Abstract
Crimean-Congo hemorrhagic fever virus (CCHFV) is a causative agent of serious hemorrhagic diseases in humans with high mortality rates. CCHFV glycoprotein Gc plays critical roles in mediating virus-host membrane fusion and has been studied extensively as an immunogen. However, the molecular mechanisms involved in membrane fusion and Gc-specific antibody-antigen interactions remain unresolved largely because structural information of this glycoprotein is missing. We designed a trimeric protein including most of the ectodomain region of Gc from the prototype CCHFV strain, IbAr10200, which enabled the cryo-electron microscopy structure to be solved at a resolution of 2.8 Å. The structure confirms that CCHFV Gc is a class II fusion protein. Unexpectedly, structural comparisons with other solved Gc trimers in the postfusion conformation revealed that CCHFV Gc adopted hybrid architectural features of the fusion loops from hantaviruses and domain III from phenuiviruses, suggesting a complex evolutionary pathway among these bunyaviruses. Antigenic sites on CCHFV Gc that protective neutralizing antibodies target were mapped onto the CCHFV Gc structure, providing valuable information that improved our understanding of potential neutralization mechanisms of various antibodies.
Keywords: Crimean-Congo hemorrhagic Fever virus (CCHFV), Glycoprotein C, Fusion protein, Bunyavirus
Highlights
-
•
Cryo-EM structure of the ectodomain of CCHFV Gc in the postfusion conformation was determined at atomic-resolution.
-
•
CCHFV Gc is a class II fusion protein and adopts hybrid architectural features of hantaviruses and phenuiviruses.
-
•
Structural mapping of Gc epitope residues targeted by neutralizing antibodies would facilitate future vaccine development.
1. Introduction
Crimean-Congo hemorrhagic fever virus (CCHFV, genus Orthonairovirus, family Nairoviridae, order Bunyavirales) is a prevalent tick-borne virus (Abudurexiti et al., 2019, Kuhn et al., 2020), which causes severe hemorrhagic fever in humans with a case fatality rate ranging from 3% to 80% (Pavel et al., 2020). CCHFV has been classified as a biosafety level 4 (BSL-4) pathogen (Weidmann et al., 2016) and has been declared by the World Health Organization (WHO) as a R&D Blueprint priority pathogen (Mehand et al., 2018). Despite being a deadly pathogen, there is currently no licensed vaccine against CCHF disease (Al-Abri et al., 2017; TipihandBurt, 2020; Tahir Ul Qamar et al., 2021).
CCHFV contains a tripartite genome composed of small (S), medium (M) and large (L) negative-stranded RNA segments (Bente et al., 2013; Zivcec et al., 2016). The M segment encodes the glycoprotein precursor that is processed by cellular proteases into several proteins, including the N-terminal mucin-like domain (MLD), two non-structural proteins (GP38 and NSm) and two structural glycoproteins (Gn and Gc) (Fig. 1A) (Bergeron et al., 2007; Guardado-Calvo and Rey, 2017). Glycoproteins Gn and Gc form a lattice of putative tetrameric projections on the viral membrane surface according to recent cryo-electron tomography studies of a nairovirus (Hazara virus) (Punch et al., 2018). The CCHFV Gc glycoprotein is predicted to be a class II membrane fusion protein (Garry and Garry, 2004) that mediates fusion of the viral envelope with the cellular membrane, and this fusion event is triggered by low pH in multivesicular bodies (Shtanko et al., 2014).
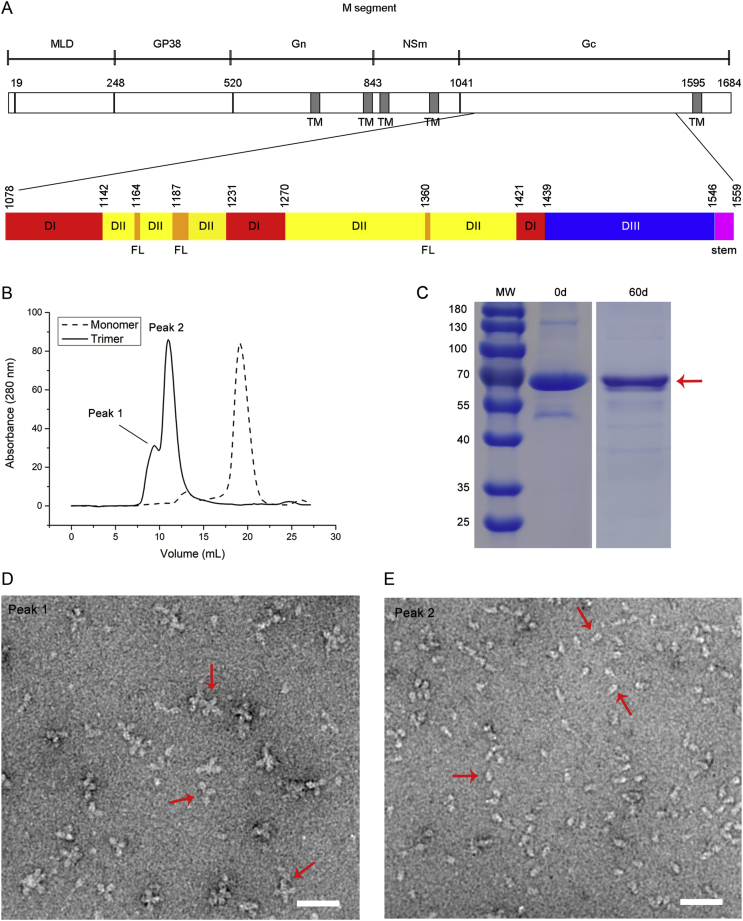
Fig. 1.
Expression of CCHFV sGc-trimers. A Domain diagram of the CCHFV M RNA segment encoding MLD, GP38, Gn, NSm and Gc and transmembrane (TM) regions, which are shown in dark gray. The first residue of each mature protein is provided above. The lower schematic outlines the ectodomain organization of Gc based on our cryo-EM structure: domain I (DI) in red, domain II (DII) in yellow with the fusion loop (FL) in orange, domain III (DIII) in blue and the stem region in magenta. The first residue of each domain is provided above. B SEC elution profiles of CCHFV Gc monomer and trimer are shown as dashed and solid lines, respectively. There are two oligomeric states for sGc-trimers, as indicated by peaks 1 and 2. C SDS-PAGE analysis of degradation of purified sGc-trimers (marked by a red arrow) at 4 °C after 60 days. D and E Negative staining analysis of purified sGc-trimers in peaks 1 and 2 from Fig. 1B. Red arrows indicate rosette-shaped particles formed by aggregation of sGc-trimers (D) and cone-shaped sGc-trimers (E). Scale bar: 50 nm. MLD, mucin-like domain; SEC, size-exclusion chromatography; CCHFV, Crimean-Congo hemorrhagic fever virus; sGc-trimer, trimer of the soluble Gc ectodomain.
Within the Bunyavirales order, most fusion proteins adopt a class II fusion fold, with the remarkable exception of arenaviruses whose GP2 is a class I fusion protein (Hulswit et al., 2021). Class II fusion proteins of bunyaviruses in their postfusion conformations are homotrimers, which have been structurally studied mainly by X-ray crystallography and are limited to the families Phenuiviridae [Rift Valley fever virus (RVFV) and severe fever with thrombocytopenia syndrome virus (SFTSV)] (Halldorsson et al., 2016; Guardado-Calvo et al., 2017) and Hantaviridae [Puumala virus (PUUV), Hantaan virus (HTNV), Andes virus (ANDV) and Maporal virus (MAPV)] (Guardado-Calvo et al., 2016; Willensky et al., 2016; Serris et al., 2020). These proteins share a common molecular architecture consisting of three characteristic β-strand-rich domains, termed I, II and III (Kielian and Rey, 2006; Harrison, 2015; Hulswit et al., 2021). Domain III is connected by a “stem” region to the C-terminal-proximal transmembrane domain, which anchors the fusion protein to the viral envelope. Exposure to acidic conditions causes the fusion loop in domain II to insert into the outer leaflet of the host-cell membrane bilayer. The fusion protein then undergoes a fusogenic conformational change and domain III moves towards the fusion loop, directing the viral membrane and the host-cell membrane to approach each other for subsequent membrane fusion. Detailed structural information on different bunyaviruses is necessary for understanding unique features of their fusion mechanisms.
