Abstract
Introduction
Integrase strand transfer inhibitors (INSTIs) are important drugs that are currently used as the first line treatment for HIV-1 patients. The aim of this study was to characterize HIV-1 INSTI mutations among ART-naive patients in Beijing from 2019–2021.
Methods
865 ART-naive patients were enrolled in this study between January 2019 and June 2021 in Beijing. The amplification of the entire pol gene containing the reverse transcriptase, protease and integrase regions was performed using a validated In-house SBS method. HIV-1 subtypes and circulating recombinant forms (CRFs) were determined using the COMET online tool (http://comet.retrovirology.lu). Stanford HIV-1 drug resistance database (HIVdb version 8.9) was used to analyze the mutations.
Results
865 HIV-1 pol sequences were successfully amplified and sequenced. Among them, no major INSTI-related mutations were identified, but 12 polymorphic accessory mutations were found. Two patients have E138A and G163R mutations respectively and both could cause low-level resistance to RAL and EVG. Furthermore, one patient having S230R mutation resulted in low-level resistance to RAL, EVG, DTG and BIC.
Conclusion
The prevalence of INSTIs mutations remains low, which demonstrated that INSTIs have good applicability currently in our city. Nevertheless, it is very important to monitor the INSTI-related mutations in Beijing.
Keywords: HIV-1, genotype, integrase strand transfer inhibitor, pre-treatment drug resistance mutation, Beijing
Introduction
Highly active antiretroviral therapy (HAART) can effectively inhibit human immunodeficiency virus (HIV) replication and achieve the goal of reducing AIDS-related morbidity and mortality.1 However, with the widespread use of HAART, the prevalence of transmitted drug resistance (TDR) mutations has gradually increased.2,3 According to the 2017 and 2019 World Health Organization (WHO) drug resistance mutation reports, the prevalence of TDR in most countries have exceeded the warning line of 5%, and even 67% of the countries have exceeded 10%.4,5 TDR mutations could reduce the choice of first line antiretroviral drugs, and it is also associated with increasing risk of virologic failure (VF).6
Integrase strand transfer inhibitors (INSTIs) are important drugs that are currently used as the first line treatment for newly diagnosed HIV-1 patients.7 In 2007, the first INSTI raltegravir (RAL) was approved by FDA of the United States and then the treatment of HIV entered a new era.8 The first generation INSTIs include raltegravir (RAL) and elvitegravir (EVG), and the second generation INSTIs include dolutegravir (DTG), bictegravir (BIC) and cabotegravir (CAB) which have better safety, tolerability, and higher genetic barriers to the emergence of drug resistance.9–11
With the widespread use of INSTIs, the INSTIs-related drug resistance mutations have gradually emerged. Previously published studies have shown that there were IN major resistance mutations in newly diagnosed HIV-1 patients in Europe, Canada, and the United States.12–14 In China, IN major resistance mutations have also been discovered.15,16 A study including 531 HIV-infected patients in Yunnan demonstrated that 9 patients had IN major resistance mutations.15 Furthermore, a study including 252 patients in Jiangsu illustrated that 4 patients had IN major resistance mutations.16 However, a study in Guangdong including 827 integrase sequences revealed that no patients have IN major resistance mutations but 12 patients had IN-related polymorphic accessory mutations.17
Conventional Sanger genotype resistance testing cannot detect low-frequency INSTI-related resistance mutations, so the prevalence of TDR to INSTIs was low in treatment naive patients. Next-generation sequencing (NGS) has improved the ability of quantitative identification of minority resistant variants (MRVs).18,19 However, few studies have focused on the application of NGS technology on TDR mutations of INSTIs. Therefore, the aim of this study was to monitor the prevalence of INSTIs-related drug resistance mutations among ART-naive patients in Beijing and to explore the Clinical implications of NGS for detecting MRVs.
Methods
Study Population and Data Collection
865 ART-naive patients were enrolled in this study between January 1, 2019 and June 1, 2021 in Beijing. The patients came from Ditan Hospital and Youan Hospital both Affiliated to Capital Medical University and were approved by the ethics committee. Demographic and clinical data including age, gender, HIV transmission route, HIV RNA viral load and CD4+ cell count at baseline were collected from the national free antiretroviral treatment database.
