Abstract
Enveloped viruses such as Coronaviridae (CoV) enter the host cell by fusing the viral envelope directly with the plasma membrane (PM) or with the membrane of the endosome. Replication of the CoV genome takes place in membrane compartments formed by rearrangement of the endoplasmic reticulum (ER) membrane network. Budding of these viruses occurs from the ER-Golgi intermediate compartment (ERGIC). The relationship between proteins and various membranes is crucial for the replication cycle of CoVs. The role of transmembrane domains (TMDs) and pre-transmembrane domains (pre-TMD) of viral proteins in this process is gaining more recognition. Here we present a thorough analysis of physico-chemical parameters, such as accessible surface area (ASA), average hydrophobicity (Hav), and contribution of specific amino acids in TMDs and pre-TMDs of single-span membrane proteins of human viruses. We focus on unique properties of these elements in CoV and postulate their role in adaptation to diverse host membranes and regulation of retention of membrane proteins during replication.
Keywords: Accessible surface area (ASA), Average hydrophobicity (Hav), Lipid rafts, Trafficking, Retention
Graphical abstract
1. Introduction
To replicate, viruses must enter the host cell and hijack the cellular machinery. Enveloped viruses consist of a lipid membrane envelope, which encloses their genome, and membrane proteins anchored to the surface of the envelope. Some of these surface proteins are essential for viral entry as they are the first to encounter the target cell membrane. The viral genome is released in the host cell and is replicated and translated to synthesize the viral proteome. The assembly of young virions occurs via budding, which can take place at any step between the endoplasmic reticulum (ER), Golgi complex (Golgi), and plasma membrane (PM), and a new envelope is acquired by pinching off of the host cell membrane (Rheinemann and Sundquist, 2021).
The first step of viral entry is the attachment of the virion to the surface of the host cell. This occurs through the noncovalent interaction of viral fusion proteins and specific surface cell receptors. Fusion proteins are anchored in the viral envelope through their transmembrane domains (TMDs) and bind to cell receptors via receptor-binding domains (RBDs). RBDs are located within the protein's ectodomain, protruding from the viral envelope to the outside of the viral particle. The second step of the viral entry is membrane fusion. Fusion between viral and host membranes is an essential step for envelope viruses to enter the cell. Membrane fusion may occur directly between the viral membrane and the host cell PM (early entry) or after internalization via endocytosis, in which the viral membrane fuses with the membrane of the endosome (late entry) (Pempler, 2012). For the membrane fusion to take place, fusion proteins must be activated. The activation requires proteolytic cleavage of fusion protein precursors (PM fusion) or an additional subsequent drop in pH (endosome fusion). Activation of these proteins results in conformational changes, which allow them to bring two membranes closer together by insertion of a fusion peptide (FP) into the membrane (White et al., 2008). FPs are formed when the preprotein undergoes its proteolytic activation. The free energy released by this conformational change is necessary to overcome the repulsive forces between the membranes, which ends in their fusion.
Certain families of enveloped viruses such as Coronaviridae, or simply coronaviruses (CoVs), can enter the host cell either by fusing directly with the PM or via internalization and endocytosis. As enveloped viruses, CoV virions contain their positive-sense, single-strand RNA genome in a membrane envelope, which harbors three to four structural proteins: the spike protein (S), the matrix protein (M), the envelope protein (E), and sometimes the hemagglutinin esterase (HE). These membrane proteins first encounter the PM at the initiation of infection and later during the replication cycle are translated and incorporated into the ER, endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and after budding, in the secretory pathway through the Golgi (Welsch et al., 2007). Interestingly, the CoV genome replication takes place in special membrane structures, which are induced via drastic modification of host membrane architecture to act as platforms for replication-transcription complexes (RTCs) and to shield newly synthesized RNA genome from the cell's immune mechanisms (reviewed in V'kovski et al., 2021). The most common membrane-derived structures formed by CoVs are double-membrane vesicles (DMVs). Additionally, RNA synthesis benefits from the association with macromolecule-rich membranes such as the intracellular membranes of ER and Golgi (Zhang et al., 2020). Therefore, CoV membrane proteins interact with a diverse array of membranes across the host cell.
The ER, ERGIC, Golgi and PM are not uniformly thick due to a different distribution of cholesterol and sphingolipids. This creates a gradient spanning from thinner and cholesterol-poor intracellular membrane compartments to thicker and cholesterol-rich PM (Bretscher and Munro, 1993). This is evident in TMDs of single-span human membrane proteins and their affinity to so-called lipid rafts. Lipid rafts are fluctuating membrane microdomains, rich in cholesterol and sphingomyelin, which have been implicated in important cell functions such as signal transduction by harboring receptor and channel proteins and association with cytoskeletal elements such as microtubules or actin filaments (Simons and Ikonen, 1997; Allen et al., 2007; Coskun and Simons, 2011). Moreover, they have been shown to play an important role in viral entry and budding (Takeda et al., 2003). It has been demonstrated that certain physico-chemical characteristics affect the association of TMDs with lipid rafts: the surface area of TMDs, their hydrophobicity, length, and palmitoylation (Lorent et al., 2017). TMDs with a smaller surface, determined by their amino acid side chains, preferentially associate with lipid rafts in PM while bulkier TMDs are more likely to reside in thinner, cholesterol-poor, intracellular membranes. This general principle, which governs the affinity of TMDs of single-span human membrane proteins to lipid rafts and different cellular membranes, begs the question of how physico-chemical properties of viral protein single-span TMDs differ between viral families and whether there is any link between viral TMD properties and viral biology.
