Abstract
The neural crest is a dynamic embryonic structure that plays a major role in the formation of the vertebrate craniofacial skeleton. Neural crest formation is regulated by a complex sequence of events directed by a network of transcription factors working in concert with chromatin modifiers. The high mobility group nucleosome binding protein 1 (Hmgn1) is a non-histone chromatin architectural protein, associated with transcriptionally active chromatin. Here we report the expression and function of Hmgn1 during Xenopus neural crest and craniofacial development. Hmgn1 is broadly expressed at the gastrula and neurula stages, and is enriched in the head region at the tailbud stage, especially in the eyes and the pharyngeal arches. Hmgn1 knockdown affected the expression of several neural crest specifiers, including sox8, sox10, foxd3 and twist1, while other genes (sox9 and snai2) were only marginally affected. The specificity of this phenotype was confirmed by rescue, where injection of Hmgn1 mRNA was able to restore sox10 expression in morphant embryos. The reduction in neural crest gene expression at the neurula stage in Hmgn1 morphant embryos correlated with a decreased number of sox10- and twist1-positive cells in the pharyngeal arches at the tailbud stage, and hypoplastic craniofacial cartilages at the tadpole stage. These results point to a novel role for Hmgn1 in the control of gene expression essential for neural crest and craniofacial development. Future work will investigate the precise mode of action of Hmgn1 in this context.
Keywords: Xenopus, neural crest, transcription, craniofacial, sox10, twist1
Introduction
The neural crest is a population of multipotent and motile cells derived from the embryonic ectoderm and unique to vertebrates. Neural crest progenitors are specified at the neural border zone at the end of gastrulation, and upon completion of neurulation these cells delaminate from the neural tube, migrate throughout the embryo and give rise to multiple cell lineages contributing to the formation of most organs of the adult. In the head region, the cranial neural crest cells migrate into the pharyngeal arches where they will form most craniofacial skeletal elements of the face and neck (Minoux and Rijli, 2010). Abnormal development of the cranial neural crest can result in severe defects affecting the orofacial complex as exemplified by mandibulofacial dysostosis (MFD) and other related craniofacial congenital disorders (Trainor and Andrews, 2013). Animal models of MFD indicate that this condition is due to defects in neural crest progenitors formation (Dixon et al., 2006, Devotta et al., 2016; Lei et al., 2017; Beauchamp et al., 2021; Park et al., 2021).
Neural crest cells are generated through a well-orchestrated sequence of events directed by a complex network of transcription factors often working in conjunction with chromatin modifiers to activate and repress gene expression over time (Hu et al. 2014; Simões-Costa and Bronner, 2015; Rogers and Nie, 2018). The high mobility group nucleosome-binding protein 1 (Hmgn1) belongs to a family of five non-histone chromatin remodelers (Hmgn1–5). These factors preferentially bind to enhancers and promoters, thereby regulating transcription (Kugler et al., 2012; Naduri et al., 2020). Hmgn proteins are vertebrate-specific and mostly ubiquitously expressed (Postnikov & Bustin, 2010; Naduri et al., 2020). Hmgn1 and Hmgn2 are especially highly expressed during early embryogenesis suggesting that it may have an important role in lineage decision and differentiation (Yang, et al., 2018). Hmgn1−/− mouse embryos are relatively normal, with impaired response to stress, and increased sensitivity to UV-induced DNA damage (Birger et al., 2005; Abuhatzira et al., 2011). The lack of a more severe embryonic phenotype in these animals is likely due to functional redundancy between Hmgn1 and Hmgn2 (Martinez de Paz and Ausio, 2016; Naduri et al., 2020).
Here, we describe the expression and function of Hmgn1 during Xenopus craniofacial development. Hmgn1 is initially broadly expressed in the embryo and then becomes progressively enriched in the head region. Morpholino-mediated knockdown of Hmgn1 affected the expression of several early neural crest genes including sox8, sox10, foxd3 and twist1. Later in development, these embryos had a decreased number of sox10- and twist1-positive cells in the pharyngeal arches, and hypoplastic craniofacial cartilages at the tadpole stage. We propose that Hmgn1 is regulating the transcription of a subset of genes essential for neural crest and craniofacial development, and in its absence neural progenitors fail to activate or maintain a neural crest developmental program.
Materials and Methods
Plasmids constructs and morpholino antisense oligonucleotides
Xenopus laevis Hmgn1.L complete coding sequence was generated by polymerase chain reaction (PCR) using the following primers: forward; 5’-ATCGATGCCACCATGCCTAAGAGAAAGCAGGTGAA-3’ and reverse; 5’- TCTAGATTATTCAGACTTGGATTCCTTGT-3’ and NF stage 11.5 cDNA using Pure Taq Ready-to-go PCR beads (ThermoFisher Scientific; Waltham, MA). The PCR product was cloned into pGEMT Easy and subsequently ligated into pCS2+ vector, using the restriction sites ClaI and XbaI. Hmgn1 translation blocking morpholino antisense oligonucleotide (Hmgn1MO; 5’-CTTTCTCTTAGGCATGGCTGCAC-3’) and control morpholino (CoMO) were purchased from GeneTools (Philomath, OR). Hmgn1MO targets both Hmgn1.L and Hmgn1.S allo-alleles. The specificity of Hmgn1MO was tested on a pCS2+ myc-tagged version of Hmgn1 containing the full Hmgn1MO target site (including 8 bp upstream to the ATG), generated using the following primers forward; 5’-GGATCCGTGCAGCCATGCCTAAGAGAA-3’ and reverse 5’-ATCGATTTCAGACTTGGATTCCTTGTCA-3’. The amplified regions were ligated into pCS2+myc-tag plasmid, using the restriction sites BamHI and ClaI.
Xenopus embryos and microinjections
Xenopus laevis embryos were staged according to Nieuwkoop and Faber (1967) and raised in 0.1X NAM (Normal Amphibian Medium; Slack and Forman, 1980). The use of animals was approved by New York University Institutional Animal Care and Use Committee (protocol #IA16–00052). Hmgn1, and β-galactosidase (LacZ) mRNAs were synthesized in vitro using the Message Machine kit (Ambion, Austin TX). CoMO, Hmgn1 MO, and Hmgn1 mRNA were injected in one blastomere at the 2-cell stage and the injected embryos were analysed by in situ hybridization at the neurula (NF stage 15) or tailbud (NF stage 25) stages. To identify the injected side, 500 pg of LacZ mRNA was co-injected as a lineage tracer. Only embryos showing co-localized expression of the lineage tracer with the marker of interest were considered for analysis.
Whole-mount in situ hybridization
Embryos were fixed in MEMFA and stained for Red-Gal (Research Organics; Cleveland, OH) to visualize the lineage tracer (LacZ mRNA) and processed for in situ hybridization (ISH). Antisense digoxigenin-labeled probes (Genius kit; Roche, Indianapolis IN) were synthesized using template cDNA encoding hmgn1, snai2 (Mayor et al., 1995), sox8 (O’Donnell et al., 2006), sox9 (Spokony et al., 2002), sox10 (Aoki et al., 2003), foxd3 (Sasai et al., 2001), dmrta1 (Huang et al., 2005), sox2 (Mizuseki et al., 1998), twist1 (Hopwood et al., 1989a), foxi4.1 (Pohl et al., 2002) pax6 (Hirsch and Harris, 1997) and foxe3 (Kenyon et al., 1999). Whole-mount ISH was performed as described (Harland, 1991; Saint-Jeannet, 2017), and the embryos imaged on a Leica M165 Stereomicroscope (Leica Microsystems Inc., Buffalo Grove, IL). For histology, NF stage 33 stained embryos were embedded in Paraplast+, sectioned on a rotary microtome (Olympus, Center Valley, PA), and briefly counterstained with Eosin Y.
