Abstract
Myogenic differentiation, the irreversible developmental process where precursor myoblast muscle stem cells become contractile myotubes, is heavily regulated by glycosylation and glycan–protein interactions at the cell surface and the extracellular matrix. The glycan-binding protein galectin-1 has been found to be a potent activator of myogenic differentiation. While it is being explored as a potential therapeutic for muscle repair, a precise understanding of its glycoprotein interactors is lacking. These gaps are due in part to the difficulties of capturing glycan–protein interactions in live cells. Here, we demonstrate the use of a proximity tagging strategy coupled with quantitative mass-spectrometry-based proteomics to capture, enrich, and identify the glycan-mediated glycoprotein interactors of galectin-1 in cultured live mouse myoblasts. Our interactome dataset can serve as a resource to aid the determination of mechanisms through which galectin-1 promotes myogenic differentiation. Moreover, it can also facilitate the determination of the physiological glycoprotein counter-receptors of galectin-1. Indeed, we identify several known and novel glycan-mediated ligands of galectin-1 as well as validate that galectin-1 binds the native CD44 glycoprotein in a glycan-mediated manner.
Graphical Abstract
INTRODUCTION
The interactions between glycan-binding proteins (GBPs), such as galectins, and their cognate glycoconjugate counter-receptors at the glycocalyx or the extracellular matrix (ECM) mediate many important biological events.1,2 Galectin-1 (14 kDa), a prototypical member of the galectin family, plays critical roles in biology,3 including myogenic differentiation, the process in which mononuclear myoblasts fuse and differentiate into multinuclear and contractile myotubes. Galectin-1 has been found to be highly upregulated during myogenesis4 and in damaged muscular tissue,5 and its knockout leads to impaired differentiation.6 Recent work has found that extracellular galectin-1 and its glycan-binding activities are directly related to these functions.7,8 Hence, galectin-1 has garnered significant interest as a potential therapeutic for muscular regeneration and muscular dystrophy.9,10 Despite its utility, however, the molecular mechanisms through which it acts are relatively unknown, and a comprehensive list of glycoprotein interactors for galectin-1 has yet to be developed. A detailed understanding of these molecular mechanisms can impact our ability to repair muscle tissues in response to injury or disease.
The study of GBP–glycan binding events presents challenges that often preclude their study in live cells, where the native expression and three-dimensional presentation of ligands are preserved. Glycan biosynthesis relies on a complex ensemble of enzymes to assemble and trim glycans in a nontemplated manner, resulting in the nonuniform glycosylation of proteins, such that a single protein may exist in myriad glycoforms, which can have differing affinities for a GBP. Conversely, a single glycan structure may be present on many other proteins. While the galectin family demonstrates similar glycan-binding preferences for β-galactoside glycans (e.g., lactose), each member can exhibit specific, fine-tuned binding differences because they can rely on cues beyond the glycan itself.11,12 Thus, it may be insufficient to define glycans or glycoproteins without the context of both its glycoforms and its anchoring protein. Moreover, GBP–glycan interactions are usually weak (dissociation constant of galectin-1 to immobilized lactose and poly-LacNAc repeats is Kd ~ 14 μM), dynamic, and require multivalent presentation to achieve sufficient avidity.13 While defined glycan arrays have significantly advanced our understanding of the glycan-binding preferences of galectin-1,14 detailed information regarding the proteins carrying these glycans has yet to be identified. This information is especially important when considering the molecular mechanisms through which GBPs function. While conventional immunoprecipitation can partially identify some of these interactors, it may not capture the entire interactome and fails to recapitulate the precise three-dimensional presentation of ligands.
The study of galectin-1 binding in live cells requires close attention to its structure. Galectin-1 contains a high number of cysteines, leaving it sensitive toward oxidation, which results in loss of glycan binding.15 It also exists in a reversible monomer–dimer equilibrium in solution, spontaneously formed by hydrophobic interactions at its N- and C-termini,16 with a dimerization constant (DC50) of ~2.6–7 μM.17,18 Whereas both monomeric and dimeric forms of galectin-1 retain the ability to bind glycans, specific glycoprotein receptors seem to favor binding to dimeric or monomeric forms, suggesting dimerization as a regulatory mechanism for binding.19,20 Importantly, in cellular studies, galectin-1 is often used at high sub-millimolar ranges to favor the formation of the dimer and enhance detection of binding events.20
We recently demonstrated the use of a proximity tagging strategy to identify the glycoprotein interactors of a similar GBP, galectin-3.21 Proximity tagging is especially apt for the study of GBP–glycoprotein interactors because it converts the weak but physiologically relevant interactions into near-covalent binding events that can be efficiently and reproducibly captured. Here, we apply the same strategy toward determining the interactors for galectin-1 in live mouse myoblasts. In this method, fusion proteins of a GBP and an engineered heme-containing ascorbate peroxidase enzyme (APEX2) are created and applied to cells to covalently tag proximally located interactors with affinity tags for capture, enrichment, and identification by quantitative mass spectrometry (MS)-based proteomics.22 We show that a fusion protein of galectin-1 and APEX2 retains the ability to dimerize, similar to the wild-type protein, and that it can be used for live cell proximity tagging. We generated a list of galectin-1 glycoprotein interactors from myoblasts and validated several proteins for binding with galectin-1. Informed by pathway-based informatic analysis, we prioritized several glycoproteins for subsequent validation of binding.
RESULTS AND DISCUSSION
Expression and Characterization of Fusion Construct.
We designed and expressed human galectin-1 fusion proteins with APEX2 fused either at the N- (NPG1) or C-terminus (CPG1) (Figure 1A and Figure S1). APEX2 was chosen because of its small size (28 kDa), potency in extracellular environments where protein–glycan interactions dominate, as well as its monomeric nature.22 Given their high sequence homology (88%), human and mouse galectin-1 have been used interchangeably in previous studies.9 Therefore, we reasoned that human galectin-1 could be used as a model for studies in mouse myoblasts. Both NPG1 and CPG1 constructs include a spacer sequence between APEX2 and galectin-1 to confer conformational flexibility, as galectin-1 can undergo significant structural changes.23–25 As expected, the expression of both constructs in Escherichia coli resulted in ~43 kDa bands by gel electrophoresis (Figure 1B and Figure S1). The proteins were expressed in high (~20 mg/L culture) yields and spontaneously multimerized, and they consistently retained high (>70%) heme occupancies.22
Figure 1.
