Abstract
Abstract
African swine fever virus (ASFV) causes a highly contagious and often lethal swine viral disease, and leads to tremendous economic losses to the swine industry. Unfortunately, there are no vaccines and effective antiviral agents available to prevent and control ASFV outbreaks. Therefore, it is necessary to develop simple and rapid strategies to monitor ASFV-infected pigs to restrain its spread. In the current study, ASFV capsid protein p72 was expressed along with its chaperone pB602L to form trimers in human embryonic kidney 293 (HEK293) cells. The p72 trimers were subsequently labeled with colloidal gold to develop a immunochromatographic strip. The strip showed high specificity to ASFV-positive serum and no cross-reactivity to other swine virus positive sera. Importantly, the strip showed a higher sensitivity of detecting ASFV antibodies in both positive standard serum and clinical serum samples than a commercial enzyme-linked immunosorbent assay (ELISA) kit. Taken together, these results demonstrate the strip as a reliable diagnostic tool against ASFV infection, which will be appropriate for application in prevention and control of ASFV.
Key points
• ASFV p72 trimers were successfully generated.
• A colloidal gold strip was developed based on ASFV p72 trimers.
• The strip is appropriate for detecting ASFV antibodies in the field.
Keywords: Antibody detection, ASFV, Colloidal gold immunochromatographic strip, P72 trimers
Introduction
African swine fever (ASF), caused by ASF virus (ASFV), is a devastating disease of swine notifiable to the World Organization for Animal Health (OIE) (2019). ASF was first reported in Kenya in 1921 and rapidly spread throughout Europe in the 1950s (Dixon et al. 2019; Montgomery 1921). In June 2007, ASF was reported in the Caucasus region of Georgia and has since become prevalent in Russia and spread to other neighboring countries (Gogin et al. 2013). In August 2018, the first ASF case emerged in China, which has half of the world’s pig population (Zhou et al. 2018). At present, ASF is epidemic in many countries burdening global swine industry (Kedkovid et al. 2020).
Unfortunately, there are no vaccines or effective treatments to restrain ASF outbreaks (Gaudreault and Richt 2019; Revilla et al. 2018). The only control strategy is to eliminate ASFV-infected pigs (Costard et al. 2009). As a consequence, ASFV antibody surveillance as an important indicator of ASFV infection is a key to prevent and control this disease (Dixon et al. 2020).
ASFV is a large enveloped DNA virus in the Asfivirus genus in the Asfarviridae family (Galindo and Alonso 2017). ASFV genome ranges from 170 to 190 kb and encodes more than 150 proteins (Wang et al. 2020). ASFV B646L gene encodes the major capsid protein p72. The amino acid sequences of p72 share high identity among different ASFV strains, including the recently identified natural mutants in China (Yu et al. 1996; Zhang et al. 2021c). The p72 is the most predominant structural component and constitutes approximately 31–33% of the total mass of ASFV virions (Revilla et al. 2018). It is also one of the first identified viral proteins responsible for induction of antibodies post infection (Carolina et al. 2013). As a consequence, p72 is well suited as an antibody detection target for ASFV infection (Heimerman et al. 2018; Jia et al. 2017; Yu et al. 1996).
In this study, we have developed a colloidal gold immunochromatographic strip with p72 trimers for ASFV antibody detection. ASFV antibodies can be detected by the naked eye in both positive standard serum and clinical serum samples using the strip without specialized equipment or professional personals. More importantly, the strip is more sensitive than a commercial enzyme-linked immunosorbent assay (ELISA) kit for clinical serum sample detection. The strip will be a potent detection tool for ASF surveillance in the field.
Materials and methods
Serum samples
Healthy swine negative serum, positive standard sera for ASFV, pseudorabies virus (PRV), classical swine fever virus (CSFV), and porcine circovirus type 2 (PCV2) were purchased from China Veterinary Culture Collection Center (Beijing, China). Positive serum samples for porcine reproductive and respiratory syndrome virus (PRRSV), porcine parvovirus (PPV), foot-and-mouth disease virus (FMDV), and 54 serum samples positive for ASFV nucleic acid were collected and stored in our laboratory. All sample treatments were strictly performed in accordance with the standard operation for ASFV by OIE.
Expression, purification, and characterization of p72
The genes of ASFV p72 and pB602L (Pig/HLJ/18 GenBank: MK333180.1) were codon optimized, synthesized and cloned into pCMV vector by Sangon Biotech (Shanghai, China) (Liu et al. 2019a). The sequences were deposited in GenBank with the p72 accession number OL698792 and the pB602L accession number OL698793. The recombinant p72 was fused with Flag-tag at its N terminus, and the recombinant pB602L was fused with His-tag at its N terminus. These two recombinant vectors were co-transfected into human embryonic kidney 293 (HEK293) cells for expression. The target protein expression was detected with anti-Flag-tag antibody (Zen BioScience, Chengdu, China) and anti-His-tag antibody (Proteintech, Wuhan, China) by western blotting (WB).
The p72 was purified with Flag affinity chromatography using anti-DYKDDDDKG affinity resin (GenScript, Nanjing, China). The concentrated p72 was further fractionated by a HiLoad 16/600 Superdex 200 pg column (GE healthcare, Uppsala, Sweden). The purified p72 was analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The purified p72 was further applied to a size exclusion chromatography column (Superdex 200 Increase 10/300 GL; GE healthcare) and then analyzed by native-PAGE. The molecular mass (MM) was determined by the calibration curve and calculated from its elution volume (Ve) and column void volume (Vo). The antigenicity of the purified p72 was tested by WB using ASFV-positive standard serum and negative serum as control. The p72 concentration was determined by BCA protein concentration determination kit (Beyotime Biotechnology, Shanghai, China) for further use.
