Abstract
The microbiome plays an important role in shaping plant growth and immunity, but few plant genes and pathways impacting plant microbiome composition have been reported. In Arabidopsis thaliana, the phosphate starvation response (PSR) was recently found to modulate the root microbiome upon phosphate (Pi) starvation through the transcriptional regulator PHR1. Here, we report that A. thaliana PHR1 directly binds to the promoters of rapid alkalinization factor (RALF) genes, and activates their expression under phosphate‐starvation conditions. RALFs in turn suppress complex formation of pathogen‐associated molecular pattern (PAMP)‐triggered immunity (PTI) receptor through FERONIA, a previously‐identified PTI modulator that increases resistance to certain detrimental microorganisms. Suppression of immunity via the PHR1‐RALF‐FERONIA axis allows colonization by specialized root microbiota that help to alleviate phosphate starvation by upregulating the expression of PSR genes. These findings provide a new paradigm for coordination of host‐microbe homeostasis through modulating plant innate immunity after environmental perturbations.
Keywords: FER, phosphate starvation response, PHR1, plant immunity, RALF
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction; Plant Biology
Arabidopsis PHOSPHATE STARVATION RESPONSE‐1 impinges on PAMP‐triggered immunity to allow for the growth of microorganisms that help alleviate phosphate stress.

Introduction
Phosphorus is an essential macronutrient for plant growth and development and is one of the major factors that limits crop production worldwide. The concentration of inorganic phosphate (Pi), the main form of phosphorus that can be absorbed by plants, is typically less than 10 μM in soils (Raghothama & Karthikeyan, 1999). To better cope with Pi‐deficient environments, plants have evolved adaptive Pi starvation responses. Common strategies for improving Pi uptake include altering root morphology, such as the rapid elongation of lateral roots (LRs) (Lambers et al, 2015) and activating Pi starvation‐related transporters and transcriptional regulators (Mudge et al, 2002; Nilsson et al, 2007; Duan et al, 2008). Another strategy involves establishing beneficial relationships with some microorganisms (Harrison, 2012; Hacquard et al, 2016; Hiruma et al, 2016). Under phosphate starvation responses (PSRs), plants shape the root microbiome to alleviate the Pi starvation in Arabidopsis thaliana (A. thaliana, Isidra‐Arellano et al, 2021; Finkel et al, 2019; Castrillo et al, 2017; Hiruma et al, 2016). In addition, genes that are involved in the Pi starvation contribute to the plant immune response and shape the root microbiome composition (Bustos et al, 2010), likely benefiting plants under Pi‐limiting conditions.
PHOSPHATE STARVATION RESPONSE 1 (PHR1) is a master transcription factor involved in the Pi starvation in A. thaliana. Under Pi starvation conditions, PHR1 binds to the PHR1‐binding site (P1BS) element in the promoters of Pi starvation‐induced (PSI) genes, activating their expression and promoting Pi uptake (Bustos et al, 2010). PHR1 also regulates plant immunity at the transcription level in A. thaliana. Transcriptomic analysis indicated that the genes related to jasmonic acid (JA) and salicylic acid (SA) are differentially expressed in a PHR1‐dependent manner during the Pi starvation. PHR1 negatively regulates immune reactions triggered by the bacterial epitope flg22 (a peptide corresponding to the most conserved domain of bacterial flagellin), a pathogen‐associated molecular pattern (PAMP) that induces PAMP‐triggered immunity (PTI) (Castrillo et al, 2017). Although PHR1 plays an important role in immunity suppression during the Pi starvation in A. thaliana, the molecular pathway underlying the PHR‐mediated inhibition of plant immunity and the understanding of how this process benefits plants in relieving Pi starvation remain unclear.
How plant innate immunity coordinates microbe‐host homeostasis is obscure. In plants, the elicitation of PTI relies on the recognition of PAMPs by cell surface‐localized receptor kinases. For instance, the receptor flagellin sensing 2 (FLS2) perceives flg22 and activates the immune responses, including the activation of mitogen‐associated protein kinase (MAPK) cascades, reactive oxygen species (ROS) bursts and the induction of defence‐responsive gene expression (Böhm et al, 2014; Macho et al, 2014). PTI triggered by flg22 also requires coreceptors such as BRASSINOSTEROID INSENSITIVE 1‐ASSOCIATED RECEPTOR KINASE 1 (BAK1) (Li et al, 2002). Furthermore, the Catharanthus roseus receptor‐like kinase 1‐like (CrRLK1L) family member FERONIA (FER) in A. thaliana can facilitate flg22‐induced FLS2‐BAK1 complex formation to initiate immune signalling (Stegmann et al, 2017). RALF (rapid alkalinization factors) peptides are ligands of CrRLK1L receptors, such as FER (Franck et al, 2018). The perception of RALF23 by FER inhibits plant immunity by disrupting the complex between FLS2 and BAK1(Stegmann et al, 2017). RALF‐FER also interferes with JA signalling to modulate plant immunity (Shen et al, 2017; Guo et al, 2018; Zhang et al, 2020). This shift in PTI allows plants to restrict the colonization of pathogenic microbes. Several studies have also investigated the impact of PTI on microbiome structure (Lebeis et al, 2015; Chen et al, 2020). Although PTI components such as immune‐related hormones affect the microbiome composition, in some cases, immune‐related mutants show relatively mild changes in root microbiota structure under natural soil conditions (Lebeis et al, 2015; Chen et al, 2020). Recently, a study showed that FER controls microbiome composition largely independent of its immune function in mediating pattern recognition receptor (PRR) complex formation under normal conditions (Song et al, 2021). At present, a direct molecular link between PTI‐associated immune pathways, microbiome composition and abiotic stress conditions of mineral starvation has yet to be identified. In this study, we established that a PHR1‐RALFs‐FER molecular pathway is used in A. thaliana to inhibit immunity under Pi‐starvation conditions and further revealed that the PHR1‐RALFs‐FER axis reduces PRR complex formation to modulate the rhizosphere microbiota composition, which in turn helps alleviate Pi stress. These results reveal a finely tuned mechanism employed by A. thaliana to balance the plant’s immune response, microbiota composition and mineral uptake under nutrient stress conditions.
Results
The RALF‐FER module is involved in low‐Pi stress‐mediated suppression of plant immunity
To determine the impact of Pi starvation on plant immune response, we compared the flg22‐induced activation of phosphorylated MAPK proteins, PAMP‐responsive marker gene expression and ROS burst in wild‐type (WT; Col‐0) A. thaliana plant roots grown under Pi‐sufficient conditions (1/2‐strength Murashige and Skoog (MS) media supplemented with 1.25 mM Pi; HP) and Pi‐deficient conditions (low‐Pi media supplemented with 10 μM Pi; LP). We observed that all tested immune hallmarks were consistently suppressed under LP conditions in roots (Appendix Fig S1A–C). To further verify these observations, we tested the growth of Pseudomonas syringae pv. tomato DC3000 (Pto DC3000) in A. thaliana roots under Pi starvation conditions. The results demonstrated that Pto DC3000 had more colonization in roots under LP conditions than under HP conditions (Appendix Fig S1D). Taken together, these results indicate that Pi starvation suppresses the plant innate immunity.
Next, we sought to determine the molecular mechanisms through which the Pi starvation suppresses the immune response. Recent studies have highlighted the role of FER in plant immunity regulation (Stegmann et al, 2017, Guo et al, 2018 and Zhang et al, 2020). Based on current research on the molecular function of FER, this receptor is also involved in nutrition stress responses (Xu et al, 2018). And interestingly, the fer‐4 (a loss‐of‐function mutant of FER) mutant (Duan et al, 2010) showed more anthocyanin accumulation (Appendix Fig S1E and F) under LP condition, which is a hallmark of Pi deficiency (Nilsson et al, 2007). So, we hypothesized that FER is involved in Pi starvation‐mediated immunity suppression. To test this hypothesis, we compared flg22‐induced MAPK activation in WT and fer‐4 mutant roots under HP and LP conditions. The results showed that the Pi starvation‐mediated MAPK inhibition observed in the WT was nearly completely abolished in the fer‐4 mutants under LP conditions (Fig 1A and Appendix Fig S1G). This suggests that FER is required for Pi starvation‐mediated immunity suppression in roots. ROS assays also confirmed a role of FER in Pi starvation‐mediated immunity suppression in roots (Fig 1B and C). FER is a receptor of RALFs and perception of RALF23 by FER inhibits immunity triggered by PAMPs such as flg22 (Stegmann et al, 2017). We questioned whether RALF23 is also involved in Pi starvation‐mediated immunity suppression. To this end, we first performed RNA sequencing (RNA‐seq) analysis with the A. thaliana roots treated with 1 μM of RALF23 or grown on LP media. Gene Ontology (GO) analysis revealed that differentially regulated genes involved in immunity were enriched under Pi‐starvation conditions (Appendix Fig S2A and B) or upon RALF23 treatment (Appendix Fig S2C and D). In total, 738 defence responsive genes (Raudvere et al, 2019) were analysed in RALF23‐treated roots and LP‐treated plants. These genes exhibited the same pattern in terms of differential expression under both treatments (Fig 1D, Table EV1). Furthermore, most of the differentially expressed genes were shifted in the same direction, and the values were highly correlated with one another (r = 0.8343, P < 0.001) (Fig 1E), suggesting that Pi starvation and RALF23 regulate similar type of plant defence response at the transcriptional level. To further confirm the role of RALF23 in Pi starvation‐mediated immunity regulation, we obtained a GFP‐labelled RALF23 overexpression transgenic line (RALF23‐OE) (Dobón et al, 2015) and compared flg22‐induced expression of the PAMP‐responsive marker gene FRK1 (Asai et al, 2002; Boudsocq et al, 2010) in RALF23‐OE and WT roots under both HP and LP conditions, respectively (Fig 1F and Appendix Fig S3A). Upon flg22 treatment, RALF23‐OE repressed FRK1 expression under HP conditions. However, the expression of FRK1 was repressed in both WT and RALF23‐OE under LP conditions, demonstrating that RALF23 in WT is involved in immunity regulation under Pi starvation (Fig 1F). We also performed bacterial growth assay on WT plants and RALF23‐OE roots. Pto DC3000 had more colonization on RALF23‐OE roots than in WT roots under HP conditions; however, there was no significant difference between WT and RALF23‐OE plants under LP conditions (Fig 1G). A ROS burst was also observed for both WT plants and RALF23 peptide treatment under LP conditions (Fig 1B and C, Appendix Fig S3B and C); however, there was no obvious ROS burst change in the fer‐4 mutants (Fig 1B and C). Taken together, these results suggest that the RALF23‐FER complex is involved in immunity suppression in A. thaliana under low‐Pi conditions.
Figure 1. The RALF‐FER module is involved in PSR‐mediated immunity suppression in Arabidopsis .
