Abstract
Although tremendous efforts have been made to prevent and treat HIV-1 infection, HIV-1/AIDS remains a major threat to global human health. The combination antiretroviral therapy (cART), although able to suppress HIV-1 replication, cannot eliminate the proviral DNA integrated into the human genome and thus requires lifelong treatment that may lead to various side effects. In recent years, clustered regularly interspaced short palindromic repeat (CRISPR)-associated nuclease 9 (Cas9) related gene-editing systems have been developed and designed as effective ways to treat HIV-1 infection. However, new gene-targeting tools derived from or functioning like CRISPR/Cas9, including base editor, prime editing, SHERLOCK, DETECTR, PAC-MAN, ABACAS, pfAGO, have been developed and optimized for pathogens detection and diseases correction. Here, we summarize recent studies on HIV-1/AIDS gene therapy and provide more gene-editing targets based on studies relating to the molecular mechanism of HIV-1 infection. We also identify the strategies and potential applications of these new gene-editing technologies for HIV-1/AIDS treatment in the future. Moreover, we discuss the caveats and problems that should be addressed before the clinical use of these versatile CRISPR-based gene targeting tools. Finally, we offer alternative solutions to improve the practice of gene targeting in HIV-1/AIDS gene therapy.
Keywords: HIV-1/AIDS, CRISPR/Cas9, Gene editing, Gene therapy
Highlights
-
•
New gene-targeting tools derived from CRISPR/Cas9 have been introduced.
-
•
Recent researches in HIV-1/AIDS gene therapy have been summarized.
-
•
The strategies and potential applications of new gene editing technologies for HIV-1/AIDS treatment have been provided.
-
•
The caveats and challenges in HIV-1/AIDS gene therapy have been discussed.
1. Introduction
1.1. Human immunodeficiency virus (HIV)
Acquired immunodeficiency syndrome (AIDS), caused by HIV-1 infection, has been a severe threat to global human health since it was first reported in the early 1980s by the Center for Disease Control and Prevention (CDC) in the USA (CDC, 1982). HIV-1 is a retrovirus with two copies of approximately 9.8-kb full-length genomic RNA consisting of two long terminal repeat (LTR) sequences and genes encoding ten viral proteins, including gag, pol, vif, vpr, vpu, env, tat, rev, nef, and antisense protein (ASP) (Liu et al., 2019). HIV-1 invades target cells by binding its envelope surface protein gp120 to the CD4 main receptor on the membrane of the target cell, and then interacts with the chemokine co-receptor CCR5 or CXCR4 (Alkhatib et al., 1996; Bleul et al., 1996). CCR5 is the main co-receptor for CCR5 (R5)-tropic HIV-1 entry into lymphocytes, monocytes, macrophages, and CD4+ T cells (Keele et al., 2008), while CXCR4 is the co-receptor for CXCR4 (X4)-tropic viral strains (Hunt et al., 2006). In the late stage of R5-tropic HIV-1 infection or when CCR5 is destroyed, HIV-1 has evolved to use CXCR4 as an alternative co-receptor for viral entry (Hütter et al., 2015; Poveda, 2015). By binding to the receptors, HIV-1 enters cells through membrane fusion and converts viral genomic RNA into double-stranded DNA (dsDNA) by RNA reverse transcriptase. Then, the dsDNA integrates into the host genome to form a provirus that can either actively transcribe RNA to generate progeny viruses or enter a latent state without viral production (Finzi et al., 1999; Zheng et al., 2005). Latent HIV reservoirs are established during the earliest stage of HIV infection in resting CD4+ T cells (Siliciano et al., 2003; Chun et al., 2008), macrophages (Smith et al., 2003), astrocytes and microglial cells (Bagasra et al., 1996; Narasipura et al., 2014), and consequently evade the immune system and antiviral drugs. Once the latent provirus is reactivated by stimuli, new viruses can be produced to infect adjacent cells, and new latent reservoirs can be established (Xiao et al., 2019b). In addition, it has been demonstrated that HIV-1 can also enter cells through endocytosis, but this is controversial and requires more evidence (Marin et al., 2019). To complete replication, packaging, and budding processes, HIV-1 must utilize some indispensable host factors, such as lens epithelium-derived growth factor (LEDGF/P75), transportin-3 (TNPO3), and nucleoporin 358 (NUP 358), which provide host drug targets against HIV-1 infection (Xiao et al., 2019b). HIV-1 mainly infects and destroys primary CD4+ T cells, leading to increased risk for opportunistic infections, other infectious diseases, and some types of cancers (Z Liu et al., 2017). HIV-1 can also infect monocytes, dendritic cells, microglia, astrocytes and perivascular macrophages in the central nervous system (Xiao et al., 2019b).
Currently, the primary treatment for HIV-1/AIDS is combined antiretroviral therapy (cART), which is a combination of multiple drugs that effectively inhibit the functions of various viral proteins during HIV-1 replication. However, cART cannot completely eradicate latent reservoirs and has many drawbacks, such as lifelong treatment, high cost, chronic liver disease, cardiovascular and cerebrovascular system damage and complications in aging and neurological disorders (Bayer et al., 1983; Durand et al., 2012; Chen et al., 2018; Guo and Buch 2019; Thakur et al., 2019). Therefore, more effective methods of removing latent HIV-1 provirus and treating HIV-1/AIDS patients must be developed.
1.2. Gene editing technology
In recent decades, gene therapy has been developed as a new strategy to improve the health of patients with genetic diseases, such as hemophilia, β-thalassemia, and other monogenic diseases (Xiao et al., 2019a). Meanwhile, clinical research reported that the "Berlin patient" with AIDS and acute myelocytic leukemia (AML), was functionally cured after accepting a CCR5 Δ32 genotype of bone marrow transplantation (Hütter, 2014). More recently, a clinical report on the "London patient" with Hodgkin's lymphoma and AIDS provided similar evidence of curative treatment (Gupta et al., 2019). In addition, Xu et al. reported the case of a "Beijing patient" with AIDS and AML who received CCR5-disrupted hematopoietic stem cell transplantation using CRISPR/Cas9 technology. The AML was alleviated; however, the replication of HIV-1 was only suppressed, and AIDS cure was not achieved due to the low gene editing efficiency (Xu et al., 2019). These clinical cases suggest that, at this time, gene editing treatment is not suitable for all AIDS patients and is only recommended as a last resort for those who also have life-threatening hematological malignancies. However, in the future, gene therapy could be a powerful strategy for HIV-1/AIDS treatment if we can overcome all limitations and ensure on-target efficiency and safety.
In HIV-1/AIDS gene therapy research, three nuclease-mediated gene-editing tools, namely transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs), and CRISPR/Cas9, have been extensively studied (Xiao et al., 2019b). Among them, ZFN and TALEN are two special nucleases that recognize genome editing sites through protein-DNA interactions. ZFN has been applied in the gene-editing of CCR5 in autologous CD4+ T cells to treat HIV-1 infected patients (Tebas et al., 2014; Xiao et al., 2019b). However, both ZFN and TALEN have limitations, such as low gene editing efficiency, high off-target rate, and costly and laborious vector construction. The CRISPR/Cas system was first identified in prokaryotic immune responses against phage infection and invading plasmids (Barrangou et al., 2007). It contains a helicase, designated as Cas, that binds to the guide RNA (gRNA) transcribed by palindrome repeat DNA and excises the target DNA hybridized with gRNA. Depending on the phylogenetic analysis of the conserved Cas proteins with comparison of gene repertoires and arrangements in CRISPR-Cas loci, CRISPR-Cas systems can be classified into three distinct types and ten subtypes (Makarova and Koonin, 2015). Specifically, in the most commonly used type II CRISPR-Cas system, the transcript of the DNA palindromes repeated sequences named the trans-activating CRISPR RNA (tracrRNA), and the transcript of spacer called CRISPR RNA (crRNA) can be fused to form a single-guide RNA (sgRNA) that directs Cas9 to break DNA located near the protospacer adjacent motifs (PAMs) (Jinek et al., 2012; Cong et al., 2013). DNA breakage can lead to non-homologous end junction (NHEJ) or homology-directed repair (HDR)-mediated genome editing, resulting in insertions or deletions in the cellular genome (Cong et al., 2013; Mali et al., 2013).
