Abstract
We have demonstrated by using an in vitro approach that interruption of the OmpK36 porin gene by insertion sequences (ISs) is a common type of mutation that causes loss of porin expression and increased resistance to cefoxitin in Klebsiella pneumoniae. This mechanism also operates in vivo: of 13 porin-deficient cefoxitin-resistant clinical isolates of K. pneumoniae, 4 presented ISs in their ompK36 gene.
Klebsiella pneumoniae is a major nosocomial pathogen, and successful chemotherapy is critical for the treatment of infections caused by this microorganism. Antibiotic-resistant strains emerge with variable frequency, particularly in the hospital environment. K. pneumoniae resistance to expanded-spectrum cephalosporins caused by plasmid-borne extended-spectrum β-lactamases is a well-documented example and has increased since the 1980s (11, 14, 17). Additionally, effects in porin expression contribute to increase the resistance levels provided by the mechanism described above (12).
We have described porins OmpK36 and OmpK35 (2, 7), the K. pneumoniae homologues of Escherichia coli porins OmpC and OmpF, respectively. Clinical isolates of this species express either OmpK36 or OmpK35 porins or both (6). Recently, we reported that loss of porins OmpK36 and OmpK35 in extended-spectrum β-lactamase-producing clinical isolates of K. pneumoniae caused increased resistance to cefoxitin and expanded-spectrum cephalosporins (3, 9). The relationship between porin loss and antimicrobial resistance has also been demonstrated in other bacterial species (1, 10). However, the mechanisms leading to porin deficiency and subsequent antimicrobial resistance are largely unknown. The purpose of the present study was to identify the mechanisms causing porin deficiency in clinical isolates of K. pneumoniae. For this purpose, we first used an in vitro approach. We selected cefoxitin-resistant mutants derived from strain LB4(pSHA2) (9). Strain LB4 is a porin-deficient, cefoxitin-resistant (MIC, 128 μg/ml) clinical isolate that reverted to cefoxitin sensitivity (MIC, 4 μg/ml) after the plasmid pSHA2 carrying the OmpK36 porin gene was cloned. Cefoxitin-resistant mutants were obtained by plating LB4(pSHA2) on increasing concentrations of cefoxitin. Mutants with increased resistance to cefoxitin (MIC, >48 μg/ml), determined according to the National Committee for Clinical Laboratory Standards recommendations, were selected and further characterized. The outer membrane proteins (OMPs) from these mutants were isolated from bacterial envelopes as sodium lauryl sarcosinate-insoluble material (5) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. We found that expression of porin OmpK36 was completely abolished in those mutants. In order to study possible mutations affecting the ompK36 gene, we isolated the pSHA2 plasmid from the cefoxitin-resistant mutants. Plasmids were analyzed by Southern blotting (4) with an ompK36 probe obtained by PCR amplification of plasmid pSHA2 with primers U228 (5′-GGTAAAAAAAACCGGATGCG-3′) and L1730 (5′-CGTGCTTAGAACTGGTAAACC-3′), which anneal to sequences 427 bp upstream and 1,097 bp downstream of the ompK36 start codon (accession no. Z33506), respectively.
These analyses revealed that different events occurred in the ompK36 gene contained in pSHA2: point mutations or small deletions, deletions of the entire gene, and insertions. These mutant plasmids derived from pSHA2 were designated pSHA followed by an arbitrary number (Table 1). As shown in Table 1, the most frequent types of changes detected in the mutant plasmids were insertions. Among 14 randomly selected mutants, 9 (64.3%) presented an insertion. Detailed characterization of the nature and position of the inserted DNA was carried out by sequencing the mutant plasmids. DNA sequencing was performed with primers U681 (5′-CGGTTACGGCCAGTGGGAATA-3′) and L1316 (5′-GACGCAGACCGAAATCGAACT-3′), which represent DNA sequences 230 and 345 bp downstream of the ompK36 start codon, and primers U228 and L1730. We detected four different insertion sequences (ISs; IS26, IS5, IS903, and IS1) located in different sites of the ompK36 gene. Furthermore, with probes specific for the identified ISs (obtained by elution of an IS internal restriction fragment from the corresponding mutant plasmid), we verified by Southern blot analysis that the ISs were present in many copies in the chromosome of the original strain LB4 (data not shown).
TABLE 1.
Analysis of randomly selected mutant plasmids conferring increased resistance to cefoxitin (MIC, >48 μg/ml) derived from strain LB4(pSHA2)
| Plasmid | Type of mutation | IS positiona |
|---|---|---|
| pSHA119 | Point mutation or small deletion | |
| pSHA124 | Point mutation or small deletion | |
| pSHA125 | Point mutation or small deletion | |
| pSHA128 | Insertion of IS1 | 87 |
| pSHA123 | Insertion of IS903 | 8 |
| pSHA103 | Insertion of IS5 | −4 |
| pSHA100 | Insertion of IS26 | 14 |
| pSHA146 | Insertion of IS1 | 95 |
| pSHA149 | Insertion of IS5 | 7 |
| pSHA151 | Insertion of IS5 | 29 |
| pSHA101 | Insertion of IS26 | 77 |
| pSHA154 | Insertion of IS903 | 28 |
| pSHA157 | Deletion | |
| pSHA104 | Deletion |
Values indicate the positions (in base pairs) of the ISs with respect to the ompK36 start codon.
