Abstract
Objective
Integrating and harmonizing disparate patient data sources into one consolidated data portal enables researchers to conduct analysis efficiently and effectively.
Materials and Methods
We describe an implementation of Informatics for Integrating Biology and the Bedside (i2b2) to create the Mass General Brigham (MGB) Biobank Portal data repository. The repository integrates data from primary and curated data sources and is updated weekly. The data are made readily available to investigators in a data portal where they can easily construct and export customized datasets for analysis.
Results
As of July 2021, there are 125 645 consented patients enrolled in the MGB Biobank. 88 527 (70.5%) have a biospecimen, 55 121 (43.9%) have completed the health information survey, 43 552 (34.7%) have genomic data and 124 760 (99.3%) have EHR data. Twenty machine learning computed phenotypes are calculated on a weekly basis. There are currently 1220 active investigators who have run 58 793 patient queries and exported 10 257 analysis files.
Discussion
The Biobank Portal allows noninformatics researchers to conduct study feasibility by querying across many data sources and then extract data that are most useful to them for clinical studies. While institutions require substantial informatics resources to establish and maintain integrated data repositories, they yield significant research value to a wide range of investigators.
Conclusion
The Biobank Portal and other patient data portals that integrate complex and simple datasets enable diverse research use cases. i2b2 tools to implement these registries and make the data interoperable are open source and freely available.
Keywords: Information storage and retrieval, data curation, data science, genomics, electronic health records, i2b2
INTRODUCTION
Observational cohort studies are an important study design to complement interventional studies and answer a wide variety of research questions.1 Prospective and retrospective cohort studies are often designed to support well-defined research hypotheses and also test new hypotheses developed after study initiation. To do so, these studies often collect data beyond what is defined in the endpoints in the analysis plan. These data may include broad electronic health records (EHRs), genomics, complex assays, imaging studies and more. Examples include large national observational cohort studies such as All of Us, Million Veteran Project, and the UK Biobank that are collecting data and biospecimens to advance biological discoveries in medicine.2–4 In addition, large disease-specific observational cohorts have also been collecting high-dimensional datasets outside of the data required to test their initial hypotheses.5–7 These cohorts link primary data collection in electronic case report forms to EHR data, questionnaires, imaging studies, and more.
Integrating these disparate and high-dimensional data types into a research data repository and making them discoverable and accessible by a diverse set of investigators is complex and requires specialized tools and informatics skills. Informatics for Integrating Biology and the Bedside (i2b2) is a software and data model initially built for cohort discovery from EHR data.8 Since its initial release over 10 years ago, the platform has been used to support an increasingly broad range of use cases including for creating networks of data registries, supporting disease-specific research registries, and aggregating data from clinical studies. The flexibility of the platform and its data model enables a wide range of customizations to support patient data. Collected data are harmonized and made available using FAIR principles of finability, accessibility, interoperability, and reusability.9
Objective
In this article, we describe an implementation of i2b2 to create the Mass General Brigham (MGB) Biobank Portal data repository. The repository integrates data from many different types and schemas for over 125 000 consented patients and is updated weekly. The resulting structured data are made available to investigators in an easy-to-use data portal where they can construct and export customized datasets for analysis.
MATERIALS AND METHODS
Data repository population
The MGB Biobank (formerly named Partners Biobank) is an ongoing observational research project that enrolls patients and employees of a multicenter health system in Eastern Massachusetts.10 Participants are enrolled using a broad-based consent process by up to 30 research coordinators located at health system practices, in public hospital locations, as part of a collaborating study (dual consent) or electronically through Patient Gateway, the MGB patient portal.11 Biobank recruitment materials are available in print and electronically at https://biobank.massgeneralbrigham.org. MGB investigators conducting their own studies also consent patients (again with the Biobank consent language) to the Biobank at the same time as they consent for their own studies. Investigators with limited resources often consent research participants into the Biobank to take advantage of centralized resources for EHR data and sample collection, management, and genotyping.