The CRISPR/Cas system can be divided into two classes according to the component of the Cas protein, and each class contains several types (Safari et al., 2020). The Class I CRISPR/Cas system comprises multiple protein complexes, including types I, III and IV (Negahdaripour et al., 2017). Class II is a mono-protein CRISPR system, including type II (Cas9), type V (Cas12a/Cpf1, Cas12b/C2c1, and C2c3), and type VI (Cas13a, Cas13b, and Cas13d) (East-Seletsky et al., 2017). The type II CRISPR/Cas system was first used in vitro to generate a double-strand break (DSB) in the target DNA at a specific site, laying the foundation for its application in genome editing (Jinek et al., 2012). Then it has been rapidly developed to target genes in mammalian cells due to its convenience, high editing efficiency, and low off-target effects (Cong et al., 2013). To date, developed CRISPR/Cas systems mainly include: (1) Traditional CRISPR-SpCas9, in which the Cas9 was obtained from Streptococcus pyogenes and its delivery vehicles such as lentivirus (LV), adenovirus (AV), and adeno-associated virus (AAV), to expand its range of applications (Chen et al., 2018). (2) CRISPR interference (CRISPRi), in which the Cas9 protein contains two important nuclease domains, RuvC and histidine-asparagine-histidine (HNH), which are essential for cleaving complementary and non-complementary strands of the target DNA. The SpCas9 RuvC (D10A) and HNH (H840A) mutations can make its structure domain convert into a DNA "nickase", which only cleaves a single strand and induces specific NHEJ and HDR. Thus, the inactive Cas9 (dCas9, SpCas9D10A/H840A) can only bind to sgRNA and target DNA without cleaving, leading to the inhibition of the expression of target genes (Chen et al., 2018). (3) CRISPR activation (CRISPRa), which is assembled by combining four copies of the transcription activator VP16 (virus protein 16 is a transcription factor encoded by the UL48 gene of Herpes simplex virus-1) or a single copy of the p65 activation domain (AD) with dCas9 (dCas9-VP64 and dCas9-p65AD), which can transactivate gene transcription in cells (Chen et al., 2018). (4) CRISPR-SaCas9 system, in which SaCas9 is obtained from Staphylococcus aureus and encoded by a 3.5- kb open reading frame (ORF) that can be delivered into cells via an AAV vector (Chen et al., 2018). (5) Enhanced Cas9 (eCas9), which is an engineered Cas9 variant first developed by two groups independently, termed enhanced Cas9-1.1 (eCas9-1.1) (Slaymaker et al., 2016) and Cas9 high-fidelity variant 1 (Cas9-HF1) (Kleinstiver et al., 2016), which has minimal or no detectable off-target effects in human cells but lose some cleaving activity. Later, several other eCas9 variants, such as hyper-accurate Cas9 variant (HypaCas9) (Chen et al., 2017) and further eCas9 (FeCas9) (Yin et al., 2019), were explored and exhibited extremely low off-target activity with no obvious loss of editing ability compared with wild-type Cas9.
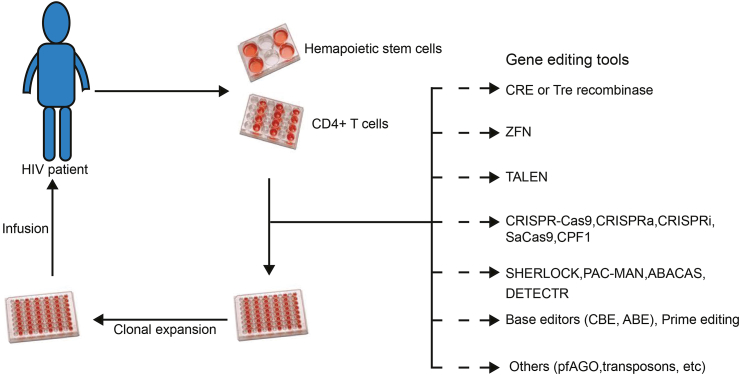
In addition to CRISPR/Cas systems, many other gene-editing tools, such as CPF1, base editor, prime editing, and pfAGO, have recently been developed (Zetsche et al., 2015; Komor et al., 2016; Anzalone et al., 2019). Moreover, CRISPR/Cas-derived SHERLOCK, DETECTR, PAC-MAN, and ABACAS have been used for pathogens detection (Safari et al., 2021). To introduce strategies for HIV-1/AIDS gene therapy and exploit these novel gene-editing and detection technologies in the future (Fig. 1), we have summarized the principles, functions and potential applications of these gene-targeting tools in HIV/AIDS treatment (Table 1).
Fig. 1.
Strategies for HIV-1/AIDS gene therapy. The allogeneic transplantation is the common method for HIV-1/AIDS gene therapy. It is performed by transplanting gene edited hematopoietic stem cells (HPSC) or CD4+ T cells derived from the bone marrow cells or peripheral-blood mononuclear cells (PBMC) of HIV-1/AIDS patient. In the process of gene editing, multi-approaches can be utilized to making cells resistant against HIV-1 infection.
Table 1.
Application of CRISPR/Cas based gene editing in HIV-1/AIDS therapy.
| CRISPR-Cas system | Delivery | Target | Cell type/organism | Animal model | AIDS patient | Ref |
|---|---|---|---|---|---|---|
| SpCas9 | Transfection, Lentivirus, Adenovirus, Electroporation | LTR, Rev, Gag, Pol, Env, CCR5, CXCR4, Restriction factors, | HEK293T, 293T-CD4-CCR5, 293 Primary T cells, Hela, TZM-b1, Jurkat, SupT1, K562, hPSC, iPSC, CHO, human CD34+ HSPCs, CHME5, U937, JLat10.6, J.Lat FL, CEMss, C8166, Ghost, pMoHIV |
Mouse | Beijing patient NCT 03164135 | (Ebina et al., 2013); (Hu et al., 2014); (Liao et al., 2015); (Wang et al., 2014); (Wang et al., 2016); (Lebbink et al., 2017); (Cho et al., 2013); (Ye et al., 2014); (Li et al., 2015); (Kang et al., 2015); (Xu et al., 2017); (Qi et al., 2018); (Hou et al., 2015); (Z Liu et al., 2017); (Bogerd et al., 2015); (Rosa et al., 2015); (Gonzalez-Enriquez et al., 2017); (Chougui et al., 2018); (Lahaye et al., 2018); (Yurkovetskiy et al., 2018); (Hultquist et al., 2016); (Xu et al., 2019); (Scott et al., 2019); (Sessions et al., 2020); |
| SaCas9 | Lentivirus, AAV, Transfection | LTR, Gag, proviral SIV DNA, CCR5, CXCR4, | HEK293T, TZM-bl, C11, Ghost, Jurkat, primary CD4+ T, human CD34+ HSPCs, J-Lat 6.3 | Tg26 transgenic mouse, SIV-infected macaques | No | (Yin et al., 2017); (Wang et al., 2018); (Wang et al., 2017); (Xiao et al., 2019a); (Olson et al., 2020); (Mancuso et al., 2020) |
| CRISPRa | Lentivirus, Transfection, Electroporation | LTR, APOBEC3G, APOBEC3B, T5, T6, | TZM-b1, J-Lat, CHME5 microglial cell, C11, HEK293T, ACH2, J1.1, CEM LChIT 3.2, Hela, U1 | No | No | (Zhang et al., 2015); (Saayman et al., 2016); (Limsirichai et al., 2016); (Ji et al., 2016); (Bogerd et al., 2015); (Kim et al., 2017); (Zhang et al., 2018) |
| CRISPRi | Lentivirus | PSMD 1, PSMD 3, PSMD 8, Spt4, Spt5, host factors (FTSJ3, TMEM178A, NICN1) | Jurkat 2D10 | No | No | (Z Li et al., 2019); (Krasnopolsky et al., 2021); (Z Li et al., 2020) |
| CPF1 | Adenovirus, Lentivirus | CCR5, LTR, Gag, Vpr, Tat, Rev | TZM-b1, SupT1-R5, Primary CD4+T cells, HEK293T | No | No | (Liu et al., 2020); (Gao et al., 2020) |
| Base editor | No | No | No | No | No | No |
| Prime editing | No | No | No | No | No | No |
| SHERLOCK | No | No | No | No | No | No |
| DETEECTR | No | No | No | No | No | No |
| PAC-MAN | No | No | No | No | No | No |
| ABACAS | No | No | No | No | No | No |
2. Application of CRISPR/Cas system in HIV-1/AIDS treatment
2.1. CRISPR/SpCas9 gene editing in HIV-1/AIDS treatment
The CRISPR/SpCas9 gene-targeting tool is the first CRISPR/Cas system that has been used in HIV-1 gene therapy research. One of the most significant challenges in the treatment of AIDS is eliminating the latent reservoir in HIV-1 infected cells. The most direct and effective gene therapy method is to remove the integrated provirus DNA from the host genome by CRISPR/SpCas9 targeting. For this purpose, in 2013, Ebina et al. reported that HIV-1 gene expression was successfully suppressed in the Jurkat cell line by CRISPR/SpCas9 mediated targeting of the NF-κB cassettes located in the U3 region and the TAR sequence in the R region of LTR. In addition to inhibiting HIV-1 proviral DNA transcription and viral replication, CRISPR/SpCas9 can eliminate integrated viral genes in the host chromosome (Ebina et al., 2013). Similarly, Hu et al. conducted a study to remove the HIV-1 genome using CRISPR/SpCas9 to target the conserved sites in the HIV-1 LTR U3 region. HIV-1 gene expression and replication were inhibited in microglial, promonocytic, and T cells, with minimal genotoxicity observed, and no off-target effects were detected (Hu et al., 2014). Zhu et al. achieved similar elimination of latent HIV-1 provirus in JLat10.6 cells by CRISPR/SpCas9 targeting of ten different sites (Zhu et al., 2015). By targeting multiple sites of the HIV-1 genome, Liao et al. improved the efficiency of gene editing and conferred primary cells with resistance to HIV-1 infection (Liao et al., 2015). It has been shown that combining two effective gRNAs targeting different regions of the HIV-1 genome can prevent viral replication and escape in primary human T cells and pluripotent stem cell (hPSC)-derived HIV reservoir cells (Lebbink et al., 2017).
