Abstract
Molecular tests assist at various stages of cancer patient management, including providing diagnosis, predicting prognosis, identifying therapeutic targets, and determining hereditary cancer risk. The current testing paradigm involves germline testing in a subset of patients determined to be at high risk for having a hereditary cancer syndrome, and tumor-only sequencing for treatment decisions in advanced cancer patients. A major limitation of tumor-only sequencing is its inability to distinguish germline versus somatic mutations. Tumor-normal sequencing has emerged as a comprehensive analysis for both hereditary cancer predisposition and somatic profiling. Here, we review recent studies involving tumor-normal sequencing, discuss its benefits in clinical care, challenges for its implementation, and novel insights it has provided regarding tumor biology and germline contribution to cancer.
Keywords: Tumor sequencing, tumor-normal sequencing, hereditary cancer predisposition
Genetic Testing in Cancer Care Beyond Tumor Sequencing
Clinical care of cancer patients today routinely includes molecular tests on tumor to detect therapeutic targets and, if hereditary risk assessment guidelines are met, germline testing to determine genetic risk and inform future management of both patient and family members. Germline testing results are also used for predicting response to certain targeted therapies. Until recently, tumor and germline testing have typically been performed separately, with patients being selected for each test based on different sets of criteria. Recent studies using unbiased paired tumor-normal testing have demonstrated several advantages of the combined analysis for clinical care of patients and expanded our understanding of cancer development and progression. In this review, we will discuss the applications and utility of tumor-normal sequencing, issues regarding its integration into clinical care, as well as lessons learned on cancer biology using this approach.
Germline Testing for Hereditary Cancer Predisposition
Hereditary contribution to cancer has long been recognized [1–4]. Over 100 cancer predisposition genes have been described to date and the majority of them are associated with cancer risk only, while certain genes, such as NF1, PTPN11, and TSC2, underlie syndromic presentations that include developmental anomalies in addition to predisposition to childhood cancers.[2] High-penetrance genes such as APC, BRCA1, BRCA2, MLH1, RB1, and TP53, were among the first genes to be discovered through observation of segregation in large families. Later, candidate gene analyses in large case-control studies have identified genes such as ATM, BRIP1, CHEK2, and PALB2 that are associated with a relatively modest increased risk and are referred to as having moderate penetrance.
Recognizing hereditary predisposition to cancer and identifying causative pathogenic germline variants are undoubtedly important to allow timely surveillance and preventative interventions for both patients and family members [5]. Germline defects in certain homologous recombination (i.e. BRCA1, BRCA2) and mismatch repair (i.e. MLH1, MSH2) pathway genes also predict response to PARP inhibitor and immune-checkpoint inhibitor therapies, respectively. With the approval of targeted treatments based on genetic biomarkers, the significance of germline testing in management of cancer patients has been more widely recognized. In addition, studies over the last two decades led to a substantial increase in the number of known hereditary cancer genes demonstrated that germline susceptibility to cancer is more prevalent than previously anticipated [6–10]. Therefore, inclusion of germline testing in management of cancer patients has become increasingly essential.
Traditional Testing Protocol for Tumor and Germline Analysis
In general, tumor and germline testing are performed separately at different time points and often by different laboratories. Germline genetic testing is done using blood or saliva as the “normal” tissue source to look for pathogenic variants that cause a known cancer predisposition syndrome. In current practice, patients are selected in a “guideline-based” manner, with only those who meet established guidelines from national and professional organizations receiving genetic testing. These guidelines and recommendations are created based on the expected the yield of genetic testing considering the tumor type, age of onset, and clinical and/or family history of patients [11–13]. Patients who do not meet the established criteria typically do not receive germline testing for cancer predisposition.
On the other hand, tumor testing is typically performed on the tumor specimens of individuals with advanced cancer who require systemic therapy. Many of these individuals show disease progression through standard treatments, and tumor testing is performed to identify somatic variants that may confer susceptibility to targeted therapies.