Demographic data and blood samples are collected at baseline and linked to EHRs data and self-reported health surveys for ongoing research. Biobank enrollment and biospecimen collection is supported through institutional funding but most data and biospecimen analyses are supported by public and private grant funding. All adult patients able to provide informed consent are eligible to participate. A small number of children have also been enrolled as part of a collaborating study with IRB-approved assent forms. Once a child turns 18, they are recontacted to consent using the adult form. If they refuse consent or are unable to be contacted, they are removed from the Biobank and their data are no longer available in the Biobank Portal. The Human Research Committee of MGB approved the Biobank research protocol (2009P002312).
i2b2 software and data model
i2b2 is both a modular software platform and a data meta-model.8 The software components include a middleware module that provides authentication and authorization based on HIPAA guidelines at a project level (named the Project Management Cell = PM). The data repository module (Clinical Research Chart) drives much of the interaction with the source data using well-defined application programming interfaces (APIs). The ontology module manages the metadata attached to the data and enables powerful query capabilities across various local and external data sources. A web user interface (webclient) is a JavaScript application that runs on users’ browser and communicates with the i2b2 APIs. The webclient is highly adaptable and extensible using plugins. The Biobank Portal web client is a customized version of the standard i2b2 webclient. i2b2 is completely open source distributed under the Mozilla Public License, version 2.012 and has an active developer and academic user group community.
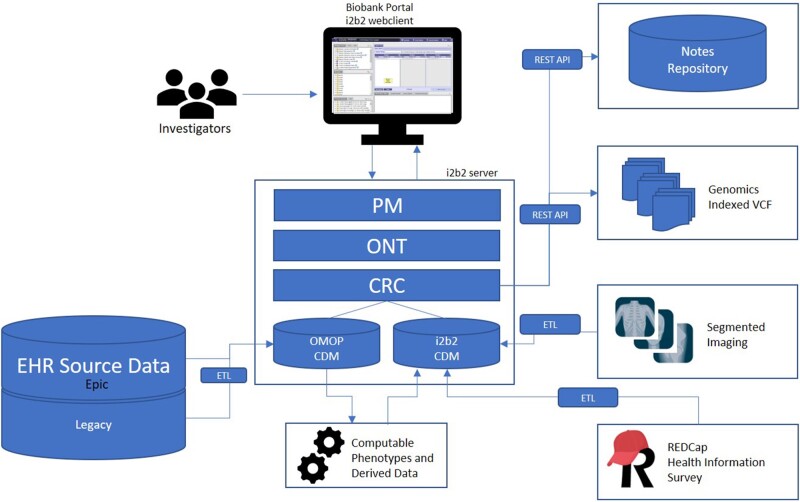
The i2b2 data model is described as a metamodel in that it defines a database schema for storing large-scale health data with the patient as the single point of reference. Beyond that there is no prescribed schema for specific types of data in the model, rather sites can customize how data are stored and define ontology rules to access that data. The data model is based on a star schema developed by Kimball and Ross13 and optimized for data warehousing use cases. The ontology is decoupled from the data, enabling a flexible and extensible data repository that can access data locally and remotely. The Biobank Portal leverages these features to allow disparate data sources to be interoperable and queryable across many different types of data which are described below. Figure 1 illustrates the overall architecture of the Biobank Portal using the i2b2 platform.
Figure 1.
The Biobank Portal architecture is based on Informatics for Integrating Biology and the Bedside (i2b2). Investigators access data through the webclient which interacts with the i2b2 application server using application programming interfaces (APIs). Most data are ingested into the data repository directly, but other data are accessed using external APIs at query time. PM: Project management cell; ONT: Ontology cell; CRC: Data repository cell; OMOP: Observational medical outcome partnership; CDM: common data model; VCF: variant call format; ETL: extract-transform-load.
Primary data
Demographic, consent, and biospecimen data
Participant identity and demographic characteristics are managed as part of the core identity management functionality of the Biobank portal platform. Demographic information is sourced from the Enterprise Master Patient Index (EMPI) and transformed to normalized age, race, gender, and ethnicity US Census categories. Each patient is mapped to one of many medical record numbers, Biobank participant ID, and other identifiers ensuring data linking to each patient is maintained across data sources and time. Patient consent information is provided by consent tracking software (CONSTRACK) which manages enrollment and withdrawal.14 Biospecimen tracking is managed by a laboratory information management system and tracks sample type, accession date, and quantity for plasma, serum, buffy coat, and other sample types. Consent and biospecimen data updates are sent via HL-7 and ingested into the main i2b2 data model tables.
EHR data
EHRs used for research have been extensively utilized for a wide range of studies. The i2b2 software began primarily as a data warehousing and query tool for EHR data before expanding to many more types of data. The Biobank Portal integrates EHR data retrieved from the MGB Research Patient Data Registry (RPDR). Data are ingested using extract-transform-load (ETL) procedures and stored in the i2b2 data model. i2b2 can also query data stored on the observational medical outcomes partnership (OMOP) common data model (CDM).15 Information on OMOP on i2b2 is available at https://community.i2b2.org/wiki/display/OMOP/OMOP+Home.
