SUMMARY
The endoplasmic reticulum (ER) is a ubiquitous organelle that is vital to the life of eukaryotic cells. It synthesizes essential lipids and proteins and initiates the glycosylation of intracellular and surface proteins. As such, the ER is necessary for cell growth and communication with the external environment. The ER is also a highly dynamic organelle, whose structure is continuously remodeled through an interaction with the cytoskeleton and the action of specialized ER shapers. Recent and significant advances in ER studies have brought to light conserved and unique features underlying the structure and function of this organelle in plant cells. In this review, exciting developments in the understanding of the mechanisms for plant ER structural and functional homeostasis, particularly those that underpin ER network architecture and ER degradation, are presented and discussed.
INTRODUCTION
“Change is the only constant in life” is a quote attributed to the Greek philosopher Heraclitus. It comes to mind when observing the endoplasmic reticulum (ER) in live plant cells and reflecting on the ability of this organelle to respond and adapt to the changing biosynthetic demands of the cell during growth and in response to stress. The ER is a pleomorphic network of interconnected membranes that are continuously remodeled while the organelle is engaged in the synthesis of at least one-third of the cellular proteome and of essential lipids and primes secretory proteins with glycan moieties. The ER also synthesizes and houses hormone receptors and is critical for cellular calcium homeostasis (Kriechbaumer and Brandizzi, 2020; Wu et al., 2002). The ER changes its structure as cells expand (Stefano et al., 2014) and tailors its ability to synthesize proteins to stress cues that alter the cell’s demands for biosynthetic cargo, including defense proteins (Pastor-Cantizano et al., 2020). Since the first visualization of the ER network in living cells using fluorescent dyes (Quader and Schnepf, 1986), the advent of fluorescent protein technology and advanced quantitative imaging in live cells has led to significant strides in the understanding of the conserved and plant-unique mechanisms underlying ER structure and dynamics. The implementation of genomics has also allowed gaining new insights on the mechanisms underlying the biosynthetic homeostasis of the ER and the maintenance of the ER network in conditions of stress. In this review, we will review recent and significant advances in the understanding of the mechanisms that control the dynamic shape of the plant ER network and connect the ER membrane with the cytoskeleton and with other cellular membranes, as well as the processes that guide ER degradation through ER-phagy.
HOMEOSTASIS OF ER NETWORK STRUCTURE
ER network structure
The plant ER network pervades the cell cortex (Figure 1), where it is sandwiched between the plasma membrane (PM) and the membrane of the large vacuole (tonoplast) and is continuous with the nuclear envelope. The plant ER traverses the cell as a bundle of membranes through invaginations of the tonoplast forming the transvacuolar strands, which are normally one or two per cell. At the cell cortex, the architecture of the ER network comprises interconvertible tubules and cisternae (Figure 2). Homotypic fusion between tubules and interconversion of cisternae into tubules are generally rapid and the most commonly seen ER reshaping events (Kriechbaumer and Brandizzi, 2020). Similar to other eukaryotes, tubules and the edges of the plant ER cisternae have positive degree of curvature. Tubules connect with each other and to the cisternae, forming the so-called “three-way junctions,” which are small, triangular sheets with negatively curved edge lines (Shemesh et al., 2014) (Figure 2). ER tubule fission is generally not observed in plant cells.
Figure 1. The ER network forms a reticulated structure at the cell cortex.
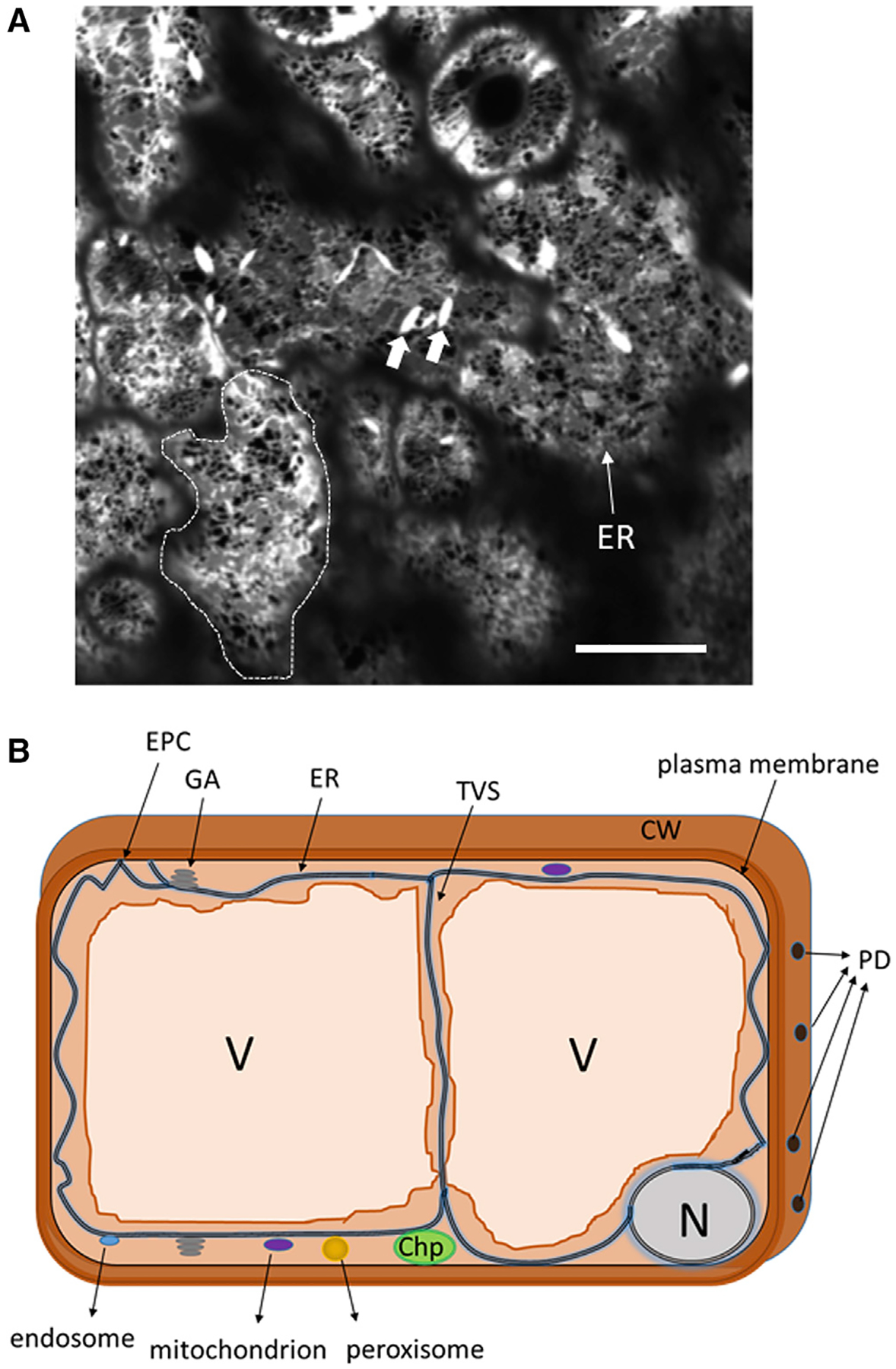
(A) Confocal microscopy image of the surface of Arabidopsis leaf epidermal cells stably expressing a fluorescent lumenal marker, ER-YK (Nelson et al., 2007), which labels the entirety of the plant ER network. The image shows the ER at the cell cortex (ER, narrow arrow). A dotted line contours the perimeter of the cortex of an epidermal cell. Arrows point to ER stress bodies, which are present in Brassicaceae species. Scale bar, 40 μm.
(B) Diagram of a cross-section of an epidermal cell to illustrate the distribution of the ER at the cell perimeter between the central vacuole (V) and the plasma membrane, and at the trans-vacuolar strands (TVS). The diagram also shows PD, organelles contacted by the ER through mechanisms discussed in the main text, and the nucleus. EPC, ER-plasma membrane contact; GA, Golgi apparatus; chp, chloroplast.
Figure 2. Plant ER structures visible through live-cell imaging and diagram of ER shapers.
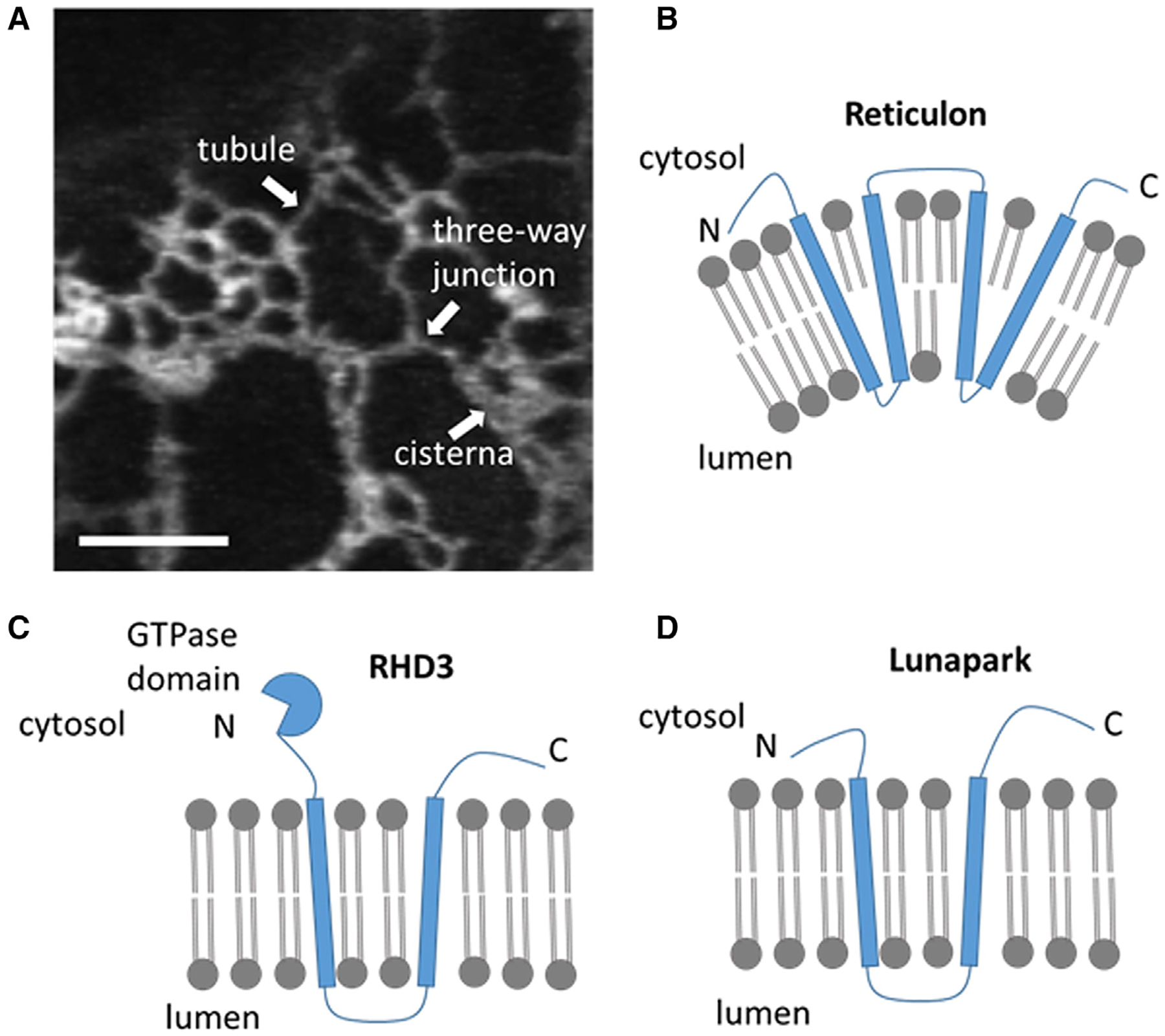
(A) Confocal microscopy image of a region of the cortical ER of a tobacco leaf epidermal cell transiently expressing the ER lumenal marker ER-YK (Nelson et al., 2007) highlighting conspicuous ER network structures: cisterna, three-way junction, and tubule. Scale bar, 5 μm.
