Abstract
Retroviruses exclusively infect vertebrates, causing a variety of diseases. The replication of retroviruses requires reverse transcription and integration into host genomes. When infecting germline cells, retroviruses become inherited vertically, forming endogenous retroviruses (ERVs). ERVs document past viral infections, providing molecular fossils for studying the evolutionary history of retroviruses. In this review, we summarize the recent advances in understanding the diversity and evolution of retroviruses from the perspectives of viral fossils, and discuss the effects of ERVs on the evolution of host biology.
Keywords: Retroviruses, Endogenous retroviruses (ERVs), Origin and evolution, Diversity
Highlights
-
•
Recent advances in understanding the diversity and evolution of retroviruses.
-
•
Methods to analyze ERVs.
-
•
The effects of ERVs on the evolution of host biology.
1. Introduction
Retroviruses (the Retroviridae family) exclusively infect vertebrates and cause a variety of diseases, including acquired immune deficiency syndrome (AIDS) and cancers (Goff, 2007; Coffin et al., 1997; Telesnitsky, 2010). Unique among RNA viruses, retrovirus replication requires reverse transcription and integration into host genomes. Retroviruses usually infect somatic cells. But when retroviruses infect germline cells, the integrated retroviruses are passed on vertically to the next generation, forming endogenous retroviruses (ERVs). ERVs were first discovered in 1960s (Weiss, 2013). In the pre-genomic era, many ERVs had been identified by Southern blot and/or PCR approaches (e.g. Repaske et al., 1985; Tristem et al., 1995; Tristem et al., 1996). Thanks to the recent development of next-generation sequencing techniques, the genomes of many vertebrates have been sequenced, providing important resources for studying the extent and distribution of ERVs with the vertebrate genomes. ERVs are widespread and highly abundant in vertebrate genomes; for example, ERVs account for ∼8% of the human genome (Hayward et al. 2013, 2015; Xu et al., 2018).
It is notoriously hard to study the deep evolution of viruses over geographic time scale due to high mutation rates of viruses (Duffy et al., 2008; Gago et al., 2009) and no physical fossils preserved. Retroviruses are no exception. Reverse transcriptase lacks the ability of proofreading, resulting in high mutation rates in retroviruses. However, ERVs evolve with their hosts as genomic loci, and thus have much lower evolutionary rates (Duffy et al., 2008; Gago et al., 2009). ERVs provide “molecular fossils” for studying the deep history of retroviruses and the ancient evolution of host-retrovirus interaction. More generally, many viruses, both RNA and DNA viruses, can integrate into host genomes, forming endogenous viral elements (EVEs) (Holmes, 2011; Feschotte and Gilbert, 2012). The discovery of EVEs in a wide range of eukaryotes lays the foundation of an emerging field, namely Paleovirology. Paleovirology is the study of the ancient evolution of viruses through analyzing endogenous viral elements or evolutionary signatures of host-virus conflicts left in the host genomes (Emerman and Malik, 2010; Katzourakis, 2013). In this review, we introduce methods to analyze ERVs, summarize the recent advances in understanding the diversity and evolution of retroviruses from the perspectives of viral fossils, and discuss the effects of ERVs on the evolution of host biology.
2. Retroviruses and endogenous retroviruses
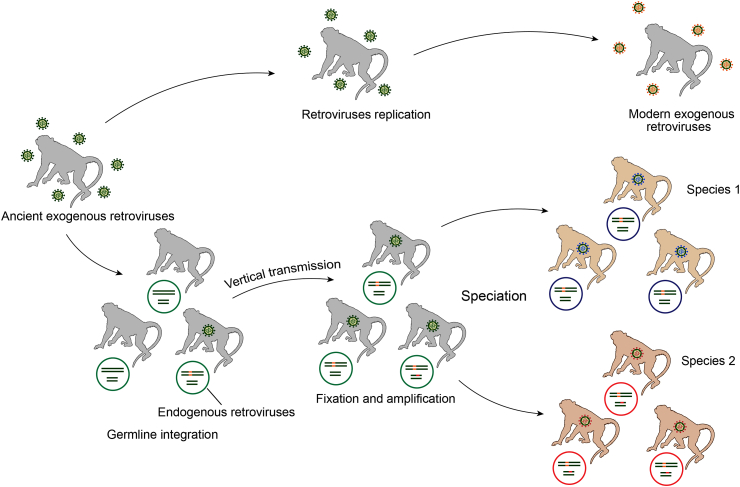
Retroviruses are enveloped viruses with RNA genomes ∼7–10 kb in length. All the retroviruses encode three common genes, namely gag, pol, and env. The gag gene encodes matrix (MA), capsid (CA), and nucleocapsid (NC) proteins, the pol gene encodes protease (PR), reverse transcriptase (RT), RNase H (RH), and integrase (IN) proteins, and the env gene encodes transmembrane (TM) and surface (SU) glycoproteins. Some complex retroviruses can also encode several accessory genes (Goff, 2007; Coffin et al., 1997; Telesnitsky, 2010). Unlike other RNA viruses, the replication of retroviruses requires reverse transcription of viral RNA genomes into double stranded DNA, and integration of the newly synthesized DNA into host genomes. Retrovirus infection typically occurs in the somatic cells of hosts. On occasion, retroviruses infect host germline cells, and the integrated retroviruses flanked by two long terminal repeats (LTRs) become vertically inherited to next generation as part of the host chromosomes, forming endogenous retroviruses (ERVs) (Stoye, 2012; Johnson, 2015) (Fig. 1).
Fig. 1.
Pattern of retrovirus transmission. Retroviruses integrate into the chromosomes of germline cells, forming endogenous retroviruses (ERVs). The new ERVs might be fixed in the host population either by positive selection or random genetic drift. Moreover, ERVs might increase their copy numbers during the evolutionary course. The colored circles represent germline cells, and green and orange lines represent host genomes and ERVs, respectively.