Case report and survey data
Each Biobank participant is asked to complete a comprehensive health information survey (available in English and Spanish) at the time of enrollment. These data are collected in REDCap, a widely used electronic data capture tool, using an online portal, either by the patient alone or together with a research coordinator.16 The data from REDCap are retrieved using a native API, transformed, and loaded into a staging table on a quarterly basis to be ingested into the core Biobank Portal data repository at each build. The latest i2b2 version provides native REDCap import functionality available at (https://community.i2b2.org/wiki/display/RM/1.7.12+Release+Notes). The most recent version of the Biobank Health Information Survey is included in the Supplementary Material. Although all participants are sent the survey only about 43% of participants complete the questionnaire. Barriers to survey completion include time and workflow issues. Many patients are enrolled in waiting rooms and do not have time to complete the 20- to 30-min survey and then do not follow-up to complete it online or return a paper version.
Genomic data
Biobank samples are genotyped using 3 versions of the Multi-Ethnic Global BeadChip SNP array offered by Illumina that is designed to capture the diversity of genetic backgrounds across the globe.17 These arrays cover over 1.7 million unphased variants which are annotated for dbSNP rs identifier, gene location, and protein and variant effect using Alamut-Batch (Interactive Biosoftware, France, https://www.interactive-biosoftware.com/alamut-batch/). Genotype calls and annotations are made available to investigators as VCF and PED file formats. Imputed genotypes are also available. To support querying by patient-level variant and zygosity, we extended the i2b2 web client to enable genomic queries by rsid and gene and allow constraints by variant effect (for gene queries) and zygosity (see Supplementary Figure S1). The VCF files are indexed using an optimized binary index and exposed as a REST API web service and integrated into the i2b2 application. Additional details and code for extending i2b2 for genomic queries is available at (https://community.i2b2.org/wiki/display/IGD)
Unstructured clinical text and reports
The MGB Notes Repository is a Microsoft SQL Server (MSSQL) relational database that hosts all clinical notes and reports since 1990. These notes are updated nightly and indexed using the MSSQL full-text service to allow text searches (Microsoft, https://docs.microsoft.com/en-us/sql/relational-databases/search/full-text-search?view=sql-server-2016). While i2b2 supports full-text searches using SQL CONTAINS statements directly in the observation_fact table, we implement another REST API query framework to search the external notes repository for patients enrolled in the Biobank. Users can enter search phrases such as “diabetes” or “atrial fibrillation” and optionally choose to exclude matches near certain negation words (eg, not, negative, denies). The API restricts searches by Census names and numbers to prevent inadvertent release of PHI. While this is not a full-featured NLP pipeline for named entity extraction (ie, negation functionality is limited to prespecified negation terms and no additional context exclusions), investigators can run exploratory queries for cohort selection or for applying broad exclusion criteria. Most text queries can run in <20 s across the full corpus of over 250 million notes and reports. Supplementary Figure S2 illustrates the user interface for querying notes in the Biobank Portal.
Derived and curated data
Computed phenotypes
EHR data are multifaceted and often complex, reflecting both disease state, healthcare processes, and data collection and processing workflows.18 As such, we devote significant effort to validating and improving EHR data quality using quality assurance, as well as machine learning computed phenotypes. Computed phenotypes are derived from both structured and unstructured EHR data and provide the ability for researchers to accurately select a disease population for genomic or other analyses. The Biobank Portal computed phenotypes (also called “Curated Disease Populations”) are trained using PheCAP, a well-defined supervised learning workflow.19 Once a model is trained, it is operationalized in the data repository build process using SQL scripts that define features based on ontology paths and run the prediction to estimate predicted probability of each patient having a disease. In the Biobank Portal webclient, users can select these phenotype patient cohorts based on different levels of precision and sensitivity depending on their use case. Supplementary Figure S3 illustrates the query interface for phenotypes. In addition, each phenotype algorithm has a dedicated Wiki page that includes information on training data and evaluation sets with information on performance on test data. Supplementary Table S1 includes disease prevalence and performance of Biobank phenotype algorithms.
Composite variables
In addition to more advanced machine learning phenotypes, we also build composite variables derived from primary data. A key example is the computation of Charlson comorbidity indexes derived from ICD-9 and ICD-10 codes to identify levels of illness and select relatively healthy controls. Another recent example is we build a composite variable of COVID-19 positive or negative status based on PCR lab test results or infection control flag status to quickly support the large number of COVID-19 studies conducted with Biobank Portal data. While these derived variables often include simple rule-based logic that could be run as a user query, they significantly increase usability of the portal.
