Abstract
As in human cells, yeast telomeres can be maintained in cells lacking telomerase activity by recombination‐based mechanisms known as ALT (Alternative Lengthening of Telomeres). A hallmark of ALT human cancer cells are extrachromosomal telomeric DNA elements called C‐circles, whose origin and function have remained unclear. Here, we show that extrachromosomal telomeric C‐circles in yeast can be detected shortly after senescence crisis and concomitantly with the production of survivors arising from “type II” recombination events. We uncover that C‐circles bind to the nuclear pore complex (NPC) and to the SAGA‐TREX2 complex, similar to other non‐centromeric episomal DNA. Disrupting the integrity of the SAGA/TREX2 complex affects both C‐circle binding to NPCs and type II telomere recombination, suggesting that NPC tethering of C‐circles facilitates formation and/or propagation of the long telomere repeats characteristic of type II survivors. Furthermore, we find that disruption of the nuclear diffusion barrier impairs type II recombination. These results support a model in which concentration of C‐circles at NPCs benefits type II telomere recombination, highlighting the importance of spatial coordination in ALT‐type mechanisms of telomere maintenance.
Keywords: alternative lengthening of telomeres, C‐circles, recombination, senescence, telomeres
Subject Categories: Chromatin, Transcription & Genomics; DNA Replication, Recombination & Repair
Extrachromosomal DNA circles, a hallmark of alternative telomere lengthening (ALT), arise also in telomerase‐deficient yeast cells and may aid recombination‐based telomere maintenance.

Introduction
Telomeres at the ends of linear chromosomes are designed for protection against DNA degradation, chromosome fusion, and homologous recombination. They form nucleoprotein structures composed of G‐rich repeated sequences ending with a 3′ single‐strand overhang and coated by a complex of proteins called shelterin (Ruis & Boulton, 2021). In Saccharomyces cerevisiae, telomeres consist of about 300 bp of TG1‐3 repeats, ending with a 3′ single‐strand overhang of 10–15 nucleotides (Wellinger et al, 1993). The double‐strand telomeric DNA is bound by Rap1 that, together with Rif1 and Rif2, plays a central role in telomere capping, while Cdc13 binds specifically to the single‐strand 3′‐overhang (Kupiec, 2014). As in mammals, telomere length homeostasis is dependent on the activity and regulation of the telomerase, which is constitutively expressed in yeast.
In metazoans, telomerase expression is usually turned off during embryogenesis and most of somatic cells do not express telomerase or do it at suboptimal level (Wright et al, 1996). In the absence of telomerase, telomeres shorten with each cell division until they reach a critical length that prevents efficient capping and induces a permanent cell cycle arrest called replicative senescence (Di Micco et al, 2021). Replicative senescence acts as a potent tumor suppressor mechanism as tumor cells need to maintain functional telomeres to proliferate continuously (Kim et al, 1994). Although most cancer cells accumulate mutation leading to de novo expression of the telomerase, some of them use homologous recombination (HR) to reconstitute functional, long telomeres in a process called Alternative Lengthening of telomeres (ALT; Shay et al, 2012).
Inactivation of telomerase in S. cerevisiae leads to severe telomere erosion in about 50 generations. Eroded dysfunctional telomeres initiate an Mec1‐dependent checkpoint cascade that arrests the cell cycle in G2 phase (Lundblad & Szostak, 1989; Enomoto et al, 2002; IJpma & Greider, 2003). This permanent G2/M arrest is defined as crisis at the cell population level. Among the population of senescent cells, few are able to recover functional telomeres through HR and hence to escape cell‐cycle arrest (Lundblad & Blackburn, 1993). Two types of survivors have been described in S. cerevisiae (Le et al, 1999; Chen et al, 2001). Type I survivors show amplification of the Y’ subtelomeric region while maintaining short terminal telomere repeats. They are dependent on the Rad52 and Rad51 recombination factors. Type II survivors, which display very long and heterogeneous terminal telomere repeats, depend on the recombination proteins Rad52 and Rad59 (Chen et al, 2001) as well as the Mre11‐Rad50‐Xrs2 complex (Tsukamoto et al, 2001), the helicase Sgs1 (Lee et al, 2007), and the nucleases Sae2 and Exo1 (Maringele & Lydall, 2004; Hardy et al, 2014). As in ALT tumor cells, maintenance of functional telomeres in both types of survivors depends on the mechanism of break‐induced replication (Lydeard et al, 2007; Dilley et al, 2016; Roumelioti et al, 2016; Min et al, 2017).
Although several components implicated in type II recombination have been identified over time (Claussin & Chang, 2015), the molecular transactions leading to type II recombination remain poorly understood. The foremost question is the nature of the template that is used to generate up to 10 kb of telomere repeats in a few generations (Teng & Zakian, 1999; Teng et al, 2000) at a time when telomere sequences are exhausted at most chromosome ends. The observation that artificial extrachromosomal circles of telomeric DNA (t‐circle) can serve as a template for telomere elongation in K. lactis (Natarajan & McEachern, 2002) raised the possibility that t‐circles might be copied through a rolling circle amplification mechanism to generate the long telomeres characteristics of ALT cells and type II survivors (McEachern & Haber, 2006; Cesare & Reddel, 2010; Kockler et al, 2021b). The presence of partially duplexed telomere circles (called C‐circles) is indeed the most robust hallmark of human ALT cancer cells (Henson et al, 2009). T‐circles have also been described in survivors after telomerase inactivation in S. cerevisiae (Larrivée & Wellinger, 2006).
Spatial localization of the DNA damage is a key determinant in the choice of the pathway used to repair double‐strand breaks (Horigome et al, 2014). We have previously demonstrated that eroded telomeres that arise in the absence of telomerase are localized to the Nuclear Pore Complexes (NPCs) in S. cerevisiae (Khadaroo et al, 2009) and that this localization favors type II survivor formation (Churikov et al, 2016). In this study, we asked whether t‐circles might also localize to the NPCs during senescence where they might serve as a substrate to either initiate telomere elongation or facilitate its spreading to all telomeres. This hypothesis was incited by the observations that noncentromeric DNA circles attach to the nuclear envelope through the NPCs and the SAGA/TREX2 complex in yeast (Shcheprova et al, 2008; Denoth‐Lippuner et al, 2014).
Here, we report detection of C‐circles in telomerase‐negative yeast cells immediately after the nadir of senescence and concurrently with the first type II recombination events. C‐circles interact with the SAGA/TREX2 components Ada2 and Sac3 and co‐immunoprecipitate with the nucleoporin Nic96 in an Ada2 and Sus1‐dependent manner. Inactivation of SAGA/TREX2 through deletion of ADA2, SUS1 or SAC3 in telomerase‐negative cells impaired the production of survivors suggesting that the localization of C‐circles to the NPCs plays a role in the formation of type II telomeres. Finally, we showed that inactivation of the nuclear diffusion barrier that restrains the segregation of noncentromeric episomes to the daughter cell (Shcheprova et al, 2008; Denoth‐Lippuner et al, 2014; Baldi et al, 2017) also impairs the formation of type II survivors. This raises the possibility that C‐circles need to concentrate in one cell for fully efficient type II recombination. Altogether, our data point out that efficient type II recombination might require spatial coordination between eroded telomeres and the template for its repair.