(B–D) Diagrams depicting the ER shapers reticulon (B), RHD3 (C) and Lunapark (D). The diagrams depict the relationship of the proteins with the ER membrane, in which they are embedded, the ER lumen and the cytosol. The diagrams are not to scale.
The nature of the plant ER cisternae is yet to be established. They may be continuous sheets of membranes and tightly packed tubules, as proposed in mammalian cells (Nixon-Abell et al., 2016), or sheets perforated by sub-diffraction holes (termed “nanoholes”) (Schroeder et al., 2019). Nonetheless, the perforations visible in confocal microscopy imaging of the plant ER cisternae, which generally enwrap other organelles, support the existence of mechanisms for localized membrane curvature, possibly through the selective distribution of the ER shapers, such as the reticulons (Rtns, see below), as shown for the nanoholes in mammalian cells (Schroeder et al., 2019).
The relative abundance of the plant ER cisternae and tubules varies during cell development. In expanding plant cells, the cisternal ER is the predominant form, while in fully expanded cells, the tubular form is the most evident (Ridge et al., 1999; Stefano et al., 2014). The machinery responsible for the maintenance of the ER cisterna space appears not to be conserved across kingdoms. In mammalian cells, CLIMP-63 serves as a spacer in the cisternae, acting through stabilization of the flat areas by oligomerization of the protein’s inter-luminal domain across the lumen of the cisternal ER (Klopfenstein et al., 2001; Shibata et al., 2010). Notably, the Arabidopsis genome does not express sequence homologs of CLIMP-63, raising the possibility that the shape of cisternae may be attained by functional homologs of CLIMP-63 with markedly divergent sequence, and/or through other structural mechanisms.
The most well-characterized and ubiquitous ER-membrane-associated proteins responsible for membrane fusion, curvature, and tubule stabilization, include the largely conserved dynamin-like proteins: atlastin (ATL) in metazoan cells, ATL-like root hair defective 3 (RHD3) in plant cells, Sey1p in yeast, the ubiquitous Rtns, and Lunapark proteins (Lnps) (Figure 2). The dynamin-like ER shapers are associated with the ER membrane via two membrane anchor domains and have a cytosolic GTPase domain (Figure 2). Protein crystallography analyses of ATLs have led to the model that, by tethering opposite ER membranes via dimerization of the GTPase domain, ATLs facilitate homotypic fusion, most likely through a GTP-hydrolysis-dependent conformational change (reviewed in Hu and Rapoport [2016]). Rtns induce and stabilize high curvature of the membrane of the tubules and cisterna edges through oligomerization and have two V-shaped transmembrane wedges joined by a cytosolic loop, with both the N and C termini facing the cytosol (Voeltz et al., 2006; Wang and Rapoport, 2019) (Figure 2). Lnp is a conserved membrane protein containing two closely spaced transmembrane segments and cytoplasmic domains (Figure 2) and is localized at the three-way junctions where it influences the stability of these ER structures (Chen et al., 2015, 2012, 2013).
The Arabidopsis genome encodes three functionally redundant isoforms of RHD3 (RHD3, RHD3-like 1, and RHD3-like 2), of which RHD3 is the most abundant (Chen et al., 2011; Stefano et al., 2012; Ueda et al., 2016; Zhang et al., 2013). The evidence that loss of RHD3-like 1 and RHD3-like 2 does not lead to visible ER structure defects (Zhang et al., 2013) indicates that RHD3 has a prevalent role in ER structure compared with the other isoforms. In support of fusogenic activity, the number of tubule fusion events per area of cortical ER is reduced in an Arabidopsis RHD3 loss-of-function mutant compared with wild type, and in vitro experiments have shown a requirement of RHD3 for ER reconstitution from microsomal fractions (Ueda et al., 2016). Although a crystal structure of RHD3 is still not available, by analogy with ATLs, RHD3 is likely to mediate membrane fusion through mechanisms similar to ATLs. This model is partially supported by the evidence that RHD3 dimerizes via the GTPase domain and that dimerization is necessary for ER fusion (Sun and Zheng, 2018). The Arabidopsis genome also encodes 21 Rtn homologs (Sparkes et al., 2009b) and two Lunapark homologs (Lnp1 and Lnp2) (Kriechbaumer et al., 2018; Ueda et al., 2018).
Despite the identification of plant ER-shaping proteins and a preliminary understanding of protein-protein interactions, there is still a significant gap in the appreciation of how these proteins function together to control plant ER network homeostasis at a mechanistic level. For example, how the interconversion of tubular and cisternal ER is controlled at a protein level is yet to be defined. We also do not know whether the mechanisms controlling the conversion of the cisternal ER into a tubular ER during cell expansion are equivalent to those underlying the dynamic remodeling of the ER in mature cells. A controlled redistribution of the ER shapers in the ER membrane might favor one ER form over the other, but how this may occur is not yet established. The presence of RHD3 blocked in the GDP or GTP-bound form in the ER leads to collapse of network structure homeostasis similar to an RHD3 loss-of-function mutant (Chen et al., 2011). Also, an over-abundance of Rtns leads to constrictions of the ER lumen and a reduction in the diffusion of ER lumenal proteins (Tolley et al., 2010). Therefore, a tight control of the activity of the ER shapers and their distribution in the ER membrane is necessary to maintain ER homeostasis.
The mechanisms for a controlled distribution of ER shapers in the ER membrane may include post-translational modifications. For example, RHD3 interacts with Rtns (Kriechbaumer et al., 2015; Lee et al., 2013). The verified phosphorylation of the C-terminal region of RHD3 (Ueda et al., 2016) may reversibly modify such an interaction, resulting in an enhanced fusion or lack of homotypic fusion. Nonetheless, successful complementation of a loss-of-function mutant of RHD3 with RHD3 protein mutants mimicking constitutively phosphorylated or dephosphorylated forms (Ueda et al., 2016) argues against this hypothesis. It cannot be excluded that other protein modifications may modulate the relative distribution of ER shapers in the ER membrane to control shape.
Similarly, protein-protein interactions may underlie a localized distribution or function of ER shapers. For example, the loss of the Arabidopsis Lnp1 and Lnp2 together leads to the appearance of an increased abundance of ER sheets with dense fenestration and ER conglomerates (Kriechbaumer et al., 2018; Sun et al., 2020a; Ueda et al., 2018), suggesting that Lnps are important for ER architecture also in plants. It has been recently established that RHD3 interacts with Lnp1 and Lnp2 in vivo and in vitro and that the interaction occurs at the three-way junctions of the cortical ER, where Lnps accumulate (Sun et al., 2020a). Interestingly, the loss of RHD3 leads to a redistribution of Lnps to the bulk ER (Sun et al., 2020a), supporting the idea that RHD3 is required for the localization of Lnps to the three-way junctions. In addition, Lnps were found to stabilize nascent three-way junctions but also to suppress the fusogenic function of RHD3 (Sun et al., 2020a). Therefore, Lnps and RHD3 together participate in the control of the dynamic architecture of ER subdomains.
Intriguingly, the RHD3 proteins can functionally replace the yeast Sey1p in complementation analyses (Zhang et al., 2013). Furthermore, in yeast, Lnp antagonizes Sey1 (Chen et al., 2012), but the Arabidopsis Lnps expressed in yeast do not antagonize the function of Sey1 (Sun et al., 2020a), supporting species-specific requirements for the control of ER shape by ATL-like proteins and Lnps.
Finally, it has been demonstrated in Arabidopsis that a double lnp1 lnp2 mutant or mutants lacking RHD3 are viable, though showing some growth defects (Chen et al., 2011; Kriechbaumer et al., 2018; Stefano et al., 2012; Sun et al., 2020a; Ueda et al., 2018, 2016; Zhang et al., 2013). On the other hand, loss of RHD3 along with either the RHD3-isoform RHD3-like 2 or RHD3-like 1 leads to unviable pollen or causes lethality, respectively (Zhang et al., 2013). These observations indicate that the control of the fusogenic activity of RHD3 by Lnps and the localized distribution of Lnps at the three-way junctions are likely not essential processes. Nonetheless, they also raise the questions whether, in addition to Lnps, other proteins control the fusogenic activity of the RHD3 family of proteins, other proteins may have fusogenic activity, and/or whether, besides ER membrane fusion, RHD3 carries out (or enables) other functions that are essential for the cell.
Recent findings shed light the role of Lnps in regulating RHD3 activity in the ER. The mammalian Lnp (mLNP) interacts with Cullin-associated and neddylation-dissociated 1 (CAND1), a regulator of Skp1-Cul1-F-box (SCF) ubiquitin ligase, which negatively controls mLnp auto-ubiquitination (Kajiho et al., 2019). Furthermore, a CAND1 knockdown mutation promotes proteosomal degradation of mLnp1 and reduces the abundance of tubular ER (Kajiho et al., 2019). Therefore, mLnp ubiquitination by CAND1 likely serves to control the three-way junction stability of the ER. It has been shown recently that, similar to mLnp, the Arabidopsis Lnp has E3 ubiquitin ligase activity (Sun et al., 2020a). Lnps can ubiquitinate RHD3, targeting the protein for proteosomal-mediated degradation, and in the absence of Lnps, RHD3 protein levels increase (Sun et al., 2020a). Therefore, Lnps are required to control the homeostatic levels of RHD3. Because the process of ubiquitination and proteasomal degradation of membrane proteins is an energy-expensive process (Grice and Nathan, 2016), the removal of Lnp from the membrane is unlikely to underlie the rapid changes in the ER morphology and may instead account for slower ER remodeling processes, such as development-associated changes in ER shape (Ridge et al., 1999; Stefano et al., 2014). Moving forward, it will be interesting to test this hypothesis and whether the stability of the plant Lnps in the membrane is also controlled by CAND1 homologs or other proteins.
Shaping plant-specific forms of the ER
Although this is yet to be tested experimentally, the plant ER is most likely a single organelle in each cell. Theoretically, the plant ER can be considered a continuous organelle across the entire organism because it passes as a modified form, known as the desmotubule, through intercellular nanopores, known as plasmodesmata (PD) (Figure 1). PD are essential for plant development as they channel cell-to-cell transport of molecules, including water, ions, metabolites, transcription factors, and RNAs (Tilsner et al., 2016). In the PD, the PM layers the boundaries of the PD pore that is traversed by the desmotubule.
Considering that they cross through a rigid cell wall, an astonishing feature of the PD is their dynamic permeability, which is regulated and responsive to several cues, including the metabolic status of cells. For example, leaf cells control PD trafficking in response to carbohydrate availability that is monitored by the highly conserved target of rapamycin (TOR) kinase (Brunkard et al., 2020). Callose deposition and removal at the PD sphincter dynamically controls the permeability of PD. However, as predicted by mathematical modeling, the ER/desmotubule complex can contribute to the PD permeability. In this model, intercellular transport of molecules in the cytoplasm surrounding the desmotubule is facilitated by a concentration gradient. A turgor pressure difference between cells can displace the ER-desmotubule complex toward the PM, thus limiting pore permeability (Nicolas et al., 2017b). While an ER/desmotubule control may facilitate regulation of rapid changes of PD permeability, callose deposition and removal at the PD sphincter control the permeability of PD for relatively slower and energetically costlier cellular responses. Indeed, in situations of stress, callose-mediated regulation of permeability of PD controls symplastic movement of viruses, pathogen-responsive proteins, and transcription factors (TFs) (Wolf et al., 1989; Wu and Gallagher, 2012; Xu et al., 2017), including the TFs responsive to proteotoxic stress in the ER (Lai et al., 2018).