Quantitative imaging
Advances in computer vision have enabled high-throughput segmentation and analysis of radiology imaging. In the Biobank Portal, we have integrated machine learning-extracted quantitative abdominal CT scan area of skeletal muscle, visceral adipose tissue, and subcutaneous adipose tissue. The data are generated by a robust, fully automated, externally validated body composition pipeline consisting of 2 deep learning algorithms.20,21 The first algorithm selects a single, axial CT slice through the third lumbar vertebral body (L3) of the spine, the second algorithm anatomically identifies the boundaries of muscle, subcutaneous adipose tissue, and visceral adipose tissue in that slice and for each quantifies the area (cm2). Absolute and normalized values for each CT scan are loaded into the data repository for querying and dataset generation.
Ontology
The i2b2 ontology of the Biobank Portal is the main mechanism of data harmonization and quality. As a flexible data model, the i2b2 CDM relies heavily on predefined ontologies. Many of these ontologies are standard to support interoperability with other data sources and network data consortium. For EHR data, the Biobank Portal primarily relies on ICD-10-CM and CPT-4 diagnosis and procedure vocabularies to provide minimal transformation from the source data.22,23 Medications use a combination of RXNORM and VA National Drug Reference and lab tests and vital signs chiefly rely on LOINC codes.24–26 Local ontologies are developed for Biobank-specific data. The Health Information Survey variables collected from participants in REDCap format are mapped to an ontology based on events, forms, and question ordering.
Because the data are completely decoupled from the ontology it is possible to have multiple ontologies covering the same data. For example, we also map ICD diagnosis to the PheCode ontology to support computable phenotype development and phenotype-genotype correlation studies. There is typically a usability and analytic trade-off between granular ontologies (eg, SNOMED-CT) and higher-level ontologies (eg, PheCode, CCS).27–29 Granular ontologies better support cohort definitions for study design however aggregated ontologies provide improved power and performance for developing high-dimensional machine learning risk prediction.30,31 i2b2 also provides the ability to share queries within study groups and within all users of this system. Users can drag over queries, ontology items and patient sets into Workplace folders for sharing.
Data updates and quality
The Biobank Portal data are rebuilt on a weekly basis. A set of ETL scripts run every Tuesday to load the CDM-ingested data sources. A set of data quality scripts is run on Wednesday to ensure all data are populated and values fall within the expected ranges and have increased or remained stable compared with the previous build. Any data quality variances are dealt with on Wednesdays and Thursdays before the new build is promoted to production on Friday mornings. Additional data quality and end-to-end testing are conducted on Friday after each build is promoted to production. The data quality workflow is integral to avoiding data errors in the setting of ingesting and harmonizing disparate data sources.
Exporting analysis datasets
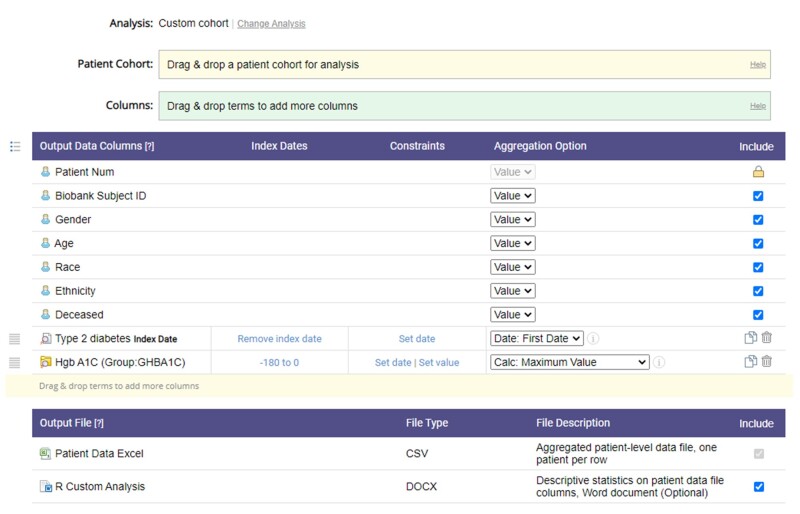
The Biobank Portal provides 2 formats for data export: a 1 row per patient “standard” analysis file (configured to be downloaded immediately as a HIPAA-coded limited dataset) or a complete and detailed data longitudinal file that includes identifiers but must have an attached IRB protocol. All users must sign a data use agreement at first login which allows access to the limited dataset available in the portal. Users can download data for any patient with data in the portal. The analysis file download tool provides many options for the user to construct an analysis file. Users select a patient cohort (defining the rows in the file) and then can drag over concepts from the ontology (columns in the file) and select aggregation options to present as values in the cells. Aggregation options vary by data type and can include count of events, count of unique dates, most recent date, mean, median, max, min, and most recent values (for numeric concepts) or simply the presence or absence of a concept. This format was used in the 2018 Biobank Disease Challenge, an open competition for developing computable phenotypes from Biobank Portal data.32
A new version of the download tool also allows users to define an index date from a concept and constrain other concepts based on the index date. For example, users can define the index date as the first diagnosis of diabetes and then define a column with the maximum hemoglobin A1C value within 6 months prior to the index date.
