To the Editor,
Among the different SARS-CoV-2 variants of concern [1], Delta and Omicron currently account for the vast majority of cases worldwide, with the latter rapidly replacing the former in many countries [2]. Notably, Omicron is the most divergent from the wild-type B.1 strain, with 15 amino acid changes in the receptor-binding domain of the spike protein, the major target of neutralizing antibodies. Indeed, Omicron partly escapes natural or vaccine-elicited immunity [3] and can decrease the efficacy of currently licensed therapeutic monoclonal antibodies (mAbs) [4,5].
We assessed the ex vivo inhibition of Omicron and Delta variants by sera obtained from unvaccinated patients treated with one of the three licensed mAb preparations, namely bamlanivimab/etesevimab, casirivimab/imdevimab, and sotrovimab. A pair of patient sera samples was collected, the first before (to exclude any prior neutralizing antibody (NtAb) activity) and the second 1 hour after mAbs infusion.
The study was conducted in accordance with the Declaration of Helsinki and approved by the local ethics committee (Neutro-COVID observational study, protocol number 4069/21). Written informed consent was obtained from all the patients enrolled.
The efficacy of the mAbs was assessed by a live virus neutralization assay against wild type (GISAID accession number EPI_ISL_2472896), Delta (EPI_ISL_2840619), the Omicron sublineage BA.1 (EPI_ISL_6777160), and the Delta sublineage AY.4.2 (EPI_ISL_6943992), recently detected in Italy [6] and carrying the additional Y145H and A222V spike substitutions (details of each viral stock are indicated in Table S1). NtAb titres were determined by a microneutralization live virus assay [7] and defined as the reciprocal value of the sample dilution that showed a 50% protection of virus-induced cytopathic effect (ID50).
Statistical analyses were performed using IBM SPSS Statistics, version 20. Sera with ID50 < 10 were defined as negative and scored as 5 for statistical analysis. Data were expressed as median (IQR), as appropriate for the distribution of data based on the Shapiro-Wilk test for normality. The Kruskal-Wallis test followed by Mann-Whitney test post hoc analysis was used to compare independent groups, and the Friedman test followed by Wilcoxon rank sum test post hoc analysis was used to compare multiple paired data. Spearman analysis was used to measure the correlation between NtAb titres against the different variants.
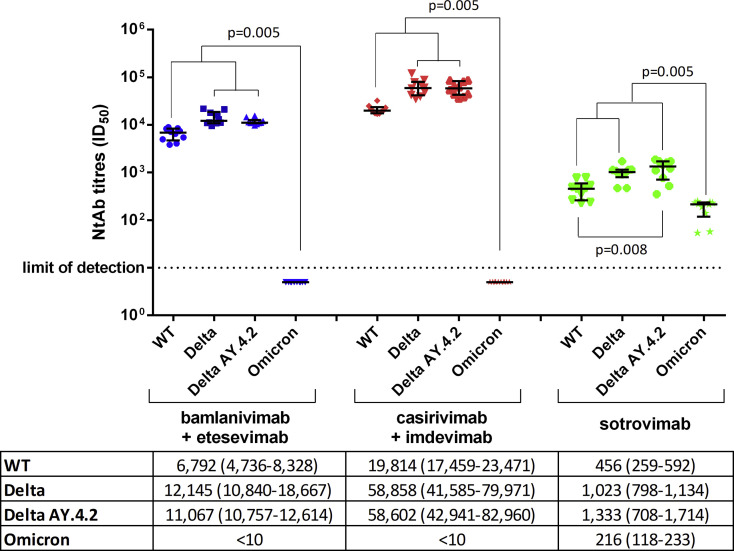
Of 30 patients studied (14 males, age 59 ± 18 years, full characteristics summarized in Table S2), one was asymptomatic and the others had mild symptoms such as cough (n = 19), fever (n = 17), headache (n = 13), gastrointestinal symptoms (n = 4), and dyspnoea (n = 2). Patients were randomly treated with bamlanivimab/etesevimab (n = 10), casirivimab/imdevimab (n = 10), or sotrovimab (n = 10), 3.5 ± 1.7 days from diagnosis. None of the patients required hospitalization. All preinfusion sera were negative for SARS-CoV-2 NtAb activity. In postinfusion sera, casirivimab/imdevimab, bamlanivimab/etesevimab, and sotrovimab showed activity against the wild-type variant (19,814 (17,459–23,471), 6792 (4736–8328), and 456 (259–592)), the Delta variant (58,858 (41,585–79,971), 12,145 (10,840–18,667), and 1023 (798–1134)), and Delta AY.4.2 (58,602 (42,941–82, 960), 11,067 (10,757–12,614), and 1333 (708–1714)). Notably, sotrovimab was the only active treatment against the Omicron variant (216 (118–233)) (Fig. 1 ).
Fig. 1.
Ex vivo anti–SARS-CoV-2 wild type, Delta, Delta AY.4.2, and Omicron neutralizing antibody (NtAb) titres measured in sera from 30 patients after infusion of the bamlanivimab/etesevimab (in blue), orcasirivimab/imdevimab (in red), or sotrovimab (in green) monoclonal antibody cocktails. Paired data were analyzed by the nonparametric Wilcoxon signed rank sum test. NtAb titre before infusion was negative against each variant tested (data not shown in figure). Median ID50 (IQR) are indicated in the table below the figure. ID50, the reciprocal value of the sera dilution showing the 50% protection of virus-induced cytopathic effect; NtAb, neutralizing antibody.
