Abstract
Resistance to lincomycin and clindamycin in the clinical isolate Enterococcus faecium HM1025 is due to a ribosomal methylase encoded by an ermAM-like gene and the plasmid-mediated inactivation of these antibiotics. We have cloned and determined the nucleotide sequence of the gene responsible for the inactivation of lincosamides, linB. This gene encodes a 267-amino-acid lincosamide nucleotidyltransferase. The enzyme catalyzes 3-(5′-adenylation) (the adenylation of the hydroxyl group in position 3 of the molecules) of lincomycin and clindamycin. Expression of linB was observed in both Escherichia coli and Staphylococcus aureus. The deduced amino acid sequence of the enzyme did not display any significant homology with staphylococcal nucleotidyltransferases encoded by linA and linA′ genes. Sequences homologous to linB were found in 14 other clinical isolates of E. faecium, indicating the spread of the resistance trait in this species.
Lincosamide antibiotics include lincomycin, naturally produced by several actinomycetes, and clindamycin, a semisynthetic derivative obtained by the chlorination of lincomycin. These antibiotics are active against many gram-positive cocci and anaerobes; they inhibit protein synthesis by blocking the peptidyltransferase activity of the 50S subunit of the bacterial ribosome (11). Resistance to lincosamides is usually due to alteration of the ribosome following the N6 dimethylation of a specific adenine in the 23S rRNA, which confers cross-resistance to macrolide, lincosamide, and streptogramin B type antibiotics, i.e., the MLSB phenotype (22, 32). In contrast to this broad-spectrum resistance, resistance specific to lincosamides, gained by bacterial modification of those antibiotics, has been reported. Phosphorylation (1) and nucleotidylation (2, 26) of the hydroxyl group in position 3 of lincosamide molecules (24, 26) have been detected in several species of Streptomyces. Inactivation of lincosamides was also observed in strains of staphylococci, streptococci, enterococci, and lactobacilli of animal origin (10, 12, 13) and in staphylococci isolated from humans (5, 20, 21). Clinical isolates of Staphylococcus haemolyticus BM4610 and Staphylococcus aureus BM4611 are highly resistant to lincomycin (MIC = 64 μg/ml) and are apparently susceptible to clindamycin (MIC = 0.12 μg/ml). In these strains, lincosamide O-nucleotidyltransferases encoded by two closely related genes named linA (lincosamide inactivation nucleotidylation) and linA′, respectively, were characterized (4, 5). These genes encode two 161-amino-acid isoenzymes that differ by 14 amino acids. These enzymes inactivate lincomycin and clindamycin by converting them to lincomycin 3-(5′-adenylate) and clindamycin 4-(5′-adenylate) by using ATP, GTP, CTP, or UTP as a nucleotidyl donor and MgCl2 as a cofactor (5). The distribution of linA and linA′ genes was studied by using DNA-DNA hybridization, and related sequences were found in strains belonging to various species of staphylococci (20).
In this paper, we report the nucleotide sequence of a new linB gene that confers resistance to lincosamides on a clinical strain of Enterococcus faecium, HM1025, by inactivating the compounds, and we further report on our study of the biochemical mechanism of the resistance.
MATERIALS AND METHODS
Bacterial strains.
Five hundred eight enterococci of various species isolated from patients at the Henri Mondor Hospital were screened for inactivation of clindamycin with Gots’ test, as previously described (17, 21). Fourteen of the 110 strains of E. faecium tested were found to inactivate clindamycin. One of these strains, E. faecium HM1025, was studied further. Species identification was based on the biochemical scheme of Facklam and Collins (15). S. haemolyticus BM4610 harboring linA, S. aureus BM4611 harboring linA′, and the recombinant plasmids pAT221 (linA) and pAT24 (linA′) were used as positive controls in hybridization experiments (5, 6). Strains were grown in brain heart infusion broth and agar (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France). All strains were incubated at 37°C.
Antibiotic susceptibility testing.
Susceptibility to antibiotics was determined by the disk diffusion technique. MICs of antibiotics were determined by the agar dilution method using Mueller-Hinton medium (Sanofi Diagnostics Pasteur) (8).
Inactivation of lincosamides.
