Abstract
Objective
The Greater Plains Collaborative (GPC) and other PCORnet Clinical Data Research Networks capture healthcare utilization within their health systems. Here, we describe a reusable environment (GPC Reusable Observable Unified Study Environment [GROUSE]) that integrates hospital and electronic health records (EHRs) data with state-wide Medicare and Medicaid claims and assess how claims and clinical data complement each other to identify obesity and related comorbidities in a patient sample.
Materials and Methods
EHR, billing, and tumor registry data from 7 healthcare systems were integrated with Center for Medicare (2011–2016) and Medicaid (2011–2012) services insurance claims to create deidentified databases in Informatics for Integrating Biology & the Bedside and PCORnet Common Data Model formats. We describe technical details of how this federally compliant, cloud-based data environment was built. As a use case, trends in obesity rates for different age groups are reported, along with the relative contribution of claims and EHR data-to-data completeness and detecting common comorbidities.
Results
GROUSE contained 73 billion observations from 24 million unique patients (12.9 million Medicare; 13.9 million Medicaid; 6.6 million GPC patients) with 1 674 134 patients crosswalked and 983 450 patients with body mass index (BMI) linked to claims. Diagnosis codes from EHR and claims sources underreport obesity by 2.56 times compared with body mass index measures. However, common comorbidities such as diabetes and sleep apnea diagnoses were more often available from claims diagnoses codes (1.6 and 1.4 times, respectively).
Conclusion
GROUSE provides a unified EHR-claims environment to address health system and federal privacy concerns, which enables investigators to generalize analyses across health systems integrated with multistate insurance claims.
Keywords: obesity, electronic health records, Centers for Medicare and Medicaid Services, PCORnet, Patient-Centered Outcomes Research Institute, cloud computing, Amazon Web Services private cloud
INTRODUCTION
In 2014, the Patient-Centered Outcomes Research Institute (PCORI) funded PCORnet to create a national research infrastructure for conducting patient-centered comparative effectiveness research (CER) using electronic health data.1 The Greater Plains Collaborative (GPC),2,3 a PCORnet Clinical Data Research Network (CDRN) currently including 12 health systems in 9 states, leverages patient engagement and informatics infrastructure developed through the National Institutes of Health (NIH) Clinical and Translational Science Award programs.4 PCORI’s funding announcement5 required CDRNs aggregate complete and comprehensive longitudinal data for a large, diverse population over 1 million individuals, and create 3 longitudinal cohorts (1 rare disease, 1 common condition, and 1 obesity). The GPC cohorts focus on amyotrophic lateral sclerosis, breast cancer, and the consequences of unhealthy versus healthy weight. In order to assess outcomes for patients who may not remain under a single health system’s care, PCORI required CDRNs to develop strategies for integrating insurance claims. Although the GPC’s breadth across 9 states is advantageous for generalizing research findings, insurance carriers vary extensively; leaving integrating Center for Medicare and Medicaid Services (CMS) claims as the most consistent initial claims strategy. CMS allows qualified organizations to access their identifiable data files, also known as research identifiable file (RIF) data, for research purposes6 and CMS funds the Research Assistance Data Center (ResDAC) to assist investigators interested in studying CMS data. The execution of this strategy was the creation of the GPC Reusable Observable Unified Study Environment (GROUSE).
In conceiving GROUSE to support PCORI’s objectives, we established an institutional review board (IRB) protocol for the 3 cohorts with broad aims to: (1) characterize the increase in data completeness and comprehensiveness provided through claims integration to provide a more “complete” picture of our patient’s health; (2) evaluate the distributions of health and care processes for the patients with our 3 conditions and their treatment patterns within the GPC versus the larger Medicare and Medicaid populations in our region to understand how studies of the GPC population generalize to the broader populations in our states; (3) use CMS claims data to enhance quality control processes for aggregating health system-derived clinical data and establish correlations with CMS claims data for health system-derived data to support trial recruitment and observational studies, that is, validating the use of EHR data for recruitment.
Although GPC supports distributed analyses via PCORnet and other national consortia (eg, ACT,7 4CE8), GROUSE provides a centralized data platform for the following reasons: (1) there are certain type of analyses that are less practical under the distributed framework, especially with time constraints,9 for example, such studies that requiring rapid feedback and frequent iterations; (2) centralizing data streamlines model validation computationally; (3) centralizing simplifies developing novel machine learning models (eg, transfer learning,10 deep learning11) which require iteration; (4) centralizing aids evaluating and minimizing the presence of “batch effects,” defined as technical or biological artifacts due to different data provenance12 that impact study generalizability;13 (5) centralizing provides consistent data management to meet federal requirements.
Cloud computing has been increasingly adopted by the academic community for cancer and genomic studies14–17 such as All-of-Us18 and more recently N3C9 and C3AI19 data lakes to support observational researches for coronavirus disease (COVID-19). Although initially deployed on premise, GROUSE was redeployed using Amazon Web Services (AWS) and the Snowflake data platform to provide scalable storage and computation for multisite collaboration, best practices for security and privacy protection, and to leverage synergies with partners using AWS and Snowflake (eg, Cerner,20 Foundation Medicine21) CMS’ data distributor (ie, NewWave-GDIT22) is migrating their data environment to AWS which may streamline data access in the future.
In this study, we outline the GROUSE cloud-based data enclave’s governance, architecture, and compliance components: including interagency agreements, facilitating health system collaboration, and ensuring security and privacy align with federal requirements and industry best practices. We highlight administrative and technical practices and report briefly GROUSE’s analytic capacity regarding obesity as a case study.
DATA GOVERNANCE AND ACQUISITION
GPC has established a governance framework, policies, and procedures at both site-level and network-level to oversee the use of electronic health record (EHR) data, with the goal of promoting collaboration while preserving data security and patient privacy.3 This framework includes:(1) a federally compliant data and analytical environment, (2) IRBs reciprocity,23 (3) a GPC data sharing agreement and data request oversight committee,24 and (4) data use agreements and security/privacy control requirements set forth by CMS for acquiring RIF files.
IRB oversight
The SMART IRB23 Master Common Reciprocal reliance agreement is used to create a central IRB for the GPC Coordinating Center (CC) at the University of Missouri. This initial IRB approval is intended to cover both contribution of data to GROUSE as well as research using GROUSE data for the 3 predefined cohorts approved by key stakeholders.
CMS data management plan
Organizations preparing a CMS Data Use Agreement (DUA) application using RIF data25 must complete a Data Management Plan Self-Attestation Questionnaire (DMP SAQ) to demonstrate compliance with CMS security and privacy requirements.26,27 This new procedure piloted by CMS in 2020 provides more structured and consistent guidance on DMP development and addresses cloud computing. The DMP SAQ better matches with the National Institute for Standards and Technology Special Publication (NIST SP 800-53),28 which dictates the necessary security and privacy controls for federal information systems and provides organizations and a process for selecting controls to protect organizational operations and assets. The DMP SAQ is reviewed by the CMS Data Privacy Safeguard Program (DPSP)29 consisting of third-party auditors from MBL technologies26,30 and its subcontractors. Our DMP SAQ process included the steps:
Risk Categorization and Data Classification: we started with classifying data based on Federal Information Processing Standards 199 and 20031,32 with our existing institutional Data Classification Level policy.33
Control Selection and Implementation: we then identified and adopted required controls specified in DMP SAQ from 18 control families established in NIST SP 800-53 Rev. 4 guidelines, as the official policy for the GROUSE infrastructure (Supplementary Appendix SA). These controls are applied to not only the system housing the sensitive data, but also enterprise functions supporting it (eg, separation of responsibility, control management).