The desmotubule is anchored to the PM via filamentous proteins and surrounded by a thin sleeve of cytoplasm, where transport occurs. Electron microscopy analyses established that the desmotubule-PM tethers are absent in post-cytokinesis PD but appear in mature PD (Nicolas et al., 2017a). Two members of the multiple C2 domains and transmembrane region protein (MCTP) family, AtMCTP3 and AtMCTP4, have been recently shown to function as desmotubule-PM tethers in PD (Brault et al., 2019). These are PD-localized proteins that insert into the ER membrane via their single-spanning transmembrane region and contact the PM via their C2 domains through an interaction with anionic phospholipids. The function of AtMCTP4 as a tether is supported by a partial complementation of ER-PM tethering defects of a yeast Δtether mutant. Furthermore, an Atmctp3/Atmctp4 double mutant showed growth defects as well as compromised PD function and composition. Since PD are essential to plants’ life (Sager and Lee, 2018), the viability of Atmctp3/Atmctp4 supports the existence of additional desmotubule-PM tethers that are yet unknown.
PD are typically 300 nm long and ~ 10 nm wide. The desmotubule is therefore constricted in the PD and virtually devoid of a lumen. The unusual arrangement of the desmotubule membrane may be supported by a distinct lipid composition forming a specialized subdomain of the ER, characterized by a non-bilayer lipid phase (Jouhet, 2013). However, it is also possible that ERlumen-constricting proteins, such as Rtns, may contribute to the stabilization of the narrow lumen of the desmotubule. Indeed, overexpression of RtnB13 leads to constriction of the ER tubules and a significantly reduced diffusion of a lumenal GFP in vivo (Tolley et al., 2008), and RtnB3 and RtnB6 have been found to reside at the PD (Kriechbaumer et al., 2015). If Rtns or other specialized proteins were responsible for the shaping of the desmotubule, a fundamental question would be how the shaping proteins accumulate to sufficient levels in the PD to constrict the ER tubule. Rtns are known to form oligomeric structures (Shibata et al., 2008), a property that may, at least in part, explain how they may accumulate in subdomains of the ER such as PD and facilitate the constriction of the desmotubule. Nonetheless, whether the processes underlying tubule shaping of the cortical ER tubules also operate at the level of the desmotubule is yet to be demonstrated.
Relationship of the ER with the cytoskeleton
A remarkable feature of the plant ER network is its movement, which refers to the flow of proteins within the ER membrane coupled with ER network remodeling, rather than the movement of entire organelles typical of other plant cell organelles (e.g., mitochondria, peroxisomes, and endosomes). Different from chemical depletion of microtubules (MTs), which only impacts a subset of ER structures (see below), chemical depletion of the actin cytoskeleton leads to a global reduction of ER remodeling (Sparkes et al., 2009a), suggesting that both cytoskeletal components contribute to ER remodeling, but that actin has a predominant role. This is markedly different from mammalian cells, in which the ER movement depends largely on MT (Waterman-Storer and Salmon, 1998). Depletion of MT in plant cells leads to an alteration of the extension of a subset of ER tubules (Hamada et al., 2014). In particular, it has been established in hypocotyl epidermal cells that some ER tubules can elongate along preexisting MT in both a plus and minus end direction and that the MT-dependent ER movement is slower than that of the actin-myosin-dependent movement (Hamada et al., 2014).
More recently, a close relationship between ER and MT has been established in the polarized growth of root hairs (Qi et al., 2016). A characteristic phenotype of the loss-of-function of RHD3 is a reduced length of the primary root and of root hair, which become visibly wavy compared with wild type (Galway et al., 1997). It has been shown that the root hair growth of rhd3 is hypersensitive to oryzalin (a MT-destabilizing agent) at 50 nM, a concentration that does not significantly affect wild type; conversely the root hair growth of rhd3 is partially rescued by taxol (a MT-stabilizing agent) at substoichiometric concentrations (0.1–0.5 μM) (Qi et al., 2016), suggesting that RHD3 is functionally connected with MT. This hypothesis is further supported by the evidence that in rhd3 the organization of the endoplasmic MT (a specific MT arrangement of tip-growing hairs in the subapex; Van Bruaene et al., 2004) is altered compared with wild type (Qi et al., 2016). Recent findings have lent further support to this notion through the characterization of two allelic enhancers of the rhd3 phenotype, named rhd3 enhancer9 (ren9), isolated through an ethyl methanesulfonate (EMS) screen of the rhd3-1 allele (Sun et al., 2020b). Ren9 alleles were mapped to missense mutations in ARK1, an armadillo-repeat-containing kinesin, which is known to promote MT catastrophe and to localize at the plus end of MT (Eng et al., 2017). When co-expressed with a fluorescent protein fusion to RHD3 (mCherry-RHD3) in epidermal leaf cells, ARK1-GFP was localized at comets/puncta that co-localize and move together with some of the RHD3 puncta along the ER tubules. Also, ARK1-GFPlocalized puncta were found to move with mCherry-RHD3 and, apparently, to pull an ER tubule toward another tubule (Sun et al., 2020b).
These findings add to the earlier discovery that plant ER tubules can elongate along preexisting MT (Hamada et al., 2014) but also support the idea that the ER can elongate at the growing ends of MT. Because RHD3 and ARK1 interact via the armadillorepeat-containing domain of the kinesin, it will be interesting to establish if a loss-of-function of ARK1 may exacerbate the RHD3 loss-of-function mutation on the ER structure and tubule fusion to test whether other proteins exist that may function redundantly with ARK1 in MT-associated ER movement. Furthermore, it will be important to validate that ARK1 functions as a bona fide kinesin for the directional dragging of the ER, which is also yet to be experimentally established. In mammalian cells, the ER membrane spanning and MT-binding protein CLIMP-63 functions as an ER-MT linker (Klopfenstein et al., 1998). Overexpression of CLIMP-63 leads to a reorganization of the ER network to overlay the MT cytoskeleton by increasing the density of interactions between the ER and the MT (Klopfenstein et al., 1998). Plants do not encode sequence homologs of CLIMP-63. Therefore, whether linkers that connect the plant ER membrane with the MT exist and what their identity may be are still open questions.
As indicated earlier, the movement and shape of the ER is mainly actin dependent. Depolymerization of actin leads to an increase in a spatiotemporal persistence of ER structures, which translates into reduced dynamics and streaming of the ER, and enlargement of the tubules (Sparkes et al., 2009a). The close relationship of the ER with actin is mediated by actin linkers that have been recently identified. These include SYP73, a plant-specific ER type-II-membrane-associated protein containing a putative soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor (SNARE) domain and NETWORKED 3B (NET3B) (Cao et al., 2016; Wang and Hussey, 2017) (Figure 3). The cytoplasmic domain of SYP73 contains actin-binding signature motifs and binds actin directly, as demonstrated by in vitro assays with a recombinant SYP73 cytosolic domain. A loss-offunction allele of SYP73 leads to reduced ER movement, similar to the reported effect of treatment with latrunculin B (LatB, an actin-depolymerizing agent) (Cao et al., 2016; Sparkes et al., 2009a). Conversely, overexpression of SYP73 leads to a restructuring of the ER network to overlay the actin cytoskeleton (Cao et al., 2016). Overexpression of SYP73 also leads to a marked reduction in cisternal ER compared with cells expressing lower levels of SYP73 (Cao et al., 2016). Therefore, the effect of overexpression of SYP73 on ER morphology mimics the reported effect of overexpression of CLIMP-63 on the ER in mammalian cells (Klopfenstein et al., 1998). Treatment of cells overexpressing SYP73 with LatB changes the ER network structure to resemble the effect of the drug on wild-type cells, further supporting the idea that SYP73 serves as a linker between the ER and the actin cytoskeleton. Together these results have led to the model that SYP73 facilitates a dynamic association of the ER and actin cables by acting as a temporary bridge between the ER and actin (Cao et al., 2016).
Figure 3. ER and cytoskeleton interactions.
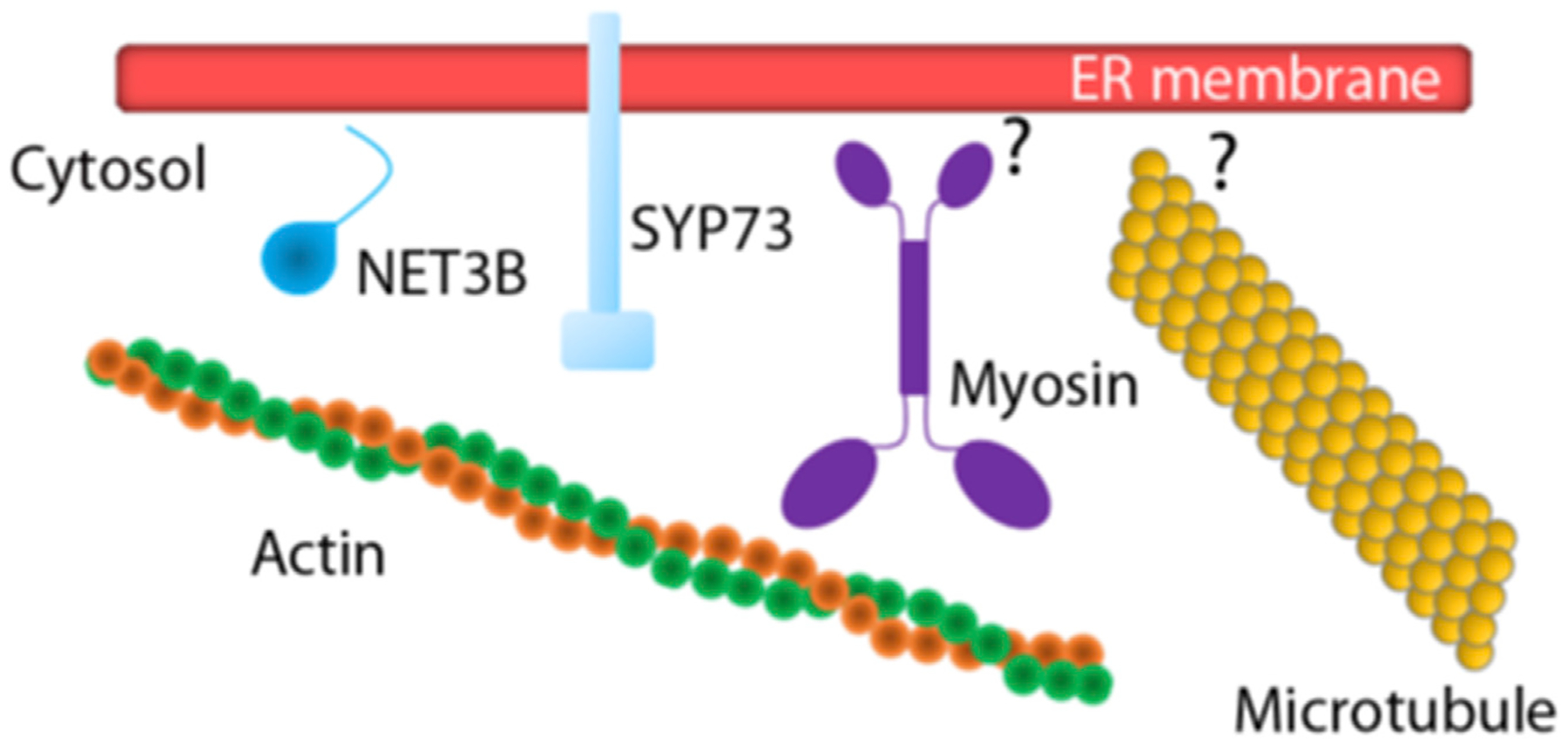
Diagram illustrating the two ER-actin anchors identified to date in plant cells: NET3B and SYP73. The current models posit that these proteins link the ER with actin cables. A direct interaction with actin has been demonstrated only for SYP73. SYP73 may facilitate a transient association of the ER membrane with actin and the propelling action of myosin motors. While myosin has been found in association with the ER in cofractionation analyses, the mechanisms for the association are yet unknown (question mark in the diagram). The ER can associate with microtubules via yet-unknown mechanisms (question mark in the diagram). As described in the text, an interaction of RHD3 with ARK1 may be one of such mechanisms. The protein diagrams are not to scale.