Western blot analysis
Embryos were injected at the 2-cell stage with 10 pg of Xenopus Hmgn1 mRNA alone or in combination with increasing doses of Hmgn1MO, and cultured to NF stage 10–15. Pools of 10 embryos were homogenized in lysis buffer (0.5% Triton X-100, 10 mM Tris–HCI at pH 7.5, 50 mM NaCl, 1 mM EDTA), containing HaltTM Protease Inhibitor Cocktail (ThermoFisher Scientific; Waltham, MA). After two successive centrifugations to eliminate lipids, the lysate was concentrated on an Amicon Ultra Centrifugal Filter (Merck Millipore; Billerica, MA), 5 μl of the lysate was resolved on a 10% NuPAGE Bis-Tris gel and transferred onto a PVDF membrane using the iBlot system (Invitrogen). The blots were incubated overnight with a monoclonal anti-myc antibody (Santa Cruz Biotechnology, Dallas TX; 1:1000 dilution) or anti α-tubulin antibody (Sigma Aldrich, St Louis, MO; 1:500 dilution). The blots were washed and incubated with anti-mouse IgG coupled to horseradish peroxidase (Santa Cruz Biotechnology, Dallas TX; 1:10,000 dilution). Peroxidase activity was detected with the Western Blotting Luminol Reagent (Santa Cruz Biotechnology, Dallas TX) and imaged on a ChemiDoc MP gel documentation system (Biorad, Hercules CA). Membranes were stripped using Restore Western Blot Stripping Buffer (ThermoFisher Scientific, Waltham MA) according to the manufacturer recommendations.
TUNEL and Proliferation Assays
TUNEL staining was conducted as described (Hensey and Gautier, 1998). Hmgn1MO- and Sf3B4MO-injected albinos embryos at stage 15 or stage 12 were fixed in MEMFA, rehydrated in PBT and washed in TdT buffer (Roche, Indianapolis IN) for 30 min. End labelling was carried out overnight at room temperature in TdT buffer containing 0.5 mM DIG-dUTP and 150 U/ml TdT (Roche, Indianapolis IN). Embryos were washed for 2 hours at 65 °C in PBS/1 mM EDTA. DIG was detected with anti-DIG Fab fragments conjugated to alkaline phosphatase (Roche, Indianapolis IN; 1:2000 dilution) and the chromogenic reaction performed using NBT/BCIP (Roche, Indianapolis IN). For phosphohistone H3 (pHH3) detection (Saka and Smith, 2001), fixed albinos embryos were incubated successively in anti-phosphohistone H3 antibody (Upstate Biotechnology, Lake Placid NY; 1 mg/ml) and an anti-rabbit IgG secondary antibody conjugated to alkaline phosphatase (ThermoFisher Scientific, Waltham MA; 1:1000). Alkaline phosphatase activity was revealed using NBT/BCIP (Roche, Indianapolis IN). To quantify changes in cell death and proliferation, embryos were individually photographed and the number of TUNEL- and pHH3-positive cells in the dorsal ectoderm counted manually on control and injected sides.
Cartilage staining
Hmgn1MO-injected embryos were cultured to NF stage 45 for cartilage analysis. Alcian blue staining of stage 45 tadpoles was performed as described (Berry et al., 1998). Tadpoles were fixed overnight in 10% formaldehyde, rinsed in tap water, skinned and eviscerated. The tadpoles were dehydrated and stained in Alcian blue (Sigma-Aldrich; St. Louis, MO) for 12 hours. After multiple rinses in 95% ethanol, tadpoles were rehydrated and macerated in 2% potassium hydroxide. Specimens were then transferred successively in 20%, 40%, 60% and 80% glycerol in 2% potassium hydroxide. The ethmoid plate was dissected out and specimens mounted flat under a coverslip in 80% glycerol, and individually imaged on a Leica M165 Stereomicroscope (Leica Microsystems Inc., Buffalo Grove, IL).
Neural crest cell migration quantification
The net distance on neural crest cells migration was measured in sox10 and twist1 stained stage 23–25 embryos using FIJI/Image J by drawing a straight line from the dorsal midline to the ventral-most edge of neural crest streams 1–3 (the 4th stream is often fused with the 3rd stream at these stages). The mean length of dorsoventral extension was calculated per embryo for each side (injected side vs. control side). The ratio of the mean dorsoventral extension is plotted after normalization to the control condition. A value of 1 represents normal dorsoventral extension.
Statistical Method
Each experiment was performed on at least three different batches of embryos obtained from different females. In MO-injected embryos we compared gene expression on the injected side to the contralateral uninjected side, and to CoMO-injected embryos. For TUNEL and pHH3 staining analyses were performed with GraphPad Prism 8 (GraphPad Software, San Diego CA) using unpaired t-test with Welch’s correction. p-value <0.05 was considered significant.
Results
Developmental expression of hmgn1
To analyse the expression pattern of hmgn1 during Xenopus laevis development, we performed in situ hybridization (ISH) on embryos at different stages. Sense (negative control) and antisense probes were used to evaluate hmgn1 expression. Hmgn1 transcripts are detected in the embryonic ectoderm at early (NF stage 10) and late (NF stage 12) gastrula stages, and appear to be ubiquitously expressed at these stages (Fig. 1A). At early neurula stage (NF stage 15), hmgn1 is still broadly expressed throughout the ectoderm. As neurulation proceeds (NF stage 18), hmgn1 expression appears to be enriched dorsally, in the prospective neural plate and neural crest territories (Fig 1A). By tailbud stages (NF stages 23 and 33), stronger signal was detected in the developing eyes, otic vesicles and the posterior pharyngeal arches (Fig. 1A). To further document hmgn1 expression we sectioned stage 33 embryos, and confirmed that hmgn1 is expressed in the skin, brain, developing retina, and to a lesser extent in the developing otic vesicle (Fig. 1B). Hmgn1 is therefore broadly expressed during Xenopus development consistent with a previous report (Korner et al., 2003), and its predicted function as a general regulator of chromatin remodeling (Nanduri et al., 2020).
Figure 1: Developmental expression of hmgn1 in Xenopus laevis embryos.
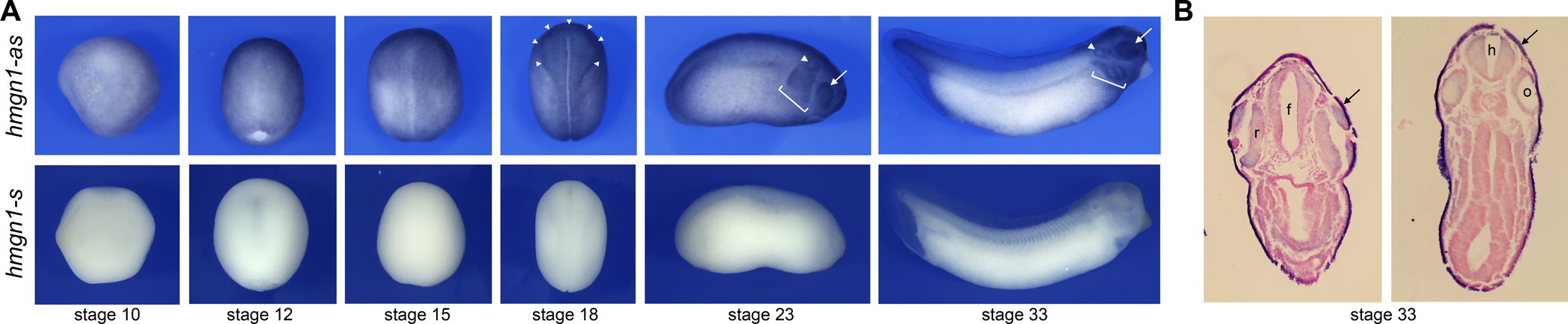
(A) At gastrula stages (NF stage 10–12), hmgn1 transcripts (hmgn1-as; upper panels) are detected throughout the embryonic ectoderm. At neurula stages (NF stage 15–18) hmgn1 appears to be enriched at the neural plate and the neural plate border (stage 18; arrowheads). At stage 23 and 33, hmgn1 is still broadly expressed with stronger expression in the pharyngeal arches (brackets) and the developing eyes (arrows). Stages 10–18 are dorsal views, anterior to top. Stages 23–33 are lateral views, dorsal to top, anterior to right. Staining with a control sense probe (hmgn1-s; lower panels) is shown for comparison. (B) Transverse sections of a stage 33 embryo at the forebrain (f; left panel) and hindbrain (h; right panel) levels, highlight hmgn1 expression in the skin (arrows), brain, retina of the eye (r) and to a lesser extent in the otic vesicle (o).