CCHFV Gc is an important neutralizing-antibody target. A panel of monoclonal antibodies (mAbs) was produced from mice immunized with glycoproteins from the CCHFV strain IbAr10200 (Bertolotti-Ciarlet et al., 2005). Many Gc-specific antibodies have neutralizing activity but only a subset of these antibodies shows protection of neonatal mice in passive-immunization experiments. In a recent report, a recombinant polyprotein bait (rGn/Gc) was used to isolate protective neutralizing antibodies from CCHF-convalescent donors (Fels et al., 2021). The resulting mAbs mainly targeted some conserved epitopes in Gc, and structural characterization of Gc was required for mapping these antigenic sites (Fels et al., 2021).
In this study, we constructed a CCHFV Gc-trimer by taking advantage of a C-terminal GCN4 trimerization motif and determined its atomic structure by cryo-electron microscopy (cryo-EM). Structure-based evolutionary studies revealed some interesting features shared by different bunyaviral families. In addition, we analyzed the distribution of critical epitope residues on the Gc structure and proposed potential neutralization mechanisms.
2. Materials and methods
2.1. Protein expression and purification
The DNA sequence encoding the soluble ectodomain of Gc (sGc, corresponding to the M segment residues 1049–1569) of CCHFV IbAr10200 strain (GenBank accession no. AAM48106) was subcloned into the pMT/BiP/V5-His vector (Invitrogen). sGc-trimers were designed by insertion of a GCN4 trimerization motif (MKQIEDKIEEILSKIYHIENEIARIKKLIGE) between sGc and the C-terminal V5-His tag. A flexible linker (GSGG) was introduced between sGc and the trimerization motif.
A mixture of expression vector and selection vector (19:1) were co-transfected into 1 mL of S2 cells (2 × 106 cells/mL) with 8 μL Cellfectin® II (10362100; Thermo Fisher Scientific Inc., Carlsbad, CA, USA). The transfection mixture was removed and replaced with 2 mL of ESF921 medium (96-001-01; Expression Systems, Davis, CA, USA) with 5% fetal bovine serum (FBS; 10099141; Gibco, Australia) after 6 h. Then, 300 μg/mL of hygromycin-B (V900372; Sigma-Aldrich, USA) was added to S2 cells every four to five days until resistant colonies (3–4 weeks) were selected. For soluble protein expression, stable resistant S2 cells were grown at 27 °C and were induced by the addition of copper sulfate when the cells reached a density of 5 × 106 cells/mL. The supernatant was harvested and clarified by centrifugation 144 h post infection. sGc and sGc-trimers were purified by nickel affinity chromatography with Ni-NTA agarose (17-5318-02; GE Healthcare, Uppsala, Sweden) followed by size-exclusion chromatography (SEC) with a Superdex 200 (10/300) GL column (17-5175-01; GE Healthcare) equilibrated with 25 mmol/L Tris-HCl, 150 mmol/L NaCl, pH 8.0.
2.2. Cryo-EM sample preparation, data collection and processing
The purified protein was first negatively stained and examined under transmission electron microscope (TEM) to evaluate sample quality. Four microliters of a specimen at a concentration of ∼ 0.01 mg/mL was applied onto glow-discharged copper grids supported by a continuous layer of carbon film for 30 s at room temperature. The grids were then submerged into 2% (w/v) uranyl acetate solution for 60 s. The air-dried grids were loaded onto a FEI Tecnai G2 20 TWIN microscope (FEI, United States) (operating at 200 kV) equipped with an Olympus SIS Cantega G2 2K × 2K CCD (Olympus, Japan).
For cryo-EM specimen preparation, 3.5 μL of the protein sample at a concentration of ∼ 0.3 mg/mL was applied onto glow-discharged holey carbon copper grids (Quantifoil R 1.2/1.3, 300-mesh). After excess sample was removed by blotting with filter paper for 5.5 s, the grid was plunge-frozen in liquid ethane using a Leica EM GP2 (Leica Microsystems, Germany) at 25 °C and 85% humidity.
Cryo-EM single particle data collection was performed using a CRYO ARM 300 electron microscope (JEOL, Japan) operated at 300 kV with an in-column energy filter (slit width 30 eV). A total of 3330 images were recorded at a nominal magnification of 80,000 ×, corresponding to 0.61 Å per pixel by a K3 direct electron detector (Gatan, United States) using Serial-EM (Li et al., 2018). Each image stack was dose-fractionated to 40 frames with an accumulated dose of ∼ 48 e/Å2 and a total exposure time of 2 s.
After manual selection, 2688 image stacks were subject to beam-induced motion correction using the MotionCor2 implementation in RELION (version 3.1) (Zivanov et al., 2019). Dose-weighted micrographs were then imported into cryoSPARC (version 3.2) for subsequent data processing procedures (Punjani et al., 2017). The contrast-transfer function parameters were fitted by the patch-based CTF estimation procedure. A total of 2514 micrographs were filtered with a CTF fit resolution better than 4.5 Å and a detected focus range of 0.5–2.5 μm.
Totally, 323,944 particles were auto-picked in 250 micrographs by the blob picker tool within cryoSPARC and were subjected to reference-free 2D classification. Five representative classes were selected as references for template-based particle picking for the whole data set. After 2D classification of the resultant 2,060,865 particles, 956,121 particles were selected from 13 good classes for ab initio generation of four volumes, which were used as initial models for heterogeneous refinement. When C3 symmetry was applied, 187,292 particles selected from the best 3D class yielded a map at an average resolution of 2.8 Å [Fourier shell correlation (FSC) = 0.143] using non-uniform refinement and CTF refinement.
2.3. Atomic model building and refinement
A starting atomic model was built using the phenix.map_to_model tool (Afonine et al., 2018). The only fragment with the correct register was N1377–T1425, including the longest α helix (α2). The crystal structure of RVFV Gc in the postfusion conformation (PDB ID: 6EGT) was used as a guide to trace the backbone of the CCHFV sGc-trimer. The coordinates were modified and the sequence was assigned using Coot (Emsley et al., 2010). The model was improved through multiple cycles of real-space refinement using phenix.real_space_refinement with geometric and secondary structure restraints, and subsequent manual adjustments in Coot. The structure was validated using the phenix.validation_cryoem tool. The model refinement statistics generated in Phenix are summarized in Table 1.
Table 1.
Cryo-EM data collection, refinement and model validation statistics.
| Item | CCHFV sGc-trimer |
|---|---|
| Data collection and processing | |
| Microscope | JEOL CRYO ARM 300 |
| Voltage (kV) | 300 |
| Magnification | 80,000 × |
| Defocus range (μm) | 0.5 to 2.5 |
| Pixel size (Å) | 0.61 |
| Image stacks (no.) | 3330 |
| Frames per stack | 40 |
| Total dose per movie (e−/Å2) | 48 |
| Final particle images (no.) | 187,292 |
| Symmetry imposed | C3 |
| Map resolution (Å) | 2.8 |
| FSC threshold | 0.143 |
| Map-sharpening B factor (Å2) | −141 |
| Model refinement | |
| Composition | |
| Chains | 3 |
| Non-hydrogen atoms | 10,143 |
| Protein residues | 1296 |
| B factors for protein (Å2) | 43.91 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.765 |
| Validation | |
| MolProbity score | 1.87 |
| Clashscore | 7.79 |
| Poor rotamers (%) | 0.00 |
| Ramachandran plot | |
| Favored (%) | 93.06 |
| Allowed (%) | 6.94 |
| Disallowed (%) | 0.00 |
CCHFV, Crimean-Congo hemorrhagic fever virus; sGc-trimer, trimer of the soluble Gc ectodomain; R.m.s, root mean square.