The kinetics of the inactivation of clindamycin by resting cells were determined in liquid medium as previously described (21). E. faecium cells suspended in 0.01 M phosphate buffer (pH 7) containing 20 μg of clindamycin or lincomycin per ml were incubated at 37°C for various periods of time. The pH of this suspension remained constant. Inactivation of lincosamides was followed by a bioassay with Micrococcus luteus ATCC 9341 as the indicator organism (21).
For preparation of modified clindamycin and lincomycin, cells of E. faecium HM1025 were treated with 500 μg of lysozyme per ml and lysed by sonication. Cell debris was removed by centrifugation at 40,000 × g for 45 min. Clindamycin and lincomycin (500 μg/ml) were then incubated in the supernatants at 37°C for 18 h in the presence of ATP (2.5 mM) and MgCl2 (50 mM). Inactivation of antibiotics was monitored as indicated above. Aliquots of inactivated clindamycin and lincomycin were freeze-dried.
High-pressure liquid chromatography–UV-MS experiments.
Samples of freeze-dried inactivated clindamycin and lincomycin, and the control samples, were each dissolved in 1 ml of 10% methanol in 2 mM ammonium acetate. A 5-μl aliquot was injected onto a 2.1- by 150-mm Waters Symmetry C18 column at 35°C in a 0.4-ml/min flow of 5% methanol in 2 mM ammonium acetate. After 1 min, the concentration of methanol was gradually increased to reach 95% at 11 min and was then held at this level for 4 min. Column effluent was monitored by mass spectrometry (MS) (unit resolution, 150 to 800 atomic mass units [amu]) with a Sciex API 300 instrument equipped with a heated nebulizer source. Under these conditions, clindamycin (424/426 amu) was eluted at 13.8 min, clindamycin B (410/412 amu) was eluted at 13.3 min, clindamycin adenylate (753/755 amu) was eluted at 11.7 min, and clindamycin B adenylate (739/741 amu) was eluted at 11.3 min. Lincomycin (406 amu) was eluted at 12.1 min, and lincomycin adenylate (735 amu) was eluted at 9.9 min. For sample purification, the inactivated solutions were concentrated to dryness, redissolved in methanol-water (1:9), evenly divided into 10 batches, and loaded into 10 SepPak (C8) cartridges prewashed with methanol and reequilibrated with water. The resins were washed with a stepwise gradient (25% increment/step) from water to methanol. It was found that the 50% methanol–water fraction contains the majority of mass for both inactivated lincomycin and inactivated clindamycin solutions. Analytical high-pressure liquid chromatography–UV-MS confirmed that the material which was eluted at 50% methanol–water was pure.
NMR experiments.
Samples for nuclear magnetic resonance (NMR) testing were prepared with approximately 40 mg of either inactivated clindamycin or inactivated lincomycin isolate dissolved in 600 μl of dimethyl sulfoxide-d6 (Isotec; 99.96% D) and placed in a standard 5-mm NMR tube (Wilmad). The proton, correlated spectroscopy, g-HMQC (gradient heteronuclear multiple quantum correlation), g-HMBC (gradient heteronuclear multiple bond correlation), and carbon spectra for each sample were recorded with a Bruker DRX 500 spectrometer operating at 499.90 MHz for 1H and 125.70 MHz for 13C. The spectrometer was equipped with a 5-mm Bruker broad-band probe with a z-axis gradient equilibrated at 27°C. Pulses for 1H and 13C were calibrated at 9.3 and 8.5 μs, respectively. The proton spectra were recorded by using 32,000 real data points, a spectral width of 7,507 Hz, and a recycle time of 5 s. The 1H-decoupled carbon spectra were recorded by using a 33° 13C pulse, WALTZ-16 decoupling, 32,000 real data points, a spectrum width of 31,446 Hz, and a recycle delay of 1 s. Two-dimensional data were typically acquired as 2,048- by 128-point matrices which were zero filled to 2,048 by 1,024 points during data processing. The chemical shifts were referenced relative to dimethyl sulfoxide-d5: δ = 2.49 for 1H and 39.5 for 13C.
Genetic techniques.
Enterococcus faecalis JH2-2 was used in mating experiments as a recipient strain. Matings were performed on filters as previously described (19). The antibiotics used for selection of transconjugants were rifampin (100 μg/ml), fusidic acid (50 μg/ml), and either erythromycin, tetracycline, chloramphenicol (each at 10 μg/ml), gentamicin (100 μg/ml), or lincomycin (50 μg/ml). In curing experiments, novobiocin or ciprofloxacin was used as described previously (3).
