Abstract
Purpose
The lncRNA MIR155 host gene (MIR155HG) plays a role in the progression of several malignant cancers. However, the specific mechanisms of MIR155HG in glioma progression have not been clearly established. The purpose of this study was to investigate the function of MIR155HG in glioma at the transcriptome level and relationship with immune infiltration.
Patients and Methods
Totally, 697 RNA-seq and 594 DNA methylation data were retrieved from The Cancer Genome Atlas (TCGA) dataset while 325 RNA-seq data were retrieved from the Chinese Glioma Genome Atlas (CGGA) dataset. The DNA methylation levels of MIR155HG CpG islands were assessed through bisulfite amplicon sequencing (BSAS). The regulatory mechanism of SP1 on MIR155HG was examined by chromatin immunoprecipitation (ChIP) and luciferase reporter assays. R language was used as the main tool for statistical analysis and graphical work.
Results
MIR155HG was predominantly expressed in the isocitrate dehydrogenase (IDH) wild-type as well as mesenchymal subtype gliomas. Promoter methylation levels of MIR155HG in glioblastoma (GBM) were remarkably decreased compared with those in lower-grade glioma (LGG). In addition, there were negative correlations between promoter methylation levels and MIR155HG expressions but positive correlations with patients’ overall survival. In vitro studies further revealed that MIR155HG expression was regulated by DNA promoter methylation and transcription factor (SP1) binding to the promoter. Moreover, there was a close association between MIR155HG expression and immune as well as stromal cell infiltrations, inflammatory activities, and immune checkpoints. Clinically, univariate and multivariate Cox analyses revealed that MIR155HG is an independent prognostic marker for glioma patients.
Conclusion
Our results established that MIR155HG is a potential biomarker for prognosis and an immunotherapeutic target in glioma.
Keywords: MIR155HG, DNA methylation, SP1, immune infiltration, glioma
Introduction
In adults, glioma is a very prevalent brain tumor of which glioblastoma (GBM) is the most aggressive, malignant subtype.1 Improvements in conventional treatment options, including surgical resection, followed by radiotherapy as well as chemotherapy have not improved the median survival time for glioma patients, which is still < 2 years.2 Given the poor outcome after standard treatment, it’s urgent to develop new therapeutic approaches. Malignant solid tumor tissues are composed of tumor cells as well as tumor-associated nontumor cells, including normal epithelial, stromal, vascular, and immune cells.3 The tumor microenvironment (TME) has a key role in glioma progression.4,5 For instance, stromal cells within the TME may promote glioma expansion and invasion by elaborating numerous compounds that stimulate angiogenesis, neoplastic cell growth, and modify the extracellular matrix (ECM).6 Meanwhile, tumor-associated macrophages (TAMs) could promote angiogenesis, maintain the glioma stem-like cell phenotypes, and secrete VEGF, PDGF, IL-6, TGFβ1, MMP2 and MMP9 to regulate ECM degradation, resulting in glioma progression.7,8 Thus, the TME-targeted therapies are promising novel approaches for glioma.
Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts whose nucleotide length exceeds 200. LncRNAs have critical, complex roles in cancer development as well as progression, including glioma.9,10 LncRNA MIR155HG is located at chromosome 21q21 and has a length of 1.5kb, which is also considered as the primary microRNA of miR-155. Previously, we have reported that MIR155HG is a prognostic factor for GBM and is involved in promoting the progression of glioma via miR-155/PCDHs/β-catenin pathway.11 Recently, two other studies further confirmed that MIR155HG contributes to GBM growth and progression by sponging miR-185 to increase ANXA2 expression, and promotes temozolomide resistance by binding to PTBP1.12,13 Additionally, MIR155HG was involved in a lncRNA signature related to ferroptosis, tumor progression, and microenvironment for stratifying the prognosis of glioma patients with adequate predictive performance.14 Moreover, MIR155HG was identified as a m6A-related prognostic lncRNA for patients with primary glioblastoma by constructing the m6A-lncRNA co-expression networks.15 Another research demonstrated that the MIR155HG/has-miR-129-5p/C1S axis is a potential marker and therapeutic target for glioblastoma.16 MIR155HG also has a vital role in facilitating the malignancy of laryngeal squamous cell carcinoma (LSCC),17 pancreatic cancer (PC),18 clear cell renal cell carcinoma (ccRCC)19 and non-small cell lung cancer (NSCLC).20 LncRNA expression is epigenetically regulated.21 Of all epigenetic modifications, DNA methylation induces heritable alterations in gene expression, but does not affect DNA sequences. It is well known that promoter hypermethylation has an association with transcriptional repression, while hypomethylation has an association with transcriptional activation.22 Currently, whether and how MIR155HG is regulated by DNA methylation in glioma remains unclear.
In addition, lncRNAs exert crucial functions in the immune and inflammatory responses.23,24 For instance, in dendritic cells and macrophages, lncRNA-Cox2 activates the toll-like receptor (TLR) signaling pathway to increase IL-23 and IL-6 secretion after microbial infection.25 Although the immunoregulatory role of miR-155 has been most extensively studied in many tumors, especially in hematologic tumors,26,27 the research on the immunoregulation of MIR155HG is still in its infancy. Recently, a bioinformatic data analysis demonstrated that MIR155HG is correlated with immune checkpoints and infiltrations in various cancers, including glioma.28 Nevertheless, the underlying function and mechanism of MIR155HG in tumor immunology still require further studies.
In the present study, we combined experimental detection with bioinformatic analysis to further explore the MIR155HG expression and clinicopathologic characteristics and its correlation with immune infiltration in glioma. The results would give us a better understanding of MIR155HG and provide a molecular basis for anti-MIR155HG treatment in glioma immunotherapy.
Materials and Methods
Study Participants and Samples
We obtained 697 RNA-seq and 685 DNA methylation data, as well as related phenotypic data from The Cancer Genome Atlas (TCGA, (http://cancergenome.nih.gov/), and an additional dataset of 325 RNA-seq data from the Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/).
In addition, 10 tissue samples, including 5 lower grade glioma (LGG) samples as well as 5 GBM samples were obtained from the Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University. This study was approved by Ethics Committee of the Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University and informed consent was obtained from all the patients. This study was conducted in accordance with the Declaration of Helsinki.
Cell Lines
Human glioblastoma cell lines (U87, U251) and the human embryonic kidney (HEK) cell line (293T) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cell lines were routinely cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100µg/mL), streptomycin (100µg/mL) at 37°C in a 5% CO2 humidified air incubator.
Extraction of RNA and Quantitative RT-PCR (qRT-PCR)
The TRIzol reagent (Invitrogen) was used for total RNA extraction from cultured cells. qRT-PCR analyses were conducted using the SYBR Green Premix Ex Taq (Takara, Tokyo, Japan) in an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). MIR155HG primer sequences were: forward 5’-CCACCCAATGGAGATGGCTC-3’, reverse 5’-AGTTGGAGGCAAAAACCCCT-3’. Target gene expression was normalized by GAPDH mRNA expressions.
Extraction of DNA and Bisulfite Amplicon Sequencing (BSAS)
DNA from glioma samples were extracted using a QIAamp DNA Mini Kit (Qiagen, Inc., Valencia, CA, USA) and subjected to bisulfite amplicon sequencing (BSAS) after DNA bisulfite conversion using the EZ DNA Methylation-Gold Kit (Zymo Research Corp., Irvine, CA, USA) for analysis of MIR155HG promoter methylation. Primer sequences for MIR155HG were: CpG island 1, forward 5’- GTAAGGAGAGAGTAGAGATATTTTTG −3’; reverse 5’- CCCTCCTACCTATTCTTAAAACCTAC −3’, and CpG island 2, forward 5’-AATTAAGGAGAYGTTTTTGGTA-3’; reverse 5’- TAAAAACAATCTCTTTTTCCACCC-5’. Then, the methylation data and average methylation level at all sites were analyzed by MethylKIT software.