RNA Extraction, PCR, and Sanger Sequencing
Viral RNA was extracted from 200ul of plasma samples using the Viral RNA Extraction Kit (Guangzhou Life Technologies Daan Diagnostics Co.,Ltd) according to the manufacturer’s instructions. The amplification of the entire pol gene containing the reverse transcriptase, protease and integrase regions was performed by a validated In-house SBS method (Guangzhou Life Technologies Daan Diagnostics Co.,Ltd). The positive PCR products were purified and the PCR products were sequenced using the 3500XL DX genetic analyzer.
Identification of HIV-1 Genotype and Drug Resistance Mutations
The BIOEDIT version 7 was used to align and edit HIV-1 pol sequences, together with reference sequences of different subtypes and circulating recombinant forms (CRFs). Reference sequences were downloaded from the Los Alamos HIV database (http://www.hiv.lanl.gov). HIV-1 subtypes and CRFs were determined using the COMET online tool (http://comet.retrovirology.lu). Stanford HIV-1 drug resistance database (HIVdb version 8.9) was used to analyze the mutations. In our study, sequences with low-level category of resistance or greater were defined as having drug resistance.
Gene Evolution Analysis
And these IN sequences from 865 HIV-1 treatment naive patients were uploaded to the Gen Bank website (https://www.ncbi.nlm.nih.gov/genbank/; Accession numbers: OM329078 - OM329942). The reference sequences of other subtypes were downloaded from Gen Bank. Phylogenetic tree was constructed using MEGA (Version 7.0, USA) software and the between group mean distance as well as bootstrap-ping test were performed.
Vela Next Generation Sequence
The HIV-1 resistance test based on Vela NGS was performed according to the manufacturer’s instructions (Vela-Dx, Germany). Plasma RNA was extracted from 730uL plasma sample and library preparation was performed on the Sentosa SX 101 workstation. The amplification of the entire pol gene was performed using one-step reverse transcription PCR. Sequencing was performed on the Sentosa SQ 301 sequencer. Preliminary analysis on the original sequencing data was performed with Sentosa® SQ Suite software, and report generation was performed by the Sentosa SQ reporter server.
Data Analysis
Continuous variables were presented as medians with interquartile ranges (IQRs) and categorical variables were expressed as number (%). The chi-square test was used to compare different groups of patients at enrollment. The above-mentioned analyses were performed using Statistical Package for the Social Sciences software (SPSS 22.0).
Ethics and Consent
In our study, all included patients provided written informed consent for the use of clinical data. This study had been approved by the ethics committee of Beijing Ditan Hospital of Capital Medical University (Approval number: 2019–037-002) and complied with the Declaration of Helsinki.
Results
Patient Characteristics
Overall, 865 treatment-naive patients with HIV infection were included in this study. 42.54% (368/865) of the patients were diagnosed in 2019, 36.76% (318/865) in 2020 and 18.61% (161/865) in 2021. 96.0% (830/865) of the patients were male and 4% (35/865) were female. The median age of the participants was 34 years old (IQR:16–79 years) at HIV diagnosis. 78.72% (681/865) of the participants were infected through homosexual transmission. The median HIV-1 RNA viral load and CD4+ cell count at baseline were 245,970 copies/mL (IQR: 13894–164049 copies/mL) and 273 cells/ul (IQR: 96–396 cells/ul) respectively. Baseline characteristics were summarized in Table 1.
Table 1.