Hmgn1 regulates the expression of a subset of neural crest genes
Because hmgn1 is enriched dorsally and in the head region, we evaluated its role during neural crest formation, a cell population that makes a major contribution to the orofacial complex. We used a translation blocking morpholino antisense oligonucleotide (Hmgn1MO; Fig. 2A) to interfere with its function. Validation of the MO was conducted by Western blot on embryos injected with mRNA encoding a tagged version of Hmgn1 (Hmgn1-Myc) alone, or in combination with increasing doses of Hmgn1MO. In these embryos, co-injection of Hmgn1MO blocked accumulation of Hmgn1 protein directed by Hmgn1-Myc mRNA (Fig. 2B). We first analyzed the expression of sox10, a gene expressed in neural crest progenitors at the neurula stage (Aoki et al., 2003). Unilateral injection of Hmgn1MO in embryos at the 2 -cell stage caused a marked reduction of sox10 expression in 78% of analyzed embryos (Figure 2C,D). To confirm the specificity of the phenotype, we performed rescue experiments by injection of a wild-type hmgn1 mRNA (Hmgn1WT) resistant to Hmgn1MO. Expression of Hmgn1WT was sufficient to restore sox10 expression in Hmgn1 morphant embryos (Figure 2C,D), while overexpression of Hmgn1WT had little to no effect on sox10 expression at this stage (Figure 2C,D). Altogether these results indicate that Hmgn1 function is required for sox10 expression, and demonstrate the specificity of the Hmgn1 knockdown phenotype.
Figure 2: Hmgn1 knockdown affects sox10 expression.
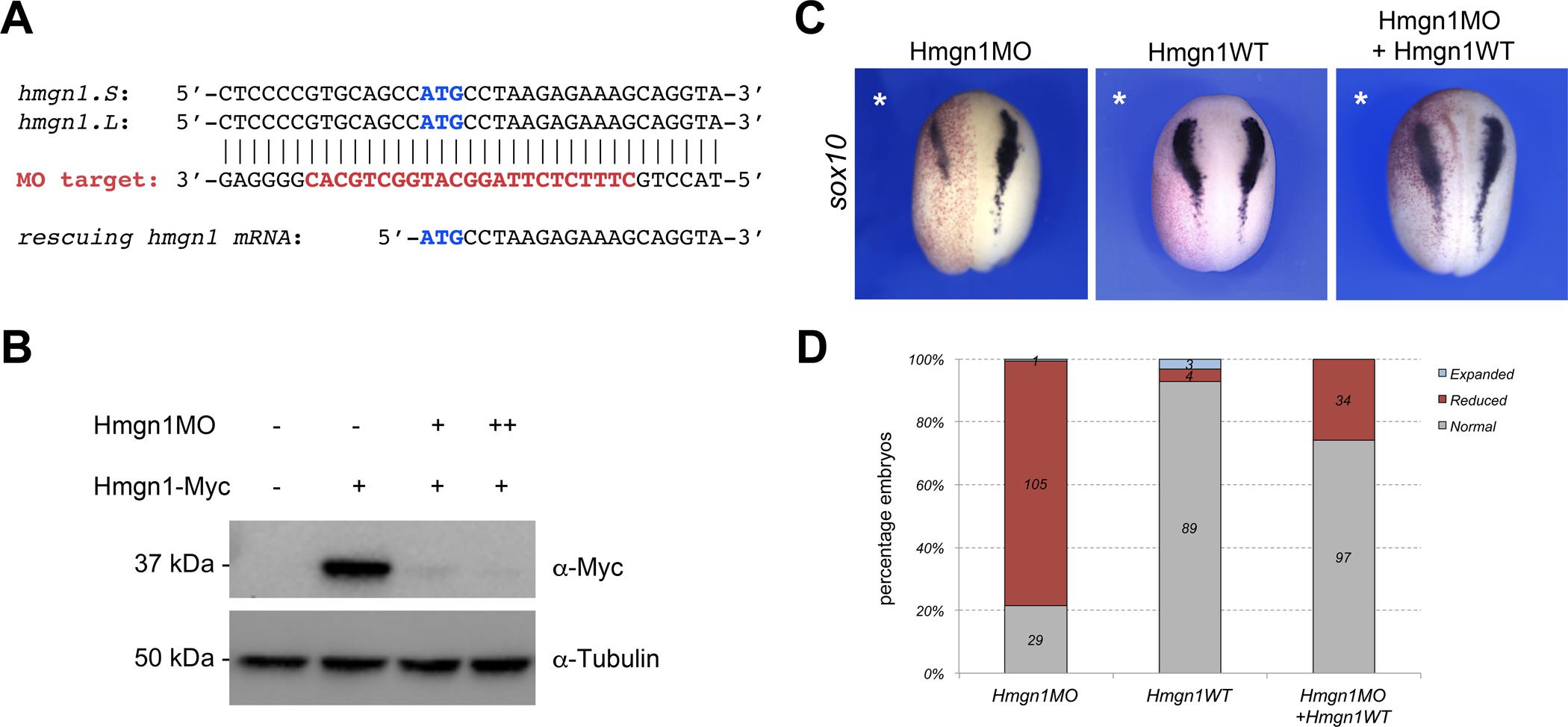
(A) The sequence targeted by the Hmgn1 translation blocking morpholino antisense oligonucleotide (Hmgn1MO) is depicted in red. Hmgn1MO targets the ATG start site (blue) of both hmgn1.S and hmgn1.L. The start sequence of the rescuing hmgn1 mRNA is shown for comparison, it lacks the 8bp upstream of the ATG targeted by Hmgn1MO. (B) Western blot analysis from lysates of embryos injected with hmgn1-Myc mRNA (100 pg) alone, or in combination with increasing doses of Hmgn1MO, 10 ng (+) and 100 ng (++). ⍺-tubulin is shown as a loading control. (C) Unilateral injection of wild-type hmgn1 mRNA (Hmgn1WT; 10 pg) restores sox10 expression in Hmgn1-depleted embryos (Hmgn1MO; 20 ng). The injected side is indicated by an asterisk. (D) Quantification of the phenotypes from 4 independent experiments. The number of embryos showing a given phenotype is indicated in each bar.