Plasmid Construction and Luciferase Reporter Assay
To construct SP1 overexpression vector, the amplified coding region of the human SP1 gene was subcloned into a pcDNA3.1 vector. The MIR155HG proximal promoter region sequences containing the CpG islands (+400 bp to −400 bp) were amplified and inserted into a pGL3-basic luciferase vector (Promega), named pGL3-MIR155HG-WT. Generation of the mutated version (pGL3-MIR155HG-Mut) was achieved by mutating the putative binding sites of SP1. For in vitro DNA methylation, pGL3-MIR155HG-WT was treated with the CpG methylase M.SssI (NEB, Ipswich, MA). Then, 48 h post-transfection, the Dual Luciferase Reporter Assay System (Promega, Madison, WI) was used to assess luciferase activities.
Chromatin Immunoprecipitation (ChIP)
ChIP assays were conducted with the Millipore Magna ChIP kit (Millipore, USA) as instructed by the manufacturer. The antibodies against SP1 and IgG were bought from Cell Signaling Technology (CST, USA). Primer sequences used in this assay were: region 1 (−490 to +105), forward 5’-GGTCTCCAGCTGATTCGGTC-3’, reverse 5’-CCAGGAGCGTCTCCTTGGTT-3’; region 2 (+109 to +480): forward 5’-TGCAGGTACGCCGACTTCAG-3’, reverse 5’-TTCTCTGGCCACTCCAACAG −3’.
Statistical Analysis
R language was used for statistical analyses and to generate figures with several publicly available packages, including survival, pheatmap, ggplot2, circlize, corrgrams, pROC, and corrplot. Comparisons of differences between groups were done by two-tailed Student’s t-tests. Kaplan-Meier curves were used to calculate overall survival differences. The statistical significance threshold was set at p < 0.05.
Results
MIR155HG Expression Was Elevated in IDH Wild-Type Glioma
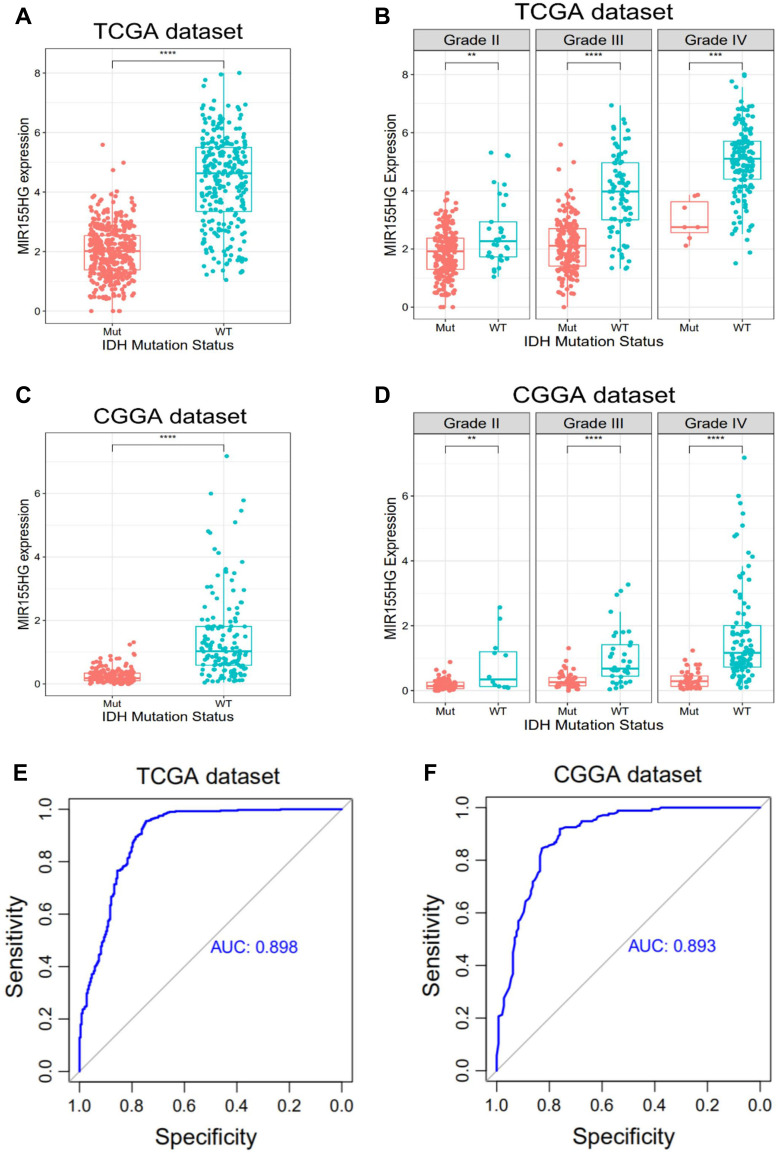
The IDH mutation status is a clinically important molecular marker for glioma progression. In glioma, especially GBM, MIR155HG is highly upregulated.11 In this study, we found that MIR155HG expressions were significantly increased in IDH wild-type glioma than that in IDH mutated glioma in both datasets (Figure 1A and C). Additionally, IDH wild-type glioma showed remarkably elevated MIR155HG expressions than IDH mutated glioma across different grades (Figure 1B and D). Then, we constructed receiver operating characteristic (ROC) curves to assess the diagnostic values of MIR155HG for the IDH wild type glioma. Interestingly, the respective areas under the curve (AUCs) were 89.8% and 89.3% for TCGA as well as CGGA datasets (Figure 1E and F). These findings suggested that MIR155HG is specifically highly expressed in IDH wild-type glioma.
Figure 1.
MIR155HG expression according to IDH status in TCGA and CGGA datasets. (A and C) MIR155HG was highly upregulated in IDH wild-type glioma of all grades. (B and D) MIR155HG was significantly increased in IDH wild-type glioma across different grades. (E and F) MIR155HG could serve as a biomarker to predict IDH wild-type glioma. **p < 0.01; ***p < 0.001; ****p < 0.0001.
MIR155HG Expressions Correlated with Mesenchymal Subtype
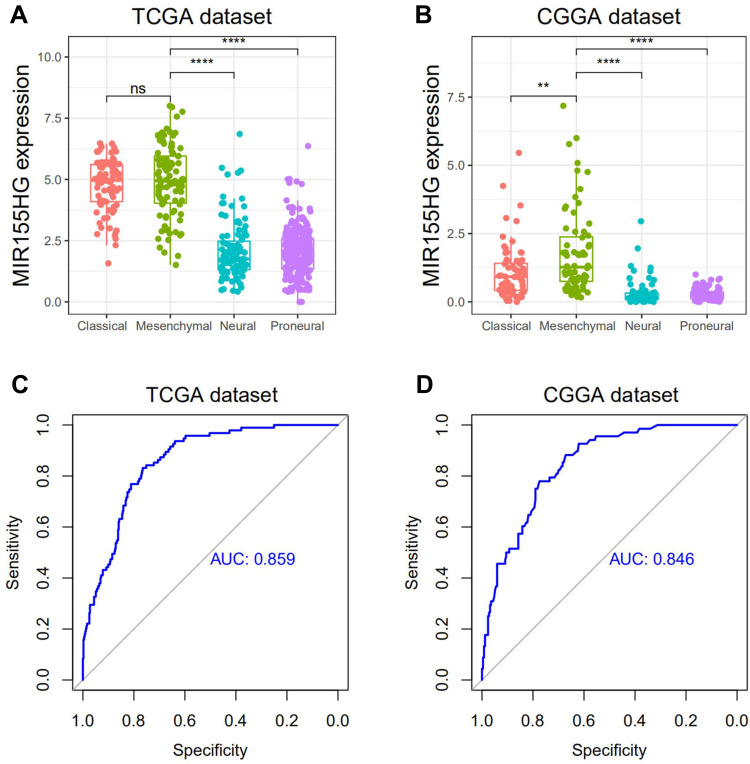
Next, we evaluated MIR155HG distributions in 4 TCGA-defined molecular subtypes. MIR155HG was apparently elevated in the mesenchymal subtype relative to other subtypes, except for the classical subtype in TCGA dataset (Figure 2A and B). To further confirm this, we performed ROC curves for MIR155HG expressions in the mesenchymal subtype. The respective AUCs were 85.9% and 84.6% for TCGA and CGGA datasets (Figure 2C and D). Therefore, these results indicated that MIR155HG was mainly enriched in the mesenchymal subtype.