There is a growing body of studies, which demonstrate that hydrophobic stretches of membrane proteins, which act as their TMD regions, have a function beyond the passive anchor they are usually associated with. A recent study has shown that TMD peptides can cause liposome membrane fusion through the mechanism of lipid binding and lipid splay (Scheidt et al., 2018). Studies have demonstrated that disruption of native TMD sequences can have consequences for correct viral entry (Broer et al., 2006) and budding (Arbely et al., 2004). An alanine scanning insertion study has shown that Mouse Hepatitis Virus (MHV) E protein TMD is crucial for correct viral assembly (Ye and Hogue, 2007). In another study, it was demonstrated that insertion of alanine residues in the predicted TMD of the SARS-CoV ORF7b protein resulted in the shift in localization of the protein from the Golgi to the PM (Schaecher et al., 2008).
We have previously looked into the physico-chemical characteristics of TMD regions of influenza A virus (IAV) hemagglutinin (HA) according to their subtypes and host organisms (Kubiszewski-Jakubiak and Worch, 2020). Our observations suggest that distinct differences in available surface area (ASA), average hydrophobicity (Hav), and the hydrophobic moment (μH) of HA TMDs could have an important effect on protein-lipid interaction, HA oligomerization, and orientation. In that study we also looked into the membrane proximal region or pre-transmembrane domain region (pre-TMD) of IAV HA, which connects the HA TMD with its ectodomain. It has been described as a flexible juxtamembrane linker and it was shown to be involved in a tilt of up to 52° of the HA TMD from the threefold axis of the HA ectodomain (Benton et al., 2018). We hypothesized that due to significant differences in TMD ASA and Hav and pre-TMD μH between H1 and H3 subtypes, it could be involved in the formation of the heterosubtypic immune response (Kubiszewski-Jakubiak and Worch, 2020). Similar pre-TMD regions have been described for other envelope viruses such as HIV-1, SARS-CoV and Ebola (Salzwedel et al., 1999; Howard et al., 2008; Lee et al., 2017). They are often referred to as the membrane proximal extended regions (MPERs) and although they differ in primary amino acid sequence they are similarly flexible, enriched in aromatic residues and contain cholesterol-binding motifs, all of which implicate that these regions may have a functional role in viral entry, specifically in the process of membrane fusion (Sainz et al., 2005; Apellániz et al., 2011).
To elucidate the importance of membrane-associated regions of viral proteins, we decided to conduct a thorough study of all available TMD and pre-TMD sequences from single-span membrane proteins of known human pathogenic viruses. We focused on various physico-chemical parameters such as the accessible surface area (ASA), average hydrophobicity (Hav), as well as the contribution of particular amino acid residues. We observed that CoV TMDs and pre-TMDs differ significantly as compared to other studied viral families, therefore we decided to focus our analysis on CoVs and discussed their unique properties in relation to human proteins and CoV replication cycle.
2. Results and discussion
2.1. CoV TMDs have larger ASA and are more hydrophobic compared to other viruses
We began by assembling a comprehensive set of sequences of TMDs and pre-TMDs of single-span membrane proteins from viruses known to infect humans. This included several enveloped viral families (Herpesviridae, Poxviridae, Retroviridae, Arenaviridae, Filoviridae, Hantavirus, Nairoviridae, Orthomyxoviridae, Paramyxoviridae, Flaviviridae, Pneumoviridae, Rhabdoviridae, Coronaviridae, Flaviviridae, Metano Virus and Togaviridae) as well as few non-enveloped viral families (Adenoviridae, Papillomaviridae, Reoviridae, Caliciviridae, Picornaviridae and Hepeviridae).
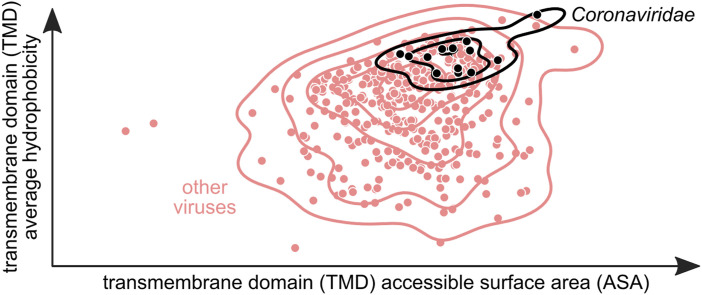
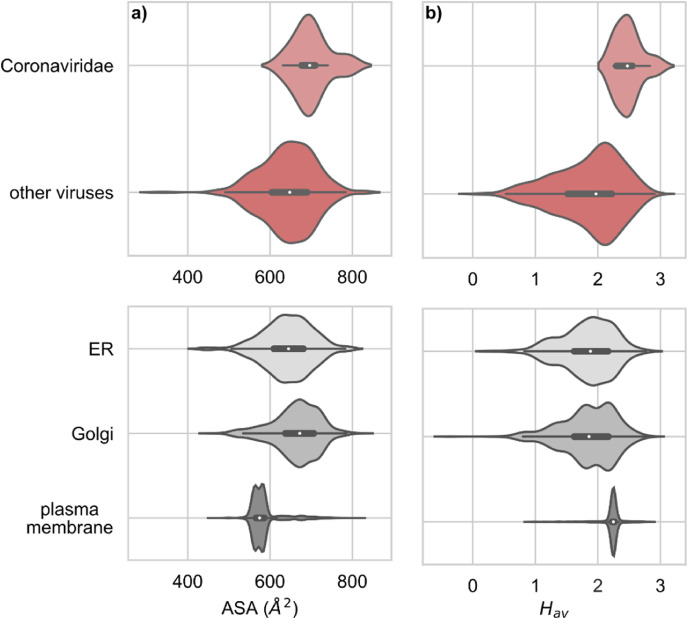
Since physico-chemical parameters of TMD regions have been shown to influence their lipid interaction properties and affinity to lipid rafts, we calculated the accessible surface area (ASA) and average hydrophobicity (Hav) of all TMD sequences in our dataset. We observed that on average, TMDs of proteins belonging to the Coronaviridae (CoV) family have a larger ASA value as compared to other viral families. The average value of ASA for CoV TMDs was 700 ± 44 Å2 (mean ± standard deviation) while that for other families was 644 ± 67 Å2 (Fig. 1 a). We also observed that ASA for human proteins from the endoplasmic reticulum (ER) (640 ± 60 Å2) and the Golgi (663 ± 56 Å2) was higher than that for the plasma membrane (PM) (585 ± 37 Å2), which was consistent with observations by Lorent et al., 2017) (Lorent et al., 2017). It seems that CoV TMD ASA has shifted towards higher values as compared to other viruses as well as human proteins located in the ER, Golgi and PM. As mentioned previously, bulkier TMDs preferentially locate in intracellular membranes such as those of ER and Golgi. As CoVs bud from the ERGIC (Ujike and Taguchi, 2015), TMDs of their membrane proteins could have evolved larger surface areas to fit in these compartments. Retention of viral membrane proteins in intracellular membranes is crucial for the correct and efficient maturation of CoV viral particles. It is hypothesized that viral proteins are retained by one of two mechanisms: the “bilayer thickness model” or the “kin recognition model”. The first one proposes that a shorter TMD would be retained in the ER and Golgi, where the bilayer is thinner than the PM (Bretscher and Munro, 1993). As mentioned above, the membrane thickness of different intracellular membranes varies due to their cholesterol and sphingolipid composition. It is hypothesized that this could play a significant role in the trafficking of membrane proteins. It is possible then, that due to the lower cholesterol and sphingolipid content of ER and Golgi compartments, as compared to the PM, CoV TMDs adapted to membranes, which are not lipid raft-rich and during trafficking would be excluded from lipid rafts destined for the PM. Additionally, raft association correlates with proteins of smaller ASA values (Lorent et al., 2017), which could explain retention in ER and Golgi of these proteins.