Generation of polyclonal antibodies (pAbs) against the purified p72
The experimental procedure for generation of pAbs was authorized and supervised by the Ethical and Animal Welfare Committee of Key Laboratory of Animal Immunology of the Ministry of Agriculture of China (Permit 2020060401). The purified p72 was used as immunogen to immunize 6-week-old New Zealand white rabbits with 300 μg in Freund’s complete adjuvant (Sigma, St. Louis, USA) per rabbit by multi-point subcutaneous injection on the back. Rabbits were immunized for another two times with 3-week interval. Two weeks after the third immunization, the immune serum was isolated from the immunized rabbit blood, and the serum antibody titers were identified by indirect ELISA. Briefly, the ELISA plate was coated with 0.2 μg/mL purified p72, and then added with two-fold serially diluted immune sera. A large amount of high-titer serum was obtained by heart blood collection method and purified by sulfate precipitation (Liu et al. 2017).
Conjugation of the purified p72 with colloidal gold
Colloidal gold was obtained by previously reported sodium citrate reduction method (Fren 1973). Briefly, 1 mL of 1.0% chloroauric acid (Aladdin, Shanghai, China) was added to 99 mL of ultrapure water and boiled for 3 min. The boiled solution was added with 1.6 mL of 1.0% trisodium citrate (Aladdin) solution and heated continuously for 5 min until the color changed to deep red with no longer changes. The colloidal gold solution was cooled to room temperature (RT) and store at 4 °C.
The optimal amount of antigen and labeling conditions was determined according to a previous report (Sun et al. 2019). The purified p72 was incubated with colloidal gold solution at RT for 30 min. The mixture was added with the 3% casein solution and further incubated at RT for 20 min. The resulting suspension was centrifuged at 12,000 × g at 4 °C for 30 min, and the colloidal gold-labeled p72 was suspended in 0.02 M sodium borate buffer containing 1% bovine serum albumin (BSA), 0.1% NaN3.
Assembly of the strip
The fiberglass pad was saturated with a buffer containing 2.0% (w/v) BSA, 20 mM sodium borate, 2.0% (w/v) sucrose and 0.1% (w/v) NaN3, and dried as the sample pad. The colloidal gold-labeled p72 was dispensed on the fiberglass pad and dried at 45 °C for 1 h as the conjugate pad. Staphylococcal protein A (SPA, Sigma) and rabbit anti-p72 pAbs were dispensed on nitrocellulose (NC) membrane (300 mm × 25 mm, Merck Millipore) as the test and control lines, respectively, and dried at 42 °C for 4 h. Pure cellulose fiber was used as the absorbent pad. These pads and NC membrane were sealed and stored at RT under dry conditions.
The assembly of the strip was performed as described previously (Li et al. 2017). Briefly, the sample pad, conjugate pad, NC membrane, and absorbent pad were assembled on a support plate sequentially with 1–2 mm overlapping each other, and then cut into individual 2.8-mm-wide strip. The strip was installed in a plastic case and sealed for further use.
Cross-reactivity evaluation of the strip
The cross-reactivity test of the strip was determined by positive sera for ASFV and other swine viruses (PRRSV, PRV, CSFV, PCV2, FMDV, PPV) and healthy swine negative serum. For testing, the serum samples were diluted with normal saline at 1:200 and added to each sample well of the strip with 100 μL. The results were observed by the naked eye 10 min later.
Detection limit evaluation of the strip
The detection limit of the strip was evaluated by ASFV-positive standard serum. The serum was diluted by two-fold serial dilutions from 1:100 to 1:819,200. The strip was tested with the diluted serum, and the test line was scanned with a TSR3000 membrane strip reader (BioDot, Carlsbad, USA) to obtain the relative optical density (ROD). The result was displayed as graph/density × area (G/D × A) of the ROD value, and it is considered positive when > 10 (Li et al. 2021). The diluted sera were tested by a commercial ELISA kit (IDVET, Marseille, France) in parallel. The results were expressed in an optical density sample/positive control (S/P) ratio. It is considered positive when S/P > 0.4 according to the manufacturer’s instruction.
Stability and repeatability evaluation of the strip
The stability and repeatability of the strip were evaluated with four-fold serially diluted ASFV-positive standard sera and a negative serum. The strips were stored at RT for 2, 4, and 6 months and tested for their stability. In the meantime, all tests were repeated three times to determine repeatability. The strip test lines were scanned by the TSR3000 membrane strip reader to obtain the ROD values. Coefficients of variation (CVs) of the strip in testing each dilution were individually calculated as the ratio of standard deviation (SD) to average value based on three independent experiments. It was considered an acceptable repeatability level for the strip when the CV value was less than 15%.
Clinical sample detection
To further evaluate the strip for detecting clinical samples, a total of 54 serum samples positive for ASFV nucleic acid were tested with the strip and the commercial IDVET ELISA kit, respectively. The seroprevalence rates were compared between these two assays.
