Abstract
Inactive carrier phases in chronic hepatitis B virus (HBV) infection present minimal liver disease and HBV replication activity suggesting partial immune reconstitution, although the mechanisms responsible remain elusive. Moreover, hepatitis B surface antigen (HBsAg) production—hypothesized to modulate the immune response—is unaltered. In the current study, we assessed the intrahepatic transcriptome in inactive carriers of HBV versus healthy liver donors, including in the context of diverse HBsAg levels (serum and liver), to better understand the phenomenon of immune control. We found a deregulated liver transcriptome in inactive carriers compared with healthy controls, despite normal liver function. Moreover, diverse HBsAg levels have minimal impact on the liver transcriptome in inactive carriers, although gene correlation analysis revealed that leukocyte activation, recruitment, and innate responses genes were correlated with liver HBsAg levels. These findings provide more insight into the mechanisms underlying anti-HBV strategies currently under development, aimed at interfering with HBsAg production or inducing a state of immune control.
Keywords: hepatitis B, HBsAg, intrahepatic transcriptome
Chronic infection with the hepatitis B virus (HBV) remains a global health burden, despite the fact that the virus was discovered >50 years ago and a highly effective prophylactic vaccine exists. According to the World Health Organization, only 9% of the estimated 257 million individuals living with chronic HBV have had their HBV infection diagnosed [1]. Untreated, chronic HBV can lead to liver fibrosis, cirrhosis, and ultimately hepatocellular carcinoma. These long-term life threatening complications are thought to be responsible for >1 million deaths annually [2, 3].
The virological characteristics in chronic HBV infection are highly diverse and categorize patients into distinct clinical phases based on the presence of the viral protein hepatitis B e antigen (HBeAg) and serum levels of HBV DNA and alanine transferase (ALT), a surrogate marker of liver damage and inflammation released by lysed hepatocytes [4]. Particularly interesting is the inactive carrier phase, marked by undetectable HBeAg and low serum HBV DNA and normal serum ALT levels, suggesting that the virus is either inactive or somehow controlled by the immune system. A detailed understanding of the immune status during this phase is still lacking, but it may provide important clues for future therapeutic strategies. Characterization of the host immune response with respect to distinct clinical phases, using systems biology approaches, has shown changes in host gene expression in phases with active hepatitis versus nonhepatitis in chronic HBV and phases with high versus low viral load [5–7].
Although low serum HBV DNA levels, normal ALT levels, and undetectable HBeAg are a hallmark of the inactive carrier phase, the viral protein hepatitis B surface antigen (HBsAg) is abundantly found in the serum of these subjects. It has been hypothesized that high levels of serum HBsAg are responsible for inducing and maintaining an impaired immune response against HBV, by inducing T-cell exhaustion [8–10]. In fact, various therapeutic approaches aimed at reducing HBsAg production and release are currently being tested in trials for clinical efficacy [9], but evidence in support of a potent immunomodulatory activity of HBsAg has not been convincingly reported, and studies using blood leukocytes from individuals with variable serum HBsAg levels showed minor differences in gene expression profiles and functional readouts in purified leukocyte subpopulations [11–13].
However, these studies failed to examine the consequence of differential HBsAg expression in the liver, where viral replication and protein synthesis takes place. Moreover, the difficulty in sampling livers of healthy individuals with core needle biopsy and the scarcity of such samples limits studies comparing healthy livers with those in patients with HBV. To the best of our knowledge, we are the first to profile the transcriptome of liver formalin-fixed paraffin-embedded (FFPE) biopsy specimens to clarify the putative differences in immune status (1) between inactive carriers and healthy individuals and (2) among inactive carriers with variable serum and liver HBsAg levels.
MATERIALS AND METHODS
Patient and FFPE Liver Samples
Core needle liver biopsy specimens from 21 patients with chronic HBV and 7 healthy controls—taken to determine their eligibility as altruistic liver donors—were collected as part of routine clinical care at the Erasmus MC and archived as FFPE tissues. Serum HBV DNA was undetectable in 10 of 21 patients with HBV. The remaining 11 had all levels <2000 IU/mL except for 3 patients (with levels of 2290, 3690, and 5160 IU/mL). All HBV patients were HBeAg negative, had normal-range ALT levels (<35 IU/mL), had no signs of hepatitis or antiviral treatment before biopsy, and had no coexisting primary liver disease or coinfection with hepatitis C virus, hepatitis E virus, hepatitis D virus, or human immunodeficiency virus, which classified them in the inactive carrier state of chronic HBV.
Histological evaluation (using the METAVIR staging system and the hepatic activity index [HAI]) was performed as described elsewhere, by a single liver pathologist in a uniform manner (Table 1) [14]. Determination of intrahepatic and peripheral virological and clinical parameters is described in the Supplementary Material and Methods. This study was conducted in accordance with the Declaration of Helsinki guidelines and the principles of Good Clinical Practice. Given the retrospective nature of this study, written informed consent was not obtained from each patient. Instead, the ethical review board of the Erasmus Medical Center approved this study as it was in accordance with the regulations of the Dutch medical scientific societies (FEDERA) guidelines, which regulate further use of coded-anonymous residual human tissue for scientific research.
Table 1.
