Abstract
Background
Immune and skeletal systems physiologically and pathologically interact with each other. Immune and skeletal diseases may share potential pleiotropic genetics factors, but the shared specific genes are largely unknown.
Objective
This study aimed to investigate the overlapping genetic factors between multiple diseases (including rheumatoid arthritis (RA), psoriasis, osteoporosis, osteoarthritis, sarcopenia, and fracture).
Methods
The canonical correlation analysis (metaCCA) approach was used to identify the shared genes for six diseases by integrating genome-wide association study (GWAS)-derived summary statistics. The versatile Gene-based Association Study (VEGAS2) method was further applied to refine and validate the putative pleiotropic genes identified by metaCCA.
Results
About 157 (p<8.19E-6), 319 (p<3.90E-6), and 77 (p<9.72E-6) potential pleiotropic genes were identified shared by two immune diseases, four skeletal diseases, and all of the six diseases, respectively. The top three significant putative pleiotropic genes shared by both immune and skeletal diseases, including HLA-B, TSBP1, and TSBP1-AS1 (p<E-300), were located in the major histocompatibility complex (MHC) region. Nineteen of 77 putative pleiotropic genes identified by metaCCA analysis were associated with at least one disease in the VEGAS2 analysis. Specifically, the majority (18) of these 19 putative validated pleiotropic genes were associated with RA.
Conclusion
The metaCCA method identified some pleiotropic genes shared by the immune and skeletal diseases. These findings help to improve our understanding of the shared genetic mechanisms and signaling pathways underlying immune and skeletal diseases.
Keywords: GWAS, metaCCA, VEGAS2, pleiotropic gene, immune diseases, skeletal diseases
1. INTRODUCTION
Bone formation and bone resorption are dynamic processes occurring in bone at all times, and its imbalance contributes to bone metabolic disorders, such as osteoporosis (OP) [1]. The immune system is one of the major systems in determining the balance of bone turnover [1]. The concept of osteoimmunology has recently been proposed to summarize novel insights into functional interdependence between immune and skeletal systems at the anatomical, vascular, cellular, and molecular levels [2]. It is well established that bone and adaptive immune systems appear at the same stage of vertebrate evolution [3]. This phenomenon suggested that the immune system is required for bone during its evolution and vice versa.
Immune and skeletal systems physiologically and pathologically interact with each other. The important role of the immune system in multiple bone disease pathologies, such as OP, osteoarthritis (OA), and rheumatoid arthritis (RA), is now well established. Osteoporosis is a disorder of decreased bone mass, microarchitectural deterioration, and fragility fractures (F) [4]. OP is a prevalent inflammatory bone loss condition in elderly people. RA is a typical chronic inflammatory joint disease characterized by persistent inflammation in multiple joints, progressive bone erosions, and loss of function. The formation of bone erosions and structural damage in inflamed joints in RA were partially attributed to osteoclasts, which were generally regarded as the dominant source of both receptor activators of NF-κB (RANK) ligand (RANKL) and osteoprotegerin (OPG) [5]. The RANKL to OPG ratio is a key determinant of osteoclast differentiation and bone resorption. Psoriasis (PsO) was associated with higher rates of pathological fractures, particularly of the vertebrae, pelvis, femur, tibia, and fibula [6, 7]. Previous studies also reported that skeletal muscle was a potent regulator of immune system function [8]. The immune system has been reported as a contributor to sarcopenia (SP), which is a syndrome of low muscle mass with low muscle strength and/or low physical performance.
Immune and skeletal systems are strongly interlinked through a quantity of shared regulatory factors, including cytokines, chemokines, transcription factors, and receptors [3]. Although previous findings have broadened our understanding of the interplay between these two systems, the shared molecular mechanism and signaling pathways between immune and skeletal diseases are still much less explored. Pleiotropy is a phenomenon in one gene that affects multiple phenotypes, leading to genetic correlation among phenotypes [9]. Identifying potential pleiotropic genes among phenotypes can help investigate the overlapping etiology of multiple diseases. The genome-wide association study (GWAS)-derived summary statistic data can be utilized to identify the shared genes for immune and skeletal diseases with canonical correlation analysis (metaCCA) framework, which was able to test the association between all SNPs located at the same gene and multiple phenotypes [10]. This method incorporates the GWAS summary statistics, linkage disequilibrium (LD) structure, and phenotypic correlations between phenotypes to enhance the statistical power for identifying novel genetic associations [11]. MetaCCA extends the statistical technique of canonical correlation analysis to the setting where original individual-level records are not available and employs a covariance shrinkage algorithm to achieve robustness [10].
This study adopted the metaCCA method to assess the overlapping genetic correlation among two immune diseases (RA and PsO), four bone skeletal disorders (OP, OA, F, and SP), and all of these six diseases using the GWAS summary statistics data, respectively. The versatile Gene-based Association Study (VEGAS2) approach was further applied to validate the putative pleiotropic genes identified by metaCCA. Our findings may provide clues for understanding the shared pathogenesis of immune and skeletal systems and may serve as a reference for further genetic research or drug development.
2. MATERIALS AND METHODS
2.1. GWAS Datasets
The summary statistics of six GWAS datasets were downloaded from GWAS Catalog (available at https://www.ebi.ac.uk/gwas/). The RA GWAS dataset comprising of ~10 million SNPs is derived from a GWAS meta-analysis with 14,361 cases and 43,923 controls of European subjects [12]. The dataset for OA GWAS included ~16 million SNPs and was downloaded from UK Biobank involving 32,970 European subjects [13]. The PsO GWAS summary data were obtained from a meta-analysis with 10,588 cases and 22,806 controls of European subjects [14]. The FNK-BMD dataset contained the association results for approximately 10 million SNPs and were obtained from a GWAS meta-analysis with 32,965 European subjects published by the Genetic Factors for Osteoporosis (GEFOS) Consortium [15]. The summary statistics for fracture (F) included ~13 million SNPs and were based on a GWAS from a European cohort consisting of 53,184 cases and 373,611 controls [16]. Many fracture sites were included but not limited to ankle, leg, hip, spine, and arm. The SP-related traits (appendicular lean body mass) GWAS dataset was estimated in 28,330 European subjects from a subset of 15 cohorts, containing summary statistics of more than 2.3 million SNPs [17]. Basic information for the GWAS studies was summarized in Table S1.
2.2. Data Preparation
The SNPs in the six summary statistics were pruned before multivariate analysis using the genotype data from the 1000 Genomes project as a reference [18]. Briefly, six datasets were combined to select overlapping SNP; then, an LD-based algorithm was adopted to prune SNPs with high pairwise correlations. Default values of the PLINK 1.9 software (50, 5, and 0.2) were set as parameters when calculating LD values (r2) between each SNP pair. The LD was calculated for windows that contained 50 SNPs. The SNP with a lower frequency of the minor allele was excluded for each pair with r2> 0.2. The calculation window was then shifted forward by 5 SNPs. Then the above process was repeated until each SNP pair was in low LD.
The SNPs were annotated to their corresponding genes using the Genome Reference Consortium Human genome build 37 (GRCh37) (available at https://www.gencodegenes.org/human/releases.html.), which contained 32,801 genes. An SNP was mapped to a certain gene when it was located between the transcription starting and ending point of that gene [11].