Within each individual treatment group, the NtAb titres to Delta and Delta AY.4.2 variants were significantly higher than those to wild type (p = 0.008 for AY.4.2 vs. wild type with sotrovimab; p = 0.005 for all other comparisons). NtAb titres to wild type, Delta, and Delta AY.4.2 variants were higher than those to Omicron within all individual treatment groups (p = 0.005 for all comparisons).
Comparing treatments, casirivimab/imdevimab neutralizing titres were significantly higher than bamlanivimab/etesevimab and sotrovimab against wild type, Delta, and the Delta AY.4.2 variants, and bamlanivimab/etesevimab neutralizing titres were significantly higher than sotrovimab for the same variants (p < 0.001 for all comparisons). When considering all 30 postinfusion sera, a significant correlation was observed between NtAb titres to any pair of wild type, Delta, and Delta AY.4.2 variants (p < 0.001 for all comparisons), whereas NtAb titres to Omicron correlated significantly only to NtAb titres to wild-type virus (p = 0.009) (Fig. S1).
A previous report documented in vitro inhibition of the Delta variant by the individual mAbs etesevimab, casirivimab, and imdevimab, but not bamlanivimab, at slightly higher levels compared with the wild-type B.1 virus [8]. Our study confirms these findings by testing the bamlanivimab/etesevimab and casirivimab/imdevimab cocktails in an ex vivo format. In addition, we demonstrated maintenance of activity against Delta AY.4.2 for the first time, implying that the additional Y145H and A222V mutations have no impact on neutralization by these mAbs preparations. Most importantly, our results support full escape of commonly used mAbs cocktails by the Omicron variant, as recently reported [4], although sotrovimab appears to retain activity against all variants tested, including Omicron (albeit reduced by an average 2.7-fold in paired measurements), similar to what has been reported in the literature [5,9]. Since Omicron has rapidly replaced the circulating variants, the mAbs arsenal should be updated accordingly. Clearly, surveillance of SARS-CoV-2 evolution over time and in different geographical areas remains a priority to adapt our defences against the pandemic.
Transparency declaration
This work was partly funded by the European Commission under HORIZON-HLTH-2021-CORONA-01, Project EuCARE, grant agreement N. 101046016. MZ reports consultancy for ViiV Healthcare, Gilead Sciences, Janssen-Cilag, Theratechnologies, and Merck Sharp and Dohme (MSD) and grants for his institution from ViiV Healthcare, Theratechnologies, and Gilead Sciences outside the submitted work. BR received support for travel to meetings from Abbvie, Gilead Sciences, Janssen-Cilag, MSD, ViiV Healthcare, and Bristol Myers Squibb and fees for attending advisory boards and speaker's honoraria from Abbvie, Gilead Sciences, Janssen-Cilag, MSD, ViiV Healthcare, and Bristol-Myers Squibb. All other authors have no conflicts to declare.
Author contributions
IV, MZ, DF, and MRG conceived the idea for this work. FD, LF, VM, AL, and ES performed the experiments. FD, LF, BR, and IV contributed to the data analysis. VM, AL, ES, GZ, MRG, BR, and DF collected the samples and provided the virus lineages for this work. IV, FD, and MZ wrote the paper. All authors contributed to the revision and approved the final version of the manuscript.
Acknowledgements
We would like to thank Dr. Cesira Nencioni and Dr. Alessandro Lanari for their support in this study.
Editor: R. Chemaly
Footnotes
The omicron variant discussed in this study is the omicron BA.1 to distinguish it from other omicron variants emerging later including BA.1.1, BA.2 and BA.3.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2022.03.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Multimedia component 2
Multimedia component 3
References
- 1.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodcroft E. CoVariants: SARS-CoV-2 mutations and variants of interest; C2021. https://covariants.org/ [cited 2022 Feb 21]. Available from:
- 3.Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S., Pollard A.J., et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet. 2022;399:234–236. doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., et al. An infectious SARS-CoV-2 B.1.1.529 omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat Med. 2022;19:1–6. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angeletti S., Giovanetti M., Fogolari M., Cella E., De Florio L., Lintas C., et al. SARS-CoV-2 AY.4.2 variant circulating in Italy: genomic preliminary insight. J Med Virol. 2022;94:1689–1692. doi: 10.1002/jmv.27451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vicenti I., Gatti F., Scaggiante R., Boccuto A., Zago D., Basso M., et al. Single-dose BNT162b2 mRNA COVID-19 vaccine significantly boosts neutralizing antibody response in health care workers recovering from asymptomatic or mild natural SARS-CoV-2 infection. Int J Infect Dis. 2021;108:176–178. doi: 10.1016/j.ijid.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 9.Tzou P.L., Tao K., Nouhin J., Rhee S.Y., Hu B.D., Pai S., et al. Coronavirus Antiviral Research Database (CoV-RDB): an online database designed to facilitate comparisons between candidate anti-coronavirus compounds. Viruses. 2020;12:1006. doi: 10.3390/v12091006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 2
Multimedia component 3