In addition to targeting the HIV-1 genome by CRISPR/SpCas9, an alternative therapeutic strategy is to edit the cellular receptors for HIV-1 entry. The main receptor, CD4, plays a critical role in the immune system; thus, it is not advisable to destroy it by gene targeting. Given the two known cases of the "Berlin Patient" and "London Patient", the co-receptor CCR5 also should be an ideal target for HIV-1/AIDS gene therapy. Indeed, Cho et al. successfully silenced CCR5 in the human embryonic kidney (HEK) 293T cells by transfecting SpCas9 and sgRNA (Cho et al., 2013). Subsequently, Ye et al. reported that CRISPR/SpCas9 and piggyBac transposase technology could be used to generate CCR5-Δ32 mutations in induced pluripotent stem cells (IPSCs) without off-target effects. The edited IPSCs could differentiate into monocytes/macrophages and showed resistance to R5-tropic HIV-1 infection in vitro (Ye et al., 2014). In addition, Li et al. used chimeric adenovirus as a carrier to deliver CCR5 silencing CRISPR/SpCas9 effectively into primary CD4+ T cells and conferred resistance to HIV-1 infection (Li et al., 2015). Furthermore, Mandal et al. produced CCR5 biallelic deletions in human primary CD34+ hematopoietic stem and progenitor cells (HSPCs) and CD4+ T cells using a dual guide approach. The CCR5-modified HSPCs retained the ability to differentiate into effector cells in NOD-PRKDCScid-IL2rgnull (NSG) mice (Mandal et al., 2014). Finally, Xu et al. generated CCR5-modified CD34+HSPCs by SpCas9 targeting of the Δ32 region, which can intrinsically differentiate and maintain CCR5 disruption in the second generation of HSCs, providing the basis for the development of HIV-1/AIDS treatment by transplantation of CCR5-modified HSPCs (Xu et al., 2017).
The other co-receptor of HIV-1, CXCR4, plays an important role in maintaining the normal physiological function of hematopoietic stem cells. A lack of CXCR4 may lead to potentially malignant diseases (Liu et al., 2018). The strategy of targeting CXCR4 to treat HIV-1 has only been conducted in CD4+ T cells. For example, we designed two sgRNAs that guide CRISPR/SpCas9 to destroy CXCR4 in primary human CD4+ T cells and confer resistance to X4-tropic HIV-1 infection without off-target effects (Hou et al., 2015). Hultquist et al. constructed tropic-dependent HIV-1 resistant primary CD4+ T cells by electroporation of the CRISPR/SpCas9 ribonucleic acid protein and sgRNAs targeting CXCR4 or CCR5, but the electroporation technique was complicated as it required large amounts of Cas9 protein and crRNA:tracrRNA to be generated in vitro (Hultquist et al., 2016). We noticed that the natural CXCR4 P191A mutant could inhibit X4 tropic HIV-1 infection without impairing HSC differentiation (Liu et al., 2014). Thus, we further used the CRISPR/SpCas9 and PiggyBac systems to construct a cell line with the CXCR4 mutant (P191A) genome that can significantly reduce HIV-1 infection, but neither affect ligand binding nor Ca2+ signaling. As the CXCR4 mutant P191A functions physically in cells and resists X4-HIV binding, it provides a new approach for HIV-1 gene therapy (Liu et al., 2018). Moreover, as HIV-1 has co-receptor conversion and evolved to use CXCR4 in the late stage of infection, we conducted a study targeting both CCR5 and CXCR4 simultaneously using CRISPR/SpCas9. The modified CD4+ T cells were resistant to X4 and/or R5-tropic HIV-1 infection and exhibited a selective advantage over unmodified cells (Liu et al., 2017). As some host factors are essential for the HIV-1 life cycle, they can be targeted to prevent HIV-1 infection. For example, Hultquist et al. introduced CRISPR/SpCas9-sgRNA targeting LEDGF and TNPO3, which play pivotal roles in HIV-1 replication in primary CD4+ T cells through electroporation, leading to non-ophilic-dependent progeny virus production (Hultquist et al., 2016). A major challenge in the knockout of certain host factors is that the disruption may affect the physiological functions of these proteins, leading to serious adverse reactions in the host. In addition to forced expression of HIV-1 restriction factors, such as TRIM5α, SAMHD-1, and NONO, by CRISPRa targeting, it is possible to produce mutants of these host factors that confer increased resistance to HIV-1 infection through a CRISPR-mediated HDR approach. For example, the introduction of R332G and R355G amino acid substitutions in the human TRIM5α (huTRIM5α) domain can effectively lead to HIV-1 restriction. Berthoux et al. used a transfection-based CRISPR/Cas9 genome editing approach to successfully mutate TRIM5α into its potentially HIV-1-restrictive version via HDR. This strategy emphasized the importance of biallelic modification of TRIM5α to achieve significant HIV-1 replication inhibition. However, due to the imprecise editing of CRISPR/Cas9 and the limitation of PAM, this strategy has low efficiency and can easily produce double-allelic targeting and undesired mutations (Dufour et al., 2018; Désaulniers et al., 2020).
2.2. CRISPR/SaCas9 and CPF1 gene editing in HIV-1/AIDS treatment
CRISPR/SaCas9 is usually delivered by AAV, which has the advantages of low toxicity, poor pathogenicity, stable gene expression, safety, efficient targeting, and a broad serotype and tropism range (Mingozzi and High, 2013). Moreover, owing to the longer PAM sequence recognized by SaCas9, the target recognition rate is improved, and the off-target effects are reduced. We first used CRISPR/SaCas9 combined with multiple gRNAs in an all-in-one lentiviral vector to efficiently target latent HIV-1 provirus in Jurkat C11 and TZM-bl cells (Wang et al., 2018). Yin et al. demonstrated that AAV-mediated SaCas9/sgRNA could be used to remove integrated HIV-1 proviral DNA from progenitor, neural stem, and C11 cells (Yin et al., 2017). To target co-receptors, we selected two gRNAs/SaCas9 packages in AAV6 to target CXCR4 in CD4+ T cells. These edited cells showed resistance to X4-tropic HIV-1 infection (Wang et al., 2017). We also successfully targeted CCR5 in primary CD4+ T cells and CD45+ HSCs with AAV6-sgRNA/SaCas9 and demonstrated that CCR5 CD45+ HSCs differentiated normally and showed HIV-1 resistance in NSG mice (Wang et al., 2017). By linking dCas9 to the transcriptional inhibitory region of the Kruppel cassette, Olson et al. could inhibit HIV-1 transcription while reactivating latent HIV-1 proviruses (Olson et al., 2020).
Anti-HIV-1 gene therapy by CRISPR/SaCas9 requires careful in vivo investigation in animal model settings. Yin et al. demonstrated the feasibility and effectiveness of using an all-in-one AAV vector containing multiple gRNAs/SaCas9 to interfere with HIV-1 provirus in three different animal models. HIV-1 Tg26 transgenic mice harboring a virus replication-deficient pNL4-3 proviral genome were intravenously injected with four gRNAs/SaCas9 AAV-DJ/8 and showed significantly reduced viral replication (Yin et al., 2017). We injected CCR5-modified CD4+ T cells into humanized mice, which showed a survival advantage over unmodified CD4+ T cells during the HIV-1 challenge (Xiao et al., 2019a). Pietro et al. further demonstrated that disease in SIV-infected rhesus monkeys intravenously injected with AAV9-CRISPR/SaCas9 was alleviated by removing the integrated proviral DNA fragments in the genome of blood cells and tissues including the lymph nodes, spleen, bone marrow and brain (Mancuso et al., 2020). The above studies lay a foundation for the design of CRISPR/SaCas9 in human HIV-1/AIDS clinical trials.
Compared with SaCas9, Cpf1 (class 2 type V) also has advantages in treating HIV-1/AIDS (Zetsche et al., 2015). Using CRISPR/AsCpf1 technology, our group performed gene editing on CCR5 of TZM-b1, Sup T1-R5, and CD4+ T cells. The CCR5 targeted cells showed resistance to R5-tropic HIV-1 infection and selective advantage over unedited cells without off-target effects or cell proliferation deficiency. This suggests that CRISPR/AsCpf1-mediated gene editing has potential for clinical applications (Liu et al., 2020).