The regression coefficient β and corresponding standard error (SE) of each SNP originatingin GWAS summary statistics of the six different phenotypes were obtained. The regression coefficient β was normalized according to:
 (1)
(1)
where SEgp is the standard error of βgp, as given by the original GWAS result, g is the number of genotypic variables, p is the number of phenotypic variables, and N is the sample number of each trait.
2.3. MetaCCA Analysis
MetaCCA analysis was performed to identify the putative pleiotropic genes among the six phenotypes. Detailed principles and formulas of this approach were described elsewhere [10]. Conducting metaCCA analysis requires a cross- covariance matrix between all genotypic SNPs (ƩXX), a phenotypic correlation structure between phenotypes (ƩYY), and the GWAS summary statistics (ƩXY). The full covariance matrix (Ʃ) can be calculated according to:

The full covariance matrix (Ʃ) was applied to obtain the final genotype-phenotype association results, which contained a significant test for each gene in the identified canonical correlation. The p-value was adjusted using the Bonferroni approach. An adjusted p-value < 0.05 was defined as the cutoff value for significant correlation. The metaCCA test was based on the script of the R package “metaCCA”.
2.4. Versatile Gene-based Association Study (VEGAS) Approach
Gene-based association analysis was performed using the Versatile Gene-based Association Study-2 (VEGAS-2) method to refine and validate the pleiotropic genes identified by metaCCA. VEGAS-2 is a gene-based approach that assesses the correlation between a phenotype and multiple SNPs within a gene and accounts for linkage disequilibrium between those SNPs to estimate the association between each SNP and each phenotype individually [19]. This method has shown higher sensitivity and lower false-positive rates compared to other gene-based approaches. Gene with a correlation p-value < 1.56E-5 (=0.05/3,209) was significantly correlated with the diseases in the VEGA-2 analysis.
2.5. Functional Annotation
For the pleiotropic genes validated by VEGAS-2, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEEG) pathway analysis using online tool DAVID (available at http://david.abcc.ncifcrf.gov/). The GO analysis contained biological processes, cellular components, and molecular function. Multiple testing corrected by FDR with a significant threshold at 0.05 was employed to assess the degree of enrichment in each GO term or KEGG pathway.
Protein-protein interaction (PPI) analysis for the genes of interest was performed using the online tool STRING (available at https://string-db.org/).
3. RESULTS
3.1. Potential Pleiotropic Genes Identified by metaCCA Analysis
3.1.1. Potential Pleiotropic Genes for Six Diseases
To investigate the overlapping genetic genes shared by the six diseases, we applied metaCCA analysis using GWAS summary statistics data. After gene annotation and SNP pruning, 17,284 SNPs located in 5,145 genes were used in metaCCA analysis. The genes with correlation p-values < 9.72E-6 (= 0.05/5,145) were significantly correlated with the six diseases. About 77 potential pleiotropic genes were significantly associated with multiple diseases (p < 9.72E-6) (Table 1). The canonical correlation r between potential pleiotropic genes and diseases ranged from 0.0363 to 0.3567. Specifically, among the 77 genes we identified, 57 were from chromosome 6.
Table 1.
Potential pleiotropic genes identified by metaCCA analysis for six traits.
| Pleiotropic Gene | Chrom | Position | Correlation Coefficient (r) | p-value |
|---|---|---|---|---|
| HLA-B | 6 | 31237268-31324965 | 0.2521 | 0 |
| TSBP1 | 6 | 32256303-32339689 | 0.3058 | 0 |
| TSBP1-AS1 | 6 | 32222417-32375540 | 0.3568 | 0 |
| HLA-DQA1 | 6 | 32595956-32614839 | 0.1728 | 4.3193E-165 |
| LINC02571 | 6 | 31261685-31269419 | 0.1733 | 2.1443E-163 |
| ABHD16A | 6 | 31654732-31671133 | 0.1525 | 1.891E-127 |
| C2 | 6 | 31865562-31913449 | 0.1483 | 3.4572E-120 |
| TAP2 | 6 | 32789610-32806547 | 0.1462 | 2.6041E-109 |
| MUC22 | 6 | 30978251-31003179 | 0.1414 | 1.7153E-104 |
| SFTA2 | 6 | 30899130-30923413 | 0.1247 | 4.6514E-74 |
| ANKRD55 | 5 | 55395507-55529157 | 0.1359 | 2.27576E-67 |
| HLA-DOB | 6 | 32780540-32788243 | 0.1149 | 1.31164E-66 |
| STK19 | 6 | 31938868-31950598 | 0.1044 | 7.52326E-58 |
| BTNL2 | 6 | 32361740-32374905 | 0.1041 | 1.54063E-57 |
| HLA-DOA | 6 | 32971959-32977368 | 0.1052 | 2.79825E-55 |
| HCG20 | 6 | 30711567-30760027 | 0.1062 | 1.01891E-51 |
| COL11A2P1 | 6 | 33071571-33075107 | 0.0982 | 5.90187E-51 |
| LINC00243 | 6 | 30766431-30798436 | 0.1018 | 8.0991E-49 |
| HIPK1-AS1 | 1 | 114466622-114472117 | 0.0950 | 1.77439E-47 |
| TRIM31 | 6 | 30070674-30080867 | 0.0908 | 6.15542E-45 |
| TRIM31-AS1 | 6 | 30073017-30082501 | 0.0939 | 1.69435E-44 |
| MICA | 6 | 31367561-31383092 | 0.0864 | 7.76903E-39 |
| TRIM39 | 6 | 30294256-30311506 | 0.0868 | 1.64094E-37 |
| TCF19 | 6 | 31126324-31134936 | 0.0836 | 5.11349E-35 |
| ATAT1 | 6 | 30594619-30614600 | 0.0821 | 6.96222E-35 |
| OR5V1 | 6 | 29321526-29399744 | 0.0884 | 1.44978E-34 |
| HLA-DMA | 6 | 32916390-32936871 | 0.0786 | 1.72345E-34 |