The detailed data format provides tables with integrated metadata and the full-time series data. It can contain full-text notes and genetic sequences. Because it is difficult to provide this kind of data as a limited dataset, we require every study to make a request only after obtaining IRB approval for the study. Users request PHI data, including clinical notes, using a detailed data request wizard that requires IRB approval and principal investigator sign-off. Each request is reviewed and validated. Data are delivered via secure, encrypted network file shares. It takes 2–3 days to put all the data together for these requests as opposed to the instantly available limited dataset requests. The detailed requests require more back-end informatics resources to fulfill.
RESULTS
As of July 2021, there are 125 645 consented patients enrolled in the MGB Biobank with data in the Biobank Portal. Enrolled patients tend to be older, more likely to be female and white non-Hispanic and have significantly higher healthcare utilization compared with the overall patient population (Table 1).
Table 1.
Mass General Brigham Biobank Participant Characteristics compared with all health system patients.
| Characteristica | Biobank Portal (N = 125 645) | All other patients (N = 3 811 544)b | P-valuec |
|---|---|---|---|
| Demographics | |||
| Gender | <.001 | ||
| Female | 71 360 (57%) | 2 094 150 (55%) | |
| Male | 54 234 (43%) | 1 ,716 594 (45%) | |
| Other/Unknown | 4 (<0.1%) | 800 (<0.1%) | |
| Age at last visit | 59 (42, 70) | 46 (26, 64) | <.001 |
| Race | <.001 | ||
| Asian | 3785 (3.0%) | 190 804 (5.0%) | |
| Black | 5960 (4.7%) | 225 ,069 (5.9%) | |
| Other | 7711 (6.1%) | 681 710 (18%) | |
| Unknown | 2251 (1.8%) | 121 ,111 (3.2%) | |
| White | 105 891 (84%) | 2 592 850 (68%) | |
| Ethnicity | <.001 | ||
| Hispanic | 3387 (2.7%) | 214 407 (5.6%) | |
| Non-Hispanic | 122 211 (97%) | 3 597 137 (94%) | |
| ACS median income4 | $70 245 ($57 313, $90 673) | $69 576 ($55 652, $88 829) | <.001 |
| Healthcare utilization | |||
| Number of visit days | 125 (44, 268) | 9 (2, 38) | <.001 |
| Number of diagnosis codes | 405 (138, 962) | 29 (7, 124) | <.001 |
| Number of clinical notes | 124 (38, 314) | 20 (6, 70) | <.001 |
| Number of diagnostic reports | 122 (47, 262) | 18 (4, 60) | <.001 |
| Available data | |||
| Electronic health records | 124 760 (99%) | 3 811 544 (100%) | — |
| Health information survey | 55 121 (44%) | — | — |
| Genomic data | 43 552 (35%) | — | — |
| Biospecimens | 88 527 (71%) | — | — |
N (%) or median (IQR).
Other patients are defined as patients with a health system visit since 2010 and not enrolled in the MGB Biobank.
Pearson’s Chi-squared test or Wilcoxon rank-sum test.
2018 American Community Survey 2018 Median income in patients zip code.
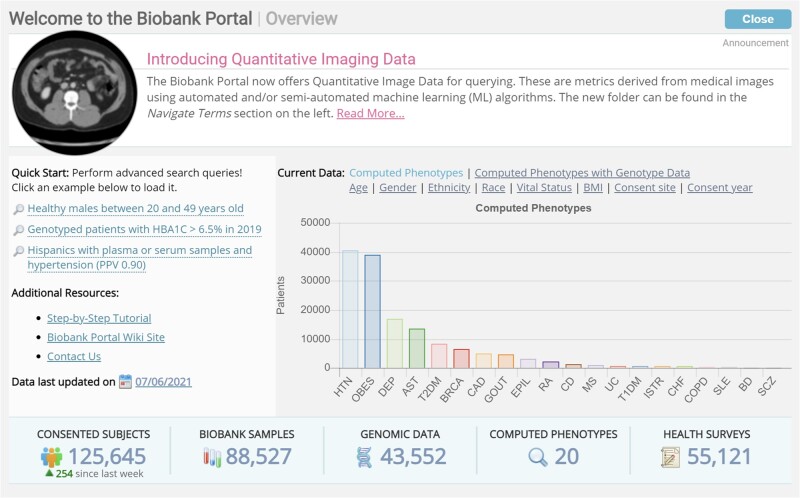
Of the Biobank-enrolled patients, 88 527 (70.5%) have a biospecimen, 55 121 (43.9%) have completed the health information survey, 43 552 (34.7%) have genomic data and 124 760 (99.3%) have linked EHR data. We have deployed 20 machine learning computed disease phenotypes that are calculated on a weekly basis. Figure 2 illustrates characteristics of the Biobank population and data status in an overview screen all users see when logging in to the Biobank Portal.