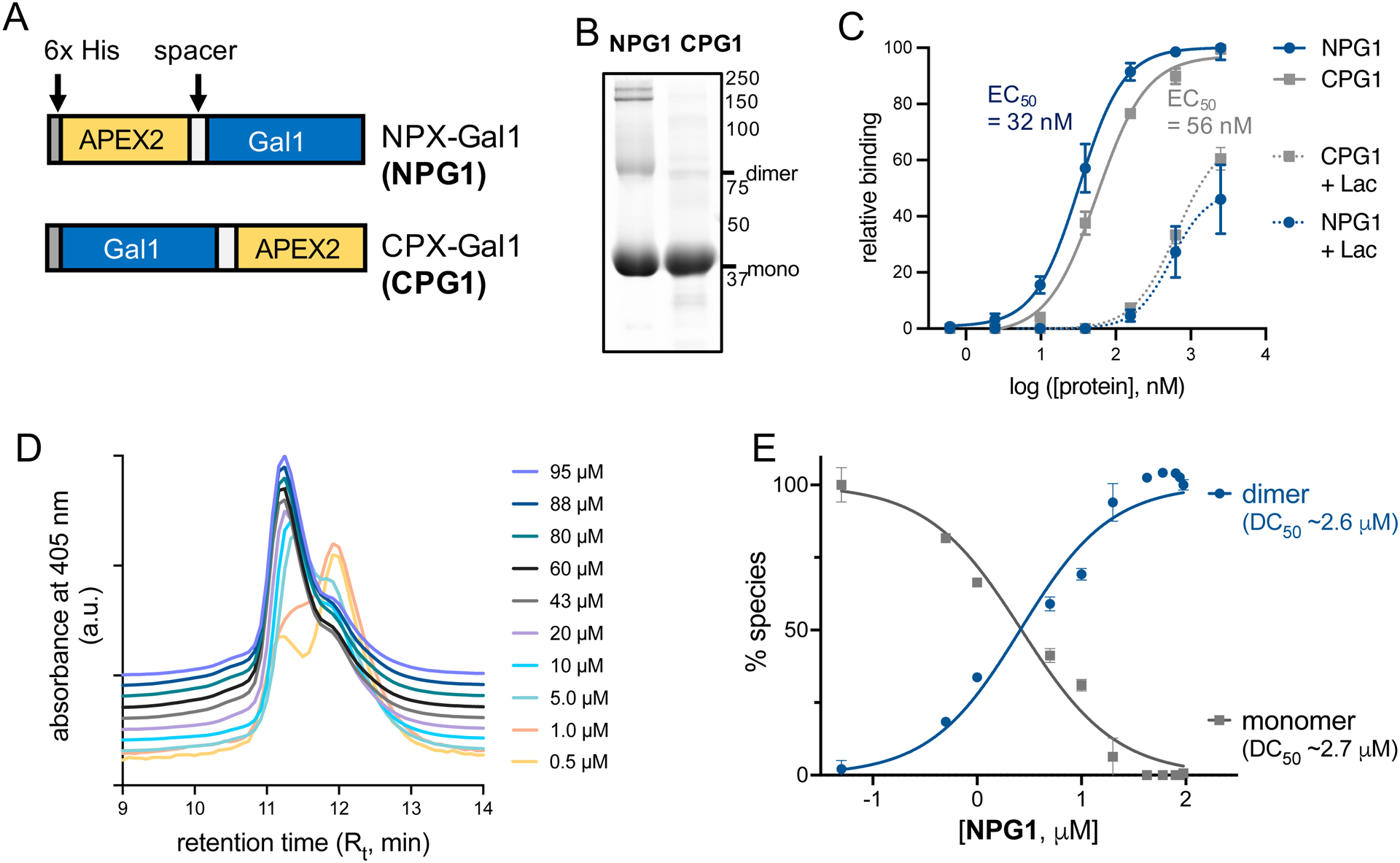
Expression and characterization of galectin-1 fusion constructs. (A) Fusion constructs of galectin-1 and the APEX2 enzyme. Constructs consist of an N-terminal 6x His tag for protein purification followed by galectin-1, with the APEX2 enzyme joined at the N- (NPG1) or C-terminus (CPG1) by a short spacer sequence. (B) SDS-PAGE analysis of purified NPG1 and CPG1 reveals spontaneous formation of higher molecular weight species at high concentrations (Figure S1). (C) ELISA using immobilized asialofetuin indicates both NPG1 and CPG1 retain their glycan binding activities with similar apparent affinity constants (EC50) measured at 32 and 56 nM, respectively. Error bars indicate standard deviation of three independent replicates. (D) Size exclusion chromatograms (offset at y-axis, overlaid) of NPG1 constructs pre-equilibrated at a range of concentrations (0.5–95 μM) and detected by absorbance at 405 nm of the APEX2 heme cofactor. The monomeric species (Rt ~ 12 min) dominates the equilibrium at lower concentrations, and the formation of the dimeric species (Rt ~ 11 min) is favored at higher concentrations. (E) Plotting the abundance of each yields a dimerization constant (DC50) ~ 2.7 μM. Errors bars represent standard deviation of two independent replicates.
To confirm that the fusion proteins retained their capacities for binding β-galactoside glycans, we performed an enzyme-linked immunosorbent assay (ELISA) with immobilized asialofetuin as a model glycoprotein (Figure 1C and Table S1).26,27 Both NPG1 and CPG1 bound asialofetuin in a dose-dependent manner and binding was substantially reduced upon co-incubation with an excess of the soluble competitor lactose (100 mM), indicating that the interactions are glycan-mediated. We observed a slight preference for NPG1 (EC50 ~ 32 nM) to bind asialofetuin over CPG1 (EC50 ~ 56 nM). Hence, NPG1 was chosen for subsequent evaluation. We further evaluated whether the presence of APEX2 altered the ability of NPG1 to interconvert between its monomeric and dimeric forms by size exclusion chromatography (SEC).18 Upon analyzing pre-equilibrated (20 h, 4 °C) NPG1 in various concentrations (0.5–95 μM), we observed both monomeric (~44 kDa) and dimeric (~88 kDa) NPG1 that migrated with different retention times (Figure 1D and Figure S2). Using this data, we determined a DC50 for NPG1 of ~2.6 μM, similar to previous reports for wild-type galectin-1.17
To initiate proximity tagging, we incubated NPG1 exogenously with live adherent C2C12 mouse myoblasts (Figure 2A). Upon binding its interactors and initiation of the tagging event with biotin–phenol (500 μM, 30 min) and H2O2 (1 mM, 1 min), APEX2 catalyzes the formation of highly reactive and short-lived (<1 ms) biotinyl radicals that diffuse within a short (<20 nm) radius of the complex and covalently react with electron-rich residues of the interactors.22,28 We observed significant and dose-dependent labeling of biotinylated interactors of NPG1 by fluorescence microscopy with as little as 200 nM of NPG1 (Figure 2B and Figure S3). Much of the fluorescent signal was localized at the cell surface, consistent with the heavy presence of glycans and glycoproteins at the glycocalyx. We also observed the appearance of extracellular and relatively diffuse fluorescence signals, which we attribute to the ECM (Figure S4). The fluorescence signals were abrogated with excess lactose, but were unaffected by the addition of excess sucrose, a nonbinding glycan29 (Figure S5), and a truncated protein construct containing only the APEX2 enzyme failed to generate significant fluorescence, suggesting that the glycan-binding activity of galectin-1 is required (Figure S5). While we observed significant fluorescence from a relatively short (30 min) incubation period with exogenous NPG1, we also observed labeling with prolonged culture (up to 24 h) and subsequent trafficking of NPG1 from the cell surface and ECM into concentrated intracellular locations (Figure S6), consistent with previous work tracing the fate of extracellular galectin-1 over time.30,31 These observations highlight the utility of APEX2 and proximity tagging toward capturing GBP–glycoprotein interactions at the cell surface and the ECM, as well as its ability to track the dynamic fate of galectin-1 across extracellular and intracellular localizations with different oxidative environments.32
Figure 2.
