Abstract
Purpose:
Capmatinib is approved for MET exon 14-altered NSCLC based on activity in targeted therapy-naïve patients. We conducted a phase II study to assess efficacy of capmatinib in patients previously treated with a MET inhibitor.
Methods:
Patients with advanced NSCLC harboring MET amplification or MET exon 14 skipping received capmatinib 400 mg twice daily. The primary endpoint was objective response rate. Secondary endpoints included progression-free survival (PFS), disease control rate (DCR), intracranial response rate, and overall survival (OS). Circulating tumor DNA was analyzed to identify capmatinib resistance mechanisms.
Results:
Twenty patients were enrolled between 5/2016 and 11/2019, including 15 patients with MET skipping alterations and 5 patients with MET amplification. All patients had received crizotinib; three had also received other MET-directed therapies. The median interval between crizotinib and capmatinib was 22 days (range 4–374). Two (10%) patients achieved an objective response to capmatinib and 14 had stable disease, yielding a DCR of 80%. Among 5 patients who discontinued crizotinib for intolerance, DCR was 83%, including two patients with best tumor shrinkage of −25% and −28%. Intracranial DCR among 4 patients with measurable brain metastases was 100%, with no observed intracranial objective responses. Overall, the median PFS and OS were 5.5 (95% CI 1.3–11.0) and 11.3 (95% CI 5.5-not reached) months, respectively. MET D1228 and Y1230 mutations and MAPK alterations were recurrently detected in post-crizotinib, pre-capmatinib plasma. New and persistent MET mutations and MAPK pathway alterations were detected in plasma at progression on capmatinib.
Conclusion:
Capmatinib has modest activity in crizotinib-pretreated MET-altered NSCLC, potentially due to overlapping resistance mechanisms.
Keywords: MET skipping, MET amplification, capmatinib, lung cancer
INTRODUCTION
The MET gene encodes a receptor tyrosine kinase that promotes cell proliferation and survival.1 In non-small cell lung cancer (NSCLC), MET dysregulation is fueled by a variety of genetic and non-genetic mechanisms. Genetic mediators of oncogenic MET activation include rare gene rearrangements, gene amplification, and an array of mutations clustered in the splice regions flanking exon 14.1 The diverse MET exon 14 splice site alterations lead to deletion of the portion of the juxtamembrane domain that contains the binding site for the CBL E3 ubiquitin ligase, thus enhancing MET signaling by impeding receptor degradation.2 The resulting reduced MET receptor turnover increases sensitivity to tyrosine kinase inhibitors (TKIs) targeting MET.3–6 In addition to activity in tumors with MET exon 14 skipping alterations, MET TKIs have shown promising activity in NSCLCs with de novo MET amplification, particularly tumors with high-level MET amplification.1
Capmatinib is a highly selective and potent MET TKI with robust central nervous system (CNS) penetration that is approved by the US FDA for treatment of NSCLC harboring MET exon 14 skipping alterations.6 In the phase II GEOMETRY mono-1 trial, capmatinib induced an objective response in 41% of chemotherapy-treated and 68% of treatment-naïve patients with MET exon 14 altered NSCLC.6 Capmatinib demonstrated promising CNS activity with an intracranial response rate of 54%.6 Although regulatory approval does not extend to tumors with MET amplification, 29–40% of patients with highly MET-amplified (gene copy number ≥ 10) NSCLC in the study achieved an objective response to capmatinib.7 In addition to capmatinib, several investigational MET TKIs have also demonstrated encouraging clinical activity in MET-altered NSCLC, including crizotinib, tepotinib, and savolitinib.3–5
In other molecular subsets of NSCLC (e.g. anaplastic lymphoma kinase rearranged), sequential treatment with increasingly potent targeted therapies has translated into dramatic improvements in outcomes.8 It is presently unknown whether a similar paradigm of sequential treatment with a more potent, selective, and brain-penetrable MET TKI can re-induce responses after failure of another MET TKI. To investigate this hypothesis and explore the potential for sequencing MET targeted therapies, we conducted a phase II trial of capmatinib in NSCLC patients with MET amplification or exon 14 skipping alterations who were previously treated with MET-directed therapy.
METHODS
Study Design and Assessments
NCT02750215 was an open-label, investigator-initiated, single institution, single arm phase II trial of capmatinib in patients with metastatic NSCLC harboring MET amplification or a MET exon 14 skipping alteration (Supplementary Figure 1). The protocol was approved by the local Institutional Review Board. Written informed consent was obtained from all patients before screening. Patients aged ≥ 18 years with stage IIIB-V (AJCC v7.0) NSCLC and measurable disease per Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 were enrolled. Patients with brain metastases who did not have progressive neurological symptoms or require escalating doses of steroids were eligible. Patients must have received prior treatment with a MET TKI, but there was no restriction on number of prior treatment regimens, including non-targeted therapy. MET exon 14 alterations were detected using next-generation sequencing (NGS) of DNA or RNA. MET amplification was defined as MET: centromere 7 ratio ≥ 1.8 (fluorescence in-situ hybridization) or ≥ 6 MET copies (FoundationOne NGS assay) in tissue or an absolute MET copy number of ≥ 2.1 in circulating tumor DNA (Guardant360 assay).9, 10
Capmatinib was administered at an initial dose of 400 mg twice daily in 21-day cycles. Safety assessments were performed at baseline and at every subsequent visit. Adverse events were graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE) version 4.0. Dose reductions and interruptions were allowed as indicated to manage toxicities. Response assessment was conducted centrally using RECIST v1.1 and RANO criteria.11 Restaging scans were performed every 2 cycles. Baseline and subsequent brain imaging were only required for patients with known brain metastases. Patients were permitted to continue treatment beyond progression at the treating investigator’s discretion. Plasma was collected for retrospective circulating tumor DNA (ctDNA) analysis using the Guardant360 assay.10 The study was conducted in accordance with the principles of the Declaration of Helsinki and Good Clinical Practice guidelines of the International Conference on Harmonization.