Figure 2.
MIR155HG expression in different molecular subtypes in TCGA and CGGA datasets. (A and B) MIR155HG was highly enriched in mesenchymal subtype glioma. (C and D) MIR155HG could serve as a biomarker to predict mesenchymal subtype glioma. **p < 0.01; ****p < 0.0001.
Abbreviations: ns, not significant; AUC, area under the curve.
MIR155HG Expression Was Associated with Promoter DNA Methylation
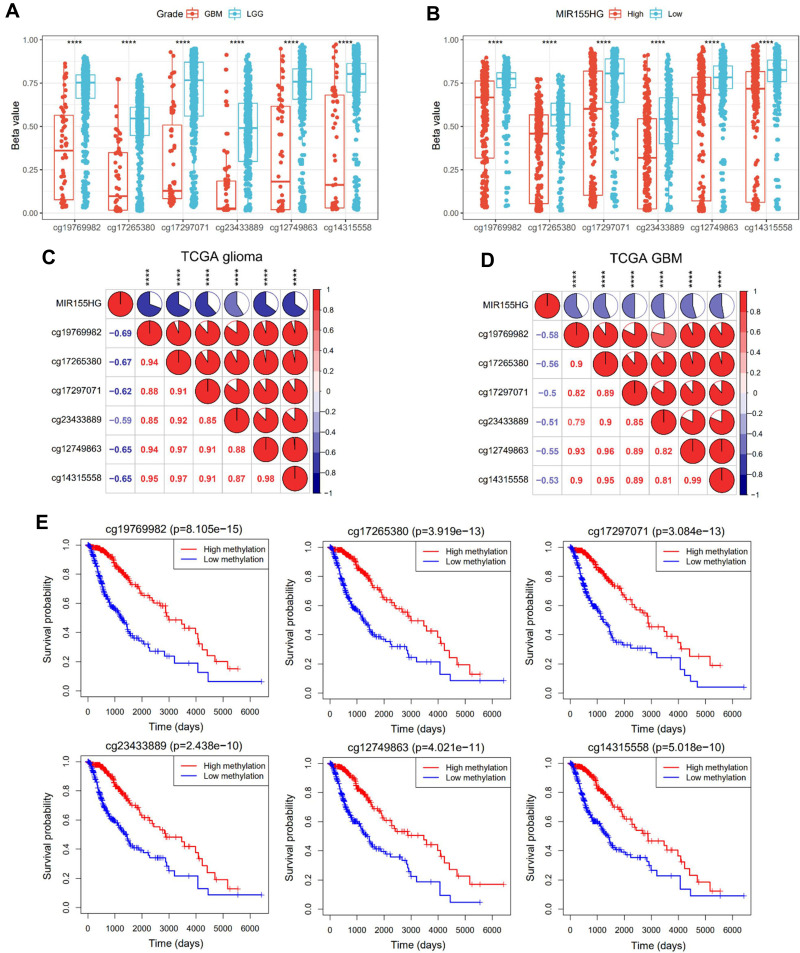
To discover the genetic determinant of MIR155HG endogenous expression, we investigated the association between MIR155HG expression levels and its DNA methylation. Integration of TCGA glioma datasets identified that 594 patients had both MIR155HG expression and DNA methylation data. Particularly, six CpG sites (cg19769982, cg17265380,cg17297071, cg23433889,cg12749863 and cg14315558) were localized in the MIR155HG promoter region. Interestingly, DNA methylation beta values at all the six sites were significantly higher in LGG than in GBM (Figure 3A) and were remarkably elevated in the low MIR155HG expression group, relative to the high MIR155HG expression group (Figure 3B). In addition, there was a negative association between promoter DNA methylation beta values and MIR155HG expressions in all gliomas as well as in GBM (Figure 3C and D). Meanwhile, patients with elevated methylation levels exhibited markedly better overall survival when compared to the low methylation level group (Figure 3E). In summary, these results suggested that MIR155HG expression is regulated by promoter DNA methylation. Therefore, this epigenetic modification may be a prognostic biomarker for glioma.
Figure 3.
Promoter DNA methylation was correlated with MIR155HG expression and glioma patients’ prognosis. (A) Comparison of DNA methylation beta values at six CpG sites (cg19769982, cg17265380, cg17297071, cg23433889, cg12749863 and cg14315558) between LGG and GBM groups. (B) Comparison of DNA methylation beta values at six CpG sites between low and high MIR155HG expression groups. (C and D) Correlation between MIR155HG expression and the promoter DNA methylation beta values in all gliomas and GBM. (E) Association between the promoter DNA methylation beta values of MIR155HG and overall survival time for glioma patients. ****p < 0.0001.
DNA Methylation Profiles of CpG Islands in MIR155HG Promoter Region
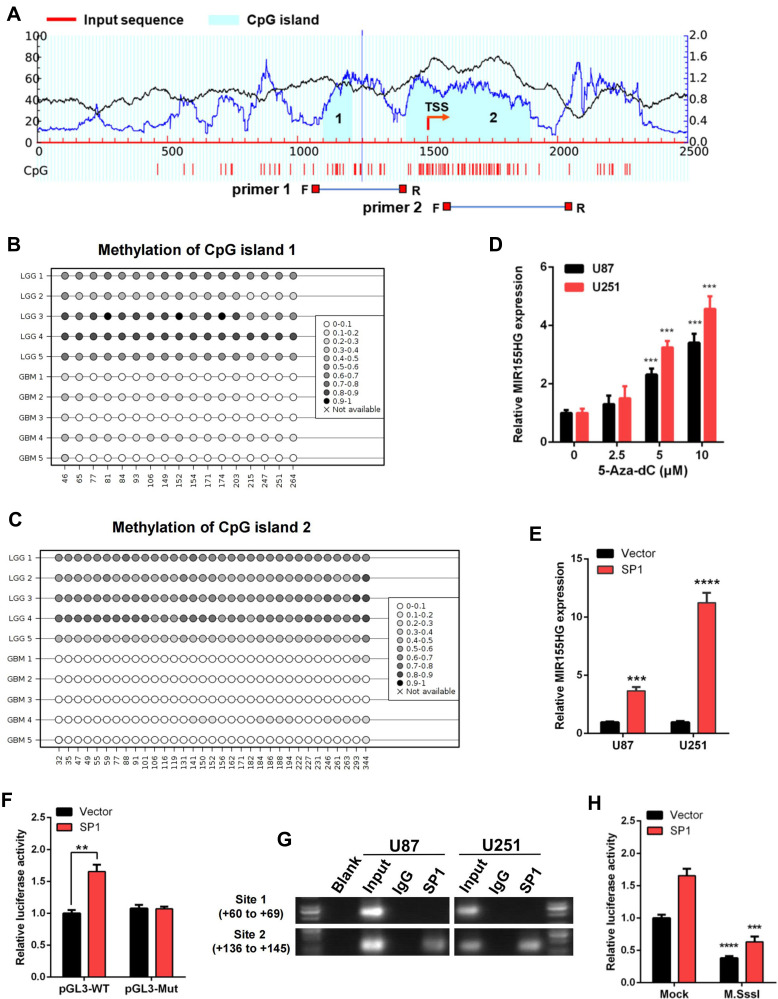
Next, we evaluated DNA sequences of the MIR155HG promoter region and identified two CpG islands via the MethPrimer 2.0 (http://www.urogene.org/methprimer2/) (Figure 4A). CpG island 1 spans −399 to −290, and CpG island 2 covers −82 to +396 when compared to the MIR155HG transcription start site (TSS). And then, we performed bisulfite amplicon sequencing (BSAS) to detect the DNA methylation levels around the CpG island region in glioma samples. The results revealed that LGG tissues exhibited a denser methylation pattern at both CpG islands in MIR155HG promoter region compared with GBM tissues (Figure 4B and C), consistent with the analysis of the TCGA data. Then for further verification of the effect of DNA methylation on MIR155HG expression, U87, as well as U251 cells, were treated with a DNA methyltransferase inhibitor (5-Aza-dC) at 0, 2.5, 5, and 10 μM, and the MIR155HG expression levels were subsequently examined by qPCR. After DNA methylation inhibition, MIR155HG expression levels were markedly elevated in both cell lines in a dose-dependent manner (Figure 4D).