2.3. Potential application of base editors and prime editing in HIV-1/AIDS treatment
Although the application of CRISPR/Cas gene editing in HIV-1 treatment has progressed and improved, there are still some inevitable defects in the process of NHEJ and HDR caused by DSB, which may lead to uncontrolled adverse reactions. To avoid DSB, Alexis et al. developed a novel genome editing technique called "base editor", which comprises a programmable DNA-binding protein, such as a catalytically impaired Cas9 that has no ability for DNA cleavage or a nickase Cas9, and a DNA-modifying enzyme, such as cytidine deaminase (Komor et al., 2016). The fused Cas9 and cytidine deaminase can bind to a target genomic locus by sgRNA recognition and directly convert cytidine to uridine, resulting in a C→T (or G→A) mutation after replication or additional DNA repair. The "base editor", cytosine base editor (CBE), has an approximate "active window", typically bases 4–8 of protospacer where PAM is counted as 21–23 bases, which can be edited efficiently. Thus, it can effectively correct various point mutations associated with human diseases (Komor et al., 2016). In addition, through a wide range of directed evolution and protein engineering strategies, David et al. generated adenine base editors (ABEs) that can effectively convert target A-T to G-C base pairs (Gaudelli et al., 2017). The CBE and ABE technologies greatly expand the application of CRISPR/Cas in genome editing research as they result in fewer off-target effects, lower genotoxicity, and broaden editing windows. Furthermore, recent studies have reported the dual-functional base editor (DBE) technology, which combines dCas9 with cytidine deaminase and adenosine deaminase to convert C to T and A to G bases simultaneously. Compared with the corresponding ABE or CBE, the base-editing efficiency of DBE is higher, and the off-target rate is lower. DBE expands the scope of DNA sequence changes and extends the application of CRISPR base editors in HIV-1 treatment (Grünewald et al., 2020; Sakata et al., 2020; Zhang et al., 2020). As we and others have found that some point mutations (S532, T534, T536, L537A/W666A, and I535A/V539G) in the HIV-1 gp41 region significantly reduced or blocked Env-mediated syncytium formation and affected viral fusion and infection, these residues could be targeted by base editors for generating dead-virus without infectivity (Freed et al., 1990; Bellamy-McIntyre et al., 2007; Lay et al., 2011; Lu et al., 2019). Therefore, the HIV-1 reservoir can be converted to noninfectious proviral DNA by key amino acid mutations that are important for viral infectivity. In addition, to target CCR5, CXCR4, and host factors, we can convert the genetic start codon or its near downstream sequence into a stop codon by base editors, leading to premature termination of protein translation (Lu et al., 2019).
Although base editors can effectively install four transition mutations (C→T, G→A, A→G, and T→C), the remaining eight conversions cannot be produced (C→A, C→G, G→C, G→T, G→T, A→C, T→A, and T→G) at this time. To solve this problem, Andrew et al. developed a novel gene-editing system called Prime Editing (PE) by fusing a catalytically damaged Cas9 (H840A) with reverse transcriptase. In the PE system, new genetic information is directly written into the specified DNA site through a prime editing guide RNA (pegRNA) that binds specifically to the target site and encodes for the required editing sequence. The pegRNA consists of three elements: a sgRNA corresponding to the target site, a primer binding sequence, and a reverse transcription template storing genetic information for targeting (Anzalone et al., 2019). PE has a wide range of applications as it causes 12 base transition mutations, small base insertions, deletions and their combinations in human cells without the need for DSB and donor DNA templates. The current PE system mainly includes PE1, PE2, PE3, and PE3b (Hsu et al., 2021). PE1 has the lowest editing efficiency by fusing reverse transcriptase with RNA programmable nickase. PE2 improves the targeting efficiency by engineering reverse transcriptase based on PE1. PE3 and PE3b co-express the sgRNA targeting the non-editing strand based on PE2 and can protect the modification information of the editing strand through the intracellular mismatch repair pathway, thus further improving the editing efficiency and enhancing indel formation (Anzalone et al., 2019; Lin et al., 2020). Compared with the CRISPR-Cas genome editing method which induces HDR, PE has fewer off-target effects and byproducts, and higher editing efficiency. In addition, compared with BE, which would cause bystander edits if the active window had multiple cytosine or adenine, PE has almost no byproduct. However, although PE has a broader range of applications and more accurate edits, the editing efficiency of it is lower than that of BE. Therefore, BE can be selected if there is a single target nucleotide in the active window, or bystander edits are acceptable. PE is more suitable for active windows with multiple cytosine/adenines or unacceptable bystander edits. However, the difficulty and complexity of pegRNA design for PE are much higher than those of sgRNA for BE. To simplify the pegRNA design, Xu et al. introduced a database to improve the practicality of PE (Hsu et al., 2021; Kim et al., 2021). For HIV-1/AIDS treatment, PE can be used as a supplement to BE. For example, we identified three highly conserved important sites (S532P, T534A and T536A) that can affect the binding of gp120 to the CD4 receptor during HIV-1 entry into cells (Lu et al., 2019). The mutation of these sites by PE may block HIV-1 infectivity.
2.4. Other CRISPR/Cas9 systems in HIV-1/AIDS therapy
In HIV-1/AIDS treatment, the "shock and kill" strategy is a promising approach to eliminate latent viruses, which is achieved by using latency-reversing agents (LRAs) to reactivate latent proviruses ("shock" phase) integrated into immune cells, and the reactivated cells could be eliminated by HIV-1 induced cytopathic effects or host immune responses ("kill" phase) (Chen et al., 2018). As the CRISPR activation (CRISPRa) system can increase gene expression, it can be considered as an LRA that involves activating viral replication (Deeks, 2012). For example, Zhang et al. demonstrated that the dCas9-SAM system could efficiently target the HIV-1 LTR promoter in TZM-b1, J-LAT, 2D10, 3C9, E4, and CHME5 microglial cells (Saayman et al., 2016). In addition, Sheena et al. explored the dCas9-VP64 system targeting the HIV-1 LTR promoter to identify the "hot spot" sites for sgRNA binding and provirus activation (Limsirichai et al., 2016). Moreover, Ji et al. used dCas9-SunTag-VP64 to target the HIV-1 LTR region and effectively reactivate the provirus in C11 and A10.6 latent cells (Ji et al., 2016).
2.5. SHERLOCK, DETECTR, PAC-MAN, ABACAS technologies in HIV-1/AIDS detection and treatment
As HIV-1 contains two copies of viral RNA and the genome RNA could be produced from the provirus, targeting RNA is another strategy for HIV-1/AIDS treatment. The invention of RNA-guided, RNA-targeting CRISPR effectors has provided the foundation for the detection and clearance of HIV-1 RNA (Freije et al., 2019; Zhang et al., 2019; Safari et al., 2021). Most RNA editing techniques are based on Cas13a and Cas12a.
Cas13a was first introduced by Shmakov et al., (2015). It can break target-RNA directed by gRNA with extremely high specificity and very low off-target effects. It has the advantage of single-base discrimination of target genes (Shmakov et al., 2015). When the first single-stranded RNA (ssRNA) is broken, Cas13a becomes highly sensitive and enzymatically "active", and performs nonspecific cleavage of other RNAs (collateral cleavage) (East-Seletsky et al., 2017; L Liu et al., 2017). Based on these properties, Cas13a can be used to target HIV-1 genomic RNA and incoming RNA in infected cells. By combing the Cas13a enzymology of RNA cleavage and nucleic acid preamplification of isothermal recombinase polymerase or reverse transcription recombinase polymerase, specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) was proposed for rapid, convenient, and sensitive target gene detection (Gootenberg et al., 2017; Kellner et al., 2019). The principle of SHERLOCK is to use the collateral cleavage of Cas13a directed by gRNA to cut the reporting ssRNA with a fluorophore and a quencher at either end. When the targeting RNA was not primed by the CRISPR/Cas13a gRNA, the reporter RNA remained intact, and the quencher quenched the fluorescence. However, once the targeting RNA was detected by the CRISPR/Cas13a gRNA, the reporter RNA could be broken to release fluorescence that a detector can measure (Kellner et al., 2019). SHERLOCK has been used to detect Zika and dengue viruses (Gootenberg et al., 2017). As HIV-1 has two copies of ssRNA, SHERLOCK could be used to screen HIV-1 in suspected cases and quantify the levels of active HIV-1 replication with high sensitivity in AIDS patients.
Similar to SHERLOCK, the Cas12a derived DNA endonuclease-targeted CRISPR trans reporter (DETECTR) can cause DSB, which induces nonspecific DNA cleavage (Li et al., 2019). Therefore, DETECTR can break the ssDNA reporter with fluorescence and quenching groups linked at either end. It may detect HIV-1 provirus DNA in AIDS patients using DETECTR with specific gRNA/Cas12a.
Direct targeting of (+) ssRNAs or RNAs responsible for virus replication and expression is an effective method for blocking RNA virus replication (Li et al., 2020). The CRISPR/Cas13d system has been developed to inhibit mRNA expression in mammalian cells or RNA viruses in plants (Konermann et al., 2018; Mahas et al., 2019; Kushawah et al., 2020; Zhou et al., 2020). In addition, Cas13d and its variants have strong collateral activity for RNA cleavage with high specificity and minimal off-target effects (Yan et al., 2018). Due to the COVID-19 pandemic caused by SARS-CoV-2 in the past year, Abbott et al. developed the prophylactic antiviral CRISPR method in human cells (PAC-MAN) based on Cas13d to target SARS-CoV-2 and all sequenced coronavirus strains (Abbott et al., 2020). By bioinformatic analyses of SARS-CoV-2 sequences, the conserved regions of SARS-CoV-2 were selected to establish a crRNA pool (library) that can directly degrade all coronaviruses. Thus, this crRNA library may serve as a preventive treatment against future pathogenic coronaviruses that evolved to escape immune surveillance (Abbott et al., 2020). Similar to SARS-CoV-2, a crRNA library may be constructed against HIV-1 strains in the PAC-MAN strategy to manage HIV-1/AIDS.