Results
Expression, purification, and characterization of recombinant p72
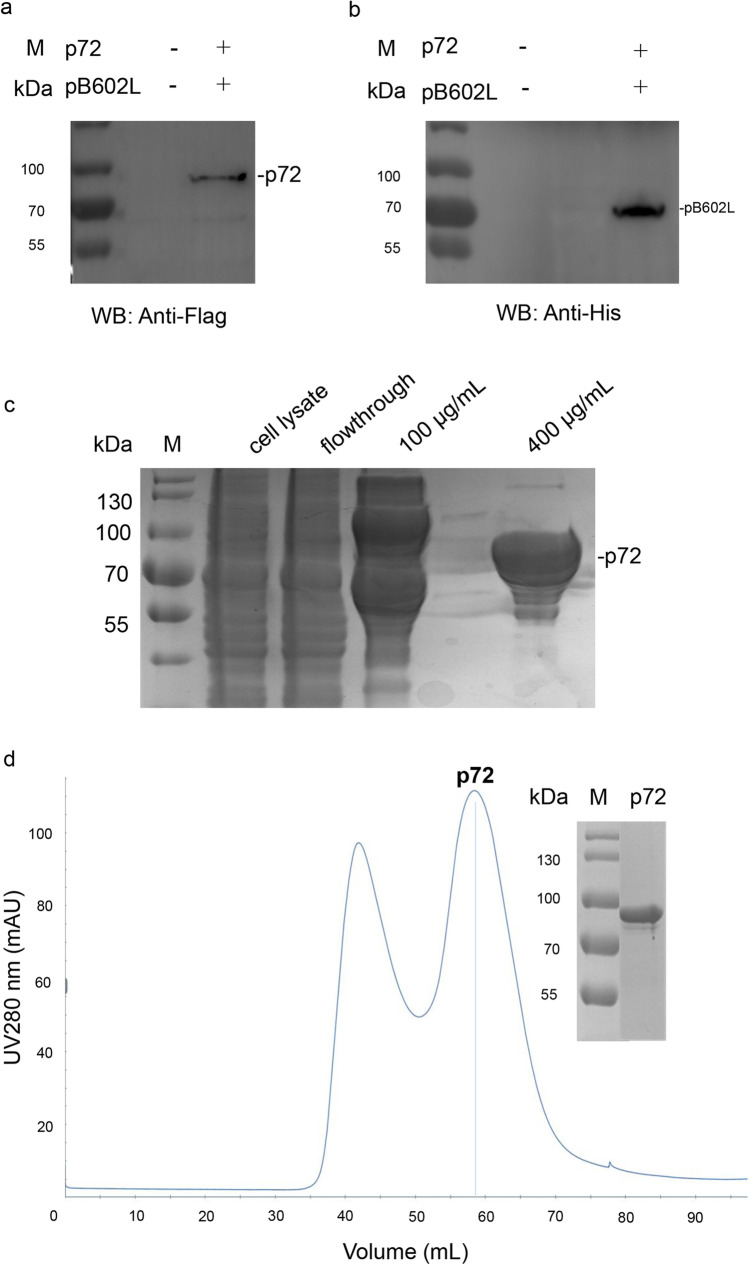
In order to obtain the trimer form of p72, we co-expressed ASFV p72 and its chaperon pB602L in HEK293 cells (Liu et al. 2019a). The tag antibodies detected that the MM of p72 was between 70 and 100 kDa, and pB602L was 70 kDa (Fig. 1a, b). The p72 was purified by Flag affinity chromatography and eluted with 400 μg/mL Flag peptide (Fig. 1c). The p72 was further obtained with high purity by gel filtration chromatography (Fig. 1d).
Fig. 1.
Expression and purification of the p72. a The expression of the p72 was verified by anti-Flag tag antibody. b The expression of the pB602L was verified by anti-His tag antibody. c SDS-PAGE analysis of the p72 purified by Flag affinity chromatography. The p72 was eluted with 400 μg/mL Flag peptide. d SDS-PAGE analyses of the p72 purified by gel filtration chromatography; c, d M protein MM marker
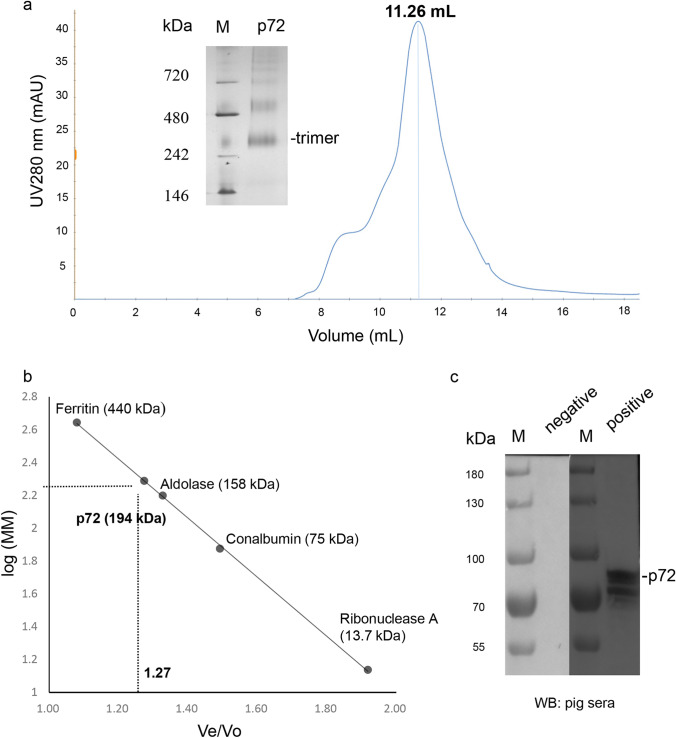
The purified p72 was applied to calibrated size exclusion chromatography and eluted at 11.26 mL (Fig. 2a), suggesting that its MM corresponds to approximately 194 kDa (Fig. 2b). The eluted p72 was further analyzed by native-PAGE (Fig. 2c) and indicated as trimers, in consistent with the previous report (Liu et al. 2019a). Importantly, the purified p72 was recognized by ASFV-positive standard serum but not negative serum (Fig. 2c), which demonstrated that p72 trimers showed good antigenicity.
Fig. 2.
Characterization of the purified p72. a Size exclusion chromatography and native-PAGE analyses of the purified p72. The purified p72 was eluted at 11.26 mL. Native-PAGE analysis of the p72 was indicated by the black line. b The calibration curve of the Superdex 200 Increase 10/300 GL column used for the purified p72 characterization. The estimated MM (194 kDa) of the p72 in solution was calculated based on the Ve/Vo value (1.27) of the p72. c Immunogenicity tests of the purified p72 by pig negative and positive sera; M: protein MM marker
Preparation of anti-p72 pAbs
The purified p72 was immunized in rabbits to prepare pAbs. After three immunizations, the titer of rabbit anti-p72 pAbs was determined by indirect ELISA. The prepared pAbs reacted well with the p72 (date not shown) and the titer reached 1:105.