Paired Tumor-Normal Testing
In paired tumor-normal analysis, DNA isolated from tumor and from nonmalignant “normal” cells of the peripheral blood, saliva, buccal swab, fibroblasts, or nails are sequenced on the same platform separately, and data from tumor and normal DNA samples from the same individual are matched to analyze together. While this method can be used solely to improve tumor sequencing results interpretation, when patient’s consent is obtained to analyze germline findings, it can also be used to determine hereditary cancer predisposition simultaneously. This approach provides several advantages over the traditional testing scheme:
1. Broad tumor-normal sequencing identifies hereditary cancer predisposition in an unbiased manner
Sequencing both tumor and normal in each patient without pre-selection based on certain criteria elucidates the hereditary contribution to cancer in an unbiased manner. Recently, multiple studies that have taken this approach demonstrated that a greater proportion of both pediatric and adult cancer patients carry germline disease-causing variants in cancer genes than previously anticipated [6–9, 14–18]. Variants indicative of hereditary cancer predisposition were identified in 3% to 12.6% of adult and 8.5–10% of pediatric patients depending on the population and genes studied [7, 9, 10, 15, 19]. It has become clear that established guidelines may miss patients with hereditary cancer predisposition who could be identified by a broad testing approach. In our MSKCC cohort of advanced cancer patients, we identified clinically actionable germline findings in 17.5% of patients by tumor-normal sequencing, over half (55.5%) of which would have been missed in guideline-driven testing [18]. While further studies are needed to understand whether the same applies to early-stage cancer, recent analyses on patients with a range of tumor types or multiple primaries also suggest that an unbiased testing approach may reveal hereditary cancer predisposition in individuals who do not meet established guidelines [14, 20–23], helping inform management and counseling of patients and their family members.
In addition to its implications for patient care, broad tumor-normal sequencing has also provided insights into the genetic landscape of cancer and germline contribution to tumorigenesis. Firstly, germline pathogenic or likely pathogenic variants have been identified in cancer genes that are not known to be associated with the patient’s particular tumor type. It largely remains unclear whether these variants contribute to tumor formation or represent unrelated incidental findings, although interesting correlations have already emerged through burden analyses, association with loss of heterozygosity (LOH) and specific mutation signatures in paired tumors [8, 9, 18]. For instance, endometrial cancer is not generally considered to be part of hereditary breast and ovarian cancer syndrome. However, through tumor-normal sequencing, one study identified 13 endometrial cancer cases with BRCA1 or BRCA2 germline mutations and showed that 8/8 BRCA1 and 2/5 BRCA2 associated endometrial cancers had LOH for the wild type allele in the tumor and genomic evidence of homologous recombination deficiency (JCO Precision Oncology, in press). Other examples include enrichment of truncating variants in RAD51C in acute myeloid leukemia and PALB2 in stomach adenocarcinoma and ovarian cancer [8]. Therefore, whether a germline mutation is playing a biological role in the tumor can be assessed using the associated tumor sequencing data. Secondly, the paired analysis is informative for determining germline and somatic interactions. Selective pressure for biallelic inactivation of tumor-suppressor genes in tumorigenesis has been repeatedly demonstrated across several cancer types [8, 9, 24, 25]. A large study of 12 cancer types reported mutual exclusivity and co-occurrence of many germline and somatic events, such as mutual exclusivity of germline ATM truncating variants and TP53 somatic mutations in lung adenocarcinoma and co-occurrence of BRCA1 germline truncating variants and TP53 somatic mutations in breast cancer [8]. Finally, tumor-normal sequencing also helps clarify the phenotypic and therapeutic relevance of germline alterations. In a recent analysis of advanced-stage cancer and germline pathogenic or somatic truncating variants in BRCA1/2, selective pressure for biallelic inactivation, phenotype penetrance and PARP inhibitor sensitivity were observed only in tumor types known to be associated BRCA1/2, and tumorigenesis was independent of mutant BRCA1/2 in patients with non-BRCA-associated cancers [26]. These findings suggested that most BRCA1/2 mutations in non-BRCA-associated cancers may be incidental and unlikely to be therapeutically relevant.