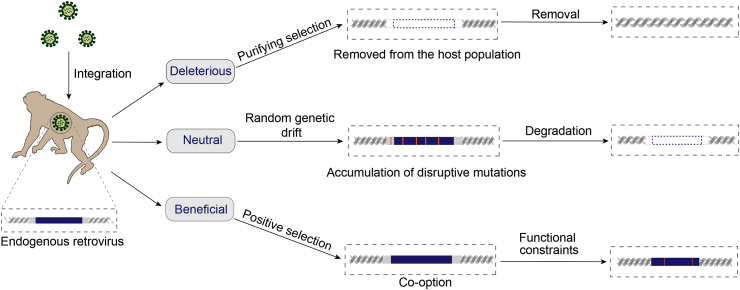
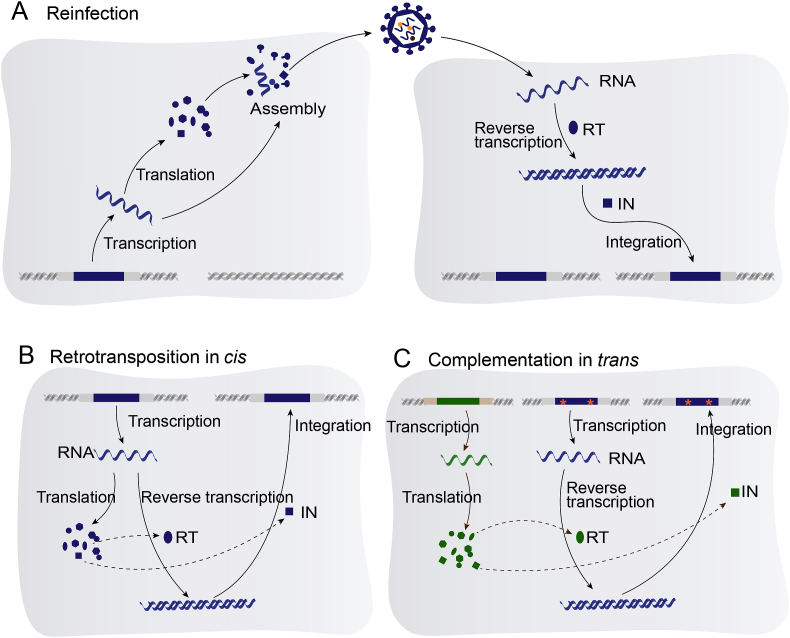
Like new mutations, the fate of novel retrovirus insertions within the host population is governed by population genetics processes. As new mutations, most of new ERV insertions are deleterious and removed from host population by purifying selection (Wang and Han, 2020). The integration of exogenous retroviruses occurs preferentially near or within host genes, but ERVs are found mainly outside host genes, suggesting that a majority of ERVs have been removed by negative selection (Brady et al., 2009; Emerman and Malik, 2010). The fates of the ERVs that have little effect on host fitness and are selectively neutral are governed by random genetic drift. The probability that a newly retroviral insertion is fixed in the host population is 1/2N, where N is the effective population size of hosts (Hartl and Clark, 2006). These neutral ERVs might accumulate disruptive mutations (premature stop codons, frameshift mutations, and insertions or deletions), which eventually lead to the degradation of these ERVs. In extreme but frequent cases, recombination occurs between 5′- and 3-LTRs, generating a solo-LTR. In the rare cases that an ERV insertion is beneficial, it will be fixed in the host population by positive selection (Fig. 2). In the meantime, ERVs retain the ability to proliferate within the host genomes by retrotransposition in cis (ERVs use their own proteins for amplification), complementation in trans (ERVs hitchhike proteins produced by other ERVs or transposable elements within the same cell), or reinfection (ERVs produce virus particles to infect other germline cells), increasing their copy numbers within the host genomes (Belshaw et al., 2005; Bannert and Kurth, 2006; Magiorkinis et al., 2012) (Fig. 3). Together, these processes generate complex insertion polymorphism pattern of ERVs within the host populations.
Fig. 2.
The evolutionary fates of new retrovirus insertions. The newly integrated ERVs are new mutations in the host population. Deleterious retrovirus insertions are eventually removed from host populations by purifying selection. The fate of neutral retrovirus insertions are governed by random genetic drift. Neutral ERVs may accumulate disruptive mutations that eventually lead to their degradation. Beneficial ERVs might be fixed in the host population by positive selection, a process known as co-option. The co-opted retrovirus genes are subject to functional constraints. Blue boxes flanked by gray boxes indicate ERVs with LTRs, and orange vertical lines indicate mutations.
Fig. 3.
Proliferation mechanisms of ERVs. A Reinfection. ERVs in germline cells or somatic cells produce intact virus particles that can be reintegrated into the chromosomes of other germline cells. B Retrotransposition in cis. Viruses use their own proteins to proliferate in germ cells, which requires ERVs to have functional gag and pol genes. C Complementation in trans. ERVs increase their copy numbers using proteins encoded by other ERVs (shown in green). ERVs that proliferate in this way do not require functional gag, pol and env genes. The asterisks represent disruptive mutations. Abbreviation: RT, reverse transcriptase; IN, integrase.
ERV insertion polymorphisms have been found in the populations of many vertebrates (Zhang et al., 2008; Turner et al., 2001; Roca et al., 2004; Polani et al., 2010; Ngo et al., 2019). An ongoing fixation of endogenous koala retrovirus (KoRV) had been observed in the wild koala (Phascolarctos cinereus) population: all the individuals in the north of Australia had endogenous KoRV, while KoRV was not detected in some individuals in the south of Australia (Tarlinton et al., 2006). Consistently, southern koala individuals carried fewer KoRV loci than northern ones (Ishida et al., 2015). The KoRV endogenization started before the late 1800s, but no more than 22,200–49,990 years ago (Ávila-Arcos et al., 2013; Ishida et al., 2015). Tens of HERV-K elements were found to be polymorphic in the human population (Wildschutte et al., 2016), and several HERV-K elements were absent from modern human genomes and present in archaic (Denisovan and Neanderthal) hominids (Lee et al., 2014), suggesting the recent activity of the HERV-K elements in the human population.
3. Mining ERVs
ERVs often accumulate a large number of mutations, making them difficult to identify. Several main strategies have been commonly used to mine ERVs within the vertebrate genomes. (i) ERVs can be identified using similarity search with representative retroviruses as queries. However, representative retroviruses might not cover the major diversity of retroviruses. (ii) Because ERVs are flanked by two LTRs, ERVs can be identified through first detecting candidate LTRs and then detecting and reconstructing retroviral proteins, as exemplified by RetroTector (Sperber et al., 2007). However, ERVs frequently accumulate large-scale deletions and become fragmented, and thus not all the ERVs are flanked by two LTRs. (iii) ERVs can also be identified through first searching homologs of retrovirus hallmark RT proteins. Because retrovirus RT proteins share detectable similarity with retrotransposon RT proteins, phylogenetic analyses are then performed to identify ERVs that cluster with representative retroviruses (Xu et al., 2018). Once again, fragmented ERVs that lack RT proteins cannot be identified using this approach. In general, the approach based on identifying RT homologs is more sensitive than the one based on detecting LTRs (Xu et al., 2018). Nevertheless, old ERVs are often more difficult to detect than young ERVs.