Figure 4.
DNA methylation and SP1 were involved in the activation of MIR155HG. (A) Schematic representation of the CpG islands and bisulfite sequencing primers. (B and C) Bisulfite amplicon sequencing (BSAS) was performed to examine methylation status of CpG island 1 and 2 at the promoter region of MIR155HG in LGG and GBM. (D) MIR155HG expression in U87 and U251 cell lines were upregulated in a dose-dependent manner after 5-Aza-dC treatment. (E) Overexpression of SP1 activated the expression of MIR155HG in U87 and U251 cell lines. (F) The luciferase reporter plasmids carrying MIR155HG promoter pGL3-WT or pGL3-Mut with the SP1 plasmid or vector were co-transfected into HEK293T cells. Relative luciferase activity in HEK293T cells were determined. (G) ChIP assay was performed to confirm the putative binding regions for SP1 in MIR155HG promoter region. (H) Luciferase reporter assay revealed that M.SssI-induced methylation could abolish the MIR155HG promoter activity activated by SP1. **p < 0.01; ***p < 0.001; ****p < 0.0001.
Transcriptional Activity of MIR155HG Promoter is Regulated by SP1 and DNA Methylation
Subsequently, we performed bioinformatic analysis using the JASPAR database (http://jaspar.genereg.net/) for the prediction of transcription factor binding sites in the MIR155HG promoter region. We identified two transcription factor SP1 putative binding sites located at site 1(+60 to +69), and site 2(+136 to +145) (Supplementary Figure S1). SP1, a DNA-binding protein, specifically binds the promoter’s GC-rich sequences. This binding capacity has a close association with CpG dinucleotide methylation. Thus, we performed qPCR assay and found that overexpression of SP1 significantly increased MIR155HG expression levels in U87 as well as U251 glioma cell lines (Figure 4E). Moreover, dual luciferase reporter assay was used to confirm SP1 binding to MIR155HG promoter. As shown in Figure 4F, SP1 overexpression dramatically induced the wild-type MIR155HG promoter activation, while having no effect on the mutated promoter activity. Furthermore, ChIP assay revealed that SP1 could bind to the MIR155HG promoter at site 2 (+136 to +145), whereas no binding was observed at site 1 (+60 to +69) (Figure 4G).
To investigate whether DNA methylation affects the MIR155HG promoter activity, the wild-type MIR155HG promoter luciferase reporter vector, containing 2 CpG islands, was methylated using CpG methyltransferase M.SssI. Luciferase reporter analysis revealed that M.SssI significantly repressed the MIR155HG promoter reporter gene activity compared with no methylase (Figure 4H). In addition, luciferase assay following cotransfection of methylated/unmethylated MIR155HG promoter constructs and SP1 expression plasmids revealed that SP1-induced activation of MIR155HG promoter was significantly abolished by M.SssI (Figure 4H). Collectively, these data indicated that SP1 transcriptionally upregulated MIR155HG expression levels by binding its proximal promoter region after DNA hypomethylation.
MIR155HG Was Closely Related to Immune Function in Glioma
GO enrichment analysis was performed to identify the biological features of MIR155HG. First, genes strongly correlated with MIR155HG expression were selected (Pearson |R|>0.5) from the TCGA as well as CGGA datasets. Biofunctions for these genes were evaluated by GO analysis on DAVID Bioinformatics Resources (https://david.ncifcrf.gov/). In biological processes analysis, genes that had a positive association with MIR155HG expressions were enriched in inflammatory and immune responses (Supplementary Figure S2A and B). Regarding cell components, the enrichment of these positive genes was mainly in the extracellular regions (Supplementary Figure S2C and D), indicating a vital role of these genes in the TME of glioma. Moreover, molecular function analysis revealed gene enrichment in protein binding (Supplementary Figure S2E and F). In summary, these results suggested that MIR155HG might participate in the regulation of the glioma immune environment.
MIR155HG Was Associated with Inflammatory Activities in Glioma
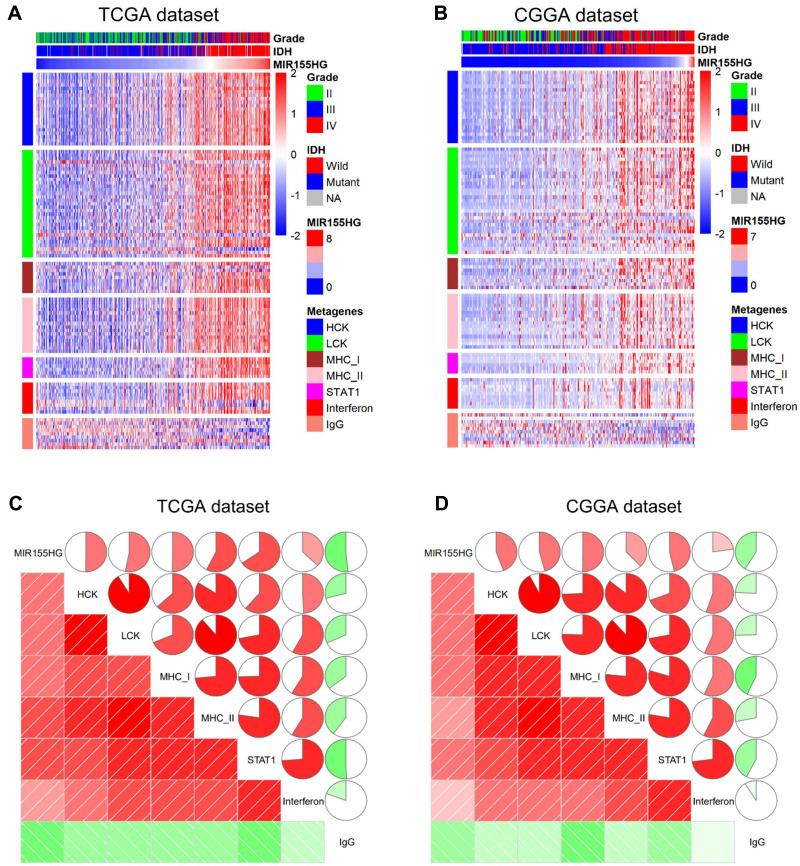
To specifically evaluate the function of MIR155HG in the glioma inflammatory response, we analyzed its relationship with seven well-established metagenes using the method described previously.29 Figure 5A and B show that MIR155HG expression levels were positively correlated with the majority of the clusters in the TCGA and CGGA datasets apart from IgG, which represented B cell activities. Furthermore, corrgrams were generated by R language based on Pearson R values between MIR155HG expression and the 7 metagenes (Figure 5C and D). There was a positive association between MIR155HG and HCK, MHC-I, LCK, MHC-II, Interferon, and STAT1, as well as negative associations with IgG, in accordance with the above findings. These findings confirm the important immune functions of MIR155HG in glioma.
Figure 5.
The relationship between MIR155HG and inflammatory activities in TCGA and CGGA datasets. (A and B) Heatmaps displaying the clinicopathological parameters, MIR155HG expression, and seven inflammatory metagenes. (C and D) Corrgrams were established based on the relationship between MIR155HG expression and seven inflammatory metagenes.