Fig. 1.
Coronaviridae (CoV) TMD have larger (a) ASA and (b) are more hydrophobic as compared to other viruses. Corresponding values for TMDs of human membrane proteins in ER, Golgi and plasma membrane (PM) are shown in the lower panel. The values for Coronaviridae differ significantly from all others: (Wilcoxon test) p-values < 0.001 in all cases, except for Coronaviridae and Golgi ASA (p < 0.02).
Similarly, we observed that the average hydrophobicity (Hav, according to GES scale) of CoV TMDs was significantly higher (2.5 ± 0.2) as compared to that of TMDs of proteins of other viral families (1.8 ± 0.5) (Fig. 1b). The distribution of hydrophobicity was also different between human proteins from different membrane systems (ER and Golgi: 1.8 ± 0.4, PM: 2.2 ± 0.2) and again CoV TMD Hav values were higher than those of other viruses and human proteins. The ASA and Hav values for particular viral families are shown in Supplementary Fig. 1. As the Hav is dependent on the number of polar versus non-polar amino acid residues within the TMD, to further analyze the detailed composition of viral TMDs we divided the amino acids into three categories: polar, strongly polar, and charged (Fig. 2 , see Materials and Methods for details).
Fig. 2.
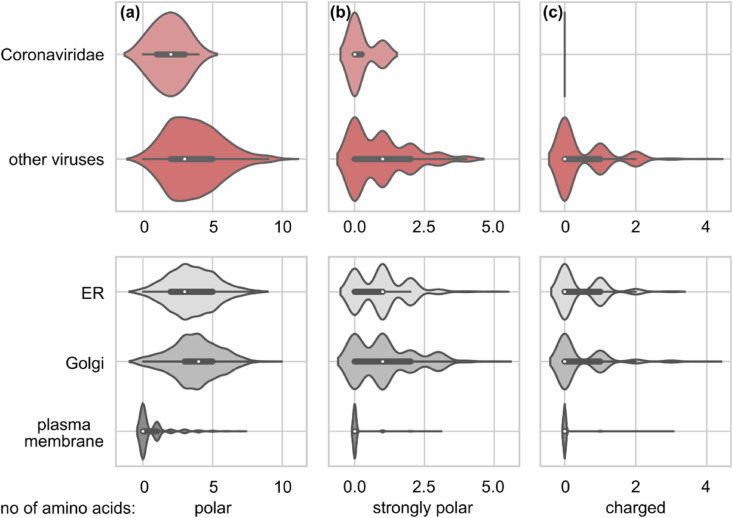
Coronaviridae (CoV) TMD have fewer (a) polar, (b) strongly polar and (c) charged amino acids as compared to other viruses. Corresponding values for human TMDs of human membrane proteins in ER, Golgi and plasma membrane (PM) are shown in the lower panel. The values for Coronaviridae differ significantly from all others: (Wilcoxon test) p-values < 0.001 in all cases, except for the numbers of charged amino acids in Coronaviridae and plasma membrane proteins (p = 0.56).
We observed that CoV TMDs contain significantly fewer polar, strongly polar, and charged amino acid residues as compared to TMDs of other viruses. In more detail, in the analyzed TMD fragments of 21 amino acids, on average CoV contained fewer polar residues (1.9 ± 1.1) as compared to other viruses (3.6 ± 2.0). In comparison to TMDs of human membrane proteins, depending on the organelle: ER: 3.5 ± 1.6, Golgi: 3.7 ± 1.7 and PM: 0.6 ± 1.2 (Fig. 2a). For strongly polar residues, CoV TMDs contained on average 0.25 ± 0.45, other viruses: 0.9 ± 1.0, whereas human proteins: 0.96 ± 0.88 in ER, 1.1 ± 1.1 in Golgi and 0.06 ± 0.28 in PM. Finally, charged residues were not found in CoV TMDs, in contrast to other viruses (0.49 ± 0.74). The average number of polar residues in TMDs showed a substantial difference between organelles of human cells: 0.44 ± 0.72 in Golgi, 0.46 ± 0.65 in ER and 0.03 ± 0.18 in plasma membrane. Probably this trend is related to avoiding unspecific, electrostatically-driven, protein-protein interactions in the membrane milieu.