Patient Characteristics
| Characteristic | Healthy Controls (n = 7) | Inactive Carriers (n = 21) | P Value |
|---|---|---|---|
| Male sex, % (no.) | 57 (4) | 47 (10) | NSa |
| Race/ethnicity | |||
| White | 6 | 7 | NA |
| African | 0 | 5 | |
| Asian | 0 | 5 | |
| Other | 1 | 4 | |
| Age, median (IQR), y | 51 (27–55) | 36 (28–42) | NSb |
| Laboratory values, median (IQR) | |||
| ALT, IU/mL | 28 (23–40) | 27 (29–33) | NSb |
| AST, IU/mL | 25 (23–04) | 24 (19–30) | NSb |
| Prothrombin time, sc | NA | 8 (10.3–12.1) | NA |
| Total bilirubin, µmol/Lc | NA | 9 (8–11.4) | NA |
| HBV DNA, median (maximum), log IU/mLd | NA | 2.3 (3.7) | NA |
| HBeAg negative | NA | 21 | NA |
| Anti-HbeAg positive | NA | 21 | NA |
| Serum HBsAg, median (IQR), IU/mL | NA | 1750 (555–9475) | NA |
| HBsAg-positive liver cells, % (IQR)e | NA | 30 (7–69) | NA |
| HBV genotype | NA | A in 4, B in 2, C in 2, D in 9, E in 1; ND in 3 | NA |
| Fibrosis score <F2 | 5 (ND in 2) | 21 | NA |
| HAI score ≤5 | ND | 21 | NA |
| NASH | |||
| No | 7 | 20 | NSa |
| Yes | 0 | 1 | |
| Steatosis | 7/0 | ||
| No | 7 | 17 | NSa |
| Yes | 0 | 4 |
Abbreviations: ALT, alanine transferase; AST, aspartate aminotransferase; HAI, hepatic activity index; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; IQR, interquartile range; NA, not applicable; NASH, non-alcoholic steatohepatitis; ND, not determined; NS, not significant.
a P values calculated with Fisher exact test.
b P values calculated with unpaired Mann-Whitney test.
cDetermined in 18 of 21 inactive carriers.
dUndetectable HBV DNA in 10 of 21 inactive carriers.
eDetermined in 16 of 21 inactive carriers.
Intrahepatic Transcriptome Data Analysis
RNA extraction from FFPE liver biopsy specimens and sequencing are described in the Supplementary Material and Methods. Raw paired-end reads were aligned to the human reference genome hg38, downloaded from the University of California Santa Cruz Genome Bioinformatics site using the STAR (version 2.7.3a) [15]. Picard tools were used to detect samples with ≥10 million reads in protein coding messenger RNA regions for downstream analysis and were otherwise excluded [16]. Gene set annotation from the Molecular Signatures Database (version 7.0) was quantified per sample as follows. Gene expression was quantified using featureCounts software (version 1.4.6-p1) and converted to log2 counts per million using the R package edgeR (version 3.32.1), and then scores were calculated using the R package gsva [17, 18]. Raw gene counts per sample were used to perform differential gene expression analysis comparing groups, using the DESeq2 package (version 1.30.1) [19]. Low-expression genes across samples were removed and normalized using the estimateSizeFactors function. The normalized gene expression matrix obtained from DESeq2 was used to performed Spearman correlations with serum and liver HBsAg levels in inactive carriers. Pathway analysis was conducted with gene set enrichment analysis [20].
Statistical Analysis
Statistical analyses were performed using GraphPad Prism software (version 9). Baseline variables between cohorts were compared χ 2 tests for categorical variables and Mann-Whitney tests for continuous variables. All reported P values are 2 sided. Significant differences were considered in all cases when P < .05. Spearman correlations were considered significant when P < .01.
RESULTS
Down-regulation of Metabolic Genes and Pathways in Livers of Inactive Carriers
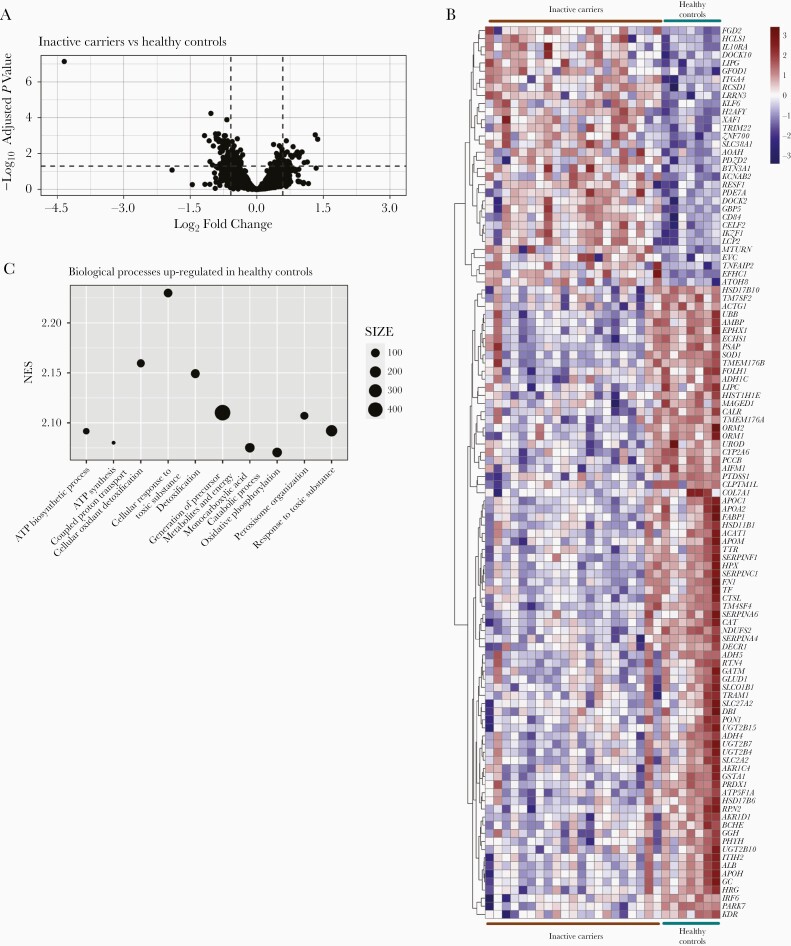