Clinical Characteristic of Patients at Enrollment
| Variable | 2019(n=161) | 2020(n=386) | 2021(n=318) | P |
|---|---|---|---|---|
| Age (years) | ||||
| <30 | 77(47.8%) | 167(43.3%) | 126(39.6%) | 0.152 |
| 30–50 | 71(44.1%) | 172(44.6%) | 164(51.6%) | |
| >50 | 13(8.1%) | 47(12.1%) | 28(8.8%) | |
| Sex | ||||
| Male | 156(96.9%) | 368(95.3%) | 306(96.2%) | 0.668 |
| Female | 5(3.1%) | 18(4.7%) | 12(3.8%) | |
| HIV transmission route | ||||
| Homosexual | 140(87.0%) | 270(69.9%) | 271(85.2%) | <0.01 |
| Heterosexual | 9(5.6%) | 44(11.4%) | 29(9.1%) | |
| Intravenous drug users | 0(0%) | 1(0.3%) | 0(0%) | |
| Former plasma donors | 1(0.6%) | 3(0.8%) | 0(0%) | |
| Unkown | 11(6.8%) | 68(17.6%) | 18(5.7%) | |
| CD4 cell count at baseline(cells/ul) | ||||
| <50 | 11(6.8%) | 90(23.4%) | 50(15.7%) | <0.01 |
| 50–199 | 20(12.4%) | 75(19.4%) | 65(20.4%) | |
| 200–349 | 52(32.3%) | 102(26.4%) | 97(30.5%) | |
| ≥350 | 77(47.8%) | 105(27.2%) | 89(28.0%) | |
| Unkown | 1(0.7%) | 14(3.6%) | 17(5.4%) | |
| CD4 count (cells/uL, IQR) | 349(317–380) | 275(240–285) | 253(221–285) | <0.01 |
| HIV-1 RNA at baseline (copies/mL) | ||||
| ≤100,000 | 115(71.4%) | 228(59.1%) | 201(63.2%) | 0.002 |
| >100,000 | 46(28.6%) | 145(37.6%) | 98(30.8%) | |
| Unkown | 0(0%) | 13(3.3%) | 19(6.0%) |
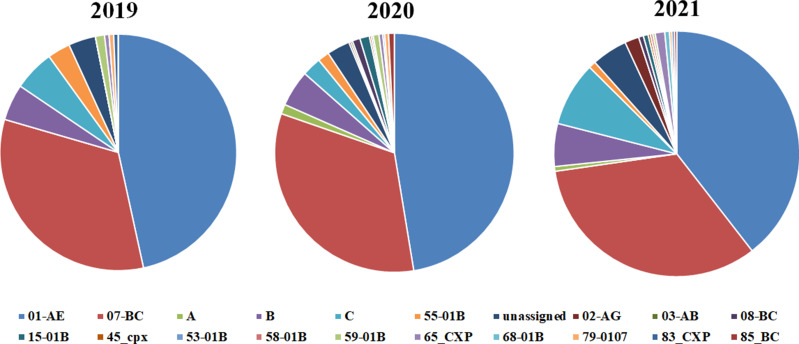
A total of 865 HIV-1 pol sequences were successfully amplified and sequenced. CRF01-AE was the genotype occurred most frequently with the proportion of 42.1%, followed by CRF07_BC (33.8%) and B (4.7%). Furthermore, the most subtype every year was CRF01-AE with the proportion of 39% (71/161), 46.1% (178/386), and 36.2% (115/ 318) in 2019, 2020, and 2021 respectively. The distribution of the subtype was shown in Figure 1. To determine the evolutionary relationship of HIV strains, phylogenetic tree analysis was performed using IN sequences (Supplementary Figure 1).
Figure 1.
Distribution of HIV-1 genotypes among 865 HIV-1 treatment naïve patients.