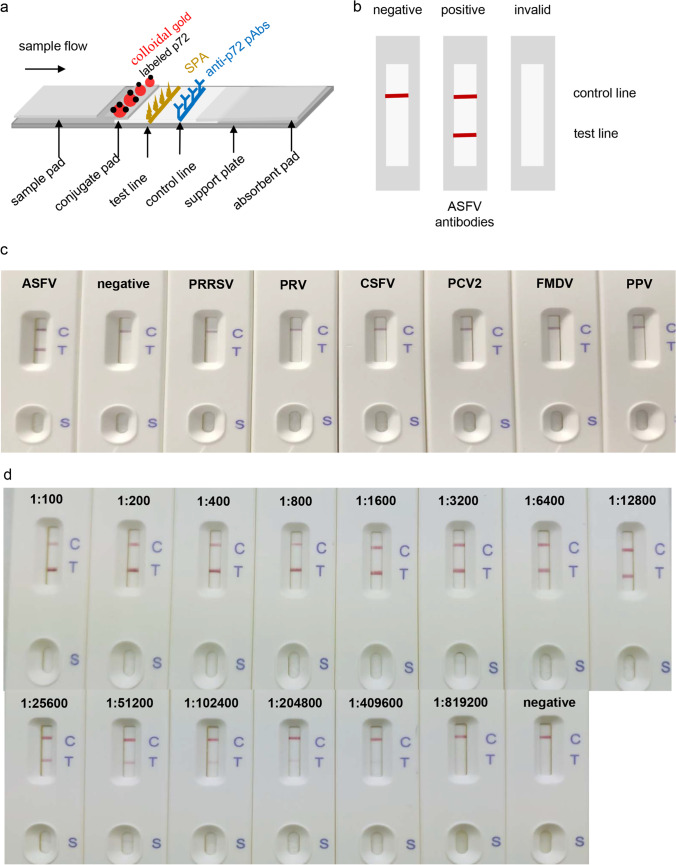
Assembly of the strip
The schematic diagram of the strip is shown in Fig. 3a. In the presence of the specific anti-p72 antibodies in the serum, they will be captured first by the colloidal gold-labeled p72 to form antibody-antigen immune complexes. The complexes will be subsequently captured by SPA, which binds to Fc fragments of mammalian IgG and certain IgM and IgA. A red band therefore appears at the test line and is visible to the naked eye. In the absence of anti-p72 antibodies, the antibody-antigen immune complexes will neither form nor show a red band at the test line. As a quality control check, the colloidal gold-labeled p72 can be captured by anti-p72 pAbs at the control line position as a visible red band whether or not the serum contains anti-p72 antibodies. Otherwise the test is considered invalid (Fig. 3b).
Fig. 3.
Schematic representation and test of the strip. a, b Schematic figures of the strip and the test results. c Cross-reactivity test of the strip. The test serum samples were diluted by 1:200 and added to the sample well of the strip. d The detection limit test of the strip. ASFV-positive standard serum was diluted by two-fold serial dilutions from 1:100 to 1:819,200 and added to the sample well of the strip. After 10 min, the results were observed by the naked eye
Cross-reactivity of the strip
Cross-reactivity of the strip was evaluated with ASFV-positive standard serum, negative serum, and sera positive for PRRSV, PRV, CSFV, PCV2, FMDV, and PPV. Serum samples with 200-fold dilution were applied to the strip for test. As shown in Fig. 3c, the strip showed only one red band at the control line when tested with negative serum and other swine virus positive sera. In contrast, the strip showed two red bands at the test and control lines, respectively, when tested with the ASFV-positive standard serum. The results demonstrate that the strip has high specificity for detection of anti-ASFV antibodies.
The detection limit of the strip
The detection limit of the strip was evaluated by two-fold serially diluted ASFV-positive standard serum with immunofluorescence assay (IFA) titer 1:4000. As shown in Fig. 3d, the red color depth of the test line is directly proportional to the antibody level within a certain range, and the strip still showed a red band at the test line with addition of 1:204,800 or even lower diluted positive serum. The strip test lines were scanned with the TSR3000 membrane strip reader, and the G/D × A of the ROD value decreased as the serum was diluted. As the sample is considered positive when the value is > 10 (Li et al. 2021), the detection limit of the strip reached up to 1:409,600 (Table 1). In parallel, we also utilized the commercial IDVET ELISA kit to perform the same test. The kit detection limit was 1:6400 (Table 1), where S/P > 0.4 is considered positive according to the instruction manual. These results provide evidence that the strip has high sensitivity for detecting anti-ASFV antibodies.
Table 1.
