Abstract
SARS-CoV-2 is currently causing an unprecedented pandemic. While vaccines are massively deployed, we still lack effective large-scale antiviral therapies. In the quest for antivirals targeting conserved structures, we focused on molecules able to bind viral RNA secondary structures. Aminoglycosides are a class of antibiotics known to interact with the ribosomal RNA of both prokaryotes and eukaryotes and have previously been shown to exert antiviral activities by interacting with viral RNA. Here we show that the aminoglycoside geneticin is endowed with antiviral activity against all tested variants of SARS-CoV-2, in different cell lines and in a respiratory tissue model at non-toxic concentrations. The mechanism of action is an early inhibition of RNA replication and protein expression mediated by direct interaction with the −1 programmed ribosomal frameshift (PRF) signal. Using in silico modeling, we have identified a potential binding site of geneticin in the pseudoknot of frameshift RNA motif. Moreover, we have selected, through virtual screening, additional RNA binding compounds, interacting with the same site with increased potency.
INTRODUCTION
Since the beginning of the SARS-CoV-2 pandemic, a huge effort has been made for the identification of effective vaccines and antivirals. The vaccines programme has been in immense success with the approval of three vaccines in less than one year, and the vaccination, at the time of writing, of 63.4% of the world population (1). The drug discovery effort has also led to the identification of three antivirals drugs, Remdesivir, Molnupiravir and Paxlovid, which have been approved by FDA (2). However, the emergence of new SARS-COV 2 variants, which can potentially escape the vaccine-mediate immunity and the effectiveness of therapies, highlights the importance to identify new potential pan antivirals agents against SARS-CoV-2.
RNA structure elements represent an attractive target for antiviral drug discovery. Viral genomes contain highly conserved RNA elements that play a critical role in gene regulation and viral replication. These RNA elements are directly involved in the viral infection process, interacting with proteins, DNA or other RNAs, modulating their activity (3). The function and activity of these RNA molecules are based on the complex three-dimensional structure they can adopt (4). Due to the conserved nature and to the well-defined structure, the RNA provides potentially unique interaction sites for selective small-molecule ligands that affect viral replication. The high conservation of untranslated regions reduces the possibility of a drug-resistant mechanism, increasing the effectiveness of potential antiviral drugs (5). Any change in nucleotide sequence can result in inactive elements through misfolding the RNA structure, as recently demonstrated with the programmed −1 ribosomal frameshifting element (−1 PRF) of SARS-CoV-2(6). Programmed ribosomal frameshifting is one of the strategies commonly used by RNA viruses, such as flaviviruses, coronaviruses, influenza A viruses, HIV, to regulate the relative expression level of two proteins encoded on the same messenger RNA (mRNA) (7–9). This strategy is rarely used by human cells, making it an attractive therapeutic target for antiviral drug development. Several studies have proposed the frameshifting element (FSE) as a target for disruption of virus replication (10–13). The SARS-CoV-2 FSE is a small region between the open reading frame (ORF) 1a and the ORF 1b. The ORF1b encodes all the enzymes necessary for viral RNA replication, including the RNA dependent RNA polymerase. The frameshifting events depend on the flexibility of the RNA structure and its ability to interact with the ribosome. A small molecule that can alter the structural organisation of the FSE can block the frameshifting event and consequently the viral replication.
Aminoglycosides are among the molecules known to interact with secondary or tertiary structures on RNA, therefore potentially inhibiting −1 PRF of SARS-CoV-2. This class of antibiotics is known to interact with the ribosomal RNA of prokaryotes and eukaryotes (14, 15) in particular with the tRNA recognition site, blocking a conformational switch of the ribosomal A site. The affinity for RNA makes this class of molecules potentially interacting with additional RNA structures as shown for RNA HIV dimerisation sites, or for a riboswitch sequence in the 5’ leader RNA of a resistance gene in bacteria(16).
Among the different aminoglycosides, geneticin is one of the few for which the cells are permeable, and it is commonly used in cell lines as a selective agent due to its alteration in eukaryotic protein synthesis when administered at high doses for prolonged time(17). However, the drug proved to be effective as well against multiple viruses (Bovine Viral Diarrhea Virus, Dengue Virus and Hepatitis C virus (HCV)) (18–20) at nontoxic concentrations. In particular, in the evaluation of the antiviral activity of geneticin against HCV, a specific interaction with a double stranded RNA switch structure in the 5’UTR of the virus was shown (18). This binding resulted in a stabilisation of the open conformation leading to inhibition of the production of non-structural protein 3 (NS3) and viral replication in cell lines.
Here we show that geneticin is active against SARS-CoV-2 through an early inhibition in its life cycle and a direct interaction with the −1 PRF region. The activity in the micromolar range is maintained against multiple variants, in different cell lines, and respiratory tissues. Importantly, we identified a putative binding site for geneticin on the −1 PRF sequence of SARS-CoV-2 through in silico modelling. After a screening of RNA binding molecules interacting with the same site, we identified compounds displaying antiviral activity at lower half-maximal effective concentrations (EC50) than geneticin, paving the road for future development of SARS-CoV-2 antivirals.
MATERIAL AND METHODS
Compounds
Geneticin was purchased from Gibco (Life Technologies), merafloxacin and MTT [1-(4,5-Dimethylthiazol-2-yl)-3,5-diphenylformazan] were purchased from Sigma.
Cells
Vero C1008 (clone E6) (ATCC CRL-1586) cells were a kind gift from Prof Gary Kobinger, Calu-3 were purchased from ATCC. Cells were propagated in DMEM High Glucose + Glutamax supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptavidin (pen/strep).
Viruses
SARS-CoV-2/Switzerland/GE9586/2020 was isolated from a clinical specimen in the University Hospital in Geneva in Vero-E6 and passaged twice before the experiments. SARS-CoV-2 GFP was a kind gift from Prof Volker Thiel (21). The other clinical strains (hCoV-19/Switzerland/VD-CHUV-GEN3159/2021, hCoV-19/Switzerland/VD-GEN3343/2021, hCoV-19/Switzerland/VD-CHUV-GEN5521/2021, hCoV-19/Switzerland/VD-CHUV-GEN8840/2021, hCoV-19/Switzerland/VD-GEN3642/2021, hCoV-19/Switzerland/VD-GEN3807/2021, hCoV-19/Switzerland/VD-GEN3770/2021) were isolated from clinical specimens from the University Hospital of Lausanne (CHUV) as described in (22). Supernatant of infected cells was collected, clarified, aliquoted, and frozen at −80°C and subsequently titrated by plaque assay in Vero-E6.
