ABSTRACT.
Highly sensitive molecular techniques for the detection of low-level Plasmodium falciparum parasitemia are highly useful for various clinical and epidemiological studies. However, differences in how blood samples are preserved, the quantity of blood stored, as well as genomic DNA extraction methods used may compromise the potential usefulness of these methodologies. This study compared diagnostic sensitivity based on microscopy and malaria rapid diagnostic tests (mRDTs), with quantitative polymerase chain reaction (qPCR) P. falciparum positivity of dried blood spots (DBS) or whole blood pellets (WBP) from pregnant women using different DNA extraction protocols (Chelex-saponin or a commercial kit). Samples from 129 pregnant women were analyzed, of which 13 were P. falciparum positive by mRDT and 5 by microscopy. By using extraction kit on WBP and on DBS, qPCR positivity was 27 (20.9%) and 16 (12.4%), respectively, whereas Chelex extraction on DBS only resulted in 4 (3.1%) P. falciparum positive samples. Thus, extraction using commercial kits greatly improve the likelihood of detecting P. falciparum infections.
INTRODUCTION
Malaria during pregnancy can have profound consequences to the mother and her fetus as the disease may result in complications such as severe anemia, hypertension, and low/reduced birth weight in newborns.1 In areas of moderate to high malaria transmission, infected pregnant women may have acquired partial immunity and may remain asymptomatic with low/undetectable parasitemia in the peripheral blood, but with higher parasitemia in the placenta as a result of placental sequestration that may render them being missed when peripheral blood is used for detection.2,3 During first visits to the antenatal clinic (ANC), pregnant women are usually screened for malaria by use of either microscopy or malaria rapid diagnostic tests (mRDTs). With these tools, however, malaria cases with low parasitemia may be missed and this may have a profound impact on pregnancy outcome.3,4
New molecular techniques of relevance for mainly epidemiological studies have been developed that are significantly more sensitive and reliable in detecting low-level parasitemia as compared with microscopy.5 However, they generally require sufficient and high-quality DNA, which is dependent on the blood sample volume, preservation method for sampled blood, and DNA extraction procedure.6 The blood for downstream molecular analysis is either stored as whole blood in tubes (frozen) or as dried blood spots (DBS) on various types of filter paper.7 DBS generally requires a small volume of blood, minimum storage space, and simple transport allowing it to be applied in remote areas with limited laboratory resources.8,9 On the other hand, blood samples preserved in tubes as pellets usually require cumbersome sampling strategies and a cold chain maintenance, limiting their use in remote settings.10
DNA extraction protocols aim to reduce potential inhibitors found in whole blood that may reduce the efficiency of the polymerase chain reaction (PCR) amplification and thus, compromise sensitivity such as hemoglobin and heme-containing components.6,11 Apart from commercial DNA extraction kits, noncommercial protocols have been developed as a cheap alternative including the Chelex100-saponin-based extraction methodology.12 Numerous molecular detection methods available for DNA-based malaria diagnosis include conventional (nested) PCR, quantitative PCR (qPCR), and loop-mediated isothermal amplification (LAMP) have been developed with varying sensitivity but generally superior to microscopy.13
In this study, we were using an ultrasensitive qPCR to compare Plasmodium falciparum detection in pregnant women when using DNA extracted by different extraction methods from either DBS by Chelex-Saponin (DBS-Chelex), DBS by kit (DBS-kit), or whole blood pellet (WBP) by kit (WBP-kit). The method with the highest number of positive samples (WBP-kit) was set as reference versus the same samples as identified by microscopy and mRDT. The study was part of a larger clinical trial entitled Improving Pregnancy Outcome by Intermittent Preventive Treatment in Africa (IMPROVE) conducted on pregnant women from Malawi, Kenya, and Tanzania. IMPROVE TRIAL-2015-1076-IMPROVE (https://www.improve-consortium.org/projects/improve-trial) with ethical approval NIMR\HQ\R.8a\vol.IX\2533. For this particular sub-study, samples obtained from Korogwe, Tanga region, Tanzania, were used.
Women attending ANC with gestational ages between 16 and 20 weeks after consent were enrolled in the IMPROVE study. Five milliliters of venous blood were collected from each woman in ethylenediaminetetraacetic acid (EDTA) collection Vacutainers. Three spots of 50 µL blood were spotted onto 903 Whatman filter paper (GE Healthcare, Whatman Grade CF 12, Sigma-Aldrich, Denmark), air-dried, and stored at −20°C in separate zip lock bags containing silica gel. The remaining blood was centrifuged, and the pellet was divided into two aliquots and cryopreserved at −80°C until use.
mRDT was done using CareStart™ (Access Bio Inc, Somerset, NJ) that detects either histidine-rich protein 2 (HRP-2) antigens from P. falciparum or parasite lactate dehydrogenase (pLDH) enzymes from the Plasmodium genus according to the manufacturer’s instructions. Giemsa-stained thick and thin smears were prepared for the detection and quantification of malaria parasites by microscopy. Parasite density was determined as the number of asexual stage parasites/200 leukocytes (500 leukocytes if < 10 parasites) and converted to number per microliter using the actual leukocytes count as estimated by using Sysmex KX-21N hematological analyzer (Sysmex Corporation, Kobe, Japan). A minimum of 100 fields was examined before a blood smear was considered negative.