Detection limit of the strip and the IDVET ELISA kit determined by ASFV-positive standard serum
| Dilution | ELISA | Strip | ||||
|---|---|---|---|---|---|---|
| OD 450 | S/P | Result | G/D × A-ROD × pixel | G/peak-ROD × pixel | Result | |
| 1:100 | 3.06 | 1.62 | + | 1035.91 | 0.79 | + |
| 1:200 | 2.83 | 1.49 | + | 984.93 | 0.68 | + |
| 1:400 | 2.74 | 1.43 | + | 866.30 | 0.60 | + |
| 1:800 | 2.53 | 1.32 | + | 794.95 | 0.51 | + |
| 1:1600 | 2.04 | 1.14 | + | 669.30 | 0.41 | + |
| 1:3200 | 1.34 | 0.66 | + | 646.25 | 0.38 | + |
| 1:6400 | 0.85 | 0.42 | + | 554.74 | 0.33 | + |
| 1:12,800 | 0.49 | 0.18 | − | 385.62 | 0.24 | + |
| 1:25,600 | 0.29 | 0.07 | − | 229.56 | 0.15 | + |
| 1:51,200 | 0.17 | 0.01 | − | 138.04 | 0.09 | + |
| 1:102,400 | 0.12 | − 0.02 | − | 87.98 | 0.06 | + |
| 1:204,800 | 0.09 | − 0.04 | − | 57.32 | 0.04 | + |
| 1:409,600 | 0.07 | − 0.06 | − | 22.45 | 0.02 | + |
| 1:819,200 | 0.06 | − 0.06 | − | 8.67 | 0.01 | − |
| N | 0.16 | 0.00 | − | 8.42 | 0.00 | − |
− negative; + positive, S/P > 0.4 is considered positive. G/D × A means density value of the sampled outline, multiplied by its area. G/peak indicates graph peak density, maximum density value of sampled line points. ROD means relative optical density, inverse of gray level value with a logarithmic transformation G/D × A-ROD × pixel > 10 is considered positive
Stability and repeatability of the strip
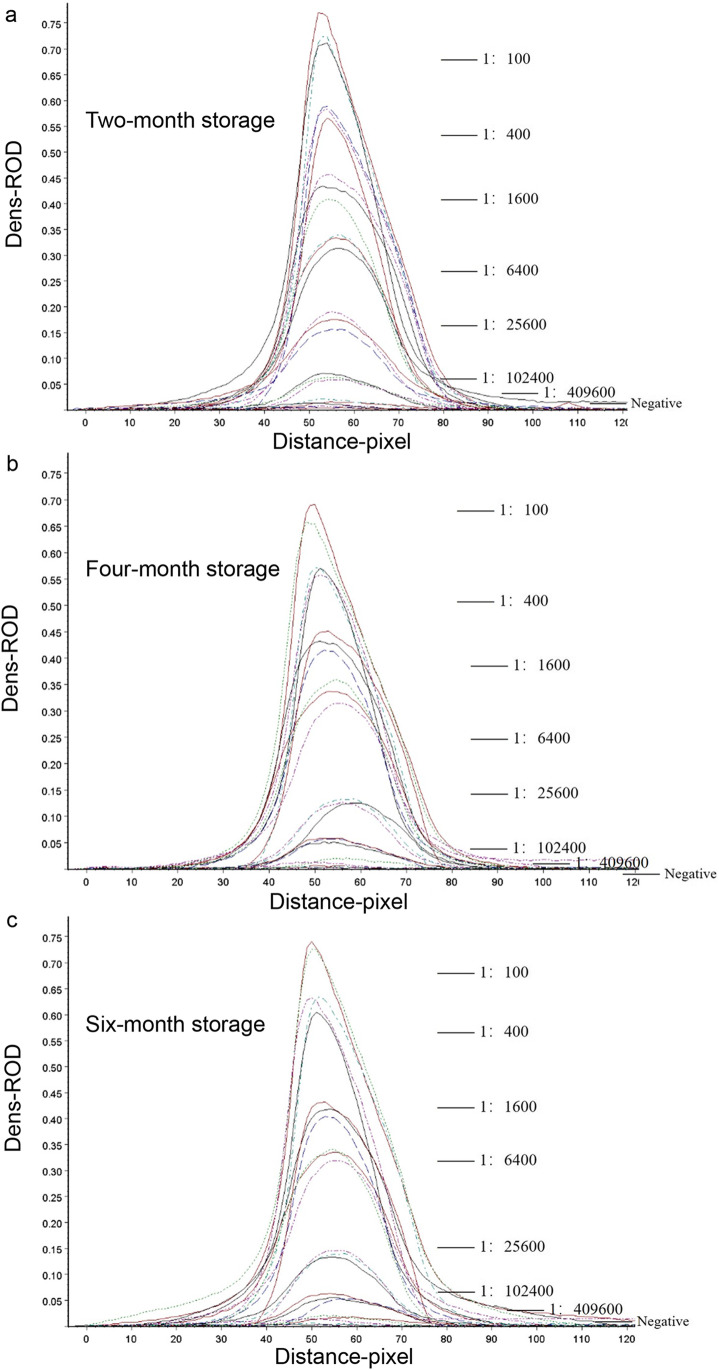
In stability assay, the strips stored for different time periods (2, 4, and 6 months) were measured by the ASFV-positive standard serum with a four-fold dilution from 1:100 to 1:409,600. The strip of 6-month storage still showed almost identical detection limit to the newly produced strip, where the detection limit reached 1:409,600 (Table 2). Additionally, the strip of each time-period storage was tested with each diluted sample for three times to assess repeatability. As shown in Fig. 4 and Table 2, all the CV values were less than 15% within the allowable range of error. These results show that the strip has good stability and repeatability within at least 6 months.
Table 2.
Repeatability of the strips with different storage time determined by CV value
| Time point (month) /Sample (dilution) | 2 | 4 | 6 | CV value (%) |
|---|---|---|---|---|
| Average G/D × A-ROD × pixel of three repeat tests ± SD | ||||
| 1:100 | 998.5±6.40 | 978.66±5.02 | 1005.31±1.21 | 1.39% |
| 1:400 | 880.89±11.56 | 749.74±11.06 | 931.53±3.63 | 10.99% |
| 1:1600 | 718.77±8.54 | 714.08±1.08 | 639.27±3.07 | 6.46% |
| 1:6400 | 542.61±1.44 | 594.63±6.51 | 535.57±7.20 | 5.79% |
| 1:25,600 | 186.79±9.32 | 185.04±3.34 | 186.78±1.51 | 0.54% |
| 1:102,400 | 105.09±1.76 | 81.44±3.96 | 88.30±6.33 | 13.28% |
| 1:409,600 | 27.78±1.05 | 23.19±3.92 | 25.89±4.76 | 9.01% |
| N | 9.07±0.76 | 7.06±0.81 | 9.21±1.17 | 14.23% |
CV value (%) = (SD/mean) × 100%
Fig. 4.
Stability and repeatability test of the strip. a The scan result of the test lines on the strips stored for 2 months by TSR3000 membrane strip reader. b. The scan result of the test lines of the strips stored for 4 months with TSR3000 membrane strip reader. c. The scan result of the test lines of the strips stored for 6 months with TSR3000 membrane strip reader. The test sample is ASFV-positive standard serum with a four-fold dilution ratio and a negative serum as control
Clinical serum sample detection of the strip
To evaluate whether the strip is suitable for ASFV antibody detection in clinical application, 54 serum samples positive for ASFV nucleic acid were simultaneously tested with the strip and the commercial IDVET ELISA kit. Table 3 shows that the detected positive sample numbers were 24 by the strip and 10 by the commercial IDVET ELISA kit, respectively. In other words, the seroprevalence rate of the strip was 44%, and that of the commercial kit was 18.5%. Consequently, the strip shows a higher sensitivity to the serum samples positive for nucleic acid and is potent for detecting ASFV antibodies in clinical serum samples.