MIR155HG Expressions Were Correlated with Glioma Immune Infiltrations
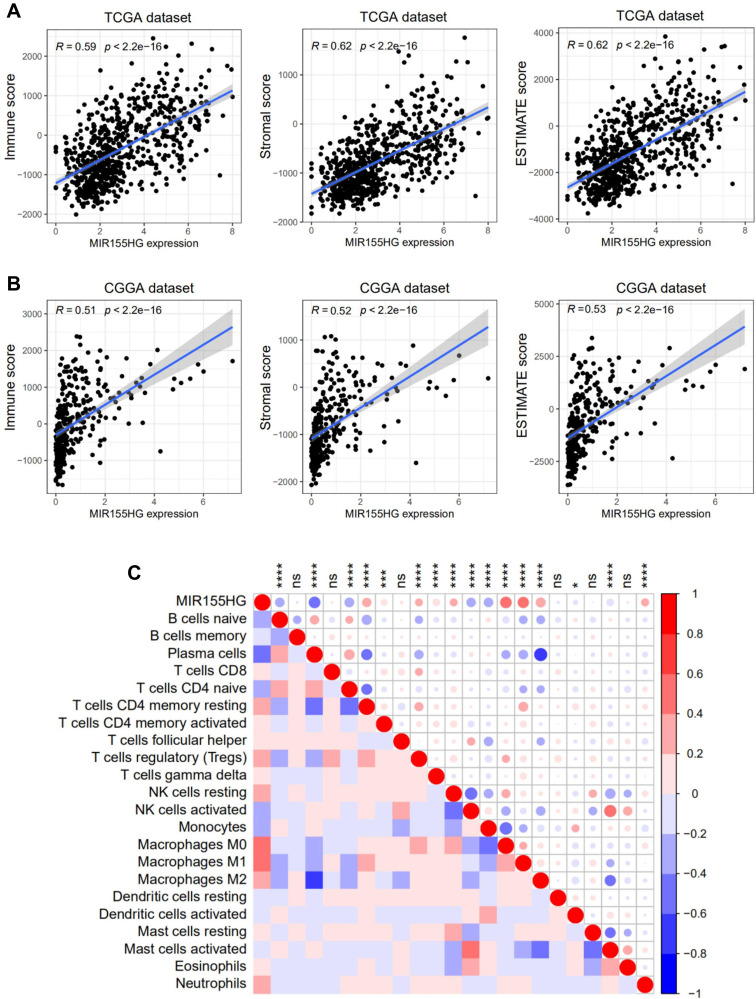
Immune as well as stromal cells are crucial constituents of the TME. Considering the results of GO analysis, we further analyzed the correlation between MIR155HG and infiltrated cells in glioma. First, we performed the ESTIMATE algorithm method as reported by Yoshihara to investigate the association between MIR155HG expression levels and ESTIMATE scores. As shown in Figure 6A and B, there were significant positive correlations between MIR155HG expressions and immune, stromal, as well as ESTIMATE scores in glioma in both datasets, indicating that it predominantly influences immune as well as stromal cell infiltrations. Furthermore, we employed the CIBERSORT algorithm to evaluate the proportions of 22 subpopulations of infiltrating immune cells in glioma and then investigated the association between MIR155HG and particular cell populations in the glioma TME. A comparative summary of immune cell proportions in various MIR155HG expression groups is shown in Supplementary Figure S3. Several kinds of immune cells were enriched in different groups. Notably, we established that M0 and M2 macrophage populations were significantly higher in the high MIR155HG expression group, while plasma cells and monocytes were significantly enriched in the low MIR155HG expression group. Furthermore, MIR155HG expression levels were significantly positively correlated with 9 cell types and negatively correlated with 7 cell types (Figure 6C). Interestingly, MIR155HG expression showed a strong correlation with M2 macrophages as well as neutrophils which were considered to be immunosuppressive cells.30 In summary, MIR155HG played a precise role in immune as well as stromal cell infiltrations in glioma.
Figure 6.
MIR155HG expression was tightly associated with ESTIMATE algorithm scores and infiltrated cells in glioma. (A and B) MIR155HG expression was positively correlated with immune score, stromal score and ESTIMATE score in glioma patients in TCGA and CGGA datasets. (C) MIR155HG expression was significantly correlated with infiltrated cells in the tumor microenvironment estimated by CIBERSORT in TCGA dataset. *p < 0.05; ***p < 0.001; ****p < 0.0001.
Abbreviation: ns, not significant.
MIR155HG Was Associated with Immune Checkpoints
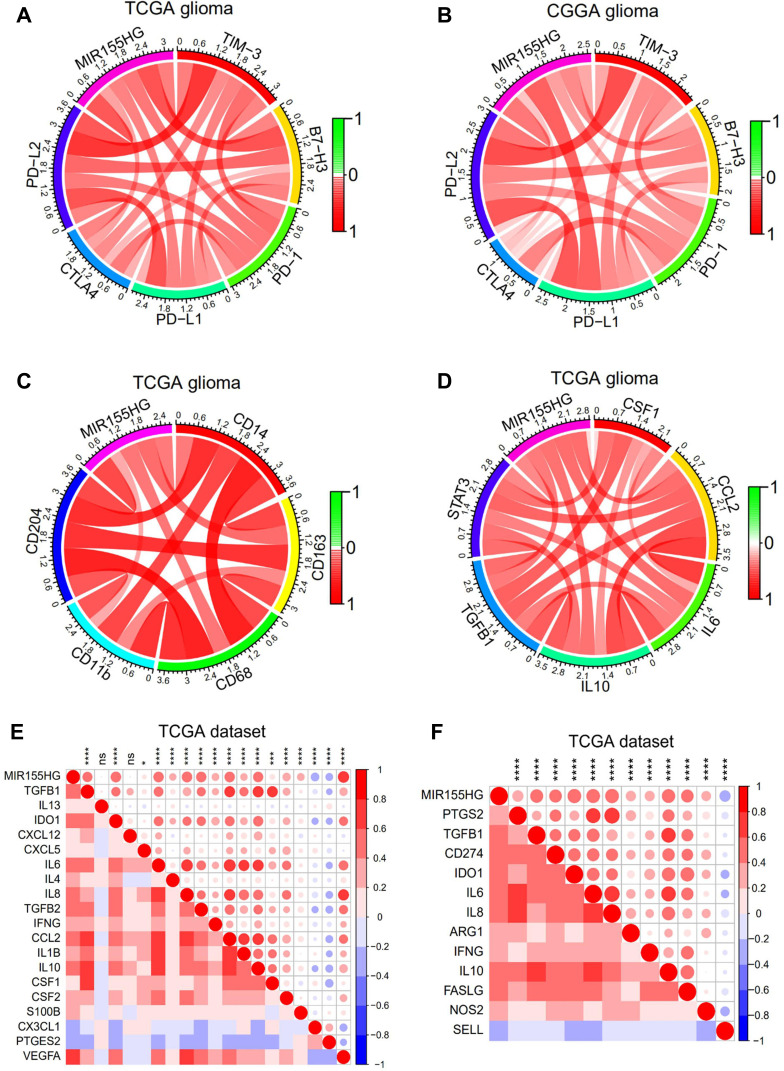
Immune checkpoint-targeting agents have been extensively evaluated in preclinical as well as clinical trials for various solid tumors, including glioma. Thus, the available immune checkpoint blocking agents, such as TIM-3, PD-1, B7-H3, PD-L1, CTLA4, and PD-L2, were enrolled in the analysis. Surprisingly, MIR155HG showed strong positive correlations with all these molecules in glioma in both datasets (Figure 7A and B). In GBM, MIR155HG exhibited a steady correlation with the above immune checkpoints, except for PD-1 and CTLA4 in TCGA dataset (Supplementary Figure S4A and B). These findings indicate the potential regulatory effects of MIR155HG on immune checkpoints in glioma.