System Security Plan: we developed a complete system security plan which entails how the NIST requirements will be met and completed the DMP SAQ.
Evidence Gathering and Independent Assessment: once the system is built based on the system security plan, we collected evidence and engaged the CMS DPSP to perform independent assessment. CMS provides an information technology concierge who provides feedback during the development of the DMP SAQ.
Privacy-preserving-data linkage
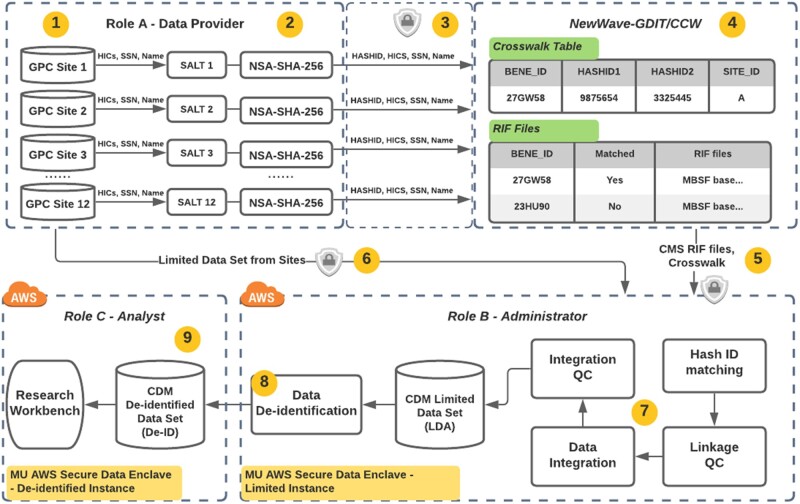
Meeting federal data management requirements, patient privacy, and reducing institutional risk while meeting PCORI contractual requirements and timelines were key considerations in the technical architecture. Claims obtained in RIF format have record linkage services provided by the CMS contractor, NewWave-GDIT. As shown in Figure 1, participating health system “sites” generate finder files which include multiple primary identifiers such as health insurance claim numbers, social security numbers, and multiple secondary identifiers such as date of birth and gender along with a 1-way hash of identifiers (hashID). It is worth noting that the multiplicity of primary and secondary identifiers is to minimize ambiguity and warranty high linkage accuracy. Finder files are then encrypted with an Advanced Encryption Standard of at least 256-bit encryption algorithm34 and sent to NewWave-GDIT but not to the GPC CC, so the integrated repository doesn’t include highly sensitive patient information. This reduces risk for organizations sharing their data and the receiving institution integrating and safeguarding data derived from millions of patients. Each GPC site sends their limited data sets (eg, PCORnet Common Data Model [CDM]35,36 and Informatics for Integrating Biology & the Bedside schemas37 holding tumor registries and additional data) along with a crosswalk file that contains the hashID, patient number, and the offset of days used for each patient to deidentify their records. GPC CC integrates the crosswalk and claims data provided by NewWave-GDIT with individual site EHR data (limited data set) by linking the hashed IDs and stages the data in databases. External investigators access the deidentified data set and not the limited data residing in the data integration environment.
Figure 1.
Privacy-Preserving Data Linkage between Center for Medicare and Medicaid Services (CMS) claims and EHR data. (1) Each participating Greater Plains Collaborative Greater Plains Collaborative (GPC) site uses its EHR data, to define patients for linkage to CMS data. (2) GPC sites generate a unique hashed ID for each patient. (3) Each GPC site sends “finder files” combining multiple primary and secondary identifiers and hashed IDs to NewWave-GDIT/Chronic Condition Data Warehouse (CCW) following a well-established encryption procedure. (4) NewWave-GDIT/CCW uses the set of identifiers from each of the GPC sites to generate a cross walk file that maps between the hashed IDs and the GPC Reusable Observable Unified Study Environment-specific BENE_ID. (5) NewWave-GDIT/CCW creates an extract of CMS data specific to the states encompassing the GPC sites. The resulting files are sent by NewWave-GDIT/CCW to the GPC CC via encrypted external media (6) GPC Coordinating Center (CC) receives Limited Data Sets containing EHR data from each of the GPC sites along with the hashed IDs sent to NewWave-GDIT/CCW. (7) GPC CC will then use the hashed IDs to link the patient records received from NewWave-GDIT/CCW with the Limited Data Sets received from each site. (8) Each merged data set is deidentified by GPC CC via dynamic views and made available to the collaborating investigators that are listed within the protocol. (9) No identifiers are retained by GPC CC after creation of the deidentified data set. The GPC site data may be refreshed over time upon agreement across sites. CMS data may be refreshed when new data becomes available. For this data refresh, individual sites will either use the same hashed IDs previously used for its patients so that they are linked automatically over time, or if a site chooses to use a different hashed ID for the refresh, then they will provide GPC CC a mapping between the previous hashed ID and the new hashed ID.
Data access request oversight
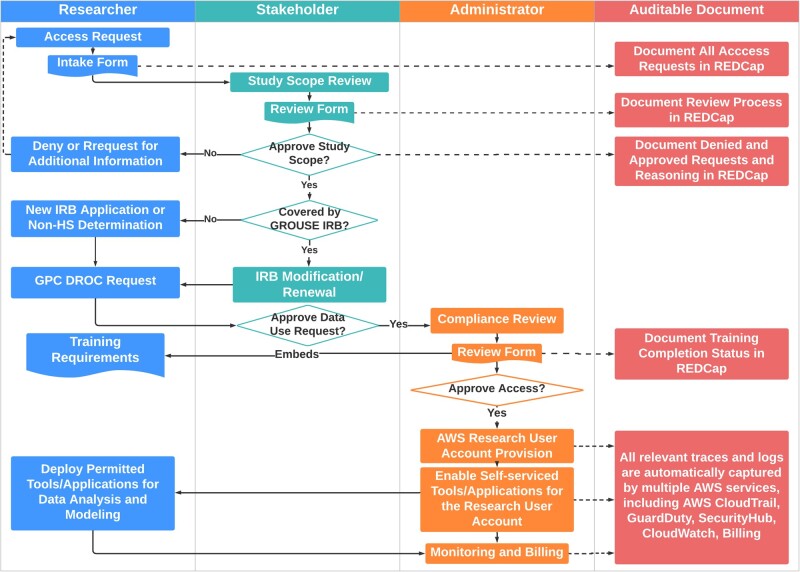
Figure 2 shows the REDCap38 supported multi-stakeholder workflow for provisioning access to GROUSE. Researchers first submit an Access Request Intake Form and trigger “Study Scope Review,” which is performed by designated stakeholders who determine: (1) whether the study can be covered by the scope of GROUSE IRB or a new IRB is needed; (2) the appropriate group/role of the requester. Upon the study scope approval, researchers will submit a GPC Data Request Oversight Committee request to acknowledge participating GPC sites. Then, a compliance review with requirements for CITI Human Subject Research and NIH security and privacy awareness training39 are checked and collected by administrators, as well as signing the data use agreement to affirm agreement to GPC terms and conditions. Finally, an AWS research user account will be provisioned with self-serviced tools and applications enabled. Each step is to support annual review and periodic auditing.
Figure 2.