Statistical Design
The primary objective of the study was to evaluate the objective response rate (ORR) of capmatinib in NSCLC patients previously treated with ≥ 1 MET TKI. Key secondary endpoints included progression-free survival, disease control rate, overall survival, and safety/tolerability. The original study design employed a Simon two-stage design to target an ORR of 40% compared to an ORR of 10%. Nine patients were enrolled in the first stage, with a plan to enroll an additional 11 patients in the second stage if at least 2 responses were observed in the first stage (Supplementary Figure 1). This design had 90% power to detect this difference in ORR, with a one-sided alpha level of 0.05. After enrollment of the first nine patients, the threshold for advancing to the second stage of the study was not met. Nonetheless, due to observed tumor shrinkage that did not meet response criteria in two patients, along with the heterogeneity of patients enrolled, the study was amended to still enroll the additional 11 patients. This was based on the hypothesis that the full cohort of 20 patients would be better positioned to identify subsets of patients who might benefit from capmatinib. The sample size of 20 patients was associated with 80% power to detect a 21% improvement in ORR (i.e., 31% vs 10%) using a one-sided binomial test with alpha level of 0.05. The data cutoff for this analysis was May 31, 2020. All analyses were done using SAS version 9.4.
RESULTS
Patient Characteristics
Between 5/2016 and 11/2019, 20 patients were enrolled (Table 1), including 15 (75%) patients with a MET exon 14 alteration and five (25%) patients with MET-amplified NSCLC (Supplementary Table 1). Most patients were female (60%), smokers (60%), with lung adenocarcinoma (80%). The median age of the study population was 70 years (range 57–88). The median number of prior lines of therapy was 2 (range 1–5). Eleven (55%) and seven (35%) patients had received chemotherapy or immunotherapy, respectively. All patients were previously treated with crizotinib. The reasons for discontinuing crizotinib included progression (n=14, 70%) and intolerance (n=6, 30%). Toxicities resulting in discontinuing crizotinib included liver test abnormalities (n=4), chest pressure/congestion (n=1), and pneumonitis (n=1). For 19 (95%) patients, crizotinib was the most recent targeted therapy, including 14 patients who transitioned directly from crizotinib to capmatinib. The median interval between discontinuing crizotinib and commencing treatment with capmatinib was 22 days (range 4–374 days).
Table 1.
Baseline Characteristics of Study Population
| Characteristic | No. (%) of Patients |
|---|---|
| All Patients (n = 20) | |
|
| |
| Age, years | |
| Median | 70 |
| Range | 57 – 88 |
|
| |
| Sex | |
| Male | 8 (40) |
| Female | 12 (60) |
|
| |
| Ethnicity | |
| White | 18 (90) |
| Asian | 1 (5) |
| Other | 1 (5) |
|
| |
| Smoking History | |
| Never | 8 (40) |
| Former | 11 (55) |
| Current | 1 (5) |
|
| |
| Histology | |
| Adenocarcinoma | 16 (80) |
| Squamous | 2 (10) |
| Sarcomatoid or Poorly Differentiated Carcinoma | 2 (10) |
|
| |
| MET Alteration | |
| Amplification | 5 (25) |
| Exon 14 Skipping | 15 (75) |
|
| |
| Prior Chemotherapy | |
| Yes | 11 (55) |
| No | 9 (45) |
|
| |
| Prior Immunotherapy | |
| Yes | 7 (35) |
| No | 13 (65) |
|
| |
| Number of Prior Lines of Therapy | |
| 1 | 9 (45) |
| 2 | 3 (15) |
| ≥3 | 8 (40) |
|
| |
| Number of Prior MET Targeted Therapies | |
| 1 | 17 (85) |
| 2 | 3 (15) |
|
| |
| Reason for Discontinuing Crizotinib | |
| Progression | 14 (70) |
| Intolerance | 6 (30) |
The median number of prior MET-directed therapies was 1 (range 1–2). In addition to crizotinib, two patients had received the MET monoclonal antibody ABBV399 and one patient was treated with glesatinib.