2.2. Contribution of specific amino acid residues and dimerization motifs in TMDs
Intrigued by differences in the content of polar, strongly polar, and charged amino acid residues, we further analyzed the contribution of specific amino acids in all TMDs (Table 1 ). We observed that 75.0% of CoV TMDs contain cysteine residues (at least one per TMD) while for other viruses it was only 40.0% (36.1% in ER and 51.0% in Golgi and only 8.7% in PM for TMDs of human membrane proteins). Cysteine residues of viral proteins are S-acylated, predominantly in the form of palmitoylation and stearation (reviewed in (Veit, 2012):). Palmitoylation is a covalent fatty acid modification, which adds a 16-carbon, palmitic acid chain to protein cysteines. It enhances protein affinity to the membrane milieu and is known to be involved in virus assembly and infection and has been proposed to have a trafficking function (Chamberlain and Shipston, 2015). Palmitoylated cysteine residues usually are located in the cytoplasmic tail of the membrane protein or the vicinity or inside of its predicted TMD. According to a recent review, many of CoV membrane protein cysteine residues are either predicted to or have been experimentally shown to be palmitoylated (Tanner and Alfieri, 2021). Perhaps due to bulkier and more hydrophobic TMDs, CoV membrane proteins contain more cysteine residues, which can be palmitoylated to increase the lipid raft association. This could act as a regulation mechanism for the retention in ER and Golgi versus loading into vesicles and trafficking via the secretion pathway. Distribution of the number of cysteine residues in CoV TMDs and other viruses is presented in Supplementary Fig. 2.
Table 1.
Percentages of TMDs containing at least one amino acid (C, G, W) or a motif for Coronaviridae, other viruses and human single-span proteins in ER, Golgi and plasma membrane (n - total number of proteins).
| amino acids |
Motifs |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | C | G | W | F | S | GxxxG | QxxS | FxxGxxxG | FxxS | WxxF | |
| Coronaviridae | 16 | 75.0 | 25.0 | 56.3 | 93.8 | 56.3 | 18.8 | 0.0 | 0.0 | 12.5 | 12.5 |
| other viruses | 472 | 40.0 | 74.6 | 28.8 | 81.4 | 67.4 | 8.9 | 0.6 | 0.6 | 9.7 | 2.5 |
| ER | 479 | 36.1 | 72.2 | 45.9 | 81.2 | 70.8 | 9.2 | 0.6 | 1.9 | 7.9 | 4.2 |
| Golgi | 553 | 51.0 | 59.0 | 36.7 | 89.2 | 75.6 | 9.2 | 1.1 | 0.4 | 9.6 | 1.3 |
| plasma membrane | 6100 | 8.7 | 95.1 | 4.7 | 28.3 | 11.5 | 37.1 | 0.0 | 0.2 | 1.0 | 0.3 |
Interestingly, only 25.0% of CoV TMDs contain glycine residues (at least one per TMD), while for other viruses it is 69.3%, 72.2% for ER, 59.0% for Golgi and 95.1% for PM TMDs (Table 1). Majority of glycine residues in CoV TMDs are present in the form of the GxxxG motif (Senes et al., 2000) but none in the extended FxxGxxxG motif (Unterreitmeier et al., 2007). It has been demonstrated that the GxxxG motif is uniquely inserted in the SARS-CoV S protein TMD, and not present in other CoV TMDs and it is important for trimerization of S protein monomers (Arbely et al., 2006) however, disputed by work from Corver et al., 2007) (Corver et al., 2007), who demonstrated that GxxxG is not involved in trimerization and is not important for S protein-mediated entry. In fact, according to Corver et al., 2007), the glycine residue G1205 alone is important for the entry of the virus. Similarly, more than 56% of CoV TMDs contain W residues, some of which (∼22%) are organized in the WxxF oligomerization motif (Johnson et al., 2007). Other motifs such as LIxxGVxxGVxxT, SxxSSxxT, and SxxxSSxxT were not observed in our viral TMD dataset. This observation is in line with the “kin recognition model” of retention of proteins, which states that membrane proteins might form large homo- or heterooligomers within the ER and Golgi membranes, which would prevent their transport into vesicles for trafficking through the secretory pathway (Nilsson et al., 1993, 1994).
Finally, we observed that a high percentage (93.8%) of CoV TMDs contained phenylalanine (F) as compared to other viruses (81.3%), ER (81.2%), Golgi (89.2%) and PM (28.3%). It has been proposed that Golgi resident proteins contain TMDs with high F content (Lundbæk et al., 2003). The F residues contain large side chains, which could be energetically unfavorable for membrane domains rich in cholesterol. That could offer another explanation for how ER and Golgi TMD and CoV TMD help the proteins to retain in the intracellular membranes.
2.3. The pre-TMD regions of CoV single-span proteins are enriched in aromatic amino acids
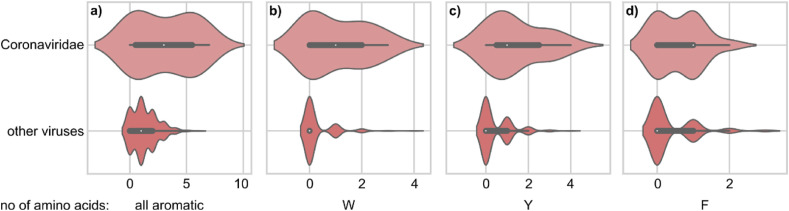
The pre-TMD regions of envelope viruses have been shown to play a role in membrane fusion and are considered the holy grail of immunization due to their conserved sequences as demonstrated for IAV HA2 (Kirkpatrick et al., 2018) and SARS-CoV-2 S2 subunits (Ng et al., 2021). We looked into these regions (11 amino acid long fragments upstream of the TMD) in single-pass viral proteins in our dataset and observed that on average CoV pre-TMD regions contain significantly more aromatic residues compared to other viral families (Fig. 5 ). Specifically, we observed that 53.3% of CoV pre-TMDs contained at least one W residue while for other viral families it was 21.0%. In case of F residues it was 53.3% in CoV and 29.7% for other viruses and for Y it was 73.3% and 35.6% for CoV and other viral families respectively (Table 2 ). Enrichment of aromatic amino acids has been previously reported for pre-TMD regions such as SARS-CoV MPER and it has been demonstrated that W residues specifically play a role in maintaining the MPER structure allowing it to form a quaternary structure with the internal fusion peptide to aid membrane fusion (Liao et al., 2015). We also observed that in all CoV pre-TMD regions in our dataset there is at least one G residue present (Table 2). This could explain the flexibility of this region, which was previously reported, and combined with aromatic enrichment explain the mechanism of pre-TMD region involvement in the membrane fusion process. Interestingly, despite a high number of G, W and F residues, we have not observed any of the mentioned dimerization motifs present in CoV pre-TMD regions.