Figure 7.
MIR155HG expression was correlated with immune checkpoints and immunosuppressive properties. (A and B) Correlation between MIR155HG and immune checkpoints in glioma in TCGA and CGGA datasets. (C) Correlation between MIR155HG and the surface markers of macrophages in TCGA dataset. (D) Correlation between MIR155HG and several key factors that drive M2 phenotype macrophages differentiation in TCGA dataset. (E) Correlation between MIR155HG and immunosuppressive cell recruitment factors in TCGA dataset. (F) Correlation between MIR155HG and immunosuppressive factors secreted by tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), and tumor-associated neutrophils in TCGA dataset. *p < 0.05; ***p < 0.001; ****p < 0.0001.
Abbreviation: ns, not significant.
MIR155HG Was Correlated with Immunosuppressive Properties
Accumulating evidence has demonstrated that increased tumor associated macrophage (TAM) infiltrations predict poor prognostic outcomes for GBM patients and correlate with upregulated glioma progression and tumor grade.31,32 Therefore, targeting TAMs is a potential therapeutic approach for intractable glioma. Herein, we further analyzed the correlation between MIR155HG and the surface markers of macrophages. Expressions of MIR155HG were markedly associated with CD14, CD163, CD68, CD11b, and CD204 in glioma in TCGA dataset (Figure 7C). Moreover, we observed that MIR155HG was closely related to several key factors that mediate M2 macrophage differentiation,33 including CSF-1, IL-6, CCL2, IL-10, STAT3, and TGFβ (Figure 7D). Similar results were shown in CGGA dataset (Supplementary Figure S4C and D). Next, correlation analysis was performed to evaluate the association between MIR155HG expression levels and critical factors involved in the recruitment of TAMs, tumor-associated neutrophils, myeloid-derived suppressor cells, and the immunosuppressive factors that are secreted by these cells. Interestingly, MIR155HG expressions were strongly positively correlated with the majority of the factors involved in immunosuppressive cell recruitment33 as well as immunosuppressive factors33,34 (Figure 7E and F). In glioma, MIR155HG is a mesenchymal transition-associated lncRNA.11 Consistently, in this study, the correlations between common EMT biomarkers and MIR155HG were significant (Supplementary Figure S5A and B). Therefore, in glioma, MIR155HG has a potential immunotherapeutic value.
Overexpression of MIR155HG Predicted Worse Survival in Glioma
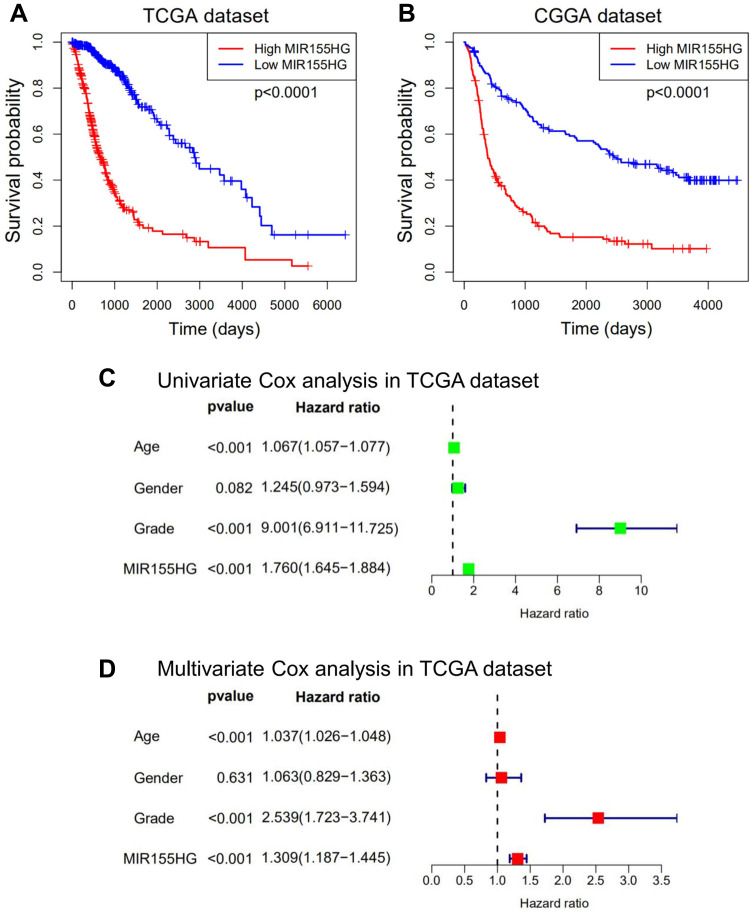
Considering that MIR155HG expression was abnormal in glioma and correlated with the histological grade as well as molecular subtypes, we explored the prognostic significance of MIR155HG in glioma. Figure 8A and B show that glioma patients with high MIR155HG expression levels exhibited significantly short overall survival times than those with low MIR155HG expressions. Moreover, univariate analysis showed that MIR155HG expression, diagnostic age, and WHO grades had a significant association with overall survival outcomes (Figure 8C). In multivariate analysis, after adjustment of the above factors, MIR155HG expression was still a significant prognostic factor (Figure 8D). Similar results were obtained in CGGA dataset (Supplementary Figure S6A and B). In summary, MIR155HG was an independent prognostic factor for glioma patients.
Figure 8.
MIR155HG predicted worse survival in glioma. (A and B) Kaplan-Meier survival analyses showed that high MIR155HG expression predicted poor prognosis for glioma patients based on the TCGA and CGGA datasets. (C and D) Univariate and multivariate Cox regression analyses revealed that MIR155HG was an independent prognostic biomarker for glioma patients in TCGA dataset.
Discussion
Gliomas are prevalent and lethal brain tumor types in adults. Despite the advances in the combination of surgery, chemotherapy, and radiotherapy, improvements in patient survival are very limited.35 Therefore, exploring new therapeutic approaches to prolong the survival of glioma patients is urgently needed. Immunotherapy is a potential option for glioma.36 In particular, immune checkpoint blockade (eg, CTLA4, PD-1)37,38 and chimeric antigen receptor T-cell immunotherapy (CAR-T)39 have exhibited potential benefits in the treatment of glioma. Nevertheless, current immunotherapies for glioma are difficult to be widely applied because of the low immunogenic response rate, therapeutic resistance, as well as autoimmune-like side effects. It is thus of great importance to develop an effective immunotherapeutic option for glioma by targeting novel molecular markers as well as molecules that have crucial roles in immunosuppression.
Previously, we found that MIR155HG is elevated in GBM and predicted a worse overall survival for GBM patients.11 Herein, we further identified that MIR155HG expression levels are elevated in IDH wild-type and mesenchymal subtype gliomas, indicating that MIR155HG was associated with more malignant biological processes. In addition, MIR155HG may serve as an indicator of IDH wild-type as well as mesenchymal subtype, and an independent prognostic factor for glioma. DNA methylation, which is the main epigenetic modification, plays various roles in the initiation and progression of different tumors.40,41 Aberrantly methylated promoters are potential markers for glioma diagnosis as well as prognosis.42,43 We took advantage of TCGA RNA-seq and DNA methylation data and demonstrated that the promoter region of MIR155HG was aberrantly hypomethylated in GBM than in LGG. The promoter methylation levels were inversely correlated with MIR155HG expression and had a predictive value for glioma patients, which was consistent with the results found for mRNA levels. In vitro experiments further confirmed that GBM tissues had markedly low methylation levels of the MIR155HG promoter region when compared to LGG tissues, and MIR155HG expression was remarkably dose-dependently upregulated after 5-Aza-dC treatment in glioma cell lines, which indicated that MIR155HG promoter region hypomethylation may be highly associated with the elevated MIR155HG expression levels in glioma.
