Abstract
Purpose
In this study, we aimed to investigate the precise relationship between hypoxia-inducible factor 1α (HIF1α), circadian clock genes, and OSA.
Methods
We recruited 21 patients with OSA and 22 age-matched controls who underwent polysomnography and had their peripheral blood collected on the evening before and the morning after sleep. OSA was defined as an apnea hypopnea index (AHI) ≥15 events/h. Patients in which T90 > 0 were defined as having nocturnal hypoxemia (NH) and were referred to as the NH group. The mRNA levels of HIF1α, HIF1β and several clock genes (Timeless, Clock, Bmal1, Per1, Per2, Per3, Cry1, Cry2, Ck1δ, Rorα, NR1D1, and NPAS2) were determined by RT-qPCR. The percentage difference in gene expression levels when compared between the morning and evening was then determined as referred to as morning-evening variation (MEV).
Results
The MEV for HIF1α mRNA expression in OSA patients increased significantly by 23% (P = 0.008) when compared to patients without OSA. The gene expression levels of Timeless (P = 0.038) and Cry2 (P = 0.012) decreased with AHI. The MEV of Bmal1, Rorα, and HIF1α mRNA levels were upregulated by 16% (P = 0.006), 14% (P = 0.027), and 25% (P = 0.005), respectively, in participants with NH when compared to those without NH. Furthermore, the MEV for HIF1α mRNA levels was positively correlated with the MEV of Bmal1, Cry1, and CK1δ mRNA levels (R = 0.638, P < 0.001; R = 0.327, P = 0.002; R = 0.332, P = 0.001, respectively) and negatively correlated with LSpO2 (R = −0.464, P =0.009) and Mean SpO2 (R = −0.500, P = 0.003).
Conclusion
Our data suggest that patients with OSA or NH tend to develop circadian rhythm disorders that may be induced by the hypoxia-mediated augmentation of HIF1α gene expression in OSA.
Keywords: obstructive sleep apnea, hypoxia-inducible factor, circadian rhythm, apnea hypopnea index
Introduction
Obstructive sleep apnea (OSA) syndrome is a partial or complete block of the airways that can cause hypopnea or apnea accompanied by a variety of clinical manifestations, including night snoring, daytime sleepiness, and a reduction in cognitive performance.1 OSA is estimated to affect 34% and 17% of middle-aged men and women in the general population;2 the prevalence of this condition is increasing coincident with the epidemic of obesity.3 Emerging data also suggest that the presence of OSA and nocturnal hypoxemia (NH) are constantly accompanied by an increased risk of hypertension,4 diabetes,5 coronary artery disease,6 and stroke.7 In OSA, the upper airway collapses repeatedly during sleep, thus causing cycles of apnea, arousal, and compensatory hyperventilation. Collectively, these events lead to alterations in day/night physiological and circadian gene expression and associated biological processes.8–10 In addition, hypoxic signals regulate the clock by slowing the circadian cycle and dampening the amplitude of oscillations in a dose-dependent manner.11 It is speculated that the disrupted expression of circadian genes induced by OSA through the hypoxia-inducible factor (HIF) pathway could result in an increased risk of a variety of metabolic diseases and cardiovascular diseases.12 Investigating the association between HIF and the expression of circadian genes could enhance our understanding of the pathophysiology of OSA and help reduce complications associated with this condition.
The internal 24-h circadian clocks are achieved in humans and most mammals by the ocular photoreception of light from the environment and the relay of signals to the suprachiasmatic nucleus (SCN) in the hypothalamus.13 Previous studies suggested that the expression of clock genes in human monocytes isolated from peripheral whole blood could be used as indicators of changes in the central circadian rhythm.14,15 A genetic network of transcriptional and translational negative feedback loops generates rhythmic 24-h oscillations in the expression levels of core clock components. The major transcription factor, Bmal1, interacts with Clock and NPAS2 (the Clock homolog) to activate the circadian gene expression of their target genes including PER (PER1, PER2, and PER3), CRY (CRY1 and CRY2), NR1D1, and RORα.16,17 Although several studies have reported changes in the expression of canonical clock genes in patients with OSA,18–21 the results derived from these studies were not consistent.
HIF1 is a heterodimer consisting of an unstable HIF1α subunit and a constitutively expressed HIF1β subunit that is ubiquitously expressed in all tissues and functions as a primary regulator of oxygen homeostasis in humans.22 Gene expression array and chromatin immunoprecipitation assays have demonstrated that more than 1000 genes are directly trans-activated by HIF1 in response to hypoxia.23,24 Several studies have reported an increase in HIF1α in patients with OSA when compared with control subjects.25,26 Furthermore, HIF1α has also been proven to exert functionality as the regulatory node that connects hypoxia signaling with the circadian clock.11 A body of evidence now suggests that HIF1 and circadian clock genes (eg, Bmal1 and Per2) undergo bidirectional interactions.27,28 According to these previous studies, we know little about the specific interactions between HIF1α and clock genes in patients with OSA; furthermore, existing information is not consistent.29
In this study, we investigated the disturbance in circadian clock gene expression in subjects experiencing different severities of OSA without any other complications. Secondly, we aimed to investigate the association between the expression of HIF1α and clock genes in patients with OSA to gain a further understanding of the effect of intermittent hypoxia induced by OSA on genes responsible for circadian rhythm.
Materials and Methods
Participants
Polysomnography (PSG) was carried out in accordance with the American Academy of Sleep Medicine (AASM) manual 2.5 in order to score sleep and associated events.30 PSG results were then used to consecutively recruit 21 males with an apnea hypopnea index (AHI) ≥ 15 events/h and 22 male controls (mean AHI: 1.6 events/h). Patients were all recruited between August 2019 and December 2019 from the Sleep Disordered Breathing Laboratory of Peking Union Medical College Hospital (PUMCH). The present study was carried out as described previously.31 The exclusion criteria were as follows: (1) patients who had been treated for sleep apnea by continuous positive airway pressure (CPAP); (2) patients with self-reported chronic diseases including uncontrolled hypertension, chronic liver diseases, chronic kidney diseases, chronic lung diseases, perivascular diseases, heart diseases, diabetes, thromboembolism diseases, tumor, nerve system diseases and mental diseases; (3) patients with acute infectious over the previous two weeks; and (4) patients who performed shift work and slept for less than 6 h. Written and informed consent was provided by all participants. The study was approved by the PUMCH ethics committee (protocol number: JS-2013) and conducted in accordance with the Declaration of Helsinki.