UCSF ChimeraX (Goddard et al., 2018) or PyMOL (The PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC) was used to visualize the 3D volume map and the atomic model, and local resolution variations were estimated using cryoSPARC. Sequences were aligned using ClustalW, and the secondary structure was analyzed by DSSP (Joosten et al., 2011) and illustrated with ESPript 3.0 (Robert and Gouet, 2014).
3. Results
3.1. Expression of CCHFV Gc
sGc, (∼ 60 kDa), which was constructed by truncating the C-terminal transmembrane domain and cytoplasmic domain (residues 1595–1684 in the polyprotein) (Fig. 1A), was expressed using a Drosophila expression system and was purified by nickel affinity chromatography. SEC analysis revealed that purified sGc was monomeric in solution (Fig. 1B). Efforts to obtain sGc crystals were hampered by the poor stability of sGc. A C-terminal trimerization motif (GCN4) with a flexible glycine-serine linker (GSGG) (Hernandez Alvarez et al., 2008; Chan et al., 2012) was appended to sGc to obtain a stable Gc candidate for structural analysis. This sGc product was purified as stable trimers (hereafter referred to as sGc-trimers) with no significant degradation over 60 days at 4 °C (Fig. 1C). SEC analysis of sGc-trimers revealed that there were two oligomeric states in solution (Fig. 1B). One (peak 1) was the aggregation of trimers, which showed a rosette pattern under TEM (Fig. 1D). Negatively stained sGc-trimers (peak 2) were homogeneous cone-shaped particles (Fig. 1E), which were used for subsequent cryo-EM studies.
3.2. Single-particle reconstruction of sGc-trimers
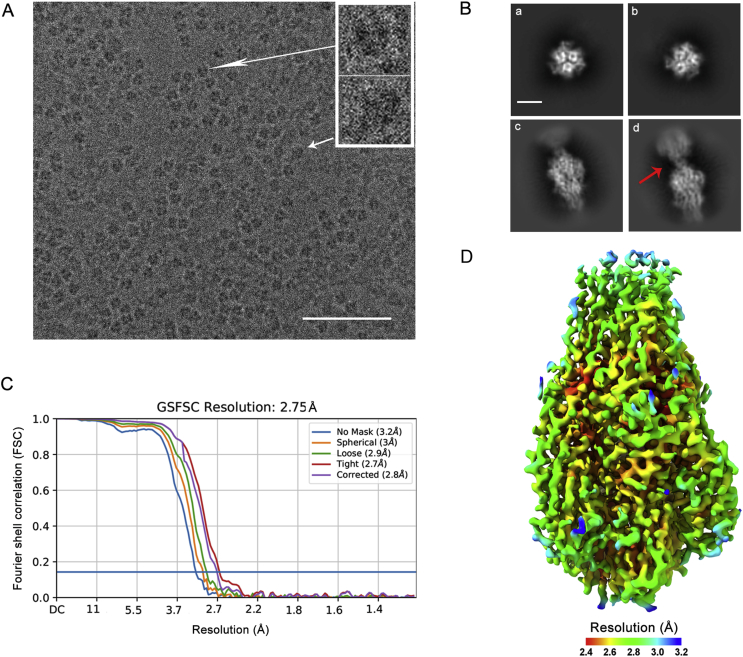
Cryo-EM images (Fig. 2A) of sGc-trimers were collected using a K3 camera mounted on a JEOL CRYO ARM 300 electron microscope. Using a template-based picking strategy, about two million particles for reference-free 2D classification in cryoSPARC were selected (Punjani et al., 2017). Dominant 2D classes showed essentially C3 symmetry (Fig. 2B, classes a and b), indicating that most of the particles oriented with their three-fold axis coinciding with the direction of the electron beams. Using heterogeneous refinement implemented in cryoSPARC, we were able to select ∼ 190,000 particles with an even orientation distribution for final reconstruction of the structure at 2.8 Å resolution when C3 symmetry was applied (Fig. 2C and D).
Fig. 2.
Cryo-EM data analysis of CCHFV sGc-trimer. A A micrograph of frozen-hydrated CCHFV sGc-trimer particles. Insets show particles in two representative orientations. Scale bar: 50 nm. B Representative 2D class averages of sGc-trimers. Classes a and b show three-fold symmetry. Class c is the side view of the particle. Class d highlights the connectivity between two adjacent particles lying flat in ice. Scale bar: 5 nm. C Gold-standard Fourier shell correlation (GSFSC) curves for the non-uniform refinement reconstructions. The 0.143 cutoff is indicated by the horizontal blue line. D Final cryo-EM map of sGc-trimers with surface coloring according to the local resolution estimated by cryoSPARC. CCHFV, Crimean-Congo hemorrhagic fever virus; sGc-trimer, trimer of the soluble Gc ectodomain.
3.3. CCHFV Gc is a class II membrane fusion protein
The cryo-EM structure of sGc-trimers at atomic resolution enabled tracing of the polypeptide chain from Lys 1078 to Ile1559 (Fig. 3A and B), except for some loop regions: Glu1094–Ile1100, Gly1123–Ser1130, Pro 1195–Gly 1202, Glu1286–Phe1289, Gly1324–His1343 and Phe1501–Thr1503. Our final model contained 482 amino acids per monomer. Interestingly, the C-terminal GCN4 trimerization motif was also missing in the density map, suggesting that the trimerization motif facilitated Gc monomer packing but was not required in forming the final stable Gc trimer structure. One or two N-linked glycosylation sites have been identified in the Gc crystal structures of RVFV, SFTSV, HTNV and PUUV (Guardado-Calvo et al., 2016, 2017; Halldorsson et al., 2016; Willensky et al., 2016), and two N-linked glycosylation sites (Asn1194 and Asn1345) were predicted in our model by NetNGlyc (Sun et al., 2007); however, no clear glycan density was observed near the two asparagine residues in our cryo-EM map. The close proximity of Asn1194 and Asn1345 to two disordered loops (Pro1195–Gly1202 and Gly1324–His1343) might contribute to the structural flexibility of the glycans. Statistics on cryo-EM data collection, 3D reconstruction and model refinement are summarized in Table 1.
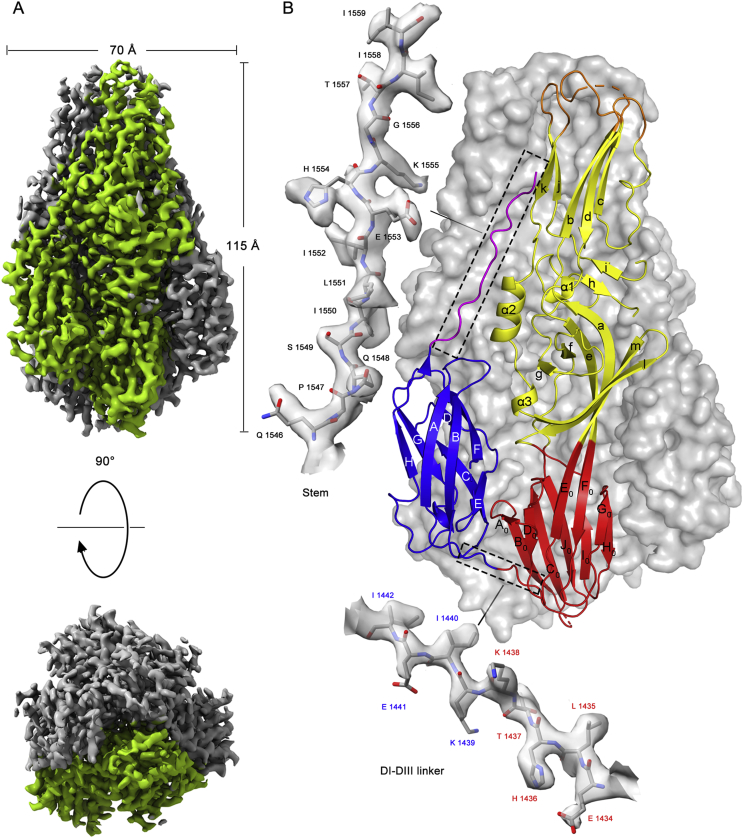
Fig. 3.