Fig. 5.
Coronaviridae (CoV) pre-TMD regions are enriched in aromatic residues as compared to other viruses. The panels show (a) all aromatic residues, (b) W, (c) Y and (d) F. p-values (Wilcoxon test): (a) p < 0.05, (b) and (c) p < 0.001, (d) p < 0.08.
Table 2.
Percentages of pre-TMDs containing at least one amino acid (C, G, W, F, S, Y) or a motif for Coronaviridae and for other viruses (n - total number of proteins).
| preTMD | N | C | G | W | F | S | Y | GxxxG | QxxS | FxxGxxxG | FxxS | WxxF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coronaviridae | 15 | 6.7 | 100.0 | 53.3 | 53.3 | 13.3 | 73.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| other viruses | 424 | 12.5 | 43.6 | 21.0 | 29.7 | 59.2 | 35.6 | 2.6 | 1.7 | 0.0 | 3.5 | 0.5 |
2.4. The role of TMD: active player versus passive hydrophobic zipper
It is becoming more evident that TMDs of viral membrane proteins play a crucial role in viral replication cycles. Previously, we have hypothesized that different physico-chemical properties of the TMD of IAV HA of different HA subtypes can have an important role in the positioning of the ectodomain and thereby influence the immunogenicity of HA protein (Kubiszewski-Jakubiak and Worch, 2020). The importance of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) ORF7b TMD was demonstrated for the retention in Golgi (Schaecher et al., 2008). SARS-CoV ORF7b is a type III integral membrane protein. The ORF7b chimera was generated by replacing the native TMD with the 21-residue TMD from the human endoprotease furin, which abolished retention in the Golgi. A scanning alanine mutagenesis was performed on the ORF7b TMD. The highest level of PM expression of the alanine mutant (i.e. lowest retention in the Golgi) was observed for mutants in the 13–15 and 19-22 section of the TMD segment, which contained all non-polar and aromatic residues.
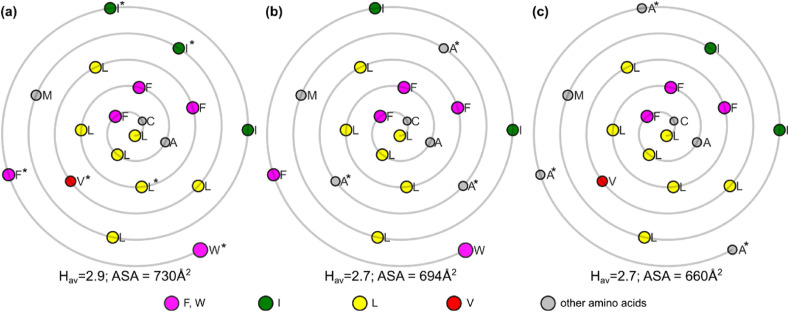
To further investigate the potential importance of TMD ASA and Hav parameters we performed the analysis of the WT ORF7b and two alanine stretch mutants (Fig. 3) by calculating the ASA and the Hav and we generated the Wenxiang diagrams of their TMDs. We observed that Hav values were slightly lower in the alanine stretch mutants (13–15: 2.7 and 19–22: 2.7) as compared to WT (2.9). Similarly, ASA was lower in the 13–15 mutant (694 Å2) and 19–22 mutant (660 Å2) as compared to the WT ASA (730 Å2). It shows that potentially a thinner TMD, with fewer hydrophobic residues, is not as likely to be retained in the Golgi. Indeed, an alanine scanning insertion mutagenesis of the TMD belonging to the MHV E protein showed that disruption in the TMD can affect virus growth (Schaecher et al., 2008). MHV E is a viroporin that plays a crucial role in the viral replication of CoV. Insertion of an alanine residue into the TMD helix causes all amino acid residues on its carboxy side to rotate by approximately 100°. In this study, eight alanine insertion mutants (designated as Ala 1–8) were constructed by positioning the residues at various places across the hydrophobic domain. All alanine insertions affected the virus growth as determined by the plaque assay but the most severe phenotype was observed for Ala 3–6 mutants. The integrity of the helix and possibly the positions of polar hydrophilic residues may be functionally important as insertions caused decreased viral growth.
Fig. 3.
Wenxiang diagrams for the (a) wild-type SARS-CoV ORF7b TMD and two mutations exhibiting impaired retention in Golgi (b) and (c), respectively. Color code: F and W: magenta, I: green, L: yellow, V: red, other: grey. Mutated residues marked with an asterisk. Sizes of the amino acids are proportional to ASA. Hav and ASA calculated for the most hydrophobic 19 amino acid fragment (see Materials and Methods for details).