Since resistance to cefoxitin in laboratory-derived mutants was dependent on mutations of the OmpK36 porin gene, predominantly insertion of ISs, we investigated whether this type of mechanism also occurred in vivo. For this purpose, we amplified the entire ompK36 gene from two pairs of strains isolated from two patients before and after antimicrobial therapy: strains LB1 and LB4 (9) and strains CMD1 and CMD2 (19). LB4 and CMD2 are porin-deficient cefoxitin-resistant clinical isolates derived from the porin-sufficient, cefoxitin-sensitive strains LB1 and CMD1, respectively. Additionally, we characterized the OmpK36 porin gene in 11 additional porin-deficient K. pneumoniae clinical isolates: strains HUSR2/94, C1, LB66, LB68, LB73, CSUB2, CSUB8R, CSUB9R, CSUB10R, CSUB11, and CSUB12.
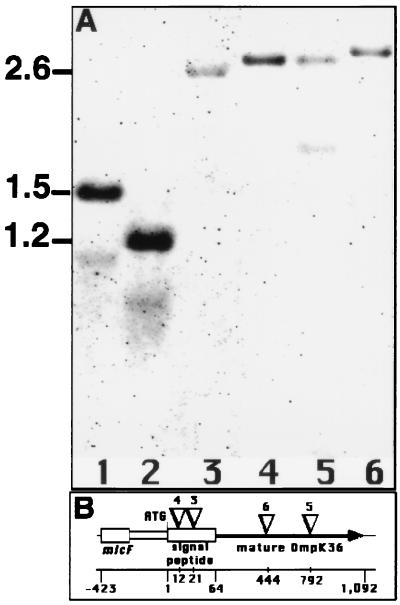
Analysis of the amplicons with the ompK36 gene probe detected insertions, seen as increases in the size of ompK36, in some of these clinical isolates (Fig. 1A). In addition, we observed in strain HUSR2/94 a deletion on the OmpK36 porin gene (Fig. 1A, lane 2). Further characterization of the above-described insertions was obtained by sequencing the PCR amplicons. We identified the presence of IS102 in strain CMD2, and we identified insertions with a high degree of sequence identity to IS5 in the amplicons from strains LB73, CSUB2, and CSUB11. These ISs were located in different positions within the ompK36 coding region (Fig. 1B). These results demonstrate that porin loss due to insertions of ISs within the porin genes is a mechanism that operates in vivo in patients undergoing antibiotic treatment.
FIG. 1.
(A) PCR and Southern blot analysis of the OmpK36 porin gene from porin-deficient clinical isolates. The entire ompK36 gene was amplified by using PCR and the U228-L1730 primer pair. Amplification products were analyzed with the ompK36 gene probe. Lanes: 1, strain CMD1; 2, strain HUSR2/94; 3, strain CMD2; 4, strain LB73; 5, strain CSUB2; 6, strain CSUB11. (B) Scheme of the ompK36 gene, its upstream micF gene, and the positions of the ISs. ISs are represented by inverted triangles and the numbers above them correspond to the lanes in panel A. The positions (in base pairs) of genes and ISs with respect to the ompK36 start codon are indicated on the bottom line.
It has been suggested that a low level of random transposition might help cells to adapt to environmental changes and to increase the survival rate (18), and it is also well known that ISs can interrupt or alter gene expression. There are many examples in the literature where ISs have been involved in resistance to certain antibiotics by creating novel promoters for antimicrobial resistance genes (8, 15). However, to our knowledge, this is the first report where antimicrobial resistance is caused by interruption of porin genes by ISs. Mechanisms of porin loss and increased resistance to imipenem were also studied in Pseudomonas aeruginosa by Yoneyama and Nakae by using an in vitro approach (20): only deletions in the OprD2 porin gene were observed. However, we have shown that, both in vitro and in vivo, ISs cause porin deficiency in K. pneumoniae. ISs are widespread in all bacterial species studied, and, although their presence has not been correlated with antimicrobial resistance, they have also been found in E. coli porins (13, 16). Thus, interruption of porin genes by ISs is therefore not an exclusive phenomenon of K. pneumoniae. Further studies will be required to determine the importance of this mechanism in other bacterial species.
Acknowledgments
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (CICYT). S.H.A. and S.A. were supported by a predoctoral fellowship and a postdoctoral contract from CICYT, respectively.