4. Classification and taxonomy of retroviruses
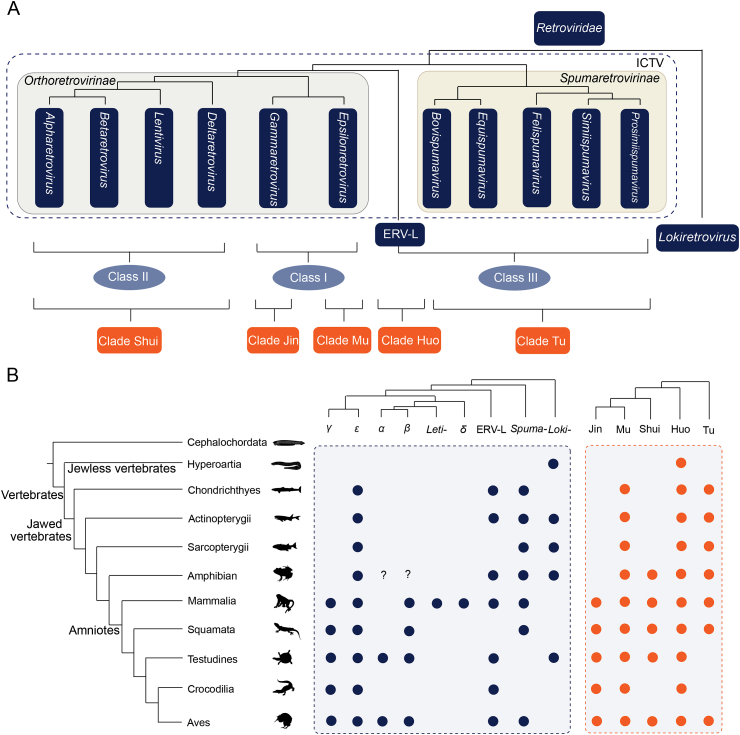
Based on the latest release of International Committee on the Taxonomy of Viruses (ICTV), the Retroviridae family is classified into two subfamilies, namely Orthoretrovirinae and Spumaretrovirinae. The Orthoretrovirinae subfamily includes six genera (Alpharetrovirus, Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Epsilonretrovirus, and Lentivirus), and the Spumaretrovirinae subfamily (also known as foamy viruses) includes five genera (Bovispumavirus, Equispumavirus, Felispumavirus, Prosimiispumavirus, Simiispumavirus) (Hendrickson et al., 2018) (Fig. 4A). However, the ICTV classification only accounts for exogenous retroviruses. ERVs have been traditionally classified into three classes based on their similarity and phylogenetic relationship with exogenous retroviruses. Class I ERVs cluster with Gammaretrovirus and Epsilonretrovirus, Class II ERVs cluster with Alpharetrovirus, Betaretrovirus, Deltaretrovirus, and Lentivirus, and Class III ERVs cluster with foamy viruses and ERV-L (Gifford and Tristem, 2003; Gifford et al., 2018) (Fig. 4A). For historical reasons, human ERVs (HERVs) had been grouped into families primarily based on the specificity of tRNA primer binding sites (PBS) (Bannert and Kurth, 2006; Jern and Coffin, 2008). However, this classification system had problems: ERVs that belong to the same phylogenetic group might have different types of PBS, while unrelated ERVs might share the same type of PBS, and mutations and deletions were present in PBS regions (Bannert and Kurth, 2006; Jern and Coffin, 2008). Therefore, the classification of exogenous and endogenous retroviruses has not been well incorporated. Most of ERVs cannot be assigned into certain genus. It is also challenging to classify and name ERVs, when ERVs arise from recent endogenization events are nested with the diversity of exogenous retroviruses (Blomberg et al., 2009).
Fig. 4.
Retrovirus taxonomy and distribution. A Classification and taxonomy of retroviruses. On the top is the phylogenetic relationship of retroviruses. The gray and yellow rectangles highlight Orthoretrovirinae and Spumaretrovirinae subfamily, respectively. The dark blue dashed rectangle represents the ICTV classification system. B Distribution of retrovirus clades in vertebrates under different taxonomic systems. On the left is the phylogenetic relationship of vertebrate groups, whereas on the top-right is the phylogenetic relationships among the major groups of retroviruses under different taxonomic systems. α, β, γ, δ, ε, Lenti-, and Spuma-represent Alpharetrovirus, Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Epsilonretrovirus, Lentivirus, and Spumaretrovirinae, respectively. The dark blue and orange circles indicate the presence of corresponding exogenous retroviruses and ERVs based on literature (Xu et al., 2018; Zheng et al., 2021; Hayward et al., 2015; Han and Worobey, 2012a, 2012b, 2012c, 2014, 2015; Chen et al. 2019, 2021a, 2021b; Wang and Han, 2021a).
It is logic to include both exogenous and endogenous retroviruses in a taxonomy of retroviruses (Blomberg et al., 2009; Gifford et al., 2018). Based on phylogenetic analyses of Pol proteins from exogenous and endogenous retroviruses sampled across vertebrates, retroviruses have been classified into five major clades, designated Jin, Mu, Shui, Huo, and Tu (Xu et al., 2018). Clades Jin and Mu include viruses related to gammaretroviruses and epsilonretroviruses, respectively, and they include class I ERVs. Clade Shui includes viruses related to alpha-, beta-, delta-retroviruses, lentiviruses, and class II ERVs. Clade Huo is related to snakehead retrovirus, ERV-L, and class III ERVs. Clade Tu is related to foamy viruses (Fig. 4A). However, RH and IN proteins were acquired multiple times in retroviruses, making phylogenetic analyses based on Pol proteins should be taken with cautions (Wang and Han, 2021a). On the other hand, based on phylogenetic analyses of the hallmark RT proteins, the positions of some retroviruses are unstable, and the support values for some groups are low. The recent discovery of lokiretroviruses makes the situation more complicated (Wang and Han, 2021a). Lokiretroviruses are sister to all the known retroviruses, and might represent a novel subfamily of retroviruses. We anticipate a classification system that fully integrates endogenous and exogenous retroviruses and follows phylogeny will be developed in the future.