| RSBN1 | 1 | 114304454-114355098 | 0.0891 | 3.54109E-34 |
| DRAIC | 15 | 69755260-70135459 | 0.0948 | 1.475E-32 |
| MUCL3 | 6 | 30902300-30921998 | 0.0787 | 1.0258E-29 |
| TMPOP1 | 6 | 30434229-30435771 | 0.0759 | 1.53914E-29 |
| HCG17 | 6 | 30201816-30293911 | 0.0837 | 3.28028E-29 |
| HLA-DPA2 | 6 | 33059530-33065072 | 0.0719 | 2.92774E-26 |
| AFF3 | 2 | 100161881-100808890 | 0.0903 | 7.18191E-25 |
| HLA-P | 6 | 29768192-29770202 | 0.0680 | 2.37849E-23 |
| HCG18 | 6 | 30254467-30295159 | 0.0734 | 3.87231E-22 |
| HLA-F-AS1 | 6 | 29694378-29716826 | 0.0662 | 7.19657E-22 |
| TNXB | 6 | 32008930-32083111 | 0.0644 | 1.22398E-21 |
| NOTCH4 | 6 | 32162620-32191844 | 0.0656 | 1.39385E-21 |
| GABBR1 | 6 | 29523406-29601753 | 0.0626 | 1.73259E-21 |
| PFDN6 | 6 | 33257079-33266178 | 0.0652 | 2.72822E-21 |
| PRKCQ | 10 | 6469105-6622263 | 0.0868 | 1.28076E-20 |
| SYNGAP1 | 6 | 33387438-33421466 | 0.0616 | 7.38002E-19 |
| HLA-DPA3 | 6 | 33098993-33111102 | 0.0606 | 3.61826E-18 |
| DCLRE1B | 1 | 114447835-114456708 | 0.0636 | 7.51786E-18 |
| MAGI3 | 1 | 113933137-114228545 | 0.0706 | 1.91163E-16 |
| HCP5 | 6 | 31430947-31446713 | 0.0560 | 4.46803E-15 |
| ZBED9 | 6 | 28538312-28583989 | 0.0553 | 6.47681E-15 |
| OLFML3 | 1 | 114522013-114578194 | 0.0575 | 3.99471E-14 |
| HLA-F | 6 | 29690552-29706305 | 0.0526 | 2.29311E-13 |
| HLA-DMB | 6 | 32902406-32908805 | 0.0567 | 6.4457E-13 |
| OR2I1P | 6 | 29518184-29525498 | 0.0502 | 4.40528E-12 |
| LINC01016 | 6 | 33835283-33864691 | 0.0575 | 6.67504E-12 |
| MICD | 6 | 29938578-29940241 | 0.0482 | 4.73015E-11 |
| ANK3 | 10 | 61786056-62493248 | 0.0702 | 3.48439E-10 |
| UQCC2 | 6 | 33662070-33679504 | 0.0458 | 7.77029E-10 |
| PRRC2A | 6 | 31588492-31605548 | 0.0455 | 1.0046E-09 |
| CCR6 | 6 | 167525295-167553184 | 0.0493 | 1.96115E-09 |
| LINC02829 | 6 | 29465252-29478335 | 0.0402 | 2.23724E-09 |
| OR10C1 | 6 | 29407083-29408754 | 0.0445 | 3.1145E-09 |
| SLC44A4 | 6 | 31830969-31846823 | 0.0440 | 5.16113E-09 |
| CCDC170 | 6 | 151815152-151942328 | 0.0478 | 8.7594E-09 |
| IRF5 | 7 | 128577511-128590092 | 0.0425 | 2.47913E-08 |
| OR11A1 | 6 | 29393281-29424848 | 0.0449 | 3.14789E-08 |
| ARID5B | 10 | 63661443-63856703 | 0.0469 | 3.39802E-08 |
| MICE | 6 | 29709508-29716746 | 0.0408 | 1.30454E-07 |
| COG6 | 13 | 40229764-40365802 | 0.0402 | 2.32812E-07 |
| ABCF1 | 6 | 30539170-30564956 | 0.0400 | 2.72518E-07 |
| IL2RA | 10 | 6052652-6104333 | 0.0498 | 1.66699E-06 |
| DAP | 5 | 10679342-10761346 | 0.0420 | 2.25497E-06 |
| LINC00824 | 8 | 129417515-129576925 | 0.0534 | 2.43457E-06 |
| MFSD4B | 6 | 111580530-111766557 | 0.0392 | 2.97114E-06 |
| PLCL2 | 3 | 16844159-17132096 | 0.0373 | 3.14587E-06 |
| FAM3C | 7 | 120988932-121036418 | 0.0368 | 4.67958E-06 |
| TRIM26BP | 6 | 30206078-30210056 | 0.0365 | 5.93577E-06 |
| ERBB3 | 12 | 56470583-56497289 | 0.0363 | 6.98855E-06 |
| PTPRC | 1 | 198607587-198726605 | 0.0428 | 9.58622E-06 |
Note: “0”: p-value <1E-300.
3.1.2. Potential Pleiotropic Genes for Two Immune Diseases
MetaCCA analysis found the pleiotropic effects shared by two immune diseases (RA and PsO). After gene annotation and SNP pruning, 19,290 SNPs located in 6,103 genes were used in metaCCA analysis. The genes with correlation p-values < 8.19E-6 (=0.05/6,103) were significantly correlated with the two autoimmune diseases. A total of 157 genes with significant threshold (p < 8.19E-6) were identified as putative pleiotropic genes (Table S2). The canonical correlation r between potential pleiotropic genes and diseases ranged from 0.0302 to 0.4769.
3.1.3. Potential Pleiotropic Genes for Four Skeletal Diseases
MetaCCA analysis was performed to identify potential pleiotropic genes for four skeletal traits (OP, OA, SP, and F). After gene annotation and SNP pruning, 67,362 SNPs located in 12,834 genes remained for metaCCA analysis. The genes with a significant threshold (p-values < 3.90E-6 (=0.05/12,833)) were identified as potential pleiotropic genes for the four skeletal diseases. We identified 319 putative pleiotropic genes that were significantly associated with multiple diseases (p < 3.90E-6) (Table S3). The canonical correlation r between potential pleiotropic gene and phenotype ranged from 0.0438 to 0.9455. These pleiotropic genes were located dispersedly at all human chromosomes.
3.2. Potential Pleiotropic Genes Validated by VEGAS Analysis
We refined and validated the 77 pleiotropic genes associated with all six diseases to investigate their associations with specific phenotypes using VEGAS-2 analysis. About 24, 102, 4, and 3 significant genes were identified for PsO, RA, OP, and F with p-values < 1.56E-5 (adjusted p < 0.05), respectively (Table S4). Notably, we found that 19 putative pleiotropic genes identified in the metaCCA analysis were associated with at least one disease in the VEGAS-2 analysis. We identified 4 genes (SLC44A4, HLA-DOB, TAP2, C2) associated with PsO, 18 (PLCL2, DAP, COG6, ARID5B, IRF5, SLC44A4, CCR6, UQCC2, MAGI3, DCLRE1B, SYNGAP1, AFF3, RSBN1, HLA-DOA, HLA-DOB, ANKRD55, TAP2, C2) genes for RA, one (CCDC170) gene for OP and one (CCDC170) gene for F (Table 2). Four (SLC44A4, HLA-DOB, TAP2, C2) genes identified by metaCCA analysis were associated with both PsO and RA.
Table 2.