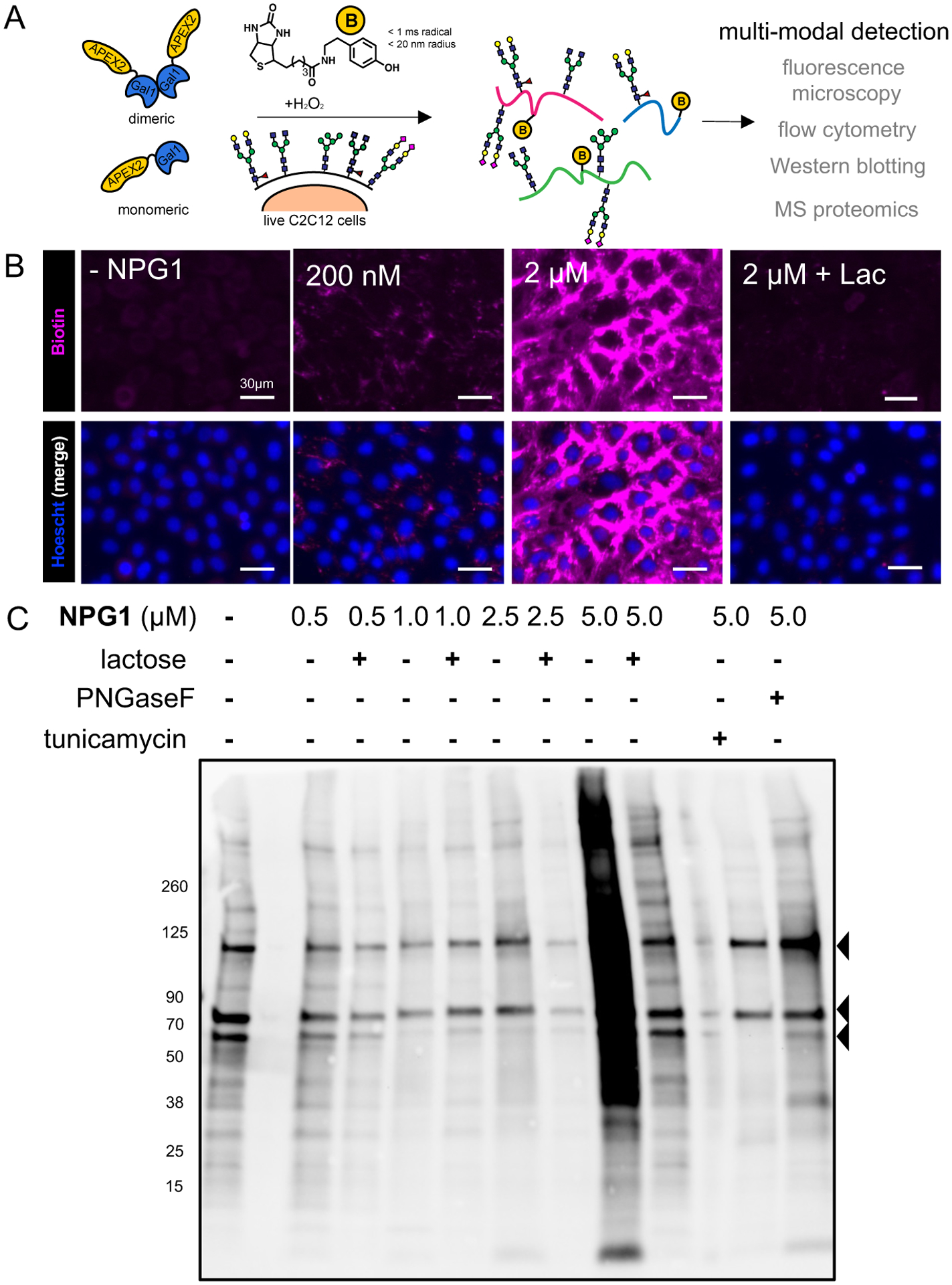
Detection of galectin-1 interactors in live myoblasts by proximity tagging. (A) Schematic illustration of the identification of galectin-1 interacting proteins in situ by proximity tagging. APEX2 catalyzes the formation of highly reactive short-lived (<1 ms) biotin–phenoxyl radicals that react within a short range (<20 nm) of diffusion. (B) Fluorescence microscopy of tagged cells demonstrates dose-dependent biotinylation (purple) with significant signal generated over the negative control (no NPG1). The addition of lactose significantly ameliorates the biotin signal. (C) Western blotting for biotin of whole cell lysates from proximity-labeled cells demonstrates dose-dependent biotinylation signals that are reduced by the addition of lactose. The addition of tunicamycin or PNGase F significantly reduces the signal, indicating that N-glycosylation is important for interaction. Arrowheads indicate endogenous biotinylated proteins, which are presumed to be pyruvate carboxylase (129 kDa), propionyl-CoA carboxylase (80 kDa), and methylcrotonoyl-CoA carboxylase (79 kDa), based on their molecular weights.
The dose-dependent increases in fluorescence was also confirmed by flow cytometry (Figure S7), as well as Western blotting of whole cell lysates (Figure 2C) and the captured biotinylated proteins (Figure S8). Similar to fluorescence imaging, we observed that many of the interactions are glycan-dependent, as competition with excess lactose, pretreatment with PNGase F (an N-glycan endoglycosidase), or with tunicamycin (an N-glycosylation inhibitor) reduced the extent of labeling. Using Western blotting permitted the observation of protein bands that remained even after glycan-mediated competition or N-glycan inhibition, suggesting that NPG1 can also interact with proteins in a non-glycan-mediated manner.33 Notably, we observed a significant increase in the number of protein bands corresponding to biotinylated proteins (Figure 2C) when cells are incubated with higher concentrations of NPG1 (5 μM), consistent with the preference for forming dimers (Figure 1C).
Determination of NPG1 Interactors by Quantitative MS.
To identify the interactors, we analyzed the captured proteins resulting from exogenous incubation of NPG1 (5 μM) with myoblasts by quantitative MS-based proteomics. To quantitatively compare protein abundances between conditions, we performed tandem mass tagging (TMT), which chemically tags tryptic peptides with unique isobaric mass tags that can be used to distinguish samples at the MS3 stage.34 As such, TMT permits the simultaneous comparison of (up to 16) multiplexed samples in a single run, ensuring high precision of the comparisons made. Across two representative biological replicates, we identified 115 (≥3 unique peptides) proteins that were highly enriched (>10-fold compared to the negative control) and statistically significant (p < 0.05; Figure 3A,B and Appendix Table 1). Of these, 74 proteins (63%) (Table S2) were glycan-mediated interactors (≥2-fold), as they were competed with excess lactose (Figure 3C). We also observed a general correlation between proteins that were enriched and those that were competed by lactose (Figure 3D). When proximity tagging was performed at the lower concentration of 1 μM NPG1, we captured a significantly smaller number (50) of glycan-mediated interactors. 47 of these proteins overlapped with the list derived from the higher concentration of NPG1 (Figure 3E) and three (mesothelin, Msln; CD166; and latent-transforming growth factor β-binding protein 1, Ltbp1) did not. Although CD166 is a known glycan-dependent galectin-1 interactor35 and galectin-1 is known to exhibit preferences for binding based on its multimerization state, it is currently unclear why these specific proteins exhibit a preference for interacting with monomeric NPG1.
Figure 3.
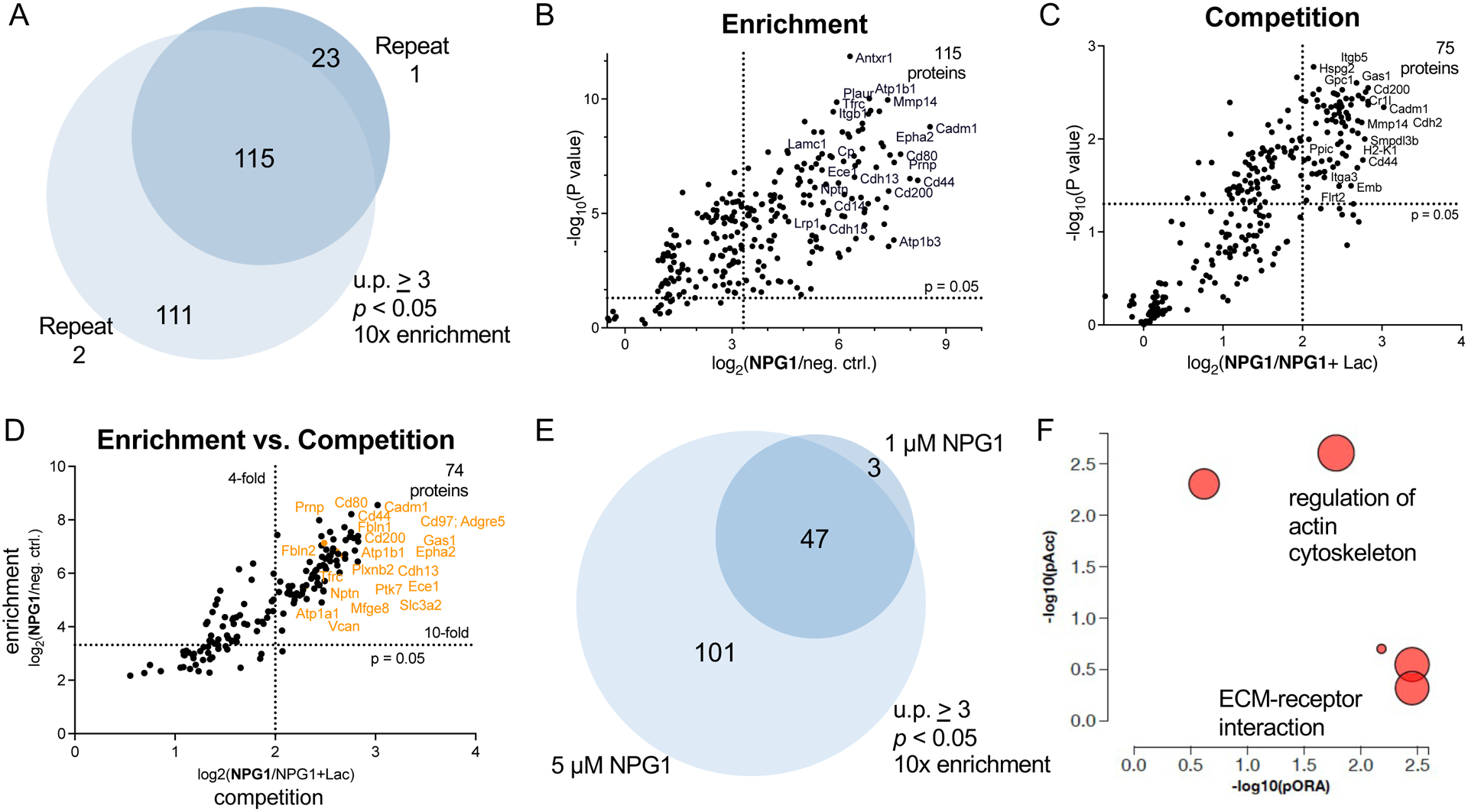
Determination of galectin-1 glycoprotein counter-receptors by quantitative MS. (A) One hundred fifteen proteins were enriched across two biological replicates. Enrichment is defined as proteins with unique peptides (u.p.) ≥ 3 and TMT ratios (5 μM NPG1/no protein) ≥ 10. The “no protein” condition indicates cells that were not treated with NPG1 but were treated with biotin–phenol and H2O2. (B) Statistically significant (p < 0.05) and enriched proteins identified. (C) Statistically significant (p < 0.05) and competed (TMT ratio 5 μM NPG1/5 μM NPG1 + lactose ≥ 4) proteins. (D) There is high agreement between proteins that were enriched and competed by lactose. (E) Fifty proteins were significantly enriched by 1 μM NPG, 47 of which were also identified by 5 μM NPG1. (F) Pathway analysis data (iPathway) based on enriched proteins represented in a perturbation vs over-representation plot. Represented pathways include the regulation of actin cytoskeleton and ECM–receptor interactions, with 14 and 17 differentially expressed members, respectively.