Figure 2.
Overview of Biobank Portal Data. Investigators see this screen at every login with information on available data, date of last update help, and quick start query examples.
Since its inception in 2015, the Biobank Portal has been rebuilt 327 times. There are currently 1220 active Biobank investigators who have run over 58 793 patient queries and exported over 10 257 analysis files (Figure 3 shows an example analysis file specification). The Biobank Portal has been used for a diverse set of research projects and use cases leading to many awarded research grants and publications. Table 2 lists example research use cases and publications generated using Biobank Portal data. Active research initiatives using Biobank and specimen are available at (https://biobank.massgeneralbrigham.org/research-initiatives).
Figure 3.
Example analysis file specification to download limited datasets.
Table 2.
Biobank Portal example use cases and publications
| Research use case | Investigator type | Data types |
|---|---|---|
| Research feasibility for grant application | All types | All |
| Multicenter genome-wide association studies33 | Clinical/bioinformatics | EHR and Genomics |
| Machine learning disease subgroup detection using NLP and genetics34 | Data scientist | EHR, Genomics, and notes |
| Polygenic risk score integration with EHR data for phenotyping35 | Psychology fellow | EHR, Genomics, and Health Survey |
| Population cohort discovery based on gene variants and laboratory results | Population epidemiologist | Genomic and EHR |
| Obtain biospecimens for control group | Basic scientist | Biospecimen |
| Developing and validating phenotype algorithms36 | All types | EHR and notes |
| Case-control association study of disease comorbidity37 | Population epidemiologist | EHR |
| Evaluating the clinical utility of polygenic risk scores38 | Clinical/bioinformatics | Genomics |
DISCUSSION
Integrating and harmonizing disparate data sources into one interoperable and consolidated data portal is critical to enabling researchers to conduct analysis efficiently and effectively. The Biobank Portal allows a wide variety of researchers to conduct study feasibility by querying across these data sources and then extract data that are most useful to them. Democratizing the data retrieval process by providing tools to bridge the data acquisition and engineering gap has proven to be an effective model for Mass General Brigham. Providing informatics infrastructure support centrally using i2b2 and avoiding data gatekeeping limited to highly technical users have scaled more effectively and generated meaningful publications, many of which the informatics team has not been directly involved with. Secondary use of EHR and other data for which there were no predefined outcomes is a key strength of this data portal and federated analysis model.
Large biobank and consortium efforts collecting similar types of data have also provided similar functionality to the Biobank Portal. The eMERGE network provides a “record counter” for submitted phenotype data that allows researchers to query demographic and diagnosis and procedure data using simple queries.39 The All of Us project also provides a data explorer to view data availability across various data types including survey and digital health data.40 These approaches rely on standard coding systems only and lack the flexibility of the ontology-driven i2b2 queries. The Observational Health Data Science Initiative provides a number of analytic tools for study cohort feasibility and generation focused primarily on EHR data with some extensions into NLP and other data.41 This approach requires a universal up-front agreement to define every element of the data model. This is less flexible than the i2b2 method of defining the data model through the ontology. The opportunity to derive queries through the ontology and quickly create complex multimodal queries has not been achieved with the OMOP model extensions and requires advanced analytic expertise unlike i2b2 where the ontology-driven queries allow any kind of extensions to be easily incorporated into query tools.
The Biobank Portal does have some limitations. First, the data integration workflow is dependent on the existence of a single master patient index. The Biobank Portal relies on the MGB EMPI but this may not be available in all cases or in multisite data repositories. Emerging work in privacy-preserving patient linking enables cross-site patient identification using hashed tokens generated from patient identifiers (names, dates of birth, address, etc.) and would enable data integration across sites and data sources.42 In addition, the Biobank population is a relatively small population compared with a large multisite study or all patients in a healthcare system. However, we have shown that the i2b2 meta-model star schema overall will scale to much larger populations. Finally, we find that the Biobank population is highly enriched for non-Hispanic white patients than the general health system patient population. This may lead to bias and poor generalizability both in data completeness and analytic findings. Additional efforts to enroll more representative participants are required for all patients to realize the benefit of research findings.