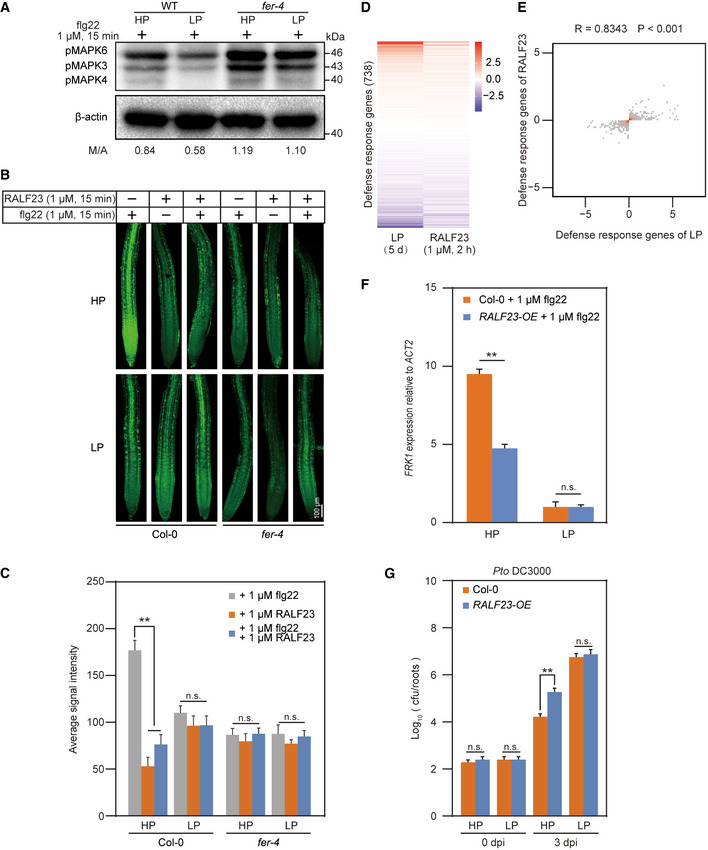
- flg22‐induced MAPK activation in Col‐0 and fer‐4 mutant roots under Pi‐starvation conditions. Seedlings grown in either HP or LP media were treated with 1 μM of flg22 for 15 min and roots were harvested. The MAPK level was determined with anti‐pMAPK, and β‐actin was used as a loading control.
- flg22‐induced ROS production in Col‐0 and fer‐4 mutant roots under Pi‐starvation conditions. Seedlings grown in either HP or LP media were treated with 1 μM of flg22, RALF23 or both flg22 and RALF23 for 15 min. Roots were harvested and stained with H2DCFDA. One representative root was shown.
- Average H2DCFDA signal intensity in roots shown in (B). The fluorescence intensity was quantified with ImageJ. The data shown indicate the means ± SDs (n = 30, n refers to the number of roots per group); n.s., not significant; **P < 0.01 (Student’s t test).
- Heatmap of differentially expressed defense‐responsive genes after LP or RALF23 treatment (n = 3, n refers to technical replicates). Seven‐day‐old Col‐0 seedlings were treated with either LP for 5 days or 1 μM of RALF23 for 2 h.
- Density figure showing the comparison of defense‐responsive genes between LP treatment and RALF23 treatment (n = 3, n refers to technical replicates). The x‐ and y‐axes represent changes in transcription level, and the red area indicates high density. Spearman correlations were used for the correlation analysis.
- RALF23 inhibits flg22‐induced MAMP‐responsive FRK1 gene expression in a Pi content‐dependent manner. Seedlings grown in either HP or LP media were treated with water or flg22 for 15 min and roots were harvested. The relative expression of FRK1 was quantified via RT‐qPCR. The data shown indicate the means ± SDs (n = 3, n refers to technical replicates); n.s., not significant; **P < 0.01 (Student’s t test).
- Growth of Pto DC3000 in Col‐0 and RALF23‐OE lines under Pi‐starvation conditions. Seedlings grown in either HP or LP media were root soaking inoculated with bacterial suspension (OD600 = 0.2) for 2 min, and the number of bacteria from individual roots was quantified at 0 dpi and 3 dpi. The data shown indicate the means ± SDs (n = 27, n refers to the number of roots per group); n.s., not significant; **P < 0.01 (Student’s t test).
Data information: All experiments were repeated three times with similar results.
PHR1 targets and activates RALF expression during the Pi starvation
To determine how RALF23‐FER is involved in Pi starvation‐mediated immunity suppression, we investigated the protein levels of RALF23 and FER, respectively in RALF23‐GFP (Dobón et al, 2015) and FER‐GFP (Duan et al, 2010) under both HP and LP conditions. Western blot analysis revealed that seedlings grown under LP conditions accumulated mature RALF23 at a much higher level than seedlings grown under HP conditions (Fig 2A), yet no differences were observed for FER (Appendix Fig S4). This indicates that the Pi starvation induces mature RALF23 accumulation. RALF23 is cleaved by SITE‐1 PROTEASE (S1P) (Srivastava et al, 2009; Stegmann et al, 2017). Sequence analysis revealed that several other RALFs, such as RALF1, RALF4, RALF22, RALF33 and RALF34, also contain the S1P cleavage site (Appendix Fig S5A) (Stegmann et al, 2017). All these RALFs inhibited flg22‐triggered immunity (Appendix Fig S6). Interestingly, we found that, except for the expression of RALF1, the expression of other RALFs was significantly induced in roots during the Pi starvation (Fig 2B). We also analysed published microarray data of A. thaliana subjected to 7 days of Pi starvation and discovered that the expression of most of the above tested RALFs was upregulated during the low‐Pi stress (Fig 2C).
Figure 2. PHR1 targets and activates RALF expression under Pi‐starvation conditions.
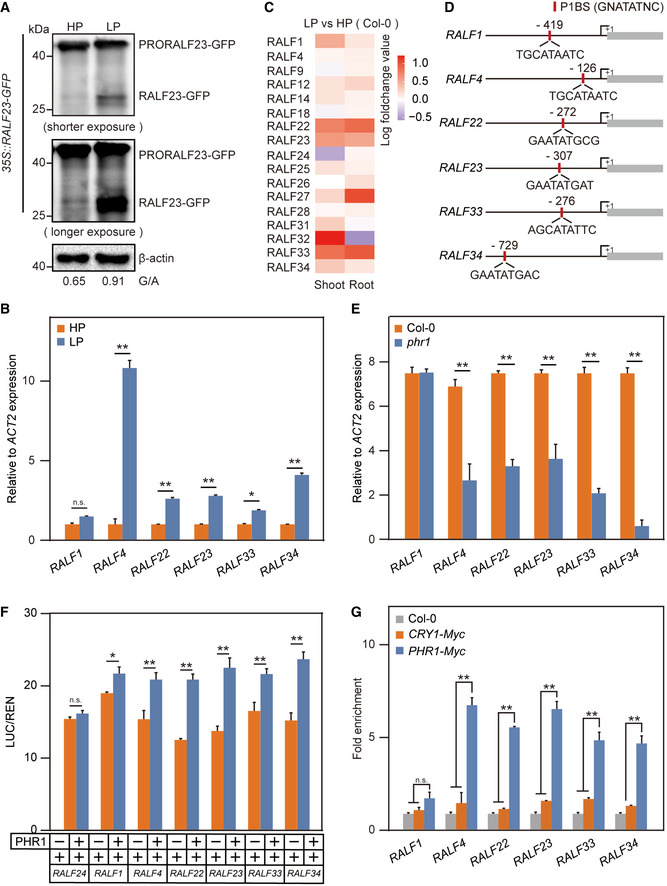
- The PSR induces the cleavage of PRORALF23‐GFP into mature RALF23‐GFP. The 35S promoter‐driven RALF23‐GFP line was grown in either HP or LP media for 5 days and roots were harvested. Premature and mature RALF23 levels were determined with anti‐GFP. β‐actin was used as a loading control.
- The PSR activates RALF expression under LP conditions. Col‐0 seedlings were grown in either HP or LP media for 5 days and roots were harvested. The relative expression of RALFs was quantified via RT‐qPCR. The data shown indicate the means ± SDs (n = 3, n refers to technical replicates); n.s., not significant; *P < 0.05; **P < 0.01 (Student’s t test).
- Expression pattern of RALFs in the shoots and roots under Pi‐starvation conditions. The microarray data were obtained from Bustos et al (2010).
- PHR1‐binding site (P1BS, GNATATNC) prediction within 1,000 bp of certain RALF promoters. The positions are relative to the start codon (+1).
- The PSR activates RALF expression in a PHR1‐dependent manner. Col‐0 and phr1 seedlings were grown in LP media for 5 days and roots were harvested. The relative expression of RALFs was quantified via RT‐qPCR. The data shown indicate the means ± SDs (n = 3, n refers to technical replicates); n.s., not significant; **P < 0.01 (Student’s t test).
- PHR1 activates RALF promoters in plants. Depicted RALF promoter‐LUC fusions (RALF24 as a negative control) were co‐expressed together with 35S promoter‐driven PHR1 or an empty vector control in Arabidopsis protoplasts. The relative LUC/REN reporter activity was quantified. The data shown indicate the means ± SDs (n = 3, n refers to technical replicates); n.s., not significant; *P < 0.05; **P < 0.01 (Student’s t test).
- PHR1 directly binds to RALF promoters, as shown via ChIP‐PCR. The Myc‐labelled PHR1 overexpression line was immunoprecipitated with anti‐Myc. ChIP analysis was performed on Col‐0, CRY1‐Myc and PHR1‐Myc transformed plants, with Col‐0 and CRY1‐Myc transformed plants as negative controls. The data shown indicate the means ± SDs (n = 3, n refers to technical replicates); n.s., not significant; **P < 0.01 (Student’s t test).
Data information: All experiments were repeated three times with similar results.
Next, we sought to determine how the expression of RALFs is induced during the Pi starvation. Based on the previously determined PHR1‐binding site sequence (P1BS, GNATATNC) (Bustos et al, 2010), we analysed the promoter regions of all 37 RALFs in A. thaliana and detected that RALF1, RALF4, RALF22, RALF23, RALF33 and RALF34 contain P1BS (Fig 2D). To test whether PHR1 directly controls RALF expression, we quantified RALF mRNA levels in the roots of WT plants and phr1 mutants. Surprisingly, the PHR1 mutation significantly reduced the expression of most tested RALFs (Fig 2E). We next cloned the native promoters of RALFs upstream of a luciferase reporter gene and co‐transfected them together with the 35S promoter‐driven PHR1 in A. thaliana protoplasts. The expression of PHR1 increased luciferase activity to a much higher extent than the vector control in all tested RALFs expect RALF24, which lacks the predicted P1BS (Fig 2F). These results suggest that PHR1 activates RALF expression. To further confirm the direct binding of PHR1 to RALF promoters, we generated a Myc‐labelled PHR1 overexpression line (PHR1‐Myc). Chromatin immunoprecipitation quantitative PCR (ChIP‐qPCR) showed that the promoters of most RALFs were highly enriched in PHR1‐Myc cells (Fig 2G, CRY1‐Myc acts as a negative control). In addition, we purified PHR1‐GST proteins and carried out electrophoretic mobility shift assays (EMSAs) in vitro; we observed that PHR1 directly binds to the RALF23 promoter (Appendix Fig S5B). Collectively, we identified several RALFs as direct targets of PHR1. To determine whether PHR1 targeting RALFs is a conserved process across different plant species, we analysed the promoter regions of RALF genes from rice, soybean and tomato. All tested RALFs contain at least one P1BS (Appendix Fig S5C–E). These results indicate that PHR1 likely targets RALFs in other plant species as well.