DNA demethylation is involved in the transcriptional reactivation of viral genes in cancer.44 Abnormal DNA methylation may also lead to abnormal lncRNA expression in tumor progression. DNA demethylation enhances the accessibility of promoter regions to transcription factors. In this study, specific SP1 binding sites were found to be localized in the CpG island region of MIR155HG promoter. SP1 is ubiquitously expressed and activates target gene transcription by binding GC-rich promoter elements, which are often interrupted by DNA methylation. We found that overexpression of SP1 enhanced the expression of MIR155HG and the transcriptional activity of MIR155HG promoter in glioma cells. However, M.SssI-induced hypermethylation of the MIR155HG promoter region abrogated this activation, indicating that SP1-associated transcriptional activation of MIR155HG promoter was suppressed by DNA methylation. MIR155HG promoter hypomethylation may result in increased SP1 binding and subsequent transactivation. This partially explained why the expression of MIR155HG in GBM with low MIR155HG promoter methylation was markedly higher than that in LGG with high methylation.
Immune checkpoint blockade, the most promising approach to achieve encouraging antitumor effects, has shown great progress for the treatment of various advanced solid malignancies.45,46 Immune checkpoint blockade takes advantage of tumor immune cell infiltration to reactivate an anti-tumor immune response that is effective. We evaluated the association between MIR155HG and immune checkpoints, such as TIM-3, PD-1, B7-H3, PD-L1, CTLA4, and PD-L2, and found that MIR155HG had a high concordance with these checkpoint members, consistent with previous findings.28 These findings implied the potential regulatory effects of MIR155HG on immune checkpoints and provided potential strategies for glioma treatment.
The TME is comprised of a diverse mixture of nontumor cells, including immune cells, stromal cells, and vascular endothelial cells, as well as ECM. The TME usually suppresses efficient lymphocytic initiation, prevents their infiltration, and inhibits effector cell infiltration, leading to tumor progression, drug resistance, immune evasion, and immune suppression.47 The composition of TME has also been shown to influence outcomes of immune checkpoint blockade therapy. Herein, we found that MIR155HG plays a role in immune and inflammatory response as well as in the TME of glioma. Furthermore, there was a significant correlation between MIR155HG expression and infiltrating stromal and immune cells, such as plasma cells, monocytes, macrophages, and neutrophils. In addition, MIR155HG was prominently correlated with a series of immunosuppressive factors in glioma, implying a vital function of MIR155HG in tumor immune regulation. Besides, MIR155HG expression levels were positively related to marker genes of macrophages and key factors that promote macrophage differentiation towards the M2 phenotype, suggesting that MIR155HG has a regulatory function in TAMs polarization. Moreover, MIR155HG expression levels were notably positively associated with chemokines or cytokines involved in immunosuppressive cell recruitment, including tumor-associated neutrophils, TAMs, and myeloid-derived suppressor cells, as well as immunosuppressive factors secreted by these cells. The EMT has a vital role in tumor immunosuppression as well as immune evasion.48 The interaction between EMT genes and the TME may facilitate tumor progression. Additionally, EMT is associated with the activation of different immune checkpoint molecules, such as PD-1, PD-L2, TIM-3, PD-L1, B7-H3, and CTLA4.49 EMT scores may also provide a novel biomarker for the prediction of clinical responses to immune checkpoint blockade.50 Previously, we have found that MIR155HG promoted the progression and EMT of glioma.11 Additionally, Cui et al showed that MIR155HG promoted laryngeal squamous cell carcinoma EMT through miR-155-5p/SOX10 axis regulation.17 In the present study, we further confirmed that MIR155HG-related genes are mainly enriched in the extracellular region and MIR155HG was significantly associated with EMT markers. Accordingly, we speculated that MIR155HG is involved in immunosuppressive microenvironment regulation and facilitates tumor immune evasion in glioma by regulating M2 polarization, recruiting immunosuppressive cells, increasing the secretion of immunosuppressive factors, and promoting EMT.
Mechanistically, MIR155HG may modulate the tumor immunity through Wnt/β-catenin signaling pathway activation. Previously, we showed that MIR155HG activated β-catenin expression via miR-155/PCDHs axis in glioma. Recently, He et al found that MIR155HG enhances temozolomide resistance through Wnt/β-catenin pathway activation by binding PTBP1 in glioma. These findings highlight the important role of Wnt/β-catenin signaling pathway in the downstream regulation of MIR155HG. Aberrant Wnt/β‐catenin pathway activation is implicated in various cancer types and is a key regulator of the TME.51 Noteworthily, Wnt/β-catenin signaling as well as its interactions with immune cells have both negative and positive effects on anticancer immunosurveillance. It is involved in leucocyte maintenance and renewal while promoting immune tolerance, immune evasion and limits the antitumor immune response.52 Furthermore, Wnt/β-catenin signaling also plays a crucial role in EMT, thus, it enhances cancer stem cell maintenance. Based on these findings, we postulate that MIR155HG modulates tumor-immune interactions via the Wnt/β-catenin signaling pathway. These findings should be verified by further molecular biology or cell experiments.
Conclusion
In conclusion, we established a novel role of MIR155HG in the promotion of malignant phenotypes which enhance glioma immune evasion. Therefore, MIR155HG is a potential novel immunotherapeutic target for glioma.
Acknowledgments
We appreciate the generosity of TCGA and CGGA networks for sharing a great amount of data.
Funding Statement
This work was supported by Jiangsu Province’s Science and Technology Project (SBK2019021295), and National Natural Science Foundation of China (82072791).
Data Sharing Statement
Publicly available datasets were analyzed in this study. TCGA dataset can be found at: https://xenabrowser.net/datapages/?cohort=TCGA%20lower%20grade%20glioma%20and%20glioblastoma%20(GBMLGG)&removeHub$=$https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443,GSE16011; CGGA dataset can be found at http://www.cgga.org.cn/download.jsp.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. These authors have contributed equally to this work and share first authorship: Xuechao Wu, Quan Wan, and Jing Wang.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Buckner JC, Brown PD, O’Neill BP, Meyer FB, Wetmore CJ, Uhm JH. Central nervous system tumors. Mayo Clin Proc. 2007;82(10):1271–1286. doi: 10.4065/82.10.1271 [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126 [DOI] [PubMed] [Google Scholar]
- 3.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quail DF, Bowman RL, Akkari L, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352(6288):aad3018. doi: 10.1126/science.aad3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutmann DH. Microglia in the tumor microenvironment: taking their TOLL on glioma biology. Neuro Oncol. 2015;17(2):171–173. doi: 10.1093/neuonc/nou346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain RK, Di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8(8):610–622. doi: 10.1038/nrn2175 [DOI] [PubMed] [Google Scholar]
- 8.Ye XZ, Xu SL, Xin YH, et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-β1 signaling pathway. J Immunol. 2012;189(1):444–453. doi: 10.4049/jimmunol.1103248 [DOI] [PubMed] [Google Scholar]