Transmitted Drug Resistance Mutation to INSTIs, NRTIs, NRTIs, and PIs
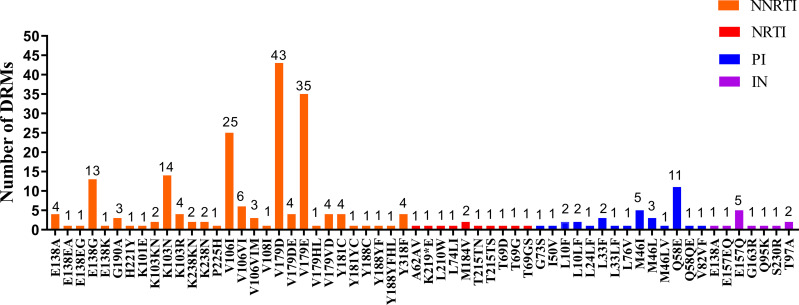
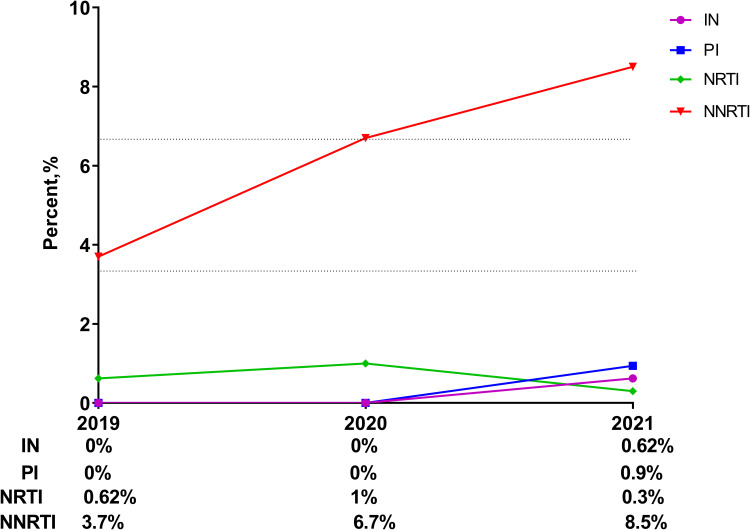
A total of 12 polymorphic accessory mutations were detected, including E157Q (0.58%, 5/865), T97A (0.23%, 2/865), E138A (0.12%, 1/865), E157EQ (0.12%, 1/865), G163R (0.12%, 1 /865), Q95K (0.12%, 1/865), and S230R (0.12%, 1/865) (as shown in Figure 2). Among 12 IN-related polymorphic accessory mutations, only E138A, S230R and G163R mutations cause low-level resistance to INSTIs. Two patients have E138A and G163R mutations respectively and either of the two lead to low-level resistance to RAL and EVG. Furthermore, one patient having S230R mutation resulted in low-level resistance to RAL, EVG, DTG and BIC (as shown in Table 2). The prevalence of TDR to INSTIs was 0.62% in 2019, 0% in 2020, and 0.62% in 2021 (as shown in Figure 3).
Figure 2.
Frequency of NRTI, NNRTI, PI and IN mutations among 865 HIV-1 treatment naïve patients.
Table 2.
Information of Samples Who Were Detected with in Resistance Mutations
| Sample ID | Year | Age | Gender | Transmission Route | Subtype | Viral Load | CD4 | Mutation | Level of Drug Resistance | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAB | BIC | DTG | EVG | RAL | |||||||||
| Patient 1 | 2019 | 54 | F | Homosexual | CRF01_AE | 20157 | 133 | E157Q | S | S | S | PL | PL |
| Patient 2 | 2019 | 32 | M | Homosexual | CRF01_AE | 11975 | 346 | E138A | PL | PL | PL | L | L |
| Patient 3 | 2020 | 25 | M | Homosexual | CRF07_BC | 331048 | 280 | T97A | S | S | S | PL | PL |
| Patient 4 | 2020 | 27 | M | Homosexual | B | 147538 | 11 | E157Q | S | S | S | PL | PL |
| Patient 5 | 2021 | 26 | M | Heterosexual | CRF01_AE | 65227 | 261 | Q95K | S | S | S | PL | PL |
| Patient 6 | 2021 | 42 | M | Homosexual | unassigned_1 | 87,194 | 12 | E157Q | S | S | S | PL | PL |
| Patient 7 | 2021 | 52 | M | Homosexual | CRF02_AG | 1334184 | 11 | G163R | S | S | S | L | L |
| Patient 8 | 2021 | 48 | F | Homosexual | CRF01_AE | 14007 | 757 | T97A | S | S | S | PL | PL |
| Patient 9 | 2021 | 19 | M | Homosexual | C | 145444 | 203 | E157Q | S | S | S | PL | PL |
| Patient 10 | 2021 | 38 | M | Homosexual | CRF01_AE | 195967 | 15 | E157Q | S | S | S | PL | PL |
| Patient 11 | 2021 | 31 | M | Homosexual | CRF07_BC | 91862 | 217 | S230R | PL | L | L | L | L |
| Patient 12 | 2021 | 35 | M | Homosexual | CRF01_AE | 47816 | 141 | E157EQ | S | S | S | PL | PL |
Abbreviations: F, Female; M, Male; CAB, Cabotegravir; BIC, Bictegravir; DTG, Dolutegravir; EVG, Elvitegravir; RAL, Raltegravir; S, Susceptible; PL, Potential low-level resistance; L, low-level resistance.