We expanded our analysis of the Hmgn1 morphant phenotype by exploring the the impact of Hmgn1 knockdown on other neural crest genes (sox8, sox9, foxd3, snai2, twist1), as well as genes expressed in other derivatives of the ectoderm including cranial placode (six1, dmrt1a, foxi4.1, sox9) and neural plate (sox2) lineages. Interestingly, we found that only a subset of neural crest genes were affected upon Hmgn1 knockdown, while the expression of sox8, foxd3 and twist1 was reduced in a majority of the embryos, sox9 and snai2 were largely unaffected (Fig. 3A,B). The pan-placodal genes foxi4.1 and six1 were only marginally affected in 11% and 26% of the embryos, respectively, whereas dmrta1, which is expressed in the prospective olfactory epithelium (Huang et al., 2005), was significantly reduced in 76% of embryos. The otic expression domain of sox9, and the neural plate and placode expression domains of sox2 were largely unperturbed in morphant embryos (Fig 3A,B). Because hgmn1 is strongly expressed in the developing eye (Fig 1) we also analyzed the expression of pax6 (prospective eye and lens) and foxe3 (lens vesicle) in morphant embryos at NF stage 23 and 30, respectively. We found that both genes were largely unaffected in these embryos (Fig 4A–C). These results suggest that Hmgn1 is necessary for the expression of a subset of neural crest genes, suggesting that it may regulate the development of these progenitors and their derivatives.
Figure 3: Hmgn1 knockdown affects neural crest formation.
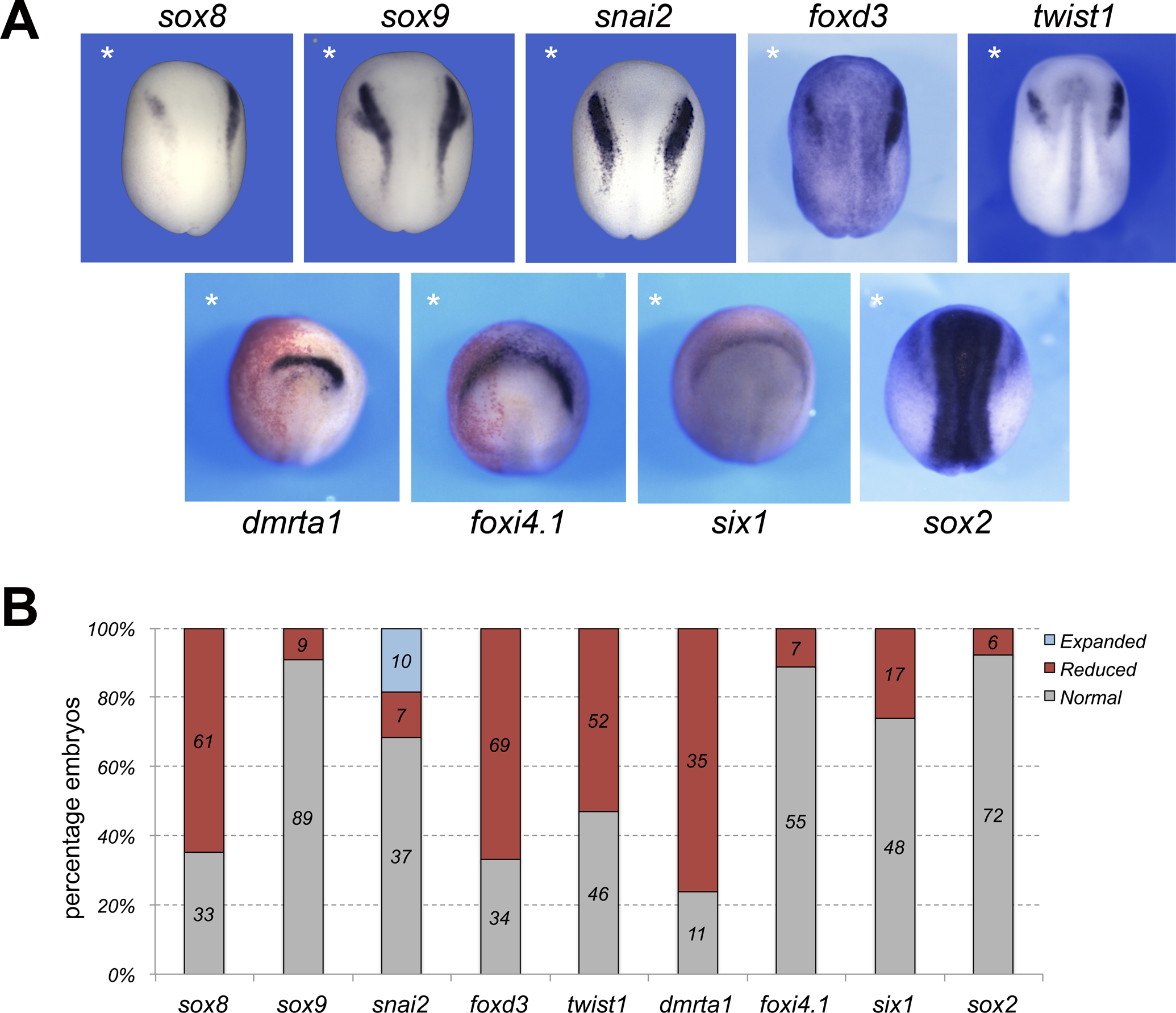
(A) Phenotype of Hmgn1MO-injected embryos (20 ng) on neural crest (upper row) and placode (lower row) gene expression at stage 15. The injected side is indicated by an asterisk. Dorsal views, anterior to top, except for dmrta1 and foxi4.1 for which anterior views are shown. (B) Quantification of the phenotypes. The number of embryos showing a given phenotype is indicated in each bar.
Figure 4: Hmgn1 depletion does not affect eye and lens development.
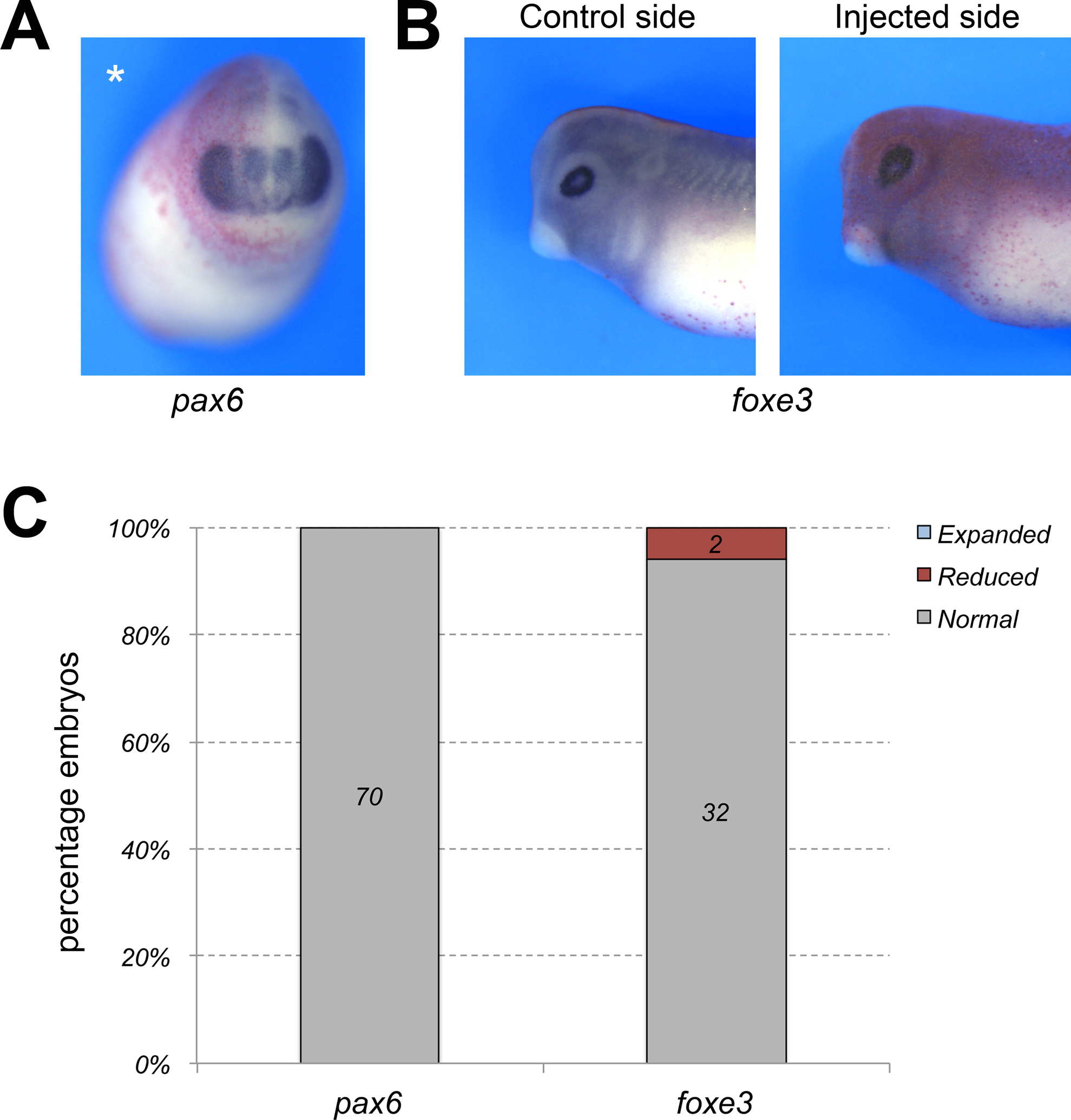
(A) Hmgn1 knockdown does not affect the expression of pax6 in the developing eye, prospective lens and forebrain at NF stage 23. Injected side (asterisk) is showing the lineage tracer Red-Gal. Frontal view, dorsal to top. (B) The expression of foxe3 in the lens vesicle is not affected in Hmgn1 morphant embryos at NF stage 30. Injected side is showing the lineage tracer Red-Gal. Lateral views, dorsal to top, anterior to left. (C) Quantification of the phenotypes. The number of embryos showing a given phenotype is indicated in each bar.