Potential pleiotropic genes identified by both metaCCA analysis and VEGAS2 analysis for six traits.
| Gene | p.meta | p.f | p.oa | p.pso | p.ra | p.sp | p.op |
|---|---|---|---|---|---|---|---|
| PLCL2 | 3.15E-06 | 6.63E-01 | 3.78E-01 | 7.59E-02 | 1.00E-06 | 8.89E-02 | 8.51E-01 |
| DAP | 2.25E-06 | 4.80E-01 | 6.72E-01 | 2.72E-01 | 1.00E-06 | 9.89E-02 | 6.24E-01 |
| COG6 | 2.33E-07 | 4.52E-02 | 9.55E-01 | 6.84E-01 | 1.00E-06 | 4.00E-01 | 3.74E-01 |
| ARID5B | 3.40E-08 | 2.86E-01 | 4.06E-01 | 2.10E-01 | 1.00E-06 | 9.30E-01 | 5.17E-01 |
| IRF5 | 2.48E-08 | 1.44E-02 | 4.86E-01 | 5.10E-02 | 1.00E-06 | 5.38E-01 | 3.56E-01 |
| CCDC170 | 8.76E-09 | 1.00E-06 | 9.87E-01 | 2.38E-01 | 7.96E-01 | 7.84E-01 | 1.00E-06 |
| SLC44A4 | 5.16E-09 | 6.77E-01 | 5.22E-01 | 1.00E-06 | 1.00E-06 | 5.98E-01 | 2.88E-03 |
| CCR6 | 1.96E-09 | 4.35E-02 | 7.30E-01 | 7.52E-01 | 1.00E-06 | 3.95E-02 | 2.38E-01 |
| UQCC2 | 7.77E-10 | 5.12E-01 | 4.12E-01 | 6.36E-03 | 1.00E-06 | 8.48E-01 | 2.54E-02 |
| MAGI3 | 1.91E-16 | 5.85E-02 | 8.26E-01 | 9.97E-01 | 1.00E-06 | 4.18E-01 | 7.49E-02 |
| DCLRE1B | 7.52E-18 | 4.87E-01 | 6.22E-01 | 9.42E-01 | 1.00E-06 | 6.83E-01 | 2.89E-02 |
| SYNGAP1 | 7.38E-19 | 8.26E-01 | 1.44E-01 | 7.84E-01 | 1.00E-06 | 4.60E-01 | 6.54E-01 |
| AFF3 | 7.18E-25 | 7.46E-01 | 5.12E-01 | 3.35E-03 | 2.00E-06 | 5.79E-01 | 1.86E-01 |
| RSBN1 | 3.54E-34 | 1.79E-01 | 1.00E+00 | 9.45E-01 | 1.00E-06 | 3.10E-01 | 6.27E-02 |
| HLA-DOA | 2.80E-55 | 9.13E-01 | 3.72E-01 | 2.22E-03 | 1.00E-06 | 5.96E-01 | 2.12E-01 |
| HLA-DOB | 1.31E-66 | 1.56E-01 | 6.16E-01 | 1.00E-06 | 1.00E-06 | 2.05E-01 | 9.83E-01 |
| ANKRD55 | 2.28E-67 | 6.29E-01 | 5.50E-01 | 2.71E-01 | 1.00E-06 | 6.08E-01 | 8.45E-01 |
| TAP2 | 2.60E-109 | 1.19E-01 | 5.89E-01 | 1.00E-06 | 1.00E-06 | 6.96E-01 | 7.53E-01 |
| C2 | 3.46E-120 | 5.97E-01 | 3.19E-01 | 1.00E-06 | 1.00E-06 | 2.68E-01 | 1.40E-03 |
Note: Bold numbers indicate a significant adjusted p-value (p < 0.05) for the VEGAS2 test, and the bold text indicates genes related to more than one trait in the VEGAS2 test.
In particular, 14 of these 19 potential pleiotropic genes (PLCL2, COG6, ARID5B, IRF5, CCDC170, CCR6, MAGI3, AFF3, RSBN1, HLA-DOA, HLA-DOB, ANKRD55, TAP2, and C2) were previously reported to be associated with more than one of these six diseases in published studies. Among the 14 confirmed pleiotropic genes, CCR6 was associated with OP, RA, PsO, and OA in previous studies, 3 genes (IRF5, TAP2, and C2) were reported to be associated with RA, PsO and OA, 3 genes (PLCL2, COG6, and AFF3) were associated with PsO and OA, CCDC170 was associated with OP and F, and 6 genes (ARID5B, MAGI3, RSBN1, HLA- DOA, HLA-DOB, and ANKRD55) were associated with RA.
3.3. Functional Annotation
GO enrichment analysis for the 77 putative pleiotropic genes identified by metaCCA indicated that these genes were significantly enriched in biological processes related to “immune response” (GO: 0006955, FDR=3.73E-3), “interferon-gamma-mediated signaling pathway” (GO: 0060333, FDR=5.39E-3), “antigen processing and presentation of peptide or polysaccharide antigen via MHC class II” (GO: 0002504, FDR=4.44E-5), “antigen processing and presentation of exogenous peptide antigen via MHC class II” (GO: 0019886, FDR=1.11E-2), “antigen processing and presentation” (GO: 0019882, FDR=3.38E-2) and “positive regulation of T cell proliferation” (GO: 0042102, FDR=3.64E-2).
Cellular components enriched by putative pleiotropic genes included “MHC class II protein complex” (GO: 0042613, FDR=3.00E-5) and “cell surface” (GO: 0009986, 2.65E-2). Molecular functions significantly enriched by these genes included “MHC class II receptor activity” (GO: 0032395, FDR=5.22E-4) and “MHC class II protein complex binding” (GO: 0023026, FDR=5.22E-4) (Table 3).
Table 3.
GO terms enriched by pleiotropic genes.
| Gene Ontology ID: Term | Genes | FDR |
|---|---|---|
| Biological process | ||
| GO: 0002504 Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 4.44E-05 |
| GO: 0006955 Immune response | HLA-DMA, HLA-DMB, IL2RA, HLA-B, CCR6, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 3.73E-03 |
| GO: 0060333 Interferon-gamma-mediated signaling pathway | HLA-B, IRF5, TRIM31, HLA-F, HLA-DQA1 | 5.39E-03 |
| GO: 0019886 Antigen processing and presentation of exogenous peptide antigen via MHC class II | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 1.11E-02 |
| GO: 0019882 Antigen processing and presentation | HLA-DMB, HLA-B, MICA, HLA-DQA1 | 3.38E-02 |
| GO: 0042102 Positive regulation of T cell proliferation | HLA-DMB, PTPRC, PRKCQ, BTNL2 | 3.64E-02 |
| Cellular component | ||
| GO:0042613 MHC class II protein complex | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 3.00E-05 |
| GO:0009986 Cell surface | HLA-DMA, PTPRC, NOTCH4, HLA-B, ANK3, CCR6, HLA-F, MICA | 2.65E-02 |
| Molecular function | ||
| GO: 0032395 MHC class II receptor activity | HLA-DMA, HLA-DOA, HLA-DOB, HLA-DQA1 | 5.22E-04 |
| GO: 0023026 MHC class II protein complex binding | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB | 5.22E-04 |
KEGG pathway analysis showed that the 77 pleiotropic genes were mainly enriched in pathways related to autoimmune/auto-inflammatory diseases and infection (Table 4).
Table 4.