To examine whether the minimal viral activity observed in the inactive carrier phase still affects liver gene expression we performed RNA sequencing on FFPE liver biopsy specimens from 21 inactive carriers with normal ALT levels, minimal to undetectable serum HBV DNA, low fibrosis and low necroinflammatory scores (METAVIR score ≤F2 and HAI ≤5, respectively) and compared findings with those in 7 healthy individuals (Table 1). Differential gene expression analysis (fold change, ≥1.5; adjusted P ≤.05) identified 110 differentially expressed genes (DEGs) (Figure 1A and Supplementary Table 1). Two-thirds (n = 78) were down-regulated in the livers of inactive carriers, while the remaining genes (n = 32) were increased compared with healthy individuals (Figure 1B). Half of the down-regulated genes were enzymatic genes, predominantly involved in lipid metabolism (HSD11B1, DECR1, and PTDSS1) and oxidative processes (GLUD1, GSTA1, and NDUFS2) (Supplementary Table 1). Interestingly, we also found liver-specific genes (FABP1 and TF), hepatocyte markers (ALB and SLC2A2) and some immune-related genes (IRF6, SOD1, CTSL, ORM1, and ORM2) to be underexpressed in livers of inactive carriers. In contrast, a third of the up-regulated genes found in inactive carriers were immune related and predominantly involved in adaptive immune cell activation (CD84, BTN3A1, IL10RA).
Figure 1.
Intrahepatic gene expression comparison in healthy control (n = 7) versus inactive carriers (n = 21). A, Volcano plot of differentially expressed genes (DEGs) in 7 healthy controls versus 21 inactive carriers. Vertical and horizontal dashed lines represent 1.5-fold change (log2 fold change, 0.58) and adjusted P value <.05 (−1.3 log10) thresholds, respectively. B, Heat map of DEGs with fold change >1.5 per patient. Color-coded values represent z scores of normalized gene counts per samples, scaled per row (gene). C, Top 10 gene set enrichment analysis biological processes up-regulated in healthy controls. Abbreviations: ATP, adenosine triphosphate; NES, Normalized enrichment score.
To better understand in which processes these DEGs participate and the level of deregulation of the host response, we conducted biological processes gene set enrichment pathway analysis. We only detected down-regulated biological processes signatures (adjusted P <.05) in inactive carriers compared with healthy individuals. These were predominantly related to liver metabolism (Supplementary Table 2), and the top 10 were involved in adenosine triphosphate synthesis and response to toxic substrates (Figure 1C). In sum, we observed a deregulated intrahepatic gene expression profile in the livers of patients with chronic HBV with controlled infection, in comparison with healthy individuals, the vast majority affecting metabolic genes and pathways with select immune genes significantly up-regulated.
Correlation Between Serum HBsAg Levels and Innate and Adaptive Liver Immune Genes
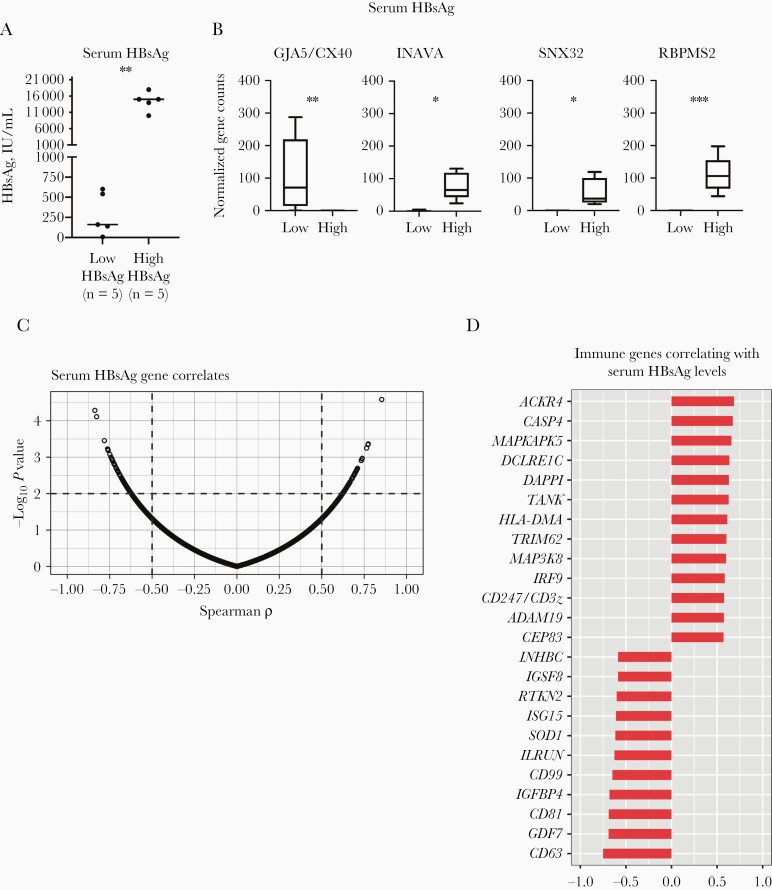
Elevated viral antigen expression has been previously shown to negatively affect immune responses, as shown in lymphocytic choriomeningitis virus-infected mouse studies [21]. Although viral replication activity and liver inflammation are low in inactive carrier, HBsAg is continuously expressed and can easily be detected in serum. A continuous presence of HBsAg has been hypothesized, which may negatively affect immune control during all phases of chronic HBV infection [9]. Inactive carriers were selected with a 10-fold difference in serum HBsAg levels (low [≤1000 IU/mL] in 5 and high [≥10 000 IU/mL] in 5) (Figure 2A). HBV genotype distribution (not shown) and age (median, 39 vs 36 years in low- and high-HBsAg groups, respectively) and were comparable between groups, albeit older age and genotype D have been associated with lower serum HBsAg levels [22, 23]. Surprisingly, we detected only 4 DEGs (INAVA, SNX32, RBPMS2, and GJA5/CX40) between the low- and high-HBsAg groups, with all except GJA5/CX40 increased in the high-HBsAg group (Figure 2B).