The postfusion structure of the CCHFV Gc ectodomain. A The cryo-EM map of CCHFV sGc-trimers. The upper map is a side view of sGc-trimers with the region corresponding to one Gc protomer shown in green and the density corresponding to the other two subunits shown in light gray. The bottom is the top view of the map along the C3 axis. Contour threshold used in ChimeraX was set to 0.44. B Ribbon diagram of one Gc ectodomain oriented roughly as shown in panel (A) and color-coded according to Fig. 1A. The β strands “A0–J0” of domain I are labeled by letters with a subscript. The β strands “a–m” of domain II are labeled with lowercase letters, whereas the three helices are labeled α1–3. The β strands “A–H” of domain III are labeled with capital letters. The C-terminal “stem” (magenta) extends along the interface between domains II from neighboring protomers towards the fusion loop (orange). Magnified view (upper left) of the cryo-EM density of the stem region (residues Gln 1546 to Ile1559) is shown as a transparent surface with the underlying atomic model in stick representation. Magnified view (bottom) highlights the connecting region between domains I and III with residues shown in stick representation under the cryo-EM density. Residues are labeled and colored to indicate the domain boundary, and the carbon, oxygen and nitrogen atoms in magnified amino acid residues are colored with gray, red and blue respectively. CCHFV, Crimean-Congo hemorrhagic fever virus; sGc-trimer, trimer of the soluble Gc ectodomain; DI, domain I; DIII, domain III.
Like other class II fusion proteins, the ectodomain of CCHFV Gc can be divided into three domains, I, II and III (Fig. 3B). Structure-based sequence alignment showed that the secondary elements of CCHFV Gc largely coincide with those of RVFV and HTNV (Fig. 4A and B), even though the total sequence identity was only 15.7% and 15.3%, respectively. sGc-trimers have a central trimeric core formed by parallel interaction of adjacent protomers involving roughly the N-terminal three-quarters of sGc (domains I and II). The remaining C-terminal portion of sGc packs laterally against the core trimer, which involves domain III and part of the stem region. CCHFV Gc trimerizes through extensive interactions between the adjacent protomers and the total surface area buried in trimer interfaces is ∼ 16,000 Å2 (compared with the ∼ 50,000 Å2 of total solvent accessible surface area of the trimer).
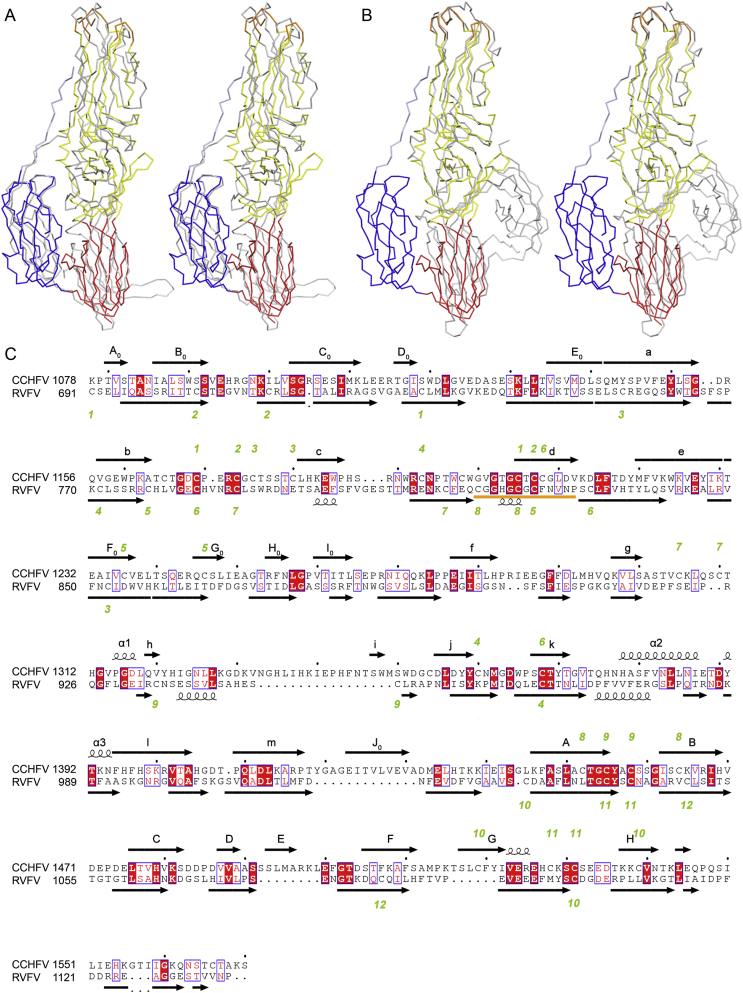
Fig. 4.
Structural comparisons between CCHFV Gc and other bunyavirual Gc. A Superposition of CCHFV Gc onto RVFV Gc (PDB ID: 6GET) is presented as stereo pairs. The CCHFV Gc monomer is colored as in Fig. 3B whereas RVFV Gc is colored gray. All three domains of CCHFV and RVFV can be roughly superpositioned. B Superposition of CCHFV Gc onto HTNV Gc (gray, PDB ID: 5LJZ). Domains I and II are superimposable, whereas domain III is on the opposite side. C Pairwise Gc sequence alignment of CCHFV and RVFV. The secondary structural elements of CCHFV Gc are shown at the top of sequences corresponding to the ribbon diagrams presented in Fig. 3B. In addition, two elongated β strands spanning domains I and II are divide into E0/a and e/F0 (corresponding to RVFV). The fusion loop of RVFV is indicated by an orange line. The disulfide bonds of CCHFV and RVFV are labeled with green letters at the top and bottom of the sequences, respectively. Conserved or similar amino acids are highlighted in light blue border, whereas fully conserved amino acids are highlighted in red. CCHFV, Crimean-Congo hemorrhagic fever virus; RVFV, Rift Valley fever virus.
CCHFV Gc has a long stem region connecting domain III to the transmembrane domain, which spans 49 residues. Electron density for most of this region is missing because of flexibility; however, the first 14 residues of the stem (1546–1559) are visible in the cryo-EM map. The last residue modeled in our structure is Ile1559, which is located ∼ 20 Å from the distal tip of domain II where the fusion loop is situated (Fig. 3B). The remaining 35 residues could readily extend towards the fusion loop to form a “hairpin” conformation with the Gc transmembrane domain and fusion loop at the same end, strongly suggesting that our structure represents CCHFV Gc in its postfusion conformation.
3.4. Distribution of disulfide bonds in Gc
There are 26 cysteine residues in the soluble Gc ectodomain and 24 of these residues are present in the cryo-EM structural model (except Cys1198 and Cys 1566). There are 11 disulfide bonds in our model with one disulfide bond in domain I, six in domain II, and the remaining four in domain III. All 11 disulfide bonds are fully conserved among different CCHFV strains (Supplementary Fig. S1).
Domain I of CCHFV Gc is composed of ten β-strands labeled A0–J0 that are arranged in two anti-parallel β-sheets to form a central β-sandwich (Fig. 3B), which is structurally similar to RVFV and HTNV (Fig. 4A and B). The B0C0J0I0H0 sheet faces the trimeric core, whereas the D0E0F0G0 sheet is located on the surface of the Gc trimer. CCHFV Gc has one disulfide bonds in domain I between strands F0 and G0 (Cys1236–Cys1246), which is also conserved in domain I of HTNV Gc (Guardado-Calvo et al., 2016).