Efficacy
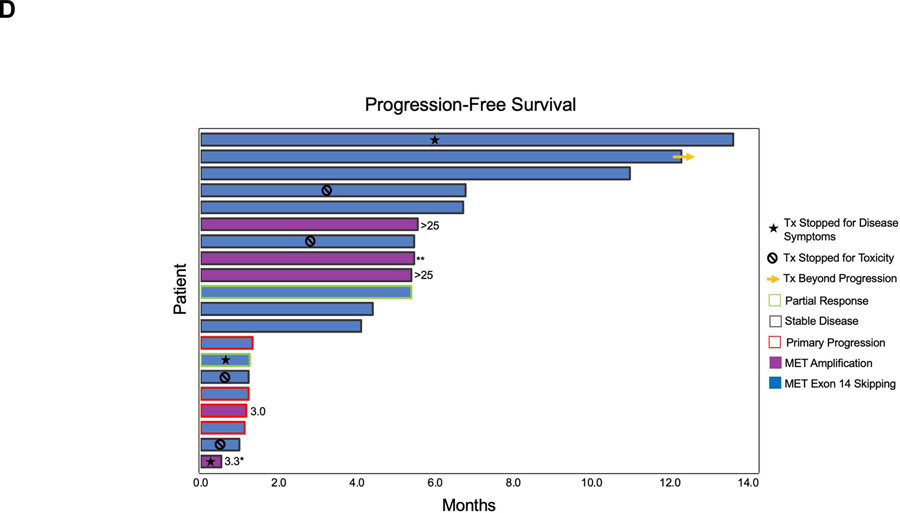
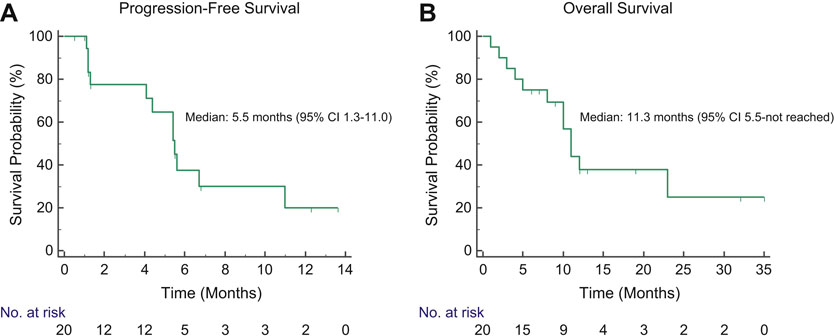
Two (10%) patients achieved a partial response (PR) to capmatinib (Figure 1A). Both had received chemotherapy + pembrolizumab as intervening therapy between crizotinib and capmatinib (Figure 1B–C). One patient with PR died from a stroke attributed to hypercoagulability of malignancy prior to subsequent restaging scans preventing confirmation of response. Of note, the patient had a history of hypercoagulable events refractory to therapeutic anticoagulation. The other patient with PR progressed after 5 months. Fourteen (70%) patients had a best response of disease stabilization, yielding a disease control rate of 80%. Two patients with stable disease had a near-PR (RECIST −28% and −25%). Several patients had stable disease lasting >6 months (Figure 1D).The ORR among patients with MET exon 14 skipping and MET amplification were 13% (n=2/15) and 0% (n=0/5), respectively. Of the 6 patients who had discontinued crizotinib for toxicity, five (83%) achieved disease control on capmatinib, including the two patients with near-PR (Figure 1A). Four patients (all with measurable intracranial disease) transitioned to capmatinib due to CNS progression on crizotinib, all of whom had previously received CNS radiation. The median interval between completion of CNS radiation and initiating capmatinib was 8.9 months (range 2.2–16.2 months). All four had stable disease as their best intracranial response to capmatinib, including two with best percent change of −29% and −22% intracranially (Supplementary Table 2). At data cutoff, three patients had ongoing CNS control lasting between 4.2 and 12.5 months. The median progression-free and overall survival of the overall population were 5.5 months (95% CI 1.3–11.0 months, Figure 2A) and 11.3 months (95% CI 5.5-not reached, Figure 2B), respectively.
Figure 1. Activity of Capmatinib in MET-Altered Lung Cancer.
(A) Waterfall plot depicts best tumor response as assessed by RECIST version 1.1. Secondary MET mutations detected in plasma analyzed immediately prior to treatment with capmatinib are listed above bars corresponding to individual patients. MET copy number is listed under the bars for patients with MET amplification, as determined by fluorescence in-situ hybridization using MET: centromeric probe 7 ratio or plasma genotyping. Absolute copy number was not provided for one patient. For patients who discontinued crizotinib for reasons other than extracranial progression, reason for stopping crizotinib is indicated by asterisk or chevron. (B) The treatment history for MGH9252 is illustrated above contrast enhanced CT images demonstrating decrease in the size of a right upper lobe mass (asterisk) after 6 weeks of treatment with capmatinib. (C) MGH9235’s treatment history is outlined above serial CT scans demonstrating decrease in a lingula mass (arrowhead), lymphangitic carcinomatosis (arrow), and pleural effusion (asterisk) after 6 weeks on capmatinib. *Capmatinib was discontinued after 8 weeks due to a stroke that was unrelated to study drug. (D) Swimmer plot illustrates progression-free survival on capmatinib for each patient (row). For patients with MET amplification, MET copy number is noted, as determined by fluorescence in-situ hybridization (ratio of MET to centromeric probe 7) or plasma next-generation sequencing (asterisk). A patient found to have MET amplification by FoundationOne next-generation sequencing assay for whom absolute copy number was not available is indicated with double asterisk (**). CNS: central nervous system; Carbo: carboplatin; pem: pemetrexed; Pembro: pembrolizumab; SD: stable disease; PR: partial response; PD: disease progression, Tx: treatment.