Figure 2.
Gene expression differences in inactive carriers based on serum hepatitis B surface antigen (HBsAg) levels. A, Serum HBsAg distribution in inactive carriers with high (n = 5; >10 000 IU/mL) or low (n = 5; <1000 IU/mL) HBsAg expression (Mann-Whitney test). B, Differentially expressed genes in inactive carriers with distinct serum HBsAg levels. Abbreviations: GJA5/CX40, gap junction protein alpha 5; INAVA, innate immunity activator; RBPMS2, RNA binding protein, messenger RNA processing factor 2; SNX32, sorting nexin 32. *P < .05; **P < .01; ***P < .005. C, Spearman correlations with serum HBsAg levels and normalized gene expression values in 21 inactive carriers. Vertical and horizontal dashed lines indicate ρ (>0.5) and P value (<.01) thresholds, respectively. D, Selected immune-related gene correlates.
To further examine the possible modulation exerted by HBsAg, we conducted correlation analysis of gene expression with serum HBsAg levels from our full cohort of 21 inactive carriers (Spearman, P < .01 and ρ >0.5). We found almost equal numbers of genes that were positively (n = 136; ρ > 0.5) or negatively (n = 135; ρ < −0.5) correlated with serum HBsAg (Supplementary Table 3). Among the extensive list of genes, we detected enzymatic genes (hydrolases, transferases, and oxidoreductases) as well as genes involved in transcriptional regulation and cell cycle (Supplementary Table 3). Furthermore, 24 correlating immune genes were identified, including some related to B-cell activation (DAPP1, DCLRE1C, CD81, and IGSF8), T-cell activation (MAP3K8 and CD247), and inflammatory response (IGFBP4, MAPKAPK5, and CASP4), chemotactic genes (ACKR4 and CD99), and genes involved in antiviral defense (ISG15 and IRF9) and antigen presentation (ADAM19 and HLA-DMA) (Figure 2D and Supplementary Table 3).
Although transcriptome comparison based on distinct serum HBsAg levels did not reveal HBsAg-induced changes in expression of immune genes, correlation analysis revealed an association between the intrahepatic gene expression levels of a few immune genes and serum HBsAg levels which suggests some degree of immune modulation due to HBsAg in livers of inactive carriers.
Percentage of HBsAg-Positive Liver Cells Positively Correlated With Serum HBsAg Expression in Inactive Carriers
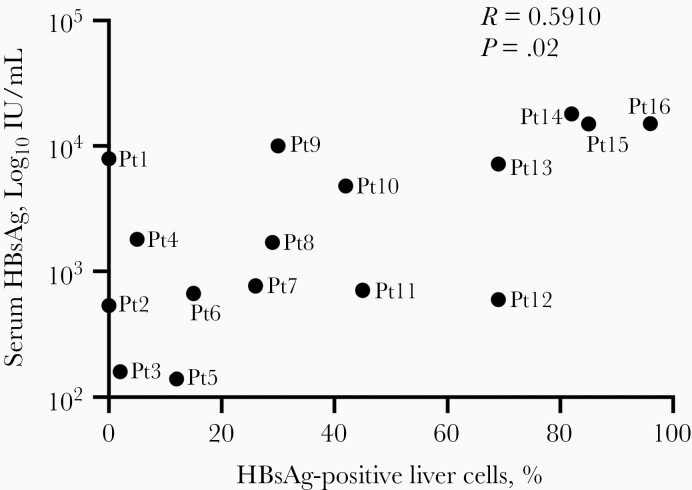
We generally use serum HBsAg levels as a proxy for HBsAg production in HBV-positive livers. However, little is known as to whether serum HBsAg levels reflect intrahepatic HBsAg levels. To study this, we performed immunofluorescence of HBsAg on FFPE liver biopsy specimens from 16 RNA sequencing–matched inactive carriers. Images of HBsAg staining in the liver are presented in Supplementary Figure 1. We found that the percentage of HBsAg-positive liver cells was significantly correlated with serum HBsAg levels (r = 0.59; P = .02) (Figure 3). However, this monotonic relationship may not be true in all patients (eg, patients 1, 4, and 12; Supplementary Figure 1), raising the question of which mechanisms are responsible for the different HBsAg secretion patterns from the liver, as well the question of clinical implications. This positive trend suggests that serum HBsAg levels may be used as a surrogate of the HBV liver burden.
Figure 3.
Spearman correlation between serum hepatitis B surface antigen (HBsAg) levels and the percentage of liver HBsAg-positive cells in inactive carriers (n = 16). Abbreviations: Pt1 (etc), patient 1 (etc).