Figure 3.
% of all categories of DRMs in different years.
The most common non-nucleoside reverse transcriptase inhibitors (NNRTIs) associated mutation was V179D (4.97%, 43/865), followed by V179E (4.05%, 35/865) and V106I (2.89%, 25/865). M184V (0.23%, 2/865) was the most common nucleoside reverse transcriptase inhibitors (NRTIs) related mutation followed by T69D (0.12%, 1/865) and T69G (0.12%, 1/865). Additionally, the most common protease inhibitors associated mutation was Q58E (0.23%, 11/865), followed by M46I (4.05%, 5/865) and L33F (2.89%, 3/865)(as shown in Figure 2). The prevalence of TDR to NNRTIs was 3.7% in 2019, 6.7% in 2020, and 8.5% in 2021; the prevalence of TDR to NRTIs was 0.62% in 2019, 1% in 2020, and 0.3% in 2021; the prevalence of TDR to PIs was 0% in 2019, 0% in 2020, and 0.9% in 2021(as shown in Figure 3).
Comparing Mutations Detected by Sanger Sequencing and NGS
NGS was performed on 12 patients with IN-related mutations, and 12 IN-related mutations were successfully detected by NGS at the same time. NGS was more sensitive in detecting low-frequency mutations and a total of 4 mutations were detected by NGS but missed by Sanger sequencing which including M184V (1.31%), K65E (3.72%), E138G (1.21%), and Y188C (1.04%). None of the three patients with low frequency RT mutations used 2NRTIs + 1NNRTI experiencing VF by week 24. The explanation of NGS results was shown in Figure 4.
Figure 4.
Interpretation results of NGS on drug resistance.
Clinical Characteristics of Patients with Integrase Gene Mutation
Among the 12 patients with IN-related mutations, HIV-1 genotypes were identified: CRF01_AE (50%, 6/12), CRF07_BC (17%, 2/12), CRF02_AG (8.3%, 1/12), B (8.3%, 1/12), C (8.3%, 1/12) and other (8.3%, 1/12). Of these patients, 83.3% (10/12) were male. The dominant transmission route was men who have sex with men 83.3% (10/12). The median age of the patients was 36 years (IQR: 26–46 years). The median HIV-1 RNA viral load at baseline were 207,701 copies/mL (IQR: 27071–183859 copies/mL) and the CD4+ cell count at baseline were 198 cells/ul (IQR: 12–275 cells/ul). The clinical characteristics of patients with IN-related mutations were shown in Table 2.
Discussion
TDRM has become an important threat to HIV treatment. In our study, there were 12 IN-related polymorphic accessory mutations among newly diagnosed HIV-1 patients in Beijing, which emphasized the importance of monitoring INSTI-related mutations. However, only 3 patients had IN-related drug resistance mutations lead to low-level resistance to INSTIs. The prevalence of TDR to INSTI was 0.34% (3/865) which suggests that INSTI have good applicability currently and the use of INSTI in our city is still at the preliminary stage.
Consistent with previous research, no major INSTI mutations were identified among ART-naive individuals in our city.20 3 IN-related drug resistance mutations were E138A, S230R and G163R. E138A mutation can cause low-level resistance to RAL and EVG. When it existed together with Q148 mutation, it will result in high-level resistance to RAL and EVG and intermediate-level resistance to DTG and BIC.21 S230R mutation can lead to potential resistance to CAB and low-level resistance to BIC, DTG, RAL and EVG.21 G163R mutation was also reported in one study in China, which could cause low-level resistance to RAL and EVG.17
With the widespread use of HARRT, the prevalence of TDR has increased in recent years.22 In our study, the overall prevalence of TDR was 4.94%, 7.7% and 10.36% in 2019, 2020 and 2021 respectively. Compared with the previous studies, the prevalence of TDR in 2020 and 2021 were both >5% in our study, which highlight the importance of genotyping resistance testing when HIV-1 patients starting antiviral treatment.23,24 Furthermore, the prevalence of TDR to NNRTIs accounts for high percentage, while the prevalence of TDR to INSTIs, NRTIs and PIs accounts for low percentage. Possible explanations for this finding include NNRTI-based regimens having low genetic barrier to resistance, and a single point mutation in the reverse transcriptase enzyme is often associated with the development of high-level resistance.25,26 The above results illustrated that we could choose the 2NRTIs+1INSTI as the first-line treatment option when genotyping resistance testing is not available.