Multi-Stakeholder Data Access Governance Model. A data access request starts from researcher submitting an Access Request Intake Form and trigger “Study Scope Review,” which is performed by designated stakeholders who determine: (1) whether the study can be covered by the scope of Greater Plains Collaborative Reusable Observable Unified Study Environment institutional review board (IRB) or a new reuse IRB is needed; (2) the appropriate group/role of the requester. Upon the study scope approval, researchers will submit a GPC Data Request Oversight Committee request to approval from participating GPC sites. Then, a compliance review with requirements for CITI Human Subject Research and NIH security and privacy awareness training are checked and collected by administrators, as well as signing the data use agreement to affirm agreement to GPC terms and conditions. Finally, an AWS research user account will be provisioned with self-serviced tools and applications enabled. Each step is to support for annual review and periodic auditing.
CLOUD ARCHITECTURE AND COMPONENTS
Software as a Service, Platform as a Service, and Infrastructure as a Service
AWS cloud as well as other cloud providers usually offer 3 types of service models—Software as a Service (SaaS), Platform as a Service (PaaS), and Infrastructure as a Service (IaaS), and customer can select 1 or a mixture to formulate the best solution for their needs. As shown in Figure 3, SaaS enables the customers to use the cloud provider’s applications that are running on a provider’s infrastructure, whereas PaaS enables consumers to create or acquire applications and tools and to deploy them on the cloud provider’s infrastructure. IaaS enables a consumer to provision processing, storage, networks and other fundamental computing resources. GROUSE was built based on a mixture of the 3 service models. Our configuration considered (sorted by importance): (1) Data Security; (2) Controllability/Auditability; (3) Operational Simplicity and Rapid Deployment; (4) Integrability; (5) Customizability/Flexibility; (6) Scalability; (7) Cost-effectiveness. Data security requires high controllability and flexibility over the underlying infrastructure. As a result, we chose the IaaS model for the architectural foundation (ie, landing zone) and security baseline.40 We also had a pressing need to rapidly deploy the environment and a preference of operational simplicity, so we chose to leverage PaaS services (eg, AWS Service Workbench,41 AWS Fargate42) and SaaS services (eg, Snowflake data warehouse,43 CloudCheckr44) to orchestrate the upper layers of the system from operating system to applications.
Figure 3.
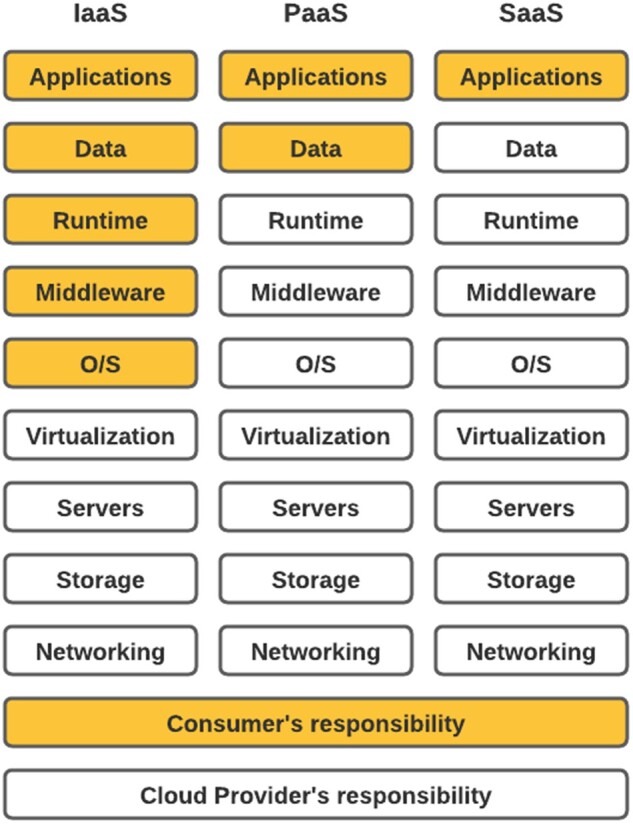
Infrastructure as a Service (IaaS) versus Platform as a Service (PaaS) versus Software as a Service (SaaS) cloud service models. The modules highlighted in yellow are consumer’s responsibility, while the white modules are cloud provider’s responsibility. SaaS enables the customers to use the cloud provider’s applications/software that are running on a provider’s infrastructure, whereas PaaS enables consumers to create or acquire applications/software and tools and to deploy them on the cloud provider’s infrastructure. IaaS enables a consumer to provision processing, storage, networks and other fundamental computing resources.
Security at scale
GROUSE is protected through a defense-in-depth architecture following the AWS “Shared Responsibility” model,45 with AWS being responsible for the “Security of the Cloud” and the GPC CC taking responsibility for the “Security in the Cloud” (SIC). To ensure SIC, we adopted the following DevSecOps46,47 best practices by leveraging tools for Infrastructure as code (IaC)48: (1) automating and centralizing Identity and Access Management; (2) automating security tasks and compliance assessments; (3) enforcing policies in a hierarchical fashion. IaC is the process of managing and provisioning information systems through machine-readable definition files, rather than physical hardware configuration or interactive configuration tools.48
Cloud architecture
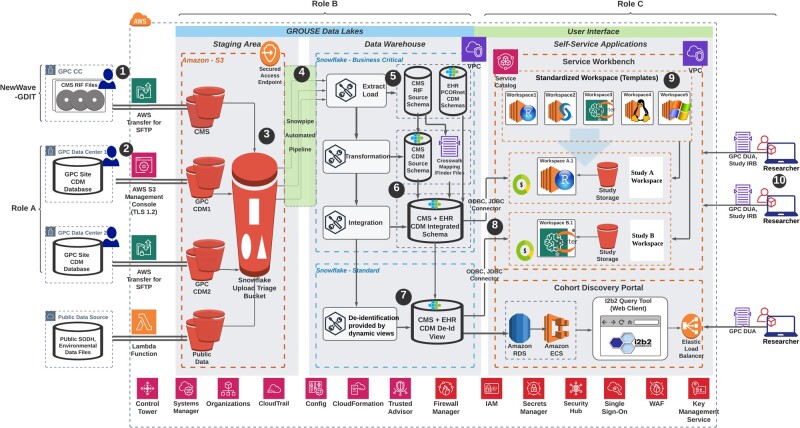
Figure 4 illustrates system architecture of the GROUSE environment, which is composed of a data lake, a data warehouse, and analytic workbenches. (1) “Data Lake”: data (including GDIT physical media) are loaded into secure S3 buckets via Secure Shell File Transfer Protocol49 or Transport Layer Security (TLS) 1.2 Protocol.36 (2) “Data Warehouse”: data are extracted and loaded into Snowflake for data transformation into the PCORnet CDM and deidentification. (3) “Analytic Workbench”: to minimize the burden on researchers of learning to navigate the cloud environment, we adopted an AWS solution—service workbench,41 where approved users can self-service to deploy either Windows or Linux analytic “workspaces” of multiple analytical applications (eg, R, Python, SAS) and varying computing power based upon their needs. From each analytical “workspace,” a dedicated connection can be created to the backend GROUSE database where researchers have full visibility to multiple schemas and can choose to either query from the original CMS schema or a transformed CDM schema.
Figure 4.