We compared P. falciparum qPCR positivity using DNA obtained from extraction of blood collected on filter paper as DBS and as WBP (250 µL) in cryo vials (stored at −80°C) using commercial DNA extraction kits (E Z 96 Tissue DNA Kit®, Omega, VWR, Denmark) and the DNA obtained from the widely used Chelex-saponin protocol14 on DBS. The sensitivity was assessed by comparing results obtained by performing P. falciparum ultrasensitive q-PCR targeting the species-specific TARE-2 gene15 on extracted DNA.
One whole DBS (50 µL) was cut and placed in a deep 96 well plate followed by the addition of 0.8 mL of 0.5% saponin in phosphate-buffered saline (PBS) to each well and the DNA was extracted by Chelex-saponin protocol as described elsewhere.16 Furthermore, a 10-fold diluted 250 µL WBP and a whole DBS were extracted with the Omega BIO-TEK E-Z-96 Tissue kit, Denmark (vacuum protocol, according to manufacturer’s protocol. (https://www.omegabiotek.com/product/e-z-96-tissue-dna-kit/)
The resulting DNA extracts were screened for the presence of P. falciparum DNA on a VIIA 7 Applied Biosystems Real-Time PCR System (Applied Biosystems, CA) using the qPCR method and conditions as described by Hofmann et al.15 targeting the high-copy telomere-associated repetitive element 2 (TARE-2,*250 copies/genome) of P. falciparum. DNA of 3 µL was added into 15 µL of PowerUp™ SYBR™ Green master mix (Applied Biosystems, CA).
Samples from 129 pregnant women residing in Korogwe, Tanga region, Tanzania, were analyzed, out of which, 5/129 (3.9%) and 13/129 (10.1%) were P. falciparum positive by microscopy and mRDT, respectively. Comparing the methodologies, 115/129 (89.1%) of the samples were negative and 4 (3.1%) were positive by both methods, respectively, whereas one sample (0.8%) was microscopy positive/mRDT negative and 9 samples (7.0%) were mRDT positive/microscopy negative. Using microscopy as the gold standard, the sensitivity and specificity of mRDT were 80.0% and 92.7%, respectively.
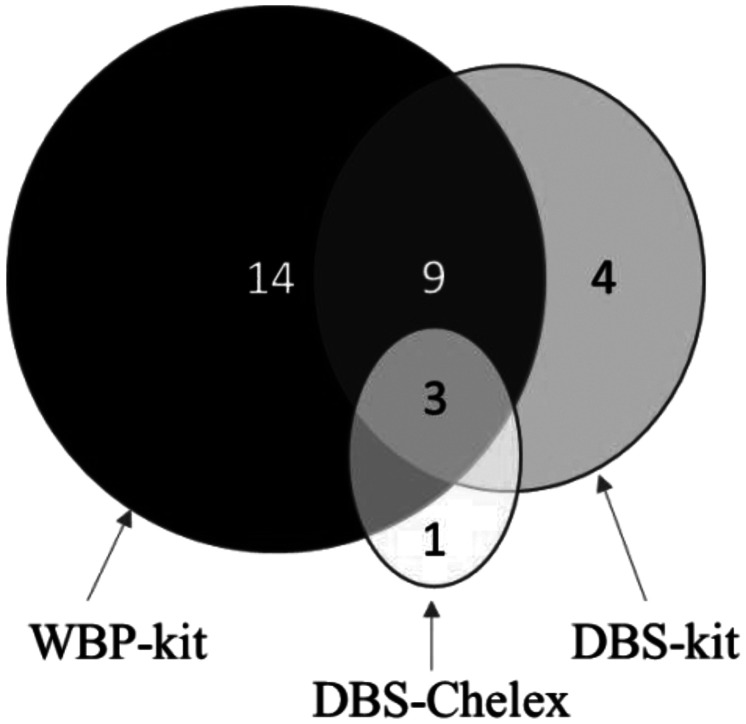
The qPCR positivity was explored using DNA obtained from the different extractions; Chelex extraction on DBS (DBS-Chelex), a commercial DNA extraction kit for DBS (DBS-kit) and WBP (WBP-kit). A total of 31 (24.0%) P. falciparum positive samples were detected by the three procedures (Figure 1): 4 (3.1%) by DBS-Chelex, 16 (12.4%) by DBS-kit, and 27 (20.9%) WBP-kit. Only 3 (9.7%) of the 31 P. falciparum positive samples were identified by all methods. Extraction from WBP showed the highest sensitivity identifying 27/31 (87.1%) of the infections and when used as reference, the sensitivity was 44.4% and 14.8% and the specificity was 96,1% and 81.6% for DBS-kit and DBS-Chelex, respectively (Supplemental Table 1).