Results
Telomeric circles are produced during senescence at the time of type II survivor formation
To detect telomeric circles in S. cerevisiae during replicative senescence, we adapted in yeast the Rolling Circle Amplification (RCA) assay previously developed in human cells (Henson et al, 2009). The in vitro RCA assay is based on the auto‐primed production of a long single‐strand DNA using the Φ29 polymerase through a rolling circle mechanism starting with either partially double‐stranded or a nicked DNA circle (Fig 1A). Haploid est2∆ clones were isolated by micromanipulation on plates of the spores originated from a heterozygous est2∆/EST2 diploid. The clones were next propagated in liquid cultures for ten days (about 120 population doublings) via serial dilutions every 24 h as previously described (Churikov et al, 2014) (Fig 1B). The growth capacity of the clonal cell populations was assessed by measurement of the cell density every day. As expected, the growth of the est2∆ clones progressively declined, in a course of five days on average, until they went through a telomere erosion‐driven crisis and formed survivors (Fig 1C). In liquid culture, type II survivors have a growth advantage over type I survivors and outcompete them completely after a single dilution (Fig 1C, middle panel). RCA assays were performed on DNA samples prepared at the different time points of the senescence. The product of the RCA assay was loaded to a membrane using a dot blotter and the RCA product was detected by hybridization with a telomeric probe in native conditions to detect only the RCA single‐strand DNA product. No signal was detected before the nadir of senescence; however, a strong signal of Φ29‐dependent amplification was observed at later time points in parallel with the appearance of type II survivors (Fig 1C and Appendix Fig S1A). The RCA signal persisted when survivors were maintained in liquid culture for up to 8 days (about 80 population doublings), suggesting a recurrent production of t‐circles (Appendix Fig S1B). Reintroduction of EST2 in est2∆ type II survivors did not abolish the RCA signal (Appendix Fig S1C). This suggests that t‐circles are byproducts of recombination at telomeres and possibly are generated during their replication as it has been proposed for ALT cancer cells (Zhang et al, 2019).
Figure 1. Production of telomeric circles during senescence and survivor formation in S. cerevisiae .

- Schematic of the RCA assay. The Φ29 polymerase produces long telomeric single‐strand concatemers of the circular DNA in a self‐primed reaction.
- Schematic of the senescence assay. Senescence assays were performed starting from the spores isolated from the heterozygous telomerase‐negative diploids. The spore colonies were inoculated at OD600 = 0.01 in 15 ml liquid YPD medium. Every 24 h, the cell density was measured and a new culture was restarted at OD600 = 0.01. Samples were collected at each time‐point for further analyses.
- Upper panel: Senescence curve of a representative est2∆ clone. DNA samples were prepared at the indicated time points to analyze telomere length (middle panel) and t‐circles production (lower panel and graph) during senescence and cell immortalization. Telomeres were analyzed by Southern blot of the XhoI cut DNA hybridized with a TG1‐3 probe. The presence of t‐circles was monitored by RCA and analyzed by TG1‐3 probed dot blot. The graph shows signal intensities of the dots measured with the Fiji software (arbitrary units). Similar results were obtained with at least 16 other est2∆ clones.
- RCA products were resolved by denaturing agarose gel electrophoresis, transferred on a membrane, and hybridized with a telomeric TG1‐3 probe.
- Dot blot of the RCA product either treated or not with the Exonuclease VII that degrades linear single‐stranded DNA in both the 3′ to 5′ and 5′ to 3′ directions (upper panel). The membrane was hybridized with a telomeric TG1‐3 probe. The signal intensities of the dots are shown (lower graph).
To further confirm that the signal was specific to the product of t‐circle amplification, the RCA reactions were loaded on a denaturing gel (Fig 1D). As expected, the single‐stranded DNA amplification product was detected close to the wells, as previously described (Henson et al, 2009; Vannier et al, 2012). This result confirmed that the dot blot analysis did not detect the presence of linear single‐stranded repeats at chromosome ends but only the product of RCA amplification. Finally, the dot blot signals dropped upon treatment of the RCA product with the exonuclease ExoVII (Fig 1E) further validating that the signals reflect the production of single‐strand DNA by the Φ29 polymerase templated by extrachromosomal circles. Since no exogenous primer was provided in the reaction, the t‐circles that serve as substrate for the RCA reaction could be either partially or completely duplexed with a nick in the G‐rich strand. Together our results show that partially single‐stranded t‐circles can be detected shortly after crisis in the absence of telomerase and this correlates with the emergence of type II survivors.
Telomeric circles are partially duplexed DNA‐DNA specific to type II survivors
We next wondered about the structure of the telomeric circles produced in S. cerevisiae. The RCA products hybridized with a telomeric CA oligonucleotide complementary to the G‐rich telomere sequence, but not with the corresponding GT probe (Fig 2A). No signal was detected with a Y′ subtelomeric probe signifying that the t‐circles did not include subtelomeric sequences (Fig 2A). Accordingly, RCA products were detected even in the absence of dCTP and dATP in the reaction mixture showing that at least some circles contain only telomeric repeats (Fig 2B). Thus, we concluded that the t‐circle backbone is formed by the C‐rich strand of the telomeric sequence (C‐circles). Adding a telomeric primer to the RCA reaction did not enhance the signal (Appendix Fig S2). To further determine whether fully single‐stranded t‐circles are also present, the DNA sample was treated with exonuclease ExoI to degrade linear single‐stranded DNA and with a duplex‐specific nuclease (DSN) that specifically degrades double‐stranded DNA (Appendix Fig S2A). RCA was then performed after addition of either CA or TG telomeric oligonucleotides to prime the reaction. No amplification was detected suggesting that type II survivors do not contain DNA circles that are entirely single‐stranded (Appendix Fig S2B). We were unable to conclude on the presence of telomeric circles carrying a nick on each DNA strand as such circles are not appropriate substrates for RCA.
Figure 2. t‐circles contain the C‐rich telomere strand without subtelomeric Y′ sequence.

- Senescence curve of a representative est2∆ clone and corresponding dot blot of the RCA product hybridized with the indicated subtelomeric or telomeric probes. t‐circle amounts were evaluated based on signal intensity of the dots and were expressed as fold increase over the signal in the absence of Φ29 (lower graph).
- Senescence curve of a representative est2∆ clone (upper panel). DNA samples were prepared at the indicated time points to analyze telomere length by Southern blot (middle panel) and t‐circles production by RCA assay performed in the absence of dCTP and dATP (lower panel). The graph shows signal intensities of the dots measured with the Fiji software (arbitrary units).
To determine whether C‐circles are specific to type II survivors, we monitored their presence in rad59Δ est2Δ and rad51Δ est2Δ cells during the course of senescence and survivor production. C‐circles were not detected in rad59Δ est2Δ cells that form only type I survivors (Fig EV1A) but were present in rad51Δ est2Δ cells concomitant with the appearance of type II survivors (Fig EV1B). Although we cannot exclude a direct role of Rad59, this result is consistent with a tight link between the presence of C‐circles and type II recombination.
Figure EV1. C‐circles are not produced in the absence of Rad59.
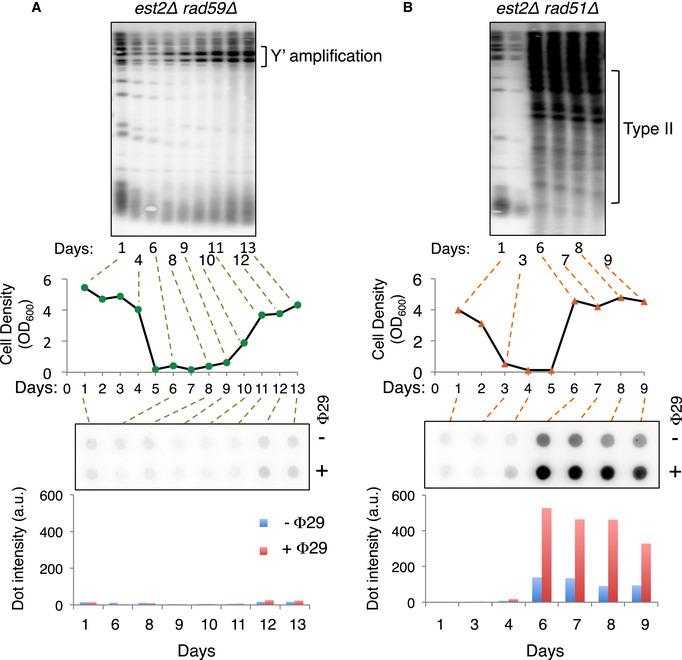
- Analyses of the telomeres and C‐circles in rad59Δ est2Δ cells during senescence. Senescence curve of one representative rad59Δ est2Δ clone is shown (middle panel). At the indicated time points, DNA was extracted and analyzed by telomere Southern blot (upper panel) and RCA assay (lower panel and graph) both hybridized with a TG1‐3 probe. The abundance of C‐circles was evaluated based on the signal intensity of the dots (lower graph). Similar results were obtained with 2 independent est2∆ rad59∆ clones.