5. Dating ERV invasion
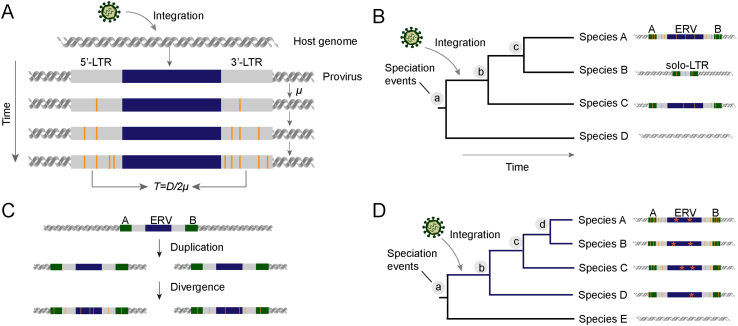
The time for ERV invasion into host genomes can help establish time scales for retrovirus evolution. Several methods have been developed to estimate the integration time of ERVs. (i) The most commonly used approach is based on the divergence of 5′- and 3′-LTRs of an ERV. When integrated into host genomes, 5′- and 3′-LTRs of an ERV are identical at sequence level (Coffin et al., 1997). Then, they accumulate mutations independently as distinct genomic loci. Therefore, the sequence divergence between 5′- and 3′-LTRs are proportional to the integration time (Johnson and Coffin, 1999). The integration time of an ERV, T = D/2μ, where D indicates the genetic distance between 5′-and 3′-LTR, and μ indicates host neutral evolutionary rate (Fig. 5A). However, the approach comes with at least two caveats: 5′- and 3′-LTRs may evolve at a rate different from host neutral evolutionary rate (Martins and Villesen, 2011); gene conversion frequently occurs in LTR regions, which decreases their genetic distance (Johnson and Coffin, 1999). Moreover, given ERVs frequently accumulate large-scale deletion and become fragmented, not all the ERVs are flanked by two LTRs and thus their ages cannot be estimated using the approach based on the divergence between 5′- and 3′-LTRs. (ii) The age of an ERV can also be estimated based on its orthologous insertions within closely related species. The possibility that two ERVs integrated independently at an orthologous position of closely related species is extremely low (Stoye, 2001). Therefore, ERV insertion should occur in the last common ancestor of species who share orthologous ERVs, and the divergence time of their last common ancestor provides a minimum time for the ERV insertion. This approach might sometimes underestimate the invasion time, if an orthologous insertion is lost in certain outgroup species (Fig. 5B). (iii) ERV insertions might undergo segmental duplication. ERVs loci generated by segmental duplication can be identified by ERVs flanking with highly similar host sequences. Calculating the divergence time of segmental duplicates provides a minimum estimate of the invasion time (Katzourakis et al., 2007) (Fig. 5C). (iv) Rarely, the invasion time of an ERV can be estimated based on the occurrence of stop codons within it. Under neutral evolution, stop codons are generated at a rate of ∼1/2310 per codon per million years in primate coding sequences (Belyi et al., 2010). However, it is unclear whether ERVs evolve neutrally after integration, and the reliability of this method should be taken with caution (Fig. 5D).
Fig. 5.
ERV integration time estimation. A The ERV integration time can be estimated based on the divergence of 5′- and 3′-LTRs of an ERV. B The age of an ERV can be estimated based on its orthologous insertions within closely related species. C ERV insertions might undergo segmental duplication. The divergence time of segmental duplicates provides a minimum estimate of the invasion time. D The integration time of an ERV can be estimated based on the occurrence of stop codons within it. The orange vertical lines indicate disruptive mutations, the asterisks (∗) represent stop codons and the green rectangles represent host genes.
6. Origin, evolution, and diversity of retroviruses
Systemic analyses of ERVs in vertebrates provide important insights into the origin, evolution, and diversity of retroviruses. Large-scale phylogenomic analyses show that ERVs are present in all the genomes of jawed vertebrates studied to date (Hayward et al. 2013, 2015; Xu et al., 2018; Zheng et al., 2021). Within jawless vertebrates, ERVs (later classified to endogenous lokiretroviruses) were also identified in sea lamprey (Petromyzon marinus) and Arctic lamprey (Lethenteron camtschaticum) (Hayward et al., 2015; Xu et al., 2018; Wang and Han, 2021a). ERVs have not been identified in any species outside vertebrates (Xu et al., 2018; Wang and Han, 2021a). The distribution of retroviruses suggests that retroviruses may have originated during the early evolution of vertebrates >450 million years ago (Xu et al., 2018). However, the distribution might also be explained by a later origin and spread across vertebrate through frequent host switching. Foamy viruses, a subfamily of retroviruses, have been found to mainly co-diverge with their jawed vertebrate hosts for >450 million years (Han and Worobey, 2012a; Aiewsakun and Katzourakis, 2017; Xu et al., 2018; Wei et al., 2019; Chen et al., 2021a). Moreover, retroviruses of tetrapods (amphibians, reptiles, birds, and mammals) nest within the diversity of fish retroviruses, and fish retrovirus occupy basal positions of all major retrovirus clades. These findings reveal an ancient aquatic origin of retroviruses >450 million years ago (Xu et al., 2018).
The isolation and characterization of retroviruses have been highly biased to human and vertebrates of agricultural or medical importance. ERVs expand our understanding of the host distribution of major retrovirus groups (Fig. 4B). Clade Jin retroviruses (including gamma-related retroviruses) infect mammals, reptiles, and birds (Xu et al., 2018; Zheng et al., 2021). Clade Mu retroviruses (including epsilon-related retroviruses) infect almost all jawed vertebrates (Xu et al., 2018). Clade Shui retroviruses (including alpha-, beta-, delta-related retroviruses and lentiviruses) infect amphibians, reptiles, birds, and mammals; among them, delta-retroviruses and lentiviruses are mammal specific (Katzourakis et al., 2007; Gifford et al., 2008; Keckesova et al., 2009; Gilbert et al., 2009; Han and Worobey, 2012b, 2015; Cui and Holmes, 2012; Hron et al. 2016, 2018; Farkašová et al., 2017; Xu et al., 2018; Chen et al. 2021a, 2021b; Zheng et al., 2021). Clade Huo retroviruses (ERV-L and snakehead retrovirus related) infect all the major lineages of vertebrates (Xu et al., 2018). Clade Tu retroviruses (or fomay viruses) infect all major linages of jawed vertebrates (Han and Worobey, 2012a; Han and Worobey, 2012c; Han and Worobey, 2014; Katzourakis et al., 2014; Ruboyianes and Worobey, 2016; Aiewsakun and Katzourakis, 2017; Xu et al., 2018; Wei et al., 2019; Aiewsakun et al., 2019; Chen et al., 2019; Aiewsakun et al., 2020; Chen et al., 2021a). Recently phylogenomics has identified a novel major retrovirus subfamily known as lokiretrovirus (Wang and Han, 2021a). Lokiretroviruses whose Env proteins share detectable similarity with fusion proteins of some negative sense single-stranded RNA viruses form a sister group to all known retroviruses. Endogenous lokiretroviruses are widely distributed in vertebrates, including lampreys, ray-finned fishes, lobe-finned fishes, amphibians, and reptiles (Fig. 4B). The number of ERVs from different retroviruses groups is dramatically different. Fossil records of delta-retroviruses, lentiviruses, and foamy viruses are extremely rare, which may reflect their inefficiency in entering germline cells, or inherent barriers of replication in germline cells (Johnson, 2015). Most of ERVs lack exogenous retrovirus counterparts, and known exogenous retroviruses cluster into several “small” groups within the large ERV phylogeny (Hayward et al. 2013, 2015; Xu et al., 2018), suggesting that an enormous number of exogenous retroviruses remain to be discovered.