Polysomnography and the Definition of Key Sleep Variables
Overnight PSG was performed using an Embla N7000 system (Natus Medical Incorporated, USA) from 10 PM to 6 AM in the sleep-disordered breathing laboratory of our hospital. The overall process and manual scoring were performed by skilled technicians who were supervised by experienced sleep physicians in accordance with the AASM manual 2.5.30 The AHI was defined as the total number of apnea and hypopnea events per hour of sleep. LSpO2 was defined as the lowest pulse oxygen saturation during sleep. The oxygen desaturation index (ODI) was defined as the frequency of instances when the blood oxygen levels decreased (desaturation) by 3% or more. T90 was defined as the percentage of time when the oxygen saturation was < 90% during the period of monitoring. An AHI ≥15 events/h was used as a cutoff to divide the patients into two groups: the control group featuring patients with an AHI <15 events/h and an OSA group of patients with an AHI ≥15 events/h based on the International Classification of Sleep Disorders (3rd Edition).31 In addition, we classified patients with a T90 > 0 into a nocturnal hypoxemia (NH) group and those with a T90 = 0 as a no NH group.
Clock Gene Expression and Biological Measurements
Samples of whole peripheral blood were collected on the evening (9–10 PM) before and early morning (6–7 AM) after PSG using PAXGene Blood RNA tubes (2.5 mL) in the sleep-disordered breathing laboratory and then stored at −80°C. Samples were processed within 6 months of acquisition. RNA was isolated with a PAXGene Blood RNA kit in accordance with the manufacturer’s instructions (QIAGEN, Germany). Total RNA was converted to cDNA using a Prime Script RT Master Mix kit (TAKARA, Japan). Relative gene expression was then quantified by quantitative real-time reverse transcription-polymerase chain reaction (RT-qPCR) performed on an Applied Biosystems StepOnePlus Real-time PCR system (Applied Biosystems, USA), using Fast SYBR Green Master Mix (Applied Biosystems, USA). Relative gene expression was analyzed by the 2−ΔΔCt method32 and normalized relative to the expression levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
All reactions were performed in duplicate, and mean expression levels for each transcript were used for analyses. The target genes were as follows: Timeless, Clock, Bmal1, Per1, Per2, Per3, Cry1, Cry2, Ck1δ, Rorα, NR1D1, NPAS2, HIF1α, and HIF1β. Forward and reverse primers specific for each gene of interest are listed in Table 1.
Table 1.
Primer Sequence Details of the Analyzed Gene (F: Forward Primer, R: Reverse Primer)
| Gene | Sequence |
|---|---|
| Human_Gapdh _F | TCGGAGTCAACGGATTTGGT |
| Human_Gapdh _R | TTCCCGTTCTCAGCCTTGAC |
| Human_Timeless _F | AGGAGCGGAGTGCAGATTTT |
| Human_Timeless _R | TCGAGAATGCCTGTTGCCTC |
| Human_Clock _F | AAGTTAGGGCTGAAAGACGACGA |
| Human_Clock _R | GAACTCCGAGAAGAGGCAGAAG |
| Human_Bmal1 _F | AGGATGGCTGTTCAGCACATGA |
| Human_Bmal1 _R | CAAAAATCCATCTGCTGCCCTG |
| Human_Per1_F | CTGAGGAGGCCGAGAGGAAAGAA |
| Human_Per1_R | AGGAGGAGGAGGCACATTTAC |
| Human_ Per 2_F | AAGCAGGTGAAAGCCAATGAAGA |
| Human_ Per 2 _R | CCACCGCAAACATATCGGCATT |
| Human_ Per 3_F | TGCAGGGCATCCTCCCTTTGA |
| Human_ Per 3_R | TCCGGCTCCAGGGATTCACAA |
| Human_ Cry1_F | CTGCGTCTACATCCTGGACC |
| Human_ Cry1_R | GAAGCAAAAATCGCCACCTGT |
| Human_ Cry2_F | TCCCTAGCATGTCAGCCCGTT |
| Human_ Cry2_R | AGGATTTGAGGCACTGTTCCGA |
| Human_ Ck1δ_F | CTCACAGAATAGCATTCCTTTCG |
| Human_ Ck1δ_R | TGCACGACAGACTGAAGACC |
| Human_ Rorα_F | TCGCAGCGATGAAAGCTCAAAT |
| Human_ Rorα_R | GTGGCATTGCTTTGCTGACT |
| Human_ NR1D1_F | ACAGCTGACACCACCCAGATC |
| Human_ NR1D1_R | CATGGGCATAGGTGAAGATTTCT |
| Human_ NPAS2_F | ATCCTGCAGGCCAATATCCG |
| Human_ NPAS2_R | CTGCTGCAGGAACATCTGGA |
| Human_ HIF1α_F | GCCGCTGGAGACACAATCAT |
| Human_ HIF1α_R | GAAGTGGCTTTGGCGTTTCA |
| Human_ HIF1β_F | GACTACTGCCAACCCCGAAA |
| Human_ HIF1β_R | CTCATCATCCGACCTGGCAA |
The levels of key biochemical parameters were determined in fasting peripheral blood within 1 h using standard techniques. Plasma was obtained after centrifugation for 30 min at 3000 g at 4°C for 15 min, stored at −20°C for less than 1 week and then transferred to −80°C. Plasma levels of HIF1α were measured in batches within 6 months of acquisition using a Human/Mouse Total HIF1α ELISA kit (R&D Systems, USA) in accordance with the manufacturer’s protocol.