KEGG pathways enriched by pleiotropic genes.
| Term | Genes | FDR |
|---|---|---|
| hsa05332 Graft-versus-host disease | HLA-DMA, HLA-DMB, HLA-B, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 4.01E-08 |
| hsa05330 Allograft rejection | HLA-DMA, HLA-DMB, HLA-B, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 4.17E-08 |
| hsa04940 Type I diabetes mellitus | HLA-DMA, HLA-DMB, HLA-B, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 4.74E-08 |
| hsa04612 Antigen processing and presentation | HLA-DMA, HLA-DMB, HLA-B, TAP2, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 4.74E-08 |
| hsa05320 Autoimmune thyroid disease | HLA-DMA, HLA-DMB, HLA-B, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 1.41E-07 |
| hsa05416 Viral myocarditis | HLA-DMA, HLA-DMB, HLA-B, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 2.08E-07 |
| hsa04514 Cell adhesion molecules (CAMs) | HLA-DMA, HLA-DMB, PTPRC, HLA-B, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 2.13E-06 |
| hsa04145 Phagosome | HLA-DMA, HLA-DMB, HLA-B, TAP2, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 2.71E-06 |
| hsa05150 Staphylococcus aureus infection | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1, C2 | 4.37E-06 |
| hsa05168 Herpes simplex infection | HLA-DMA, HLA-DMB, HLA-B, TAP2, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 8.33E-06 |
| hsa05310 Asthma | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 1.12E-05 |
| hsa05166 HTLV-I infection | HLA-DMA, HLA-DMB, IL2RA, HLA-B, HLA-DOA, HLA-F, HLA-DOB, HLA-DQA1 | 5.94E-05 |
| hsa04672 Intestinal immune network for IgA production | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 5.94E-05 |
| hsa05321 Inflammatory bowel disease (IBD) | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 1.89E-04 |
| hsa05322 Systemic lupus erythematosus | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1, C2 | 2.30E-04 |
| hsa05140 Leishmaniasis | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 2.49E-04 |
| hsa05323 Rheumatoid arthritis | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 5.40E-04 |
| hsa05145 Toxoplasmosis | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 0.001199 |
| hsa05164 Influenza A | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 0.006251 |
| hsa05152 Tuberculosis | HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DQA1 | 0.006318 |
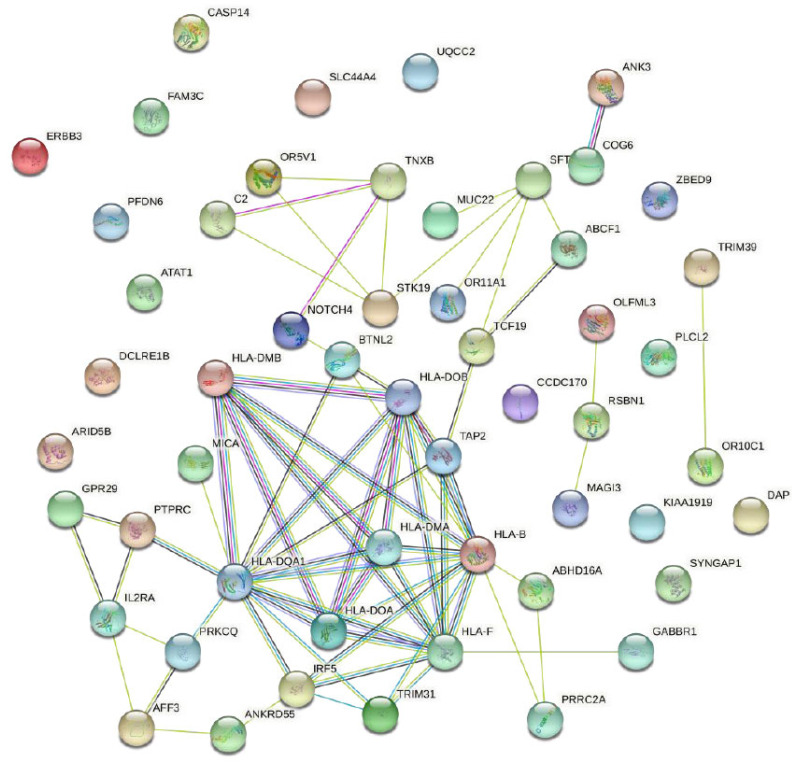
The complex protein-protein interaction network, constituted by the above 77 putative pleiotropic genes, highlighted a network containing 31 genes (Fig. 1). Among these 31 genes, eleven genes (BTNL2, HLA-DMB, HLA-DQA1, HLA-DMA, HLA-DOA, HLA-DOB, TAP2, HLA-B, HLA-F, IRF5, and TRIM31) constructed a very complex network.
Fig. (1).
PPI network of 77 putative pleiotropic genes.
Notes: The nodes represent pleiotropic genes-encoded protein(s).
Node content: empty nodes mean unknown 3D structure.
Edges: different colors of edges represent various types of associations (interaction). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
4. DISCUSSION
The present study performed multivariable analytical metaCCA by combining six available independent GWAS summary statistics datasets to identify the genes shared by immune and skeletal diseases. To refine and confirm the genes identified in metaCCA analysis, the gene-based VEGAS-2 analysis was conducted for these six diseases, respectively. We identified 77 potential pleiotropic genes in the metaCCA analysis. Among these putative pleiotropic genes, 19 were associated with at least one disease in the VEGAS-2 analysis. In particular, 14 of these 19 potential pleiotropic genes were previously reported to be associated with more than one of these six diseases. By trimming the multiple testing burden at the initial filtering step, the metaCCA approach allows for the detection of more phenotype-related loci than what would be identified by VEGAS-2 alone. The identification of putative pleiotropic genes and the associated biological pathways may provide a better understanding of the shared genetic factors involved in the development of immune and skeletal diseases.
Among the 19 confirmed pleiotropic genes, 18 genes (PLCL2, DAP, COG6, ARID5B, IRF5, SLC44A4, CCR6, UQCC2, MAGI3, DCLRE1B, SYNGAP1, AFF3, RSBN1, HLA-DOA, HLA-DOB, ANKRD55, TAP2, C2) were associated with RA in the VEGAS-2 analysis. Notably, 13 of these genes were involved in the pathogenesis of RA in previous studies. Several genes, such as PLCL2, COG6, RIF5, ARID5B, and CCR6, were reported to be the susceptibility loci for RA [20-22]. Chemokine (C-C motif) receptor 6 gene (CCR6) encodes an important protein that is expressed in memory T-cells and immature dendritic cells and plays a critical role in B-cell maturation and differentiation [23]. A variant of the CCR6 rs3093024 variant was significantly associated with RA-risk both in a Pakistani and Chinese population [24]. Univariate SNP-multivariate phenotype analysis in the MetaCCA method indicated that rs3093026 was significantly associated with the six diseases (p=5.91E-08) (Table S5). Interestingly, the SNPs rs3093024 and rs3093026 were in strong LD (r2=0.7). This is probably because the real associated SNP was removed during the process of data preparation using an LD-based pruning method, which was performed to remove one SNP of pairs with an R2 value greater than 0.2. Removing the real associated SNPs in the LD-based pruning method may also prevent the pleiotropic genes that are truly associated with multiple diseases from being detected. We noticed that cytokines such as RANKL and TNF-α, which play an important role both in immune and skeletal systems, were insignificantly associated with these six diseases in the metaCCA analysis. This may also result from the absence of SNPs in the gene region. As expected, there was no SNP mapped to the gene regions of RANKL and TNF-α.