Hmgn1 depletion does not affect cell death or proliferation in the ectoderm
To determine the mechanism underlying the reduction of neural crest gene expression in Hmgn1-depleted embryos we analyzed cell death in the ectoderm of Hmgn1 morphant embryos at the neurula stage by TUNEL staining. We observed no significant change in the number of TUNEL-positive cells in the ectoderm of Hmgn1-depleted embryos (Fig. 5A,B). By comparison injection of Sf3b4MO resulted in increased TUNEL positivity, as previously reported (Devotta et al., 2016). We next examined whether changes in neural crest gene expression could be the result of a defect in neural crest progenitor proliferation rate by performing phospho-histone H3 (pHH3) immunostaining at neurula stage (NF stage 15). We found that the number of pHH3-positive cells was not significantly affected in Hmgn1MO-injected embryos as compared to controls (Fig. 5C,D). To ascertain that defects in cell death and proliferation did not occur at an earlier time point (prior to neurula stage), we also performed TUNEL and pHH3 staining in late gastrula stage embryos (NF stage 12), and observed no significant differences (Supplementary Figure S1; not shown). Based on these observations we can speculate that reduced neural crest gene expression in Hmgn1-depleted embryos may indicate that these cells fail to differentiate and/or maintain an undifferentiated/pluripotent state.
Figure 5: Hmgn1 knockdown does not affect cell death or proliferation in the ectoderm.
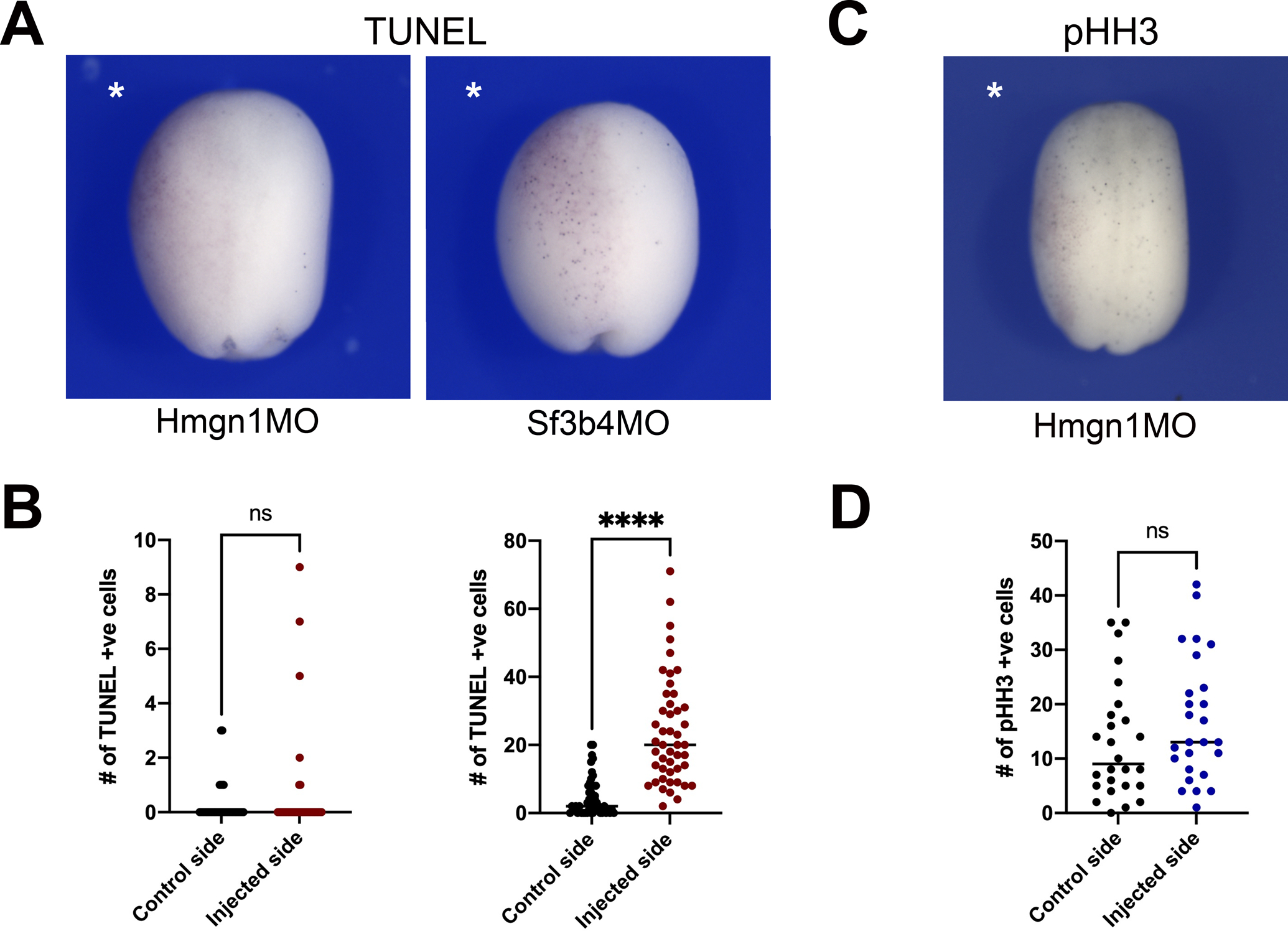
(A) TUNEL staining of representative Hmgn1 and Sf3b4 morphant embryos at stage 15. The injected side is indicated by an asterisk. Dorsal view, anterior to top. (B) Quantification of the number of TUNEL-positive cells in control and injected sides of Hmgn1MO (n=40; left graph) and Sf3b4MO (n=47; right graph) injected embryos at stage15. (C) pHH3 immunostaining of a representative Hmgn1 morphant embryos at stage 15. (D) Quantification of the number of pHH3-positive cells in control and injected sides of Hmgn1MO injected embryos (n=26). (B, D) Each dot represents one embryo. p-values were calculated using unpaired t-test with Welch’s correction, **** p<0.0001; ns: not significant.