Cell toxicity assay
Cell viability was measured by the MTT assay or MTS assay (Promega) for tissues. Confluent cell cultures seeded in 96-well plates were incubated with different concentrations of geneticin in duplicate under the same experimental conditions described for the antiviral assays. Absorbance was measured using a Microplate Reader at 570 nm. The effect on cell viability at different concentrations of geneticin and additional compounds was expressed as a percentage, by comparing the absorbance of treated cells with the one of cells incubated with equal concentrations of solvent in medium. The 50 % cytotoxic concentrations (CC50) and 95 % confidence intervals (CIs) were determined using Prism software (Graph-Pad Software, San Diego, CA).
Antiviral assay in Vero-E6 cells
Vero-E6 cells (105 cells per well) were seeded in 24-well plate. Cells were infected with SARS-CoV-2 (MOI, 0.001 PFU/cell) for 1 hour at 37°C. The monolayers were then washed and overlaid with medium supplemented with 5% FBS containing serial dilutions of compounds for the experiments with SARS-CoV-2 expressing GFP. For experiments with the different SARS-CoV-2 variant and analogues of geneticin, Vero-E6 cells were overlaid instead with 0.4% avicel gp3515 in medium containing 2.5% FBS. Two days after infection, cells were fixed with 4% formaldehyde and stained with crystal violet solution containing ethanol. Plaques were counted, and the percent inhibition of virus infectivity was determined by comparing the number of plaques in treated wells with the number in untreated control wells. 50% effective concentration (EC50) was calculated with Prism 8 (GraphPad).
Antiviral assay in Calu3 cells
Calu-3 cells (4 × 104 cells per well) were seeded in 96-well plate. Cells were infected with SARS-CoV-2 (MOI 0.1 PFU/cell) for 1 hour at 37°C. The monolayers were then washed and overlaid with medium containing serial dilutions of geneticin. At 24 hpi, supernatant was collected and viral RNA was extracted with EZNA total RNA kit (Omega Bio-tek). SARS-CoV-2 RNA was quantified by qPCR with the QuantiTect Kit (Qiagen, 204443) with Sarbeco E gene primers and probe in a QuantStudio 3 thermocycler (Applied Biosystems). Percent inhibition of virus infectivity was determined by comparing viral load in treated wells with the viral load in untreated control wells. EC50 was calculated with Prism 8 (GraphPad).
Kinetics of RNA expression
Vero-E6 cells were seeded in 24-well plates at a density of 105 cells per well and infected in duplicate with SARS-CoV-2 at an MOI of 0.1 PFU/cell for 1 hour at 37°C. After the removal of the inoculum the treatment was started and cells were lysed with TRK buffer (Omega Biotech) at 0, 4, 8 and 24 hours post infection. RNA was extracted with the Total RNA kit (Omega Biotech) and amplified with the E-sarbeco primers for SARS-CoV-2.
Flow cytometry analysis
Vero-E6 cells were seeded in 24-well plates at a density of 105 cells per well and infected in duplicate with SARS-CoV-2 at an MOI of 0.01 PFU/cell for 1 hour at 37°C. The cells were then treated with geneticin and incubated at 37 °C for additional 24 or 48 hours. Supernatant was collected, cells washed once and detached with trypsin. Once in suspension cells were pelleted and then fixed with paraformaldehyde 4% in PBS. Percentages of GFP positive cells and mean GFP value for each positive cell was evaluated with an Accuri C6 cytometer (BD biosciences).
Dual luciferase
pSGDLuc v3.0 was modified to include the −1 PRF signal of SARS-CoV-2 as described in (23). Vero-E6 cells were seeded 24 hours in advance in 96-well plates (104 cells per well), treated with geneticin, merafloxacin or geneticin analog and transfected with Lipofectamine 3000 (Thermofisher) and the plasmid containing the −1PRF sequence or the in frame control. Luciferase was evaluated 24 hours post transfection with the Dual Glo Kit (Promega). The percentage of ribosomal frameshift was calculated as described in (23).
PRF sequencing
RNA was extracted from isolated clinical SARS-CoV-2 with E.Z.N.A total RNA (Omega Bio-Tek). Maxima H Minus cDNA Synthesis (Thermofisher) and Platinium II Taq (Thermofisher) were used as RT-PCR kits with designed primers (Fwd 5’-GCC ACA GTA CGT CTA CAA GC-3’, Rev 5’-GGC GTG GTT TGT ATG AAA TC-3’). PCR products were Sanger sequenced by Microsynth.
MucilAir antiviral assays
Tissues were obtained from Epithelix (Geneva, Switzerland). For all experiments, epithelia were prepared with different single donor’s biopsies. Before inoculation with the viruses, MucilAir tissues were incubated in 250 μL of PBS Ca2+Mg2+ (PBS++) for 45 min at 37°C. Infection was done with 106 RNA copies/tissue with ancestral SARS-CoV-2 or 105 RNA copies with SARS-CoV-2 B.1.1.529 (omicron). At 4 hours after incubation at 33°C, tissues were rinsed three times with MucilAir medium to remove non-adsorbed virus and cultures were continued in the air-liquid interface. Every 24 hours, 200 μL of MucilAir medium was applied to the apical face of the tissue for 20 min at 33°C for sample collection, followed by apical treatment with geneticin (30 μg/tissue) starting at 24 hpi. Viral load was determined by qPCR as described previously. At the same time point, the basal medium was replaced with 500 μL of fresh MucilAir medium. At the end of the experiments, tissues were fixed and subjected to immunofluorescence.
Statistics and data analysis
Experiments were performed in duplicate and from two to four independent experiments as stated in the figure legends. Results are shown as mean and SEM. The EC50 and CC50 values for inhibition curves were calculated by regression analysis using the program GraphPad Prism version 8.0 (GraphPad Software, California, USA) to fit a variable slope sigmoidal dose-response curve. One-way Anova followed by multiple comparison analysis was used as statistical tests to compare grouped analysis. Unpaired t test was used to compare two different conditions. Area under the curve analysis followed by unpaired t test or one-way ANOVA was done to compare curves.