Total cellular DNA and plasmids were isolated from enterococcal and staphylococcal strains, as described previously (14, 23). In hybridization experiments, total-DNA extracts from E. faecium and staphylococci were immobilized on nylon membranes (Hybond-N; Amersham France, Les Ullis, France) in 50% formamide at 42°C under stringent conditions (23). The linA, linA′, and ermAM probes, previously described (20, 24), were labeled with digoxigenin (Boehringer Mannheim France, Meylan, France), and hybrids were detected by using an antidigoxigenin-alkaline phosphatase conjugate with a chromogenic enzyme substrate.
DNA fragments were cloned by using plasmids pUC18, pCR 2.1 (Invitrogen, Carlsbad, Calif.), and the shuttle vector pJIM2246 (30). S. aureus RN4220 and Escherichia coli DB10 were transformed by electroporation. DNA was sequenced in an automated ABI PRISM 310 system (Perkin-Elmer Corp., Norwalk, Conn.).
The proteins specified by the linear templates amplified by PCR were synthesized with an E. coli in vitro transcription-translation system (Promega, Madison, Wis.). Electrophoresis of proteins labeled with l-[α-35S]methionine was performed in a sodium dodecyl sulfate–15% polyacrylamide gel. For study of the distribution of the linB gene in E. faecium, DNA sequences specific for the gene were amplified by PCR by using the primers LINB1 ([nucleotides 3 to 22] 5′ CCTACCTATTGTTTGTGGAA 3′) and LINB2 ([nucleotides 947 to 928] 5′ ATAACGTTACTCTCCTATTC 3′) through a precycle of 5 min at 94°C followed by 35 cycles of 45 s at 94°C, 45 s at 54°C, and 1 min at 72°C and a final cycle of 5 min at 72°C.
Pulsed-field gel electrophoresis of the enterococcal total-DNA extract digested with SmaI or SfiI was performed as previously described (29). Restricted DNA was transferred to nylon membranes under a vacuum and hybridized to digoxigenin-labeled DNA fragments (Boehringer Mannheim SA, Meylan, France) obtained after amplification of the linB gene with the specific primers LINB1 and LINB2.
Nucleotide sequence accession number.
The nucleotide sequence of the linB gene from E. faecium HM1025 has been deposited in the GenBank data library under accession no. AF110130.
RESULTS AND DISCUSSION
Properties of E. faecium strains.
Resistance to lincosamides gained by the inactivation of the antibiotics was detected by Gots’ test in 14 E. faecium strains and was associated with cross-resistance to macrolide, lincosamide, and streptogramin B type antibiotics (MLSB phenotype). Hybridization experiments failed to detect the genes related to linA or linA′ which are responsible for the nucleotidylation of lincosamides in staphylococci. E. faecium HM1025 was resistant to chloramphenicol, penicillin G (MIC = 32 μg/ml), tetracycline, and high levels of aminoglycosides (streptomycin, kanamycin, and gentamicin). In this strain, resistance to macrolide, lincosamide, and streptogramin B type antibiotics was due to ribosomal methylation mediated by an ermAM-like gene, as revealed by hybridization experiments (data not shown).
Localization of the determinant responsible for lincosamide inactivation.
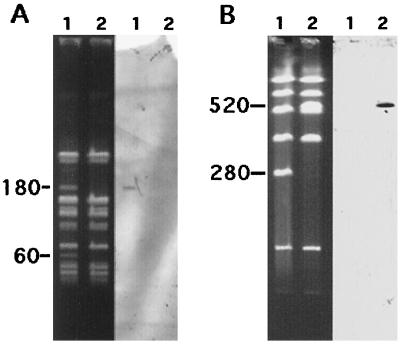
Resistance to high levels of streptomycin and gentamicin, tetracycline, macrolide, lincosamide, and streptogramin B type antibiotics, and lincosamides by inactivation was cured in the presence of novobiocin or ciprofloxacin in approximately 10% of colonies. The resistances were transferred (en bloc or without the resistance to gentamicin) by conjugation to E. faecalis JH2-2 at a frequency of 10−5 per donor CFU after the mating period. The susceptibilities to lincosamides of the E. faecium strains, the transconjugant E. faecalis JH2-2/1025 (resistant to erythromycin, lincomycin, and high levels of gentamicin and streptomycin), and the cured derivative E. faecium HM1025-1 are listed in Table 1. The total-DNA extracts of E. faecium HM1025 and E. faecium HM1025-1 were digested with SmaI and analyzed by pulsed-field gel electrophoresis (Fig. 1). Comparison of fragment patterns revealed the presence of two faint SmaI bands of nearly 180 and 60 kb in E. faecium HM1025 which were absent in HM1025-1. Hybridization with a probe specific for linB showed that this gene was borne by the 180-kb SmaI fragment (Fig. 1). Analysis of the SfiI-generated fragment patterns of the total-DNA extracts of E. faecalis JH2-2 and JH2-2/1025 showed the insertion of an approximately 240-kb DNA segment bearing the linB gene into a 280-kb SmaI fragment from E. faecalis JH2-2 (Fig. 1). These results suggested that the linB gene was borne by a ca. 240-kb plasmid containing two SmaI restriction sites which could be integrated into the chromosome of E. faecalis JH2-2 after conjugative transfer.