Statistical Analysis
All calculations were performed using SPSS version 22.0 (IBM, USA) and GraphPad Prism 5.0 (GraphPad, USA). The normality of variables was assessed by the Kolmogorov–Smirnov normality test. We used t-tests or Chi-squared tests to compare between groups for normally distributed variables and categorical variables, respectively. The non-parametric Mann–Whitney U-test was used when data were not normally distributed. An ANOVA or Kruskal–Wallis test was used for non-parametric multi-group comparisons. For each sample of whole blood, CT data were examined, and samples in which the expression level of GAPDH was ≥23, thus indicating unreliable; these samples were excluded from our analyses. Gene expression data were analyzed by comparing the mean fold change of transcript levels between groups. The percentage difference between morning and evening (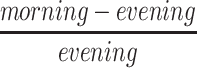 ) was used to describe morning-evening variation (MEV). Pearson’s or Spearman’s methods were used to analyze the correlation between gene expression and PSG results. P < 0.05 was considered statistically significant.
) was used to describe morning-evening variation (MEV). Pearson’s or Spearman’s methods were used to analyze the correlation between gene expression and PSG results. P < 0.05 was considered statistically significant.
Results
Characteristics of Participants
Demographic variables, clinical characteristics, PSG parameters, and laboratory data are shown in Table 2. The mean age of the cohort was 39 years (range, 26–56 years); there was no significant difference between the control and OSA groups in terms of age. The mean body mass index (BMI) was 26.6 kg/m2 and was significantly higher in OSA (P = 0.001) or NH (P < 0.001) subjects than those in the control group. In addition, 49% of the study cohort were diagnosed as NH because T90 (percent of time spent with SpO2 below 90%) was > 0. After adjustment for age and BMI, the serum HbA1c (hemoglobin A1C) levels in OSA patients were significantly higher than those of the control participants (P = 0.03). Moreover, the levels of glucose, HbA1c, and ALT (Alanine aminotransferase) in patients with NH were significantly higher than the subjects in the no NH group after adjusting for age and BMI (P = 0.022, P = 0.033 and P = 0.039, respectively). AHI, ODI, mean SpO2, LSpO2 (%, lowest pulse oxygen saturation), and T90 (%) are greatly increased both in patients with OSA or NH.
Table 2.
General Characteristics of the Study Participants
| Control (AHI<15 Events/h, n=22) | OSA (AHI≥15 Events/h, n=21) | P value | no NH (T90<0, n=22) | NH (T90>0, n=21) | P value | |
|---|---|---|---|---|---|---|
| Age (years) | 39.0±7.8 | 38.9±8.3 | 0.969 | 37.5±6.9 | 40.4±8.8 | 0.238 |
| BMI (kg/m2) | 25.1±3.1 | 28.1±2.4 | 0.001 | 25.0±3.1 | 28.2±2.3 | <0.001 |
| Current smokers(n) | 3 | 5 | 0.457 | 4 | 4 | 1.000 |
| Hypertension (n) | 5 | 9 | 0.203 | 4 | 10 | 0.055 |
| AHI (events/h) | 1.6 (0.6, 6.9) | 42.4 (21.1, 63.1) | <0.001 | 1.6 (0.6, 8.6) | 40.4 (18.2, 63.2) | <0.001 |
| ODI (events/h) | 1.4(0.3,4.9) | 38.6(19.1,60.1) | <0.001 | 1.4(0.3,4.9) | 38.6(17.0,60.1) | <0.001 |
| Mean SpO2(%) | 97.3 (95.6, 98.1) | 95.8 (94.0, 97.4) | 0.029 | 97.7 (95.8, 98.4) | 95.1 (94.0, 96.7) | <0.001 |
| LSpO2(%) | 91.6±4.0 | 81.7±6.7 | <0.001 | 92.4±2.6 | 80.8±5.8 | <0.001 |
| T90(%) | 0(0, 0) | 0.6(0.2,1.6) | <0.001 | 0(0, 0) | 0.6(0.3, 1.6) | <0.001 |
| ALT(U/L) | 25(19, 35.5) | 46(27.5, 56) | 0.017 | 25(19, 33.2) | 44(27.5, 62.5) | 0.011 |
| Cr (umol/L) | 80.1±8.2 | 75.4±11.5 | 0.128 | 80.3±7.8 | 75.2±11.7 | 0.099 |
| Glucose (mmol/L) | 4.9±0.6 | 5.2±0.6 | 0.082 | 4.8±0.6 | 5.3±0.6 | 0.008 |
| HBA1C(%) | 5.2(5.0, 5.5) | 5.5(5.3, 5.6) | 0.025 | 5.2(5.0, 5.5) | 5.5(5.4, 5.6) | 0.006 |
| TG (mmol/L) | 1.1(0.8, 1.8) | 2.4(1.5, 4.2) | 0.004 | 1.1(0.8, 1.8) | 2.4(1.5, 4.2) | 0.004 |
| TC (mmol/L) | 4.7±0.7 | 4.8±0.8 | 0.722 | 4.7±0.7 | 4.8±0.8 | 0.965 |
| LDL (mmol/L) | 3.2±0.7 | 2.9±0.8 | 0.357 | 3.2±0.7 | 2.9±0.8 | 0.200 |
| HDL (mmol/L) | 1.2±0.3 | 1.0±0.2 | 0.026 | 1.2±0.3 | 1.0±0.2 | 0.005 |
| UA (umol/L) | 391.6±74.8 | 425.4±93.5 | 0.197 | 410.1±95.8 | 406.0±74.8 | 0.874 |
| hsCRP (mg/L) | 0.8(0.6, 1.1) | 2.0(0.8, 2.6) | 0.002 | 0.9(0.6, 1.8) | 1.2(0.8, 2.4) | 0.194 |
Notes: Data are presented as mean values and SD, counts, or medians and interquartile ranges, as appropriate. Differences tested by t-test for each OSA subset against Controls.
Abbreviations: AHI, Apnea-hypopnea index; ALT, Alanine aminotransferase; BMI, Body mass index; Cr, Creatinine; HBA1C, hemoglobin A1C; HDL, high-density lipoprotein; hsCRP, high-sensitivity C-reactive protein; LDL, low-density lipoprotein; LSpO2, Lowest pulse arterial oxygen saturation; T90 (%), percent of time spent with SpO2 below 90%; TC, Total cholesterol; TG, Triglyceride; UA, Uric acid.