Molecular Modelling
All molecular modelling experiments were performed on Asus WS X299 PRO Intel® i9–10980XECPU @ 3.00GHz × 36 running Ubuntu 18.04 (graphic card: GeForce RTX 2080 Ti). Molecular Operating Environment (MOE, 2019.10, Montreal, QC, Canada)(24); Maestro (Schrödinger Release 2021–1, New York, NY, USA)(25); GROMACS (2020.4) (26); Dock6 (27); Annapurna (28); RNAsite(29), Barnaba(30) were used as molecular modelling software. A library of commercially available RNA-targeting compounds was downloaded from Enamine and ChemDiv website.
Molecular dynamic simulations
MD simulations were performed with Gromacs software package. The ff99+bsc0+χOL3 force field was used for MD simulation since this is the most validated and recommended FFs for RNA system (31). The cryo-EM of the SARS-COV-2 FSE was download from PDB (http://www.rcsb.org/; PDB entry 6xrz).
The structure was solvated with 14,0812 TIP4P-Ew waters and 87 Na+ counterions to neutralise the charge on the RNA. All the molecular dynamics simulations were performed for 100ns on the isothermal-isobaric ensemble, using the stochastic velocity rescaling thermostat at 300 K and the Berendsen barostat with an isotropic pressure coupling of 1 bar. The eRMSD, an RNA-specific metric distance based on relative orientation and position of nucleobases, was used to verify the stability of the simulated systems during the MD simulation. The conformations obtained after 40 ns were extracted, and eRMSD between all structures was used to perform a cluster analysis to group the different RNA conformations and to select a representative structure.
Binding site identification and molecular docking
The refined cryo-EM structure was prepared for further refinement with the Schrödinger Protein Preparation Wizard. Protonation states of RNA nucleotides were calculated considering a temperature of 300 K and a pH of 7.4, and restrained energy minimisation of the added hydrogens using the OPLS4 force field was performed. The Geneticin and the RNA-targeting compounds were prepared using the Maestro LigPrep tool by energy minimising the structures (OPLS4 force filed), generating possible ionisation states at pH 7 ± 2 (Epik), tautomers and stereoisomers per each ligand. RNAsite was employed to identify a potential binding site using the refined structure(32). An 11 Å docking grid was prepared using as the centroid the predicted binding pocket previously identified by RNAsite. A Glide XP precision was employed to screen the compounds keeping the default parameters and setting 3 as the number of output poses per input ligand. The best-docked poses were then refined using PRIME/MM-GBSA module. The docking poses obtained were then rescored using Annapurna and amber DOCK6 scoring functions. The values of the three different scoring functions for each docking pose were then analysed together (consensus score) and only the Docking poses falling in the top 25% of the score value range in all the three scoring functions were selected for the final visual inspection. The visual inspection process, conducted as the last step of the structure-based virtual screening, was performed using MOE 2019.10.
RESULTS
Geneticin is active against different variants of SARS-CoV-2
Antiviral activity of geneticin against several variants of SARS-CoV-2 was assessed in Vero-E6 cells with addition of the molecule post-infection in parallel with merafloxacin, a molecule previously shown to inhibit SARS-CoV-2(33). Importantly the seven different variants tested, including the alpha (B.1.1.7), the beta (B.1.135), the delta (B.1.617.2) and the omicron (B.1.1.529) were directly isolated from clinical specimens at the University Hospital of Lausanne with minimal passaging in cell lines to avoid any cell adaptation. We observed dose-response activity in the micromolar range for all the variants tested (Table 1) with no evidence of toxicity at the tested doses. Analysis of the sequences did not reveal any particular cell adaptation, nor common changes in the variants showing higher EC50s if compared to the others (Supplementary Figure 1).
Table 1.
Antiviral activity of geneticin against SARS-CoV-2
| Variant | EC50 (95% CI) [μM] | EC90 (95% CI) [μM] | CC50 [μM] | |
|---|---|---|---|---|
| Geneticin | B. 1.1.7 (Alpha) | 47.2 (35.1 – 62.4) | 245 (149 – 460) | 3951 |
| B. 1.351 (Beta) | 129 (84.7 – 188) | 542 (273 – 1537) | 3951 | |
| B.1.617.2 (Delta) | 32.8 (21.3 – 48.2) | 201 (100 – 505) | 3951 | |
| B.1.1.529 (Omicron) | 25.7 (16.9 – 38.9) | 155 (60 – 528) | 934 | |
| B. 1.258 | 42.4 (29.7 – 60.4) | 163 (86 – 377) | 3951 | |
| B.1.160 | 31.6 (21.2 – 44.8) | 150 (74 – 368) | 3951 | |
| B.1.177 | 101.7 (74.8 – 138) | 407 (222 – 852) | 3951 | |
| Merafloxacin | B. 1.1.7 (Alpha) | 23.7 (16.8 – 33.8) | 116 (52.9 – 458) | > 100 |
EC50: half-maximal effective concentration. EC90: 90% effective concentration. 95% CI: confidence interval 95%
The antiviral activity is maintained in human respiratory cell lines and in tissues
To assess the antiviral activity in more relevant cell models we evaluated the antiviral activity in dose response of geneticin in Calu3 cells, a lung adenocarcinoma cell line, which was previously shown to mimic faithfully SARS-CoV-2 infection into respiratory cell line(21). The results evidenced a sustained antiviral activity (EC50 179.6 μM) also in this cellular model in absence of toxicity (Figure 1A). We then tested the activity in a pseudostratified model of human respiratory tract (Mucilair, Epithelix). This tissue model is composed by the typical cells of the human upper respiratory tract, namely ciliated, goblet and basal cells. In this infection model, we aimed to mimic a possible treatment with the molecule by starting the treatment at 24hpi when the infection of the tissue was already well established and we used viral stocks produced in the same tissue and never passaged in cell lines to exclude any adaptation. The treatment was performed apically and the infection monitored up to 4 days post infection by collecting an apical wash and performing either a ta qPCR or a titration in VeroE6 (Figure 1B). The results evidenced a significant protection from viral infection with both ancestral (B) and omicron (C) variants, in absence of decrease of viability for the tissues (Supplementary Figure 2).
Figure 1.