NGS was successfully performed on 12 samples containing IN-related polymorphic accessory mutations and 12 IN-related mutations were successfully detected by NGS at the same time. Meanwhile, consistent with previous studies, NGS can detect more low-frequency mutations that are missed by Sanger sequencing.18,27 But the clinical consequences of low-frequency mutations detected by NGS are still controversial. Studies have shown that NNRTI-related low-frequency resistance mutations are closely related to VF.28,29 However, in this study, 2 patients with NNRTI-related low frequency mutations started NNRTI-based antiretroviral therapy and none experienced VF by week 24. Of course, our sample size is too small to explain the problem.
Our study has some limitations. First of all, this study adopted a random sampling method, rather than all treatment naive patients in Beijing. Secondly, Because of economic constraints, vast majority of patients in this study used Sanger sequencing to identify drug-resistant mutations, the possibility of under-estimating the prevalence of minor resistance-associated variants by NGS still cannot be totally excluded given the small number of patients who underwent NGS studies.
Conclusion
The overall prevalence of INSTI mutations remains low, which suggests that INSTI have good applicability currently and the use of INSTI in our city is still at the preliminary stage. Nevertheless, there were 12 IN-related polymorphic accessory mutations among newly diagnosed HIV-1 patients in Beijing, which emphasized the importance of monitoring INSTI-related polymorphic accessory mutations in Beijing.
Acknowledgments
We would like to thank Mr. Ruolei Xin of Beijing CDC for his participation. We are grateful to the Mr Xin for agreeing to help us make a phylogenetic tree in this study.
Funding Statement
This work was funded in part by Beijing Municipal Administration of Hospitals’ Ascent Plan (DFL20191802) and Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX202126).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Sterne JAC, Hernán MA, Ledergerber B, et al. Long-term effectiveness of potent antiretroviral therapy in preventing AIDS and death: a prospective cohort study. Lancet. 2005;366(9483):378–384. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, Zhang M, Shi CX, et al. Mortality and virological failure among HIV-infected people who inject drugs on antiretroviral treatment in China: an observational cohort study. Drug Alcohol Depend. 2017;170:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Godfrey C, Bobkova M, Boucher C, et al. Regional challenges in the prevention of human immunodeficiency virus drug resistance. J Infect Dis. 2017;216(suppl_9):S816–S819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. HIV Drug Resistance Report 2017. Geneva: World Health Organization; 2017. [Google Scholar]
- 5.World Health Organization. HIV Drug Resistance Report 2019. Geneva: World Health Organization; 2019. [Google Scholar]
- 6.Wittkop L, Günthard HF, de Wolf F, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011;11(5):363–371. [DOI] [PubMed] [Google Scholar]
- 7.AIDS and Hepatitis C Professional Group SoID, Chinese Medical Association, Chinese Center for Disease Control and Prevention. Chinese guidelines for diag-nosis and treatment of HIV/AIDS (version 2018). Chin J Intern Med. 2018;57:867–884. [Google Scholar]
- 8.Blanco J, Whitlock G, Milinkovic A, Moyle G. HIV integrase inhibitors: a new era in the treatment of HIV. Expert Opin Pharmacother. 2015;16:1313–1324. [DOI] [PubMed] [Google Scholar]
- 9.Mbhele N, Chimukangara B, Gordon M. HIV-1 integrase strand transfer inhibitors: a review of current drugs, recent advances and drug resistance. Int J Antimicrob Agents. 2021;57(5):106343. [DOI] [PubMed] [Google Scholar]
- 10.Gallant J, Lazzarin A, Mills A, et al. Bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir, Abacavir, and lamivudine for initial treatment of HIV-1 infection (GS-US-380-1489): a double-blind, multicentre, Phase 3, randomised controlled non-inferiority trial. The Lancet. 2017;390(10107):2063–2072. [DOI] [PubMed] [Google Scholar]