Correlation Between Liver HBsAg Expression and Liver Leukocyte Activation Genes
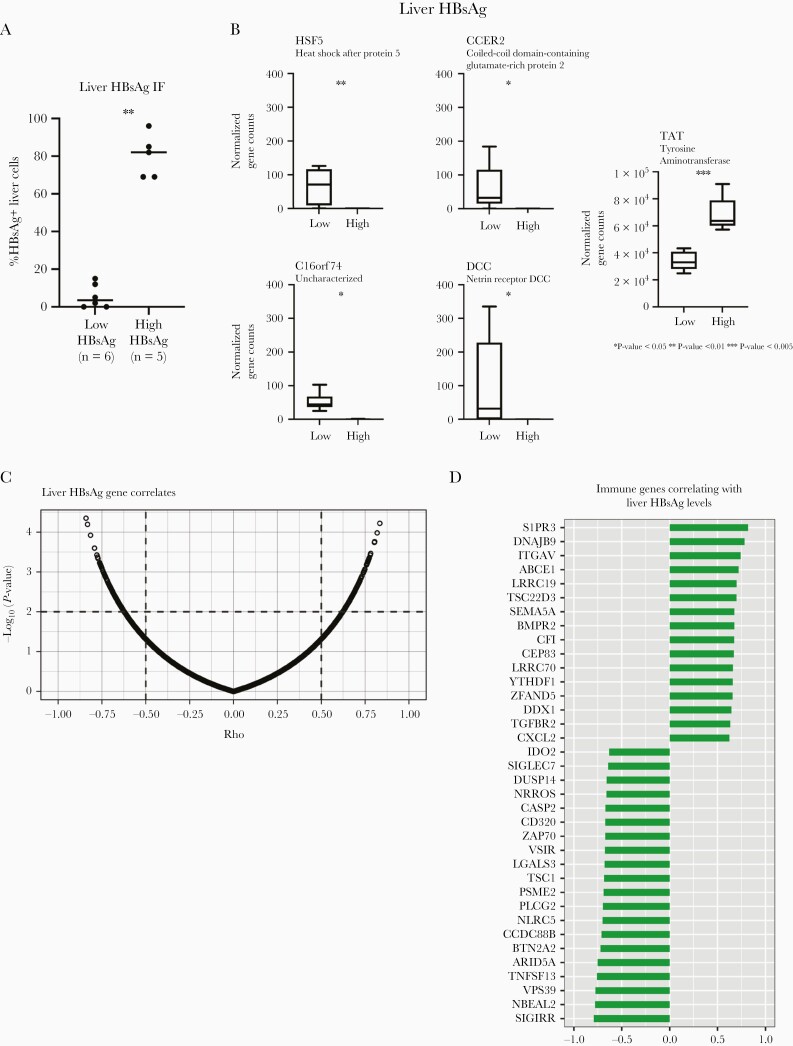
Although we observed a positive correlation between the HBsAg levels detected in serum and liver, not all patients adhered to this pattern (patients 1, 4 and 12; Supplementary Figure 1), and therefore we decided to also examine the liver transcriptome with respect to the number of intrahepatic HBsAg-expressing cells. For this, 5 inactive carriers with >60% of cells positive for HBsAg were compared with 6 inactive carriers whose livers had <20% of cells positive for HBsAg by immunofluorescence (Figure 4A). This analysis resulted in only 5 DEGs (HSF5, C16orf74, CCER2, DCC, and TAT). All except TAT were increased in the low-HBsAg group (Figure 4B).
Figure 4.
Gene expression differences in inactive carrier based on the percentage of intrahepatic hepatitis B surface antigen (HBsAg) expression. A, Percentage distribution of HBsAg-positive liver cells in inactive carriers with high (n = 5; >60%) or low (n = 6; <20%) percentages (Mann-Whitney test). Abbreviation: IF, immunofluorescence. B, Differentially expressed genes in inactive carriers with distinct liver HBsAg expression. Abbreviations: CCER2, coiled-coil domain–containing glutamate-rich protein 2; DCC, netrin receptor DCC; HSF5, heat shock factor protein 5; TAT, tyrosine aminotransferase. *P < .05; **P < .01; ***P < .005. C, Spearman correlations between liver HBsAg levels and normalized gene expression values in 16 inactive carriers. Vertical and horizontal dashed lines indicate ρ (>0.5) and P value (<0.01) thresholds, respectively. D, Selected immune-related gene correlates.
Again, we further conducted correlation analysis of gene expression with the percentage of liver HBsAg-positive cells in the cohort of 16 inactive carriers, whose livers were subjected to HBsAg immunofluorescence. Correlation analysis revealed a total of 270 genes correlated with the percentage of HBsAg positivity in the liver (Supplementary Table 4). The numbers of genes positively (ρ > 0.5) or negatively (ρ < −0.5) correlated with the levels of HBsAg in the liver were almost identical (128 and 142 genes, respectively; Figure 4C). Among these we found a large number of genes acting as transcription factors/regulators and DNA binding, cell cycle, enzymatic (oxidoreductase, transferase and hydrolase functions), and lipid and protein transport genes, involved in cytoskeleton arrangement, lipid metabolism, autophagy, and apoptotic programs (Supplementary Table 4).