The activity of geneticin in maintained in Calu3 cells and in human derived tissues. A) Calu3 cells were infected with SARS-CoV-2 (alpha variant) for 1 hour at 37°C. After the removal of the inoculum, the cells were treated with serial dilutions of geneticin. At 24 hpi supernatant was collected and viral RNA copies evaluated with qPCR. B) and C) Mucilair tissues were infected with B) ancestral SARS-CoV-2 106 RNA copies or C) SARS-CoV-2 Omicron 105 RNA copies, the following day the apical treatment with 30 μg/tissue started. Every 24 hours an apical wash was performed and collected after 20 minutes at 37°C. The supernatant was then used for viral RNA quantification (solid lines) or for plaque assay (dashed lines). The results are mean and SEM of two to three independent experiments performed in duplicate. P values <0.0332 (*), <0.0021 (**), <0.0002 (***), < 0.0001 (****)
Geneticin inhibits the −1PRF of SARS-CoV-2
In order to assess the mechanism of action and the stage of viral replication of SARS-CoV-2 inhibited by geneticin, we first assessed viral protein expression. We exploited a GFP expressing SARS-CoV-2 previously generated (21) evaluating the GFP expression in presence or absence of the drug at 24h and 48hpi (Fig 2A). The results evidenced, as expected, a marked reduction in the number of infected cells (Supplementary Figure 3). In addition, in the infected cells treated with geneticin the GFP intensity was significantly reduced, if compared to the untreated control (Figure 2A). These results show a decrease of viral protein production, therefore a block of infection at an initial stage of the viral life cycle. To evaluate if the inhibition of protein expression was related to a block of translation or of viral replication we then monitored viral replication through a RT-qPCR measuring the viral RNA replication at different time points. In this case, we included as control, a compound previously shown to interfere with −1 PRF signal, merafloxacin. As shown in Figure 2B, the addition or geneticin or merafloxacin, results in inhibition of viral RNA replication at 4h, 8h and 24h post infection, showing a very rapid inhibition of viral replication of the two drugs. Finally, we assessed the ability of geneticin, in comparison with merafloxacin, to interfere with the programmed ribosomal frameshifting element of SARS-CoV-2 with a dual luciferase assay. The −1 PRF signal was cloned between Renilla and Firefly luciferase and the relative expression of the luciferases was evaluated in presence or absence of the drugs as described in(23). The results of figure 2C show a reduction in the −1 PRF efficiency in presence of both compounds suggesting a direct interaction of the drug with the −1PRF sequence resulting in impaired replication (Figure 2B) and protein production (Figure 2A). The effect of geneticin on the frameshift is more marked for the lower dose (50 μg/ml) because we observe an overall reduction of luciferase expression with the in frame control at 300 μg/ml that is partially affecting the results (Supplementary File 1).
Figure 2.
Mechanism of action of geneticin. A) Vero-E6 cells were infected with SARS-CoV-2 expressing GFP at MOI 0.01 and treated post-infection with geneticin (300 μg/mL) at 24 and 48 hpi cells were detached and analysed with flow cytometer, results are expressed as percentage of untreated control. B) Vero-E6 were infected with SARS-CoV-2 at MOI 0.1 for 1 hour at 37°C. After the removal of the inoculum, geneticin (300 μg/mL) or merafloxacin (100 μM) were added into the well. At 0, 4, 8 and 24 hours post.-infection cells were lysed and viral RNA was quantified. C) Dual luciferase evaluation was performed at 24 hours post-transfection in Vero-E6 cells treated with geneticin (300 μg/mL or 50 μg/mL) or merafloxacin (50 μM). The frameshift efficiency was normalized compared to untreated. The results are mean and SEM of at least three independent experiments performed in duplicate. P values <0.0332 (*), <0.0021 (**), <0.0002 (***), < 0.0001 (****)
In silico modelling and prediction of geneticin binding site
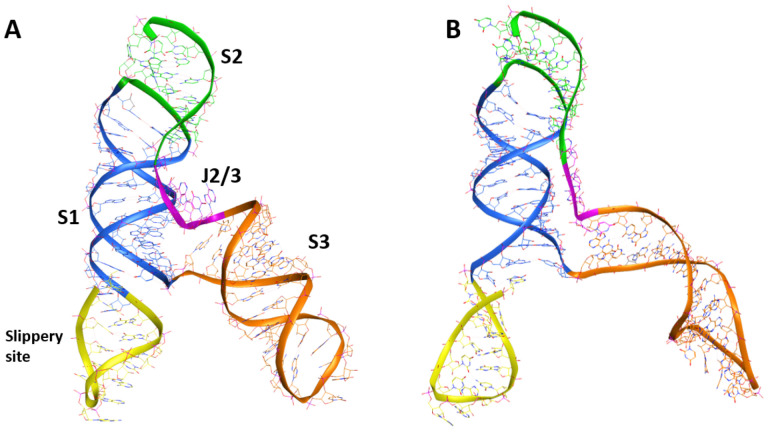
To further rationalise the results obtained by dual-luciferase and antiviral assays, the cryo-EM structure of the RNA frameshift-stimulatory element (FSE) was used to investigate the Geneticin-FSE binding complex (34).The cryo-EM RNA structure shows a λ-like tertiary arrangement composed of a three-stemmed H-type pseudoknot structure with three loops. Starting from the 5’-end and proceeding to the 3’-end, the cryo-EM structure begins with a slippers site, followed by a first strand (S1), which lead to the Loop (L1), and it continues to a second strand (S2) (Figure 3A). Stem 1 and 2 are intertwined via basepair region, which ties the 5’-end of the RNA to the 3’-end. From the second stem (S2), the RNA strands continue to form a hairpin region (S3), followed by an unpaired segment J3/2, which leads back to the Stem 2 and close the Stem 1-Stem 2 pseudoknot (Figure 3A). The cryo-EM data also suggested alternative conformations due to the structural flexibility at the 5’-ends, which appeared poorly resolved (34). Moreover, the cryo-EM structure was resolved at low-mid resolution 6.9 Å, which can affect the assignment of the atom position with high certainty. Molecular dynamic (MD) simulations have proven useful in refining macromolecular structures, particularly unveiling the atomic details for low-resolution regions of the cryo-EM map(35–37). Thus, in this study, we initially refined the cryo-EM FSE structure by 100 ns molecular dynamics simulation using the GROMACS software package(26). Overall, after an initial 40 ns of equilibration, the structural fluctuation of the RNA diminished, with the simulation system converging around a fixed eRMSD value of 1.5 Å. This eRMSD value was chosen as a cut-off for selecting a series of different conformers, which were successively clustered to select a representative structure (Figure 3B). The comparison between the cryo-EM and our model showed a similar structure rearrangement with a minimum eRMSD variations in nucleotides position, except for the slipper site and S3 region, which displayed a higher level of flexibility (Figure 3B). These results are in line with previous studies conducted by Omar et al. and Rangan et al., showing that stem 3 could adopt multiple conformations (38, 39).