To target more specifically of SARS-CoV-2 by Cas13, antibody and CAS fusion (ABACAS) method was developed by fusing Cas13 to a specific antibody of main virus structural proteins, such as S protein. The combination of viral antigens by ABACAS can suppress virus entry into cells. Additionally, when the virus enters cells by endocytosis, Cas13 can cleave viral RNA in infected cells, thus inhibiting viral replication (Joyce et al., 2020; Yuan et al., 2020). As HIV-1 can also enter target cells through endocytosis, the virus selectivity and RNA cleavage efficiency could be improved by assembling ABACAS with Cas13 and an HIV-1 antibody.
3. Conclusions and future directions
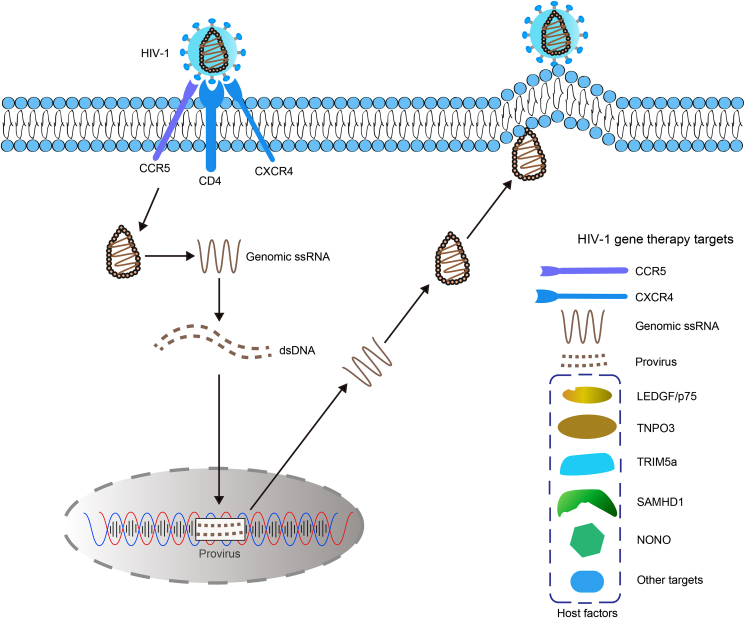
At present, the common approach of treating HIV-1/AIDS by gene therapy can be divided into four categories (as shown in Fig. 2): (1) Gene editing of HIV-1 co-receptors. As HIV-1 enters immune cells through co-receptors CCR5 and CXCR4, the most effective method is using gene-editing technology to silent or mutate co-receptors on cell surface against HIV-1 infection. (2) Targeting the HIV-1 RNA genome or provirus. HIV-1 RNA can be detected and/or degraded by RNA-guided RNA-targeting CRISPR effectors derived from Cas12a and Cas13a. In addition, HIV-1 provirus could either be cleaved by gene-editing tools or activated by CRISPRa for the “shock and kill” strategy. (3) Targeting indispensable host factors, such as LEDGF/p75 and TNPO3, which are essential in the HIV-1 life cycle. Mutating or knockout of these genes by CRISPR-Cas will restrict virus replication in cells. (4) Forced expression of HIV-1 restriction factors, such as TRIM5α, SAMHD-1, and NONO, by CRISPRa targeting, or gene editing to develop mutants that confer increased resistance to HIV-1 infection.
Fig. 2.
Gene editing targets in the life cycle of HIV-1. The potential targets for HIV-1/AIDS gene therapy can be divided into four categories: (1) HIV-1 co-receptors. As HIV-1 enters immune cells through co-receptors CCR5 and CXCR4, the most effective method is using gene-editing technology to silent or mutate co-receptors and confer cell resistance against HIV-1 infection. (2) HIV-1 RNA genome or provirus. HIV-1 RNA can be detected and/or degraded by RNA-guided RNA-targeting CRISPR effectors derived from Cas12a and Cas13a. HIV-1 provirus could either be cleaved by gene-editing tools or activated by CRISPRa for the “shock and kill” strategy. (3) Indispensable host factors, such as LEDGF/p75 and TNPO3, which are essential in the HIV-1 life cycle. Mutating or knockout of these genes by CRISPR-Cas will restrict virus replication in cells. (4) Forced expression of HIV-1 restriction factors, such as TRIM5α, SAMHD-1, and NONO, by CRISPRa targeting, or gene editing to develop mutants that confer increased resistance to HIV-1 infection.
The ZFN genome-editing tool has been applied to the clinical treatment of HIV-1/AIDS (Tebas et al., 2014). Although the first CRISPR/Cas9-mediated clinical trial was conducted in 2016, in which modified human T cells were re-injected into an individual with metastatic non-small cell lung cancer (Lu et al., 2020), the technology for HIV-1/AIDS therapy has mostly been used in mice or primary cells. Clinical trials of CRISPR/Cas9 for HIV-1 treatment have been conducted in the "Beijing patient", who suffered both from HIV-1/AIDS and AML (Xu et al., 2019). To consider safety in the clinical use of CRISPR-based gene editing for HIV/AIDS therapy, some caveats must be addressed before widespread use. First, the off-target effects of CRISPR/Cas9 cause nonspecific cleavage and result in cytotoxicity. A previous report demonstrated the tolerance of mismatching by SpCas9/sgRNA targeting (Lin et al., 2014). In addition, lentiviral vector-mediated permanent gRNA/Cas9 expression would lead to undesirable off-target effects and promote DSBs by increasing the tolerability of mismatches in the PAM and guide-matching region (Ortinski et al., 2017). This can be improved by using alternative Cas9 delivery vehicles, such as transient expression vectors, electroporation of mRNA or Cas9 proteins, and nanoparticles. Second, SpCas9 and SaCas9 can cause DSB, which would induce NHEJ or HDR, resulting in adverse reactions by uncontrollable deletion and insertion. Third, CRISPR/Cas9 targeting relies on the recognition of the PAM sequence, which limits the application of this method. Fourth, the in vivo delivery efficiency of CRISPR/Cas9 components still needs to be improved. Fifth, for HIV-1/AIDS targeting, the escape of the virus increases the difficulty of eliminating the reservoir in HIV-1 infected cells. Recently, the above CRISPR/Cas9 defects have been partially addressed and improved by researchers. For example, Jordan et al. designed a YE1 variant that retains the substrate-targeting scope of high-activity CBEs while maintaining minimal Cas9-independent off-target editing (Doman et al., 2020). Shannon et al. showed that targeting range of SpCas9 and its engineering variants was strongly restricted to its recognition sequence PAM to contain the guanines. They adopted phage-assisted non-continuous evolution (PANCE) and three new phage-assisted continuous evolution (PACE) selection strategies to identify three new SpCas9 variants with NRNH types of PAMs (R = A or G and H = A, C, or T). These new Cas9 variants with multiple PAMs have extended the application of CRISPR/Cas9 targeting (Miller et al., 2020) and allow certain strategies to be improved and generate mutants of host proteins (Dufour et al., 2018). In addition, Youdiil et al. successfully inhibited HIV-1 replication in the long term and restricted the generation of escape mutants by diverting an all-in-one lentiviral vector containing six gRNAs targeting the HIV-1 genome in T cells (Ophinni et al., 2020).
Gene editing technologies have broad application prospects, particularly for treating many diseases caused by gene mutations or pathogen infections. For HIV-1/AIDS gene therapy in clinical research using CRISPR/Cas9, this remains a challenge. To improve the practice in future applications, we should: (1) design more specific gRNA and Cas9 mutants to reduce the off-target effects; (2) develop single- or multi-functional base editors that can directly and accurately edit bases without DSB-induced NHEJ or HDR; (3) design Cas9 variants with flexible PAM sequences to expand the application of CRISPR/Cas9; (4) explore new vehicles for the safe and efficient delivery of gene-editing tools; and (5) optimize the HIV-1/AIDS animal models and perform more in vivo gene therapy experiments, which will lay a foundation for clinical use. With the above progress, the CRISPR/Cas9 and other genome editing technologies may bring exciting and promising advances in HIV-1/AIDS gene therapy.
Declaration of competing interest
The authors declare that they have no potential conflict of interests.
Acknowledgments
This study was supported by the Fundamental Research Funds for the Central Universities (2042021kf0195), love creates future research funding, and Hubei natural science foundation (2021CFB483). Research in the authors' laboratory was supported by grants from China National Special Program for Major Infectious Diseases (2014ZX10001003 and 2017ZX10202102), the National Natural Science Foundation of China (81401659, 82172258) and China Postdoctoral Science Foundation (2015T80838 and 2014M560622). We apologize authors whose work has not been cited owing to the limited space of the present study.
Contributor Information
Wei Hou, Email: houwei@whu.edu.cn.