2. Somatic versus germline variants can be differentiated in tumor sequencing results
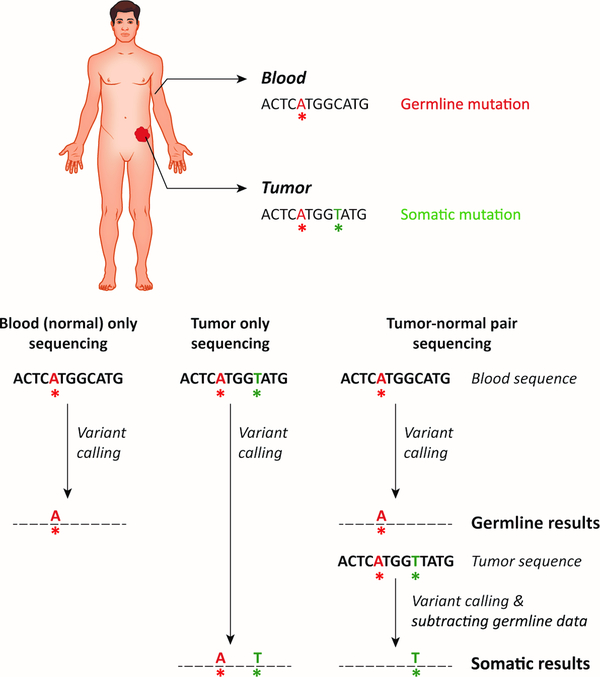
Tumor sequencing data include a mixture of variants that are of germline origin and those that have arisen somatically during tumor formation. From a clinician’s perspective, it is critical to determine whether a pathogenic variant identified in tumor sequencing represents a somatic versus a germline finding. Because of the implications of germline results for the patient and their family members, identification of a pathogenic variant in a cancer syndrome-related gene during tumor-only sequencing often initiates a cascade of genetic consultation and follow-up testing of a nonmalignant tissue to determine whether the variant is of germline origin. Therefore, tumor-only sequencing may frequently lead to additional genetic tests, which would be avoided with paired normal tissue analysis. One of the major advantages of tumor-normal sequencing is the ability to subtract variants in matched normal tissue from tumor data, thus revealing those that are truly of somatic origin (Figure 1).
Figure 1. Mutations Reported in Blood-only, Tumor-only, and Paired Tumor-Normal Sequencing.
In blood-only sequencing, only germline variants detected in the blood (see limitations about mosaic and clonal hematopoiesis-related variants) are reported, informing about hereditary cancer predisposition. Tumor-only sequencing detects and reports both germline and somatic mutations, and is unable to distinguish between the two sources. Paired tumor-normal sequencing allows one to accurately both report germline variants related to hereditary cancer predisposition and somatic mutations in the tumor. Mutations called in the tumor that are not present in the paired normal sample are annotated as somatic and are reported in the somatic report.
In the absence of a “normal” control, various approaches have been undertaken to predict and filter out variants that are likely to be germline. One approach filters out variants that are present in large population databases such as dbSNP, 1000 Genomes, or ExAC for likely having germline origin [19, 27–29]. While this approach can filter out common variants, those that are rare in the general population will remain in the sequencing data. This is particularly problematic for individuals of non-White ancestries and results in a higher rate of false positive germline findings in these individuals, as the majority of current large population databases consist of White/European ancestries [27]. Another approach looks at variant allele fractions in tumor sequencing data. Allele fractions close to ~50% and ~100% are assumed to likely represent heterozygous or homozygous germline variants, respectively, and lower allele fractions are predicted to represent somatic mutations [19]. However, this method gets complicated by varying tumor purity, normal tissue contamination, and changes in tumor allele fractions due to loss of heterozygosity or deletion/amplification events. In addition, the fraction of variants in NGS data may differ from the expected 50% or 100% depending on their genomic location or their size, such that those in regions of high homology or large deletion and insertion variants may have lower than expected representation in the total mapped reads in NGS. Often, a combination of the above methods is used to maximize the prediction and filtration of germline variants from tumor-only sequencing results. During this filtration process, true somatic variants that may be identical to germline variants in population databases may inadvertently get removed, resulting in false-negatives. In order to reduce the likelihood of removing cancer-related somatic mutations this way, many groups rescue variants that have been reported in the COSMIC (Catalogue of Somatic Mutations in Cancer) database [19, 27, 28]. One study has shown that molecular pathologist review of tumor-only sequencing results following germline prediction/filtering steps classified the majority of true germline variants as having uncertain significance for cancer and thereby reduced the rate of false positive germline cancer-related calls [27]. Overall, despite germline prediction and filtering processes, tumor-only sequencing led to false positive and/or false negative mutation calls, including those in potentially actionable genes [19, 27]. For example, a recent study of 17,152 matched tumor-normal pairs found of the 17,075 pathogenic variants detected in tumor sequencing, 1494 (8.7%) were actually of germline origin [30]. These variants would either be misattributed as somatic in nature or require identification as potentially germline by the oncologist and subsequent germline testing under the tumor-only sequencing approach. Therefore, matched tumor-normal sequencing is the ideal approach to differentiate somatic versus germline variants and for precise interpretation of both results.