Figure 1.
Number of samples that are Plasmodium falciparum positive as identified by use of quantitative polymerase chain reaction (qPCR) targeting the high-copy telomere-associated repetitive element 2 (TARE-2) of P. falciparum in 129 blood samples from Tanzanian women. 27, 16, and 4 samples were positive extracted from whole blood pellets (WBP) kit, dried blood spots (DBS) kit, and DBS-Chelex, respectively. Three samples were identified positive by all methods.
Figure 2 shows the distribution of P. falciparum positive samples as identified by microscopy, mRDT, and using qPCR on the WBP-kit as a reference. Four samples were positive by all methods and 99 samples were negative by all methods. Using qPCR, 16 additional P. falciparum samples that were negative by microscopy and mRDT were identified as positive whereas all samples identified as P. falciparum positive by microscopy were all qPCR positive. On the other hand, three samples that were mRDT positive were negative by qPCR.
Figure 2.
Number of Plasmodium falciparum positive identified by use of microscopy, malaria rapid diagnostic tests (mRDT), and quantitative polymerase chain reaction (qPCR) in blood samples from Tanzanian women. 27, 13, and 5 were positive by whole blood pellets (WBP) kit, mRDT, and Microscopy, respectively. Four samples were identified positive by all methods, six were both WBP-kit and mRDT positive. A total of 22 (17.1%) women were positive for P. falciparum by PCR, but negative by microscopy, 17 (13.2%) were positive for P. falciparum by PCR and negative by mRDT. However, 9 (7%) women were positive for malaria by RDT but negative by microscopy.
Infections by P. falciparum during pregnancy have a significant impact on pregnancy outcomes if not correctly detected and treated. This study explored P. falciparum positivity by qPCR of DNA extracted by different extraction procedures from blood preserved on DBS or WBP. Studies from elsewhere have shown the importance of optimal quality and quantity of genomic DNA for the subsequent molecular diagnostic analyses.11,17 Other studies have reported that the use of different volumes of blood material from participants (either 50 µL of DBS or 250 µL of WBP) and DNA extraction protocols could have a significant effect on the P. falciparum positivity when applying an ultrasensitive qPCR. We hereby show that the qPCR positivity was 20.9%, 12.4%, and 3.1% for WBP-kit, DBS-kit, and DBS-Chelex, respectively, indicating choice of extraction method and DNA preservation could have a direct effect on parasite DNA yield and hence detection efficiency. Similar observations to ours was reported from another study that found Chelex extraction was less effective as compared with commercial DNA extraction kits,12 but contrary to a study by Strøm et al. that showed that extracting DNA from DBS using either Chelex or various DNA extraction kits performed equally well.16 The conflicting results between these studies possibly arise as a consequence of how the DBS are stored, for how long the DBS are stored and subsequently handled. Moreover, a low resulting DNA concentration in a solution that contains suspended impurities can inhibit the qPCR, which may be a particular challenge when applying Chelex extraction.18 The average cycle threshold (Ct) was 30,7, 34,8, and 39,2 for WBP-kit, DBS-kit, and DBS-Chelex, respectively (Supplemental Table 2) indicating that higher concentration of DNA is achieved by use of WBP kit as compared with DBS-kit and DBS-Chelex and as well when comparing DBS-kit with DBS-Chelex.
When comparing the gold standard microscopy and mRDT, out of 129 samples 5 samples were positive by microscopy and 13 were positive by mRDT. Microscopy is not efficient for diagnosing subpatent parasites in asymptomatic Plasmodium infections even though it is the gold standard.10 However, despite the inability in identifying several submicroscopic infections, the use of microscopy and mRDT is applied for routine and early case detection of the majority of P. falciparum infections and the resulting management of the disease.19 Generally using molecular technique like PCR is found to be more sensitive to identify a significant proportion of submicroscopic infections irrespective of the preservation method for blood samples and extraction methods used.
Well aware that this is a limited study, it nevertheless clearly shows when identifying all malaria-infected women is crucial, like for instance in clinical trials evaluating intermittent preventative treatment in pregnancy or the like, it is essential to explore the efficiency of protocols for extracting malaria parasite DNA to increase the likelihood of identifying P. falciparum positive cases. No one method will have all the attributes of being cost-effective, require little time, and avoid the use of hazardous chemicals. Using a higher volume of blood increases the chance of detecting very low parasite density. Thus, when financial resources are available the use of commercial kits and WBP should be applied. Chelex should not be used for submicroscopic detection in pregnant women or other low parasitemia samples. DNA extracted by kit on DBS is recommended for molecular diagnostic method by PCR in malaria research in endemic countries.