Figure 3.
Comparison of the cryo-EM RNA structure (A) and the refined RNA structure by molecular dynamic simulation (B).
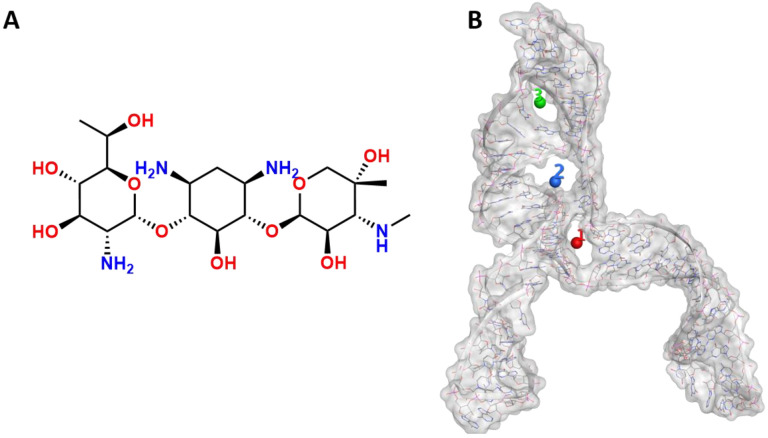
Mutational studies showed that the virus replication is highly sensitive to any conformational change in the pseudoknots region, as evident by the point mutation of guanidine (G13486) to adenine in the S1-J3/2 region, which reduced the frameshifting efficiency to 30%(6). According to the mutation results and the uncertainty of the S3 region, we hypothesised that geneticin could significantly alter and disrupt the FSE conformational plasticity and consequently the viral replication, directly binding the S1/S2-J3/2 pseudoknots region. A previous study showed that geneticin can interact with tertiary RNA structures through hydrogen bond and electrostatic interaction(40, 41). The binding affinity of geneticin for the RNA structures is mainly due to the presence of four amino groups which are positively charged at physiological pH and can form strong electrostatic interaction with the negatively charged phosphates in the nucleic acid backbone (Figure 4A). Furthermore, the presence of seven hydroxyl groups can stabilise the RNA-binding complex through a series of hydrogen bonds with the bases atoms and phosphate oxygen atoms of the nucleic acid. Several studies demonstrated the preference of aminoglycoside compounds to bind RNA helix and junction sites (42). We first investigated if there were potential geneticin-binding sites using the refined cryo-EM structure. The binding site analyses, performed by the RNAsite module(29), identified 3 different potential active sites situated between stem 1, stem 2 and junction site (figure 4B), which partially confirmed the results obtained by Zhang et collaborator, which reported the presence of a ‘ring site’, a ‘J3/2 site’ and the ‘slippery hairpin binding site(34). Our results showed that two potential binding sites, 1 and 2, were located in close proximity from one another, sharing 3 nucleotide residues (G18, G19 and G20), similar to the ring site and J3/2 site reported by Zhang et collaborator (34); meanwhile, the binding site 3 was located at the beginning of the stem 2 instead of the slippery site (figure 4B).
Figure 4.
2D Geneticin molecular structure (A) and the 3 binding sites indetified by RNAsite (B). The binding site 1 (ring site); 2 (J3/2) and 3 (stem 2) are highlighted in red, blue and green, respectively.
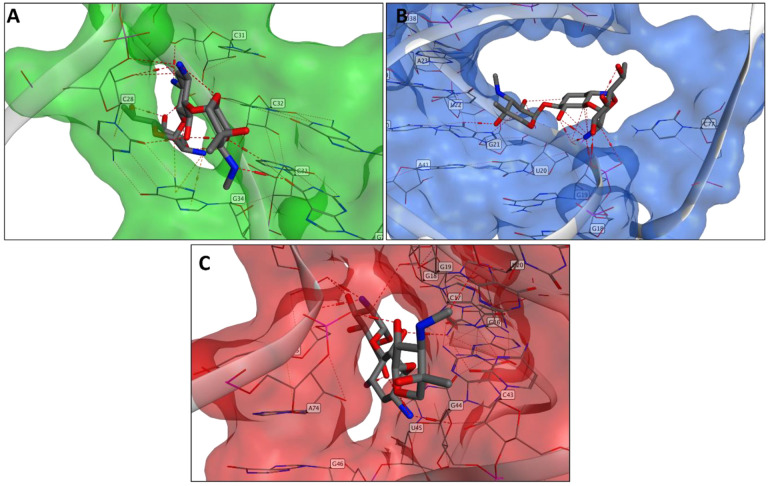
The geneticin-binding affinity was evaluated against all the three potential binding sites using an in silico protocol, which comprises three steps: firstly, the compound was docked using XP GLIDE module (Maestro, Schrodinger), then the docked poses were refined using MM-GBSA module, and lastly the refined poses were rescored using two scoring functions optimised specifically for RNA-ligand complex, Annapurna and Amber score function (DOCK6). The purpose of multiple scoring functions was to ascertain the most potentially accurate ligand poses and avoid any possible bias associated with using a single docking program/scoring function. The docking results showed that although geneticin can be well accommodated inside all three binding sites in different rational configurations, it has a slighter higher affinity for site 1 compared to site 2 and 3 (ΔGmm-gbsa −102.98, −90.34, −80.77 kcal/mol, respectively). Site 2 and 3 showed the largest surface area, but are solvent-exposed, which affect the ligand-RNA interaction: geneticin was only partially in contact with the RNA surface while the rest of the molecule was exposed to solvent (figure 5A and 5B). On the other hand, site 1 showed a smaller surface area, but it was surrounded by nucleotides (G18, G19, G20, G43, G44, G46, U75 and A76), which form a tunnel-like binding site. Geneticin can well occupy the active site with the streptamine core inside the tunnel cavity and amino group chain close to phosphate groups of U75 and A76 (Figure 5). To confirm the results obtained by the MM-GBSA analysis, the refined docked poses were rescored using Annapurna and DOCK6 score function. In both software, the top-ranked binding poses were predicted to site 1 (Supplementary Table 1), suggesting that this site might be more accessible and druggable than the other two binding sites.