- Analyses of the telomeres and C‐circles in rad51Δ est2Δ cells during senescence. Middle panel: Senescence curve of one representative rad51Δ est2Δ clone. At the indicated time points, DNA was extracted and analyzed by Southern blot (upper panel) and RCA assay (lower panel), both hybridized with a TG1‐3 probe. The abundance of C‐circles was evaluated based on the signal intensity of the dots (lower graph).
Because telomere recombination during senescence is enhanced by TERRA transcription and the presence of R‐loops at telomeres (Balk et al, 2013; Yu et al, 2014; Graf et al, 2017), we questioned whether C‐circles are DNA/RNA hybrids. Prolonged incubation of the DNA samples with high concentrations of RNaseA or RNaseH before RCA assay did not prevent RCA product formation (Fig EV2A and B). Furthermore, overexpression of RNH1 in est2∆ cells during the course of senescence did not prevent the production of C‐circles in type II survivors (Fig EV2C).
Figure EV2. C‐circles do not contain RNA–DNA hybrid.

- Senescence curve of one est2Δ clone (upper panel). DNA extracted at the indicated time points was treated with 0.5 mg/ml RNaseA before RCA assay. The dot blot was hybridized with a telomeric TG1‐3 probe (lower panel).
- Senescence curve of one est2Δ clone (upper panel). DNA extracted at the indicated time points was treated with 5 U of RNaseH1 before RCA assay. The dot blot was hybridized with a specific telomeric TG1‐3 probe (middle panel). The abundance of t‐circles evaluated based on the signal intensity of the dots (minus the intensity of the signal in the absence of Φ29) (lower graph).
- Upper panel: Senescence curve of one representative pGal‐RNH1 est2Δ clone grown in the presence of galactose. DNA samples were prepared at the indicated time points to analyze telomere length by TG1‐3 probed Southern blot of the XhoI cut DNA (middle panel) and t‐circle production by RCA followed by dot blot probed with a telomeric TG1‐3 probe (lower panel). The abundance of t‐circles was evaluated based on the signal intensity of the dots (lower graph). Two pGal‐RNH1 est2Δ clones were analyzed with similar results.
Taken together, our data showed that S. cerevisiae type II survivors carry C‐circles similar to those found in human ALT cancer cells (Henson et al, 2009).
C‐circles bind to the nuclear pore complexes
We next addressed the possibility that C‐circles localize or are enriched at the NPCs as previously reported for other types of extrachromosomal DNA circles (Shcheprova et al, 2008). To this purpose, we used the nucleoporin Nic96, which is part of the inner ring of the NPCs, to set up the C‐circles co‐IP experiment. We isolated independent haploid est2∆ Nic96‐RFP cells from a heterozygous diploid as described above and propagated them in liquid culture until the appearance of type II survivors (Fig 3A and Appendix Fig S3A). est2∆ clones isolated from the same diploid were used as a control showing that the NIC96‐RFP allele did not impact the kinetics of senescence and the capacity of est2∆ cells to form type II survivors (Fig 3A and B). Type II survivor cells either expressing or not Nic96‐RFP were fixed with para‐formaldehyde and the NPCs were immunoprecipitated from cell extracts. After crosslink reversal, RCA assays were performed on the purified DNA that co‐immunoprecipitated with Nic96‐RFP. No C‐circles were detected in immunoprecipitates from either telomerase positive control cells expressing Nic96‐RFP or in est2∆ cells with untagged Nic96 (Fig 3C). In contrast, Nic96‐RFP immunoprecipitates from type II survivors displayed RCA signal (Fig 3C and D).
Figure 3. C‐circles bind to the NPCs.

- Senescence curves of four est2∆ and six est2∆ NIC96‐RFP independent clones used to generate type II survivors for C‐circles co‐IP experiments. The time when the cells were collected for the Co‐IP is indicated by an arrow. The results are shown for one of the three independent experiments with similar outcomes.
- Telomere length and recombination were analyzed by TG1‐3 probed Southern blot of XhoI‐digested DNA prepared from samples of the replicative senescence. Two representative clones are shown.
- Detection of the t‐circles co‐immunoprecipitated with Nic96‐RFP from the extracts obtained from several type II survivors. The DNA co‐immunoprecipitated with Nic96‐RFP was amplified by RCA assay, loaded on a dot blot and detected with a telomeric TG1‐3 probe. The numbers above the blot refer to the growth curves in (A) and correspond to type II survivors produced in independent cultures. The lower panel shows the efficiency of Nic96‐RFP IP that was evaluated by western blot with anti‐mCherry antibody.
- Quantification of the RCA product after co‐IP of the C‐circles with Nic96‐RFP showing the effect of ADA2 deletion. Dot signal intensities were quantified using the Fiji software and were calculated as the percentage of INPUT that was immunoprecipitated normalized to est2∆ signals. Relative signals were calculated by setting the mean of est2∆ signals from each independent experiment to 1. Means and s.e.m. of the six independent co‐IPs experiments each with 2–6 clones per genotype are shown. Outliers were removed using ROUT robust outlier test. The p‐values from one‐way ANOVA were adjusted using the Bonferroni’s correction for multiple comparisons. est2∆ versus est2∆ NIC96‐RFP (P = 0.000007), est2∆ NIC96‐RFP versus est2∆ ada2∆ NIC96‐RFP (P = 0.00012). ***P < 0.0001.
- Quantification of the RCA product after co‐IP of the C‐circles with Nic96‐RFP showing the effect of SUS1 deletion. Quantification was done as in D. The graph shows means and s.e.m. of the three independent co‐IPs with 2–5 clones per genotype. Outliers were removed using ROUT robust outlier test. The p‐values from one‐way ANOVA were adjusted using the Bonferroni’s correction for multiple comparisons. est2∆ versus est2∆ NIC96‐RFP (P = 0.002), est2∆ versus est2∆ sus1∆ NIC96‐RFP (P = 0.0006). **P < 0.001, ***P < 0.0001.
The NPC‐associated complex TREX2 and the transcriptional co‐activator SAGA are part of the gene gating mechanism that couples transcription and mRNA export (García‐Oliver et al, 2012). SAGA‐TREX2 is also implicated in the retention of extrachromosomal DNA circles at the NPCs (Denoth‐Lippuner et al, 2014). We previously reported that deletion of ADA2, a component of the SAGA complex, impairs survivor formation with some est2∆ ada2∆ clones either unable to form type II survivors or being greatly delayed in producing them (Churikov et al, 2016). Deletion of ADA2 did not prevent the production of C‐circles in est2∆ cells when they eventually formed type II survivors. The same holds true for the deletion of SAC3, a component of the TREX2 complex and the deletion of SUS1, which is part of both the SAGA and TREX2 complexes (Appendix Fig S3B) suggesting that SAGA/TREX2 is not required for the production of the circles per se. We thus asked whether the localization of C‐circles to the NPCs depends on SAGA/TREX2. We observed a significant drop of C‐circle amplification in Nic96‐RFP immunoprecipitates in type II survivors arising from the est2∆ ada2∆ and est2∆ sus1∆ clones (Figs 3 D and E, and EV3 A and B) suggesting that retention of C‐circles at the NPCs requires a functional SAGA/TREX2 complex.