The mRNA Expression Levels of Clock Genes and HIF1α According to OSA Status
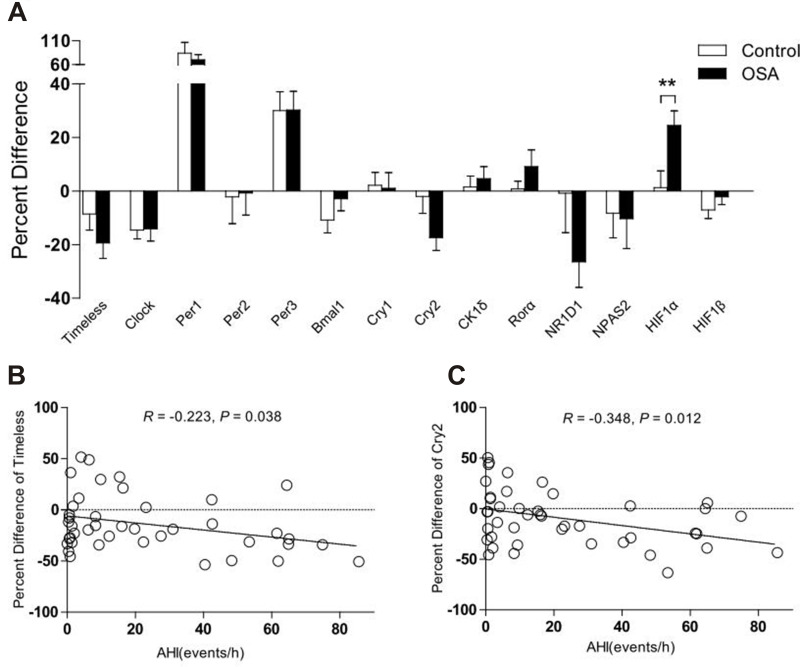
No significant differences were detected between the OSA and control group with regards to the mRNA expression levels of clock genes and HIF1a; this was the case for both the evening and morning samples (P > 0.05). No significant differences were identified between the evening and morning samples of both groups (P > 0.05) in terms of the mRNA levels of clock genes and HIF1α (P > 0.05). The MEVs of each target gene (the percentage difference for each gene) is depicted in Figure 1A. Of these, the MEV for HIF1α mRNA expression increased by 23% in subjects with OSA when compared to those without OSA (P = 0.008) after one night of sleep. However, no significant changes were detected in the expression levels of other clock genes when compared to control subjects. Further analysis detected a significant trend towards the downregulation of MEV for the Timeless gene (R = −0.223, P = 0.038) and Cry2 gene (R = −0.348, P = 0.012) as AHI values increased (Figure 1B and C).
Figure 1.
The percentage difference of morning-evening variation in clock gene expression for subjects with and without OSA. (A) expression of HIF1α increased by 23% in subjects with OSA when compared with those without OSA (P = 0.008) (B and C) Relative gene expression of Timeless (R = −0.223, P = 0.038) and Cry2 (R= −0.348, P = 0.012) in subjects with OSA decreased with increasing AHI. Data represent means ± SEM. **P < 0.01.
The mRNA Expression Levels of Clock Genes and HIF1α According to NH Status
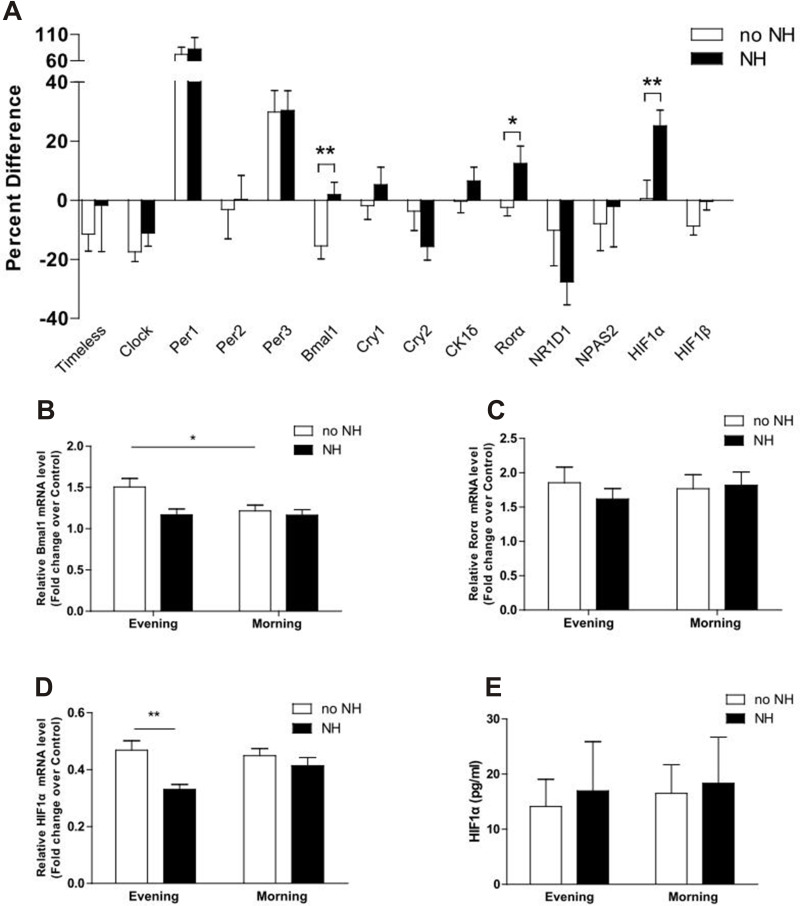
In the NH group, the MEVs of Bmal1, Rorα, and HIF1α were upregulated by 16% (P = 0.006), 14% (P = 0.027), and 25% (P = 0.005), respectively, when compared with the no NH group (Figure 2A). The mRNA expression of Bmal1 mRNA in the morning decreased significantly after sleep in the no NH group (P = 0.039), but not in the NH group (Figure 2B). A similar trend was also observed for the expression of Rorα, although without statistical significance (Figure 2C). The mRNA levels of HIF1α increased after a single night of sleep in the NH group but this change was not statistically significant. Analyses also showed that the evening levels of HIF1α mRNA were significantly higher in the no NH group than in the NH group (P = 0.003) (Figure 2D). Consequently, the protein levels of HIF1α in both groups had increased the following morning; however, these changes were not statistically significant due to wide variation and the small sample size in the NH and no NH groups at both time points (Figure 2E). No statistical differences were found for the mRNA expression levels of other clock genes when comparing the NH and no NH group at both time points or when comparing evening and morning samples for both groups.