We observed many N-linked or O-linked glycoproteins (93%) within the 115 interactors of NPG1 (Table 1 and Table S2), some of which have yet to be assigned as galectin-1 ligands. Among these proteins are integrins, including integrin-α5/β1 (Itgα−5), which regulates cellular proliferation by adhering to ECM proteins and has been identified as an interactor of dimeric galectin-1.36 Several cadherins were also identified, including cadherin-2 (N-Cadherin), which is differentially expressed with galectin-1 and is implicated in cell–cell adhesion events.37 Other known ligands of galectin-1 found in our dataset include CD44,38 epidermal growth factor receptor (Egfr),39 integrin-αv (Itgαv),40 and vascular cell adhesion protein (Vcam-1).41 Consistent with our fluorescence microscopy data (Figure 2B) and the importance of the ECM in myogenesis,42 we identified several ECM proteins, including vasorin (Vasn), fibulin-2 (Fbln-2), Notch-2, lactadherin (Mfgm), and versican (Cspg-2). Furthermore, our dataset contains many surface N-linked glycoproteins on C2C12 myoblasts, including ephrin receptor type-A 2 (Epha-2), adhesion G-protein-coupled receptor E5 (CD97), glypican 1 (Gpc1), and Egfr.43 We also identified a number of novel galectin-1 interactors, including growth arrest specific protein 1 (Gas-1),44,45 Epha-2,46,47 CD97, endothelin converting enzyme 1 (Ece-1), integrin-β3 (Itgβ-3), and the proteoglycan Gpc1,48 which have all previously been implicated in myoblast proliferation or differentiation. Upon analysis of our dataset using iPathway49 (AdvaitaBio), we discovered pathways involved in the regulation of actin cytoskeleton and in ECM–receptor interactions, as defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Figure 3F).
Table 1.
Abbreviated Glycoprotein Interactors Identified by MS-Proteomics (Glycosylation State and Gene Ontology (GO) Localization Assigned by Uniprot)
| gene name | protein name | Uniprot ID | glycosylation state | GO localization | known interactor? |
|---|---|---|---|---|---|
| ECE1 | endothelin-converting enzyme 1 | Q4PZA2 | 10 N-linked sites | CM | no |
| CD97 | adhesion-G protein-coupled receptor E5 | Q9Z0M6 | 7 N-linked sites | CM | no |
| CD44 | CD44 antigen | P15379 | 10 N-linked sites | CM | yes |
| NOTC2 | notch 2 | O35516 | 5 N-linked, 2 O-linked sites | CM, ECM | no |
| GAS1 | growth arrest-specific protein 1 | Q01721 | 2 N-linked sites | CM | no |
| VCAM1 | vascular cell adhesion protein 1 | P29533 | 5 N-linked sites | CM | yes |
| EPHA2 | ephrin type-A receptor 2 | Q03145 | 2 N-linked sites | CM | no |
| ADGRL2 | adhesion G-protein-coupled receptor L2 | Q8JZZ7 | 3 N-linked sites | CM | no |
| CADM1 | cell adhesion molecule 1 | Q8R5M8 | 6 N-linked sites | CM | no |
| VASN | vasorin | Q9CZT5 | 6 N-linked sites | CM, ECM | yes |
| ITA5 | integrin α−5 | P11688 | 14 N-linked sites | CM | no |
| ITB5 | integrin β−5 | O70309 | 7 N-linked sites | CM | no |
| EGFR | epidermal growth factor receptor | Q01279 | 10 N-linked sites | CM | yes |
| CSPG4 | chondroitin sulfate proteoglycan 4 | Q8VHY0 | 16 N-linked sites, 1 O-linked site | CM, ECM | no |
| FBLN2 | fibulin-2 | P37889 | 4 N-linked sites | Secreted, ECM | no |
| DAG1 | dystroglycan | Q62165 | 4 N-linked sites, 3 O-linked sites | CM, Secreted | no |
| GPC1 | glypican-1 | Q9QZF2 | 2 N-linked sites, 3 O-linked sites | CM, ECM | no |
| CDH2 | cadherin-2 | P15116 | 7 N-linked sites | CM | yes |
| CDH13 | cadherin-13 | Q9WTR5 | 7 N-linked sites | CM | no |
| CDH15 | cadherin-15 | P33146 | 5 N-linked sites | CM | no |
Validation of Galectin-1 Interactors.
Within this dataset of interactors are ligands of galectin-1. A key challenge in validating whether proteins are true physiological and glycan-mediated counter-receptors for galectin-1 is recapitulating the correct glycosylation patterns of the native glycoproteins. A common approach used to validate protein–protein interactions entails interrogating the isolated recombinant proteins in an ELISA.21 Hence, we sourced recombinant glycoproteins (Table S3) and evaluated binding against NPG1. It is important to note that these glycoproteins may not accurately reflect the glycosylation patterns present in myoblasts. We immobilized integrin-β3, Epha-2 and Ece-1, Notch-2, and Gas1 and found that NPG1 binds the three former glycoproteins, with apparent EC50 values of 42, 14, and 156 nM, respectively (Figure 4A and Table S4). The addition of excess lactose mildly reduced binding, and a greater excess (200 vs 100 mM) of neither lactose (Figure S9A) nor the galectin-1 inhibitor TD13950 completely abrogated binding (Figure S9B). The lack of competition was also observed with recombinant galectin-1 binding with integrin-β3 and Epha-2 (Figure S9C). These observations perhaps suggest that, although glycosylation plays a role in mediating these interactions, additional protein–protein interactions could be large contributors to binding. Of course, we cannot exclude the possibilities that the recombinant glycoproteins do not match the native protein glycoforms found in myoblasts or other indirect effects that glycosylation might play to alter binding, including modulating protein conformation or causing potential steric effects on protein–protein binding events.
Figure 4.