PHR1‐mediated RALF activation suppresses FLS2‐BAK1 complex formation to inhibit immunity during the Pi starvation
To clarify whether PHR1 is involved in mediating immune repression under Pi starvation condition, we analysed flg22‐induced MAPK activation in WT plants and phr1 mutants grown in HP and LP media. The MAPK level was suppressed in the WT plants during the Pi starvation; however, the inhibitory effect was reduced in the phr1 mutants (Fig 3A). This result, consistent with previous work (Castrillo et al, 2017), indicates PHR1‐mediated immunity suppression. In addition, our results showed that PHR1 activates RALF expression (Fig 2). Moreover, RALF23 and RALF34 have been shown to inhibit immune responses by disrupting flg22‐induced FLS2 and BAK1 complex formation (Stegmann et al, 2017). We postulated that PHR1‐mediated RALF expression during the Pi starvation interferes with immune complex formation between FLS2 and BAK1. To test this hypothesis, we immunoprecipitated the FLS2‐BAK1 complex together with anti‐FLS2. LP treatment reduced the flg22‐induced FLS2‐BAK1 complex association in the WT but not in the phr1 mutants protoplasts (Fig 3B), suggesting that PHR1 is required for Pi starvation‐mediated immune complex inhibition. Further, the expression of PHR1‐Myc in phr1 protoplasts partially recovered Pi starvation‐mediated suppression of immune complex formation (Fig 3B). Next, we performed bacterial growth assay with WT plants, phr1 mutants and PHR1‐Myc plants. Pto DC3000 had more colonies in the WT and PHR1‐Myc roots under LP conditions than under HP conditions (Fig 3C). However, a similar trend was also observed for the phr1 mutants (Fig 3C), suggesting the potential existence of a PHR1‐independent pathway regulating immunity (e.g. PHR1 has a weakly redundant paralogue PHL1, Castrillo et al, 2017). Since RALFs are direct downstream targets of PHR1, we speculated that exogenous application of RALF peptides would rescue PHR1‐mediated FLS2‐BAK1 complex disruption in phr1 mutants under LP conditions. Indeed, RALF23 and RALF34 treatment significantly reduced the flg22‐triggered FLS2‐BAK1 complex association in phr1 mutants during Pi starvation (Fig 3D). Consistent with the above observations, Pto DC3000 colonized phr1 mutants treated with RALF23 and RALF34 better than it colonized phr1 mutants under the mock treatment (Fig 3E). As FER is the receptor of RALFs, we hypothesized that PHR1‐RALF‐ mediated immune suppression under low‐Pi stress is FER‐dependent. We immunoprecipitated the FLS2‐BAK1 complex with anti‐FLS2 in fer‐4 mutants and FER/fer‐4 complementation lines and found that LP treatment inhibited the flg22‐induced FLS2‐BAK1 complex association in the WT and FER/fer‐4 complementation lines (Fig 3F), while the inhibitory effect was reduced under both HP and LP condition in the fer‐4 mutants (Fig 3F). These results suggest that FER is involved in the PHR1‐RALF mediated immune inhibition under low‐Pi stress. To further confirm the role of FLS2 and BAK1 in Pi starvation‐mediated immunity suppression, we compared Pto DC3000 colonization in the roots of fls2 and bak1 mutants. Pto DC3000 had more colonization in roots of fls2 and bak1 mutants compared to WT plants under LP conditions (Fig 3G, Appendix Fig S7). Notably, no colonization difference was observed for Pto DC3000 between WT, fls2 and bak1 mutants under HP conditions (Fig 3G, Appendix Fig S7). This finding suggests that Pto DC3000 colonization in roots under HP conditions was independent of FLS2 and BAK1. Taken together, these results suggest that PHR1 inhibits PAMP‐triggered FLS2‐BAK1 complex formation by activating RALF expression to suppress immunity under Pi‐deficient conditions.
Figure 3. PHR1‐mediated RALF activation disrupts FLS2‐BAK1 complex formation to inhibit immunity.
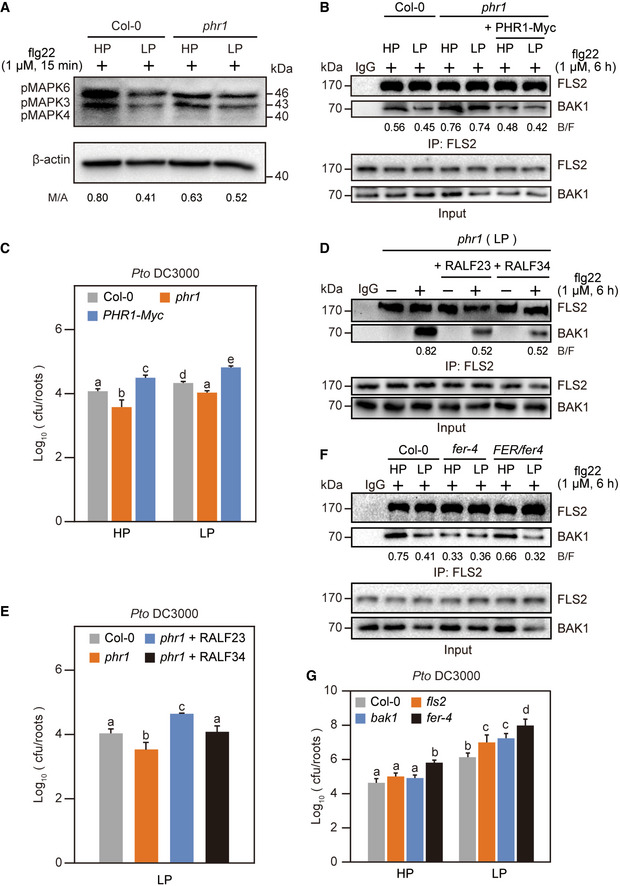
- The PSR inhibits flg22‐induced MAPK activation in a PHR1‐dependent manner. Col‐0 and phr1 mutant seedlings grown in either HP or LP media were treated with flg22 for 15 min and roots were harvested. The MAPK level was determined with anti‐pMAPK, and β‐actin was used as a loading control.
- PHR1 negatively regulates FLS2‐BAK1 complex formation under Pi‐starvation conditions. Col‐0, phr1 mutant seedlings were grown in either HP or LP media for 5 days, then lysing roots into protoplasts, transformed with water or PHR1‐Myc plasmid and treated with flg22 for 6 h. The amount of immunoprecipitated FLS2 and coimmunoprecipitated BAK1 were determined using anti‐FLS2 and anti‐BAK1 antibodies, respectively. Preimmune serum (IgG) was used as negative control.
- PHR1 enhances the growth of Pto DC3000 under Pi‐starvation conditions. Col‐0, phr1 mutant and PHR1‐Myc overexpression seedlings grown in either HP or LP media were root inoculated (by soaking) with a bacterial suspension (OD600 = 0.2) for 2 min, and the number of bacteria from individual roots was quantified at 5 dpi. The data shown indicate the means ± SDs (n = 18, n refers to the number of roots per group). The different lowercase letters indicate statistical significance (one‐way ANOVA).
- Application of RALF23/34 disrupted FLS2‐BAK1 complex formation in phr1 mutants. phr1 mutants grown in LP media were treated with water, 1 μM of flg22, 1 μM of RALF23, 1 μM of RALF34 or both flg22 and RALF for 6 h. The amount of immunoprecipitated FLS2 and coimmunoprecipitated BAK1 were determined using anti‐FLS2 and anti‐BAK1 antibodies, respectively. IgG was used as a negative control.
- RALF23/34 promoted the growth of Pto DC3000 in phr1 mutants. Col‐0 and phr1 mutant seedlings grown in LP media were root inoculated (by soaking) with a bacterial suspension (OD600 = 0.2) for 2 min, and the number of bacteria from individual root was quantified at 5 dpi. The data shown indicate the means ± SDs (n = 18, n refers to the number of roots per group). The different lowercase letters indicate statistical significance (one‐way ANOVA).
- FER negatively regulates FLS2‐BAK1 complex formation under Pi‐starvation conditions. Col‐0, fer‐4 mutant and FER/fer‐4 complementation lines seedlings were grown in either HP or LP media for 5 days and treated with flg22 for 6 h. The amount of immunoprecipitated FLS2 and coimmunoprecipitated BAK1 was determined using anti‐FLS2 and anti‐BAK1 antibodies, respectively. Preimmune serum (IgG) was used as a negative control.
- LP promoted the growth of Pto DC3000 in fls2 and bak1 mutants. Col‐0, fls2, bak1 and fer‐4 mutant seedlings grown in either HP or LP media were inoculated (by soaking) with a GFP‐labelled bacterial suspension (OD600 = 0.2) for 2 min, and the number of bacteria from individual root was quantified at 5 dpi. The data shown indicate the means ± SDs (n = 18, n refers to the number of roots per group). The different lowercase letters indicate statistical significance (one‐way ANOVA).
Data information: All experiments were repeated three times with similar results.
LP and RALF23 treatment promotes the colonization of rhizosphere bacteria that help plants resist LP stress
Having ascertained that PHR1 targets RALFs to suppress immunity during Pi starvation, we sought to understand how this would benefit plants under LP conditions. Col‐0 and PSR mutants such as phr1 assemble different root microbiome communities, even under normal Pi conditions (Castrillo et al, 2017). To determine the effect of low‐Pi stress on the composition of overall rhizosphere microbiome composition, Col‐0, fer‐4 and phr1 mutants grown under HP and LP conditions were inoculated with a natural microbiome population collected from the local region (Tianma Road 19, Changsha, Hunan, China). Three weeks post inoculation, roots were harvested and subjected to 16S rRNA‐based sequencing. We observed no obvious shift at the phylum level in Col‐0 between HP and LP conditions (Appendix Fig S8A); however, the relative abundances of several bacterial genera, such as Pseudomonas, Bacillus, Geodermatophilus and Methylobacillus, were highly enriched in Col‐0 after low‐Pi stress (Fig 4A, C, E and G, Appendix Figs S8E and S9E), suggesting that plants recruit a specialized root microbiota under LP conditions. Notably, no significant difference was observed for these bacterial groups in fer‐4 and phr1 mutants between HP and LP conditions (Fig 4A, C, E and G). This suggests that FER and PHR1 play a key role in shaping root microbiota structure under LP conditions. We further determined the effect of RALF23 on root microbiota composition by using an engineered strain Bacillus subtilis (Zhang & Gleason, 2020) that delivers RALF23. Bacterial groups that were enriched in Col‐0 under LP conditions (e.g. Pseudomonas and Bacillus) were also significantly enriched in RALF23‐treated roots in Col‐0 (Fig 4B, D, F, H, Appendix Fig S9A–D and F). This suggests that LP and RALF23 treatment act similarly to recruit specific microbial lineages. RALF23‐regulated root microbiota changes were also observed in phr1 but not in fer‐4 mutants, which confirms that RALF23 is a downstream regulator of PHR1.