Shuliang Chen, Email: chen-shuliang@whu.edu.cn.
References
- Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., Pande T., Endy D., La Russa M.F., Lewis D.B., Qi L.S. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–876. doi: 10.1016/j.cell.2020.04.020. e812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib G., Combadiere C., Broder C.C., Feng Y., Kennedy P.E., Murphy P.M., Berger E.A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., Liu D.R. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagasra O., Lavi E., Bobroski L., Khalili K., Pestaner J.P., Tawadros R., Pomerantz R.J. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS. 1996;10:573–585. doi: 10.1097/00002030-199606000-00002. [DOI] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bayer R., Levine C., Murray T.H. Guidelines for confidentiality in research on AIDS. AIDS Res. 1983;1:275–297. doi: 10.1089/aid.1.1983.1.275. [DOI] [PubMed] [Google Scholar]
- Bellamy-McIntyre A.K., Lay C.S., Baär S., Maerz A.L., Talbo G.H., Drummer H.E., Poumbourios P. Functional links between the fusion peptide-proximal polar segment and membrane-proximal region of human immunodeficiency virus gp41 in distinct phases of membrane fusion. J. Biol. Chem. 2007;282:23104–23116. doi: 10.1074/jbc.M703485200. [DOI] [PubMed] [Google Scholar]
- Bleul C.C., Farzan M., Choe H., Parolin C., Clark-Lewis I., Sodroski J., Springer T.A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- Bogerd H.P., Kornepati A.V., Marshall J.B., Kennedy E.M., Cullen B.R. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E7249–E7256. doi: 10.1073/pnas.1516305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Update on acquired immune deficiency syndrome (AIDS)--United States. MMWR Morb. Mortal. Wkly. Rep. 1982;31 507-508, 513-504. [PubMed] [Google Scholar]
- Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Yu X., Guo D. CRISPR-cas targeting of host genes as an antiviral strategy. Viruses. 2018;10:40. doi: 10.3390/v10010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Chougui G., Munir-Matloob S., Matkovic R., Martin M.M., Morel M., Lahouassa H., Leduc M., Ramirez B.C., Etienne L., Margottin-Goguet F. HIV-2/SIV viral protein X counteracts HUSH repressor complex. Nat. Microbiol. 2018;3:891–897. doi: 10.1038/s41564-018-0179-6. [DOI] [PubMed] [Google Scholar]
- Chun T.W., Nickle D.C., Justement J.S., Meyers J.H., Roby G., Hallahan C.W., Kottilil S., Moir S., Mican J.M., Mullins J.I., Ward D.J., Kovacs J.A., Mannon P.J., Fauci A.S. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks S.G. HIV: shock and kill. Nature. 2012;487:439–440. doi: 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- Désaulniers K., Ortiz L., Dufour C., Claudel A., Plourde M.B., Merindol N., Berthoux L. Editing of the TRIM5 gene decreases the permissiveness of human T lymphocytic cells to HIV-1. Viruses. 2020;13:24. doi: 10.3390/v13010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour C., Claudel A., Joubarne N., Merindol N., Maisonnet T., Masroori N., Plourde M.B., Berthoux L. Editing of the human TRIM5 gene to introduce mutations with the potential to inhibit HIV-1. PLoS One. 2018;13 doi: 10.1371/journal.pone.0191709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand C.M., Blankson J.N., Siliciano R.F. Developing strategies for HIV-1 eradication. Trends Immunol. 2012;33:554–562. doi: 10.1016/j.it.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A., O'Connell M.R., Burstein D., Knott G.J., Doudna J.A. RNA targeting by functionally orthogonal type VI-A CRISPR-cas enzymes. Mol. Cell. 2017;66:373–383. doi: 10.1016/j.molcel.2017.04.008. e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H., Misawa N., Kanemura Y., Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D., Blankson J., Siliciano J.D., Margolick J.B., Chadwick K., Pierson T., Smith K., Lisziewicz J., Lori F., Flexner C., Quinn T.C., Chaisson R.E., Rosenberg E., Walker B., Gange S., Gallant J., Siliciano R.F. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Freed E.O., Myers D.J., Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije C.A., Myhrvold C., Boehm C.K., Lin A.E., Welch N.L., Carter A., Metsky H.C., Luo C.Y., Abudayyeh O.O., Gootenberg J.S., Yozwiak N.L., Zhang F., Sabeti P.C. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Fan M., Das A.T., Herrera-Carrillo E., Berkhout B. Extinction of all infectious HIV in cell culture by the CRISPR-Cas12a system with only a single crRNA. Nucleic Acids Res. 2020;48:5527–5539. doi: 10.1093/nar/gkaa226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Enriquez G.V., Escoto-Delgadillo M., Vazquez-Valls E., Torres-Mendoza B.M. SERINC as a restriction factor to inhibit viral infectivity and the interaction with HIV. J. Immunol. Res. 2017;2017:1548905. doi: 10.1155/2017/1548905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald J., Zhou R., Lareau C.A., Garcia S.P., Iyer S., Miller B.R., Langner L.M., Hsu J.Y., Aryee M.J., Joung J.K. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020;38:861–864. doi: 10.1038/s41587-020-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M.L., Buch S. Neuroinflammation & pre-mature aging in the context of chronic HIV infection and drug abuse: role of dysregulated autophagy. Brain Res. 2019;1724:146446. doi: 10.1016/j.brainres.2019.146446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.K., Abdul-Jawad S., McCoy L.E., Mok H.P., Peppa D., Salgado M., Martinez-Picado J., Nijhuis M., Wensing A.M.J., Lee H., Grant P., Nastouli E., Lambert J., Pace M., Salasc F., Monit C., Innes A.J., Muir L., Waters L., Frater J., Lever A.M.L., Edwards S.G., Gabriel I.H., Olavarria E. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019;568:244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P., Chen S., Wang S., Yu X., Chen Y., Jiang M., Zhuang K., Ho W., Hou W., Huang J., Guo D. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Grünewald J., Szalay R., Shih J., Anzalone A.V., Lam K.C., Shen M.W., Petri K., Liu D.R., Joung J.K., Pinello L. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat. Commun. 2021;12:1034. doi: 10.1038/s41467-021-21337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., Luo B., Alvarez-Carbonell D., Garcia-Mesa Y., Karn J., Mo X., Khalili K. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist J.F., Schumann K., Woo J.M., Manganaro L., McGregor M.J., Doudna J., Simon V., Krogan N.J., Marson A. A Cas9 ribonucleoprotein platform for functional genetic studies of HIV-host interactions in primary human T cells. Cell Rep. 2016;17:1438–1452. doi: 10.1016/j.celrep.2016.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt P.W., Harrigan P.R., Huang W., Bates M., Williamson D.W., McCune J.M., Price R.W., Spudich S.S., Lampiris H., Hoh R., Leigler T., Martin J.N., Deeks S.G. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J. Infect. Dis. 2006;194:926–930. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- Hütter G. More on shift of HIV tropism in stem-cell transplantation with CCR5 delta32/delta32 mutation. N. Engl. J. Med. 2014;371:2437–2438. doi: 10.1056/NEJMc1412279. [DOI] [PubMed] [Google Scholar]
- Hütter G., Bodor J., Ledger S., Boyd M., Millington M., Tsie M., Symonds G. CCR5 targeted cell therapy for HIV and prevention of viral escape. Viruses. 2015;7:4186–4203. doi: 10.3390/v7082816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Jiang Z., Lu P., Ma L., Li C., Pan H., Fu Z., Qu X., Wang P., Deng J., Yang X., Wang J., Zhu H. Specific reactivation of latent HIV-1 by dCas9-SunTag-VP64-mediated guide RNA targeting the HIV-1 promoter. Mol. Ther. 2016;24:508–521. doi: 10.1038/mt.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce M.G., Sankhala R.S., Chen W.H., Choe M., Bai H., Hajduczki A., Yan L., Sterling S.L., Peterson C.E., Green E.C., Smith C., de Val N., Amare M., Scott P., Laing E.D., Broder C.C., Rolland M., Michael N.L., Modjarrad K. A cryptic site of vulnerability on the receptor binding domain of the SARS-CoV-2 spike glycoprotein. bioRxiv. 2020 doi: 10.1101/2020.03.15.992883. [DOI] [Google Scholar]
- Kang H., Minder P., Park M.A., Mesquitta W.T., Torbett B.E., Slukvin CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol. Ther. Nucleic Acids. 2015;4:e268. doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G., Sun C., Grayson T., Wang S., Li H., Wei X., Jiang C., Kirchherr J.L., Gao F., Anderson J.A., Ping L.H., Swanstrom R., Tomaras G.D., Blattner W.A., Goepfert P.A., Kilby J.M., Saag M.S., Delwart E.L., Busch M.P., Cohen M.S., Montefiori D.C., Haynes B.F., Gaschen B., Athreya G.S., Lee H.Y., Wood N., Seoighe C., Perelson A.S., Bhattacharya T., Korber B.T., Hahn B.H., Shaw G.M. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.K., Yu G., Park J., Min S., Lee S., Yoon S., Kim H.H. Predicting the efficiency of prime editing guide RNAs in human cells. Nat. Biotechnol. 2021;39:198–206. doi: 10.1038/s41587-020-0677-y. [DOI] [PubMed] [Google Scholar]