Table 3.
The detection results of the sera positive for ASFV nucleic acid by the strip and the IDVET ELISA kit
| ELISA | Strip positive rate | |||
|---|---|---|---|---|
| Positive | Negative | |||
| Strip | Positive | 10(A) | 14(B) | 44% |
| Negative | 0(C) | 30(D) | ||
| ELISA positive rate | 18% | |||
Strip positive rate (%) = (A + B) / (A + B + C + D), ELISA positive rate (%) = (A + C) / (A + B + C + D)
Discussion
At present, ASF cases have been reported in more than 60 countries around the world (Clemmons et al. 2021). Most ASFV strains cause up to 100% mortality in domestic swine (Wormington et al. 2019). China is the largest pig-producing and pork-consuming country in the world. ASF was first reported in China in 2018 and will continue to circulate for a long time based on reports and experience in other countries (Tao et al. 2020; Terrestrial code OIE 2019). Therefore, ASFV detection is crucial for prevention and control of ASF (Blome et al. 2020).
Polymerase chain reaction (PCR), real-time PCR, and ELISA are conventional diagnostic methods for ASFV antigens/antibodies recommended by OIE (2019). However, these methods require specialized equipment and professional personals. The colloidal gold immunochromatographic strip does not require specialized equipment or professional personals. Hence, it is easy to operate, and the detection results can be rapidly judged within 20 min by the naked eye in field (Bahadır and Sezgintürk. 2016). The colloidal gold immunochromatographic strips have been combined with recombinase polymerase amplification (RPA) to detect ASFV nucleic acid by other scholars (Wang et al. 2021; Zhai et al. 2020; Zhang et al. 2021b). In addition, ASFV protein detection has been developed based on monoclonal antibody-mediated colloidal gold test strip (Zhang et al. 2021a). Due to the presence of natural variant strains, the virulence of ASFV was weakened to some extent, and therefore ASFV infection is not easy to be identified by nucleic acid/protein diagnosis. As a consequence, these ASFV stains are more likely to result in wide spread, making eradication more difficult (Zhang et al. 2021c). Under these situations, ASFV diagnosis by detection of the specific antibodies is of great importance.
For ideal antibody diagnosis candidates, Kollnberger et al. identified several immunogenic proteins of ASFV, including both structural and non-structural proteins (Kollnberger et al. 2002). Among them, the p72 is considered as one of the most immunogenic and suitable candidates for antibody detection (Yu et al. 1996). The p72 has been reported to exist as trimers in ASFV capsid by cryo-electron microscopy (cryo-EM) (Liu et al. 2019a, b; Wang et al. 2019). Therefore, preparation of the p72 in its native form is the key for antibody test to accurately distinguish positive and negative sera and reduce false positive reactions (Carolina et al. 2013). Previous studies expressed the p72 as insoluble inclusion bodies in Escherichia coli expression system (Freije et al. 1993; Garcia et al. 1998). Additionally, Wan et al. expressed the truncated p72 through E.coli expression system (Wan et al. 2022). Other reports produced the p72 alone in baculovirus expression system (Heimerman et al. 2018). However, the p72 was shown to fail in forming trimers when expressed alone (Cobbold et al. 2001). Interestingly, Zhu et al. reported to express the 72 alone in yeast competent cells and obtain p72 trimers (Zhu et al. 2022). In fact, the pB602L, an ASFV non-structural protein, has been described as a chaperone facilitating the correct folding of the p72 (Cobbold et al. 2001; Epifano et al. 2006; Meng et al. 2020). In the current study, the p72 was co-expressed with the chaperone pB602L in the eukaryotic expression system (Fig. 1). The MM of purified p72 is approximately 200 kDa as analyzed by size exclusion chromatography and native-PAGE (Fig. 2), showing that the p72 monomers form into trimers, in consistent with the previous study (Liu et al. 2019a).
Next, a colloidal gold immunochromatographic strip was developed based on p72 trimers in this work. Our strip is highly specific to ASFV-positive serum, without cross-reactivity to other swine virus positive sera. During the review of our article, Wan et al. have developed a colloidal-gold dual immunochromatographic strip with recombinant p30 and truncated p72 as described above. The sensitivity of the strip is 1:256, which is equivalent to a commercial ELISA kit (Wan et al. 2022). Zhu et al. have also utilized gold nanoparticle–labeled and acid-treated p72 to establish a lateral flow strip. The sensitivity of the strip is 1:10,000 for clinical positive sera (Zhu et al. 2022). The sensitivity of our strip is 1:204,800, which is more sensitive than the commercial ELISA kit when testing positive standard serum. As a consequence, it is not surprising that the strip has a higher seroprevalence rate in the clinical serum samples positive for ASFV nucleic acid. In fact, we need ASFV-challenged pig sera to confirm the time point when the strip can detect antibodies. In the future, the detection of large-scale clinical samples is necessary for promotion and application of the strip.
In conclusion, a colloidal gold immunochromatographic strip was developed based on p72 trimers in this study and showed a high diagnostic sensitivity, specificity, and repeatability. As a supplement to nucleic acid detection, the strip can accurately identify ASFV antibodies in swine serum, providing a reliable tool in elimination of infected pigs and epidemiological investigations.
Author contribution
R G, S-L Q, and G-P Z conceived and designed the research. R G, Y-N S, H-F M, J-F Y, R L, Z-X Q, and Q-X L conducted the experiments and analyzed the data. R G wrote the manuscript. R L, S-L Q, and G-P Z revised the manuscript. All the authors read and approved the manuscript.