- 9.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi: 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C, Wei Y, Wang X, et al. DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer. 2020;19(1):28. doi: 10.1186/s12943-020-1137-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Wang Y, Yu T, et al. Blocking MIR155HG/miR-155 axis inhibits mesenchymal transition in glioma. Neuro Oncol. 2017;19(9):1195–1205. doi: 10.1093/neuonc/nox017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu W, Yu T, Wu Y, Tian W, Zhang J, Wang Y. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J Exp Clin Cancer Res. 2019;38(1):133. doi: 10.1186/s13046-019-1132-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X, Sheng J, Yu W, Wang K, Zhu S, Liu Q. LncRNA MIR155HG promotes temozolomide resistance by activating the wnt/beta-catenin pathway via binding to PTBP1 in glioma. Cell Mol Neurobiol. 2021;41:1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Ye Y, Tian W, Qiu H. A novel lncRNA panel related to ferroptosis, tumor progression, and microenvironment is a robust prognostic indicator for glioma patients. Front Cell Dev Biol. 2021;9:788451. doi: 10.3389/fcell.2021.788451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Li J, Lin F, Guo J, Zhao J. Identification of N(6)-methyladenosine-related lncRNAs for patients with primary glioblastoma. Neurosurg Rev. 2021;44(1):463–470. doi: 10.1007/s10143-020-01238-x [DOI] [PubMed] [Google Scholar]
- 16.Yu K, Yang H, Lv QL, et al. Construction of a competitive endogenous RNA network and analysis of potential regulatory axis targets in glioblastoma. Cancer Cell Int. 2021;21(1):102. doi: 10.1186/s12935-021-01789-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui W, Meng W, Zhao L, Cao H, Chi W, Wang B. TGF-β-induced long non-coding RNA MIR155HG promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the miR-155-5p/SOX10 axis. Int J Oncol. 2019;54(6):2005–2018. doi: 10.3892/ijo.2019.4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin Y, Liu X, Pan L, Zhou R, Zhang X. Long noncoding RNA MIR155HG facilitates pancreatic cancer progression through negative regulation of miR-802. J Cell Biochem. 2019;120(10):17926–17934. doi: 10.1002/jcb.29060 [DOI] [PubMed] [Google Scholar]
- 19.Tao M, Zhou Y, Jin Y, Pu J. Blocking lncRNA MIR155HG/miR-155-5p/-3p inhibits proliferation, invasion and migration of clear cell renal cell carcinoma. Pathol Res Pract. 2020;216(2):152803. doi: 10.1016/j.prp.2019.152803 [DOI] [PubMed] [Google Scholar]
- 20.Ren XY, Han YD, Lin Q. Long non-coding RNA MIR155HG knockdown suppresses cell proliferation, migration and invasion in NSCLC by upregulating TP53INP1 directly targeted by miR-155-3p and miR-155-5p. Eur Rev Med Pharmacol Sci. 2020;24(9):4822–4835. doi: 10.26355/eurrev_202005_21171 [DOI] [PubMed] [Google Scholar]
- 21.Amin V, Harris RA, Onuchic V, et al. Epigenomic footprints across 111 reference epigenomes reveal tissue-specific epigenetic regulation of lincRNAs. Nat Commun. 2015;6:6370. doi: 10.1038/ncomms7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230 [DOI] [PubMed] [Google Scholar]
- 23.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–419. doi: 10.1016/j.it.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores-Concha M, Onate AA. Long non-coding RNAs in the regulation of the immune response and trained immunity. Front Genet. 2020;11:718. doi: 10.3389/fgene.2020.00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. doi: 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Wang L, Fan J, et al. Host miR155 promotes tumor growth through a myeloid-derived suppressor cell-dependent mechanism. Cancer Res. 2015;75(3):519–531. doi: 10.1158/0008-5472.CAN-14-2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Z, Sun R, Zhao HJ, et al. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol Cancer. 2019;18(1):54. doi: 10.1186/s12943-019-0977-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng L, Chen Z, Chen Y, Wang X, Tang N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019;8(17):7161–7173. doi: 10.1002/cam4.2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu X, Hou P, Qiu Y, Wang Q, Lu X. Large-scale analysis reveals the specific clinical and immune features of DGCR5 in glioma. Onco Targets Ther. 2020;13:7531–7543. doi: 10.2147/OTT.S257050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi: 10.1038/nn.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prosniak M, Harshyne LA, Andrews DW, et al. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin Cancer Res. 2013;19(14):3776–3786. doi: 10.1158/1078-0432.CCR-12-1940 [DOI] [PubMed] [Google Scholar]
- 32.Lu-Emerson C, Snuderl M, Kirkpatrick ND, et al. Increase in tumor-associated macrophages after antiangiogenic therapy is associated with poor survival among patients with recurrent glioblastoma. Neuro Oncol. 2013;15(8):1079–1087. doi: 10.1093/neuonc/not082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Q, Long W, Xing C, et al. Cancer stem cells and immunosuppressive microenvironment in glioma. Front Immunol. 2018;9:2924. doi: 10.3389/fimmu.2018.02924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motz GT, Coukos G. Deciphering and reversing tumor immune suppression. Immunity. 2013;39(1):61–73. doi: 10.1016/j.immuni.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woehrer A, Bauchet L, Barnholtz-Sloan JS. Glioblastoma survival: has it improved? Evidence from population-based studies. Curr Opin Neurol. 2014;27(6):666–674. doi: 10.1097/WCO.0000000000000144 [DOI] [PubMed] [Google Scholar]
- 36.Dunn GP, Cloughesy TF, Maus MV, Prins RM, Reardon DA, Sonabend AM. Emerging immunotherapies for malignant glioma: from immunogenomics to cell therapy. Neuro Oncol. 2020;22(10):1425–1438. doi: 10.1093/neuonc/noaa154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11(9):504–514. doi: 10.1038/nrneurol.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genoud V, Marinari E, Nikolaev SI, et al. Responsiveness to anti-PD-1 and anti-CTLA-4 immune checkpoint blockade in SB28 and GL261 mouse glioma models. Oncoimmunology. 2018;7(12):e1501137. doi: 10.1080/2162402X.2018.1501137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, You Y, Shen Z, Shi L. EGFRvIII-CAR-T cells with PD-1 knockout have improved anti-glioma activity. Pathol Oncol Res. 2020;26(4):2135–2141. doi: 10.1007/s12253-019-00759-1 [DOI] [PubMed] [Google Scholar]
- 40.Jia D, Lin W, Tang H, et al. Integrative analysis of DNA methylation and gene expression to identify key epigenetic genes in glioblastoma. Aging. 2019;11(15):5579–5592. doi: 10.18632/aging.102139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–786. doi: 10.1016/S0140-6736(18)31268-6 [DOI] [PubMed] [Google Scholar]
- 42.Su J, Ma Q, Long W, et al. LCTL is a prognostic biomarker and correlates with stromal and immune infiltration in gliomas. Front Oncol. 2019;9:1083. doi: 10.3389/fonc.2019.01083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong X, Deng Q, Nie X, et al. Downregulation of HTATIP2 expression is associated with promoter methylation and poor prognosis in glioma. Exp Mol Pathol. 2015;98(2):192–199. doi: 10.1016/j.yexmp.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 44.Tian HP, Lun SM, Huang HJ, et al. DNA methylation affects the SP1-regulated transcription of FOXF2 in breast cancer cells. J Biol Chem. 2015;290(31):19173–19183. doi: 10.1074/jbc.M114.636126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–1355. doi: 10.1126/science.aar4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadreddini S, Baradaran B, Aghebati-Maleki A, et al. Immune checkpoint blockade opens a new way to cancer immunotherapy. J Cell Physiol. 2019;234(6):8541–8549. doi: 10.1002/jcp.27816 [DOI] [PubMed] [Google Scholar]
- 47.Tang H, Qiao J, Fu YX. Immunotherapy and tumor microenvironment. Cancer Lett. 2016;370(1):85–90. doi: 10.1016/j.canlet.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dongre A, Rashidian M, Reinhardt F, et al. Epithelial-to-mesenchymal transition contributes to immunosuppression in breast carcinomas. Cancer Res. 2017;77(15):3982–3989. doi: 10.1158/0008-5472.CAN-16-3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lou Y, Diao L, Cuentas ER, et al. Epithelial-mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin Cancer Res. 2016;22(14):3630–3642. doi: 10.1158/1078-0432.CCR-15-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mak MP, Tong P, Diao L, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res. 2016;22(3):609–620. doi: 10.1158/1078-0432.CCR-15-0876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel S, Alam A, Pant R, Chattopadhyay S. Wnt signaling and its significance within the tumor microenvironment: novel therapeutic insights. Front Immunol. 2019;10:2872. doi: 10.3389/fimmu.2019.02872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan Y, Ogana H, Gang E, Kim HN, Kim YM. Wnt signaling in the tumor microenvironment. Adv Exp Med Biol. 2021;1270:107–121. [DOI] [PMC free article] [PubMed] [Google Scholar]