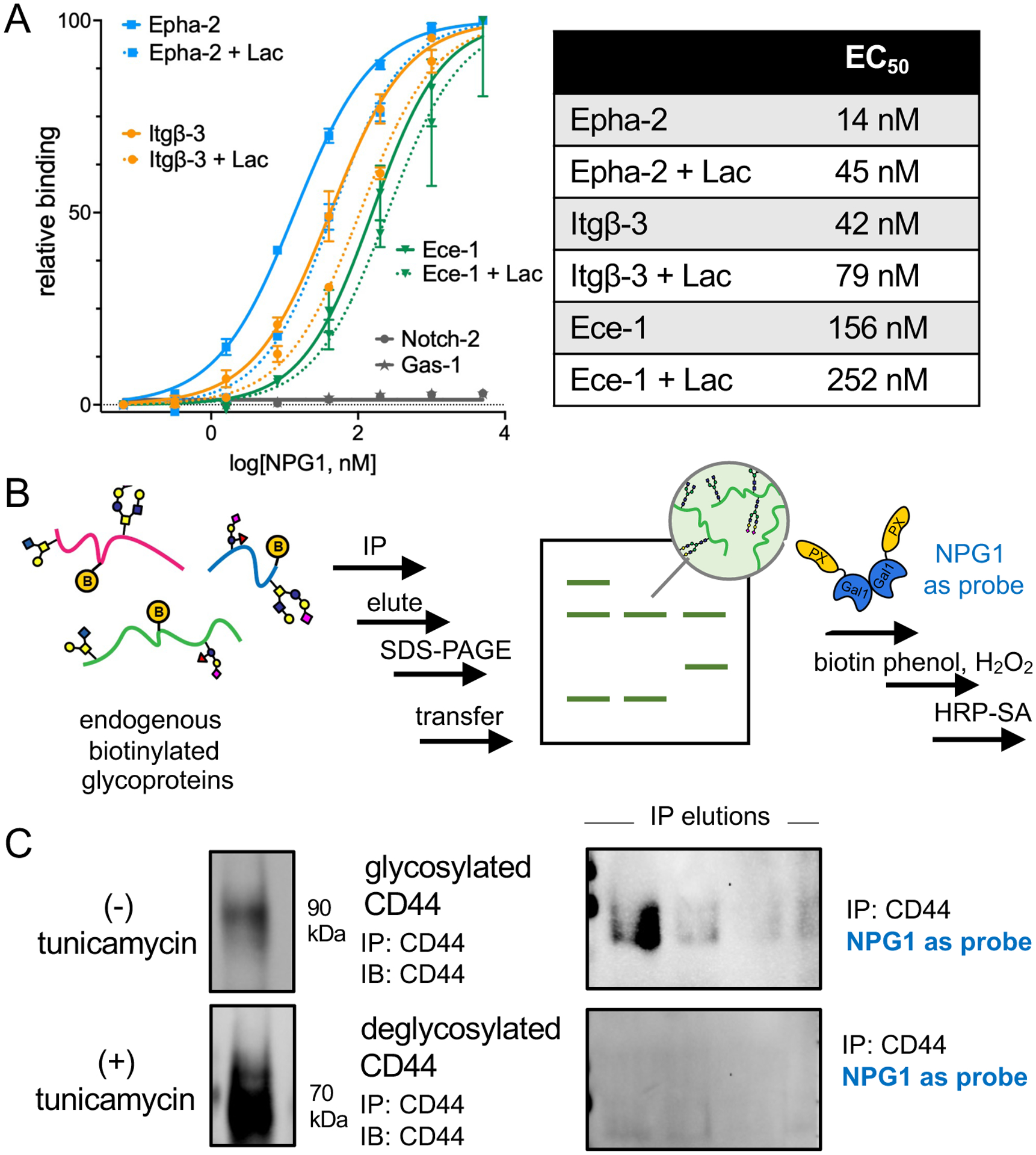
Validation of glycoprotein interactors by ELISA and direct blotting with NPG1. (A) Recombinant glycoproteins were evaluated for binding to NPG1 by ELISA. Epha-2, Itgβ-3, and Ece-1 demonstrated dose-dependent binding to NPG1 and partial competition with excess lactose (100 mM). Errors bars indicate standard deviation derived from three independent replicates. (B) Native CD44 was captured by immunoprecipitation (IP). Following proximity tagging with 5 μM NPG1 on membranes, biotinylation was detected by HRP-streptavidin (HRP-SA). (C) Probing for CD44 revealed successful IP of the endogenous protein from both nontreated and cells pretreated with tunicamycin (left panel). The bands in tunicamycin-treated conditions are of a lower molecular weight, indicative of deglycosylation. Probing with NPG1 and HRP-SA revealed a band corresponding to CD44 only in the nontreated condition, indicating that endogenous N-linked glycosylation is necessary for the interaction between NPG1 and CD44.
To move toward validating interactions with native glycoproteins, we used NPG1 as a probe in a direct blot assay upon immunoprecipitation from cell lysates (Figure 4C and Figure S10). We immunoprecipitated the highly abundant glycoprotein CD44 using an antibody that displays broad reactivity toward multiple glycoforms of the antigen (Figure S9), and upon successive incubation of the membrane with NPG1, biotin–phenol and H2O2, and HRP-streptavidin (HRP-SA), we observed the presence of bands corresponding to CD44 (Figure 4C). Signals were abrogated with tunicamycin, lactose, and TD139 but not with sucrose (Figure S11), and no signals were observed with a truncated APEX2 construct (Figure S11), implying further that the N-linked glycans of CD44 contribute to binding.
To support the presence of correct native protein glycoforms for mediating glycan-dependent interactions with galectin-1, we profiled the cell surface N-linked glycans on C2C12 myoblasts using MS-based glycomics. Although mucin-type O-linked glycans have been observed to bind galectin-1,4,12 a majority of glycan-dependent activity that occurs with galectin-1 have been found to be N-linked.4,14 The most abundant glycans found on C2C12 myoblast cell surfaces consisted mostly of complex and oligomannose N-glycans (Figure 5 and Table S5), consistent with previous analyses of rat myoblasts.4 The complex N-glycans were either capped with N-acetyl (Neu5Ac) or N-glycolyl (Neu5Gc) neuraminic acids or were uncapped, such that they present a terminal galactose residue. Although we have yet to profile the linkages in these residues (galectin-1 can tolerate α(2–3)Neu5Ac substitutions), the abundance of the terminally galactosylated N-glycans strongly indicates that appropriate glycan motifs are present on cell surface glycoproteins for glycan-mediated galectin-1 interactions.
Figure 5.
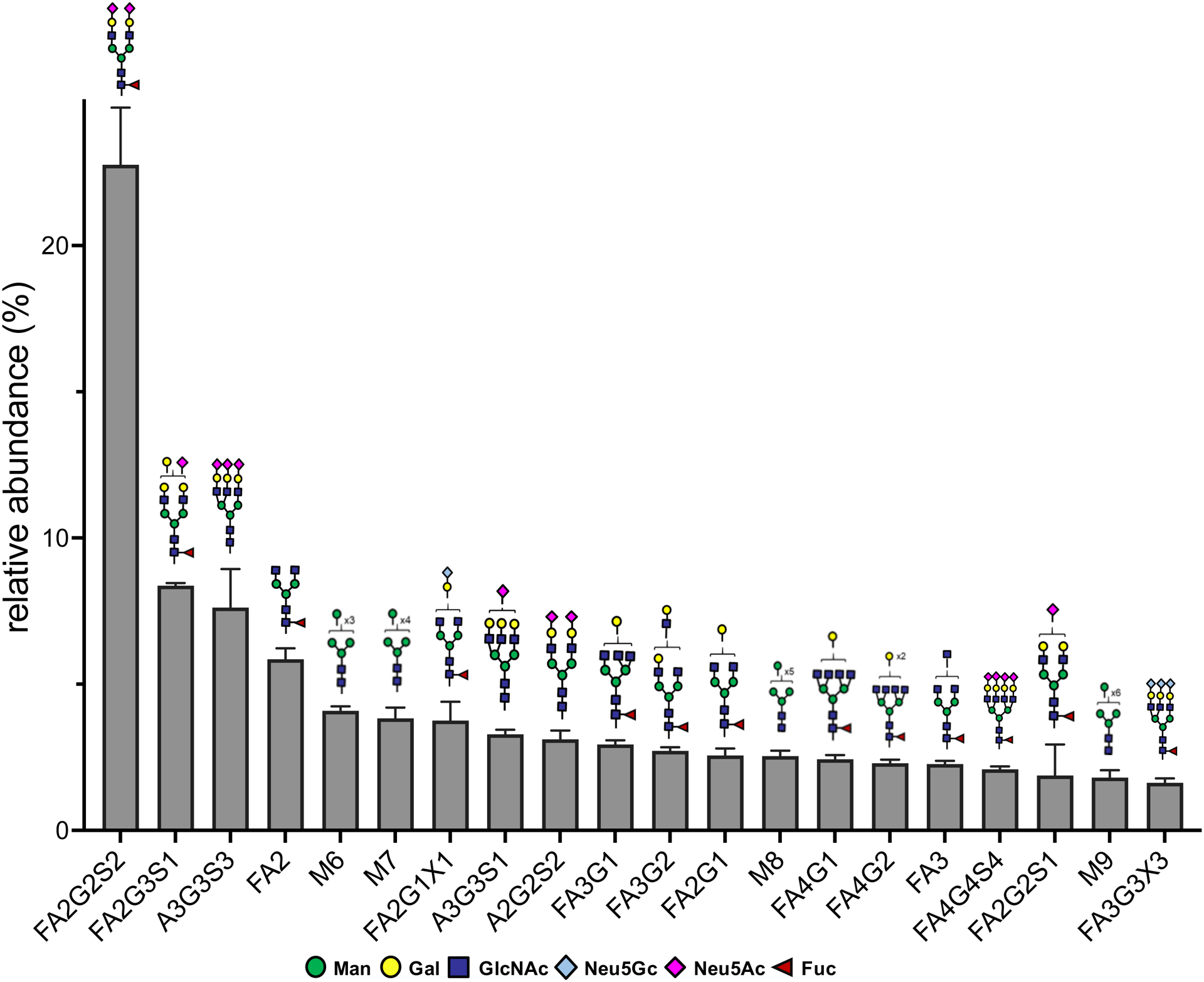
MS-based determination of N-glycan abundance and composition of C2C12 cell surfaces. Composition and relative amounts of the top 20 most abundant N-glycans found on cell surfaces. Error bars indicate standard deviation derived from three replicates.