Figure 2. Progression-Free and Overall Survival of Patients Treated with Capmatinib.
Kaplan-Meier curves depict progression-free (A) and overall survival (B) on capmatinib among study participants.
Safety
The safety analysis included all 20 patients (Table 2). Treatment-related adverse events (AEs) occurring in ≥ 15% of patients included lower extremity edema (n=13, 65%), fatigue (n=7, 35%), nausea (n=7, 35%), myalgia (n=4, 20%), amylase elevation (n=4, 20%), creatinine elevation (n=4, 20%), lipase elevation (n=4, 20%), ALT elevation (n=3, 15%), and neuropathy (n=3, 15%). Lower extremity edema was primarily grade 2 (n=8/13, 62%). Eight (40%) grade 3 treatment-related AEs were observed, including asymptomatic lipase elevation (n=3, 15%), asymptomatic amylase elevation (n=2, 10%), asymptomatic AST and ALT elevation in a single patient (n=1 each, 5%), and dyspnea (n=1, 5%). No grade 4 AEs were seen. In general, AEs were ameliorated by dose modification and interruption. Eight (40%) patients required treatment interruption for toxicities. Seven (35%) patients had toxicities necessitating dose reduction. Three patients discontinued treatment for toxicities, specifically intolerable edema (after 8 cycles), elevated transaminases (after 10 cycles), and combined toxicities of nausea, congestion, and fatigue (after 1 cycle). The mean capmatinib dose and median duration of capmatinib exposure were 360 mg BID and 5.5 months (range 0.5–13.9), respectively.
Table 2.
Treatment-Related Adverse Events Occurring in ≥ 10% of Patients
| Adverse Event | No. (%) of Patients with Treatment-Related Adverse Event by Grade | |||||
|---|---|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | Grade 4 | Grade 5 | All Grades | |
| Lower Extremity Edema | 5 (25) | 8 (40) | 0 (0) | 0 (0) | 0 (0) | 13 (65) |
| Fatigue | 7 (35) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (35) |
| Amylase Elevation | 1 (5) | 1 (5) | 2 (10) | 0 (0) | 0 (0) | 4 (20) |
| Creatinine Elevation | 4 (20) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (20) |
| Lipase Elevation | 1 (5) | 0 (0) | 3 (15) | 0 (0) | 0 (0) | 4 (20) |
| Myalgia | 4 (20) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (20) |
| ALT Elevation | 2 (10) | 0 (0) | 1 (5) | 0 (0) | 0 (0) | 3 (15) |
| Neuropathy | 3 (15) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (15) |
| AST Elevation | 1 (5) | 0 (0) | 1 (5) | 0 (0) | 0 (0) | 2 (10) |
| Diarrhea | 2 (10) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (10) |
| Hypophosphatemia | 1 (5) | 1 (5) | 0 (0) | 0 (0) | 0 (0) | 2 (10) |
| Pneumonitis | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (10) | 2 (10) |
| Vomiting | 2 (10) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (10) |
AST: aspartate aminotransferase, ALT: alanine aminotransferase
Two (10%) patients developed grade 5 pneumonitis. The first patient experienced grade 3 pneumonitis on crizotinib and started capmatinib two months later when dyspnea and inflammatory changes had resolved. This patient developed recurrent dyspnea and opacities after 3 months on capmatinib. Although the AE was designated as pneumonitis, respiratory failure due to progression of disease or pneumonia could not be excluded. The second patient developed pneumonitis after two months on capmatinib, a timepoint corresponding to two weeks after completing palliative thoracic spine radiation. Capmatinib had been held while the patient received radiation during cycle 1 and was resumed the day after radiation. Neither patient had received immunotherapy.
As mentioned above, six patients discontinued crizotinib due to toxicity. Of these patients, three experienced recurrence of the same toxicity on capmatinib, including one case each of chest congestion/pressure, hepatitis, and pneumonitis (as described above).
Analysis of Plasma Specimens
Analysis of Pre-Capmatinib Plasma
Plasma was collected from all patients prior to initiating capmatinib. Sixteen (80%) patients had detectable ctDNA pre-capmatinib. The four non-diagnostic specimens were from patients relapsing only in the CNS or thorax. For six of the 16 specimens with detectable ctDNA, plasma genotyping did not detect the MET driver alteration despite identifying alterations in other genes, potentially reflecting decreased sensitivity of DNA-based analyses for detecting exon 14 skipping and challenges of estimating copy number from ctDNA.12 Among the 16 patients with detectable ctDNA, five (31%) had secondary MET mutations, 3 (19%) had MAPK pathway alterations, and 2 (13%) had ERBB pathway alterations. Detailed below is a summary of the alterations found among patients with detectable ctDNA pre-capmatinib who stopped crizotinib due to intolerance versus those who discontinued crizotinib for progression.