Moreover, 36 immune genes were identified. Most of which were involved in B-cell (CD320, IDO2, TNFSF13, DNAJB9, PLCG2, and ZAP70), T-cell (LGALS3, VSIR, TSC22D3/GILZ, TSC1, PLCG2, ZAP70, BTN2A2, and ITGAV), and natural killer (NK) cell (SIGLEC7, NBEAL2, CCD88B, and SEMA5A) activation or function. Furthermore, a small fraction of detected genes was related to antigen presentation (NLRC5 and YTHDF1), transforming growth factor b signaling (TGFBR2 and VPS39), leukocyte recruitment (S1PR3 and ITGAV), interferon-lambda 3 stimulation (PSME2), and inflammation (CXCL2, LRRC70, and ARID5A) (Figure 4D). Interestingly, none of these genes were found among the list of genes correlating with serum HBsAg levels; <2% (RPL41, SUPV3L1, CNPPD1, CEP83, MON2, and USP37) of the genes found to be correlated with liver HBsAg levels were also correlated with serum HBsAg levels (not shown). These findings point toward changes in liver leukocyte activation profiles among inactive carriers, associated with liver HBsAg levels.
Discussion
The present study examines the putative mechanisms underlying immune control during the inactive carrier phase of chronic HBV. Through intrahepatic transcriptome analysis in inactive carriers and healthy liver donors, we found down-regulation of metabolic genes and pathways and a minor increase in immune genes among inactive carriers compared with healthy individuals. Moreover, whereas gene expression differences were almost nonexistent in a subset of inactive carriers based on the HBsAg level in serum or liver, correlation analysis identified a subset of immune genes that were correlated with serum or liver HBsAg levels.
To gain insight into the mechanisms that act during the inactive carrier phase in patients with chronic HBV, we demonstrated that the expression of metabolic genes in liver is down-regulated in these patients compared with healthy individuals. Thus, although viral replication and liver damage are minimal in these patients, metabolic dysregulation at the genetic level was observed, including the gene encoding albumin. Given the key role of hepatocytes in metabolic processes and the abundance of hepatocytes relative to immune cells, it is likely that down-regulated gene expression is the consequence of either continued translation of viral protein from covalently closed circular DNA in infected hepatocytes or low HBV replicative activity in hepatocytes. To note, the observed transcriptome differences were unlikely to be driven by HBV DNA levels because no DEGs were identified between inactive carriers on the basis of HBV DNA (detectable vs undetectable; not shown). Nonetheless, despite the reduced expression of metabolic genes in the liver, prothrombin and total bilirubin levels were within the normal range in the majority of inactive carriers, suggesting a conserved synthetic liver function, as often observed in more advanced forms of liver disease.
Our findings observed in livers of inactive carriers differ from those from earlier studies in which an increase in fatty acid synthesis, gluconeogenesis, and aerobic oxidation during this phase of chronic HBV was observed [24, 25]. In this respect, it is important to note that our study is unique, in that we compared core needle liver biopsy specimens obtained from patients and healthy donors, which were collected and processed using identical procedures. Most other studies used healthy liver material from surgical procedures or obtained results from in vitro studies using hepatoma tumor cell lines, in which the persistence of the HBV infection and the inactive carrier phase are difficult to interpret [26–29].
In contrast, a recent study evaluating the transcriptomic changes over time in distinct liver-injury mice models revealed down-regulation of metabolic genes and pathways during peak up-regulation of inflammatory genes and pathways, which were controlled by the same upstream regulators, suggesting that these changes participate in liver regeneration and liver-injury [30]. Furthermore, the same study also showed that these transcriptomic signatures were present in different liver conditions in human livers, including HBV infection [30]. Whether these findings can be translated to our data set, composed of patients with HBV with no signs of hepatitis and minimal liver inflammation (HAI <5) warrants further investigation. However, it is important to mention that we previously demonstrated that serum metabolomics conducted in chronic HBV showed deregulated lipid metabolite patterns in inactive carriers [31], which might be the consequence of altered expression of metabolic genes.
Besides an effect on expression levels of metabolic genes, transcriptome comparison showed up-regulation of a small number of immune genes when we compared livers of inactive carriers with those of healthy individuals. Most of these genes are involved in leukocyte activation (DOCK2 and ITGA4) and T-cell responses (BTN3A1 and CD84). Whether these signatures reflect inflammatory activity during chronic hepatitis or contribute to partial immune control in the livers of inactive carriers is difficult to ascertain and warrants functional studies. However, enhanced expression of DOCK2 and IL10RA, in addition to the previously mentioned genes, suggests either infiltration of immune cells, pronounced activation, or regulation of intrahepatic immune responses during the inactive phase of chronic HBV infection. Nonetheless, we detected an increased expression of TRIM22, a potent HBV replication and transcription suppressor, in infected patients, suggesting an active anti-HBV response [32].
Although signs of immune control in our cohort of inactive carriers are limited, a previous study in which NanoString analysis was performed on FFPE core needle liver biopsy specimens from patients with chronic HBV observed a general gene expression down-regulation of innate antiviral effectors and Toll-like receptor– and interferon-related genes in HBeAg-negative patients, compared with uninfected controls [33]. These discrepancies might be explained by differences in cohort composition between studies, since a third of the HBeAg-negative patients showed signs of active hepatitis, and no matching details were provided for the uninfected cohort. Furthermore, HBeAg-negative patients with minimal HBV DNA and normal ALT levels displayed a comparable gene expression in some of the studied ISGs compared with healthy controls, supporting the findings in our cohort of inactive carriers [33].