We thank J. A. M. van de Klundert (University Hospital Leiden), G. Jacoby (Lahey Hitchcock Clinic), and J. Liñares and C. Ardanuy (Hospital de Bellvitge) for clinical isolates, the Servicio de Secuenciación del Centro de Investigaciones Biológicas for DNA sequencing, and J. Casadesús (Universidad de Sevilla) for helpful discussions throughout the work.
REFERENCES
- 1.Aggeler R, Then R, Ghosh R. Reduced expression of outer-membrane proteins in β-lactam-resistant mutants of Enterobacter cloacae. J Gen Microbiol. 1987;133:3383–3392. doi: 10.1099/00221287-133-12-3383. [DOI] [PubMed] [Google Scholar]
- 2.Albertí S, Rodríguez-Quiñones F, Schirmer T, Rummel G, Tomás J M, Rosenbusch J P, Benedí V J. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional structure, and complement binding. Infect Immun. 1995;63:903–910. doi: 10.1128/iai.63.3.903-910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ardanuy C, Liñares J, Domínguez M A, Hernández-Allés S, Benedí V J, Martínez-Martínez L. Outer membrane profiles of clonally related Klebsiella pneumoniae isolated from clinical samples and activity of cephalosporins and carbapenems. Antimicrob Agents Chemother. 1998;42:1636–1640. doi: 10.1128/aac.42.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1997. [Google Scholar]
- 5.Filip C, Fletcher G, Wulf J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernández-Allés S, Albertí S, Alvarez D, Doménech-Sánchez A, Martínez-Martínez L, Gil J, Tomás J M, Benedí V J. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology. 1999;145:673–679. doi: 10.1099/13500872-145-3-673. [DOI] [PubMed] [Google Scholar]
- 7.Hernández-Allés S, Albertí S, Rubirés X, Merino S, Tomás J M, Benedí V J. Isolation of FC3-11, a bacteriophage specific for the Klebsiella pneumoniae porin OmpK36, and its use for the isolation of porin-deficient mutants. Can J Microbiol. 1996;41:399–406. [Google Scholar]
- 8.Jaurin B, Normark S. Insertion of IS2 creates a novel ampC promoter in Escherichia coli. Cell. 1983;32:809–816. doi: 10.1016/0092-8674(83)90067-3. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Martínez L, Hernández-Allés S, Albertí S, Tomás J M, Benedí V J, Jacoby G A. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1996;40:342–348. doi: 10.1128/aac.40.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medeiros A A, O’Brien T F, Rosenberg E Y, Nikaido H. Loss of OmpC porin in a strain of Salmonella typhimurium causes increased resistance to chephalosporins during therapy. J Infect Dis. 1987;156:751–757. doi: 10.1093/infdis/156.5.751. [DOI] [PubMed] [Google Scholar]
- 11.Meyer K S, Urban C, Eagan J A, Berger B J, Rahal J J. Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Ann Intern Med. 1993;119:353–358. doi: 10.7326/0003-4819-119-5-199309010-00001. [DOI] [PubMed] [Google Scholar]
- 12.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozawa Y, Mizushima S, Mizuno T. Osmoregulatory expression of the ompC gene in Escherichia coli K-12; IS1 insertion in the upstream regulatory region results in constitutive activation of the promoter. FEMS Microbiol Lett. 1990;68:295–300. doi: 10.1016/s0378-1097(05)80057-6. [DOI] [PubMed] [Google Scholar]
- 14.Philippon A, Labia R, Jacoby G A. Extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podglajen I, Breuil J, Collatz E. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol Microbiol. 1994;12:105–114. doi: 10.1111/j.1365-2958.1994.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 16.Prilipov A, Phale P S, Koebnik R, Widmer C, Rosenbusch J P. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J Bacteriol. 1998;180:3388–3392. doi: 10.1128/jb.180.13.3388-3392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice L B, Willey S H, Papanicolau A, Medeiros A A, Eliopoulos G M, Moellering R C, Jacoby G A. Outbreak of ceftazidime resistance caused by extended-spectrum β-lactamases at a Massachusetts chronic-care facility. Antimicrob Agents Chemother. 1990;34:2193–2199. doi: 10.1128/aac.34.11.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syvanen M. The evolutionary implications of mobile genetic elements. Annu Rev Genet. 1984;18:271–293. doi: 10.1146/annurev.ge.18.120184.001415. [DOI] [PubMed] [Google Scholar]
- 19.van de Klundert J A M, van Gestel M H, Meerdink G, de Marie S. Emergence of bacterial resistance to cefamandole in vivo due to outer membrane protein deficiency. Eur J Clin Microbiol Infect Dis. 1988;7:776–777. doi: 10.1007/BF01975046. [DOI] [PubMed] [Google Scholar]
- 20.Yoneyama H, Nakae T. Mechanism of efficient elimination of protein D2 in outer membrane of imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993;37:2385–2390. doi: 10.1128/aac.37.11.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]