CONCLUSION
The Biobank Portal and other patient data portals that integrate complex and simple datasets enable a diverse set of research use cases. i2b2 tools to implement these registries are open source and freely available. While institutions require substantial informatics resources to establish and maintain these types of data registries, they yield significant research value to a wide range of investigators.
FUNDING
This study was supported by Mass General Brigham institutional funds. The work was supported by the National Human Genome Research Institute grants R01-HG009174 and U01HG008685-05S1 and the National Heart, Lung, and Blood Institute award 1OT2HL161841-01. The authors had the final responsibility for the decision to submit for publication.
AUTHOR CONTRIBUTIONS
VMC was the primary author of this article. VSG, NW, and SNM also contributed substantially to the article. BB, AC, BG, SG, RM, HP, DW, MM, MR, and CH all contributed intellectual value, technical support, and text to the article.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the contributions of Mass General Brigham Biobank participants—without them this work would not be possible. We’d also like to acknowledge the research coordinators and the Biobank team for their work in enrolling patients and collecting data. Finally, we’d like to acknowledge the RPDR team for their work maintaining the enterprise research patient data warehouse.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The limited dataset underlying this article cannot be shared publicly due to privacy of individuals that participated in the study. Deidentified data and samples may be shared in collaboration with a Mass General Brigham investigator. A subset of genotype and phenotype data is available on dbGAP at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001584.v2.p2.
REFERENCES
- 1. Thiese MS. Observational and interventional study design types; an overview. Biochem Med (Zagreb) 2014; 24 (2): 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 2016; 70: 214–23. [DOI] [PubMed] [Google Scholar]
- 3.All of Us Research Program Investigators. The “All of Us” Research Program. N Engl J Med 2019; 381: 668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018; 562 (7726): 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oelsner EC, Allen NB, Ali T, et al. Collaborative Cohort of Cohorts for COVID-19 Research (C4R) Study: Study Design. medRxiv Published Online First: March 20, 2021. doi: 10.1101/2021.03.19.21253986. [DOI] [PMC free article] [PubMed]
- 6. Bild DE, Detrano R, Peterson D, et al. Ethnic differences in coronary calcification: the Multi-Ethnic Study of Atherosclerosis (MESA). Circulation 2005; 111 (10): 1313–20. [DOI] [PubMed] [Google Scholar]
- 7. Yamanaka H, Tanaka E, Nakajima A, et al. A large observational cohort study of rheumatoid arthritis, IORRA: providing context for today’s treatment options. Mod Rheumatol 2020; 30 (1): 1–6. [DOI] [PubMed] [Google Scholar]
- 8. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 17 (2): 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016; 3: 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karlson EW, Boutin NT, Hoffnagle AG, et al. Building the partners healthcare biobank at partners personalized medicine: informed consent, return of research results, recruitment lessons and operational considerations. J Pers Med 2016; 6 (1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boutin NT, Mathieu K, Hoffnagle AG, et al. Implementation of electronic consent at a Biobank: an opportunity for precision medicine research. J Pers Med 2016; 6 (2): 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mozilla Public License, version 2.0. https://www.mozilla.org/en-US/MPL/2.0/ Accessed July 13, 2021.
- 13. Kimball R, Ross M.. The Data Warehouse Toolkit: The Complete Guide to Dimensional Modeling. Indianapolis, IN: John Wiley & Sons; 2011. [Google Scholar]
- 14. Boutin N, Holzbach A, Mahanta L, et al. The information technology infrastructure for the translational genomics core and the Partners Biobank at Partners Personalized Medicine. J Pers Med 2016; 6 (1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hripcsak G, Duke JD, Shah NH, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
- 16. Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42 (2): 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Infinium Multi-Ethnic Global-8 Kit. https://www.illumina.com/products/by-type/microarray-kits/infinium-multi-ethnic-global.html Accessed July 12, 2021.