Figure 4. Analysis of bacterial colonization after LP stress and delivery of RALF23 by B. subtilis treatment.
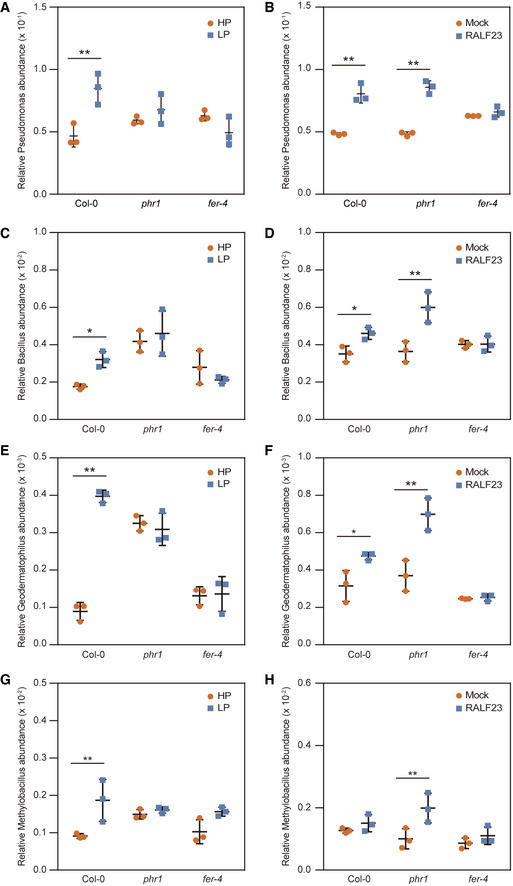
-
A, BRelative Pseudomonas abundance in rhizosphere samples after (A), Pi‐stress (LP) or (B) treatment with B. subtilis expressing RALF23 (RALF23). The data shown indicate the means ± SDs (n = 3, n refers to biological replicates). **P < 0.01 (Student’s t test).
-
C, DRelative Bacillus abundance in rhizosphere samples after (C), Pi‐stress (LP) or (D) treatment with B. subtilis expressing RALF23 (RALF23). The data shown indicate the means ± SDs (n = 3, n refers to biological replicates). *P < 0.05; **P < 0.01 (Student’s t test).
-
E, FRelative Geodermatophilus abundance in rhizosphere samples after (E), Pi‐stress (LP) or (F) treatment with B. subtilis expressing RALF23 (RALF23). The data shown indicate the means ± SDs (n = 3, n refers to biological replicates). *P < 0.05; **P < 0.01 (Student’s t test).
-
G, HRelative Methylobacillus abundance in rhizosphere samples after (G), Pi‐stress (LP) or (H) treatment with B. subtilis expressing RALF23 (RALF23). The data shown indicate the means ± SDs (n = 3, n refers to biological replicates). **P < 0.01 (Student’s t test).
Enrichment of certain bacterial genera after LP treatment indicates that these microorganisms might be involved in the alleviation of Pi starvation. To test this hypothesis, we selected Pto DC3000 and B. subtilis as the representative of Pseudomonas and Bacillus, respectively. Pto DC3000 typically colonizes leaf tissue as a pathogenic bacterium. A recent study showed that this bacterium can also colonize plant roots to regulate LR formation and development (Kong et al, 2020), and it cannot develop disease upon root inoculation in a natural soil community (Song et al, 2021). Using the soil‐soak inoculation method, we inoculated A. thaliana seedlings grown under HP and LP conditions with water, Pto DC3000 or B. subtilis strain RIK1285 and used the shoot fresh weight as the alleviated Pi‐starvation phenotypic characteristic. We found that Pto DC3000 and B. subtilis colonization significantly increased the shoot fresh weight of WT plants and phr1 mutants (Fig 5A and B). In addition, Pto DC3000 and B. subtilis colonization also promoted LR growth of WT plants and phr1 mutants under LP conditions (Appendix Fig S10), suggesting that bacterial colonization by members of these lineages can indeed alleviate low‐Pi stress by promoting LR growth during the Pi starvation. However, bacterial inoculation did not increase the shoot fresh weight in the fer‐4 mutants (Fig 5B). These results are consistent with the idea that LP‐ and RALF23‐regulated microorganisms can alleviate Pi starvation. Next, we were interested in how bacterial colonization alleviates Pi starvation. We checked the effect of bacterial inoculation on the expression of Pi uptake‐associated genes expression. qPCR analysis showed that the Pto DC3000 and B. subtilis inoculation significantly upregulated PHT1:1 and PHT1:4 expression in WT and phr1 mutants, but fer‐4 attenuates almost all these response (Fig 5C). Altogether, these results indicate that LP and RALF23 treatment shape a specialized microbiota that relieves Pi starvation, and a possible strategy that this specialized microbiota used to relief Pi starvation is induction of Pi uptake gene expression.
Figure 5. Bacterial colonization alleviates LP stress.
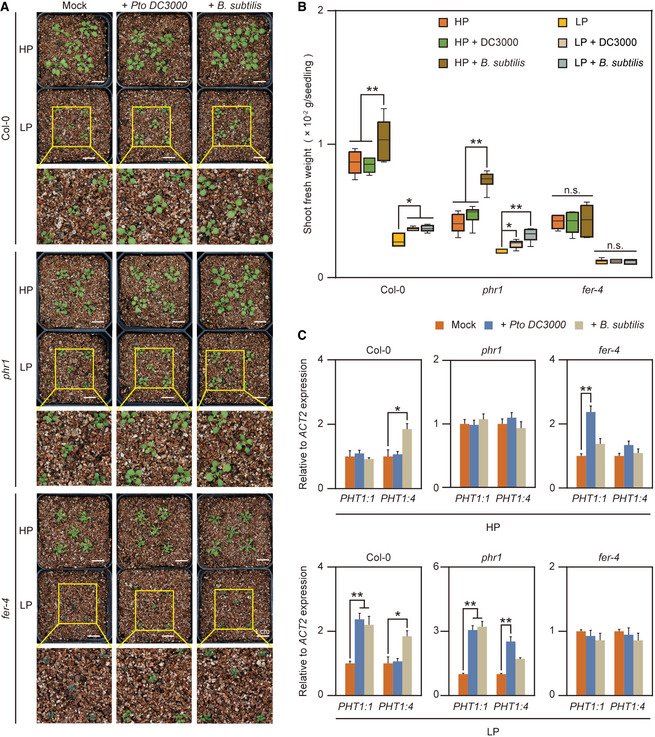
-
A, BBacterial inoculation promotes plant growth and alleviates LP stress. Arabidopsis seeds were germinated on 1/2‐strength MS media for 3 days and then transplanted to either HP or LP media for another 5 days. The seedlings were then transferred to vermiculite and inoculated with Pto DC3000 or B. subtilis. (A), Images were taken 4 weeks after inoculation. (B), The seedlings were harvested, and the shoot fresh weight was measured 4 weeks after inoculation. The data shown indicate the means ± SDs (n = 30, n refers to the number of seedlings per group), Central bands: median, boxes: interquartile range, whiskers: max./min; n.s., not significant; *P < 0.05, **P < 0.01 (Student’s t test).
-
CPto DC3000 and B. subtilis upregulate phosphate absorption gene expression. Col‐0, phr1 and fer‐4 mutant roots were harvested 48 h after inoculation with Pto DC3000 or B. subtilis in Pi‐sufficient or Pi‐deficient condition, and the relative expression of Pi‐absorbing genes was quantified via RT‐qPCR. The data shown indicate the means ± SDs (n = 3, n refers to technical replicates); *P < 0.05; **P < 0.01 (Student’s t test).
Data information: All experiments were repeated three times with similar results.
The root microbiota of fer‐4 mutant alleviates the Pi starvation
We further determined the effect of FER on the overall rhizosphere microbiome composition. The fer‐4 microbiome had lower richness (number of operational taxonomic units, OTUs) and Shannon diversity (a metric of species richness and evenness) than Col‐0 (Appendix Fig S8C and D), which is consistent with a previous report (Song et al, 2021), the relative abundance of the phylum Bacteroidota was significantly increased in fer‐4 mutants (Appendix Fig S8B). Further analysis revealed that the genera Flavobacterium, Pseudomonas and Delftia were also more abundant in fer‐4 mutants (Appendix Fig S8F). Previous studies have shown that the genera Flavobacterium, Pseudomonas and Delftia were among the most‐predominant genera of plant‐beneficial bacteria (Lebeis et al, 2015; Rolli et al, 2015; Bernal et al, 2017; Lally et al, 2017; Melnyk et al, 2019). Notably, the high abundance of Pseudomonas in the Col‐0 was also observed under LP treatment (Fig 4A). Therefore, we compared the FER‐regulated and LP‐regulated microbiota at the genus level. We found a large overlap in bacterial genera that were regulated by FER and LP (Fig 6A), suggesting a functional link between the FER‐ and LP‐ regulated microbiomes. Song et al reported that FER regulates ROS levels in roots to control Pseudomonas under normal condition, which is largely independent of its immune function in mediating FLS2‐BAK1 complex formation (Song et al, 2021). However, we found that the immune receptors such as FLS2 and BAK1 can change specific microbe under LP conditions (Fig 3G and Appendix Fig S7). Additionally, we found that the expression of genes that are involved in synthesis of the secondary metabolites glucosinolates, are induced in fer‐4 mutants (Appendix Fig S11). These metabolites are produced in plants upon perception of PAMPs by pattern recognition receptors and are needed for broad‐spectrum defence to restrict the growth of pathogens (Bednarek et al, 2009; Clay et al, 2009). Therefore, we propose that FER‐mediated root microbiota change is probably through regulation of the FLS2‐BAK1 complex formation and biosynthesis of secondary metabolites in plants roots. We hypothesized that, similar to the LP microbiome, the fer‐4 microbiome is involved in the alleviation of Pi starvation. To test this hypothesis, we collected the microbiome population from the rhizosphere of 3‐week‐old WT and fer‐4 plants by shaking the soil in sterile water and applied them to WT, fer‐4 and phr1 mutants grown in HP and LP conditions, respectively. Indeed, treatment with the fer‐4 microbiome significantly enhanced plant growth in WT and phr1 mutants compared to the WT microbiome treatment (Fig 6B and C), suggesting that the mutation of FER assembles a rhizosphere microbiome that is beneficial to Pi starvation alleviation. We further checked the impact of microbiome treatment on the expression of Pi uptake‐associated gene expression. qPCR analysis showed that the fer‐4 microbiome application significantly upregulated several PSI genes, such as PHL1, WRKY6, WRKY45, WRKY75 and PHT1 families in WT plants (Fig 6D). Altogether, these results indicate that the fer‐4 microbiome can alleviate Pi starvation and induce the expression of PSI genes.