Patients who Discontinued Crizotinib for Intolerance
Five of the 16 patients with detectable ctDNA had discontinued crizotinib for intolerance rather than progression; of these, four had stable disease on capmatinib and one had primary progression. One patient had a MET G110A mutation of unknown significance in pre-capmatinib plasma. The other four did not have secondary MET mutations. Two of the five cases had MAPK pathway alterations, both of which were BRAF V600E. BRAF V600E was not detected in tissue at diagnosis for either patient. BRAF V600E overlapped with MET G110A in one case and ERBB2 amplification in the other. The patient with BRAF V600E (0.05% allelic frequency) and MET G110A (0.37% allelic frequency) experienced primary progression on capmatinib, whereas the patient with BRAF V600E (0.06% allelic frequency) and ERBB2 amplification had stable disease.
Patients who Discontinued Crizotinib for Progression
Eleven patients with detectable pre-capmatinib ctDNA had progressed on crizotinib. Crizotinib was the last therapy for seven patients. A secondary MET mutation was detected in plasma from 4 (36%) patients with crizotinib-resistant NSCLC. The detected mutations included MET D1228H (n=2), Y1230H (n=1), and D1228N +Y1230H (n=1). The best response to capmatinib was progressive disease in one patient with D1228H, stable disease in 2 patients (one with D1228H; the other with Y1230H), and partial response in the patient with D1228N+Y1230H (Figure 1A, Supplementary Table 3). The patient with partial response discontinued treatment after 5 weeks due to death from stroke as discussed above. One patient with stable disease progressed after 5.5 months whereas the other discontinued treatment soon after first restaging (3 weeks on treatment) due to persistent disease-related symptoms.
Among the seven patients who progressed on crizotinib that did not have secondary MET mutations in pre-capmatinib plasma (Supplementary Table 3), one had MAPK alterations (KRAS V14I and a truncating NF1 mutation) and another had a non-kinase domain ERBB2 P1241S mutation of unknown significance. Five (71%) of the seven patients achieved disease control on capmatinib, including one patient with partial response. The patient with KRAS V14I and a truncating NF1 mutation, both of which activate MAPK signaling,13 had primary progression on capmatinib. The patient with ERBB2 P1241S had stable disease lasting 4 months.
When all patients with pre-capmatinib ctDNA were considered (including those with crizotinib intolerance), the DCR among patients with MET kinase domain mutations, MAPK alterations, and ERBB2 alterations was 75% (n=3/4), 33% (n=2/3), and 100% (n=2/2), respectively.
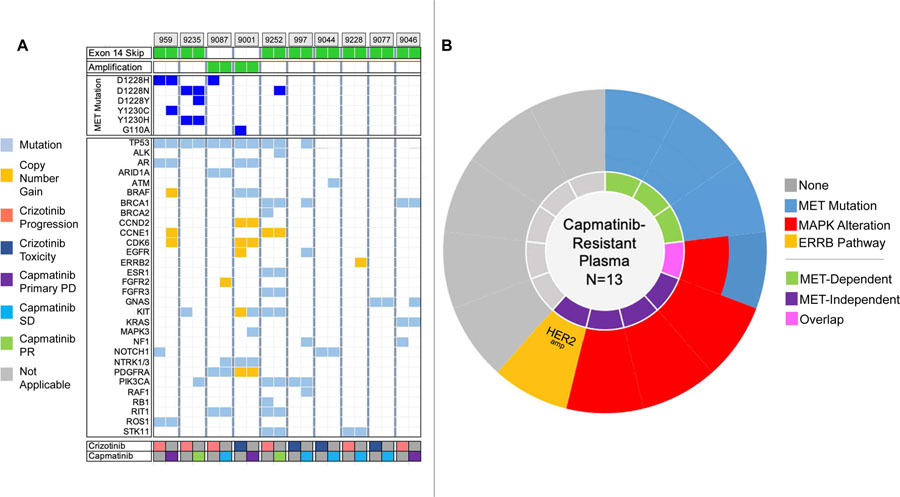
Analysis of Post-Capmatinib Plasma
Thirteen patients had detectable post-capmatinib ctDNA and are described in Figure 3 and Supplementary Table 3. Ten of these patients had paired pre- and post-capmatinib plasma specimens (Figure 3A). In total, four (31%) of 13 post-capmatinib specimens harbored secondary MET mutations and four (31%) had MAPK alterations (Figure 3B). One specimen had ERBB2 amplification and two others had EGFR mutations of unknown significance (R958H and T259M).
Figure 3. Genetic Alterations in Pre- and Post-Capmatinib Plasma Specimens.
(A) Heatmap illustrates findings in pre-capmatinib and post-capmatinib plasma specimens from 10 patients with paired specimens. Colors in legend correspond to reason for discontinuing crizotinib (crizotinib row) and best response to capmatinib (capmatinib row). Gray box: not applicable (crizotinib and capmatinib rows). PD: disease progression; PR: partial response; SD: stable disease; tox: toxicity. (B) Molecular alterations detected in plasma from 13 patients relapsing on capmatinib. Each slice corresponds to a single patient. Amp: amplification; del: deletion; mut: mutation.
Among the patients with secondary MET mutations, three patients had pre-capmatinib MET D1228 and/or Y1230 mutations. For two of these patients (one with PR and D1228N +Y1230H; one with primary progression and D1228H), the same MET mutations were detected in the post-capmatinib specimen. Both patients also had a new D1228 or Y1230 mutation in the post-capmatinib plasma. The third patient “lost” MET D1228H after 6 months of stable disease on capmatinib. One additional patient who had not had a secondary MET mutation in pre-capmatinib plasma acquired MET D1228N after 5 months of partial response to capmatinib.