- 11.Acosta RK, Willkom M, Martin R, et al. Resistance Analysis of Bictegravir-Emtricitabine-Tenofovir Alafenamide in HIV-1 Treatment-Naive Patients through 48 Weeks. Antimicrob Agents Chemother. 2019;63:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casadella M, van Ham PM, Noguera-Julian M, et al. Primary resistance to integrase strand-transfer inhibitors in Europe. J Antimicrob Chemother. 2015;70(10):2885–2888. [DOI] [PubMed] [Google Scholar]
- 13.Ji H, Patterson A, Taylor T, et al. Prevalence of Primary Drug Resistance Against HIV-1 Integrase Inhibitors in Canada. J Acquir Immune Defic Syndr. 2018;78(1):e1–e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stekler JD, McKernan J, Milne R, et al. Lack of resistance to integrase inhibitors among antiretroviral-naive subjects with primary HIV-1 infection, 2007-2013. Antivir Ther. 2015;20(1):77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng XM, Liu JF, Zhang M. Mutations of primary integrase gene resistance of HIV/AIDS patients in Yunnan province. Clin J AIDS STD. 2019;25(4):327–341. [Google Scholar]
- 16.Yin YQ, Lu J, Zhou Y, et al. Drug Resistance to HIV-1 Integrase Inhibitors among Treatment-naive Patients in Jiangsu, China. Biomed Environ Sci. 2021;34(5):400–403. [DOI] [PubMed] [Google Scholar]
- 17.Lan Y, Li L, Chen W, et al. Absence of Integrase Inhibitor-Associated Resistance Among Antiretroviral Therapy-Naive HIV-1-Infected Adults in Guangdong Province, China, in 2018. Infect Drug Resist. 2020;13:4389–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan L, Yu F, Zhang H, et al. Transmitted and Acquired HIV-1 Drug Resistance from a Family: a Case Study. Infect Drug Resist. 2020;13:3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Liang S, Zhou C, et al. HIV Drug Resistance Mutations Detection by Next-Generation Sequencing during Antiretroviral Therapy Interruption in China. Pathogens. 2021;10:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Dai L, Yao J, et al. Lack of HIV-1 integrase inhibitor resistance among 392 antiretroviral-naïve individuals in a tertiary care hospital in Beijing, China. AIDS. 2019;33:1945–1947. [DOI] [PubMed] [Google Scholar]
- 21.Standard University HIV Drug Resistance Database. HIVdb Program.Available from: https://hivdb.stanford.edu/hivdb/by-mutations/.
- 22.Zhang F, Maria A, Haberer J, Zhao Y. Overview of HIV drug resistance and its implications for China. Chin Med J. 2007;119:1999–2004. [PubMed] [Google Scholar]
- 23.Zhao S, Feng Y, Hu J, et al. Prevalence of Transmitted HIV drug resistance in antiretroviral treatment naive newly diagnosed individuals in China. Sci Rep. 2018;8(1):12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao L, Qing Y, Shang H, et al. The Prevalence of Transmitted Antiretroviral Drug Resistance in Treatment-Naive HIV-Infected Individuals in China. J Acquir Immune Defic Syndr. 2010;53(Suppl 1):S10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sluis-Cremer N. The emerging profile of cross-resistance among the nonnucleoside HIV-1 reverse transcriptase inhibitors. Viruses. 2014;6(8):2960–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams J, Patel N, Mankaryous N, Tadros M, Miller CD. Nonnucleoside reverse transcriptase inhibitor resistance and the role of the second-generation agents. Ann Pharmacother. 2010;44(1):157–165. [DOI] [PubMed] [Google Scholar]
- 27.Fogel JM, Bonsall D, Cummings V, et al. Performance of a high-throughput next-generation sequencing method for analysis of HIV drug resistance and viral load. J Antimicrob Chemother. 2020;75(12):3510–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simen BB, Simons JF, Hullsiek KH, et al. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis. 2009;199(5):693–701. [DOI] [PubMed] [Google Scholar]
- 29.Johnson J, Li J-F, Wei X, et al. Minority HIV-1 Drug Resistance Mutations Are Present in Antiretroviral Treatment–Naïve Populations and Associate with Reduced Treatment Efficacy. PLoS Med. 2008;5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]