Funding
This work was supported by Program for Central Plains Youth Top Talent (ZYYCYU202012148), China Agriculture Research System of MOF and MARA (CARS-35), the Special Fund for Henan Agriculture Research System (S2012-06), and the Science-Technology Foundation for Outstanding Young Scientists of Henan Academy of Agricultural Sciences (2022YQ25). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Declarations
Ethics approval and consent to participate
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to and the appropriate ethical review committee approval has been received. The animal experimental procedures have been reviewed and approved by the Ethical and Animal Welfare Committee of Key Laboratory of Animal Immunology of the Ministry of Agriculture of China (Permit 2020060401).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rui Geng and Yaning Sun contributed equally to this work.
Contributor Information
Songlin Qiao, Email: cdj565@gmail.com.
Gaiping Zhang, Email: zhanggaip@126.com.
References
- Blome S, Franzke K, Beer M. African swine fever - a review of current knowledge. Virus Res. 2020;287:198099. doi: 10.1016/j.virusres.2020.198099. [DOI] [PubMed] [Google Scholar]
- Bahadır E, Sezgintürk K. Lateral flow assays: principles, designs and labels. Trends Analyt Chem. 2016;82:286–306. doi: 10.1016/j.trac.2016.06.006. [DOI] [Google Scholar]
- Clemmons EA, Alfson KJ, Dutton JR (2021) Transboundary animal diseases, an overview of 17 diseases with potential for global spread and serious consequences. Animals (Basel) 11. 10.3390/ani11072039 [DOI] [PMC free article] [PubMed]
- Cobbold C, Windsor M, Wileman T. A virally encoded chaperone specialized for folding of the major capsid protein of african swine fever. Virus J Virol. 2001;75:7221–7229. doi: 10.1128/JVI.75.16.7221-7229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costard S, Wieland B, Glanville W, Jori F, Rowlands R, Vosloo W, Roger F, Pfeiffer DU, Dixon LK. African swine fever: how can global spread be prevented? Philos Trans R Soc B Biol Sci. 2009;364:2683–2696. doi: 10.1098/rstb.2009.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carolina C, Silvia GS, Noelia M, María CN, Leopold KMM, Carlos JQ, Livio H, Eric MC, Ferran J, Jose ME, Esther B. African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res. 2013;173:159–167. doi: 10.1016/j.virusres.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Dixon LK, Stahl K, Jori F, Vial L, Pfeiffer DU. African swine fever epidemiology and control. Annu Rev Anim Biosci. 2020;8:221–246. doi: 10.1146/annurev-animal-021419-083741. [DOI] [PubMed] [Google Scholar]
- Dixon LK, Sun H, Roberts H. African swine fever. Antiviral Res. 2019;165:34–41. doi: 10.1016/j.antiviral.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Epifano C, Krijnse-Locker J, Salas ML, Rodriguez JM, Salas J. The African swine fever virus nonstructural protein pB602L is required for formation of the icosahedral capsid of the virus particle. J Virol. 2006;80:12260–12270. doi: 10.1128/JVI.01323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije JM, Munoz M, Vinuela E, Lopez-Otin C. High-level expression in Escherichia coli of the gene coding for the major structural protein (p72) of African swine fever virus. Gene. 1993;123:259–262. doi: 10.1016/0378-1119(93)90134-o. [DOI] [PubMed] [Google Scholar]
- Fren G. Preparation of gold dispersions of varying particle size: controlled nucleation for the regulation of the particle size in monodisperse gold suspension. Nat Phys. 1973;241:20–22. doi: 10.1038/physci241020a0. [DOI] [Google Scholar]
- Galindo I, Alonso C. African swine fever virus A Review. Viruses. 2017;9:103. doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia ER, Andres G, Almazan F, Vinuela E. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J Virol. 1998;72:3185–3195. doi: 10.1128/JVI.72.4.3185-3195.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault NN, Richt JA (2019) Subunit vaccine approaches for African swine fever virus. Vaccines (Basel) 7. 10.3390/vaccines7020056 [DOI] [PMC free article] [PubMed]
- Gogin A, Gerasimov V, Malogolovkin A, Kolbasov D. African swine fever in the North Caucasus region and the Russian Federation in years 2007–2012. Virus Res. 2013;173:198–203. doi: 10.1016/j.virusres.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Heimerman ME, Murgia MV, Wu P, Lowe AD, Jia W, Rowland RR. Linear epitopes in African swine fever virus p72 recognized by monoclonal antibodies prepared against baculovirus-expressed antigen. J Vet Diagn Invest. 2018;30:406–412. doi: 10.1177/1040638717753966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia N, Ou Y, Pejsak Z, Zhang Y, Zhang J. Roles of African swine fever virus structural proteins in viral infection. J Vet Res. 2017;61:135–143. doi: 10.1515/jvetres-2017-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedkovid R, Sirisereewan C, Thanawongnuwech R. Major swine viral diseases: an Asian perspective after the African swine fever introduction. Porcine Health Manag. 2020;6:20. doi: 10.1186/s40813-020-00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollnberger SD, Gutierrez-Castañeda B, Foster-Cuevas M, Corteyn A, Parkhouse RME. Identification of the principal serological immunodeterminants of African swine fever virus by screening a virus cDNA library with antibody. J Gen Virol. 2002;83:1331–1342. doi: 10.1099/0022-1317-83-6-1331. [DOI] [PubMed] [Google Scholar]