Among the patients with MAPK alterations, one had KRAS V14I (described above) in pre-capmatinib plasma which was maintained after exposure to capmatinib. Three patients—two with a best response of primary progression and one with stable disease—had MAPK pathway alterations that were only detected after treatment with capmatinib (Supplementary Table 3).
DISCUSSION
Capmatinib is a potent MET-selective TKI that is approved for treatment of NSCLCs that harbor MET exon 14 skipping.6 Several other MET TKIs have also demonstrated robust clinical activity in MET-altered NSCLC.3, 4 As clinical trials for these MET TKIs occurred in parallel and prior MET-directed therapy was prohibited, it remains to be established whether patients can benefit from sequential treatment with multiple MET TKIs.
Our study demonstrates that the anti-tumor activity of capmatinib is modest in crizotinib-resistant MET-altered NSCLC. We observed a signal of activity among patients who discontinued crizotinib for intolerance and those receiving intervening non-MET directed therapies. Interestingly, the ORR and disease control rate among 4 patients who received chemotherapy and/or immunotherapy as an intervening therapy after progression on crizotinib were 50% and 100%, respectively. The sensitivity to capmatinib in these settings may reflect absence of drug-resistant clones or retreatment effect due to repopulation with MET TKI-sensitive clones in the absence of selective pressure during treatment with intervening therapies. Stabilization of CNS metastases was seen in all four patients with CNS progression on crizotinib, with two having significant shrinkage of disease, albeit not meeting criteria for RANO PR. Still, despite the overall disease control rate of 80%, the ORR of 10% and median PFS of 5.5 months underscore the need for alternative therapeutic strategies that can induce deeper and more durable responses.
In the GEOMETRY mono-1 study that led to capmatinib’s approval, the response rate was 68% in patients treated in the first-line setting and 41% among patients who had previously received chemotherapy and/or immunotherapy.6 These data in combination with the modest efficacy of capmatinib when sequenced after crizotinib suggest that the optimal position of capmatinib is as first-line treatment of patients with NSCLC that harbors MET exon 14 skipping.
MET TKIs are divided into several classes based on receptor binding mechanics. Type I inhibitors (e.g., capmatinib and crizotinib) bind the receptor in its active conformation whereas type II inhibitors target the receptor’s inactive state.1 Type I binding relies on critical interactions with the MET receptor hinge region and π-stacking interactions between the TKI and the Y1230 residue in the kinase activation loop.14 A salt bridge involving D1228 and K1110 stabilizes the activation loop in the optimal configuration for binding.14 Models predict that destabilization of the salt bridge or re-positioning of Y1230 will weaken the binding affinity of all type I TKIs for the MET receptor.14 Thus, capmatinib shares vulnerabilities with crizotinib that may compromise its activity in a crizotinib pre-treated population.
Early analyses have identified substitutions involving D1228 and Y1230 in clinical specimens from patients with crizotinib-resistant MET-altered NSCLC.15, 16 Among the four patients with these mutations at initiation of capmatinib, we observed only one partial response, albeit a disease control rate of 75%. Notably, the allelic frequency (AF) of D1228 (0.33) and Y1230 (0.5) mutations for the patient who achieved a partial response was much lower than the AF of the MET splice variant (6.5%), suggesting that the majority of tumor cells did not harbor the secondary mutations. We found that MET D1228 and Y1230 mutations persisted in plasma during treatment with capmatinib and identified a subset of patients who acquired these mutations at progression on capmatinib, suggesting that—consistent with preclinical studies—capmatinib cannot easily overcome these secondary mutations.14 Considering the small number of patients in our study with these mutations, larger datasets are needed to rigorously explore the correlation between MET activating loop mutations and capmatinib response.
Despite disease stabilization, patients in our study did not have robust responses to capmatinib. Indeed, two patients with stable disease per RECIST v1.1 withdrew from the study due to persistent disease-related symptoms, underscoring the need for alternative therapeutic strategies. In preclinical studies, class II MET TKIs (e.g., cabozantinib, merestinib, glesatinib) that bind to MET in a configuration that does not rely on interactions with the activation loop retain activity against MET D1228 and Y1230 mutations.14 These observations have been confirmed by clinical case reports.14 Studies of merestinib (NCT02920996) and cabozantinib (NCT03911193) in MET TKI-pretreated patients may help answer whether this preclinical sensitivity consistently translates into clinical responses. Given the variation in ability of distinct MET TKIs to overcome MET secondary mutations, there is rationale for pursuing repeat molecular profiling to inform selection of subsequent MET therapies.