TABLE 1.
MICs of erythromycin and lincosamides against bacterial strains
| Organism | MICs (μg/ml) of:
|
Origin of strain or reference | ||
|---|---|---|---|---|
| Erythromycin | Lincomycin | Clindamycin | ||
| E. faecium HM1025 | >128 | >128 | >128 | This study |
| E. faecium HM1025-1 | 0.25 | 0.25 | 0.12 | Cured derivative of E. faecium HM1025 |
| E. faecalis JH2-2 | 0.12 | 8 | 2 | Recipient strain |
| E. faecalis JH2-2/1025 | >128 | >128 | >128 | Transconjugant from E. faecium HM1025 |
| E. coli DB10 | 4 | 8 | 0.25 | Datta et al. (9) |
| E. coli DB10/pVMM25 | 4 | 64 | 8 | This study |
| E. coli DB10/pVMM27 | 4 | 64 | 8 | This study |
| S. aureus RN4220/pJIM2246 | 0.25 | 0.5 | 0.06 | This study |
| S. aureus RN4220/pVMM26 | 0.25 | 64 | 0.5 | This study |
FIG. 1.
Analysis of genomic DNA from E. faecium HM1025 and HM1025-1, digested with SmaI (A and B, left) and from E. faecalis JH2-2 and a transconjugant digested with SfiI (A and B, right) by pulsed-field gel electrophoresis and hybridization. (A) Lanes 1, E. faecium HM1025; lanes 2, E. faecium HM1025-1. (B) Lanes 1, E. faecalis JH2-2; lanes 2, E. faecalis JH2-2/L1 (transconjugant resistant to erythromycin, lincomycin, gentamicin, streptomycin, and tetracycline). The digested fragments were transferred to a nylon sheet and hybridized to an in vitro digoxigenin-labeled linB probe. Numbers on the left of the gels are molecular sizes in kilobases.
E. faecium HM1025 produces a 3-lincosamide O-nucleotidyltransferase.
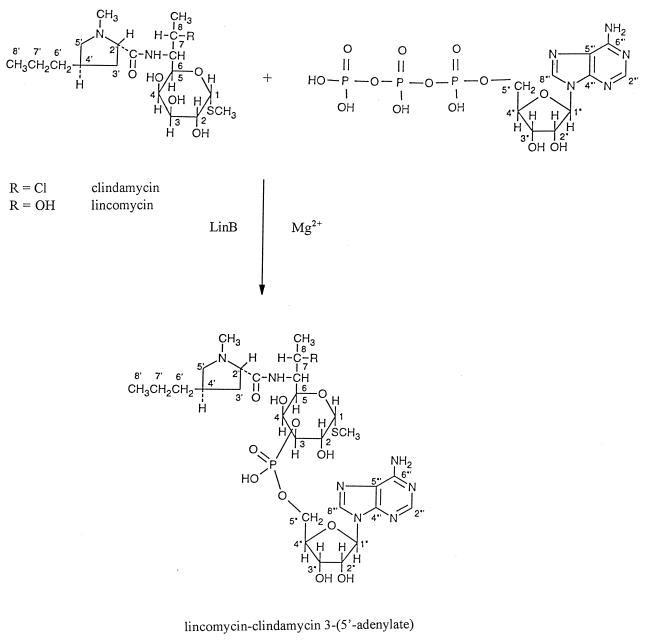
Inactivation of 20 μg of lincomycin and clindamycin per ml by resting E. faecium HM1025 cells was achieved within 6 h. Inactivation of 500 μg of lincomycin and clindamycin per ml was also observed when crude extracts of the strains were incubated with ATP and MgCl2 but not when cells were incubated in the absence of ATP. After purification, the inactivated materials were shown to have masses of 735 and 753/755 amu for lincomycin and clindamycin, respectively. Although these results suggested that the inactivation products are adenylated lincomycin and clindamycin, it was not possible, to assign the site of adenylation with certainty by MS results. The purified material was therefore subjected to two-dimensional correlated spectroscopy, g-HMQC, and g-HMBC NMR experiments. With the exception of a few hydroxyl protons, all of the remaining proton and carbon NMR resonances were rigorously assigned. It is important to note that C-3 of the hexose ring was found to have a significant shift from the control (2). The chemical shift of the clindamycin C-3 appears at 71.2 ppm, while the chemical shifts of adenylated clindamycin and lincomycin were 74.5 and 74.6 ppm, respectively. Further, several confirming vicinal C-O-P couplings were also observed for both inactivated materials (31). These data clearly show that the 3 position is the site of