Filtering out germline variants from tumor sequencing data also yields more reliable information to estimate tumor phenotypic features such as mutation burden, microsatellite instability, and other mutation signatures [31, 32]. These attributes often are informative about underlying molecular processes for tumor formation, help determine its site of origin, and serve as biomarkers for targeted therapies, such as immune checkpoint inhibitors for tumors with high mutation burden or microsatellite instability [33–35]. Somatic mutation calling using a matched normal sample is the ideal approach for accurate mutation signature analysis.
3. Comparing tumor and normal sequencing results allows to identify loss-of heterozygosity and “second-hits”
Somatic biallelic inactivation through loss of heterozygosity (LOH) or a second mutation (“second hit”) on the opposite allele are the most common cancer initiation mechanisms in individuals with germline pathogenic variants in tumor suppressor genes [36, 37]. Matched tumor-normal analysis in individuals with hereditary cancer predisposition allows assessing LOH and second-hits in tumors, which is informative for both somatic evaluation to elucidate molecular mechanism of tumor formation and in germline assessment to identify cancer predisposition genes [38–40] and assess clinical significance of variants [39, 41]. While tumor mutation data including LOH needs to be reviewed carefully in addition to other types of evidence regarding a variant’s pathogenicity [41], it is useful for variant classification in cancer predisposition genes. For instance, while evaluating a germline RB1 variant in an individual with retinoblastoma, loss of the allele carrying this variant in the tumor would suggest that the variant is not disease-related. Similarly, in an individual with renal cell carcinoma and a rare VHL germline variant, identification of biallelic truncating somatic mutations in VHL in the tumor would be suggestive of a sporadic cancer and be considered as evidence supporting that the germline VHL variant is not causative for disease.
4. Matched analysis of a second tissue specimen from the same individual allows detection of mosaicism and clonal hematopoiesis
Blood is the standard source of nonmalignant DNA used for germline analyses. However, DNA isolated from blood does not solely represent true germline variants, but may also include those that are mosaic or arose as a somatic mutation in the hematopoietic lineage, referred to as clonal hematopoiesis (CH) [42, 43] (Figure 2). Generally, CH is common in cancer patients and is associated with aging, smoking, or radiation therapy [44]. Clonal hematopoiesis may be detected in the absence of any evidence for a hematopoietic neoplasm; however, it is a risk factor for the development of hematologic cancer [44–46].
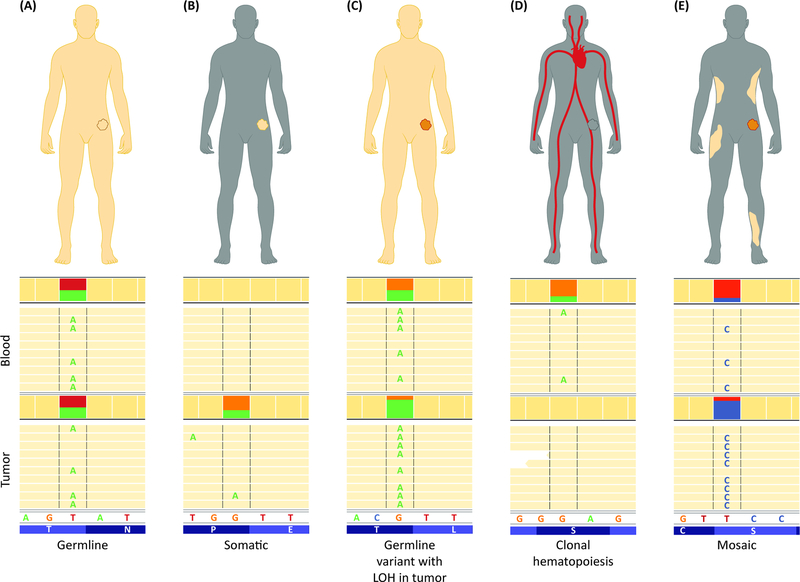
Figure 2: Paired Tumor-Normal Analysis Distinguishes Variants of Various Origins.