- Kim V., Mears B.M., Powell B.H., Witwer K.W. Mutant Cas9-transcriptional activator activates HIV-1 in U1 cells in the presence and absence of LTR-specific guide RNAs. Matters (Zur) 2017;2017 doi: 10.19185/matters.201611000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676. doi: 10.1016/j.cell.2018.02.033. e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnopolsky S., Novikov A., Kuzmina A., Taube R. CRISPRi-mediated depletion of Spt4 and Spt5 reveals a role for DSIF in the control of HIV latency. Biochim. Biophys. Acta Gene Regul. Mech. 2021;1864:194656. doi: 10.1016/j.bbagrm.2020.194656. [DOI] [PubMed] [Google Scholar]
- Kushawah G., Hernandez-Huertas L., Abugattas-Nuñez Del Prado J., Martinez-Morales J.R., DeVore M.L., Hassan H., Moreno-Sanchez I., Tomas-Gallardo L., Diaz-Moscoso A., Monges D.E., Guelfo J.R., Theune W.C., Brannan E.O., Wang W., Corbin T.J., Moran A.M., Sánchez Alvarado A., Málaga-Trillo E., Takacs C.M., Bazzini A.A., Moreno-Mateos M.A. CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev. Cell. 2020;54:805–817. doi: 10.1016/j.devcel.2020.07.013. e807. [DOI] [PubMed] [Google Scholar]
- Lahaye X., Gentili M., Silvin A., Conrad C., Picard L., Jouve M., Zueva E., Maurin M., Nadalin F., Knott G.J., Zhao B., Du F., Rio M., Amiel J., Fox A.H., Li P., Etienne L., Bond C.S., Colleaux L., Manel N. NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell. 2018;175:488–501. doi: 10.1016/j.cell.2018.08.062. e422. [DOI] [PubMed] [Google Scholar]
- Lay C.S., Ludlow L.E., Stapleton D., Bellamy-McIntyre A.K., Ramsland P.A., Drummer H.E., Poumbourios P. Role for the terminal clasp of HIV-1 gp41 glycoprotein in the initiation of membrane fusion. J. Biol. Chem. 2011;286:41331–41343. doi: 10.1074/jbc.M111.299826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbink R.J., de Jong D.C., Wolters F., Kruse E.M., van Ham P.M., Wiertz E.J., Nijhuis M. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 2017;7:41968. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Guan X., Du T., Jin W., Wu B., Liu Y., Wang P., Hu B., Griffin G.E., Shattock R.J., Hu Q. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J. Gen. Virol. 2015;96:2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- Li H., Wang S., Dong X., Li Q., Li M., Li J., Guo Y., Jin X., Zhou Y., Song H., Kou Z. CRISPR-Cas13a cleavage of dengue virus NS3 gene efficiently inhibits viral replication. Mol. Ther. Nucleic Acids. 2020;19:1460–1469. doi: 10.1016/j.omtn.2020.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- Li Z., Hajian C., Greene W.C. Identification of unrecognized host factors promoting HIV-1 latency. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wu J., Chavez L., Hoh R., Deeks S.G., Pillai S.K., Zhou Q. Reiterative Enrichment and Authentication of CRISPRi Targets (REACT) identifies the proteasome as a key contributor to HIV-1 latency. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.K., Gu Y., Diaz A., Marlett J., Takahashi Y., Li M., Suzuki K., Xu R., Hishida T., Chang C.J., Esteban C.R., Young J., Izpisua Belmonte J.C. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- Limsirichai P., Gaj T., Schaffer D.V. CRISPR-mediated activation of latent HIV-1 expression. Mol. Ther. 2016;24:499–507. doi: 10.1038/mt.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A.V., Raguram A., Doman J.L., Liu D.R., Gao C. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020;38:582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Li X., Wang J., Wang M., Chen P., Yin M., Li J., Sheng G., Wang Y. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168:121–134. doi: 10.1016/j.cell.2016.12.031. e112. [DOI] [PubMed] [Google Scholar]
- Liu S., Wang Q., Yu X., Li Y., Guo Y., Liu Z., Sun F., Hou W., Li C., Wu L., Guo D., Chen S. HIV-1 inhibition in cells with CXCR4 mutant genome created by CRISPR-Cas9 and piggyBac recombinant technologies. Sci. Rep. 2018;8:8573. doi: 10.1038/s41598-018-26894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhou J., Pan J.A., Mabiala P., Guo D. A novel approach to block HIV-1 coreceptor CXCR4 in non-toxic manner. Mol. Biotechnol. 2014;56:890–902. doi: 10.1007/s12033-014-9768-7. [DOI] [PubMed] [Google Scholar]
- Liu Z., Chen S., Jin X., Wang Q., Yang K., Li C., Xiao Q., Hou P., Liu S., Wu S., Hou W., Xiong Y., Kong C., Zhao X., Wu L., Li C., Sun G., Guo D. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4(+) T cells from HIV-1 infection. Cell Biosci. 2017;7:47. doi: 10.1186/s13578-017-0174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Liang J., Chen S., Wang K., Liu X., Liu B., Xia Y., Guo M., Zhang X., Sun G., Tian G. Genome editing of CCR5 by AsCpf1 renders CD4(+)T cells resistance to HIV-1 infection. Cell Biosci. 2020;10:85. doi: 10.1186/s13578-020-00444-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Torresilla C., Xiao Y., Nguyen P.T., Caté C., Barbosa K., É Rassart, Cen S., Bourgault S., Barbeau B. HIV-1 antisense protein of different clades induces autophagy and associates with the autophagy factor p62. J. Virol. 2019;93 doi: 10.1128/JVI.01757-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Chen S., Yu J., Behrens R., Wiggins J., Sherer N., Liu S.L., Xiong Y., Xiang S.H., Wu L. The polar region of the HIV-1 envelope protein determines viral fusion and infectivity by stabilizing the gp120-gp41 association. J. Virol. 2019;93 doi: 10.1128/JVI.02128-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y., Shen H., Tong R., Wang W., Li L., Song J., Li J., Su X., Ding Z., Gong Y., Zhu J., Wang Y., Zou B., Zhang Y., Li Y., Zhou L., Liu Y., Yu M., Wang Y., Zhang X., Yin L., Xia X., Zeng Y., Zhou Q., Ying B., Chen C., Wei Y., Li W., Mok T. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- Mahas A., Aman R., Mahfouz M. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 2019;20:263. doi: 10.1186/s13059-019-1881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Koonin E.V. Annotation and classification of CRISPR-cas systems. Methods Mol. Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso P., Chen C., Kaminski R., Gordon J., Liao S., Robinson J.A., Smith M.D., Liu H., Sariyer I.K., Sariyer R., Peterson T.A., Donadoni M., Williams J.B., Siddiqui S., Bunnell B.A., Ling B., MacLean A.G., Burdo T.H., Khalili K. CRISPR based editing of SIV proviral DNA in ART treated non-human primates. Nat. Commun. 2020;11:6065. doi: 10.1038/s41467-020-19821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P.K., Ferreira L.M., Collins R., Meissner T.B., Boutwell C.L., Friesen M., Vrbanac V., Garrison B.S., Stortchevoi A., Bryder D., Musunuru K., Brand H., Tager A.M., Allen T.M., Talkowski M.E., Rossi D.J., Cowan C.A. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M., Kushnareva Y., Mason C.S., Chanda S.K., Melikyan G.B. HIV-1 fusion with CD4+ T cells is promoted by proteins involved in endocytosis and intracellular membrane trafficking. Viruses. 2019;11:100. doi: 10.3390/v11020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.M., Wang T., Randolph P.B., Arbab M., Shen M.W., Huang T.P., Matuszek Z., Newby G.A., Rees H.A., Liu D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020;38:471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasipura S.D., Kim S., Al-Harthi L. Epigenetic regulation of HIV-1 latency in astrocytes. J. Virol. 2014;88:3031–3038. doi: 10.1128/JVI.03333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negahdaripour M., Nezafat N., Hajighahramani N., Rahmatabadi S.S., Ghasemi Y. Investigating CRISPR-Cas systems in Clostridium botulinum via bioinformatics tools. Infect. Genet. Evol. 2017;54:355–373. doi: 10.1016/j.meegid.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Olson A., Basukala B., Lee S., Gagne M., Wong W.W., Henderson A.J. Targeted chromatinization and repression of HIV-1 provirus transcription with repurposed CRISPR/Cas9. Viruses. 2020;12:1154. doi: 10.3390/v12101154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophinni Y., Miki S., Hayashi Y., Kameoka M. Multiplexed tat-targeting CRISPR-cas9 protects T cells from acute HIV-1 infection with inhibition of viral escape. Viruses. 2020;12:1223. doi: 10.3390/v12111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinski P.I., O'Donovan B., Dong X., Kantor B. Integrase-deficient lentiviral vector as an all-in-one platform for highly efficient CRISPR/Cas9-Mediated gene editing. Mol. Ther. Methods Clin. Dev. 2017;5:153–164. doi: 10.1016/j.omtm.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda E. HIV tropism shift: new paradigm on cell therapy strategies for HIV cure. AIDS Rev. 2015;17:65. [PubMed] [Google Scholar]