adenylation for both the lincomycin and clindamycin inactivated materials (Fig. 2). The biochemical mechanism of resistance differs from that reported for staphylococcal enzymes which catalyze the conversion of lincomycin to its 3-(5′-ribonucleotide) and clindamycin to its 4-(5′-ribonucleotide) (5). By contrast, the mechanism is similar to that reported for Streptomyces coelicolor Müller NRRL 3532 (25). It has been proposed that genes for antibiotic resistance originated in antibiotic producers. However, the producer of lincomycin, Streptomyces lincolnensis, resists lincomycin by the monomethylation of an adenine residue of the 23S rRNA but not by nucleotidylation, and S. coelicolor does not produce this antibiotic (16). To test the hypothesis that the enzyme found in enterococci derives from a Streptomyces enzyme, determination of the sequence of the gene encoding the nucleotidyltransferase from Streptomyces and comparing it with that of E. faecium HM1025 would be of interest.
FIG. 2.
Mechanism whereby clindamycin and lincomycin are converted to lincomycin and clindamycin 3-(5′-adenylate) by LinB in the presence of ATP and MgCl2+.
Characterization of the linB gene and its product.
The total-DNA extracts from E. faecium HM1025 and plasmid pUC18 were digested with HindIII and mixed, ligated, and introduced by electrotransformation into E. coli DB10, a mutant susceptible to lincosamides. Transformants selected on lincomycin (20 μg/ml) were screened for their plasmid content by agarose gel electrophoresis. The smallest hybrid plasmid, pVMM25, was found to contain a 2.6-kb HindIII fragment, and this plasmid conferred resistance to lincomycin by inactivating the antibiotic in E. coli DB10 but did not confer resistance to erythromycin (Table 1). The nucleotide sequence of the 2,684-bp insert of plasmid pVMM25 was determined. Analysis of the sequence revealed two open reading frames (ORFs), ORF1 and ORF2. To identify the gene responsible for resistance to lincosamide, ORF1 (774 bp), ORF2 (804 bp), and the corresponding putative ribosome-binding sites were separately amplified by PCR and subcloned into the vector plasmid pCR2.1. Only the recombinant plasmid containing ORF2, pVMM27, conferred resistance to lincosamides when introduced into E. coli DB10. The cloned gene, named linB, was amplified by PCR, and the product of the amplification was used as a template in a cell-free coupled transcription-translation system. One band of ca. 31 kDa, a mass that closely approximates the 31,195-Da predicted mass of LinB, was encoded by this fragment (Fig. 3). The deduced amino acid sequence of linB did not display any significant homology with the sequences of lincosamide nucleotidyltransferases encoded by linA and linA′ or those of the aminoglycoside nucleotidyltransferases ANT(2"), ANT(3")(9), ANT(4′)(4"), ANT(6), and ANT(9), (6, 7, 18, 27, 28).
FIG. 3.