In contrast to the 74 glycan-mediated interactors we identified for NPG1, our previous work identified a larger number (248) for galectin-3.21 Although this disparity could be explained by the change in cell types, previous work using affinity-based techniques also reports a substantially lower number of galectin-1 compared to galectin-3 interactors (15 vs 131).40 While the molecular basis of this disparity is unclear, our dataset currently represents the largest list of galectin-1 interactors. Most importantly, this list is derived from interactions of galectin-1 with live cells, providing relevance for subsequent work profiling their roles of the enriched proteins in myogenic differentiation. Given the dynamic changes that the glycocalyx undergoes during myogenic differentiation,51,4 it will be interesting to profile the corresponding changes in interactors for NPG1 as it traffics into the cell. Our ability to detect these interactions at lower concentrations consistent with physiological levels further increases the likelihood that they are biologically relevant. The use of engineered monomeric galectin-1 that binds specific glycoproteins16,17 in proximity tagging will enable us to further dissect why some glycoproteins exhibit this preference.
CONCLUSIONS
Galectin-1 is an important GBP that promotes myogenic differentiation. While the glycan structures responsible for binding galectin-1 have been assigned, the interacting glycoproteins that ultimately contribute to its function and the roles of these interactions in myogenesis remain poorly understood. We demonstrate that proximity tagging enables the determination of the glycan-mediated glycoprotein interactors of galectin-1 in mouse myoblasts and that this dataset of interactors can reveal galectin-1 counter-receptors. This dataset of 74 interactors represents a significant increase in the known list of glycoprotein interactors for galectin-1.3 It reveals both known and novel glycoprotein interactors of galectin-1, and it highly implicates the regulation of cytoskeleton as a relevant pathway, consistent with the required substantial remodeling of the cytoskeletal structure in myogenic differentiation.52,53 We further show that proximity tagging to capture glycan-mediated binding partners is amenable toward multiple modes of detection (fluorescence microscopy, Western blotting, flow cytometry). Consistent with the glycan-binding requirements for galectin-1, we find that mouse C2C12 myoblasts are decorated with β-galactoside and sialoside N-glycans, and that the native CD44 glycoprotein binds galectin-1. Our dataset of interactors can serve as resource for future work exploring the functional role of these glycoproteins in myogenic differentiation.
METHODS
In Situ Proximity Labeling of Live C2C12 Cells.
C2C12 cells (6 × 106) were seeded on poly-D-lysine-coated plates and incubated with NPG1 (30 min, 37 °C, Dulbecco’s modified Eagle medium, DMEM). For controls, cells were either pretreated with PNGase F (5 μg/mL, 4 h, 37 °C, DMEM), tunicamycin (10 μg/mL, 24 h, 37 °C, DMEM + 10% fetal bovine serum, FBS), or NPG1 was preincubated (5 min, DMEM) with lactose (100 mM), sucrose (100 mM), or TD139 (62 μM). Excess NPG1 was removing by washing three times with phosphate-buffered saline (PBS) before biotin–phenol (500 μM, 15% FBS/DMEM, 30 min, 37 °C) and H2O2 (1 mM, 1 min, RT) were added, followed by quenching (three washes with 10 mM sodium ascorbate, 10 mM sodium azide, and 5 mM Trolox in PBS).
Size Exclusion Chromatography.
Pre-equilibrated (0.25 mL/min, PBS, pH 7.14, 30 min) samples of NPG1 were monitored on a SEC column (4 × 400 mm, 5 μm MAbPac, Thermofisher #074696) at 280 and 405 nm.
Preparation of Samples for Proteomics.
Following proximity tagging, adherent cells were scraped, resuspended in 400 μL of PBS, and lysed by sonication. After precipitation in cold MeOH (overnight, −20 °C), the resulting pellet was resuspended in 6 M urea in PBS and 10 μL of 10% (w/v) sodium dodecyl sulfate (SDS). Dissolved proteins were reduced by adding 1:1 solution of 20 mM (tris(2-carboxyethyl)phosphine (50 μL, PBS) and 600 mM K2CO3 (PBS) and further incubated (shaking, 30 min, 37 °C). Samples were treated with iodoacetamide, followed by 10% SDS before incubation with streptavidin agarose beads (1.5 h, room temperature). After being washed to remove unbound materials, beads were resuspended in 100 mM triethylammonium bicarbonate and sequencing grade porcine trypsin and digested overnight (37 °C). Supernatants were harvested and labeled with a tandem mass tag (Thermo Scientific cat# A34808; 1 h, room temperature). The reaction was quenched with hydroxylamine and acidified with formic acid before being dried by vacuum centrifugation. Dried samples were redissolved in 5% (v/v) MeCN, 0.1% formic acid, combined, desalted, dried by vacuum centrifugation, and stored at −80 °C until ready for injection.
Proteomics Liquid Chromatography-Mass Spectrometry (LC/MS) Analysis.
Samples were solubilized in 1% trifluoroacetic acid and desalted using ZipTips (Millipore), according to the manufacturer’s instructions. Peptides were online eluted into a Fusion Tribrid mass spectrometer (Thermo Scientific) from an EASY PepMapTM RSLC C18 column (2 mm, 100 Å, 75 mm × 50 cm, Thermo Scientific). The precise LC gradient is available in the Supporting Information. Ions were created at 1.9 kV using an EASY Spray source (Thermo Scientific) held at 50 °C. A synchronous precursor selection (SPS)-MS3 mass spectrometry method was used based on the work of Ting et al. (see Supporting Information).34
Proteomics Data Analysis.
Proteomics data were processed using Proteome Discoverer 2.4 (Thermo Scientific). Precise settings are available in the Supporting Information. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE54 partner repository with the dataset identifier PXD026718.
Cell Surface Glycomics.
Confluent C2C12 cells were harvested, washed in PBS, and trypsinized. After heat inactivation (100 °C, 5 min), glycans were released from glycopeptides by PNGase F (7.5 μg/μL in 25 mM Tris, pH 7.2, 37 °C, overnight). Released glycans were purified with a C18 column (Thermo Scientific HyperSep, #03251257), and permethylated samples were dissolved in 20% (v/v) MeCN prior to injection. Experiments were repeated across two independent biological replicates. Chromatography was performed on an Ultimate 3,000 UHPLC (Thermo Scientific) with a reversed-phase Waters Acquity Peptide BEH C18 column (150 mm × 2.1 mm inner diameter, 130 Å particle size). The UHPLC was interfaced with an LTQ XL ETD Hybrid Ion Trap-Orbitrap ESI (Thermo Scientific). Precise LC/MS conditions are available in the Supporting Information. Data were processed with XCalibur 2.1 (Thermo Scientific). Glycan compositions and structures were identified using Simglycan and GlycoWorkbench.55
Immunoprecipitation from C2C12 Cells.