Findings from our study and others suggest that MET-independent resistance mechanisms (including MAPK alterations and EGFR/ERBB2 amplification) are present in post-treatment specimens and can overlap with MET mutations.16–18 In other series, de novo concurrent MAPK alterations mediated primary resistance to type I MET TKIs.17, 18 In support of this, we observed primary progression on capmatinib in two of three patients who had MAPK pathway alterations in their pre-capmatinib specimen. In preclinical models, pairing a MEK inhibitor with a MET TKI re-sensitizes MET-altered tumors with MAPK alterations to treatment.17, 18 Thus, while it is important to investigate sensitivity of acquired MET resistance mutations to type II MET TKIs, it is equally essential to characterize and target molecular drivers of MET-independent resistance to develop successful post-capmatinib combination strategies.17, 18
Our study has several important limitations. The sample size in this single institution study was intentionally small. While the safety profile was reassuringly overall consistent with the larger GEOMETRY Mono-1 study,6 the rate of pneumonitis in our study was higher (10% vs 4.5%). The increased frequency of pneumonitis may reflect patient-specific factors (e.g., prior history of pneumonitis) that would have been prohibited in the larger study. With respect to efficacy, the limited number of patients and the heterogeneous makeup of their tumors may have led us to underestimate the potential for patients to benefit from sequential type I MET TKIs. We only enrolled five patients with MET amplification, in part due to the rarity of de novo MET amplification in NSCLC. As response to MET TKIs may be proportional to level of MET amplification and capmatinib is a more potent MET TKI than crizotinib,1 it is possible that the response rate to capmatinib may have been more robust if the MET copy number cutoff for eligibility were higher. We did not mandate baseline and serial CNS imaging. As a result, the intracranial activity of capmatinib after crizotinib was incompletely characterized. Our correlative analyses suffered from small numbers, a lack of post-crizotinib specimens, and the limited number of paired pre- and post-capmatinib specimens with detectable ctDNA. Finally, the lack of tumor biopsy samples limits our ability to fully assess all resistance mechanisms, including amplification events and histologic transformation.
In summary, in this phase II study, we observed modest anti-tumor activity of capmatinib in crizotinib pre-treated patients, potentially due to emergence of resistance mechanisms on crizotinib that conferred cross-resistance to capmatinib. Together with data demonstrating differential efficacy of capmatinib in the first-line vs later-line setting, our findings support prioritizing capmatinib as initial therapy for metastatic MET exon 14 altered NSCLC. Furthermore, our ctDNA analysis suggests that effective post-capmatinib therapeutic strategies will need to address secondary MET mutations and off-target resistance mechanisms involving MAPK pathway.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank all participating patients, their families and caregivers, and the co-investigators, study staff, and research coordinators whose contributions were invaluable to the conduct of this study.
CONFLICTS OF INTEREST/DISCLOSURES:
IDJ has received honoraria from Foundation Medicine, CEC, and American Lung Association, consulting fees from Boehringer-Ingelheim, Catalyst, AstraZeneca, Novocure, Pfizer, Syros, and Xcovery, research support from Array, Genentech, Novartis, Pfizer, and Guardant Health, and travel support from Array and Pfizer. JFG: has served as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Loxo/Lilly, Blueprint, Oncorus, Regeneron, Gilead, Helsinn, AstraZeneca, Pfizer, Incyte, Novartis, Merck, Agios, Amgen, and Array; research support from Novartis, Genentech/Roche, and Ariad/Takeda; institutional research support from Bristol-Myers Squibb, Tesaro, Moderna, Blueprint, Jounce, Array Biopharma, Merck, Adaptimmune, Novartis, and Alexo; and has an immediate family member who is an employee of Ironwood Pharmaceuticals. ZP has served as a compensated consultant for AstraZeneca, Jazz Pharmaceuticals, Blueprint Medicines, C4 Therapeutics, InCyte, Eli Lilly, Medtronic, Genentech and received research funding from AstraZeneca, Novartis, Takeda, Spectrum, Tesaro and Cullinan Oncology. JJL has served as a compensated consultant or received honorarium from Chugai Pharma, Boehringer-Ingelheim, Pfizer, C4 Therapeutics, Nuvalent, Turning Point Therapeutics, Blueprint Medicines, and Genentech; received institutional research funds from Hengrui Therapeutics, Turning Point Therapeutics, Neon Therapeutics, Relay Therapeutics, Roche/Genentech, Pfizer, and Novartis; received CME funding from OncLive, MedStar Health, and Northwell Health; and received travel support from Pfizer. LVS has served as a compensated consultant for AstraZeneca, Janseen, and Genentech; received research funding from Boehringer Ingelheim, AstraZeneca, Genentech, LOXO, and Blueprint. ATS has served as a compensated consultant or received honoraria from Achilles, Archer, Ariad/Takeda, Bayer, Blueprint Medicines, Chugai, Daiichi-Sankyo, EMD Serono, Foundation Medicine, Guardant, Ignyta, KSQ Therapeutics, Loxo Oncology, Natera, Novartis, Pfizer, Roche-Genentech, Servier, Syros, Taiho Pharmaceutical, and TP Therapeutics; received institutional research funding from Ariad, Ignyta, Novartis, Pfizer, Roche-Genentech, and TP Therapeutics; received travel support from Genentech and Pfizer; and is currently employed by and owns stock in Novartis. SRD provides independent image analysis for hospital contracted clinical research programs for Merck, Pfizer, Bristol Meyers Squibb, Novartis, Roche, Polaris, Cascadian, Abbvie, Gradalis, Clinical Bay, and Zai laboratories; has received honoraria from Siemens Healthineers and research funding from Lunit Inc. KP is an employee of Guardant Health. ANH: has received grant/research funding from Amgen, Pfizer, Roche/Genentech, Blueprint Medicines, Novartis, Eli Lilly, Relay Therapeutics. RSH: has served as a compensated consultant or received honoraria from Novartis, Merck Healthcare KGaA, Pfizer, Roche, Apollomics, Tarveda, Boehringer Ingelheim, Daichii Sankyo; institutional research support from Novartis, Genentech Roche, Corvus, Incyte, Exelixis, Abbvie, Daichii Sankyo, Agios, Mirati, Turning Point, Lilly. The remaining authors have no conflicts of interest to disclose.