The putative HBsAg immunomodulatory effect on the immune response in chronic HBV remains under continuous debate as human data fail to recapitulate in vitro and animal models observations. In the current study, we showed that liver gene expression differences in inactive carriers with variable serum and liver HBsAg levels are minimal, with >5 genes found to be differentially expressed. These results mirror the findings from the previous study in which Montanari et al [11] showed that variations in serum HBsAg levels resulted in minimal gene expression changes in different cell-sorted peripheral leukocytes populations (ie, monocytes, CD4 and CD8 T cells, B cells, NK cells, and dendritic cells). Likewise, the phenotypic characterization and function of peripheral T and NK cells in patients with chronic HBV have been shown to be comparable irrespective of serum HBsAg levels [34].
However, in mouse models, knockdown of HBV antigen expression using small interfering RNA increased CD8+ T-cell responses against HBV after therapeutic vaccination [35]. In contrast, a different study showed that the frequency and function of HBsAg-specific T cells in HBV transgenic mice were independent of the level of circulating serum HBsAg levels [36]. In line with this study, Le Bert et al [34] showed that the duration of HBV infection, not circulating HBsAg levels, is associated with the frequency of HBsAg-specific T cells in patients with chronic HBV. These findings could not be extrapolated to our study, since we found that the intrahepatic transcriptome variance was not associated with patient age (not shown), albeit that the HBsAg burden in serum and particularly in the liver was negatively associated with patient age (Supplementary Figure 2). Likewise, infection with HBV genotype D, which was predominant in our cohort and has been shown to be associated with lower serum HBsAg levels [23], showed no transcriptomic differences compared with samples from patients with non-D genotypes (not shown).
Additional correlation analysis on a continuous scale of serum and liver HBsAg levels showed a fraction of the correlating genes to be immune related. For instance, the antiviral effector ISG15 was found to be negatively correlated with serum HBsAg, an observation in line with a study by Lebossé et al [33], which showed that HBeAg-negative patients with lower HBsAg levels displayed increased ISG15 expression. Although we found a set of immune genes related to innate and adaptive immune responses to be correlated with serum HBsAg levels, it is unclear whether these changes have consequences for the functionality of the immune response.
Interestingly, the positive correlation between liver HBsAg levels and expression of innate immune genes (CFI, DDX1 and LRRC19), chemotactic genes (CXCL2), and genes involved in leukocyte recruitment (S1PR3 and ITGAV) may reflect signs of HBsAg-induced immune activation. Similarly, the negative correlations observed between low liver HBsAg levels and the increased expression of T- and B-cell–activated genes may point toward a limited HBsAg-induced immune exhaustion and enhanced immune control and/or a higher proportion of leukocytes in the livers of these patients.
In summary, profiling of liver transcriptomes in healthy individuals versus inactive carriers with variable HBsAg levels revealed limited immune activation signals in livers of inactive carriers and a previously unreported down-regulated metabolic profile. Furthermore, our results underline the limited but detectable signs of immunomodulation in the livers of inactive carriers, and mildly more active T-cell and B-cell gene signatures in patients with low liver HBsAg levels. Our findings are highly relevant in the current search for novel anti-HBV strategies that aim at reinvigorating the antiviral response by interfering with HBsAg production or inducing a state of immune control.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank Gertine Van Oord for her valuable contribution organizing the operational procedures for sample shipment and all research participants for their contribution to the research.
Author contributions. N. R. M. and A. B. designed the study. T. P. P. v. d. B., and M. D. provided liver formalin-fixed paraffin-embedded tissues and conducted histological examination and liver hepatitis B surface antigen staining. R. R. and N. V. B. overlooked and organized RNA sequencing. N. R. M. and R. R. conducted NA-seq analysis. N. R. M. and A. B. wrote the manuscript. N. R. M., R. R., N. V. B., T. P. P. v. d. B., M. D., J. D. D., B. F., and A. B. critically revised the manuscript. B. F. and A. B. secured funding. All authors approved the final version of the manuscript.
Financial support. This work was supported by the Foundation for Liver and Gastrointestinal Research.
Potential conflicts of interest. A. B. received grant support from Janssen, GSK, Fujirebio, and Gilead Sciences. At the time this study was conducted, R. R., N. V. B., and B. F. were employees and stockholders of Gilead Sciences. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lampertico P, Agarwal K, Berg T, et al. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol 2017; 67:370–98. [DOI] [PubMed] [Google Scholar]
- 2. Akinyemiju T, Abera S, Ahmed M, et al. Global Burden of Disease Liver Cancer Collaboration. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the Global Burden of Disease Study 2015. JAMA Oncol 2017; 3:1683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang H, Naghari M, Allen C, et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016; 388:1459–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gish RG, Given BD, Lai CL, et al. Chronic hepatitis B: virology, natural history, current management and a glimpse at future opportunities. Antiviral Res 2015; 121:47–58. [DOI] [PubMed] [Google Scholar]
- 5. Liu H, Li F, Zhang X, et al. Differentially expressed intrahepatic genes contribute to control of hepatitis B virus replication in the inactive carrier phase. J Infect Dis 2018; 217:1044–54. [DOI] [PubMed] [Google Scholar]