The discoveries of ERVs also help establish or refine the time scales of retrovirus evolution. Molecular clock analyses of exogenous lentiviruses and delta-retroviruses indicate their evolutionary time scales in the unit of hundred years, and both have been thought to be young viral groups (Van Dooren et al., 2001; Wertheim and Worobey, 2009). Endogenous lentiviruses have been identified in lagomorphs, primates, carnivores, and Dermoptera (Katzourakis et al., 2007; Keckesova et al., 2009; Gifford et al., 2008; Gilbert et al., 2009; Han and Worobey, 2012b, 2015; Cui and Holmes, 2012). These endogenous lentiviruses invaded host genomes from 1.9 to 3.8 million years ago (Mya) (in Malagasy lemurs) to 21–40 Mya (in Colugo) (Han and Worobey, 2012b, 2015; Cui and Holmes, 2012; Hron et al., 2016). Endogenous delta-retroviruses have been identified in bats, carnivores, cetaceans, and insectivores (Farkašová et al., 2017; Hron et al., 2018; Zheng et al., 2021). A delta-retrovirus was estimated to integrate into carnivore genomes >24.6 Mya (Hron et al., 2018). These findings strongly support that both lentiviruses and delta-retroviruses are not young viral groups, and they have been circulating in mammals for millions of years.
7. Biological function of endogenous retroviruses
Upon infecting host germ cells, ERVs are inherited in the Mendelian way. ERVs have various effects on their hosts: deleterious, neutral, and beneficial (Fig. 2). If ERVs are inserted into host genes, they are likely to interrupt the normal open reading frames and function of the host genes, which are generally detrimental. Indeed, ERVs are mainly found outside of the host genes (Brady et al., 2009; Emerman and Malik, 2010). ERVs have been proposed to be responsible for a wide range of diseases, such as cancer, autoimmune diseases, and neurodegenerative diseases, but these hypotheses remain controversial and unproven (Voisset et al., 2008; Stoye, 2012; Bhardwaj and Coffin, 2014; Küry et al., 2018). On the other hand, the transcripts or proteins of HERVs might represent biomedicine markers for the diagnosis of those diseases (Song et al., 2021). The deleterious ERV insertions will be removed from the host population by purifying selection. Most of the remaining ERVs are neutral and will become degraded due to the accumulation of disruptive mutations, and insertions or deletions. Beneficial ERVs will be recruited by hosts for host biological functions, a process known as co-option or exaptation (Wang and Han, 2020, 2021b) (Fig. 2).
Broadly, the functionality of ERVs can be divided into three categories, namely physiological, immunological, and regulatory functions (Frank and Feschotte, 2017; Feschotte, 2008; Chuong et al. 2016, 2017). (i) Physiological roles. Some ERV Env proteins, termed as syncytins, were repurposed to function in placenta formation and development (Mi et al., 2000; Blaise et al., 2003). Synctins originated tens of times in mammals and a viviparous lizard, which might be pivotal in the emergence of mammals (Dupressoir et al., 2005; Lavialle et al., 2013; Cornelis et al., 2017). Moreover, Synctins might also contribute to myoblast fusion and muscle sexual dimorphism in mice (Redelsperger et al., 2016). (ii) Immunological roles. Some ERV-derived proteins can protect the hosts from exogenous retrovirus infection. Expression of the ERV env gene is known to inhibit exogenous retrovirus infection by saturating receptors recognized by exogenous retroviruses (Aswad and Katzourakis, 2012; Kozak et al., 1984; McDougall et al., 1994; Palmarini et al., 2004; Tikhonenko and Lomovskaya, 1990). Hosts also employ the ERV-derived proteins to block the replication of exogenous retroviruses. Some endogenous jaagsiekte sheep retroviruses (JSRVs) express defective Gag proteins, which prevent virion release and interfere with exogenous JSRV (Arnaud et al., 2007; Palmarini et al., 2004; Mura et al., 2004). Fv1 gene was derived from the gag gene of a murine ERV-L that was inserted into the genomes of species in Muroidae ∼45 million years ago, and serves as the restriction factor to protect the hosts against infections of various retroviruses (Boso et al., 2018; Young et al., 2018; Best et al., 1996; Yap et al., 2014; Yan et al., 2009). Moreover, the level of ERV expression controls host's respond to the microbiota in homeostatic or inflammatory manner (Lima-Junior et al., 2021). (iii) Regulatory roles. Given that ERVs contain specific regulatory sequences modulating their RNA synthesis and processing, they may provide new regulatory elements and thus influence the expression pattern of nearby host genes (Feschotte, 2008; Stoye, 2012). ERVs have been found to shape the evolution of transcriptional networks underlying immune response and placental function (Finnegan, 2012; Chuong et al. 2016, 2017; Pasquesi et al., 2020; Sun et al., 2021). Moreover, ERV-derived long noncoding RNA, designated lnc-EPAV, serves as a positive regulator of innate immune responses (Zhou et al., 2019). HERVH might act as an enhancer associated lncRNA that interacts with mediator kinase proteins and pluripotency-associated transcription factors to regulate the transcription of pluripotency-associated genes (Sexton et al., 2022). Besides, homologous recombination of ERVs at different chromosome regions can mediates chromosome rearrangement, which might play a major role in vertebrate genome evolution (Hughes, 2001).
Recently, a comprehensive analysis of vertebrate genomes identified 177 independent retroviral gene (gag and env) co-option events in vertebrates, suggesting ERV genes had been frequently co-opted during the evolution of vertebrates (Wang and Han, 2020). The evolution of retroviral gene co-option followed a birth and death model, in which retrovirus genes were frequently co-opted, and co-opted retroviruses were frequently lost (Wang and Han, 2020, 2021b). These co-opted retroviral genes were subject to different selection pressure, implying potentially diverse cellular functionalities, but the specific cellular functionality remained to be studied (Wang and Han, 2020).