Figure 6. The fer‐4 rhizosphere microbiome promotes growth in the next generation of plants under LP conditions.
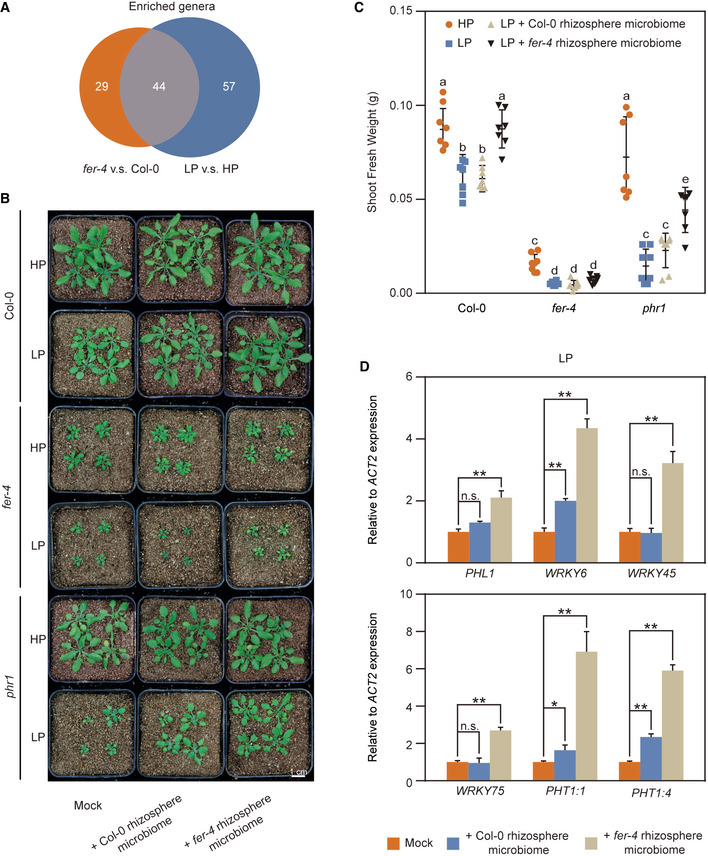
-
AVenn diagram showing the total and overlapping numbers of genera enriched in the fer‐4 mutant and under Pi starvation.
-
B, CThe fer‐4 rhizosphere microbiome promotes plant growth and alleviates LP stress. Arabidopsis seeds were germinated on 1/2‐strength MS media for 3 days and then transplanted to either HP or LP media for another 5 days. The seedlings were then transferred to vermiculite and inoculated with Col‐0 or fer‐4 rhizosphere microbiomes. (B), Images were taken 4 weeks after inoculation. (C), The seedlings were harvested, and the shoot fresh weight was measured 4 weeks after inoculation. The data shown indicate the means ± SDs (n = 7, n refers to the number of flowerpots (4 seedlings per flowerpot) per group). The different lowercase letters indicate statistical significance (one‐way ANOVA).
-
DThe fer‐4 rhizosphere microbiome upregulated phosphate absorption gene expression. The roots of Col‐0 were harvested after 4 weeks of inoculation with the Col‐0 or fer‐4 rhizosphere microbiome under LP condition, and the relative expression of Pi‐absorbing genes was quantified via RT‐qPCR. The data shown indicate the means ± SDs (n = 3, n refers to technical replicates); n.s., not significant; *P < 0.05; **P < 0.01 (Student’s t test).
Data information: All experiments were repeated three times with similar results.
Discussion
This study identified a PHR1‐RALFs‐FER pathway that intersects with FLS2/BAK1 to shape the root microbiome during the Pi starvation in A. thaliana. We show that PHR1, the key transcriptional regulator that coordinates the Pi starvation and defence (Castrillo et al, 2017), directly targets and activates the expression of several RALF genes under LP conditions (Fig 2). PHR1‐mediated RALF activation inhibits plant immunity by suppressing PAMP‐triggered FLS2‐BAK1 complex formation in an FER‐dependent manner (Fig 3), which is consistent with the known function of RALF‐FER in plant immunity regulation (Stegmann et al, 2017). We found that suppression of plant immunity during the Pi starvation facilitates colonization of certain rhizosphere microorganisms activating PSI gene expression and alleviating the Pi starvation (Figs 4 and 5C). Furthermore, the microbiota from the fer‐4 mutants significantly enhanced plant growth and relieved Pi starvation. Altogether, our work revealed PHR1‐RALFs/FER‐FLS2/BAK1 as a novel signalling pathway that plants use to suppress immunity, shape a specialized microbiota and alleviate the Pi starvation under low‐Pi conditions (Fig 7). Though recent studies revealed that individual Pi starvation or a combination of Pi starvation and chemical elicitation promote defence responses against insect herbivory or bacteria (Khan et al, 2016; Morcillo et al, 2020), our work showed that Pi starvation suppresses PTI. This discrepancy is likely due to the fact that different immune eliciting organisms or chemicals were used.
Figure 7. Model of the function of PHR1 in the balance between plant immunity and the PSR.
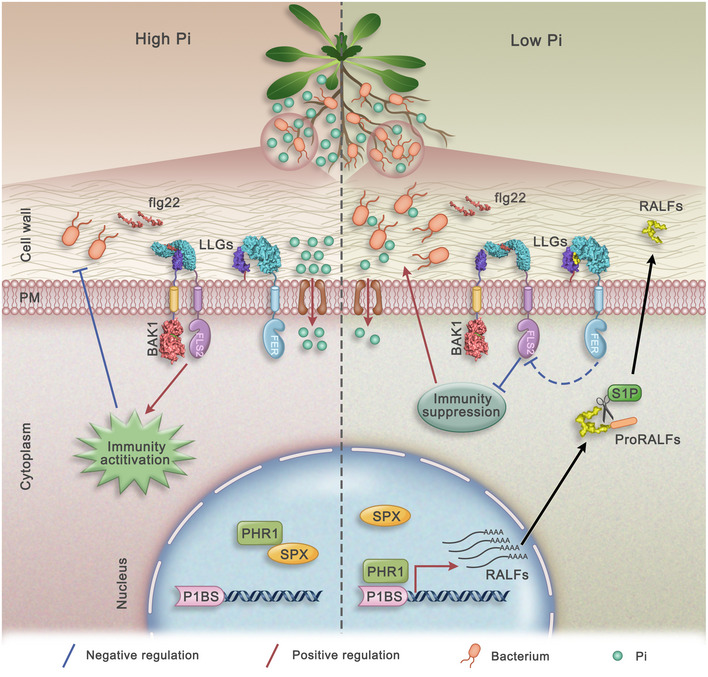
On the left side, under HP conditions, PHR1 is bound by SPX proteins and thus remains inactive. Bacterial flg22 is perceived by the plant receptor FLS2, triggering the immune response and inhibiting bacterial growth. On the right side, under LP conditions, PHR1 is released from the SPX protein complex, binds to the promoters of RALF genes and activates RALF expression. RALF mRNA is translated and further processed into mature RALF peptides that interfere with flg22‐induced immunity complex formation between FLS2 and BAK1 through FER. Suppression of plant immunity by RALF peptides enhances bacterial growth, which helps plants increase Pi uptake under Pi‐limiting conditions and thus promotes plant growth.
Interestingly, all of the studies published until now support the idea that FER mutation increases resistance to certain harmful microorganisms, including bacteria (Keinath et al, 2010), fungi (Kessler et al, 2010; Masachis et al, 2016) and nematodes (Zhang et al, 2020). However, a recent study (Song et al, 2021) showed that the RALF23/FER pathway can also recruit a specific beneficial taxon that helps maximize plant fitness [i.e. growth in Song et al (2021) and alleviated Pi starvation in this work (Fig 6B)]. Furthermore, our Pto DC3000 infection and 16S rRNA microbiome sequencing data support that the FER mutation recruits beneficial Pseudomonas under normal growth conditions, which is consistent with Song et al’s observation. Interestingly, we further found that LP treatment also significantly enriched Pseudomonas, Bacillus, Geodermatophilus and Methylobacillus in a FER‐dependent manner during the Pi starvation (Fig 4A, C, E and G). Importantly, application of the fer‐4 microbiome can significantly relieve the Pi starvation and promote the growth of WT and phr1 mutant plants under LP conditions. Together with the results reported by Song et al (2021), our work indicates the role of the RALF‐FER pathway in recruiting beneficial microbes under both normal conditions and during the Pi starvation. Of note, the regulation of microbes by FER under normal growth conditions is largely independent of its immune function as a scaffold protein of FLS2 and BAK1 (Song et al, 2021). Mutation of some immune‐associated genes only shows relatively subtle changes in root microbiota when tested in natural soil (Lebeis et al, 2015). This is probably because of the complexity of most root microbiomes that can contain both the immune suppressive and immune non‐suppressive microbes (Ma et al, 2021). However, our data showed that PHR1‐mediated RALF‐FER inhibits the immune response and regulates specific microbe composition by suppressing FLS2 and BAK1 complex formation under LP conditions. This finding is important as it indicates that plants might use different mechanisms to regulate microbiota structure under normal and stress conditions, respectively. Lastly, in addition to Pseudomonas, we found that the FER mutation also recruits other beneficial bacteria, such as Bacillus, under LP conditions to increase plant fitness. In support of this finding, B. subtilis inoculation also promoted plant growth under LP conditions. Consistent with these findings, the delivery of RALF23 by B. subtilis also significantly enriched Pseudomonas and Bacillus in the A. thaliana root microbiota. Based on these observations, we propose that plants suppress immunity during the Pi starvation through the RALFs/FER‐FLS2/BAK1 pathway to recruit a specialized microbiota, including Pseudomonas and Bacillus, which may be critical for alleviating Pi starvation.
Additionally, we used Pto DC3000 as a model microbe to study immunity suppression in response to Pi starvation in roots. Pto DC3000 is a well‐known bacterial pathogen that infects leaves. However, certain pathogenic microbes can change their pathogenicity or even become beneficial under specific nutritional conditions (Kessler et al, 2010). For example, the fungal pathogen Colletotrichum tofieldiae (Ct) becomes beneficial and transfers phosphorus macronutrients to A. thaliana under Pi‐deficient conditions (Hiruma et al, 2016). Recent studies have shown that Pto DC3000 can also colonize A. thaliana LRs to promote plant growth and fitness (Mang et al, 2017; McClerklin et al, 2018; Kong et al, 2020). As a leaf pathogen, Pto DC3000 might promote Pi absorption in host roots under low‐Pi stress, thus enhancing LR growth. Induction of LRs is typically associated with increased uptake of nutrients such as Pi (Péret et al, 2011). We observed that Pto DC3000 inoculation significantly reduced the Pi‐starvation phenotypic characteristics, promoted LR growth in WT plants as well as the phr1 mutants under LP conditions and it did not show negative effects on A. thaliana (Fig 5A and Appendix Fig S10). Inoculation of Pto DC3000 on roots can stunt plant growth at early stages; however, we checked for phenotypes 4 weeks post Pto DC3000 treatment. Plants may have recovered from early stage stunting caused by Pto DC3000. Additionally, a recent report showed that the effects of soil phosphorus content on plant microbiota are changed by a bacterial synthetic community (SynCom) (Finkel et al, 2019). The authors also showed that in the absence of Burkholderia from the SynCom, plant shoots accumulated higher Pi levels than shoots colonized with the full SynCom under Pi starvation conditions (Finkel et al, 2019). That shows that rhizosphere microbiomes can alleviate Pi starvation under LP condition; similarly, we found the Pto DC3000 and B. subtilis can promote LR growth and Pi‐uptake genes expression to alleviate Pi starvation under LP condition (Fig 5C and Appendix Fig S10).