Supplemental Material
ACKNOWLEDGMENTS
We thank the National Institute of Medical Research (NIMR) for providing a permit for the study and NIMR-Tanga Centre for providing the samples. The Kilimanjaro Clinical Research Institute (KCRI) for providing the facility for the analysis of the sample.
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1.Fried M Muehlenbachs A Duffy PE , 2012. Diagnosing malaria in pregnancy: an update. Expert Rev Anti Infect Ther 10: 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azizi SC Chongwe G Chipukuma H Jacobs C Zgambo J Michelo C , 2018. Uptake of intermittent preventive treatment for malaria during pregnancy with sulphadoxine-pyrimethamine (IPTp-SP) among postpartum women in Zomba District, Malawi: a cross-sectional study. BMC Pregnancy Childbirth 18: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy SC Shott JP Parikh S Etter P Prescott WR Stewart VA , 2013. Review article: malaria diagnostics in clinical trials. Am J Trop Med Hyg 89: 824–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tedla M , 2019. A focus on improving molecular diagnostic approaches to malaria control and elimination in low transmission settings: review. Parasite Epidemiol Control 6: e00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zainabadi K Nyunt MM Plowe CV , 2019. An improved nucleic acid extraction method from dried blood spots for amplification of Plasmodium falciparum kelch13 for detection of artemisinin resistance. Malar J 18: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seesui K Imtawil K Chanetmahun P Laummaunwai P Boonmars T , 2018. An alternative method for extracting Plasmodium DNA from EDTA whole blood for malaria diagnosis. Korean J Parasitol 56: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi T Gamboa D Ndiaye D Cui L Sutton PL Vinetz JM , 2015. Malaria diagnosis across the International Centers of Excellence for Malaria Research: platforms, performance, and standardization. Am J Trop Med Hyg 93(Suppl): 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth JM Korevaar DA Leeflang MMG Mens PF , 2016. Molecular malaria diagnostics: a systematic review and meta-analysis. Crit Rev Clin Lab Sci 53: 87–105. [DOI] [PubMed] [Google Scholar]
- 9.Choi E-H Lee SK Ihm C Sohn Y-H , 2014. Rapid DNA extraction from dried blood spots on filter paper: potential applications in biobanking. Osong Public Health Res Perspect 5: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miguel RB Coura JR Samudio F Suarez-Mutis MC , 2013. Evaluation of three different DNA extraction methods from blood samples collected in dried filter paper in Plasmodium subpatent infections from the Amazon region in Brazil. Rev Inst Med Trop São Paulo 55: 205–208. [DOI] [PubMed] [Google Scholar]
- 11. Taylor BJ, Martin KA, Arango E, Agudelo OM, Maestre A, Yanow SK, 2011. Real-time PCR detection of Plasmodium directly from whole blood and filter paper samples. Malar J 1: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyan Hera Wiluyaningtias Y , 2015. Evaluation of five methods used to extract deoxyribonucleic acid (DNA) from human malaria parasitized blood spotted on the filter paper. Asian J Appl Sci 3: 429–436. [Google Scholar]
- 13.Moyeh MN et al. 2019. Comparison of the accuracy of four malaria diagnostic methods in a high transmission setting in coastal Cameroon. J Parasitol Res 2019. Available at: 10.1155/2019/1417967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon N Shallat J Williams-Weitzikoski C Harrington WE , 2020. Optimization of Chelex 100 resin-based extraction of genomic DNA from dried blood spots. Biol Methods Protoc 5: bpaa009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann N Mwingira F Shekalaghe S Robinson LJ Mueller I Felger I , 2015. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med 12: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strøm GEA Tellevik MG Hanevik K Langeland N Blomberg B , 2014. Comparison of four methods for extracting DNA from dried blood on filter paper for PCR targeting the mitochondrial Plasmodium genome. Trans R Soc Trop Med Hyg 108: 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canier L et al. 2015. Malaria PCR detection in Cambodian low-transmission settings: dried blood spots versus venous blood samples. Am J Trop Med Hyg 92: 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh UA Kumari M Iyengar S , 2018. Method for improving the quality of genomic DNA obtained from minute quantities of tissue and blood samples using Chelex 100 resin. Biol Proced Online 20. Available at: 10.1186/S12575-018-0077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minja DTR et al. 2013. Plasmodium falciparum mutant haplotype infection during pregnancy associated with reduced birthweight, Tanzania. Emerg Infect Dis 19: 1446–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.