3.5. Putative fusion loops are structurally similar to those of hantaviruses
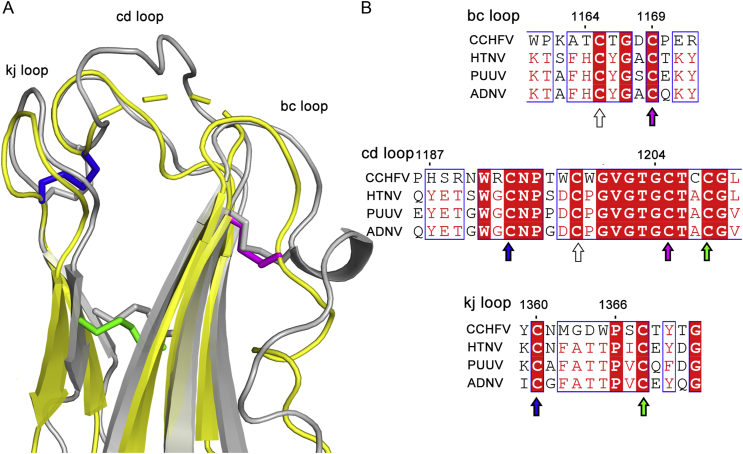
Elongated domain II consists of two insertions in domain I (one is between strands E0 and F0 and the other is between strands I0 and J0). Domain II is composed of thirteen β-strands (labeled a through m) and three short helices (α1, α2 and α3). Like other class II fusion proteins, domain II of CCHFV Gc can be divided into two subdomains. One subdomain is proximal to domain I, folding into an open β-barrel composed of six antiparallel β-strands “aefglm”. In comparison to RVFV and HTNV Gc structures, CCHFV Gc has a longer hairpin structure formed by “lm” β-strands that extend to an adjacent protomer (Fig. 3B). The other subdomain resides at the distal tip of the Gc ectodomain, forming a β-sandwich between the “bdc” β-sheet and the “kj” β-hairpin that projects the “bc” (Thr1164–Cys1169), “cd” (His1187–Gly1204) and “jk” (Cys1360–Pro1366) loops at its outmost end. In our postfusion Gc model, several residues of the “cd” loop are missing. Pairwise sequence alignment shows that the fusion loop of RVFV (Cys818–Pro830) corresponds to a region of CCHFV Gc spanning part of the “cd” loop and complete “d” β-strand (Fig. 4C), suggesting the fusion loop in the two bunyaviral families are not conserved.
Five disulfide bonds in domain II are distributed near the fusion loop region and are essential for structural integrity. The top and middle moieties of the CCHFV Gc “bc” loop are covalently connected with the “d” β-strand by two disulfide bonds (Cys1169–Cys1205 and Cys1173–Cys1207), and the “bc” loop is also stabilized by an internal disulfide bond (Cys1175–Cys1180). There is a disulfide bond bridging “cd” and “jk” loops (Cys1193–Cys1360), and a disulfide bond stapling together β-strands “k” and “d” (Cys1208–Cys1368) (Fig. 5).
Fig. 5.
Structure and sequence alignment of fusion loops and conserved disulfide bonds between CCHFV Gc and HTNV Gc. A Superposition of Gc fusion loops of CCHFV (yellow) onto HTNV (gray; PDB ID: 5LJZ) reveals highly similar conformations. Residues from Pro 1195 to Gly 1202 in the cd loop of CCHFV Gc are missing in the density map and are depicted as a dashed line. Three conserved disulfide bonds of CCHFV are shown as sticks and in magenta (Cys1169–Cys1205), blue (Cys1193–Cys1360) and green (Cys1208–Cys1368), respectively. Three conserved disulfide bonds of HTNV are shown as gray sticks. B Sequence alignment of bc, cd and kj loops between CCHFV and three hantaviruses. The first and last residues of each loop are numbered on the top of the sequences. Residues highlighted in red are strictly conserved. The cysteine residues of conserved disulfide bonds are labeled by arrows that match the coloring used in (A). Conserved or similar amino acids are highlighted in light blue border, whereas fully conserved amino acids are highlighted in red. It is worthy to note that a cysteine pair (Cys1165-Cys1198, labeled as empty arrows) is highly conserved. Although the cysteine pair forms a disulfide bond in hantaviruses (Guardado-Calvo et al., 2016), further experimental evidence is needed to confirm whether this is also the case in CCHFV Gc, since the density for Cys1198 is missing in the cryo-EM model. CCHFV, Crimean-Congo hemorrhagic fever virus; HTNV, Hantaan virus; PUUV, Puumala virus; ADNV, Andes virus.
HTNV Gc and CCHFV Gc share three conserved disulfide bonds in the tip region of domain II (Cys1169–Cys1205, Cys1193–Cys1360 and Cys1208–Cys1368), and the overall structure of the fusion loops of HTNV is also similar to that of CCHFV (Fig. 5). It has been reported that instead of a single “fusion loop”, all three loops at the tip of domain II from HTNV Gc insert into the target membrane (Guardado-Calvo et al., 2016). Structural conservation of the fusion loops between the two families suggests that nairoviruses could also use a tri-partite membrane targeting strategy to facilitate viral membrane fusion (Guardado-Calvo et al., 2016). The functional roles of individual loops need to be experimentally determined.
3.6. Organization of domain III resembles its phenuivirus counterpart
Domain III has an immunoglobulin superfamily C2 subtype fold formed with an eight-stranded (ABCDEFGH) β-barrel-like module (Halaby et al., 1999) (Fig. 3A). The AB, CD and FG loops are close to the β-sheets of the adjacent protomer domain II. In contrast to domain III from RVFV or HTNV Gc with seven β-strands, domain III from CCHFV Gc has eight β-strands with the extra E β-strand, resulting in a more closed β-barrel structure. The four disulfide bonds in domain III function to stabilize the AB and GH loops (Fig. 4C).
Compared with other Gc structures in their postfusion states from the Phenuiviridae and Hantaviridae families, all three domains of CCHFV Gc can be roughly aligned to those of RVFV Gc, whereas superimposable regions of HTNV Gc are limited to domains I and II (Fig. 4A and B). The polypeptide sequence linking domains I and III is well defined in the cryo-EM map (Fig. 3B), validating that this domain arrangement is distinct from “the swapping of domain III” observed in hantaviruses (Guardado-Calvo et al., 2016). There are 23 conserved residues in domain III between CCHFV and RVRF, especially for the A β-strand (5 of the 10 residues being conserved) (Fig. 4C), whereas only 11 residues are conserved between CCHFV and HTNV (Supplementary Fig. S2). In addition, there is one conserved disulfide bond between RVFV and CCHFV in domain III (Cys1455–Cys1458), whereas no conserved disulfide bond is present between HTNV and CCHFV in this region. All these structural observations strongly indicate that nairoviruses and phenuiviruses share a possible common evolutionary origin for domain III.
3.7. Epitope sites on CCHFV Gc
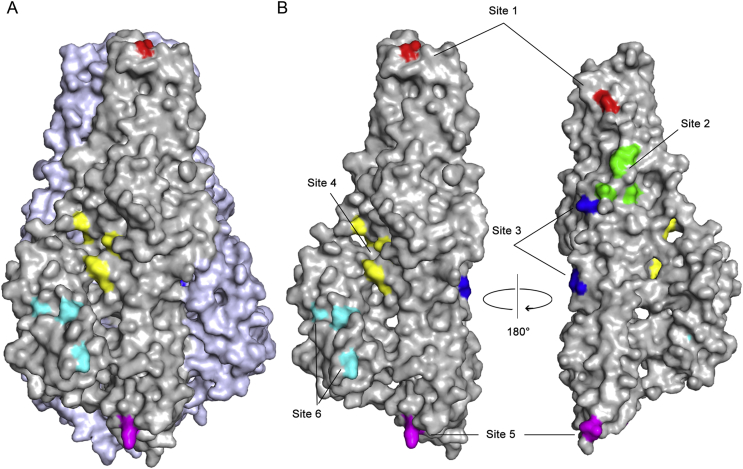
Recent research has identified six antigenic sites on CCHFV Gc that protective neutralizing antibodies target: site 1 (Trp 1185, Trp 1199), site 2 (Ser 1309, Val 1314, Asp 1352, Gly 1353), site 3 (Leu 1307, Ile 1229), site 4 (Lys 1297, Asn 1386, Lys 1393), site 5 (Val 1124, Ser 1128, Val 1093) and site 6 (Glu 1500, Asp 1504, Leu 1450, Ile 1522, Leu 1518, Phe 1520) (Fels et al., 2021). We mapped these residues onto the CCHFV Gc trimer and protomer structures (Fig. 6).