2.5. TMD ASA of viruses budding by cellular exocytosis is higher compared to other viral families
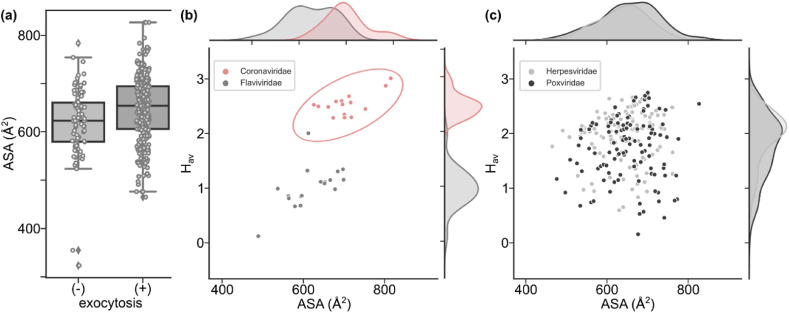
The process of budding from ER, ERGIC, and Golgi, which takes place during the maturation of enveloped viruses, implies that young viral particles are exported by cellular exocytosis. Therefore, we decided to check whether the TMD ASA of exocytotic viruses in our database is different when compared to non-exocytotic viruses. Indeed we observed that on average TMDs of proteins belonging to exocytotic human pathogenic viruses had larger ASA values in comparison to non-exocytotic pathogens (Fig. 4a). As CoVs are known to replicate in previously mentioned membrane structures, such as ER DMVs, and have been shown to bud from ERGIC (Ujike and Taguchi, 2015), it seems likely that their TMDs would have physico-chemical characteristics adapted to these membranes.
Fig. 4.
Physico-chemical parameters for TMDs of viruses budding by cellular exocytosis. (a) ASA of TMDs is larger for viruses budding by exocytosis (Wilcoxon test, p-value <0.05), (b) Coronaviridae and Flaviviridae appear in clusters on Hav vs ASA map in contrast to Herpesviridae and Poxviridae (c).
As we have observed that both ASA and Hav are significantly higher in CoV TMDs as compared with other human viral pathogens, we wanted to see how these two parameters correlate. Therefore, we checked the correlation between ASA and Hav for individual viral families, which are known to exit the cell via exocytosis. Interestingly, we observed that values for ASA versus Hav cluster for CoV and Flaviviridae (Fig. 4b). TMDs of CoV membrane proteins have higher ASA values (700 ± 44 Å2) and are more hydrophobic (2.5 ± 0.2) as compared to Flaviviridae (626 ± 50 Å2 and 1.0 ± 0.0, ASA and Hav, respectively). Both of these families have been shown to bud from specialized membrane structures; however, CoVs replicate in DMVs while Flaviviridae form ER spherules for their genome replication. Moreover, CoV virions bud from ERGIC while Flaviviridae from the ER (Ujike and Taguchi, 2015; Welsch et al., 2009). Perhaps there is a very specific and discrete combination of ASA and Hav of TMD segments, which allows CoVs to traffic and retain their membrane proteins for precise assembly and budding from the ERGIC compartment. The other viral family known to bud from ERGIC, namely the Arteriviridae, was not included in our dataset as there are no known human pathogens in this family according to the ViralZone database.
Noticeably, there is no clear clustering of ASA vs Hav for other exocytotic viral families such as Herpesviridae and Poxviridae. Both families showed a wide spread of ASA and Hav parameters (Fig. 4c). These viral families have been shown to bud from the Golgi and the trans-Golgi network (TGN) respectively (Mettenleiter et al., 2006; Smith and Law, 2004). Moreover, they do not employ modified host cell membrane structures for genome replication. Instead, they replicate their genome within the host cell nucleus (Herpesviridae) or viroplasm (Poxviridae), which is a cytoplasmic inclusion compartment unrelated to membranes. This could suggest that there is a functional link between the ASA and Hav of viral membrane TMDs and the way viruses coordinate the genome replication in modified membrane structures, assembly of properly targeted membrane proteins, and budding within the complex membrane network of the host cell. As for other exocytotic viral families, our database contains only one entry for Reoviridae (on average ASA = 631.5 Å2; Hav = 1.1) while there is no representation from Bunyaviridae and Hepadnaviridae since there are no predicted membrane proteins in those families.
3. Conclusion
Evidently, TMD and pre-TMD regions of viral proteins have essential functions in viral replication cycles. Our analysis of human pathogenic viral membrane-associated segments has shown that CoV TMDs and pre-TMDs have distinct features when compared to other viral families. We demonstrated that CoV TMDs have a higher accessible surface area (ASA) and higher average hydrophobicity (Hav), which might play an important role in protein-lipid interactions and affinity to lipid rafts. We observed that CoV TMDs amino acid sequences shifted towards dimerizing motifs as calculated by the contribution of G and W residues. Moreover, CoV TMDs have a high number of large side chain-containing phenylalanine residues as well as cysteine residues, which can be potentially S-acylated, all of which could act as a retention/secretion regulation mechanism. We postulate that CoV TMDs have acquired these features to adapt to varying membrane milieu across the cell which facilitates efficient CoV replication, budding, and exocytosis.
4. Materials and methods
4.1. Source of sequences
We assembled the set of viral proteins based on ViralZone (https://viralzone.expasy.org/) (Hulo et al., 2011) entries (human viruses, reference stains). In the case of polyproteins, the sequences were compared with Uniprot (https://www.uniprot.org/) (Bateman et al., 2021) PTM/Processing annotations and manually divided into sequences of processed proteins (in total 39 entries). Their protein sequence fragments resulting from cleavage were added manually to the dataset. The sequences of human proteins were downloaded from UniProt. Organelle location was obtained by adding a corresponding keyword to the query (keyword: “Endoplasmic reticulum” or “Golgi apparatus” or “Cell membrane”). Viral proteins were excluded by the keyword NOT host: "Homo sapiens (Human) [9606]".