8. Outstanding questions
The discoveries of ERVs have greatly refined our understanding of the origin, deep evolution, and diversity of retroviruses. ERVs also shape the evolution of the genome complexity of their hosts. However, many outstanding questions remain: (i) Retroviruses have been thought to originate during the early evolution of vertebrates >450 Mya. However, it remains unclear how retroviruses originated (Hayward, 2017). (ii) Multiple classification systems of retroviruses have been developed, but all these systems have their own shortcomings. A classification system that fully integrates endogenous and exogenous retroviruses and follows phylogeny is eagerly needed. (iii) Retrovirus genes or regulatory sequences have been frequently co-opted in vertebrates, but their functionalities need to be experimentally characterized. (iv) The relationship between ERVs and human diseases should be fully evaluated. (v) ERVs are ubiquitously distributed in the vertebrate genomes, but only tens of exogenous retroviruses have been isolated. More efforts should be paid to isolate exogenous retroviruses in wild animals and assess their potential pathogenicity, which are crucial for predicting and controlling zoonotic disease outbreaks in the future. (vi) ERVs are highly abundant in vertebrates, but some vertebrate genomes harbor more ERVs than others (Hayward et al. 2013, 2015; Xu et al., 2018; Zheng et al., 2021). It remains obscure about the factors that shape the landscape of ERVs within a certain genome.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgement
This work was supported by National Natural Science Foundation of China (31922001).
References
- Aiewsakun P. Avian and serpentine endogenous foamy viruses, and new insights into the macroevolutionary history of foamy viruses. Virus. Evol. 2020;6:vez057. doi: 10.1093/ve/vez057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiewsakun P., Katzourakis A. Marine origin of retroviruses in the early Palaeozoic Era. Nat. Commun. 2017;8:13954. doi: 10.1038/ncomms13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiewsakun P., Simmonds P., Katzourakis A. The first Co-opted endogenous foamy viruses and the evolutionary history of reptilian foamy viruses. Viruses. 2019;11:641. doi: 10.3390/v11070641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud F., Caporale M., Varela M., Biek R., Chessa B., Alberti A., Golder M., Mura M., Zhang Y.P., Yu L., Pereira F., Demartini J.C., Leymaster K., Spencer T.E., Palmarini M. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad A., Katzourakis A. Paleovirology and virally derived immunity. Trends Ecol. Evol. 2012;27:627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Ávila-Arcos M.C., Ho S.Y., Ishida Y., Nikolaidis N., Tsangaras K., Hönig K., Medina R., Rasmussen M., Fordyce S.L., Calvignac-Spencer S., Willerslev E., Gilbert M.T., Helgen K.M., Roca A.L., Greenwood A.D. One hundred twenty years of koala retrovirus evolution determined from museum skins. Mol. Biol. Evol. 2013;30:299–304. doi: 10.1093/molbev/mss223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N., Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genom. Hum. Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- Belshaw R., Katzourakis A., Paces J., Burt A., Tristem M. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol. Biol. Evol. 2005;22:814–817. doi: 10.1093/molbev/msi088. [DOI] [PubMed] [Google Scholar]
- Belyi V.A., Levine A.J., Skalka A.M. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S., Le Tissier P., Towers G., Stoye J.P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- Bhardwaj N., Coffin J.M. Endogenous retroviruses and human cancer: is there anything to the rumors? Cell Host Microbe. 2014;15:255–259. doi: 10.1016/j.chom.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Blaise S., de Parseval N., Bénit L., Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg J., Benachenhou F., Blikstad V., Sperber G., Mayer J. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene. 2009;448:115–123. doi: 10.1016/j.gene.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Boso G., Buckler-White A., Kozak C.A. Ancient evolutionary origin and positive selection of the retroviral restriction factor Fv1 in muroid rodents. J. Virol. 2018;92 doi: 10.1128/JVI.00850-18. e00850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady T., Lee Y.N., Ronen K., Malani N., Berry C.C., Bieniasz P.D., Bushman F.D. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 2009;23:633–642. doi: 10.1101/gad.1762309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Guo X., Zhang L. Unexpected discovery and expression of Amphibian class II endogenous retroviruses. J. Virol. 2021;95:e01806–e01820. doi: 10.1128/JVI.01806-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wei X., Zhang G., Holmes E.C., Cui J. Identification and evolution of avian endogenous foamy viruses. Virus. Evol. 2019;5:vez049. doi: 10.1093/ve/vez049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang Y.Y., Wei X., Cui J. Multiple infiltration and cross-species transmission of foamy viruses across the paleozoic to the cenozoic era. J. Virol. 2021;95 doi: 10.1128/JVI.00484-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong E.B., Elde N.C., Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong E.B., Elde N.C., Feschotte C. Regulatory activities of trans- posable elements: from conflicts to benefits. Nat. Rev. Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J.M., Hughes S.H., Varmus H.E. In: Retroviruses. Cold Spring Harbor. Coffin J.M., Hughes S.H., Varmus H.E., editors. Cold Spring Harbor Laboratory Press; (NY): 1997. The interactions of retroviruses and their hosts. [PubMed] [Google Scholar]
- Cornelis G., Funk M., Vernochet C., Leal F., Tarazona O.A., Meurice G., Heidmann O., Dupressoir A., Miralles A., Ramirez-Pinilla M.P., Heidmann T. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E10991–E11000. doi: 10.1073/pnas.1714590114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Holmes E.C. Endogenous lentiviruses in the ferret genome. J. Virol. 2012;86:3383–3385. doi: 10.1128/JVI.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Dupressoir A., Marceau G., Vernochet C., Bénit L., Kanellopoulos C., Sapin V., Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. U.S.A. 2005;102:725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerman M., Malik H.S. Paleovirology--modern consequences of ancient viruses. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkašová H., Hron T., Pačes J., Hulva P., Benda P., Gifford R.J., Elleder D. Discovery of an endogenous Deltaretrovirus in the genome of long-fingered bats (Chiroptera: miniopteridae) Proc. Natl. Acad. Sci. U. S. A. 2017;114:3145–3150. doi: 10.1073/pnas.1621224114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- Finnegan D.J. Retrotransposons. Curr. Biol. 2012;22:R432–R437. doi: 10.1016/j.cub.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Frank J.A., Feschotte C. Co-option of endogenous viral sequences for host cell function. Curr. Opin. Virol. 2017;25:81–89. doi: 10.1016/j.coviro.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gago S., Elena S.F., Flores R., Sanjuán R. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323:1308. doi: 10.1126/science.1169202. [DOI] [PubMed] [Google Scholar]