The existence of ER-actin linkers has been supported by the investigation NET3B, a protein that can be found in association with the actin cables in live cells, and whose overexpression leads to overlay of the ER with actin (Wang and Hussey, 2017). Similar to SYP73 overexpression, the phenotype of NET3B overexpression is reverted by LatB treatment (Wang and Hussey, 2017). Differently from SYP73, however, NET3B is a cytosolic protein. Therefore, the bridging of ER with the actin cytoskeleton is dependent on multiple types of linkers, which likely function in a redundant fashion and contribute to the shape and movement of the ER. However, unlikely SYP73, whose loss hampers root growth and ER morphology in early stages of seedling development (Cao et al., 2016), the loss of NET3B does not affect ER morphology and plant growth (Wang and Hussey, 2017), indicating that, individually, some of these linkers may make a more significant contribution to the homeostasis of ER structure than others, or that they may have overlapping roles with other paralogs.
Through a spatial association of the ER with actin, the ER-actin bridging provided by linkers may facilitate the propelling action of actin-associated motors that drive the movement of the ER. The most likely motors are members of the plant-specific myosin XI family, whose genetic depletion, primarily of myosin XI-K (Peremyslov et al., 2010; Ueda et al., 2010), or overexpression of the tail domain alone can alter the ER organization and movement (Griffing et al., 2014; Sparkes et al., 2009a). Despite a demonstrated cofractionation of myosin with the ER fraction in Arabidopsis cell extracts (Ueda et al., 2010), the mechanisms for an association of myosin with the ER are unknown.
ER contact sites with other organelles, a two-way connection
Similar to a spider web hanging from a wall, the cortical ER contacts the PM at the so-called ER-PM contact sites (EPCs). These sites have been involved in a large number of processes, including signaling in immune response, controlling viral movement, responses to environmental clues, endocytosis, autophagy, and stabilizing the cortical ER network (Kim et al., 2016;Lee et al., 2019;Levy et al., 2015; Lewis and Lazarowitz, 2010;Pérez-Sancho et al., 2015; Schapire et al., 2008; Stefano et al., 2018; Uchiyama et al., 2014; Wang et al., 2014, 2019; Yamazaki et al., 2008). The EPCs represent regions of juxtaposed cortical ER and PM, which are tethered by ER proteins and/or electrostatic interactions with PM phospholipids and interactions with PM proteins (Bayer et al., 2017). The ER components of the EPCs include the evolutionary conserved synaptotagmins (SYTs) (Ishikawa et al., 2018; McFarlane et al., 2017; Pérez-Sancho et al., 2015; Siao et al., 2016) and the vesicle-associated membrane-associated protein 27s (VAP27s) and VAP27-related proteins (Stefano et al., 2018; Wang et al., 2017, 2016). The distribution of SYT1 and VAP27 at EPCs is not always coincidental, in the sense that while SYT1-enriched EPCs (S-EPCs) coincide with VAMP27-enriched EPCs (V-EPCs), S-EPCs can also be distinct from V-EPCs (Siao et al., 2016). The existence of EPCs has been documented in earlier electron microscopy studies (Staehelin, 1997), but the implementation of fluorescent protein fusions in live-cell analyses has provided opportunities to gain insights on the function and structural requirements of these sites. VAP27s are type II membrane proteins that reside in the bulk ER and concentrate at the EPCs where they show limited diffusion in and out of the EPCs, in a manner that is dependent upon cellular availability of SYT1 (Siao et al., 2016). At the EPCs, VAP27s interact with the plant-specific actin-binding protein NETWORK 3C (NET3C), MT, heavy and light clathrin chains, clathrin adaptor proteins, and AtEH1/Pan1, a component of the endocytic TPLATE complex (TPC) and a regulator of autophagosome formation (Stefano et al., 2018; Wang et al., 2014, 2019). VAP27-1 and VAP27-3 also interact directly with PI(3)P, PI(4)P, PI(5)P, PI(3,5)P2, PI(4,5)P2, and PI(3,4)P2, lipids that are abundant in membranes associated with endocytosis, such as early endosomes and the PM (Stefano et al., 2018). Furthermore, NET3C has been recently found to interact with two microtubule binding proteins: kinesin-light-chain-related protein 1 (KLCR1) and IQ67-domain 2 (IQD2), and it has proposed that the NET3C-KLCR1-IQD2 forms an actin-microtubule bridging complex at the cell cortext (Zang et al., 2021).
EPCs are generally immobile (Ishikawa et al., 2018; Wang et al., 2014), but their number is influenced by a number of factors. For example, a correlation has been established between the developmental stage of cells of roots and hypocotyls, with the appearance of more EPCs per area of young cells than mature cells of the same tissues (McFarlane et al., 2017). Despite a change in number during cell development, during physiological growth conditions the size of EPCs does not change and is generally within the 50–300-nm range (McFarlane et al., 2017). Nonetheless, it has been recently demonstrated that ionic stress (e.g., NaCl stress) causes enlargement of EPCs with a redistribution of SYT1 along the tubules of the cortical ER and an increase in ER-PM connectivity. The evidence that osmotic stress generators (i.e., mannitol or sorbitol) do not have a similar effect supports the hypothesis that ionic imbalance is the cause of changes in EPC morphology under conditions of salt stress (Lee et al., 2019).
Recent findings in maize have indicated that the ZmVAP27-1 homolog interacts with a water channel, the PM aquaporin ZmPIP2;5, an interaction that was also replicated with the respective Arabidopsis homolog VAP27 and aquaporin proteins (Fox et al., 2020). Interestingly, when co-expressed in Xenopus oocytes, ZmVAP27-1 and ZmPIP2;5 increase the water permeability coefficient of the PM. Therefore, at least in vitro, these proteins functionally impact water transport across the membrane. Because of the involvement of EPCs in ionic stress, verified at least for SYT1, it is possible that the interaction of VAP27 with PIPs may contribute to salt stress responses.
The mechanisms for tethering of ER-EPCs components to the PM are not fully understood. VAP27 proteins interact with lipids that are present in endocytic membranes (Stefano et al., 2018), and it has been shown that the NaCl-induced expansion of S-EPCs correlates with an increase of PI(4,5)P2 accumulation at the PM (Lee et al., 2019). Based on these results, it is possible that the tethering of ER proteins to the PM occurs via electrostatic interactions of ER proteins with phosphoinositides at the PM, if these are present in sufficient quantity. Because phosphoinositides are generally in low abundance in the PM, it is possible that these phospholipids preferentially accumulate at the EPCs to favor an ER-PM interaction. It also cannot be excluded that protein-phosphoinositide interactions contribute only partially to EPC establishment or stability and operate in parallel or additively with protein-based tethers embedded within the PM. The verified interaction of the aquaporin ZmPIP2;5 with VAP27 (Fox et al., 2020), for example, lends support to protein-protein interactions as a mechanism to anchor EPCs with PM-embedded proteins. Because S-EPCs and the V-EPCs do not completely overlap spatially (Siao et al., 2016), it is possible that the mechanisms underlying the ER-PM association are different for distinct EPCs subpopulations.
Whether V-EPCs and S-EPCs are functionally equivalent is yet unknown. The evidence that a double mutant of VAP27-1 and VAP27-3 shows reduced endocytosis compared with wild type and that VAP27s interact with endocytic machinery components (Stefano et al., 2018) supports a direct role of V-EPCs in endocytosis, most likely through the recruitment of endocytic machinery at the EPCs through VAP27s (Stefano et al., 2018). Similarly, SYT1 is also required for endocytosis (Lewis and Lazarowitz, 2010), but it is unclear whether this role is executed at spatially coincidental V-EPCs and S-EPCs, and/or at S-EPCs that are not coincident with V-EPCs.
Spatiotemporal analyses of the S-EPCs provide new insights into formation of the EPCs. Using three-dimensional imaging in super-resolution confocal live imaging microscopy (SCLIM), SYT1 was specifically localized to the ER at the ER-PM interface on immobile ER tubules only. In addition, SYT1 was found to be distributed to edges of ER sheets that eventually morphed into immobile ER tubules. Loss of functional SYT1 led to a reduction of the immobile tubules, an enlargement of the ER into a more sheeted network structure, and possibly a detachment of the ER from the PM (Ishikawa et al., 2018). Therefore, by providing a contact with the PM, SYT1 is likely to be involved in stabilizing an ER subdomain that leads to the formation of the immotile EPCs on the tubular ER at the interface with the PM. This hypothesis is further supported by evidence that loss of SYT1 compromises the stability of V-EPCs (Siao et al., 2016). Interestingly, treatment with the acting depolymerizing agent LatB does not alter the distribution of SYT1-EPCs. Conversely, LatB treatment and MT-depolymerization alters the mobility of NET3c and VAP27 at the EPCs, respectively (Wang et al., 2014). These results suggest that EPC components have different structural requirements and support the idea that the steady-state composition of the EPCs depends, at least in part, on the cytoskeleton.
The vicinity of the ER to the tonoplast in the cell cortex and at the transvacuolar strands most likely facilitates the existence of points of contacts between the ER and the tonoplast, but these have not been reported as functional entities yet. Nonetheless, the ER provides the membrane for the biogenesis of the central vacuole (Viotti et al., 2013).
The ER has been observed to associate with chloroplasts, Golgi, peroxisomes, mitochondria, and endosomes (Barton et al., 2013; Stefano et al., 2014, 2015, 2018). The identity of the machinery facilitating these contacts is beginning to emerge, at least for mitochondria and Golgi. For example, optical laser tweezer experiments (White et al., 2020) have shown that in tobacco leaf epidermal cells about 80% of the mitochondria trapped by a laser tweezer remain attached to the ER. The activity of Miro2, a membrane-anchored GTPase presumably localized in the mitochondria outer envelope, was found to influence the ER-mitochondria association (White et al., 2020). Specifically, as revealed by optical laser trapping, expression of dominant mutants of Miro2 affecting the GTPase domains (i.e., residues K23V and K434V) led to significant changes in the number of mitochondria attached to the ER. In particular, putatively inactivating GTPase mutations led to about 20% reduction of the mitochondria attached to the ER compared with wild type and a putatively active variant of Miro2. These results support the hypothesis that Miro influences the association of the ER with mitochondria, and further studies will help clarify the underlying mechanisms supporting the role of Miro2 in ER-mitochondria association. It will also be interesting to test the functional and physiological role of the association of the ER and mitochondria in plants for which not much is currently known.