An enduring challenge in this field is to understand how plants can recruit beneficial microbes and restrict pathogens at the same time. There are some paradigms in arbuscular mycorrhizal fungi and rhizobia (Zipfel & Oldroyd, 2017; Thoms et al, 2021; Zhang et al, 2021) that demonstrate how selection of symbionts is achieved, but the life history and recruitment mechanisms of the majority of plant‐microbiome associations are poorly understood. At the ETI level, plants can reach this aim by using a “gene for gene” strategy, but at the PTI level, it seems more complex and challenging. Here, we show that RALF/FER partially targets the PTI‐related FLS2/BAK1 complex to recruit microbes that are beneficial for phosphate uptake and plant growth. Furthermore, recent work showed that plant roots can discriminate harmful and beneficial microorganisms by sensing cell wall integrity (Atkinson et al, 2013; Zhou et al, 2020; Rebaque et al, 2021). Interestingly, RALF/FER is well‐known for its role in sensing cell wall integrity to modulate cell growth and the stress response (Feng et al, 2018; Zhao et al, 2018). In the future, it would be worthwhile to study whether the RALF/FER pathway may act through its ability to sense the cell wall integrity to further regulate ROS production (Lebeis et al, 2015) and help select beneficial microorganisms.
Materials and Methods
Plant growth conditions
A. thaliana seeds were sterilized and then vernalized at 4°C for 3 days before being grown on 1/2 MS with 1% (w/v) sucrose solidified with 1% (w/v) agar (Sigma‐Aldrich, Missouri, USA) at 23°C, at 65% relative humidity under 16/8 h (long days).
For phosphate stress treatment assay, seeds were first grown in 1/2 MS medium for 3 days and then transferred to either 1/2 MS medium containing 1.25 mM of Pi (HP) or low‐Pi medium containing 10 μM of Pi (LP) for another 5 days. The standard HP medium was 1/2 MS medium with 1% (w/v) sucrose and 1% (w/v) agarose (Biowest Regular Agarose G‐10). For the LP medium, KH2PO4 in the HP medium was replaced with K2SO4. The pH was adjusted to 5.8 with 1 M of NaOH for both HP and LP media (Zheng et al, 2019).
Western blot
For flg22‐induced MAPK phosphorylation assay, seedlings were grown in either HP or LP medium for 5 days and treated with water or flg22 (Phytotech, Lenexa, USA) for 15 min, and then roots were harvested. Protein extraction was performed by adding Laemmli sample buffer and incubated for 10 min at 95°C. Analysis was carried out by SDS‐PAGE and western blot using anti‐pMAPK (a phospho‐p44/42 MAPK (Erk1/2) (Thr202/ Tyr204)) (Cell Signaling Technology, Boston, USA). β‐actin (Abmart, Shanghai, China) was used as loading control.
For FER accumulation assay, seedlings were grown in either HP or LP medium for 5 days and roots were harvested. Protein extraction was performed by adding Laemmli sample buffer and incubating for 10 min at 95°C. Analysis was carried out by SDS‐PAGE and western blot using anti‐GFP (Abmart, Shanghai, China). β‐actin was used as loading control.
For RALF23 cleavage assay, RALF23‐GFP seedlings were grown in either HP or LP medium for 5 days and roots were harvested. Protein extraction was performed by adding Laemmli sample buffer and incubating for 10 min at 95°C. Analysis was carried out by SDS‐PAGE and western blot using anti‐GFP. β‐actin was used as loading control.
For Co‐immunoprecipitation assay, Co‐IP was performed as previously described with some modifications (Du et al, 2016; Li et al, 2018). Briefly, seedlings were grown in either HP or LP medium for 2 weeks, transferred to water (pH = 5.8) and incubated for 2 h. Then, 1 µM of flg22 and/or 1 µM of RALF23/34 were added and incubated for 6 h. Seedlings were then frozen in liquid nitrogen. For protoplasts preparation, protoplasts were isolated from seedlings, which were grown in either HP or LP medium for 2 weeks. Seedling strips were incubated in the cell wall–degrading enzyme solution (0.4 M of Mannitol, 20 mM of MES (pH 5.7), 20 mM of KCl, 10 mM of CaCl2, 0.1% BSA, 1.25% Cellulase and 0.1% Pectolyase) in the dark for 3 h. Protoplasts were purified and transfected with 100 μg of PHR1‐Myc plasmid DNA and an equal volume of PEG solution. The transfected protoplasts were incubated in the dark at 23°C for 16 h to allow expression of the PHR1‐Myc proteins. Then seedlings or protoplasts solubilized with NEBT buffer (1% Triton X‐100, 20 mM of HEPES [pH = 7.5], 40 mM of KCl, 1 mM of EDTA, 1 mM of PMSF (Selleck Chemicals, Houston, USA) and 1% protease inhibitor (Selleck Chemicals, Houston, USA)). For immunoprecipitation A/G magnetic beads (Bimake, Houston, USA) coupled with anti‐FLS2 (Zhao et al, 2018) were used and incubated with the crude extract overnight at 4°C. Subsequently, beads were washed five times with NEB‐T buffer before adding Laemmli sample buffer and incubating for 10 min at 95°C. Analysis was carried out by SDS‐PAGE and western blot using anti‐FLS2 and anti‐BAK1 (Stegmann et al, 2017).
Measurement of flg22‐induced ROS burst
For H2DCFDA staining assay, all genotypes seedlings were germinated and grown on 1/2 MS medium for 3 days and then transferred to either HP or LP medium for another 5 days then treat with 1 µM of flg22, RALF23 or both flg22 and RALF23 for 15 min. H2DCFDA was dissolved in dimethyl sulfoxide (10 mg/ml), then diluted to a 500‐mM stock (10×) in 0.1 M of PB buffer (pH 7.0) and stored at −20°C. Before use, H2DCFDA aliquots were thawed in the dark, stored on ice and diluted to a 1× working concentration in 2 ml 1/2× MS media with 0.1% 2‐(N‐morpholino) ethanesulfonic acid sodium salt. Whole seedlings were transferred to a 12‐well plate with 2 ml staining solution per well and stained for 15 min at room temperature in the dark. Confocal images were acquired with a 10×/0.45 numerical aperture objective on a Leica SP8 laser scanning confocal microscope using a white‐light laser. H2DCFDA was excited with a 514‐nm laser and a HyD detector was used to capture emissions between 511 and 611 nm.
For total ROS measurement, the ROS burst assay was performed as previously described (Stegmann et al, 2017). Briefly, 4‐week‐old leaves were cut into 4 × 4‐mm pieces that were transferred to 96‐well plates containing 100 µl of sterile water and then covered and incubated in the dark overnight. After 24 h, the water was replaced with 100 µl of a solution containing 20 µM of luminol L‐012 (Sigma‐Aldrich, Missouri, USA), 1 µg/ml of horseradish peroxidase (HRP, Sigma‐Aldrich), 100 nM of flg22 and the different RALFs (1 µM) described above or with elution buffer. Luminescence was measured using a Fluoroskan Ascent FL microplate fluorometer and luminometer (Thermo Scientific, USA). ROS production was reported as either the increase in photon counts or the sum of total photon counts.
Gene expression analysis
For RT‐PCR assay, the total RNAs were extracted from roots and qPCR was performed using the CFX96 Touch Real‐Time PCR Detection System (Bio‐Rad, USA) with SYBR Premix ExTaqII (Takara, Japan). Reactions were performed using a PCR System (C1000, Bio‐Rad) with initial incubation at 95°C for 5 min followed by 40 cycles of 15 s at 95°C, 15 s at 58°C and 1 min/kb at 72°C. ACTIN2 was used as a reference in qPCR analysis. Data are shown as the mean expression ± SD. When indicated, the fold change is shown for the treated sample in comparison to the control sample. The PCR‐sequences are listed in Appendix Table S1.
Microscopy imaging
Confocal images of the pFER::FER‐GFP and Pto DC3000‐GFP accumulation in LP‐stress were obtained on a Zeiss confocal laser scanning microscope using a 488 nm band‐pass filter. Quantification of the fluorescence intensity was carried out with ImageJ software.
Bacterial treatment
For Pto DC3000 infection assay, Pseudomonas syringae pv. tomato DC3000 strain was grown overnight in King’s B medium (10 g/l of proteose peptone, 1.5 g/l of anhydrous K2HPO4, 5 g/l of MgSO4) with shaking at 28°C. Bacteria were collected and resuspended in deionized water to an OD600 = 0.2. Seven‐day‐old seedlings grown in either HP or LP medium were soak inoculated with bacterial suspension for 2 min and then transfer to HP or LP medium (only put the roots on the medium and the shoot should stand in the air. One can achieve this by discarding about 1/4 or 1/3 agar medium from one side of the petri dish) without sucrose, and then quantifying the bacterial amount from individual seedlings at 3 and 5 days after inoculation (dpi). Eighteen seedling roots were collected and ground with a drill‐adapted pestle. Serial dilutions were plated on LB agar and colonies were counted 2 days later.
For bacteria relieve Pi starvation assay, seeds were first phosphate stress treatment for 5 days and then transferred to vermiculite soils. After 3 days transfer, the B. subtilis (OD600 = 0.2) and Pto DC3000 (OD600 = 0.2) suspensions were used to treat the A. thaliana plants in pots, with 1 ml per seedling and co‐cultured for 3 weeks. Four‐week‐old seedlings were harvested and the shoot fresh weight were measured previously described.
RNA‐seq sample preparation
For LP treatment, Col‐0 seedlings were grown on 1/2 MS medium for 3 days under long‐day conditions (16 h light/8 h dark) then transferred to either MS or LP medium for another 5 days, with three independent groups used for one treatment. Seedlings were then frozen in liquid nitrogen.
For RALF23 treatment, to avoid an effect of the manipulation of the seedlings during the transfer from solid to liquid 1/2 MS medium for RALF treatment, we first removed the 7‐day‐old A. thaliana Col‐0 seedlings from the solid 1/2 MS medium. Then, we transferred the seedling roots into liquid 1/2 MS medium and pre‐incubated them for 1 h before treatment. Subsequently, Col‐0 seedlings were treated with 1 µM of RALF 23 or elution buffer for 2 h, with three independent groups used for one treatment. Seedlings were then frozen in liquid nitrogen.