C‐circles bind to SAGA/TREX2 complexes
We next asked if SAGA/TREX2 components interact with C‐circles. The C‐circles co‐IP assay followed by RCA was conducted in type II survivors expressing a Myc‐tagged Ada2. For that, est2∆ ADA2‐Myc clones isolated from a heterozygous diploid were propagated in liquid cultures until the formation of type II survivors (Fig 4A and B) and RCA assays were performed on DNA immunoprecipitated with Ada2‐Myc. C‐circles were detected exclusively in the immunoprecipitates from type II survivors expressing Ada2‐Myc (Fig 4C and D). To demonstrate that the signal on the dot blots results from amplification of the C‐circles and not of the long terminal repeats present in type II survivors, products of the RCA reaction were separated by denaturing agarose gel electrophoresis. As expected, the long single‐stranded DNA produced by the amplification of C‐circles with the Φ29 polymerase was detected close to the wells while no gross amplification of the chromosomal terminal repeats was detected (Fig EV3C). Remarkably, C‐circles also co‐immunoprecipitated with Sac3 (Fig EV3D). Together, these data suggest that C‐circles produced slightly before or during type II recombination localize to the NPCs through interaction with the SAGA/TREX2 complex as described for other noncentromeric episomes (Denoth‐Lippuner et al, 2014).
Figure 4. C‐circles interact with SAGA/TREX2.

- Senescence curves of two est2∆ and two est2∆ ADA2‐Myc clones used to generate type II survivors for Co‐IP‐RCA. This experiment is representative out of the 4 independent experiments performed during this study.
- Telomere length and recombination were analyzed by TG1‐3‐probed Southern blot of XhoI‐digested DNA prepared from samples of the replicative senescence.
- Detection of the C‐circles co‐immunoprecipitated with Ada2‐Myc in type II survivors. The DNA co‐immunoprecipitated with Ada2‐Myc was amplified by RCA, loaded on a dot blot and detected with a telomeric TG1‐3 probe. The numbers above the blot refer to clones in (A) and correspond to type II survivors produced in independent cultures. The lower panel shows the efficiency of Ada2‐Myc IP analyzed by western blot with anti‐Myc antibody.
- Quantification of the dot blot signals in Ada2‐Myc co‐IP. Dot signals were quantified as in Fig 3D. The graph shows means and s.e.m. from n = 3 independent co‐IP experiments each performed with 3–4 independent clones per genotype. The p‐value is obtained for the Mann–Whitney two‐tailed test (***P = 0.00000005).
Figure EV3. Association of the C‐circles with NPCs is SAGA/TREX2‐dependent.

- Interaction between the C‐circles and Nic‐96‐RFP is decreased in the absence of Ada2 (related to Fig 3D). The DNA co‐immunoprecipitated with Nic96‐RFP was amplified by RCA assay, loaded on a dot blot and detected with a telomeric TG1‐3 probe. The genotypes of the survivors are indicated, and the numbers refer to independent clones. Dot signals were quantified using the Fiji software and are represented as the percentage of INPUT that was immunoprecipitated after normalization to the est2∆ signals set to one.
- Interaction between the C‐circles and Nic96‐RFP is decreased in the absence of Sus1 (related to Fig 3E). The DNA co‐immunoprecipitated with Nic96‐RFP was amplified by RCA assay, loaded on a dot blot and detected with a telomeric TG1‐3 probe. The genotypes of the survivors are indicated, and the numbers refer to independent cultures. Dot signals were quantified as described in A.
- Analysis of the RCA product by denaturing agarose gel electrophoresis (related to Fig. 4). Whole cell extracts were prepared from the indicated strains. Numbers correspond to independent type II survivors. The DNA co‐immunoprecipitated with Ada2‐Myc was amplified by RCA. The RCA product from the Co‐IP and INPUT samples was resolved by denaturing agarose gel electrophoresis and hybridized with a telomeric TG1‐3 probe.
- RCA of the C‐circles co‐immunoprecipitated with Sac3‐Myc. Type II survivors derived from senescence of est2∆ and est2∆ SAC3‐myc clones were isolated. Whole cell extracts from the indicated strains were IP with anti‐Myc antibodies, and the co‐immunoprecipitated DNA was amplified by RCA. Products of the RCA were loaded on a dot blot and detected with a telomeric TG1‐3 probe. Dot signals were quantified as described in A. The lower panel shows the efficiency of Sac3‐Myc IP analyzed by western blot with anti‐Myc antibodies. These results are representative of two independent experiments.
SAGA/TREX2 favors type II recombination
We next addressed whether TREX2 inactivation also impacts type II recombination. SUS1 and SAC3 deletions were combined to est2∆ as described above, and liquid senescence assays were conducted with several independent est2∆ sus1∆ and est2∆ sac3∆ clones (Fig 5 A and B and Appendix Fig S4). The deletion of SUS1 extended the crisis period to the extent similar to that of ADA2 (Churikov et al, 2016), indicating that formation of survivors was compromised in est2∆ sus1∆ cells (Fig 5C). Accordingly, telomere length analysis by Southern blot of individual clones showed that sus1∆ impaired type II survivor formation (Fig 5D and Appendix Fig S5). The delay in forming survivors was much more pronounced in est2∆ sac3∆ cells as compared to est2∆ ada2∆ and est2∆ sus1∆ cells (Fig 5C). However, type II survivors were eventually detected at late time points in most of the clones that were analyzed (Fig 5D and Appendix Fig S6). The prolonged crisis in the absence of Sac3 suggests that its deletion also affects type I survivor formation leaving more time for the cells to perform type II recombination even though its efficiency is decreased. In favor of this possibility, Sac3 is a scaffold protein with three regions that interact with various partners including Sus1 (Gordon et al, 2017).
Figure 5. Inactivation of SAGA/TREX‐2 affect type II telomere recombination.
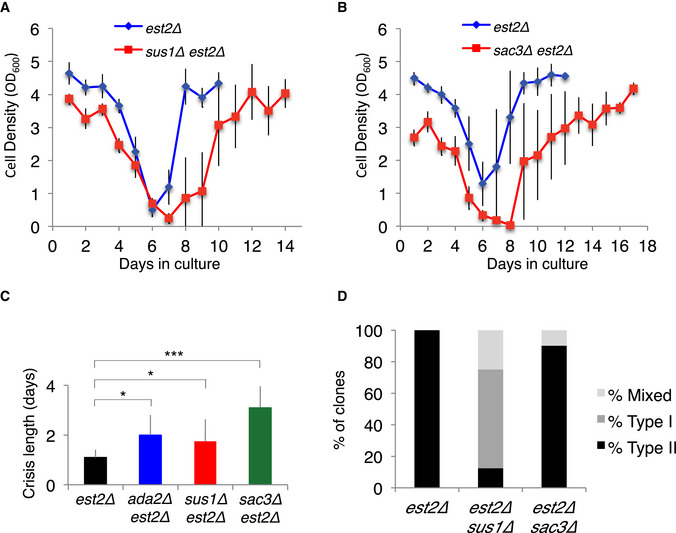
- Mean senescence profiles of the est2Δ (n = 7) and est2Δ sus1Δ (n = 8) clones isolated from the same heterozygous diploid. The error bars are SDs.
- Mean senescence profiles of the est2Δ (n = 10) and est2Δ sac3Δ (n = 10) clones isolated from one heterozygous diploid. The error bars are SDs.