Data that flow from multiple sources, including (1) NewWave-GDIT physical media and (2) other Greater Plains Collaborative sites will be load into secured S3 bucket via Secure File Transfer Protocol or using AWS S3 management console (TLS 1.2). (3) Raw files are externally staged in S3 buckets and then loaded into Snowflake data warehouse via (4) the Snowpipe automated pipeline (a Snowflake functionality). (5) Data in source Center for Medicare and Medicaid Services (CMS) research identifiable file or site Common Data Model (CDM) schema are first extracted as they are in 1 database. (6) CMS data will then be transformed into PCORnet CDM and integrated with electronic health record data using the finder file provided by CMS. (7) The integrated CDM will be deidentified using the built-in dynamic view functionality provided by Snowflake. (8) Both the limited and deidentified view can be accessed via ODBC or JDBC connector with researchers’ service workbench workspaces. (9) Service workbench provides templated and reusable workspaces (AWS EC2 instances) with various computing power, operating systems and prepackaged software that can satisfy most of the research needs. (10) Approved researchers can deploy the self-serviced applications to perform either advanced analysis using the service workbench or simply discover study cohort using an integrated Informatics for Integrating Biology & the Bedside query tool. Various underlying Amazon Web Services are marked at each step described above as well as at the bottom of the figure.
CASE STUDY: OBESITY COHORT
As the largest population defined in our approved CMS DUA, we provide our obesity cohort as a case study to demonstrate how integrating claims with EHRs can improve data completeness for cross-walked populations and provide comparison with the broader population outside the GPC participating health systems. We define the group of Medicare or Medicaid beneficiaries as the “CMS Cohort”; those patients with at least 1 valid diagnosis code observable from the GPC EHR data as the “Observable Cohort”; and the subpopulation of the observable cohort with cross-walked Medicare of Medicaid claims available as the “Crosswalk Cohort.”
Study population and covariates
The body weight cohort consisted of yearly patient cross-sections with at least 1 diagnosis code during 2011–2016 from 7 GPC sites participating in this study. At the time of this study, GROUSE incorporated fee-for-service Medicare claims from 2011 to 2016 but only Medicaid claims from 2011 to 2012 were included due to delays in several states providing their Medicaid claims via ResDAC50 (CMS file types available are detailed in Supplementary Appendix SB). Site-level EHR data analyses leveraged PCORnet CDM databases. Individuals under 2 years of age were excluded because obesity is not typically diagnosed in this population. Obesity is identified either by physical measurement, i.e., body mass index (BMI) or BMI percentile, or International Classification of Disease (ICD) diagnosis codes:
Physical obesity. Following the recommended Centers for Disease Control and Prevention (CDC) growth charts which defined for children and teens ages 2 through 19 years, we identified obesity for population under 19 years old by BMI Percentile ≥ 95 for population under 19 years old.51 For population above 19 years old, we defined obesity as BMI ≥ 30 kg/m2.
Obesity diagnosis codes. ICD-9-CM (278.00, 278.01, 278.03) or ICD-10-CM (E66.0, E66.01, E66.09, E66.1, E66.2, E66.8, E66.9, and Z68.30-Z68.45).
Physical measurement is only contained in EHR from participating healthcare systems, whereas diagnosis codes are available from the participating GPC sites’ EHRs as well as from Medicare and Medicaid claims. In addition, we also collect demographic information (available in both EHR and claims) as well as multiple comorbidities for the obese cohort using ICD codes.
Analysis
Yearly prevalence rates of obesity were estimated and compared between the overall claim population and the cross-walked GPC cohort. We also estimated and compared the prevalence rates of obesity using different markers: physical measurement (“BMI-defined obesity”), diagnosis code assignment in claims, or code assignment in sites’ EHRs (“Code-defined Obesity”). Obesity coding relative to physical measurement was calculated using the sites EHR data. For the Crosswalk Cohort, we calculated the rates of Code-defined Obesity (1) within the sites’ EHR only, (2) within CMS claims only, and (3) within both EHR and CMS claims. Trends in physical and coded obesity were presented annually from 2011 to 2016. For patients with multiple physical measurements within the same year, we chose to use the highest value to calculate the population-level rates. Finally, we explored claims data augmentation of site EHR data for detecting the presence of sleep apnea and type II diabetes using ICD codes (Supplementary Appendix SC) in obese populations by comparing the prevalence rates of each condition estimated from (1) EHR only, (2) CMS claims only, and (3) either EHR or CMS claims.
RESULTS
GROUSE contained a CMS Cohort with more than 24 million beneficiaries and an Observable Cohort of over 6.6 million patients, as well as a Crosswalk Cohort of 1 674 134 patients. As shown in Table 1, there was a steady increasing trend of both the CMS cohort and Crosswalk Cohort over time and more than 50% of CMS cohort did not have any enrollment gap over the study period (ie, continuously enrolled in Medicare from 2011 to 2016, or Medicaid from 2011 to 2012).
Table 1.
Center for Medicare and Medicaid Services (CMS) Cohort and Crosswalk Cohort sizes by source year and years of continuous coverage
| CMS Cohort |
Crosswalk Cohort |
|||
|---|---|---|---|---|
| Calendar year | Medicare (N) | Medicaid (N) | Medicare (N) | Medicaid (N) |
| 2011 | 8 756 666 | 11 437 745 | 759 438 | 661 435 |
| 2012a | 9 076 119 | 12 287 716a | 802 840 | 607 159a |
| 2013 | 9 353 333 | 837 145 | ||
| 2014 | 9 564 777 | 859 538 | ||
| 2015 | 9 824 974 | 878 290 | ||
| 2016a | 10 684 220a | 831 310a | ||
| At least 1 year enrollment | 12 902 644 | 13 997 184 | 993 396 | 680 738 |
| Years of continuous coverage | ||||
| 1 Year coverage | 1 788 714 (14%) | 4 268 907 (30%) | 21 343 (2%) | 92 882 (14%) |
| 2 (maximum for Medicaid) | 1 066 092 (8%) | 9 728 278 (70%) | 79 547 (8%) | 587 856 (86%) |
| 3 | 1 302 144 (10%) | 99 781 (10%) | ||
| 4 | 987 096 (8%) | 86 124 (9%) | ||
| 5 | 1 067 213 (8%) | 95 321 (10%) | ||
| 6 (maximum for Medicare) | 6 691 385 (52%) | 611 280 (62%) | ||
Due to random data shifting for data deidentification, the population size of the year 2012 for Medicaid population and year 2016 for Medicare population are slightly lower than the actual accounts.
Within the GPC Observable Cohort, there is a total of 3,471,533 (52%) patients with at least 1 observation of physical measurement (ie, weight and height or BMI or BMI percentile), which we define it as the “GPC Weight Cohort.” In Table 2, we reported the demographic changes of GPC Weight Cohort over calendar years. We defined 3 age groups: “age group 1” includes people aged 2–19, which covers the child and teenager group with obesity defined by BMI percentile; for people above 19 years old whose BMI becomes the defining metric for obesity, we further broke the range down into “age group 2,” 20–64 years old, including adult population of working age who are not Medicare eligible; “age group 3”, 65 years old and older, who are Medicare eligible.
Table 2.