Hmgn1 is necessary for cranial neural crest cells migration
At the tailbud stage, cranial neural crest cells migrate ventrally as individual streams towards the pharyngeal arches. We next analysed the dorsoventral extension of neural crest streams using ISH for twist1 and sox10, two genes expressed in migratory neural crest cells. We found that in both cases the vast majority of the Hmgn1 morphant embryos had defects in neural crest streams formation as compared to CoMO-injected embryos (Fig 6A; Supplementary Figure S2). To further quantify this defect we measured the dorsoventral extension of the first three neural crest streams (Fig. 6B) of CoMO- and Hmgn1MO-injected embryos after ISH with sox10 and twist1. We observed a severe reduction of the net distance migrated in the posterior cranial neural crest streams (2nd and 3rd) while the first stream was only marginally affected (Fig 6C–D).
Figure 6: Hmgn1 knockdown affects neural crest streams formation.
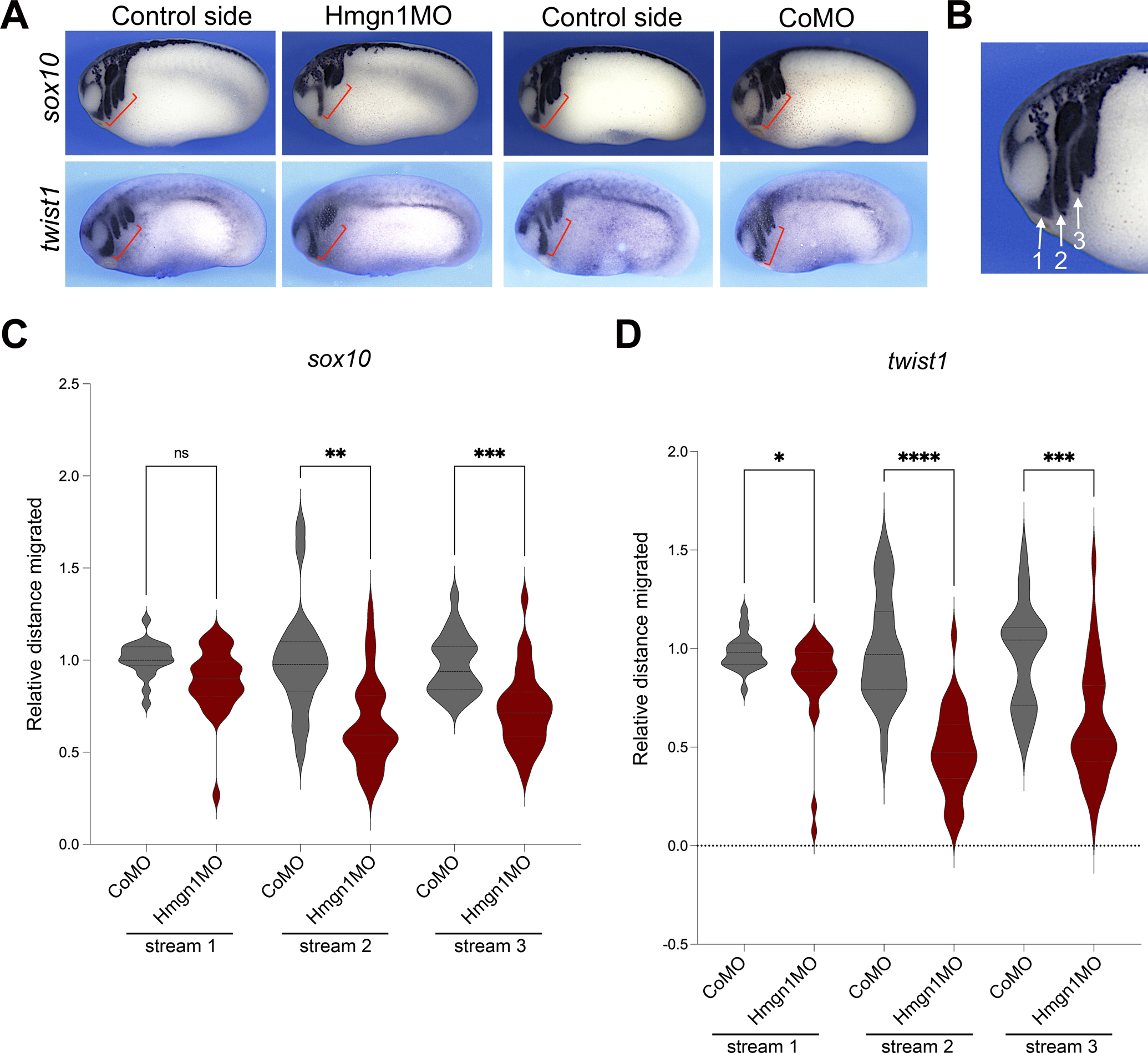
(A) Phenotype of CoMO (20 ng) and Hmgn1MO (20 ng) injected embryos at NF stage 23–25 analyzed for sox10 and twist1 expression. The red brackets indicate the extent of dorso-ventral extension of neural crest streams in control and MO-injected sides. (B) Higher magnification of an embryo stained with sox10 showing the position of the three neural crest streams. Lateral view, dorsal to top, anterior to left. (C-D) Graph plotting the distance migrated by neural crest cells in stream 1, 2 and 3 of CoMO- and Hmgn1MO-injected embryos stained by sox10 (C) or twist1 (D). A similar number of embryos were analyzed for each gene (CoMO, n=21 and Hmgn1MO, n=30). Brown-Forsyth and welch ANOVA test, with multiple comparisons test (Dunnett) with individual variances computed for each comparison. *, p=0.0141; **, p=0.0013; ***, p=0.0003; ****, p<0.0001; ns: not significant.
Hmgn1-depleted embryos display abnormal craniofacial cartilages
To evaluate the long-term effects of Hmgn1 depletion on neural crest derived craniofacial structures, embryos with unilateral injection of Hmgn1MO were cultured to the tadpole stage (NF stage 45) to analyze craniofacial cartilage formation using alcian blue staining. Morphant tadpoles exhibited size reduction of craniofacial cartilage, with the posterior cartilages being the most consistently affected when compared to the uninjected side (Fig. 7A,B). This is consistent with the observation that the extent of dorsoventral cranial neural crest cells migration is more severely affected in the posterior pharyngeal arches (Fig 6).
Figure 7: Hmgn1 knockdown affects craniofacial cartilage formation.
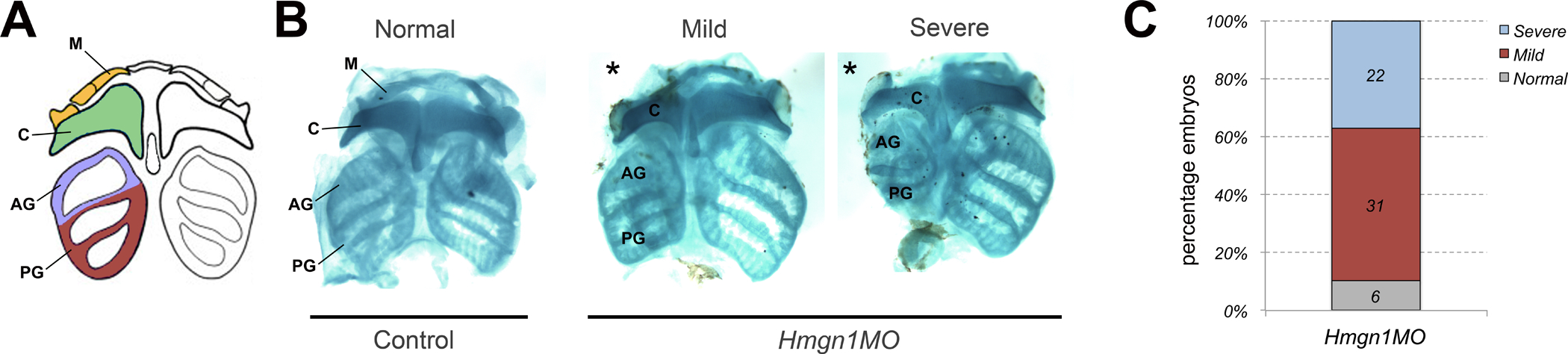
(A) Schematic representation of neural crest derived cranial cartilages at the tailbud stage, (modified from Sadaghiani and Thiebaud; 1987). Neural crest cells in stream 1 contribute to Meckel’s cartilage (M), in stream 2 they contribute to ceratohyal cartilage (C), and in streams 3 and 4 they contribute to the anterior (AG) and posterior (PG) gill cartilages, respectively. (B) Alcian blue staining of dissected craniofacial cartilages of control and Hmgn1MO (20 ng), injected tadpoles at NF stage 45. The injected side is indicated by an asterisk. (C) Quantification of the phenotypes. The number of embryos showing a given phenotype is indicated in each bar.
