. . Antimicrob. Agents Chemother. 1967. [PubMed] [Google Scholar]
- 49.Oethinger M, Conrad S, Kaifel K, Cometta A, Bille J, Klotz G, Glauser M P, Marre R, Kern W V. Molecular epidemiology of fluoroquinolone-resistant Escherichia coli bloodstream isolates from patients admitted to European cancer centers. Antimicrob Agents Chemother. 1996;40:387–392. doi: 10.1128/aac.40.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oram M, Fisher L M. An Escherichia coli DNA topoisomerase I mutant has a compensatory mutation that alters two residues between functional domains of the DNA gyrase A protein. J Bacteriol. 1992;174:4175–4178. doi: 10.1128/jb.174.12.4175-4178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavitt G D, Higgins C F. Chromosomal domains of supercoiling in Salmonella typhimurium. Mol Microbiol. 1993;10:685–696. doi: 10.1111/j.1365-2958.1993.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 52.Piddock L J V, Hall M C, Walters R N. Phenotypic characterization of quinolone-resistant mutants of Enterobacteriaceae selected from wild type, gyrA type and multiply-resistant (marA) type strain. J Antimicrob Chemother. 1991;28:185–198. doi: 10.1093/jac/28.2.185. [DOI] [PubMed] [Google Scholar]
- 53.Pruss G J, Franco R J, Chevalier S G, Manes S H, Drlica K. Effects of DNA gyrase inhibitors in Escherichia coli topoisomerase I mutants. J Bacteriol. 1986;168:276–282. doi: 10.1128/jb.168.1.276-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reece R J, Maxwell A. DNA gyrase: structure and function. Crit Rev Biochem Mol Biol. 1991;26:335–375. doi: 10.3109/10409239109114072. [DOI] [PubMed] [Google Scholar]
- 55.Ruiz J, Marco F, Goni P, Gallardo F, Mensa J, Trilla A, Jimenez de Anta T, Vila J. High frequency of mutations at codon 83 of the gyrA gene of quinolone-resistant clinical isolates of Escherichia coli. J Antimicrob Chemother. 1995;36:737–738. doi: 10.1093/jac/36.4.737. [DOI] [PubMed] [Google Scholar]
- 56.Simon R U, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 57.Steck T R, Franco R J, Wang J Y, Drlica K. Topoisomerase mutations affect the relative abundance of many Escherichia coli proteins. Mol Microbiol. 1993;10:473–481. doi: 10.1111/j.1365-2958.1993.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 58.Sternglanz R, DiNardo S, Voelkel K A, Nishimura Y, Hirota Y, Becherer K, Zumstein L, Wang J C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affect transcription and transposition. Proc Natl Acad Sci USA. 1981;78:2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 61.Vila J, Ruiz J, Goni P, De Anta M T. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:491–493. doi: 10.1128/aac.40.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]