Figure 5.
Binding pose of geneticin in the PRF binding site 3(A), 2(B) and 1(C).
Identification of −1PRF binding compounds
To test the druggability of the binding site, we screened an RNA targeted library (Enamine, ChemDIV), which contains 44520 commercially available RNA-binding compounds, against site 1. The virtual screening was performed using the previously described protocol. Firstly, the XP glide docking mode was employed to virtually screen the RNA-target library. The best 10% of docked poses to this initial screening were refined and rescored through MM-GBSA. To validate the top-scored docking results, the compounds were rescored using Annapurna and DOCK6 scoring functions. After applying a consensus score procedure, 132 molecules were chosen, and their potential interactions with the RNA binding site were visually inspected. Twenty virtual hits were selected, purchased and evaluated in antiviral assays. Among them, three compounds could inhibit the virus replication with an EC50 in the micromolar range, with higher potency than geneticin (Table 2).
Table 2.
Antiviral activity of site 1 −1 PRF binders against SARS-CoV-2.
| Analog | Structure | EC50 [μM] | CC50 [μM] |
|---|---|---|---|
| 1 (AB-3234) |

|
13.0 | >100 |
| 2 (AB-3241) |

|
25.2 | >100 |
| 3 (AB-3285) |
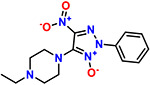
|
12.0 | >100 |
EC50 : half-maximal effective concentration, CC50: half-maximal cytotoxic concentration
The most potent compound was further analysed for kinetics of RNA expression (Figure 6A) and dual luciferase (Figure 6B) confirming a similar activity to geneticin. The in-silico results showed that 3 could completely occupy the tunnel-binding site, forming a cation-pi with G19 and 2 H-bonds with G19 and G18 (Figure 6C). We hypothesize that the NO-G18 interaction is crucial for the RNA-binding capacity of these compounds: the N-nitroso compounds group are known to alkylate RNA and DNA, mediated by transfer of the nitroso group to nucleotides bases(19).
Figure 6.
Mechanism of action of AB-3285. A) Vero-E6 were infected with SARS-CoV-2 at MOI 0.1 for 1 hour at 37°C. After the removal of the inoculum, AB-3285 (250 μM) were added into the well. At 0, 4, 8 and 24 hours post.-infection cells were lysed and viral RNA was quantified. C) Dual luciferase evaluation was performed at 24 hours post-transfection in Vero-E6 cells treated with AB-3285 (500 μM). The frameshift efficiency was normalized compared to untreated. The results are mean and SEM of at least two independent experiments performed in duplicate. C) Binding pose of AB-3285 in the PRF binding site 1. P values <0.0332 (*), <0.0021 (**), <0.0002 (***), < 0.0001 (****)
More recently, the SARS-COV-2 FSE structure has been solved by x-ray, which confirmed the cryo-EM three-stemmed H-type pseudoknot structure, but it showed different tertiary arrangements: the cryo-EM structure has a λ-like tertiary arrangement, meanwhile, the x-ray adopts a vertical conformation (20). Although the x-ray shows a higher resolution of 2.09 Å, it lacks the 5’-slippery site sequence, which might affect the tertiary arrangement. These different arrangements of the FSE have also been supported by previous chemical probing, mutational, and NMR studies demonstrating that the arrangement of stem 1 and stem 2 relative to stem 3 can be flexible (45). The superposition of the x-ray structure and our model showed a similar binding site in the experimental structure, as also revealed by RNAsite, which overlaps our identified binding site 1 (supplementary Figure 4).
DISCUSSION
The alteration of the flexibility of the FSE of SARS-CoV-2 is detrimental to the replication of the virus (23, 46–49). If the viral RNA cannot interact correctly with the ribosome, the −1 PRF is altered and ORF 1ab cannot be expressed at the correct ratio, with lack of production of the viral polymerase and consequent reduction of the replication(23). The FSE of SARS-CoV-2 was previously shown to be a possible target for antiviral development with basic modelling(12) or with empiric screening with dual luciferase assays(50). The precise druggable pockets of the FSE however were not previously identified.
With the aim of identifying new molecules interacting with viral RNA we tested geneticin, an aminoglycoside known to interact with RNA secondary structures.
The compound proved to be effective against multiple variants of SARS-CoV-2 (Table 1). The range of EC50s determined (Table 1) might be linked to the fitness of the variants in the VeroE6 and their plaque forming ability. To exclude any bias, we verified as well the activity of geneticin in Calu-3 cells, and we tested both the ancestral and the omicron variant in a respiratory airway model. In all conditions we confirmed the antiviral activity of geneticin (Figure1).
We then verified an early inhibition in the life cycle with reduced viral protein expression and RNA replication (Figure 2A–B), and we tested the activity on the PRF through dual luciferase assays (Figure 2C). Finally, we studied the interaction of geneticin with the FSE through molecular dynamic simulations (Figure 4–5).
The high flexibility and plasticity of the FSE is an essential requirement for its biological activity(23, 46–49). This unique characteristic is also supported by cryo-EM and x-ray structures recently published(34, 44, 51). In particular, the pseudoknot structure seems to be highly dynamic before encountering the ribosome. However, the unique 3-stem architecture of the FSE (Figure 3) and its mechanism made the FSE a viable target for small molecules. Our computational studies confirmed the presence of a suitable binding site pseudoknot, originally identified by Zhang et collaborator (34). This binding site is located between J2/3 and stem 3 regions, and it is large enough to accommodate geneticin and small ligands. Interestingly, this pocket is close to the S3 region of the FSE. Our molecular dynamic simulation studies revealed that the S3 region is particularly flexible, showing higher fluctuations than the other regions. According to these results, we hypothesised that the S3 region might play a critical role in the conformational change of the FSE, necessary for the frameshifting event. Hence, geneticin could exert its antiviral activity by altering the flexibility of this region, and consequently interfering with the conformational changes between the two main FSE structures.
The resulting antiviral activity is however linked to several limitations: the activity is in the micromolar range, and further studies should focus on the identification of more potent compounds. The antiviral activity is at nontoxic concentrations, also in human derived respiratory tissues (Supplementary Figure 2), however the selectivity index of geneticin is narrow since it is known to bind eukaryotic ribosome and it is associated with toxicity in cell culture. Although the administration in a viral infection is most likely to be for a short duration, future work should be directed to the identification of compounds devoid of interaction with ribosomal RNA. Moreover, aminoglycosides are associated with nephrotoxicity and ototoxicity when administered systemically, therefore a topical administration should be envisaged for compounds similar to geneticin.