(A) Germline heterozygous variants are typically detected at ~50% allele fraction in both normal and tumor sample (in the absence of additional copy number-altering or copy-neutral LOH events). (B) Somatic variants are absent from the normal tissue and can be detected at various fractions in the tumor, depending on their time of occurrence. (C) Variants with ~50% allele fraction in the normal sample and at much higher fraction in the tumor are likely germline variants with LOH at that region in the tumor. (D) Variants that are detected in the blood but are absent from the tumor, without the presence of LOH at that locus, likely represent clonal hematopoiesis. (E) Variants detected with low allele fraction in the blood and are present in the tumor may represent mosaic alterations, that can be confirmed by testing of additional non-malignant tissue samples.
Blood-only sequencing is often unable to distinguish true germline variants from those that are mosaic or representative of CH [47, 48]. Typically, a variant’s fraction in the blood specimen is used as a metric to suspect mosaicism or CH in blood-only sequencing [49]. Most true heterozygous germline variants are detected at a fraction of ~50% in the blood, while mosaic and CH variants are expected to be detected at lower fractions. However, various technical reasons may cause a true germline variant to be detected at a lower fraction than expected in NGS, including problems with mapping of sequencing reads in high homology regions or nearby homopolymers, and those that involve indels. On the other hand, high-level mosaic variants or CH variants that arose early in differentiation or amplified during clonal expansion may be detected at high fractions that are close to the expected range for germline variants. These issues complicate results analysis in blood-only sequencing and may result in inaccurate interpretation of their origin [43].
An incorrect assumption of CH variants as germline has significant implications for the patient’s clinical management [50]. For instance, a pathogenic TP53 variant of CH origin may indicate risk for hematologic cancer and would necessitate clinical follow-up accordingly. On the other hand, if interpreted as a germline variant, it would be considered as diagnostic of Li Fraumeni syndrome and would initiate a life-time long series of comprehensive and often invasive screening tests for a broad range of cancers, including sarcomas, breast cancer, and brain tumors [51]. Identification of a germline variant also has implications for reproductive planning and risk to family members. Since somatic mutations in the blood are not inherited and cannot be transmitted to future generations, distinguishing them from germline variants is also important to avoid unnecessary genetic tests in extended family members.
Mosaicism is the presence of a variant in some but not all cells of an individual that are derived from the same zygote [52, 53]. A mosaic pathogenic variant may be present at varying levels in different tissues and may be clinically significant or inconsequential if it is restricted to tissues not pertinent to the gene’s function, occurred after a time frame that is relevant for the gene’s function, or is present in a number of cells that is lower than a critical threshold to impact tissue function [52, 53]. Therefore, predicting the clinical consequence of a mosaic variant is challenging and differentiating them from true heterozygous germline variants is important for recommendations regarding surveillance, prophylactic treatments and reproductive planning.
In order to confirm whether a suspected variant detected in peripheral blood cells is germline, mosaic, or CH-associated, testing a separate tissue specimen is required. For this purpose, the variant is typically tested in fibroblasts from the individual isolated via a skin biopsy. A matched tumor analysis is often informative to identify potential mosaic or CH-associated variants, as the tumor represents another tissue specimen from the tested individual (Figure 2). Because other events that may result in lower variant fraction in the tumor, such as copy number-altering structural rearrangements or copy-neutral loss of heterozygosity events are common in tumor genomes, one should assess the presence of such alterations before considering a variant as a potential CH-associated mutation. Similarly, a variant detected at a fraction lower than the expected ~50% for a heterozygous germline variant in blood but was also detected in the tumor may be suspected of being mosaic. Nonetheless, a non-malignant tissue specimen should be tested to clarify the status of suspected CH or mosaic variants.
5. With patient’s consent, both tumor data and hereditary cancer predisposition can be assessed simultaneously with a single test
Tumor sequencing is performed for diagnostic and therapeutic purposes; however, it frequently provides incomplete information to guide patient management. For instance, in a patient with unilateral breast cancer and a germline pathogenic BRCA1 variant, surgery decisions may be made based on germline versus somatic origin of mutation, such that the patient may choose to pursue prophylactic bilateral mastectomy and oophorectomy if the variant is germline instead of a unilateral mastectomy only. In a traditional testing scheme, tumor sequencing may be performed first and germline testing is pursued for variants suspected of having germline origin, such as known founder mutations or those predicted to be germline based on their allele fraction. However, as discussed above, distinguishing germline versus somatic mutations in tumor-only sequencing has limitations, leading to high number of variants that will fail to confirm as germline, or incorrect calling of a pathogenic germline variant as somatic and thus missing the diagnosis of a hereditary cancer predisposition syndrome. From a clinician’s perspective, in addition to accurate calling of somatic and germline variants, a simultaneous testing approach also has several practical advantages over separate analyses at different time points. Imagine a Lynch syndrome patient with a germline pathogenic MLH1 variant. In such patients, reaching a diagnosis often takes several rounds of sequential tumor and germline testing that starts with tumor immunohistochemistry and/or microsatellite instability analysis indicating a deficiency in one of the mismatch-repair (MMR) genes, followed by tumor MLH1 hypermethylation and/or tumor-only sequencing of MMR genes, and finally a germline test to identify/confirm the pathogenic MLH1 variant. This approach requires coordinating multiple clinic visits and sample collections from the patient, as well as ordering and gathering results of tests that are often performed by different laboratories. A combined testing approach eliminates this long cascade, allowing timely and accurate diagnosis.