Demographic characteristic changes of Greater Plains Collaborative Weight cohort over time
| 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | |
|---|---|---|---|---|---|---|
| N (Total) | 1 951 306 | 2 209 608 | 2 343 943 | 2 490 952 | 2 668 068 | 2 734 073 |
| Age | ||||||
| Age group 1 (2–19) | 460 283 (23.6%) | 532 646 (24.6%) | 555 133 (23.7%) | 565 557 (22.7%) | 581 869 (21.8%) | 600 160 (22%) |
| Age group 2 (20–64) | 1 148 037 (58.8%) | 1 287 471 (59.5%) | 1 365 400 (58.3%) | 1 463 500 (58.8%) | 1 591 222 (59.6%) | 1 592 967 (58.3%) |
| Age group 3 (65) | 342 986 (17.6%) | 342 986 (15.9%) | 423 410 (18.1%) | 461 895 (18.5%) | 494 977 (18.6%) | 540 946 (19.8%) |
| Gender | ||||||
| Female | 1 109 376 (56.9%) | 1 254 592 (56.8%) | 1 336 098 (57%) | 1 373 698 (56.4%) | 1 536 637 (57.6%) | 1 562 339 (57.1%) |
| Male | 841 882 (43.1%) | 954 915 (43.2%) | 1 007 732 (43%) | 1 060 995 (43.6%) | 1 131 297 (42.4%) | 1 171 536 (42.8%) |
| Other | 57 (0%) | 101 (0%) | 113 (0%) | 109 (0%) | 134 (0%) | 198 (0%) |
| Race | ||||||
| White | 1 368 825 (70.1%) | 1 609 826 (72.9%) | 1 735 676 (74%) | 1 840 661 (73.9%) | 1 967 581 (73.7%) | 2 008 078 (73.4%) |
| African American | 196 668 (10.1%) | 242 416 (11%) | 267 422 (11.4%) | 281 333 (11.3%) | 298 807 (11.2%) | 299 407 (11%) |
| Other | 174 331 (8.9%) | 146 986 (6.7%) | 147 143 (6.3%) | 165 797 (6.7%) | 186 941 (7%) | 190 526 (7%) |
| Unknown | 211 482 (10.8%) | 210 380 (9.5%) | 193 702 (8.3%) | 203 161 (8.2%) | 214 739 (8%) | 236 062 (8.6%) |
| Ethnicity | ||||||
| Hispanic | 230 213 (10.8%) | 283 422 (12.8%) | 318 597 (13.6%) | 334 192 (13.4%) | 349 944 (13.1%) | 363 022 (13.3%) |
| Non-Hispanic | 1 342 151 (63.1%) | 1 548 366 (70.1%) | 1 673 609 (71.4%) | 1 794 564 (72%) | 1 928 727 (72.3%) | 1 965 167 (71.9%) |
| Other | 4984 (0.2%) | 5250 (0.2%) | 5877 (0.3%) | 6147 (0.2%) | 6587 (0.2%) | 6552 (0.2%) |
| Unknown | 549 465 (25.8%) | 372 570 (16.9%) | 345 860 (14.8%) | 356 049 (14.3%) | 382 810 (14.3%) | 399 332 (14.6%) |
Across our 7 participating health systems, the GPC Weight Cohort increased from 2011 to 2016, with age group 3 (65 and older) the highest relative growth from 342 986 to 540 946 (58%), resulting in an increase of its proportion from 17.6% to 19.8%. Proportions of the “Unknown” category for race and ethnicity reduced significantly over time from 10.8–8.6% to 25.8–14.6%, respectively. Gender distribution remains consistent over the years with a higher ratio of male within age group 1 (male > 50%) and high ratio of female for age group 2 and 3 (female > 50%; Supplementary Appendix SD).
EHR-based rates of obesity
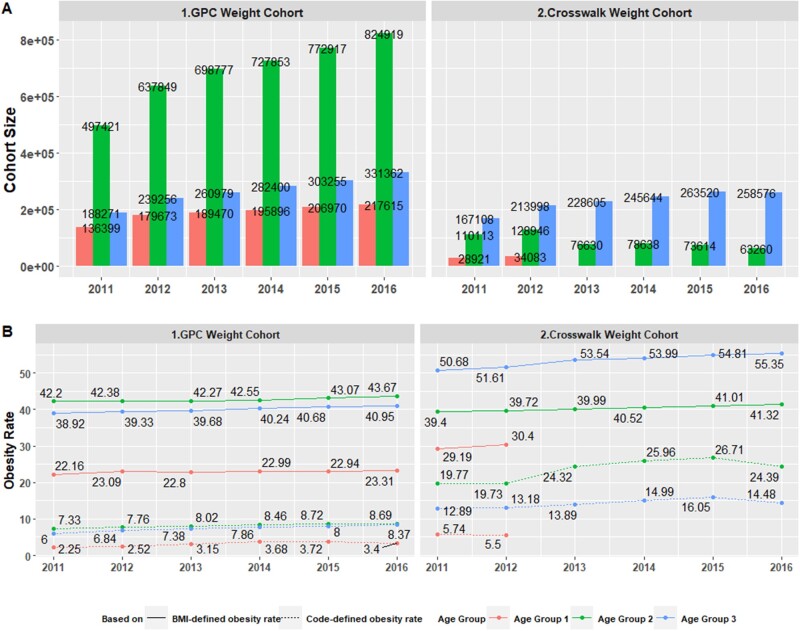
Figure 5A2 provides obesity rates using the physical measurements available for the GPC Weight Cohort. 22.16% of age group 1 (2–19), 42.20% of age group 2 (20–64), and 38.92% of age group 3 (65 and older) were obese in 2011, increasing to 23.31%, 43.67%, and 40.95%, respectively, in 2016. The obesity rates using diagnosis codes in EHR were an order of magnitude lower: 2.25% for age group 1, 7.33% of age group 2, and 6.00% of age group 3 were coded as obese in 2011. These rates increased for all 3 age groups to 3.40%, 8.69%, and 8.37%, respectively, by 2016.
Integrating claims and EHR to obtain obesity rates
Within the Crosswalk Cohort, 983 450 out of the 1 674 134 (58.7%) patients had at least 1 valid physical measurement in EHR (“Crosswalk Weight Cohort”) with 478 300 (48.6%) noted as being obese in at least 1 year. The prevalence rates of obesity estimated by combining CMS and EHR data were always significantly higher than estimations based on either source. For example, for age group 3, obesity rates estimated by CMS diagnosis codes and EHRs were 8.92–11.08% and 8.04–11.34%, respectively, whereas the combined rates were from 12.89% to 16.05% (Figure 5B2). More adults (age groups 2 and 3) were more likely to be assigned with obesity diagnosis codes than children and teenagers (age group 1), regardless of data source (Figure 5). For the rest 505 150 (51.4%) of Crosswalk Weight Cohort who had some BMI records which suggesting nonobese, there were 34 826 (7%) patients had at least 1 obesity diagnosis and majority of such inconsistent events were identified by CMS claims. In addition, there were 80 291 patients who were within GPC Cohort, had an obesity diagnosis but missing physical measurement. Among them, 10 389 (13%) even had a second obesity diagnosis from CMS.
Figure 5.
Electronic health record and claims based obesity rates for different age groups. (A1) and (A2) plot Greater Plains Collaborative Weight Cohort and Crosswalk Weight Cohort sizes stratified by 3 age groups (2–19, 20–64, 65, and older) by calendar year. (B1) and (B2) plot obesity rates where the solid lines correspond to body mass index-defined obesity, whereas the dotted line correspond Code-defined obesity.