Fig. 6.
Critical residues for six epitope sites are mapped onto the surface of CCHFV Gc. A Surface representation of Gc trimer with one protomer colored in dark gray and the other two in light blue. B Two views of the Gc protomer illustrate the outer surface [left, in the same orientation as in (A)] and buried surface (right).
Site 1 is located near the fusion loop “cd”, and the antibodies targeting this site can effectively block the insertion of the fusion loop into the target membrane. Sites 2 and 3 are embedded in the interior of the trimer, preventing the conformational transition of Gc by blocking the formation of the postfusion homotrimer. Site 4 is situated between the α2 helix and “g” β-strand. The binding of an antibody to this position probably prohibits the complete folding back of domain III. Site 5 is located at the bottom of domain III and near the linker region between domains I and III, and antibody binding may hinder the movement of domain III. Critical residues in site 6 are located in domain III and binding of antibodies to this site may also affect the free movement of domain III, which is required for the prefusion-to-postfusion transition.
4. Discussion
In this report, we presented a cryo-EM reconstruction of the CCHFV Gc ectodomain at atomic resolution, which enabled us to build a model with clear structural details comparable to previously reported crystal structures of postfusion Gc trimers from other bunyaviral families. During 2D classification of selected sGC-trimer particles, it was observed that most particles in vitreous ice had their three-fold axes oriented perpendicular to the vitreous ice layer, which might be caused by exposure of hydrophobic fusion loops to the air-water interface. Even particles lying flat in the vitreous ice layer showed a head-to-tail arrangement (Fig. 2B, class d), in which the fusion loops of one sGC-trimer might insert into the base region of domain I (near missing residues Glu1094–Ile1100, Gly1123–Ser1130) in the adjacent homotrimer to harbor their hydrophobic fusion loops. Testing with different surfactants to tackle the problem of preferential orientation (Drulyte et al., 2018) may facilitate expansion of cryo-EM studies to other bunyaviral Gc.
In recent years, the range of CCHFV circulation has increased (Bente et al., 2013; Arteaga et al., 2021). This increase has been accompanied by reports of multiple CCHFV strains co-circulating within a geographic region (Goedhals et al., 2014). Our results suggest that CCHFV may have “borrowed” different structural elements from various bunyaviral families, showing a complex evolutionary strategy within this large virus group. Studying the possible transfer of genetic material among bunyaviral families represents an interesting future study.
5. Conclusions
This report describes the atomic-resolution structure of CCHFV Gc in the postfusion conformation using cryo-EM and sheds light on how CCHFV Gc combines structural features from two other bunyaviral families. Our cryo-EM structure of CCHFV Gc provides a structure-based blueprint for developing new neutralizing antibodies that target the glycoprotein surface of this group of important emerging pathogens.
Data availability
The Cryo-EM map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-31579, and the corresponding atomic model has been deposited in the Protein Data Bank (PDB) under accession code 7FGF.
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Na Li: conceptualization, formal Analysis, investigation, writing-original draft, writing-review and editing. Guibo Rao: data curation, methodology, writing-original draft. Zhiqiang Li: methodology, visualization. Jiayi Yin: data Curation. Tingting Chong: methodology. Kexing Tian: formal Analysis. Yan Fu: project administration. Sheng Cao: conceptualization, funding acquisition, resources, supervision, writing-review and editing.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank Profs. Zhihong Hu, Rui Gong and Manli Wang for important discussions. We thank the Center for Instrumental Analysis and Metrology of Wuhan Institute of Virology for providing technical assistance. This work was supported by the National Natural Science Foundation of China (31570161).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.01.015.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Sequence alignment of CCHFV Gc from representative strains. The disulfide bonds of CCHFV are labeled with green letters at the bottom of sequences. Viral strains used for the alignment are IbAr10200 (GenBank ID: AAM48106), SPU 115/87 (ARB51462), Caceres 2014 (ASV45881), Sudan AB1-2009 (AEI70589), Badajoz 2018 (QQL93572), SK/2015/human (AOF43039), Baghdad 12 (CAD61343), Afg09-2990 (ADQ57289), Iran-53 (ABD98125), 813040 UAE (ASW20657), 61T Pakistan (QIQ51272), MCL-18-H-1780 (QCY54392), NIV1311122 (QBP34417), U2/2-002/U 6415 (AAW84283), FK16116 (QEQ12756), Hodzha (AAP29978), SCT ex Afghanistan (KC344856), Oman (ABB30032), BA66019 (AF350448), Kosova Hoti (EU037902) and YL04057 (ACM78471). CCHFV, Crimean-Congo hemorrhagic fever virus.
Pairwise Gc sequence alignment of CCHFV and HTNV. The secondary structural elements of CCHFV Gc are shown at the top of sequences corresponding to the ribbon diagrams presented in Fig. 3B. The three fusion loops of CCHFV and HTNV are indicated by orange lines. The disulfide bonds of CCHFV and HTNV are labeled with green letters at the top and bottom of the sequences, respectively. CCHFV, Crimean-Congo hemorrhagic fever virus; HTNV, Hantaan virus.