- Duration of the short telomere‐induced crisis of the clones analyzed in (A). The crisis period was determined as the number of days the cell population stayed arrested without notable increase in cell density. The average crisis period is plotted for each genotype. est2∆ ada2∆ data are from (Churikov et al, 2016) and are shown for comparison. The error bars are SD of n independent clones. est2Δ (n = 17), est2Δ ada2Δ (n = 11), est2Δ sus1Δ (n = 8) and est2Δ sac3Δ (n = 10). The p values are from two‐tailed Student’s tests. P = 0.000087 for ada2∆, P = 0.047 for sus1∆, P = 0.049 for ada2∆. *P < 0.05, ***P < 0.0001.
- Frequencies of the telomerase‐independent survivor types formed by the clones shown in A.
As mentioned above, the SAGA complex acts as a transcriptional co‐activator that plays a role genome‐wide (Bonnet et al, 2014) and TREX2 mediates the anchoring of transcribed genes to the NPCs through a direct interaction between Nup1 and Sac3 (Jani et al, 2014) to favor mRNA export (Fischer et al, 2002; Rodríguez‐Navarro et al, 2004). Furthermore, inactivation of TREX2 components interferes with transcription of long GC‐rich genes and induces high levels of recombination consecutive to conflicts between the replication and transcription machineries (Santos‐Pereira et al, 2014). Because of the role of TERRA in telomere recombination (Balk et al, 2013), we addressed the possibility that the defect of type II survivor formation observed in SAGA/TREX2 mutant relies on TERRA metabolism. RT‐qPCR analysis of TERRA RNA production at single telomeres during the course of senescence and survivor formation did not reveal a defect of TERRA transcription in est2∆ ada2∆ as compared to est2∆ control cells (Fig EV4A and B). Thus, it seems unlikely that the failure of type II formation observed in est2∆ ada2∆ cells resulted from an alteration of TERRA‐induced recombination.
Figure EV4. TERRA transcription and telomere localization in est2Δ ada2Δ cells.

- Senescence curves of the est2Δ and est2Δ ada2Δ clones, one of each. Cell populations were maintained in the exponential phase of growth by diluting the cultures to OD600 = 0.01 as soon as they reached OD600 = 0.8. The generation time is shown as a function of the number of population doublings.
- TERRA levels at telomere 15L (upper graph) and 10R (lower graph) during replicative senescence in est2Δ and est2Δ ada2Δ clones shown in (A). TERRA levels were analyzed by qPCR after reverse transcription of the total RNA. TERRA levels were normalized to the ACT1 RNA levels and are represented as fold change relative to the first time point of the est2∆ control that is set to one for each telomere. Two technical replicates are shown for each genotype.
- Localization of eroded telomeres in est2∆ ada2∆ cells during senescence and at crisis. Eroded telomeres were detected as foci containing Cdc13‐YFP and Rad52‐RFP. Percentages of the Cdc13‐YFP/Rad52‐RFP foci that co‐localize with CFP‐Nup49 are shown in the nup133ΔN background in which NPCs cluster at one side of the nucleus (Charifi et al, 2021). Three independent est2∆ ada2∆ clones were analyzed. The error bars are SDs.
Previously, we showed that recombinogenic eroded telomeres localize to the NPCs (Churikov et al, 2014, 2016). Potentially, TREX2‐dependent gene gating of the transcribed telomeres could contribute to their localization to the NPCs. Therefore, we monitored the localization of dysfunctional telomeres detected by colocalization of Cdc13‐YFP and Rad52‐RFP (Fig EV4C). Cdc13‐YFP/Rad52‐RFP foci localized to the NPC marked by CFP‐Nup49 in est2∆ ada2∆ cells (Fig EV4C), excluding the possibility that gene gating through TREX2 participates in the relocalization of eroded telomeres.
Overall, these data suggest that the defect in survivor formation observed upon inactivation of the SAGA/TREX2 complex is linked to its role in tethering C‐circles to the NPCs and that this function may facilitate the formation and/or propagation of type II survivors.
Inactivation of components of the nuclear diffusion barrier impairs type II recombination
Although long controversial, it is now clear that retention of at least a subset of NPCs in the mother cell during anaphase is part of the mechanisms leading to the accumulation of extrachromosomal ribosomal circles (ERCs) with age (Shcheprova et al, 2008; Gehlen et al, 2011; Khmelinskii et al, 2011; Denoth‐Lippuner et al, 2014). Localization of the C‐circles to the NPCs raised a possibility that fully efficient type II survivor formation and/or propagation might depend on the nuclear diffusion barrier (NDB). In this scenario, the NDB would help to reach a threshold level of C‐circles necessary to facilitate their interaction with the eroded telomeres that transiently relocalize to the NPCs. To challenge this hypothesis, we evaluated the capacity of the NDB mutants to produce type II survivors. Two proteins with known function in NDB are Shs1 and Bud6, a septin localized at the mother bud neck and an actin binding protein, respectively (Shcheprova et al, 2008). Deletions of SHS1 and BUD6 were combined to est2∆ and est1∆ respectively and liquid senescence assays conducted with several independently isolated clones as previously described (Fig 6A and B, Appendix Fig S7). The deletions of SHS1 and BUD6 extended the crisis period indicating a defect in survivor formation (Fig 6C). Accordingly, telomere length analysis by Southern blot revealed a defect in type II survivor formation upon inactivation of either Shs1 or Bud6 (Fig 6D, Appendix Figs S8 and S9).
Figure 6. The nuclear diffusion barrier promotes type II recombination.

- Mean senescence profiles of the est2Δ (n = 10) and est2Δ shs1Δ (n = 16) clones. The error bars are SDs.
- Mean senescence profiles of the est1Δ (n = 6) and est1Δ bud6Δ (n = 7) clones. Est1, similarly to Est2, is essential for telomere maintenance. est1∆ has been used in this experiment instead of est2∆ because EST2 and BUD6 genes are linked on chromosome XII. The error bars are SDs.
- Duration of the short telomere‐induced crisis for the clones analyzed in (A) and B). The crisis period was determined as the number of days the cell population stayed arrested without notable increase in cell density. The average crisis period is plotted for each genotype. The error bars are SDs of n independent clones: est2Δ (n = 10), est2Δ shs1Δ (n = 16), est1Δ (n = 6) and est1Δ bud6Δ (n = 7). P values are from two‐tailed Student’s tests (*P = 0.014).
- Frequencies of the survivor types formed by the clones used in the senescence assays shown in A and B.
It has recently been demonstrated that the extended lifespan observed in yeast upon mild temperature stress results from the downregulation of the nuclear diffusion barrier and hence relaxation of the retention of DNA circles in the mother cell (Baldi et al, 2017). Strikingly, performing a parallel senescence assay at 30°C and 37°C on est2∆ cells issued from the same spore showed that elevated temperature not only accelerated senescence but also dramatically interfered with type II recombination (Fig EV5A and B).
Figure EV5. Type II recombination is prevented at 37°C.

-
A, B(A) Left panel: An entire est2∆ spore colony was diluted in YPD and the cell suspension split to inoculate two cultures at OD600 = 0.01. One was grown at 30°C and the other at 37°C. The senescence curves are shown. Right Panel: DNA samples were prepared at the different time points, and telomere length analysis was performed by TG1‐3 probed Southern blot after XhoI digestion. (B) Same as in (A), with a second independently isolated clone.