For these reasons, and to validate the druggability of the binding pocket identified, we used a virtual screening simulation to identify additional molecules, from a library of RNA binders. Our in silico screening against the “J2/3-stem 3” site revealed that the architecture of the pocket might be sufficiently complex to be targeted by more specific ligands. Through our simulations, we have identified molecules that might engage the FSE targeting the J2/3-stem 3 pocket, enhancing or reducing the pseudoknot stability. The identification of compound 3 with increased potency, reduction of RNA replication, and alteration of the −1PRF (Figure 6) demonstrates the feasibility of our approach. Future work will be directed on the identification of analogues with increased potency, retaining the same mechanism of action, with suitable pharmacological properties.
Supplementary Material
ACKNOWLEDGEMENT
We thank the diagnostics of the Institute of Microbiology of Lausanne for providing the clinical specimens of SARS-CoV-2.
FUNDING
This work was supported by the National Institutes of Health [AA123456 to A.B., BB123456 to C.D.], the Swiss National Science Foundation [PZ00P3_193289 to V.C.] and the University Hospital of Geneva PRD financing. Funding for open access charge: Swiss National Science Foundation and National Institutes of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest
REFERENCES
- 1.Statistics and Research Coronavirus Vaccinations (https://ourworldindata.org/covid-vaccinations) access october 2021.
- 2.Parums D. v. (2022) Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Patients. Medical Science Monitor, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Embarc-Buh A., Francisco-Velilla R. and Martinez-Salas E. (2021) Rna-binding proteins at the host-pathogen interface targeting viral regulatory elements. Viruses, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ganser L.R., Kelly M.L., Herschlag D. and Al-Hashimi H.M. (2019) The roles of structural dynamics in the cellular functions of RNAs. Nature Reviews Molecular Cell Biology, 20, 474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner K.D., Hajdin C.E. and Weeks K.M. (2018) Principles for targeting RNA with drug-like small molecules. Nature Reviews Drug Discovery, 17, 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatt P.R., Scaiola A., Loughran G., Leibundgut M., Kratzel A., Meurs R., Dreos R., O’Connor K.M., McMillan A., Bode J.W., et al. (2021) Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. Science, 372, 1306–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penn W.D., Harrington H.R., Schlebach J.P. and Mukhopadhyay S. (2020) Annual Review of Virology Regulators of Viral Frameshifting: More Than RNA Influences Translation Events. 10.1146/annurev-virology-012120. [DOI] [PMC free article] [PubMed]
- 8.Brierley I. and dos Ramos F.J. (2006) Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Research, 119, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth A.E., Jagger B.W., Wise H.M., Nelson C.C., Parsawar K., Wills N.M., Napthine S., Taubenberger J.K., Digard P. and Atkins J.F. (2012) Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC-UUU-CGU and is in the +1 direction. Open Biology, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wit E., van Doremalen N., Falzarano D. and Munster V.J. (2016) SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews Microbiology, 14, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haniff H.S., Tong Y., Liu X., Chen J.L., Suresh B.M., Andrews R.J., Peterson J.M., O’Leary C.A., Benhamou R.I., Moss W.N., et al. (2020) Targeting the SARS-COV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RiboTAC) degraders. ACS Central Science, 6, 1713–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park S.J., Kim Y.G. and Park H.J. (2011) Identification of rna pseudoknot-binding ligand that inhibits the - 1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. Journal of the American Chemical Society, 133, 10094–10100. [DOI] [PubMed] [Google Scholar]
- 13.Ahn D.G., Lee W., Choi J.K., Kim S.J., Plant E.P., Almazán F., Taylor D.R., Enjuanes L. and Oh J.W. (2011) Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antiviral Research, 91, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garreau De Loubresse N., Prokhorova I., Holtkamp W., Rodnina M. v., Yusupova G. and Yusupov M. (2014) Structural basis for the inhibition of the eukaryotic ribosome. Nature, 513, 517–522. [DOI] [PubMed] [Google Scholar]
- 15.Vicens Q. and Westhof E. (2003) RNA as a drug target: The case of aminoglycosides. ChemBioChem, 4, 1018–1023. [DOI] [PubMed] [Google Scholar]
- 16.Jia X., Zhang J., Sun W., He W., Jiang H., Chen D. and Murchie A.I.H. (2013) Riboswitch control of aminoglycoside antibiotic resistance. Cell, 152, 68–81. [DOI] [PubMed] [Google Scholar]
- 17.Davies J. and Jimenez A. (1980) A new selective agent for eukaryotic cloning vectors. American Journal of Tropical Medicine and Hygiene, 29, 1089–1092. [DOI] [PubMed] [Google Scholar]
- 18.Ariza-Mateos A., Díaz-Toledano R., Block T.M., Prieto-Vega S., Birk A. and Gómez J. (2016) Geneticin stabilizes the open conformation of the 5′ region of hepatitis C virus RNA and inhibits viral replication. Antimicrobial Agents and Chemotherapy, 60, 925–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X.G., Mason P.W., Dubovi E.J., Xu X., Bourne N., Renshaw R.W., Block T.M. and Birk A. v. (2009) Antiviral activity of geneticin against dengue virus. Antiviral Research, 83, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birk1 Alexander V, Dubovi Edward J, Zhang Xianchaoand Szeto Hazel H(2008) Antiviral activity of geneticin against bovine viral diarrhoea virus. Antiviral Chemistry & Chemotherapy, 19, 33–40. [DOI] [PubMed] [Google Scholar]
- 21.Thi Nhu Thao T., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., et al. (2020) Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature, 582, 561–565. [DOI] [PubMed] [Google Scholar]
- 22.Mathez G. and Cagno V. (2021) Clinical severe acute respiratory syndrome coronavirus 2 isolation and antiviral testing. Antiviral Chemistry and Chemotherapy, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhatt P.R., Scaiola A., Loughran G., Leibundgut M., Kratzel A., Meurs R., Dreos R., O K.M., McMillan A., Bode J.W., et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. [DOI] [PMC free article] [PubMed]