References
- Abudurexiti A., Adkins S., Alioto D., Alkhovsky S.V., Avsic-Zupanc T., Ballinger M.J., Bente D.A., Beer M., Bergeron E., Blair C.D., Briese T., Buchmeier M.J., Burt F.J., Calisher C.H., Chang C., Charrel R.N., Choi I.R., Clegg J.C.S., de la Torre J.C., de Lamballerie X., Deng F., Di Serio F., Digiaro M., Drebot M.A., Duan X., Ebihara H., Elbeaino T., Ergunay K., Fulhorst C.F., Garrison A.R., Gao G.F., Gonzalez J.J., Groschup M.H., Gunther S., Haenni A.L., Hall R.A., Hepojoki J., Hewson R., Hu Z., Hughes H.R., Jonson M.G., Junglen S., Klempa B., Klingstrom J., Kou C., Laenen L., Lambert A.J., Langevin S.A., Liu D., Lukashevich I.S., Luo T., Lu C., Maes P., de Souza W.M., Marklewitz M., Martelli G.P., Matsuno K., Mielke-Ehret N., Minutolo M., Mirazimi A., Moming A., Muhlbach H.P., Naidu R., Navarro B., Nunes M.R.T., Palacios G., Papa A., Pauvolid-Correa A., Paweska J.T., Qiao J., Radoshitzky S.R., Resende R.O., Romanowski V., Sall A.A., Salvato M.S., Sasaya T., Shen S., Shi X., Shirako Y., Simmonds P., Sironi M., Song J.W., Spengler J.R., Stenglein M.D., Su Z., Sun S., Tang S., Turina M., Wang B., Wang C., Wang H., Wang J., Wei T., Whitfield A.E., Zerbini F.M., Zhang J., Zhang L., Zhang Y., Zhang Y.Z., Zhang Y., Zhou X., Zhu L., Kuhn J.H. Taxonomy of the order Bunyavirales: update 2019. Arch. Virol. 2019;164:1949–1965. doi: 10.1007/s00705-019-04253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Abri S.S., Abaidani I.A., Fazlalipour M., Mostafavi E., Leblebicioglu H., Pshenichnaya N., Memish Z.A., Hewson R., Petersen E., Mala P., Nhu Nguyen T.M., Rahman Malik M., Formenty P., Jeffries R. Current status of crimean-Congo haemorrhagic fever in the World Health organization eastern mediterranean region: issues, challenges, and future directions. Int. J. Infect. Dis. 2017;58:82–89. doi: 10.1016/j.ijid.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga L.M., Bellido J.L.M., Negredo A.I., Criado J.G., Lista M.C.V., Serrano J.A.S., Santiago M.B.V., Bernus A.L., Manchon F.D., Seco M.P.S., Leralta N., Sardon M.A., Muro A., Belhassen-Garcia M. New circulation of genotype V of Crimean-Congo haemorrhagic fever virus in humans from Spain. PLoS Neglected Trop. Dis. 2021;15:e0009197. doi: 10.1371/journal.pntd.0009197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bente D.A., Forrester N.L., Watts D.M., McAuley A.J., Whitehouse C.A., Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antivir. Res. 2013;100:159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Bergeron E., Vincent M.J., Nichol S.T. Crimean-Congo hemorrhagic fever virus glycoprotein processing by the endoprotease SKI-1/S1P is critical for virus infectivity. J. Virol. 2007;81:13271–13276. doi: 10.1128/JVI.01647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A., Smith J., Strecker K., Paragas J., Altamura L.A., McFalls J.M., Frias-Staheli N., Garcia-Sastre A., Schmaljohn C.S., Doms R.W. Cellular localization and antigenic characterization of crimean-Congo hemorrhagic fever virus glycoproteins. J. Virol. 2005;79:6152–6161. doi: 10.1128/JVI.79.10.6152-6161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.P., Lu M., Dutta S., Yan L., Barr J., Flora M., Feng Y.R., Xu K., Nikolov D.B., Wang L.F., Skiniotis G., Broder C.C. Biochemical, conformational, and immunogenic analysis of soluble trimeric forms of henipavirus fusion glycoproteins. J. Virol. 2012;86:11457–11471. doi: 10.1128/JVI.01318-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drulyte I., Johnson R.M., Hesketh E.L., Hurdiss D.L., Scarff C.A., Porav S.A., Ranson N.A., Muench S.P., Thompson R.F. Approaches to altering particle distributions in cryo-electron microscopy sample preparation. Acta Crystallographica Section D-Structural Biology. 2018;74:560–571. doi: 10.1107/S2059798318006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels J.M., Maurer D.P., Herbert A.S., Wirchnianski A.S., Vergnolle O., Cross R.W., Abelson D.M., Moyer C.L., Mishra A.K., Aguilan J.T., Kuehne A.I., Pauli N.T., Bakken R.R., Nyakatura E.K., Hellert J., Quevedo G., Lobel L., Balinandi S., Lutwama J.J., Zeitlin L., Geisbert T.W., Rey F.A., Sidoli S., McLellan J.S., Lai J.R., Bornholdt Z.A., Dye J.M., Walker L.M., Chandran K. Protective neutralizing antibodies from human survivors of Crimean-Congo hemorrhagic fever. Cell. 2021 doi: 10.1016/j.cell.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry C.E., Garry R.F. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of Bunyaviruses are class II viral fusion protein (beta-penetrenes) Theor. Biol. Med. Model. 2004;1:10. doi: 10.1186/1742-4682-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhals D., Bester P.A., Paweska J.T., Swanepoel R., Burt F.J. Next-generation sequencing of southern African Crimean-Congo haemorrhagic fever virus isolates reveals a high frequency of M segment reassortment. Epidemiol. Infect. 2014;142:1952–1962. doi: 10.1017/S0950268814000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardado-Calvo P., Atkovska K., Jeffers S.A., Grau N., Backovic M., Pérez-Vargas J., de Boer S.M., Tortorici M.A., Pehau-Arnaudet G., Lepault J., England P., Rottier P.J., Bosch B.J., Hub J.S., Rey F.A. A glycerophospholipid-specific pocket in the RVFV class II fusion protein drives target membrane insertion. Science. 2017;358:663–667. doi: 10.1126/science.aal2712. [DOI] [PubMed] [Google Scholar]
- Guardado-Calvo P., Bignon E.A., Stettner E., Jeffers S.A., Perez-Vargas J., Pehau-Arnaudet G., Tortorici M.A., Jestin J.L., England P., Tischler N.D., Rey F.A. Mechanistic insight into bunyavirus-induced membrane fusion from structure-function analyses of the hantavirus envelope glycoprotein Gc. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardado-Calvo P., Rey F.A. The envelope proteins of the Bunyavirales. Adv. Virus Res. 2017;98:83–118. doi: 10.1016/bs.aivir.2017.02.002. [DOI] [PubMed] [Google Scholar]
- Halaby D.M., Poupon A., Mornon J. The immunoglobulin fold family: sequence analysis and 3D structure comparisons. Protein Eng. 1999;12:563–571. doi: 10.1093/protein/12.7.563. [DOI] [PubMed] [Google Scholar]
- Halldorsson S., Behrens A.-J., Harlos K., Huiskonen J.T., Elliott R.M., Crispin M., Brennan B., Bowden T.A. Structure of a phleboviral envelope glycoprotein reveals a consolidated model of membrane fusion. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:7154–7159. doi: 10.1073/pnas.1603827113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Alvarez B., Hartmann M.D., Albrecht R., Lupas A.N., Zeth K., Linke D. A new expression system for protein crystallization using trimeric coiled-coil adaptors. Protein Eng. Des. Sel. 2008;21:11–18. doi: 10.1093/protein/gzm071. [DOI] [PubMed] [Google Scholar]