Discussion
Extrachromosomal circular DNAs from chromosomal origin are common in yeast and human cells (Møller et al, 2015, 2018; Qiu et al, 2021). However, how they are produced and whether they interfere with DNA metabolism or DNA damage repair remain elusive. One of the best studied examples is the extrachromosmal ribosomal DNA circles that arise from recombination between the rDNA repeats and accumulate in the mother cells where they contribute to aging (Sinclair & Guarente, 1997). This asymmetric segregation of ERCs at anaphase relies on both the binding of ERCs to the NPCs and the nuclear diffusion barrier that is thought to limit the diffusion of the NPC‐bound circles into the bud (Shcheprova et al, 2008; Denoth‐Lippuner et al, 2014; Baldi et al, 2017). Extrachromosomal circles are also a hallmark of ALT cancer cells that maintain functional telomeres through recombination instead of telomerase activity (Henson et al, 2009). The most abundant telomeric circles in ALT cells are composed of partially duplexed C‐rich strand (C‐circles) that might be byproducts of the repair of the DNA replication forks stalled in long telomere sequences (Henson et al, 2009; Rivera et al, 2017). Here, we show that C‐circles are also produced in S. cerevisiae at the time of type II survivor formation. The presence of t‐circles has already been revealed in type II survivors using 2D‐gel analysis (Lin et al, 2005; Larrivée & Wellinger, 2006). However, although this approach detected partially double‐stranded circles, these circles contained the G‐rich rather than C‐rich strand (Larrivée & Wellinger, 2006). We did not detect G‐circles using the RCA assay. This suggests that G‐circles might be nicked on both strands preventing their amplification by the Φ29 polymerase. A single‐stranded TG‐rich telomeric probe has not been used in this founding study likely explaining why the C‐circles described here were not detected by 2D‐gel electrophoresis. The mechanism of C‐circle production remains elusive. It has been proposed that they arise from processing of DNA structures generated upon replication stress (Rivera et al, 2017). Accordingly, telomeres are an obstacle to replication fork progression in both yeast and mammals (Simon et al, 2016; Özer et al, 2018; Bonnell et al, 2021).
Here, we show that C‐circles in yeast localize to the NPCs in a pathway dependent on the SAGA/TREX2 components Ada2 and Sus1. We cannot exclude that this pathway is part of a detoxification process as it has been suggested for ERCs (Sinclair & Guarente, 1997; Shcheprova et al, 2008). Alternatively and not exclusively, we propose that retention of C‐circles at the NPCs might be part of the process that ultimately results in the production of type II survivors. We have previously shown that in the absence of telomerase, eroded telomeres bound by SUMOylated proteins are recognized by the SUMO‐targeted ubiquitin ligase Slx5‐Slx8 and relocate to the NPCs (Khadaroo et al, 2009). Relocalization to the NPCs favors type II survivor recombination, which involves BIR although the identity of the template used to produce elongated telomeres remains elusive (McEachern & Haber, 2006; Churikov et al, 2016). We propose that C‐circles may provide a template for telomere elongation through single strand annealing and rolling circle amplification (Fig 7) as it has been suggested in the yeast K. lactis (McEachern & Haber, 2006). In this hypothesis, the proximity of eroded telomeres and C‐circles at the NPCs would spatially favor their interaction and hence telomere elongation. In line with this possibility, our data show that inactivation of SAGA/TREX2 components impacts type II recombination.
Figure 7. Model of type II telomere recombination that relies on telomeric circles.

C‐circles localize at the NPCs in a SAGA/TREX2‐dependent process. We previously showed that eroded telomeres also localize to the NPCs in a SUMO and Slx5/Slx8‐dependent manner, which favors type II recombination (Churikov et al, 2016). Co‐localization of the C‐circles and eroded telomeres at NPCs may favor SSA between the telomeric 3’‐overhang and the telomeric circles, thereby promoting telomere elongation. In this scheme C‐circles serve as a template for elongation of the G‐strand of telomeres which is followed by synthesis of the complementary strand by Polα (not shown).
Unfortunately, we were unable to detect C‐circles before the detection of the first type II recombination events. This prevented us to directly test our model and to determine the genetic requirements for the formation of C‐circles. This may be due to technical constraints related to the fact that the C‐circles would be below the threshold of RCA detection in the population of senescing cells of which only a few (on average one in 105 cells) succeed in restoring their telomeres via recombination and become type II survivors (McEachern & Haber, 2006; Kockler et al, 2021a). Alternatively, C‐circles might be produced during the first recombination events that generate long telomeres and then used to facilitate spreading of telomere repeats to other eroded telomeres. In line with this possibility, it has been recently proposed that the production of type II survivors might involve mixed BIR processes, each with specific genetic requirements (Kockler et al, 2021a, 2021b). It is conceivable that this complexity actually reflects different stages of survivor maturation, from the elongation of one or a few telomeres to the regeneration of a complete set of functional telomeres allowing resumption of growth, and finally to the maintenance of telomeres recurrently dependent on HR.
Our data also implicate components of the nuclear diffusion barrier in facilitating type II survivor formation. Whether NDB plays its role by concentrating C‐circles in the mother cells to facilitate telomere elongation or to facilitate the spreading of telomere repeats among the chromosome ends will need further investigation. It is noteworthy that although maintenance of telomeres through type II recombination affects neither the growth rate nor the genome integrity of the cells, it significantly reduces their replicative life span (Chen et al, 2009).
Overall, our study suggests that formation of the type II survivors depends on multiple spatially organized events with a central role of the NPCs in recruiting both eroded telomeres and C‐circles. This multifactorial constrains might explain the very low efficiency of type II survivor formation in S. cerevisiae and, by extension, the low frequency of ALT in cancer cells.
Materials and Methods
Yeast strains and primers
Strains and primers used in this study are listed in the Appendix Tables S1 and S2, respectively. All strains were constructed using standard genetic methods.
Senescence assays
Senescence assays were performed in liquid culture starting from haploid spores obtained by dissection of the tetrads produced by sporulation of diploid cells heterozygous for EST2 (EST2/est2Δ) and for the gene(s) of interest. Diploids obtained by crossing two haploid mutants were grown in YPD plates for about 50 PDs before sporulation to ensure homogeneous telomere length. After dissection, spores were grown for 3 days on plate at 30°C (about 25–30 PDs). The entire colonies were inoculated and then propagated in 15 ml of YPD at 30°C via serial dilutions to OD600 = 0.01 every 24 h until the appearance of survivors. The duration of the crisis was estimated as the number of days the culture remains at the lowest OD (with no increase in cell density). As soon as the culture gained some growth capacity, even intermediate, we considered that survivors started to appear to avoid any putative side effect of the mutation on the growth rate. Determination of the type of survivors has been done at the second day after stabilization of the growth rate.
Telomere Southern blot analysis
Genomic DNA (25 μg estimated with a nanodrop) was digested overnight with XhoI at 37°C. Digested DNA was resolved in 0.9% agarose gel and transferred onto the nylon membrane (Hybond‐XL) in alkaline conditions. The blotted DNA was hybridized with a probe prepared by random priming of the DNA fragment composed of TG1‐3 repeats in the presence of [α‐32P]dCTP.
Chromatin immunoprecipitation
Yeast cells were grown in YPD at 30°C to OD600 = 1 and fixed at RT for 10 min with 1% formaldehyde (Sigma; F8775). Fixation was quenched with 0.25 M of glycine for 5 min. After three washes with cold 1× TBS, the cell pellets were frozen and stored at −80°C. Thawed were resuspended in the lysis buffer (50 mM of HEPES–KOH pH 7.5, 150 mM of NaCl, 1 mM of EDTA, 1% Triton X‐100, 0.1% of Na‐deoxycholate) containing 1 mM of PMSF and a protease inhibitor cocktail (cOmplete tablet, EDTA‐free, Roche) and disrupted in a bead‐beater for 3 × 30 s at 5,000 rpm (Precellys 24, Bertin). Lysates were cleared by centrifugation (2,000 g, 4°C) and supernatants were collected. Aliquots were saved to check the protein level and for RCA experiments (Inputs). The cleared lysates were then incubated with either a mouse monoclonal anti‐Myc 9E10 antibody (Santa Cruz Biotechnology, sc‐40) or a rabbit polyclonal anti‐pHis‐mCherry antibody (Home‐made) for 2 h 30 min at 4°C. Magnetic Dynabeads Protein G (Invitrogen Dynal AS, Oslo, Norway) were then added for another 2 h 30 min incubation at 4°C. The beads were washed consecutively with lysis buffer, lysis Buffer + 500 mM of NaCl, Wash Buffer (10 mM of Tris–HCl pH 8, 250 mM of LiCl, 0.5% NP40, 0.5% Na‐deoxycholate, 1 mM of EDTA) and with TE 1X. The protein–DNA complexes were eluted from the beads in 1× TE 1% SDS solution. An aliquot of the elution was saved to check the level of the immunoprecipitated proteins. The cross‐links were reversed by incubation at 65°C overnight. The samples were treated with proteinase K (Roche), and DNA was purified through phenol‐chloroform extraction and resuspended in 25 µl of H2O. The samples were analyzed by RCA assay.