RNA‐seq analysis
RNA‐Seq was performed by OE Biotech (Shanghai, China). Total RNA was extracted using a Universal Plant Total RNA Extraction Kit (BioTeke, Beijing, China) followed by treatment with a TURBO DNA‐free Kit (Ambion, USA). RNA integrity was evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Samples with RNA integrity numbers (RINs) ≥ 7 were subjected to downstream analysis. Libraries were constructed using a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, USA) according to the manufacturer’s instructions and then sequenced on the Illumina sequencing platform (HiSeqTM 2500 or Illumina HiSeq X Ten, USA), to generate 125‐bp/150‐bp paired‐end reads. Raw data (raw reads) were processed using the NGS QC Toolkit. Reads containing poly‐N sequences and low‐quality reads were removed to obtain high‐quality reads. The clean reads were mapped to the reference A. thaliana genome using Bowtie2 or TopHat (http://tophat.cbcb.umd.edu/). The fragments per kilobase of exon model per million reads mapped (FPKM) value of each gene was calculated using cufflinks (Cole et al, 2012), and the read counts of each gene were obtained using htseq‐count. DEGs were identified using the DESeq estimating function and nbinom Test. Notably, P < 0.05 and fold changes > 2 or < 0.5 were used as the criteria for identifying significant DEGs. A hierarchical clustering analysis of the DEGs was performed to explore gene expression patterns. The GO enrichment of the DEGs was estimated using the DESeq R package based on the hypergeometric distribution.
Dual‐luciferase assay
The dual‐LUC assay was performed as described (Liu et al, 2008), with modified steps. The RALFs promoter was cloned into the pGreen‐0800‐LUC vector as the reporter plasmid (pRALF‐pGreen II). The effector plasmid 35S::PHR1 was constructed using the pEGAD vector (PHR1‐pEGAD). The reporter plasmid and effector plasmid were transferred into A. thaliana protoplasts. Samples were incubated in the dark at 23°C (16 h) for the dual‐LUC assay using the Dual Luciferase Reporter Gene Assay Kit (Beyotime, Shanghai, China). The LUC and REN signals were detected using a Modulus microplate multimode reader (Turner Biosystem). Three biological repeats were measured for each sample, and similar results were obtained.
Chromatin immunoprecipitation assay
The ChIP assay was performed as described (Liu et al, 2008). Two‐week‐old seedlings were harvested and then treated with 1% formaldehyde under vacuum for 15 min at room temperature. Glycine was added to a final concentration of 0.125 M to stop cross‐linking. The seedlings were washed twice with sterile water, frozen in liquid nitrogen, ground to a fine powder and homogenized in the nuclear extraction buffer 1 (10 mM of Tris–HCl [pH = 8.0], 0.4 M sucrose, 10 mM of MgCl2, 0.1 mM of PMSF and protease inhibitor). Nuclei were precipitated by centrifugation in a centrifuge at 4,000 g for 20 min, washed with nuclear extraction buffer 2 (10 mM of Tris–HCl [pH = 8.0], 0.25 M of sucrose, 10 mM of MgCl2, 1% Triton X‐100, 0.1 mM of PMSF and protease inhibitor) and lysed in the nucleus lysis buffer (50 mM of Tris–HCl [pH = 8.0], 10 mM of EDTA, 1% SDS, 0.1 mM of PMSF and protease inhibitor). Chromatin was sheared by sonication to approximately 500 bp. The chromatin solution was diluted 10‐fold with ChIP dilution buffer (16.7 mM of Tris–HCl [pH = 8.0], 167 mM of NaCl, 1.1% Triton X‐100, 1.2 mM of EDTA, 0.1 mM of PMSF and protease inhibitor). Myc magnetic beads (Bimake, Houston, USA, 30 ml per IP) were washed with ChIP dilution buffer two times and then mixed with the chromatin solution and incubated at 4°C overnight. Immunocomplexes were precipitated and washed with four different buffers: low‐salt buffer (20 mM of Tris–HCl [pH = 8.0], 150 mM of NaCl, 0.2% SDS, 0.5% Triton X‐100, 2 mM of EDTA), high‐salt buffer (20 mM of Tris–HCl [pH 8.0], 500 mM of NaCl, 0.2% SDS, 0.5% Triton X‐100, 2 mM of EDTA), LiCl washing buffer (20 mM of Tris–HCl [pH 8.0], 0.25 M LiCl, 1% NP‐40, 1% sodium deoxycholate, 1 mM of EDTA) and TE washing buffer (10 mM of Tris–HCl [pH = 8.0], 1 mM of EDTA). The bound chromatin fragments were eluted with the elution buffer (50 mM of Tris–HCl [pH = 8.0], 10 mM of EDTA, 1% SDS) and the cross‐links were reversed by incubating at 65°C overnight. The mixture was treated with Proteinase K for 1 h at 45°C to remove proteins. DNA was extracted with phenol/chloroform/isoamyl alcohol and precipitated with a two‐fold volume of 100% ethanol at −80°C for 4 h. To recover the DNA, the sample was spun at 16,000 g for 20 min at 4°C. The pellet was dried briefly and resuspended in 25 ml of TE buffer for further real‐time PCR analysis.
DNA‐EMSA assay
The EMSA was performed as described (Liu et al, 2008), with some modified steps. PHR1‐GST protein and GST protein were used for EMSA assay. The primer sequences were synthesized and labelled with FITC fluorescence probe (TsingKe Biological Technology, Beijing, China). The DNA‐PHR1‐binding reaction contained 100 pg probe, 100 ng PHR1‐GST protein, 10 mM of Tris (pH 7.5), 5% glycerol, 1 mM of MgCl2 50 mM of KCl, 0.2 mg/ml of bovine serum albumin (BSA), 0.5 mM of DTT, 0.5 mg/ml of polyglutamate and the indicated amount of unlabelled competitor. The reactions were incubated at room temperature for 20 min and fractioned by electrophoresis in a 6% native polyacrylamide gel (acrylamide: bisacrylamide, 29:1) containing 10% glycerol, 89 mM of Tris (pH 8.0), 89 mM of boric acid and 2 mM of EDTA. The FITC signal was detected after electrophoresis using KODAK 4000MM Image Station.
RALFs treatment
For RALF‐induced root growth inhibition assay, Seeds were surface‐sterilized and grown on 1/2 MS agar plates for 3 days before transferring individual seedlings in each well of a 48‐well plate containing 1/2 MS medium with 1 µM of RALFs. Seedlings root were measured 3 days after transfer.
Processing of 16S sequencing data
Col‐0, fer‐4 and phr1 mutants grown under HP and LP conditions were inoculated with a natural microbiome population collected from the top 8 cm of soils from the local region (Tianma Road 19, Changsha, Hunan, China (+112°94’54, +28°17’57)), which is free of pesticide and heavy mental contamination for many years. The soil was diluted with sterile water in a 1:1 ration (w/v) and shook for 10 min. And then watered onto the roots of Arabidopsis seedlings. For B. subtilis delivered RALF23 pre‐treatment assay, we cloned codon‐optimized mature RALF23 peptide region into the pBE‐S vector and introduced it into B. subtilis RIK1285 by electroporation. The bacterial cultures were spun down and pellets were resuspended in deionized water (OD600 = 0.6). Three millilitres of the B. subtilis suspensions were used to treat the roots 3 days before inoculating with a natural microbiome. Three weeks post inoculation, roots were harvested and total genomic DNA from samples was extracted using the FastDNA SPIN Kit for soil (MP Biomedicals) according to the manufacturer’s instructions. DNA concentration and purity were monitored on 1% agarose gels. According to the concentration, DNA was diluted to 1 ng/μl using sterile water. 16S rRNA genes of distinct regions V5‐V7 (799F and 1193R) were amplified used specific primer with the barcode. All PCR reactions were carried out with 15 µl of Phusion® High‐Fidelity PCR Master Mix (New England Biolabs); 2 µM of forward and reverse primers and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s and elongation at 72°C for 30 s. Finally, 72°C for 5 min. Sequencing libraries were generated using TruSeq® DNA PCR‐Free Sample Preparation Kit (Illumina, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina NovaSeq platform and 250 bp paired‐end reads were generated.
Sequence analysis was performed by Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/). Sequences with ≥ 97% similarity were assigned to the same OTUs. For each representative sequence, the Silva Database (https://www.arb‐silva.de/) was used based on Mothur algorithm to annotate taxonomic information. Representative sequence for each OTU was screened for further annotation. In order to study phylogenetic relationship of different OTUs, and the difference of the dominant species in different samples (groups), multiple sequence alignment was conducted using the MUSCLE software (Version 3.8.31, http://www.drive5.com/muscle/). OTUs abundance information was normalized using a standard of sequence number corresponding to the sample with the least sequences. Subsequent analysis of alpha diversity and beta diversity were all performed basing on this output normalized data.
Alpha diversity is applied in analysing complexity of species diversity for a sample through 6 indices, including Observed‐species, Chao1, Shannon, Simpson, ACE, Good‐coverage. All this indices in our samples were calculated with QIIME (Version 1.7.0) and displayed with R software (Version 4.0.2).
Statistical analysis
Statistical significance was determined by conducting Student’s t tests or one‐way ANOV A using SPSS 23.0 software (SPSS).
Author contributions
Jing Tang: Data curation; Software; Formal analysis; Investigation; Methodology; Writing—original draft. Dousheng Wu: Data curation; Formal analysis; Funding acquisition; Writing—original draft; Project administration; Writing—review & editing. Xiaoxu Li: Data curation; Project administration. Lifeng Wang: Project administration. Ling Xu: Software; Formal analysis. Yi Zhang: Data curation. Fan Xu: Data curation. Hongbin Liu: Data curation. Qijun Xie: Data curation. Shaojun Dai: Project administration; Writing—review & editing. Devin Coleman‐Derr: Project administration; Writing—review & editing. Sirui Zhu: Data curation. Feng Yu: Funding acquisition; Methodology; Writing—original draft; Writing—review & editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
FY, JT, DW, XL, DC and FX designed the research; JT, FX, YZ, LW, HL, QX, SZ and DW performed the experiments; JT, LX and DW analysed the data. YF, JT, DW, DC, XL, SD and LX wrote the paper with input from other co‐authors.
Supporting information
Appendix
Table EV1
Acknowledgements
The authors thank Dr. Yang Bai (Chinese Academy of Sciences), Dr. Ertao Wang (Chinese Academy of Sciences), Dr. Cyril Zipfel (University of Zurich), Dr. Feng Feng, Dr. Yi Song and Dr. Liang Wang for helpful discussions. They thank Dr. Haiyang Wang for providing the phr1 mutant, Dr. Pablo Vera and Cyril Zipfel for RALF23‐GFP overexpression plants, Dr. Dong Liu for PHR1‐OX overexpression plants, and Dr. Zhaojun Ding for providing the Pto DC3000‐GFP strain. They thank Mei Xia Hu and Shufeng Song for their technical support on confocal microscope. Jia Chen, Weijun Chen and other members in the laboratory are thanked for critical reading of the manuscript and helpful discussions. This work was financially supported by the Natural Science Foundation of China (32001974, 32070769, 31871396 and 31571444), and the Science and Technology Innovation Program of Hunan Province (2020WK2014).