- Li G, Wang AP, Chen YM, Sun YN, Du YK, Wang X, Ding PY, Jia R, Wang YW, Zhang GP. Development of a colloidal gold-based immunochromatographic strip for rapid detection of severe acute respiratory syndrome coronavirus 2 spike protein. Front Immunol. 2021;12:635677. doi: 10.3389/fimmu.2021.635677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HW, Yang JF, Bao DK, Hou J, Zhi YB, Yang YY, Ji PC, Zhou EM, Qiao SL, Zhang GP. Development of an immunochromatographic strip for detection of antibodies against porcine reproductive and respiratory syndrome virus. J Vet Sci. 2017;18:307–316. doi: 10.4142/jvs.2017.18.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ma B, Qian N, Zhang F, Tan X, Lei J, Xiang Y. Structure of the African swine fever virus major capsid protein p72. Cell Res. 2019;29:953–955. doi: 10.1038/s41422-019-0232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Luo YZ, Wang YJ, Li SH, Zhao ZN, Bi YH, Sun JQ, Peng RC, Song H, Zhu DJ, Sun Y, Li S, Zhang L, Wang W, Sun YP, Qi JX, Yan JH, Shi Y, Gao F. Cryo-EM structure of the African swine fever virus. Cell Host Microbe. 2019;26:836–843. doi: 10.1016/j.chom.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Liu S, Li S, Zhang Y, Wang Y, Zhu Y, Wang B, Chen ZN. Purification of a polyclonal antibody against CD147 for ELISA using antigenimmunoaffinity chromatography. Mol Med Rep. 2017;15:4035–4040. doi: 10.3892/mmr.2017.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng KW, Zhang YP, Zhu WZ, Xiang Y, Meng G (2020) Expression and purification of p72 trimers as subunit vaccine candidate. 10.1101/2020.01.30.926808
- Montgomery RE. On a form of swine fever occurring in British East Africa (Kenya Colony) J Comp Pathol Ther. 1921;34:159–191. doi: 10.1016/S0368-1742(21)80031-4. [DOI] [Google Scholar]
- Revilla Y, Perez-Nunez D, Richt JA. African swine fever virus biology and vaccine approaches. Adv Virus Res. 2018;100:41–74. doi: 10.1016/bs.aivir.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Sun YN, Wang L, Yang SZ, Xx Chen, Yw Liu, Ll Zhang, Rg Deng, Gp Zhang. Research and development of swine fever antibody detection test paper and optimization of preparation process. Northwest Agricultural Journal. 2019;28(02):169–175. [Google Scholar]
- Tao DP, Sun DP, Liu YM, Wei S, Yang ZF, An TQ, Shan FP, Chen ZL, Liu JL. One year of African swine fever outbreak in China. Acta Trop. 2020;211:105602. doi: 10.1016/j.actatropica.2020.105602. [DOI] [PubMed] [Google Scholar]
- Terrestrial code OIE (2019) Chapter 15.1. Infection with Africa swine fever virus. In: Terrestrial Animal Health Code. 28th ed. Pairs, France: World Organisation for Animal Health. http://www.oie.int/index.php?id=169&L=0&htmfile=chapitre_asf.htm. Accessed 6 May 2019
- Wan Y, Shi Z, Peng G, Wang L, Luo J, Ru Y, Zhou G, Ma Y, Song R, Yang B, Cao L, Tian H, Zheng H. Development and application of a colloidal-gold dual immunochromatography strip for detecting African swine fever virus antibodies. Appl Microbiol Biotechnol. 2022;106(2):799–810. doi: 10.1007/s00253-021-11706-z. [DOI] [PubMed] [Google Scholar]
- Wang N, Zhao D, Wang J, Zhang Y, Wang M, Gao Y, Wang J, Bu Z, Wang X. Architecture of African swine fever virus and implications for viral assembly. Science. 2019;366(6465):640–644. doi: 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jia L, Li J, Liu H, Liu D. Pan-Genomic analysis of African swine fever virus. Virol Sin. 2020;35:662–665. doi: 10.1007/s12250-019-00173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yu W, Xie R, Yang S, Chen A. A strip of lateral flow gene assay using gold nanoparticles for point-of-care diagnosis of African swine fever virus in limited environment. Anal Bioanal Chem. 2021;413:4665–4672. doi: 10.1007/s00216-021-03408-2. [DOI] [PubMed] [Google Scholar]
- Wormington JD, Golnar A, Poh KC, Kading RC, Martin E, Hamer SA, Hamer GL. Risk of African swine fever virus sylvatic establishment and spillover to domestic swine in the United States. Vector Borne Zoonotic Dis. 2019;19:506–511. doi: 10.1089/vbz.2018.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Morrissy CJ, Westbury HA. Strong sequence conservation of African swine fever virus p72 protein provides the molecular basis for its antigenic stability. Arch Virol. 1996;141:1795–1802. doi: 10.1007/BF01718302. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Ma P, Fu X, Zhang L, Cui P, Li H, Yan W, Wang H, Yang X. A recombinase polymerase amplification combined with lateral flow dipstick for rapid and specific detection of African swine fever virus. J Virol Methods. 2020;285:113885. doi: 10.1016/j.jviromet.2020.113885. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu X, Wu X, Ren W, Zou Y, Xia X, Sun H. A colloidal gold test strip assay for the detection of African swine fever virus based on two monoclonal antibodies against P30. Arch Virol. 2021;166:871–879. doi: 10.1007/s00705-020-04915-w. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li Q, Guo J, Li D, Wang L, Wang X, Xing G, Deng R, Zhang G. An isothermal molecular point of care testing for African swine fever virus using recombinase-aided amplification and lateral flow assay without the need to extract nucleic acids in blood. Front Cell Infect Microbiol. 2021;11:633–763. doi: 10.3389/fcimb.2021.633763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang JY, Yang JJ, Yang JM, Han X, Mi LJ, Zhang F, Qi Y, Zhang SF, Wang Y, Zhou XT, Yue HX, Wang SC, Chen T, Hu RL (2021c) Identification of a natural varian of African swine fever virus in China. Chin J Vet Sci 199–207. 10.16303/j.cnki.1005-4545.2021.02.01
- Zhou X, Li N, Luo Y, Liu YE, Miao F, Chen T, Hu R. Emergence of African swine fever in China, 2018. Transbound Emerg Dis. 2018;65:1482–1484. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
- Zhu W, Meng K, Zhang Y, Bu Z, Zhao D, Meng G. Lateral flow assay for the detection of African swine fever virus antibodies using gold nanoparticle-labeled acid-treated p72. Front Chem. 2022;9:804–981. doi: 10.3389/fchem.2021.804981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.