Similar to mitochondria, chloroplasts are in close association with the ER. Using trans-organellar complementation assays based on complementation of biosynthetic mutants of the vitamin E pathway, which occurs in the plastids, it has been possible to show that the ER-chloroplast interactions are functional (Mehrshahi et al., 2013). In these assays, plastidial enzymes were shown to complement the respective chloroplast mutants with proteins retargeted to the ER. These results are consistent with the notion that, together, chloroplasts and ER produce essential lipids whose biosynthesis requires non-overlapping steps that occur in both organelles (Xu et al., 2008). The identity of the molecular machinery connecting chloroplasts and ER is unknown, but it is most likely protein mediated. This is supported by data showing that optical laser tweezers applying to chloroplasts forces of up to 400 pN were insufficient to pull chloroplasts away from the ER in disrupted protoplasts (Andersson et al., 2007). In the future, it will be interesting to establish the identity of the proteins underlying the ER-chloroplasts connections and test their relevance to lipid biosynthesis and, possibly, protein shuttling between the two organelles.
The plant ER is also in close association with the Golgi apparatus, which is dispersed in mini stacks that are mobile within the cell (Brandizzi et al., 2002; daSilva et al., 2004). It has been demonstrated that a Golgi stack trapped by an optical tweezer can be repositioned in proximity of an ER tubule and anchored to the ER (Sparkes et al., 2009c), supporting the existence of contacts between the two organelles. The ER-Golgi connection is likely mediated by a golgin-based tethering complex. Golgins are coiled-coil domain proteins that have been implicated in Golgi integrity and membrane traffic (Muschalik and Munro, 2018). In dual color optical laser tweezer experiments based on the co-expression of a fluorescent protein fusion to AtCASP, a Golgi-localized golgin, and a Golgi marker, Golgi stacks labeled by a truncated AtCASP-DCC (i.e., without the coiled-coil domains) were easier to trap than Golgi stacks labeled by wild-type AtCASP (Osterrieder et al., 2017). The interaction of AtCASP-ΔCC-labeled Golgi stacks with the ER was easier to disrupt compared with wild-type AtCASP-labeled Golgi stacks (Osterrieder et al., 2017). Given that the Golgi apparatus is the first receiving site of cargo produced by the ER in conventional secretion and that it is a mobile organelle in plant cells, a connection secured by golgins between the ER and the Golgi stacks likely facilitates efficient ER-Golgi membrane traffic. This hypothesis can be tested in experiments based on loss-of-function mutants of golgins like AtCASP.
Similar to Golgi, chloroplasts, and mitochondria, the ER is also in close association with endosomes, as demonstrated by electron tomography and time-lapse microscopy analyses (Stefano et al., 2015). Dual-fluorochrome analyses of the distribution and movement of early and late endosomes in cells coexpressing an ER marker, established that 85% and 75% of late and early endosomes, respectively, were in continuous association with the ER within a 1 min of observation period (Stefano et al., 2015). While the machinery underlying ER-endosome association in plant cells is unknown, functional studies on the relationship between ER structure homeostasis and endosomal function have revealed that homeostasis of the ER structure is required for the spatial distribution of endosomes, their velocity as well as endocytosis (Stefano et al., 2015). This was demonstrated in experiments based on alteration of ER structure upon overexpression of the Rtn RTNLB3 or a loss of RHD3 (Stefano et al., 2015). In plant cells, early endosomes receive endocytic cargo. Therefore, the verified reduction in endocytosis efficiency in an rhd3 background coupled with a reduced endosome mobility led to the suggestion that the ER-endosome association is likely necessary for maintaining the endocytic function of endosomes, possibly by ensuring an appropriate subcellular distribution of endosomes and their movement (Stefano et al., 2015). An ER-endosome association also facilitates other processes, for example the movement of TFs. The root patterning TF SHORT-ROOT (SHR) moves intracellularly, associates with endosomes, and interacts with SHR-INTERACTING EMBRYONIC LETHAL (SIEL) (Gallagher et al., 2004; Koizumi et al., 2011). SIEL is a MT-binding protein associated with endosomes. It is required for the intercellular movement of SHR (Koizumi et al., 2011) and is known to interact with the MT-binding protein KINESIN G (KinG), a calponin homology kinesin, which also regulates the movement of SHR (Spiegelman et al., 2018). It has been demonstrated that both SHR and KinG localize to endosomes in close proximity to the ER and that defects in ER structure in loss-of-function mutants of RHD3 or the ER-membrane-associated SYT1 reduce intercellular movement of SHR (Spiegelman et al., 2019). Based on these results and the evidence that SHR-associated endosomes pause on MT where KinG is present (Spiegelman et al., 2018), it has been hypothesized that the sites of the ER where the interaction between SHR and KinG occurs at MT-associated junctions are the locations where endosomes pause to facilitate protein-protein interactions necessary to ensure cell-to-cell SHR movement (Spiegelman et al., 2019). Under this light, the ER provides a surface to direct the spatial organization of organelles and facilitate their functional interactions.
Homeostasis of ER function
ER function: foundations and impact on cell growth and homeostasis.
The ER is the biosynthetic organelle of the cell and its productivity is continuously challenged by internal and external cues, such as growth factors and environmental stresses that impact the cell. Indeed, accumulation of misfolded proteins in the ER leads to a potentially lethal condition, known as ER stress. ER homeostasis is monitored and maintained by a surveillance system that is called the unfolded protein response (UPR). The UPR is an ancestral signaling pathway that senses changes in ER protein folding status and alteration of ER membrane equilibrium and reprograms nuclear gene expression to mitigate the load of unfolded proteins (Hetz et al., 2020; Pastor-Cantizano et al., 2020). The regulators of the plant UPR are the bifunctional ribonuclease and kinase IRE1 and the membrane-tethered-transcription factor, bZIP28. IRE1 is functionally conserved in yeast, plants, and mammals. Metazoan genomes encode a functional equivalent of bZIP28, termed ATF6. Plants and yeast do not encode protein kinase RNA-like endoplasmic reticulum kinase (PERK), which guides the third arm of the UPR in mammalian cells. Terminally misfolded proteins are cleared by the ER-associated degradation (ERAD) pathway that facilitates translocation of ER proteins into the cytosol for degradation via the ubiquitinproteasome system. Because the plant UPR and ERAD have been recently reviewed, they will not be discussed in this review (Chen et al., 2020; Pastor-Cantizano et al., 2020). Here, we will focus on ER-phagy an important phenomenon for which exciting new findings are emerging in plants.
While the UPR and ERAD monitor ER proteostasis, autophagy processes facilitate the selective turnover of ER domains, a process termed ER-phagy (Chino and Mizushima, 2020). In general, autophagy is a cellular process designed to reshape cell content during growth and in conditions of stress. Autophagy is highly selective and relies on the recognition of cargo, such as damaged organelles or protein aggregates, by specific receptors (Johansen and Lamark, 2020). There are two major types of autophagy described thus far in mammalian, plant, and yeast cells: macroautophagy and microautophagy (Schuck, 2020;Sieńko et al., 2020; Yang and Bassham, 2015), with the former being the most extensively studied. Macroautophagy and microautophagy are mechanistically distinct. In microautophagy, cellular components are engulfed directly into the degrading compartments. Specifically, in plants, the tonoplast engulfs cargo directly and pinches off inward to form autophagic bodies that contain cytoplasmic cargo for storage or degradation in the vacuolar lumen (Sieńko et al., 2020). Macroautophagy is initiated by the formation of the autophagosome, a double-membrane cup-shaped structure, which originates from a membrane structure, the phagophore. The ER is a membrane source for the autophagosome (Yamamoto and Noda, 2020). The phagophore enwraps the autophagy cargo to form a sealed autophagic body that is delivered to the vacuole and fuses with the tonoplast to release the cargo for degradation. The largely conserved AUTOPHAGY-RELATED (Atg) proteins mediate the various macroautophagy steps. For example, upon activation of autophagy, Atg8, a ubiquitin-like protein, is conjugated to the phagophore (Stolz et al., 2014;Zaffagnini and Martens, 2016). In non-plant systems, macroand microphagy contribute to ER-phagy along with the degradation of ER-derived vesicles, an additional ER degradation pathway (Chino and Mizushima, 2020). The documented existence of ER-phagy adaptors that act as linkers connecting the ER with autophagic membranes support the idea that ER-phagy is based, at least in part, on the selective recognition of the ER as cargo by proteins on the autophagosomal membrane. Several ER-phagy receptors are known to date in mammalian cells (C53, FAM134B, RTN3L [a long isoform of Rtn3], CCPG1, SEC62, TEX264, CALCOCO1, and ATL3) and yeast cells (Epr1, the Lst1-Sec23 complex, Atg39, and Atg40) (Chino and Mizushima, 2020; Cui et al., 2019; Nthiga et al., 2020a, 2020b; Zhao et al., 2020). In vertebrates, TEX264 is expressed more ubiquitously than the other mammalian ER-phagy receptors (An et al., 2019; Chino et al., 2019) and is considered a master receptor for ER-phagy under both basal and starvation conditions (Delorme-Axford et al., 2019). The apparent lack of TEX264 homologs in non-vertebrate species suggests that the mechanisms of ER-phagy have assumed some unique features across kingdoms, at least for what concerns the identity of the receptors.
The yeast and mammalian ER-phagy receptors in non-plant species have been ascribed distinct roles. For example, yeast Atg39 interacts with Atg8 directly to mediate ER-phagy of the nuclear envelope, which is continuous with the ER, while Atg40 interacts with Atg8 and Lnp1 for the degradation of the cortical ER (Mochida et al., 2015). The interaction of Atg40 with Lnp1 brings Atg40-containing ER regions to the sites of phagophore initiation (Chen et al., 2018). In mammalian cells, the long isoform of the reticulon Rtn3 (RTN3L) (Grumati et al., 2017) and FAM134B (Bhaskara et al., 2019; Khaminets et al., 2015) are ER-phagy receptors that function in conjunction with four other ER-resident proteins, namely CCPG1 (Smith et al., 2018), SEC62 (Chino et al., 2019; Fumagalli et al., 2016), ATL3 (Chen et al., 2019), and TEX264 (An et al., 2019; Chino et al., 2019) for cargo selection during ER-phagy, and for ER-phagy via an interaction with Atg8. RTN3L and ATL3 appear to mediate constitutive clearance of ER tubules, while FAM134B removes ER sheets (Grumati et al., 2017; Khaminets et al., 2015). CCPG1 protects against aggregation of ER lumenal proteins (Smith et al., 2018), while the translocon complex component SEC62 controls ER turnover after transient ER stress to adjust ER volume homeostasis (Fumagalli et al., 2016). Unlikely the other known ER-phagy receptors, C53 is a cytosolic protein that is connected to the ER quality control system via the ufmylation pathway, a post-translational protein modification that occurs in plants and metazoans, and is functionally specialized in resolving ribosome stalling, a situation that can occur during co-translational protein translocation via the signal recognition particle (SRP) pathway (Banerjee et al., 2020; Stephani et al., 2020).
Compared with mammalian cells, the number of identified ER-phagy receptors in plants is smaller and includes SEC62, the reticulons Rtn3, and Rtn2, and C53 (Figure 4). In plants, it has been suggested that similar to the mammalian counterpart, the Arabidopsis ER integral membrane protein SEC62 serves a role as an ER-phagy receptor (Hu et al., 2020). This suggestion is based on the evidence that SEC62 is required for resistance to ER stress and that, in cells challenged by ER stress agents, relative to controls, a larger percentage of YFP-AtSec62 labeled ring-like structures, which were distributed throughout the ER network, were induced by ER stress and co-localized with the autophagy marker mCh-Atg8e (Hu et al., 2020). Furthermore, it was shown that SEC62 and Atg8e interact directly and that this interaction is necessary for the ER-stress-induced degradation of SEC62 in the vacuole. Also, the formation of SEC62 ring-like structures and the delivery of SEC62 to the vacuole were reduced in lossof-function backgrounds of ATG7 and ATG5, two ATG-proteins required for ER stress resistance and autophagosome biogenesis (Hu et al., 2020). Therefore, Sec62 is likely an ER-phagy receptor whose function depends on the core autophagic machinery.