4.2. Transmembrane region prediction
Reviewed sequences were downloaded in FASTA format and were used as an input for TMD prediction using TMHMM 2.0 server. Single-span membrane proteins were selected by PredHel = 1 notation in the output for both viral and human datasets to exclude the multi-span protein bias. To further confirm the presence of a single-span TMD we have manually scanned all sequences and added Uniprot single-span annotated proteins, which were missed. We applied the procedure of TMD selection performed in other studies (Senes et al., 2000; Unterreitmeier et al., 2007). In short, the sequences predicted by the TMHMM 2.0 were extended by 5 amino acids from both ends, and the region of 21 amino acids of the highest hydrophobicity according to the GES scale was assigned as TMD. Only 6.4% of the entries with predicted TMDs did not have “transmembrane” annotation for the corresponding UniProt ID. To avoid potential bias, the set was reduced to 40% homology (separately for viral and human proteins). The pre-TMD regions were chosen as an 11 amino acid fragment upstream of the predicted TMD while fragments shorter than 11 amino acids were not included in the analysis. For the TMDs of the normalized lengths average hydrophobicity (GES scale) and accessible surface area (ASA) were calculated. The ASA values for individual amino acids (a set of values for membranous regions of membrane proteins) were taken from (Yuan et al., 2006). For the composition analysis, we divided the amino acids into the following categories, similarly to Worch at al. (Worch et al., 2010): polar (S, T, Y, N, Q, H, R, K, D, E), strongly polar (for which S, T and Y were excluded) and charged (R, K, D, E; H was treated as neutral). The analysis was performed using home-written Python 3.7 scripts. Supplementary data 1 and Supplementary data 2 contain the records for viral and human TMDs, respectively.
CRediT authorship contribution statement
Szymon Kubiszewski-Jakubiak: Data curation, Validation, Writing – review & editing. Remigiusz Worch: Conceptualization, Methodology, Visualization, Software, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Sonata Bis 8 grant from the National Centre of Science, Poland (UMO-2018/30/E/NZ1/00257).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2022.03.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Allen J.A., Halverson-Tamboli R.A., Rasenick M.M. Lipid raft microdomains and neurotransmitter signalling. Nat. Rev. Neurosci. 2007;8:128–140. doi: 10.1038/nrn2059. [DOI] [PubMed] [Google Scholar]
- Apellániz B., Ivankin A., Nir S., Gidalevitz D., Nieva J.L. Membrane-proximal external HIV-1 gp41 motif adapted for destabilizing the highly rigid viral envelope. Biophys. J. 2011;101:2426–2435. doi: 10.1016/j.bpj.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbely E., Khattari Z., Brotons G., Akkawi M., Salditt T., Arkin I.T. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 2004;341:769–779. doi: 10.1016/j.jmb.2004.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbely E., Granot Z., Kass I., Orly J., Arkin I.T. A trimerizing GxxxG Motif is uniquely inserted in the severe acute respiratory syndrome (SARS) coronavirus spike protein transmembrane domain. Biochemistry. 2006;45:11349–11356. doi: 10.1021/bi060953v. [DOI] [PubMed] [Google Scholar]
- Bateman A., Martin M.J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bursteinas B., Bye-A-Jee H., Coetzee R., Cukura A., Da Silva A., Denny P., Dogan T., Ebenezer T.G., Fan J., Castro L.G., Garmiri P., Georghiou G., Gonzales L., Hatton-Ellis E., Hussein A., Ignatchenko A., Insana G., Ishtiaq R., Jokinen P., Joshi V., Jyothi D., Lock A., Lopez R., Luciani A., Luo J., Lussi Y., MacDougall A., Madeira F., Mahmoudy M., Menchi M., Mishra A., Moulang K., Nightingale A., Oliveira C.S., Pundir S., Qi G., Raj S., Rice D., Lopez M.R., Saidi R., Sampson J., Sawford T., Speretta E., Turner E., Tyagi N., Vasudev P., Volynkin V., Warner K., Watkins X., Zaru R., Zellner H., Bridge A., Poux S., Redaschi N., Aimo L., Argoud-Puy G., Auchincloss A., Axelsen K., Bansal P., Baratin D., Blatter M.C., Bolleman J., Boutet E., Breuza L., Casals-Casas C., de Castro E., Echioukh K.C., Coudert E., Cuche B., Doche M., Dornevil D., Estreicher A., Famiglietti M.L., Feuermann M., Gasteiger E., Gehant S., Gerritsen V., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., Hyka-Nouspikel N., Jungo F., Keller G., Kerhornou A., Lara V., Le Mercier P., Lieberherr D., Lombardot T., Martin X., Masson P., Morgat A., Neto T.B., Paesano S., Pedruzzi I., Pilbout S., Pourcel L., Pozzato M., Pruess M., Rivoire C., Sigrist C., Sonesson K., Stutz A., Sundaram S., Tognolli M., Verbregue L., Wu C.H., Arighi C.N., Arminski L., Chen C., Chen Y., Garavelli J.S., Huang H., Laiho K., McGarvey P., Natale D.A., Ross K., Vinayaka C.R., Wang Q., Wang Y., Yeh L.S., Zhang J. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton D.J., Nans A., Calder L.J., Turner J., Neu U., Lin Y.P., Ketelaars E., Kallewaard N.L., Corti D., Lanzavecchia A., Gamblin S.J., Rosenthal P.B., Skehel J.J. Influenza hemagglutinin membrane anchor. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115:10112–10117. doi: 10.1073/pnas.1810927115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M.S., Munro S. Cholesterol and the golgi apparatus. Science. 1993;261(80-):1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Broer R., Boson B., Spaan W., Cosset F.-L., Corver J. Important role for the transmembrane domain of severe acute respiratory syndrome coronavirus spike protein during entry. J. Virol. 2006;80:1302–1310. doi: 10.1128/jvi.80.3.1302-1310.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain L.H., Shipston M.J. The physiology of protein s-acylation. Physiol. Rev. 2015;95:341–376. doi: 10.1152/physrev.00032.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver J., Broer R., van Kasteren P., Spaan W. GxxxG motif of severe acute respiratory syndrome coronavirus spike glycoprotein transmembrane domain is not involved in trimerization and is not important for entry. J. Virol. 2007;81:8352–8355. doi: 10.1128/jvi.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun Ü., Simons K. Cell membranes: the lipid perspective. Structure. 2011;19:1543–1548. doi: 10.1016/j.str.