Autoradiogram of l-[35S]methionine-labeled polypeptide specified in vitro by the amplification product of linB. (A) Protein electrophoresis in a 15% polyacrylamide gel containing sodium dodecyl sulfate. After separation, proteins were stained with Coomassie blue. (B) Visualization of labeled protein bands in the gel by autoradiography. Lanes 1, control without template; lanes 2, protein products synthesized from PCR-generated linB DNA; lanes 3, molecular mass markers (masses on the left are expressed in kilodaltons). The position of LinB (31,195 Da) is indicated.
Heterospecific expression of lincosamide resistance.
The 2,684-bp HindIII fragment of pVMM25 was excised after further digestion with the restriction enzymes BamHI and PstI and was subcloned into the shuttle plasmid pJIM2246. The resulting recombinant plasmid, pVMM26, was introduced into E. coli DB10 and S. aureus RN4220. In both backgrounds, the transformants inactivated clindamycin and lincomycin. As shown in Table 1, resistance to lincomycin and clindamycin was expressed differently in the gram-negative and gram-positive hosts. MICs of lincomycin were increased by factors of 8 and 128 and the MICs of clindamycin were increased by factors of 32 and 8 in the E. coli and S. aureus backgrounds, respectively. Similar differences in resistances were found for nucleotidyltransferases encoded by linA and linA′ genes in staphylococci (5). These discrepancies could be hypothetically explained by differences in the affinity of lincosamide for either the nucleotidyltransferase or the E. coli and S. aureus ribosomes.
Distribution of linB in clinical isolates of E. faecium.
Inactivation of lincosamides following the acquisition of linB appears to have already spread in E. faecium, since DNA could be amplified by using specific primers in all 14 of the clinical isolates of E. faecium which inactivated lincosamides. By contrast, the inactivation of lincosamides was not detected in E. faecalis. The apparent specificity of the gene for E. faecium is surprising. This specificity does not appear to be due to a narrow spectrum of transferability of the resistance, since we could readily transfer inactivation of lincosamides together with resistance to erythromycin and aminoglycosides from E. faecium HM1025 to E. faecalis JH2-2. Possibly, because of the intrinsic resistance to lincomycin of E. faecalis, the acquisition of an additional resistance to lincosamides did not provide any selective advantage for this species (22).
REFERENCES
- 1.Argoudelis A D, Coats J H. Microbial transformation of antibiotics. II. Phosphorylation of lincomycin by streptomyces species. J Antibiot. 1969;22:341–343. doi: 10.7164/antibiotics.22.341. [DOI] [PubMed] [Google Scholar]
- 2.Argoudelis A D, Coats J H, Mizsak S A. Microbial transformation of antibiotics. Clindamycin ribonucleotides. J Antibiot. 1977;30:474–487. doi: 10.7164/antibiotics.30.474. [DOI] [PubMed] [Google Scholar]
- 3.Bouanchaud H C, Scavizzi M R, Chabbert Y A. Elimination by ethidium bromide of antibiotic resistance in enterobacteria and staphylococci. J Gen Microbiol. 1969;54:417–425. doi: 10.1099/00221287-54-3-417. [DOI] [PubMed] [Google Scholar]
- 4.Brisson-Noël A, Courvalin P. Nucleotide sequence of gene linA encoding resistance to lincosamides in Staphylococcus haemolyticus. Gene. 1986;43:247–253. doi: 10.1016/0378-1119(86)90213-1. [DOI] [PubMed] [Google Scholar]
- 5.Brisson-Noël A, Delrieu P, Samain D, Courvalin P. Inactivation of lincosaminide antibiotics in Staphylococcus. Identification of lincosaminide O-nucleotidyltransferases and comparison of the corresponding resistance genes. J Biol Chem. 1988;263:15880–15887. [PubMed] [Google Scholar]
- 6.Cameron F, Groot Obbink D J, Ackerman V P, Hall R M. Nucleotide sequence of the AAD(2") aminoglycoside adenyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 1986;14:8625–8635. doi: 10.1093/nar/14.21.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlier C, Courvalin P. Emergence of 4′,4"-aminoglycoside nucleotidyltransferase in enterococci. Antimicrob Agents Chemother. 1990;34:1565–1569. doi: 10.1128/aac.34.8.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comité de l’Antibiogramme de la Société Française de Microbiologie. 1996 report of the Comité de l’Antibiogramme de la Société Française de Microbiologie. Technical recommendations for in vitro susceptibility testing. Clin Microbiol Infect. 1996;2S1:11–25. [Google Scholar]