FUNDING
The study was funded by Novartis. Plasma analysis was performed through a research collaboration with Guardant Health. Funding support for this work was also provided by U54CA224068 and a SPORE Developmental Project Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guo R, Luo J, Chang J, Rekhtman N, Arcila M, Drilon A. MET-dependent solid tumours - molecular diagnosis and targeted therapy. Nat Rev Clin Oncol. Jun 2020;doi: 10.1038/s41571-020-0377-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. Jan 2006;66(1):283–9. doi: 10.1158/0008-5472.CAN-05-2749 [DOI] [PubMed] [Google Scholar]
- 3.Drilon A, Clark JW, Weiss J, et al. Antitumor activity of crizotinib in lung cancers harboring a MET exon 14 alteration. Nat Med. 01 2020;26(1):47–51. doi: 10.1038/s41591-019-0716-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paik PK, Felip E, Veillon R, et al. Tepotinib in Non-Small-Cell Lung Cancer with. N Engl J Med. May 2020;doi: 10.1056/NEJMoa2004407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gan HK, Millward M, Hua Y, et al. First-in-Human Phase I Study of the Selective MET Inhibitor, Savolitinib, in Patients with Advanced Solid Tumors: Safety, Pharmacokinetics, and Antitumor Activity. Clin Cancer Res. Aug 2019;25(16):4924–4932. doi: 10.1158/1078-0432.CCR-18-1189 [DOI] [PubMed] [Google Scholar]
- 6.Wolf J, Seto T, Han JY, et al. Capmatinib in. N Engl J Med. Sep 2020;383(10):944–957. doi: 10.1056/NEJMoa2002787 [DOI] [PubMed] [Google Scholar]
- 7.Wolf J, Overbeck TR, Han J-Y, et al. Capmatinib in patients with high-level MET-amplified advanced non-small cell lung cancer (NSCLC): results from the phase 2 GEOMETRY mono-1 study. Journal of Clinical Oncology 2020. [Google Scholar]
- 8.Solomon BJ, Besse B, Bauer TM, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol. Dec 2018;19(12):1654–1667. doi: 10.1016/S1470-2045(18)30649-1 [DOI] [PubMed] [Google Scholar]
- 9.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. Nov 2013;31(11):1023–31. doi: 10.1038/nbt.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res. Aug 2018;24(15):3539–3549. doi: 10.1158/1078-0432.CCR-17-3831 [DOI] [PubMed] [Google Scholar]
- 11.Lin NU, Lee EQ, Aoyama H, et al. Response assessment criteria for brain metastases: proposal from the RANO group. Lancet Oncol. Jun 2015;16(6):e270–8. doi: 10.1016/S1470-2045(15)70057-4 [DOI] [PubMed] [Google Scholar]
- 12.Benayed R, Offin M, Mullaney K, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res. Aug 2019;25(15):4712–4722. doi: 10.1158/1078-0432.CCR-19-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hernández-Porras I, Schuhmacher AJ, Garcia-Medina R, et al. K-Ras(V14I) - induced Noonan syndrome predisposes to tumour development in mice. J Pathol. 06 2016;239(2):206–17. doi: 10.1002/path.4719 [DOI] [PubMed] [Google Scholar]
- 14.Engstrom LD, Aranda R, Lee M, et al. Glesatinib Exhibits Antitumor Activity in Lung Cancer Models and Patients Harboring. Clin Cancer Res. Nov 2017;23(21):6661–6672. doi: 10.1158/1078-0432.CCR-17-1192 [DOI] [PubMed] [Google Scholar]
- 15.Heist RS, Sequist LV, Borger D, et al. Acquired Resistance to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol. 08 2016;11(8):1242–1245. doi: 10.1016/j.jtho.2016.06.013 [DOI] [PubMed] [Google Scholar]
- 16.Recondo G, Bahcall M, Spurr LF, et al. Molecular Mechanisms of Acquired Resistance to MET Tyrosine Kinase Inhibitors in Patients with MET Exon 14-Mutant NSCLC. Clin Cancer Res. Jun 2020;26(11):2615–2625. doi: 10.1158/1078-0432.CCR-19-3608 [DOI] [PubMed] [Google Scholar]
- 17.Rotow JK, Gui P, Wu W, et al. Co-occurring Alterations in the RAS-MAPK Pathway Limit Response to MET Inhibitor Treatment in MET Exon 14 Skipping Mutation-Positive Lung Cancer. Clin Cancer Res. Jan 2020;26(2):439–449. doi: 10.1158/1078-0432.CCR-19-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzawa K, Offin M, Lu D, et al. Activation of KRAS Mediates Resistance to Targeted Therapy in MET Exon 14-mutant Non-small Cell Lung Cancer. Clin Cancer Res. 02 2019;25(4):1248–1260. doi: 10.1158/1078-0432.CCR-18-1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.