- Hulswit R.J.G., Paesen G.C., Bowden T.A., Shi X. Recent advances in bunyavirus glycoprotein research: precursor processing, receptor binding and structure. Viruses. 2021;13:353. doi: 10.3390/v13020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten R.P., Beek T.A.H.T., Krieger E., Hekkelman M.L., Hooft R.W.W., Schneider R., Sander C., Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011;39:D411–D419. doi: 10.1093/nar/gkq1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielian M., Rey F.A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Adkins S., Alioto D., Alkhovsky S.V., Amarasinghe G.K., Anthony S.J., Avsic-Zupanc T., Ayllon M.A., Bahl J., Balkema-Buschmann A., Ballinger M.J., Bartonicka T., Basler C., Bavari S., Beer M., Bente D.A., Bergeron E., Bird B.H., Blair C., Blasdell K.R., Bradfute S.B., Breyta R., Briese T., Brown P.A., Buchholz U.J., Buchmeier M.J., Bukreyev A., Burt F., Buzkan N., Calisher C.H., Cao M., Casas I., Chamberlain J., Chandran K., Charrel R.N., Chen B., Chiumenti M., Choi I.R., Clegg J.C.S., Crozier I., da Graca J.V., Dal Bo E., Davila A.M.R., de la Torre J.C., de Lamballerie X., de Swart R.L., Di Bello P.L., Di Paola N., Di Serio F., Dietzgen R.G., Digiaro M., Dolja V.V., Dolnik O., Drebot M.A., Drexler J.F., Durrwald R., Dufkova L., Dundon W.G., Duprex W.P., Dye J.M., Easton A.J., Ebihara H., Elbeaino T., Ergunay K., Fernandes J., Fooks A.R., Formenty P.B.H., Forth L.F., Fouchier R.A.M., Freitas-Astua J., Gago-Zachert S., Gao G.F., Garcia M.L., Garcia-Sastre A., Garrison A.R., Gbakima A., Goldstein T., Gonzalez J.J., Griffiths A., Groschup M.H., Gunther S., Guterres A., Hall R.A., Hammond J., Hassan M., Hepojoki J., Hepojoki S., Hetzel U., Hewson R., Hoffmann B., Hongo S., Hoper D., Horie M., Hughes H.R., Hyndman T.H., Jambai A., Jardim R., Jiang D., Jin Q., Jonson G.B., Junglen S., Karadag S., Keller K.E., Klempa B., Klingstrom J., Kobinger G., Kondo H., Koonin E.V., Krupovic M., Kurath G., Kuzmin I.V., Laenen L., Lamb R.A., Lambert A.J., Langevin S.L., Lee B., Lemos E.R.S., Leroy E.M., Li D., Li J., Liang M., Liu W., Liu Y., Lukashevich I.S., Maes P., Marciel de Souza W., Marklewitz M., Marshall S.H., Martelli G.P., Martin R.R., Marzano S.L., Massart S., McCauley J.W., Mielke-Ehret N., Minafra A., Minutolo M., Mirazimi A., Muhlbach H.P., Muhlberger E., Naidu R., Natsuaki T., Navarro B., Navarro J.A., Netesov S.V., Neumann G., Nowotny N., Nunes M.R.T., Nylund A., Okland A.L., Oliveira R.C., Palacios G., Pallas V., Palyi B., Papa A., Parrish C.R., Pauvolid-Correa A., Paweska J.T., Payne S., Perez D.R., Pfaff F., Radoshitzky S.R., Rahman A.U., Ramos-Gonzalez P.L., Resende R.O., Reyes C.A., Rima B.K., Romanowski V., Robles Luna G., Rota P., Rubbenstroth D., Runstadler J.A., Ruzek D., Sabanadzovic S., Salat J., Sall A.A., Salvato M.S., Sarpkaya K., Sasaya T., Schwemmle M., Shabbir M.Z., Shi X., Shi Z., Shirako Y., Simmonds P., Sirmarova J., Sironi M., Smither S., Smura T., Song J.W., Spann K.M., Spengler J.R., Stenglein M.D., Stone D.M., Strakova P., Takada A., Tesh R.B., Thornburg N.J., Tomonaga K., Tordo N., Towner J.S., Turina M., Tzanetakis I., Ulrich R.G., Vaira A.M., van den Hoogen B., Varsani A., Vasilakis N., Verbeek M., Wahl V., Walker P.J., Wang H., Wang J., Wang X., Wang L.F., Wei T., Wells H., Whitfield A.E., Williams J.V., Wolf Y.I., Wu Z., Yang X., Yang X., Yu X., Yutin N., Zerbini F.M., Zhang T., Zhang Y.Z., Zhou G., Zhou X. Taxonomic update for phylum Negarnaviricota (Riboviria: orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020;165:3023–3072. doi: 10.1007/s00705-020-04731-2. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Liu J., Xiao C., Deng H., Xie Q., Han H. A fast forward 3D connection algorithm for mitochondria and synapse segmentations from serial EM images. BioData Min. 2018;11:24. doi: 10.1186/s13040-018-0183-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehand M.S., Al-Shorbaji F., Millett P., Murgue B. The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antivir. Res. 2018;159:63–67. doi: 10.1016/j.antiviral.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavel S.T.I., Yetiskin H., Kalkan A., Ozdarendeli A. Evaluation of the cell culture based and the mouse brain derived inactivated vaccines against Crimean-Congo hemorrhagic fever virus in transiently immune-suppressed (IS) mouse model. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punch E.K., Hover S., Blest H.T.W., Fuller J., Hewson R., Fontana J., Mankouri J., Barr J.N. Potassium is a trigger for conformational change in the fusion spike of an enveloped RNA virus. J. Biol. Chem. 2018;293:9937–9944. doi: 10.1074/jbc.RA118.002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serris A., Stass R., Bignon E.A., Muena N.A., Manuguerra J.C., Jangra R.K., Li S., Chandran K., Tischler N.D., Huiskonen J.T., Rey F.A., Guardado-Calvo P. The hantavirus surface glycoprotein lattice and its fusion control mechanism. Cell. 2020;183:442–456. doi: 10.1016/j.cell.2020.08.023. e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtanko O., Nikitina R.A., Altuntas C.Z., Chepurnov A.A., Davey R.A. Crimean-Congo hemorrhagic fever virus entry into host cells occurs through the multivesicular body and requires ESCRT regulators. PLoS Pathog. 2014;10:e1004390. doi: 10.1371/journal.ppat.1004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Liu Y., Lu W., Cheng G., Zhou H., Zhou X., Wei L., Dai Z., Guo K., Lu H. Con A affinity glycoproteomics of normal human liver tissue. Sci. China C Life Sci. 2007;50:403–411. doi: 10.1007/s11427-007-0027-z. [DOI] [PubMed] [Google Scholar]
- Tahir Ul Qamar M., Ismail S., Ahmad S., Mirza M.U., Abbasi S.W., Ashfaq U.A., Chen L.L. Development of a novel multi-epitope vaccine against crimean-Congo hemorrhagic fever virus: an integrated reverse vaccinology, vaccine informatics and biophysics approach. Front. Immunol. 2021;12:669812. doi: 10.3389/fimmu.2021.669812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipih T., Burt F.J. Crimean-Congo hemorrhagic fever virus: advances in vaccine development. Biores Open Access. 2020;9:137–150. doi: 10.1089/biores.2019.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann M., Avsic-Zupanc T., Bino S., Bouloy M., Burt F., Chinikar S., Christova I., Dedushaj I., El-Sanousi A., Elaldi N., Hewson R., Hufert F.T., Humolli I., Jansen van Vuren P., Kocak Tufan Z., Korukluoglu G., Lyssen P., Mirazimi A., Neyts J., Niedrig M., Ozkul A., Papa A., Paweska J., Sall A.A., Schmaljohn C.S., Swanepoel R., Uyar Y., Weber F., Zeller H. Biosafety standards for working with Crimean-Congo hemorrhagic fever virus. J. Gen. Virol. 2016;97:2799–2808. doi: 10.1099/jgv.0.000610. [DOI] [PubMed] [Google Scholar]
- Willensky S., Bar-Rogovsky H., Bignon E.A., Tischler N.D., Modis Y., Dessau M. Crystal structure of glycoprotein C from a hantavirus in the post-fusion conformation. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Scheres S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M., Scholte F.E.M., Spiropoulou C.F., Spengler J.R., Bergeron E. Molecular insights into crimean-Congo hemorrhagic fever virus. Viruses-Basel. 2016;8:106. doi: 10.3390/v8040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of CCHFV Gc from representative strains. The disulfide bonds of CCHFV are labeled with green letters at the bottom of sequences. Viral strains used for the alignment are IbAr10200 (GenBank ID: AAM48106), SPU 115/87 (ARB51462), Caceres 2014 (ASV45881), Sudan AB1-2009 (AEI70589), Badajoz 2018 (QQL93572), SK/2015/human (AOF43039), Baghdad 12 (CAD61343), Afg09-2990 (ADQ57289), Iran-53 (ABD98125), 813040 UAE (ASW20657), 61T Pakistan (QIQ51272), MCL-18-H-1780 (QCY54392), NIV1311122 (QBP34417), U2/2-002/U 6415 (AAW84283), FK16116 (QEQ12756), Hodzha (AAP29978), SCT ex Afghanistan (KC344856), Oman (ABB30032), BA66019 (AF350448), Kosova Hoti (EU037902) and YL04057 (ACM78471). CCHFV, Crimean-Congo hemorrhagic fever virus.
Pairwise Gc sequence alignment of CCHFV and HTNV. The secondary structural elements of CCHFV Gc are shown at the top of sequences corresponding to the ribbon diagrams presented in Fig. 3B. The three fusion loops of CCHFV and HTNV are indicated by orange lines. The disulfide bonds of CCHFV and HTNV are labeled with green letters at the top and bottom of the sequences, respectively. CCHFV, Crimean-Congo hemorrhagic fever virus; HTNV, Hantaan virus.
Data Availability Statement
The Cryo-EM map has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-31579, and the corresponding atomic model has been deposited in the Protein Data Bank (PDB) under accession code 7FGF.