- Gifford R., Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Gene. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- Gifford R.J., Blomberg J., Coffin J.M., Fan H., Heidmann T., Mayer J., Stoye J., Tristem M., Johnson W.E. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology. 2018;15:59. doi: 10.1186/s12977-018-0442-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R.J., Katzourakis A., Tristem M., Pybus O.G., Winters M., Shafer R.W. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C., Maxfield D.G., Goodman S.M., Feschotte C. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.P. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 2007;5:253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- Han G.Z., Worobey M. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.Z., Worobey M. Endogenous lentiviral elements in the weasel family (Mustelidae) Mol. Biol. Evol. 2012;29:2905–2908. doi: 10.1093/molbev/mss126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.Z., Worobey M. An endogenous foamy virus in the aye-aye (Daubentonia madagascariensis) J. Virol. 2012;86:7696–7698. doi: 10.1128/JVI.00650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.Z., Worobey M. Endogenous viral sequences from the Cape golden mole (Chrysochloris asiatica) reveal the presence of foamy viruses in all major placental mammal clades. PLoS One. 2014;9 doi: 10.1371/journal.pone.0097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.Z., Worobey M. A primitive endogenous lentivirus in a colugo: insights into the early evolution of lentiviruses. Mol. Biol. Evol. 2015;32:211–215. doi: 10.1093/molbev/msu297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D., Clark A. Sinauer Associates; 2006. Principles of Population Genetics. [Google Scholar]
- Hayward A. Origin of the retroviruses: when, where, and how? Curr. Opin. Virol. 2017;25:23–27. doi: 10.1016/j.coviro.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A., Cornwallis C.K., Jern P. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc. Natl. Acad. Sci. U. S. A. 2015;112:464–469. doi: 10.1073/pnas.1414980112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A., Grabherr M., Jern P. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20146–20151. doi: 10.1073/pnas.1315419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson R.C., Orton R.J., Siddell S.G., Smith D.B. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV) Nucleic Acids Res. 2018;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C. The evolution of endogenous viral elements. Cell Host Microbe. 2011;10:368–377. doi: 10.1016/j.chom.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hron T., Farkašová H., Gifford R.J., Benda P., Hulva P., Görföl T., Pačes J., Elleder D. Remnants of an ancient Deltaretrovirus in the genomes of horseshoe bats (rhinolophidae) Viruses. 2018;10:185. doi: 10.3390/v10040185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hron T., Farkašová H., Padhi A., Pačes J., Elleder D. Life history of the oldest lentivirus: characterization of ELVgv integrations in the dermopteran genome. Mol. Biol. Evol. 2016;33:2659–2669. doi: 10.1093/molbev/msw149. [DOI] [PubMed] [Google Scholar]
- Hughes D.C. Alternative splicing of the human VEGFGR-3/FLT4 gene as a consequence of an integrated human endogenous retrovirus. J. Mol. Evol. 2001;53:77–79. doi: 10.1007/s002390010195. [DOI] [PubMed] [Google Scholar]
- Ishida Y., Zhao K., Greenwood A.D., Roca A.L. Proliferation of endogenous retroviruses in the early stages of a host germ line invasion. Mol. Biol. Evol. 2015;32:109–120. doi: 10.1093/molbev/msu275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P., Coffin J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Johnson W.E. Endogenous retroviruses in the genomics era. Annu. Rev. Virol. 2015;2:135–159. doi: 10.1146/annurev-virology-100114-054945. [DOI] [PubMed] [Google Scholar]
- Johnson W.E., Coffin J.M. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A. Paleovirology: inferring viral evolution from host genome sequence data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120493. doi: 10.1098/rstb.2012.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Aiewsakun P., Jia H., Wolfe N.D., LeBreton M., Yoder A.D., Switzer W.M. Discovery of prosimian and afrotherian foamy viruses and potential cross species transmissions amidst stable and ancient mammalian co-evolution. Retrovirology. 2014;11:61. doi: 10.1186/1742-4690-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A., Tristem M., Pybus O.G., Gifford R.J. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z., Ylinen L.M., Towers G.J., Gifford R.J., Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;384:7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C.A., Gromet N.J., Ikeda H., Buckler C.E. A unique sequence related to the ecotropic murine leukemia virus is associated with the Fv-4 resistance gene. Proc. Natl. Acad. Sci. U. S. A. 1984;81:834–837. doi: 10.1073/pnas.81.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küry P., Nath A., Créange A., Dolei A., Marche P., Gold J., Giovannoni G., Hartung H.P., Perron H. Human endogenous retroviruses in neurological diseases. Trends Mol. Med. 2018;24:379–394. doi: 10.1016/j.molmed.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavialle C., Cornelis G., Dupressoir A., Esnault C., Heidmann O., Vernochet C., Heidmann T. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A., Huntley D., Aiewsakun P., Kanda R.K., Lynn C., Tristem M. Novel denisovan and neanderthal retroviruses. J. Virol. 2014;88:12907–12909. doi: 10.1128/JVI.01825-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima-Junior D.S., Krishnamurthy S.R., Bouladoux N., Collins N., Han S.J., Chen E.Y., Constantinides M.G., Link V.M., Lim A.I., Enamorado M., Cataisson C., Gil L., Rao I., Farley T.K., Koroleva G., Attig J., Yuspa S.H., Fischbach M.A., Kassiotis G., Belkaid Y. Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell. 2021;184:3794–3811. doi: 10.1016/j.cell.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorkinis G., Gifford R.J., Katzourakis A., De Ranter J., Belshaw R. Env-less endogenous retroviruses are genomic superspreaders. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7385–7390. doi: 10.1073/pnas.1200913109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins H., Villesen P. Improved integration time estimation of endogenous retroviruses with phylogenetic data. PLoS One. 2011;6 doi: 10.1371/journal.pone.0014745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall A.S., Terry A., Tzavaras T., Cheney C., Rojko J., Neil J.C. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J. Virol. 1994;68:2151–2160. doi: 10.1128/jvi.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S., Keith J.C., Jr., McCoy J.M. Syncytin is a captive retroviral envelope protein involved in human placenta morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- Mura M., Murcia P., Caporale M., Spencer T.E., Nagashima K., Rein A., Palmarini M. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11117–11122. doi: 10.1073/pnas.0402877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo M.H., Arnal M., Sumi R., Kawasaki J., Miyake A., Grant C.K., Otoi T., Fernández de Luco D., Nishigaki K. Tracking the fate of endogenous retrovirus segregation in wild and domestic cats. J. Virol. 2019;93 doi: 10.1128/JVI.01324-19. e01324-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmarini M., Mura M., Spencer T.E. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J. Gen. Virol. 2004;85:1–13. doi: 10.1099/vir.0.19547-0. [DOI] [PubMed] [Google Scholar]