Approximately 1 mg of preclarified and precleared lysate from C2C12 cells was incubated with 10 μg of α-CD44 antibody (overnight, 4 °C) to form the immunocomplex, which was then captured with protein G Sepharose Fast Flow beads (overnight, 4 °C). Beads were washed with PBS, and bound protein was eluted in glycine (0.2 M, pH 2.6). Elutions were quenched with an equal volume Tris buffer (1 M, pH 9.0).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to T. O’Leary, the Scripps Research Florida Proteomics (G. Tsaprailis) and Bioinformatics (P. Natarajan and G. Crynen) Core Facilities for their assistance with data analysis. We thank M. Caffrey for their gift of the OmpA3-PNGaseF-TEV-His6 plasmid (Addgene plasmid #114274; http://n2t.net/addgene:114274; RRID:Addgene_114274). In this work, Z.V., E.J., M.C., and M.L.H. are supported by the NIH K99/R00 Pathway to Independence Award to M.L.H. (R00HD090292) and the NIGMS (R35GM142462). E.J. is supported by a Skaggs Graduate Fellowship, enabled by the Henry and Jennifer Luttrell Foundation. We are also grateful for additional support from the Joe W. & Dorothy Dorsett Brown Foundation.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acschembio.1c00313.
Appendix Table 1 (XLSX)
Additional and extended methods, amino acid sequences of protein constructs, NPG1 characterization, asialofetuin ELISA analysis parameters, NPG1 dimerization characterization, high-magnification and dose-dependent NPG1 microscopy, evaluation of APEX activity and localization by microscopy, flow cytometry of NPG1-labeled cells, Western blot of enriched NPG1 interactors, significantly enriched/competed proteins by MS proteomics, glycoproteins and ELISA analysis parameters for NPG1 interaction validation, ELISA of interactors with additional controls, Western blot of CD44 immunoprecipitation, direct blot of CD44 with additional controls, glycans identified by glycomics (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acschembio.1c00313
The authors declare no competing financial interest.
Contributor Information
Zak Vilen, Department of Molecular Medicine, Scripps Research, Jupiter, Florida 33458-5284, United States.
Eugene Joeh, Department of Molecular Medicine, Scripps Research, Jupiter, Florida 33458-5284, United States.
Meg Critcher, Department of Molecular Medicine, Scripps Research, Jupiter, Florida 33458-5284, United States.
Christopher G. Parker, Department of Chemistry, Scripps Research, Jupiter, Florida 33458-5284, United States.
Mia L. Huang, Department of Molecular Medicine, Scripps Research, Jupiter, Florida 33458-5284, United States; Department of Chemistry, Scripps Research, Jupiter, Florida 33458-5284, United States;.
REFERENCES
- (1).Crocker PR, and Feizi T (1996) Carbohydrate recognition systems: functional triads in cell-cell interactions. Curr. Opin. Struct. Biol 6, 679–691. [DOI] [PubMed] [Google Scholar]
- (2).Critcher M, O’Leary T, and Huang M (2021) Glycoengineering: scratching the surface. Biochem. J 478, 703–719. [DOI] [PubMed] [Google Scholar]
- (3).Camby I, Le Mercier M, Lefranc F, and Kiss R (2006) Galectin-1: a small protein with major functions. Glycobiology 16, 137R–157R. [DOI] [PubMed] [Google Scholar]
- (4).Blazev R, Ashwood C, Abrahams J, Chung L, Francis D, Yang P, Watt K, Qian H, Quaife-Ryan G, Hudson J, Gregorevic P, Thaysen-Andersen M, and Parker B (2020) Integrated glycoproteomics identifies a role of N-glycosylation and galectin-1 on myogenesis and muscle development. Mol. Cell Proteomics 20, 100030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Cerri DG, Rodrigues LC, Stowell SR, Araujo DD, Coelho MC, Oliveira SR, Bizario JC, Cummings RD, Dias-Baruffi M, and Costa MC (2008) Degeneration of dystrophic or injured skeletal muscles induces high expression of Galectin-1. Glycobiology 18, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Georgiadis V, Stewart HJ, Pollard HJ, Tavsanoglu Y, Prasad R, Horwood J, Deltour L, Goldring K, Poirier F, and Lawrence-Watt DJ (2007) Lack of galectin-1 results in defects in myoblast fusion and muscle regeneration. Dev. Dyn 236, 1014–1024. [DOI] [PubMed] [Google Scholar]
- (7).Goldring K, Jones G, Thiagarajah R, and Watt D (2002) The effect of galectin-1 on the differentiation of fibrobalsts and myoblasts in vitro. J. Cell Sci 115, 355–366. [DOI] [PubMed] [Google Scholar]
- (8).Chan J, O’Donoghue K, Gavina M, Torrente Y, Kennea N, Mehmet H, Stewart H, Watt DJ, Morgan JE, and Fisk NM (2006) Galectin-1 induces skeletal muscle differentiation in human fetal mesenchymal stem cells and increases muscle regeneration. Stem Cells 24, 1879–1891. [DOI] [PubMed] [Google Scholar]
- (9).Van Ry PM, Wuebbles RD, Key M, and Burkin DJ (2015) Galectin-1 Protein Therapy Prevents Pathology and Improves Muscle Function in the mdx Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther 23, 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wuebbles RD, Cruz V, Van Ry P, Barraza-Flores P, Brewer PD, Jones P, and Burkin DJ (2019) Human Galectin-1 Improves Sarcolemma Stability and Muscle Vascularization in the mdx Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther.–Methods Clin. Dev 13, 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Horlacher T, Oberli MA, Werz DB, Krock L, Bufali S, Mishra R, Sobek J, Simons K, Hirashima M, Niki T, and Seeberger PH (2010) Determination of carbohydrate-binding preferences of human galectins with carbohydrate microarrays. ChemBioChem 11, 1563–1573. [DOI] [PubMed] [Google Scholar]
- (12).Nielsen MI, Stegmayr J, Grant OC, Yang Z, Nilsson UJ, Boos I, Carlsson MC, Woods RJ, Unverzagt C, Leffler H, and Wandall HH (2018) Galectin binding to cells and glycoproteins with genetically modified glycosylation reveals galectin-glycan specificities in a natural context. J. Biol. Chem 293, 20249–20262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Collins BE, and Paulson JC (2004) Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr. Opin. Chem. Biol 8, 617–625. [DOI] [PubMed] [Google Scholar]
- (14).Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, and Cummings RD (2008) Galectin-1, −2, and −3 exhibit differential recognition of sialylated glycans and blood group antigens. J. Biol. Chem 283, 10109–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Guardia CM, Caramelo JJ, Trujillo M, Mendez-Huergo SP, Radi R, Estrin DA, and Rabinovich GA (2014) Structural basis of redox-dependent modulation of galectin-1 dynamics and function. Glycobiology 24, 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cho M, and Cummings RD (1996) Characterization of Monomeric Forms of Galectin-1 Generated by Site-Directed Mutagenesis. Biochemistry 35, 13081–13088. [DOI] [PubMed] [Google Scholar]
- (17).Salomonsson E, Larumbe A, Tejler J, Tullberg E, Rydberg H, Sundin A, Khabut A, Frejd T, Lobsanov YD, Rini JM, Nilsson UJ, and Leffler H (2010) Monovalent interactions of galectin-1. Biochemistry 49, 9518–9532. [DOI] [PubMed] [Google Scholar]
- (18).Cho M, and Cummings RD (1995) Galectin-1, a beta-galactoside-binding lectin in Chinese hamster ovary cells. I. Physical and chemical characterization. J. Biol. Chem 270, 5198–5206. [DOI] [PubMed] [Google Scholar]
- (19).Dias-Baruffi M, Zhu H, Cho M, Karmakar S, McEver RP, and Cummings RD (2003) Dimeric galectin-1 induces surface exposure of phosphatidylserine and phagocytic recognition of leukocytes without inducing apoptosis. J. Biol. Chem 278, 41282–41293. [DOI] [PubMed] [Google Scholar]
- (20).Leppanen A, Stowell S, Blixt O, and Cummings RD (2005) Dimeric galectin-1 binds with high affinity to alpha2,3-sialylated and non-sialylated terminal N-acetyllactosamine units on surface-bound extended glycans. J. Biol. Chem 280, 5549–5562. [DOI] [PubMed] [Google Scholar]