Figure 2.
The percentage difference of morning-evening variation in clock gene expression for subjects with and without NH. (A-D) The percentage difference of Bmal1, Rorα, and HIF1α were up-regulated by 16% (P = 0.006), 14% (P = 0.027), and 25% (P = 0.005), respectively, in subjects with nocturnal hypoxemia. (E) The protein concentration of HIF1α was increased in patients with NH after one-night of hypoxia but did not reach a significant statistical difference. Data represent means ± SEM. * P < 0.05, ** P < 0.01.
Correlations Between the MEVs of HIF1α and the MEVs of Clock Gene and PSG Parameters
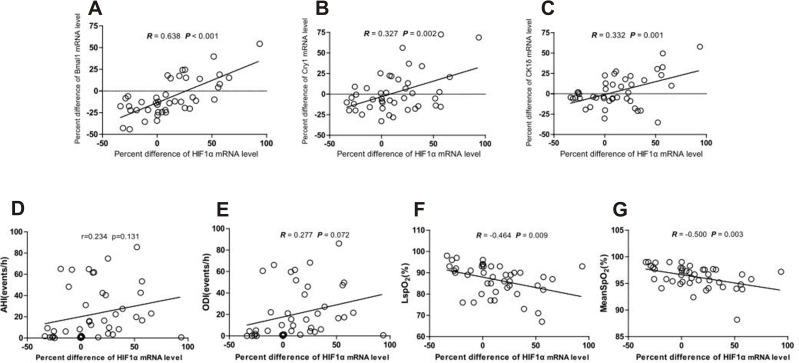
Next, we attempted to identify correlations between the MEVs of HIF1α and clock genes mRNA expression levels and found that the MEV of HIF1α mRNA levels was positively correlated with the MEVs of Bmal1, Cry1, and CK1δ mRNA levels (R = 0.638, P < 0.001; R = 0.327, P = 0.002; R = 0.332, P = 0.001, respectively, Figure 3A–C); the MEV of HIF1α did not correlate with the expression of any of the other clock genes. We also investigated the MEVs of HIF1α mRNA levels and PSG parameters. As shown in Figure 3F and G, the MEV of HIF1α mRNA expression was negatively correlated with LSpO2 (%) and mean SpO2 (%) (R = −0.46, P = 0.009; R = −0.5, P = 0.003; respectively). There was no significant correlation between AHI, ODI, and the MEV for HIF1α mRNA.
Figure 3.
Correlations between morning-evening variation of HIF-1α and circadian genes mRNA levels (Bmal1, Cry1, and CK1δ), PSG parameters (AHI, ODI LSpO2, and Mean SpO2). (A–C) Correlation between morning-evening variation for HIF1α and Bmal1, Cry1, and CK1δ mRNA level (R = 0.638, P < 0.001; R = 0.327, P = 0.032; R = 0.332, P = 0.030, respectively). (D–G) Correlation between morning-evening variation for HIF1α mRNA level and PSG parameters (AHI, ODI, LSpO2, and Mean SpO2) (R = 0.234, P = 0.131; R = 0.277, P = 0.072; R = −0.464, P = 0.009; R = −0.500, P = 0.003, respectively).
Discussion
In this preliminary study, we showed that patients with OSA or NH had a significantly higher MEV for HIF1α mRNA expression. We also found that the MEVs for Bmal1 and Rorα mRNA expression were significantly increased in patients with NH. In addition, we identified positive correlations between the MEVs for Bmal1, Cry1, CK1δ, and HIF1α. Intermittent hypoxia is known to be the main contributor to OSA-related pathologies and place patients at a higher risk of developing multiple comorbidities.33 Our findings suggest that an important interrelationship may exist between OSA-induced hypoxia and clock gene disturbance.
We also observed a remarkable increase in the amplitude of HIF1α mRNA expression levels in the mornings in patients from the OSA or NH groups after one night of sleep. A similar trend, albeit not significant, was observed for the levels of HIF1α protein in subjects from the NH group. This may be explained by changes in post-translational regulation rather than transcriptional control which leads to a mismatch between HIF1α expression at the mRNA and protein levels in blood serum. In addition, the MEV for HIF1α mRNA level was negatively correlated with LSpO2 and mean SpO2 but not AHI. These results further implied that more HIF1α were upregulated in response to acute hypoxia in one night than the chronic intermittent hypoxia induced by OSA. In fact, previous studies on the relationship between HIF1α and OSA are inconsistent.25,29,34 Our current findings are similar to those reported by Kaczmarek et al, who showed that the mRNA expression of HIF1α in skin biopsies in severely hypoxemic OSA patients was significantly higher than those in control subjects or those who had mildly hypoxemic OSA.34 However, other studies found that the serum levels of HIF1α protein were chronically increased in patients with OSA when compared with control patients.25,29 In another study, Gabryelska et al found that the serum levels of HIF1α protein were higher in patients with OSA in both the evening and the morning; moreover, there were no significant differences between the OSA and control group with regards to HIF1α mRNA expression.25 In comparison, we believe our findings might be due to the fact that a single night, with a reduction in the oxygen saturation of hemoglobin, may have directly affected the mRNA expression of HIF1α in the peripheral blood cells. However, this changes were not sufficient to cause a significant upregulation in the protein levels of HIF1α in our cohort of patients, who were relatively young OSA patients with a small BMI, and without significant complications. Another interpretation is that, during hypoxia, the overexpressed HIF1α protein translocate from the cytoplasm to the nucleus to form a transcriptionally active HIF1 complex via dimerization with HIF-1β to induce gene expression. Thus, the increase in serum HIF-1α protein was not so evident.