RCA assay
2.5 μg of genomic DNA (estimated with a nanodrop) was digested with PvuII at 37°C prior Phi29 amplification reaction. For inputs and co‐immunoprecipitated samples, 5 µl were used for RCA per condition. For all experiments, samples were incubated with 7.5 U of Φ29 polymerase (NEB) in the 1× Φ29 buffer supplemented with 1 mM of dNTPs, 0.2 mg/ml of BSA, 0.1% Tween overnight at 30°C. The Φ29 polymerase was inactivated 10 min at 65°C. Samples were diluted with 200 μl 2X SSC and loaded onto 6X SSC‐soaked Hybond‐XL membrane (GE Healthcare) using a dot blotter. When indicated, the RCA products were loaded on a denaturing 0.8% agarose gel (adapted from Vannier et al, 2012) and transferred to a Hybond‐XL membrane (GE Healthcare). DNA was UV‐crosslinked onto the membrane and hybridized either with the end‐labeled single‐stranded probes at 37°C (CA probe: [5′‐32P]CACCACACCCACACACA/GT probe: [5′‐32P] TGTGTGTGGGTGTGGTG ) or at 55°C with radiolabelled double‐stranded probes. When indicated, treatment with ExoVII (NEB) was performed for 4 h at 37°C to degrade linear ssDNA. Treatment with ExoI (NEB) was performed for 1 h at 37°C followed by DSN (NEB) treatment for 2 h at 37°C. The samples were then denatured and a 3′ 2‐thio‐U primer was added before treatment with Phi29 polymerase.
Membranes were exposed to a phosphorimager screen and the signal was detected via a Typhoon scanner (GE Healthcare). For co‐IP/RCA, quantification was done with the Fiji software by measuring the integrated intensity of the signals. Background signal obtained without Φ29 treatment was subtracted from the Φ29‐treated samples. The percentage of the INPUT that was immunoprecipitated was normalized to the est2∆ samples. Relative signals were calculated by setting the mean of the est2∆ signals from each independent experiment to 1. The mean values of the signal for independent clones used in each experiment were represented as a bar graph with the error bars indicating (s.e.m.).
TERRA transcription
Cells were grown in 25 ml of YPD to mid‐exponential phase (OD: 0.6–0.8). Total RNA was extracted with hot acidic phenol as described (Graf et al, 2017). Three DNase I treatments were done with the RNase‐free DNase set from Qiagen to completely remove telomeric DNA from RNA. Reverse transcription was done using 200 U of SuperScript III per reaction and TERRA was reverse‐transcribed using the CA oligonucleotide as a primer. All primer sequences are listed in Appendix Table S2. TERRA levels were normalized to the ACT1 RNA levels and converted to relative TERRA expression by normalization to the first time point of the control est2Δ strain.
Author contributions
Paula Aguilera: Conceptualization; Formal analysis; Validation; Investigation; Methodology; Writing — original draft. Marion Dubarry: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Writing — original draft. Julien Hardy: Investigation; Methodology. Michael Lisby: Investigation. Marie‐Noelle Simon: Conceptualization; Formal analysis; Supervision; Funding acquisition; Validation; Investigation; Methodology; Writing — original draft; Project administration. Vincent Geli: Conceptualization; Formal analysis; Supervision; Funding acquisition; Project administration; Writing — review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
PA, MD, JH, ML, and MNS performed experiments. M‐NS and VG conceived this study. PA, MD, VG, and M‐NS designed the experiments and discussed the results. MD analyzed the results and contributed to the writing of the manuscript. PA, VG, and M‐NS wrote the manuscript.
Supporting information
Appendix
Expanded View Figures PDF
Acknowledgments
The authors thank Brian Luke for the vector overexpressing RNH1 and Dmitri Churikov for comments on the manuscript. P.A. was supported by the Région Provence‐Alpes‐Côte d’Azur/Inserm and the Fondation ARC pour la Recherche sur le Cancer. M.D was supported by the Agence Nationale de Recherche (ANR‐19‐CE12‐0023 NIRO). V.G. is supported by the Ligue Nationale Contre le Cancer (Equipe labellisée).
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
The EMBO Journal (2022) 41: e108736.
Contributor Information
Marie‐Noëlle Simon, Email: marie-noelle.simon@inserm.fr.
Vincent Géli, Email: vincent.geli@inserm.fr.
Data availability
This study includes no data deposited in external repositories.
References
- Baldi S, Bolognesi A, Meinema AC, Barral Y (2017) Heat stress promotes longevity in budding yeast by relaxing the confinement of age‐promoting factors in the mother cell. eLife 6: e28329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke‐Glaser S, Bender K, Luke B (2013) Telomeric RNA‐DNA hybrids affect telomere‐length dynamics and senescence. Nat Struct Mol Biol 20: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Bonnell E, Pasquier E, Wellinger RJ (2021) Telomere replication: solving multiple end replication problems. Front Cell Dev Biol 9: 668171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet J, Wang C‐Y, Baptista T, Vincent SD, Hsiao W‐C, Stierle M, Kao C‐F, Tora L, Devys D (2014) The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev 28: 1999–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11: 319–330 [DOI] [PubMed] [Google Scholar]
- Charifi F, Churikov D, Eckert‐Boulet N, Minguet C, Jourquin F, Hardy J, Lisby M, Simon M‐N, Géli V (2021) Rad52 SUMOylation functions as a molecular switch that determines a balance between the Rad51‐ and Rad59‐dependent survivors. iScience 24: 102231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Ijpma A, Greider CW (2001) Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol Cell Biol 21: 1819–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X‐F, Meng F‐L, Zhou J‐Q (2009) Telomere recombination accelerates cellular aging in Saccharomyces cerevisiae . PLoS Genet 5: e1000535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Charifi F, Simon M‐N, Géli V (2014) Rad59‐facilitated acquisition of Y’ elements by short telomeres delays the onset of senescence. PLoS Genet 10: e1004736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Charifi F, Eckert‐Boulet N, Silva S, Simon M‐N, Lisby M, Géli V (2016) SUMO‐dependent relocalization of eroded telomeres to nuclear pore complexes controls telomere recombination. Cell Rep 15: 1242–1253 [DOI] [PubMed] [Google Scholar]
- Claussin C, Chang M (2015) The many facets of homologous recombination at telomeres. Microb Cell 2: 308–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoth‐Lippuner A, Krzyzanowski MK, Stober C, Barral Y (2014) Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. eLife 3: e03790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Krizhanovsky V, Baker D, d’Adda di Fagagna F (2021) Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat Rev Mol Cell Biol 22: 75–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA (2016) Break‐induced telomere synthesis underlies alternative telomere maintenance. Nature 539: 54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S, Glowczewski L, Berman J (2002) MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae . Mol Biol Cell 13: 2626–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Strässer K, Rácz A, Rodriguez‐Navarro S, Oppizzi M, Ihrig P, Lechner J, Hurt E (2002) The mRNA export machinery requires the novel Sac3p‐Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J 21: 5843–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Oliver E, García‐Molinero V, Rodríguez‐Navarro S (2012) mRNA export and gene expression: the SAGA‐TREX‐2 connection. Biochim Biophys Acta 1819: 555–565 [DOI] [PubMed] [Google Scholar]
- Gehlen LR, Nagai S, Shimada K, Meister P, Taddei A, Gasser SM (2011) Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr Biol 21: 25–33 [DOI] [PubMed] [Google Scholar]