- 6. Hou J, Brouwer WP, Kreefft K, et al. Unique intrahepatic transcriptomics profiles discriminate the clinical phases of a chronic HBV infection. PLoS One 2017; 12:e0179920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vanwolleghem T, Hou J, van Oord G, et al. Re-evaluation of hepatitis B virus clinical phases by systems biology identifies unappreciated roles for the innate immune response and B cells. Hepatology 2015; 62:87–100. [DOI] [PubMed] [Google Scholar]
- 8. Fisicaro P, Barili V, Rossi M, et al. Pathogenetic mechanisms of T cell dysfunction in chronic HBV infection and related therapeutic approaches. Front Immunol 2020; 11:849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tout I, Loureiro D, Mansouri A, Soumelis V, Boyer N, Asselah T. Hepatitis B surface antigen seroclearance: immune mechanisms, clinical impact, importance for drug development. J Hepatol 2020; 73:409–22. [DOI] [PubMed] [Google Scholar]
- 10. Boni C, Laccabue D, Lampertico P, et al. Restored function of HBV-specific T cells after long-term effective therapy with nucleos(t)ide analogues. Gastroenterology 2012; 143:963–73.e9. [DOI] [PubMed] [Google Scholar]
- 11. Montanari NR, Conceição-Neto N, Van Den Wyngaert I, et al. Differential gene expression, irrespective of circulating hepatitis B surface antigen levels, between inactive carrier and nucleos(t)ide analogue-treated hepatitis B virus patients. J Infect Dis doi: 10.1093/infdis/jiaa614. Published 3 October 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aliabadi E, Mix C, Manns MP, Kraft A, Cornberg M. FRI-131-impact of HBsAg level on cellular immune responses in HBeAg negative patients with chronic hepatitis B virus infection. J Hepatol 2019; 70:e445. [Google Scholar]
- 13. Gill US, Hansi N, Le Bert N, et al. HBV-specific T cell responses in low replicating inactive carrier patients are independent of hepatitis B surface antigen load. J Hepatol 2018;68:S793–S794. [Google Scholar]
- 14. Brouwer WP, van der Meer AJ, Boonstra A, et al. The impact of PNPLA3 (rs738409 C > G) polymorphisms on liver histology and long-term clinical outcome in chronic hepatitis B patients. Liver Int 2015; 35:438–47. [DOI] [PubMed] [Google Scholar]
- 15. Haeussler M, Zweig AS, Tyner C, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res 2019; 47:D853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Broad Institute. Picard toolkit. Broad Institute, GitHub Repos; 2019. http://broadinstitute.github.io/picard/. Accessed 03 February 2020. [Google Scholar]
- 17. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013; 14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102: 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mueller SN, Ahmed R. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc Natl Acad Sci U S A 2009; 106:8623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taniguchi H, Iwasaki Y, Aimi M, Shimazaki G, Moriya A. Clinical features of chronic hepatitis B patients with low hepatitis B surface antigen levels and determinants of hepatitis B surface antigen seroclearance. JGH Open 2020; 4:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Salpini R, Battisti A, Piermatteo L, et al. Key mutations in the C-terminus of the HBV surface glycoprotein correlate with lower HBsAg levels in vivo, hinder HBsAg secretion in vitro and reduce HBsAg structural stability in the setting of HBeAg-negative chronic HBV genotype-D infection. Emerg Microbes Infect 2020; 9:928–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masson JJ, Billings HW, Palmer CS. Metabolic reprogramming during hepatitis B disease progression offers novel diagnostic and therapeutic opportunities. Antivir Chem Chemother 2017; 25:53–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi YX, Huang CJ, Yang ZG. Impact of hepatitis B virus infection on hepatic metabolic signaling pathway. World J Gastroenterol 2016; 22:8161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang MD, Wu H, Huang S, et al. HBx regulates fatty acid oxidation to promote hepatocellular carcinoma survival during metabolic stress. Oncotarget 2016; 7:6711–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim KH, Shin HJ, Kim K, et al. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPARgamma. Gastroenterology 2007; 132:1955–67. [DOI] [PubMed] [Google Scholar]
- 28. Shin HJ, Park YH, Kim SU, et al. Hepatitis B virus X protein regulates hepatic glucose homeostasis via activation of inducible nitric oxide synthase. J Biol Chem 2011; 286:29872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yue D, Zhang Y, Cheng L, et al. Hepatitis B virus X protein (HBx)-induced abnormalities of nucleic acid metabolism revealed by 1H-NMR-based metabonomics. Sci Rep 2016; 6:24430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campos G, Schmidt-Heck W, De Smedt J, et al. Inflammation-associated suppression of metabolic gene networks in acute and chronic liver disease. Arch Toxicol 2020; 94:205–17. [DOI] [PubMed] [Google Scholar]
- 31. Schoeman JC, Hou J, Harms AC, et al. Metabolic characterization of the natural progression of chronic hepatitis B. Genome Med 2016; 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao B, Duan Z, Xu W, Xiong S. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 2009; 50:424–33. [DOI] [PubMed] [Google Scholar]
- 33. Lebossé F, Testoni B, Fresquet J, et al. Intrahepatic innate immune response pathways are downregulated in untreated chronic hepatitis B. J Hepatol 2017; 66:897–909. [DOI] [PubMed] [Google Scholar]
- 34. Le Bert N, Gill US, Hong M, et al. Effects of hepatitis B surface antigen on virus-specific and global T cells in patients with chronic hepatitis B virus infection. Gastroenterology 2020; 159:652–64. [DOI] [PubMed] [Google Scholar]
- 35. Michler T, Kosinska AD, Festag J, et al. Knockdown of virus antigen expression increases therapeutic vaccine efficacy in high-titer hepatitis B virus carrier mice. Gastroenterology 2020; 158:1762–1775.e9. [DOI] [PubMed] [Google Scholar]
- 36. Fumagalli V, Di Lucia P, Venzin V, et al. Serum HBsAg clearance has minimal impact on CD8+ T cell responses in mouse models of HBV infection. J Exp Med 2020; 217:e20200298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.