- 9.Datta N, Hedges R W, Becker D, Davies J. Plasmid-determined fusidic acid resistance in Enterobacteriaceae. J Gen Microbiol. 1974;83:191–196. doi: 10.1099/00221287-83-1-191. [DOI] [PubMed] [Google Scholar]
- 10.Devriese L A. Two new types of resistance to lincomycin in pathogenic staphylococci from animals. Ann Inst Pasteur (Paris) 1980;131B:261–266. [PubMed] [Google Scholar]
- 11.Dhawan V K, Thadepalli H. Clindamycin: a review of fifteen years of experience. Rev Infect Dis. 1982;4:1133–1153. doi: 10.1093/clinids/4.6.1133. [DOI] [PubMed] [Google Scholar]
- 12.Dutta G N, Devriese L A. Degradation of macrolide-lincosamide-streptogramin antibiotics by Lactobacillus strains from animals. Ann Inst Pasteur (Paris) 1981;132A:51–57. [PubMed] [Google Scholar]
- 13.Dutta G N, Devriese L A. Resistance to macrolide, lincosamide and streptogramin antibiotics and degradation of lincosamide antibiotics in streptococci from bovine mastitis. J Antimicrob Chemother. 1982;10:403–408. doi: 10.1093/jac/10.5.403. [DOI] [PubMed] [Google Scholar]
- 14.Ehrenfeld E E, Clewell D B. Transfer functions of the Streptococcus faecalis plasmid pAD1: organization of plasmid DNA encoding response to sex pheromone. J Bacteriol. 1987;169:3473–3481. doi: 10.1128/jb.169.8.3473-3481.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujisawa Y, Weisblum B. A family of r-determinants in Streptomyces spp. that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J Bacteriol. 1981;146:621–631. doi: 10.1128/jb.146.2.621-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gots J S. The detection of penicillinase production properties of microorganisms. Science. 1945;102:309. doi: 10.1126/science.102.2647.309. [DOI] [PubMed] [Google Scholar]
- 18.Hollingshead S, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 19.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclercq R, Brisson-Noël A, Duval J, Courvalin P. Phenotypic expression and genetic heterogeneity of lincosamide inactivation in Staphylococcus spp. Antimicrob Agents Chemother. 1987;31:1887–1891. doi: 10.1128/aac.31.12.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclercq R, Carlier C, Duval J, Courvalin P. Plasmid-mediated resistance to lincomycin by inactivation in Staphylococcus haemolyticus. Antimicrob Agents Chemother. 1985;28:421–424. doi: 10.1128/aac.28.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leclercq R, Derlot E, Weber M, Duval J, Courvalin P. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob Agents Chemother. 1989;33:10–15. doi: 10.1128/aac.33.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mabilat C, Courvalin P. Gene heterogeneity for resistance to macrolides, lincosamides, and streptogramins in Enterobacteriaceae. Ann Inst Pasteur Microbiol. 1988;139:677–681. doi: 10.1016/0769-2609(88)90072-5. [DOI] [PubMed] [Google Scholar]
- 25.Marshall V P, McGee J E, Cialdella J I, Baczynskyj L, Chirby D G, Yurek D A, Liggett W F, Kuo M S. Purification of lincosaminide O-nucleotidyltransferase from Streptomyces coelicolor Müller. J Antibiot. 1991;44:895–900. doi: 10.7164/antibiotics.44.895. [DOI] [PubMed] [Google Scholar]
- 26.Marshall V P, Liggett W F, Cialdella J I. Enzymic inactivation of lincosaminide and macrolide antibiotics: divalent metal cation and coenzyme specificities. J Antibiot. 1989;42:826–830. doi: 10.7164/antibiotics.42.826. [DOI] [PubMed] [Google Scholar]
- 27.Matsumura M, Katakura Y, Imanaka T, Aiba S. Enzymatic and nucleotide sequence studies of a kanamycin-inactivating enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J Bacteriol. 1984;160:413–420. doi: 10.1128/jb.160.1.413-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy E. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3")(9) Mol Gen Genet. 1985;200:33–39. doi: 10.1007/BF00383309. [DOI] [PubMed] [Google Scholar]
- 29.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for Gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 31.Stothers J B. Carbon-13 spectroscopy. New York, N.Y: Academic Press; 1972. p. 378. [Google Scholar]
- 32.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]