6. Testing two independent samples from the same individual allows sample identity check and prevents sample mix-ups
Despite many quality check steps including DNA barcoding and SNP identity testing in independent extractions or aliquots, sample mix-ups still occur and pose a major problem for clinical laboratories. Additionally, typical practices only allow checking mix-ups that happen after the DNA extraction step at the laboratory, although mix-ups may happen at the phlebotomy, surgical pathology, or sample accessioning steps, as well. By comparing SNPs identified in matched tumor and blood samples, one can ensure that they belong to the same individual, thus preventing sample mix-ups that may happen at any stage of the testing process.
Challenges associated with tumor-normal sequencing
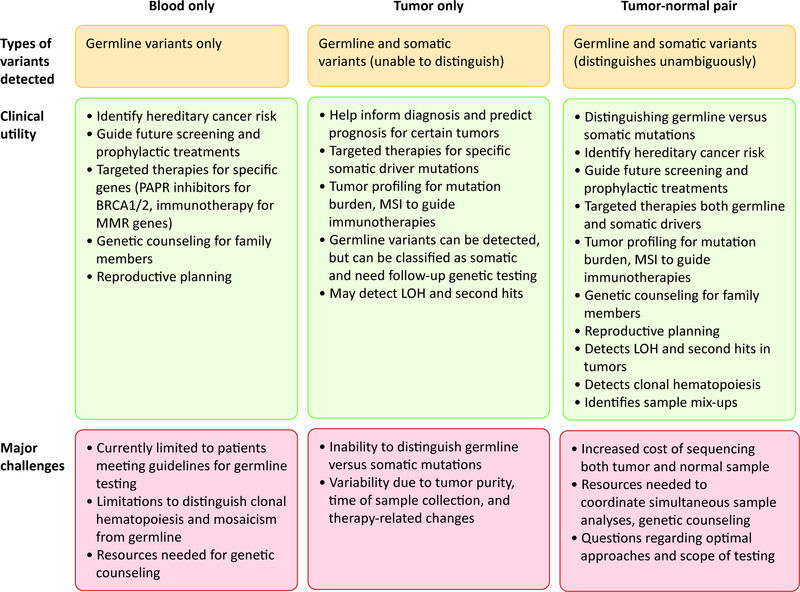
Matched tumor-normal testing is an efficient and advantageous approach for both somatic and germline analyses (Figure 3). However, there are several challenges to its implementation at a large scale in any institution. First, building an infrastructure to have both blood and tumor specimen sent to the laboratory in an orderly manner requires an institution-wide organization, which may be difficult to establish. Second, costs associated with sequencing and interpreting both tumor and germline DNA may be prohibitive, especially because the germline testing component may not be covered by insurances at present, if the patient does not meet current guidelines for clinical germline testing. In addition, germline testing requires pre- and post-test genetic counseling and therefore significant amount of resources should be dedicated to providing access to genetic consultation for each patient to be tested. Finally, expanded germline analyses in individuals who do not meet current testing guidelines is likely to identify more variants of uncertain significance compared to targeted tests in high-risk individuals, and therefore may result in inconclusive results for a higher number of patients. These factors need to be considered to take advantage of the benefits of tumor-normal sequencing.