- (21).Joeh E, O’Leary T, Li W, Hawkins R, Hung JR, Parker CG, and Huang ML (2020) Mapping glycan-mediated galectin-3 interactions by live cell proximity labeling. Proc. Natl. Acad. Sci. U. S. A 117, 27329–27338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, and Ting AY (2015) Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Earl LA, Bi S, and Baum LG (2011) Galectin multimerization and lattice formation are regulated by linker region structure. Glycobiology 21, 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Garner OB, and Baum LG (2008) Galectin-glycan lattices regulate cell-surface glycoprotein organization and signalling. Biochem. Soc. Trans 36, 1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Diehl C, Engström O, Delaine T, Håkansson M, Genheden S, Modig K, Leffler H, Ryde U, Nilsson UJ, and Akke M (2010) Protein Flexibility and Conformational Entropy in Ligand Design Targeting the Carbohydrate Recognition Domain of Galectin-3. J. Am. Chem. Soc 132, 14577–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tamura M, Watanabe T, Igarashi T, Takeuchi T, Kasai K, and Arata Y (2014) Crosslinking of Cys-Mutated Human Galectin-1 to the Modeal Glycoprotein Ligands Asialofetuin and Laminin by Using a Photoactivatable Bifunctional Reagent. Biol. Pharm. Bull 37, 877–882. [DOI] [PubMed] [Google Scholar]
- (27).Joeh E, O’Leary T, Li W, Hawkins R, Hung J, Parker C, and Huang M (2020) Mapping glycan-mediated galectin-3 interactions by live cell proximity labeling. Proc. Natl. Acad. Sci. U. S. A 117, 27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Trinkle-Mulcahy L (2019) Recent advances in proximity-based labeling methods for interactome mapping. F1000Research 8, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Tinari N, Kuwabara I, Huflejt ME, Shen PF, Iacobelli S, and Liu F-T (2001) Glycoprotein 90K/MAC-2BP Interacts with Galectin-1 and Mediates Galectin-1-Induced Cell Aggregation. Int. J. Cancer 91, 167–172. [DOI] [PubMed] [Google Scholar]
- (30).Cooper D, and Barondes SH (1990) Evidence for Export of a Muscle Lectin from Cytosol to Extracellular Matrix and for a Novel Secretory Mechanism. J. Cell Biol 110, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Harrison L, and Wilson TJ (1992) The 14 kDa β-galactoside binding lectin in myoblast and myotube cultures: localization by confocal microscopy. J. Cell Sci 101, 635–646. [DOI] [PubMed] [Google Scholar]
- (32).May EA, Kalocsay M, D’Auriac IG, Schuster PS, Gygi SP, Nachury MV, and Mick DU (2021) Time-resolved proteomics profiling of the ciliary Hedgehog response. J. Cell Biol 220, e202007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Adams L, Kenneth Scott G, and Weinberg CS (1996) Biphasic modulation of cell growth by recombinant human galectin-1. Biochim. Biophys. Acta, Mol. Cell Res 1312, 137–144. [DOI] [PubMed] [Google Scholar]
- (34).Ting L, Rad R, Gygi SP, and Haas W (2011) MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods 8, 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Escoda-Ferran C, Carrasco E, Caballero-Banos M, Miro-Julia C, Martinez-Florensa M, Consuegra-Fernandez M, Martinez VG, Liu FT, and Lozano F (2014) Modulation of CD6 function through interaction with Galectin-1 and −3. FEBS Lett. 588, 2805–2813. [DOI] [PubMed] [Google Scholar]
- (36).Fischer C, Sanchez-Ruderisch H, Welzel M, Wiedenmann B, Sakai T, Andre S, Gabius HJ, Khachigian L, Detjen KM, and Rosewicz S (2005) Galectin-1 interacts with the {alpha}5-{beta}1 fibronectin receptor to restrict carcinoma cell growth via induction of p21 and p27. J. Biol. Chem 280, 37266–37277. [DOI] [PubMed] [Google Scholar]
- (37).Zhu J, Zheng Y, Zhang H, Liu Y, Sun H, and Zhang P (2019) Galectin-1 induces metastasis and epithelial-mesenchymal transition (EMT) in human ovarian cancer cells via activation of the MAPK JNK/p38 signalling pathway. Am. J. Transl. Res 11, 3862–3878. [PMC free article] [PubMed] [Google Scholar]
- (38).Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, Szpyt J, Tam S, Zarraga G, Pontano-Vaites L, Swarup S, White AE, Schweppe DK, Rad R, Erickson BK, Obar RA, Guruharsha KG, Li K, Artavanis-Tsakonas S, Gygi SP, and Harper JW (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Foerster S, Kacprowski T, Dhople VM, Hammer E, Herzog S, Saafan H, Bien-Moller S, Albrecht M, Volker U, and Ritter CA (2013) Characterization of the EGFR interactome reveals associated protein complex networks and intracellular receptor dynamics. Proteomics 13, 3131–3144. [DOI] [PubMed] [Google Scholar]
- (40).Obermann J, Priglinger CS, Merl-Pham J, Geerlof A, Priglinger S, Gotz M, and Hauck SM (2017) Proteome-wide Identification of Glycosylation-dependent Interactors of Galectin-1 and Galectin-3 on Mesenchymal Retinal Pigment Epithelial (RPE) Cells. Mol. Cell Proteomics 16, 1528–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Humphries JD, Byron A, Bass MD, Craig SE, Pinney JW, Knight D, and Humphries MJ (2009) Proteomic Analysis of Integrin-Associated Complexes Identifies RCC2 as a Dual Regulator of Rac1 and Arf6. Sci. Signaling 2, ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Wang PY, Thissen H, and Tsai WB (2012) The roles of RGD and grooved topography in the adhesion, morphology, and differentiation of C2C12 skeletal myoblasts. Biotechnol. Bioeng 109, 2104–2115. [DOI] [PubMed] [Google Scholar]
- (43).Gundry RL, Raginski K, Tarasova Y, Tchernyshyov I, Bausch-Fluck D, Elliott ST, Boheler KR, Van Eyk JE, and Wollscheid B (2009) The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Mol. Cell Proteomics 8, 2555–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Li L, Rozo M, Yue S, Zheng X, Tan FJ, Lepper C, and Fan C-M (2019) Muscle stem cell renewal suppressed by Gas1 can be reversed by GDNF in mice. Nat. Metab 1, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Leem YE, Han JW, Lee HJ, Ha HL, Kwon YL, Ho SM, Kim BG, Tran P, Bae GU, and Kang JS (2011) Gas1 cooperates with Cdo and promotes myogenic differentiation via activation of p38MAPK. Cell. Signalling 23, 2021–2029. [DOI] [PubMed] [Google Scholar]
- (46).Zhang X, Wang L, Qiu K, Xu D, and Yin J (2019) Dynamic membrane proteome of adipogenic and myogenic precursors in skeletal muscle highlights EPHA2 may promote myogenic differentiation through ERK signaling. FASEB J. 33, 5495–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Menges CW, and McCance DJ (2008) Constitutive activation of the Raf-MAPK pathway causes negative feedback inhibition of Ras-PI3K-AKT and cellular arrest through the EphA2 receptor. Oncogene 27, 2934–2940. [DOI] [PubMed] [Google Scholar]
- (48).Gutiérrez J, Cabrera D, and Brandan E (2014) Glypican-1 regulates myoblast response to HGF via Met in a lipid raft-dependent mechanism: effect on migration of skeletal muscle precursor cells. Skeletal Muscle 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, and Romero R (2007) A systems biology approach for pathway level analysis. Genome Res. 17, 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Hsieh TJ, Lin HY, Tu Z, Lin TC, Wu SC, Tseng YY, Liu FT, Hsu ST, and Lin CH (2016) Dual thiodigalactoside-binding modes of human galectins as the structural basis for the design of potent and selective inhibitors. Sci. Rep 6, 29457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Janot M, Audfray A, Loriol C, Germot A, Maftah A, and Dupuy F (2009) Glycogenome expression dynamics during mouse C2C12 myoblast differentiation suggests a sequential reorganization of membrane glycoconjugates. BMC Genomics 10, 483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Komati H, Naro F, Mebarek S, De Arcangelis V, Adamo S, Lagarde M, Prigent A-F, and Nemoz G (2005) Phospholipase D Is Involved in Myogenic Differentiation through Remodeling of Actin Cytoskeleton. Mol. Biol. Cell 16, 1232–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Formigli L, Meacci E, Sassoli C, Squecco R, Nosi D, Chellini F, Naro F, Francini F, and Zecchi-Orlandini S (2007) Cytoskeleton/stretch-activated ion channel interaction regulates myogenic differentiation of skeletal myoblasts. J. Cell. Physiol 211, 296–306. [DOI] [PubMed] [Google Scholar]
- (54).Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, and Vizcaino JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Ceroni A, Maass K, Geyer H, Geyer R, Dell A, and Haslam SM (2008) GlycoWorkbench: A Tool for the Computer-Assisted Annotation of Mass Spectra of Glycans. J. Proteome Res 7, 1650–1659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