As a chemokine receptor, CCR6 is not only the susceptibility gene for RA, through binding to a specific chemokine, CCL20, to exert its function on the maturation, differentiation, and migration of immune cells, but also plays an important role in the pathogenesis of OA, PsO and OP [23]. The CCL20-CCR6 axis is upregulated in the lesional skin of human psoriasis, and psoriasiform dermatitis can be suppressed by blocking this axis using a specific antibody [25]. In addition, CCL20 chemokine could induce osteoclast differentiation. Previous studies demonstrated that the expression of RANK on CD14+ cells was positively correlated with that of CCR6. Monocytes expressing both RANK and CCR6 differentiate into osteoclasts [26]. The above findings may help drive force for understanding the osteoimmune axis.
Of the 77 putative pleiotropic genes identified by the metaCCA approach, several (HLA-B, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DOB, HLA-DPA2, HLA-DPA3, HLA-DQA1, HLA-F, C2, TAP2) were HLA genes, which played a critical role in the processing and presentation of antigens [27]. It has been well established that the HLA region was strongly associated with autoimmune disease (AID). For instance, the association of components of the HLA class II -encoded HLA-DRB1-DQA1-DQB1 haplotype has been determined with several AIDs such as RA. Molecules encoded by this region play a key role in exogenous antigen presentation to CD4+ Th cells, indicating the importance of this pathway in AID initiation and progression. However, the association between the HLA genes and skeletal diseases has been poorly studied. A previous study aimed to investigate the relationships between polymorphisms of the HLA-B gene and postmenopausal osteoporosis showed that the frequency of the HLA-B* 3501 allele was significantly higher in postmenopausal osteoporosis patients than in the control group in a Chinese Han population and likely an important risk factor for postmenopausal osteoporosis [28]. Our results hinted that HLA genes' role in skeletal diseases might not be adequately explored.
GO analyses suggested that the identified putative pleiotropic genes for two immune diseases were significantly enriched in biological processes related to “positive regulation of T cell proliferation” (GO: 0042102, FDR=2.51E-2), “immune response” (GO: 0006955, FDR=2.51E-2), “interferon-gamma-mediated signaling pathway” (GO: 0060333, FDR=2.51E-2) and “antigen processing and presentation of peptide or polysaccharide antigen via MHC class II” (GO: 0002504, FDR=3.33E-2). These processes were reported to be associated with inflammatory autoimmune disorders. It suggested that the result of metaCCA should be reliable and reasonable. The metaCCA method is an effective technique to systematically and comprehensively identify pleiotropic genes associated with multiple complex diseases. By leveraging large GWAS summary statistics of six diseases, the metaCCA approach could increase the sample size and statistical power of the study compared to the univariate GWAS analysis based on a cross-sectional population [10]. However, the proportions of variability could not be assessed due to no such data being available. Additionally, biological experimental verification of these pleiotropic genes identified in this study is required.
CONCLUSION
In summary, by applying metaCCA and VEGAS2 methods, we identified several putative pleiotropic genes associated with two immune diseases (e.g., RA and PsO) four skeletal disorders (e.g., OP, OA, F, and SP), and all of these six diseases by using GWAS summary statistics data, respectively. These findings may provide a novel molecular basis for understanding the interaction of the pathogenesis between immune diseases and skeletal diseases.
ACKNOWLEDGEMENTS
Declared none.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No animals/humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The study was supported by Natural Science Foundation of China (81872681, 31401079 and 81473046), the Science and Technology Project of Suzhou (SS202050), and a Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article.
REFERENCES
- 1.Caetano-Lopes J., Canhão H., Fonseca J.E. Osteoimmunology-the hidden immune regulation of bone. Autoimmun. Rev. 2009;8(3):250–255. doi: 10.1016/j.autrev.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 2.Geusens P., Lems W.F. Osteoimmunology and osteoporosis. Arthritis Res. Ther. 2011;13(5):242. doi: 10.1186/ar3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto K., Takayanagi H. Osteoimmunology. Cold Spring Harb. Perspect. Med. 2019;9(1):a031245. doi: 10.1101/cshperspect.a031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane J.M., Russell L., Khan S.N. Osteoporosis. Clin. Orthop. Relat. Res. 2000;(372):139–150. doi: 10.1097/00003086-200003000-00016. [DOI] [PubMed] [Google Scholar]
- 5.Asagiri M., Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40(2):251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Ramot Y. Psoriasis and osteoporosis: The debate continues. Br. J. Dermatol. 2017;176(5):1117–1118. doi: 10.1111/bjd.15437. [DOI] [PubMed] [Google Scholar]
- 7.Kathuria P., Gordon K.B., Silverberg J.I. Association of psoriasis and psoriatic arthritis with osteoporosis and pathological fractures. J. Am. Acad. Dermatol. 2017;76(6):1045–1053.e3. doi: 10.1016/j.jaad.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Nelke C., Dziewas R., Minnerup J., Meuth S.G., Ruck T. Skeletal muscle as potential central link between sarcopenia and immune senescence. EBioMedicine. 2019;49:381–388. doi: 10.1016/j.ebiom.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auge G.A., Penfield S., Donohue K. Pleiotropy in developmental regulation by flowering-pathway genes: Is it an evolutionary constraint? New Phytol. 2019;224(1):55–70. doi: 10.1111/nph.15901. [DOI] [PubMed] [Google Scholar]