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
The EMBO Journal (2022) 41: e109102.
Data availability
The RNA‐seq raw sequence data reported in this paper have been deposited (PRJCA005011) in the Genome Sequence Archive in the BIG Data Center (Xu et al, 2018), Chinese Academy of Sciences under accession codes CRA004168 (https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=PRJCA005011). The microbiome sequencing data reported in this paper have been deposited (PRJCA005003) in the Genome Sequence Archive in the BIG Data Center (Xu et al, 2018), Chinese Academy of Sciences under accession codes CRA004166 (https://ngdc.cncb.ac.cn/search/?dbId=gsa&q=PRJCA005003).
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez‐Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Lilley CJ, Urwin PE (2013) Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol 162: 2028–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Pislewska‐Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez‐Vallet A et al (2009) A glucosinolate metabolism pathway in living plant cells mediates broad‐spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Bernal P, Allsopp LP, Filloux A, Llamas MA (2017) The pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J 528: 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H, Albert I, Fan L, Reinhard A, Nürnberger T (2014) Immune receptor complexes at the plant cell surface. Curr Opin Plant Biol 20: 47–54 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez‐Pérez J, Solano R, Leyva A, Paz‐Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis . PLoS Genet 6: e1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJPL, Herrera Paerdes S, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield N, Mieczkowski P, Jones CD et al (2017) Direct integration of phosphate starvation and immunity in response to a root microbiome. Nature 543: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ullah C, Reichelt M, Beran F, Yang ZL, Gershenzon J, Hammerbacher A, Vassão DG (2020) The phytopathogenic fungus sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat Commun 11: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 347–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole T, Adam R, Loyal G, Geo P, Daehwan K, Kelley DR, Harold P, Salzberg SL, Rinn JL, Lior P (2012) Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobón A, Canet JV, García‐Andrade J, Angulo C, Neumetzler L, Persson S, Vera P (2015) Novel disease susceptibility factors for fungal necrotrophic pathogens in Arabidopsis . PLoS Pathog 11: 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Li X, Chen J, Chen W, Li B, Li C, Wang L, Li J, Zhao X, Lin J et al (2016) Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis . Proc Natl Acad Sci USA 113: E8326–E8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P (2008) Characterization of a sub‐family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Duan Q, Kita D, Li C, Cheung AY, Wu HM (2010) FERONIA receptor‐like kinase regulates RHO GTPase signaling of root hair development. Proc Natl Acad Sci USA 107: 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu M‐C, Maman J, Steinhorst L, Schmitz‐Thom I et al (2018) The FERONIA receptor kinase maintains cell‐wall integrity during salt stress through Ca2+ signaling. Curr Biol 28: 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel OM, Salas‐González I, Castrillo G, Spaepen S, Law TF, Teixeira PJPL, Jones CD, Dangl JL (2019) The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol 17: e3000534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck CM, Westermann J, Boisson‐Dernier A (2018) Plant malectin‐like receptor kinases: from cell wall integrity to immunity and beyond. Annu Rev Plant Biol 69: 301–328 [DOI] [PubMed] [Google Scholar]
- Guo H, Nolan TM, Song G, Liu S, Xie Z, Chen J, Schnable PS, Walley JW, Yin Y (2018) FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana . Curr Biol 28: 3316–3324 [DOI] [PubMed] [Google Scholar]
- Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido‐Oter R, Thon MR, Weimann A, Damm U, Dallery J‐F, Hainaut M et al (2016) Survival trade‐offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun 7: 11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MJ (2012) Cellular programs for arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 15: 691–698 [DOI] [PubMed] [Google Scholar]
- Hiruma K, Gerlach N, Sacristán S, Nakano R, Hacquard S, Kracher B, Neumann U, Ramírez D, Bucher M, O’Connell R et al (2016) Root endophyte colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidra‐Arellano MC, Delaux P‐M, Valdés‐López O (2021) The phosphate starvation response system: its role in the regulation of plant–microbe interactions. Plant Cell Physiol 62: 392–400 [DOI] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato‐Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R (2010) PAMP (Pathogen‐associated Molecular Pattern)‐induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SA, Shimosato‐Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, Grossniklaus U (2010) Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- Khan GA, Vogiatzaki E, Glauser G, Poirier Y (2016) Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol 171: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Zhang C, Zheng H, Sun M, Zhang F, Zhang M, Cui F, Lv D, Liu L, Guo S et al (2020) Antagonistic interaction between auxin and SA signaling pathways regulates bacterial infection through lateral root in Arabidopsis . Cell Rep 32: 108060 [DOI] [PubMed] [Google Scholar]
- Lally RD, Galbally P, Moreira AS, Spink J, Ryan D, Germaine KJ, Dowling DN (2017) Application of endophytic pseudomonas fluorescens and a bacterial consortium to brassica napus can increase plant height and biomass under greenhouse and field conditions. Front Plant Sci 8: 2193–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Martinoia E, Renton M (2015) Plant adaptations to severely phosphorus‐impoverished soils. Curr Opin Plant Biol 25: 23–31 [DOI] [PubMed] [Google Scholar]
- Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Glavina del Rio T, Jones CD, Tringe SG et al (2015) PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349: 860–864 [DOI] [PubMed] [Google Scholar]
- Li C, Liu X, Qiang X, Li X, Li X, Zhu S, Wang L, Wang Y, Liao H, Luan S et al (2018) EBP1 nuclear accumulation negatively feeds back on FERONIA‐mediated RALF1 signaling. PLoS Biol 16: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, An Arabidopsis LRR receptor‐like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liu HT, Yu XH, Li KW, Klejnot J, Yang HY, Lisiero D, Lin CT (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis . Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Ma K‐W, Niu Y, Jia Y, Ordon J, Copeland C, Emonet A, Geldner N, Guan R, Stolze SC, Nakagami H et al (2021) Coordination of microbe–host homeostasis by crosstalk with plant innate immunity. Nat Plants 7: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, Roy S, Kadota Y, Oh M‐H, Sklenar J, Derbyshire P et al (2014) A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Mang H, Feng B, Hu Z, Boisson‐Dernier A, Franck CM, Meng X, Huang Y, Zhou J, Xu G, Wang T et al (2017) Differential regulation of two‐tiered plant immunity and sexual reproduction by ANXUR receptor‐like kinases. Plant Cell 29: 3140–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, Leon‐Ruiz M, Fürst U, el Ghalid M, Leonard G, López‐Berges MS, Richards TA, Felix G et al (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat Microbiol 1: 1–8 [DOI] [PubMed] [Google Scholar]
- McClerklin SA, Lee SG, Harper CP, Nwumeh R, Jez JM, Kunkel BN (2018) Indole‐3‐acetaldehyde dehydrogenase‐dependent Auxin synthesis contributes to virulence of pseudomonas syringae strain DC3000. PLoS Pathog 14: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk RA, Hossain SS, Haney CH (2019) Convergent gain and loss of genomic islands drive lifestyle changes in plant‐associated Pseudomonas. ISME J 13: 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo RJL, Singh SK, He D, An G, Vílchez JI, Tang K, Yuan F, Sun Y, Shao C, Zhang S et al (2020) Rhizobacterium‐derived diacetyl modulates plant immunity in a phosphate‐dependent manner. EMBO J 39: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis . Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB‐related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana . Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T (2011) Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Raghothama KG, Karthikeyan AS (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists. Nucleic Acids Res 47: W191–W198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebaque D, Hierro I, López G, Bacete L, Vilaplana F, Dallabernardina P, Pfrengle F, Jordá L, Sánchez‐Vallet A, Pérez R et al (2021) Cell wall‐derived mixed‐linked β‐1,3/1,4‐glucans trigger immune responses and disease resistance in plants. Plant J 106: 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolli E, Marasco R, Vigani G, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R et al (2015) Improved plant resistance to drought is promoted by the root‐associated microbiome as a water stress‐dependent trait. Environ Microbiol 17: 316–331 [DOI] [PubMed] [Google Scholar]
- Shen Q, Bourdais G, Pan H, Robatzek S, Di T (2017) Arabidopsis glycosylphosphatidylinositol‐anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc Natl Acad Sci USA 114: 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YI, Wilson AJ, Zhang X‐C, Thoms D, Sohrabi R, Song S, Geissmann Q, Liu Y, Walgren L, He SY et al (2021) FERONIA restricts Pseudomonas in the rhizosphere microbiome via regulation of reactive oxygen species. Nat Plants 5: 644–654 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Guo H, Yin Y, Howell SH (2009) Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis . Plant J 59: 930–939 [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska‐Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF‐regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Thoms D, Liang Y, Haney CH (2021) Maintaining symbiotic homeostasis: How do plants engage with beneficial microorganisms while at the same time restricting pathogens? Mol Plant Microbe Interact 34: 462–469 [DOI] [PubMed] [Google Scholar]
- Xu X, Hao L, Zhu J, Tang B, Zhou Q, Chen T, Zhang S, Dong L, Lan L et al (2018) Database Resources of the BIG Data Center in 2018. Nucleic Acids Res 46: D14–D20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, He J, Dai H, Wang G, Zhang X, Wang C, Shi J, Chen X, Wang D, Wang E (2021) Discriminating symbiosis and immunity signals by receptor competition in rice. Proc Natl Acad Sci USA 118: e2023738118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gleason C (2020) Enhancing potato resistance against root‐knot nematodes using a plant‐defence elicitor delivered by bacteria. Nat Plants 6: 625–629 [DOI] [PubMed] [Google Scholar]
- Zhang X, Peng H, Zhu S, Xing J, Li X, Zhu Z, Zheng J, Wang L, Wang B, Chen J et al (2020) Nematode‐encoded RALF peptide mimics facilitate parasitism of plants through the FERONIA receptor kinase. Mol Plant 13: 1434–1454 [DOI] [PubMed] [Google Scholar]
- Zhao C, Zayed O, Yu Z, Jiang W, Zhu P, Hsu CC, Zhang L, Andy Tao W, Lozano‐Durán R, Zhu JK (2018) Leucine‐rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis . Proc Natl Acad Sci USA 115: 13123–13128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Wang Z, Wang X, Liu D (2019) Blue light triggered‐chemical reactions underlie phosphate deficiency‐induced inhibition of root elongation of Arabidopsis seedlings grown in petri dishes. Mol Plant 11: 1515–1523 [DOI] [PubMed] [Google Scholar]
- Zhou F, Emonet A, Dénervaud Tendon V, Marhavy P, Wu D, Lahaye T, Geldner N (2020) Co‐incidence of damage and microbial patterns controls localized immune responses in roots. Cell 180: 440–453.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Oldroyd GED (2017) Plant signalling in symbiosis and immunity. Nature 543: 328–336 [DOI] [PubMed] [Google Scholar]