Figure 3:
Comparison of blood-only, tumor-only, and tumor-normal sequencing
Concluding Remarks
Paired tumor-normal sequencing undeniably has many advantages over blood-only and tumor-only approaches. However, questions remain regarding the benefit and cost-effectiveness of a comprehensive genomic analysis in different types of cancers and early-stage patients (Outstanding Questions). One option to minimize these issues while still deriving the benefits of tumor-normal sequencing might be to restrict the test to a targeted set of highly actionable genes and/or variants. In addition, several other methods are currently under development and can be useful to assist both germline and tumor studies. RNA testing is informative for identifying structural alterations and the impact of variants that affect splicing. Cell-free DNA analysis can be a noninvasive test for tumor profiling. Ultimately, further studies on broader cohorts are needed to understand the optimal approach of testing for diverse patient populations. Future efforts should also be dedicated to providing patients equal access to the broad range of testing options that become available and provide benefit for their clinical care.
Highlights.
Clinical care of cancer patients increasingly involves germline and tumor molecular analyses.
Paired tumor-normal sequencing is being adapted at various institutions and provides many advantages to tumor-only or blood-only sequencing, such as simultaneous detection of both hereditary cancer predisposition and somatic mutations to guide targeted therapies, distinguishing between germline and somatic mutations, and identifying potential clonal hematopoiesis-related variants.
Combined tumor-normal analysis has revealed novel associations between hereditary cancer predisposition genes and tumor formation, and provided insights into germline contribution to cancer.
Several challenges exist regarding the implementation of paired tumor-normal analyses in patient care, including technical, practical and cost-related considerations.
Outstanding Questions.
What is the optimal testing approach in patients with various tumor types and at different stages of cancer?
In what circumstances should a comprehensive versus a targeted testing approach be pursued?
Do the benefits of paired tumor-normal sequencing justify the practical and cost-related challenges that need to be undertaken in all patients? Is universal tumor-normal sequencing necessary?
What is the optimal non-malignant “normal” tissue specimen? Blood is the current gold-standard, but could saliva, nail, or non-malignant biopsy specimens be used as a matched “normal”?
Can cell-free DNA and RNA testing be integrated into tumor-normal analysis methods?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garber JE and Offit K (2005) Hereditary cancer predisposition syndromes. J Clin Oncol 23, 276–292 [DOI] [PubMed] [Google Scholar]
- 2.Rahman N (2014) Realizing the promise of cancer predisposition genes. Nature 505, 302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lichtenstein P, et al. (2000) Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343, 78–85 [DOI] [PubMed] [Google Scholar]
- 4.Stadler ZK, et al. (2014) Cancer genomics and inherited risk. J Clin Oncol 32, 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robson ME, et al. (2015) American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. J Clin Oncol 33, 3660–3667 [DOI] [PubMed] [Google Scholar]
- 6.AlDubayan SH, et al. (2018) Inherited DNA-Repair Defects in Colorectal Cancer. Am J Hum Genet 102, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsons DW, et al. (2016) Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol 2, 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, et al. (2015) Patterns and functional implications of rare germline variants across 12 cancer types. Nat Commun 6, 10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang KL, et al. (2018) Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 173, 355–370 e314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, et al. (2015) Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med 373, 2336–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampel H, et al. (2015) A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med 17, 70–87 [DOI] [PubMed] [Google Scholar]
- 12.Daly MB, et al. (2017) NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast and Ovarian, Version 2.2017. J Natl Compr Canc Netw 15, 9–20 [DOI] [PubMed] [Google Scholar]
- 13.Gupta S, et al. (2017) NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Colorectal, Version 3.2017. J Natl Compr Canc Netw 15, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 14.Carlo MI, et al. (2018) Prevalence of Germline Mutations in Cancer Susceptibility Genes in Patients With Advanced Renal Cell Carcinoma. JAMA Oncol 4, 1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrader KA, et al. (2016) Germline Variants in Targeted Tumor Sequencing Using Matched Normal DNA. JAMA Oncol 2, 104–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seifert BA, et al. (2016) Germline Analysis from Tumor-Germline Sequencing Dyads to Identify Clinically Actionable Secondary Findings. Clin Cancer Res 22, 4087–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meric-Bernstam F, et al. (2016) Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol 27, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandelker D, et al. (2017) Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. JAMA 318, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones S, et al. (2015) Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med 7, 283ra253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitworth J, et al. (2018) Comprehensive Cancer-Predisposition Gene Testing in an Adult Multiple Primary Tumor