Contribution of claims to EHR in identifying comorbidity
Although claims integration had modest improvement in identifying additional obese patients relative to physical measurements, we hypothesized that claims data would increase detection of related comorbidities that might be underreported during specialty or acute care at tertiary academic medical centers. Among 478 300 obese patients (measured physically) with available claims, 52 441 had obstructive sleep apnea diagnosis codes found in the EHR, 83 649 had diagnosis codes found in claims, and 35 791 in both EHR and claims. Claims identified an additional 47 858 (91% increase) patients. For type II diabetes, 104 307 of the 478 300 patients had a diagnosis code recorded in the EHR, 149 894 in claims, and 82 100 with codes in both EHR and claims. For diabetes, claims identified an additional 67 974 (65% increase) patients.
DISCUSSION
Creating GROUSE required extensive coordination across GPC organizations and legal teams, ResDAC staff, data management plan approval, CMS approval, and PCORI’s program officers and leadership. PCORnet’s vision of reusable infrastructure leveraging integrated claims is aligned with the data collection purpose envisioned by Congress’ legislation52 authorizing PCORI:
(3) DATA COLLECTION.—(A) IN GENERAL.—Secretary shall, with appropriate safeguards for privacy, make available to the Institute such data collected by the Centers for Medicare & Medicaid Services under the programs under titles XVIII, XIX, and XXI, as well as provide access to the data networks developed under section 937(f) of the Public Health Service Act, as the Institute and its contractors may require to carry out this section…
However, the processes for obtaining data from Medicare are designed for single studies. At the regional level, the GPC was acting as a distributed research network and had conducted only moderately sized cohort studies across its member organizations. There were concerns about data transfer from members as well as the institutional risk assumed by the GPC CC in managing organizations protected health information. It took over 2 years from inception to full integration and deidentification of CMS and EHR data to be available for analysis across multiple GPC member organizations. Trust was facilitated by (1) transparent communication and collaboration tools, (2) leveraging established CMS procedures for record linkage obviating the need for GPC CC to consolidate identifiable health information, and (3) designing GROUSE as a deidentified environment to support external investigators. Maintaining focus was strengthened by an additional cancer research study funded by PCORI where primary analysis was conducted by investigators at the University of Iowa.53 Since this study period, we have incorporated additional years of Medicare Claims and extended GPC membership to additional organizations. Currently, the timing required for data integration both organizationally and technically is considerable and at best accomplished annually.
In the case study, we found that the rates of BMI-Defined Obesity for the GPC network patients were consistently higher than national estimates with a similar increasing trend compared with national statistics.54–56 For adults (age groups 2 and 3), CDC reports obesity prevalence as 34.9%, 37.7%, and 39.6% for year intervals 2011–2012, 2013–2014, 2015–2016, respectively. Comparable numbers of GPC network data were 40.7%, 41.2%, and 42.1%. For age group 1, CDC-based obesity prevalence statistics were 16.9%, 17.2%, 18.5%, whereas GPC statistics were 22.6%, 22.8%, and 22.9%. The difference between these prevalence statistics is plausible and could be explained given that our study population is a restricted subpopulation of individuals who seek medical attention and our use of the highest average measurement. The discrepancy between BMI-defined and Code-defined obesity rates further confirmed the underdiagnosis issue of obesity especially among the children and adolescents (age group 1), echoing a prior study with similar conclusion.57 Our study’s Code-defined obesity rate (13.5%) based on the Medicaid and Medicare claims (averaged over the complete time period for combined middle aged and elderly populations), is consistent with the 14.6% estimated in the study by Ammann et al,58 based on commercial insurance data.
For supporting CER use cases, leveraging height and weight measurements recorded in EHRs is the dominant approach to selecting obese cohorts for a trial recruitment. CMS claims were linked to EHR data for the purpose of examining a more complete picture of care received, and especially to capture obesity and comorbidity diagnoses given outside the GPC network healthcare systems. Although the additional gain of obesity diagnoses from CMS claims was not substantial, claims significantly increased cohort size for sleep apnea and type II diabetes. This difference between the impact of claims integration to detect obesity versus comorbidity diagnosis codes for cohort selection can partially be explained by the case-mix at our sites. Most GPC members are academic medical centers with a greater percentage of specialty providers relative to community health systems. A minority of GPC members (eg, Marshfield Clinic) provides extensive regional primary care and has a more balanced provider mix. In claims data, diagnosis codes are included to indicate medical necessity. Thus obesity-related codes can be underrepresented because obesity-related comorbidity (eg, sleep apnea and diabetes) offer stronger and/or sufficient justification of medical necessity. Few services/procedures are provided for, and only for, obesity.
Integrating Medicare claims with health systems EHRs is especially beneficial for analyzing patients over 65 as over 98% of these United States citizens are covered by Medicare59,60 and since 2017 processes are also established for obtaining claims for those beneficiaries covered by Advantage managed care programs.61 Additionally, the summary statistics provided by Table 1 indicate majority of Medicare-eligible beneficiaries are continuously enrolled in the program over at least 5 years. During our study period, studying multistate longitudinal Medicaid populations was complicated as state Medicaid programs transitioned to the new Transformed Medicaid Statistical Information System (T-MSIS) in different years.62 As a result, the appropriate Medicaid RIF claims format and costs varied by state and year.63
As we move forward in support of all 3 cohorts and additional research requesting reuse of GROUSE, we anticipate value in part D medication and Durable Medical Equipment claims to support strong computable phenotyping of the existence of comorbidities but also severity of comorbidities (eg, amyotrophic lateral sclerosis disease progression from assistive devices to powered wheelchairs). We also plan to incorporate additional years of Medicare claims in support of NIH funded research programs reusing GROUSE and revisit incorporating additional Medicaid claims as states have largely completed transition to T-MSIS.
CONCLUSION
GROUSE facilitates data-driven research reproducibility by leveraging multiple health systems data integrated with Medicare and Medicaid claims; allowing investigators to understand model performance across health systems with different populations and explore how performance varies when claims incorporate care received outside largely tertiary health systems. Additionally, researchers can use the health characteristics reflected in the state-wide claims as a baseline. Our findings indicate the EHR-only measures of obesity are more than sufficient for supporting prospective and observational studies while observational studies incorporating comorbidities suffer if only diagnosis codes from within the health systems are included. As PCORI’s reauthorization expands its research to consider costs and healthcare utilization as outcomes, GROUSE’s integrated claims provide direct access to consistent financial information for procedures and encounters as well as medications and home health supplies; providing a new dimension for PCORnet to support CER at increased scale. We anticipate that increased adoption of cloud infrastructure supporting direct querying across enterprise data assets versus transferring files will catalyze data sharing across health systems, federal agencies, external laboratories, and increased consumer- and patient-generated data.
FUNDING
This work is supported through a Patient-Centered Outcomes Research Institute award No. RI-CRN-2020-003-IC and a National Institutes of Health Clinical and Translational Science Award grant from NCATS awarded to the University of Kansas for Frontiers: University of Kansas Clinical and Translational Science Institute No. UL1TR002366.
AUTHOR CONTRIBUTIONS
LRW and AMD designed and conceptualized the overall study. DLW and XS performed cohort extraction, data cleaning, descriptive analysis. XS contributed to the development of cloud environment and GROUSE database migration. LRW, MCS, AMD, and XS contributed to the evaluation of experimental results. LRW, JJV, ASM, DCC, and LPP contributed in data collection and extraction. AMD advised regarding motivation of the clinical design. ETA advised on system security of the cloud environment. All authors reviewed the article critically for scientific content, and all authors gave final approval of the article for publication. LRW and XS contributed equally to this work.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We want to acknowledge AWS professional service team and DLT reseller for their technical support and delivery of the Guardrail Accelerator Program. We also want to acknowledge the close involvement of University of Kansas Medical Center Information Resources security team and General Counsel’s office in developing the initial DMP and data agreements and then University of Missouri Health Care Information Security team during the development and review of DMP SAQ. We are also grateful to be supported by University of Missouri Department of IT, especially the Information Security & Access Management (ISAM) team.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The clinical data used for training and validation in this study are not publicly available and restrictions apply to its use. The deidentified dataset may be available from the Greater Plains Collaborative clinical data network, subjective to individual institution and network-wide ethical approvals.