- Qi C., Li D., Jiang X., Jia X., Lu L., Wang Y., Sun J., Shao Y., Wei M. Inducing CCR5Δ32/Δ32 homozygotes in the human Jurkat CD4+ cell line and primary CD4+ cells by CRISPR-cas9 genome-editing technology. Mol. Ther. Nucleic Acids. 2018;12:267–274. doi: 10.1016/j.omtn.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Chande A., Ziglio S., De Sanctis V., Bertorelli R., Goh S.L., McCauley S.M., Nowosielska A., Antonarakis S.E., Luban J., Santoni F.A., Pizzato M. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature. 2015;526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S.M., Lazar D.C., Scott T.A., Hart J.R., Takahashi M., Burnett J.C., Planelles V., Morris K.V., Weinberg M.S. Potent and targeted activation of latent HIV-1 using the CRISPR/dCas9 activator complex. Mol. Ther. 2016;24:488–498. doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari F., Afarid M., Rastegari B., Borhani-Haghighi A., Barekati-Mowahed M., Behzad-Behbahani A. CRISPR systems: novel approaches for detection and combating COVID-19. Virus Res. 2021;294:198282. doi: 10.1016/j.virusres.2020.198282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari F., Hatam G., Behbahani A.B., Rezaei V., Barekati-Mowahed M., Petramfar P., Khademi F. CRISPR system: a high-throughput toolbox for research and treatment of Parkinson's disease. Cell. Mol. Neurobiol. 2020;40:477–493. doi: 10.1007/s10571-019-00761-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata R.C., Ishiguro S., Mori H., Tanaka M., Tatsuno K., Ueda H., Yamamoto S., Seki M., Masuyama N., Nishida K., Nishimasu H., Arakawa K., Kondo A., Nureki O., Tomita M., Aburatani H., Yachie N. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020;38:865–869. doi: 10.1038/s41587-020-0509-0. [DOI] [PubMed] [Google Scholar]
- Scott T., Urak R., Soemardy C., Morris K.V. Improved Cas9 activity by specific modifications of the tracrRNA. Sci. Rep. 2019;9:16104. doi: 10.1038/s41598-019-52616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions K.J., Chen Y.Y., Hodge C.A., Hudson T.R., Eszterhas S.K., Hayden M.S., Howell A.L. Analysis of CRISPR/Cas9 guide RNA efficiency and specificity against genetically diverse HIV-1 isolates. AIDS Res. Hum. Retrovir. 2020;36:862–874. doi: 10.1089/aid.2020.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., Zhang F., Koonin E.V. Discovery and functional characterization of diverse class 2 CRISPR-cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano J.D., Kajdas J., Finzi D., Quinn T.C., Chadwick K., Margolick J.B., Kovacs C., Gange S.J., Siliciano R.F. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P.D., Meng G., Salazar-Gonzalez J.F., Shaw G.M. Macrophage HIV-1 infection and the gastrointestinal tract reservoir. J. Leukoc. Biol. 2003;74:642–649. doi: 10.1189/jlb.0503219. [DOI] [PubMed] [Google Scholar]
- Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G., Holmes M.C., Gregory P.D., Ando D.G., Kalos M., Collman R.G., Binder-Scholl G., Plesa G., Hwang W.T., Levine B.L., June C.H. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur K.T., Boubour A., Saylor D., Das M., Bearden D.R., Birbeck G.L. Global HIV neurology: a comprehensive review. AIDS. 2019;33:163–184. doi: 10.1097/QAD.0000000000001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Chen S., Xiao Q., Liu Z., Liu S., Hou P., Zhou L., Hou W., Ho W., Li C., Wu L., Guo D. Genome modification of CXCR4 by Staphylococcus aureus Cas9 renders cells resistance to HIV-1 infection. Retrovirology. 2017;14:51. doi: 10.1186/s12977-017-0375-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu S., Liu Z., Ke Z., Li C., Yu X., Chen S., Guo D. Genome scale screening identification of SaCas9/gRNAs for targeting HIV-1 provirus and suppression of HIV-1 infection. Virus Res. 2018;250:21–30. doi: 10.1016/j.virusres.2018.04.002. [DOI] [PubMed] [Google Scholar]
- Wang W., Ye C., Liu J., Zhang D., Kimata J.T., Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Pan Q., Gendron P., Zhu W., Guo F., Cen S., Wainberg M.A., Liang C. CRISPR/Cas9-Derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- Xiao Q., Chen S., Wang Q., Liu Z., Liu S., Deng H., Hou W., Wu D., Xiong Y., Li J., Guo D. CCR5 editing by Staphylococcus aureus Cas9 in human primary CD4(+) T cells and hematopoietic stem/progenitor cells promotes HIV-1 resistance and CD4(+) T cell enrichment in humanized mice. Retrovirology. 2019;16:15. doi: 10.1186/s12977-019-0477-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Guo D., Chen S. Application of CRISPR/Cas9-Based gene editing in HIV-1/AIDS therapy. Front. Cell Infect. Microbiol. 2019;9:69. doi: 10.3389/fcimb.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Wang J., Liu Y., Xie L., Su B., Mou D., Wang L., Liu T., Wang X., Zhang B., Zhao L., Hu L., Ning H., Zhang Y., Deng K., Liu L., Lu X., Zhang T., Xu J., Li C., Wu H., Deng H., Chen H. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- Xu L., Yang H., Gao Y., Chen Z., Xie L., Liu Y., Liu Y., Wang X., Li H., Lai W., He Y., Yao A., Ma L., Shao Y., Zhang B., Wang C., Chen H., Deng H. CRISPR/Cas9-Mediated CCR5 ablation in human hematopoietic stem/progenitor cells confers HIV-1 resistance in vivo. Mol. Ther. 2017;25:1782–1789. doi: 10.1016/j.ymthe.2017.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018;70:327–339. doi: 10.1016/j.molcel.2018.02.028. e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Wang J., Beyer A.I., Teque F., Cradick T.J., Qi Z., Chang J.C., Bao G., Muench M.O., Yu J., Levy J.A., Kan Y.W. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C., Zhang T., Qu X., Zhang Y., Putatunda R., Xiao X., Li F., Xiao W., Zhao H., Dai S., Qin X., Mo X., Young W.B., Khalili K., Hu W. In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol. Ther. 2017;25:1168–1186. doi: 10.1016/j.ymthe.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Liu M., Liu Y., Wu J., Gan T., Zhang W., Li Y., Zhou Y., Hu J. Optimizing genome editing strategy by primer-extension-mediated sequencing. Cell Discov. 2019;5:18. doi: 10.1038/s41421-019-0088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Guney M.H., Kim K., Goh S.L., McCauley S., Dauphin A., Diehl W.E., Luban J. Primate immunodeficiency virus proteins Vpx and Vpr counteract transcriptional repression of proviruses by the HUSH complex. Nat. Microbiol. 2018;3:1354–1361. doi: 10.1038/s41564-018-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., Koonin E.V., Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhao Y., Ye J., Cao X., Xu C., Chen B., An H., Jiao Y., Zhang F., Yang X., Zhou G. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019;17:1185–1187. doi: 10.1111/pbi.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhu B., Chen L., Xie L., Yu W., Wang Y., Li L., Yin S., Yang L., Hu H., Han H., Li Y., Wang L., Chen G., Ma X., Geng H., Huang W., Pang X., Yang Z., Wu Y., Siwko S., Kurita R., Nakamura Y., Yang L., Liu M., Li D. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 2020;38:856–860. doi: 10.1038/s41587-020-0527-y. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Arango G., Li F., Xiao X., Putatunda R., Yu J., Yang X.F., Wang H., Watson L.T., Zhang L., Hu W. Comprehensive off-target analysis of dCas9-SAM-mediated HIV reactivation via long noncoding RNA and mRNA profiling. BMC Med. Genom. 2018;11:78. doi: 10.1186/s12920-018-0394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yin C., Zhang T., Li F., Yang W., Kaminski R., Fagan P.R., Putatunda R., Young W.B., Khalili K., Hu W. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 2015;5:16277. doi: 10.1038/srep16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.H., Lovsin N., Peterlin B.M. Newly identified host factors modulate HIV replication. Immunol. Lett. 2005;97:225–234. doi: 10.1016/j.imlet.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Zhou H., Su J., Hu X., Zhou C., Li H., Chen Z., Xiao Q., Wang B., Wu W., Sun Y., Zhou Y., Tang C., Liu F., Wang L., Feng C., Liu M., Li S., Zhang Y., Xu H., Yao H., Shi L., Yang H. Glia-to-Neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell. 2020;181:590–603. doi: 10.1016/j.cell.2020.03.024. e516. [DOI] [PubMed] [Google Scholar]
- Zhu W., Lei R., Le Duff Y., Li J., Guo F., Wainberg M.A., Liang C. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]