- 24.Molecular Operating Environment (MOE), Montreal, QC, Canada, H3A 2R7, 2019. http://www.Chemcomp.Com. [Google Scholar]
- 25.Schrödinger L. Schrödinger, LLC, New York, NY, 2019. https://www.Schrodinger.Com. [Google Scholar]
- 26.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B. and Lindah E. (2015) Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. [Google Scholar]
- 27.Lang P.T., Brozell S.R., Mukherjee S., Pettersen E.F., Meng E.C., Thomas V., Rizzo R.C., Case D.A., James T.L. and Kuntz I.D. (2009) DOCK 6: Combining techniques to model RNA-small molecule complexes. RNA, 15, 1219–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefaniak F. 4 and Bujnicki J.M. (2020) AnnapuRNA: a scoring function for 2 predicting RNA-small molecule 3 interactions. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su H., Peng Z. and Yang J. (2021) Recognition of small molecule–RNA binding sites using RNA sequence and structure. Bioinformatics, 37, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bottaro S., Bussi G., Pinamonti G., Reißer S., Boomsma W. and Lindorff-Larsen K. (2019) Barnaba: software for analysis of nucleic acid structures and trajectories. 10.1261/rna. [DOI] [PMC free article] [PubMed]
- 31.Aytenfisu A.H., Spasic A., Grossfield A., Stern H.A. and Mathews D.H. (2017) Revised RNA Dihedral Parameters for the Amber Force Field Improve RNA Molecular Dynamics. Journal of Chemical Theory and Computation, 13, 900–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su H., Peng Z. and Yang J. (2021) Recognition of small molecule–RNA binding sites using RNA sequence and structure. Bioinformatics, 37, 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C. and Pöhlmann S. (2020) Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature, 585, 588–590. [DOI] [PubMed] [Google Scholar]
- 34.Zhang K., Zheludev I.N., Hagey R.J., Haslecker R., Hou Y.J., Kretsch R., Pintilie G.D., Rangan R., Kladwang W., Li S., et al. (2021) Cryo-EM and antisense targeting of the 28-kDa frameshift stimulation element from the SARS-CoV-2 RNA genome. Nature Structural & Molecular Biology, 28, 747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nierzwicki Ł. and Palermo G. (2021) Molecular Dynamics to Predict Cryo-EM: Capturing Transitions and Short-Lived Conformational States of Biomolecules. Frontiers in Molecular Biosciences, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bissaro M., Sturlese M. and Moro S. (2020) Exploring the RNA-Recognition Mechanism Using Supervised Molecular Dynamics (SuMD) Simulations: Toward a Rational Design for Ribonucleic-Targeting Molecules? Frontiers in Chemistry, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGreevy R., Teo I., Singharoy A. and Schulten K. (2016) Advances in the molecular dynamics flexible fitting method for cryo-EM modeling. Methods, 100, 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omar S.I., Zhao M., Sekar R.V., Moghadam S.A., Tuszynski J.A. and Woodside M.T. (2021) Modeling the structure of the frameshift-stimulatory pseudoknot in SARS-CoV-2 reveals multiple possible conformers. PLoS Computational Biology, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rangan R., Watkins A.M., Chacon J., Kretsch R., Kladwang W., Zheludev I.N., Townley J., Rynge M., Thain G. and Das R. (2021) De novo 3D models of SARS-CoV-2 RNA elements from consensus experimental secondary structures. Nucleic Acids Research, 49, 3092–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vicens Q. and Westhof E. (2003) Crystal structure of geneticin bound to a bacterial 16 S ribosomal RNA A site oligonucleotide. Journal of Molecular Biology, 326, 1175–1188. [DOI] [PubMed] [Google Scholar]
- 41.Prokhorova I., Altman R.B., Djumagulov M., Shrestha J.P., Urzhumtsev A., Ferguson A., Chang C.W.T., Yusupov M., Blanchard S.C., Yusupova G., et al. (2017) Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proceedings of the National Academy of Sciences of the United States of America, 114, E10899–E10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aradi K., di Giorgio A. and Duca M. (2020) Aminoglycoside Conjugation for RNA Targeting: Antimicrobials and Beyond. Chemistry - A European Journal, 26, 12273–12309. [DOI] [PubMed] [Google Scholar]
- 43.Montesano R., Pegg A.E. and Margison G.P. (1980) Alkylation of dna and carcinogenicity of w-nitroso compounds. Journal of Toxicology and Environmental Health, 6, 81–88. [DOI] [PubMed] [Google Scholar]
- 44.Roman C., Lewicka A., Koirala D., Li N.-S. and Piccirilli J.A. (2021) The SARS-CoV-2 Programmed −1 Ribosomal Frameshifting Element Crystal Structure Solved to 2.09 Å Using Chaperone-Assisted RNA Crystallography. ACS Chemical Biology, 10.1021/acschembio.1c00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlick T., Zhu Q., Dey A., Jain S., Yan S. and Laederach A. (2021) To Knot or Not to Knot: Multiple Conformations of the SARS-CoV-2 Frameshifting RNA Element. Journal of the American Chemical Society, 143, 11404–11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huston N.C., Wan H., Strine M.S., de Cesaris Araujo Tavares R., Wilen C.B. and Pyle A.M. (2021) Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Molecular Cell, 81, 584–598.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlick T., Zhu Q., Dey A., Jain S., Yan S. and Laederach A. (2021) To Knot or Not to Knot: Multiple Conformations of the SARS-CoV-2 Frameshifting RNA Element. Journal of the American Chemical Society, 143, 11404–11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omar S.I., Zhao M., Sekar R.V., Moghadam S.A., Tuszynski J.A. and Woodside M.T. (2021) Modeling the structure of the frameshift-stimulatory pseudoknot in SARS-CoV-2 reveals multiple possible conformers. PLoS Computational Biology, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manfredonia I., Nithin C., Ponce-Salvatierra A., Ghosh P., Wirecki T.K., Marinus T., Ogando N.S., Snijder E.J., van Hemert M.J., Bujnicki J.M., et al. (2020) Genome-wide mapping of SARS-CoV-2 RNA structures identifies therapeutically-relevant elements. Nucleic Acids Research, 48, 12436–12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y., Abriola L., Surovtseva Y. v, Lindenbach B.D. and Guo J.U. (2020) Restriction of SARS-CoV-2 Replication by Targeting Programmed −1 Ribosomal Frameshifting In Vitro. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatt P.R., Scaiola A., Loughran G., Leibundgut M., Kratzel A., Meurs R., Dreos R., O K.M., McMillan A., Bode J.W., et al. Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.