REFERENCES
- 1. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS.. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 2014; 21 (4): 578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Greater Plains Collaborative (GPC). Secondary The Greater Plains Collaborative. 2015. http://www.gpcnetwork.org/ Accessed July 1, 2021.
- 3. Waitman LR, Aaronson LS, Nadkarni PM, Connolly DW, Campbell JR.. The Greater Plains Collaborative: a PCORnet Clinical Research Data Network. J Am Med Inform Assoc 2014; 21 (4): 637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NIH. National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Science Award (CTSA) Program. Secondary National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Science Award (CTSA) Program. 2013. https://ncats.nih.gov/ctsa.
- 5.PCORI. Patient-Centered Outcomes Research Institute Cooperative Agreement Funding Announcement: Improving Infrastructure for Conducting Patient-Centered Outcomes Research. The National Patient-Centered Clinical Research Network: Clinical Data Research Networks (CDRN)—Phase One. Secondary Patient-Centered Outcomes Research Institute Cooperative Agreement Funding Announcement: Improving Infrastructure for Conducting Patient-Centered Outcomes Research. The National Patient-Centered Clinical Research Network: Clinical Data Research Networks (CDRN)—Phase One. 2013. https://www.pcori.org/sites/default/files/PCORI-PFA-CDRN-071713.pdf.
- 6.Services CoMaM. Identifiable Data Files. Secondary Identifiable Data Files. https://www.cms.gov/Research-Statistics-Data-and-Systems/Files-for-Order/IdentifiableDataFiles.
- 7. Visweswaran S, Becich MJ, D’Itri VS, et al. Accrual to Clinical Trials (ACT): a Clinical and Translational Science Award Consortium Network. JAMIA Open 2018; 1 (2): 147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brat GA, Weber GM, Gehlenborg N, et al. International electronic health record-derived COVID-19 clinical course profiles: the 4CE consortium. NPJ Digit Med 2020; 3: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haendel MA, Chute CG, Bennett TD, et al. ; N3C Consortium. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc 2021; 28 (3): 427–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Estiri H, Vasey S, Murphy SN.. Generative transfer learning for measuring plausibility of EHR diagnosis records. J Am Med Inform Assoc 2021; 28 (3): 559–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rank N, Pfahringer B, Kempfert J, et al. Deep-learning-based real-time prediction of acute kidney injury outperforms human predictive performance. NPJ Digit Med 2020; 3 (1): 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010; 11 (10): 733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song X, Yu ASL, Kellum JA, et al. Cross-site transportability of an explainable artificial intelligence model for acute kidney injury prediction. Nat Commun 2020; 11 (1): 5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Afgan E, Baker D, Coraor N, et al. Harnessing cloud computing with Galaxy Cloud. Nat Biotechnol 2011; 29 (11): 972–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heath AP, Greenway M, Powell R, et al. Bionimbus: a cloud for managing, analyzing and sharing large genomics datasets. J Am Med Inform Assoc 2014; 21 (6): 969–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Madduri RK, Sulakhe D, Lacinski L, et al. Experiences building Globus Genomics: a next-generation sequencing analysis service using Galaxy, Globus, and Amazon Web Services. Concurr Comput 2014; 26 (13): 2266–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau JW, Lehnert E, Sethi A, et al. ; Seven Bridges CGC Team. The Cancer Genomics Cloud: collaborative, reproducible, and democratized-a new paradigm in large-scale computational research. Cancer Res 2017; 77 (21): e3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denny J. All of Us Research Program Begins Beta Testing of Data Platform. Secondary All of Us Research Program Begins Beta Testing of Data Platform. 2020. https://allofus.nih.gov/news-events-and-media/announcements/all-us-research-program-begins-beta-testing-data-platform.
- 19.C3AI. C3 AI COVID-19 Data Lake. Secondary C3 AI COVID-19 Data Lake. 2020. https://c3.ai/products/c3-ai-covid-19-data-lake/.
- 20.Cerner. Cerner—Strategic Innovation in Healthcare for Today and Tomorrow. Secondary Cerner—Strategic Innovation in Healthcare for Today and Tomorrow. https://www.cerner.com/about.
- 21.Medicine F. Foundation Medicine—Advancing Cancer Care Together. Secondary Foundation Medicine—Advancing Cancer Care Together. https://www.foundationmedicine.com/.
- 22.NewWave. NewWave-GDIT, LLC—Awarded Centers for Medicare & Medicaid Services (CMS) Chronic Condition Warehouse (CCW) and Virtual Research Data Center (VRDC) Contract. Secondary NewWave-GDIT, LLC—Awarded Centers for Medicare & Medicaid Services (CMS) Chronic Condition Warehouse (CCW) and Virtual Research Data Center (VRDC) Contract. 2015. https://blog.newwave.io/newwave-gdit-llc-awarded-centers-for-medicare-medicaid-services-cms-chronic-condition-warehouse-ccw-and-virtual-research-data-center-vrdc-contract.
- 23. Cobb N, Witte E, Cervone M, et al. The SMART IRB platform: a national resource for IRB review for multisite studies. J Clin Transl Sci 2019; 3 (4): 129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Network G. Greater Plains Collaborative CDRN Collaboration. Secondary Greater Plains Collaborative CDRN Collaboration. 2015. https://www.gpcnetwork.org/?q=Collaboration.
- 25.Services CfMM. Data Disclosures and Data Use Agreements (DUAs)—Identifiable Data Files. Secondary Data Disclosures and Data Use Agreements (DUAs)—Identifiable Data Files. 2021. https://www.cms.gov/Research-Statistics-Data-and-Systems/Files-for-Order/Data-Disclosures-Data-Agreements/Researchers.
- 26.Govsurf. GS35F0475X—MBL TECHNOLOGIES, INC. Secondary GS35F0475X—MBL TECHNOLOGIES, INC. 2021. https://usaspending.gov.surf/Spending?awardId=GS35F0475X.
- 27.ResDAC. CMS Research Identifiable Request Process & Timeline. Secondary CMS Research Identifiable Request Process & Timeline. https://resdac.org/cms-research-identifiable-request-process-timeline.
- 28.NIST. NIST-800-53—Security and Privacy Controls for Federal Information Systems and Organizations. Secondary NIST-800-53—Security and Privacy Controls for Federal Information Systems and Organizations. 2015. https://csrc.nist.gov/publications/detail/sp/800-53/rev-4/final.
- 29.ResDAC. CMS’s Data Privacy Safeguard Program (DPSP). Secondary CMS’s Data Privacy Safeguard Program (DPSP). 2014. https://resdac.org/articles/cmss-data-privacy-safeguard-program-dpsp.
- 30.MBLtechnologies. MBL Technologies. Secondary MBL Technologies. 2021. https://www.mbltechnologies.com/about-us/#.
- 31.NIST. FIPS 199—Standards for Security Categorization of Federal Information and Information Systems. Secondary FIPS 199—Standards for Security Categorization of Federal Information and Information Systems. 2004. https://csrc.nist.gov/publications/detail/fips/199/final.