- 18. Hripcsak G, Albers DJ.. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2013; 20 (1): 117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Cai T, Yu S, et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019; 14 (12): 3426–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bridge CP, Rosenthal M, Wright B, et al. Fully-automated analysis of body composition from ct in cancer patients using convolutional neural networks In: Or 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis. Cham, Switzerland: Springer International Publishing; 2018: 204–13. [Google Scholar]
- 21. Magudia K, Bridge CP, Bay CP, et al. Population-scale CT-based body composition analysis of a large outpatient population using deep learning to derive age-, sex-, and race-specific reference curves. Radiology 2021; 298 (2): 319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ICD—ICD-10-CM—International Classification of Diseases, Tenth Revision, Clinical Modification. 2021. https://www.cdc.gov/nchs/icd/icd10cm.htm Accessed July 13, 2021.
- 23.American Medical Association. CPT® (Current Procedural Terminology). https://www.ama-assn.org/amaone/cpt-current-procedural-terminology Accessed July 13, 2021.
- 24. Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc 1998; 5 (3): 276–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu S, Ma W, Moore R, et al. RxNorm: prescription for electronic drug information exchange. IT Prof 2005; 7: 17–23. [Google Scholar]
- 26. Smith MW, Joseph GJ.. Pharmacy data in the VA health care system. Med Care Res Rev 2003; 60 (3 Suppl): 92S–123S. [DOI] [PubMed] [Google Scholar]
- 27. Wu P, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform 2019; 7 (4): e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical Classifications Software Refined (CCSR). https://www.hcup-us.ahrq.gov/toolssoftware/ccsr/ccs_refined.jsp Accessed July 13, 2021. [DOI] [PMC free article] [PubMed]
- 29. Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud Health Technol Inform 2006; 121: 279–90. [PubMed] [Google Scholar]
- 30. Hong C, Rush E, Liu M, et al. Clinical Knowledge Extraction via Sparse Embedding Regression (KESER) with Multi-Center Large Scale Electronic Health Record Data. medRxiv Published Online First: 2021. https://www.medrxiv.org/content/10.1101/2021.03.13.21253486v1.abstract. [DOI] [PMC free article] [PubMed]
- 31. Rasmy L, Tiryaki F, Zhou Y, et al. Representation of EHR data for predictive modeling: a comparison between UMLS and other terminologies. J Am Med Inform Assoc 2020; 27 (10): 1593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winners Announced from the first “Biobank Disease Challenge.” 2018. https://rc.partners.org/news-events/announcements/winners-announced-first-biobank-disease-challenge Accessed July 13, 2021.
- 33. Bonde A, Gaitanidis A, Breen K, et al. Identification of a new genetic variant associated with cholecystitis: a multicenter genome-wide association study. J Trauma Acute Care Surg 2020; 89 (1): 173–8. [DOI] [PubMed] [Google Scholar]
- 34. McCoy TH Jr, Castro VM, Hart KL, et al. Genome-wide association study of dimensional psychopathology using electronic health records. Biol Psychiatry 2018; 83 (12): 1005–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheutlin AB, Dennis J, Karlsson Linnér R, et al. Penetrance and pleiotropy of polygenic risk scores for Schizophrenia in 106,160 patients across four health care systems. Am J Psychiatry 2019; 176 (10): 846–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu SH, Wan ES, Cho MH, et al. An independently validated, portable algorithm for the rapid identification of COPD patients using electronic health records. Sci Rep 2021; 11 (1): 19959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kronzer VL, Huang W, Zaccardelli A, et al. Association of sinusitis and upper respiratory tract diseases with incident rheumatoid arthritis: a case-control study [published online ahead of print October 15, 2021]. J Rheumatol 2021; doi: 10.3899/jrheum.210580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vassy J, Hao L, Kraft P, et al. Clinical validation, implementation, and reporting of polygenic risk scores for common diseases. Research Square Preprint 2021; doi: 10.21203/rs.3.rs-743779/v1. [Google Scholar]
- 39. McCarty CA, Chisholm RL, Chute CG, et al. ; eMERGE Team. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramirez AH, Gebo KA, Harris PA.. Progress with the All of Us research program: opening access for researchers. JAMA 2021; 325 (24): 2441–2. [DOI] [PubMed] [Google Scholar]
- 41.OHDSI. The Book of OHDSI: Observational Health Data Sciences and Informatics. OHDSI; 2019. https://ohdsi.github.io/TheBookOfOhdsi/ Accessed July 13, 2021. [Google Scholar]
- 42. Bian J, Loiacono A, Sura A, et al. Implementing a hash-based privacy-preserving record linkage tool in the OneFlorida clinical research network. JAMIA Open 2019; 2 (4): 562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The limited dataset underlying this article cannot be shared publicly due to privacy of individuals that participated in the study. Deidentified data and samples may be shared in collaboration with a Mass General Brigham investigator. A subset of genotype and phenotype data is available on dbGAP at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001584.v2.p2.