Our results showed no significant changes in the expression of circadian rhythm genes in OSA patients, irrespective of whether we considered evening or morning time points. These findings are somewhat different from those described in previous studies, in which the changes of circadian genes are inconsistent neither. A cross-sectional analysis of 49 veterans with chronic kidney disease found that the expression of Per3 was reduced in sleep apnea, and that the expression levels of NPAS2, Per1, and NR1D1 were reduced in subjects with NH.18 Similarly, another study described the disruption of Per1 mRNA expression levels in subjects with OSA and that CPAP treatment reversed this condition in 16 subjects.20 However, Moreira et al reported statistically significant differences for the expression of clock genes in 13 patients with severe OSA when compared to 8 healthy subjects; this condition was not reversed by CPAP therapy.21
Furthermore, our results showed that the MEVs for Cry2 and Timeless mRNA levels decreased with the increasing severity of sleep apnea. Cry1 and Cry2 are necessary for the maintenance of circadian behavioral rhythms; both circadian rhythmicity and light-induced suppression of pineal melatonin concentration were disturbed in Cry1 and Cry2 double-deficient mice.35,36 Meanwhile, the human Timeless protein interacts with both Cry2 and cell cycle checkpoint proteins and plays an important role in the DNA damage checkpoint response.37 We speculated that OSA may influence the DNA repair process, or potentially apoptosis. Moreover, we identified a positive correlation between the mRNA levels of HIF1α and Cry1; this was similar to previous findings in which the evening levels of Cry1 proteins were positively related to the levels of HIF1α protein.38 According to our current study, the two isoforms of Cry protein may play opposing roles in OSA and NH.
After a single night in the presence of apnea and hypoxia, we found that the gene expression of Bmal1, Rorα, and HIF1α were enhanced in participants with NH when compared to the control group. In addition, the MEV for Bmal1 was also positively correlated with the expression of HIF1α. Previous reports showed that Rorα can directly activate the transcription of Bmal1, thereby maintaining a robust circadian rhythm in the peripheral tissues,39 and the murine SCN.40 Simultaneously, transcription of the Rorα gene appeared to be upregulated by hypoxia in a panel of cell lines derived from different human tissues.41 Moreover, it is known that Bmal1 and Rorα form a regulatory loop and increase the transcriptional activity of HIF1α within the nucleus.42,43 Consistent with this view, our data showed that the mRNA expression of Rorα and HIF1α were both upregulated in the morning after one night of hypoxia. Therefore, our results provide a new insight for the function of Rorα in the amplification of hypoxia signaling. Further research is now needed to elucidate the potential role of Rorα in OSA. CK1δ is a serine/threonine-specific kinase that phosphorylates HIF1α at S247 to reduce its affinity for its hetero-dimerization partner HIF-1β.44,45 Studies also showed that the silencing of CK1δ enhanced the activity of HIF1 under hypoxic conditions while the overexpression of CK1δ inhibited HIF1 activity under such conditions; these data suggest the importance of the CK1δ-dependent phosphorylation of HIF1α.46 In the present study, the enhancement of CK1δ expression under hypoxic conditions may contribute to the accumulation of HIF1α protein in patients with OSA.
The current study had key advantages that need to be highlighted. For example, only a very limited number of human studies have been published relating to the potential association between HIF1α and clock gene expression in patients with OSA. Secondly, our methodology was clinically accessible and implemented efficiently. In our study, we only included males without any severe comorbidities; in this manner, we were able to reduce the potential effects of confounding factors because OSA is a heterogeneous disease with different clinical phenotypes.47 However, there are some limitations that need to be considered. First, we only analyzed the effect of hypoxia on HIF1α and clock genes after one night; we did not consider data over longer periods of time. Further research is now needed to investigate the mRNA and protein levels of HIF1α at more time points during the day and determine the precise correlations between HIF1α expression at the mRNA and protein levels. Secondly, the relatively small sample size limits the generalizability of our conclusions; future studies should be conducted in a larger OSA cohort to investigate the expression of HIF1α and clock genes in a more comprehensive manner.
Conclusions
In conclusion, the current study indicated that OSA was associated with some alterations in the expression of several clock genes, including Timeless, Cry1, Cry2, Bmal1, Rorα, and CK1δ. Interestingly, these disruptions in clock gene expression may be mediated by HIF1α; the mRNA morning–evening variation for HIF1α was positively correlated with the expression of Bmal1, Cry1, and CK1δ. Our findings suggest that OSA and OSA-related nocturnal hypoxia may be associated with the altered expression of clock genes during the response to hypoxia. A better understanding of the direction and magnitude of the association between OSA and 24-h gene expression patterns of key clock genes may open the door for the development of novel therapeutic targets to treat OSA-related complications.
Acknowledgments
The help from Hairong Zhang, Shuning Li in collecting data is greatly appreciated. Informed consent was obtained from all participants involved in the study. All authors have seen and approved the submitted version of this manuscript.
Funding Statement
This study was supported by the National Natural Science Foundation of China (81570085) and CAMS Innovation Fund for Medical Sciences(CIFMS)(2021-I2M-C&T-B-013).
Brief Summary
There is a significant lack of literature relating to circadian clock disruption in patients experiencing OSA. This study aimed to investigate the relationship between HIF1α, a series of circadian clock genes, and polysomnography variables.
Data highlight the positive association between the mRNA expression of a series of clock genes and HIF1α: a trend towards the downregulation of Timeless and Cry2 with increasing OSA severity, and an increase in the morning-evening variation of Bmal1 and Rorα mRNA expression in patients with NH. Findings enhance our understanding of the complex relationship between intermittent hypoxia, circadian rhythms, and OSA.