- 10.Cichonska A., Rousu J., Marttinen P., Kangas A.J., Soininen P., Lehtimäki T., Raitakari O.T., Järvelin M-R., Salomaa V., Ala-Korpela M., Ripatti S., Pirinen M. metaCCA: Summary statistics-based multivariate meta-analysis of genome-wide association studies using canonical correlation analysis. Bioinformatics. 2016;32(13):1981–1989. doi: 10.1093/bioinformatics/btw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Greenbaum J., Qiu C., Li K., Wang Q., Tang S-Y., Deng H-W. Identification of pleiotropic genes between risk factors of stroke by multivariate metaCCA analysis. Mol. Genet. Genomics. 2020;295(5):1173–1185. doi: 10.1007/s00438-020-01692-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., Graham R.R., Manoharan A., Ortmann W., Bhangale T., Denny J.C., Carroll R.J., Eyler A.E., Greenberg J.D., Kremer J.M., Pappas D.A., Jiang L., Yin J., Ye L., Su D-F., Yang J., Xie G., Keystone E., Westra H-J., Esko T., Metspalu A., Zhou X., Gupta N., Mirel D., Stahl E.A., Diogo D., Cui J., Liao K., Guo M.H., Myouzen K., Kawaguchi T., Coenen M.J.H., van Riel P.L.C.M., van de Laar M.A.F.J., Guchelaar H-J., Huizinga T.W.J., Dieudé P., Mariette X., Bridges S.L., Jr, Zhernakova A., Toes R.E.M., Tak P.P., Miceli-Richard C., Bang S-Y., Lee H-S., Martin J., Gonzalez-Gay M.A., Rodriguez-Rodriguez L., Rantapää-Dahlqvist S., Arlestig L., Choi H.K., Kamatani Y., Galan P., Lathrop M., Eyre S., Bowes J., Barton A., de Vries N., Moreland L.W., Criswell L.A., Karlson E.W., Taniguchi A., Yamada R., Kubo M., Liu J.S., Bae S-C., Worthington J., Padyukov L., Klareskog L., Gregersen P.K., Raychaudhuri S., Stranger B.E., De Jager P.L., Franke L., Visscher P.M., Brown M.A., Yamanaka H., Mimori T., Takahashi A., Xu H., Behrens T.W., Siminovitch K.A., Momohara S., Matsuda F., Yamamoto K., Plenge R.M. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zengini E., Hatzikotoulas K., Tachmazidou I., Steinberg J., Hartwig F.P., Southam L., Hackinger S., Boer C.G., Styrkarsdottir U., Gilly A., Suveges D., Killian B., Ingvarsson T., Jonsson H., Babis G.C., McCaskie A., Uitterlinden A.G., van Meurs J.B.J., Thorsteinsdottir U., Stefansson K., Davey Smith G., Wilkinson J.M., Zeggini E. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat. Genet. 2018;50(4):549–558. doi: 10.1038/s41588-018-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsoi L.C., Spain S.L., Knight J., Ellinghaus E., Stuart P.E., Capon F., Ding J., Li Y., Tejasvi T., Gudjonsson J.E., Kang H.M., Allen M.H., McManus R., Novelli G., Samuelsson L., Schalkwijk J., Ståhle M., Burden A.D., Smith C.H., Cork M.J., Estivill X., Bowcock A.M., Krueger G.G., Weger W., Worthington J., Tazi-Ahnini R., Nestle F.O., Hayday A., Hoffmann P., Winkelmann J., Wijmenga C., Langford C., Edkins S., Andrews R., Blackburn H., Strange A., Band G., Pearson R.D., Vukcevic D., Spencer C.C.A., Deloukas P., Mrowietz U., Schreiber S., Weidinger S., Koks S., Kingo K., Esko T., Metspalu A., Lim H.W., Voorhees J.J., Weichenthal M., Wichmann H.E., Chandran V., Rosen C.F., Rahman P., Gladman D.D., Griffiths C.E.M., Reis A., Kere J., Nair R.P., Franke A., Barker J.N.W.N., Abecasis G.R., Elder J.T., Trembath R.C. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 2012;44(12):1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H-F., Forgetta V., Hsu Y-H., Estrada K., Rosello-Diez A., Leo P.J., Dahia C.L., Park-Min K.H., Tobias J.H., Kooperberg C., Kleinman A., Styrkarsdottir U., Liu C-T., Uggla C., Evans D.S., Nielson C.M., Walter K., Pettersson-Kymmer U., McCarthy S., Eriksson J., Kwan T., Jhamai M., Trajanoska K., Memari Y., Min J., Huang J., Danecek P., Wilmot B., Li R., Chou W-C., Mokry L.E., Moayyeri A., Claussnitzer M., Cheng C-H., Cheung W., Medina-Gómez C., Ge B., Chen S-H., Choi K., Oei L., Fraser J., Kraaij R., Hibbs M.A., Gregson C.L., Paquette D., Hofman A., Wibom C., Tranah G.J., Marshall M., Gardiner B.B., Cremin K., Auer P., Hsu L., Ring S., Tung J.Y., Thorleifsson G., Enneman A.W., van Schoor N.M., de Groot L.C.P.G.M., van der Velde N., Melin B., Kemp J.P., Christiansen C., Sayers A., Zhou Y., Calderari S., van Rooij J., Carlson C., Peters U., Berlivet S., Dostie J., Uitterlinden A.G., Williams S.R., Farber C., Grinberg D., LaCroix A.Z., Haessler J., Chasman D.I., Giulianini F., Rose L.M., Ridker P.M., Eisman J.A., Nguyen T.V., Center J.R., Nogues X., Garcia-Giralt N., Launer L.L., Gudnason V., Mellström D., Vandenput L., Amin N., van Duijn C.M., Karlsson M.K., Ljunggren Ö., Svensson O., Hallmans G., Rousseau F., Giroux S., Bussière J., Arp P.P., Koromani F., Prince R.L., Lewis J.R., Langdahl B.L., Hermann A.P., Jensen J-E.B., Kaptoge S., Khaw K-T., Reeve J., Formosa M.M., Xuereb-Anastasi A., Åkesson K., McGuigan F.E., Garg G., Olmos J.M., Zarrabeitia M.T., Riancho J.A., Ralston S.H., Alonso N., Jiang X., Goltzman D., Pastinen T., Grundberg E., Gauguier D., Orwoll E.S., Karasik D., Davey- Smith G., Smith A.V., Siggeirsdottir K., Harris T.B., Zillikens M.C., van Meurs J.B.J., Thorsteinsdottir U., Maurano M.T., Timpson N.J., Soranzo N., Durbin R., Wilson S.G., Ntzani E.E., Brown M.A., Stefansson K., Hinds D.A., Spector T., Cupples L.A., Ohlsson C., Greenwood C.M.T., Jackson R.D., Rowe D.W., Loomis C.A., Evans D.M., Ackert-Bicknell C.L., Joyner A.L., Duncan E.L., Kiel D.P., Rivadeneira F., Richards J.B. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature. 2015;526(7571):112–117. doi: 10.1038/nature14878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris J.A., Kemp J.P., Youlten S.E., Laurent L., Logan J.G., Chai R.C., Vulpescu N.A., Forgetta V., Kleinman A., Mohanty S.T., Sergio C.M., Quinn J., Nguyen-Yamamoto L., Luco A-L., Vijay J., Simon M-M., Pramatarova A., Medina-Gomez C., Trajanoska K., Ghirardello E.J., Butterfield N.C., Curry K.F., Leitch V.D., Sparkes P.C., Adoum A-T., Mannan N.S., Komla-Ebri D.S.K., Pollard A.S., Dewhurst H.F., Hassall T.A.D., Beltejar M.G., Adams D.J., Vaillancourt S.M., Kaptoge S., Baldock P., Cooper C., Reeve J., Ntzani E.E., Evangelou E., Ohlsson C., Karasik D., Rivadeneira F., Kiel D.P., Tobias J.H., Gregson C.L., Harvey N.C., Grundberg E., Goltzman D., Adams D.J., Lelliott C.J., Hinds D.A., Ackert-Bicknell C.L., Hsu Y-H., Maurano M.T., Croucher P.I., Williams G.R., Bassett J.H.D., Evans D.M., Richards J.B. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019;51(2):258–266. doi: 10.1038/s41588-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zillikens M.C., Demissie S., Hsu Y-H., Yerges-Armstrong L.M., Chou W-C., Stolk L., Livshits G., Broer L., Johnson T., Koller D.L., Kutalik Z., Luan J., Malkin I., Ried J.S., Smith A.V., Thorleifsson G., Vandenput L., Hua Zhao J., Zhang W., Aghdassi A., Åkesson K., Amin N., Baier L.J., Barroso I., Bennett D.A., Bertram L., Biffar R., Bochud M., Boehnke M., Borecki I.B., Buchman A.S., Byberg L., Campbell H., Campos Obanda N., Cauley J.A., Cawthon P.M., Cederberg H., Chen Z., Cho N.H., Jin Choi H., Claussnitzer M., Collins F., Cummings S.R., De Jager P.L., Demuth I., Dhonukshe-Rutten R.A.M., Diatchenko L., Eiriksdottir G., Enneman A.W., Erdos M., Eriksson J.G., Eriksson J., Estrada K., Evans D.S., Feitosa M.F., Fu M., Garcia M., Gieger C., Girke T., Glazer N.L., Grallert H., Grewal J., Han B.G., Hanson R.L., Hayward C., Hofman A., Hoffman E.P., Homuth G., Hsueh W.C., Hubal M.J., Hubbard A., Huffman K.M., Husted L.B., Illig T., Ingelsson E., Ittermann T., Jansson J.O., Jordan J.M., Jula A., Karlsson M., Khaw K.T., Kilpeläinen T.O., Klopp N., Kloth J.S.L., Koistinen H.A., Kraus W.E., Kritchevsky S., Kuulasmaa T., Kuusisto J., Laakso M., Lahti J., Lang T., Langdahl B.L., Launer L.J., Lee J.Y., Lerch M.M., Lewis J.R., Lind L., Lindgren C., Liu Y., Liu T., Liu Y., Ljunggren Ö., Lorentzon M., Luben R.N., Maixner W., McGuigan F.E., Medina-Gomez C., Meitinger T., Melhus H., Mellström D., Melov S., Michaëlsson K., Mitchell B.D., Morris A.P., Mosekilde L., Newman A., Nielson C.M., O’Connell J.R., Oostra B.A., Orwoll E.S., Palotie A., Parker S.C.J., Peacock M., Perola M., Peters A., Polasek O., Prince R.L., Räikkönen K., Ralston S.H., Ripatti S., Robbins J.A., Rotter J.I., Rudan I., Salomaa V., Satterfield S., Schadt E.E., Schipf S., Scott L., Sehmi J., Shen J., Soo Shin C., Sigurdsson G., Smith S., Soranzo N., Stančáková A., Steinhagen-Thiessen E., Streeten E.A., Styrkarsdottir U., Swart K.M.A., Tan S.T., Tarnopolsky M.A., Thompson P., Thomson C.A., Thorsteinsdottir U., Tikkanen E., Tranah G.J., Tuomilehto J., van Schoor N.M., Verma A., Vollenweider P., Völzke H., Wactawski-Wende J., Walker M., Weedon M.N., Welch R., Wichmann H.E., Widen E., Williams F.M.K., Wilson J.F., Wright N.C., Xie W., Yu L., Zhou Y., Chambers J.C., Döring A., van Duijn C.M., Econs M.J., Gudnason V., Kooner J.S., Psaty B.M., Spector T.D., Stefansson K., Rivadeneira F., Uitterlinden A.G., Wareham N.J., Ossowski V., Waterworth D., Loos R.J.F., Karasik D., Harris T.B., Ohlsson C., Kiel D.P. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nat. Commun. 2017;8(1):80. doi: 10.1038/s41467-017-00031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia X., Shi N., Feng Y., Li Y., Tan J., Xu F., Wang W., Sun C., Deng H., Yang Y., Shi X. Identification of 67 pleiotropic genes associated with seven autoimmune/autoinflammatory diseases using multivariate statistical analysis. Front. Immunol. 2020;11:30. doi: 10.3389/fimmu.2020.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J.Z., McRae A.F., Nyholt D.R., Medland S.E., Wray N.R., Brown K.M., Hayward N.K., Montgomery G.W., Visscher P.M., Martin N.G., Macgregor S. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 2010;87(1):139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myrthianou E., Zervou M.I., Budu-Aggrey A., Eliopoulos E., Kardassis D., Boumpas D.T., Kougkas N., Barton A., Sidiropoulos P., Goulielmos G.N. Investigation of the genetic overlap between rheumatoid arthritis and psoriatic arthritis in a Greek population. Scand. J. Rheumatol. 2017;46(3):180–186. doi: 10.1080/03009742.2016.1199734. [DOI] [PubMed] [Google Scholar]
- 21.Márquez A., Vidal-Bralo L., Rodríguez-Rodríguez L., González-Gay M.A., Balsa A., González-Álvaro I., Carreira P., Ortego-Centeno N., Ayala-Gutiérrez M.M., García-Hernández F.J., González-Escribano M.F., Sabio J.M., Tolosa C., Suárez A., González A., Padyukov L., Worthington J., Vyse T., Alarcón-Riquelme M.E., Martín J. A combined large-scale meta-analysis identifies COG6 as a novel shared risk locus for rheumatoid arthritis and systemic lupus erythematosus. Ann. Rheum. Dis. 2017;76(1):286–294. doi: 10.1136/annrheumdis-2016-209436. [DOI] [PubMed] [Google Scholar]
- 22.Cheng P., Zhang Y., Huang H., Zhang W., Yang Q., Guo F., Chen A. Association between CCR6 and rheumatoid arthritis: A meta-analysis. Int. J. Clin. Exp. Med. 2015;8(4):5388–5396. [PMC free article] [PubMed] [Google Scholar]
- 23.Kulkarni N., Meitei H.T., Sonar S.A., Sharma P.K., Mujeeb V.R., Srivastava S., Boppana R., Lal G. CCR6 signaling inhibits suppressor function of induced-Treg during gut inflammation. J. Autoimmun. 2018;88:121–130. doi: 10.1016/j.jaut.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Chang W-C., Woon P-Y., Wei J.C-C., Chang C-M., Hsu Y-W., Guo Y-C., Hwang D-Y., Kochi Y., Yen J-H. A single-nucleotide polymorphism of CCR6 (rs3093024) is associated with susceptibility to rheumatoid arthritis but not ankylosing spondylitis, in a Taiwanese population. J. Rheumatol. 2012;39(8):1765–1766. doi: 10.3899/jrheum.120040. [DOI] [PubMed] [Google Scholar]
- 25.Furue K., Ito T., Tsuji G., Nakahara T., Furue M. The CCL20 and CCR6 axis in psoriasis. Scand. J. Immunol. 2020;91(3):e12846. doi: 10.1111/sji.12846. [DOI] [PubMed] [Google Scholar]
- 26.Nanke Y., Kobashigawa T., Yago T., Kawamoto M., Yamanaka H., Kotake S. RANK expression and osteoclastogenesis in human monocytes in peripheral blood from rheumatoid arthritis patients. BioMed Res. Int. 2016;2016:4874195. doi: 10.1155/2016/4874195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosaad Y.M. Clinical role of human leukocyte antigen in health and disease. Scand. J. Immunol. 2015;82(4):283–306. doi: 10.1111/sji.12329. [DOI] [PubMed] [Google Scholar]
- 28.Li S.M., Zhou D.X., Liu M.Y. Associations between polymorphisms of HLA-B gene and postmenopausal osteoporosis in Chinese Han population. Int. J. Immunogenet. 2014;41(4):324–329. doi: 10.1111/iji.12130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s website along with the published article.
Data Availability Statement
Not applicable.