- Gordon JMB, Aibara S, Stewart M (2017) Structure of the Sac3 RNA‐binding M‐region in the Saccharomyces cerevisiae TREX‐2 complex. Nucleic Acids Res 45: 5577–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf M, Bonetti D, Lockhart A, Serhal K, Kellner V, Maicher A, Jolivet P, Teixeira MT, Luke B (2017) Telomere length determines TERRA and R‐loop regulation through the cell cycle. Cell 170: 72–85 [DOI] [PubMed] [Google Scholar]
- Hardy J, Churikov D, Géli V, Simon M‐N (2014) Sgs1 and Sae2 promote telomere replication by limiting accumulation of ssDNA. Nat Commun 5: 5004 [DOI] [PubMed] [Google Scholar]
- Henson JD, Cao Y, Huschtscha LI, Chang AC, Au AYM, Pickett HA, Reddel RR (2009) DNA C‐circles are specific and quantifiable markers of alternative‐lengthening‐of‐telomeres activity. Nat Biotechnol 27: 1181–1185 [DOI] [PubMed] [Google Scholar]
- Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Dion V, Harata M, Gasser SM (2014) SWR1 and INO80 chromatin remodelers contribute to DNA double‐strand break perinuclear anchorage site choice. Mol Cell 55: 626–639 [DOI] [PubMed] [Google Scholar]
- IJpma AS, Greider CW (2003) Short telomeres induce a DNA damage response in Saccharomyces cerevisiae . Mol Biol Cell 14: 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani D, Valkov E, Stewart M (2014) Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res 42: 6686–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadaroo B, Teixeira MT, Luciano P, Eckert‐Boulet N, Germann SM, Simon MN, Gallina I, Abdallah P, Gilson E, Géli V et al (2009) The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol 11: 980–987 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A, Meurer M, Knop M, Schiebel E (2011) Artificial tethering to nuclear pores promotes partitioning of extrachromosomal DNA during yeast asymmetric cell division. Curr Biol 21: R17–R18 [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Kockler ZW, Comeron JM, Malkova A (2021a) A unified alternative telomere‐lengthening pathway in yeast survivor cells. Mol Cell 81: 1816–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockler ZW, Osia B, Lee R, Musmaker K, Malkova A (2021b) Repair of DNA breaks by break‐induced replication. Annu Rev Biochem 90: 165–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M (2014) Biology of telomeres: lessons from budding yeast. FEMS Microbiol Rev 38: 144–171 [DOI] [PubMed] [Google Scholar]
- Larrivée M, Wellinger RJ (2006) Telomerase‐ and capping‐independent yeast survivors with alternate telomere states. Nat Cell Biol 8: 741–747 [DOI] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE, Greider CW (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kozak M, Martin JD, Pennock E, Johnson FB (2007) Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol 5: e160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C‐Y, Chang H‐H, Wu K‐J, Tseng S‐F, Lin C‐C, Lin C‐P, Teng S‐C (2005) Extrachromosomal telomeric circles contribute to Rad52‐, Rad50‐, and polymerase delta‐mediated telomere‐telomere recombination in Saccharomyces cerevisiae . Eukaryot Cell 4: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1‐ senescence. Cell 73: 347–360 [DOI] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break‐induced replication and telomerase‐independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2004) EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics 166: 1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE (2006) Break‐induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75: 111–135 [DOI] [PubMed] [Google Scholar]
- Min J, Wright WE, Shay JW (2017) Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break‐induced replication processes. Mol Cell Biol 37: e00226‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller HD, Mohiyuddin M, Prada‐Luengo I, Sailani MR, Halling JF, Plomgaard P, Maretty L, Hansen AJ, Snyder MP, Pilegaard H et al (2018) Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun 9: 1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller HD, Parsons L, Jørgensen TS, Botstein D, Regenberg B (2015) Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci USA 112: E3114–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan S, McEachern MJ (2002) Recombinational telomere elongation promoted by DNA circles. Mol Cell Biol 22: 4512–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özer Ö, Bhowmick R, Liu Y, Hickson ID (2018) Human cancer cells utilize mitotic DNA synthesis to resist replication stress at telomeres regardless of their telomere maintenance mechanism. Oncotarget 9: 15836–15846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu G‐H, Zheng X, Fu M, Huang C, Yang X (2021) The decreased exclusion of nuclear eccDNA: From molecular and subcellular levels to human aging and age‐related diseases. Ageing Res Rev 67: 101306 [DOI] [PubMed] [Google Scholar]
- Rivera T, Haggblom C, Cosconati S, Karlseder J (2017) A balance between elongation and trimming regulates telomere stability in stem cells. Nat Struct Mol Biol 24: 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez‐Navarro S, Fischer T, Luo M‐J, Antúnez O, Brettschneider S, Lechner J, Pérez‐Ortín JE, Reed R, Hurt E (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore‐associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Roumelioti F‐M, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD, Gagos S (2016) Alternative lengthening of human telomeres is a conservative DNA replication process with features of break‐induced replication. EMBO Rep 17: 1731–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruis P, Boulton SJ (2021) The end protection problem‐an unexpected twist in the tail. Genes Dev 35: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos‐Pereira JM, García‐Rubio ML, González‐Aguilera C, Luna R, Aguilera A (2014) A genome‐wide function of THSC/TREX‐2 at active genes prevents transcription‐replication collisions. Nucleic Acids Res 42: 12000–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Reddel RR, Wright WE (2012) Cancer. Cancer and telomeres–an ALTernative to telomerase. Science 336: 1388–1390 [DOI] [PubMed] [Google Scholar]
- Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y (2008) A mechanism for asymmetric segregation of age during yeast budding. Nature 454: 728–734 [DOI] [PubMed] [Google Scholar]
- Simon M‐N, Churikov D, Géli V (2016) Replication stress as a source of telomere recombination during replicative senescence in Saccharomyces cerevisiae . FEMS Yeast Res 16: fow085 [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L (1997) Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91: 1033–1042 [DOI] [PubMed] [Google Scholar]
- Teng SC, Chang J, McCowan B, Zakian VA (2000) Telomerase‐independent lengthening of yeast telomeres occurs by an abrupt Rad50p‐dependent, Rif‐inhibited recombinational process. Mol Cell 6: 947–952 [DOI] [PubMed] [Google Scholar]
- Teng SC, Zakian VA (1999) Telomere‐telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae . Mol Cell Biol 19: 8083–8093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA (2001) The role of the Mre11‐Rad50‐Xrs2 complex in telomerase‐ mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol 11: 1328–1335 [DOI] [PubMed] [Google Scholar]
- Vannier J‐B, Pavicic‐Kaltenbrunner V, Petalcorin MIR, Ding H, Boulton SJ (2012) RTEL1 dismantles T loops and counteracts telomeric G4‐DNA to maintain telomere integrity. Cell 149: 795–806 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA (1993) Saccharomyces telomeres acquire single‐strand TG1‐3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW (1996) Telomerase activity in human germline and embryonic tissues and cells. Dev Genet 18: 173–179 [DOI] [PubMed] [Google Scholar]
- Yu T‐Y, Kao Y, Lin J‐J (2014) Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc Natl Acad Sci USA 111: 3377–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhang Z, Shengzhao G, Li X, Liu H, Zhao Y (2019) Strand break‐induced replication fork collapse leads to C‐circles, C‐overhangs and telomeric recombination. Plos Genet 15: e1007925 [DOI] [PMC free article] [PubMed] [Google Scholar]