Abbreviations
AASM, American Academy of Sleep Medicine; AHI, apnea-hypopnea index; ALT, alanine aminotransferase; BMI, body mass index; CK1, casein kinase 1; CPAP, continuous positive airway pressure; Cr, creatinine; Cry, cryptochrome; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HBA1C, hemoglobin A1C; HDL, high-density lipoprotein; HIF1α, hypoxia-inducible factor 1α; hsCRP, high-sensitivity C-reactive protein; LDL, low-density lipoprotein; LSpO2, Lowest pulse arterial oxygen saturation; MEV, morning-evening variation; NH, nocturnal hypoxemia; OSA, obstructive sleep apnea; Per, period; PSG, polysomnography; PUMCH, Peking Union Medical School of Hospital; RT-qPCR, quantitative real-time polymerase chain reaction; SCN, suprachiasmatic nucleus; SpO2, oxygen saturation; T90 (%), Sleep time spent with oxygen saturation < 90%; TBA, total bile acid; TC, total cholesterol; TG, triglyceride; UA, uric acid.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Gottlieb DJ, Punjabi NM. Diagnosis and management of obstructive sleep apnea: a review. JAMA J Am Med Assoc. 2020;323(14):1389. doi: 10.1001/jama.2020.3514 [DOI] [PubMed] [Google Scholar]
- 2.Peppard PE, Young T, Barnet JH, Palta M, Hagen EW, Hla KM. Increased prevalence of sleep-disordered breathing in adults. Am J Epidemiol. 2013;177(9):1006–1014. doi: 10.1093/aje/kws342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garvey JF, Pengo MF, Drakatos P, Kent BD. Epidemiological aspects of obstructive sleep apnea. J Thorac Dis. 2015;7(5):920–929. doi: 10.3978/j.issn.2072-1439.2015.04.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bangash A, Wajid F, Poolacherla R, Mim FK, Rutkofsky IH. Obstructive Sleep Apnea and Hypertension: a Review of the Relationship and Pathogenic Association. Cureus. 2020;12(5):e8241–e8241. doi: 10.7759/cureus.8241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doumit J, Prasad B. Sleep Apnea in Type 2 Diabetes. Diabetes Spectrum. 2016;29(1):14. doi: 10.2337/diaspect.29.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent BD, Garvey JF, Ryan S, Nolan G, Dodd JD, McNicholas WT. Severity of obstructive sleep apnoea predicts coronary artery plaque burden: a coronary computed tomographic angiography study. Eur Respir J. 2013;42(5):1263. doi: 10.1183/09031936.00094812 [DOI] [PubMed] [Google Scholar]
- 7.Yaggi HK, Concato J, Kernan WN, Lichtman JH, Brass LM, Mohsenin V. Obstructive Sleep Apnea as a Risk Factor for Stroke and Death. N Eng J Med. 2005;353(19):2034–2041. doi: 10.1056/NEJMoa043104 [DOI] [PubMed] [Google Scholar]
- 8.Butler MP, Thosar SS, Smales C, et al. Effects of obstructive sleep apnea on endogenous circadian rhythms assessed during relaxed wakefulness; an exploratory analysis. Chronobiol Int. 2020;37(6):856–866. doi: 10.1080/07420528.2020.1740723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manella G, Aviram R, Bolshette N, Muvkadi S, Asher G. Hypoxia induces a time- And tissue-specific response that elicits intertissue circadian clock misalignment. Proc National Acad Sci. 2019;117(1):201914112. doi: 10.1073/pnas.1914112117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connell EJ, Martinez CA, Liang YG, Cistulli PA, Cook KM. Out of Breath, Out of Time: interactions between HIF and Circadian Rhythms. AJP Cell Physiology. 2020;319(3):9857. doi: 10.1152/ajpcell.00305.2020 [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Tang D, Liu N, et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017;25(1):73–85. doi: 10.1016/j.cmet.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 12.Gabryelska A, Turkiewicz S, Karuga FF, Sochal M, Strzelecki D, BiaBasiewicz P. Disruption of Circadian Rhythm Genes in Obstructive Sleep Apnea Patient“ Possible Mechanisms Involved and Clinical Implication. Int J Mol Sci. 2022;23. doi: 10.3390/ijms23020709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer A, Merrow M. Circadian Clocks. Handb Exp Pharmacol. 2013;148(5):S29. [Google Scholar]
- 14.Burioka N, Takata M, Endo M, et al. Treatment with β 2 ‐Adrenoceptor Agonist in Vivo Induces Human Clock Gene, Per1, mRNA Expression in Peripheral Blood. Chronobiol Int. 2007;24:183–189. doi: 10.1080/07420520601140043 [DOI] [PubMed] [Google Scholar]
- 15.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102(12):4143–4145. doi: 10.1182/blood-2003-03-0779 [DOI] [PubMed] [Google Scholar]
- 16.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965 [DOI] [PubMed] [Google Scholar]
- 17.Schibler U. The daily timing of gene expression and physiology in mammals. Dialogues Clin Neurosci. 2007;9(3):257–272. doi: 10.31887/DCNS.2007.9.3/uschibler [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang M-Y, Lin P-W, Lin H-C, et al. Alternations of Circadian Clock Genes Expression and Oscillation in Obstructive Sleep Apnea. J Clin Med. 2019;8(10):987. doi: 10.3390/jcm8101634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canales MT, Holzworth M, Bozorgmehri S, et al. Clock gene expression is altered in veterans with sleep apnea. Physiol Genomics. 2019;51(3):77–82. doi: 10.1152/physiolgenomics.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burioka N, Koyanagi S, Endo M, et al. Clock gene dysfunction in patients with obstructive sleep apnoea syndrome. Eur Respir J. 2008;32(1):105. doi: 10.1183/09031936.00138207 [DOI] [PubMed] [Google Scholar]
- 21.Moreira S, Rodrigues R, Barros AB, et al. Changes in Expression of the CLOCK Gene in Obstructive Sleep Apnea Syndrome Patients Are Not Reverted by Continuous Positive Airway Pressure Treatment. Front Med. 2017;4:187. doi: 10.3389/fmed.2017.00187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc National Acad Sci. 1995;92(12):5510. doi: 10.1073/pnas.92.12.5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manalo D, Rowan A, Lavoie T, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958 [DOI] [PubMed] [Google Scholar]