Series Shows a Broad Range of Deleterious Variants and Atypical Tumor Phenotypes. Am J Hum Genet 103, 3–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowery MA, et al. (2018) Prospective Evaluation of Germline Alterations in Patients With Exocrine Pancreatic Neoplasms. J Natl Cancer Inst 110, 1067–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You YN, et al. (2019) Detection of Pathogenic Germline Variants Among Patients With Advanced Colorectal Cancer Undergoing Tumor Genomic Profiling for Precision Medicine. Dis Colon Rectum 62, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beitsch PD, et al. (2019) Underdiagnosis of Hereditary Breast Cancer: Are Genetic Testing Guidelines a Tool or an Obstacle? J Clin Oncol 37, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nones K, et al. (2019) Whole-genome sequencing reveals clinically relevant insights into the aetiology of familial breast cancers. Ann Oncol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latham A, et al. (2019) Microsatellite Instability Is Associated With the Presence of Lynch Syndrome Pan-Cancer. J Clin Oncol 37, 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson P, et al. (2019) Tumour lineage shapes BRCA-mediated phenotypes. Nature 571, 576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garofalo A, et al. (2016) The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med 8, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sukhai MA, et al. (2019) Somatic Tumor Variant Filtration Strategies to Optimize Tumor-Only Molecular Profiling Using Targeted Next-Generation Sequencing Panels. J Mol Diagn 21, 261–273 [DOI] [PubMed] [Google Scholar]
- 29.Hiltemann S, et al. (2015) Discriminating somatic and germline mutations in tumor DNA samples without matching normals. Genome Res 25, 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandelker D, et al. (2019) Germline-Focused Analysis of Tumour-Only Sequencing: Recommendations from the ESMO Precision Medicine Working Group. Ann Oncol 30, 1221–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Hoeck A, et al. (2019) Portrait of a cancer: mutational signature analyses for cancer diagnostics. BMC Cancer 19, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teer JK, et al. (2017) Evaluating somatic tumor mutation detection without matched normal samples. Hum Genomics 11, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandal R, et al. (2019) Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science 364, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samstein RM, et al. (2019) Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51, 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abida W, et al. (2018) Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knudson AG (2001) Two genetic hits (more or less) to cancer. Nat Rev Cancer 1, 157–162 [DOI] [PubMed] [Google Scholar]
- 37.Thiagalingam S, et al. (2002) Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: molecular basis of its occurrence. Curr Opin Oncol 14, 65–72 [DOI] [PubMed] [Google Scholar]
- 38.Park S, et al. (2018) Systematic discovery of germline cancer predisposition genes through the identification of somatic second hits. Nat Commun 9, 2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diaz-Gay M, et al. (2019) Integrated Analysis of Germline and Tumor DNA Identifies New Candidate Genes Involved in Familial Colorectal Cancer. Cancers (Basel) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanchi KL, et al. (2014) Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun 5, 3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh MF, et al. (2018) Integrating somatic variant data and biomarkers for germline variant classification in cancer predisposition genes. Hum Mutat 39, 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busque L, et al. (2012) Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 44, 1179–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ptashkin RN, et al. (2018) Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors. JAMA Oncol 4, 1589–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coombs CC, et al. (2017) Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 21, 374–382 e374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaiswal S, et al. (2014) Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371, 2488–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genovese G, et al. (2014) Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371, 2477–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coombs CC, et al. (2018) Identification of Clonal Hematopoiesis Mutations in Solid Tumor Patients Undergoing Unpaired Next-Generation Sequencing Assays. Clin Cancer Res 24, 5918–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weitzel JN, et al. (2018) Somatic TP53 variants frequently confound germ-line testing results. Genet Med 20, 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slavin TP, et al. (2019) Prevalence and characteristics of likely-somatic variants in cancer susceptibility genes among individuals who had hereditary pan-cancer panel testing. Cancer Genet 235-236, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell RL, et al. (2018) Misdiagnosis of Li-Fraumeni Syndrome in a Patient With Clonal Hematopoiesis and a Somatic TP53 Mutation. J Natl Compr Canc Netw 16, 461–466 [DOI] [PubMed] [Google Scholar]
- 51.Kratz CP, et al. (2017) Cancer Screening Recommendations for Individuals with Li-Fraumeni Syndrome. Clin Cancer Res 23, e38–e45 [DOI] [PubMed] [Google Scholar]
- 52.Biesecker LG and Spinner NB (2013) A genomic view of mosaicism and human disease. Nat Rev Genet 14, 307–320 [DOI] [PubMed] [Google Scholar]
- 53.Forsberg LA, et al. (2017) Mosaicism in health and disease - clones picking up speed. Nat Rev Genet 18, 128–142 [DOI] [PubMed] [Google Scholar]