- Pasquesi G.I.M., Perry B.W., Vandewege M.W., Ruggiero R.P., Schield D.R., Castoe T.A. Vertebrate lineages exhibit diverse patterns of transposable element regulation and expression across tissues. Genome. Biol. Evol. 2020;12:506–521. doi: 10.1093/gbe/evaa068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polani S., Roca A.L., Rosensteel B.B., Kolokotronis S.O., Bar-Gal G.K. Evolutionary dynamics of endogenous feline leukemia virus proliferation among species of the domestic cat lineage. Virology. 2010;405:397–407. doi: 10.1016/j.virol.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Redelsperger F., Raddi N., Bacquin A., Vernochet C., Mariot V., Gache V., Blanchard-Gutton N., Charrin S., Tiret L., Dumonceaux J., Dupressoir A., Heidmann T. Genetic evidence that captured retroviral envelope syncytins contribute to myoblast fusion and muscle sexual dimorphism in mice. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repaske R., Steele P.E., O'Neill R.R., Rabson A.B., Martin M.A. Nucleotide sequence of a full-length human endogenous retroviral segment. J. Virol. 1985;54:764–772. doi: 10.1128/jvi.54.3.764-772.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca A.L., Pecon-Slattery J., O'Brien S.J. Genomically intact endogenous feline leukemia viruses of recent origin. J. Virol. 2004;78:4370–4375. doi: 10.1128/JVI.78.8.4370-4375.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruboyianes R., Worobey M. Foamy-like endogenous retroviruses are extensive and abundant in teleosts. Virus. Evol. 2016;2:vew032. doi: 10.1093/ve/vew032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton C.E., Tillett R.L., Han M.V. The essential but enigmatic regulatory role of HERVH in pluripotency. Trends Genet. 2022;38:12–21. doi: 10.1016/j.tig.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Li X., Wei X., Cui J. Human endogenous retroviruses as biomedicine markers. Virol. Sin. 2021;36:852–858. doi: 10.1007/s12250-021-00387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber G.O., Airola T., Jern P., Blomberg J. Automated recognition of retroviral sequences in genomic data--RetroTector. Nucleic Acids Res. 2007;35:4964–4976. doi: 10.1093/nar/gkm515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J.P. Endogenous retroviruses: still active after all these years? Curr. Biol. 2001;11:R914–R916. doi: 10.1016/s0960-9822(01)00553-x. [DOI] [PubMed] [Google Scholar]
- Stoye J.P. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat. Rev. Microbiol. 2012;10:395–406. doi: 10.1038/nrmicro2783. [DOI] [PubMed] [Google Scholar]
- Sun M.A., Wolf G., Wang Y., Senft A.D., Ralls S., Jin J., Dunn-Fletcher C.E., Muglia L.J., Macfarlan T.S. Endogenous retroviruses drive lineage-specific regulatory evolution across primate and rodent placentae. Mol. Biol. Evol. 2021;38:4992–5004. doi: 10.1093/molbev/msab223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlinton R.E., Meers J., Young P.R. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- Telesnitsky A. Retroviruses: molecular biology, genomics and pathogenesis. Future Virol. 2010;5:539–543. doi: 10.2217/fvl.10.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonenko A.T., Lomovskaya O.L. Avian endogenous provirus (ev-3) env gene sequencing: implication for pathogenic retrovirus origination. Virus Gene. 1990;3:251–258. doi: 10.1007/BF00393184. [DOI] [PubMed] [Google Scholar]
- Tristem M., Herniou E., Summers K., Cook J. Three retroviral sequences in amphibians are distinct from those in mammals and birds. J. Virol. 1996;70:4864–4870. doi: 10.1128/jvi.70.7.4864-4870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristem M., Myles T., Hill F. A highly divergent retroviral sequence in the tuatara (Sphenodon) Virology. 1995;210:206–211. doi: 10.1006/viro.1995.1333. [DOI] [PubMed] [Google Scholar]
- Turner G., Barbulescu M., Su M., Jensen-Seaman M.I., Kidd K.K., Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Van Dooren S., Salemi M., Vandamme A.M. Dating the origin of the African human T-cell lymphotropic virus type-i (HTLV-I) subtypes. Mol. Biol. Evol. 2001;18:661–671. doi: 10.1093/oxfordjournals.molbev.a003846. [DOI] [PubMed] [Google Scholar]
- Voisset C., Weiss R.A., Griffiths D.J. Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol. Mol. Biol. Rev. 2008;72:157–196. doi: 10.1128/MMBR.00033-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Han G.Z. Frequent retroviral gene Co-option during the evolution of vertebrates. Mol. Biol. Evol. 2020;37:3232–3242. doi: 10.1093/molbev/msaa180. [DOI] [PubMed] [Google Scholar]
- Wang J., Han G.Z. A sister lineage of sampled retroviruses corroborates the complex evolution of retroviruses. Mol. Biol. Evol. 2021;38:1031–1039. doi: 10.1093/molbev/msaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Han G.Z. Unearthing LTR retrotransposon gag genes Co-opted in the deep evolution of eukaryotes. Mol. Biol. Evol. 2021;38:3267–3278. doi: 10.1093/molbev/msab101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Chen Y., Duan G., Holmes E.C., Cui J. A reptilian endogenous foamy virus sheds light on the early evolution of retroviruses. Virus. Evol. 2019;5:vez001. doi: 10.1093/ve/vez001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.A. On the concept and elucidation of endogenous retroviruses. Phil. Trans. Biol. Sci. 2013;368:20120494. doi: 10.1098/rstb.2012.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J.O., Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte J.H., Williams Z.H., Montesion M., Subramanian R.P., Kidd J.M., Coffin J.M. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E2326–E2334. doi: 10.1073/pnas.1602336113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhao H., Gong Z., Han G.Z. Endogenous retroviruses of non-avian/mammalian vertebrates illuminate diversity and deep history of retroviruses. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Buckler-White A., Wollenberg K., Kozak C.A. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3259–3263. doi: 10.1073/pnas.0900181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap M.W., Colbeck E., Ellis S.A., Stoye J.P. Evolution of the retroviral restriction gene Fv1: inhibition of non-MLV retroviruses. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G.R., Yap M.W., Michaux J.R., Steppan S.J., Stoye J.P. Evolutionary journey of the retroviral restriction gene Fv1. Proc. Natl. Acad. Sci. U. S. A. 2018;115:10130–10135. doi: 10.1073/pnas.1808516115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Maksakova I.A., Gagnier L., van de Lagemaat L.N., Mager D.L. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wang J., Gong Z., Han G.Z. Molecular fossils illuminate the evolution of retroviruses following a macroevolutionary transition from land to water. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Qi F., Wu F., Nie H., Song Y., Shao L., Han J., Wu Z., Saiyin H., Wei G., Wang P., Ni T., Qian F. Endogenous retrovirus-derived long noncoding RNA enhances innate immune responses via derepressing RELA expression. mBio. 2019;10 doi: 10.1128/mBio.00937-19. e00937-19. [DOI] [PMC free article] [PubMed] [Google Scholar]