Discussion
In recent years, important advances have been made in the characterization of the gene regulatory network (GRN) underlying cranial neural crest formation (Simões-Costa and Bronner, 2015; Chong-Morrison and Sauka-Spengler, 2021). Here, we describe the role of Hmgn1, a novel player in the neural crest GRN, and its requirement during neural crest and craniofacial development in Xenopus. Hmgn1 is a ubiquitously expressed protein unique to vertebrates that binds specifically to chromatin nucleosome core particles. The interaction of Hmgn1 with nucleosomes modulates the organization of the chromatin and histone modifications, and as such Hmgn1 is considered an epigenetic regulator of transcription (Kugler et al., 2012; Martinez de Paz and Ausio, 2016, Naduri et al., 2020). Other reported functions of Hmgn1 include DNA replication and repair (Vestner et al., 1998; Birger et al., 2003).
We confirmed that hmgn1 is broadly expressed during Xenopus early embryogenesis (Gawantka et al., 2008; Korner et al., 2003), however we also found that hmgn1 transcripts were enriched in the pharyngeal arches and developing eyes. This expression in the head region propounded a possible role for Hmgn1 in the regulation of craniofacial development. Hmgn1 morpholino-mediated knockdown resulted in decreased expression of a subset of neural crest specifier genes (sox8, sox10, foxd3 and twist1). Importantly, this phenotype could be rescued by restoring Hmgn1 expression in morphant embryos, demonstrating the specificity of the knockdown phenotype. At the tailbud stage, cranial neural crest cells migrate as streams in a stereotypical manner towards the pharyngeal arches where they will eventually differentiate into the unique skeletal elements of the face (Sadaghiani and Thiebaud, 1987; Minoux and Rijli, 2010). At this stage, the dorsoventral extension of neural crest streams was significantly reduced in Hmgn1 morphant as compared to control embryos, resulting in severe craniofacial cartilage defects at the tadpole stage. We propose that the craniofacial phenotype observed at these tadpoles is likely due to the inability of neural crest cells to reach the pharyngeal arches in sufficient numbers to generate full size cartilages.
In our hands, Hmgn1 overexpression did not significantly affect sox10 expression at the neurula stage. However a previous study has shown that injection in Xenopus embryos of Hmgn1 proteins, or its related factor Hmgn2, caused malformations affecting primarily the head region and the body axis, a phenotype that was not observed in embryos overexpressing a mutant form of Hmgn2 lacking a functional nucleosomal binding domain (Korner et al., 2003). Hmgn2 knockdown had a very similar phenotype to the gain of function phenotype, suggesting that changes in Hmgn protein levels are broadly detrimental to embryonic development. The phenotype of Hmgn1 knockdown has not been previously reported by Korner et al. (2003).
Unlike other genes that have been linked to increased apoptosis and craniofacial defect, such as Sf3b4 in Nager syndrome (Devotta et al., 2016), Eftud2 in mandibulofacial dysostosis with microcephaly (Lei et al., 2017; Beauchamp et al., 2021) or Tcof1 in Treacher Collins syndrome (Dixon et al, 2006), we did not observed any changes in the rate of cell death or proliferation in the embryonic ectoderm at the late gastrula or neurula stage upon Hmgn1 knockdown. This result signifies that reduced neural crest gene expression cannot be explained by a targeted depletion of neural crest progenitors through apoptosis or by a reduction in the proliferative capacity of these progenitors. One possibility is that these cells may have adopted another fate, however this is not supported by our results showing that neural plate (sox2) and placode (six1 and foxi4.1) genes expression were largely unaffected or reduced (dmrta1) in Hmgn1 morphant embryos. An alternative interpretation is that these cells in the absence of Hmgn1 function may lose the ability to differentiate and rather maintain an undifferentiated/multipotent state. Consistent with this possibility, a recent study using reprogrammed mouse embryonic fibroblasts has shown that HMGN proteins are required at cell-type specific regulatory regions to maintain cell fate, by optimizing and stabilizing the epigenetic landscape that ensures maintenance of cell identity (He et al., 2018). HMGN proteins have been shown to regulate the expression of cell-type specific genes in other contexts as well. For example, mouse embryonic stem cells lacking both HMGN1 and HMGN2 down-regulate the expression of genes involved in oligodendrocyte differentiation (OLIG1 and OLIG2). In vivo, mouse lacking HMGN1 and HMGN2 have a reduced number of oligodendrocytes in the spinal cord, impaired myelination and neurological defects (Deng et al., 2017). Therefore the presence of HMGN proteins is essential for the expression of neural differentiation genes. Future studies will determine whether this is also the case in the context of the neural crest.
In conclusion, we present evidence of a novel role of Hmgn1 in the regulation of neural crest and craniofacial development in Xenopus. Whilst Hmgn proteins regulate global transcription, there is evidence that these proteins can regulate the expression of cell-type specific genes either directly (West et al., 2004; Rubinstein et al., 2005) or indirectly by modulating the binding of accessory transcription factors at the cis-regulatory regions of these genes (Amen et al., 2008; Ueda et al., 2009). Future lines of investigation will address the precise molecular mechanisms underlying Hmgn1 function that ensure proper development of the cranial neural crest and its derivatives.
Supplementary Material
Acknowledgements
We are grateful to Drs. Aditi Dubey and Nadege Gouignard for assistance with the GraphPad analysis, and to members of the lab for discussions. The work benefited from the support of Xenbase (http://www.xenbase.org/-RRID: SCR_003280) and the National Xenopus Resource (http://mbl.edu/xenopus/-RRID: SCR_013731).
Funding
The work was supported by a grant from the National Institutes of Health to J-P-S-J (R01DE025468).
Footnotes
Declaration of competing interest
The authors declare no competing interests.