Figure 4. Plant ER-phagy receptors.
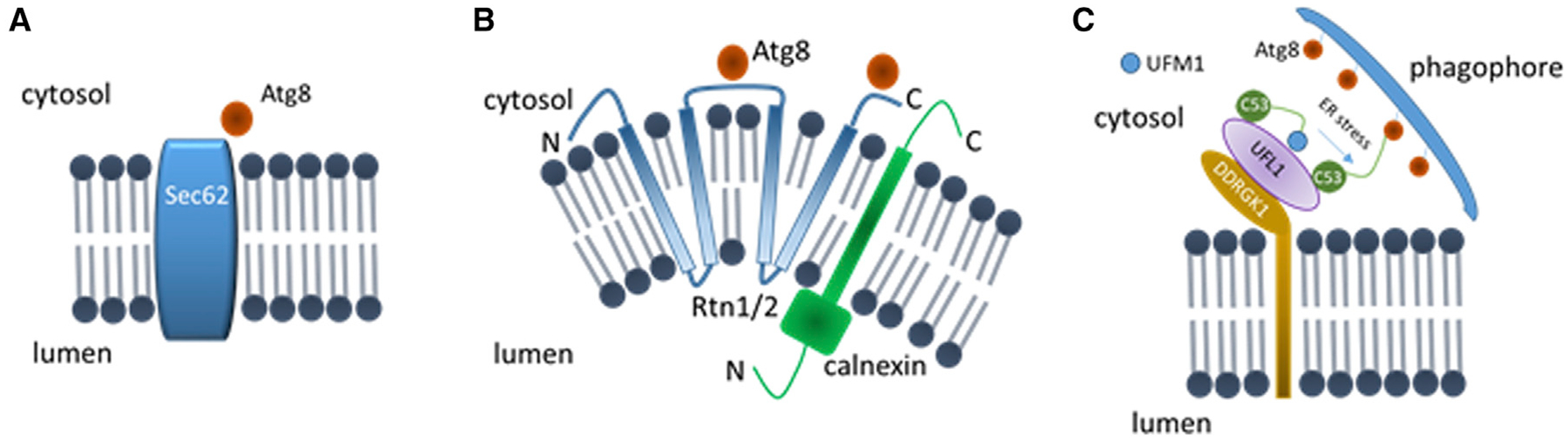
(A–C) Diagrams depicting the plant ER-phagy receptors identified to date: Arabidopsis Sec62 (A), maize Rtn1/2 (B), and Arabidopsis C53 (C). The interacting proteins or complexes are also illustrated. C53 is in a heteromeric receptor complex with UFL1 and DDRGK1. The strength of the interaction between C53 and Atg8 is enhanced by ER stress induced by tunicamycin, while the interaction between C53 and UMF1 is reduced in these conditions. See main text for details. Diagrams not in scale. Orientation of the ER-phagy receptors is presented with respect to the ER membrane, ER lumen, and cytosol. N and C, N and C terminus of the proteins.
In addition to SEC62, a recent study identified maize Rtn1 (Zm00001d043551) and Rtn2 (Zm00001d012776), two members of the cereal-restricted subgroup of reticulons (Clade 1-1), as ER-phagy receptors (Zhang et al., 2020). Rtn1 and Rtn2 are expressed in the starchy endosperm and aleurone at 18–22 days after pollination. Rtn1 and Rtn2 localize to the ER, and, when overexpressed, cause a marked ER architecture reorganization, supporting their role as reticulon proteins (Zhang et al., 2020). Rtn1 and Rtn2 interact with Atg8a via specific Rtn domains localized at the C-terminal region, one at the cytoplasmic loop and two within the predicted transmembrane regions close to the cytoplasmic face of the ER membrane (Zhang et al., 2020). Rnt2-Atg8a binding increases during ER stress, and in these conditions, a fluorescent protein fusion of Rtn2 was found to associate with autophagosome-like structures decorated with a fluorescent protein fused to Atg8a (Zhang et al., 2020). These structures were stabilized in the vacuole by treatment with concanamycin A (ConA), a drug that suppresses protein turnover in the vacuole (Zhang et al., 2020). Therefore, Rtn2 actively undergoes autophagic degradation during ER stress. Based on these results and the evidence that in a rtn2 loss-of-function mutant bulk autophagy is increased, it was concluded that Rnt2 is an ER shaper that functions as an ER-phagy receptor by interacting with autophagy effectors like Atg8a. Indeed, it has been proposed that loss of Rtn2 leads to elevated bulk autophagy likely as a compensatory mechanism when ER-phagy is impaired (Zhang et al., 2020).
Moving forward in plant ER-phagy studies, it will be interesting to establish whether the plant ER-phagy receptors have a general role in ER degradation or if they are specialized in selective clearance of ER subdomains similar to some of the mammalian ER-phagy receptors. Also, it will be interesting to establish whether the plant ER-phagy receptors function in response to specific stress or growth cues that alter ER homeostasis and how these response pathways communicate with ER-phagy. For example, the Arabidopsis C53 was identified in a screen designed to discover Atg8-binding proteins via a peptide array (Stephani et al., 2020). C53 is closely associated with the ER, and in conditions of ER stress it is recruited into autophagosomes. Furthermore, quantitative proteomics assays on wild type and an Arabidopsis c53 mutant suggested that C53 is necessary for the degradation of ER-resident and -secreted proteins as well as cell wall proteins and lipid droplet proteins (Stephani et al., 2020). The Arabidopsis C53 interacts with the ER-associated ufmylation ligase UFL1, the membrane adaptor DDRGK1, in addition to Atg8 (Stephani et al., 2020) (Figure 4). Notably, the association between Atg8 and C53 was strengthened by tunicamycin, an ER stress inducer; conversely, the interaction between the ubiquitin-like modifier UFM1 and C53 was weakened by this chemical (Stephani et al., 2020). UFM1 may shield the C53-heteromeric complex from Atg8 in normal conditions, a role that is reduced during ER stress. Therefore, UFM1 and Atg8 most likely compete with each other for an association with the C53 to modulate its cellular activity (Figure 4). The Arabidopsis c53, ddrgk1, and ufl1 loss-of-function alleles were found to be hypersensitive to chemically induced ER stress compared with wild type. These results, along with the evidence that unlike the core autophagy mutants atg5 and atg2, c53 did not show hypersensitivity to nitrogen or carbon starvation, support the idea that C53 is specifically responsive to ER stress (Stephani et al., 2020). The evidence that C53 is not activated by associating with UPR sensors (Stephani et al., 2020) raises the question of how C53 may sense ER stress. Interestingly, despite the high level of conservation of mammalian and plant C53, the loss of C53 in Arabidopsis does not lead to a visible plant phenotype (Stephani et al., 2020). This is markedly different from mammalian cells, where the loss of C53 and ufmylation causes cell dysfunction and disease (Gerakis et al., 2019), indicating fundamental differences in ER-phagy strategies and possible relevance to cell homeostasis across kingdoms.
An additional connection between ER-phagy and stress pathways was suggested when the ER-phagy receptor Rnt2 was found to bind to the ER lectin calnexin and two other ER proteins involved in protein folding, binding protein (BiP) and protein disulfide isomerase (PDI). The binding depended on the level of ER stress (Zhang et al., 2020). Consistent with an interaction of Rnt2 with this ER folding machinery, a rtn2 loss-of-function mutation led to an increase in expression of the ER stress biomarker BiP2 in cells challenged by ER stress (Zhang et al., 2020). These results prompted the suggestion that a reduction in Rtn-mediated functions, including ER-phagy, causes ER stress increases, promoting general autophagy as a compensatory mechanism for ER turnover (Zhang et al., 2020). Although Rtn2 is required for attenuating ER stress, a loss of RHD3 in Arabidopsis causes a functional impairment of IRE1, a UPR master regulator, and reduces the expression of ER stress biomarkers in ER stress conditions compared with wild type (Lai et al., 2014). Genetic and functional analyses of an ire1 rhd3 high-order mutant compared with ire1 and rhd3 single mutants have indicated that RHD3 functions upstream of IRE1 (Lai et al., 2014). The evidence that Rnt2 and RHD3 contribute to ER stress responses are consistent with the idea that ER shape is correlated with ER function in plants. However, the differences in contribution of Rnt2 and RHD3 to ER stress suggest either the existence of species-specific mechanisms underlying ER stress responses (i.e., maize versus Arabidopsis) or the possibility that the ER-shaping machine does not equally contribute to ER function. The nature of the mechanisms underlying the role of Rnt2 in ER-phagy is an open question. ER-phagy requires severing of the ER tubules. Overexpression of plant reticulons leads to constrictions of ER tubules but not fragmentation. Therefore, even though it is possible that during ER-phagy reticulons accumulate at ER subdomains to modify tubule shape, the process of tubule severing would require additional machinery. In mammalian cells, human atlastin 2 (ATL2), a functional homolog of RHD3, is required for FAM134B-mediated ER-phagy (Liang et al., 2018), but a role of RHD3 in ER-phagy is yet to be reported. An increase in the total level of phospholipids and proteins has been reported in rhd3 alleles (Maneta-Peyret et al., 2014). By analogy with the reported functional interaction of ATL2 and FAM134B in ER-phagy in mammalian cells (Liang et al., 2018), a loss in RHD3 may be conducive to a missed clearance of ER membranes through ER-phagy in rhd3 and an increase in phospholipids.
CONCLUSIONS
Although any organelle of a cell is remarkable for their function and relevance to the life of the cell, the ER is a pervasive organelle that stands out for its combination of unique shape, functions, and ability to adapt to growth and environmental cues. In the last decade, important milestones in our understanding of the mechanisms underlying ER morphological and functional integrity have been reached. These include identification of ER shapers, description of the proteome that connects the ER with other membranes and controls the proteostasis and membrane abundance of this organelle, and insights into mechanisms that in several instances have assumed unique features in plants. Moving forward, it will be important to understand how the ER shapers and anchors are regulated during development and their role in facilitating responses to stress. Furthermore, it will be interesting to adopt other cell models to study the ER besides expanded cells (e.g., epidermal cells) at interphase or growing cells (e.g., root hairs) that are commonly adopted for the ease of to visualize the ER in light microscopy. For example, fascinating questions revolve around how the ER partitions in dividing plant cells undergoing mitosis and what the role and regulation of the ER shapers and cytoskeleton may be in the process. During plant cell division, the ER undergoes extensive reorganization (Gupton et al., 2006). During late cytokinesis, the phragmoplast, a scaffold for cell plate assembly, is formed prior to the generation of a new cell wall separating the two daughter cells. Live-cell imaging has shown that ER reorganization during mitosis is maintained mainly by MT (Gupton et al., 2006), tipping the relevance of the cytoskeleton in ER structural homeostasis away from actin. As mitosis progresses, the ER tubules are entrapped in the forming cell wall to give rise to PD (Hepler, 1982); however, it cannot be excluded that processes of severing of the ER membrane may occur to complete cell separation. Therefore, mechanistically, cell division may offer new unexpected insights in plant ER homeostasis.