- 24.Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117(23):e207–17. doi: 10.1182/blood-2010-10-314427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabryelska A, Szmyd B, Panek M, Szemraj J, Biaasiewicz P. Serum Hypoxia-Inducible Factor-1α protein level as a diagnostic marker of obstructive sleep apnea. Polskie Archiwum Medycyny Wewn. 2019;130(2):287. doi: 10.20452/pamw.15104 [DOI] [PubMed] [Google Scholar]
- 26.Gabryelska A, Szmyd B, Szemraj J, Stawski R, Sochal M, Białasiewicz P. Patients with obstructive sleep apnea present with chronic upregulation of serum HIF-1α protein. J Clin Sleep Med. 2020;16(10):1761–1768. doi: 10.5664/jcsm.8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi M, Morinibu A, Koyasu S, Goto Y, Hiraoka M, Harada H. A circadian clock gene, PER2, activates HIF-1 as an effector molecule for recruitment of HIF-1α to promoter regions of its downstream genes. FEBS J. 2017;284(22):3804–3816. doi: 10.1111/febs.14280 [DOI] [PubMed] [Google Scholar]
- 28.Peek CB, Levine DC, Cedernaes J, et al. Circadian Clock Interaction with HIF1α Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab. 2017;25(1):86–92. doi: 10.1016/j.cmet.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabryelska A, Stawski R, Sochal M, Szmyd B, Bialasiewicz P. Influence of one‐night CPAP therapy on the changes of HIF‐1α protein in OSA patients: a pilot study. J Sleep Res. 2020. doi: 10.1111/jsr.12995 [DOI] [PubMed] [Google Scholar]
- 30.Berry RB, Albertario CL, Harding SM, et al. The AASM Manual for the Scoring of Sleep and Associated Events: rules, Terminology and Technical Specifications. Original Res. 2018:2018;76. [Google Scholar]
- 31.Luo J, Wang X, Guo Z, et al. Endothelial Function and Arterial Stiffness Should Be Measured to Comprehensively Assess Obstructive Sleep Apnea in Clinical Practice. Original Research. Front Cardiovascular Med. 2021;2021:8. doi: 10.3389/fcvm.2021.716916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 33.American Academy of Sleep Medicine. International Classification of Sleep Disorders. 3rd ed. Darien. IL: American Academy of Sleep Medicine; 2014. [Google Scholar]
- 34.Kaczmarek E, Bakker JP, Clarke DN, et al. Molecular biomarkers of vascular dysfunction in obstructive sleep apnea. PLoS One. 2013;8(7):e70559. doi: 10.1371/journal.pone.0070559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horst G, Muijtjens M, Kobayashi K, Takano R, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627–630. doi: 10.1038/19323 [DOI] [PubMed] [Google Scholar]
- 36.Yamanaka Y, Suzuki Y, Todo T, Honma K, Honma S. Loss of circadian rhythm and light-induced suppression of pineal melatonin levels in Cry1 and Cry2 double-deficient mice. Genes to Cells. 2010;15(10):1063–1071. doi: 10.1111/j.1365-2443.2010.01443.x [DOI] [PubMed] [Google Scholar]
- 37.Unsal-Kaçmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25(8):3109–3116. doi: 10.1128/mcb.25.8.3109-3116.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gabryelska A, Sochal M, Turkiewicz S, Biaasiewicz P. Relationship between HIF-1 and Circadian Clock Proteins in Obstructive Sleep Apnea Patients—Preliminary Study. J Clin Med. 2020;9(5):1599. doi: 10.3390/jcm9051599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12(5):441–448. doi: 10.1038/nsmb925 [DOI] [PubMed] [Google Scholar]
- 40.Sato TK, Panda S, Miraglia LJ, Reyes TM, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018 [DOI] [PubMed] [Google Scholar]
- 41.Hongyu L, Longjian Z, Jun D. Retinoic acid receptor-related orphan receptor ROR regulates differentiation and survival of keratinocytes during hypoxia. J Cell Physiol. 2018;233(1):641–650. doi: 10.1111/j.1471-4159.2008.05739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suyama K, Silagi ES, Choi H, et al. Circadian factors BMAL1 and RORα control HIF-1α transcriptional activity in nucleus pulposus cells: implications in maintenance of intervertebral disc health. Oncotarget. 2014;7(17):786. doi: 10.18632/oncotarget.8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chauvet C, Bois-Joyeux B, Berra E, Pouyssegur J, Danan JL. The gene encoding human retinoic acid-receptor-related orphan receptor alpha is a target for hypoxia-inducible factor 1. Biochem J. 2004;384(Pt 1):84. doi: 10.1042/BJ20040709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalousi A, Mylonis I, Politou AS, Chachami G, Simos G. Casein kinase 1 regulates human hypoxia-inducible factor HIF-1. J Cell Sci. 2010;123(Pt 17):2976–2986. doi: 10.1242/jcs.068122 [DOI] [PubMed] [Google Scholar]
- 45.Kourti M, Ikonomou G, Giakoumakis -N-N, et al. CK1δ restrains lipin-1 induction, lipid droplet formation and cell proliferation under hypoxia by reducing HIF-1α/ARNT complex formation. Cell Signal. 2015;27(6):1129–1140. doi: 10.1016/j.cellsig.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koyasu S, Kobayashi M, Goto Y, Hiraoka M, Harada H. Regulatory mechanisms of hypoxia-inducible factor 1 activity: two decades of knowledge. Cancer Sci. 2018;109(3):560–571. doi: 10.1111/cas.13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keenan BT, Kim J, Singh B, et al. Recognizable clinical subtypes of obstructive sleep apnea across international sleep centers: a cluster analysis. Sleep. 2018;41(3):zsx214. doi: 10.1093/sleep/zsx214 [DOI] [PMC free article] [PubMed] [Google Scholar]