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Howard M.W., Travanty E.A., Jeffers S.A., Smith M.K., Wennier S.T., Thackray L.B., Holmes K.V. Aromatic amino acids in the juxtamembrane domain of severe acute respiratory syndrome coronavirus spike glycoprotein are important for receptor-dependent virus entry and cell-cell fusion. J. Virol. 2008;82:2883–2894. doi: 10.1128/jvi.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo C., De Castro E., Masson P., Bougueleret L., Bairoch A., Xenarios I., Le Mercier P. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res. 2011;39:576–582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.M., Hecht K., Deber C.M. Aromatic and cation-π interactions enhance helix-helix association in a membrane environment. Biochemistry. 2007;46:9208–9214. doi: 10.1021/bi7008773. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick E., Qiu X., Wilson P.C., Bahl J., Krammer F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-28706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiszewski-Jakubiak S., Worch R. Influenza a h1 and h3 transmembrane domains interact differently with each other and with surrounding membrane lipids. Viruses. 2020;12:1–13. doi: 10.3390/v12121461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Nyenhuis D.A., Nelson E.A., Cafiso D.S., White J.M., Tamm L.K. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E7987–E7996. doi: 10.1073/pnas.1708052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Zhang S.M., Neo T.L., Tam J.P. Tryptophan-dependent membrane interaction and heteromerization with the internal fusion peptide by the membrane proximal external region of SARS-CoV spike protein. Biochemistry. 2015;54:1819–1830. doi: 10.1021/bi501352u. [DOI] [PubMed] [Google Scholar]
- Lorent J.H., Diaz-Rohrer B., Lin X., Spring K., Gorfe A.A., Levental K.R., Levental I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017;8:1–9. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundbæk J.A., Andersen O.S., Werge T., Nielsen C. Cholesterol-induced protein sorting: an analysis of energetic feasibility. Biophys. J. 2003;84:2080–2089. doi: 10.1016/S0006-3495(03)75015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T.C., Klupp B.G., Granzow H. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Ng K.T., Mohd-Ismail N.K., Tan Y.J. vol. 9. 2021. Spike S2 Subunit: the Dark Horse in the Race for Prophylactic and Therapeutic Interventions against Sars-Cov-2, Vaccines; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Slusarewicz P., Hoe M.H., Warren G. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-B. [DOI] [PubMed] [Google Scholar]
- Nilsson T., Hoe M.H., Slusarewicz P., Rabouille C., Watson R., Hunte F., Watzele G., Berger E.G., Warren G. Kin recognition between medial golgi enzymes in HeLa cells. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pempler K.R. Cell enter of enveloped viruses. Curr. Opin. Virol. 2012;1:92–100. doi: 10.1016/j.coviro.2011.06.002.Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinemann L., Sundquist W.I. Virus Budding. Encycl. Virol. 2021:519–528. doi: 10.1016/b978-0-12-814515-9.00023-0. [DOI] [Google Scholar]
- Sainz B., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. The aromatic domain of the coronavirus class I viral fusion protein induces membrane permeabilization: putative role during viral entry. Biochemistry. 2005;44:947–958. doi: 10.1021/bi048515g. [DOI] [PubMed] [Google Scholar]
- Salzwedel K., West J.T., Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for env-mediated fusion and virus infectivity. J. Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Diamond M.S., Pekosz A. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the golgi complex. J. Virol. 2008;82:9477–9491. doi: 10.1128/jvi.00784-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidt H.A., Kolocaj K., Veje Kristensen J., Huster D., Langosch D. Transmembrane helix induces membrane fusion through lipid binding and splay. J. Phys. Chem. Lett. 2018;9:3181–3186. doi: 10.1021/acs.jpclett.8b00859. [DOI] [PubMed] [Google Scholar]
- Senes A., Gerstein M., Engelman D.M. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and association with β-branched residues at neighboring positions. J. Mol. Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. S. K, I. E. [DOI] [PubMed] [Google Scholar]
- Smith G.L., Law M. The exit of Vaccinia virus from infected cells. Virus Res. 2004;106:189–197. doi: 10.1016/j.virusres.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Takeda M., Leser G.P., Russell C.J., Lamb R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner J.E., Alfieri C. vol. 13. 2021. pp. 1–18. (The Fatty Acid Lipid Metabolism Nexus in COVID-19, Viruses). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7:1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterreitmeier S., Fuchs A., Schäffler T., Heym R.G., Frishman D., Langosch D. Phenylalanine promotes interaction of transmembrane domains via GxxxG motifs. J. Mol. Biol. 2007;374:705–718. doi: 10.1016/j.jmb.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Veit M. Palmitoylation of virus proteins. Biol. Cell. 2012;104:493–515. doi: 10.1111/boc.201200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Müller B., Kräusslich H.G. More than one door - budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K.E., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320.Structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worch R., Bökel C., Höfinger S., Schwille P., Weidemann T. Focus on composition and interaction potential of single-pass transmembrane domains. Proteomics. 2010;10:4196–4208. doi: 10.1002/pmic.201000208. [DOI] [PubMed] [Google Scholar]
- Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 2007;81:3597–3607. doi: 10.1128/jvi.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Zhang F., Davis M.J., Bodén M., Teasdale R.D. Predicting the solvent accessibility of transmembrane residues from protein sequence. J. Proteome Res. 2006;5:1063–1070. doi: 10.1021/pr050397b. [DOI] [PubMed] [Google Scholar]
- Zhang J., Lan Y., Sanyal S. Membrane heist: coronavirus host membrane remodeling during replication. Biochimie. 2020;179:229–236. doi: 10.1016/j.biochi.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.