ACKNOWLEDGMENTS
We apologize to those colleagues whose work was not included in this review due to space limitations. This work was funded primarily by the National Institutes of Health (GM136637), National Science Foundation (MCB1727362), the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy (award number DE-FG02-91ER20021), and AgBioResearch (MICL02598) to F.B.
REFERENCES
- An H, Ordureau A, Paulo JA, Shoemaker CJ, Denic V, and Harper JW (2019). TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol. Cell 74, 891–908.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Goksör M, and Sandelius AS (2007). Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J. Biol. Chem 282, 1170–1174. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Kumar M, and Wiener R (2020). Decrypting UFMylation: how proteins are modified with UFM1. Biomolecules 10, 1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton K, Mathur N, and Mathur J (2013). Simultaneous live-imaging of peroxisomes and the ER in plant cells suggests contiguity but no luminal continuity between the two organelles. Front. Physiol 4, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Sparkes I, Vanneste S, and Rosado A (2017). From shaping organelles to signalling platforms: the emerging functions of plant ER-PM contact sites. Curr. Opin. Plant Biol 40, 89–96. [DOI] [PubMed] [Google Scholar]
- Bhaskara RM, Grumati P, Garcia-Pardo J, Kalayil S, Covarrubias-Pinto A, Chen W, Kudryashev M, Dikic I, and Hummer G (2019). Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat. Commun 10, 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, and Hawes C (2002). Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14, 1293–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault ML, Petit JD, Immel F, Nicolas WJ, Glavier M, Brocard L, Gaston A, Fouché M, Hawkins TJ, Crowet J-M, et al. (2019). Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep 20, e47182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard JO, Xu M, Scarpin MR, Chatterjee S, Shemyakina EA, Goodman HM, and Zambryski P (2020). TOR dynamically regulates plant cell-cell transport. Proc. Natl. Acad. Sci. USA 117, 5049–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Renna L, Stefano G, and Brandizzi F (2016). SYP73 anchors the ER to the actin cytoskeleton for maintenance of ER integrity and streaming in Arabidopsis. Curr. Biol 26, 3245–3254. [DOI] [PubMed] [Google Scholar]
- Chen J, Stefano G, Brandizzi F, and Zheng H (2011). Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci 124, 2241–2252. [DOI] [PubMed] [Google Scholar]
- Chen Q, Xiao Y, Chai P, Zheng P, Teng J, and Chen J (2019). ATL3 is a tubular ER-Phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol 29, 846–855.e6. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yu F, and Xie Q (2020). Insights into endoplasmic reticulum-associated degradation in plants. New Phytol 226, 345–350. [DOI] [PubMed] [Google Scholar]
- Chen S, Cui Y, Parashar S, Novick PJ, and Ferro-Novick S (2018). ER-phagy requires Lnp1, a protein that stabilizes rearrangements of the ER network. Proc. Natl. Acad. Sci. USA 115, E6237–E6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Desai T, McNew JA, Gerard P, Novick PJ, and Ferro-Novick S (2015). Lunapark stabilizes nascent three-way junctions in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 112, 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, and Ferro-Novick S (2012). ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol 14, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, and Ferro-Novick S (2013). ER structure and function. Curr. Opin. Cell Biol 25, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chino H, Hatta T, Natsume T, and Mizushima N (2019). Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell 74, 909–921.e6. [DOI] [PubMed] [Google Scholar]
- Chino H, and Mizushima N (2020). ER-Phagy: quality control and turnover of endoplasmic reticulum. Trends Cell Biol 30, 384–398. [DOI] [PubMed] [Google Scholar]
- Cui Y, Parashar S, Zahoor M, Needham PG, Mari M, Zhu M, Chen S, Ho HC, Reggiori F, Farhan H, et al. (2019). A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 365, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, and Brandizzi F (2004). Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16, 1753–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E, Popelka H, and Klionsky DJ (2019). TEX264 is a major receptor for mammalian reticulophagy. Autophagy 15, 1677–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng RC, Halat LS, Livingston SJ, Sakai T, Motose H, and Wasteneys GO (2017). The ARM Domain of ARMADILLO-REPEAT kinesin 1 is not required for microtubule catastrophe but can negatively regulate Nima-RELATED kinase 6 in Arabidopsis thaliana. Plant Cell Physiol 58, 1350–1363. [DOI] [PubMed] [Google Scholar]
- Fox AR, Scochera F, Laloux T, Filik K, Degand H, Morsomme P, Alleva K, and Chaumont F (2020). Plasma membrane aquaporins interact with the endoplasmic reticulum resident VAP27 proteins at ER–PM contact sites and endocytic structures. New Phytol 228, 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Soldà T, et al. (2016). Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol 18, 1173–1184. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, and Benfey PN (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol 14, 1847–1851. [DOI] [PubMed] [Google Scholar]
- Galway ME, Heckman JW Jr., and Schiefelbein JW (1997). Growth and ultrastructure of Arabidopsis root hairs: the rhd3 mutation alters vacuole enlargement and tip growth. Planta 201, 209–218. [DOI] [PubMed] [Google Scholar]
- Gerakis Y, Quintero M, Li H, and Hetz C (2019). The UFMylation system in proteostasis and beyond. Trends Cell Biol 29, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice GL, and Nathan JA (2016). The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci 73, 3497–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing LR, Gao HT, and Sparkes I (2014). ER network dynamics are differentially controlled by myosins XI-K, XI-C, XI-E, XI-I, XI-1, and XI-2. Front. Plant Sci 5, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Hölper S, Mari M, Harwardt MI, Yan R, Muller S, Reggiori F, Heilemann M, and Dikic I (2017). Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife 6, e25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton SL, Collings DA, and Allen NS (2006). Endoplasmic reticulum targeted GFP reveals ER organization in tobacco NT-1 cells during cell division. Plant Physiol. Biochem 44, 95–105. [DOI] [PubMed] [Google Scholar]
- Hamada T, Ueda H, Kawase T, and Hara-Nishimura I (2014). Microtubules contribute to tubule elongation and anchoring of endoplasmic reticulum, resulting in high network complexity in Arabidopsis. Plant Physiol 166, 1869–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK (1982). Endoplasmic reticulum in the formation of the cell plate and plasmodesmata. Protoplasma 111, 121–133. [Google Scholar]
- Hetz C, Zhang K, and Kaufman RJ (2020). Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol 21, 421–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, and Rapoport TA (2016). Fusion of the endoplasmic reticulum by membrane-bound GTPases. Semin. Cell Dev. Biol 60, 105–111. [DOI] [PubMed] [Google Scholar]
- Hu S, Ye H, Cui Y, and Jiang L (2020). AtSec62 is critical for plant development and is involved in ER-phagy in Arabidopsis thaliana. J. Integr. Plant Biol 62, 181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Tamura K, Ueda H, Ito Y, Nakano A, Hara-Nishimura I, and Shimada T (2018). Synaptotagmin-associated endoplasmic reticulum-plasma membrane contact sites are localized to immobile ER tubules. Plant Physiol 178, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, and Lamark T (2020). Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol 432, 80–103. [DOI] [PubMed] [Google Scholar]
- Jouhet J (2013). Importance of the hexagonal lipid phase in biological membrane organization. Front. Plant Sci 4, 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiho H, Yamamoto Y, and Sakisaka T (2019). CAND1 regulates lunapark for the proper tubular network of the endoplasmic reticulum. Sci. Rep 9, 13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. [DOI] [PubMed] [Google Scholar]
- Kim H, Kwon H, Kim S, Kim MK, Botella MA, Yun HS, and Kwon C (2016). Synaptotagmin 1 negatively controls the two distinct immune secretory pathways to powdery mildew fungi in Arabidopsis. Plant Cell Physiol 57, 1133–1141. [DOI] [PubMed] [Google Scholar]
- Klopfenstein DR, Kappeler F, and Hauri HP (1998). A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J 17, 6168–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, and Hauri HP (2001). Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J. Cell Biol 153, 1287–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K, Wu S, MacRae-Crerar A, and Gallagher KL (2011). An essential protein that interacts with endosomes and promotes movement of the SHORT-ROOT transcription factor. Curr. Biol 21, 1559–1564. [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Botchway SW, Slade SE, Knox K, Frigerio L, Oparka K, and Hawes C (2015). Reticulomics: protein-protein interaction studies with two plasmodesmata-localized Reticulon family proteins identify binding partners enriched at plasmodesmata, endoplasmic reticulum, and the plasma membrane. Plant Physiol 169, 1933–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, and Brandizzi F (2020). The plant endoplasmic reticulum: an organized chaos of tubules and sheets with multiple functions. J. Microsc 280, 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Breeze E, Pain C, Tolmie F, Frigerio L, and Hawes C (2018). Arabidopsis Lunapark proteins are involved in ER cisternae formation. New Phytol 219, 990–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YS, Stefano G, and Brandizzi F (2014). ER stress signaling requires RHD3, a functionally conserved ER-shaping GTPase. J. Cell Sci 127, 3227–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YS, Stefano G, Zemelis-Durfee S, Ruberti C, Gibbons L, and Brandizzi F (2018). Systemic signaling contributes to the unfolded protein response of the plant endoplasmic reticulum. Nat. Commun 9, 3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Vanneste S, Pérez-Sancho J, Benitez-Fuente F, Strelau M, Macho AP, Botella MA, Friml J, and Rosado A (2019). Ionic stress enhances ER-PM connectivity via phosphoinositide-associated SYT1 contact site expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 116, 1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sparkes I, Gattolin S, Dzimitrowicz N, Roberts LM, Hawes C, and Frigerio L (2013). An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol 197, 481–489. [DOI] [PubMed] [Google Scholar]
- Levy A, Zheng JY, and Lazarowitz SG (2015). Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol 25, 2018–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, and Lazarowitz SG (2010). Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA 107, 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Ahmed S, and Corn JE (2018). Atlastins remodel the endoplasmic reticulum for selective autophagy. J. Cell Biol 217, 3354–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneta-Peyret L, Lai YS, Stefano G, Fouillen L, Brandizzi F, and Moreau P (2014). Phospholipid biosynthesis increases in RHD3-defective mutants. Plant Signal. Behav 9, e29657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HE, Lee EK, van Bezouwen LS, Ross B, Rosado A, and Samuels AL (2017). Multiscale structural analysis of plant ER-PM contact sites. Plant Cell Physiol 58, 478–484. [DOI] [PubMed] [Google Scholar]
- Mehrshahi P, Stefano G, Andaloro JM, Brandizzi F, Froehlich JE, and DellaPenna D (2013). Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc. Natl. Acad. Sci. USA 110, 12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, and Nakatogawa H (2015). Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362. [DOI] [PubMed] [Google Scholar]
- Muschalik N, and Munro S (2018). Golgins. Curr. Biol 28, R374–R376. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, and Nebenführ A (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nicolas WJ, Grison MS, and Bayer EM (2017a). Shaping intercellular channels of plasmodesmata: the structure-to-function missing link. J. Exp. Bot 69, 91–103. [DOI] [PubMed] [Google Scholar]
- Nicolas WJ, Grison MS, Trépout S, Gaston A, Fouché M, Cordelières FP, Oparka K, Tilsner J, Brocard L, and Bayer EM (2017b). Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat. Plants 3, 17082. [DOI] [PubMed] [Google Scholar]
- Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, Pasolli HA, Harvey K, Hess HF, Betzig E, et al. (2016). Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nthiga TM, Kumar Shrestha B, Sjøttem E, Bruun JA, Bowitz Larsen K, Bhujabal Z, Lamark T, and Johansen T (2020a). CALCOCO1 acts with VAMP-associated proteins to mediate ER-phagy. EMBO J 39, e103649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nthiga TM, Shrestha BK, Lamark T, and Johansen T (2020b). CALCOCO1 is a soluble reticulophagy receptor. Autophagy 16, 1729–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder A, Sparkes IA, Botchway SW, Ward A, Ketelaar T, de Ruijter N, and Hawes C (2017). Stacks off tracks: a role for the golgin AtCASP in plant endoplasmic reticulum-Golgi apparatus tethering. J. Exp. Bot 68, 3339–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Cantizano N, Ko DK, Angelos E, Pu Y, and Brandizzi F (2020). Functional diversification of ER stress responses in Arabidopsis. Trends Biochem. Sci 45, 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Prokhnevsky AI, and Dolja VV (2010). Class XI myosins are required for development, cell expansion, and F-actin organization in Arabidopsis. Plant Cell 22, 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sancho J, Vanneste S, Lee E, McFarlane HE, Esteban Del Valle A, Valpuesta V, Friml J, Botella MA, and Rosado A (2015). The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol 168, 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Sun J, and Zheng H (2016). A GTPase-dependent fine ER is required for localized secretion in polarized growth of root Hairs1. Plant Physiol 171, 1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quader H, and Schnepf E (1986). Endoplasmic reticulum and cytoplasmic streaming: fluorescence microscopical observations in adaxial epidermis cells of onion bulb scales. Protoplasma 131, 250–252. [Google Scholar]
- Ridge RW, Uozumi Y, Plazinski J, Hurley UA, and Williamson RE (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol 40, 1253–1261. [DOI] [PubMed] [Google Scholar]
- Sager RE, and Lee JY (2018). Plasmodesmata at a glance. J. Cell Sci 131. [DOI] [PubMed] [Google Scholar]
- Schapire AL, Voigt B, Jasik J, Rosado A, Lopez-Cobollo R, Menzel D, Salinas J, Mancuso S, Valpuesta V, Baluska F, and Botella MA (2008). Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20, 3374–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder LK, Barentine AES, Merta H, Schweighofer S, Zhang Y, Baddeley D, Bewersdorf J, and Bahmanyar S (2019). Dynamic nanoscale morphology of the ER surveyed by STED microscopy. J. Cell Biol 218, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S (2020). Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J. Cell Sci 133. [DOI] [PubMed] [Google Scholar]
- Shemesh T, Klemm RW, Romano FB, Wang S, Vaughan J, Zhuang X, Tukachinsky H, Kozlov MM, and Rapoport TA (2014). A model for the generation and interconversion of ER morphologies. Proc. Natl. Acad. Sci. USA 111, E5243–E5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, and Rapoport TA (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, and Voeltz GK (2008). The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J. Biol. Chem 283, 18892–18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siao W, Wang P, Voigt B, Hussey PJ, and Baluska F (2016). Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27-1-enriched endoplasmic reticulum-plasma membrane contact sites. J. Exp. Bot 67, 6161–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieńko K, Poormassalehgoo A, Yamada K, and Goto-Yamada S (2020). Microautophagy in plants: consideration of its molecular mechanism. Cells 9, 887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, and Wilkinson S (2018). CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell 44, 217–232.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, and Griffing L (2009a). Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21, 3937–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Frigerio L, Tolley N, and Hawes C (2009b). The plant endoplasmic reticulum: a cell-wide web. Biochem. J 423, 145–155. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Ketelaar T, de Ruijter NC, and Hawes C (2009c). Grab a Golgi: laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic 10, 567–571. [DOI] [PubMed] [Google Scholar]
- Spiegelman Z, Lee CM, and Gallagher KL (2018). KinG is a plant-specific kinesin that regulates both intra- and intercellular movement of SHORT-ROOT. Plant Physiol 176, 392–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman Z, Wu S, and Gallagher KL (2019). A role for the endoplasmic reticulum in the cell-to-cell movement of SHORT-ROOT. Protoplasma 256, 1455–1459. [DOI] [PubMed] [Google Scholar]
- Staehelin LA (1997). The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J 11, 1151–1165. [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, and Brandizzi F (2014). The endoplasmic reticulum exerts control over organelle streaming during cell expansion. J. Cell Sci 127, 947–953. [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, Lai Y, Slabaugh E, Mannino N, Buono RA, Otegui MS, and Brandizzi F (2015). ER network homeostasis is critical for plant endosome streaming and endocytosis. Cell Discov 1, 15033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G, Renna L, Moss T, McNew JA, and Brandizzi F (2012). In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J 69, 957–966. [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, Wormsbaecher C, Gamble J, Zienkiewicz K, and Brandizzi F (2018). Plant endocytosis requires the ER membrane-anchored proteins VAP27-1 and VAP27-3. Cell Rep 23, 2299–2307. [DOI] [PubMed] [Google Scholar]
- Stephani M, Picchianti L, Gajic A, Beveridge R, Skarwan E, Sanchez de Medina Hernandez V, Mohseni A, Clavel M, Zeng Y, Naumann C, et al. (2020). A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife 9, e58396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ernst A, and Dikic I (2014). Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol 16, 495–501. [DOI] [PubMed] [Google Scholar]
- Sun J, Movahed N, and Zheng H (2020a). LUNAPARK is an E3 ligase that mediates degradation of ROOT HAIR DEFECTIVE3 to maintain a Tubular ER Network in Arabidopsis. Plant Cell 32, 2964–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang M, Qi X, Doyle C, and Zheng H (2020b). Armadillo-repeat kinesin1 interacts with Arabidopsis atlastin RHD3 to move ER with plus-end of microtubules. Nat. Commun 11, 5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, and Zheng H (2018). Efficient ER fusion requires a dimerization and a C-terminal tail mediated membrane anchoring of RHD3. Plant Physiol 176, 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J, Nicolas W, Rosado A, and Bayer EM (2016). Staying tight: plasmodesmal membrane contact sites and the control of cell-to-cell connectivity in plants. Annu. Rev. Plant Biol 67, 337–364. [DOI] [PubMed] [Google Scholar]
- Tolley N, Sparkes I, Craddock CP, Eastmond PJ, Runions J, Hawes C, and Frigerio L (2010). Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J 64, 411–418. [DOI] [PubMed] [Google Scholar]
- Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, and Frigerio L (2008). Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9, 94–102. [DOI] [PubMed] [Google Scholar]
- Uchiyama A, Shimada-Beltran H, Levy A, Zheng JY, Javia PA, and Lazarowitz SG (2014). The Arabidopsis synaptotagmin SYTA regulates the cellto-cell movement of diverse plant viruses. Front. Plant Sci 5, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Ohta N, Kimori Y, Uchida T, Shimada T, Tamura K, and Hara-Nishimura I (2018). Endoplasmic reticulum (ER) membrane proteins (LUNAPARKs) are required for proper configuration of the cortical ER network in plant cells. Plant Cell Physiol 59, 1931–1941. [DOI] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kutsuna N, Shimada T, Tamura K, Shimmen T, Hasezawa S, Dolja VV, and Hara-Nishimura I (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107, 6894–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yokota E, Kuwata K, Kutsuna N, Mano S, Shimada T, Tamura K, Stefano G, Fukao Y, Brandizzi F, et al. (2016). Phosphorylation of the C terminus of RHD3 has a critical role in homotypic ER membrane fusion in Arabidopsis. Plant Physiol 170, 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bruaene N, Joss G, and Van Oostveldt P (2004). Reorganization and in vivo dynamics of microtubules during Arabidopsis root hair development. Plant Physiol 136, 3905–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M, Neubert C, Fink F, Lupanga U, Scheuring D, Boutté Y, Frescatada-Rosa M, Wolfenstetter S, et al. (2013). The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell 25, 3434–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, and Rapoport TA (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. [DOI] [PubMed] [Google Scholar]
- Wang N, and Rapoport TA (2019). Reconstituting the reticular ER network - mechanistic implications and open questions. J. Cell Sci 132, jcs227611. [DOI] [PubMed] [Google Scholar]
- Wang P, Hawes C, and Hussey PJ (2017). Plant endoplasmic reticulum-plasma membrane contact sites. Trends Plant Sci 22, 289–297. [DOI] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Richardson C, Cummins I, Deeks MJ, Sparkes I, Hawes C, and Hussey PJ (2014). The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr. Biol 24, 1397–1405. [DOI] [PubMed] [Google Scholar]
- Wang P, and Hussey PJ (2017). NETWORKED 3B: a novel protein in the actin cytoskeleton-endoplasmic reticulum interaction. J. Exp. Bot 68, 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Pleskot R, Zang J, Winkler J, Wang J, Yperman K, Zhang T, Wang K, Gong J, Guan Y, et al. (2019). Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun 10, 5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Richardson C, Hawkins TJ, Sparkes I, Hawes C, and Hussey PJ (2016). Plant VAP27 proteins: domain characterization, intracellular localization and role in plant development. New Phytol 210, 1311–1326. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer CM, and Salmon ED (1998). Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol 8, 798–806. [DOI] [PubMed] [Google Scholar]
- White RR, Lin C, Leaves I, Castro IG, Metz J, Bateman BC, Botchway SW, Ward AD, Ashwin P, and Sparkes I (2020). Miro2 tethers the ER to mitochondria to promote mitochondrial fusion in tobacco leaf epidermal cells. Commun. Biol 3, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Deom CM, Beachy RN, and Lucas WJ (1989). Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Science 246, 377–379. [DOI] [PubMed] [Google Scholar]
- Wu S, and Gallagher KL (2012). Transcription factors on the move. Curr. Opin. Plant Biol 15, 645–651. [DOI] [PubMed] [Google Scholar]
- Wu Z, Liang F, Hong B, Young JC, Sussman MR, Harper JF, and Sze H (2002). An endoplasmic reticulum-bound Ca(2+)/Mn(2+) pump, ECA1, supports plant growth and confers tolerance to Mn(2+) stress. Plant Physiol 130, 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Cheval C, Laohavisit A, Hocking B, Chiasson D, Olsson TSG, Shirasu K, Faulkner C, and Gilliham M (2017). A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol 215, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Cornish AJ, and Benning C (2008). Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 20, 2190–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YH, and Noda T (2020). Autophagosome formation in relation to the endoplasmic reticulum. J. Biomed. Sci 27, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Kawamura Y, Minami A, and Uemura M (2008). Calciumdependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20, 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, and Bassham DC (2015). New insight into the mechanism and function of autophagy in plant cells. Int. Rev. Cell Mol. Biol 320, 1–40. [DOI] [PubMed] [Google Scholar]
- Zaffagnini G, and Martens S (2016). Mechanisms of selective autophagy. J. Mol. Biol 428, 1714–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang J, Klemm S, Pain C, Duckney P, Bao Z, Stamm G, Kriechbaumer V, Bürstenbinder K, Hussey PJ, and Wang P (2021). A novel plant actin-microtubule bridging complex regulates cytoskeletal and ER structure at ER-PM contact sites. Curr. Biol 10.1016/j.cub.2020.12.009. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wu F, Shi J, Zhu Y, Zhu Z, Gong Q, and Hu J (2013). ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol 163, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ding X, Marshall RS, Paez-Valencia J, Lacey P, Vierstra RD, and Otegui MS (2020). Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. eLife 9, e51918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Zou CX, Liu XM, Jiang ZD, Yu ZQ, Suo F, Du TY, Dong MQ, He W, and Du LL (2020). A UPR-induced soluble ER-Phagy receptor acts with VAPs to confer ER stress resistance. Mol. Cell 79, 963–977.e3. [DOI] [PubMed] [Google Scholar]