References
- Abuhatzira L, Shamir A, Schones DE, Schaffer AA & Bustin M (2011). The chromatin-binding protein HMGN1 regulates the expression of methyl CpG-binding protein 2 (MECP2) and affects the behavior of mice. J. Biol. Chem 286: 42051–42062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amen M, Espinoza HM, Cox C, Liang X, Wang J, Link TM, Brennan RG, Martin JF, & Amendt BA (2008). Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/beta-catenin signaling, Nucleic Acids Res 36, 462–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee Y-H, Credidio C & Saint-Jeannet J-P (2003). Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev. Biol 259, 19–33. [DOI] [PubMed] [Google Scholar]
- Beauchamp M-C, Djedid A, Bareke E, Mekuri F Aber R, Tam AS, Lines MA, Boycott KM, Sterling PC, Fish JL, Majewski J & Jerome-Majewska LA (2021). Mutation in Eftud2 causes craniofacial defects in mice via mis-splicing of Mdm2 and increased P53. Human Molecular Genetics, doi: 10.1093/hmg/ddab051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Rose CS, Remo BF & Brown DD (1998). The expression pattern of thyroid hormone response genes in remodeling tadpole tissues defines distinct growth and resorbtion gene expression programs. Dev. Biol 203, 24–35. [DOI] [PubMed] [Google Scholar]
- Birger Y, West KL, Postnikov YV, Lim JH, Furusawa T, Wagner JP, Laufer CS, Kraemer KH & Bustin M (2003) Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J, 22, 1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birger Y, Catez F, Furusawa T, Lim JH, Prymakowska-Bosak M, West KL, Postnikov YV, Haines DC & Bustin M (2005). Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1, Cancer Res 65, 6711–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong-Morrison V & Sauka-Spengler T (2021). The cranial neural crest in a mutiomics era. Front Physiol 12:634440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Postnikov Y, Zhang S, Garrett L, Becker L, Racz I, Holter SM, Wurst W, Fuchs H, Gailus-Durner V, et al. (2017). Interplay between H1 and HMGN epigenetically regulates OLIG1&2 expression and oligodendrocyte differentiation. Nucleic Acids Res 45, 3031–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devotta A, Juraver-Geslin H, Gonzalez JA, Hong C-S & Saint-Jeannet J-P (2016) Sf3b4-depleted Xenopus embryos: a model to study the pathogenesis of craniofacial defects in Nager syndrome. Dev Biol 415, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey J-P, Dixon M & Trainor PA (2006). Tcof1/Treacle is required for NC cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA 103, 13403–13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, et al. , (1998). Gene expression screening in Xenopus identifies molecular pathways, predicts gene function and provides a global view of embryonic patterning. Mech. Dev 77, 95–141. [DOI] [PubMed] [Google Scholar]
- Harland RM (1991). In situ hybridization: an improved whole-mount method for Xenopus embryos. Meth. Cell Biol 36, 685–695. [DOI] [PubMed] [Google Scholar]
- He B, Deng T, Zhu I, Furusawa T, Zhang S, Tang W, Postnikov Y, Ambs S, Li CC, Livak F, et al. (2018). Binding of HMGN proteins to cell specific enhancers stabilizes cell identity. Nat. Commun 9, 5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensey C & Gautier J (1998). Programmed cell death during Xenopus development: a spatio-temporal analysis. Developmental Biology, 203, 36–48. [DOI] [PubMed] [Google Scholar]
- Hirsch N & Harris WA (1997). Xenopus pax6 and retinal development. J. Neurobiol 32, 45–61. [PubMed] [Google Scholar]
- Hock R, Furusawa T, Ueda T, Bustin M (2007). HMG chromosomal proteins in development and disease. Trends Cell Biol 17, 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Strobl-Mazzulla PH & Bronner ME (2014). Epigenetic regulation in neural crest development. Dev Biol 396,159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood ND, Pluck A & Gurdon JB (1989). A Xenopus mRNA related to Drosophila twist is expressed in response to induction in the mesoderm and the neural crest. Cell 59, 893–903. [DOI] [PubMed] [Google Scholar]
- Huang X, Hong CS, O’Donnell M & Saint-Jeannet J-P. (2005). The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl Acad. Sci. USA 102, 11349–11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KL, Moody SA & Jamrich M (1999). A novel forkhead gene mediates early steps during Xenopus lens formation. Development 126, 5107–5116. [DOI] [PubMed] [Google Scholar]
- Korner U, Bustin M, Scheer U & Hock R (2003). Developmental role of HMGN proteins in Xenopus laevis. Mech. Dev 120, 1177–1192. [DOI] [PubMed] [Google Scholar]
- Kugler JE, Deng T & Bustin M (2012). The HMGN family of chromatin-binding proteins: Dynamic modulators of epigenetic processes. Biochim. Biophys. Acta 1819, 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Yan SY, Yang R, Chen JY, Li Y, Bu Y, Chang N, Zhou Q, Zhu X, Li CY et al. (2017) Spliceosomal protein eftud2 mutation leads to p53-dependent apoptosis in zebrafish neural progenitors. Nucleic acids research, 45, 3422–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Paz A & Ausio J (2016). HMGNs: The enhancer charmers. Bioessay 38, 226–231. [DOI] [PubMed] [Google Scholar]
- Mayor R, Morgan R & Sargent MG (1995). Induction of the prospective neural crest of Xenopus. Development 121, 767–777. [DOI] [PubMed] [Google Scholar]
- Minoux M, & Rijli FM (2010). Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development, 137(16), 2605–2621. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S & Sasai Y (1998). Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development 125, 579–587. [DOI] [PubMed] [Google Scholar]
- Naduri R, Furusawa T & Bustin M (2020). Biological functions of HMGN chromosomal proteins. Int. J. Mol. Sci 21, 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD & Faber J (1956). Normal table of Xenopus laevis (Daudin). Amsterdam: North-Holland Publishing Co. [Google Scholar]
- O’Donnell M, Hong CS, Huang X, Delnicki RJ & Saint-Jeannet JP (2006). Functional analysis of Sox8 during neural crest development in Xenopus. Development 133, 3817–3826. [DOI] [PubMed] [Google Scholar]
- Park B-Y, Tachi-Duprat M, Devotta A & Saint-Jeannet J-P (2021). The core splicing factors EFTUD2, SNRPB and TXNL4a are essential for neural crest and craniofacial development. Hum. Mol. Genet submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl BS, Knochel S Dillinger K & Knochel W (2002). Sequence and expression of FoxB2 (XFD-5) and FoxI1c (XFD-10) in Xenopus embryogenesis, Mech. Dev 117, 283–287. [DOI] [PubMed] [Google Scholar]
- Postnikov YV & Bustin M (2010). Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta, 1799, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD & Nie S (2018). Specifying neural crest cells: from chromatin to morphogens and factors in between. WIREs Dev. Biol 2018;7:e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein YR, Furusawa T, Lim JH, Postnikov YV, West KL, Birger Y, Lee S, Nguyen P, Trepel JB, & Bustin M (2005). Chromosomal protein HMGN1 modulates the expression of N-cadherin, FEBS J 272, 5853–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani B & Thiebaud CH (1987). Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev. Biol 124, 91–110. [DOI] [PubMed] [Google Scholar]
- Saint-Jeannet J-P (2017). Whole-mount in situ hybridization of Xenopus embryos. Cold Spring Harb. Protoc 2017, pdb.prot097287. [DOI] [PubMed] [Google Scholar]
- Saka Y, & Smith JC (2001). Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Developmental Biology 229, 307–318. [DOI] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K & Sasai Y (2001). Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development 128, 2525–2536. [DOI] [PubMed] [Google Scholar]
- Simões-Costa M & Bronner ME (2015). Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM & Forman D (1980). An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos. Journal of Embryology and Experimental Morphology, 56, 283–299. [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E & Saint-Jeannet J-P (2002). The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development 129, 421–432. [DOI] [PubMed] [Google Scholar]
- Trainor PA, & Andrews BT (2013). Facial dysostoses: Etiology, pathogenesis and management. American Journal of Medical Genetics Part C: Seminars in Medical Genetics, 163, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Furusawa T, Kurahashi T, Tessarollo L, & Bustin M (2009). The nucleosome binding protein HMGN3 modulates the transcription profile of pancreatic β-cells and affects insulin secretion, Mol. Cell. Biol 29, 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestner B, Bustin M & Gruss C (1998) Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J. Biol. Chem, 273, 9409–9414. [DOI] [PubMed] [Google Scholar]
- West KL, Castellini MA, Duncan MK & Bustin M (2004). Chromosomal proteins HMGN3a and HMGN3b regulate the expression of glycine transporter 1. Mol. Cell. Biol 24, 3747–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