- 32.NIST. FIPS 200—Minimum Security Requirements for Federal Information and Information Systems. Secondary FIPS 200—Minimum Security Requirements for Federal Information and Information Systems. 2006. https://csrc.nist.gov/publications/detail/fips/200/final.
- 33.Security UoMI. Data Classification System. Secondary Data Classification System. 2019. https://www.umsystem.edu/ums/is/infosec/classification-definitions.
- 34.(NIST). USNIoSaT. Federal Information Processing Standards Publication 197—Announcing the Advanced Encryption Standard (AES). 2001.
- 35. Carnahan RM, Waitman LR, Charlton ME, et al. Exploration of PCORnet data resources for assessing use of molecular-guided cancer treatment. JCO Clin Cancer Inform 2020; (4): 724–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.PCORI. The National Patient-Centered Clinical Research Network (PCORnet) Common Data Model v6.0. Secondary The National Patient-Centered Clinical Research Network (PCORnet) Common Data Model v6.0. 2020. https://pcornet.org/data/.
- 37. Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC.. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annu Symp Proc 2006; 2006: 1040. [PMC free article] [PubMed] [Google Scholar]
- 38. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG.. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42 (2): 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.NIH. NIH Information Security and Information Management Training Courses—Information Security and Management Refresher. Secondary NIH Information Security and Information Management Training Courses—Information Security and Management Refresher. https://irtsectraining.nih.gov/publicUser.aspx.
- 40.AWS. AWS Well-Architected Framework. Secondary AWS Well-Architected Framework. 2020. https://docs.aws.amazon.com/wellarchitected/latest/framework/wellarchitected-framework.pdf.
- 41.Service AW. Service Workbench on AWS—A web portal for researchers to accelerate their time to science. Secondary Service Workbench on AWS—A web portal for researchers to accelerate their time to science. https://aws.amazon.com/government-education/research-and-technical-computing/service-workbench/.
- 42.AWS. AWS Fargate—Serverless compute for containers. Secondary AWS Fargate—Serverless compute for containers. 2021. https://aws.amazon.com/fargate/.
- 43.Snowflake. The Snowflake Platform. Secondary The Snowflake Platform. https://www.snowflake.com/cloud-data-platform/.
- 44.CloudCheckr. CloudCheckr for Cost Management. Secondary CloudCheckr for Cost Management. https://cloudcheckr.com/solutions/cloud-cost-optimization/.
- 45.AWS. Whitepaper—Shared Responsibility Model.
- 46. Garbis J, Chapman JW.. Zero trust scenarios. Zero Trust Security 2020; 4: 239–65. [Google Scholar]
- 47.Myrbakken H, Colomo-Palacios R. DevSecOps: a multivocal literature review. In: Mas A, Mesquida A, O’Connor RV, Rout T, Dorling A, eds. Software Process Improvement and Capability Determination. Cham, Switzerland: Springer International Publishing; 2017: 17–29.
- 48.AWS. Infrastructure as Code. Secondary Infrastructure as Code. https://docs.aws.amazon.com/whitepapers/latest/introduction-devops-aws/infrastructure-as-code.html.
- 49.Corp SCS. The Secure Shell (SSH) Connection Protocol. Secondary The Secure Shell (SSH) Connection Protocol 2006. https://www.hjp.at/doc/rfc/rfc4254.html.
- 50.Services CfMM. Research Data Assistance Center (ResDAC). Secondary Research Data Assistance Center (ResDAC) 2018. https://www.cms.gov/Research-Statistics-Data-and-Systems/Research/ResearchGenInfo/ResearchDataAssistanceCenter.
- 51.Centers for Disease Control and Prevention HW, Nutrition, and PHysical Activity. BMI Percentile Calculator for Child and Teen. Secondary BMI Percentile Calculator for Child and Teen. 2021. https://www.cdc.gov/healthyweight/bmi/calculator.html.
- 52.United States C. Public Law 111-148. Patient Protection and Affordable Care Act, Section 6301: Patient-Centered Outcomes Research, Subsection (d)(3)(A) – DATA COLLECTION.730. U.S.: Government Publishing Office.
- 53.PCORI. Using PCORnet to Understand Use of Molecular Tests and Treatments for Cancerous Tumors. Secondary Using PCORnet to Understand Use of Molecular Tests and Treatments for Cancerous Tumors 2017. https://www.pcori.org/research-results/2017/using-pcornet-understand-use-molecular-tests-and-treatments-cancerous-tumors.
- 54.Centers for Disease Control and Prevention DoN, Physical Activity, and Obesity, National Center for Chronic Disease Prevention and Health Promotion. Adult Obesity Prevalence Maps and Animated Maps 2011 to 2018. Secondary Adult Obesity Prevalence Maps and Animated Maps 2011 to 2018. https://www.cdc.gov/obesity/data/prevalence-maps.html#overall.
- 55.Environment KDoHa. Behavioral Risk Factor Surveillance System. Percentage of Adults Who are Obese. Secondary Behavioral Risk Factor Surveillance System. Percentage of Adults Who are Obese 2015. http://www.kdheks.gov/brfss/Survey2015/ct2015_obesity.html.
- 56. Hales CM, Carroll MD, Fryar CD, Ogden CL.. Prevalence of obesity among adults and youth: United States, 2015-2016. NCHS Data Brief 2017 (288): 1–8. [PubMed] [Google Scholar]
- 57. Martin B-J, Chen G, Graham M, Quan H.. Coding of obesity in administrative hospital discharge abstract data: accuracy and impact for future research studies. BMC Health Serv Res 2014; 14 (1): 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ammann EM, Kalsekar I, Yoo A, Johnston SS.. Validation of body mass index (BMI)-related ICD-9-CM and ICD-10-CM administrative diagnosis codes recorded in US claims data. Pharmacoepidemiol Drug Saf 2018; 27 (10): 1092–100. [DOI] [PubMed] [Google Scholar]
- 59.AoA. 2020 Profile of Older Americans: The Administration for Community Living. 2021.
- 60.CMS. Total Medicare Enrollment: Part A and/or Part B Enrollees, by Demographic Characteristics, Calendar Year 2019. Secondary Total Medicare Enrollment: Part A and/or Part B Enrollees, by Demographic Characteristics, Calendar Year 2019. 2019. https://www.cms.gov/files/document/2019cpsmdcrenrollab5.pdf.
- 61.Center RDA. 2017 Medicare Advantage (Part C) Encounter Data Now Available. Secondary 2017 Medicare Advantage (Part C) Encounter Data Now Available. 2020. https://resdac.org/cms-news/2017-medicare-advantage-part-c-encounter-data-now-available.
- 62.Medicaid.gov. Transformed Medicaid Statistical Information System (T-MSIS). Secondary Transformed Medicaid Statistical Information System (T-MSIS). https://www.medicaid.gov/medicaid/data-systems/macbis/transformed-medicaid-statistical-information-system-t-msis/index.html.
- 63.Center RDA. Fee List for RIFs: Physical Research Data Request. Secondary Fee List for RIFs: Physical Research Data Request. 2021. https://resdac.org/sites/datadocumentation.resdac.org/files/2021-08/CMS%20Fee%20List%20for%20Research%20Files_0.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The clinical data used for training and validation in this study are not publicly available and restrictions apply to its use. The deidentified dataset may be available from the Greater Plains Collaborative clinical data network, subjective to individual institution and network-wide ethical approvals.